Abstract
We isolated a temperature-sensitive mutant, hrd4–1, deficient in ER-associated degradation (ERAD). The HRD4 gene was identical to NPL4, a gene previously implicated in nuclear transport. Using a diverse set of substrates and direct ubiquitination assays, our analysis revealed that HRD4/NPL4 is required for a poorly characterized step in ERAD after ubiquitination of target proteins but before their recognition by the 26S proteasome. Our data indicate that this lack of proteasomal processing of ubiquitinated proteins constitutes the primary defect in hrd4/npl4 mutant cells and explains the diverse set of hrd4/npl4 phenotypes. We also found that each member of the Cdc48p-Ufd1p-Npl4p complex is individually required for ERAD.
INTRODUCTION
The endoplasmic reticulum is a major site for protein degradation in the cell (Arias et al., 1969; Fra and Sitia, 1993; Brodsky and McCracken, 1999). This endoplasmic reticulum-associated degradation (ERAD) serves several functions: ERAD removes aberrant, misfolded proteins from the ER as a means of “quality control” for ER proteins (Hammond and Helenius, 1995; Wickner et al., 1999). ERAD is also used in the regulation of HMG-CoA reductase (HMGR), a rate-limiting enzyme in cholesterol biosynthesis (Hampton et al., 1994). We have identified HRD genes required for Hmg-CoA Reductase Degradation (Hampton et al., 1996). The characterization of these HRD genes along with other studies has revealed that ERAD proceeds via the ubiquitin-proteasome pathway (Sommer and Latterich, 1993; Hiller et al., 1996; Hampton and Bhakta, 1997). Several Hrd proteins are required for the ubiquitination of ERAD substrates, including the ubiquitin-protein ligase (E3) Hrd1p (Bays et al., 2001), its associated membrane protein Hrd3p (Gardner et al., 2000), and the ubiquitin-conjugating enzymes (E2s) Ubc7p and Ubc1p (Hampton and Bhakta, 1997; Bays et al., 2001). Other Hrd proteins, like Hrd2p, are components of the 26S proteasome itself (Tsurumi et al., 1996). To extend the range of our genetic studies, we modified the original hrd selection (Hampton et al., 1996) to allow the recovery of temperature-sensitive hrd mutants. We isolated strains expressing the hrd allele hrd4–1, which grew normally at 30°C but were inviable at 35°C. The wild-type allele corresponding to the hrd4–1 mutation was cloned and found to be identical to the gene NPL4.
NPL4 is an essential gene (null alleles are not viable), and was previously identified in a selection for mutants deficient in nuclear import/export (DeHoratius et al., 1996). npl4 mutants fail both to import nuclear localization signal (NLS)-bearing proteins into the nucleus and to export poly(A)+ RNA from the nucleus when shifted to the restrictive temperature. npl4 mutants also exhibit defects in nuclear structure at the restrictive temperature. These nuclear abnormalities include herniations of the nuclear envelope, separation of the inner and outer nuclear membranes, and large membrane protusions containing accumulations of poly(A)+ RNA (DeHoratius et al., 1996).
NPL4 has also been implicated in unsaturated fatty acid (UFA) biosynthesis. Hitchcock et al. (2001) isolated the OLE1 gene as a partial high-copy suppressor of npl4–1 and npl4–2 mutant growth phenotypes. OLE1 is an essential gene encoding the sole Δ9-fatty acid desaturase in yeast (Stukey et al., 1990). MGA2 and SPT23 were also isolated as partial high-copy suppressors of npl4 (Hitchcock et al., 2001). The products of these genes were previously identified as functionally redundant transcription factors required for the production of OLE1 transcript (Zhang et al., 1999). Recently, it has been shown that Mga2p and Spt23p reside in the endoplasmic reticulum as inactive membrane-bound transcription factors (Hoppe et al., 2000). When cells are deprived of fatty acids, the Mga2 and Spt23 proteins are cleaved from their membrane anchors in a ubiquitin- and proteasome-dependent process, liberating the transcription factor domains to enter the nucleus and promote transcription of the OLE1 gene. The proteasome-dependent processing of Mga2p and Spt23p requires NPL4. In npl4 mutants, Mga2p and Spt23p cleavage is defective (Hitchcock et al., 2001). Furthermore, at least some of the nuclear defects in npl4 mutants can also be suppressed by unsaturated fatty acids and increased OLE1 expression (Hitchcock et al., 2001).
Npl4p physically associates with Cdc48p via Ufd1p to form a Cdc48p-Ufd1p-Npl4p complex (Meyer et al., 2000; Hitchcock et al., 2001). Ufd1p was previously identified in a screen for mutants that fail to degrade a fusion protein with a nonremovable ubiquitin moiety (Johnson et al., 1995). Cdc48p is a AAA ATPase required for a variety of cellular processes including cell division, protein degradation, and ER membrane fusion (Moir et al., 1982; Latterich et al., 1995; Ghislain et al., 1996). Cdc48p actually associates with two other Ufd proteins, Ufd2p and Ufd3p/Doa1p, as well as several other proteins with no known function in protein degradation (Ghislain et al., 1996; Koegl et al., 1999). This ability to bind multiple proteins has prompted models of an adapter function for Cdc48p (Patel and Latterich, 1998).
Here, we unite these diverse phenotypes for HRD4/NPL4 by describing the role of Hrd4p/Npl4p in the proteasomal processing of ubiquitinated proteins at the ER. hrd4/npl4 mutants are defective in the degradation of several ER proteins, but HRD4/NPL4 is not required for the actual ubiquitination of ERAD substrates. Importantly, general proteasome function is not impaired in hrd4/npl4 mutant cells. Therefore, we conclude that Hrd4p/Npl4p functions at a postubiquitination but preproteasomal step in ERAD. Our analysis also shows that the primary defect in hrd4/npl4 cells is a lack of proteasomal processing of ubiquitinated proteins, not a defect in nuclear transport or fatty acid biosynthesis. These diverse phenotypes apparently arise from the loss of Hrd4p/Npl4p function in the ubiquitin-proteasome pathway.
MATERIALS AND METHODS
Identification of HRD4 as NPL4
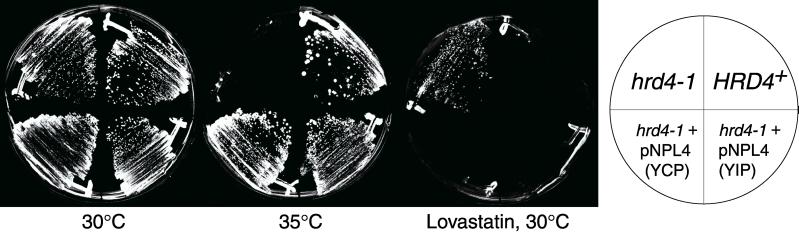
The hrd4–1 mutant allele was recovered from a lovastatin-resistant colony isolated in the hrd selection described previously (Hampton et al., 1996). The hrd4–1 allele yielded a yeast strain that grew well at 30° but was inviable at 35°C — failing to complete any further cell division after the temperature shift. This strain bred true for lovastatin resistance (Lovr) and so was crossed to the wild-type parent strain. A wild-type diploid resulted from the cross, indicating that recessive allelle(s) were conferring lovastatin-resistance. This diploid was then sporulated. Tetrad analysis revealed that the Lovr and Ts− phenotypes were the result of a single recessive mutant allele, hrd4–1. The hrd4–1 allele defined a new HRD complementation group, as crosses to other haploid hrd mutant strains always resulted in a Hrd+ Ts+ diploid. When sporulated and dissected, these diploids yielded progeny with Hrd and Ts phenotypes in the expected ratios for two unlinked hrd alleles.
We cloned the wild-type allele for HRD4 using a yeast genomic library bearing the URA3 prototrophy marker (Rose et al., 1987). A Ura− Ts− Lovr hrd4–1 strain was transformed with library DNA, and Ura+ Ts+ colonies were then selected from the transformants by incubation at 35°C. Ts+ colonies were analyzed for lovastatin resistance. Those colonies that showed both Ts+ and Lovs phenotypes were subjected to URA3 counterselection using 5-fluoroorotic acid (Guthrie and Fink, 1991). This selection for loss of the library plasmid resulted in Ura− Ts− Lovr colonies displaying the original hrd4–1 phenotypes. Plasmid DNA was extracted from the original Ura+ Ts+ Lovs transformants, bacterially amplified, and retransformed into a hrd4–1 mutant strain. Plasmids capable of reversing the hrd4–1 Ts− Lovr phenotypes were sequenced. Two different plasmids containing a common region of yeast chromosome II were isolated. Further subcloning of the library plasmids revealed that only one orf (open reading frame) was required for complementation. That orf corresponded to the previously identified gene NPL4. To test whether HRD4 was indeed NPL4, the NPL4 gene was cloned into a yeast integrating plasmid bearing the URA3 gene. This URA3-marked NPL4 was integrated at the NPL4 locus in both HRD4+ and hrd4–1 Ura− strains. After transformation, both strains were Ura+ Ts+ Lovs (Figure 1). These strains were then crossed to a Ura− hrd4–1 strain. The resulting diploid was sporulated. Every tetrad showed 2:2 segregation of the Ura+/Ura−, Ts+/Ts−, and Lovr/Lovs phenotypes; and no Ura+ segregant was ever Ts− or Lovr. These results indicated that hrd4–1 was indeed a mutant allele of NPL4.
Figure 1.
hrd4–1 phenotypes and complementation by NPL4. The indicated yeast strains were incubated as designated by each labeled plate. (YCP) indicates the hrd4–1 strain was transformed with an ARS/CEN yeast genomic library plasmid, isolated for its ability to complement the hrd4–1 mutation and containing a genomic fragment bearing the NPL4 gene. (YIP) indicates a plasmid bearing only the NPL4 and URA3 genes was integrated at the ura3–52 locus in a hrd4–1 strain. “Lovastatin” indicates the addition of 200 μg/ml lovastatin to the indicated plate.
Yeast Strains
Genotypes of yeast strains used in this study are listed in Table 1. The following alleles were described previously: hrd1–1, hrd2–1, hrd1Δ::TRP1, ubc6Δ::KanMX, ubc7Δ::HIS3. (Hampton et al., 1996; Hampton and Bhakta, 1997; Wilhovsky et al., 2000). The nup116Δ allele was introduced into RHY400 by transformation with a nup116Δ deletion construct described previously to create RHY877 (Wente and Blobel, 1993). Deletion of NUP116 was verified by immunoblotting with anti-Nup116p antibody. OLE1+ and ole1Δ strains were described previously, as were the NPL4+/npl4–1/npl4–2, CDC48+/cdc48–2, and UFD1/ufd1–1 strains (Stukey et al., 1990; Johnson et al., 1995; Latterich et al., 1995; DeHoratius et al., 1996). All strains were constructed according to standard techniques (Guthrie and Fink, 1991). Yeast strains were grown in yeast minimal media, as described previously (Hampton et al., 1994), except plates in Figure 4, which used synthetic complete media (-uracil in panel C) prepared as described (Guthrie and Fink, 1991). Oleic and palmitoleic acids were added to media where indicated from 10% stocks in absolute ethanol to a final concentration of 0.5 mM each unsaturated fatty acid (UFA). One percent Tergitol NP-40 (Sigma, St. Louis, MO) was added to plates containing UFA for solubilization. (Tergitol NP-40 is not the same as Nonidet NP-40 [sigma], which is somewhat toxic to yeast [our unpublished results and personal communication, Charles Martin])
Table 1.
