Abstract
Predictive testing to characterise substances for their skin sensitisation potential has historically been based on animal models such as the Local Lymph Node Assay (LLNA) and the Guinea Pig Maximisation Test (GPMT). In recent years, EU regulations, have provided a strong incentive to develop non-animal alternatives, such as expert software systems. Here we selected three different types of expert systems: VEGA (statistical), Derek Nexus (knowledge based), TIMES-SS (hybrid) and evaluated their performance using two large sets of animal data, one of 1249 substances from eChemportal and a second of 515 substances from NICEATM. A model was considered successful at predicting skin sensitisation potential if it had at least the same balanced accuracy as the LLNA and the GPMT had in predicting the others’ outcomes, which ranged from 79% to 86%. We found that highest balanced accuracy of any of the expert systems evaluated was 65% when making global predictions. However, for substances within the domain of TIMES-SS, balanced accuracies were found to be 79% and 82%, for the two datasets. In cases where a chemical was within the domain of TIMES-SS, the TIMES-SS skin sensitisation hazard prediction had the same confidence as the result from the LLNA or the GPMT.
Keywords: (Q)SAR, expert system, TIMES-SS, VEGA, Derek Nexus, Skin Sensitisation
Introduction
Allergic contact dermatitis (ACD) is clinically defined as the presence of skin erythema and oedema that result specifically from delayed type IV T-cell-mediated immune skin hypersensitivity [1]. ACD is estimated to constitute about 10–15% of all occupational diseases [2]. In Europe, about 20% of the general population suffers from allergy to at least one contact allergen [3]. The clear social and economic impact of ACD is reflected by the requirement for skin sensitisation potential of a substance entering commerce to be assessed world-wide, the EU REACH regulation being one example [4]. Traditionally animal tests such as the Guinea Pig Maximisation Test (GPMT) and the Local Lymph Node Assay (LLNA) (described in OECD Test Guidelines 406 and 429 ([5, 6])) have been used to identify and characterise skin sensitising substances. However, in response to EU regulations overseeing both the cosmetic [7] and chemicals sectors [4], there has been a concerted effort to develop non-animal approaches and anchor these to key events in the associated adverse outcome pathway (AOP) for skin sensitisation [8–10]. Significant progress has been made in developing and evaluating non-animal test methods. Several methods have undergone OECD validation or have been included in the OECD Test Guideline work programme.[11] Under the EU REACH regulation [15] to address information requirements for the skin sensitisation endpoint, the following non-animal test methods are now considered acceptable: the direct peptide reactivity assay (DPRA) [8]; the KeratinoSens™ [9], the LuSens[12], the SENS-IS[13]; the human Cell Line Activation Test (h-CLAT) [11]; the U-SENS™ [14]; and the IL-8 Luc assay [11]. There are parallel efforts to develop and evaluate the utility of in silico models for skin sensitisation including local and global (Q)SARs as well as expert systems (as discussed in references [15] and [16]).
In this study, we evaluated the performance of three different expert systems: VEGA 1.1.4; Derek Nexus 5.0.2; and, TIMES-SS 2.27.19.13 using two large datasets of substances that had been assessed for their skin sensitisation potential in animal models. Similar evaluations have been performed in the past (e.g. [17, 18]), however they were based on much smaller datasets of up to 200 substances. Here we sought to exploit, the large amount of skin sensitisation information collected under the EU REACH regulation [4]. One dataset (comprising 1249 substances with 354 sensitisers and 895 non-sensitisers) was compiled by querying OECD’s eChemPortal (https://www.echemportal.org/) to identify substances that had been assessed for their skin sensitisation potential under REACH. A second dataset of 515 substances (329 sensitisers and 186 non-sensitisers) was taken from the NTP Interagency Center for the Evaluation of Alternative Toxicological Methods (NICEATM) LLNA database, a resource that had been collected as a reference set for the development and evaluation of non-animal approaches (https://ntp.niehs.nih.gov/pubhealth/evalatm/test-method-evaluations/immunotoxicity/index.html).
Three different types of expert systems were used: statistical, knowledge-based, and hybrid. The typenames ‘statistical’, ‘knowledge-based’, ‘hybrid’ have been discussed in more detail in [19]. The skin sensitisation model within the VEGA platform (https://www.vegahub.eu/) is an example of a statistical expert system, based on LLNA data from Gerberick et al. [20]. This model was developed under the EU funded project CAESAR (http://www.caesar-project.eu/) using an adaptive fuzzy partitioning algorithm based on eight descriptors. The algorithm assigns substances into two classes; sensitisers and non-sensitisers. An assessment of the applicability domain of the prediction was performed to provide a qualitative measure of reliability (low, moderate and good) [21]. The low, moderate, and good rankings are based on the structural similarity of the compound being predicted to compounds in the training set [21]. For a full description, the reader is referred to the model guide for skin sensitisation within VEGA [21].
Derek Nexus is an example of a knowledge based expert system, developed by Lhasa Ltd (www.lhasalimited.org). In version 5.0.2 of Nexus v2.1.1, there are over 850 alerts covering 72 different toxicological endpoints. An alert consists of a toxicophore, a substructure known or thought to be responsible for the toxicity alongside associated literature references, comments and examples. The skin sensitisation knowledge base in Derek Nexus was initially developed in collaboration with Unilever in 1993 using its historical database of GPMT data for 294 substances and contained approximately forty alerts [22]. Since that time, the knowledge base has undergone extensive improvements as more data and knowledge have become available [23, 24]. The current version contains about 89 alerts for skin sensitisation.
The TImes MEtabolism Simulator platform for predicting skin sensitisation (TIMES-SS) is an example of a hybrid commercial expert system that was developed by the Laboratory of Mathematical Chemistry (LMC) at University As. Zlatarov, Bulgaria using funding and data initially from a Consortium comprising industry and regulators [25]. TIMES-SS encodes structure-toxicity and structure-skin metabolism relationships through a number of transformations, some of which are underpinned by mechanistic 3D QSARs. It was developed on a training set of substances assessed for their skin sensitisation potential and potency from three main data sources – LLNA (several published sources, the largest of which was from Gerberick et al., [20]), GPMT [26] and the BgVV1 [27]. The BgVV data source comprised 264 substances evaluated by an expert group that was established by the BgVV in 1985. The expert group included dermatologists from universities, chemical industry, and regulatory authorities. The group collected and evaluated data from the literature on substances with documented contact allergenic properties in humans (from clinical data and experimental studies) and animals. The evaluation listed chemicals as belonging to one of three categories (A–C): Category A represented significant contact allergens; B, a solid-based indication for contact allergenic potential; and C, insignificant or questionable contact-allergenic potential [27]. A unifying potency scale derived from these 3 data sources results in TIMES-SS predicting skin sensitisation potency as significant, weak, or non-sensitising. Characterisation and evaluation of TIMES-SS with respect to the OECD Validation Principles for (Q)SARs is described in more detail in references [22] and [26]. An updated review of TIMES-SS since then have been summarised in Patlewicz et al., [28].
Materials and Methods
Skin sensitisation Dataset construction
Two datasets of chemicals that had been assessed for their skin sensitisation potential in animal models were constructed. One dataset exploited the large amount of skin sensitisation information submitted under REACH [4] that have been made publicly available on the European Chemical Agency (ECHA) website (https://echa.europa.eu/) and which is searchable through the OECD eChemPortal (https://www.echemportal.org/). The REACH data comprises information submitted by industry to satisfy their registration requirements, hence it is subject to limited quality control by ECHA prior to publication (only 5% of dossiers within each tonnage are evaluated for technical compliance, see https://echa.europa.eu/regulations/reach/evaluation/compliance-checks). In view of this, a second dataset extracted from the NICEATM LLNA Database was used to compare the performance characteristics derived, from the ECHA data.
eChemPortal skin sensitisation dataset
OECD’s eChemPortal (http://www.echemportal.org/echemportal/index.action) was used to search for substances with reported in vivo skin sensitisation experimental outcomes. The chemical property data search option was selected to query for skin sensitisation information. The search parameters were as follows: ‘Study result type’ was set to experimental, ‘reliability’ was set to 1 or 2 (where reliability is defined by the criteria described by Klimisch et al. [29], ‘year’ was set to greater than 1, ‘type of study’ was set to GPMT or LLNA and, ‘interpretation of results’ was set to ‘sensitising’ or ‘not sensitising’. All other search criteria were left empty. The results generated were exported as a csv file. This was processed for subsequent analysis using a python script which is available in the supplemental information.
