Purpose and appropriate sample types
This 21-color flow cytometry-based OMIP[1] enables simultaneous quantification of monocytes, basophils, granulocytes, dendritic cells, natural killer cells, B cells, and all well-defined T and T helper cell subsets in the human peripheral blood. This panel captures the major phenotypes described in the NIH Human Immunology Project [2, 3] with additional markers for deep T cell analysis [4]. We specifically designed this panel for analysis of peripheral blood from patients involved in our clinical trials of novel agents for the treatment of graft versus host disease (GVHD) after allogeneic hematopoietic stem cell transplantation (alloHSCT). We have optimized this panel for the analysis of 1×10^6 fresh or previously frozen peripheral blood mononuclear cells (PBMCs).
Background
We initially designed this panel for the analysis of the PBMCs from patients who have undergone alloHSCT, particularly those enrolled in drug studies for the prevention and treatment of GVHD. Prior studies in humans and animal models have implicated many immune cell types in the initiation and progression of GVHD, and the data have, at times, conflicted, depending on species, model, and individual laboratories. In particular, prior studies have identified imbalances in T regulatory cells (Tregs), T follicular helper (Tfh) cells, T helper type 1 (Th1), type 2 (Th2), type 17 (Th17), myeloid derived suppressor cells (MDSCs), natural killer cells (NKs), dendritic cells (DCs), and others in modulating engraftment, GVHD, and treatment responses [5, 6]. Accordingly, we aimed to develop a standardized panel to capture all major human lymphoid and myeloid populations with deep T cell phenotyping in a single analysis, thus reducing experimental variability, redundancy, and the need for a high quantity of input cells. As to the last point, post-HSCT patients typically have few circulating leukocytes until hematopoietic engraftment and reconstitution. Thus, multiple flow cytometry panels and/or CyTOF analyses pose a greater challenge than a single, comprehensive flow-based panel. Beyond our HSCT-focused studies, this panel should find broad application in the study of many inflammatory and neoplastic conditions. Of note, this panel uses antibodies targeting exclusively surface receptors, making fixation and permeabilization unnecessary.
After gating on FSC/SSC, single, live cells (Figure 1A), PBMCs broadly segregate into T cells (CD3+CD20-), B cells (CD3-CD20+), and non-B/T cells (CD3-CD20-), the latter of which includes dendritic cells, natural killer cells, myeloid, and progenitor populations (Figure 1B; full gating strategy Online Table 3). To further define non-lymphoid phenotypes described in the Human Immunology Project, we included CD14, CD16, HLADR, CD56, CD123, and CD11c surface markers. First, CD14 and HLADR distinguish monocytes (CD14+HLADR+/−) and dendritic cells (CD14-HLADR+) from other granulocytes and NKs (CD14-HLADR-) (Figure 1C, Non-B/T). Within this latter NK/granulocyte population, CD123 expression denotes basophils and CD56 identifies natural killer cells (Figure 1C, NK/Granulocytes). NK cells further segregate into at least three populations according to CD56 and CD16 density (Figure 1C, NKs) [7]. Within the CD16-HLADR+ DC population, CD11c and CD123 distinguish plasmacytoid DCs and monocytic DCs (Figure 1C, DCs). Finally, within the CD14+ monocyte population, CD16 and HLADR identify at least three populations: classical monocytes (HLADR+CD16-), non-classical monocytes (HLADR+CD16+), and a subset containing myeloid derived suppressor cells (MDSCs; HLADR-CD16-) (Figure 1C, Monocytes). Of note, further analyses of chemokine receptor expression can be performed on any non-B/T subset, which may have particular relevance in diseased states (data not shown).
Figure 1.
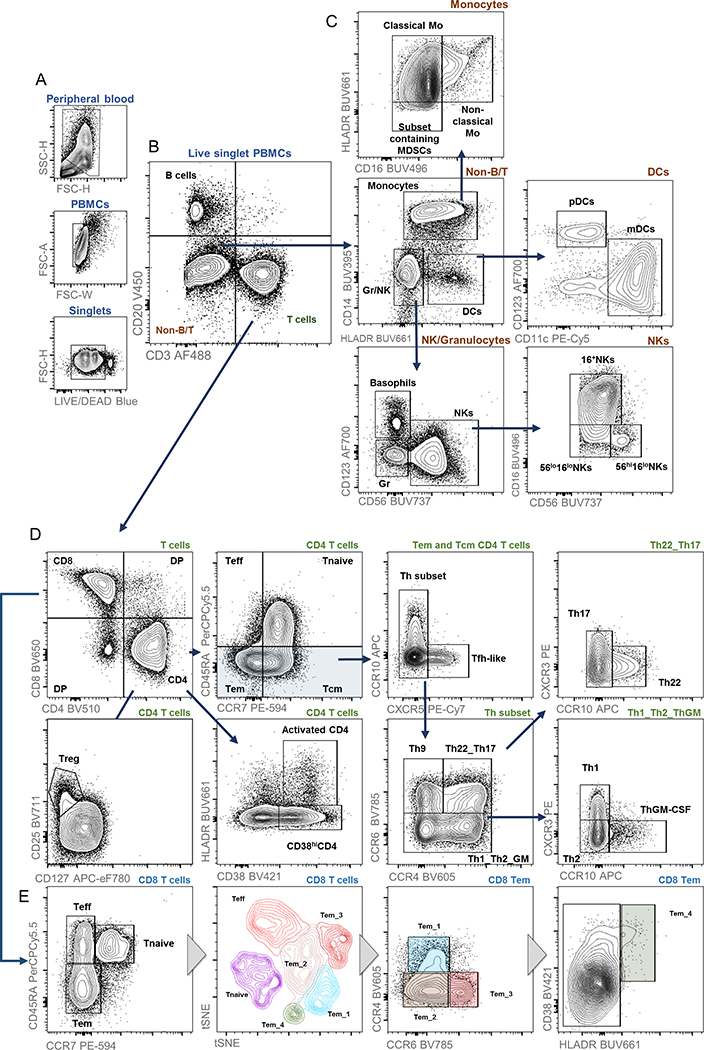
Example gating strategy for major immune cell subsets on stained PBMCs from healthy donors.
