Abstract
Purpose
We previously demonstrated an association between decreased SMAD4 expression and cetuximab resistance in head and neck squamous cell carcinoma (HNSCC). The purpose of this study was to further elucidate the clinical relevance of SMAD4 loss in HNSCC.
Experimental Design
SMAD4 expression was assessed by immunohistochemistry in 130 newly diagnosed and 43 recurrent HNSCC patients. Correlative statistical analysis with clinicopathological data was also performed. OncoFinder, a bioinformatics tool, was used to analyze molecular signaling in TCGA tumors with low or high SMAD4 mRNA levels. The role of SMAD4 was investigated by shRNA knockdown and gene reconstitution of HPV-negative HNSCC cell lines in vitro and in vivo.
Results
Our analysis revealed that SMAD4 loss was associated with an aggressive, HPV-negative, cetuximab-resistant phenotype. We found a signature of pro-survival and anti-apoptotic pathways that were commonly dysregulated in SMAD4-low cases derived from TCGA-HNSCC dataset and an independent oral cavity squamous cell carcinoma (OSCC) cohort obtained from GEO. We show that SMAD4 depletion in a HNSCC cell line induces cetuximab resistance and results in worse survival in an orthotopic mouse model in vivo. We implicate JNK and MAPK activation as mediators of cetuximab resistance and provide the foundation for the concomitant EGFR and JNK/MAPK inhibition as a potential strategy for overcoming cetuximab resistance in HNSCCs with SMAD4 loss.
Conclusions
Our study demonstrates that loss of SMAD4 expression is a signature characterizing the cetuximab-resistant phenotype and suggests that SMAD4 expression may be a determinant of sensitivity/resistance to EGFR/MAPK or EGFR/JNK inhibition in HPV-negative HNSCC tumors.
Keywords: SMAD4, epithelial–mesenchymal transition (EMT), head and neck squamous cell carcinoma (HNSCC), cetuximab, human papillomavirus (HPV)
INTRODUCTION
Head and neck squamous cell carcinoma (HNSCC) is the sixth most common cancer in the world, with an annual incidence of over 50,000 cases in the United States alone (1). HNSCC is a heterogeneous disease that includes squamous cell carcinomas (SCCs) of the oral cavity, pharynx, and larynx. The disease is more prevalent in males, and those individuals who smoke or chew tobacco and/or consume alcohol are at much higher risk for HNSCC (2). Human papillomavirus (HPV) is also associated with oropharyngeal primary sites with significantly better prognosis than patients with HPV-negative tumors (3, 4). In particular, HPV-negative HNSCC is notorious for poor prognosis, which reflects the propensity of the tumor to be treatment resistant. Although patients with early stage disease (stage I or II) have better prognosis, these patients are still at risk for developing locoregional recurrences, distant metastases, and second primary tumors. Despite improvements in molecular diagnosis, cetuximab is the only FDA-approved targeted therapy available for this disease (5). Cetuximab, a monoclonal antibody targeting epidermal growth factor receptor (EGFR), has been the standard of care in the treatment of HNSCC for many years. However, the response rate to cetuximab monotherapy is low (10–13%), and the majority of patients develop resistance even after an initial response (6, 7). Thus, HNSCC continues to represent a challenging disease to treat.
SMAD4 is a well-known tumor suppressor and the central signal transduction component of transforming growth factor beta (TGFβ), a multi-functional cytokine that regulates cell growth and differentiation and is frequently upregulated in many human cancers including HNSCC (8–11). Although TGF-β/SMAD signaling exerts a suppressive effect on normal epithelial cells, tumor cells frequently become refractory to the growth inhibitory effect of TGFβ and acquire an ability to increase expression and secretion of TGFβ (11–13). Previous studies have demonstrated that in tumor cells, TGFβ-activated SMAD4–SMAD2/3 complex stimulates the expression of SNAIL1 and TWIST1, which cooperate with SMAD proteins to repress the expression of epithelial genes such as CDH1 (encoding E-cadherin) (14, 15). This switch enables tumor cells to leverage the tumor-promoting effects of TGFβ in the tumor microenvironment to facilitate tumor progression, invasion, and metastasis (10–15). The nature of the switch that determines whether TGFβ acts as a tumor promoter or as a tumor suppressor has been the subject of intense research (14).
While mutant TP53 has been suggested to be one potential explanation (16), more recent studies have shown that loss of SMAD4 activity may also result in such a switch (17, 18). Supporting this concept, the formation and subsequent progression of various solid malignancies was strongly associated with loss of SMAD4 activity via inactivating mutations or loss of heterozygosity within the 18q21 locus, containing SMAD4 (19–22). For example, loss of SMAD4 promotes pancreatic and colorectal tumor progression and is strongly associated with increased metastatic potential, lower overall survival and poor chemotherapeutic responses (23–25). Likewise, loss of SMAD4 activity was reported to cause spontaneous HNSCC formation, to induce epithelial-to-mesenchymal transition (EMT) and cetuximab resistance, and to result in genomic instability through the downregulation of DNA repair-related genes (18, 26, 27). Although the link between SMAD4 loss or inactivation and tumorigenesis is strong and compelling, mechanisms underlying the resistance to cetuximab, an FDA approved HNSCC targeted therapy, in tumors with SMAD4 loss has not been reported.
Previously, we demonstrated that cetuximab resistant clones of an HPV-negative HNSCC cell line had reduced SMAD4 gene expression (27). Interestingly, HPV-negative tumors in HNSCC patients also had lower SMAD4 expression compared to HPV-positive tumors, suggesting that the worse overall survival of HPV-negative HNSCC patients may be in part due to low SMAD4 expression. In this study, we demonstrate that loss of SMAD4 in a cohort of 130 newly diagnosed and 43 recurrent HNSCC patients is associated with poor outcome. Our work also aimed to discover mechanisms of cetuximab resistance in SMAD4-low HNSCC that can be targeted with combination therapy. Using OncoFinder, a bioinformatics software suite for qualitative analysis of intracellular signaling pathway activation using transcriptomic data (28), we uncovered commonly upregulated pathways in the TCGA HNSCC dataset and an independent oral cavity squamous cell carcinoma cohort with low SMAD4 expression. We find that SMAD4 expression is associated with cetuximab resistance in HNSCC cell lines and that SMAD4-low tumors are associated with worse overall survival of tumor-bearing mice in vivo. Furthermore, our data indicates that JNK and MAPK activation may be potential mediators of this process, and demonstrates the potential utility of JNK or MAPK inhibition as a novel strategy for overcoming cetuximab resistance in HNSCC tumors with loss of SMAD4 expression.
MATERIALS AND METHODS
Expression data processing
Raw RNA-Seq or microarray data had been retrieved from publicly available TCGA or NCBI GEO repository databases. RNA-Seq data preprocessing and normalization steps were performed in R version 3.1.0 using DEseq package from Bioconductor. The resulting matrix contained mRNA expression information for over 20K genes across all analyzed samples. Normalized gene expression data were loaded into Oncofinder pathway activation scoring platform (28). The software enables calculation of the Pathway Activation Score (PAS) for each of the 271 pathways analyzed, a value which serves as a quantitative measure of differential pathway activation between the two states. The signalome knowledge base developed by SABiosciences (http://www.sabiosciences.com/pathwaycentral.php) was used to determine structures of intracellular pathways, which was used for the computational algorithm as described previously (29, 30). Pathways with positive PAS values are upregulated, while negative PAS values correspond to down-regulated pathways. The algorithm used to calculate AS is as follows:
Here, CNRn is the ratio of the expression level of a gene n in the tumor sample and in the control; BTIFn is a value of beyond tolerance interval flag, which equals 0 or 1; and ARRn is an activator/repressor role equal to −1, −0.5, 0, 0.5 or 1, defined by the role of protein n in the pathway. More information can be found in previous publications (29–31). Pathway activation strengths were determined using the default parameters of OncoFinder, a sigma filter of 2 and a CNR value less than 0.67 or higher than 1.5. PAS heat map generation and hierarchical clustering were performed using R package gplots. Statistical tests and correlation analysis were done with the MS Excel software.
Tissue microarrays
Two tissue microarrays (TMA) containing tumor cores from 133 newly diagnosed HNSCC patients were obtained under Institutional Review Board-approved protocols from Stanford University. Detailed clinical description of the TMAs has been published (32, 33). Cores from three patients lacked tumor and were excluded from the study. In addition, forty-three unstained formalin-fixed paraffin-embedded tumor slides from retrospectively selected patients for having recurrent HNSCC with available tumors were also obtained under Institutional Review Board-approved protocols at Vanderbilt University.
Formalin-Fixed Cell Pellet Blocks
FaDu, SCC25 and SCC61 cells were harvested by trypsinization when 70% confluent, washed twice in PBS, resuspended in 10% neutral buffered formalin (NBF) and pipetted into a 0.6ml microfuge tube containing 200μl of solidified agarose (2% in PBS). The microfuge tube was centrifuged at 200g (5 min at room temperature) to form a uniform layer on the agarose bed. Supernatant was aspirated and pellet was carefully overlaid with 200μl 10% NBF. After repeating centrifugation, the microfuge tube was carefully submerged in 10ml 10% NBF solution. Cell pellet was allowed to fix for 24–48 hours and submitted for processing and paraffin embedding.
Immunohistochemistry
Slides were stained at the Johns Hopkins Oncology Tissue Immunostaining Core Facility, a Clinical Laboratory Improvement Amendments (CLIA) certified lab, with SMAD4 antibody at 1:500 dilution (Santa Cruz Biotechnology, Inc., Santa Cruz, CA) in Universal IHC Blocking Diluent (Leica Biosystems, Wetzlar, Germany) for 15 minutes using an automated IHC system (Leica BOND-MAX, Wetzlar, Germany), and a p16 antibody (pre-dilute, mtm-CINtech, E6H4) using iView DAB detection and Ventana BenchMark XT staining system (Roche, Basel, Switzerland) as previously described (4, 24, 34). Formalin-fixed paraffin-embedded (FFPE) cell pellet blocks generated from FaDu cells with complete loss of SMAD4 and stained under the same conditions as patient samples served as a negative control, and blocks from SCC25 and SCC61 cells served as a positive control. Scoring of the IHC staining was performed by trained head and neck cancer pathologist. Scoring was based upon the percentage of tumor cells stained in an overall assessment of all tumor tissue available. Cases were scored for SMAD4 and p16 as dichotomous variables based on the presence or absence of protein expression in the tumor. p16 was used as a surrogate marker for high-risk HPV and positive was defined as >70% nuclear or nuclear and cytoplasmic expression. SMAD4 loss was defined as <5% tumor cells in order to minimize confounding effects introduced by varying degrees of SMAD4 downregulation.
