Xeroderma pigmentosum (XP), Cockayne syndrome (CS), and trichothiodystrophy (TTD) constitute a family of sun-sensitive human diseases (1). They are caused by mutations in the components of the nucleotide excision repair (NER) system and in the postreplication repair system involving the low-fidelity polymerase H (also known as XP variant or XPV). XP was initially recognized as a disease involving a genetic predisposition to extremely high levels of skin carcinogenesis from solar exposure. The relationship between DNA damage from solar UVB exposure and deficiencies in repair, leading to mutagenesis, genetic instability, and eventual carcinogenesis, provides a deceptively simple explanation for the major clinical features of XP and has provided a paradigm for environmentally induced human cancer. These diseases are, however, much more complex at all levels (1, 2), as the new report in this issue by Murai et al. (3) indicates (Fig. 1). NER has two main pathways: GGR and TCR. Global repair involves the genes XPC and XPE and is generally slower and modulated by p53 (4), which also modulates the pol H pathway (5, 6). TCR results in a more rapid repair of the transcribed strand of expressed genes, and involves the two CS genes (CSA, CSB), several of the XP genes (especially XPA, -B, and -D), and the mismatch repair system (1). Some of the genes are involved in both pathways, especially XPA. The XPG endonuclease is also a cofactor for a glycosylase (endonuclease III, encoded by nth) that acts on oxidative damage (7). Mutations in XPB and XPD also can give rise to diseases that combine one or more of the three main disorders, XP, CS, and TTD, depending in part on the precise site of the mutations in the genes.
Figure 1.
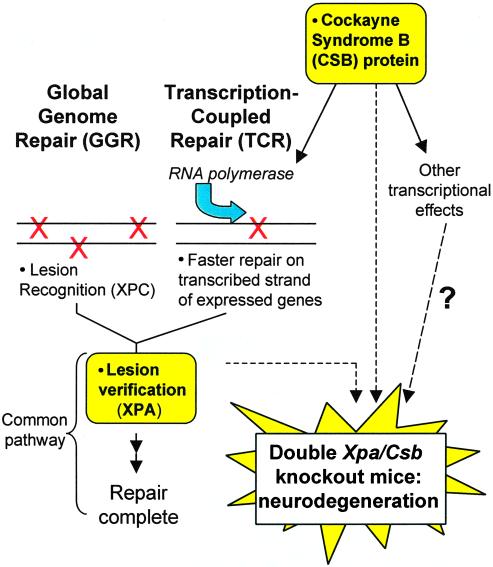
The DNA-repair pathway of NER involves two subpathways: global genome repair (GGR) and transcription-coupled repair (TCR). The xeroderma pigmentosum A (XPA) protein plays a role in both pathways. The Cockayne syndrome B (CSB) protein is involved in TCR and also has additional effects related to transcription. Murai et al. (in this issue) demonstrate neurodegeneration in Xpa/Csb double-knockout mice.
Clinically, the situation is exceedingly complex, because cancer is only part of the symptomology and is not even found in CS and TTD that are primarily disorders of development and neural functions (1). Patients with CS and TTD suffer from progressive mental deterioration, dysmeyelination, growth defects, wizened facies, and hypogonadism. TTD also has characteristic sulfur deficiencies in hair and nails, resulting in a banding pattern identifiable in polarized light. Certain of the XP groups also show a spectrum of neurodegeneration involving neuronal loss and mental deterioration that can be evident from earliest ages or that sets in slowly over time. In general, those genes that are involved only in GGR and postreplication repair (XPC, XPE, XPV) are not associated with neurodegeneration, hinting that the failure of TCR is more closely correlated with neurodegeneration. Fibroblasts from patients with neurological disorders are generally the more UV-sensitive of the XP groups (8). An understanding of the clinical situation could, it was hoped, be addressed by developing mouse models with the various genes knocked out (KOs; ref. 9), but initial results were disappointing. The first KOs of Xpa and a mouse model of a TTD mutation in Xpd did not demonstrate neurological disorders, whereas a KO of Csb demonstrated only a mild neurological phenotype in older mice. Instead, these animals were more cancer-prone than the corresponding human disorder (10–13).
The work of Murai et al. shows that global genome repair and transcription coupled repair have important nonoverlappng functions in rodents.
