Abstract
Precision medicine initiatives come amid the rapid growth in quantity and variety of biomedical data, which exceeds the capacity of matrix-oriented data representations and many current analysis algorithms. Tensor factorizations extend the matrix view to multiple modalities and support dimensionality reduction methods that identify latent groups of data for meaningful summarization of both features and instances. In this opinion article, we analyze the modest literature on applying tensor factorization to various biomedical fields including genotyping and phenotyping. Based on the cited work including work of our own, we suggest that tensor applications could serve as an effective tool to enable frequent updating of medical knowledge based on the continually growing scientific and clinical evidence. We encourage extensive experimental studies to tackle challenges including design choice of factorizations, integrating temporality and algorithm scalability.
Keywords: tensor factorization, precision medicine, biomedical data mining, multiple data modalities
Introduction
The collection of electronic medical data, while growing rapidly, poses technical challenges owing to large volume, uncertainty from noise and missing data and the fact that it draws from multiple modalities including clinical and genomic profiles, medication prescriptions and environmental exposures. Precision medicine aims to harness information from all modalities, develop a comprehensive view of a patient’s pathophysiologic progression and administer personalized therapies. Existing efforts are often based on only a few biomarkers, and their generalization demands new computational solutions, particularly to address the growing volume, uncertainty and number of modalities of data.
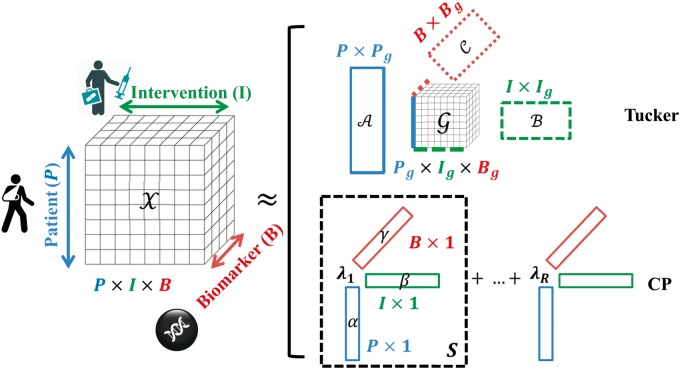
Tensor factorization has emerged as a promising solution for the computational challenges of precision medicine. A tensor is a multidimensional array where each modality spans one dimension (mode of a tensor). Figure 1 shows the tensor for modeling interactions among patients, biomarkers and interventions. Various factorization schemes have been proposed to decompose a tensor into factor matrices, which not only reduces dimensionality but also helps discover latent groups in each modality and identify group-wise interactions (see [1] for a general review). Typical matrix factorization approaches concatenate multiple data modalities into a single second dimension of the matrix, thus disallowing explicit representation of interactions among these modalities. In contrast to matrix factorization [2], different tensor factorizations can also integrate additional domain-specific prior knowledge to constrain the tensor structure. Figure 1 shows a visualization of two types of factorization: Tucker [3] and CANDECOMP/PARAFAC (CP) [4].
Figure 1.
Tensor modeling and factorization schemes. The data tensor x models the interactions among modes including patient, biomarker and medical intervention. The Tucker factorization (above, [3]) decomposes x into three factor matrices specifying groups in each mode and a core tensor 𝒢 specifying levels of interaction between the groups from different modes. In general, number of groups in each mode is less than the dimensionality of that mode and the core tensor 𝒢 can be thought of as a compression of x. The CP factorization (below, [4]) decomposes x as a weighted sum of rank-1 sub-tensors, each of which is the outer-product () of a patient factor vector (α), an intervention factor vector (β) and a biomarker factor vector (γ). The weights indicate relative importance of sub-tensors. Compared with Tucker, the structural hypothesis of CP requires the same number of groups for each mode.
Tensor factorization in biomedical informatics
Applying tensor factorization to biomedical informatics has gained traction over the past decade. Earlier applications focused on DNA microarray or sequencing data. Tucker and/or CP factorizations have been frequently applied to subjects including functionally related gene sets regarding protein/gene locus links and responses to stimulants [5], bacteria sub-lineage structure characterized by multiple types (modalities) of biomarkers [6], mouse brain genetic organizations across three-dimensional anatomical voxel positions [7] and relations between genes and transcription factors extracted from scientific literature [8]. To account for uncertainty, multiple authors proposed probabilistic Tucker and/or CP factorizations to incorporate priors on tensor structural parameters. Those priors can specify dependence between exposure to environmental chemicals and single nucleotide polymorphism level differences [9], or probability of gene sequence conditioned on the composing nucleotides and chromosomal positions [10, 11].
