ABSTRACT
All multicellular organisms must properly pattern cell types to generate functional tissues and organs. The organized and predictable cell lineages of the Brachypodium leaf enabled us to characterize the role of the MAPK kinase kinase gene BdYODA1 in regulating asymmetric cell divisions. We find that YODA genes promote normal stomatal spacing patterns in both Arabidopsis and Brachypodium, despite species-specific differences in those patterns. Using lineage tracing and cell fate markers, we show that, unexpectedly, patterning defects in bdyoda1 mutants do not arise from faulty physical asymmetry in cell divisions but rather from improper enforcement of alternative cellular fates after division. These cross-species comparisons allow us to refine our understanding of MAPK activities during plant asymmetric cell divisions.
KEY WORDS: Asymmetric cell division, MAPK pathway, Stomata, Brachypodium, Comparative development
Highlighted Article: Analysis of Brachypodium leaf epidermis development reveals that the MAPKKK BdYODA1 regulates asymmetric divisions by enforcing resultant cell fates rather than driving initial physical asymmetries.
INTRODUCTION
The correct establishment of cell types during development is essential for the generation of cellular diversity and patterning of tissues, organs and organisms. Asymmetric cell division, the process that gives rise to daughter cells with different physical appearance and/or developmental fate, is a crucial mechanism that most eukaryotes employ to generate diverse but organized cell populations. Asymmetric cell divisions are necessary from the very first to the very last events in plant development, and have been studied extensively during embryogenesis and root, shoot and reproductive development in flowering plants (Van Norman, 2016; Marzec et al., 2015; Abrash and Bergmann, 2009) as well as in basal lineages (Harrison et al., 2009; De Smet and Beeckman, 2011), albeit in a more limited fashion. A number of regulatory players and mechanisms have been identified in specific cellular contexts; however, unifying players and modules used repeatedly among these different asymmetric cell division contexts are only just beginning to come to light (Abrash and Bergmann, 2009; De Smet and Beeckman, 2011).
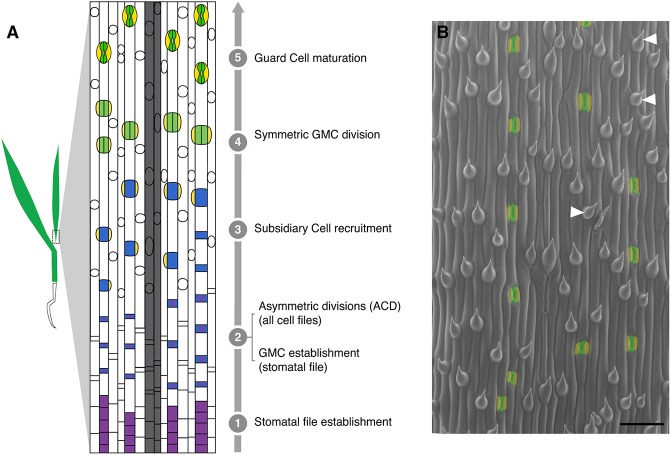
The stomatal lineages of various flowering plants offer particularly rich and accessible model systems for the study of asymmetric division regulation. Stomata are epidermal valves consisting of paired guard cells (GCs) flanking a central pore, and they contract and relax to regulate gas exchange between the plant and its environment. In many plant species, asymmetric cell divisions produce one daughter cell that acts as a stomatal precursor and another that differentiates as an epidermal pavement cell (Fryns-Claessens and Van Cotthem, 1973; Dong and Bergmann, 2010; Lau and Bergmann, 2012). In broadleaf dicots like Arabidopsis, the asymmetric divisions are self-renewing, i.e. once a cell undergoes an asymmetric division, its progeny may also continue dividing asymmetrically, in a situation somewhat analogous to the transit-amplifying cells in animal lineages (Matos and Bergmann, 2014). In the grasses, by contrast, the stomatal lineage exhibits a less flexible pattern of divisions in which stomatal precursors undergo a single asymmetric division that yields a terminal precursor (the guard mother cell, or GMC) and a larger sister cell fated to become a pavement cell. Also, notably in grasses, the lateral neighbors of the GMC undergo unique additional asymmetric divisions, parallel to the long axis of the leaf, to form the subsidiary cells (SCs) that act to facilitate stomatal function (Hepworth et al., 2017). In Brachypodium distachyon, a forage grass related to wheat, asymmetric cell divisions accompany the creation of many epidermal cell types (Raissig et al., 2016). Here, both stomatal and hair cell lineage development are subject to tight temporal regulation and occur in a developmental progression from the base (younger tissue) to the tip (older tissue) of the leaf blade (Fig. 1A,B). The common deployment of asymmetric divisions in multiple epidermal lineages in grasses requires that stomatal fate be later superimposed in particular cell files to specify the smaller daughter cells as stomatal precursors (Raissig et al., 2016).
Fig. 1.
Stomatal development in Brachypodium as model to study the progression of asymmetric divisions. (A) Simplified model of leaf blade epidermal development in Brachypodium. Specific cell files at predictable distances from veins (gray files) acquire stomatal lineage fate (stage 1) and undergo stomatal differentiation in a tip-to-base gradient. All cells in the epidermis then divide asymmetrically (ACD). In stomatal files, the smaller daughter cell of each division becomes a GMC (blue, stage 2). In all other files, these cells develop into hair cells (white circles in non-stomatal files). GMCs then recruit SCs (yellow, stage 3), divide once symmetrically to form two GCs (green, stage 4), and mature as four-celled complexes (stage 5). (B) Scanning electron micrograph of WT (Bd21-3) leaf epidermis. GCs and SCs are false-colored green and yellow, respectively. Co-existence of stomatal and hair fates in a single file is highlighted by white arrowheads. Scale bar: 50 μm.
Asymmetric divisions during development are guided by fate, polarity, and signaling inputs. In Arabidopsis stomatal production, basic helix-loop-helix (bHLH) transcription factors, the polarity protein BASL and a signaling pathway comprising ligands, receptors and a mitogen-activated protein kinase (MAPK) cascade have been connected to these roles (Lau and Bergmann, 2012; Pillitteri et al., 2016). The MAPK pathway is headed by the MAPK kinase (MAPKKK) YODA (AtYDA) (Bergmann et al., 2004). Genetic and biochemical data indicate that AtYDA responds to positional information provided by peptide ligands of the EPIDERMAL PATTERNING FACTOR (EPF) family via transmembrane receptors TOO MANY MOUTHS (TMM) and members of the ERECTA (ER) family (ERf) (Nadeau and Sack, 2002; Kim et al., 2012; Hunt and Gray, 2009; Kondo et al., 2010; Sugano et al., 2010; Hara et al., 2007; Shpak et al., 2005). AtYDA then relays this information through downstream kinases MKK4/5 and MAPK3/6 (Lampard et al., 2009; Wang et al., 2007) to phosphorylate and inhibit the bHLH transcription factor SPEECHLESS (AtSPCH), the primary regulator of entry into the stomatal lineage pathway (Lampard et al., 2008). Loss of AtYDA activity results in the production of excess stomata arranged in large clusters, whereas overactivity suppresses development of stomata (Bergmann et al., 2004), phenotypes opposite of those ascribed to loss and gain of AtSPCH function (Lampard et al., 2009; Macalister et al., 2007).
In grasses, the stomatal lineage requires homologs of the bHLH transcription factors known from Arabidopsis, though sometimes employed in different ways (Raissig et al., 2016; Raissig et al., 2017; Liu et al., 2009). BdICE1, BdSPCH1 and BdSPCH2 are needed for stomatal lineage initiation, and the two SPCH homologs are expressed in epidermal cells before they undergo asymmetric cell divisions, but their expression becomes restricted to the smaller daughter cells after division (Raissig et al., 2016). EPF overexpression has been shown to arrest stomatal production in barley (Hughes et al., 2017), potentially acting upstream of these transcription factors, but whether EPFs enforce stereotyped asymmetric divisions and the ‘every other cell’ epidermal pattern in grasses is not yet known.
