Abstract
Herpes Simplex Virus type 2 (HSV-2) is a neurotropic human pathogen. Upon de novo infection, the viral infected cell protein 0 (ICP0) is immediately expressed and interacts with various cellular components during the viral replication cycle. ICP0 is a multifunctional regulatory protein that has been shown to be important for both efficient viral replication and virus reactivation from latency. In particular, as previously demonstrated in transfected tissue culture models, ICP0 interacts with the cellular E3 ubiquitin ligase SIAH-1, which targets ICP0 for proteasomal degradation. However, the consequence of this virus-host interaction during the establishment of HSV-2 infection in vivo has not yet been elucidated. Here we confirmed that ICP0 of HSV-2 interacts with SIAH-1 via two conserved PxAxVxP amino acid binding motifs. We also demonstrate in vitro that a SIAH-1 binding-deficient HSV-2 strain, constructed by homologous recombination technology, exhibits an attenuated growth curve and impaired DNA and protein synthesis. This attenuated phenotype was also confirmed in an in vivo ocular infection mouse model. Specifically, viral load of the SIAH-1 binding-deficient HSV-2 mutant was significantly reduced in the trigeminal ganglia and brain stem at day 5 and 7 post infection. Our findings indicate that the interplay between ICP0 and SIAH-1 is important for efficient HSV-2 replication in vivo, thereby affecting viral dissemination kinetics in newly infected organisms, and possibly revealing novel targets for antiviral therapy.
Introduction
Herpes Simplex Virus type 2 (HSV-2) is the causative agent of genital herpes and a ubiquitous human virus present in about 10–20% of the human population in Europe and the US [1]. HSV-2 initially infects and replicates in epithelial cells and invades the nervous system by entering the innervating sensory neurons [2]. The virus then undergoes retrograde transport along the axon to the neuronal cell bodies, where a lifelong latency is established [2,3]. Due to a variety of stimuli, HSV can occasionally reactivate from its latent state. In this case, the virus replicates in the neuron and is transported anterograde to or near the side of primary infection to re-infect epithelial cells [2]. While HSV-2 is mostly benign in hosts with an intact immune system, it can cause severe disease in immunocompromised patients, for instance when the central nervous system is infected, or when HSV is transmitted to a newborn during birth [4–6]. Moreover, it has been reported that genital HSV infection significantly increases the risk of HIV-1 infection [7].
HSV gene expression is regulated by host and viral transcription factors and takes place in a cascade-like manner [5,8]. One of the first viral genes transcribed is the immediate early gene RL2, which encodes infected cell protein 0 (ICP0). ICP0 is a multifunctional trans-activator of viral gene expression during lytic infection [9–16] and is said to play a role in reactivating the virus from latency [15,17–19]. ICP0 acts as an E3 ubiquitin ligase through its N-terminal RING domain, an activity essential for many functions ascribed to ICP0 [9,20]. Thus, ICP0-mediated polyubiquitination leads to proteasomal degradation of cellular targets [9,12]. Examples are the ICP0-mediated degradation of SUMOylated isoforms of PML and Sp100 in the host cell nucleus that occurs shortly after HSV infection [12,21–24].
The importance of ICP0 for the virus can be further demonstrated with ICP0-deficient HSV mutants. These mutants are severely defective in establishing infection and high MOIs are necessary for these mutant viruses to overcome intrinsic antiviral cellular defenses [9,12,25–29]. Therefore, studying the interaction of ICP0 with cellular targets is crucial to understand the molecular pathology of HSV-2 and to elucidate possible novel targets for antiviral therapy.
Previously, we described a novel cellular interaction partner of ICP0, the human E3 ubiquitin ligase seven in absentia homolog 1 (SIAH-1) [30]. Initially identified as seven in absentia (SINA) in Drosophila, the family of SIAH proteins contains two human homologs, SIAH-1 and SIAH-2 [31]. Although they differ at their N-terminus, both human SIAH homologs share a RING domain that confers E3 ubiquitin ligase activity, two zinc-finger domains, as well as a C-terminal substrate-binding domain [32,33].
Here, we investigated the effect of the ICP0:SIAH-1 interaction on HSV-2 infection and replication in vitro and in vivo employing various mutants generated by homologous recombination from the parental HSV-2 strain MS. Our results revealed that SIAH-1 binding-deficient HSV-2 strains are growth attenuated and synthesize significantly less that viral protein and DNA, indicating that the ICP0:SIAH-1 interaction is required for efficient HSV-2 replication in vivo.
Materials and methods
Cell lines
African Green monkey kidney epithelial cells (Vero; ATCC#: CCL-81), baby hamster kidney epithelial cells (BHK-21, ATCC CCL-10), human embryonic kidney cells (HEK293T; ATCC#: CRL-11268), and human osteosarcoma cells (U2OS; ATCC#: HTB-96) were cultured in DMEM supplemented with 10% fetal calf serum (FCS), 2 mM L-glutamine, 1 mM sodium pyruvate, and 3.75 mg/ml sodium bicarbonate. Human hepatic cells (HepaRGTM; Life Technologies, Carlsbad, CA) were cultured in full DMEM supplemented with 5 μg/ml bovine insulin and 50 μM hydrocortisone.
Mouse strains
C57Bl/6J mice were originally obtained from the Jackson Laboratory and bred and maintained under specific pathogen-free (SPF) conditions at the animal facility of the Heinrich-Pette-Institute, Hamburg. Animals were daily monitored for signs of obvious suffering, such as weight loss, back arches and prostrated behavior.
Animal experiments were performed according to the guidelines of the German Animal Protection Law. The experimental protocols were reviewed and approved by the relevant German authority, the Freie und Hansestadt Hamburg, Behörde für Gesundheit und Verbraucherschutz (Nr.: 144/15).
Viruses
Unmodified plaque purified [34] HSV-2 strain MS (original non-purified virus received from ATCC#: VR-540) served as the initial virus for homologous recombination. Construction of HSV-2 ICP0GFPΔ19162 (HSV-2 0ΔRING) is described elsewhere [28].
Reagents and antibodies
Protein G sepharose and glutathione sepharose were obtained from GE Healthcare (Freiburg, Germany). GFPtrapA beads comprise a single domain GFP antibody immobilized on agarose beads (Chromotek, Martinsried, Germany). Proteasome inhibitor MG132 was purchased from Sigma (Munich, Germany). The following antibodies were used: anti-tubulin, mouse IgG (Sigma); anti-GFP (Novus Biologicals, CO); anti-PML H-238, anti-SIAH-1 N-15 (Santa Cruz), anti-HSV1/2 VP5 H1.4 (Acris, Herford, Germany); anti-ICP0 R147 and anti-ICP27 R152 (Biomedizinische Forschungsgesellschaft mbH Vienna, Austria), and anti-GST-DX700 (IRDye® labeled antibodies; Rockland Immunochemicals, Gilbertsville, CA). For infrared fluorescence detection (Li-COR Biosciences, Bad Homburg, Germany) secondary antibodies coupled to IRDye® fluorophores (Rockland) were used. Glutathione sepharose for GST-pulldown experiments was obtained from GE Healthcare (Freiburg, Germany).
Expression constructs
A eukaryotic expression plasmid encoding a codon-optimized and intron-less HSV-2 ICP0 sequence with a 3’ in-frame fusion to the coding sequence of green fluorescent protein (eGFP), as well as an expression plasmid encoding ICP0 with a deletion of the RING domain (ΔRING, Δaa126-166) were described previously [30]. Using the QuikChange site-directed mutagenesis kit (Agilent), point mutations in the two SIAH binding motifs were introduced using following oligonucleotide primers: 5’-GCACGTCCACGCGCTGCTAATGCAAATCGCGTGC GCTCTCCACC-3’ and 5’-GGTGGAGAGCGCACGCGATTTGCATTAGCAGCGCGTGGACGTGC-3’ for the SIAH binding motif at aa position 414–416 (NxN1); 5’-GGCCCAGCAGTGGCAGCTAAT GTGAATCGTGTGGCATCTCTGCCAC-3’ and 5’-GTGGCAGAGATGCCACACGATTCACATTAGCT GCCACTGCTGGGCC-3’ for the motif at aa position 316–318 (NxN2). Eukaryotic expression plasmids encoding HA-SIAH-1, HA-SIAH-1C44S, as well as a plasmid for bacterial expression of GST-SIAH-1 were described elsewhere [30,35].
Plasmid precursors of HSV-2 recombinant viruses
To construct the SIAH-binding deficient HSV-2 ICP0 mutants, transfer plasmids were generated. First, a homology plasmid was constructed by subcloning two DNA fragments derived from wild-type HSV-2 MS genomic DNA into a pUC19 plasmid vector (New England Biolabs), encompassing sequences homologous to 517 bp upstream of the HSV-2 ICP0 start codon and 545 bp downstream of the ICP0 stop codon. During cloning, XbaI and BamHI restriction sites were introduced in-between the two homology arms. Next, pICP0-GFP plasmids (see above) and the homology plasmid were treated with the enzymes XbaI and BamHI and fragments were separated via gel electrophoresis. The linearized homology plasmid as well as the XbaI-BamHI DNA fragment (ICP0-GFP sequence) from the pICP0-GFP plasmids, were extracted and the ICP0-GFP sequence was ligated into the homology plasmid using the DNA Rapid Dephosphorylation and Ligation Kit (Roche). The resulting transfer plasmids, where the wild-type or mutated ICP0-GFP sequence is flanked by the homology arms, are shown in S1 Table.
HSV-2 mutagenesis by homologous recombination
U2OS cells were transfected with 2.5 μg of the linearized transfer plasmid. After 24 hours, transfected cells were infected with plaque-purified HSV-2 strain MS at an MOI of 5 pfu/cell. Recombinant viruses were identified by GFP fluorescence and isolated via plaque picking and up to four cycles of subsequent plaque purification by limiting dilution [34]. Prior to further characterization, two independent recombinant viruses were routinely isolated from parallel experiments.
DNA sequencing
The sequences encoding ICP0-derived SIAH binding motifs in the various constructs were amplified by using following oligonucleotides: forward primer, 5’-GATGAAGATGATGACCTG-3’; reverse primer, 5’-AAGATGCAGAACTAGACG-3’. The resulting 850 bp amplification products were ligated into a TOPO vector using the TOPO PCR cloning kit (Life Technologies) and transformed into the bacterial strain DH5α. Selected clones were sequenced with standard M13 forward and reverse primers to exclude PCR-derived point mutations as false negatives. Positive mutants were subsequently transfected into U2OS cells and plaque-purified three to four times.