Murai et al. have made significant progress in developing a neurological model by crossing Xpa and Csb KO mice to produce double-KO mice lacking GGR and TCR with major neurological defects that are similar in many respects to the human situation. This cross combined a total block in both GGR and TCR by elimination of XPA, which is involved in a common damage recognition and binding step for both GGR and TCR, with a block to the additional, TCR-specific CSB branch (Fig. 1). These double-KO mice showed ataxia and abnormal locomotor activity, as well as postnatal growth retardation and lethality at ≈3 weeks of age. There seemed to be a selective reduction in size of the cerebella of these mice, which the authors attributed both to decreased neurogenesis and increased apoptosis. In addition, Purkinje cells displayed abnormal dendritic morphology. To appreciate the significance of the current study of Murai et al., we should consider the general problem of correlating DNA-repair deficiency with developmental and neurological defects, and the unique problems of the differences between human and mouse NER.
To understand the cause of neurodegeneration in the TCR-deficient disorders requires answering questions such as: is it a direct consequence of the deficiency and not an alternative purely transcription function; what is the origin and nature of the damage; and what confers the specificity to the symptoms. Much of the work on XP has related the cancer aspect of this disease to the effects of NER deficiencies on mutation frequencies. However, mutations may be irrelevant for the neurological aspects of these syndromes. Although mice deficient in Xpa have an increased mutation frequency in the liver, mutation frequency is unaltered in brain (14), probably because most cells there are postmitotic. An alternate explanation, therefore, must be put forward for the involvement of NER in neurodegeneration.
Endogenous oxidative stress, resulting from leakage of electron transport in mitochondria, is an obvious culprit as a source of both bulky and structurally minor DNA lesions that involve, at least in part, the NER pathway. The brain is also a tissue with one of the highest levels of oxidative metabolism. Although NER is defective in general for mitochondria in normal cells, XPA is required for the repair of oxidative damage to mitochondrial DNA (15). At least three hypotheses, which are not mutually exclusive, can be put forward linking persistent DNA damage to neurodegeneration.
First, decreased DNA repair and persistent DNA damage may cause neurodegeneration by impairing the transcription of undefined critical neural genes. Several candidate lesions that are produced by oxidative stress and are processed at least in part by the NER pathway (5′,8-purine cyclodeoxynucleosides, purine dimers, 8-oxoguanine, thymine glycol; refs. 16–19) can block or delay transcription (20, 21).
Second, unrepaired DNA damage can be a trigger for apoptosis, and the consequent loss of neurons by apoptosis could result in the neurodegeneration seen in XP and related patients. There is a contrasting role for apoptosis in cancer and neurodegeneration: whereas an efficient apoptotic pathway can remove genetically unstable cells from a pool of potentially malignant proliferating cells, such losses from nondividing neuronal population may result in functional deficits. Increased apoptosis has been observed in several mouse models of DNA-repair deficiency. Mice deficient in homologous recombination repair (Xrcc2), nonhomologous end-joining (Ku 70, Ku 80, Xrcc4, DNA-ligase IV gene) or base-excision repair (DNA polymerase β gene) all show increased apoptosis under conditions of endogenous oxidative stress (22–26). The DNA-damage checkpoints Ataxia-telangiectasia mutated (Atm) and p53 also play a role. Atm is required for central nervous system (CNS) apoptosis after ionizing radiation (27). Atm and p53 are required for the neuronal apoptosis in at least two of the DNA-repair KO mice listed above, because crossbreeding to Atm-null or p53-null mice can rescue the embryolethality of those strains (28–30). In the current study, Murai et al. found increased TUNEL labeling (a marker for DNA breaks generated during apoptosis) in the cerebellum, a result similar to that recently observed in Xpg-null mice (31) that are deficient in both NER (common pathway) and TCR of the oxidative lesion thymine glycol (7).
A third hypothesis is that persistent DNA damage results in defective neurogenesis. Abnormal neurogenesis has been reported for mice deficient in double-strand break repair (Xrcc2, Ku 70, Ku 80) (22, 25), base excision repair (DNA polymerase β gene) (26) and the DNA-damage checkpoint Atm (32). In the current study, Murai et al. observed reduced cell proliferation, as measured by BrdUrd labeling, in the developing cerebellum, providing further support for this hypothesis.
The work of Murai et al. shows that GGR and TCR have important nonoverlapping functions in rodents. These results are important because much early work on rodent cells in culture showed that they were often innately low in DNA repair, because of cell culture-derived GGR deficiencies. A cross between Xpc and Csb KO animals would be exceedingly interesting, because cross-breeding would determine whether more limited deficiencies in GGR combined with TCR also could give rise to neurological deficiencies.