In a screen aimed at identifying mutations that affect stomatal patterning in Brachypodium, we have identified a mutation in BdYDA1 that displays clustered stomata similar to loss of AtYDA in Arabidopsis. The mutation in BdYDA1 also led to patterning and fate defects in other epidermal cell types and changes in overall plant morphology. These additional phenotypes were interesting in light of previous findings that AtYDA is not exclusively a stomatal lineage regulator; it was originally characterized for its role in asymmetric divisions of the zygote (Lukowitz et al., 2004) and was subsequently shown to act during development of the inflorescence (Meng et al., 2012) and anthers (Hord et al., 2008), in defense (Sopeña-Torres et al., 2018) and in specifying division plane orientation in the root (Smékalová et al., 2014). Based on work in Arabidopsis, YDA was surmised to establish physically asymmetric divisions that lead to different cell fates in the daughters. By taking advantage of the highly spatially and temporally organized development of the Brachypodium leaf, however, we show that patterning defects do not arise from a fault in the physical asymmetry of cell divisions, but from improper enforcement of alternative cellular fates in these tissues. This comparative work supports a role for YDA as a conserved regulator of asymmetric cell divisions and expands our understanding of its role within those divisions.
RESULTS
Mutations in BdYDA1 lead to severe disruptions in stomatal pattern
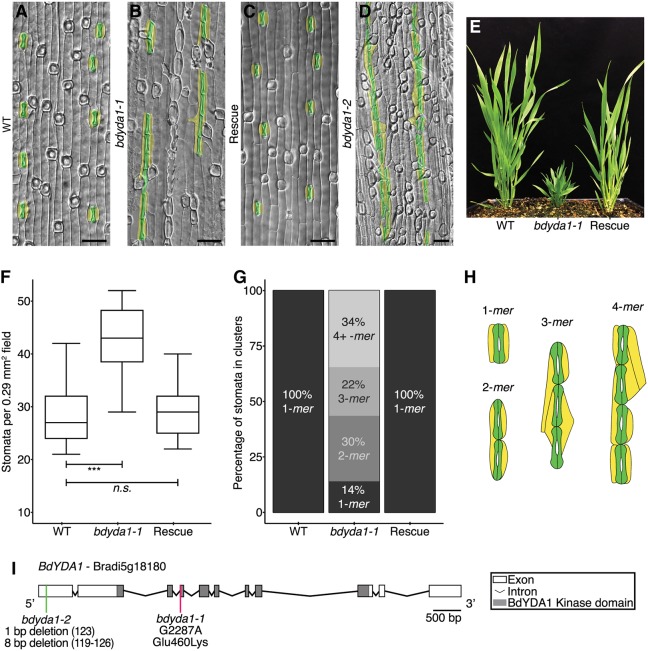
In wild-type Brachypodium distachyon Bd21-3 (WT), stomata are separated by at least one intervening non-stomatal cell (Fig. 1A,B). From an ethyl methanesulfonate (EMS)-mutagenized population of WT plants (described by Raissig et al., 2016), we identified a line segregating plants that, unlike WT, bore large groups of stomata in contact in a single row (Fig. 2A,B,H). The mutant did not exhibit ectopic stomatal rows, suggesting that the mutation affected processes occurring after specification of stomatal row identity. In addition to the stomatal patterning defects, mutants displayed whole plant growth defects, including compressed internodes and lateral organs, reduced overall stature, dark green coloration, and sterility (Fig. 2E). This combination of phenotypes was similar to that previously described for Arabidopsis plants bearing mutations in the MAPKKK-encoding gene AtYDA (Bergmann et al., 2004; Lukowitz et al., 2004). YDA orthologs can be identified in many plant species; in the grasses, YDA has been duplicated (Fig. S1). We sequenced the gene with the higher sequence similarity to AtYDA [BdYDA1 (Bradi5g18180)] and found a missense mutation (G2287A) predicted to confer a charge change (E460>K) in a conserved residue of the kinase domain (Fig. 2I).
Fig. 2.
BdYDA1 is required for proper spacing of stomata. (A-D) DIC images of cleared WT (Bd21-3) (A), bdyda1-1 mutant (B), bdyda1-1 complemented with BdYDA1pro:BdYDA1-YFP:Yt transgene (C), and bdyda1-2 (D) abaxial leaf epidermis. GCs and SCs are false-colored green and yellow, respectively. Images are of the sixth leaf from base (third from main tiller) 27 days post-germination (dpg) with WT and rescued line images being cleared leaves and bdyda1-1 images epidermal peels. bdyda1-2 image shows cleared leaf from T0 regenerant. Scale bars: 40 μm. (E) Whole-plant phenotype of bdyda1-1 mutant (middle), WT (left) and rescue (right) (5 weeks post-germination). (F) Stomatal density of bdyda1-1 mutants compared with that of WT and rescued bdyda1-1 [sixth leaf from base (third from main tiller) at 27 dpg]. n=4 individuals for WT control and n=5 for rescued plants. For each sample, five different regions of the leaf were imaged and quantified. n=5 for bdyda1-1 mutants for which four different regions of the leaf were peeled, imaged and quantified. ***P<0.001; n.s., not significant (based on Kruskal–Wallis test followed by Dunn's multiple comparisons test). In boxplot, the black horizontal line indicates the median; hinges (upper and lower edges of the box) represent versions of the upper and lower quartiles; whiskers extend to the largest observation within 1.5 interquartile ranges of the box. (G) Stomatal cluster profile as percentage of clustering of quantified stomata in F (n=566 stomata for WT controls, n=729 stomata for rescue, n=835 stomata for bdyda1-1). Clusters of four or more stomata were grouped in last category ‘4+ -mer’. (H) Schematics of representative patterns of GC and SC clusters in bdyda1-1. (I) Gene/protein diagram of BdYDA1. The vertical magenta bar indicates the bdyda1-1 EMS mutation and the green bar indicates the bdyda1-2 CRISPR/Cas9-induced mutation. Model generated in Gene Structure Display Server (Hu et al., 2015).
To determine whether the identified mutation was indeed causal for the phenotype, we generated BdYDA1pro:BdYDA1-YFP:Yt, a complementation construct consisting of ∼5.1 kb of upstream sequence, the BdYDA1 genomic region (including introns), a YFP tag and ∼1.5 kb of downstream sequence. The lateral organ and internode elongation defects of bdyda1-1 were rescued by this construct (Fig. 2E), as were the stomatal patterning defects (Fig. 2C, quantified in 2F,G). This was strong evidence that the mutation in BdYDA1 was responsible for the phenotype. Previous work on AtYDA and other MAPKKKs demonstrated that mutations in the kinase domain could lead to hypomorphic or null alleles (Sopeña-Torres et al., 2018; Lukowitz et al., 2004). We therefore also created an additional clustered regularly interspaced short palindromic repeat (CRISPR)/CRISPR-associated protein 9 (Cas9) mutation-based allele in the first exon of BdYDA1 to attempt to eliminate the protein altogether (Fig. S2A). bdyda1-2 is heteroallelic for frameshift mutations predicted to encode truncated proteins of 41 and 95 amino acids in contrast to the normal BdYDA1 protein of 896 amino acids (Fig. S2C) and resulted in the same suite of stomatal and morphological phenotypes as bdyda1-1; however, the magnitude of the clustering was increased (Fig. 2D, Fig. S2B). The difference between the phenotypes produced by the E460>K missense allele compared with the truncation allele suggests that bdyda1-1 is a hypomorphic allele. CRISPR/Cas9-induced mutations in the second YDA homolog [BdYDA2 (Bradi3g51380)] did not result in any obvious stomatal or overall growth phenotypes (Fig. S3). To test whether there might be redundancy between BdYDA1 and BdYDA2, we created early truncation bdyda2 CRISPR alleles in a bdyda1-1/+ background. In progeny homozygous for bdyda1-1 and bdyda2, we did not see any novel leaf epidermal phenotypes, nor did we see enhancement of the bdyda1 phenotype (data not shown).
