Abstract
Degenerative diseases of organs lead to their impaired function. The cellular and molecular mechanisms underlying organ degeneration are therefore of great research and clinical interest but are currently incompletely characterized. Here, using a forward-genetic screen for genes regulating liver development and function in zebrafish, we identified a cq5 mutant that exhibited a liver-degeneration phenotype at 5 days postfertilization, the developmental stage at which a functional liver develops. Positional cloning revealed that the liver degeneration was caused by a single point mutation in the gene zc3h8 (zinc finger CCCH-type containing 8), changing a highly conserved histidine to glutamine at position 353 of the Zc3h8 protein. The zc3h8 mutation–induced liver degeneration in the mutant was accompanied by reduced proliferation, increased apoptosis, and macrophage phagocytosis of hepatocytes. Transcriptional profile analyses revealed up-regulation and activation of both proinflammatory cytokines and the NF-κB signaling pathway in the zc3h8 mutant. Suppression of NF-κB signaling activity efficiently rescued the proinflammatory cytokine response, as well as the inflammation-mediated liver degeneration phenotype of the mutant. Of note, the zc3h8 mutation-induced degeneration of several other organs, including the gut and exocrine pancreas, indicating that Zc3h8 is a general repressor of inflammation in zebrafish. Collectively, our findings demonstrate that Zc3h8 maintains organ homeostasis by inhibiting the NF-κB–mediated inflammatory response in zebrafish and that Zc3h8 dysfunction causes degeneration of multiple organs, including the liver, gut, and pancreas.
Keywords: cell signaling, inflammation, NF-κ B (NF-κB), zebrafish, zinc finger, degeneration, digestive organ
Introduction
The liver is a crucial digestive organ with a central role in metabolic homeostasis. Hepatocytes constitute the majority of the liver and perform several critical functions, including the production of bile, storage of glycogen, detoxification, and regulation of blood homeostasis (1–3). Degenerative diseases involve a continuous process of cell degeneration, which affects tissue or organ function, with increasing deterioration occurring over time. Nonreversible damage to liver function results in the death of the affected organism. Thus, a better understanding of the molecular mechanisms involved in liver degeneration is essential for the development of novel drug therapies and cures for these diseases.
Inflammation is triggered by microbial pathogens or danger signals derived from the host. The receptors of antigen-presenting cells sense microbial products (4) and endogenous signals released by damaged cells (5), leading to their activation, and subsequently initiate an inflammatory response. Previous studies (6, 7) have shown that inflammation is a major cause of most chronic liver diseases, and inhibition of the activation of inflammasomes (multiprotein complexes that recognize danger signals from damaged cells and pathogens (8)) could ameliorate inflammation-related diseases. The transcription factor NF-κB is a major factor that mediates and regulates the expression of many genes involved in inflammation (9). Inactive NF-κB is located in the cytoplasm. Once it becomes phosphorylated and activated, it translocates to the nucleus, where it binds to the promoters of specific target genes and can up-regulate the transcription of cytokines and chemokines.
Zebrafish have been recognized as a powerful model organism for the study of liver development and regeneration because of their embryological and genetic advantages (10–14). In this study, we used forward genetic screening to identify genes that regulate liver development and regeneration in zebrafish. Interestingly, we found that the cq5 mutant exhibited severe liver degeneration defects at later stages of development (after 5 days postfertilization (dpf))2 and a shorter life span. Positional cloning revealed that the zinc finger protein Zc3h8 (zinc finger CCCH-type containing 8) was mutated and contributed to the cq5 mutant degenerative phenotype. Further analysis showed that liver degeneration in the cq5 mutant was accompanied by a proinflammatory response and activation of the NF-κB pathway, whose inhibition efficiently rescued hepatocyte degeneration in the mutant. These findings suggest a repressor role of Zc3h8 in liver homeostasis during the inflammatory response in zebrafish and may provide a new avenue for research on and treatment of degenerative diseases in humans.
Results
The cq5 mutant exhibits liver degeneration
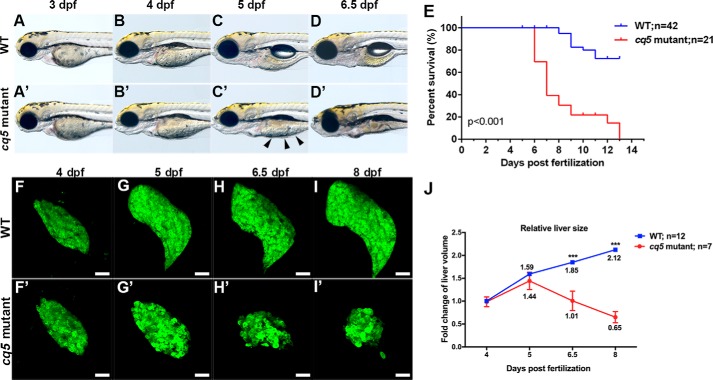
Abnormal liver development of the cq5 mutant was identified in a zebrafish N-ethyl-N-nitrosourea forward genetic screen under the Tg(fabp10a:Dendra2-NTR)cq1 background (10) for genes regulating liver development and regeneration. Homozygous cq5 mutants did not show any observable phenotype before 4 dpf (Fig. 1, A, A′, B, and B′) but exhibited a large amount of unabsorbed yolk at 5 dpf (Fig. 1, C and C′). Survival curves showed that the survival rate of the mutants decreased dramatically from 6 dpf on, with all the mutants dead at 13 dpf (Fig. 1E).
Figure 1.
Cq5 is a liver degeneration mutant. A–D and A′–D′, WT (n = 14) and cq5 mutant (n = 18) embryos were imaged from 3 to 6.5 dpf; unabsorbed yolk is indicated by black arrowheads. E, cumulative survival of cq5 mutant larvae (n = 21) and WT siblings (n = 42). p < 0.001 by the log-rank Mantel-Cox test. F–I and F′–I′, the cq5 mutants showed significant liver degeneration (57 of 59), as normalized to the WT (62 of 62). Scale bar, 50 μm. J, the relative fold change of liver size was measured in WT (n = 12) and mutant (n = 7) larvae from 4 to 8 dpf. Liver volume was measured with Imaris software and normalized to the liver size of WT larvae at 4 dpf. The error bars represent S.E. ***, p < 0.001 by Student's t test.
At 4 dpf, the liver of the mutant remained normal (Fig. 1, F and F′). From 4 to 5 dpf, although the mutant liver continued growing, its outgrowth rate was slower than the WT fish (Fig. 1, F, F′, G, and G′). The cq5 mutant subsequently exhibited degeneration of hepatocytes (Fig. 1, H, H′, I, and I′), displaying a one-third volume of the WT liver at 8 dpf (Fig. 1J). These results demonstrate that mutation of the cq5 mutation is ineffective to the early liver development but does impair liver growth and homeostasis, thus leading to liver degeneration after 4 dpf.