Yeast Strains
| Strain | Genotype |
|---|---|
| RHY244 | MATa ade2-101 lys2-801 his3Δ200 hmg1Δ∷LYS2 hmg2Δ∷HIS3 met2 ura3-52∷PTDH3-6MYC-HMG2∷URA3 |
| RHY400 | MATa ade2-101 lys2-801 his3Δ200 hmg1Δ∷LYS2 hmg2Δ∷HIS3 met2 ura3-52∷PTDH3-6MYC-HMG2 |
| RHY554 | MATa ade2-101 lys2-801 his3Δ200 hmg1Δ∷LYS2 hmg2Δ∷HIS3 met2 ura3-52∷PTDH3-6MYC-HMG2 hrd4-1 |
| RHY587 | MATa ade2-101 lys2-801 his3Δ200 hmg1Δ∷LYS2 hmg2Δ∷HIS3 met2 ura3-52∷PTDH3-6MYC-HMG2 hrd1Δ∷URA3 |
| RHY877 | MATa ade2-101 lys2-801 his3Δ200 hmg1Δ∷LYS2 hmg2Δ∷HIS3 met2 ura3-52∷PTDH3-6MYC-HMG2 nup116Δ∷URA3 |
| RHY1172 | MATalpha ade2-101 lys2-801 his3Δ200 hmg1Δ∷LYS2 hmg2Δ∷HIS3 met2 ura3-52∷PTDH3-1MYC-HMG |
| RHY1216 | MATa ade2-101 lys2-801 his3Δ200 hmg1Δ∷LYS2 hmg2Δ∷HIS3 met2 ura3-52∷PTDH3-6MYC-HMG2 pRH696a[fur4-430∷URA3] |
| RHY1220 | MATa ade2-101 lys2-801 his3Δ200 hmg1Δ∷LYS2 hmg2Δ∷HIS3 met2 ura3-52∷PTDH3-6MYC-HMG2 hrd4-1 pRH696a[fur4-430∷URA3] |
| RHY1221 | MATalpha ade2-101 lys2-801 his3Δ200 hmg1Δ∷LYS2 hmg2Δ∷HIS3 met2 ura3-52∷PTDH3-6MYC-HMG2 ubc7Δ∷HIS3 pRH696a[fur4-430∷URA3] |
| RHY1234 | MATalpha ade2-101 lys2-801 his3Δ200 hmg1Δ∷LYS2 hmg2Δ∷HIS3 met2 trp1∷hisG ura3-52∷PTDH3-1MYC-HMG2 hrd1Δ∷TRP1 |
| RHY1246 | MATa ade2-101 lys2-801 his3Δ200 hmg1Δ∷LYS2 hmg2Δ∷HIS3 met2 ura3-52∷PTDH3-1MYC-HMG2 hrd4-1 |
| RHY1252 | MATa ade2-101 lys2-801 his3Δ200 hmg1Δ∷LYS2 hmg2Δ∷HIS3 met2 ura3-52∷PTDH3-6MYC-HMG2∷URA3 hrd4-1 |
| RHY1389 | MATa ade2-101 lys2-801 his3Δ200 hmg1Δ∷LYS2 hmg2Δ∷HIS3 met2 ura3-52∷PTDH3-1MYC-HMG2 hrd2-1 |
| RHY1628 | MATa ade2-101 lys2-801 his3Δ200 hmg1Δ∷LYS2 hmg2Δ∷HIS3 met2 ura3-52∷PTDH3-1MYC-HMG2∷URA3 hrd2-1 |
| RHY2458 | MATalpha leu2-3,112 ura3-52∷hmg2-GFP∷URA3 |
| RHY2461 | MATalpha ura3-52∷hmg2-GFP∷URA3 cdc48-2 |
| RHY2472 | MATa ade2-101 lys2-801 his3Δ200 hmg1Δ∷LYS2 hmg2Δ∷HIS3 met2 ura3-52∷PTDH3-1MYC-HMG2∷URA3∷PKAR2-GFP |
| RHY2473 | MATa ade2-101 lys2-801 his3Δ200 hmg1Δ∷LYS2 hmg2Δ∷HIS3 met2 trp1∷hisG ura3-52∷PTDH3-1MYC-HMG2∷URA3∷PKAR2-GFP hrd1Δ∷TRP1 |
| RHY2474 | MATa ade2-101 lys2-801 his3Δ200 hmg1Δ∷LYS2 hmg2Δ∷HIS3 met2 ura3-52∷PTDH3-1MYC-HMG2∷URA3∷PKAR2-GFP hrd4-1 |
| RHY2476 | MATa ade2-101 lys2-801 his3Δ200 hmg1Δ∷LYS2 hmg2Δ∷HIS3 leu2Δ met2 trp1∷hisG ura3-52∷PTDH3-1MYC-HMG2∷URA3∷PTDH3-Deg1-GFP |
| RHY2477 | MATa ade2-101 lys2-801 his3Δ200 hmg1Δ∷LYS2 hmg2Δ∷HIS3 leu2Δ met2 trp1∷hisG ura3-52∷PTDH3-1MYC-HMG2∷URA3∷PTDH3-Deg1-GFP hrd4-1 |
| RHY2478 | MATa ade2-101 lys2-801 his3Δ200 hmg1Δ∷LYS2 hmg2Δ-4 leu2Δ met2 trp1∷hisG ura3-52∷PTDH3-1MYC-HMG2∷URA3∷PTDH3-Deg1-GFP ubc6Δ∷KanMX ubc7Δ∷HIS3 |
| RHY2479 | MATa ade2-101 lys2-801 his3Δ200 hmg1Δ∷LYS2 hmg2Δ∷HIS3 met2 ura3-52∷PTDH3-1MYC-HMG2∷URA3∷PTDH3-Deg1-GFP hrd2-1 |
| RHY2485 | MATa ade2-101 lys2-801 his3Δ200 hmg1Δ∷LYS2 hmg2Δ∷HIS3 leu2Δ met2 trp1∷hisG ura3-52∷PTDH3-1MYC-HMG2 pRH1377a[HA-prc1-1∷URA3] |
| RHY2486 | MATa ade2-101 lys2-801 his3Δ200 hmg1Δ∷LYS2 hmg2Δ∷HIS3 leu2Δ met2 trp1∷hisG ura3-52∷PTDH3-1MYC-HMG2 pRH1377a[HA-prc1-1∷URA3] hrd4-1 |
| RHY2488 | MATa ade2-101 lys2-801 his3Δ200 hmg1Δ∷LYS2 hmg2Δ-4 leu2Δ met2 trp1∷hisG ura3-52∷PTDH3-1MYC-HMG2 pRH1377a[HA-prc1-1∷URA3] ubc6Δ∷KanMX ubc7Δ∷HIS3 |
| RHY2494 | MATa ade2-101 lys2-801 his3Δ200 hmg1Δ∷LYS2 hmg2Δ∷HIS3 met2 ura3-52∷PTDH3-6MYC-HMG2∷URA3 |
| RHY2495 | MATa ade2-101 lys2-801 his3Δ200 hmg1Δ∷LYS2 hmg2Δ∷HIS3 met2 ura3-52∷PTDH3-6MYC-HMG2∷URA3 hrd4-1 |
| RHY2496 | MATa ade2-101 lys2-801 his3Δ200 hmg1Δ∷LYS2 hmg2Δ∷HIS3 met2 ura3-52∷PTDH3-6MYC-HMG2∷URA3∷NPL4 hrd4-1 |
| RHY2497 | MATa ade2-101 lys2-801 his3Δ200 hmg1Δ∷LYS2 hmg2Δ∷HIS3 leu2Δ met2 trp1∷hisG ura3-52∷PTDH3-1MYC-HMG2∷URA3 |
| RHY2498 | MATa ade2-101 lys2-801 his3Δ200 hmg1Δ∷LYS2 hmg2Δ∷HIS3 leu2Δ met2 trp1∷hisG ura3-52∷PTDH3-1MYC-HMG2∷URA3 hrd4-1 |
| RHY2499 | MATa ade2-101 lys2-801 his3Δ200 hmg1Δ∷LYS2 hmg2Δ∷HIS3 leu2Δ met2 trp1∷hisG ura3-52∷PTDH3-1MYC-HMG2 hrd4-1∷URA3∷NPL4 |
| RHY2659 | MATa ade2-101 lys2-801 his3Δ200 hmg1Δ∷LYS2 hmg2Δ∷HIS3 met2 ura3-52∷PTDH3-6MYC-HMG2 hrd4-1 pRH540b[URA3] |
| RHY2660 | MATa ade2-101 lys2-801 his3Δ200 hmg1Δ∷LYS2 hmg2Δ∷HIS3 met2 ura3-52∷PTDH3-6MYC-HMG2 hrd4-1 pRH1443b[OLE1∷URA3] |
| RHY2661 | MATa ade2-101 lys2-801 his3Δ200 hmg1Δ∷LYS2 hmg2Δ∷HIS3 met2 ura3-52∷PTDH3-6MYC-HMG2 pRH540b[URA3] |
| RHY2662 | MATa ade2-101 lys2-801 his3Δ200 hmg1Δ∷LYS2 hmg2Δ∷HIS3 met2 ura3-52∷PTDH3-6MYC-HMG2 pRH1443b[OLE1∷URA3] |
| RHY2669/DTY11A | MATalpha ade2-1 can1-100 leu2-3,112 trp1-1 |
| RHY2670 | MATalpha ade2-1 can1-100 leu2-3,112 trp1-1 ole1Δ∷LEU2 |
| RHY2673 | MATalpha ade2-1 can1-100 leu2-3,112 trp1-1 pRH540b[URA3] |
| RHY2674 | MATalpha ade2-1 can1-100 leu2-3,112 trp1-1 ole1Δ∷LEU2 pRH1443b[OLE1∷URA3] |
| RHY2675 | MATalpha his3Δ200 leu2Δ1 ura3-52∷hmg2-GFP∷URA3 |
| RHY2676 | MATa leu2Δ1 ura3-52∷hmg2-GFP∷URA3 npl4-1 |
| RHY2677 | MATa his3Δ200 ura3-52∷hmg2-GFP∷URA3 npl4-2 |
| RHY2679/BWG-7a | MATa ade1-100 his4-519 leu 2-3,112 ura3-52 ufd1-1 |
| RHY2680 | MATa ade1-100 his4-519 leu 2-3,112 ura3-52 |
| RHY2681 | MATa ade2-101 lys2-801 his3Δ200 hmg1Δ∷LYS2 hmg2Δ∷HIS3 met2 ura3-52∷PTDH3-6MYC-HMG2 hrd4-1 pRH590a[NPL4∷URA3] |
| RHY2702 | MATa ade1-100 his4-519 leu 2-3,112 ura3-52∷hmg2-GFP∷URA3 ufd1-1 |
| RHY2703 | MATa ade1-100 his4-519 leu 2-3,112 ura3-52∷hmg2-GFP∷URA3 |
| RHY2704 | MATalpha ade2-101 lys2-801 his3Δ200 hmg1Δ∷LYS2 hmg2Δ∷HIS3 met2 ura3-52∷PTDH3-1MYC-HMG pRH1562a[ubiquitin-R-GFP∷URA3] |
| RHY2705 | MATa ade2-101 lys2-801 his3Δ200 hmg1Δ∷LYS2 hmg2Δ∷HIS3 met2 ura3-52∷PTDH3-1MYC-HMG hrd4-1 pRH1562a[ubiquitin-R-GFP∷URA3] |
| RHY2706 | MATa ade2-101 lys2-801 his3Δ200 hmg1Δ∷LYS2 hmg2Δ∷HIS3 met2 ura3-52∷PTDH3-1MYC-HMG hrd2-1 pRH1562a[ubiquitin-R-GFP∷URA3] |
| RHY2707 | MATalpha ade2-101 lys2-801 his3Δ200 hmg1Δ∷LYS2 hmg2Δ∷HIS3 met2 ura3-52∷PTDH3-1MYC-HMG pRH1563a[ubiquitin-L-GFP∷URA3] |
| RHY2708 | MATa ade2-101 lys2-801 his3Δ200 hmg1Δ∷LYS2 hmg2Δ∷HIS3 met2 ura3-52∷PTDH3-1MYC-HMG hrd4-1 pRH1563a[ubiquitin-L-GFP∷URA3] |
| RHY2709 | MATa ade2-101 lys2-801 his3Δ200 hmg1Δ∷LYS2 hmg2Δ∷HIS3 met2 ura3-52∷PTDH3-1MYC-HMG hrd2-1 pRH1563a[ubiquitin-L-GFP∷URA3] |
| RHY2710 | MATalpha ade2-101 lys2-801 his3Δ200 hmg1Δ∷LYS2 hmg2Δ∷HIS3 met2 ura3-52∷PTDH3-1MYC-HMG pRH1564a[ubiquitin-P-GFP∷URA3] |
| RHY2711 | MATa ade2-101 lys2-801 his3Δ200 hmg1Δ∷LYS2 hmg2Δ∷HIS3 met2 ura3-52∷PTDH3-1MYC-HMG hrd4-1 pRH1564a[ubiquitin-P-GFP∷URA3] |
| RHY2712 | MATa ade2-101 lys2-801 his3Δ200 hmg1Δ∷LYS2 hmg2Δ∷HIS3 met2 ura3-52∷PTDH3-1MYC-HMG hrd2-1 pRH1564a[ubiquitin-P-GFP∷URA3] |
| RHY2713 | MATalpha ade2-101 lys2-801 his3Δ200 hmg1Δ∷LYS2 hmg2Δ∷HIS3 met2 ura3-52∷PTDH3-1MYC-HMG pRH1565a[ubiquitinG76V-GFP∷URA3] |
| RHY2714 | MATa ade2-101 lys2-801 his3Δ200 hmg1Δ∷LYS2 hmg2Δ∷HIS3 met2 ura3-52∷PTDH3-1MYC-HMG hrd4-1 pRH1565a[ubiquitinG76V-GFP∷URA3] |
| RHY2715 | MATa ade2-101 lys2-801 his3Δ200 hmg1Δ∷LYS2 hmg2Δ∷HIS3 met2 ura3-52∷PTDH3-1MYC-HMG hrd2-1 pRH1565a[ubiquitinG76V-GFP∷URA3] |
ARS/CEN plasmid.