As an alternative to Tucker or CP factorizations, another vein of work viewed tensor factorization as a series of matrix factorizations with shared structural constraints, and termed their models Generalized Singular Value Decomposition (GSVD) or Higher-Order Singular Value Decomposition (SVD) (HOSVD). Some authors performed comparative analysis using ‘organism × gene × experimental condition’ tensors [12–14], or ‘nucleotide × sequence position × organism’ tensors [15]; others studied the effect and regulation of targeted pathways [16, 17] and further predicted treatment responses [18, 19]. When two of the tensor modalities are symmetric, eigenvalue decomposition replaces SVD, as seen in gene network functional grouping using binary/weighted ‘network × gene × gene’ tensors [20, 21]. However, it is difficult to extend GSVD/HOSVD to probabilistic versions to account for uncertainty.
In other biomedical fields, CP and Tucker factorizations have been used to localize and extract artifacts from Electroencephalogram (EEG) data to analyze epileptic seizures [22–24], where tensor modes include time points, electrodes of the multi-channel EEG and subjects (see [25] for a brief review). Probabilistic CP was shown to improve EEG classification accuracy when missing data are present [26]. In image analysis, HOSVD was applied to factorize a ‘patient × voxel × fMRI (functional Magnetic Resonance Imaging) mode’ tensor and to classify cognitive normal or declining status [27]. Wang et al. [28] demonstrated the potential of using tensor modeling to generalize sparse logistic regression to multiple modalities on fMRI data. In Electronic Health Record (EHR) phenotyping, CP has been adapted to enforce sparsity constraints [29], to explicitly account for interactions among groups of the same modality [30] and to incorporate medical knowledge via customized regularization terms [31], all with the goal of extracting clinically meaningful groups of patients. Both Tucker and CP seem to have broader adoptions than GSVD/HOSVD in non-genomic biomedical fields, perhaps owing to the relative ease of imposing probabilistic and other regularizations. Although CP produces summation of rank-1 sub-tensors (Figure 1) and leads to simplified interpretation, Tucker provides a more flexible and sometimes more realistic factorization by allowing varying number of groups in different modalities. Selecting a type of factorization is largely a design choice dependent on both data and outcome, and deserves extensive experimental studies and characterizations.
Toward precision medicine—discussion and future work
The advent of precision medicine initiatives, coupled with the welcome growth of new modes of data, suggests that medical knowledge needs continuous update. The current revision process, often involving meta-analysis of multiple studies and agreement of consensus groups, has difficulty in keeping up with the pace of change. An interesting alternative is to allow data-driven processes to suggest nimble and timely updates. Toward this goal, Luo et al. [32, 33] aimed to automatically identify from pathology reports a panel of test results that are diagnostic of lymphoma subtypes. Compared with a conventional ‘patient × word’ matrix, they composed a ‘patient × test result × word’ tensor and used non-negative Tucker factorization to identify diagnostic panels of test results. One can use such panels to suggest amendment to diagnostic guidelines in a format understandable to clinicians. However, extending tensor factorization to enable frequent updating in other fields such as genomics and biomedical signal processing remains an open question.
Another big challenge concerns how to properly model temporality within tensor factorization. Most existing work treats time points as independent, thus losing significant information [16, 17, 22–24]. Although we can add temporal locality constraints as an additional regularizer, this imposes new computational complexity and still lacks constraints on temporal ordering. Integrating stochastic processes into tensor factorization represents a theoretically appealing approach toward modeling temporality, but related work with biomedical applications is still in its infancy [26]. Specifically, it remains a major challenge to select appropriate stochastic processes based on consistency with biologic knowledge instead of mathematical convenience, yet maintain efficient inference procedures. Tensor factorization also needs to address data sparsity and algorithm scalability, which are more broadly recognized challenges in general domains. Only successfully answering all these challenges can lead to breakthroughs in supporting personalized medicine by properly drawing evidence with uncertainty from multi-modal, longitudinal and constantly evolving medical big data and the medical knowledge base.