The cq5 mutant phenotypes are caused by the mutation of zc3h8 encoding a CCCH-type zinc finger protein
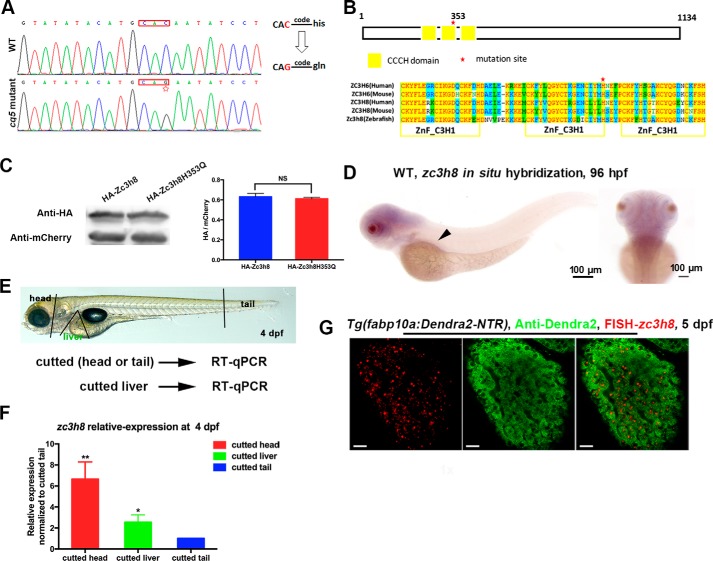
Positional cloning identified that the mutation was localized in three candidate genes, fbln7, zc3h8, and itga9, flanked by the markers 1006 and 1023 on the linkage group 13 (LG13) (Fig. S1, A and B). Through sequence analyses of these three candidate cDNAs, we identified a single point mutation in zc3h8 (Fig. 2A). This C-to-G transition generated an amino acid switch from histidine to glutamine at position 353 (Fig. 2A). ZC3H8 is a member of the CCCH-type zinc finger-containing protein family (15, 16), which exhibit broadly reported functions in maintaining the physiological metabolism of organisms (17–22). Collectively, the results of synteny analyses and homologous protein sequence alignment of Zc3h8 in different species (Fig. S1, B and C) revealed that the proteins in all species contained three conserved CCCH-type functional domains, and the mutated histidine in the cq5 mutant was evolutionarily conserved (Fig. 2B).
Figure 2.
zc3h8 is required for the maintenance of liver homeostasis. A, sequencing of zc3h8 from cq5 mutant and WT embryos identified a C-to-G point mutation (red star marking the mutation site), resulting in a coding change from histidine to glutamine at position 353. B, amino acid sequence alignment of Zc3h8 homologs/orthologs according to the three conserved zinc finger domains (red star marking the cq5 amino acid mutation site, ZnF_C3H1 domain indicated by yellow frame) in human, mouse, and zebrafish. C, Western blotting of FASC sorted mCherry-positive cells showing that mutation of cq5 (Zc3h8H353Q) does not alter protein stability in zebrafish embryos. mCherry levels were used as loading controls. Ratio of the anti-HA with anti-mCherry densitometry was used as the relative repression level. The error bars represent S.D. N.S. represents no significance and p value by Student's t test. D, zc3h8 expression pattern in WT embryos determined via whole-mount in situ hybridization at 96 hpf. Arrowheads indicate the region of digestive organ expression. E, scheme for determining the expression levels of zc3h8 in different regions in WT larvae via qPCR. F, quantitative real-time PCR analysis of zc3h8 expression in different areas (head, liver, and tail) in WT embryos at 4 dpf. The error bars indicate S.E. *, p < 0.05; **, p < 0.01 by Student's t test. G, double fluorescence labeling of zc3h8 RNA probe and anti-Dendra2 antibody staining at 5 dpf. Note the zc3h8 transcripts expressed in the hepatocytes. Scale bars, 20 μm.
To validate that the cq5 liver degeneration phenotype was caused by the zc3h8 mutation, we generated a Tg(hsp70l:HA-zc3h8-T2A-mCherry)cq43 transgenic line (hereafter referred to as hs:zc3h8) (Fig. S2A), which permitted visualization of red fluorescence after heat shock. Heat shock–induced overexpression of Zc3h8 in the cq5 mutant efficiently rescued the degenerated liver at 6.5 dpf (Fig. S2, B and C). Furthermore, hepatocyte-specific replenishment of WT Zc3h8 using the Tg(fabp10a:HA-zc3h8-T2A-mCherry)cq44 (referred to as fabp10a:zc3h8) transgenic background also efficiently rescued the degenerated liver in the cq5 mutant (Fig. S2, D–H). These results confirm that the liver phenotypes of the cq5 mutant caused by the zc3h8 mutation.
Further confirmation was carried out using the transcription activator-like effector nuclease (TALEN) technology (23) to generate a zc3h8 mutant. Exon 7 was selected as the target of TALEN nuclease, where the cq5 mutation is located (Fig. S3A). Two independent zc3h8 mutant alleles were generated. One allele harbored a 3-bp deletion and a 8-bp insertion (referred to as zc3h8inscq45), leading to disrupted protein sequence from amino acid 351 and a premature stop codon at amino acid 382. The other allele harbored a 10-bp deletion (referred to as zc3h8Δ10cq46), leading to disrupted protein sequence from amino acid 351 and a premature stop codon at amino acid 377 (Fig. S3, B and C). Both zc3h8ins5cq45 and zc3h8Δ10cq46 showed liver degeneration at 6.5 dpf, similar to the cq5 mutant (Fig. S3, D–G). These data validate that the liver degeneration phenotype of cq5 mutant is caused by the zc3h8 mutation.
To determine whether the histidine to glutamine switch of Zc3h8 affects the protein stability, the heat shock promoter-driven HA-zc3h8H353Q-T2A-mCherry plasmid that contained the same mutation as the cq5 mutant, as well as the WT HA-zc3h8-T2A-mCherry plasmid, were generated. Similar expression levels of the WT and mutated proteins (Fig. 2C) indicated that the point mutation was ineffective to protein stability of Zc3h8. In situ hybridization and qPCR of zc3h8 showed strong expressions in the brain and eyes and relatively weak expressions in the digestive system at 96 hpf (Fig. 2, D–F). Fluorescent in situ hybridizations (FISHs) confirmed the expression of zc3h8 in hepatocytes (Fig. 2G). These data indicate that zc3h8 is required for the maintenance of liver homeostasis.