The eChemportal search results contained some duplicates which were removed from the dataset. A result was considered a duplicate if it had the same CAS number, the same test type (LLNA or GPMT), and the same result (sensitising or not sensitising). We could not determine if these were duplicates or additional studies on the same compound of the same type with the same results. This reduced the set from 3622 entries to 2795. Since CAS numbers were used to systematically search for structures within the US EPA Chemistry Dashboard (https://comptox.epa.gov/dashboard), all substances that did not have a CAS number were removed from the dataset, resulting in 2574 unique substances. The resulting set of CAS numbers were queried within the Chemistry Dashboard using the ‘Batch Search’ feature in order to download a tsv file of the substances with their associated molecular formulas. Any substances which were not matches based on a CAS search in the Dashboard (i.e. did not form part of the 747,000 inventory of chemicals) were removed from the dataset, leaving 1587 compounds. Substances that were organometallic or inorganic were also excluded since the expert systems being evaluated are not able to make reliable predictions for these types of substances, further reducing the set to 1348 substances. These were identified by removing substances that had molecular formulas which included elements other than the following: C, H, O, N, F, Cl, Br, I, S, P, Ca, Na, K. The remaining unique CAS numbers were used to query the Chemistry dashboard to download a structure data file (sdf). The structures were desalted (e.g. ions like Na+ were removed) and converted into a compatible sdf format (V2000) using Spectrus DB (ACD Labs) for processing within the three expert systems. ACD Lab’s Spectrus DB was used to check for duplicate structures. Duplicate structures can occur because a single chemical may be associated with multiple CAS numbers. The final dataset used for the performance evaluation (referred herein as the eChemPortal dataset) comprised 1295 unique structures with associated skin sensitisation potential data. See figure 1 for a diagram of how the eChemportal data used in the evaluation was selected.
Figure 1.
Schematic for processing data from eChemportal.
NICEATM LLNA Database
A set of 515 organic substances all with Simplified Molecular Input Line Entry System (SMILES) codes taken from the NICEATM LLNA Database (https://ntp.niehs.nih.gov/pubhealth/evalatm/test-method-evaluations/immunotoxicity/index.html) was supplied by NICEATM scientists (personal communication). The SMILES were converted to a V2000 sdf using the online CACTUS translator (https://cactus.nci.nih.gov/translate/), for processing within Derek Nexus because it would not process the SMILES file. This NICEATM dataset comprised 329 sensitisers and 186 non-sensitisers.
VEGA
Predictions of skin sensitisation were made with CAESAR (v. 2.1.6 within VEGA version 1.1.4) and exported out as a text (.txt) file. Substances from the eChemPortal dataset were loaded in CAESAR using the sdf file, for details on how the sdf file was created, see the eChemPortal skin sensitisation dataset section. The NICEATM dataset was loaded into CAESAR using the available SMILES codes.
Derek Nexus
Predictions of skin sensitisation potential were made in batch mode using Derek Nexus v.5.0.2 within the Nexus v.2.1.1 platform. Structures from the eChemportal and NICEATM datasets were first loaded into Derek Nexus by importing the sdf files of each. Under the prediction tab in Nexus “Derek Prediction”, “Derek Batch Setup…” was selected to customise the prediction settings as follows: mouse and guinea pig were selected as species; and endpoint predictions were limited to skin sensitisation. The batch predictions were exported as tsv files. Under the “Report Display Options” tab “Show predictions of at least” was set to “IMPOSSIBLE”. All other settings were left as default. Although there is a model to predict LLNA sensitisation potency predictions (i.e. EC3 values) within Derek Nexus, this functionality was not used as part of this study.
TIMES-SS
Skin sensitisation potency predictions were made using the Skin Sensitisation with Autoxidation v. 21.26 model within TIMES-SS v2.27.19.13. However only the prediction of potential was used. Substances from the eChemPortal dataset were loaded into TIMES-SS from the sdf file created whereas the NICEATM dataset was loaded using the available SMILES codes. Results were exported by selecting “Summary” under the “Report” tab. The settings were left to default for the export results files which were subsequently saved as text (.txt) files.
Performance assessment of data underlying the eChemportal set
The performance of the GPMT predicting the LLNA and vice versa was assessed using a subset of 84 substances from the eChemPortal dataset where there was a reported GPMT and LLNA study for the same substance. A confusion matrix (an example is in table 1) was constructed from which the accuracy, sensitivity, specificity and balanced accuracy were calculated. The metrics balanced accuracy, accuracy, sensitivity, specificity, positive predictive value, negative predictive value are calculated as follows:
Table 1.
An example confusion matrix.
| Actual | |||
|---|---|---|---|
| Sensitising | Non-Sensitising | ||
| Predicted | Sensitising | True Positives (TP) | False Positives (FP) |
| Non-Sensitising | False Negatives (FN) | True Negatives (TN) | |
The positive predictive value and the negative predictive value are reported in the supplemental any time the other metrics are presented in the manuscript.
Performance assessment of the expert systems
The performance characteristics of the three expert systems were assessed by creating a confusion matrix and then calculating the accuracy, sensitivity, specificity and balanced accuracy for the two datasets individually. This assumed a binary outcome – i.e. sensitising or non-sensitising. VEGA’s skin sensitisation model has been trained to provide a binary outcome with a confidence metric. Derek Nexus identifies alerts and assigns a level of confidence to the prediction from nine possible options starting from: certain, probable, plausible, equivocal, doubted, improbably, impossible, open, through to contradicted. Substances that were predicted as plausible or higher were considered for the purposes of this evaluation as sensitisers. Equivocal substances were considered indeterminate predictions whereas a substance featuring no alert was considered a non-sensitiser. No alert could also be indicative of ‘no knowledge’ but for the purposes of this evaluation, it was interpreted as non-sensitising. There were no substances reported by Derek Nexus as doubted, improbable, impossible, open or contradicted. The TIMES-SS skin sensitisation model predicts a semi-quantitative potency which was converted into a binary outcome, by considering all weak and strong sensitisers to be sensitisers.
Substances’ belonging to the training sets of any of the expert systems (as far as this could be determined), were excluded from the performance comparison. This was straightforward to do in the case of the TIMES-SS and VEGA since both systems provide an indication of whether a substance is part of the underlying training set. In the case of Derek Nexus, some assumptions were made since the underlying training sets underpinning each of the alerts are not provided. Example chemicals that are included to illustrate the scope of a given alert are provided as part of the mechanistic justification for an alert but are additionally assigned a higher confidence level by the reasoning engine when a prediction is made. If a prediction resulted in a “certain” or “probable” level of confidence, it was assumed that experimental data was available within the Derek Nexus knowledge base for that substance and therefore it likely formed part of the underlying training set.
Results
Skin sensitisation datasets
The search of skin sensitisation data collected under REACH using the OECD eChemPortal resulted in 4189 non-unique records. Removal of duplicates left 2795 unique records linked to 2575 unique substances. Structures for 1587 substances were retrieved by searching the US EPA Chemistry Dashboard by CAS number, of the ~1000 we did not retrieve a structure for, most were mixtures or the CAS number was not linked to a specific substance in DSSTox. It should be noted that no searching was conducted by substance name nor was any curation performed to verify the association between a given CAS number and a given substance name. After removal of inorganic and organometallic substances, the resulting dataset comprised 1295 unique structures with in vivo skin sensitisation data. This set consisted of 354 sensitisers, 895 non-sensitisers and 46 substances with conflicting results between LLNA and GPMT. Table 2 summarises the comparison of the GPMT and LLNA outcomes for the 1285 substances with unique structures. For example, there were 174 cases where the only LLNA results reported were in agreement with each other, whereas there were 37 cases where the GPMT results were in conflict with the LLNA results. A comparison table of the substances that had 3 conflicting outcomes is provided in the supplemental information.
Table 2.
1285 substances from eChemPortal with LLNA and GPMT results are compared against each other.
| Sensitising LLNA result | Non-sensitising LLNA result | Sensitising GPMT result | Non-Sensitising GPMT result | |
|---|---|---|---|---|
| Sensitising LLNA result | 174 | |||
| Non-Sensitising LLNA result | 8 | 385 | ||
| Sensitising GPMT result | 37 | 3 | 143 | |
| Non-Sensitising GPMT result | 9 | 35 | 16 | 475 |
Metrics to compare the predictive performance of the LLNA relative to the GPMT are provided in Table 3. The performance metrics of the animal models served as a reasonable and convenient benchmark from which to compare the performance of the three expert systems. When making the comparisons of the expert systems to the animal test results, we used the highest result for the animal values predicting one another. For example, if the balanced accuracy of the LLNA predicting the GPMT was higher than the balanced accuracy of the GPMT predicting the LLNA we used the greater value as our bench mark.
Table 3.
Performance metrics of the LLNA and GPMT against each another. Metrics taking into account the 10 additional records which had three conflicting outcomes are shown in parenthesis. An example of a record with three conflicting results would be a compound with sensitising and non-sensitising LNNA results and a non-sensitising GPMT result.
| Accuracy | Balanced Accuracy | Sensitivity | Specificity | |
|---|---|---|---|---|
| LLNA is predictive of GPMT | 86% (79%) | 86% (79%) | 93% (86%) | 80% (72%) |
| GPMT is predictive of LLNA | 86% (79%) | 86% (80%) | 80% (75%) | 92% (84%) |
The NICEATM LLNA dataset contained 515 substances, with 329 sensitisers and 186 non-sensitisers. Seventy-three substances overlapped between the eChemPortal and NICEATM datasets. The majority of the seven-three substances that the two sets had in common may well have originated from the same study but we could not verify this entirely given the lack of meta data provided in the NICEATM dataset. The eChemportal dataset used for the analysis comprised 1249 substances. The two datasets were not combined because it could not be assured that they were of the same quality due to the fore mentioned lack of meta data.