The integrity of HSV-2 genomes was verified by next generation sequencing. For this, purified viral DNA isolated from infected BHK cells was sheared by sonification. Library preparation was conducted according to the Protocol for use with NEB Next Ultra DNA Library Prep Kit for Illumina (E7370) (NEB) and libraries were sequenced using a MiSeq (Illumina) device. SPAdes v3.10.1 was used for in silico assembly of the respective sequencing reads, which were subsequently mapped to the reference HSV-2 strain HG52 (NC_001798.2) using the blast v2.7.1.
DNA transfection
U2OS cells were transfected using TransIT (Mirus) according to the manufacturer’s protocol. HEK293T cells were transfected using Polyethylenimine (PEI; Polysciences, Eppelheim, Germany) at a mass ratio of PEI and DNA of 6:1.
Virus production
BHK-21 cells (ATCC#: CCL-10) were used for preparation of virus stocks from infected cell culture supernatants as described previously [36,37]. Virus stocks of passage 2 or 3 were used for all subsequent experiments.
Quantification of viral DNA
Viral DNA was isolated from infected cell pellets using the Qiagen Blood Mini Kit (Hilden, Germany) or from infected tissue using the peqGOLD TriFast reagent (Thermo Fisher Scientific, Waltham, MA) and DNA was quantified by Taqman real-time PCR (Applied Biosystems 7500 Real-Time PCR System; Life Technologies) using oligonucleotide-primers specific for the HSV-2 UL27 gene sequence encoding glycoprotein B [38] and for the GFP-sequence as follows: UL27-forward primer, 5’-TGCAGTTTACGTATAACCACATACAGC-3´; UL27-reverse primer, 5´-AGCTTGCGGGCCTCGTT-3´; UL27-probe, 5’-FAM-CGCCCCAGCATGTCATTCACGT-TAMRA-3´; GFP-forward primer, 5´-ACAAGCTGGAGTACAACTA-3´; GFP-reverse primer, 5’-TGTTCTGCTGGTAGTGGTC-3’; and GFP-probe, 5’-FAM-CAGCCACAACGTCTATATCATG-TAMRA-3´. In human cell lines, viral DNA copies were normalized to sequences of the human beta-globin (HBG) gene [39]; HBG-forward primer, 5’-CTTAATGCCTTAACATTGTGTATAA-3’; HBG-reverse primer, 5’-GAATATGCAAATAAGCACA CATATAT-3’; HBG-probe, 5’-FAM-ACTTTACACAGTCTGCCTAGT ACATTAC-TAMRA-3’. In murine cells (tissue), viral DNA copies were normalized to beta-actin (BAK) gene sequences. BAK-forward primer 5´-TCCTGAGACTCCCAGCACAC-3´; BAK-reverse primer, 5’-ACACTCAG GGCAGGTGAAACT-3´; BAK-probe, 5’-FAM-TGCACTCCTTGCAT GTCTCAGA-TAMRA-3’.
Southern blot analysis
Viral DNA was digested using the restriction endonucleases HindIII and NcoI. DNA fragments were separated via standard gel electrophoresis and transferred to nitrocellulose membranes by capillary blotting. The utilized probes were directed against a 726 bp sequence in GFP and against a 860 bp sequence within HSV-2 wild-type ICP0 that did not recognize the codon-adjusted ICP0-GFP sequence in the recombinant viruses. Probes were synthesized via PCR using following GFP-specific oligonucleotide-primers: forward, 5’-ATTTACTCGAGGCTAGCATGGCCACAACCATGGTGAG-3’ containing an XhoI and NheI restriction site; reverse, 5’-TAAATCTCGAGTTACTTGTACAGCTCGTCCATG-3’ containing an XhoI restriction site. The wild-type ICP0 primers used were as follows: forward, 5’-ATTTACTGCAGCACGGTGAGAGGGCGA-3’; reverse, 5’-ATTTACTGCAGACTATCAGGTACGC CACC-3’ both with PstI restriction sites. Probes were radioactively labelled with 32P-dCTP (Hartmann Analytic, Braunschweig, Germany). Hybridization was conducted using the ExpressHyp hybridization solution (TAKARA/CLONTECH). After hybridization, the membrane was subjected to autoradiography.
Protein-protein interaction analysis
Preparation of cell lysates from transfected cells and immunoprecipitation using GFPtrapA beads or Protein G Sepharose-coupled antibodies were performed as described previously [30]. For infected cell lysates, U2OS cells were infected with wild-type HSV-2 strain MS, HSV-2-ICP0-GFP or the respective SIAH binding-deficient ICP0-GFP mutants at an MOI of 0.01 pfu/cell. At 48 hours post infection, cells were harvested, washed with PBS, centrifuged at 20,000 x g for 20 sec and resuspended in 650 μl RIPA lysis buffer +1x protease inhibitors. Lysates were incubated on ice for 30 min and afterwards centrifuged at 20,000 x g for 15 min at 4°C. 10% of the supernatant was resuspended in 10 μl 6x protein sample buffer (PP) (350 mM Tris/HCL pH 6.8, 10% [w/v] SDS, 36% [v/v] glycerol, 6% [v/v] beta-mercaptoethanol, 0.01% [w/v] bromophenol blue in distilled water) as control and the cell pellet was resuspended in 650 μl 1x PP.
For GST-pulldown assays, E.coli BL21(DE3)pLysS (Promega), carrying pGEX-SIAH-1 or the empty GST plasmid pGEX-5T were grown overnight in LB medium at 37°C. The next day, 250 ml LB medium with 100 μg/ml ampicillin were inoculated with 100 μl of overnight culture and shaken at 37°C until the OD600 reached 0.5. For induction, IPTG was added at 1 μg/ml (GST-SIAH-1) or 5 ng/ml (GST) and the cultures were shaken further at 30°C for 4 h, 0.1 μM ZnCl2 were included in the GST-SIAH-1 culture during induction. The cell pellets were lysed for 10 min on ice with 3 mg/ml lysozyme in 50 mM Tris-Cl, pH 8.0, 150 mM NaCl, 5 mM EDTA, 1% TX-100, sonicated and centrifuged at 20,000 × g for 30 min at 4°C. The lysates were shaken for 30 min at room temperature with 1 ml glutathion sepharose beads equilibrated in NETN buffer (20 mM Tris-HCl pH 8.0, 100 mM NaCl, 1 mM EDTA, 0.4% NP-40). For preclearing, the GST-loaded beads were washed three times with NETN and then added to the cell lysates containing the potential SIAH interacting proteins and shaken on an overhead mixer for 1 h at 4°C. Afterwards, the beads were removed by pelleting and the precleared lysates were incubated with GST-SIAH-1 coupled beads overnight at 4°C. The beads were washed three times with 50 mM HEPES pH 7.5, 150 mM NaCl, 5 mM EDTA, 0.5% (v/v) NP-40 and the pellets were resuspended in 2×SDS-PAGE sample buffer (see below).
Western blot analysis
For preparation of cellular extracts, cells were washed with PBS and dissolved in 2xSDS-PAGE sample buffer (116 mM Tris-HCl pH 6.8, 3.3% [w/v] SDS, 12% [v/v] glycerol, 2% [v/v] beta-mercaptoethanol, bromophenol blue) and 1x complete cOmplete™ protease inhibitor cocktail (Roche). After separation by SDS-PAGE, proteins were transferred to nitrocellulose membranes (Schleicher & Schuell) and subjected to immune blot analysis.
Virological assays
Standard plaque assays were conducted in a 6-well format to determine the titers (plaque formation efficiency) of wild-type and mutant HSV-2. Duplicate dilution series were set up in RPMI (RPMI 1640 containing 10% [v/v] FCS) and were used to inoculate confluent U2OS, HepaRG or Vero cells. After 1 h of incubation on a rocking platform, the inoculum was removed and cells were covered with 2 ml complete DMEM containing 25 μg/ml HSV neutralizing human polyclonal IgG (Sigma-Aldrich, St. Louis, MO) and incubated at 36°C and 5% CO2. Procedures were according to the protocol in [40]. After 48 hours, culture medium was removed and cells were fixed with 0.5 ml/well ice cold methanol and stained with 0.1% crystal violet.
Ocular infection of mice
Six to eight week old female C57Bl/6J mice were anesthetized by intraperitoneal (i.p.) injection of ketamine (100 mg/kg of body weight) and xylazine (5 mg/kg of body weight). Each cornea was scarified in a crosshatch pattern with a 26-gauge needle and subsequently inoculated with 5x105 plaque forming units (PFU) of HSV-2-ICP0-GFP or HSV-2-ICP0NxN1/2-GFP strain MS, or with DMEM as control. The virus was suspended in 4 μl of complete cell culture medium and administered as an eye drop. Viral titers were determined at the indicated time points post inoculation on Vero cell monolayers.
Determination of viral titers in the tear film and tissue of C57Bl/6J mice
For analysis of ocular virus shed, tear film swabs were taken from both eyes on day 1, 2, 3, 5 and 7 post infection using a cotton-tipped applicator. Each tip was placed in 0.4 ml of complete cell culture medium, vortexed and surplus liquid was removed by squeezing. The amount of virus was determined by a microtiter plate plaque assay (96-well format) on Vero cell monolayers [41].
Mice were euthanized and the trigeminal ganglia (TG) and brain stem were isolated and placed in 0.6 ml of complete cell culture medium on day 3, 5 and 7 post infection. TGs from the left and the right eye were pooled. The isolated tissues were homogenized with a sonifier (Branson B-12) and freeze/thawed afterwards. The cell debris was removed by centrifugation at 3,000 x g for 5 min and the supernatant was taken for virus titer determination using standard plaque assay on Vero cell monolayers.
Statistical tests
GraphPad Prism 5.03 was used to perform statistical analysis for the qRT-PCR data and viral growth analysis in vitro and in vivo. For in vivo analysis, mean ± standard error of the mean (SEM) are reported for each group. T-test was used for two-group comparison with Mann-Whitney post-test when Gaussian distribution was not assumed. 2-way analysis of variance (ANOVA) with Bonferroni post-test was used to compare multiple groups and time points and replicate means. Significance was determined with p-values <0.001 defining *** significance and p-values >0.05 defining no significance.