Statistical analysis of SMAD4 and p16 immunohistochemical staining and clinical outcomes
Patient characteristics were analyzed by SMAD4 status using Fisher’s exact test for categorical factors and Wilcoxon test for continuous variables. To evaluate the prognostic effect of SMAD4, the clinical outcome of overall survival, defined as the elapsed time from diagnosis to death due to any cause, were considered. We used the Cox regression model to correlate pertinent markers with overall survival. For multivariate analysis, backward selection technique was used to select the variables to include in the model, while SMAD4 status and known prognostic factors were restricted within the model. The significance level for removing any variable was 0.2. All p-values were based on two-sided tests.
Cell lines and reagents
FaDu, SCC61 and SCC25 HNSCC cell lines were used for these studies. FaDu and SCC25 cell lines were obtained from American Type Culture Collection (ATCC), and SCC61 was received from Dr. Ralph Weichselbaum (University of Chicago). Short tandem repeat analysis (Identifiler, Applied Biosystems, Foster City, CA) authenticated cell lines before use. The cells were periodically monitored for mycoplasma at Johns Hopkins Genetic Resources Core Facility using the MycoDtect kit (Greiner Bio-One, Kremsmunster, Austria). All experiments were performed within 6 months of the mycoplasma screen. Cell lines were cultured in DMEM/F12 medium supplemented with 10% FBS, Penn-Strep, and 0.4ug/mL hydrocortisone. Stable cell lines were maintained in culture medium with selection antibiotics Puromycin (5 μg/mL FaDu, 4 μg/mL SCC61), Hygromycin (400 μg/mL FaDu, 100 μg/mL SCC61). Cetuximab (Bristol-Myers Squibb, Princeton, NJ) was purchased from the Johns Hopkins Pharmacy.
Antibodies
The following primary antibodies were from Cell Signaling Technology (Boston, MA): phospho-SMAD2Ser465/467 (138D4), SMAD2/3 (D7G7), phospho-STAT3Tyr705 (D3A7), STAT3 (79D7), phospho-AKTSer473 (D9E), AKT(C67E7), Phospho-EGF Receptor Tyr1045, Tyr1068 (D7A5), and Tyr1173 (53A5), EGF Receptor (D38B1), Phospho-p44/42 MAPKThr202/Tyr204, p44/p42 MAPK (137F5), GAPDH (14C10), Phospho-JNKThr183/Tyr185 (81E11), JNK (56G8), BCL-2 (50E3), BCL-XL (54H6), BIM (C34C5), beta-Actin (13E5). SMAD4 (B-8) was from Santa Cruz Biotechnology (Dallas, TX).
Immunoblotting
Protein lysates were prepared in RIPA lysis buffer and protein concentration was measured by the bicinchoninic acid method (Thermo scientific, Waltham, MA). Proteins from each sample were resolved under reducing conditions using NuPAGE gels (Novex; Invitrogen) according to manufacturer’s instructions and transferred to PVDF membrane (EMD Millipore). Membranes were blocked in 5% BSA in TBS and incubated with primary antibodies overnight at 4°C followed by incubation with HRP-linked secondary antibodies. Protein bands were visualized by chemiluminescence using the ECL Western blotting Detection System (GE Healthcare, Piscataway, NJ) or SuperSignal West Femto Maximum Sensitivity Substrate (Thermo Scientific/ Pierce Biotechnology, Rockford, IL).
Generation of SMAD4 stable knockdown cells
SCC61 or SCC25 cells were seeded in 6-well plates 24 hours prior to transduction. Per manufacturer’s instructions, an appropriate amount of lentiviral transduction particles of Smad4 (Clone Number: TRCN0000010321, Sigma, NY) or control transduction particles (#SHC-002V, Sigma) were added into the media in the presence with hexadimethrine bromide (Sigma). After puromycin selection for 2 weeks, surviving cells were pooled and knockdown was confirmed by real time PCR using primers for Smad4 (Hs00929647-m1) and β-actin (ACTB, Hs00357333-g1), or by immunoblotting.
Generation of SMAD4 expressing cells
To establish the SMAD4 expression vector, pLenti-CMV puro lentiviral vector was used as the backbone. pLenti-CMV was a kind gift from Dr. Charles Rudin and Dr. John Poirer (Oncology, Johns Hopkins University). The SMAD4 gene sequence was obtained from the pDPC-WT vector, a kind gift from Dr. Scott Kern (Oncology, Johns Hopkins University), and pLenti-Smad4-CMV was made by Gateway Technology (Life Technologies). All vectors were confirmed to have correct gene sequence. Viral particles were produced by co-transfecting 900 ng of each expression vector, 100 ng of psPAX2 as packaging vector and 1 μg pMD 2G as envelope vector into HEK-293T cells using 10 μl of Lipofectamine2000. Virus culture supernatants were obtained 24–48 hours after transfection. FaDuL cells were exposed to virus containing media for 24 hours, and were selected using puromycin (Life Technologies) 5 μg/ml for at least two weeks. For confirmation of Smad4 expression, total RNA was extracted from transduced cells using Qiagen RNeasy Mini kit (Qiagen, Valencia, CA) according to the manufacturer’s protocol. Smad4 over-expression was confirmed by conventional PCR using forward primer; 5′-CCATTTCCAATCATCCTGCT-3′ and reverse primer; 5′-ACCTTTGCCTATGTGCAACC -3′, or by immunoblotting.
Colony formation assay in Matrigel
1x103 cells with 100 μl media were seeded to each well of a 96-well dish coated with 45 μl of Matrigel (BD Bioscience, San Jose, CA) and reagents were added next day. The media and reagents were replaced every 3 days. Colonies were scanned and analyzed with GelCountTM (Oxford Optronix Ltd; UK) on day 7 after MTT (4 mg/ml 3-(4,5-Dimethythiazol-2-yl)-2,5-diphenyltetrazolium bromide, Sigma) staining for 2 hours. The total area of colonies was calculated by average colony area multiplied by colony number.
Cell viability assays
Cells were counted with trypan blue staining using a TC20 Automated Cell Counter (Bio-Rad) and 3000 cells in 100 μl were seeded in 96-well culture plates in triplicate. DMSO, SP600125 or U0126 was added 24 hours later. Cell survival rates were calculated upon measuring absorbance following 72 hours using the CellTiter 96® AQueous One Solution Cell Proliferation Assay (Promega) or alamarBlue® (ThermoFisher) according to the manufacturer’s recommendations. Percent viability was determined by comparing DMSO treatment to inhibitor treatment.
Bioluminescent Imaging
Bioluminescent images of mice orthotopically implanted with luciferase-transduced cells were acquired using a Xenogen IVIS Spectrum system (Xenogen Corporation, Berkley, USA). Mice were anesthetized with 2% isoflurane (Abbott Laboratories, Chicago, USA), and images were acquired at 10 min post injection of 50 mg/kg i.p. dose of luciferin (Xenogen Corporation).
Animal care and Orthotopic sublingual injection
Female athymic nude mice, 6–10 weeks of age, were kept in a specific pathogen-free animal facility. The animals were fed clean and autoclaved food and water. All of the animal procedures were performed in accordance with a protocol approved by the Institutional Animal Care and Usage Committee. All mice were anesthetized with Ketamine (Sigma) 80–100 mg/kg and Xylazine (Sigma) 5–10 mg/kg before injection. Tumor cells (5x105) were prepared in 50 μl of Hanks’ balanced buffered solution (Life Technologies), and injected submucosally directly into the anterior tongue with a 27-gauge needle attached to a 1-ml tuberculin syringe. Mice were then examined 1 or 2 times a week for the development of tongue tumors and weight loss. The mice were euthanized by CO2 inhalation when they had 25% body weight loss as when compared with their pre-injection weight. After euthanasia, mouse tongue, cervical lymph node, salivary gland, lungs and liver were resected. All tissues were fixed in formalin, embedded in paraffin, and stained with hematoxylin and eosin.
RESULTS
SMAD4 loss is associated with cetuximab resistance and poor survival in HPV-negative recurrent HNSCC patients
To further determine the clinical significance of SMAD4 loss, 130 tumors obtained from patients with newly diagnosed HNSCCs were analyzed for SMAD4 protein expression by IHC (Supplemental Table 1, Supplemental Figure 1). The average age of patients was 59.3 years with 73.8% of the population being male. The primary tumor site was the oropharynx for 48.5% of the patients and 50.8% of those were p16 positive. SMAD4 expression was detected in 86.9% of patients, but was undetectable in 6 (4.6%) tumors (Supplemental Table 1). This is consistent with 7% of HNSCC having SMAD4 alteration via homozygous deletion, and nonsense, missense, and silent mutations resulting in complete loss, while relative downregulation of SMAD4 expression is more commonly seen in 44.8% of HNSCC based on the gene expression analysis of TCGA dataset as previously published by our group (27). Of the 6 SMAD4 negative patients, 5 patients had a tumor site other than the oropharynx and were HPV negative.
We next evaluated SMAD4 status in a cohort of 43 recurrent HNSCC patients (Supplemental Table 2). Similar to the newly diagnosed patients, the average age was 55.1 years with 72.1% of the population being male. Unlike the newly diagnosed patients, a larger subset of recurrent HNSCCs was SMAD4 negative. Approximately 26% of the recurrent HNSCC samples lacked SMAD4 protein expression and were likely HPV-negative given the primary tumor site and p16 staining status. Univariate analysis of SMAD4 expression in these patients showed that SMAD4 loss is associated with a trend toward worse survival. A strong association between SMAD4 loss and distant metastasis development was also observed (Table 1). Multivariate analysis showed that patients with SMAD4-negative tumors had more than a 5-fold increased risk of developing distant metastases (HR: 5.85, 95% CI 1.83–18.67, p-value = 0.003; Table 2).