2μ plasmid.
Figure 4.
Unsaturated fatty acids or OLE1 overexpression do not suppress the degradation defect in hrd4–1 cells. (A-C) Serial dilutions of the indicated strains were grown at the conditions labeling each panel. “+ UFA” indicates 0.5 mM each of oleic acid and palmitoleic acid were added to the labeled plates. “Lovastatin” indicates the addition of 200 μg/ml lovastatin.
Plasmids
The yeast centromeric plasmid pRH590 (YCpNPL4) was constructed by PCR amplification of the NPL4 locus from the genomic library plasmid pRH562, using Vent DNA polymerase (New England Biolabs, Beverly, MA) and the primers oRH571:5′-AAGCTTATGTGATTTTTGGTAAGGGGACG-3′ and oRH572:5′-GGTACCGGCAA-ACTCAAGTAGTTGTGCGTAC-3′. The product was then digested with HindIII and KpnI and ligated into pRS416 digested with the same enzymes. The integrating form of pNPL4 (pRH617) was created by ligating the KpnI-SacI fragment of pRH590 containing NPL4 into pRS406. The PTDH3-Deg1-GFP (green fluorescent protein) fusion was created by PCR amplification of the gene coding for GFPS65T (contained in pRH465) with the following primers: oRH1219 5′-CGCGGGGATCCAAATGGGTAAAGGAGAAGAACTT-3′ and oRH1220 5′-CAAATGTGGTATGGCTGAT-3′. The product was digested with BamHI and SalI and ligated into pRH421 (containing the Deg1 sequence) cut with BglII and SalI.
N-end rule and UFD pathway substrates were constructed as follows. Ubiquitin-X-GFP fusions (where X indicates amino acid residue following Glycine 76 of ubiquitin) were previously constructed for expression in mammalian cells (Dantuma et al., 2000). The pEGFP-N1-based (commercial vector: Clontech) ubiquitin-M-GFP construct was digested with NheI and BsaHI. The 2.4 kb fragment containing the ubiquitin-M-GFP orf was then ligated into SpeI-ClaI digested pRH1556, a previously described ARS/CEN plasmid driving expression of cloned genes from the TDH3 promoter (Mumberg et al., 1995). The resulting plasmid, pRH1561, served as the recipient vector for the ubiquitin-R-GFP, ubiquitin-L-GFP, ubiquitin-P-GFP, and ubiquitinG76V-GFP orfs previously described (Dantuma et al., 2000). Each orf was extracted from the mammalian expression vector by digestion with EcoRI and NotI. The 1000 base pair fragment was then ligated into the 7.3-kbp backbone of pRH1561 to form pRH1562 (ubiquitin-R-GFP), pRH1563 (ubiquitin-L-GFP), pRH1564 (ubiquitin-P-GFP), and pRH1565 (ubiquitinG76V-GFP). The following plasmids were described previously: CPY*-HA, PKAR2-GFP, nup116Δ::URA3, and 2 μ OLE1 (Wente and Blobel, 1993; Pollard et al., 1998; Zhang et al., 1999; Ng et al., 2000).
Assays for Protein Degradation
Stationary chase and cycloheximide chases were performed as previously described (Hampton et al., 1994; Cronin and Hampton, 1999) and methods are summarized in figure legends. Flow cytometry was performed with the use of a BectonDickinson FACScalibur™ instrument. Statistical analysis of flow cytometry data was performed as described (Young, 1977). The graph for Figures 6B and 7D was created to quantify loss of immunoreactivity in the experiments shown in Figure 6A and 7C, respectively. Different exposures of x-ray film were obtained after immunoblotting and were scanned at 600 dpi resolution with the use of NIH Image 1.61 software for MacOS (National Institutes of Health, Bethesda, MD), a UMAX PowerLook 1100 scanner, and a Power Macintosh G4 computer. These scanned images were then subjected to densitometric analysis using the program NIH Image 1.61 for MacOS according to the supplied instructions. This analysis determines the shade of gray (degree of x-ray film exposure) for each pixel and determines the total degree of exposure for a single band. A “threshold” test was used for each band to ensure that no pixel was saturated and therefore that a valid linear comparison could be made between timepoints. These data were then expressed as loss of immunoreactivity over time with the exposure at time 0 set as 100% immunoreactivity.
Figure 6.
Degradation of Deg1-GFP is unaffected by the hrd4–1 mutation. (A) Cycloheximide chase of Deg1-GFP was performed by the addition of cycloheximide to log-phase cells at the indicated times before collection. Equal numbers of cells were lysed and analyzed by SDS-PAGE, followed by immunoblotting for GFP with the use of anti-GFP antibody. (B) Loss of Deg1-GFP immunoreactivity during cycloheximide chase. Densitometric analysis of data obtained in (A) was used to determine loss of GFP immunoreactivity in each indicated strain at 0, 30, and 60 min. (C) Steady-state fluorescence of the Deg1-GFP protein was determined for the indicated strains by flow cytometry. Fluorescence was identical for the following strains: HRD4+ Deg1-GFP, hrd4–1 Deg1-GFP, and a HRD4+ strain transformed with no GFP construct at all (cell autofluorescence). All experiments were performed at the permissive temperature of 30°C.
Figure 7.
Degradation of several cytosolic proteins requires the 26S proteasome, but not Hrd4p. (A) Steady-state fluorescence of GFP fusions bearing different N-terminal amino acids was measured by flow cytometry. (B) Degradation of the UFD substrate, ubiquitinG76V-GFP, was examined by cycloheximide chase followed by flow cytometry. (C) Cycloheximide chase of ubiquitinG76V-GFP followed by immunoblotting with the use of anti-GFP antibody. (D) Loss of ubiquitinG76V-GFP immunoreactivity during cycloheximide chase was determined by densitometric analysis of data obtained in (C).
Antibodies, Immunoprecipitation, and Immunoblotting
Monoclonal anti-myc antibodies were produced as cell-culture supernatant from 9e10 hybridomas obtained from ATCC. Monoclonal anti-HA antibodies (clone 12CA5) were obtained from Babco as purified antibody derived from mouse ascites fluid. Anti-GFP antisera was a gift from Charles Zuker (University of California, San Diego, CA). Anti-Nup116p antisera (to verify deletion of NUP116) was a gift from Susan Wente (Washington University, St. Louis, MO). Anti-Hmg2p antisera was described previously (Hampton et al., 1994). Antiubiquitin antibodies were purchased from Zymed (So. San Francisco, CA). SDS-PAGE was performed using 8% Tris-glycine gels, except for the experiment in Figure 5, which used 3–8% Tris-acetate gels (Invitrogen, Carlsbad, CA). Immunoprecipitation was performed as described in Hampton and Rine, 1994, but with additional protease inhibitors (n-ethylmaleimide, AEBSF, E-64, benzamidine, and ε-amino-n-caproic acid). Immunoblotting was also performed as described in Hampton and Rine, 1994, except that Tris-buffered saline contained 0.45% Tween 20, and 20% heat-inactivated bovine calf serum was used as the blocking agent. Antiubiquitin blots were also processed as previously described (Swerdlow et al., 1986).
Figure 5.
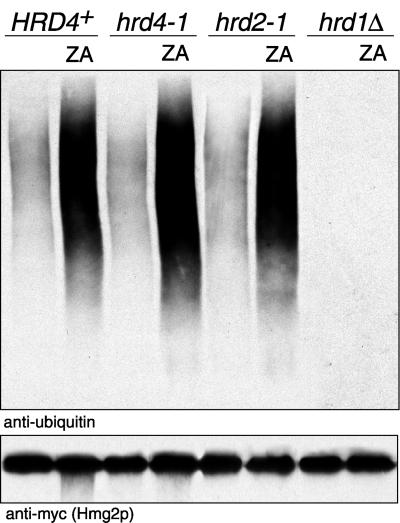
Hmg2p is fully ubiquitinated in a hrd4–1 mutant strain. Ubiquitination of 1myc-Hmg2p was determined by immunoprecipitation of 1myc-Hmg2p from lysates obtained from log-phase cells with the use of anti-Hmg2p antisera. Immunoprecipitated material was separated by SDS-PAGE and followed by immunoblotting for ubiquitin and the myc epitope tag as indicated.
RESULTS
HRD4/NPL4 Is Required for ERAD
We previously described a selection strategy to identify HRD genes required for HMG-CoA reductase (HMGR) degradation (Hampton et al., 1996). This selection used the drug lovastatin, a specific inhibitor of HMGR's essential catalytic activity, to select for mutants with elevated HMGR due to slowed degradation. We modified the previously described hrd selection to allow the isolation of lovastatin-resistant hrd mutants that were also temperature-sensitive for growth. The selection yielded mutations in several genes, including a previously unidentified HRD gene, HRD4. The gene corresponding to the hrd4–1 mutation was cloned and found to be identical to NPL4, an essential gene previously identified in a selection for genes involved in nuclear transport (Figure 1). We have characterized the HRD4/NPL4 gene and found it essential for the ubiquitin-mediated degradation of a diverse group of ER proteins.