Basic T cell markers include CD4 and CD8 (Figure 1D, T cells). Next, a combination of cell surface markers, including multiple chemokine receptors, identifies T cell activation, T regulatory cells (Tregs), T cell memory status, and all major Th subsets [3, 4, 8, 9]. Of note, HLADR and CD38 expression identifies T cell activation status within any subset [10], with an example shown for all CD4+ cells. Within the CD4+ T cell population, Tregs identify as CD25+CD127-/lo, a population highly correlated with Tregs traditionally defined as FOXP3+ CD4+ [8, 11, 12]. CD45RA and CCR7 further define CD4 and CD8 T cells into four major subsets: T effector cells (Teff; CD45RA+CCR7-), naïve T cells (Tnaive; CD45RA+CCR7+), T effector memory cells (Tem; CD45RA-CCR7-), and T central memory cells (Tcm; CD45RA-CCR7+) (Figure 1D, second panel and Figure 1E, first panel). Within the CD4+ T memory population (i.e. all cells that are CD20-CD3+CD4+CD45RA-), various chemokine receptors distinguish Th1, Th2, Th9, Th22, a subset containing T follicular helper cells (Tfh), and T GM-CSF-secreting (ThGM-CSF) cells [4]. First, within the T memory population, CCR10 and CXCR5 expression identify the subset containing Tfh cells (CCR10-CXCR5+) (Figure 1D, Tem and Tcm CD4 cells). Within the CCR10-CXCR5-Th subset, Th9 cells can be identified as CCR6+CCR4-(Figure 1D, Th subset). Further gating on CCR6, CCR4, CXCR3, and CCR10 distinguishes the remaining Th subsets: Th1 (CXCR5-CCR6-CXCR3+CCR10-), Th2 (CXCR5-CCR6-CXCR3-CCR10-), Th17 (CXCR5-CCR6+CCR4+CXCR3-CCR10-), and Th22 (CXCR5-CCR6+CCR4+CXCR3-CCR10+) [3, 4] (Figure 1D, Th22_Th17 and The1_Th2_ThGM).
Although further subsets of CD8+ T cells are not rigorously defined, high-dimensional analysis with t stochastic neighbor embedding (tSNE) revealed differences in normal human PBMCs according to chemokine and Fc receptor expression (Figure 1E). In this example, tSNE discriminated distinct populations of CD8+ Tem cells, which on further examination, segregated according to CCR6, CCR4, and HLADR/CD38 expression. Interestingly, a single prior report has postulated this CD8+ CCR6+ Tem subset as a modulator of mucosal immunity [13], and another report identified CD8+CCR4+ cells as potential mediators of synovial inflammation in rheumatoid arthritis [14]. Thus, this high-color flow panel allows high-dimensional data visualization techniques to uncover unknown and/or poorly-defined cell types in both normal and diseased states.
In summary, our 21-color panel provides a powerful tool for in-depth analysis of lymphoid and myeloid cells in the human peripheral blood with deep T cell analysis and coverage of most populations defined in the NIH’s Human Immunology Project. Future panels could substitute certain T cell markers (e.g. CCR4, CXCR5, CCR10) in favor of increased B cell discrimination (e.g. CD19, CD27, IgD). Of note, by comparison to CyTOF, which can simultaneously detect 20–40 antigens, this panel requires fewer input cells, less acquisition time, and less money, while still permitting worthwhile high-dimensional analysis.
Human Subjects
Peripheral blood mononuclear cells were obtained from healthy donors. The use of human tissue in this study was approved by the Institutional Review Board at Washington University in St. Louis.