Zc3h8 mutant liver degeneration is accompanied by reduced proliferation and increased apoptosis of hepatocytes
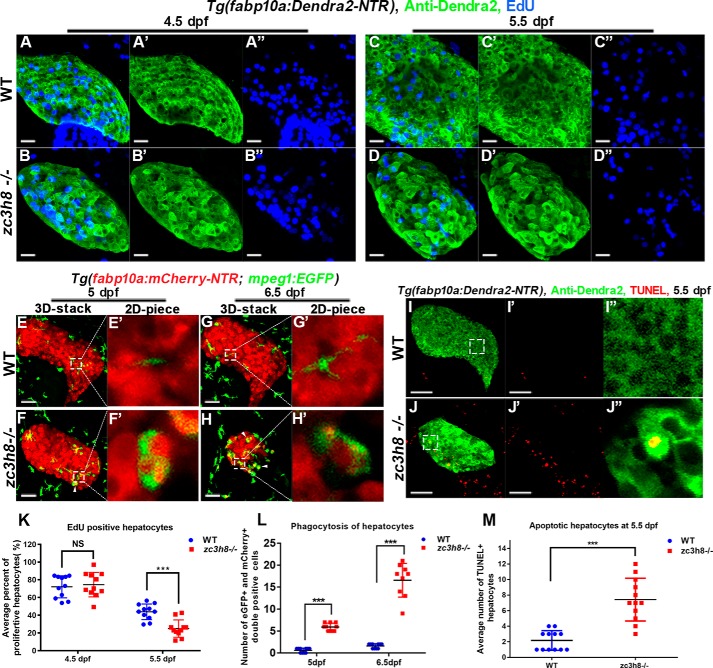
The balance of cell proliferation and apoptosis affects tissue development and determines organ size (24). Many previously reported mutants exhibiting smaller livers show low proliferation of hepatocytes (25–27). To determine whether the liver-degenerative phenotype of the cq5 mutant was the result of abnormal cell proliferation or not, EdU incorporation ratios were analyzed. The proliferation rates of mutant hepatocytes remained similar to the control at 4.5 dpf (Fig. 3, A, A′, A″, B, B′, and B″) but were significantly reduced at 5.5 dpf (Fig. 3, C, C′, D, D″, and K).
Figure 3.
Reduced proliferation and increased apoptosis of hepatocytes are associated with zc3h8 mutation. A–D and A′–D″, confocal images of EdU and anti-Dendra2 antibody labeling in WT and mutant larvae from 4.5 (29 of 29, 25 of 27) to 5.5 dpf (28 of 29, 25 of 28). Scale bars, 20 μm. E–H and E′–H′, Tg(fabp10a:mCherry-NTRcq2;mpeg1:EGFP) double-transgenic line fish were visualized via confocal microscopy from 5 (E-E′ and F-F′; 53 of 53, 48 of 52) to 6.5 dpf (G-G′ and H-H′; 54 of 54, 55 of 59) for WT and zc3h8 mutant larvae. White arrowheads highlight macrophages engulfing degenerated hepatocytes. Scale bars, 20 μm. I, I′, J, and J″, confocal images of TUNEL-labeled apoptotic cells in zc3h8 mutant and WT livers at 5.5 dpf (38 of 40, 42 of 45). Scale bars, 50 μm. K, quantification of EdU-labeled hepatocytes in the zc3h8 mutant and the WT at 4.5 dpf (n = 5, n = 9) and 5.5 dpf (n = 8, n = 8). The error bars indicate S.D. N.S. represents no significance. ***, p < 0.001 by Student's t test. L, quantification of macrophages engulfing degenerated hepatocytes in zc3h8 mutant and WT larvae at 5 dpf (n = 10, n = 10) and 6.5 dpf (n = 9, n = 10). The error bars indicate S.D. ***p < 0.001 by Student's t test. M, quantification of TUNEL-positive hepatocytes in zc3h8 mutant and WT (n = 12) larvae at 5.5 dpf. The error bars indicate S.D. ***, p < 0.001 by Student's t test.
Macrophages play important roles in inflammation, recognition, and phagocytosis of apoptotic cells, as well as tissue repair and regeneration (28). To investigate roles of macrophages in the liver degeneration process, the Tg(fabp10a:mCherry-NTR)cq2 and Tg(mpeg1:EGFP) transgenic backgrounds were applied. Macrophage recruitment to the mutant liver and engulfment of hepatocytes became observed at 5 dpf (Fig. 3, E, E′, F, and F′), which were significantly increased at 6.5 dpf (Fig. 3, G, G′, H, H′, and L). TUNEL assays showed that apoptotic hepatocytes significantly increased in the mutant at 5.5 dpf (Fig. 3, I, I′, I″, J, J′, J″, and M). These data suggest that liver degeneration in the cq5 mutant is contributed by both of the decreased hepatocyte proliferation and increased hepatocyte apoptosis.
Blockade of inflammation and NF-κB signaling rescue liver degeneration in the mutant
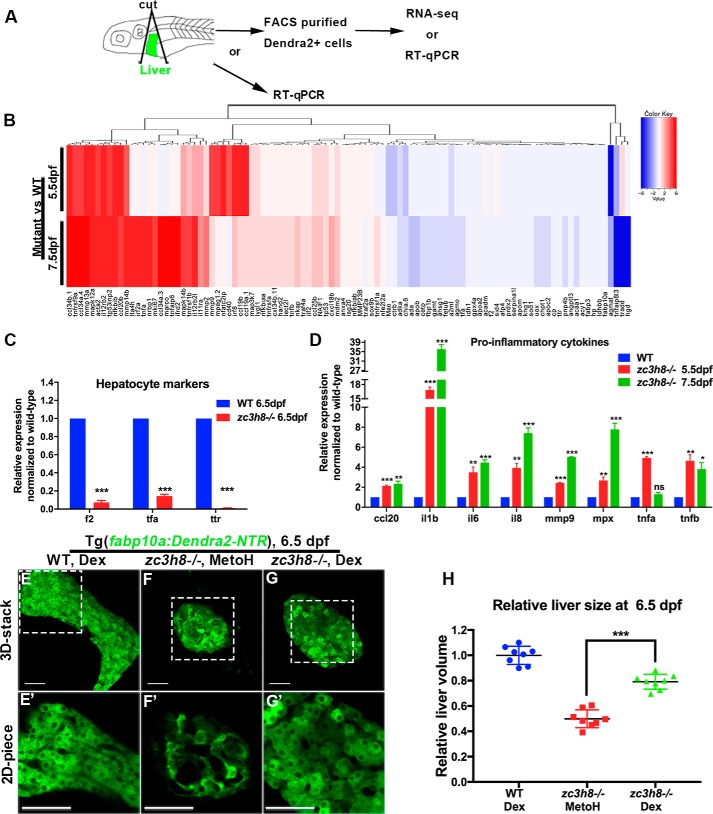
To explore the regulatory molecules and signaling pathways involved in mutant liver degeneration, we dissected the liver and sorted the Dendra2-positive hepatocytes at two different stages (middle stage at 5.5 dpf and late stage at 7.5 dpf) using the Tg(fabp10a:Dendra2-NTR)cq1 transgenic background (Fig. 4A). Transcriptional profile analyses revealed up-regulation of inflammation-related molecules in the mutant hepatocytes at both middle and late degenerative stages (Fig. 4B). qPCRs verified that expressions of hepatocyte-specific markers (f2, tfa, and ttr) significantly decreased in the zc3h8 mutant (Fig. 4C), whereas expressions of inflammation-related genes increased at both 5.5 and 7.5 dpf (Fig. 4D). These data demonstrated activation of inflammatory responses in the mutant. Treatment of ani-inflammatory drug dexamethasone (Dex) from 4 to 6 dpf partially rescued liver degeneration in the mutant (Fig. 4, E–H), suggesting the inflammatory response as one of the inducers of liver degeneration.