Table 1.
Univariate analysis of SMAD4 expression and patient characteristics for overall survival in patients with recurrent HNSCC.
| Overall Survival | |||
|---|---|---|---|
| Characteristic | HR | 95% CI | P value |
| SMAD4 | |||
| Negative vs. positive | 1.99 | [0.95,4.16] | 0.069 |
| Age | 1.02 | [0.99, 1.06] | 0.154 |
| Gender | |||
| Female vs. male | 0.85 | [0.39, 1.86] | 0.694 |
| Race | |||
| AA/other vs. caucasian | 1.16 | [0.40, 3.34] | 0.789 |
| Primary tumor site | |||
| Oropharynx vs. other | 3.21 | [1.44, 7.15] | 0.004 |
| T stage | |||
| T3/T4 vs. T1/T2 | 1.47 | [0.69, 3.13] | 0.313 |
| N stage | |||
| N2/N3 vs. N0/N1 | 2.52 | [1.19, 5.31] | 0.015 |
| M stage | |||
| M1 vs. M0 | 5.53 | [1.95, 15.68] | 0.001 |
Table 2.
Multivariate analysis of SMAD4 expression adjusted for other patient characteristics in patients with recurrent HNSCC.
| Overall Survival | |||
|---|---|---|---|
| Characteristic | HR | 95% CI | P value |
| SMAD4 | |||
| Negative vs. positive | 3.05 | [1.34, 6.95] | 0.008 |
| M stage | |||
| M1 vs. M0 | 5.85 | [1.83, 18.67] | 0.003 |
| Primary tumor site | |||
| Oropharynx vs. other | 4.07 | [1.65, 10.06] | 0.002 |
| Age | 1.008 | [0.98, 1.04] | 0.64 |
Of the 43 recurrent and/or metastatic disease patients, 15 were treated with cetuximab monotherapy. Treatment was given off-protocol limiting the ability to assess response or survival. However, cetuximab treatment duration can be used as a surrogate for potential clinical benefit as treatment was halted at time of disease progression. Eight patients with SMAD4-positive tumors received cetuximab treatment significantly longer than the seven patients with SMAD4-negative lesions (mean length of cetuximab treatment, 140.5 days vs. 40.4 days; median length of cetuximab treatment, 63 days vs. 35 days; p-value=0.02). The clinical data suggest that SMAD4 loss is associated with recurrence and cetuximab resistance in HNSCC.
SMAD4 expression correlates with dysregulation of cancer promoting pathways
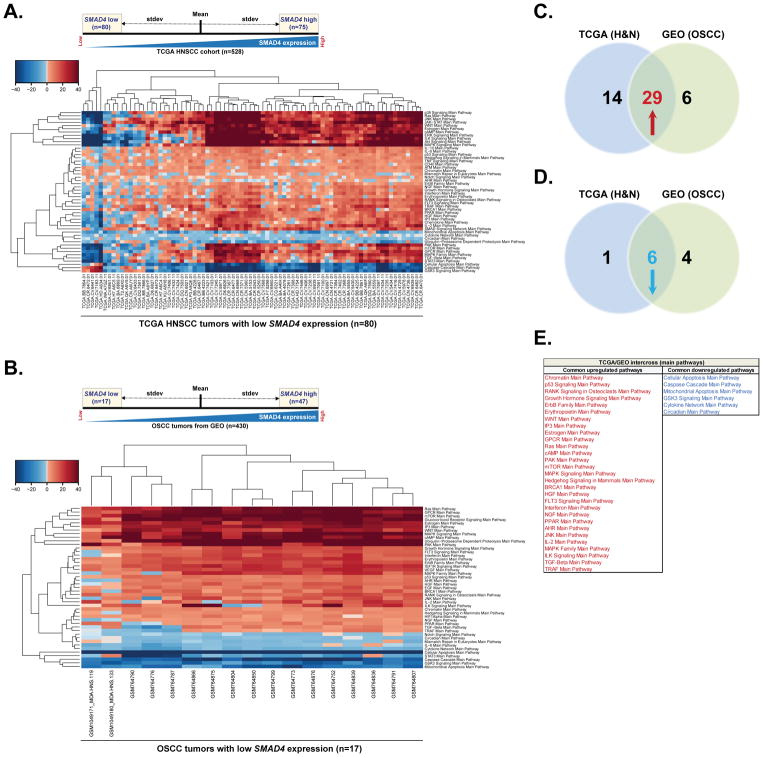
To assess differentially expressed pathways in HNSCC tumors based on the SMAD4 expression levels, we used RNA-sequencing data from The Cancer Genome Atlas (TCGA) HNSCC dataset (n=528) and categorized SMAD4 mRNA expression as high level (one standard deviation above the mean of the TCGA tumor group) or low level (one standard deviation below the mean of the TCGA tumor group) (Figure 1A). We then used OncoFinder, a new bioinformatic software suite for qualitative analysis of intracellular signaling pathway activation (SPA) based on transcriptomic data (28–30), to analyze pathway activation in TCGA tumors with low (n=80) or high (n=75) SMAD4 mRNA levels. Analysis revealed that pathways involved in cancer initiation, progression and maintenance, such as those associated with AKT, JNK, JAK/STAT, ILK, RAS, MAPK/ERK, p38, and WNT signaling, were significantly upregulated in most of the patients in the SMAD4-low expression group (Figure 1A), whereas pathways associated with apoptosis were significantly downregulated in this cohort (Figure 1A).
Figure 1. SMAD4 expression correlates with dysregulation of cancer promoting pathways.
(A). SMAD4 RNA-Seq mRNA expression in the TCGA HNSCC cohort (n=528) was categorized as high level (one standard deviation above the mean) or low level (one standard deviation below the mean) and pathway activation strength (PAS) values have been calculated according to OncoFinder algorithm. Hierarchically clustered heatmap represents differentially activated pathways in 80 tumors with low SMAD4 mRNA expression (75 tumors with high SMAD4 expression were used as a reference). Downregulated PAS values for each sample/pathway are indicated in blue, whilst upregulated PAS values are shaded in red. (B) Gene expression microarray data for 430 OSCC patients was retrieved from publicly available NCBI GEO repository database. Tumors were categorized based on the SMAD4 mRNA expression level and PAS values were then calculated as described in (A). Hierarchically clustered heatmap represents differentially activated pathways in 17 tumors with low SMAD4 mRNA expression. 47 tumors with high SMAD4 expression were used as a reference. Downregulated PAS values for each sample/pathway are indicated in blue, whilst upregulated PAS values are shaded in red. (C) Venn diagram summarizing the number of upregulated pathways predicted in tumors with low SMAD4 expression form TCGA HNSCC cohort, GEO database or both. (D) Venn diagram summarizing the number of downregulated pathways predicted in tumors with low SMAD4 expression form TCGA HNSCC cohort, GEO database or both. (E) List of commonly up- or downregulated pathways predicted in tumors with low SMAD4 expression in both TCGA and GEO datasets.
Although most of the HNSCC TCGA samples for whom HPV-status is available are HPV-negative (35, 36), HPV-status has not yet been evaluated for many patients in this dataset. Our SMAD4 IHC dataset suggests SMAD4 loss predominantly occurs in HPV-negative tumors. Therefore, to further validate the role of SMAD4 in regulating signaling networks in HPV-negative patients, we applied a similar analysis to a separate set of transcriptomic data from 430 oral squamous cell carcinoma (OSCC) tumors, the most common subtype of HNSCC (5), derived from the well annotated publicly available Gene Expression Omnibus (GEO) datasets (GSE9844, GSE41613, GSE30784, GSE42743, GSE31056 and GSE6791). All the datasets were obtained using the microarray platform Affymetrix Human Genome U133 Plus 2.0 Array. HPV infection does not appear to be a significant risk factor for OSCC and the prevalence of HPV-positive oral cavity cancers is relatively low, at less than 6% (37). After categorizing samples based on their SMAD4 mRNA level, we used OncoFinder suite to analyze pathway activation in tumors with low (n=17) or high (n=47) SMAD4 expression. Consistent with the results observed in TCGA cohort, well-known cancer driving signaling axes such as RAS, PAK/p38, MAPK, EGFR, HGFR, JNK and TGFβ pathways were shown to be upregulated in virtually all SMAD4-low tumors, whereas apoptotic pathways had been significantly downregulated (Figure 1B). Notably, 29 pathways were simultaneously upregulated (Figure 1C) and six pathways were commonly downregulated (Figure 1D) among the SMAD4-low tumors in both datasets (Figure 1E). Commonly upregulated pathways reflected activation of ERBB family members and receptors important in EMT phenotype, including WNT, hGH, and TGFβ as well as downstream pathway modulators such as RAS, MAPK, JNK, and mTOR (Figure 1E). Conversely, commonly downregulated pathways consisted of predominantly decreased apoptosis and cytokine regulation (Figure 1E). Taken together, these data suggest that SMAD4 may regulate genes associated with key cancer pathways and further support the role of SMAD4 as a potent tumor suppressor in head and neck cancers.