Key Points
Precision medicine demands new computational solutions generalizing from limited number of biomarkers to address the growing volume, uncertainty and number of modalities of electronic medical data.
Tensor factorizations can easily integrate multiple data modalities, reduce dimensionality and identify latent groups in each mode for meaningful summarization of both features and instances in medical data.
Tensor factorizations demonstrated successes in genotyping and phenotyping applications, and showed promises in enabling frequent updating of medical knowledge out of continuously growing scientific and clinical evidence.
Challenges including design choices of factorization schemes, integrating temporality, addressing data sparsity and algorithm scalability pose exciting research opportunities to bioinformatics community, toward fully harnessing tensor factorization in the emerging horizon of precision medicine.
Funding
The work described was supported in part by Grants U54LM008748 and U54HG007963 from the National Library of Medicine.
Biographies
Yuan Luo is an assistant professor at Northwestern University, Department of Preventive Medicine. He works in the area of natural language processing, time series analysis and computational genomics, with a focus on medical and clinical applications.
Fei Wang is an associate professor at University of Connecticut, Department of Computer Science and Engineering. His research interests include data mining, machine learning algorithms and their applications in health informatics.
Peter Szolovits is a professor at Massachusetts Institute of Technology, Department of Electrical Engineering and Computer Science. His research centers on the application of Artificial Intelligence methods to problems of medical decision making, natural language processing to extract meaningful data from clinical narratives to support translational medicine and the design of information systems for health care institutions and patients.
References
- 1. Kolda TG, Bader BW.. Tensor decompositions and applications. SIAM Rev 2009;51:455–500. [Google Scholar]
- 2. Lee DD, Seung HS.. Learning the parts of objects by non-negative matrix factorization. Nature 1999;401:788–91. [DOI] [PubMed] [Google Scholar]
- 3. Tucker LR. Some mathematical notes on three-mode factor analysis. Psychometrika 1966;31:279–311. [DOI] [PubMed] [Google Scholar]
- 4. Carroll JD, Chang JJ.. Analysis of individual differences in multidimensional scaling via an N-way generalization of Eckart-Young decomposition. Psychometrika 1970;35: 283–319. [Google Scholar]
- 5. Yener B, Acar E, Aguis P, et al. Multiway modeling and analysis in stem cell systems biology. BMC Syst Biol 2008;2:63.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ozcaglar C, Shabbeer A, Vandenberg S, et al. Sublineage structure analysis of Mycobacterium tuberculosis complex strains using multiple-biomarker tensors. BMC Genomics 2011;12:S1.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ji S. Computational network analysis of the anatomical and genetic organizations in the mouse brain. Bioinformatics 2011;27:3293–9. [DOI] [PubMed] [Google Scholar]
- 8. Roy S, Homayouni R, Berry MW, et al. Nonnegative tensor factorization of biomedical literature for analysis of genomic data In Data Mining for Service, Springer: Berlin, 2014, 97–110. [Google Scholar]
- 9. Kessler DC, Taylor J, Dunson DB.. Learning phenotype densities conditional on many interacting predictors. Bioinformatics 2014;30:1562–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Yang Y, Dunson DB.. Bayesian conditional tensor factorizations for high-dimensional classification. J Am Stat Assoc 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhou J, Bhattacharya A, Herring AH, et al. Bayesian factorizations of big sparse tensors. J Am Stat Assoc 2015;110:1562–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sankaranarayanan S, Schomay T, Aiello K, et al. Tensor GSVD of patient- and platform-matched tumor and normal DNA copy-number profiles uncovers chromosome arm-wide patterns of tumor-exclusive platform-consistent alterations encoding for cell transformation and predicting ovarian cancer survival. PLoS One 2015;10:e0121396.