Results
ICP0 interacts with SIAH-1 via two minimal interaction motifs
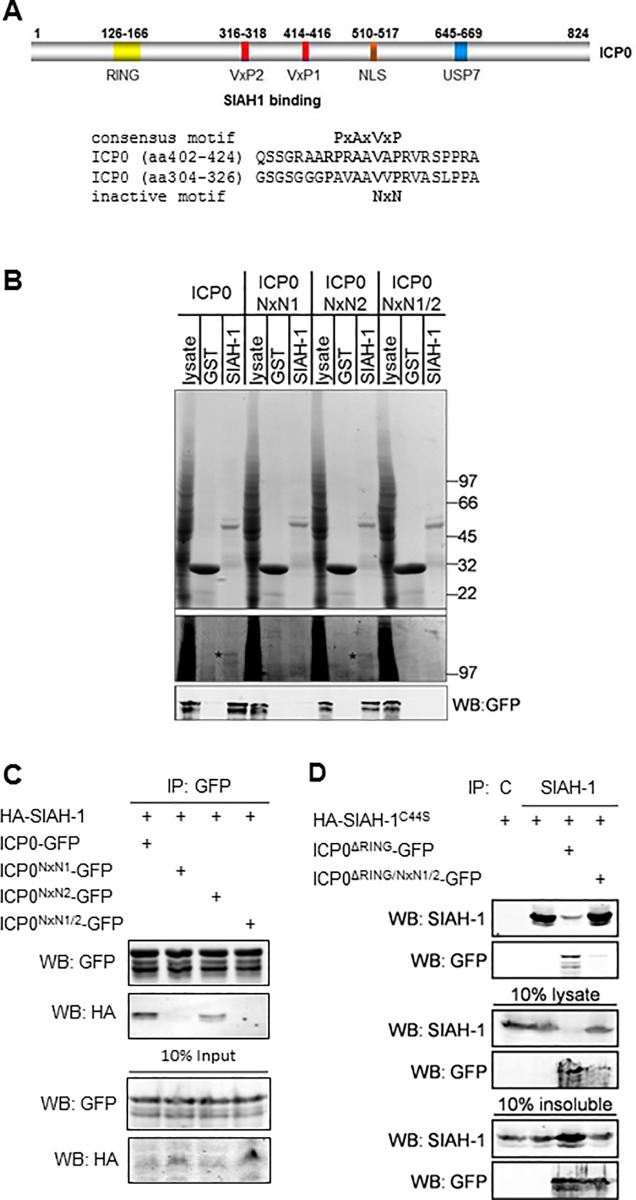
Previously, we identified a SIAH-1 binding consensus motif PxAxVxP in HSV-2 ICP0 [30]. Its most important core residues are located at ICP0 amino acid positions 414–416 (VxP1; Fig 1A). Co-immunoprecipitation studies with several ICP0 truncation and internal deletion mutants demonstrated that deletion of residues 410–420 greatly reduced the ICP0:SIAH-1 functional interaction. We also identified a second putative SIAH-1 binding motif around residues 316–318 (VxP2, Fig 1A), which does not perfectly overlap with the consensus PxAxVxP sequence. In many proteins targeted by SIAH ubiquitin ligases, a VxP motif is essential for efficient binding and mutation of the core residues to NxN inactivates SIAH-binding [42,43]. Therefore, to analyze in vitro whether a NxN mutation is sufficient to abolish the ICP0:SIAH-1 interaction, we first introduced point mutations into a previously described HSV-2-ICP0-GFP expression plasmid [30].
Fig 1. ICP0 interacts with SIAH-1 via two minimal interaction motifs.
(A) Schematic representation of HSV-2 ICP0 indicating the position of the RING domain (yellow), the USP7 interaction domain (blue), the nuclear localization signal (brown) and the two SIAH interaction motifs VxP1 and VxP2 (red). The primary amino acid sequence surrounding the predicted SIAH binding motifs is depicted together with the consensus motif and the position of the inactivating NxN mutation. (B) HEK293T cells were transfected with plasmids encoding GFP-tagged ICP0 and its mutants and the cell lysates were incubated with GST or GST-SIAH-1-loaded glutathione sepharose beads. The upper SDS-PAGE gel shows a Coomassie staining of the respective input control (lysate) and the eluates from the GST or GST-SIAH-1 beads. Below, a contrast enhanced section of the gel with putative ICP0 bands indicated by asterisks. ICP0 was detected by Western blotting using an antibody against the C-terminal GFP tag. Size markers in kDa. (C) HEK293T cells were transfected with plasmids encoding GFP-tagged ICP0 and its mutants and HA-tagged SIAH-1. The ICP0-GFP proteins were immunoprecipitated from the lysates, ICP0 and SIAH-1 were detected by SDS-PAGE and Western blotting using antibodies against the GFP and HA-tags. (D) HEK293T cells were transfected with plasmids encoding the inactive mutant HA-SIAH-1C44S and GFP-ICP0ΔRING or GFP-ICP0ΔRING/NxN1/2 as indicated. Immunoprecipitation from the cell lysates was performed using control mouse IgG or anti-SIAH-1. ICP0 and SIAH-1 were detected by SDS-PAGE and Western blotting using antibodies against SIAH-1 or the GFP-tag. The lower panel shows the analysis of the RIPA buffer-insoluble pellet after cell lysis.
ICP0-GFP or its NxN mutants were transiently expressed in HEK293T cells and the cell lysates were incubated with GST-SIAH-1-loaded glutathione sepharose beads, or beads loaded with GST alone as a control. Pulldown assays and subsequent Western blotting demonstrated that ICP0 and ICP0NxN2 were bound to SIAH-1-loaded beads (Fig 1B, lower panel), whereas ICP0NxN1 and the double mutant ICP0NxN1/2 were no longer bound. Subsequently, the ICP0:SIAH-1 interaction was confirmed by immunoprecipitation of GFP from lysates of cells co-transfected with hemagglutinin tagged SIAH-1 (HA-SIAH-1) and ICP0-GFP (Fig 1C). Consistent with the previous experiment, ICP0, and to a lesser extent ICP0NxN2, bound to SIAH-1, while ICP0NxN1 binding was barely detectable and ICP0NxN1/2 showed no binding. In reverse, SIAH-1 immunoprecipitation confirmed the loss of interaction of the ICP0NxN1/2 mutant (Fig 1D). In this experiment we employed RING-deficient ICP0 and SIAH-1 mutants to increase protein stability. Less SIAH-1 was present in the cell lysates co-transfected with ICP0ΔRING than in the absence of ICP0 or when ICP0ΔRING/NxN1/2 was present. This might be due to the formation of buffer-insoluble SIAH-1:ICP0 aggregates (Fig 1D, lower panels).
In summary, mutation of the two VxP core motifs resulted in reduced binding of HSV-2 ICP0 to SIAH-1. The affinity of the VxP1 motif appears to be stronger than that of the VxP2 motif.
Construction and validation of SIAH-binding deficient ICP0 mutant virus
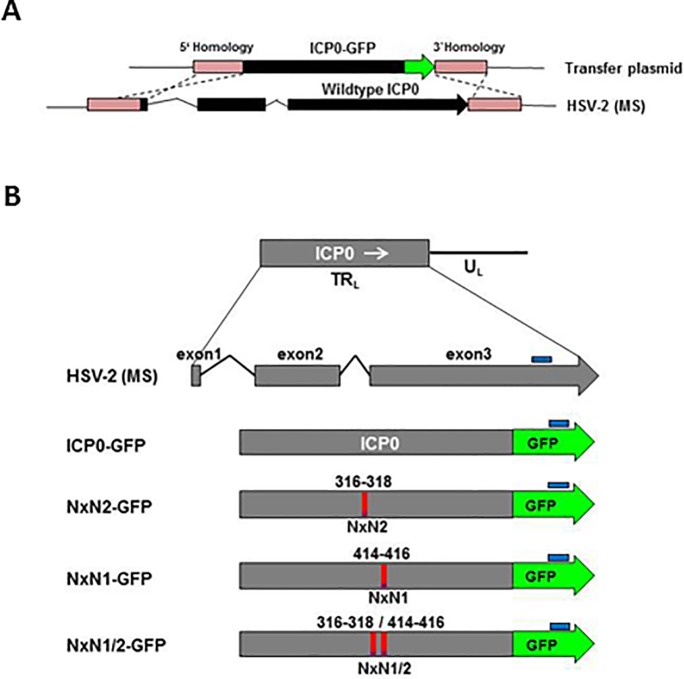
To analyze the relevance of the ICP0:SIAH-1 interaction in the context of viral infection we introduced the VxP1 and/or VxP2 mutations (Fig 1A) into both ICP0 coding regions of a full-length HSV-2 genome. HSV-2 mutants with ICP0 genes additionally fused 3’ in-frame to the GFP coding region were generated by homologous recombination (Fig 2A; for list of transfer plasmids see S1 Table). The viruses expressing either wild-type ICP0-GFP or ICP0-GFP carrying mutated VxP core residues in the two SIAH binding domains, either individually mutated (NxN1 or NxN2) or mutated in combination (NxN1/2; Fig 2B) were analyzed by restriction endonuclease digestion and subsequent Southern blot analysis (S1 Fig). Furthermore, mutations, as well as overall genome integrity were confirmed by Illumina MiSeq™ sequencing of the full-length viral constructs.
Fig 2. Construction of HSV-2 mutant viruses with mutated SIAH binding motifs in ICP0.
(A) Depiction of homologous recombination. The native ICP0 sequence in HSV-2 strain MS was exchanged by a sequence encoding ICP0-eGFP fusion protein. (B) Maps of the various ICP0 constructs. The wild-type gene is located in the HSV-2 TRL region. The various ICP0-GFP DNA sequences were accordingly adjusted to reduce their GC content. The NxN1 and NxN2 mutations in the SIAH-1 binding domains are marked in red. Sequences recognized by Southern blot probes are marked in blue.
Subsequent in vitro control experiments revealed that no significant growth differences exist between wild-type HSV-2 strain MS and HSV‐2‐ICP0‐GFP (S2 Fig). Furthermore, wild-type and mutant constructs (i.e. HSV-2-ICP0-GFP and HSV-2-ICP0NxN1/2-GFP, respectively) displayed comparable phenotypes with respect to PML degradation (S3 Fig) and timely translocation of ICP0 to the cytoplasm (S4 Fig).