SMAD4 downregulation leads to cetuximab resistance in HNSCC cell lines
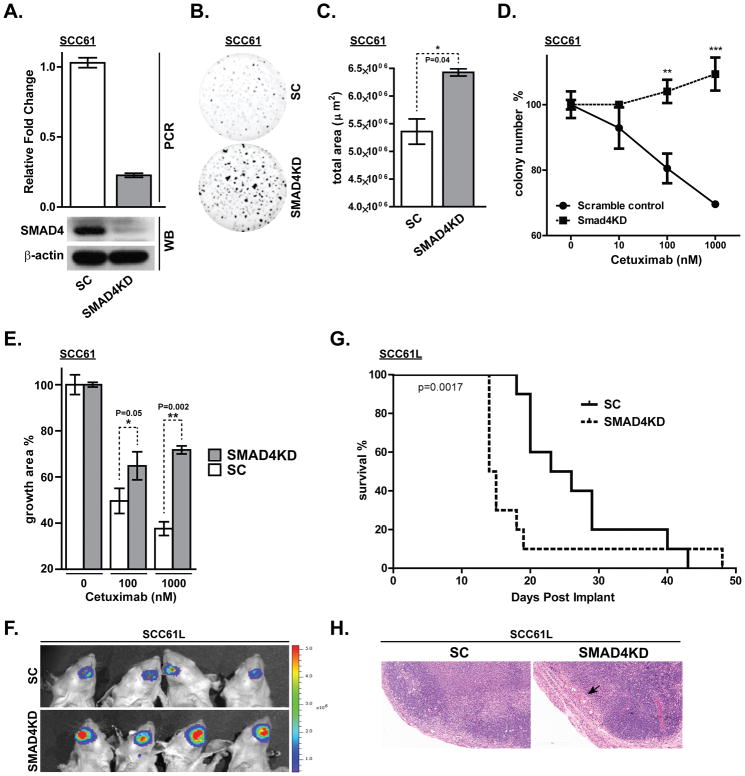
As an in vitro model system to validate our computational findings and to further elucidate the functional consequences of SMAD4 loss, we used SMAD4-specific shRNA under a lentiviral promoter to selectively knock down its expression in the HPV-negative SCC61 HNSCC cell line, known to express high endogenous SMAD4 level. Cells infected with the scrambled shRNA were used as a control. The shRNA-mediated knockdown was confirmed by western blot and real time PCR analysis and resulted in 80% reduction of SMAD4 protein and mRNA levels (Figure 2A). SMAD4 depletion in SCC61 cells did not affect SMAD2/3 levels or SMAD2 phosphorylation (Supplemental Figure 2). To assess the effect of SMAD4 depletion on the long-term cell-growth, Matrigel colony formation assays were performed with either SMAD4-depleted (SCC61-SMAD4KD) or control (SCC61-SC) cell lines. SCC61-Smad4KD cells displayed significantly higher clonogenic survival over the SCC61-SC cell line (Figure 2B and 2C). Moreover, SMAD4 knockdown cells acquired a potent resistance to cetuximab, as demonstrated by markedly increased number and size of the colonies under cetuximab treatment, compared to the SCC61-SC cell line (Figure 2D and 2E). To validate that this effect is SMAD4 loss-dependent, we have knocked down SMAD4 expression in additional HPV-negative cell line, SCC25 (Supplemental Figure 3A). Consistently with our results in SCC61 cells, depletion of SMAD4 in SCC25 cell line rendered them less sensitive to cetuximab (Supplemental Figure 3B), confirming that modulation of SMAD4 expression levels may mediate cetuximab resistance in HNSCC.
Figure 2. SMAD4 depletion in an HPV (−) HNSCC cell line increases clonogenic survival and cetuximab resistance.
(A) Real time PCR and western blot (WB) confirming knockdown of SMAD4 expression by stable expression of short-hairpin RNA in the SCC61 cell line. (SC: scrambled control shRNA, SMAD4KD: shRNA against SMAD4). (B, C) Clonogenic survival was determined by Matrigel colony formation assay for 7 days and total area was quantified (*p<0.05). Matrigel colony formation assay was performed for 7 days in PBS, 10nM, 100nM or 1000nM cetuximab showing (D) colony number (**p<0.01. ***p<0.0001) and (E) percent growth area. (F) SCC61L-SC and SCC61L-SMAD4KD cells that stably expresses firefly luciferase were submucosally injected into oral tongue of athymic nude mice. Bioluminescence images were taken at 13 days after tumor cell inoculation. (G) Kaplan-Meier survival curves for orthotopic xenograft mouse models of SCC61L-SC and SCC61L-SMAD4KD cells. (H) Representative H&E staining of tumor free lymph node derived from SCC61L-SC xenograft model and lymph node harvested from SCC61L-SMAD4KD model that shows metastatic tumor nodules (black arrow). Bar, 200 μm.
To test whether SMAD4 depletion affects tumorigenicity in vivo, SCC61L-SC and SCC61L-SMAD4KD cells that stably express firefly luciferase were injected into the oral tongue of athymic nude mice (10 mice per group) to generate orthotopic xenograft tumors. Mice were then examined for the development of oral cavity tumors and weight loss. Mice engrafted with the bioluminescent SCC61L-SMAD4KD cells exhibited enhanced tumor growth compared to the animals inoculated with the SCC61L-SC cell line (Figure 2G). The mice were euthanized when they reached 25% body weight loss compared to their pre-injection weight. Kaplan-Meier survival curve indicated that SMAD4 knockdown significantly reduces the overall survival (p=0.0017) in our orthotopic HNSCC model (average days until euthanasia was 26.8 and 18.5 for SCC61L-SC and SCC61L-SMAD4KD, respectively) (Figure 2F). Notably, metastasis to cervical lymph nodes were detected in 2 (20%) mice injected with SCC61L-SMAD4KD cells, whereas lymph nodes harvested from SCC61L-SC xenografts showed no signs of metastatic lesion (Figure 2H). Collectively, these findings further support our hypothesis that low SMAD4 expression may contribute to poor clinical outcomes seen in HPV-negative HNSCC patients.
SMAD4 loss induces JNK and MAPK pathway activation
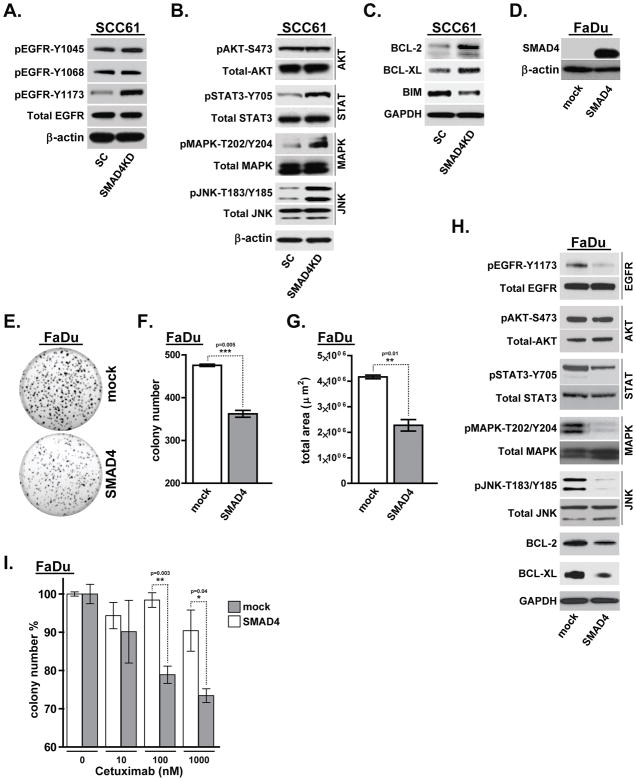
Multiple pathways predicted to be upregulated in SMAD4-low HNSCC tumors are principally related to proliferative and pro-survival EGFR signaling (Figure 1E). Since EGFR regulates activation of several tyrosine kinase pathways such as MAPK, AKT and JNK, we next evaluated the activation status of EGFR and several key downstream signaling components (AKT, STAT3, MAPK and JNK), following the depletion of SMAD4 in SCC61 cells. We first tested the phosphorylation of three major tyrosine autophosphorylation sites within the extreme carboxyl-terminal region of EGFR (Y1045, Y1068 and Y1173). SMAD4 depletion did not affect autophosphorylation in the position Y1048 and Y1068, whereas phosphorylation at residue Y1173 was significantly increased in SCC61-SMAD4KD cells compared to SCC61-SC cells (Figure 3A). While there were no significant changes in AKT phosphorylation in SCC61-SMAD4KD cells, the levels of pSTAT3, p42/44 MAPK and pJNK were significantly upregulated following SMAD4 depletion (Figure 3B). Consistently, depletion of SMAD4 in SCC25 cell line has also resulted in a substantial upregulation of pMAPK and pJNK levels (Supplemental Figure 3C).
Figure 3. SMAD4 downregulation induces JNK and MAPK pathway activation.
SCC61-SC and SCC61-SMAD4KD cell lysates were analyzed by western blot for (A) phosphorylation status of key EGFR tyrosine residues, (B) AKT, STAT3, p42/44 MAPK, JNK activation, and (C) pro-survival (BCL-2, BCL-XL) and pro-apoptotic (BIM) markers. (D) Western blots confirming SMAD4 overexpression in FaDuL-SMAD4 cell line. (E) Colony formation of FaDu-mock vs. FaDu-SMAD4 in Matrigel colony formation assay quantified as (F) colony number and (G) total area. (H) Western blot showing EGFR, AKT, STAT3, MAPK and JNK activity as well as BCL-2 and BCL-XL proteins expression levels in FaDu-SMAD4 cells compared with control FaDu-mock cell line. Actin and GAPDH were probed as loading controls where indicated. (I) Matrigel colony formation assay was performed for 7 days with FaDu-SMAD4 or control FaDu-mock cells treated with PBS, 10nM, 100nM or 1000nM cetuximab (*p<0.05. **p<0.01).
Since computational analysis predicted the downregulation of pro-apoptotic signaling in SMAD4-low HNSCC tumors, we next tested whether SMAD4 depletion in SCC61 alters expression of pro- and anti-apoptotic markers. In accordance with our computational analysis, SMAD4 knockdown resulted in an increase of anti-apoptotic proteins BCL-2 and BCL-XL (Figure 3C), whereas protein expression level of BIM, a promoter of apoptosis, was significantly decreased in SCC61-SMAD4KD cells compared to control SCC61-SC cell line (Figure 3C).