Performance of VEGA, Derek Nexus, and TIMES-SS
Table 4 provides an overview of the predictive performance of VEGA, Derek Nexus, and TIMES-SS sensitisation predictions for the eChemPortal and NICEATM datasets. Substances that formed part of the training set of any of the models had been excluded from this performance comparison. This reduced the eChemPortal dataset to 903 substances, with 220 sensitisers and 683 non-sensitisers and the NICEATM set to 180 substances, with 85 sensitisers and 95 non-sensitisers. The reduced number of substances in each set meant that there were only 13 common substances between the two datasets.
Table 4.
Performance characteristics for the eChemPortal and NICEATM datasets excluding any training set substances.
| eChemPortal Substances | NICEATM Substances | |||||||
|---|---|---|---|---|---|---|---|---|
| Accuracy | Balanced Accuracy | Sensitivity | Specificity | Accuracy | Balanced Accuracy | Sensitivity | Specificity | |
| VEGA | 44% | 56% | 80% | 32% | 57% | 58% | 76% | 40% |
| Derek Nexus | 71% | 65% | 53% | 76% | 61% | 61% | 55% | 66% |
| TIMES-SS | 67% | 63% | 55% | 71% | 61% | 61% | 65% | 57% |
In the eChemPortal dataset, 182 substances were predicted correctly by all three models whereas 67 substances were predicted incorrectly. In the NICEATM dataset, 43 substances were predicted correctly by all three models and 16 substances were predicted incorrectly. The substances predicted incorrectly every time by all three systems are discussed in more detail below.
Overall, Derek Nexus and TIMES-SS had a higher accuracy and balanced accuracy than VEGA on both datasets. The sensitivity of VEGA was much higher than either TIMES-SS or Derek Nexus but it had a much lower specificity on account of its many false positives. The performance metrics were also assessed when the applicability domain of the expert systems were accounted for (see Table 5). Since Derek Nexus does not have a structural applicability domain characterised for its entire sensitisation knowledge base, it was not included in Table 5. A substance was considered within the domain of the TIMES-SS model if it fell within the same four subdomains (general parametric requirements, structural domain, mechanistic domain, and interpolation space) as the correctly predicted substances within the training set [30]. The accuracy of TIMES-SS was much improved when predictions were restricted to substances lying within its structural applicability domain. For VEGA, the low, moderate, and good rankings are based on the structural similarity of the substance of interest being compared to substances in the training set. For a full description, see the model guide for the skin sensitisation endpoint within VEGA [21]. A marked increase was noted in the sensitivity for VEGA predictions with ‘good reliability’, but the specificity decreased even further when the domain was not accounted for. A complete assessment of how VEGA, Derek Nexus, and TIMES-SS performed individually on the two datasets is provided in the supplemental information.
Table 5.
Results from TIMES-SS and VEGA on substances which are not in their training sets. Derek Nexus is excluded from the table due to its lack of domain. Substances within the domain of TIMES-SS are those which have fragments completely contained in the model. The low, moderate, and good rankings are based on the structural similarity of the substance being predicted to substances in the training set [21].
| eChemPortal Chemicals | NICEATM Chemicals | |||||||
|---|---|---|---|---|---|---|---|---|
| Accuracy | Balanced Accuracy | Sensitivity | Specificity | Accuracy | Balanced Accuracy | Sensitivity | Specificity | |
| TIMES-SS | ||||||||
| In Domain | 80% | 79% | 75% | 82% | 85% | 82% | 86% | 77% |
| Out of Domain | 66% | 63% | 57% | 69% | 58% | 58% | 63% | 53% |
| VEGA | ||||||||
| Good | 40% | 59% | 98% | 20% | 71% | 66% | 91% | 40% |
| Moderate | 31% | 51% | 82% | 19% | 62% | 54% | 96% | 11% |
| Low | 51% | 57% | 71% | 42% | 55% | 55% | 75% | 36% |
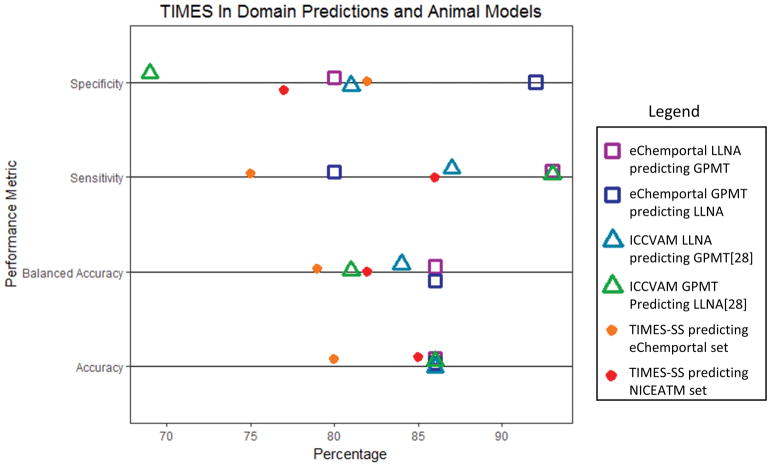
When comparing the results of VEGA, Derek Nexus, and TIME-SS, to the results of the animal data we find that only the in domain predictions of TIMES-SS in table 5 are similar to the results of the animal test predicting one another in table 3. This is illustrated in figure 2 where the accuracy, balanced accuracy, sensitivity, and specificity of the TIMES-SS predictions are compared to the same metrics of GPMT predicting the LLNA and vice versa.
Figure 2.
Results of TIMES-SS compared to animal studies.
Discussion
Currently the most widely accepted animal test for assessing the sensitising potential of a substance is the LLNA, which has largely replaced the GPMT, at least in the EU.[4] Since these two tests are considered to be the most comprehensive means of predicting the sensitising potential of chemicals, their balanced accuracies were used as a benchmark for comparison with the performance of the 3 expert systems being evaluated.
LLNA and GPMT data
A comparison of the results of the LLNA to the GPMT and vice versa resulted in an accuracy and balanced accuracy of 86% (shown in table 3). These metric values reduced to ~79% when accounting for 3 different outcomes e.g. a substance tested with a sensitising and a non-sensitising outcome in the LLNA and with a sensitising outcome in the GPMT. Substances with three different outcomes disproportionately affected the confusion matrix, hence the metrics were calculated with and without them. The performance metrics derived were comparable to a study performed by Interagency Coordinating Committee on the Validation of Alternative Methods (ICCVAM) who reported an accuracy of 86% for the GPMT and LLNA predicting one another on the basis of a dataset of 126 substances, a balanced accuracy of 84% for the GPMT predicting LLNA results and a balanced accuracy of 81% for the LLNA predicting the GPMT [31]. Based on our results and the previously published ICCVAM results, a balanced accuracy range of 79% to 86% was used as a benchmark for the expert systems.
Global performance assessment
The highest balanced accuracy (65%) was found for Derek Nexus on the basis of the eChemPortal dataset. However, this was still 14 points lower than the balanced accuracies when comparing the 2 animal tests. Amongst the expert systems themselves, Derek Nexus and TIMES-SS performed significantly better than VEGA. VEGA is the most conservative in its predictions, as evidenced by its high sensitivities and very low specificities. This is not altogether surprising since the VEGA developers rationalised all of the incorrect test set predictions as false positives [21].
Analysis of incorrect predictions from the 3 systems
There were 67 substances from the eChemportal data set that were incorrectly predicted by all 3 models (Table 6). The alerts from TIMES-SS and Derek Nexus are provided in each case along with suggested rationales on why the predictions were incorrect. There were 16 substances from the NICEATM data set that were consistently incorrectly predicted by all three expert systems (Table 7). Rationales are proposed to account for the incorrect predictions in each case.
Table 6.