We first examined the requirement for HRD4/NPL4 in ERAD by testing the effect of the hrd4–1 mutation on the degradation of 6myc-Hmg2p, the unregulated version of HMGR used in the hrd selection. 6myc-Hmg2p is a constitutively degraded ER membrane protein and is targeted for proteasomal degradation by the ERAD E3 complex, Hrd1p-Hrd3p (Hampton et al., 1996; Gardner et al., 2000; Bays et al., 2001). To examine the degradation of 6myc-Hmg2p, we performed a “stationary chase” of hrd4–1 strains (Hampton et al., 1994). In this assay, cells are grown into stationary phase, where protein synthesis slows while degradation continues. As a result, hrd mutants display a considerably higher level of 6myc-Hmg2p in stationary phase compared with wild-type cells. hrd4–1 cells showed significantly more 6myc-Hmg2p immunoreactivity than wild-type cells in a stationary chase, and this increase was reversed upon the addition of the NPL4 gene to hrd4–1 strains (Figure 2A). We then determined the effect of the hrd4–1 mutation on the native Hmg2 protein. Unlike 6myc-Hmg2p, 1myc-Hmg2p undergoes mevalonate-pathway regulated degradation. When mevalonate and its derivatives are abundant, 1myc-Hmg2p degradation is fast. Conversely, when these same molecules are scarce, 1myc-Hmg2p degradation is slow. We found that regulated 1myc-Hmg2p also required HRD4/NPL4 for degradation. When a stationary chase was performed with 1myc-Hmg2p in a hrd4–1 strain, degradation of 1myc-Hmg2p was impaired (Figure 2A). 1myc-Hmg2p immunoreactivity was identical in wild-type cells and in hrd4–1 cells transformed with a single copy of the NPL4 gene (Figure 2a). Degradation of 6myc-Hmg2p and 1myc-Hmg2p was also assessed with the use of a “cycloheximide chase” (Hampton et al., 1994). In this assay, cycloheximide is added to log-phase cells to stop protein synthesis. However, protein degradation continues and can be measured by detecting a loss of immunoreactivity (for myc-tagged Hmg2p) or a loss of fluorescence (for Hmg2-GFP). When examined by cycloheximide chase, wild-type cells showed a decrease in immunoreactivity over the chase period, while hrd4–1 and hrd1–1 strains showed little loss over the same period (our unpublished results). Therefore, HRD4/NPL4 was required for the degradation of normally regulated 1myc-Hmg2p and its constitutively degraded variant, 6myc-Hmg2p.
Figure 2.
Hrd4p/Npl4p is required for degradation of ER proteins. (A) Both panels are stationary chase assays of protein degradation. The indicated yeast strains were grown into early stationary phase (12 h after strains reached an OD600 of 0.1). Equal numbers of cells were then lysed and subjected to SDS-PAGE, followed by immunoblotting for the myc-epitope tag. (B) Degradation of Hmg2-GFP was measured by cycloheximide chase, followed by flow cytometry. Cycloheximide was added to log-phase (OD600 < 0.2) cells 3 h before collection and analysis by flow cytometry. Each histogram represents 20,000 cells. (C) CPY* degradation in the indicated strains was assayed by cycloheximide chase. Equal numbers of log-phase (OD600 < 0.5) cells were collected at the indicated times post-cycloheximide addition, lysed and analyzed by SDS-PAGE, followed by immunoblotting for the HA-epitope tag present on the CPY*-HA protein. (D) Cycloheximide chase as in C, except that immunoblotting with the use of anti-Fur4p antibodies was performed to detect UP* (E) Steady-state cell fluorescence for the indicated strains expressing the UPR reporter construct, PKAR2-GFP, was analyzed by flow cytometry during log-phase growth. All experiments were performed at the permissive temperature of 30°C.
Because NPL4 has been implicated in a phenotype quite distinct from ERAD, we asked whether the ERAD defect seen in hrd4 mutants was a general feature of losing HRD4/NPL4 function or if the ERAD phenotype was unique to the hrd4/npl4 allele we isolated. npl4–1 and npl4–2 alleles were isolated in the original npl selection for mutants that fail to properly localize an NLS (nuclear localization signal)-bearing protein (DeHoratius and Silver, 1996). We tested whether npl4–1 and npl4–2 strains were defective in ERAD by assaying their ability to degrade the reporter protein Hmg2-GFP. Hmg2-GFP is degraded via the Hrd pathway just like wild-type Hmg2p, and the GFP fusion allows detection of Hmg2-GFP levels by flow cytometry. When wild-type NPL4+ cells expressing Hmg2-GFP were subjected to a cycloheximide chase, loss of fluorescence was observed as the Hmg2-GFP protein was degraded (Figure 2B). However, very little loss of fluorescence was observed during a cycloheximide chase of npl4–1 and npl4–2 strains (Figure 2B), indicating that npl4–1 and npl4–2 mutant strains were indeed deficient in Hmg2-GFP degradation. Therefore, several different hypomorphic alleles of NPL4 isolated in very different genetic studies were deficient in ERAD.
We tested the degradation of several other known ERAD substrates in hrd4–1 mutant strains. CPY* is a mutant, misfolded form of carboxypeptidase Y that is retained in the endoplasmic reticulum and degraded by the Hrd pathway (Finger et al., 1993; Bordallo et al., 1998). To test the effect of the hrd4–1 mutation on the degradation of CPY*, we performed a cycloheximide chase of CPY* in HRD4+ and hrd4–1 cells. CPY* was rapidly degraded in HRD4+ cells, but degradation in hrd4–1 cells was clearly impaired (Figure 2C). Loss of Ubc7p, the principle ubiquitin conjugating enzyme in ERAD, also blocked the degradation of CPY* (Figure 2C).
The degradation of some ERAD substrates does not require the Hrd1p-Hrd3p ubiquitin ligase complex (Wilhovsky et al., 2000). For example, a mutant form of uracil permease (UP*) does not require either Hrd1p or Hrd3p for its ER-associated degradation (Wilhovsky et al., 2000). We decided to test whether the degradation of UP* required Hrd4p/Npl4p to see how broadly Hrd4p/Npl4p affected ERAD. In a cycloheximide chase, UP* was degraded in wild-type cells (Figure 2D). However, the degradation of UP* was slowed in hrd4–1 cells (Figure 2D). Loss of Ubc7p also caused a defect in UP* degradation (Figure 2D). Therefore, Hrd4p function was required for the degradation of a larger set of ERAD substrates than those ubiquitinated by the action of the Hrd1p-Hrd3p ubiquitin ligase complex.
As Hrd4p/Npl4p appeared to function broadly in ERAD, we tested hrd4 mutants for an elevated unfolded protein response (UPR). The UPR is a coordinated regulation of gene expression induced upon an increase of unfolded proteins in the ER (Sidrauski et al., 1998). The genes up-regulated by the UPR include those coding for protein folding machinery, like the chaperone Kar2p, and ERAD components like the E3 Hrd1p (Kohno et al., 1993; Travers et al., 2000). The UPR serves as sensitive indicator of conditions that increase the abundance of misfolded proteins in the ER. For instance, mutations that block ERAD often lead to an elevated UPR, as cells are unable to remove misfolded proteins from the ER (Friedlander et al., 2000; Travers et al., 2000). To assess the UPR in hrd4–1 cells, we introduced a UPR reporter construct (PKAR2-GFP) into wild-type, hrd4–1, and hrd1Δ cells. Flow cytometry was then used to measure cell fluorescence. When hrd4–1 cells were analyzed, they showed a clear increase in cell fluorescence compared with wild-type cells –indicating that, indeed, hrd4–1 cells exhibited an elevated UPR (Figure 2E). As previously described, hrd1Δ cells also displayed an increased UPR compared with wild-type cells (Friedlander et al., 2000; Travers et al., 2000). These results are consistent with a loss of Hrd4p/Npl4p that leads to an increased burden of unfolded proteins in the ER.
HRD4/NPL4 has been implicated in both nuclear transport and fatty acid biosynthesis (DeHoratius et al., 1996; Hitchcock et al., 2001). Therefore, we asked whether one or both of these defects was the cause of ERAD deficiency of hrd4/npl4 mutants. The following results indicated that neither nuclear transport nor fatty acid biosynthesis explained the defects in ERAD.
A Block in Nuclear Import/Export Does Not Affect ERAD
npl4 mutants are defective in nuclear import/export and were originally isolated by virtue of this deficiency. However, the npl4 mutant defect in nuclear import/export is seen after a shift to the restrictive temperature of 37°C (DeHoratius et al., 1996). There appears to be no significant deficit in nuclear transport at 30°C in npl4–1, npl4–2 or hrd4–1 cells. In contrast, the same npl4/hrd4 cells show a substantial defect in ERAD at the permissive temperature of 30°C, with almost no detectable growth deficit. (Note: All degradation experiments in this paper were performed at the permissive temperature of 30°C with the exception of those in Figure 3.) Because nuclear import and export were not significantly compromised at 30°C in npl4/hrd4 strains, a deficiency in nuclear import/export would have been insufficient to explain the ERAD defect in npl4/hrd4 cells. After all, the ERAD defect was present in hrd4/npl4 cells at 30°C, while the nuclear transport defect was not present at 30°C. Nonetheless, we tested whether blocking nuclear import/export with a distinct mutation would had any effect on ERAD. To achieve a block in nucleocytoplasmic traffic, we used strains bearing the temperature-sensitive nup116Δ allele. Loss of the bona fide nuclear pore protein Nup116p (Ho et al., 2000; Rout et al., 2000; Strawn et al., 2000) leads to a profound block in nuclear transport after a shift to the restrictive temperature of 37°C (Wente and Blobel, 1993). We chose the nup116Δ allele for several reasons. First, we could achieve a complete loss of function in a nuclear pore protein and an apparently complete block in nuclear traffic by using nup116Δ cells, providing a stringent test of any requirement for nuclear transport in ERAD. Second, the block in nuclear traffic and the presence of nuclear membrane herniations in nup116Δ cells are strikingly similar to the phenotypes of npl4 mutant cells at the nonpermissive temperature (Wente and Blobel, 1993; DeHoratius et al., 1996). Therefore, nup116Δ offered a type of block in nuclear traffic equivalent to npl4. Consistent with this, NUP116 and NPL4 also show genetic indications of affecting similar functions, as overexpression of NPL4 can partially suppress the structural and growth defects of nup116Δ strains (DeHoratius et al., 1996). These numerous similarities make nup116Δ especially appropriate for testing whether a block in nuclear traffic underlies the ERAD phenotypes of npl4 mutants.
Figure 3.
A block in nuclear transport has no effect on the degradation of an ER membrane protein. (A) The indicated strains expressing the constitutively-degraded 6MYC-Hmg2p were grown at the permissive temperature of 23°C. Log phase (OD600 = 0.1) cells were then shifted to the restrictive temperature of 37°C for 3.5 h before the addition of cycloheximide and subsequent chase at the indicated times. During this time, wild-type cells doubled at the normal rate, while the temperature-sensitive hrd4–1 and nup116Δ cells failed to complete even one additional doubling after temperature shift. Equal numbers of cells were collected at the indicated times following addition of cycloheximide and lysed. These lysates were then separated by SDS-PAGE, and immunoblotting for the myc epitope tag was performed. (B) Nup116 protein is absent in nup116Δ cells. Lysates from the indicated strains were subjected to SDS-PAGE and immunoblotting using anti-Nup116p antibodies.