The large clusters of stomata in Arabidopsis yda mutants arise from aberrant asymmetric divisions in the self-renewing precursor cells before they commit to becoming GMCs (Bergmann et al., 2004). Such self-renewing asymmetric divisions, however, are absent in grasses. An advantage of the more streamlined and rigid developmental trajectory in grasses is that we could generate a pseudo-timecourse by imaging a single Brachypodium leaf from base to tip and observing the cells at different ontological stages in bdyda1-1 and WT (Fig. 3A-H). To our surprise, early epidermal asymmetric divisions appeared normal in the bdyda1-1 mutant (Fig. 3A). By the SC recruitment stage (Fig. 3B), however, it was evident that the smaller daughters of the previous asymmetric division were not the only cells that had acquired stomatal fate. The larger daughter cells appeared to undergo extra divisions (Fig. 3B), and unusual SC morphologies, such as the spanning of two GC complexes by a single SC (Fig. 3B,C), were consistent with supernumerary stomatal-row cells taking on GC identity. The later stages (Fig. 3C,D) of stomatal differentiation, including the symmetric GMC division and subsequent stomatal pore formation, occurred fairly normally, as they do in atyda mutants, suggesting that BdYDA1 acts primarily in the early fate decisions and not in stomatal GC differentiation.
Fig. 3.
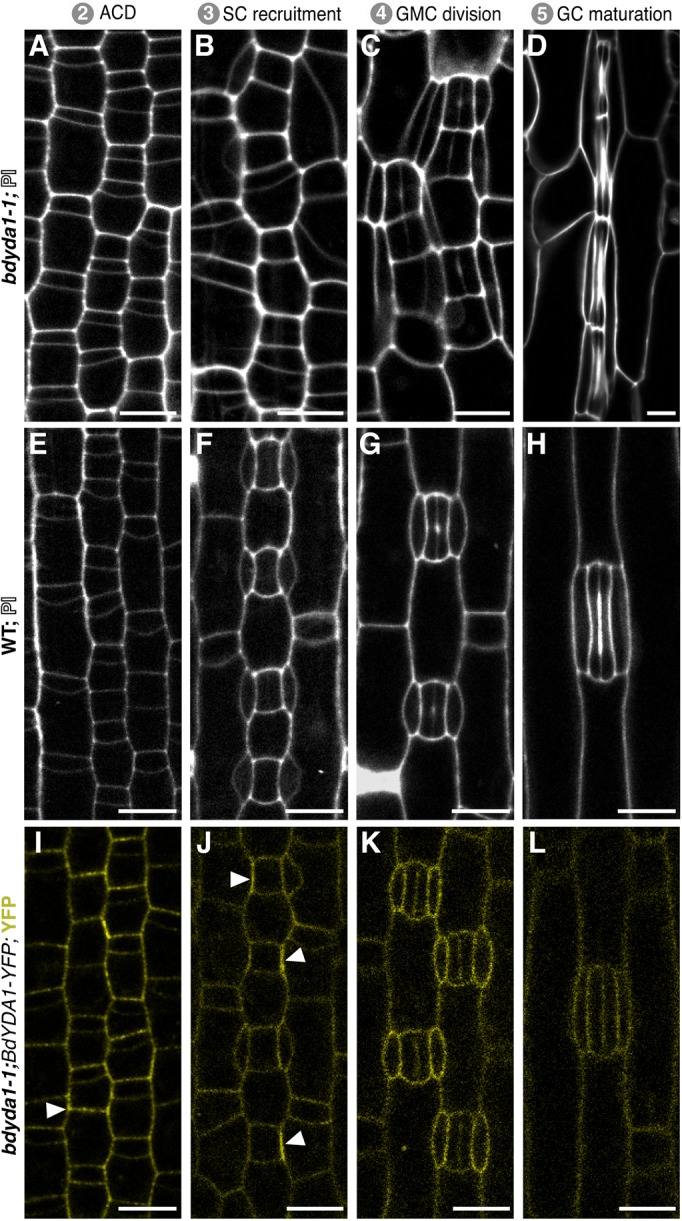
Imaging early development indicates that BdYDA1 is expressed throughout the stomatal lineage and that the initial defect in bdyda1-1 appears to be improper enforcement of non-stomatal fates. (A-H) Confocal images of progression of cells though four stages (as defined in Fig. 1A) of stomatal development in bdyda1-1 mutants (A-D) and WT (Bd21-3) (E-H) (emerging second leaf at 6 dpg, stained with PI). (I-L) Expression of rescuing BdYDA1pro:BdYDA1-YFP:Yt in bdyda1-1 (T1 plant; emerging second leaf at 6 dpg; YFP channel only). Arrowheads in I and J indicate accumulations of transgene signal. Scale bars: 10 μm. All images are oriented with the base of the leaf blade (younger cells) towards the bottom and the tip of the leaf (older cells) towards the top.
To address how activity of BdYDA1 might enforce stomatal patterning in Brachypodium, we visualized the expression pattern of the complementing BdYDA1pro:BdYDA1-YFP:Yt reporter. BdYDA1-YFP signal was not specific to the stomatal lineage and appeared to be present at roughly comparable levels in all the different leaf epidermal cell types we monitored (Fig. 3I-L). Fluorescence was strongest in the cytoplasm and/or at the cell periphery and was not detectable in the nucleus. In some cells, BdYDA1-YFP fluorescence appeared to be concentrated at a single face of a cell, most frequently at the interface between a GMC and its neighbor cell that will give rise to an SC (Fig. 3I,J, arrowheads; Fig. S4). This broad expression pattern is similar to that of AtYDA, suggesting that, like in Arabidopsis, it is the presence of appropriate upstream activating signals and downstream targets that provides specificity to YDA-mediated signaling activity.
Cell fate marker expression suggests defects in fate reinforcement in bdyda1-1
To further dissect the defects during GMC specification and SC recruitment observed in bdyda1-1, we examined the expression and behavior of stomatal lineage fate markers. BdSCRM2pro:YFP-BdSCRM2 is a pan-stomatal lineage marker (Raissig et al., 2016). It can be visualized in nuclei from the time stomatal rows are specified; when these cells start dividing asymmetrically, BdSCRM2 is confined to the smaller daughter of these divisions (Fig. 4A) and remains restricted to stomatal complexes as they mature (Fig. 4B-D). In bdyda1-1, however, restriction of signal to the smaller daughter of an asymmetric division is lost (Fig. 4E,F). Specifically, we observed signal in larger cells between stomatal precursors (Fig. 4E, arrowheads). In some cases, these presumed pavement cell precursors underwent an ectopic asymmetric division generating a stomatal precursor positive for YFP-BdSCRM2 expression adjacent to the earlier-specified stomatal precursor (Fig. 4F, arrows).
Fig. 4.