To further evaluate the role of SMAD4 in HNSCC, we investigated the effect of stable SMAD4 overexpression in HNSCC FaDuL cell-line, which has a complete loss of SMAD4 due to a de novo truncation mutation (Figure 3D). A control cell line containing vector only was also created. SMAD4 overexpression in FaDu cells conferred potent long-term anti-proliferative effect, as seen by a marked reduction in the colony-forming ability of SMAD4 overexpressing FaDu-SMAD4 cells, compared to control FaDu-mock cell line (Figure 3E, 3F and 3G). Consistent with our OncoFinder result that SMAD4 depletion induces MAPK and JNK activation, reconstitution of the SMAD4 expression in FaDu cell line resulted in significantly lower p42/44 MAPK, pJNK and pSTAT3 phosphorylation (Figure 3H). Furthermore, FaDu-SMAD4 cells displayed lower pEGFR (Y1173), BCL-2 and BCL-XL levels compared to the control cell line (Figure 3H).
To test the effect of SMAD4 overexpression in vivo, FaDu-mock and FaDu-SMAD4 cells were injected into the oral tongue of athymic nude mice (5 mice per group) to generate orthotopic xenograft tumors. Engraftment of control FaDu-mock cell-line resulted in successful implantation in 100% of the animals, with metastasis to lymph node detected in 2 (40%) mice. Conversely, injection of FaDu-SMAD4 cells demonstrated a much lower take rate, with only 2 of 5 animals successfully developing tumors and showing no signs of metastatic lesion (Supplemental Table 3). Although the ability to conduct survival analysis was limited by low tumorigenicity of SMAD4 overexpressing cells, our data further supports the role of SMAD4 as a potent tumor suppressor gene in HNSCC. Notably, SMAD4 overexpression resulted in a significant increase in cetuximab sensitivity, as demonstrated by Matrigel colony formation assays performed with either FaDu-SMAD4 or control cell lines (Figure 3I). Taken together with the data revealed by pathway activation analysis, our observations suggest that SMAD4 may regulate cellular survival and proliferation dynamics by modulating activation of key positive regulators of HNSCC.
Targeted inhibition of JNK and MAPK pathways contributes to overcome cetuximab resistance
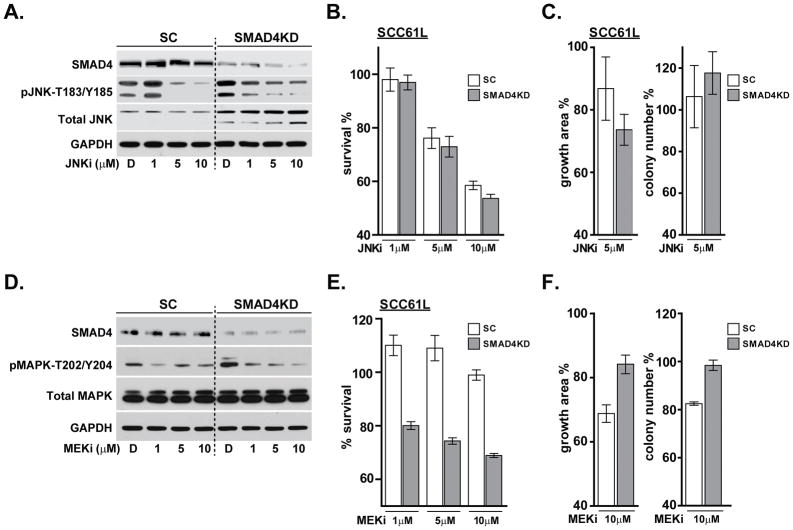
Based on the increased JNK and MAPK activation in SMAD4-depleted SCC61 cells, we targeted these kinases with small molecule inhibitors in SMAD4 knockdown and control cell lines. Cells were serum starved and then exposed to JNK inhibitor (SP600125) for 24 hours with serum stimulation. Western blot analysis demonstrated that SMAD4-depleted cells express higher total JNK protein compared to the SCC61-SC cells (Figure 4A). Notably, while JNK phosphorylation was efficiently inhibited by 5 μM and 10 μM SP600125 in SC cells, SMAD4KD cells displayed only a modest decrease in phospho-JNK levels even when treated with 10 μM SP600125 (Figure 4A). In order to examine the effect of JNK inhibition on cell viability, SCC61-SC and SCC61-SMAD4KD cells were cultured in the presence of JNK inhibitor or DMSO for 48 hours and MTS cell viability assay was performed. Although JNK inhibition resulted in a short term anti-proliferative effect in both cell lines (Figure 4B), a 7-day treatment with SP600125 did not significantly affect colony formation in a Matrigel colony growth assay (Figure 4C).
Figure 4. Small molecule inhibition of JNK and MAPK pathways in SCC61 cells.
SCC61-SC and SMAD4KD cells were serum starved for 24 hours and then incubated with DMSO or JNK inhibitor (SP600125) for 24 hours. Cell lysates were analyzed by (A) western blot for SMAD4, total and phosphorylated JNK (T183/Y185), and GAPDH as a loading control. (B) AlamarBlue assay measuring cell viability with JNKi or DMSO treatment for 48 hours. (C) Matrigel colony growth assay with 5uM JNK inhibitor for 7 days. SCC61-SC and SMAD4KD cells were serum starved and then incubated with either DMSO or MEK inhibitor (U0126) for 24 hours. Cell lysates were analyzed by (D) western blot for SMAD4, total and phosphorylated MAPK (T202/Y204), and GAPDH as a loading control. (E) AlamarBlue assay measuring cell viability with MEKi or DMSO treatment for 48 hours. (F) Clonogenic survival in response to 10uM MEKi was examined by the Matrigel colony growth assay for 7 days.
Similarly, to evaluate the effect of inhibiting the MAPK pathway, SCC61-SC and Smad4KD cells were synchronized by serum starvation and treated with a selective MAPK/MEK inhibitor U0126 or DMSO for 24 hours. Western blot analysis confirmed the increased phospho-MAPK level in SCC61-SMAD4KD cells compared to control (Figure 4D). U0126 treatment significantly lowered MAPK phosphorylation in control cells at 1 μM, whereas in SMAD4KD cells the best response was observed at 10 μM of U0126 (Figure 4D). While 48-hour exposure to U0126 selectively reduced survival of SMAD4KD cells over control at all inhibitor concentrations (Figure 4E), the long-term clonogenic survival of U0126-treated SCC61-SMAD4KD cells was no different or even higher than SCC61-SC (Figure 4F). Although SMAD4 reconstitution in FaDu cell line confirmed the decrease in pMAPK and pJNK levels and restored dose-dependent inhibition of JNK and MAPK activity in response to SP600125 or U0126 respectively (Supplemental Figure 4A and 4D), it failed to induce short term anti-proliferative benefit (Supplemental Figure 4B and 4E) and its impact on long-term clonogenic survival was significant, but not egregious (Supplemental Figure 4C and 4F). Multifaceted EGFR signaling orchestrates numerous pro-survival cellular processes (38). Therefore, activation of alternative EGFR downstream pathway targets that compensate for the loss of MAPK or JNK activity (i.e. adaptive rewiring) may provide one possible explanation for non-effectiveness of JNK and MAPK inhibitors as a monotherapy.
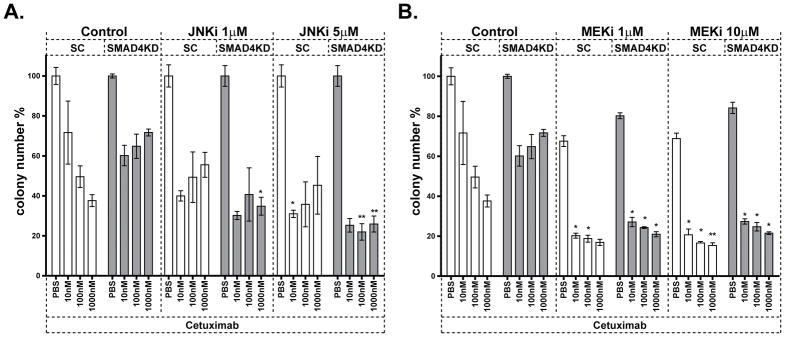
To examine if JNK or MAPK inhibitors may enhance the anti-proliferative effect of cetuximab, we treated SCC61-SC and SCC61-SMAD4KD cells with cetuximab in combination with either SP600125 or U0126. As expected, SCC61-SMAD4KD cells displayed resistance to cetuximab as a single agent compared to control cells (Figure 5). Notably, while both JNK and MAPK inhibitors successfully sensitized SMAD4KD cells to cetuximab, JNK inhibitor was considerably more effective in SCC61-KD cells compared to control SCC61-SC cell line (Figure 5). In alignment with these observations, inhibition of JNK and MAPK signaling effectively enhanced cetuximab sensitivity in SMAD4 null cell line (FaDu-mock), and further augmented its anti-proliferative activity in SMAD4 expressing FaDu-SMAD4 cells (Supplemental Figure 5). Taken together, our data indicate JNK and MAPK activation as potential mediators of cetuximab resistance and suggest the utility of concomitant EGFR/JNK or EGFR/MAPK inhibition as a novel strategy for overcoming cetuximab resistance in HNSCC tumors with low levels of SMAD4 expression.
Figure 5. JNK or MEK inhibitor sensitized SMAD4KD cells to cetuximab.
Matrigel colony formation assay of SCC61-SC and SMAD4KD cells treated with PBS or Cetuximab in combination with (A) JNKi (SP600125) or (B) MEKi (U0126) at indicated concentrations for 7 days. (*p<0.05 and **p<0.01 compared to SCC61-SC and SMAD4KD without MAPK/JNK inhibitor).
DISCUSSION
TGFβ is a multifunctional cytokine that is overexpressed in a majority of cancers (8–11). The high-affinity binding of TGFβ to TGFβ receptor II recruits TGFβ receptor I into a heterotetrameric complex that initiates SMAD-mediated transcriptional activation or repression of a network of genes that control cell growth, differentiation, and migration (14). While dual effects of TGFβ signaling on tumor initiation and progression are cell-specific and have yet to be determined under particular contexts, a number of studies have indicated that SMAD4 (a key downstream mediator of TGFβ signaling) plays crucial roles in maintaining tissue homeostasis and suppressing tumorigenesis (39–41). While a causal link between somatic inactivation of SMAD4 and tumor progression was observed in various solid malignancies, including HNSCC (23, 26, 42, 43), the molecular mechanisms underlying SMAD4 loss–associated HNSCC tumorigenesis are not yet fully elucidated.