Mutation of ICP0 in HSV-2 abolishes SIAH-1 binding
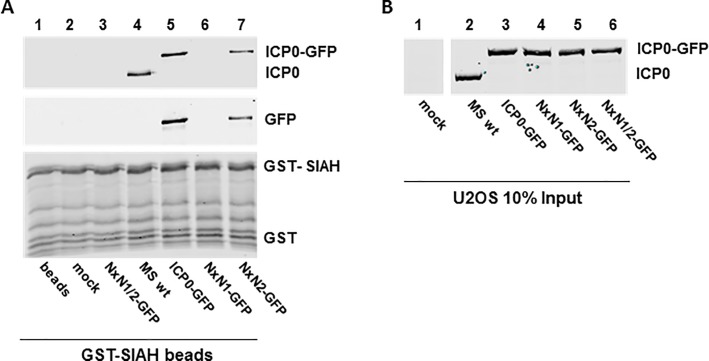
To test SIAH-1 binding efficiency by the various viruses, U2OS cells were infected for 48 hours with an MOI of 0.01 pfu/cell before performing GST pull-down assays (Fig 3). Cell lysates were harvested and incubated with GST-SIAH-1-loaded glutathione sepharose beads. ICP0 expressed by wild-type HSV-2 strain MS (MS wt) or the HSV-2-ICP0-GFP (ICP0-GFP) clearly bound to GST-SIAH-1 loaded beads, whereas the mutated HSV-2-ICP0NxN1/2-GFP (NxN1/2-GFP) showed no detectable binding (Fig 3). Interestingly, ICP0 still interacted with SIAH-1 when the VxP2 binding motif was disrupted, although the binding affinity seemed weaker than with wild-type ICP0. In agreement with our transfection experiments, inactivation of the VxP1 motif was sufficient to almost completely abolish SIAH-1-binding in these in vitro experiments, suggesting that particularly the VxP1 motif is critical for the interaction between HSV-2 ICP0 and cellular SIAH-1.
Fig 3. Virally expressed ICP0NxN1/2 does not bind to SIAH-1.
(A) U2OS cells were infected for 48 h with the indicated mutants and HSV-2 strain MS (MS wt) at an MOI 0.01 pfu/cell. Cell lysates were incubated with GST-SIAH-1-loaded glutathione sepharose beads. Eluates were analyzed by SDS-PAGE and Western blotting using antibodies directed against HSV-2 ICP0 and GFP. (B) Input controls (10%) of the GST-pulldown were analyzed as before using ICP0-specific antibody.
The NxN1/2 mutation in ICP0 attenuates viral growth
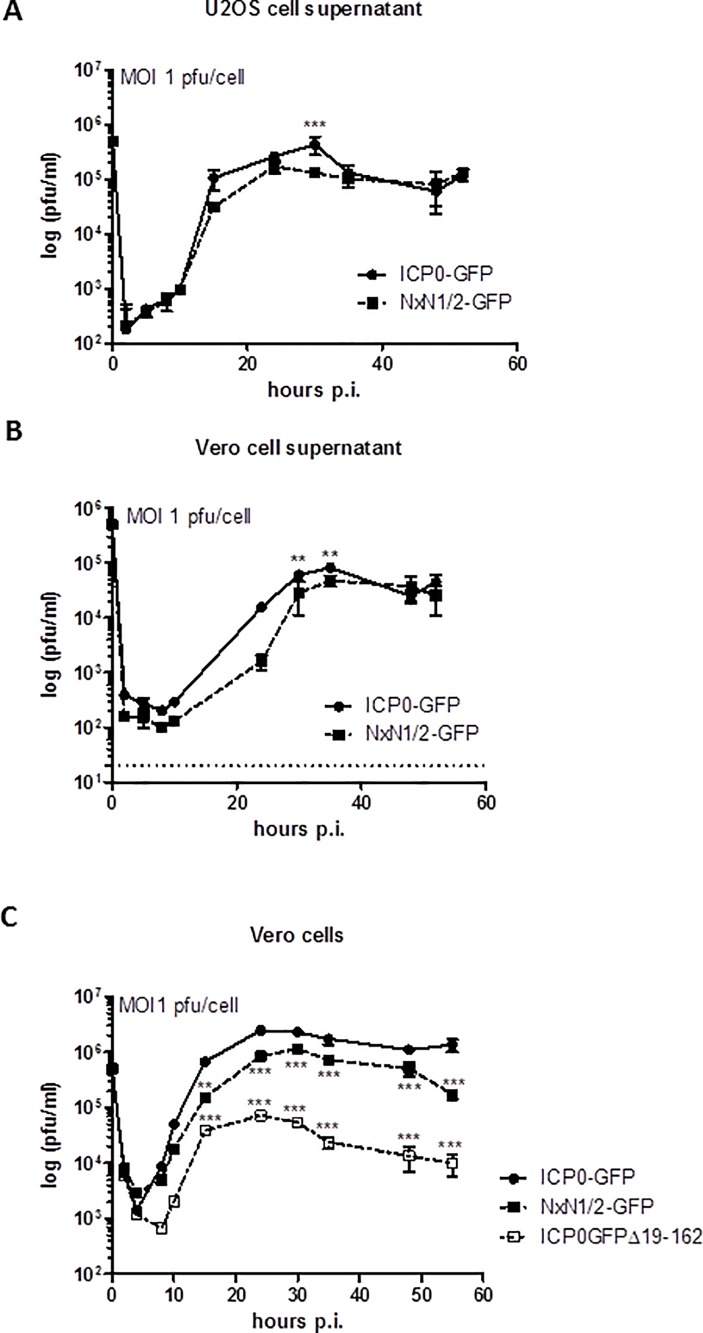
Compared to wild-type HSV, significantly fewer plaques are produced by ICP0null mutants on many cell lines, including Vero cells [16,26,27]. In sharp contrast, deletion of ICP0 does not affect plaque formation in U2OS cells [38,44,45]. However, the previously reported RING-finger-deficient HSV-2-GFP-ICP0Δ19–162 (HSV-2 0ΔRING) mutant [28] produced severely reduced vital titers, by almost two logs, on Vero cells compared to U2OS cells (S2 Table and S5 Fig). However, plaque formation of the SIAH-1 binding-deficient HSV-2 ICP0 mutant was clearly not affected on these cell lines (S2 Table and S6 Fig). Notwithstanding, during preparation of virus stocks we noted delayed growth of our HSV-2-ICP0NxN1/2-GFP mutant. To confirm this observation, we compared the growth kinetics of HSV-2-ICP0NxN1/2-GFP and HSV-2-ICP0-GFP in U2OS and Vero cells using an MOI of 1 pfu/cell. Again, HSV-2-GFP-ICP0Δ19–162 was included as a control in some experiments. Of note, analysis of HSV-2-ICP0NxN1/2-GFP and HSV-2-ICP0-GFP particle preparations by major capsid protein VP5-specific immunoblotting and UL27-specific droplet digital PCR demonstrated comparable virion/pfu ratios (not shown).
After infecting U2OS or Vero cells, supernatants were harvested at regular intervals for subsequent titration using Vero cell monolayers (Fig 4A and 4B). In another set-up, infected Vero cell cultures were lysed and titrated (Fig 4C). Independently of the procedure and cell line used, the titers of the ICP0NxN1/2-GFP mutant were significantly lower at 30 to 35 hours post infection than the titers of HSV-2-ICP0-GFP. By about 40 to 48 hours post infection, both viruses reached approximately comparable titers when analyzing virus-containing supernatants (Fig 4A and 4B). However, when viral particles isolated from Vero cell pellets were analyzed, the observed growth differences between the NxN1/2 mutant and HSV-2-ICP0-GFP were more pronounced (Fig 4C). At 55 hours post infection, when this experiment was terminated, the titer of the SIAH-1 binding-deficient mutant (NxN1/2) was clearly lower than the ICP0-GFP virus titer. Indeed, titer differences between the two viruses were already significantly different at about 15 hours and increased at 25 hours post infection.
Fig 4. The NxN1/2 mutation in ICP0 of HSV-2 results in delayed viral growth.
(A) U2OS cells were infected in duplicates with an MOI of 1 pfu/cell of HSV-2-ICP0-GFP (ICP0-GFP) and HSV-2-ICP0NxN1/2-GFP (NxN1/2-GFP). At the indicated time points supernatants were harvested and titrated on Vero cell monolayers. Mean ± SD of viral titers (n = 2 per data point) were plotted as a function of time. (B) Vero cells were infected in duplicates and evaluated as above. (C) Vero cells were infected in duplicates with HSV-2-ICP0-GFP, HSV-2-ICP0NxN1/2-GFP and HSV-2-GFP-ICP0Δ19–162 (i.e. ICP0ΔRING mutant) at an MOI of 1 pfu/cell. At the indicated time points supernatants were harvested, titrated and evaluated as above. Titer differences were analyzed using two-way ANOVA, with ** p<0.01 and *** p<0.001. The detection limits are shown by a dashed line.
As expected, the RING-finger-deficient mutant (i.e. HSV-2-GFP-ICP0Δ19–162) produced significantly diminished viral titers, also starting at 15 hours post infection (Fig 4C). By 35 hours post infection, the titers of this ΔRING mutant were more than one log below the titer of the NxN1/2-GFP mutant and almost two logs below the titer of wild-type virus (Fig 4C). These results suggest that preventing interaction between ICP0 and SIAH-1 attenuates growth of HSV-2, although clearly this growth disadvantage is not as striking as the reduced growth behavior of the RING-finger-deficient (ICP0ΔRING) mutant (Fig 4C).