Figure 4.
zc3h8 mutant hepatocyte degeneration is inflammatory response–dependent. A, schematic representation of the process of transcriptome sequencing or qPCR analysis of sorted hepatocytes and direct qPCR analysis of liver tissues from WT and zc3h8 mutants. B, heat map of the log10-transformed expression of statistically significant, differentially expressed genes between sorted hepatocytes of WT and zc3h8 mutants. Red indicates up-regulation, and blue indicates down-regulation of mRNA compared with WT. C, quantitative real-time PCR of hepatocyte-specific markers for sorted hepatocytes from zc3h8 mutant and WT larvae at 6.5 dpf. The error bars represent S.E. *, p < 0.05; **, p < 0.01; ***, p < 0.001 by Student's t test. D, quantitative real-time PCR analysis of proinflammatory cytokine expression in liver tissues at 5.5 and 7.5 dpf. The error bars represent S.E. ns represents no significance. *, p < 0.05; **, p < 0.01; ***, p < 0.001 by Student's t test. E–G and E′–G′, fluorescence confocal microscopy of Tg(fabp10a:Dendra2-NTR)cq1 in zc3h8 mutant (31 of 44) and WT (49 of 56) livers treated with Dex or methanol (56 of 56) at 6.5 dpf. Note that the zc3h8 mutant degeneration phenotype could be partially rescued by treatment with the anti-inflammatory drug Dex. Scale bars, 50 μm. H, quantification effects of Dex on zc3h8 mutant degeneration liver at 6.5 dpf. The error bars represent S.D. ***, p < 0.001 by Student's t test.
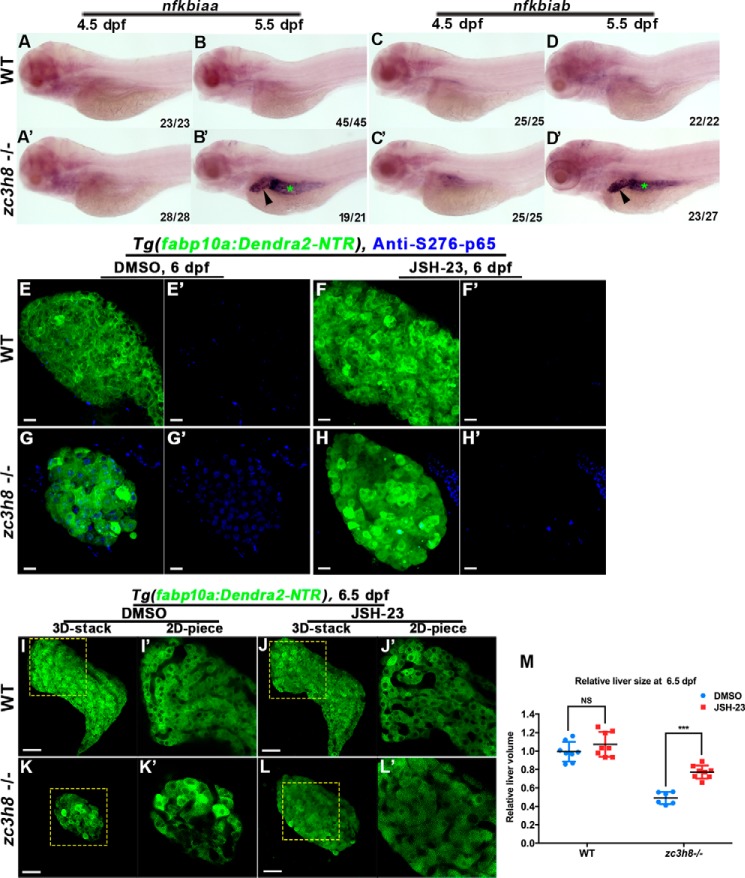
NF-κB plays important roles in innate and adaptive responses, inflammation, proliferation, and cellular differentiation (29–31). Inactive NF-κB is located in the cytoplasm. Once it is phosphorylated and activated, NF-κB translocates into the nucleus and up-regulates the transcription of cytokines and chemokines (32, 33). To investigate whether the inflammation-induced liver degeneration is mediated by the NF-κB activities, expressions of the active NF-κB components nfkbiaa and nfkbiab were first analyzed. Transcriptional activations of nfkbiaa and nfkbiab were undetectable at 4.5 dpf but became significantly increased at 5.5 dpf (Fig. 5, A, A′, B, B′, C, C′, D, and D′). To obtain additional evidences for the increased presence of activated NF-κB in degenerating hepatocytes, immunostainings of the active NF-κB component, phosphorylated p65 (S276-p65), were performed. Although the hepatic S276-p65 signals in the mutant at 4 dpf remained similar to the WT, nuclear accumulation of S276-p65 steadily increased from 4 to 6 dpf in the mutant liver (Fig. S4, A–F and A′–F′, and Fig. 5, G and G′). Because the number of apoptotic hepatocytes remained unchanged in the mutant at 4.5 dpf (Fig. S4, G–M), activation of the NF-κB signaling occurred prior to the hepatocyte apoptosis in the mutant.
Figure 5.
The NF-κB signaling pathway is activated and contributes to liver degeneration in zc3h8 mutants. A–D and A–D′, whole-mount in situ hybridization showed increased expression of NF-κB components (nfkbiaa and nfkbiab) in the livers (arrowhead) and intestines (green asterisks) of zc3h8 mutant larvae at 4.5 and 5.5 dpf. E–H and E′–H′, antibody staining of phosphorylated p65 (S273-p65) in Tg(fabp10a:Dendra2-NTR)cq1 zc3h8 mutant (38 of 43) and WT (45 of 47) larvae treated with DMSO and an NF-κB inhibitor (JSH-23) (zc3h8 mutant (28 of 33) and WT (32 of 32)) at 6 dpf. Scale bars, 20 μm. I–L and I′–L′, fluorescence confocal microscopy of Tg(fabp10a:Dendra2-NTR)cq1 in zc3h8 mutant (17 of 20) and WT (24 of 24) livers treated with an NF-κB inhibitor (JSH-23) and DMSO (zc3h8 mutant (17 of 19) and WT (15 of 17)) at 6.5 dpf. Note that the mutant degeneration phenotype could also be partially rescued by treatment with an NF-κB signaling pathway inhibitor. Scale bars, 50 μm. M, quantification effects of JSH-23 on zc3h8 mutant degeneration liver at 6.5 dpf. The error bars represent S.D. NS represents no significance. ***, p < 0.001 by Student's t test.