We introduced the nup116Δ allele into a strain expressing 6myc-Hmg2p. The absence of Nup116 protein was verified by immunoblotting. Nup116p immunoreactivity was present in the wild-type strain but not in the nup116Δ strain (Figure 3B). As expected from the previous characterization of the nup116Δ allele (Wente and Blobel, 1993), strains lacking Nup116p were temperature-sensitive (our unpublished results). nup116Δ cells were not lovastatin-resistant, indicating that loss of Nup116p did not cause a significant defect in 6myc-Hmg2p degradation (our unpublished results). Furthermore, using biochemical assays of 6myc-Hmg2p, we found no effect of nup116Δ on 6myc-Hmg2p stability. In these assays, wild-type, hrd4–1 and nup116Δ cells were grown at the permissive temperature of 23°C. Cells were shifted to 37°C for 3.5 h before the addition of cycloheximide and subsequent chase of 6myc-Hmg2p immunoreactivity. This shift to 37°C initiated a rapid and profound block in nuclear import/export in nup116Δ cells (Wente and Blobel, 1993). Despite this block in nucleocytoplasmic traffic, nup116Δ cells showed no detectable deficit in 6myc-Hmg2p degradation (Figure 3A). hrd4–1 cells showed little degradation of the 6myc-Hmg2 protein under the same conditions (Figure 3A). Therefore, ERAD proceeded normally despite the block in nuclear import/export caused by the nup116Δ allele and despite the numerous phenotypic similarities between nup116Δ and npl4/hrd4 mutants.
Unsaturated Fatty Acids and Npl4p
In yeast, unsaturated fatty acids (UFA) are synthesized by the action of Ole1p, a Δ9 fatty acid desaturase that catalyzes the formation of palmitoleic acid (16:1) and oleic acid (18:1), from palmitoyl-CoA (16:0) and stearolyl-CoA (18:0), respectively (Stukey et al., 1990). The OLE1 gene is essential, but the addition of either palmitoleic acid or oleic acid can restore growth to ole1Δ cells (Zhang et al., 1999). Transcription of the OLE1 gene requires two related and redundant transcription factors: Spt23p and Mga2p (Zhang et al., 1999). Recently, it has been shown that Spt23p and Mga2p are made as inactive proteins residing in the ER/nuclear membrane. To become active transcription factors, Spt23p and Mga2p are processed in a UFA-regulated process requiring the 26S proteasome (Hoppe et al., 2000). This processing of Mga2p and Spt23p requires HRD4/NPL4 (Hitchcock et al., 2001). Because UFAs are critical to the function of membranes throughout the cell, and because ERAD occurs at the ER membrane, we were compelled to ask whether the degradation defect in hrd4/npl4 mutant strains was the result of a lack of unsaturated fatty acids (UFAs). We began our analysis by testing whether UFAs could reverse the temperature-sensitivity and lovastatin-resistance of hrd4 mutant strains.
To test the reversal of temperature sensitivity, HRD4+ and hrd4–1 cells were plated on media with and without unsaturated fatty acids. These plates were then incubated at a series of permissive and restrictive temperatures. At 35°C, hrd4–1 cells failed to grow even on media containing unsaturated fatty acids (Figure 4A). HRD4+ cells grew at 35°C on both media (Figure 4A). Using a series of restrictive temperatures, we found that colony size could increase slightly when hrd4–1 cells were plated on UFA media at 33°C (our unpublished results), but at no temperature did we observe growth of hrd4–1 cells exclusively on UFA media, with no growth on media lacking unsaturated fatty acids. Therefore, the addition of unsaturated fatty acids had very little, if any, effect on the Ts− phenotype of hrd4–1 cells. In the same experiments, the growth phenotype of ole1Δ cells was completely reversed upon the addition of unsaturated fatty acids. We also tested the npl4–1 and npl4–2 alleles for suppression by unsaturated fatty acids, noting that these alleles were isolated in a different parent strain than the hrd4–1 allele. However, both npl4–1 and npl4–2 cells failed to grow at 37°C, even when UFA were added to the media (our unpublished results). Although UFA failed to restore wild-type growth to npl4 mutant strains, the addition of UFA did allow npl4 mutant strains to tolerate an increase of 1°C in restrictive temperature, as npl4–1 and npl4–2 cells could grow at 34°C only when plated on media containing UFA (our unpublished results). Therefore, there does appear to be a partial suppression of npl4 temperature sensitivity by UFA, but not a complete suppression. These results are consistent with those reported by Hitchcock et al. (2001) who found that UFA can partially suppress the temperature sensitivity of npl4–1 and npl4–2 strains.
We also looked for suppression of npl4/hrd4 temperature sensitivity with the use of an OLE1 2 μ (multicopy) plasmid. Suppression of npl4/hrd4 mutants by an OLE1 2 μ plasmid is an important test of the genetic relationship between Npl4p and Spt23p/Mga2p, because the OLE1 2 μ plasmid completely suppresses the growth and UFA phenotypes of a mga2Δ spt23ts double mutant strain (Zhang et al., 1999). Although mga2Δ spt23ts mutant strains fail to produce OLE1 transcript at the restrictive temperature, transformation with an OLE1 2 μ plasmid produces an abundance of OLE1 mRNA, allowing full complementation (Zhang et al., 1999). If the sole effect of losing Npl4p/Hrd4p function were an inability to process (and thus activate) Mga2p and Spt23p, then npl4/hrd4 loss-of-function mutants should be phenotypically identical to mga2 spt23 loss-of-function mutants and thus would be completely suppressed by overexpression of OLE1. We tested this by transformation of HRD4+ and hrd4–1 strains with the 2 μ OLE1 plasmid isolated as a high-copy suppressor of the mga2Δ spt23ts mutation. When plated at 35°C, hrd4–1 strains failed to survive even when bearing the OLE1 2 μ plasmid, although a slight increase in colony size before death was seen in strains with the OLE1 plasmid. (Figure 4C). For the npl4–1 and npl4–2 alleles, we saw only the partial suppression seen with UFA supplementation — OLE1 overexpression allowed a 1°C increase in the restrictive temperature but did not restore wild-type growth to npl4–1 or npl4–2 strains (our unpublished results). In contrast, transformation with the OLE1 2 μ plasmid was able to completely restore growth to ole1Δ cells.
We also tested the ability of unsaturated fatty acids and OLE1 overexpression to suppress the degradation defect in hrd4–1 strains. HRD4+ and hrd4–1 cells were plated on media with and without lovastatin. hrd4–1 cells showed the same plating efficiency on lovastatin even when supplemented with UFA (Figure 4B). Likewise, hrd1–1 cells also showed lovastatin resistance, whether the plates were supplemented with UFA or not (Figure 4B). HRD4+ cells were lovastatin-sensitive in both cases. Likewise, transformation of hrd4–1 strains with a 2 μ OLE1 plasmid failed to restore degradation and thus reverse the lovastatin-resistance phenotype (Figure 4C). HRD4+ cells were identically lovastatin-sensitive, whether transformed with an empty or OLE1 2 μ plasmid. These results indicated that the hrd4/npl4 defect in degradation was not due to an indirect effect of low unsaturated fatty acid concentrations in the cell.
Hrd4p Functions at a Postubiquitination But Preproteasomal Step in the Ubiquitin-Proteasome Pathway
ERAD of Hmg2p and other proteins proceeds by the ubiquitin-proteasome pathway. Previously described Hrd proteins function at two distinct steps in this pathway: they are components of either the E2/E3 ubiquitination machinery (e.g., Hrd1p, Ubc7p, Ubc1p) or the 26S proteasome itself (e.g., Hrd2p/Rpn1p) (Hampton et al., 1996; Bays et al., 2001). E2 and E3 proteins are required for ubiquitination of proteins, such that their loss abrogates ubiquitination. In contrast, deficiencies in components of the 26S proteasome leave proteins fully ubiquitinated. We examined where Npl4p/Hrd4p functions in ubiquitin-mediated degradation by testing the ubiquitination of Hmg2p in npl4/hrd4 mutant strains.
The ubiquitination of Hmg2p is subject to feedback regulation, and drugs that alter the Hmg2p (mevalonate) pathway also alter the ubiquitination of Hmg2p. Zaragozic acid, an inhibitor of the pathway enzyme squalene synthase, increases ubiquitination of Hmg2p by allowing the accumulation of a natural signal for Hmg2p degradation. (Figure 5 and Hampton and Bhakta, 1997; Gardner and Hampton, 1999). When ubiquitination was assayed in a wild-type HRD4+ strain, Hmg2p ubiquitination was increased dramatically by a 5-min addition of zaragozic acid (Figure 5, “ZA”). This was also the case for hrd4–1 and hrd2–1 strains, which showed full regulated ubiquitination of Hmg2p (Figure 5). In contrast, a hrd1Δ strain showed no ubiquitination of Hmg2p even when maximally stimulated by zaragozic acid (Figure 5). Therefore, Hrd4p/Npl4p was not required for the actual ubiquitination of Hmg2p, but instead, like Hrd2p/Rpn1p, Hrd4p/Npl4p was required for the degradation of fully-ubiquitinated Hmg2p.
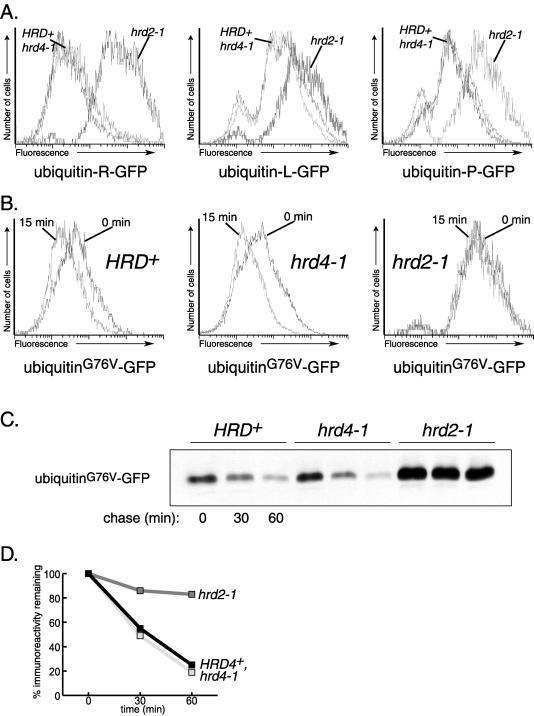
Because both hrd4 and the proteasomal hrd2/rpn1 mutants allowed ubiquitination but not degradation of Hmg2p, we asked whether Hrd4p/Npl4p was required for general proteasomal degradation. To test the effect of the hrd4–1 allele on the activity of the 26S proteasome, we evaluated the degradation of several cytosolic proteins. The first protein, Deg1-GFP, is a variant of GFP bearing the Deg1 sequence at its N terminus (Deg1-GFP). The Deg1 sequence originates from the rapidly degraded Matα2 protein and meets the classic definition of a “degron,” in that it can be transferred to a number of different stable proteins to target them for degradation by the ubiquitin-proteasome pathway (Hochstrasser et al., 1991; Chen et al., 1993). Indeed, the addition of the Deg1 sequence to GFP caused the protein to be rapidly degraded (Figure 6A and our unpublished results). In fact, this Deg1-GFP fusion was so rapidly degraded that its steady-state fluorescence in flow cytometry was barely detectable above normal cellular autofluorescence of wild-type cells (Figure 6C and our unpublished results). Only when Deg1-GFP degradation was impaired by the ubc6Δ ubc7Δ double mutation were Deg1-GFP expressing cells bright (Figure 6C). No difference in fluorescence was seen between HRD4+ and hrd4–1 cells, indicating that HRD4 was not required for degradation of Deg1-GFP (Figure 6C). We confirmed this by examining Deg1-GFP stability with a cycloheximide chase followed by immunoblotting rather than flow cytometry. Deg1-GFP was indeed rapidly degraded (Figure 6A and 6B). This degradation was impaired in a ubc6Δ ubc7Δ strain (Figure 6A and 6B), an observation consistent with the requirement for these UBC genes in Deg1-mediated degradation (Chen et al., 1993). Similarly, loss of function in the 26S proteasome gene HRD2/RPN1 also impaired degradation of Deg1-GFP (Figure 6A and 6B), which is also consistent with the previously described requirement for the 26S proteasome in Deg1-mediated degradation (DeMarini et al., 1995). In striking contrast, loss of Hrd4p/Npl4p function had no detectable effect on the degradation rate of Deg1-GFP (Figure 6A and 6B). We therefore concluded that proteasome function was not impaired in hrd4–1 strains because the degradation of the proteasome-dependent substrate Deg1-GFP was unaffected by the hrd4–1 mutation. We extended this observation by testing the effect of hrd4–1 on two other ubiquitin-mediated routes to proteasomal degradation: the N-end rule and UFD (ubiquitin-fusion degradation) pathways (Johnson et al., 1995; Varshavsky, 1996). To test effects on the N-end rule pathway, ubiquitin-R-GFP, ubiquitin-L-GFP, and ubiquitin-P-GFP fusions were expressed in HRD+, hrd4–1 and hrd2–1 strains. As expected, ubiquitin-R-GFP was destabilized by the presence of the arginine amino-terminal residue resulting from in vivo cleavage of the ubiquitin moiety (Figure 7A and our unpublished results). The degradation of ubiquitin-R-GFP was equally rapid in both HRD+ and hrd4–1 strains, but it was severally impaired in a hrd2–1 strain (Figure 7A and our unpublished results). The same result was seen for the other N-end rule substrates, ubiquitin-L-GFP and ubiquitin-P-GFP (Figure 7A and our unpublished results; note that ubiquitin-P-GFP is also subject to UFD degradation due to slow cleavage of ubiquitin moiety).