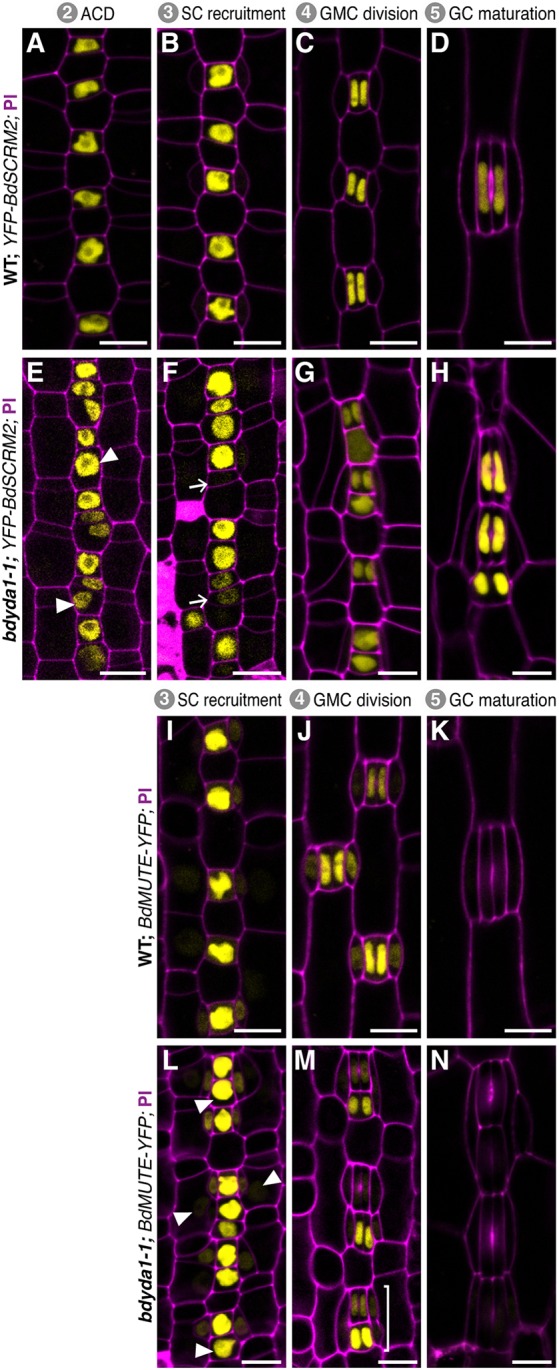
Misexpression of stomatal fate reporters in bdyda1-1 mutants is consistent with the terminal fate specification defects. Confocal images of emerging second leaf of 6 dpg T1 plants. (A-H) BdSCRM2pro:YFP-BdSCRM2 reporter in WT (Bd21-3) (A-D) and bdyda1-1 mutant (E-H) during stomatal development. Early in WT development, BdSCRM2pro:YFP-BdSCRM2 appears only in the smaller daughter of an asymmetric division (A,B). However, at the same stage in the bdyda1-1 mutant, signal is also present in mis-specified larger daughter cells (E,F). Arrowheads in E indicate examples of improper re-enforcement of non-stomatal fate in larger daughter of asymmetric division. Arrows in F point to examples of improper inhibition of division potential. (I-N) BdMUTEpro:BdMUTE-YFP reporter in WT (I-K) and bdyda1-1 mutant (L-N) during SC recruitment and GMC and SC specification. Reporter expression in WT is present only in GMCs, subsidiary mother cells (SMCs), and SCs as stomata mature. In bdyda1-1, the same reporter also marks mis-specified and clustered GMCs, SMCs and SCs. Arrowheads in L and bracket in M indicate ectopic marker expression during the SC recruitment and GMC division, respectively. Scale bars: 10 μm. Cell outlines are visualized with PI. All images are oriented with the base of the leaf (younger cells) towards the bottom and the tip of the leaf (older cells) towards the top.
To investigate later specification events and SC recruitment, we analyzed the expression pattern of BdMUTEpro:BdMUTE-YFP, which, in WT, starts in young GMCs and shows strong signal in mature GMCs and weak signal in adjacent subsidiary mother cell files (Fig. 4I). BdMUTE expression is maintained until after GMC division in both GCs and SCs and disappears during complex maturation (Fig. 4J,K). In bdyda1-1, young and mature GMCs showed marked patterning defects, with numerous BdMUTE-positive cells positioned adjacent to one another (Fig. 4L). Marker expression was also present in SCs even if they were recruited and formed abnormally. BdMUTE persisted in stomatal clusters through GMC division, but was extinguished rapidly as the GCs matured, as observed in WT (Fig. 4M,N).
Taken together, the reporter results agree with the morphological assessments and suggest that in bdyda1-1 mechanisms in place to control fate establishment and/or division potential early in the lineage are disrupted, but stomatal differentiation and morphogenesis are largely unaffected.
BdYDA1 regulates cell patterning in other cell lineages
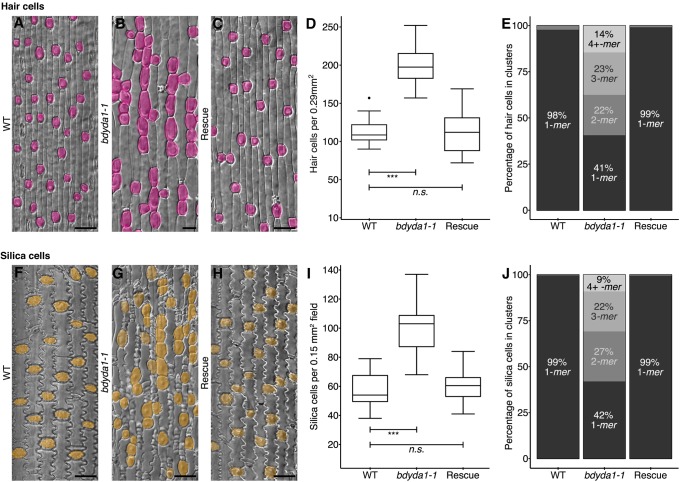
In studying the effects of bdyda1-1 on the stomatal lineage, it became apparent that this was not the only epidermal cell lineage disrupted by the mutation. Brachypodium also produces regularly spaced hair cells in the leaf epidermis. These hairs arise via asymmetric divisions in a manner similar to stomata, and the two fates appear to be closely related and somewhat interchangeable [e.g. a cell file may contain a number of stomata, then a hair, then more stomata (Raissig et al., 2016); for an example, see Fig. 1B]. In bdyda1-1 plants, the hair cell lineage displays defects very similar to the stomatal lineage, producing hair cells at higher density than in WT and often in longitudinal clusters (Fig. 5A,B, rescued in 5C, and quantified in 5D,E). In addition, the epidermis of the leaf sheath contains crenellated pavement cells and round silica cells sometimes accompanied by a small lens-shaped cell and a small triangular cell (Fig. 5F). Patterning and proliferation defects were evident in all of these cell types in bdyda1-1, and were complemented by the BdYDA1 reporter (Fig. 5G,H). We quantified patterning and proliferation defects in the large, round silica cells because, among the sheath cell types, they were most unambiguously identified. Like stomata and hair cells, the silica cells were produced at a greater density than in WT and were found in numerous longitudinal clusters (Fig. 5I,J). Cross-sections of mature leaves revealed that the overall organization of the leaf was similar to WT, but bulliform, photosynthetic and vascular cells were present in slightly higher numbers (Fig. S5).
Fig. 5.
bdyda1-1 mutants exhibit disruption of cell fates in other asymmetrically dividing epidermal lineages. (A-C) DIC images of cleared WT (Bd21-3) (A), bdyda1-1 mutant (B), and bdyda1-1 rescued with BdYDA1pro:BdYDA1-YFP:Yt (C) leaf epidermis. Hair cells are false-colored magenta. WT and complemented bdyda1-1 images show sixth leaf from base (third from main tiller) at 27 dpg. bdyda1-1 images show epidermal peels of sixth leaf from base (third from main tiller) at 27 dpg. Scale bars: 40 μm. (D) Hair cell density of bdyda1-1 mutants compared with that of WT and rescued bdyda1-1 [sixth leaf from base (third from main tiller) at 27 dpg]. n=4 individuals for WT control and n=5 for rescued plants. For each sample, five different regions of the leaf were imaged and quantified. n=5 for bdyda1-1 mutants for which four different regions of the leaf were peeled, imaged, and quantified. ***P<0.001; n.s., not significant (based on Kruskal–Wallis test followed by Dunn's multiple comparisons test). (E) Hair cell cluster profile as percentage of clustering of quantified hair cells in bdyda1-1 mutants, WT, and rescued bdyda1-1 (n=2286 hair cells for WT controls, n=3988 hair cells for rescue, n=2789 hair cells for bdyda1-1). Clusters of four or more hair cells were grouped in last category ‘4+ -mer’. (F-H) DIC images of cleared WT (F), bdyda1-1 mutant (G), and rescued bdyda1-1 (H) sheath epidermis. Silica cells are false-colored orange. For all genotypes, images show the sheath of the sixth leaf from base (third from main tiller) at 27 dpg. Scale bars: 40 μm. (I) Silica cell density of bdyda1-1 compared with that of WT and rescued bdyda1-1 [sheath of the sixth leaf from base (third from main tiller) at 27 dpg]. n=4 individuals for WT control, n=5 for rescued plants, and n=5 for bdyda1-1 mutants. For all, four to six different regions of the sheath were imaged and quantified. ***P<0.001; n.s., not significant (based on Kruskal–Wallis test followed by Dunn's multiple comparisons test). (J) Silica cell cluster profile as percentages of clustering of quantified silica cells in bdyda1-1 mutants, WT, and rescued bdyda1-1 (n=1328 silica cells for WT controls, n=2995 silica cells for rescue, n=1691 silica cells for bdyda1-1). Clusters of four or more silica cells were grouped in last category ‘4+ -mer’.