Several additional considerations may help explain the difference between human and mouse phenotypes resulting from single-gene deficiencies or KOs. One possibility is that the actual level of oxidative DNA damage is higher in humans. Accordingly, the levels of DNA damage may need to be increased in mice before a phenotype becomes apparent. Apoptosis is dramatically increased in cultured Xpa-null neurons by exposure to UV light relative to wild-type controls (33), suggesting that, in vivo Xpa-null mice might exhibit a neurological phenotype under conditions of exogenous genotoxic stress. Mice deficient in nonhomologous end-joining (DNA-PKcs) exhibit no endogenous phenotype, but do show an increased susceptibility to exogenous agents in vitro and in vivo (34).
One factor that might contribute to differences between levels of endogenous DNA damage between mice and humans could be the activity of antioxidative enzymes. Cultured cells from some XP patients are also deficient in catalase (35), although activities of catalase or other antioxidative enzymes in NER-deficient mice have yet to be measured. In human patients, the increased endogenous DNA damage in the CNS could result from dysregulated glutamate, because reduced expression of the glial glutamate transporters EATT1 and GLT-1 and an increase in oxidatively modified proteins were observed to a greater extent in brains of CS than XPA patients (36), and brains from these patients differed in the regions most affected by cell loss (37). In support of such a theory, knockdown of these glutamate transporters by antisense techniques can exacerbate neuronal loss in vivo in animal models (38, 39).
Another possibility may be that the existing mouse models of NER and TCR deficiencies have not been examined in sufficient detail in the optimal mouse strains. Use of strains with a known propensity to develop neurological deficits might increase our ability to detect subtle or overt neurological and behavioral defects, and even identify modifier genes that influence neurological development. Examination of embryos and neonates might also reveal defects no longer evident in adult animals. For example, several strains of KO mice deficient in nonhomologous end-joining repair (Ku 70, Ku 80) were originally thought to have no neurological phenotype when adult animals were studied (40–42), but when embryos were analyzed, neural deficits became apparent (24).
XP, CS, and TTD are extremely rare hereditary disorders, but their study by numerous investigators has been exceedingly informative about the causes of human cancer. Similar study of the neurodegeneration in these disorders promises to be equally informative about the causes of major human neurodegenerative conditions. A growing number of studies support a role for endogenous DNA damage, including lesions handled by the NER pathway, in the mechanism of several major human neurodegenerative conditions: Alzheimer's, Parkinson's, and amyotrophic lateral sclerosis (ALS). Clarification of the mechanisms of neurodegeneration of the Xpa/Csb double-KO mice, and extension to other crosses, will be greatly assisted by the findings of Murai et al. in the current issue of this journal. This work may help clarify the mechanisms of common human neurodegenerative conditions and provide model systems for the development of treatment methods for human patients.
Acknowledgments
We thank Mr. and Mrs. Dan Mahar of the XP Society and Mr. and Mrs. Kevin O'Brien of the Luke O'Brien Foundation for their interest and support.
Footnotes
See companion article on page 13379.
References
- 1.Bootsma D, Kraemer K H, Cleaver J E, Hoeijmakers J H J. In: The Metabolic and Molecular Bases of Inherited Disease. Scriver C R, Beaudet A L, Sly W S, Valle D, editors. Vol. 1. San Francisco: McGraw–Hill; 2001. pp. 677–703. [Google Scholar]