Along with ubiquitin-P-GFP, we examined the degradation of another UFD substrate, ubiquitinG76V-GFP. (The mutation of the carboxy-terminal glycine in ubiquitin blocks cleavage of the ubiquitin moiety and renders the entire fusion unstable.) UbiquitinG76V-GFP was rapidly degraded in both HRD+ and hrd4–1 strains (Figure 7B-7D), but its degradation was dramatically slowed by the hrd2–1 mutation. Although ubiquitinG76V-GFP degradation was quite sensitive to a defect in the 26S proteasome, its degradation was not affected by loss of Hrd4p/Npl4p function. Taken together, these studies on three distinct classes of soluble proteasomal substrates indicated that loss of Hrd4p/Npl4p does not affect general proteasome function. Furthermore, hrd4–1 strains showed no altered sensitivity to the amino acid analog canavanine (our unpublished results), unlike most proteasome mutants previously tested (Hilt et al., 1993; Heinemeyer et al., 1994; Fu et al., 1998; Rubin et al., 1998).
CDC48, UFD1, and HRD4/NPL4 Are Each Required for ER Protein Degradation
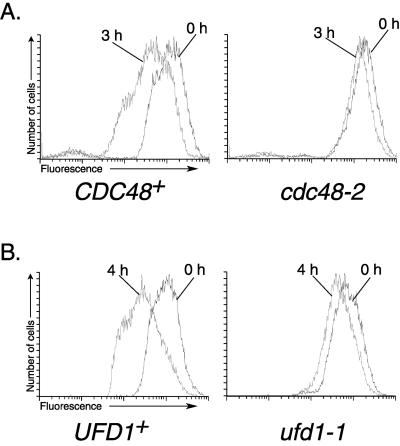
Hrd4p/Npl4p is present in a complex with Ufd1p and Cdc48p in the cell (Meyer et al., 2000). Cdc48p is an AAA ATPase capable of interacting with an impressive array of proteins, including three proteins in the Ufd degradation pathway: Ufd1p, Ufd2p, and Ufd3p/Doa1p (Ghislain et al., 1996; Koegl et al., 1999; Meyer et al., 2000). Cdc48p also physically interacts with the 26S proteasome in an ATP-dependent manner (Verma et al., 2000). Therefore, we decided to test whether CDC48 was required for ERAD. We transformed CDC48+ and cdc48–2 mutant strains with Hmg2-GFP and performed cycloheximide chase analysis of Hmg2-GFP degradation. Although Hmg2-GFP was degraded in wild-type CDC48+ cells, cdc48–2 showed little degradation over the chase period (Figure 8A, performed at the permissive temperature of 30°C). This result indicated that CDC48 was indeed required for ERAD.
Figure 8.
CDC48 and UFD1 are required for ERAD. Hmg2-GFP was transformed into strains with the indicated CDC48 (A) and UFD1 (B) alleles. Degradation of Hmg2-GFP was then measured by cycloheximide chase followed by flow cytometry. All experiments were performed at the permissive temperature of 30°C.
We also tested the requirement for the Cdc48p-interacting protein, Ufd1p, in ERAD. The Hmg2-GFP reporter protein was expressed in both UFD1+ and ufd1–1 strains. The loss of fluorescence during a cycloheximide chase of Hmg2-GFP protein was measured by flow cytometry. ufd1–1 strains showed only a minor loss of fluorescence over the chase period, while UFD1+ strains displayed typical Hmg2-GFP degradation (Figure 8B). Therefore, UFD1 was required for ERAD. Furthermore, each member of the Hrd4p/Npl4p-Ufd1p-Cdc48p complex was required for ERAD, but not for the degradation of UFD pathway substrates.
DISCUSSION
Hrd4p/Npl4p and the Endoplasmic Reticulum
Hrd4p/Npl4p is required for ERAD of diverse substrates including 6myc-Hmg2p, native Hmg2p, CPY*, and UP*. HRD4/NPL4-dependent substrates include at least one protein, UP*, whose degradation does not require the Hrd1p ubiquitin ligase (Wilhovsky et al., 2000). The processing of the ER-membrane-bound transcription factors Spt23p and Mga2p also requires NPL4. hrd4/npl4 mutant strains fail to liberate the ER membrane–bound Spt23 or Mga2 proteins when cells are deprived of unsaturated fatty acids (Hitchcock et al., 2001). This processing of Spt23p and Mga2p is a ubiquitin- and proteasome-dependent process requiring the E3 Rsp5p (Hoppe et al., 2000). Thus, loss of Hrd4p/Npl4p function appears to create a broad lesion in the ER-localized processing of ubiquitinated proteins as the defect in hrd4/npl4 mutant cells crosses several E3 and E2 boundaries.
The accumulation of misfolded proteins at the ER leads to an elevated gene expression known as the unfolded protein response (UPR). Genes elevated in the UPR include those coding for protein folding machinery like the chaperone Kar2p as well as ERAD machinery like the E3 Hrd1p (Travers et al., 2000). The UPR serves as a sensitive and useful indicator of the protein folding state at the ER, because the loss of genes required for protein folding and protein degradation at the ER leads to a constitutively elevated UPR (Friedlander et al., 2000; Travers et al., 2000). We tested hrd4/npl4 mutant strains for an elevated UPR and found that hrd4-/npl4- cells showed an unfolded protein response, indicating that Hrd4p/Npl4p is centrally involved in ER protein maintenance.
These instances of Hrd4p/Npl4p function at the endoplasmic reticulum do not preclude the involvement of Hrd4p/Npl4p in cytoplasmic processes. It is possible that Hrd4p/Npl4p exerts some cytosolic effect, as the majority of Npl4 protein is found in the cytosol (Hitchcock et al., 2001). Nevertheless, the “ER-centric” behavior of Hrd4p/Npl4p is striking and prompts further study into Hrd4p/Npl4p function at the endoplasmic reticulum. Given the striking similarity of Hrd4p/Npl4p sequence among divergent species, these studies will undoubtedly illuminate an essential process conserved among eukaryotes.
Hrd4p/Npl4p Functions at a Novel Step in ERAD
Our previous genetic studies in ERAD identified two broad classes of mutants: mutants that block the actual ubiquitination of a target protein and mutants affecting proteasome function. For example, Hrd1p and Hrd3p are components of an ERAD-dedicated ubiquitin ligase, whereas Hrd2p/Rpn1p is a subunit of the 26S proteasome (Hampton et al., 1996; Gardner et al., 2000; Bays et al., 2001). HRD4/NPL4 is not required for the ubiquitination of Hmg2p, consistent with the observation that HRD4/NPL4 is required for the degradation of substrates with different E3 and E2 requirements. Because npl4 mutants did not fall into the first class of mutants, we tested whether hrd4/npl4 mutants were defective in proteasome function. We found that the proteasome could function normally in hrd4/npl4 mutant cells as the degradation of a variety of cytosolic, proteasome-dependent substrates was normal in hrd4/npl4 mutant cells. We also observed that hrd4/npl4 mutants had no altered sensitivity to canavanine, unlike other proteasome mutants (Hilt et al., 1993; Heinemeyer et al., 1994; Fu et al., 1998; Rubin et al., 1998). Furthermore, several ambitious and thorough efforts, biochemical and genetic, to identify all of the components of the 26S proteasome have never implicated Hrd4p/Npl4p as a proteasome subunit or even as a proteasome-interacting protein. (Heinemeyer et al., 1994; Glickman et al., 1998; Verma et al., 2000) It appears that neither ubiquitination or proteasomal function are deficient in hrd4/npl4 mutant cells, indicating that hrd4/npl4 mutants are blocked in degradation at a step between ER ubiquitination of the target protein and its degradation by the 26S proteasome. These results suggest that Hrd4p/Npl4p has an interesting and novel role in proteasomal processing of ubiquitinated proteins at the ER.
We placed the function of Hrd4p/Npl4p between ubiquitination and proteasomal degradation, but the possibility does exist that Hrd4p/Npl4p may act at the ubiquitination step in the construction and/or editing of the multiubiquitin chains added to target proteins. In this case, the loss of Hrd4p/Npl4p function would somehow cause the formation of multiubiquitin chains that are not competent for recognition by the 26S proteasome. The observation that the rate and pattern of Hmg2p ubiquitination is not noticeably affected in a hrd4–1 mutant strain may argue somewhat against this model, but it cannot rule out the possibility that Hrd4p/Npl4p may somehow affect the structure of multi-ubiquitin chain.
One continuing area of investigation in protein degradation concerns whether the proteasome alone can directly recognize ubiquitinated proteins. Although a fundamental question, a definitive answer has remained elusive. In npl4- cells, the 26S proteasome fails to recognize ubiquitinated ER proteins, although the proteasome is fully capable of degrading a model cytosolic substrate. This suggests, at least for ERAD substrates, that the proteasome alone is not capable of recognizing ubiquitinated proteins and requires some function that is lost in hrd4/npl4 mutant cells. Whether Hrd4p/Npl4p directly presents ubiquitinated ER proteins to the proteasome is not yet clear, but further study of the function lost in hrd4-/npl4- cells will likely provide much needed insight into how ubiquitinated proteins are recognized by the proteasome.