We have previously shown that in HNSCC obtained from TCGA dataset, HPV-negative tumors are more likely to have lower SMAD4 expression than HPV-positive HNSCC (27). Furthermore, we showed that decreased SMAD4 expression induces HPV-negative tumor cells to undergo EMT and renders them resistant to the EGFR inhibitor cetuximab (27). Consistent with these observations, our analyses of HNSCC tumors obtained from newly diagnosed or recurrent patients revealed that lack of SMAD4 expression is highly associated with HPV-negative tumor status. Notably, the frequency of SMAD-negative tumors was significantly higher among recurrent HNSCC patients and strongly associated with distant metastasis development, further supporting the observation that in some cases loss of SMAD4 may facilitate a rapidly progressing and aggressive tumor phenotype (44–46). Moreover, recurrent and/or metastatic patients with SMAD4 negative tumors underwent significantly shorter cetuximab treatment than patients whose tumors expressed markedly higher SMAD4 levels, suggesting that loss of SMAD4 expression is a molecular signature that characterizes aggressive, cetuximab-resistant tumors. Further studies in a larger cohort of clinical samples are necessary to address the clinical significance of varying degrees of SMAD4 downregulation and functional SMAD4 insufficiency at a less stringent staining criterion.
The response to EGFR-targeted agents is inversely correlated with EMT in multiple types of tumors without known EGFR mutations, including NSCLC, head and neck, bladder, colorectal, pancreas and breast carcinomas (47–50). These data suggest that EMT is a common denominator of tumors that are resistant to EGFR inhibitors. Although the significance of SMAD4 downregulation in the induction of EMT was recently described in our previous study (27), a recent work in pancreatic cells reported a paradigm in which TGFβ-induced Smad4-dependent EMT perturbs a pro-tumorigenic transcriptional network resulting in apoptosis, which is prevented by Smad4 loss (51). While this study further confirms a pivotal role of SMAD4 in mediating EMT-associated signaling, the molecular mechanisms underlying this association in HNSCC have not yet been defined, and remain the subject of future research.
In the current investigation, we used the new bioinformatics software suite for the analysis of intracellular signaling pathway activation using transcriptomic data (29–31), for quantitative and qualitative comparison of the signaling pathway activation between SMAD4-low and SMAD4-high HNSCC tumors. Our comprehensive computational analysis has predicted that several oncogenic signaling pathways important for induction of the EMT phenotype in various solid malignancies, such as WNT (52), hGH (53, 54), and TGFβ (14, 52), as well as downstream pathway modulators such as RAS (52), MAPK (52), JNK (52), PAK/p38 (52), and mTOR (52), were significantly upregulated among the SMAD4-low tumors in both TCGA-HNSCC dataset and a subset of HPV-negative OSCC tumors obtained from GEO. These pro-survival signaling axes play a crucial role in cancer initiation, progression and maintenance, and may contribute to survival and promotion of cetuximab resistance via inhibition of apoptosis and induction of cell proliferation. In line with these observations, pro-apoptotic pathways were predicted to be significantly downregulated in most of the SMAD4-low tumors.
In accordance with the in-silico analysis, SMAD4 depletion induced cetuximab resistance in HPV-negative HNSCC tumor-derived human cells and was able to promote tumorigenicity in both in vitro and orthotopic in vivo models, further supporting the role of SMAD4 as a potent tumor suppressor in HNSCC, especially in HPV-negative tumors (27). Interestingly, the intracellular p53 regulatory network was predicted to be upregulated in tumors with decreased SMAD4 level. Since the viral oncoprotein E6 inactivates p53 in HPV-positive HNSCC tumors (55) and low SMAD4 expression was predominantly seen in HPV-negative cases (27), one possible explanation for the increased p53 signaling in SMAD4-low samples is that these patients may be TP53 WT, HPV-negative HNSCC, and SMAD4 loss is an alternative tumor suppressor functional loss to TP53. It was reported that mutation of TP53 occurs in 50% of HNSCC (21, 56), resulting in altered p53 expression and function. However, the differing nature and effects of these alterations on p53 expression and activation are frequently discordant. Therefore, additional studies are warranted to further define the role of SMAD4 functional loss in context of p53 signaling in tumorigenesis and therapeutic resistance of HNSCCs.
While dysregulation of signaling pathways downstream of EGFR is a common feature of EMT progression (57–59), many studies focusing on elucidating the role of EGFR signaling in EMT regulation often do not distinguish the specific phosphorylation site differences. Our data reveal that while Y1048 and Y1068 positions of EGFR are constitutively phosphorylated independent of SMAD4 status, phosphorylation at residue Y1173 was significantly upregulated in SMAD4-depleted cetuximab-resistant SCC61 cells, compared to SMAD4 expressing cetuximab-sensitive control. Subsequently, reconstitution of SMAD4 expression in FaDu cells resulted in a substantial decrease in Y1173 level. Interestingly, it was recently reported that phosphorylation at Y1173, which is a major target of EGFR kinase inhibitors (60), leads to the activation of MAPK/ERK signaling cascade and correlates with poor prognosis in patients with OSCC tumors (61). In accordance with our in-silico pathway activation analysis, phosphorylation of STAT3, MAPK and JNK was significantly upregulated following SMAD4 depletion. Although these results are consistent with EGFR activation at Y1173, which has been implicated in downstream activation of the MAPK/ERK cascade (62), additional studies are warranted to better understand how SMAD4 loss might influence site-specific phosphorylation changes of activated EGFR. Furthermore, a recent study reporting that SMAD4 may execute its tumor suppressive function via attenuation of JNK/MAPK activation (63), parallels our observation that pJNK level was significantly upregulated in SMAD4 depleted cells. Moreover, reconstitution of the SMAD4 expression in a SMAD4-negative cell line resulted in marked inhibition of cell growth and concomitant reduction in MAPK and JNK phosphorylation, suggesting that dysregulation of MAPK and JNK signaling may contribute to cetuximab resistance in subset of HPV-negative HNSCC tumors expressing low SMAD4 level.
Chemoresistance often ensues as a result of the concomitant activation of multiple, often overlapping signaling pathways. Therefore, for some cancers, inhibition of multiple, cross-talking pathways involved in cell growth and survival control with combination therapy is usually more effective in decreasing the likelihood that cancer cells will develop therapeutic resistance than with single agent therapy. Interestingly, while JNK or MEK inhibitors as standalone agents had limited effects on growth and survival of SMAD4-depleted cetuximab-resistant cells, dual targeted inhibition of the MEK or JNK signaling combined with cetuximab, significantly reduced colony formation ability of SMAD4 knockdown cells, highlighting the potential therapeutic benefit of concomitant EGFR and JNK or MAPK inhibition in HNSCC patients with low SMAD4 expression. Recent pre-clinical studies in patients with colorectal cancer and pancreatic ductal adenocarcinoma (64), have demonstrated that treatment with MEK inhibitors combined with EGFR-targeting agents are well tolerated and display modest antitumor activity. Since low SMAD4 expression is more prevalent in HPV-negative HNSCCs (27, 65), it is tempting to assume that the level of SMAD4 expression may act as a determinant of sensitivity/resistance to EGFR/MAPK or EGFR/JNK inhibition in HPV-negative tumors. Nevertheless, further exploration in a wider range of cell lines and tumor samples of HPV subgroups is warranted to determine the ability of SMAD4 to stratify the subset of patients whose tumors could respond and who would clinically benefit from the combination therapy.
In summary, our data implicate JNK and MAPK activation as mediators of cetuximab resistance and provide the foundation for the concomitant EGFR/JNK or EGFR/MAPK inhibition as a novel strategy for overcoming cetuximab resistance in a subtype of HNSCC tumors with loss of SMAD4 expression.
Supplementary Material
TRANSLATIONAL RELEVANCE.
HPV-negative HNSCC has poor prognosis with treatment resistance. Cetuximab, an anti-EGFR monoclonal antibody and the only FDA-approved targeted therapy for HNSCC, has limited efficacy due to development of resistance. There is an imminent need for new therapeutic strategies to circumvent the de novo or acquired resistance. We show that SMAD4 loss is associated with an aggressive, cetuximab-resistant phenotype in HPV-negative HNSCC patients, and that SMAD4 depletion results in cetuximab resistance in vitro and worse overall survival of tumor bearing mice in vivo. Using a bioinformatic analysis, we reveal a signature of pro-survival and anti-apoptotic pathways specifically dysregulated in SMAD4-low HNSCCs and indicate JNK and MAPK activation as potential mediators of cetuximab resistance. Our data suggest that SMAD4-low, HPV-negative HNSCCs will likely have poor cetuximab response and demonstrate the utility of concurrent EGFR and JNK/MAPK inhibition as a novel strategy for overcoming cetuximab resistance in tumors with loss of SMAD4 expression.
Acknowledgments
Financial support: This project was supported by NIH grants 5R21DE023430 and RO1 DE017982 to C. H. Chung, K25 CA141053 to E. J. Fertig, and SPORE P50 DE019032 to D. Sidransky. The terms of this arrangement are being managed by Johns Hopkins University in accordance with its conflict of interest policies.