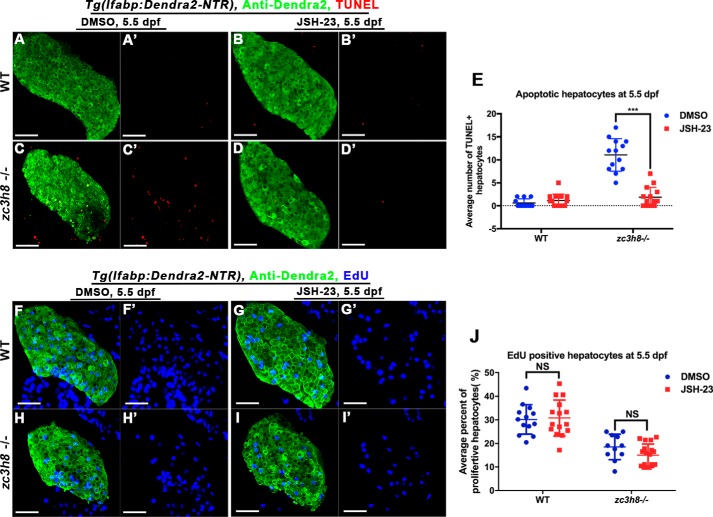
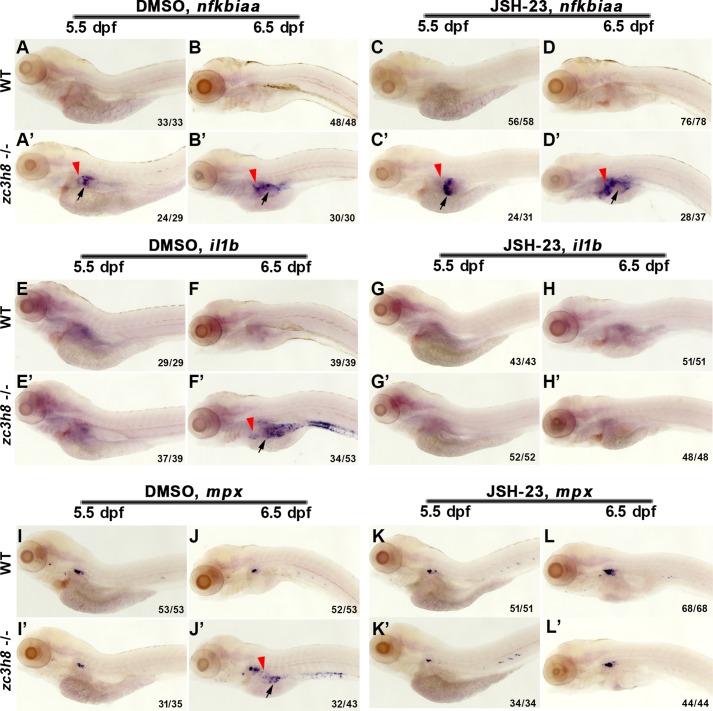
Blockade of NF-κB signaling using a chemical inhibitor JSH-23 (34) reduced the nuclear accumulation of S276-p65 (Fig. 5, F–H and F′–H′), resulting in partial rescue of degenerated liver and even full rescue of the increased hepatocyte apoptosis in the mutant (Figs. 5, I–M, and 6, A–E). The reduced proliferation of mutant hepatocytes was not rescued by the treatment of JSH-23 (Fig. 6, F–J). JSH-23 treatment was ineffective to the activation of NF-κB component nfkbiaa in the mutant (Fig. 7, C, C′, D, and D′). However, activations of il1b and mpx at 6.5 dpf in the mutant were efficiently repressed by the JSH-23 (Fig. 7, G, G′, H, H′, K, K′, L, and L′). These data indicate that the NF-κB–mediated inflammation acts as one of the inducers of liver degeneration caused by zc3h8 deficiency.
Figure 6.
JSH-23 rescues zc3h8 mutant hepatocyte apoptosis, but not proliferation. A–D and A′–D′, double fluorescence labeling with TUNEL and anti-Dendra2 antibody staining in control larvae (n = 23) or larvae treated with JSH-23 (n = 25) at 5.5 dpf. Scale bars, 50 μm. E, quantification of apoptotic hepatocytes in livers from zc3h8 mutant and WT larvae (n = 13) treated with JSH-23. The error bars indicate S.D. ***, p < 0.001 by Student's t test. F–I and F′–I′, double labeling with EdU and anti-Dendra2 antibody staining in WT and zc3h8 mutant larvae (28 of 32 and 24 of 28) or larvae treated with JSH-23 (23 of 28 and 19 of 22) at 5.5 dpf. Scale bars, 50 μm. J, quantification of hepatocyte proliferation in livers from zc3h8 mutant and WT larvae (n = 13) treated with JSH-23. The error bars indicate S.D. NS represents no significance, p value test by Student's t test.
Figure 7.
Uncontrolled inflammation is induced by activated NF-κB signaling. A–D and A′–D′, whole-mount in situ hybridization of an NF-κB component gene (nfkbiaa) in WT and zc3h8 mutant larvae at 5.5 and 6.5 dpf after pretreatment with DMSO or JSH-23. Red arrowheads point to the liver, and the arrows point to the intestine. E–L and E′–L′, whole-mount in situ hybridization of proinflammatory cytokines (il1b and mpx) in WT and zc3h8 mutant larvae at 5.5 and 6.5 dpf after pretreatment with DMSO or JSH-23. Red arrowheads point to the liver, and the black arrows point to the intestine.
Zc3h8 acts as a general inflammation repressor primarily through NF-κB signaling
Activations of nfkbiaa and nfkbiab were also observed in the gut of the cq5 mutant (Fig. 5, B′ and D′), implicating that the degeneration phenotype is not only restricted to the liver. Expressions of digestive organ markers (fabp10a, insulin, trypsin, and ifabp) indicated normal liver, pancreas, and gut at 3 dpf (Fig. S5A). At 4 dpf, expressions of trypsin and ifabp started to become weaker in the mutant than in the WT (Fig. S5B). Applications of the Tg(mpeg1:EGFP), Tg(cdh17:EGFP), and Tg(p48:GFP) transgenic backgrounds revealed degenerations of gut and exocrine pancreas from 5 to 7 dpf (Fig. S5, C, D, E–E″, F–F″, G–G″, and H–H″). The degenerated pancreas was partially rescued by the treatment of Dex or JSH-23 (Fig. S6, A and B), suggesting that degenerations of other digestive organs caused by zc3h8 deficiency involve mechanisms similar to liver degeneration. To investigate whether Zc3h8 could act as a general repressor of inflammation, we microinjected Escherichia coli into the circulation at 2 dpf to induce the inflammatory response (35). qPCR analysis indicated that the highly expressed proinflammatory cytokines were partially rescued by Zc3h8 overexpression (Fig. S6C), suggesting Zc3h8 as a general repressor of inflammation.