The NxN1/2 mutation impairs viral protein and DNA synthesis in vitro
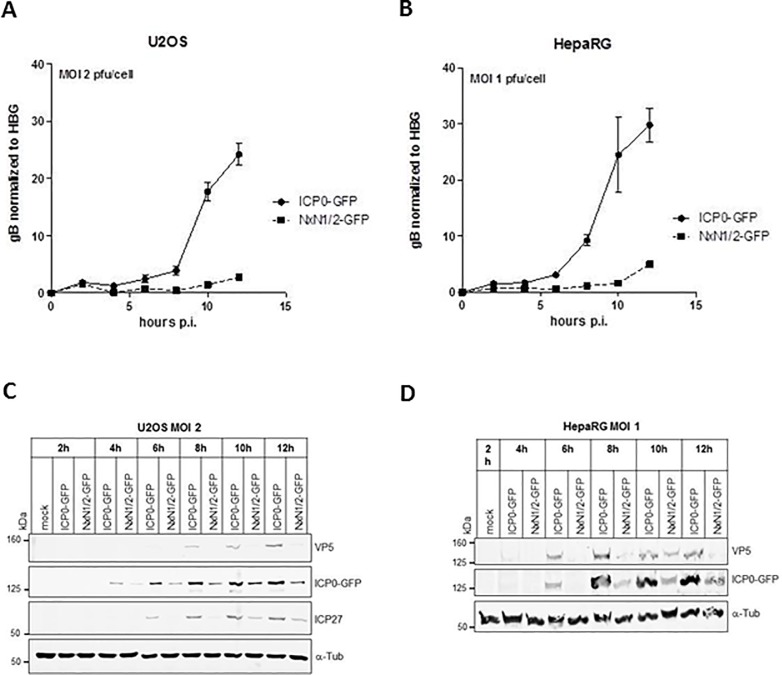
Next, we compared DNA and protein expression in vitro mediated by the HSV-2NxN1/2-GFP and HSV-2-ICP0-GFP virus constructs. After infecting U2OS or HepaRG cells with both viruses at an MOI 1 or 2 pfu/cell, respectively, cell extracts were prepared at different time points post infection for subsequent analysis of viral DNA levels by quantitative real-time PCR, and viral protein expression by Western blotting. Viral DNA synthesis commenced shortly after infection (Fig 5A and 5B). However, starting at 5 hours post infection viral DNA copy numbers in HSV-2-ICP0NxN1/2-GFP infected cells were significantly lower than copy numbers produced by HSV-2-ICP0-GFP at each time point analyzed. (Fig 5A and 5B). This difference remained during the entire experimental period of 12 hours and became even more distinct as time progressed.
Fig 5. NxN1/2 mutation in ICP0 results in delayed and reduced vital DNA and protein synthesis.
(A) U2OS cells, and (B) HepaRG cells were infected with HSV-2-ICP0-GFP (ICP0-GFP) or HSV-2-ICP0NxN1/2-GFP (NxN1/2-GFP) at an MOI of 2 or 1 pfu/cell, respectively. Infected cells were harvested at the indicated time points post infection (p.i.) for isolation of genomic and viral DNA. Viral DNA copies were determined by quantitative real-time PCR using oligonucleotide-primers and probes directed against the UL27 gene encoding glycoprotein gB. Copy numbers were normalized to the sequence of the human single-copy beta-globin gene (HBG). (C) U2OS cells, and (D) HepaRG cells were infected with HSV-2-ICP0-GFP or HSV-2-ICP0NxN1/2-GFP at an MOI of 2 for up to 24 hours, or 1 pfu/cell for 12 hours, respectively. At the indicated time points, cells were harvested. Whole cell lysates were analyzed by SDS-PAGE and Western blot against the indicated proteins. Signals were quantified in S2 Fig.
The relative attenuation of HSV-2-ICP0NxN1/2-GFP was also reflected at the level of viral de novo protein expression (Fig 5C and 5D). In immunoblot analyses, immediate-early gene synthesis was detected by ICP0- and ICP27-specific antibodies, while viral late gene expression was monitored detecting the major capsid protein VP5. Within the first 12 hours post infection protein abundance was lower in cells infected with the SIAH-1 binding-deficient virus mutant HSV-2-ICP0NxN1/2-GFP (Fig 5C and 5D). This was observed with both immediate early (ICP0 and ICP27) and late proteins (VP5), although synthesis of the latter was clearly more impaired (Fig 5C and 5D). The differences in viral protein expression became even more obvious when the quantified Western blot signals were normalized to the fluorescence intensity of the loading control α-tubulin (S7 Fig). Specifically, between 8 and 12 hours post infection all viral protein signals, including those of ICP0, were clearly higher in cells infected with wild-type virus than in cells infected with SIAH-1 binding-deficient virus (S7A and S7B Fig).
Inactivation of the SIAH binding motif in ICP0 results in impaired HSV-2 replication in trigeminal ganglia and the brain stem in vivo
We next challenged C57Bl/6J mice via the ocular route (Fig 6A) [41,46]. After infection, viruses replicate in the eye, enter the innervating sensory neurons of the trigeminal ganglia (TG) and travel in a retrograde direction towards neuronal cell bodies and even further to the brain stem [4,5,47,48]. In our experimental set-up, 105 plaque forming units (pfu) of the respective virus preparation (HSV-2-ICP0NxN1/2-GFP or HSV-2-ICP0-GFP) were applied to the cornea of anesthetized animals and virus replication was detected in the tear film obtained by ocular swabs, initially taken at 5 hours and at day 1, 2, 3, 5 and 7 post infection (Fig 6A). When comparing the viral load, particularly at 1 day post infection titers of the SIAH-1 binding-deficient mutant were significantly lower than the titers of the virus expressing ICP0-GFP protein (Fig 6B). Clearly, at later time points both viral constructs reached comparable titers (Fig 6B). At day 7 post infection, virus could no longer be detected in the tear film, indicating a complete clearance of virus in the cornea.
Fig 6. HSV-2 replication in the cornea of C57Bl/6J mice.
(A) Schematic representation of the murine ocular HSV-2 infection model. Both corneas of anesthetized mice were scarified. Virus was placed on the eyes using 105 pfu/eye. Ocular swabs were taken at the indicated time points post infection (p.i.). Animals were sacrificed at day 3, 5 and 7 post infection, and trigeminal ganglia (TG) and the brain stem were isolated for further analyses. (B) Ocular swabs from HSV-2-ICP0-GFP (ICP0-GFP) or HSV-2-ICP0NxN1/2-GFP (NxN1/2-GFP) infected animals taken at the indicated time points p.i. were analyzed for viral titers. For every experiment a group of n = 10 mice were infected and experiments were repeated four times. Each symbol represents the mean ± SEM of the titers, determined on Vero cell monolayers. The limit of detection is indicated by a dashed line. Statistical differences were calculated using two-way ANOVA with *** p< 0.001.
Following corneal infection, virus particles can be detected in the TG as early as 2 to 3 days post infection [49] and virus clearance commences at day 6 post infection [50]. Therefore, we analyzed virus dissemination to the innervating neurons, and virus replication in the infected tissue at 3, 5 and 7 days post infection by isolating and homogenizing the TG and brain stem of acutely infected mice (Fig 7). Half of the tissue samples were used for determining viral titers and the other half were saved for isolating DNA to quantify viral genome copy numbers by quantitative real-time PCR.
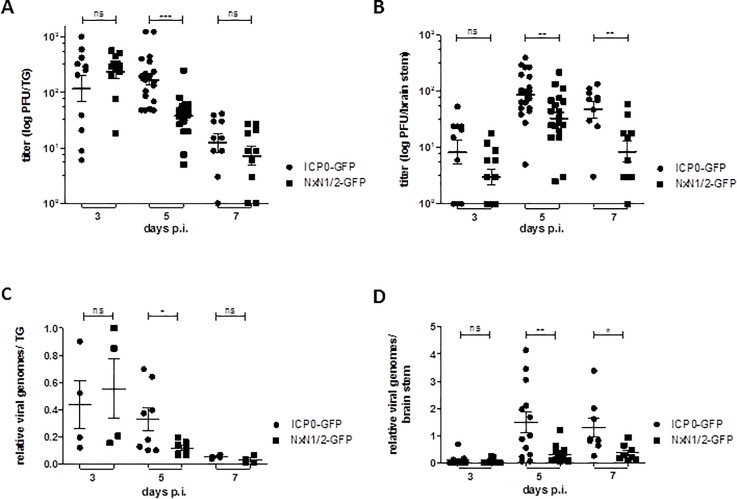
Fig 7. The NxN1/2 mutation in ICP0 leads to impaired replication in the TG and brain stem of C57Bl/6J mice.
At day 3, 5 and 7 post infection (p.i.), (A) the trigeminal ganglia (TG), and (B) brain stem was extracted from HSV-2-ICP0-GFP and HSV-2-ICP0NxN1/2-GFP infected mice, homogenized and viral titers were determined as in Fig 6. Symbols represent individual animals; floating bars represent the mean ± SEM. Titer differences were determined by the Mann-Whitney test. (C) Half of the homogenized TG tissue from panel A, and (D) half of the tissue from panel B was lysed and total DNA was isolated to determine viral genome copy numbers. Oligonucleotide-primers specific for the HSV-2 UL27 gene were used and copy numbers were normalized to murine β-actin sequences. Each symbol represents the relative genome number of an individual animal, relative to the genome copy number of a single HSV-2-ICP0-GFP infected mouse, which was set as 100%. Floating bars represent the mean ± SEM. Statistical differences were determined as above.
Titer measurements revealed that comparable amounts of viral wild-type and NxN1/2 mutant particles were detected in the TG at day 3 post infection (Fig 7A), suggesting that the ICP0:SIAH-1 interaction is not required for virus dissemination from the eye to the innervating neurons. However, on day 5 post infection titers of the NxN1/2 virus construct were significantly lower in the TG when compared to the titers reached by the ICP0 wild-type virus. This was also seen when the brain stem was analyzed (Fig 7B). In the TG, virus titers were clearly reduced at day 7 post infection and no significant difference was detected between the NxN1/2 mutant and the ICP0 wild-type construct; however comparably higher average virus titers were observed in the brain stem (Fig 7B).
Although less sensitive due to limited TG and brain stem samples, analysis of total viral DNA in these tissues confirmed the results above, showing significantly elevated levels of viral genome copies of the ICP0 wild-type virus as opposed to the NxN1/2 virus mutant, particularly in the brain stem (Fig 7C and 7D). These results suggest that the SIAH-binding deficient mutation significantly impairs HSV-2 replication in the TG and brain stem, thereby possibly affecting virus dissemination kinetics.
Discussion
In a previous study we identified SIAH-1 as a host cell factor that specifically interacts with the viral regulatory protein ICP0 [30]. Here, we show that abolishing the ICP0:SIAH-1 interaction affects the HSV-2 phenotype during infection. In cell culture, the SIAH-binding deficient mutant synthesized significantly less DNA and protein, and virus growth was clearly attenuated compared to wild-type virus (Fig 5). In an animal model, HSV-2 carrying the SIAH-binding deficient mutation caused an acute infection in the murine eye and was able to spread to the innervating neurons (Fig 6B and Fig 7). However, the titers of the SIAH-1 binding-deficient virus detected in the TG and brain stem at day 5 post infection were much lower than the titers of wild-type virus (Fig 7A and 7B). Also, the viral DNA load in these tissues was clearly lower in case of the SIAH-1 binding-deficient virus (Fig 7C and 7D). Of note, the mouse model was considered a suitable system to study ICP0:SIAH-1 interaction, since, due to its evolutionary conservation, the murine genome encodes a SIAH-1 homolog called SIAH1A [51]. Human SIAH-1 and murine SIAH1A only differ in a single amino acid, and apparently fulfill identical functions [31].