Hrd4−/Npl4− Phenotypes Result from a Defect in Protein Degradation
Because Hrd4p/Npl4p is required for protein degradation, nuclear transport, and unsaturated fatty acid biosynthesis, we were compelled to ask which primary function was actually being lost in an hrd4/npl4 mutant and whether loss of that one function could explain the other observed hrd4/npl4 phenotypes. We first considered whether nuclear transport was the primary function lost in hrd4/npl4 mutants, but we found this model unconvincing, as hrd4/npl4 mutants showed a profound defect in protein degradation at the permissive temperature, where nuclear transport functioned normally. Furthermore, we found that an equally severe block in nucleocytoplasmic traffic caused by the nup116Δ mutant had no effect on ERAD. Thus, loss of nuclear transport did not cause general ERAD defects. Conversely, several reports have indicated a requirement for protein degradation in nuclear transport. For example, a mutation in the gene coding for the proteasome subunit Rpn2p causes a profound block in nuclear transport, as do mutations in the ubiquitin-protein ligase Tom1p (Yokota et al., 1996; Utsugi et al., 1999). Taken together, the reasonable model is that the original nuclear defects of npl4 mutants are the result of the degradation defect in these cells.
Recent studies of Hrd4p/Npl4p involvement in unsaturated fatty acid (UFA) biosynthesis prompted another investigation into the cause of hrd4/npl4 phenotypes. Hitchcock et al. (2001) show that npl4 mutant strains are deficient in the UFA-regulated processing of two transcription factors (Spt23p and Mga2p) required for the production of Ole1p, a Δ9-fatty acid desaturase (Zhang et al., 1999). Because UFA levels can have profound effects on functions throughout the cell and especially at cell membranes like the ER membrane (Stukey et al., 1990; Stewart and Yaffe, 1991; Zhang et al., 1999), we tested the effects of UFAs on our hrd4–1 strain but found only a slight suppression of the Ts− phenotype by UFA supplementation and no suppression of the lovastatin-resistance phenotype. Although Hrd4p/Npl4p clearly plays an important role in the processing of the transcription factors Spt23p and Mga2p, processing of Spt23p and/or Mga2p cannot be the sole function of Hrd4p/Npl4p, because even mga2Δ spt23ts mutants are completely suppressed by overexpression of OLE1 at their restrictive temperature. hrd4/npl4 mutants are not completely suppressed by OLE1 and thus are not phenocopies of cells that simply lack functional Mga2p and/or Spt23p. Furthermore, we could find no role for UFA or OLE1 in the ERAD defect of hrd4/npl4 mutants.
A defect in proteasomal processing of ubiquitinated proteins at the ER best explains the reported hrd4/npl4 phenotypes–hrd4/npl4 mutants are deficient in the proteasome-dependent processing of ER-bound transcription factors required for unsaturated fatty acid synthesis. This reduction in cell UFA levels leads to some growth defect and appears to be at least partially responsible for the nuclear transport defect in hrd4/npl4 mutants (Hitchcock et al., 2001). The npl4/hrd4 defect in proteasomal processing also affects the degradation of ER proteins because ERAD proceeds by action of the ubiquitin-proteasome pathway. Therefore, we offer that protein degradation is the primary function lost in hrd4/npl4 mutants and that the reported hrd4/npl4 phentoypes are best explained by this loss of ubiquitin-mediated degradation.
The Mechanism of Hrd4p/Npl4p Function
In hrd4/npl4 mutants, ubiquitinated ER proteins fail to be processed by a functional 26S proteasome. This phenotype suggests several models of Hrd4p/Npl4p function. In one model, Hrd4p/Npl4p may physically mediate interaction between the 26S proteasome and ERAD substrates. This mediation may also require Cdc48p, which has been shown to associate physically with Npl4p via Ufd1p (Meyer et al., 2000). Cdc48p has also been shown to associate with the proteasome in an ATP-dependent manner (Verma et al., 2000). It is intriguing to speculate that the Cdc48p-Ufd1p-Hrd4p complex actually anchors or recruits the 26S proteasome to the ER. Because a specific population of ER-bound proteasomes exists in the cell (Hori et al., 1999), it will be enlightening to determine whether cdc48 and/or npl4 mutants have any effect on the abundance or distribution of these ER-bound proteasomes. As Cdc48p is required for protein degradation at the both the cytosol (Ghislain et al., 1996) and the endoplasmic reticulum (this report), Cdc48p may mediate the physical association of ubiquitinated proteins and the proteasome at both locations with Hrd4p/Npl4p acting as the “ER-specific adapter.” This model of Cdc48p function is consistent with recent data suggesting that Cdc48p acts in the recognition of multiubiquitin chains (Dai and Li, 2001). Determining the effect of Hrd4p/Npl4p on the recognition of multiubiquitinated proteins may well lend important insight into Hrd4p/Npl4p function as well as the “ER-centric” behavior of Hrd4p/Npl4p.
ACKNOWLEDGMENTS
The authors thank Susan Wente (Washington University, St. Louis, MO) for strains, plasmids, antibodies, and advice; Martin Latterich (Diversa Corp., San Diego, CA) for strains, plasmids, and advice; Charles Martin (Rutgers University, Piscataway, NJ) for strains, plasmids, and advice about UFA supplementation and detergents in growth media; Pamela Silver (Harvard University, Cambridge, MA), Heike Krebber (Phillips-University of Marburg, Germany), and Amy Hitchcock (Harvard University, Cambridge, MA) for strains, plasmids, advice, and sharing invaluable unpublished data; Davis Ng (Pennsylvania State University, University Park, PA) for the CPY*-HA plasmid; Michael Yaffe (University of California, San Diego, CA) for strains, reagents, and advice; Douglass Forbes (University of California, San Diego, CA) for instruction; members of the Hampton laboratory for valuable discussion. This work was supported by the American Heart Association (N.B.), ARCS Foundation, San Diego, CA (N.B.), the National Institutes of Health, Bethesda, MD (R.H.), and a Searle Scholarship (R.H.).
REFERENCES
- Arias IM, Doyle D, Schimke RT. Studies on the synthesis and degradation of proteins of the endoplasmic reticulum of rat liver. J Biol Chem. 1969;244:3303–3315. [PubMed] [Google Scholar]
- Bays NW, Gardner RG, Seelig LP, Joaezerio C, Hampton RY. Hrd1p/Der3p is a membrane-anchored ubiquitin ligase required for ER-associated degradation. Nat Cell Biol. 2001;3:24–29. doi: 10.1038/35050524. [DOI] [PubMed] [Google Scholar]
- Bordallo J, Plemper RK, Finger A, Wolf DH. Der3p-Hrd1p is required for endoplasmic reticulum-associated degradation of misfolded lumenal and integral membrane proteins. Mol Biol Cell. 1998;9:209–222. doi: 10.1091/mbc.9.1.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodsky JL, McCracken AA. ER protein quality control and proteasome-mediated protein degradation. Semin Cell Dev Biol. 1999;10:507–513. doi: 10.1006/scdb.1999.0321. [DOI] [PubMed] [Google Scholar]
- Chen P, Johnson P, Sommer T, Jentsch S, Hochstrasser M. Multiple Ubiquitin-Conjugating Enzymes Participate in the In Vivo Degradation of the Yeast Mat-α-2 Repressor. Cell. 1993;74:357–369. doi: 10.1016/0092-8674(93)90426-q. [DOI] [PubMed] [Google Scholar]
- Cronin SR, Hampton RY. Measuring Protein Degradation with Green Fluorescent Protein. In: Conn PM, editor. Methods in Enzymology. 302, Green Fluorescent Protein. San Diego, CA: Academic Press; 1999. pp. 58–73. [DOI] [PubMed] [Google Scholar]
- Dai RM, Li CH. Valosin-containing protein is a multi-ubiquitin chain-targeting factor required in ubiquitin-proteasome degradation. Nat Cell Biol. 2001;3:740–744. doi: 10.1038/35087056. [DOI] [PubMed] [Google Scholar]
- Dantuma NP, Lindsten K, Glas R, Jellne M, Masucci MG. Short-lived green fluorescent proteins for quantifying ubiquitin/proteasome-dependent proteolysis in living cells. Nat Biotechnol. 2000;18:538–543. doi: 10.1038/75406. [DOI] [PubMed] [Google Scholar]
- DeHoratius C, Silver PA. Nuclear transport defects and nuclear envelope alterations are associated with mutation of the Saccharomyces cerevisiae NPL4 gene. Mol Biol Cell. 1996;7:1835–1855. doi: 10.1091/mbc.7.11.1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeMarini DJ, Papa FR, Swaminathan S, Ursic D, Rasmussen TP, Culbertson MR, Hochstrasser M. The yeast SEN3 gene encodes a regulatory subunit of the 26S proteasome complex required for ubiquitin-dependent protein degradation in vivo. MolCell Biol. 1995;15:6311–6321. doi: 10.1128/mcb.15.11.6311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finger A, Knop M, Wolf DH. Analysis of two mutated vacuolar proteins reveals a degradation pathway in the endoplasmic reticulum or a related compartment of yeast. Eur J Biochem. 1993;218:565–574. doi: 10.1111/j.1432-1033.1993.tb18410.x. [DOI] [PubMed] [Google Scholar]
- Fra A, Sitia R. The endoplasmic reticulum as a site of protein degradation. In: Borgese N, Harris JR, editors. Subcellular Biochemistry. 21. Endoplasmic reticulum. New York, NY: Plenum Publishing; 1993. pp. 143–168. [DOI] [PubMed] [Google Scholar]
- Friedlander R, Jarosch E, Urban J, Volkwein C, Sommer T. A regulatory link between ER-associated protein degradation and the unfolded-protein response. Nat Cell Biol. 2000;2:379–384. doi: 10.1038/35017001. [DOI] [PubMed] [Google Scholar]
- Fu H, Sadis S, Rubin DM, Glickman M, van Nocker S, Finley D, Vierstra RD. Multiubiquitin chain binding and protein degradation are mediated by distinct domains within the 26 S proteasome subunit Mcb1. J Biol Chem. 1998;273:1970–1981. doi: 10.1074/jbc.273.4.1970. [DOI] [PubMed] [Google Scholar]
- Gardner RG, Foss GM, Bays NW, Cronin SR, Wilhovsky SK, Seelig LP, Kim CM, Hampton RY. ER degradation requires lumen to cytosol signaling: transmembrane control of Hrd1p by Hrd3p. J Cell Biol. 2000;151:69–82. doi: 10.1083/jcb.151.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner RG, Hampton RY. A highly conserved signal controls degradation of 3-hydroxy-3-methylglutaryl-coenzyme A (HMG-CoA) reductase in eukaryotes. J Biol Chem. 1999;274:31671–31678. doi: 10.1074/jbc.274.44.31671. [DOI] [PubMed] [Google Scholar]
- Ghislain M, Dohmen RJ, Levy F, Varshavsky A. Cdc48p interacts with Ufd3p, a WD repeat protein required for ubiquitin- mediated proteolysis in Saccharomyces cerevisiae. EMBO J. 1996;15:4884–4899. [PMC free article] [PubMed] [Google Scholar]
- Glickman MH, Rubin DM, Fried VA, Finley D. The regulatory particle of the Saccharomyces cerevisiae proteasome. Mol Cell Biol. 1998;18:3149–3162. doi: 10.1128/mcb.18.6.3149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guthrie C, Fink GR. Guide to Yeast Genetics and Molecular Biology. San Diego, CA: Academic Press Inc; 1991. [Google Scholar]