Footnotes
Conflict of interest: CHC has received honoraria from Bristol Myers Squibb and Lilly Oncology for serving scientific advisory boards. All other authors declare no conflict of interest.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA: a cancer journal for clinicians. 2015;65:5–29. doi: 10.3322/caac.21254. [DOI] [PubMed] [Google Scholar]
- 2.Maier H, Dietz A, Gewelke U, Heller WD, Weidauer H. Tobacco and alcohol and the risk of head and neck cancer. Clin Investig. 1992;70:320–7. doi: 10.1007/BF00184668. [DOI] [PubMed] [Google Scholar]
- 3.D’Souza G, Kreimer AR, Viscidi R, Pawlita M, Fakhry C, Koch WM, et al. Case-control study of human papillomavirus and oropharyngeal cancer. N Engl J Med. 2007;356:1944–56. doi: 10.1056/NEJMoa065497. [DOI] [PubMed] [Google Scholar]
- 4.Ang KK, Harris J, Wheeler R, Weber R, Rosenthal DI, Nguyen-Tan PF, et al. Human papillomavirus and survival of patients with oropharyngeal cancer. The New England journal of medicine. 2010;363:24–35. doi: 10.1056/NEJMoa0912217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Leemans CR, Braakhuis BJ, Brakenhoff RH. The molecular biology of head and neck cancer. Nature reviews Cancer. 2011;11:9–22. doi: 10.1038/nrc2982. [DOI] [PubMed] [Google Scholar]
- 6.Burtness B. The role of cetuximab in the treatment of squamous cell cancer of the head and neck. Expert opinion on biological therapy. 2005;5:1085–93. doi: 10.1517/14712598.5.8.1085. [DOI] [PubMed] [Google Scholar]
- 7.Vermorken JB, Trigo J, Hitt R, Koralewski P, Diaz-Rubio E, Rolland F, et al. Open-label, uncontrolled, multicenter phase II study to evaluate the efficacy and toxicity of cetuximab as a single agent in patients with recurrent and/or metastatic squamous cell carcinoma of the head and neck who failed to respond to platinum-based therapy. J Clin Oncol. 2007;25:2171–7. doi: 10.1200/JCO.2006.06.7447. [DOI] [PubMed] [Google Scholar]
- 8.Derynck R, Goeddel DV, Ullrich A, Gutterman JU, Williams RD, Bringman TS, et al. Synthesis of messenger RNAs for transforming growth factors alpha and beta and the epidermal growth factor receptor by human tumors. Cancer research. 1987;47:707–12. [PubMed] [Google Scholar]
- 9.Dickson RB, Kasid A, Huff KK, Bates SE, Knabbe C, Bronzert D, et al. Activation of growth factor secretion in tumorigenic states of breast cancer induced by 17 beta-estradiol or v-Ha-ras oncogene. Proceedings of the National Academy of Sciences of the United States of America. 1987;84:837–41. doi: 10.1073/pnas.84.3.837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xu J, Lamouille S, Derynck R. TGF-beta-induced epithelial to mesenchymal transition. Cell research. 2009;19:156–72. doi: 10.1038/cr.2009.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bedi A, Chang X, Noonan K, Pham V, Bedi R, Fertig EJ, et al. Inhibition of TGF-beta enhances the in vivo antitumor efficacy of EGF receptor-targeted therapy. Molecular cancer therapeutics. 2012;11:2429–39. doi: 10.1158/1535-7163.MCT-12-0101-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gregory PA, Bracken CP, Smith E, Bert AG, Wright JA, Roslan S, et al. An autocrine TGF-beta/ZEB/miR-200 signaling network regulates establishment and maintenance of epithelial-mesenchymal transition. Molecular biology of the cell. 2011;22:1686–98. doi: 10.1091/mbc.E11-02-0103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu Q, Wang L, Li H, Han Q, Li J, Qu X, et al. Mesenchymal stem cells play a potential role in regulating the establishment and maintenance of epithelial-mesenchymal transition in MCF7 human breast cancer cells by paracrine and induced autocrine TGF-beta. International journal of oncology. 2012;41:959–68. doi: 10.3892/ijo.2012.1541. [DOI] [PubMed] [Google Scholar]
- 14.Massague J. TGFbeta signalling in context. Nature reviews Molecular cell biology. 2012;13:616–30. doi: 10.1038/nrm3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vincent T, Neve EP, Johnson JR, Kukalev A, Rojo F, Albanell J, et al. A SNAIL1-SMAD3/4 transcriptional repressor complex promotes TGF-beta mediated epithelial-mesenchymal transition. Nature cell biology. 2009;11:943–50. doi: 10.1038/ncb1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Adorno M, Cordenonsi M, Montagner M, Dupont S, Wong C, Hann B, et al. A Mutant-p53/Smad complex opposes p63 to empower TGFbeta-induced metastasis. Cell. 2009;137:87–98. doi: 10.1016/j.cell.2009.01.039. [DOI] [PubMed] [Google Scholar]
- 17.Zhang B, Halder SK, Kashikar ND, Cho YJ, Datta A, Gorden DL, et al. Antimetastatic role of Smad4 signaling in colorectal cancer. Gastroenterology. 2010;138:969–80. e1–3. doi: 10.1053/j.gastro.2009.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.White RA, Malkoski SP, Wang XJ. TGFbeta signaling in head and neck squamous cell carcinoma. Oncogene. 2010;29:5437–46. doi: 10.1038/onc.2010.306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim SK, Fan Y, Papadimitrakopoulou V, Clayman G, Hittelman WN, Hong WK, et al. DPC4, a candidate tumor suppressor gene, is altered infrequently in head and neck squamous cell carcinoma. Cancer research. 1996;56:2519–21. [PubMed] [Google Scholar]
- 20.Takebayashi S, Ogawa T, Jung KY, Muallem A, Mineta H, Fisher SG, et al. Identification of new minimally lost regions on 18q in head and neck squamous cell carcinoma. Cancer research. 2000;60:3397–403. [PubMed] [Google Scholar]
- 21.Agrawal N, Frederick MJ, Pickering CR, Bettegowda C, Chang K, Li RJ, et al. Exome sequencing of head and neck squamous cell carcinoma reveals inactivating mutations in NOTCH1. Science. 2011;333:1154–7. doi: 10.1126/science.1206923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xie W, Aisner S, Baredes S, Sreepada G, Shah R, Reiss M. Alterations of Smad expression and activation in defining 2 subtypes of human head and neck squamous cell carcinoma. Head & neck. 2013;35:76–85. doi: 10.1002/hed.22924. [DOI] [PubMed] [Google Scholar]
- 23.Voorneveld PW, Kodach LL, Jacobs RJ, Liv N, Zonnevylle AC, Hoogenboom JP, et al. Loss of SMAD4 alters BMP signaling to promote colorectal cancer cell metastasis via activation of Rho and ROCK. Gastroenterology. 2014;147:196–208. e13. doi: 10.1053/j.gastro.2014.03.052. [DOI] [PubMed] [Google Scholar]
- 24.Tascilar M, Skinner HG, Rosty C, Sohn T, Wilentz RE, Offerhaus GJ, et al. The SMAD4 protein and prognosis of pancreatic ductal adenocarcinoma. Clinical cancer research : an official journal of the American Association for Cancer Research. 2001;7:4115–21. [PubMed] [Google Scholar]
- 25.Yan P, Klingbiel D, Saridaki Z, Ceppa P, Curto M, McKee TA, et al. Reduced Expression of SMAD4 Is Associated with Poor Survival in Colon Cancer. Clinical cancer research : an official journal of the American Association for Cancer Research. 2016;22:3037–47. doi: 10.1158/1078-0432.CCR-15-0939. [DOI] [PubMed] [Google Scholar]
- 26.Bornstein S, White R, Malkoski S, Oka M, Han G, Cleaver T, et al. Smad4 loss in mice causes spontaneous head and neck cancer with increased genomic instability and inflammation. The Journal of clinical investigation. 2009;119:3408–19. doi: 10.1172/JCI38854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cheng H, Fertig EJ, Ozawa H, Hatakeyama H, Howard JD, Perez J, et al. Decreased SMAD4 expression is associated with induction of epithelial-to-mesenchymal transition and cetuximab resistance in head and neck squamous cell carcinoma. Cancer biology & therapy. 2015;16:1252–8. doi: 10.1080/15384047.2015.1056418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ozerov IV, Lezhnina KV, Izumchenko E, Artemov AV, Medintsev S, Vanhaelen Q, et al. In silico Pathway Activation Network Decomposition Analysis (iPANDA) as a method for biomarker development. Nature communications. 2016;7:13427. doi: 10.1038/ncomms13427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Buzdin AA, Zhavoronkov AA, Korzinkin MB, Venkova LS, Zenin AA, Smirnov PY, et al. Oncofinder, a new method for the analysis of intracellular signaling pathway activation using transcriptomic data. Frontiers in genetics. 2014;5:55. doi: 10.3389/fgene.2014.00055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Buzdin AA, Zhavoronkov AA, Korzinkin MB, Roumiantsev SA, Aliper AM, Venkova LS, et al. The OncoFinder algorithm for minimizing the errors introduced by the high-throughput methods of transcriptome analysis. Front Mol Biosci. 2014;1:8. doi: 10.3389/fmolb.2014.00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhu Q, Izumchenko E, Aliper AM, Makarev E, Paz K, Buzdin AA, et al. Pathway activation strength is a novel independent prognostic biomarker for cetuximab sensitivity in colorectal cancer patients. Human genome variation. 2015;2:15009. doi: 10.1038/hgv.2015.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen Y, Shi G, Xia W, Kong C, Zhao S, Gaw AF, et al. Identification of hypoxia-regulated proteins in head and neck cancer by proteomic and tissue array profiling. Cancer research. 2004;64:7302–10. doi: 10.1158/0008-5472.CAN-04-0899. [DOI] [PubMed] [Google Scholar]