Analysis of substances from the eChemportal set that were predicted incorrectly by all three in silico models, the justification for the TIMES-SS and Derek predictions are given, VEGA does not give a mechanistic description. The authors’ proposed rationales for the incorrect predictions are also given. Mechanistic domains – Michael Acceptor (MA), Schiff Base former (SB), Acyl transfer agent, SN2 etc. make reference to the reaction pathways described in Aptula and Roberts [33]
| Structure | DSSTox Substance Identifier (DTXSID) |
CASRN | Result | TIMES-SS | Derek Nexus | Author proposed rationales/comments |
|---|---|---|---|---|---|---|
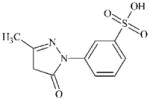
|
DTXSID0044785 | 119-17-5 | LLNA-NS | Pyrazolones and pyrazolidinones | Hydrazine or precursor | Hydrazine assignment appears incorrect. TIMES-SS alert is more likely but probably deactivated by the SO3H. |
|
|
DTXSID4020371 | 80-08-0 | LLNA-NS | Metabolite C-Nitroso compounds | Aromatic primary or secondary amine | NH2 oxidation deactivated by the SO2 moiety hence neither alert seems plausible. |
|
|
DTXSID5044784 | 1189-08-8 | GPMT-NS | alpha, beta-Carbonyl compounds with polarised double bonds | alpha, beta-Unsaturated ester or precursor | Not a sufficiently reactive Michael acceptor (MA). Substance would tend to be a borderline sensitiser or non-sensitiser. Probably polymerises upon air exposure. |
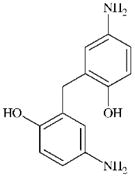
|
DTXSID10181758 | 27311-52-0 | GPMT-NS | Quinone methide(s)/imines, Quinoide oxime structure, Nitroquinones, | Ortho or para amino- or hydroxy-aniline | Unexpected outcome. On paper, the quinone imine could de-activate itself by intramolecular addition of the OH group to the C=C-C=O of the oxidised ring. However, based on the GPMT results, this substance was found to be an irritant which may account for the lack of sensitisation observed. Test substance was treated at 10% and 25% induction concentrations. Substance was irritating at 25% causing erthymea, oedema, necrotising dermatitis, encrustation and black discolouration. |
|
|
DTXSID7029168 | 141-10-6 | GPMT-NS | alpha, beta-Carbonyl compounds with polarised double bondsMetabolite: Di-substituted a,b-unsaturated aldehydes | alpha, beta-Unsaturated ketone or precursor | Not sufficiently reactive as a MA. Deactivated by the 6-Me group. A soft nucleophile (Nu) such as cysteine may preferentially attack at the 6 position which has the Me substituent. |
|
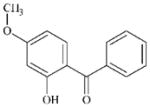
DTXSID3022405 | 131-57-7 | GPMT-LLNA-NS | Aromatic carbonyl compounds | Resorcinol or precursor | Similar to 190965-45-8. Would require demethylation of the OMe and then activation of the resorcinol which is deactivated by the COAr group. |
|
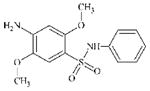
DTXSID2044686 | 52298-44-9 | LLNA-NS | Metabolite: Quinone methide(s)/imines, Quinoide oxime structure, Nitroquinones, | Aromatic primary or secondary amine | SO2NH deactivates Ar-NH2 activation. |
|
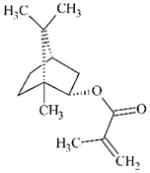
DTXSID9027653 | 7534-94-3 | GPMT-NS | alpha, beta-Carbonyl compounds with polarised double bonds | alpha, beta-Unsaturated ester or precursor | Insufficiently reactive as MA. |
|
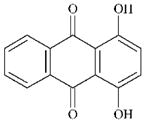
DTXSID8044464 | 81-64-1 | GPMT-NS | Aromatic carbonyl compoundsMetabolite: Quinone methide(s)/imines, Quinoide oxime structure, Nitroquinones, | Hydroquinone or derivative | Unexpected outcome. Would expect it to sensitise by oxidation of the p-diOH to the quinone structure in the right hand side of the ring which would make for a good MA. Oxidation might be disfavoured by the dicarbonyl groups. |
|
|
DTXSID2028303 | 67124-09-8 | LLNA-S | No Alert | No Alert | Not obvious. A possible explanation may be oxidation of the sulphide to sulphoxide or sulphone which can then lose water to give a reactive MA RSO2CH=CHMe. The reported EC3 value was 16% (a weak sensitiser) but the test substance actually comprised a mixture of branched forms of the sulphide structure which may account for the discrepancy.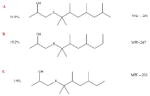
|
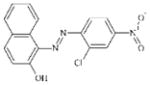
|
DTXSID7044637 | 2814-77-9 | LLNA-NS | Metabolite: Quinone methide(s)/imines, Quinoide oxime structure, Nitroquinones, | Aromatic azo compound | Skin metabolism not sufficiently significant. Would be surprising to be more than a weak sensitiser on account of this pathway as N=N reductive cleavage is bacterial. |
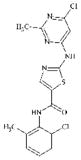
|
DTXSID20460723 | 302964-08-5 | LLNA-NS | Activated aryl and heteroaryl compounds | Activated N-heterocycle | Unexpected, potentially deactivated by hydrolysis? |

|
DTXSID6074332 | 895-85-2 | GPMT-NS | Diacyl peroxides, anhydrides (sulphur analogues of diacyl peroxides) | Acyl peroxide | Unexpected experimental outcome in view of other diacyl peroxides. |
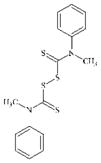
|
DTXSID90147413 | 10591-84-1 | LLNA-NS (non standard LLNA called the IMDS – integrated model for the differentiation of skin reactions. Cell counts are used in lieu of radiolabels. The threshold for a positive outcome is a cell count index of 1.4 or greater. Cell count indices for the test substances did not reach or exceed 1.4 when tested up to 50% concentration | Thiols and disulfide compounds | Thiuram mono- or di-sulphide | Unexpected experimental outcome in view of other disulphides. |
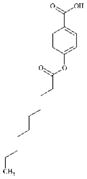
|
DTXSID60394134 | 86960-46-5 | GPMT-LLNA-NS | Activated aryl esters | Phenyl ester | Weak acyl transfer agent – relatively low reactivity and surface active |
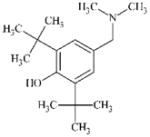
|
DTXSID0044997 | 88-27-7 | LLNA-S | No Alert | No Alert | Unexpected experimental result, possible free radical mechanism. Sterically constrained OH loses a H resulting in a radical which can lose another H from CH2NMe2 giving a quinone-methide. |
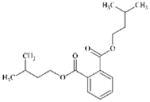
|
DTXSID80209236 | 605-50-5 | LLNA-S | No Alert | No Alert | Possibly a false positive (FP). |
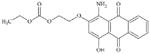
|
DTXSID5068235 | 40530-60-7 | LLNA-NS | Aromatic carbonyl compoundsMetabolite: Quinone methide(s)/imines, Quinoide oxime structure, Nitroquinones, | Resorcinol or precursor | Could form quinone-imine by oxidation but this is disfavoured by the electronegative substituents (CO and ether groups). |
|
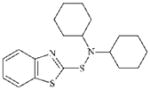
DTXSID3027584 | 4979-32-2 | GPMT-NS | Heteroarene sulfenamidesMetabolite: Thiols and disulfide compounds | Thiol or thiol exchange agent | Not sufficiently reactive. |
|
|
DTXSID9052343 | 6600-31-3 | LLNA-NS | Metabolite: Hydroperoxides | Alkyl aldehyde precursor | Could hydrolyse to cyclohex-3-ene-1-aldehyde but ring structure of the di-acetal would make this more resistant and the aldehyde would not be particularly potent. NS outcome is not surprising. |
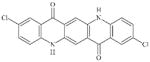
|
DTXSID1029253 | 3089-17-6 | GPMT-LLNA-NS | Metabolite: Generated free radicals | Ortho or para amino- or hydroxy-aniline | NH groups are more amide like so oxidation of the central ring to a di-imine is not likely to happen. |
|
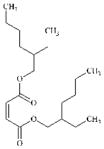
DTXSID2027094 | 142-16-5 | LLNA-NS | alpha, beta-Carbonyl compounds with polarised double bonds | alpha, beta-Unsaturated ester or precursor | False negative in the LLNA. Unbranched isomer dioctyl maleate is a sensitiser in man and in the GPMT. |
|
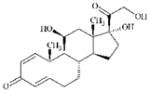
DTXSID9021184 | 50-24-8 | LLNA-NS | alpha, beta-Carbonyl compounds with polarised double bonds | 1,2-Dicarbonyl compound or precursor | Skin metabolism is not significant in the mouse but a similar reaction can occur in guinea pigs and man [32]. |
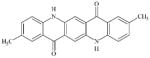