DISCUSSION
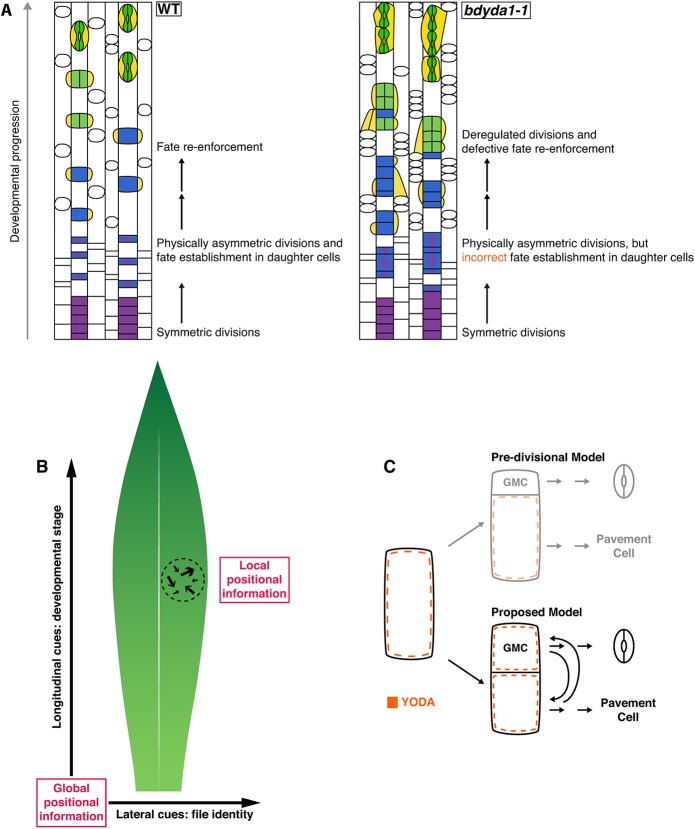
By characterizing a mutation that disrupted stomatal patterning in Brachypodium distachyon, we identified the BdYDA1 gene, a Brachypodium ortholog of the Arabidopsis MAPKKK gene AtYDA. Like atyda (Bergmann et al., 2004; Lukowitz et al., 2004), the bdyda1 mutant produces excess stomata arranged in clusters and displays a stunted growth phenotype characterized by compressed internodes and compact lateral organs. Strikingly, we could show that clusters in bdyda1 arise from mis-specification of alternative fates in the epidermis, rather than being caused by alterations in the physical asymmetry of the divisions themselves. In addition, other non-stomatal epidermal cell types are also affected. Taken together, our results demonstrate that BdYDA1 is a general regulator of cell fate establishment and enforcement, two processes that are crucial for the correct execution of asymmetric divisions during epidermal patterning of the grass leaf (Fig. 6A).
Fig. 6.
Summary of YDA's proposed role in asymmetric divisions. (A) Schematic of the bdyda1-1 phenotype and interpretations of the role of BdYDA1 in epidermal patterning of Brachypodium leaves. (B) Global and local positional information feed into developmental decisions that orient and position stomatal precursors. Global positional information in the form of lateral and longitudinal cues direct cell file identities and developmental progression of lineages, respectively. Local positional information controls fate re-enforcement to establish the correct pattern and distribution of stomata and their precursors. The relative influence of global versus local sources of positional information is likely to be species specific, i.e. longitudinally growing grass leaves are more heavily influenced by global cues and radially growing leaves with self-renewing stem-like divisions, such as in Arabidopsis, by local cues. (C) In contrast to Arabidopsis-derived pre-divisional models, which suggest that YDA mainly acts to establish physical asymmetry prior to fate establishment, we propose that YDA is primarily a post-divisional fate re-enforcer. This requires that YDA be present in both daughters of an asymmetric cell division and be available for reciprocal (and continuous) signal transduction downstream of cell-cell communication systems.
Asymmetric divisions are produced and oriented by a combination of extrinsic and intrinsic cues. AtYDA plays roles in the transduction of both sources of developmental information in Arabidopsis stomatal development. Positional (extrinsic) information conveyed by EPFL family ligands interacting with ERf/TMM receptors can activate the AtYDA MAPK cascade, leading to AtSPCH inhibition and repression of stomatal identity (reviewed by Pillitteri and Torii, 2012). More recently, a role in intrinsic polarity via a physical association between AtYDA and the polarity factor BASL emerged (Zhang et al., 2015). The cortical AtYDA/BASL complex is preferentially inherited by the larger daughter cell of an asymmetric division, leading to differential signaling capacity; the cell with higher AtYDA has lower AtSPCH levels and consequently loses stomatal identity (Zhang et al., 2016).
Could BdYDA1 participate in similar signaling or intrinsic polarity modules? The observation of non-uniform BdYDA1-YFP distribution around cells might hint to it being localized by a differentially inherited polarity complex. However, two observations make this solution unlikely. First, the specific location of the BdYDA1-YFP enrichment, at the boundary between GC and prospective SC, is not consistent with it being preferentially segregated to the larger (non-stomatal) cell during normal development. Moreover, polar localization would require an as-yet-unknown polarity partner, as BASL homologs have not been detected in the grasses.
In terms of the cell-cell signaling response, Arabidopsis YDA and Brachypodium YDA1 proteins show appreciable sequence similarity (65% overall and 90% in kinase domain; Fig. S6) and components of the YDA MAPK pathway have clear orthologs in Brachypodium, including downstream kinases MKK4 (Bradi1g46880), MKK5 (Bradi3g53650), MPK3 (Bradi3g53650) and MPK6 (Bradi1g49100). Upstream signaling components include multiple EPFL family members, TMM (Bradi2g43940), and ERECTA, although the ERECTA family consists of only two members (Bradi1g46450 and Bradi1g49950). Although the presence of homologous signaling pathway genes makes participation in parallel signaling cascades possible for AtYDA and BdYDA1, there remains the issue of the distinct ontogeny of dicot and grass stomata. With no self-renewing divisions, mature complexes restricted to specific cell files, and consistent orientation of each complex along the leaf's proximal-distal axis, the positional information required in grasses is very different from that in Arabidopsis in which distributed ‘point sources’ of signals and extensive neighbor-to-neighbor signaling are dominant patterning mechanisms (Torii, 2015) (Fig. 6B). As disorderly as epidermal identities become within bdyda1 cell files, they still obey tissue-wide fate arrangements, indicating that different factors control lateral (and proximal-distal) positional information in the leaf. Although none of the work with grass EPF homologs has yet demonstrated an effect on lateral patterning (Hughes et al., 2017; Yin et al., 2017), expanded expression of SHORTROOT, a factor normally involved in internal cell fates, leads to production of supernumerary stomatal rows in rice (Schuler et al., 2018), suggesting that lateral information may be provided through different pathways.