Discussion
Declining organ function is a common disease (36, 37), and research on this topic has important implications for human health. CCCH-type zinc finger proteins are important members of the zinc finger family proteins (18, 22, 38). Many previous studies on these proteins have been limited to mammals, and almost no studies have described the functions of CCCH-type zinc finger proteins or the phenotypes of mutants in zebrafish. This report is the first to identify a liver defect-related mutant, cq5, harboring a mutation in zc3h8, which encodes a typical zinc finger protein containing three conserved CCCH-type domains. This mutation resulted in degeneration of the zebrafish liver, gut, and pancreas. Further investigation revealed roles of Zc3h8 in maintaining hepatocyte metabolism through inhibition of proinflammatory responses in zebrafish.
To determine the function of the zc3h8 gene in zebrafish, the results of systemic gene analysis and homologous protein sequence alignment were evaluated, which indicated that zc3h8 of fish may exhibit the conserved functions of zc3h6 and zc3h8 in mammals (humans and mice). However, no clear function has been reported for zc3h6 in humans and mice. Another homologous gene of zc3h8 in mice (fetal liver zinc-finger protein 1, Fliz1) has been reported to be expressed in the fetal mouse liver (39) and plays a role in regulating thymocyte homeostasis (40). Additionally, overexpression of fliz1 represses the expression of gata3 (GATA-binding protein 3) (41). However, the expression of gata3 does not differ between WT and zc3h8-overexpressing zebrafish (data not shown). This observation indicates that zc3h8 of zebrafish may have a different function than fliz1 in the mouse.
All CCCH-type zinc finger proteins function through the characteristic CCCH zinc finger domain. The cq5 mutation site is located in the second CCCH zinc finger domain, where an amino acid coding change from histidine to glutamine occurs. We produced two lines of TALEN-induced knockout fish, zc3h8ins5cq45 and zc3h8Δ10cq46 (not containing the third CCCH zinc finger domain-truncated proteins), both of which showed the same degeneration phenotype as the cq5 mutant. Although we did not determine the molecules that could directly bind to the CCCH domain, these results suggest the importance of the second and third CCCH zinc finger domains for the function of the Zc3h8 protein.
In our analysis of the mutant phenotype, we noted that cq5 mutant larvae exhibited gradual digestive organ loss and degradation after 4 dpf, especially for the liver. However, the expression levels of organ markers and morphological phenotypes before 4 dpf were comparable between the mutant and WT larvae. The relatively stable expression of zc3h8 in hepatocytes demonstrated by FISH analysis could likely explain the degenerative phenotype observed in the mutant liver, and hepatocyte specific expression of Zc3h8 could absolutely rescue the liver defect of zc3h8 mutant (Figs. S2, D–H, S7E) and partly reduce the proinflammatory response (Fig. S7, F–H). Although the other defect phenotypes of gut, pancreas, retina, swim bladder, and brain not rescued by the liver overexpression Zc3h8 (Fig. S7, A–D), this indicated that the degeneration process of the mutant hepatocytes was autonomous and independent of the other type cells induced.
Our transcriptome analysis revealed that NF-κB signaling and inflammatory cytokines were both up-regulated during liver degeneration. This finding is consistent with previous studies showing that NF-κB signaling–mediated inflammatory responses are important for liver homeostasis and wound healing (42–45). At the RNA level, in situ hybridization of NF-κB component genes and qPCR analysis of inflammatory cytokine levels showed increased expression of NF-κB signaling-related genes and inflammatory cytokines in the degenerated mutant liver. At the protein level, the results of antibody staining for phosphorylated p65 (RelA) combined with treatment using inhibitors indicated that blocking the NF-κB signal or inflammatory response could rescue the mutant phenotype. Further confirming the Zc3h8 role as a repressor of inflammatory response, bacteria-induced inflammatory model was established, and its high expression of inflammation cytokines could be partially rescued by the overexpression of Zc3h8 (Fig. S6C). These observations are consistent with studies analyzing other CCCH-type zinc finger proteins, such as TTP (18, 21, 22, 46).
In summary, although the zc3h8 mutant exhibited the nonliver specific abnormal phenotype, but our results indicated the degeneration process and mechanism in the liver were similar with the other digestive organs. Our work reveals that Zc3h8 plays important roles in maintaining digestive organs homeostasis, and mutation of this gene causes their degeneration. Mechanistically, this degeneration process depends on the NF-κB pathway and related inflammatory responses. This study expands our understanding of CCCH-type zinc finger protein function in protecting digestive system cells including the hepatocytes from degeneration and maintaining organ development in zebrafish and provides new insight for future research and the development of degenerative organ disease therapeutics.
Experimental procedures
Ethics statement
All experimental protocols were approved by the School of Life Sciences, Southwest University (Chongqing, China), and the experiments were carried out in accordance with the approved guidelines. The zebrafish facility and study were approved by the Institutional Review Board of Southwest University (Chongqing, China). Zebrafish were maintained in accordance with the Guidelines for Experimental Animal Welfare from the Ministry of Science and Technology of the People's Republic of China (2006) and the Institutional Animal Care and Use Committee protocols of Southwest University (2007).
Zebrafish lines and embryo culture
Adult fish and embryos were raised and maintained under standard laboratory conditions according to Institutional Animal Care and Use Committee protocols. They were staged as previously described (47). The following transgenic lines were used: Tg(fabp10a:Dendra2-NTR)cq1, Tg(fabp10a:mCherry-NTR)cq2, Tg(sox17:GFP)s870, Tg(p48:GFP), Tg(cdh17:EGPF), and Tg(mpeg1:EGFP). Fish embryos were treated with 0.003% 1-phenyl-2-thiourea (Sigma–Aldrich) from 24 hpf to inhibit pigmentation.
Positional cloning of the cq5 mutation gene
Heterozygous cq5 fish were crossed with the polymorphic line SJD to generate the mapping population. A total of 240 SSLP (simple sequence length polymorphism) Luo lab Z markers were used in bulked segregant analysis. The amplification reactions of positional cloning were performed with 2× Taq Master Mix (Novoprotein, Shanghai, China). SSLP markers z6657 and z30283 from chromosome 13 were found to be linked to the cq5 mutation. After testing 560 mutant embryos, the location of the cq5 mutation was further narrowed to between z55656 (14 recombinants) and z30283 (47 recombinants). New SSLP markers were designed. Two markers, 1006 and 1023, identified three and two recombinants, respectively, within genomic DNA fragments of 0.31 million bp, containing three genes (fbln7, zc3h8 and itga9). The mutant genotype was determined by sequencing the PCR fragment containing the mutated bases.