- 33.Kong CS, Cao H, Kwok S, Nguyen CM, Jordan RC, Beaudry VG, et al. Loss of the p53/p63 target PERP is an early event in oral carcinogenesis and correlates with higher rate of local relapse. Oral surgery, oral medicine, oral pathology and oral radiology. 2013;115:95–103. doi: 10.1016/j.oooo.2012.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wilentz RE, Su GH, Dai JL, Sparks AB, Argani P, Sohn TA, et al. Immunohistochemical labeling for dpc4 mirrors genetic status in pancreatic adenocarcinomas : a new marker of DPC4 inactivation. The American journal of pathology. 2000;156:37–43. doi: 10.1016/S0002-9440(10)64703-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gross AM, Orosco RK, Shen JP, Egloff AM, Carter H, Hofree M, et al. Multi-tiered genomic analysis of head and neck cancer ties TP53 mutation to 3p loss. Nature genetics. 2014;46:939–43. doi: 10.1038/ng.3051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Parfenov M, Pedamallu CS, Gehlenborg N, Freeman SS, Danilova L, Bristow CA, et al. Characterization of HPV and host genome interactions in primary head and neck cancers. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:15544–9. doi: 10.1073/pnas.1416074111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lingen MW, Xiao W, Schmitt A, Jiang B, Pickard R, Kreinbrink P, et al. Low etiologic fraction for high-risk human papillomavirus in oral cavity squamous cell carcinomas. Oral oncology. 2013;49:1–8. doi: 10.1016/j.oraloncology.2012.07.002. [DOI] [PubMed] [Google Scholar]
- 38.Kandasamy K, Mohan SS, Raju R, Keerthikumar S, Kumar GS, Venugopal AK, et al. NetPath: a public resource of curated signal transduction pathways. Genome biology. 2010;11:R3. doi: 10.1186/gb-2010-11-1-r3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Takaku K, Miyoshi H, Matsunaga A, Oshima M, Sasaki N, Taketo MM. Gastric and duodenal polyps in Smad4 (Dpc4) knockout mice. Cancer Res. 1999;59:6113–7. [PubMed] [Google Scholar]
- 40.Tan X, Weng T, Zhang J, Wang J, Li W, Wan H, et al. Smad4 is required for maintaining normal murine postnatal bone homeostasis. J Cell Sci. 2007;120:2162–70. doi: 10.1242/jcs.03466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schwarte-Waldhoff I, Klein S, Blass-Kampmann S, Hintelmann A, Eilert C, Dreschers S, et al. DPC4/SMAD4 mediated tumor suppression of colon carcinoma cells is associated with reduced urokinase expression. Oncogene. 1999;18:3152–8. doi: 10.1038/sj.onc.1202641. [DOI] [PubMed] [Google Scholar]
- 42.Bardeesy N, Cheng KH, Berger JH, Chu GC, Pahler J, Olson P, et al. Smad4 is dispensable for normal pancreas development yet critical in progression and tumor biology of pancreas cancer. Genes Dev. 2006;20:3130–46. doi: 10.1101/gad.1478706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ding Z, Wu CJ, Chu GC, Xiao Y, Ho D, Zhang J, et al. SMAD4-dependent barrier constrains prostate cancer growth and metastatic progression. Nature. 2011;470:269–73. doi: 10.1038/nature09677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Inamoto S, Itatani Y, Yamamoto T, Minamiguchi S, Hirai H, Iwamoto M, et al. Loss of SMAD4 Promotes Colorectal Cancer Progression by Accumulation of Myeloid-Derived Suppressor Cells through the CCL15-CCR1 Chemokine Axis. Clinical cancer research : an official journal of the American Association for Cancer Research. 2016;22:492–501. doi: 10.1158/1078-0432.CCR-15-0726. [DOI] [PubMed] [Google Scholar]
- 45.Boone BA, Sabbaghian S, Zenati M, Marsh JW, Moser AJ, Zureikat AH, et al. Loss of SMAD4 staining in pre-operative cell blocks is associated with distant metastases following pancreaticoduodenectomy with venous resection for pancreatic cancer. Journal of surgical oncology. 2014;110:171–5. doi: 10.1002/jso.23606. [DOI] [PubMed] [Google Scholar]
- 46.Haeger SM, Thompson JJ, Kalra S, Cleaver TG, Merrick D, Wang XJ, et al. Smad4 loss promotes lung cancer formation but increases sensitivity to DNA topoisomerase inhibitors. Oncogene. 2016;35:577–86. doi: 10.1038/onc.2015.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yauch RL, Januario T, Eberhard DA, Cavet G, Zhu W, Fu L, et al. Epithelial versus mesenchymal phenotype determines in vitro sensitivity and predicts clinical activity of erlotinib in lung cancer patients. Clin Cancer Res. 2005;11:8686–98. doi: 10.1158/1078-0432.CCR-05-1492. [DOI] [PubMed] [Google Scholar]
- 48.Thomson S, Buck E, Petti F, Griffin G, Brown E, Ramnarine N, et al. Epithelial to mesenchymal transition is a determinant of sensitivity of non-small-cell lung carcinoma cell lines and xenografts to epidermal growth factor receptor inhibition. Cancer Res. 2005;65:9455–62. doi: 10.1158/0008-5472.CAN-05-1058. [DOI] [PubMed] [Google Scholar]
- 49.Adam L, Zhong M, Choi W, Qi W, Nicoloso M, Arora A, et al. miR-200 expression regulates epithelial-to-mesenchymal transition in bladder cancer cells and reverses resistance to epidermal growth factor receptor therapy. Clin Cancer Res. 2009;15:5060–72. doi: 10.1158/1078-0432.CCR-08-2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Barr S, Thomson S, Buck E, Russo S, Petti F, Sujka-Kwok I, et al. Bypassing cellular EGF receptor dependence through epithelial-to-mesenchymal-like transitions. Clin Exp Metastasis. 2008;25:685–93. doi: 10.1007/s10585-007-9121-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.David CJ, Huang YH, Chen M, Su J, Zou Y, Bardeesy N, et al. TGF-beta Tumor Suppression through a Lethal EMT. Cell. 2016;164:1015–30. doi: 10.1016/j.cell.2016.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lamouille S, Xu J, Derynck R. Molecular mechanisms of epithelial-mesenchymal transition. Nature reviews Molecular cell biology. 2014;15:178–96. doi: 10.1038/nrm3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Subramani R, Lopez-Valdez R, Salcido A, Boopalan T, Arumugam A, Nandy S, et al. Growth hormone receptor inhibition decreases the growth and metastasis of pancreatic ductal adenocarcinoma. Experimental & molecular medicine. 2014;46:e117. doi: 10.1038/emm.2014.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang W, Qian P, Zhang X, Zhang M, Wang H, Wu M, et al. Autocrine/Paracrine Human Growth Hormone-stimulated MicroRNA 96-182-183 Cluster Promotes Epithelial-Mesenchymal Transition and Invasion in Breast Cancer. The Journal of biological chemistry. 2015;290:13812–29. doi: 10.1074/jbc.M115.653261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Scheffner M, Werness BA, Huibregtse JM, Levine AJ, Howley PM. The E6 oncoprotein encoded by human papillomavirus types 16 and 18 promotes the degradation of p53. Cell. 1990;63:1129–36. doi: 10.1016/0092-8674(90)90409-8. [DOI] [PubMed] [Google Scholar]
- 56.Poeta ML, Manola J, Goldwasser MA, Forastiere A, Benoit N, Califano JA, et al. TP53 mutations and survival in squamous-cell carcinoma of the head and neck. N Engl J Med. 2007;357:2552–61. doi: 10.1056/NEJMoa073770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yadav A, Kumar B, Datta J, Teknos TN, Kumar P. IL-6 promotes head and neck tumor metastasis by inducing epithelial-mesenchymal transition via the JAK-STAT3-SNAIL signaling pathway. Mol Cancer Res. 2011;9:1658–67. doi: 10.1158/1541-7786.MCR-11-0271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mulholland DJ, Kobayashi N, Ruscetti M, Zhi A, Tran LM, Huang J, et al. Pten loss and RAS/MAPK activation cooperate to promote EMT and metastasis initiated from prostate cancer stem/progenitor cells. Cancer Res. 2012;72:1878–89. doi: 10.1158/0008-5472.CAN-11-3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lo HW, Hsu SC, Xia W, Cao X, Shih JY, Wei Y, et al. Epidermal growth factor receptor cooperates with signal transducer and activator of transcription 3 to induce epithelial-mesenchymal transition in cancer cells via up-regulation of TWIST gene expression. Cancer Res. 2007;67:9066–76. doi: 10.1158/0008-5472.CAN-07-0575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Morgan MA, Parsels LA, Kollar LE, Normolle DP, Maybaum J, Lawrence TS. The combination of epidermal growth factor receptor inhibitors with gemcitabine and radiation in pancreatic cancer. Clinical cancer research : an official journal of the American Association for Cancer Research. 2008;14:5142–9. doi: 10.1158/1078-0432.CCR-07-4072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Monteiro L, Ricardo S, Delgado M, Garcez F, do Amaral B, Lopes C. Phosphorylated EGFR at tyrosine 1173 correlates with poor prognosis in oral squamous cell carcinomas. Oral diseases. 2014;20:178–85. doi: 10.1111/odi.12087. [DOI] [PubMed] [Google Scholar]
- 62.Sturla LM, Amorino G, Alexander MS, Mikkelsen RB, Valerie K, Schmidt-Ullrichr RK. Requirement of Tyr-992 and Tyr-1173 in phosphorylation of the epidermal growth factor receptor by ionizing radiation and modulation by SHP2. The Journal of biological chemistry. 2005;280:14597–604. doi: 10.1074/jbc.M413287200. [DOI] [PubMed] [Google Scholar]
- 63.Zhang X, Cao J, Pei Y, Zhang J, Wang Q. Smad4 inhibits cell migration via suppression of JNK activity in human pancreatic carcinoma PANC-1 cells. Oncology letters. 2016;11:3465–70. doi: 10.3892/ol.2016.4427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ko AH, Bekaii-Saab T, Van Ziffle J, Mirzoeva OM, Joseph NM, Talasaz A, et al. A Multicenter, Open-Label Phase II Clinical Trial of Combined MEK plus EGFR Inhibition for Chemotherapy-Refractory Advanced Pancreatic Adenocarcinoma. Clin Cancer Res. 2016;22:61–8. doi: 10.1158/1078-0432.CCR-15-0979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Baez A, Cantor A, Fonseca S, Marcos-Martinez M, Mathews LA, Muro-Cacho CA, et al. Differences in Smad4 expression in human papillomavirus type 16-positive and human papillomavirus type 16-negative head and neck squamous cell carcinoma. Clinical cancer research : an official journal of the American Association for Cancer Research. 2005;11:3191–7. doi: 10.1158/1078-0432.CCR-04-1299. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.