When considering downstream targets of a BdYDA1-mediated MAPK cascade in cell fate reinforcement roles, the stomatal initiation module could be targeted to inhibit stomatal fate establishment in larger daughter cells of asymmetric divisions, much as it is in Arabidopsis, but the precise protein target in this complex may be different. In Arabidopsis, AtSPCH is the demonstrated target of MAPK regulation (Lampard et al., 2008), but in Brachypodium, BdICE1 may play a more dominant role as it possesses predicted high-fidelity MAPK target sites within the protein degradation-associated PEST domain, whereas BdSPCH1 and BdSPCH2 do not (Raissig et al., 2016). Furthermore, expression of ubiquitin promoter-driven YFP-BdICE1 accumulates only in the stomatal lineage cells of the leaf, indicating that this protein is subject to post-translational regulation, as one would expect from a target of a MAPK cascade (Raissig et al., 2016).
BdYDA1 also regulates fate re-enforcement in other non-stomatal epidermal cell linages; however, the means by which it does so remain to be explored. It is likely that BdYDA1 has targets that could enforce fate asymmetry in all of these decisions, whereas the specific fate of the cells (stomata, hair or silica) would be determined by other information. Such is the case with Arabidopsis embryos and stomata in which signaling can work when swapped between these developmental contexts (Bayer et al., 2009), but downstream transcription factors provide unique cell identities (Jeong et al., 2011; Ueda et al., 2017). We noticed that in the early truncation allele bdyda1-2 the degree of clustering is less in non-stomatal epidermal cells than in stomatal files (Fig. 2D, Fig. S2B) suggesting that BdYDA1-independent fate-determining mechanisms also exist in these lineages.
In Arabidopsis embryos, roots, and the stomatal lineage, loss or overactivity of AtYDA changes an asymmetric division into one for which daughters exhibit equalized cell fates and cell sizes. From these phenotypes, it is intuitive to imagine AtYDA's role as one initiating asymmetry in the mother cell of these formative divisions (Fig. 6C). Furthermore, considering that YDA's downstream effectors are the microtubule-regulating kinases AtMPK3 and AtMPK6, AtYDA was suggested to mediate cytoskeletal behaviors leading to the placement of division planes and creation of asymmetric divisions (Smékalová et al., 2014). In our present study, however, we showed that the physical asymmetry of divisions is not affected in the absence of BdYDA1, prompting us to re-evaluate whether AtYDA actually generates pre-divisional, physical asymmetry, or whether the failure to create different-sized cells in atyda mutants stems from a post-divisional failure in cell identity. High-resolution time-lapse imaging of developmental decisions in Arabidopsis would be needed to distinguish between YDA playing primarily a pre- or post-divisional role.
The work presented here emphasizes the value of comparative developmental studies, as the bdyda1 mutations reveal roles for the YDA pathway in numerous cell fate decisions, some of which represent cell types not present in Arabidopsis. This work also provokes a conceptual shift in our emphasis on MAPK signaling as required for the creation of asymmetry to MAPK signaling required for the post-divisional enforcement of asymmetric fates. In a kingdom devoid of Notch-Delta lateral inhibition systems with scant evidence for any segregated fate determinants, and one characterized by exceedingly flexible development, it may be that plant cell fate decisions are rarely made in advance, but are subject to multiple rounds of confirmation through post-divisional communication and re-enforcement.
MATERIALS AND METHODS
Plant material
The bdyda1-1 mutant was recovered from the M3 generation of an EMS mutagenesis of the Bd21-3 ecotype (seeds provided by Dr John Vogel, Joint Genome Institute, CA, USA; Raissig et al., 2016), and maintained through selection of heterozygous individuals that segregated the mutations in typical 3:1 Mendelian recessive ratios. Bd21-3 was used as WT for all experiments described in this paper (Vogel and Hill, 2007).
Plants were initially grown on half-strength MS agar plates in a 22°C chamber with a 16-h light/8-h dark cycle (110 μmol m−2 s−1), then subsequently transferred to soil and placed in a greenhouse with a 20-h light/4-h dark cycle (250-300 μmol m−2 s−1; day temperature: 28°C; night temperature: 18°C). Seeds were stratified for at least 2 days at 4°C before transferred to light.
Generation of constructs
All primer sequences are provided in Table S1. BdSCRM2:YFP-BdSCRM2 was described by Raissig et al. (2016). To create BdYDA1pro:BdYDA1-YFP:Yt and BdMUTEpro:BdMUTE-YFP, sequences were amplified from BACs BD_ABa0027F22 and BD_ABa0042O15/BD_AB0008G12 (Arizona Genomics Institute; http://www.genome.arizona.edu/), respectively. For BdYDA1pro:BdYDA1-YFP-Yt, 5.1 kb upstream sequence (primers BdYDA1pro5.1kb-1F and BdYDA1pro_AscI-1R) and 3′ sequence (BdYDA1term-1F and BdYDA1term-1R) of the BdYDA1 gene were cloned into pIPKb001 (Himmelbach et al., 2007). This then was recombined with a BdYDA1-YFP fusion in pENTR-D, composed of the BdYDA1 genomic region (primers BdYDA1proPacI-1F and BdYDA1cDNAnscAscI-1R) followed by AscI-flanked Citrine YFP.
For BdMUTEpro:BdMUTE-YFP, 1.1 kb upstream sequence of the BdMUTE gene (Bradi1g18400) (primers BdMUTEpro-FWD and BdMUTEpro-REV) was cloned into pIPKb001t (Raissig et al., 2016). Separately, the BdMUTE genomic sequence (primers BdMUTE-CDS-FWD and BdMUTE-CDS-REV) was cloned into pENTR-D with a poly-alanine linker (annealed primers Ala_linker-F and Ala_linker-R) and an AscI-flanked Citrine YFP inserted 3′ of the gene by AscI digest. Finally, the entry clone was recombined into the pIPKb001t vector.
CRISPR constructs were designed using the vectors pH-Ubi-cas9-7 and pOs-sgRNA (vectors and protocol described by Miao et al., 2013). The online server, CRISPR-P, was used to identify candidate spacer sequences (Lei et al., 2014). Spacers were generated by annealing oligo duplexes priMXA38F+39R for BdYDA1 CRISPR_sgRNA_6 (which generated bdyda1-2) and priMXA30F+31R for BdYDA2 CRISPR_sgRNA_11. Primers priMXA50 and 52 were used to genotype bdyda1-2 and primers priMXA48 and 49 were used to genotype bdyda2-1 and bdyda2-2. BdYDA1 CRISPRs were transformed into WT and BdYDA2 CRISPRs into both WT and bdyda1-1/+ genotypes as described below.
Generation of transgenic lines
Brachypodium calli derived from Bd21-3 and bdyda1-1/+ parental plants were transformed with AGL1 Agrobacterium tumefaciens, selected and regenerated according to standard protocols (https://jgi.doe.gov/our-science/science-programs/plant-genomics/brachypodium/). Plants and calli from bdyda1-1/+ parents were genotyped using primers priMXA25, 26 and 27 in a single PCR reaction as described by Gaudet et al. (2009).
Microscopy and phenotypic analysis
For imaging on a Leica SP5 confocal microscope, leaves were carefully taken out from the surrounding sheath and stained with propidium iodide (PI; 1:100 dilution of 1 mg/ml stock) to visualize cell walls, then mounted in water. For differential interference contrast (DIC) imaging on a Leica DM6 B microscope of WT and rescue plant leaf (1.5-2 cm distal of the sixth leaf blade of 22 dpg leaves) and sheath (topmost centimeter of sheath tissue surrounding seventh leaf of 22 dpg plants), tissue was collected into 7:1 ethanol:acetic acid and incubated overnight to remove chlorophyll, then rinsed with water, and mounted in Hoyer's medium to clear. The same was done for DIC imaging of bdyda1-2, except no sheath tissue was collected for it. For DIC imaging of bdyda1-1, epidermal peels were collected from the tissue of interest into Hoyer's medium, mounted on slides, and examined. Cell numbers and extent of stomatal clusters were counted directly on a computer display attached to the microscope (0.29 mm2 field of view for stomata and hair cell counts; 0.15 mm2 field of view for silica cell counts). Only cells fully contained within the image frame were included in the respective analysis. In cases when a cluster expanded beyond the image frame, cells outside the image frame were included to correctly represent the number of cells part of the cluster. For DIC imaging of WT and bdyda1-1 leaf cross-sections, fully emerged adult leaves were taken directly from a growing plant (∼6 weeks old) then hand-sectioned and immediately mounted in Hoyer's medium. For scanning electron microscope (SEM) images, WT leaves were taken directly from a growing plant, then introduced into an Environmental SEM (FEI Quanta 200) without any further treatments.