The cellular/proteomic consequences evoked in the infected host cell by disrupting the interaction between ICP0 and SIAH-1 are currently not known. Both proteins belong to the RING-finger domain-containing family of E3 ubiquitin ligases, which induce proteasome-dependent degradation of various cellular factors [52,53]. For example, ICP0 and SIAH are both capable of mediating promyelocytic leukemia protein (PML) degradation [24,54,55]; PML structures are considered to be the sites where viral transcription and replication take place [56,57]. However, the dispersal of PML bodies appeared not to depend on the ICP0:SIAH-1 interaction. In fact, PML bodies were still degraded to comparable degrees in cells infected with either SIAH-1 binding-deficient virus or the ICP0 wild-type virus (data not shown). These results confirmed previous experiments obtained in cell cultures transiently transfected with ICP0 expression constructs [30]. However at this point, it cannot be completely ruled out that ICP0-mediated proteasomal degradation of other PML-unrelated cellular targets might be hampered when the ICP0:SIAH-1 interaction is compromised.
A single PxAxVxP-type binding motif for SIAH recognition is also located at amino acid positions 401–407 in ICP0 from Herpes Simplex Virus type 1 (HSV-1) [30]. This ICP0 region was previously analyzed for its ability to promote, for example, transcription from viral promoters or to affect HSV-1 growth. For instance, plasmids encoding ICP0 with a five aa insertion at residue 406 (thus disrupting the VxP motif), or deletion of the VxP motif, both displayed a reduced capability (7–22%) to induce transcription from the gD promoter in cooperation with ICP4 transcription [58], although the intracellular localization of both regulators, ICP0 and ICP4, was unaltered [59]. Furthermore, another independent study using promoter activation assays showed that deletion of the aa residues 393–448, or aa residues at positions 400–462 in HSV-1 ICP0 reduced its transcriptional activity to 34–81% of wild-type activity, depending on the reporter construct used [60]. Moreover, depending on MOI and cell type, an HSV-1 mutant encoding ICP0Δaa400-462 yielded only 4–17% of total viral particles compared to wild-type virus [61]. Finally, the aforementioned HSV-1 mutant containing a five aa insertion at ICP0 position 406 [58] was severely attenuated in BHK cells and displayed reduced protein synthesis, although in human fetal lung cells this defect was not as pronounced [25].
It may be conceivable that the mutant phenotypes described above can be explained, at least in part, by the lack of ICP0:SIAH interaction, thereby compromising ICP0’s ability to efficiently induce viral transcription, and in consequence, virus replication. This notion is in good accordance with the attenuated phenotype observed with our HSV-2-ICP0NxN1/2-GFP mutant virus. Although the RING domain probably constitutes the region of ICP0 most important for initiation and promotion of HSV infection [6,38,50], the inability of ICP0 to bind SIAH also proved to be detrimental to virus growth. Clearly, structural impairment of ICP0 could also result in a loss of ICP0 function. The two amino acid exchanges in our NxN1/2-mutant could result in small structural alterations, which could interfere with other ICP0 functions apart from SIAH-1 binding. It is noted, however, that localization of ICP0 during HSV-2 infection was unaffected by this mutation (data not shown).
So why does HSV-2 require ICP0 binding to SIAH-1 for efficient replication? One could envision three scenarios: i) SIAH-1 acquisition could expand ICP0’s target range in its role of degrading cellular proteins; ii) ICP0 could inactivate SIAH-1 by ubiquitination, or iii) ICP0 may function as a decoy to sequester SIAH-1 away from its own targets. Such mechanisms would increase the abundance of SIAH-1 target proteins, some of which might be required for efficient viral replication.
It was previously noticed that after cotransfection SIAH-1 is stabilized by ICP0, which may be explained by the inhibition of SIAH-1 auto-ubiquitination [30]. Altogether, interaction with ICP0 may have multiple consequences that could functionally inactivate SIAH-1 (for a model see S8 Fig): i) Both E3 RING ubiquitin ligases have the potential for mutual polyubiquitination; ii) ICP0 inhibits SIAH auto-ubiquitination; iii) both proteins interact in low-soluble aggregates; iv) ICP0 might inhibit ubiquitination of SIAH target proteins [30]. Although SIAH-1 appears to negatively affect the stability of ICP0 by ubiquitination [30], the net outcome of the ICP0:SIAH-1 interaction might be that SIAH-1 is captured by ICP0, which results in SIAH-1 depletion from the active cellular pool. Although SIAH ubiquitin ligases have so far not been regarded as a general intrinsic antiviral defense mechanism, their inactivation by ICP0 surely contributes to a cellular environment favorable to virus infection.
Of the many SIAH targets, some are involved in cellular pathways that might be crucial at different stages of infection. For example, SIAH’s involvement in DNA damage signaling [35,62] or in maintaining presynaptic proteostasis [63] may be of particular relevance for replication or the spread of a neurotropic DNA virus. An interesting speculation is driven by the observation that SIAH-1 mediates the polyubiquitination and degradation of the ELL2 protein, a crucial component of super-elongation complexes (SEC) [64]. SECs contain several transcription factors required for elongation by RNA polymerase II, and were originally identified by their roles in HIV-1 transcription and gene regulation through mixed lineage leukemia protein [65]. Interestingly, inhibition of SIAH-1 resulted not only in increased intracellular ELL2 levels, but also in increased transcription from the HIV-1 LTR promoter, which was shown to depend on functional SECs [64,66]. It is therefore tempting to speculate that interfering with SIAH-1 function is a viral strategy adopted by ICP0 to promote efficient HSV-2 gene expression at the level of transcriptional elongation. Future experiments may therefore focus on the effect of ICP0 on the abundance of SIAH-1 target proteins. These data might reveal valuable information about virus-induced changes in cellular protein interaction networks, and broaden the base for developing novel antiviral therapies.
Supporting information
(PDF)
Passage 2 (P2) virus of the listed HSV-2 constructs was produced in BHK cells (see Methods). The P2 inoculum was subsequently titrated on U2OS, Vero and HepaRG cells to compare the growth efficiencies on different cell lines.
(PDF)
Viral DNA was digested with HindIII and NcoI and subjected to Southern blot analyses using ICP0- (left panel) and GFP-specific (right panel) radioactively-labelled probes. Constructs marked in green were selected for further analysis.
(PDF)
Vero cells were infected in duplicates with HSV‐2‐ICP0‐GFP (ICP0‐GFP) or HSV-2 MS (wt) using (A) an MOI of 0,01 pfu/cell or (B) an MOI of 1 pfu/cell. At the indicated time points, cells were harvested and titrated on Vero cell monolayers. Mean ± SD of viral titers (n = 2 per data point) were plotted as a function of time.
(PDF)
Vero cells were infected with HSV‐2‐ICP0‐GFP or HSV‐2‐ICP0NxN1/2‐GFP at an MOI of 1 for up to 24 hours. Cells were harvested at the indicated time points and whole cell lysates were analyzed by SDS‐PAGE and Western blot using antibodies raised against the indicated proteins.
(PDF)
U2OS cells (2x105) were infected with an MOI of 5 pfu/cell of the indicated virus construct. At 2, 4 and 6 h post infection, cells were fixed for 20 min with 4% PFA. Nuclei were stained with 2 μM Hoechst 33342 and plasma membrane structures with Alexa Fluor®647 wheat germ agglutinin (WGA). Localization of ICP0-GFP was visualized by confocal fluorescence microscopy using a Nikon Eclipse Ti-E system.
(PDF)
Graphical comparison of viral titers as listed in S2 Table.
(PDF)
Dilution series were performed to determine the relative plaque formation efficiency of the indicated HSV-2 constructs on Vero and HepaRG cells. Obtained titers were used to calculate the ratio of plaques on Vero cells to U2OS cells, and HepaRG cells to U2OS cells, independently for every HSV-2 construct. The titer on U2OS cells was arbitrarily set as 100%; wt, wild-type.
(PDF)
(A) HepaRG cells were infected with the indicated constructs as described in Fig 5C and (B) U2OS cells were infected with the indicated constructs as described in Fig 5D. The fluorescent signal intensities of the secondary antibodies detecting the indicated viral proteins were quantified and normalized to the signal intensities of the loading control α-tubulin. Relative intensities are shown in pixels.
(PDF)
ICP0 and SIAH-1 are E3 ubiquitin ligases that are potentially able to ubiquitinate each other and, mostly in absence of a target protein, can also auto-ubiquitinate. It is hypothesized that ICP0 ubiquitinates SIAH-1 in vivo leading to the proteasomal degradation of the latter and to the stabilization of a so far unknown target protein that is needed for efficient progression of HSV-2 infection. When ICP0 auto-ubiquitinates or is ubiquinated by SIAH-1, it can be de-ubiquitinated by USP7. In turn, ICP0 can also ubiquitinate USP7, leading to the proteasomal degradation of the latter.