- 2.Volker M, Mone M J, Karmakar P, van Hoffen A, Schul W, Vermeulen W, Hoeijmakers J H, van Driel R, van Zeeland A A, Mullenders L H. Mol Cell. 2001;8:213–224. doi: 10.1016/s1097-2765(01)00281-7. [DOI] [PubMed] [Google Scholar]
- 3.Murai M, Enokido Y, Inamura N, Yoshino M, Nakatsu Y, van der Horst G T J, Hoeijmakers J H J, Tanaka K, Hatanaka H. Proc Natl Acad Sci USA. 2001;98:13379–13384. doi: 10.1073/pnas.231329598. . (First Published October 30, 2001; 10.1073/pnas.231329598) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ford J M, Hanawalt P C. J Biol Chem. 1997;272:28073–28080. doi: 10.1074/jbc.272.44.28073. [DOI] [PubMed] [Google Scholar]
- 5.Cleaver J E, Afzal V, Feeney L, McDowell M, Sadinski W, Volpe J P, Busch D B, Coleman D M, Ziffer D W, Yu Y, et al. Cancer Res. 1999;59:1102–1108. [PubMed] [Google Scholar]
- 6.Limoli C L, Giedzinski E, Morgan W F, Cleaver J E. Proc Natl Acad Sci USA. 2000;97:7939–7946. doi: 10.1073/pnas.130182897. . (First Published June 20, 2000; 10.1073/pnas.130182897) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cooper P K, Nouspikel T, Clarkson S G, Leadon S A. Science. 1997;275:990–993. doi: 10.1126/science.275.5302.990. [DOI] [PubMed] [Google Scholar]
- 8.Robbins J H, Kraemer K H. Mutat Res. 1972;15:92–97. doi: 10.1016/0027-5107(72)90098-x. [DOI] [PubMed] [Google Scholar]
- 9.Friedberg E C, Meira L B. Mutat Res. 2000;459:243–274. doi: 10.1016/s0921-8777(00)00006-9. [DOI] [PubMed] [Google Scholar]
- 10.de Vries A, van Oostrom C T, Hofhuis F M, Dortant P M, Berg R J, de Gruijl F R, Wester P W, van Kreijl C F, Capel P J, van Steeg H, et al. Nature (London) 1995;377:169–173. doi: 10.1038/377169a0. [DOI] [PubMed] [Google Scholar]
- 11.Nakane H, Takeuchi S, Yuba S, Saijo M, Nakatsu Y, Murai H, Nakatsuru Y, Ishikawa T, Hirota S, Kitamura Y, et al. Nature (London) 1995;377:165–168. doi: 10.1038/377165a0. [DOI] [PubMed] [Google Scholar]
- 12.de Boer J, van Steeg H, Berg R J, Garssen J, de Wit J, van Oostrum C T, Beems R B, van der Horst G T, van Kreijl C F, de Gruijl F R, et al. Cancer Res. 1999;59:3489–3494. [PubMed] [Google Scholar]
- 13.van der Horst G T J, van Steeg H, Berg R J W, van Gool A J, de Wit J, Weeda G, Morreau H, Beems R B, van Kreijl C F, de Gruijl F R, et al. Cell. 1997;89:425–435. doi: 10.1016/s0092-8674(00)80223-8. [DOI] [PubMed] [Google Scholar]
- 14.Giese H, Dolle M E, Hezel A, van Steeg H, Vijg J. Oncogene. 1999;18:1257–1260. doi: 10.1038/sj.onc.1202404. [DOI] [PubMed] [Google Scholar]
- 15.Driggers W J, Grishko V I, LeDoux S P, Wilson G L. Cancer Res. 1996;56:1262–1266. [PubMed] [Google Scholar]
- 16.Brooks P J, Wise D S, Berry D A, Kosmoski J V, Smerdon M J, Somers R L, Mackie H, Spoonde A Y, Ackerman E J, Coleman K, et al. J Biol Chem. 2000;275:22355–22362. doi: 10.1074/jbc.M002259200. [DOI] [PubMed] [Google Scholar]
- 17.Kuraoka I, Bender C, Romieu A, Cadet J, Wood R D, Lindahl T. Proc Natl Acad Sci USA. 2000;97:3832–3837. doi: 10.1073/pnas.070471597. . (First Published April 4, 2000; 10.1073/pnas.070471597) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reardon J T, Bessho T, Kung H C, Bolton P H, Sancar A. Proc Natl Acad Sci USA. 1997;94:9463–9468. doi: 10.1073/pnas.94.17.9463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Satoh M S, Jones C J, Wood R D, Lindahl T. Proc Natl Acad Sci USA. 1993;90:6335–6339. doi: 10.1073/pnas.90.13.6335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Randerath K, Zhou G D, Somers R L, Robbins J H, Brooks P J. J Biol Chem. 2001;276:36051–36057. doi: 10.1074/jbc.M105472200. [DOI] [PubMed] [Google Scholar]
- 21.Hatahet Z, Purmal A A, Wallace S S. Ann N Y Acad Sci. 1994;726:346–348. doi: 10.1111/j.1749-6632.1994.tb52847.x. [DOI] [PubMed] [Google Scholar]