Zc3h8 TALEN assembly, targeting, and allele analysis
A TALEN targeting zc3h8 exon7 was designed using TAL effector Nucleotide Targeter 2.0 (TALE-NT 2.0) (48) and was assembled using the modified Golden TALEN scaffold (49). After mixing the pair of target-site TALEN mRNAs, a final concentration of ∼150–200 pg was injected into one-cell-stage Tg(fabp10a:Dendra2-NTR)cq1 WT embryos. Genomic DNA was subsequently extracted from the embryos, and the PCR products were sequenced. The primers used for amplification of the zc3h8 exon7 are listed in Table 1; a 524-bp DNA fragment containing the mutant target site was amplified via PCR and sequenced. After the embryos were grown to maturity, they were outcrossed to identify F1 progeny in which infidelity during repair by nonhomologous end joining introduced indels at the cleavage site that could disrupt the zc3h8 reading frame.
Table 1.
PCR primers used in this study
| Target | Forward (5′ → 3′) | Reverse (5′ → 3′) | Used for |
|---|---|---|---|
| 1006 | CCATACCGTGACCACTGCTA | TCTTCATGTTGACCTCTTCTAC | PCR |
| 1023 | GCACATGGTTGCACAAAAGCT | TACTGTCCTGACCATCTCAC | PCR |
| zc3h8(exon7) | AAAGGCCCACCCATTTTCCA | AGGCTCATGGGAGAACTTGC | PCR |
| zc3h8-HA | ATGTACCCATACGATGTTCCAGATTACGCTATGGCTTTGTGAGCCTCTT | GCTGTGTATTTAGTGTGAGTTTA | PCR |
| zc3h8-L | GGATCCACCGGTGCTAGCCATGTACCCATACGATGTTC | GAGGCAACGATATCCTGTAGTCACTGTATTTGTC | PCR |
| zc3h8-R | GACTACAGGATATCGTTGCCTCCGTATGCACCAC | CTCCACTGCCCTGACAGAATGGTGATGC | PCR |
| T2A-mcherry | CCATTCTGTCAGGGCAGTGGAGAGGGCAG | CTCGAGAAGCTTGTTTAGTTACTTGTACAGCTC | PCR |
| f2 | CTGTGATTTAGAGTTGTGCG | GTCTGCTTTGTTGATCTTCTC | qPCR |
| tfa | TCTGCTCCGTCCAATCATTG | GCATCTCATTAGGACCCTCC | qPCR |
| ttr | CTCTGTAGATCTGCTCCAGTG | GTGCACTTCACCAGTCATGTC | qPCR |
| ccl20 | CATTTTCCACACCGTGACAGG | GCTCATCGTCTTCGTCTTCATTG | qPCR |
| il1b | ATCAAACCCCAATCCACAGAGT | GGCACTGAAGACACCACGTT | qPCR |
| il6 | GCAGTATGGGGGAACTATCCG | GCTGATCCTGACCCCTTCAA | qPCR |
| il8 | TGTTTTCCTGGCATTTCTGACC | TTTACAGTGTGGGCTTGGAGGG | qPCR |
| mmp9 | CATTAAAGATGCCCTGATGTATCCC | AGTGGTGGTCCGTGGTTGAG | qPCR |
| mpx | TCAATATGAGGACGCCGTTTCT | GAATGCGATTGGAAACCAGTCT | qPCR |
| tnfa | GGCAATTTCACTTCCAAGGCT | GCTGATGTGCAAAGACACCTG | qPCR |
| tnfb | AAGCCAAACGAAGAAGGTCA | AACCCATTTCAGCGATTGTC | qPCR |
| nfbkiaa | AGTCATGCCAGAGAGCGAAT | CAGAGCCGGATGTCATCATA | qPCR |
| fabp10a | CGCTCAGGAGAACTACGAGGAG | GACGATGCACTTGAGCTTCTTG | qPCR |
| b-actin | TACAATGAGCTCCGTGTTGC | ACATACAATGGCAGGGGTGTT | qPCR |
| zc3h8 | AGGAGACAACTGCAAGTTCTC | GCCAAGTCCACAGTAGGCTG | qPCR |
| nfkbiaa | ATTGTAATACGACTCACTATAGGCTATCTCCCACAGAGCCGGATG | ATGGATTTACACAGAGCCGCAAT | WISH |
| nfkbiab | ATTGTAATACGACTCACTATAGGCTACTGCCCCATCACTTTAATATCATC | ATGGAGCTTTACCGAGGCACCA | WISH |
| zc3h8 | ATTGTAATACGACTCACTATAGGGGATGAGGAGAATGTGAAG | GCGTCCAATCATGAGTCAGGA | WISH |
| insulin | GGTCGTGTCCAGTGTAAGCA | CAGGTGTTTCTGGCATCGG | WISH |
| fabp10a | GTGTTGAGCTTCTCCAGAAAG | GAGTTATGGTGAAACGCTTCAG | WISH |
| ifabp | ATGACCTTCAACGGGACCTG | GGAGTGCAGATAACAGTTTAAG | WISH |
| Mpx | CATTAGCACTTCGAGAAACC | CCAGGCAAGTATAATGCATG | WISH |
| Il1b | CCAGCTCTGAAATGATGGC | CTGAAGAACAGCAGCTGGTC | WISH |
Construction of the zc3h8-overexpressing transgenic plasmid and lines
To generate the Tg(hsp70l:HA-zc3h8-T2A-mCherry)cq43 and Tg(fabp10a:HA-zc3h8-T2A-mCherry)cq44 transgenic lines of zebrafish, we first constructed the transgenic hsp70l:HA-zc3h8-T2A-mCherry plasmid (Fig. S8A). An HA-tag–encoding sequence was incorporated into the forward primer (Table 1). The full-length cDNA of zebrafish zc3h8 (XM_682645) was amplified from 24-hpf ABGO cDNA using high-fidelity Prime STAR DNA polymerase (TaKaRa, Shiga, Japan), and the PCR products were cloned into the pGEM-T easy vector (Promega, Madison, WI) to generate pGEMT-HA-zc3h8. Positive clones were subsequently picked and sequenced. Next, we amplified fragments with no mutations in the HA-zc3h8 fragment and inserted them into the pDsRed2-vector, which had a Kana-resistant backbone, to generate pDsRed2-HA-zc3h8 by replacing DsRed2 via NheI/NotI digestion. We separately amplified the left and right arms (∼500 bp) of the HA-zc3h8 fragment from the pGEMT-HA-zc3h8 and T2A-mcherry fragments from the pLentilox3.7 shRNA GFP vector plasmid. Two cycles of overlapping PCR were performed to generate HA-zc3h8 arm-T2A-mcherry, which was then cloned into the hsp70l:Cre plasmid after the excision of Cre with BamHI/NotI. Recombineering was accomplished via the following steps. We transformed a mixture of plasmid pDsRed-HA-zc3h8 with linearized hsp70l:HA-zc3h8 arm-T2A-mcherry fragments into electrocompetent SW102 cells, which can produce the recombinase used for the recombination reaction, to generate hsp70l:HA-zc3h8-T2A-mcherry. The HA-zc3h8-T2A-mcherry fragment was cloned into fabp10a:Dendra2NTR plasmid after excision of Dendra2NTR with NheI/NotI to generate fabp10a: HA-zc3h8-T2A-mcherry. The plasmids were coinjected with I-SceI meganuclease into one-cell stage WT embryos as previously described (50). Acceptable adult founders were isolated and propagated. The plasmid carrying heat shock promoter-driven hsp70l:HA-zc3h8H353Q-T2A-mcherry was mutated similarly to the cq5 mutant via site-directed mutagenesis as previously described (51).