(PDF)
Acknowledgments
We thank Bettina Abel and Britta Weseloh (Heinrich-Pette-Institute, Hamburg, Germany) for technical assistance, Daniela Indenbirken for next generation sequencing reactions, and Andreas Möller (Peter MacCallum Cancer Research Center, Melbourne, Australia) for helpful discussions. The authors are indebted to William P. Halford (Southern Illinois University, Springfield IL, USA) for assistance in the study and, in particular, for teaching the murine ocular HSV-2 infection model. This study was supported by the Landesforschungsförderung Hamburg, Graduiertenkolleg “Degradation For Life Graduate School Hamburg (DELIGRAH)” and the “Viral Latency” program at the Heinrich-Pette-Institute—Leibniz Institute for Experimental Virology, Hamburg.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This study was funded by the Free and Hanseatic City of Hamburg, Graduiertenkolleg DELIGRAH #LFF-GK06. An author of this study [MME] is employed by Biomedizinische Forschungsgesellschaft mbH, Vienna, Austria. Biomedizinische Forschungsgesellschaft mbH did not serve as a funder of this study. This study was supported by the Landesforschungsförderung Hamburg, Graduiertenkolleg “Degradation For Life Graduate School Hamburg (DELIGRAH)” and the “Viral Latency” program at the Heinrich-Pette-Institute - Leibniz Institute for Experimental Virology, Hamburg. These funders provided support in the form of salaries for authors [JSC,C-HN, MV] and research materials, but did not have any additional role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Roizman B, Knipe DM (2001) Herpes Simplex viruses and their replication In: Knipe DM, Howley P, Griffin DE, Lamb RA, Martin MA et al. , editors. Fields Virology. Philadelphia: Lippincott Williams & Wilkins; pp. 2399–2459. [Google Scholar]
- 2.Roizman B, Whitley RJ (2013) An inquiry into the molecular basis of HSV latency and reactivation. Annu Rev Microbiol 67: 355–374. 10.1146/annurev-micro-092412-155654 [DOI] [PubMed] [Google Scholar]
- 3.Thellman NM, Triezenberg SJ (2017) Herpes Simplex Virus Establishment, Maintenance, and Reactivation: In Vitro Modeling of Latency. Pathogens 6 pathogens6030028 [pii]; 10.3390/pathogens6030028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Smith G (2012) Herpesvirus transport to the nervous system and back again. Annu Rev Microbiol 66: 153–176. 10.1146/annurev-micro-092611-150051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sandri-Goldin RM (2007) Initiation of transcription and RNA synthesis, processing and transport in HSV and VZV infected cells. NBK47363 [bookaccession]. [PubMed]
- 6.Whitley R, Kimberlin DW, Prober CG (2007) Pathogenesis and disease. NBK47449 [bookaccession]. [PubMed]
- 7.Barnabas RV, Celum C (2012) Infectious co-factors in HIV-1 transmission herpes simplex virus type-2 and HIV-1: new insights and interventions. Curr HIV Res 10: 228–237. CHIVR-EPUB-20120229-004 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rajcani J, Andrea V, Ingeborg R (2004) Peculiarities of herpes simplex virus (HSV) transcription: an overview. Virus Genes 28: 293–310. 5269258 [pii]. [DOI] [PubMed] [Google Scholar]
- 9.Boutell C, Everett RD (2013) Regulation of alphaherpesvirus infections by the ICP0 family of proteins. J Gen Virol 94: 465–481. vir.0.048900–0 [pii]; 10.1099/vir.0.048900-0 [DOI] [PubMed] [Google Scholar]
- 10.Roizman B (2011) The checkpoints of viral gene expression in productive and latent infection: the role of the HDAC/CoREST/LSD1/REST repressor complex. J Virol 85: 7474–7482. JVI.00180-11 [pii]; 10.1128/JVI.00180-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Roizman B, Zhou G, Du T (2011) Checkpoints in productive and latent infections with herpes simplex virus 1: conceptualization of the issues. J Neurovirol 17: 512–517. 10.1007/s13365-011-0058-x [DOI] [PubMed] [Google Scholar]
- 12.Everett RD, Rechter S, Papior P, Tavalai N, Stamminger T, Orr A (2006) PML contributes to a cellular mechanism of repression of herpes simplex virus type 1 infection that is inactivated by ICP0. J Virol 80: 7995–8005. 10.1128/JVI.00734-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roizman B, Gu H, Mandel G (2005) The first 30 minutes in the life of a virus: unREST in the nucleus. Cell Cycle 4: 1019–1021. 10.4161/cc.4.8.1902 [DOI] [PubMed] [Google Scholar]
- 14.Everett RD (2000) ICP0, a regulator of herpes simplex virus during lytic and latent infection. Bioessays 22: 761–770. [DOI] [PubMed] [Google Scholar]
- 15.Cai W, Astor TL, Liptak LM, Cho C, Coen DM, Schaffer PA (1993) The herpes simplex virus type 1 regulatory protein ICP0 enhances virus replication during acute infection and reactivation from latency. J Virol 67: 7501–7512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cai W, Schaffer PA (1992) Herpes simplex virus type 1 ICP0 regulates expression of immediate-early, early, and late genes in productively infected cells. J Virol 66: 2904–2915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Efstathiou S, Preston CM (2005) Towards an understanding of the molecular basis of herpes simplex virus latency. Virus Res 111: 108–119. S0168-1702(05)00117-6 [pii]; 10.1016/j.virusres.2005.04.017 [DOI] [PubMed] [Google Scholar]
- 18.Halford WP, Kemp CD, Isler JA, Davido DJ, Schaffer PA (2001) ICP0, ICP4, or VP16 expressed from adenovirus vectors induces reactivation of latent herpes simplex virus type 1 in primary cultures of latently infected trigeminal ganglion cells. J Virol 75: 6143–6153. 10.1128/JVI.75.13.6143-6153.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Halford WP, Schaffer PA (2001) ICP0 is required for efficient reactivation of herpes simplex virus type 1 from neuronal latency. J Virol 75: 3240–3249. 10.1128/JVI.75.7.3240-3249.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Everett R, O'Hare P, O'Rourke D, Barlow P, Orr A (1995) Point mutations in the herpes simplex virus type 1 Vmw110 RING finger helix affect activation of gene expression, viral growth, and interaction with PML-containing nuclear structures. J Virol 69: 7339–7344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chelbi-Alix MK, de The H (1999) Herpes virus induced proteasome-dependent degradation of the nuclear bodies-associated PML and Sp100 proteins. Oncogene 18: 935–941. 10.1038/sj.onc.1202366 [DOI] [PubMed] [Google Scholar]
- 22.Müller S, Dejean A (1999) Viral immediate-early proteins abrogate the modification by SUMO-1 of PML and Sp100 proteins, correlating with nuclear body disruption. J Virol 73: 5137–5143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maul GG, Everett RD (1994) The nuclear location of PML, a cellular member of the C3HC4 zinc-binding domain protein family, is rearranged during herpes simplex virus infection by the C3HC4 viral protein ICP0. J Gen Virol 75: 1223–1233. 10.1099/0022-1317-75-6-1223 [DOI] [PubMed] [Google Scholar]
- 24.Maul GG, Guldner HH, Spivack JG (1993) Modification of discrete nuclear domains induced by herpes simplex virus type 1 immediate early gene 1 product (ICP0). J Gen Virol 74: 2679–2690. 10.1099/0022-1317-74-12-2679 [DOI] [PubMed] [Google Scholar]
- 25.Everett RD (1989) Construction and characterization of herpes simplex virus type 1 mutants with defined lesions in immediate early gene 1. J Gen Virol 70 (Pt 5): 1185–1202. [DOI] [PubMed] [Google Scholar]
- 26.Everett RD, Boutell C, Orr A (2004) Phenotype of a herpes simplex virus type 1 mutant that fails to express immediate-early regulatory protein ICP0. J Virol 78: 1763–1774. 10.1128/JVI.78.4.1763-1774.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stow ND, Stow EC (1986) Isolation and characterization of a herpes simplex virus type 1 mutant containing a deletion within the gene encoding the immediate early polypeptide Vmw110. J Gen Virol 67 (Pt 12): 2571–2585. [DOI] [PubMed] [Google Scholar]
- 28.Halford WP, Puschel R, Rakowski B (2010) Herpes simplex virus 2 ICP0 mutant viruses are avirulent and immunogenic: implications for a genital herpes vaccine. PLoS One 5: e12251 10.1371/journal.pone.0012251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mossman KL, Saffran HA, Smiley JR (2000) Herpes simplex virus ICP0 mutants are hypersensitive to interferon. J Virol 74: 2052–2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nagel CH, Albrecht N, Milovic-Holm K, Mariyanna L, Keyser B, Abel B, Weseloh B, Hofmann TG, Eibl MM, Hauber J (2011) Herpes simplex virus immediate-early protein ICP0 is targeted by SIAH-1 for proteasomal degradation. J Virol 85: 7644–7657. 10.1128/JVI.02207-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hu G, Chung YL, Glover T, Valentine V, Look AT, Fearon ER (1997) Characterization of human homologs of the Drosophila seven in absentia (sina) gene. Genomics 46: 103–111. 10.1006/geno.1997.4997 [DOI] [PubMed] [Google Scholar]
- 32.Polekhina G, House CM, Traficante N, Mackay JP, Relaix F, Sassoon DA, Parker MW, Bowtell DD (2002) Siah ubiquitin ligase is structurally related to TRAF and modulates TNF-alpha signaling. Nat Struct Biol 9: 68–75. 10.1038/nsb743 [pii]. [DOI] [PubMed] [Google Scholar]
- 33.Santelli E, Leone M, Li C, Fukushima T, Preece NE, Olson AJ, Ely KR, Reed JC, Pellecchia M, Liddington RC, Matsuzawa S (2005) Structural analysis of Siah1-Siah-interacting protein interactions and insights into the assembly of an E3 ligase multiprotein complex. J Biol Chem 280: 34278–34287. M506707200 [pii]; 10.1074/jbc.M506707200 [DOI] [PubMed] [Google Scholar]
- 34.Marconi P, Manservigi R (2014) Herpes simplex virus growth, preparation, and assay. Methods Mol Biol 1144: 19–29. 10.1007/978-1-4939-0428-0_2 [DOI] [PubMed] [Google Scholar]
- 35.Winter M, Sombroek D, Dauth I, Moehlenbrink J, Scheuermann K, Crone J, Hofmann TG (2008) Control of HIPK2 stability by ubiquitin ligase Siah-1 and checkpoint kinases ATM and ATR. Nat Cell Biol 10: 812–824. 10.1038/ncb1743 [DOI] [PubMed] [Google Scholar]