|
DTXSID2052655 | 980-26-7 | GPMT-LLNA-NS | Metabolite: Quinone methide(s)/imines, Quinoide oxime structure, Nitroquinones, | Ortho or para amino- or hydroxy-aniline | NH groups are more amide like so oxidation of the central ring to a di-imine is not likely to happen. |
|
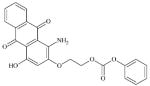
|
DTXSID60403376 | 106461-41-0 | GPMT-NS | Metabolite: Quinone methide(s)/imines, Quinoide oxime structure, Nitroquinones, | Hydrazine or precursor | Unexpected result. Would have expected possible oxidation of the bottom ring to a quinone imine (quaternised) structure. |
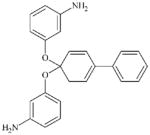
|
DTXSID8067361 | 28173-59-3 | LLNA-NS | Aromatic carbonyl compoundsMetabolite: Quinone methide(s)/imines, Quinoide oxime structure, Nitroquinones, | Phenyl carbonate | Phenyl ester is an acyl transfer agent but not very reactive. |
|
DTXSID70668237 | 105112-76-3 | LLNA-NS | Metabolite: Quinone methide(s)/imines, Quinoide oxime structure, Nitroquinones, | Aromatic primary or secondary amine | This will hydrolyse to 4-phenylphenol (unreactive) and meta amino phenol which needs further activation. Probably just not potent enough to register in the LLNA. |
|
|
DTXSID1059319 | 105-74-8 | GPMT-NS | Diacyl peroxides, anhydrides (sulphur analogues of diacyl peroxides) | Acyl peroxide | Unexpected result. Could be a bioavailability issue given how ultra hydrophobic this is. |
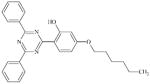
|
DTXSID4051299 | 147315-50-2 | GPMT-NS | Metabolite: Polarised alkene - alkenyl pyridines, pyrazines, pyrimidines or triazines | Resorcinol or precursor | Would need de-alkylation of the OR group to give a resorcinol. When R>Me this doesn’t happen to a significant extent. |
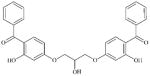
|
DTXSID20361327 | 23911-85-5 | GPMT-NS | Aromatic carbonyl compounds | Resorcinol or precursor | Cleavage of the aromatic ether groups would be needed for resorcinol formation and this is only really significant for OMe. |
|
|
DTXSID5062475 | 2500-88-1 | LLNA-NS | Thiols and disulfide compounds | Thiol or thiol exchange agent | Probably too hydrophobic. |
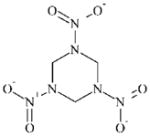
|
DTXSID9024142 | 121-82-4 | GPMT-NS | Aldehydes | Formaldehyde donor | Probably too hydrophilic and not reactive enough. |
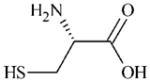
|
DTXSID8022876 | 52-90-4 | LLNA-NS | Thiols and disulfide compounds | Thiol or thiol exchange agent | SH would need to ionise for it to be reactive and this is unlikely as CO2H will ionise preferentially. |
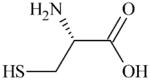
|
DTXSID0020367 | 52-89-1 | LLNA-NS | Thiols and disulfide compounds | Thiol or thiol exchange agent | SH would need to ionise for it to be reactive and this is unlikely as CO2H will ionise preferentially. |

|
DTXSID1044853 | 17766-26-6 | GPMT-NS | Metabolite: Azlactones and unsaturated lactone derivatives | Thiourea | Not reactive enough, particularly in Na salt form and not hydrophobic enough (acyl transfer logP-dependent). |
|
|
DTXSID4041107 | 110-60-1 | GPMT-NS | Aldehydes | Diamine | Apparent unexpected result but if QMM for SB is used (Roberts et al 2006)[33] an EC3 >100 is predicted which would equate with a non-sensitising outcome. |
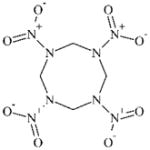
|
DTXSID3024237 | 2691-41-0 | GPMT-NS | Aldehydes | Formaldehyde donor | Probably insufficiently reactive. |
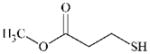
|
DTXSID6027509 | 2935-90-2 | LLNA-NS | Thiols and disulfide compounds | Thiol or thiol exchange agent | Would need the SH to ionise for nucleophilic sensitisation, but not acidic enough (1-C further away from CO than cysteine SH is. |
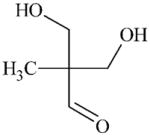
|
DTXSID4066381 | 18516-18-2 | LLNA-NS | Aldehydes | Aldehyde | LogP is too low for significant potency per QMM in Roberts et al (2006)[33]. |
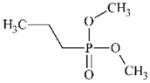
|
DTXSID0066406 | 18755-43-6 | LLNA-NS | Phosphonates | Alkyl ester of phosphoric or phosphonic acid | Phosphonate may be too poor a leaving group, hence insufficiently reactive. |
|
|
DTXSID50721910 | 38172-91-7 | LLNA-NS | Epoxides, Aziridines and Sulfuranes | Epoxide | Not reactive/hydrophobic enough. |
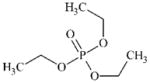
|
DTXSID8026228 | 78-40-0 | LLNA-NS | (Thio)Phosphates | Alkyl ester of phosphoric or phosphonic acid | Phosphate may be too poor a leaving group, hence insufficiently reactive. |
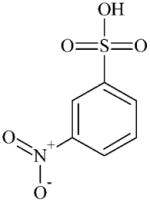
|
DTXSID2027048 | 127-68-4 | GPMT-S | No Alert | No Alert | Surprising outcome. Impurities in the substance tested could be implicated. |
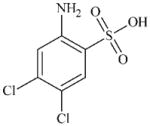
|
DTXSID5074921 | 6331-96-0 | LLNA-NS | No Alert | Aromatic primary or secondary amine | Would expected SO3H to deactivated NH2 oxidation |
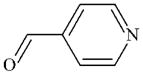
|
DTXSID1061237 | 872-85-5 | LLNA-S | No Alert | No Alert | Schiff Base (SB) formation is possibly activated sufficiently by the ring N. |
|
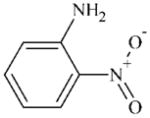
DTXSID1025726 | 88-74-4 | GPMT-LLNA-NS | Metabolites: Quinone methide(s)/imines, Quinoide oxime structure, Nitroquinones, | Aromatic primary or secondary amine | NO2 should deactivate NH2 oxidation. |
|
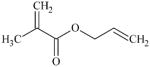
DTXSID2021816 | 96-05-9 | GPMT-NS | alpha, beta-Carbonyl compounds with polarized double bonds | alpha, beta-Unsaturated ester or precursor | MA too unreactive to occur. |
|
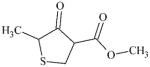
DTXSID70505936 | 66319-07-1 | LLNA-S | No Alerts | No Alerts | Maybe the ketone group is sufficiently activated by the CO2Me and ring S groups to be an SB electrophile. |
|
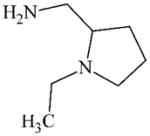
DTXSID3057698 | 26116-12-1 | LLNA-NS | Metabolite: Aldehydes | Diamine | Possibly multiple amine oxidation to give rise to a reactive SB. |
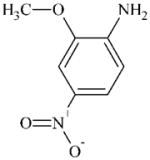
|
DTXSID0038700 | 97-52-9 | GPMT-NS | Metabolite: Quinone methide(s)/imines, Quinoide oxime structure, Nitroquinones, | Aromatic primary or secondary amine | Single aromatic amino isn’t a “confident alert” – afterall aniline is very borderline. Demethylation of the ortho (to NH2) OMe group would give an ortho aminophenol alert for strong sensitiser (via further oxidation). Demethylation isn’t a “confident alert” and in this case could be disfavoured by the electronegative meta NO2 group. |
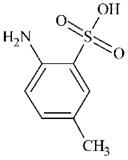
|
DTXSID7026526 | 88-44-8 | LLNA-NS | Metabolite: Quinone methide(s)/imines, Quinoide oxime structure, Nitroquinones, | Aromatic primary or secondary amine | As for 97-52-9. |
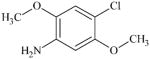
|
DTXSID9040718 | 6358-64-1 | GPMT-NS | Metabolite: Quinone methide(s)/imines, Quinoide oxime structure, Nitroquinones, | Aromatic primary or secondary amine | As for 97-52-9. |
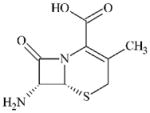
|
DTXSID4057687 | 22252-43-3 | LLNA-NS | beta-Lactams | Ring-strained amide, ester, thioamide or thioester | Probably not hydrophobic enough – acyl domain is logP-dependent. Penicillin, which contains this sub-structure is more hydrophobic, yet was only weakly sensitising in the LLNA [34]. |
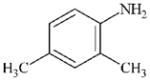
|
DTXSID8026305 | 95-68-1 | LLNA-NS | Metabolite: Quinone methide(s)/imines, Quinoide oxime structure, Nitroquinones, | Aromatic primary or secondary amine | As for 97-52-9. |
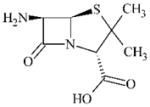
|
DTXSID7046097 | 551-16-6 | LLNA-NS | beta-Lactams | Ring-strained amide, ester, thioamide or thioester | As for 22252-43-3. |
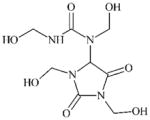
|
DTXSID0029559 | 78491-02-8 | GPMT-NS | Metabolite: Aldehydes | Formaldehyde donor | Probably too hydrophilic and maybe not reactive enough. |
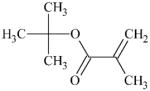
|