Statistical analysis and plotting
Statistical analysis was performed in R (R Core Team, 2017). The Shapiro–Wilk test (shapiro.test function) was used to check samples for normality; as many samples displayed non-normal distributions, the Kruskal–Wallis test (a nonparametric analog of ANOVA; kruskal.test function) was used, followed by Dunn's Multiple Comparisons tests (dunn.test, dunnTest) to assess significance of pairwise comparisons.
Supplementary Material
Acknowledgements
We thank Akhila Bettadapur for technical assistance, Dr John Vogel (JGI) for creating Brachypodium resources, Dr Kathryn Barton (Carnegie, DPB) for use of her SEM, Emanuel Schorsch for consultation on statistics, and Dr Michael Raissig, Dr Annika Weimer, and other members of the Bergmann lab for discussions and comments on the manuscript.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: E.A., M.X.A.G., D.C.B.; Methodology: E.A., M.X.A.G.; Formal analysis: E.A., M.X.A.G.; Investigation: E.A., M.X.A.G., J.L.M.; Resources: D.C.B.; Writing - original draft: E.A., M.X.A.G., D.C.B.; Writing - review & editing: E.A., M.X.A.G., J.L.M., D.C.B.; Visualization: E.A., M.X.A.G.; Supervision: D.C.B.; Project administration: D.C.B.; Funding acquisition: D.C.B.
Funding
E.A. was supported by an National Science Foundation graduate research fellowship and a Stanford graduate fellowship. D.C.B. is an investigator of the Howard Hughes Medical Institute. Deposited in PMC for release after 6 months.
Supplementary information
Supplementary information available online at http://dev.biologists.org/lookup/doi/10.1242/dev.165860.supplemental
References
- Abrash E. B. and Bergmann D. C. (2009). Asymmetric cell divisions: a view from plant development. Dev. Cell 16, 783-796. 10.1016/j.devcel.2009.05.014 [DOI] [PubMed] [Google Scholar]
- Bayer M., Nawy T., Giglione C., Galli M., Meinnel T. and Lukowitz W. (2009). Paternal control of embryonic patterning in Arabidopsis thaliana. Science 323, 1485-1488. 10.1126/science.1167784 [DOI] [PubMed] [Google Scholar]
- Bergmann D. C., Lukowitz W. and Somerville C. R. (2004). Stomatal development and pattern controlled by a MAPKK kinase. Science 304, 1494-1497. 10.1126/science.1096014 [DOI] [PubMed] [Google Scholar]
- De Smet I. and Beeckman T. (2011). Erratum: asymmetric cell division in land plants and algae: the driving force for differentiation. Nat. Rev. Mol. Cell Biol. 12, 273-273. 10.1038/nrm3094 [DOI] [PubMed] [Google Scholar]
- Dong J. and Bergmann D. C. (2010). Stomatal patterning and development. Curr. Top. Dev. Biol. 91, 267-297. 10.1016/S0070-2153(10)91009-0 [DOI] [PubMed] [Google Scholar]
- Fryns-Claessens E. and Van Cotthem W. (1973). A new classification of the ontogenetic types of stomata. Bot. Rev. 39, 71-138. 10.1007/BF02860071 [DOI] [Google Scholar]
- Gaudet M., Fara A.-G., Beritognolo I. and Sabatti M. (2009). Allele-specific PCR in SNP genotyping. Methods Mol. Biol. 578, 415-424. 10.1007/978-1-60327-411-1_26 [DOI] [PubMed] [Google Scholar]
- Hara K., Kajita R., Torii K. U., Bergmann D. C. and Kakimoto T. (2007). The secretory peptide gene EPF1 enforces the stomatal one-cell-spacing rule. Genes Dev. 21, 1720-1725. 10.1101/gad.1550707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison C. J., Roeder A. H. K., Meyerowitz E. M. and Langdale J. A. (2009). Local cues and asymmetric cell divisions underpin body plan transitions in the moss Physcomitrella patens. Curr. Biol. 19, 461-471. 10.1016/j.cub.2009.02.050 [DOI] [PubMed] [Google Scholar]
- Hepworth C., Caine R. S., Harrison E. L., Sloan J. and Gray J. E. (2017). Stomatal development: focusing on the grasses. Curr. Opin. Plant Biol. 41, 1-7. 10.1016/j.pbi.2017.07.009 [DOI] [PubMed] [Google Scholar]
- Himmelbach A., Zierold U., Hensel G., Riechen J., Douchkov D., Schweizer P. and Kumlehn J. (2007). A set of modular binary vectors for transformation of cereals. Plant Physiol. 145, 1192-1200. 10.1104/pp.107.111575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hord C. L. H., Sun Y.-J., Pillitteri L. J., Torii K. U., Wang H., Zhang S. and Ma H. (2008). Regulation of arabidopsis early anther development by the Mitogen-Activated Protein Kinases, MPK3 and MPK6, and the ERECTA and related receptor-like kinases. Mol. Plant 1, 645-658. 10.1093/mp/ssn029 [DOI] [PubMed] [Google Scholar]
- Hu B., Jin J., Guo A.-Y., Zhang H., Luo J. and Gao G. (2015). GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics 31, 1296-1297. 10.1093/bioinformatics/btu817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes J., Hepworth C., Dutton C., Dunn J. A., Hunt L., Stephens J., Waugh R., Cameron D. D. and Gray J. E. (2017). Reducing stomatal density in barley improves drought tolerance without impacting on yield. Plant Physiol. 174, 776-787. 10.1104/pp.16.01844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt L. and Gray J. E. (2009). The signaling peptide EPF2 controls asymmetric cell divisions during stomatal development. Curr. Biol. 19, 864-869. 10.1016/j.cub.2009.03.069 [DOI] [PubMed] [Google Scholar]
- Jeong S., Palmer T. M. and Lukowitz W. (2011). The RWP-RK factor GROUNDED promotes embryonic polarity by facilitating YODA MAP kinase signaling. Curr. Biol. 21, 1268-1276. 10.1016/j.cub.2011.06.049 [DOI] [PubMed] [Google Scholar]
- Kim T.-W., Michniewicz M., Bergmann D. C. and Wang Z.-Y. (2012). Brassinosteroid regulates stomatal development by GSK3-mediated inhibition of a MAPK pathway. Nature 482, 419-422. 10.1038/nature10794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo T., Kajita R., Miyazaki A., Hokoyama M., Nakamura-Miura T., Mizuno S., Masuda Y., Irie K., Tanaka Y., Takada S. et al. (2010). Stomatal density is controlled by a mesophyll-derived signaling molecule. Plant Cell Physiol. 51, 1-8. 10.1093/pcp/pcp180 [DOI] [PubMed] [Google Scholar]
- Kumar S., Stecher G. and Tamura K. (2016). MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870-1874. 10.1093/molbev/msw054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampard G. R., Macalister C. A. and Bergmann D. C. (2008). Arabidopsis stomatal initiation is controlled by MAPK-mediated regulation of the bHLH SPEECHLESS. Science 322, 1113-1116. 10.1126/science.1162263 [DOI] [PubMed] [Google Scholar]
- Lampard G. R., Lukowitz W., Ellis B. E. and Bergmann D. C. (2009). Novel and expanded roles for MAPK signaling in arabidopsis stomatal cell fate revealed by cell type-specific manipulations. Plant Cell 21, 3506-3517. 10.1105/tpc.109.070110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau O. S. and Bergmann D. C. (2012). Stomatal development: a plant's perspective on cell polarity, cell fate transitions and intercellular communication. Development 139, 3683-3692. 10.1242/dev.080523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei Y., Lu L., Liu H.-Y., Li S., Xing F. and Chen L.-L. (2014). CRISPR-P: a web tool for synthetic single-guide RNA design of CRISPR-system in plants. Mol. Plant 7, 1494-1496. 10.1093/mp/ssu044 [DOI] [PubMed] [Google Scholar]