- 36.Döhner K, Radtke K, Schmidt S, Sodeik B (2006) Eclipse phase of herpes simplex virus type 1 infection: Efficient dynein-mediated capsid transport without the small capsid protein VP26. J Virol 80: 8211–8224. 80/16/8211 [pii]; 10.1128/JVI.02528-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sodeik B, Ebersold MW, Helenius A (1997) Microtubule-mediated transport of incoming herpes simplex virus 1 capsids to the nucleus. J Cell Biol 136: 1007–1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Namvar L, Olofsson S, Bergstrom T, Lindh M (2005) Detection and typing of Herpes Simplex virus (HSV) in mucocutaneous samples by TaqMan PCR targeting a gB segment homologous for HSV types 1 and 2. J Clin Microbiol 43: 2058–2064. 43/5/2058 [pii]; 10.1128/JCM.43.5.2058-2064.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sarkar I, Hauber I, Hauber J, Buchholz F (2007) HIV-1 proviral DNA excision using an evolved recombinase. Science 316: 1912–1915. 10.1126/science.1141453 [DOI] [PubMed] [Google Scholar]
- 40.Döhner K, Wolfstein A, Prank U, Echeverri C, Dujardin D, Vallee R, Sodeik B (2002) Function of dynein and dynactin in herpes simplex virus capsid transport. Mol Biol Cell 13: 2795–2809. 10.1091/mbc.01-07-0348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Halford WP, Schaffer PA (2000) Optimized viral dose and transient immunosuppression enable herpes simplex virus ICP0-null mutants To establish wild-type levels of latency in vivo. J Virol 74: 5957–5967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.House CM, Frew IJ, Huang HL, Wiche G, Traficante N, Nice E, Catimel B, Bowtell DD (2003) A binding motif for Siah ubiquitin ligase. Proc Natl Acad Sci U S A 100: 3101–3106. 10.1073/pnas.0534783100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.House CM, Hancock NC, Möller A, Cromer BA, Fedorov V, Bowtell DD, Parker MW, Polekhina G (2006) Elucidation of the substrate binding site of Siah ubiquitin ligase. Structure 14: 695–701. 10.1016/j.str.2005.12.013 [DOI] [PubMed] [Google Scholar]
- 44.Everett RD, Parada C, Gripon P, Sirma H, Orr A (2008) Replication of ICP0-null mutant herpes simplex virus type 1 is restricted by both PML and Sp100. J Virol 82: 2661–2672. 10.1128/JVI.02308-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Everett RD, Parsy ML, Orr A (2009) Analysis of the functions of herpes simplex virus type 1 regulatory protein ICP0 that are critical for lytic infection and derepression of quiescent viral genomes. J Virol 83: 4963–4977. 10.1128/JVI.02593-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Arbuckle JH, Turner AM, Kristie TM (2014) Analysis of HSV Viral Reactivation in Explants of Sensory Neurons. Curr Protoc Microbiol 35: 14E–21. 10.1002/9780471729259.mc14e06s35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Webre JM, Hill JM, Nolan NM, Clement C, McFerrin HE, Bhattacharjee PS, Hsia V, Neumann DM, Foster TP, Lukiw WJ, Thompson HW (2012) Rabbit and mouse models of HSV-1 latency, reactivation, and recurrent eye diseases. J Biomed Biotechnol 2012: 612316 10.1155/2012/612316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pennington MR, Ledbetter EC, Van de Walle GR (2017) New paradigms for the study of ocular alphaherpesvirus infections: Insights into the use of non-traditional host model systems. Viruses 9 v9110349 [pii]; 10.3390/v9110349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nicoll MP, Proenca JT, Efstathiou S (2012) The molecular basis of herpes simplex virus latency. FEMS Microbiol Rev 36: 684–705. 10.1111/j.1574-6976.2011.00320.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Divito S, Cherpes TL, Hendricks RL (2006) A triple entente: virus, neurons, and CD8+ T cells maintain HSV-1 latency. Immunol Res 36: 119–126. IR:36:1:119 [pii]; 10.1385/IR:36:1:119 [DOI] [PubMed] [Google Scholar]
- 51.Della NG, Senior PV, Bowtell DD (1993) Isolation and characterisation of murine homologues of the Drosophila seven in absentia gene (sina). Development 117: 1333–1343. [DOI] [PubMed] [Google Scholar]
- 52.Deshaies RJ, Joazeiro CA (2009) RING domain E3 ubiquitin ligases. Annu Rev Biochem 78: 399–434. 10.1146/annurev.biochem.78.101807.093809 [DOI] [PubMed] [Google Scholar]
- 53.Jackson PK, Eldridge AG, Freed E, Furstenthal L, Hsu JY, Kaiser BK, Reimann JD (2000) The lore of the RINGs: substrate recognition and catalysis by ubiquitin ligases. Trends Cell Biol 10: 429–439. S0962-8924(00)01834-1 [pii]. [DOI] [PubMed] [Google Scholar]
- 54.Everett RD, Maul GG (1994) HSV-1 IE protein Vmw110 causes redistribution of PML. EMBO J 13: 5062–5069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fanelli M, Fantozzi A, De Luca P, Caprodossi S, Matsuzawa S, Lazar MA, Pelicci PG, Minucci S (2004) The coiled-coil domain is the structural determinant for mammalian homologues of Drosophila Sina-mediated degradation of promyelocytic leukemia protein and other tripartite motif proteins by the proteasome. J Biol Chem 279: 5374–5379. 10.1074/jbc.M306407200 [DOI] [PubMed] [Google Scholar]
- 56.Maul GG (1998) Nuclear domain 10, the site of DNA virus transcription and replication. Bioessays 20: 660–667. [DOI] [PubMed] [Google Scholar]
- 57.Everett RD (2001) DNA viruses and viral proteins that interact with PML nuclear bodies. Oncogene 20: 7266–7273. 10.1038/sj.onc.1204759 [DOI] [PubMed] [Google Scholar]
- 58.Everett RD (1987) A detailed mutational analysis of Vmw110, a trans-acting transcriptional activator encoded by herpes simplex virus type 1. EMBO J 6: 2069–2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Everett RD (1988) Analysis of the functional domains of herpes simplex virus type 1 immediate-early polypeptide Vmw110. J Mol Biol 202: 87–96. [DOI] [PubMed] [Google Scholar]
- 60.Chen JX, Zhu XX, Silverstein S (1991) Mutational analysis of the sequence encoding ICP0 from herpes simplex virus type 1. Virology 180: 207–220. [DOI] [PubMed] [Google Scholar]
- 61.Chen J, Silverstein S (1992) Herpes simplex viruses with mutations in the gene encoding ICP0 are defective in gene expression. J Virol 66: 2916–2927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dimitrova YN, Li J, Lee YT, Rios-Esteves J, Friedman DB, Choi HJ, Weis WI, Wang CY, Chazin WJ (2010) Direct ubiquitination of beta-catenin by Siah-1 and regulation by the exchange factor TBL1. J Biol Chem 285: 13507–13516. 10.1074/jbc.M109.049411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Waites CL, Leal-Ortiz SA, Okerlund N, Dalke H, Fejtova A, Altrock WD, Gundelfinger ED, Garner CC (2013) Bassoon and Piccolo maintain synapse integrity by regulating protein ubiquitination and degradation. EMBO J 32: 954–969. emboj201327 [pii]; 10.1038/emboj.2013.27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Liu M, Hsu J, Chan C, Li Z, Zhou Q (2012) The ubiquitin ligase Siah1 controls ELL2 stability and formation of super elongation complexes to modulate gene transcription. Mol Cell 46: 325–334. S1097-2765(12)00214-6 [pii]; 10.1016/j.molcel.2012.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Luo Z, Lin C, Shilatifard A (2012) The super elongation complex (SEC) family in transcriptional control. Nat Rev Mol Cell Biol 13: 543–547. nrm3417 [pii]; 10.1038/nrm3417 [DOI] [PubMed] [Google Scholar]
- 66.He N, Liu M, Hsu J, Xue Y, Chou S, Burlingame A, Krogan NJ, Alber T, Zhou Q (2010) HIV-1 Tat and host AFF4 recruit two transcription elongation factors into a bifunctional complex for coordinated activation of HIV-1 transcription. Mol Cell 38: 428–438. S1097-2765(10)00312-6 [pii]; 10.1016/j.molcel.2010.04.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF)
Passage 2 (P2) virus of the listed HSV-2 constructs was produced in BHK cells (see Methods). The P2 inoculum was subsequently titrated on U2OS, Vero and HepaRG cells to compare the growth efficiencies on different cell lines.
(PDF)
Viral DNA was digested with HindIII and NcoI and subjected to Southern blot analyses using ICP0- (left panel) and GFP-specific (right panel) radioactively-labelled probes. Constructs marked in green were selected for further analysis.
(PDF)
Vero cells were infected in duplicates with HSV‐2‐ICP0‐GFP (ICP0‐GFP) or HSV-2 MS (wt) using (A) an MOI of 0,01 pfu/cell or (B) an MOI of 1 pfu/cell. At the indicated time points, cells were harvested and titrated on Vero cell monolayers. Mean ± SD of viral titers (n = 2 per data point) were plotted as a function of time.
(PDF)
Vero cells were infected with HSV‐2‐ICP0‐GFP or HSV‐2‐ICP0NxN1/2‐GFP at an MOI of 1 for up to 24 hours. Cells were harvested at the indicated time points and whole cell lysates were analyzed by SDS‐PAGE and Western blot using antibodies raised against the indicated proteins.
(PDF)
U2OS cells (2x105) were infected with an MOI of 5 pfu/cell of the indicated virus construct. At 2, 4 and 6 h post infection, cells were fixed for 20 min with 4% PFA. Nuclei were stained with 2 μM Hoechst 33342 and plasma membrane structures with Alexa Fluor®647 wheat germ agglutinin (WGA). Localization of ICP0-GFP was visualized by confocal fluorescence microscopy using a Nikon Eclipse Ti-E system.
(PDF)
Graphical comparison of viral titers as listed in S2 Table.
(PDF)
Dilution series were performed to determine the relative plaque formation efficiency of the indicated HSV-2 constructs on Vero and HepaRG cells. Obtained titers were used to calculate the ratio of plaques on Vero cells to U2OS cells, and HepaRG cells to U2OS cells, independently for every HSV-2 construct. The titer on U2OS cells was arbitrarily set as 100%; wt, wild-type.
(PDF)
(A) HepaRG cells were infected with the indicated constructs as described in Fig 5C and (B) U2OS cells were infected with the indicated constructs as described in Fig 5D. The fluorescent signal intensities of the secondary antibodies detecting the indicated viral proteins were quantified and normalized to the signal intensities of the loading control α-tubulin. Relative intensities are shown in pixels.
(PDF)
ICP0 and SIAH-1 are E3 ubiquitin ligases that are potentially able to ubiquitinate each other and, mostly in absence of a target protein, can also auto-ubiquitinate. It is hypothesized that ICP0 ubiquitinates SIAH-1 in vivo leading to the proteasomal degradation of the latter and to the stabilization of a so far unknown target protein that is needed for efficient progression of HSV-2 infection. When ICP0 auto-ubiquitinates or is ubiquinated by SIAH-1, it can be de-ubiquitinated by USP7. In turn, ICP0 can also ubiquitinate USP7, leading to the proteasomal degradation of the latter.
(PDF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.