DTXSID3060405 | 585-07-9 | GPMT-NS | alpha, beta-Carbonyl compounds with polarised double bonds | alpha, beta-Unsaturated ester or precursor | A MA but not a sufficiently reactive one. |
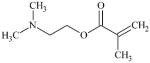
|
DTXSID1027504 | 2867-47-2 | GPMT-NS | alpha, beta-Carbonyl compounds with polarised double bonds | alpha, beta-Unsaturated ester or precursor | Insufficiently reactive as MA. |
|
DTXSID4041573 | 867-13-0 | LLNA-NS | PhosphonatesMetabolite: (Thio)Phosphates | Alkyl ester of phosphoric or phosphonic acid | Alkyl phosphonate is probably a poor leaving group hence not reactive enough. OEt is also a poor leaving group which would thwart reaction as an acyl transfer agent. |
|
|
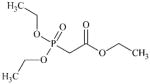
DTXSID4041238 | 2579-20-6 | GPMT-NS | Metabolite: Aldehydes | Diamine | Corresponding di-aldehyde would not be particularly reactive. Roberts et al (2006) SB QMM estimates a EC3 of 91%, which is consistent with the non-sensitising outcome. |
|
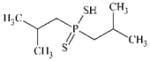
DTXSID9044798 | 13360-78-6 | LLNA-S | No Alerts | No Alerts | Potentially a nucleophilic sensitiser. EC3 reported as 18.1%. |
|
DTXSID3038694 | 91-56-5 | LLNA-S | No Alerts | No Alerts | Schiff base former. |
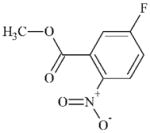
|
DTXSID60408886 | 393-85-1 | GPMT-S | No Alerts | No Alerts | SNAr electrophile with F as the leaving group. Per the QMM in Roberts and Aptula [35], this would be a moderate sensitiser. |
|
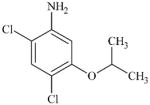
DTXSID90194124 | 41200-96-8 | LLNA-NS | Metabolite: Quinone methide(s)/imines, Quinoide oxime structure, Nitroquinones, | Aromatic primary or secondary amine | As for 97-52-9. |
|
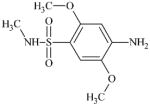
DTXSID2068507 | 49701-24-8 | LLNA-NS | Metabolite: Quinone methide(s)/imines, Quinoide oxime structure, Nitroquinones, | Aromatic primary or secondary amine | As for 97-52-9. Demethylation of the ortho OMe to the NH2 may be feasible but activation to form a o-orthoquinone imine may be deactivated by the SO2NHMe. |
|
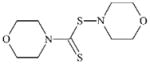
DTXSID5021095 | 13752-51-7 | LLNA-NS | Dithiocarbamate esters | Thiol or thiol exchange agent | Probably insufficiently reactive. |
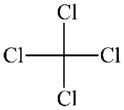
|
DTXSID8020250 | 56-23-5 | LLNA-S | No Alerts | No Alerts | False positive in the LLNA |
Table 7.
Analysis of substances from the NICEATM set that were predicted incorrectly by all three in silico models, the justification for the TIMES-SS and Derek predictions are given, VEGA does not give a mechanistic description. The authors’ proposed rationales for the incorrect predictions are also given. Mechanistic domains – Michael Acceptor (MA), Schiff Base former (SB), Acyl transfer agent, SN2 etc. make reference to the reaction pathways described in Aptula and Roberts [33]
| Structure | DSSTox Substance Identifier (DTXSID) |
CAS | Experimental sensitisation outcome |
TIMES-SS | Derek Nexus |
Author proposed rationales/comments |
|---|---|---|---|---|---|---|
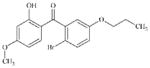
|
DTXSID60697617 | 190965-45-8 | LLNA-NS | Aromatic carbonyl compounds | Resorcinol or precursor | SB would be more plausible if COAr was ortho to OH was an aldehyde. Resorcinol formation is deactivated by the COAr which is electron withdrawing and therefore deactivated resorcinol’s oxidation. |
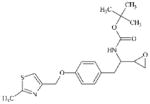
|
DTXSID90469546 | 313680-92-1 | LLNA-NS | Epoxides, Aziridines and Sulfuranes | Epoxide | Epoxide is probably insufficiently reaction (see [36]). |
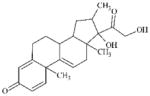
|
13504-15-9 | LLNA-NS | alpha, beta-Carbonyl compounds with polarised double bonds | 1,2-Dicarbonyl compound or precursor | This metabolic activation is probably not significant in mouse skin vs guinea pig or man (see 50-24-8). | |
|
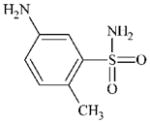
DTXSID40284367 | 6973-09-7 | LLNA-NS | Metabolite: Quinone methide(s)/imines, Quinoide oxime structure, Nitroquinones, | Aromatic primary or secondary amine | Oxidation is deactivated by SO2Me. |
|
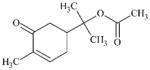
DTXSID60337127 | 87578-93-6 | LLNA-NS | alpha, beta-Carbonyl compounds with polarised double bonds | alpha, beta-Unsaturated ketone or precursor | Low reactivity as a methacrylate. |
|
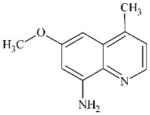
DTXSID70345475 | 57514-21-3 | LLNA-NS | Metabolites: Hydroperoxides | Aromatic primary or secondary amine | Oxidation of the aromatic amine would be deactivated by OMe and the fused pyridine ring. |
|
DTXSID9020112 | 1912-24-9 | LLNA-NS | Activated aryl and heteroaryl compounds | Activated N-heterocycle | The negative experimental outcome is surprising. Would expect this substance to be reactive as a SNAr. Hydrolysis might account for the NS outcome reported. |
|
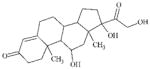
DTXSID9020112 | 50-23-7 | LLNA-NS | Ketones | 1,2-Dicarbonyl compound or precursor | Metabolic activation in mouse skin is probably not significant to lead to a sensitising outcome. (see 50-24-8). |
|
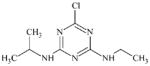
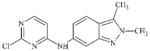
DTXSID50469840 | 444731-74-2 | LLNA-NS | Activated aryl and heteroaryl compounds | Activated N-heterocycle | Surprising outcome. Would expect this substance to react as a SNAr electrophile, hydrolysis could be a competing factor. |
|
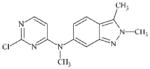
DTXSID70469403 | 444731-75-3 | LLNA-NS | Activated aryl and heteroaryl compounds | Activated N-heterocycle | Surprising outcome. Would expect this substance to react as a SNAr electrophile, hydrolysis could be a competing factor. |
|
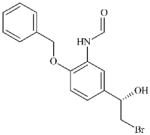
DTXSID40456769 | 201677-59-0 | LLNA-NS | Epoxides, Aziridines and Sulfuranes | Alkyl halide | OH relative to the Br leaving group has a deactivating effect. Not sufficiently reactive to act as a SN2 electrophile. |
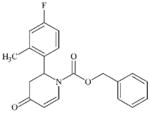
|
DTXSID10620233 | 414909-98-1 | LLNA-NS | Carbamates | alpha, beta-Unsaturated ketone or precursor | Poor acyl transfer agent, weak SN2 and deactivated as MA by beta Nitrogen. |
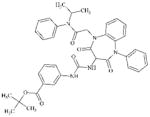
|
DTXSID70725319 | 305366-94-3 | LLNA-S | No Alerts | No Alerts | Right hand side benzene ring has 2 ortho N substituents. Hydrolysis of the amides in the 7 membered ring would give rise to a ortho-di(aryl)amino benzene which is oxidisable to a quinone imine that could act as a MA. |
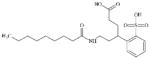
|
168151-92-6 | LLNA-S | No Alerts | No Alerts | Anionic with surfactant features, likely false positive (FP) result. | |
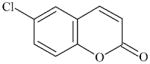
|
2051-59-4 | LLNA-S | No Alerts | No Alerts | MA – lactone; phenyl ester. | |
|
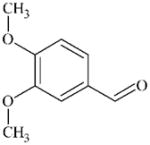
DTXSID7026285 | 120-14-9 | LLNA-S | No Alerts | No Alerts | Aromatic aldehyde SB former that happens to be sufficiently reactive to sensitise. |
Identifying why a prediction is incorrect across all three programs can be used to help improve models in the future, by modifying the structural alerts contained within them. Specific examples include the alert for resorcinol or precursors, if the alkyl group on the oxygen is an ethyl or longer, then the substance is likely to be non-sensitising or the alert for aromatic primary and secondary amines – presence of an electronegative group such as SO2NH- or NO2 or SO3H will deactivate oxidation of the aromatic amine group. It also demonstrates that in some cases the models are actually better than the animal test, take for example the three substances with CAS numbers 605-50-5, 56-23-5, and 168151-92-6 which are all predicted to be false positives in the LLNA. All three programs correctly predicted them to be non-sensitisers.