- Liu T., Ohashi-Ito K. and Bergmann D. C. (2009). Orthologs of Arabidopsis thaliana stomatal bHLH genes and regulation of stomatal development in grasses. Development 136, 2265-2276. 10.1242/dev.032938 [DOI] [PubMed] [Google Scholar]
- Lukowitz W., Roeder A., Parmenter D. and Somerville C. (2004). A MAPKK kinase gene regulates extra-embryonic cell fate in Arabidopsis. Cell 116, 109-119. 10.1016/S0092-8674(03)01067-5 [DOI] [PubMed] [Google Scholar]
- Macalister C. A., Ohashi-Ito K. and Bergmann D. C. (2007). Transcription factor control of asymmetric cell divisions that establish the stomatal lineage. Nat. Publ. Group 445, 537-540. 10.1038/nature05491 [DOI] [PubMed] [Google Scholar]
- Marzec M., Melzer M. and Szarejko I. (2015). Root hair development in the grasses: what we already know and what we still need to know. Plant Physiol. 168, 407-414. 10.1104/pp.15.00158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matos J. L. and Bergmann D. C. (2014). Convergence of stem cell behaviors and genetic regulation between animals and plants: insights from the Arabidopsis thaliana stomatal lineage. F1000Prime Rep. 6, 53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng X., Wang H., He Y., Liu Y., Walker J. C., Torii K. U. and Zhang S. (2012). A MAPK cascade downstream of ERECTA receptor-like protein kinase regulates Arabidopsis inflorescence architecture by promoting localized cell proliferation. Plant Cell 24, 4948-4960. 10.1105/tpc.112.104695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao J., Guo D., Zhang J., Huang Q., Qin G., Zhang X., Wan J., Gu H. and Qu L.-J. (2013). Targeted mutagenesis in rice using CRISPR-Cas system. Cell Res. 23, 1233-1236. 10.1038/cr.2013.123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadeau J. A. and Sack F. D. (2002). Stomatal development in Arabidopsis. The Arabidopsis Book 1, e0066 10.1199/tab.0066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillitteri L. J. and Torii K. U. (2012). Mechanisms of stomatal development. Annu. Rev. Plant Biol. 63, 591-614. 10.1146/annurev-arplant-042811-105451 [DOI] [PubMed] [Google Scholar]
- Pillitteri L. J., Guo X. and Dong J. (2016). Asymmetric cell division in plants: mechanisms of symmetry breaking and cell fate determination. Cell. Mol. Life Sci. 73, 4213-4229. 10.1007/s00018-016-2290-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team (2017). R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; https://www.R-project.org/. [Google Scholar]
- Raissig M. T., Abrash E., Bettadapur A., Vogel J. P. and Bergmann D. C. (2016). Grasses use an alternatively wired bHLH transcription factor network to establish stomatal identity. Proc. Natl. Acad. Sci. USA 113, 8326-8331. 10.1073/pnas.1606728113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raissig M. T., Matos J. L., Gil M. X. A., Kornfeld A., Bettadapur A., Abrash E., Allison H. R., Badgley G., Vogel J. P., Berry J. A. et al. (2017). Mobile MUTE specifies subsidiary cells to build physiologically improved grass stomata. Science 355, 1215-1218. 10.1126/science.aal3254 [DOI] [PubMed] [Google Scholar]
- Schuler M. L., Sedelnikova O. V., Walker B. J., Westhoff P. and Langdale J. A. (2018). SHORTROOT-mediated increase in stomatal density has no impact on photosynthetic efficiency. Plant Physiol. 176, 757-772. 10.1104/pp.17.01005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shpak E. D., McAbee J. M., Pillitteri L. J. and Torii K. U. (2005). Stomatal patterning and differentiation by synergistic interactions of receptor kinases. Science 309, 290-293. 10.1126/science.1109710 [DOI] [PubMed] [Google Scholar]
- Sievers F., Wilm A., Dineen D., Gibson T. J., Karplus K., Li W., Lopez R., McWilliam H., Remmert M., Söding J. et al. (2011). Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7, 539-539. 10.1038/msb.2011.75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smékalová V., Luptovčiak I., Komis G., Šamajová O., Ovečka M., Doskočilová A., Takáč T., Vadovič P., Novák O., Pechan T. et al. (2014). Involvement of YODA and mitogen activated protein kinase 6 in Arabidopsis post-embryogenic root development through auxin up-regulation and cell division plane orientation. New Phytol. 203, 1175-1193. 10.1111/nph.12880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sopeña-Torres S., Jordá L., Sánchez-Rodríguez C., Miedes E., Escudero V., Swami S., López G., Piślewska-Bednarek M., Lassowskat I., Lee J. et al. (2018). YODA MAP3K kinase regulates plant immune responses conferring broad-spectrum disease resistance. New Phytol. 19, 1665. [DOI] [PubMed] [Google Scholar]
- Sugano S. S., Shimada T., Imai Y., Okawa K., Tamai A., Mori M. and Hara-Nishimura I. (2010). Stomagen positively regulates stomatal density in Arabidopsis. Nat. Publ. Group 463, 241-244. 10.1038/nature08682 [DOI] [PubMed] [Google Scholar]
- Torii K. U. (2015). Stomatal differentiation: the beginning and the end. Curr. Opin. Plant Biol. 28, 16-22. 10.1016/j.pbi.2015.08.005 [DOI] [PubMed] [Google Scholar]
- Ueda M., Aichinger E., Gong W., Groot E., Verstraeten I., Vu L. D., De Smet I., Higashiyama T., Umeda M. and Laux T. (2017). Transcriptional integration of paternal and maternal factors in the Arabidopsis zygote. Genes Dev. 31, 617-627. 10.1101/gad.292409.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Norman J. M. (2016). Asymmetry and cell polarity in root development. Dev. Biol. 419, 165-174. 10.1016/j.ydbio.2016.07.009 [DOI] [PubMed] [Google Scholar]
- Vogel J. and Hill T. (2007). High-efficiency Agrobacterium-mediated transformation of Brachypodium distachyon inbred line Bd21-3. Plant Cell Rep. 27, 471-478. 10.1007/s00299-007-0472-y [DOI] [PubMed] [Google Scholar]
- Wang H., Ngwenyama N., Liu Y., Walker J. C. and Zhang S. (2007). Stomatal development and patterning are regulated by environmentally responsive mitogen-activated protein kinases in Arabidopsis. Plant Cell 19, 63-73. 10.1105/tpc.106.048298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin X., Biswal A. K., Dionora J., Perdigon K. M., Balahadia C. P., Mazumdar S., Chater C., Lin H.-C., Coe R. A., Kretzschmar T. et al. (2017). CRISPR-Cas9 and CRISPR-Cpf1 mediated targeting of a stomatal developmental gene EPFL9 in rice. Plant Cell Rep. 36, 745-757. 10.1007/s00299-017-2118-z [DOI] [PubMed] [Google Scholar]
- Zhang Y., Wang P., Shao W., Zhu J.-K. and Dong J. (2015). The BASL polarity protein controls a MAPK signaling feedback loop in asymmetric cell division. Dev. Cell 33, 136-149. 10.1016/j.devcel.2015.02.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Guo X. and Dong J. (2016). Phosphorylation of the polarity protein BASL differentiates asymmetric cell fate through MAPKs and SPCH. Curr. Biol. 26, 2957-2965. 10.1016/j.cub.2016.08.066 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.