- Hammond C, Helenius A. Quality control in the secretory pathway. Curr Opin Cell Biol. 1995;7:523–529. doi: 10.1016/0955-0674(95)80009-3. [DOI] [PubMed] [Google Scholar]
- Hampton RY, Bhakta H. Ubiquitin-mediated regulation of 3-hydroxy-3-methylglutaryl-CoA reductase. ProcNatl Acad SciUSA. 1997;94:12944–12948. doi: 10.1073/pnas.94.24.12944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampton RY, Gardner RG, Rine J. Role of 26S proteasome and HRD genes in the degradation of 3-hydroxy-3-methylglutaryl-CoA reductase, an integral endoplasmic reticulum membrane protein. Mol Biol Cell. 1996;7:2029–2044. doi: 10.1091/mbc.7.12.2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampton RY, Rine J. Regulated degradation of HMG-CoA reductase, an integral membrane protein of the endoplasmic reticulum, in yeast. J Cell Biol. 1994;125:299–312. doi: 10.1083/jcb.125.2.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinemeyer W, Troendle N, Albrecht G, Wolf DH. PRE5 and PRE6, the last missing gene encoding 20S proteasome subunits from yeast? Indication for a set of 14 different subunits in the eukaryotic proteasome core. Biochemistry. 1994;33:12229–12237. doi: 10.1021/bi00206a028. [DOI] [PubMed] [Google Scholar]
- Hiller MM, Finger A, Schweiger M, Wolf DH. ER degradation of a misfolded luminal protein by the cytosolic ubiquitin-proteasome pathway. Science. 1996;273:1725–1728. doi: 10.1126/science.273.5282.1725. [DOI] [PubMed] [Google Scholar]
- Hilt W, Enenkel C, Gruhler A, Singer T, Wolf DH. The PRE4 gene codes for a subunit of the yeast proteasome necessary for peptidylglutamyl-peptide-hydrolyzing activity. Mutations link the proteasome to stress- and ubiquitin-dependent proteolysis. J Biol Chem. 1993;268:3479–3486. [PubMed] [Google Scholar]
- Hitchcock, A.L., Krebber, H., Frietze, S., Lin, A., Latterich, M., and Silver, P.A. (2001). The Conserved Npl4 Protein Complex Mediates Proteasome-Dependent Membrane-Bound Transcription Factor Activation. Mol. Biol. Cell (in press). [DOI] [PMC free article] [PubMed]
- Ho AK, Shen TX, Ryan KJ, Kiseleva E, Levy MA, Allen TD, Wente SR. Assembly and preferential localization of Nup116p on the cytoplasmic face of the nuclear pore complex by interaction with Nup82p. Mol Cell Biol. 2000;20:5736–5748. doi: 10.1128/mcb.20.15.5736-5748.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochstrasser M, Ellison MJ, Chau V, Varshavsky A. The short-lived MATα2 transcriptional regulator is ubiquitinated in vivo. Proc Natl Acad SciUSA. 1991;88:4606–4610. doi: 10.1073/pnas.88.11.4606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoppe T, Matuschewski K, Rape M, Schlenker S, Ulrich HD, Jentsch S. Activation of a membrane-bound transcription factor by regulated ubiquitin/proteasome-dependent processing. Cell. 2000;102:577–586. doi: 10.1016/s0092-8674(00)00080-5. [DOI] [PubMed] [Google Scholar]
- Hori H, Nembai T, Miyata Y, Hayashi T, Ueno K, Koide T. Isolation and characterization of two 20S proteasomes from the endoplasmic reticulum of rat liver microsomes. J Biochem (Tokyo) 1999;126:722–730. doi: 10.1093/oxfordjournals.jbchem.a022509. [DOI] [PubMed] [Google Scholar]
- Johnson ES, Ma PCM, Ota IM, Varshavsky A. A proteolytic pathway that recognizes ubiquitin as a degradation signal. J Biol Chem. 1995;270:17442–17456. doi: 10.1074/jbc.270.29.17442. [DOI] [PubMed] [Google Scholar]
- Koegl M, Hoppe T, Schlenker S, Ulrich HD, Mayer TU, Jentsch S. A novel ubiquitination factor, E4, is involved in multiubiquitin chain assembly. Cell. 1999;96:635–644. doi: 10.1016/s0092-8674(00)80574-7. [DOI] [PubMed] [Google Scholar]
- Kohno K, Normington K, Sambrook J, Gething MJ, Mori K. The promoter region of the yeast kar2 bip gene contains a regulatory domain that responds to the presence of unfolded proteins in the endoplasmic reticulum. Mol Cell Biol. 1993;13:877–890. doi: 10.1128/mcb.13.2.877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latterich M, Frohlich KU, Schekman R. Membrane fusion and the cell cycle: Cdc48p participates in the fusion of ER membranes. Cell. 1995;82:885–893. doi: 10.1016/0092-8674(95)90268-6. [DOI] [PubMed] [Google Scholar]
- Meyer HH, Shorter JG, Seemann J, Pappin D, Warren G. A complex of mammalian ufd1 and npl4 links the AAA-ATPase, p97, to ubiquitin and nuclear transport pathways. EMBO J. 2000;19:2181–2192. doi: 10.1093/emboj/19.10.2181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moir D, Stewart SE, Osmond BC, Botstein D. Cold-sensitive cell-division-cycle mutants of yeast: isolation, properties, and pseudoreversion studies. Genetics. 1982;100:547–563. doi: 10.1093/genetics/100.4.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mumberg D, Muller R, Funk M. Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene. 1995;156:119–122. doi: 10.1016/0378-1119(95)00037-7. [DOI] [PubMed] [Google Scholar]
- Ng DT, Spear ED, Walter P. The unfolded protein response regulates multiple aspects of secretory and membrane protein biogenesis and endoplasmic reticulum quality control. J Cell Biol. 2000;150:77–88. doi: 10.1083/jcb.150.1.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel S, Latterich M. The AAA team: related ATPases with diverse functions. Trends Cell Biol. 1998;8:65–71. [PubMed] [Google Scholar]
- Pollard MG, Travers KJ, Weissman JS. Ero1p: A novel and ubiquitous protein with an essential role in oxidative protein folding in the endoplasmic reticulum. Mol Cell. 1998;1:171–182. doi: 10.1016/s1097-2765(00)80018-0. [DOI] [PubMed] [Google Scholar]
- Rose MD, Novick P, Thomas JH, Botstein D, Fink GR. A Saccharomyces cerevisiae genomic plasmid bank based on a centromere-containing shuttle vector. Gene. 1987;60:237–243. doi: 10.1016/0378-1119(87)90232-0. [DOI] [PubMed] [Google Scholar]
- Rout MP, Aitchison JD, Suprapto A, Hjertaas K, Zhao Y, Chait BT. The yeast nuclear pore complex: composition, architecture, and transport mechanism. J Cell Biol. 2000;148:635–651. doi: 10.1083/jcb.148.4.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin DM, Glickman MH, Larsen CN, Dhruvakumar S, Finley D. Active site mutants in the six regulatory particle ATPases reveal multiple roles for ATP in the proteasome. EMBO J. 1998;17:4909–4919. doi: 10.1093/emboj/17.17.4909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidrauski C, Chapman R, Walter P. The unfolded protein response: an intracellular signaling pathway with many surprising features. Trends Cell Biol. 1998;8:245–249. doi: 10.1016/s0962-8924(98)01267-7. [DOI] [PubMed] [Google Scholar]
- Sommer T, Jentsch S. A protein translocation defect linked to ubiquitin conjugation at the endoplasmic reticulum. Nature (London) 1993;365:176–179. doi: 10.1038/365176a0. [DOI] [PubMed] [Google Scholar]
- Stewart LC, Yaffe MP. A role for unsaturated fatty acids in mitochondrial movement and inheritance. J Cell Biol. 1991;115:1249–1257. doi: 10.1083/jcb.115.5.1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strawn LA, Shen T, Wente SR. The GLFG regions of Nup116p and Nup100p serve as binding sites for both Kap95p and Mex67p at the nuclear pore complex. J Biol Chem. 2000;4:4. doi: 10.1074/jbc.M008311200. [DOI] [PubMed] [Google Scholar]
- Stukey JE, McDonough VM, Martin CE. The OLE1 gene of Saccharomyces cerevisiae encodes the delta 9 fatty acid desaturase and can be functionally replaced by the rat stearoyl-CoA desaturase gene. J Biol Chem. 1990;265:20144–20149. [PubMed] [Google Scholar]
- Swerdlow PS, Finley D, Varshavsky A. Enhancement of immunoblot sensitivity by heating of hydrated filters. Anal Biochem. 1986;156:147–153. doi: 10.1016/0003-2697(86)90166-1. [DOI] [PubMed] [Google Scholar]
- Travers KJ, Patil CK, Wodicka L, Lockhart DJ, Weissman JS, Walter P. Functional and genomic analyses reveal an essential coordination between the unfolded protein response and ER-associated degradation. Cell. 2000;101:249–258. doi: 10.1016/s0092-8674(00)80835-1. [DOI] [PubMed] [Google Scholar]
- Tsurumi C, et al. CDNA cloning and functional analysis of the p97 subunit of the 26S proteasome, a polypeptide identical to the type-1 tumor-necrosis-factor-receptor-associated protein-2–55.11. Eur J Biochem. 1996;239:912–921. doi: 10.1111/j.1432-1033.1996.0912u.x. [DOI] [PubMed] [Google Scholar]
- Utsugi T, Hirata A, Sekiguchi Y, Sasaki T, Toh-e A, Kikuchi Y. Yeast tom1 mutant exhibits pleiotropic defects in nuclear division, maintenance of nuclear structure and nucleocytoplasmic transport at high temperatures. Gene. 1999;234:285–295. doi: 10.1016/s0378-1119(99)00197-3. [DOI] [PubMed] [Google Scholar]
- Varshavsky A. The N-end rule: functions, mysteries, uses. Proc Natl Acad Sci USA. 1996;93:12142–12149. doi: 10.1073/pnas.93.22.12142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma R, Chen S, Feldman R, Schieltz D, Yates J, Dohmen J, Deshaies RJ. Proteasomal proteomics: identification of nucleotide-sensitive proteasome-interacting proteins by mass spectrometric analysis of affinity-purified proteasomes. Mol Biol Cell. 2000;11:3425–3439. doi: 10.1091/mbc.11.10.3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wente SR, Blobel G. A temperature-sensitive NUP116 null mutant forms a nuclear envelope seal over the yeast nuclear pore complex thereby blocking nucleocytoplasmic traffic. J Cell Biol. 1993;123:275–284. doi: 10.1083/jcb.123.2.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickner S, Maurizi MR, Gottesman S. Posttranslational quality control: folding, refolding, and degrading proteins. Science. 1999;286:1888–1893. doi: 10.1126/science.286.5446.1888. [DOI] [PubMed] [Google Scholar]
- Wilhovsky SK, Gardner RG, Hampton RY. HRD gene dependence of ER-associated degradation. Mol Biol Cell. 2000;11:1697–1708. doi: 10.1091/mbc.11.5.1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokota KY, et al. CDNA cloning of p112, the largest regulatory subunit of the human 26S proteasome, and functional analysis of its yeast homologue, Sen3p. Mol Biol Cell. 1996;7:853–870. doi: 10.1091/mbc.7.6.853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young IT. Proof without prejudice: use of the Kolmogorov-Smirnov test for the analysis of histograms from flow systems and other sources. J Histochem Cytochem. 1977;25:935–941. doi: 10.1177/25.7.894009. [DOI] [PubMed] [Google Scholar]
- Zhang S, Skalsky Y, Garfinkel DJ. MGA2 or SPT23 is required for transcription of the delta9 fatty acid desaturase gene, OLE1, and nuclear membrane integrity in Saccharomyces cerevisiae. Genetics. 1999;151:473–483. doi: 10.1093/genetics/151.2.473. [DOI] [PMC free article] [PubMed] [Google Scholar]