Comparing our Evaluation to Previous Evaluations of in Silico Models
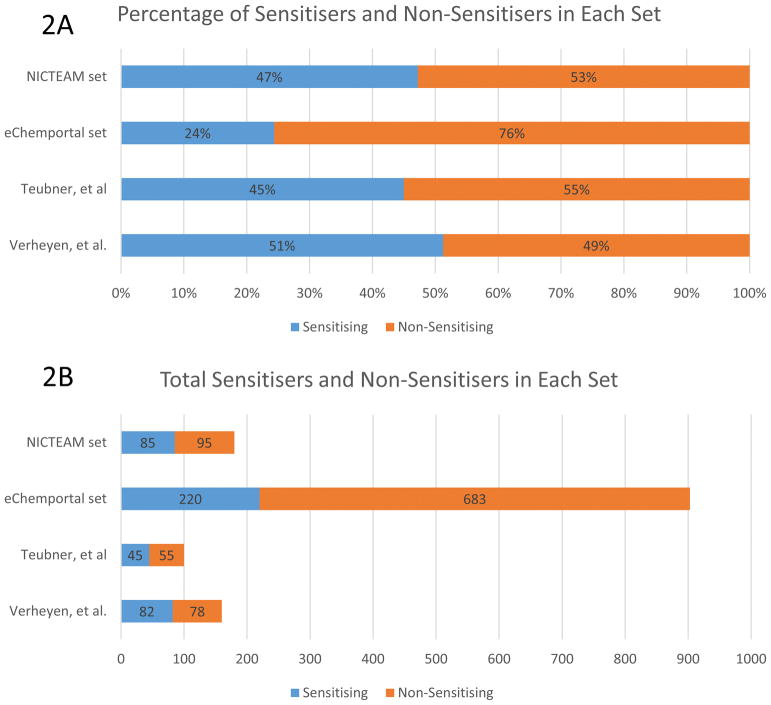
There have been other evaluations of computational skin sensitisation models. However, these evaluations had more limited datasets. Verheyen et al. [37] used a dataset of 160 substances whereas Teubner et al.[18] used a data set of 100 substances. By contrast, our eChemportal evaluation set contained 903 substances excluding substances within the training set of the three models and the NICEATM set contained 180 substances excluding substances within the training set of the three models, see figure 3 for a comparison of the four data sets used in the evaluations.
Figure 3.
Comparison of the four data sets.
Not only do the number of substances in the eChemportal set exceed all other sets, it’s proportion of sensitisers and non-sensitisers are very different as well. The eChemportal set contains approximately 25% sensitisers, while the other three sets contain about 50% sensitisers. This is an important distinction given that the scope of the eChemportal data is much larger than the others and likely a better representation of chemical universe. The large and diverse data set also allowed us to make a comparison of GPMT to LLNA, and then to compare that result to the in silico models, something that had not been done on such a large scale until now.
Conclusions
On the basis of the datasets evaluated in this study, TIMES-SS gave rise to similar performance metrics as the animal tests when a prediction was being made for a substance within its domain. i.e. for substances within the domain, TIMES-SS predictions could be used in lieu of animal methods. For substances outside of domain of TIMES-SS, both Derek Nexus and TIMES-SS performed similarly to each other but significantly poorer than the animal tests. The expert systems evaluated could be extended in light of the additional data collected as part of this study. The incorrect predictions offer new suggestions for how the existing alerts within these expert systems could be improved. Suggestions are provided for the majority of cases where either a modification could be made for an existing alert flagged or where a new alert might be warranted. Indeed, 903 substances (220 sensitisers and 683 non-sensitisers) from the eChemPortal set and 180 substances (85 sensitisers and 95 non-sensitisers) from the NICEATM set had not been used in the development of these 3 expert systems. These datasets also offer exciting opportunities for the development of new models.
Supplementary Material
Footnotes
BgVV = Bundesinstitut für Gesundheitlichen Verbraucherschutz Und Veterinärmedizin, the Federal Institute for Health Protection of Consumers and Veterinary Medicine
Disclaimer
The views expressed in this article are those of the authors and do not necessarily represent the views or policies of the U.S. Environmental Protection Agency. Mention of trade names or commercial products does not constitute endorsement or recommendation for use. The authors alone are responsible for the content and writing of this article. The datasets have been downloaded using eChemPortal on February 21, 2017 or were donated by NICEATM and are being provided as is. They have not been curated on a study level, nor has any chemical mapping curation been undertaken to verify CAS to given name associations.
References
- 1.Smith Pease CK. From xenobiotic chemistry and metabolism to better prediction and risk assessment of skin allergy. Toxicology. 2003;192:1–22. doi: 10.1016/s0300-483x(03)00246-4. [DOI] [PubMed] [Google Scholar]
- 2.Dotson AMGS, Siegel PD, Anderson SE, Green BJ, Stefaniak AB, Codispoti CD, Kimber I. Setting Occupational Exposure Limits for Chemical Allergens Understanding the Challenges. J Occup Environ Hyg. 2015;12(sup1):S82–S89. doi: 10.1080/15459624.2015.1072277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Peiser M, Tralau T, Heidler J, Api AM, Arts JHE, Basketter DA, English J, Diepgen TL, Fuhlbrigge RC, Gaspari AA, Johansen JD, Karlberg AT, Kimber I, Lepoittevin JP, Liebsch M, Maibach HI, Martin SF, Merk HF, Platzek T, Rustemeyer T, Schnuch A, Vandebriel RJ, White IR, Luch A. Allergic contact dermatitis: epidemiology, molecular mechanisms, in vitro methods and regulatory aspects: Current knowledge assembled at an international workshop at BfR, Germany. Cell Mol Life Sci. 2012;69:763–781. doi: 10.1007/s00018-011-0846-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.The European Parliament and the Council of the European Union, Corrigendum to Regulation (EC) No 1907/2006 of the European Parliament and of the Council of 18 December 2006 concerning the Registration, Evaluation, Authorisation and Restriction of Chemicals (REACH), establishing a European Chemicals Agency, amending Directive 1999/45/EC and repealing Council Regulation (EEC) No 793/93 and Commission Regulation (EC) No 1488/94 as well as Council Directive 76/769/EEC and Commission Directives 91/155/EEC, 93/67/EEC, 93/105/EC and 2000/21/EC, OJEU. L136(2007), pp. 3–280. Communities.