Similarity to Published OMIPs
This panel builds upon OMIPs −024, −015, and −030, which identify pan-leukocytes, T regulatory cells without intracellular staining, and all major T helper subsets, respectively. This single 21-color panel identifies the majority of subsets described in these three OMIPs, captures the major lymphoid and myeloid immunophenotypes defined in the NIH’s Human Immunology Project [3], and uniquely allows for detailed chemokine receptor analysis on non-B/T cell subsets.
Supplementary Material
Table 1.
| Purpose | Myeloid and lymphoid comprehensive immunophenotyping |
| Cell types | Human PBMCs |
| Cross-reference | OMIP-030, OMIP-015, OMIP-024 |
Table 2.
| Specificity | Fluorochrome | Ab Clone | Purpose |
|---|---|---|---|
| CD14 | BUV395 | MΦP9 | monocytes |
| Live/Dead | n/a | n/a | viability |
| CD16 | BUV496 | 3G8 | monocytes |
| HLADR | BUV661 | G46–6 | DCs |
| CD56 | BUV737 | NCAM16.2 | NKs |
| CD38 | BV421 | HIT2 | activation |
| CD20 | BV450 | L27 | B cells |
| CD4 | BV510 | SK3 | CD4 |
| CD194/CCR4 | BV605 | L291H4 | Th subset |
| CD8 | BV650 | RPA-T8 | CD8 |
| CD25 | BV711 | 2A3 | Treg |
| CD196/CCR6 | BV785 | G034 | Th subset |
| CD3 | AF488 | UCHT1 | T cells |
| CD45RA | PerCP-Cy5.5 | H1100 | naïve/memory |
| CD183/CXCR3 | PE | 1C6 | Th subset |
| CD197/CCR7 | PE-CF594 | 150503 | central/effector |
| CD11c | PE-Cy5 | Bly6 | mDCs |
| CD185/CXCR5 | PE-Cy7 | RF8B2 | Th subset |
| CCR10 | APC | 314305 | Th subset |
| CD123 | AF700 | 32703 | pDCs |
| CD127 | APC-eF780 | RDR5 | Treg |
Footnotes
The authors have no conflict of interest to declare.
References
- 1.Mahnke Y, Chattopadhyay P, and Roederer M Publication of optimized multicolor immunofluorescence panels. Cytometry A, 2010. 77A(9): p. 814–8. [DOI] [PubMed] [Google Scholar]
- 2.Finak G, et al. , Standardizing Flow Cytometry Immunophenotyping Analysis from the Human ImmunoPhenotyping Consortium. Sci Rep, 2016. 6: p. 20686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Maecker HT, McCoy JP, and Nussenblatt R, Standardizing immunophenotyping for the Human Immunology Project. Nat Rev Immunol, 2012. 12(3): p. 191–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wingender G and Kronenberg M, OMIP-030: Characterization of human T cell subsets via surface markers. Cytometry A, 2015. 87A(12): p. 1067–9. [DOI] [PubMed] [Google Scholar]
- 5.Cooke KR, et al. , The Biology of Chronic Graft-versus-Host Disease: A Task Force Report from the National Institutes of Health Consensus Development Project on Criteria for Clinical Trials in Chronic Graft-versus-Host Disease. Biol Blood Marrow Transplant, 2017. 23(2): p. 211–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zeiser R, Socie G, and Blazar BR, Pathogenesis of acute graft-versus-host disease: from intestinal microbiota alterations to donor T cell activation. Br J Haematol, 2016. 175(2): p. 191–207. [DOI] [PubMed] [Google Scholar]
- 7.Michel T, et al. , Human CD56bright NK Cells: An Update. J Immunol, 2016. 196(7): p. 2923–31. [DOI] [PubMed] [Google Scholar]
- 8.Mahnke YD, Beddall MH, and Roederer M, OMIP-015: human regulatory and activated T-cells without intracellular staining. Cytometry A, 2013. 83A(2): p. 179–81. [DOI] [PubMed] [Google Scholar]
- 9.Zhu J, Yamane H, and Paul WE, Differentiation of effector CD4 T cell populations (*). Annu Rev Immunol, 2010. 28: p. 445–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meditz AL, et al. , HLA-DR+ CD38+ CD4+ T lymphocytes have elevated CCR5 expression and produce the majority of R5-tropic HIV-1 RNA in vivo. J Virol, 2011. 85(19): p. 10189–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Seddiki N, et al. , Expression of interleukin (IL)-2 and IL-7 receptors discriminates between human regulatory and activated T cells. J Exp Med, 2006. 203(7): p. 1693–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu W, et al. , CD127 expression inversely correlates with FoxP3 and suppressive function of human CD4+ T reg cells. J Exp Med, 2006. 203(7): p. 1701–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kondo T, Takata H, and Takiguchi M, Functional expression of chemokine receptor CCR6 on human effector memory CD8+ T cells. Eur J Immunol, 2007. 37(1): p. 54–65. [DOI] [PubMed] [Google Scholar]
- 14.Cho BA, et al. , Characterization of effector memory CD8+ T cells in the synovial fluid of rheumatoid arthritis. J Clin Immunol, 2012. 32(4): p. 709–20. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