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Alter O, Brown PO, Botstein D.. Generalized singular value decomposition for comparative analysis of genome-scale expression data sets of two different organisms. Proc Natl AcadSci 2003;100:3351–6. p [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ponnapalli SP, Saunders MA, Van Loan CF, et al. A higher-order generalized singular value decomposition for comparison of global mRNA expression from multiple organisms. PloS One 2011;6:e28072.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Muralidhara C, Gross AM, Gutell RR, et al. Tensor decomposition reveals concurrent evolutionary convergences and divergences and correlations with structural motifs in ribosomal RNA. PloS One 2011;6:e18768.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Omberg L, Golub GH, Alter O.. A tensor higher-order singular value decomposition for integrative analysis of DNA microarray data from different studies. Proc Natl Acad Sci 2007;104:18371–6. p [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Omberg L, Meyerson JR, Kobayashi K, et al. Global effects of DNA replication and DNA replication origin activity on eukaryotic gene expression. Mol Syst Biol 2009;5:312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Li Y, Ngom A.. Non-negative matrix and tensor factorization based classification of clinical microarray gene expression data. In: 2010 IEEE International Conference on Bioinformatics and Biomedicine, 2010, pp. 438–43. [Google Scholar]
- 19. Li X, Ye Y, Ng M, et al. MultiFacTV: module detection from higher-order time series biological data. BMC Genomics 2013;14:S2.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Alter O, Golub GH.. Reconstructing the pathways of a cellular system from genome-scale signals by using matrix and tensor computations. Proc Natl Acad Sci USA 2005;102:17559–64. p [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Li W, Liu CC, Zhang T, et al. Integrative analysis of many weighted co-expression networks using tensor computation. PLoS Comput Biol 2011;7:e1001106.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Acar E, Aykut-Bingol C, Bingol H, et al. Multiway analysis of epilepsy tensors. Bioinformatics 2007;23:i10–8. [DOI] [PubMed] [Google Scholar]
- 23. Mørup M, Hansen LK, Herrmann CS, et al. Parallel factor analysis as an exploratory tool for wavelet transformed event-related EEG. NeuroImage 2006;29:938–47. [DOI] [PubMed] [Google Scholar]
- 24. Lee H, Kim YD, Cichocki A, et al. Nonnegative tensor factorization for continuous EEG classification. Int J Neural Syst 2007;17:305–17. p [DOI] [PubMed] [Google Scholar]
- 25. Cong F, Lin QH, Kuang LD, et al. Tensor decomposition of EEG signals: a brief review. J Nneurosci Methods 2015;248:59–69. [DOI] [PubMed] [Google Scholar]
- 26. Rai P, Wang Y, Guo S, et al. Scalable Bayesian low-rank decomposition of incomplete multiway tensors. In: Proceedings of the 31st International Conference on Machine Learning (ICML-14), 2014, Beijing, China, on June 21–June 26, 2014c, p. 180008.
- 27. Batmanghelich N, Dong A, Taskar B, et al. Regularized tensor factorization for multi-modality medical image classification In Medical Image Computing and Computer-Assisted Intervention–MICCAI 2011. Springer: Berlin, 2011, 17–24. [DOI] [PubMed] [Google Scholar]
- 28. Wang F, Zhang P, Qian B, et al. Clinical risk prediction with multilinear sparse logistic regression. In Proceedings of the 20th ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, ACM, 2014, 145–54.
- 29. Ho JC, Ghosh J, Steinhubl SR, et al. Limestone: high-throughput candidate phenotype generation via tensor factorization. J Biomed Inform 2014;52:199–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ho JC, Ghosh J, Sun J. Marble: high-throughput phenotyping from electronic health records via sparse nonnegative tensor factorization. In Proceedings of the 20th ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, ACM, 2014, 115–24.
- 31. Wang Y, Chen R, Ghosh J, et al. Rubik: Knowledge Guided Tensor Factorization and Completion for Health Data Analytics. In Proceedings of the 21th ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, Sydney, NSW: ACM, 2015, 1265–74. [DOI] [PMC free article] [PubMed]
- 32. Luo Y, Sohani A, Hochberg E, et al. Automatic lymphoma classification with sentence subgraph mining from pathology reports. J Am Med Inform Assoc 2014;21:824–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Luo Y, Xin Y, Hochberg E, et al. Subgraph augmented non-negative tensor factorization (SANTF) for modeling clinical text. J Am Med Inform Assoc 2015;22:1009–19. [DOI] [PMC free article] [PubMed] [Google Scholar]