- 5.OECD. Test No 429: Skin Sensitisation. OECD Publishing; 2010. pp. 1–20. [Google Scholar]
- 6.OECD. Test No 406: Skin Sensitisation. OECD Publishing; 1992. pp. 1–9. [Google Scholar]
- 7.The European Parliament and the Council of the European Union. Regulation (EC) No 1223/2009 of the European Parliament and of the Council of 30 November 2009 on Cosmetic Products, OJEU L342. 2009:59–209. [Google Scholar]
- 8.OECD. The Adverse Outcome Pathway for Skin Sensitisation Initiated by Covalent Binding to Proteins Part 1: Scientific Evidence. 2012a. Series on Testing and Assessment No. 168 ENV/JM/MONO(2012)10/PART1. [Google Scholar]
- 9.OECD. The Adverse Outcome Pathway for Skin Sensitisation Initiated by Covalent Binding to Proteins Part 2: Use of the AOP to develop chemical categories and Integrated Assessment and Testing Approaches. 2012b. Series on Testing and Assessment No. 168 ENV/JM/MONO(2012)10/PART2. [Google Scholar]
- 10.Reisinger K, Hoffmann S, Alépée N, Ashikaga T, Barroso J, Elcombe C, Gellatly N, Galbiati V, Gibbs S, Groux H, Hibatallah J, Keller D, Kern P, Klaric M, Kolle S, Kuehnl J, Lambrechts N, Lindstedt M, Millet M, Martinozzi-Teissier S, Natsch A, Petersohn D, Pike I, Sakaguchi H, Schepky A, Tailhardat M, Templier M, van Vliet E, Maxwell G. Systematic evaluation of non-animal test methods for skin sensitisation safety assessment. Toxicol in Vitro. 2015;29:259–270. doi: 10.1016/j.tiv.2014.10.018. [DOI] [PubMed] [Google Scholar]
- 11.Casati S, Aschberger K, Barroso J, Casey W, Delgado I, Kim TS, Kleinstreuer N, Kojima H, Lee JK, Lowit A, Park HK, Régimbald-Krnel MJ, Strickland J, Whelan M, Yang Y, Zuang V. Standardisation of defined approaches for skin sensitisation testing to support regulatory use and international adoption: position of the International Cooperation on Alternative Test Methods. Arch Toxicol. 2017 doi: 10.1007/s00204-017-2097-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ramirez T, Stein N, Aumann A, Remus T, Edwards A, Norman KG, Ryan C, Bader JE, Fehr M, Burleson F, Foertsch L, Wang X, Gerberick F, Beilstein P, Hoffmann S, Mehling A, van Ravenzwaay B, Landsiedel R. Intra- and inter-laboratory reproducibility and accuracy of the LuSens assay: A reporter gene-cell line to detect keratinocyte activation by skin sensitizers. Toxicol in Vitro. 2016;32:278–286. doi: 10.1016/j.tiv.2016.01.004. [DOI] [PubMed] [Google Scholar]
- 13.Cottrez F, Boitel E, Ourlin J-C, Peiffer J-L, Fabre I, Henaoui I-S, Mari B, Vallauri A, Paquet A, Barbry P, Auriault C, Aeby P, Groux H. SENS-IS, a 3D reconstituted epidermis based model for quantifying chemical sensitization potency: Reproducibility and predictivity results from an inter-laboratory study. Toxicol in Vitro. 2016;32:248–260. doi: 10.1016/j.tiv.2016.01.007. [DOI] [PubMed] [Google Scholar]
- 14.OECD. Test No 442E: In Vitro Skin Sensitisation. OECD Publishing; 2017. pp. 1–68. [Google Scholar]
- 15.ECHA. Guidance on Information Requirements and Chemical Safety Assessment Chapter R7a: Endpoint Specific Guidance. ECHA; 2017. pp. 271–314. [Google Scholar]
- 16.Patlewicz G, Aptula AO, Roberts DW, Uriarte E. A Minireview of Available Skin Sensitization (Q)SARs/Expert Systems. QSAR Comb Sci. 2008;27:60–76. [Google Scholar]
- 17.Patlewicz G, Aptula AO, Uriarte E, Roberts DW, Kern PS, Gerberick GF, Kimber I, Dearman RJ, Ryan CA, Basketter DA. An evaluation of selected global (Q)SARs/expert systems for the prediction of skin sensitisation potential. SAR and QSAR in Environ Res. 2007;18:515–541. doi: 10.1080/10629360701427872. [DOI] [PubMed] [Google Scholar]
- 18.Teubner W, Mehling A, Schuster PX, Guth K, Worth A, Burton J, van Ravenzwaay B, Landsiedel R. Computer models versus reality: How well do in silico models currently predict the sensitization potential of a substance. Regul Toxicol Pharm. 2013;67:468–485. doi: 10.1016/j.yrtph.2013.09.007. [DOI] [PubMed] [Google Scholar]
- 19.Patlewicz G, Fitzpatrick JM. Current and Future Perspectives on the Development, Evaluation, and Application of in Silico Approaches for Predicting Toxicity. Chem Res Toxicol. 2016;29:438–451. doi: 10.1021/acs.chemrestox.5b00388. [DOI] [PubMed] [Google Scholar]
- 20.Gerberick FG, Ryan CA, Kern PS, Schlatter H, Dearman RJ, Kimber I, Patlewicz GY, Basketter DA. Compilation of Historical Local Lymph Node Data for Evaluation of Skin Sensitization Alternative Methods. Dermatitis. 2005;16:157–202. [PubMed] [Google Scholar]
- 21.Chaudhry Q, Piclin N, Cotterill J, Pintore M, Price NR, Chrétien JR, Roncaglioni A. Global QSAR models of skin sensitisers for regulatory purposes. Chem Cent J. 2010;4:S5. doi: 10.1186/1752-153X-4-S1-S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barratt MD, Basketter DA, Chamberlain M, Payne MP, Admans GD, Langowski JJ. Development of an expert system rulebase for identifying contact allergens. Toxicol in Vitro. 1994;8:837–839. doi: 10.1016/0887-2333(94)90081-7. [DOI] [PubMed] [Google Scholar]
- 23.Payne MP, Walsh PT. Structure-activity relationships for skin sensitization potential: Development of structural alerts for use in knowledge-based toxicity prediction systems. J Chem Inf Comp Sci. 1994;34:154–161. doi: 10.1021/ci00017a019. [DOI] [PubMed] [Google Scholar]
- 24.Langton K, Patlewicz GY, Long A, Marchant CA, Basketter DA. Structure–activity relationships for skin sensitization: recent improvements to Derek for Windows. Contact Dermatitis. 2006;55:342–347. doi: 10.1111/j.1600-0536.2006.00969.x. [DOI] [PubMed] [Google Scholar]
- 25.Patlewicz G, Dimitrov SD, Low LK, Kern PS, Dimitrova GD, Comber MI, Aptula AO, Phillips RD, Niemela J, Madsen C, Wedebye EB, Roberts DW, Bailey PT, Mekenyan OG. TIMES-SS--a promising tool for the assessment of skin sensitization hazard. A characterization with respect to the OECD validation principles for (Q)SARs and an external evaluation for predictivity. Regul Toxicol Pharm. 2007;48:225–39. doi: 10.1016/j.yrtph.2007.03.003. [DOI] [PubMed] [Google Scholar]
- 26.Cronin MTD, Basketter DA. Multivariate Qsar Analysis of a Skin Sensitization Database. SAR QSAR Environ Res. 1994;2:159–179. doi: 10.1080/10629369408029901. [DOI] [PubMed] [Google Scholar]
- 27.Schlede E, Aberer W, Fuchs T, Gerner I, Lessmann H, Maurer T, Rossbacher R, Stropp G, Wagner E, Kayser D. Chemical substances and contact allergy—244 substances ranked according to allergenic potency. Toxicology. 2003;193:219–259. doi: 10.1016/s0300-483x(03)00266-x. [DOI] [PubMed] [Google Scholar]
- 28.Patlewicz G, Kuseva C, Mehmed A, Popova Y, Dimitrova G, Ellis G, Hunziker R, Kern P, Low L, Ringeissen S, Roberts DW, Mekenyan O. TIMES-SS – Recent refinements resulting from an industrial skin sensitisation consortium. SAR and QSAR in Environ Res. 2014;25:367–391. doi: 10.1080/1062936X.2014.900520. [DOI] [PubMed] [Google Scholar]
- 29.Klimisch HJ, Andreae M, Tillmann U. A Systematic Approach for Evaluating the Quality of Experimental Toxicological and Ecotoxicological Data. Regul Toxicol Pharm. 1997;25:1–5. doi: 10.1006/rtph.1996.1076. [DOI] [PubMed] [Google Scholar]
- 30.Dimitrov S, Dimitrova G, Pavlov T, Dimitrova N, Patlewicz G, Niemela J, Mekenyan O. A Stepwise Approach for Defining the Applicability Domain of SAR and QSAR Models. J Chem Inf Model. 2005;45:839–849. doi: 10.1021/ci0500381. [DOI] [PubMed] [Google Scholar]
- 31.ICCVAM. The Murine Local Lymph Node Assay: A Test Method for Assessing the Allergic Contact Dermatitis Potential of Chemicals/Compounds. National Institute of Environmental Health Sciences, National Institutes of Health, U.S. Public Health Service, Department of Health and Human Services; 1999. p. 13. [PubMed] [Google Scholar]
- 32.Bundgaard H. The possible implication of steroid-glyoxal degradation products in allergic reactions to corticosteroids. Arch Pharm Chem Sci. 1980;8:83–90. [Google Scholar]
- 33.Roberts DW, Aptula AO, Patlewicz G. Mechanistic Applicability Domains for Non-Animal Based Prediction of Toxicological Endpoints. QSAR Analysis of the Schiff Base Applicability Domain for Skin Sensitization. Chem Res Toxicol. 2006;19:1228–1233. doi: 10.1021/tx060102o. [DOI] [PubMed] [Google Scholar]
- 34.Roberts DW, Aptula AO, Patlewicz G. Electrophilic Chemistry Related to Skin Sensitization. Reaction Mechanistic Applicability Domain Classification for a Published Data Set of 106 Chemicals Tested in the Mouse Local Lymph Node Assay. Chem Res Toxicol. 2007;20:44–60. doi: 10.1021/tx060121y. [DOI] [PubMed] [Google Scholar]
- 35.Roberts DW, Aptula AO. Electrophilic Reactivity and Skin Sensitization Potency of SNAr Electrophiles. Chem Res Toxicol. 2014;27:240–246. doi: 10.1021/tx400355n. [DOI] [PubMed] [Google Scholar]
- 36.Roberts DW, Aptula A, Api AM. Structure–Potency Relationships for Epoxides in Allergic Contact Dermatitis. Chem Res Toxicol. 2017;30:524–531. doi: 10.1021/acs.chemrestox.6b00241. [DOI] [PubMed] [Google Scholar]
- 37.Verheyen GR, Braeken E, Van Deun K, Van Miert S. Evaluation of in silico tools to predict the skin sensitization potential of chemicals. SAR and QSAR in Environ Res. 2017;28:59–73. doi: 10.1080/1062936X.2017.1278617. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.