- 22.Deans B, Griffin C S, Maconochie M, Thacker J. EMBO J. 2000;19:6675–6685. doi: 10.1093/emboj/19.24.6675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barnes D E, Stamp G, Rosewell I, Denzel A, Lindahl T. Curr Biol. 1998;8:1395–1398. doi: 10.1016/s0960-9822(98)00021-9. [DOI] [PubMed] [Google Scholar]
- 24.Gao Y, Sun Y, Frank K M, Dikkes P, Fujiwara Y, Seidl K J, Sekiguchi J M, Rathbun G A, Swat W, Wang J, et al. Cell. 1998;95:891–902. doi: 10.1016/s0092-8674(00)81714-6. [DOI] [PubMed] [Google Scholar]
- 25.Gu Y, Sekiguchi J, Gao Y, Dikkes P, Frank K, Ferguson D, Hasty P, Chun J, Alt F W. Proc Natl Acad Sci USA. 2000;97:2668–2673. doi: 10.1073/pnas.97.6.2668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sugo N, Aratani Y, Nagashima Y, Kubota Y, Koyama H. EMBO J. 2000;19:1397–1404. doi: 10.1093/emboj/19.6.1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Herzog K H, Chong M J, Kapsetaki M, Morgan J I, McKinnon P J. Science. 1998;280:1089–1091. doi: 10.1126/science.280.5366.1089. [DOI] [PubMed] [Google Scholar]
- 28.Lee Y, Barnes D E, Lindahl T, McKinnon P J. Genes Dev. 2000;14:2576–2580. doi: 10.1101/gad.837100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Frank K M, Sharpless N E, Gao Y, Sekiguchi J M, Ferguson D O, Zhu C, Manis J P, Horner J, DePinho R A, Alt F W. Mol Cell. 2000;5:993–1002. doi: 10.1016/s1097-2765(00)80264-6. [DOI] [PubMed] [Google Scholar]
- 30.Gao Y, Ferguson D O, Xie W, Manis J P, Sekiguchi J, Frank K M, Chaudhuri J, Horner J, DePinho R A, Alt F W. Nature (London) 2000;404:897–900. doi: 10.1038/35009138. [DOI] [PubMed] [Google Scholar]
- 31.Sun X Z, Harada Y N, Takahashi S, Shiomi N, Shiomi T. J Neurosci Res. 2001;64:348–354. doi: 10.1002/jnr.1085. [DOI] [PubMed] [Google Scholar]
- 32.Allen D M, van Praag H, Ray J, Weaver Z, Winrow C J, Carter T A, Braquet R, Harrington E, Ried T, Brown K D, et al. Genes Dev. 2001;15:554–566. doi: 10.1101/gad.869001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Enokido Y, Inamura N, Araki T, Satoh T, Nakane H, Yoshino M, Nakatsu Y, Tanaka K, Hatanaka H. J Neurochem. 1997;69:246–251. doi: 10.1046/j.1471-4159.1997.69010246.x. [DOI] [PubMed] [Google Scholar]
- 34.Culmsee C, Bondada S, Mattson M P. Brain Res Mol Brain Res. 2001;87:257–262. doi: 10.1016/s0169-328x(01)00008-0. [DOI] [PubMed] [Google Scholar]
- 35.Hoffschir F, Daya-Grosjean L, Petit P X, Nocentini S, Dutrillaux B, Sarasin A, Vuillaume M. Free Radical Biol Med. 1998;24:809–816. doi: 10.1016/s0891-5849(97)00350-x. [DOI] [PubMed] [Google Scholar]
- 36.Hayashi M, Itoh M, Araki S, Kumada S, Shioda K, Tamagawa K, Mizutani T, Morimatsu Y, Minagawa M, Oda M. J Neuropathol Exp Neurol. 2001;60:350–356. doi: 10.1093/jnen/60.4.350. [DOI] [PubMed] [Google Scholar]
- 37.Itoh M, Hayashi M, Shioda K, Minagawa M, Isa F, Tamagawa K, Morimatsu Y, Oda M. Brain Dev. 1999;21:326–333. doi: 10.1016/s0387-7604(99)00033-9. [DOI] [PubMed] [Google Scholar]
- 38.Rao V L, Dogan A, Todd K G, Bowen K K, Kim B T, Rothstein J D, Dempsey R J. J Neurosci. 2001;21:1876–1883. doi: 10.1523/JNEUROSCI.21-06-01876.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rao V L, Dogan A, Bowen K K, Todd K G, Dempsey R J. Eur J Neurosci. 2001;13:119–128. [PubMed] [Google Scholar]
- 40.Nussenzweig A, Chen C, da Costa Soares V, Sanchez M, Sokol K, Nussenzweig M C, Li G C. Nature (London) 1996;382:551–555. doi: 10.1038/382551a0. [DOI] [PubMed] [Google Scholar]
- 41.Gu Y, Seidl K J, Rathbun G A, Zhu C, Manis J P, van der Stoep N, Davidson L, Cheng H L, Sekiguchi J M, Frank K, et al. Immunity. 1997;7:653–665. doi: 10.1016/s1074-7613(00)80386-6. [DOI] [PubMed] [Google Scholar]
- 42.Ouyang H, Nussenzweig A, Kurimasa A, Soares V C, Li X, Cordon-Cardo C, Li W, Cheong N, Nussenzweig M, Iliakis G, et al. J Exp Med. 1997;186:921–929. doi: 10.1084/jem.186.6.921. [DOI] [PMC free article] [PubMed] [Google Scholar]