Whole-mount in situ hybridization, fluorescent in situ hybridization, and antibody staining
Whole-mount in situ hybridization and fluorescent in situ hybridization were performed as previously described (12). Probes for the insulin, fabp2, fabp10a, mpx, il1b, nfkbiaa, nfkbiab, and zc3h8 transcripts were generated from linearized plasmids (insulin, fabp2, fabp10a, trypsin, mpx, and il1b) and PCR products (nfkbiaa, nfkbiab, and zc3h8) using the digoxigenin RNA labeling kit (Roche Applied Science). Primers for trypsin (52) were designed based on the available data. The embryos were imaged using a Zeiss microscope (SteREO DiscoveryV20) equipped with Axio Vision Rel 4.8.2 software. Antibody staining was performed as previously described (10) using the following antibodies: anti-Dendra2 (1:100; Evrogen, Moscow, Russia), anti-GFP (1:100; Santa Cruz Biotechnology, Santa Cruz, CA), S276-p65 (1:100; Abcam, Cambridge, MA), 2F11 (1:1000; Abcam, Cambridge, MA), and mCherry (1:100; Abcam, Cambridge, MA). ZEN2010 software equipped on an LSM780 confocal microscope (Carl Zeiss) was used for the imaging of antibody-stained larvae.
EdU staining and TUNEL assays
The EdU and TUNEL assays were performed as previously described (10).
Western blotting assays
Western blotting assays were performed as previously described (12). The hsp70l:HA-zc3h8-T2A-mCherry and hsp70l:HA-zc3h8H353Q-T2A-mCherry plasmids were injected into WT embryos, and the mCherry-positive cells detectable at 54 hpf were sorted by FACS for Western blotting analysis. Quantification of the Western blotting densitometry band was analyzed by ImageJ software.
Heat shock and chemical inhibitor treatment
For the induction of Zc3h8 overexpression from Tg(hsp70l:HA-zc3h8-T2A-mCherry)cq43, embryos were placed in culture medium and then transferred to a 38.5 °C water bath for 35 min at the indicated stage. Larvae at 4 dpf were incubated with 15 mg/liter dexamethasone (Sangon Biotech, Shanghai, China) in the egg water for 2 consecutive days, and dexamethasone supplementation was performed every 24 h. Control larvae were incubated in egg water containing methanol (same volume as Dex). Larvae at 4 dpf were incubated with JSH-23 (5 μm; Selleck Chemicals, Houston, TX) in the egg water from 4 to 6 dpf, with the water being changed every 24 h. Control larvae were incubated in egg water containing DMSO (Sangon Biotech, Shanghai, China) (same volume as JSH-23).
Cell sorting, transcriptome sequencing, and quantitative real-time PCR
Approximately 150–200 transgenic Tg(fabp10a:Dendra2-NTR)cq1 larvae at 5.5 or 7.5 dpf were dissected, and their hepatocytes were then dissociated and sorted as previously described (12). The Dendra2-positive hepatocytes separated from the control or sample groups were subjected to transcriptome sequencing and analysis by the Annoroad Gene Technology Company. Quantitative real-time PCR was performed for hepatocyte markers (f2, tfa, and ttr) and proinflammatory cytokines (ccl20, il1b, il6, li8, mmp9, mpx, tnfa, and tnfb) using Fast-Start Universal SYBR Green Master Mix (Roche) following the manufacturer's protocols, and their levels were normalized to that of β-actin. The primers used for these analyses are listed in Table 1.
E. coli–induced inflammation assays
The E. coli–induced inflammatory response assay was performed as previously described (35).
Quantification and statistical analysis
The liver volume of confocal Z-stack images was measured by Imaris version 9.0.2 for Windows. All statistical tests were performed with GraphPad Prism version 7.0 for Windows (GraphPad Software). The data were analyzed with Student's t test, and multiple comparisons performed with analysis of variance tests were used to determine statistical significance. Statistical significance was defined as p < 0.05 (*), p < 0.01 (**), and p < 0.001 (***).
Author contributions
L. L. X. L. H. L., and Q. Z. designed the experimental strategy, analyzed the data, and wrote the manuscript. K. G. performed the FISH experiments. Q. Y., X. T., and J. H. performed the hepatocyte sorting experiments. Q. Z. performed all other experiments in this study.
Supplementary Material
Acknowledgments
We thank Dr. Li Li for providing us the Tg(mpeg1:EGFP) transgenic line and the JSH-23 inhibitor, Dr. Huang Honghui for providing Tg(cdh17:EGFP), and Dr. Wensheng Wei for kindly providing the pLentilox3.7 shRNA GFP vector plasmids.
This work was supported by National Key Basic Research Program of China Grant 2015CB942800; National Natural Science Foundation of China Grants 31330051, 31730060, 31371483, and 91539201; Natural Science Foundation Project of Chongqing Grants cstc2014jcyjA10062 and cstc2014jcyjA10088; Fundamental Research Funds for the Central Universities Grant XDJK2015B011; and 111 Program B14037. The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1–S8.
- dpf
- days postfertilization
- hpf
- hours postfertilization
- TALEN
- transcription activator-like effector nuclease
- EdU
- 5-ethynyl-2′-deoxyuridine
- TUNEL
- terminal deoxynucleotidyltransferase-mediated dUTP nick end labeling
- qPCR
- quantitative PCR
- Dex
- dexamethasone
- SSLP
- simple sequence length polymorphism
- FISH
- fluorescent in situ hybridization.
References
- 1. Kawamoto S., Matsumoto Y., Mizuno K., Okubo K., and Matsubara K. (1996) Expression profiles of active genes in human and mouse livers. Gene 174, 151–158 10.1016/0378-1119(96)00512-4 [DOI] [PubMed] [Google Scholar]
- 2. Pack M., Solnica-Krezel L., Malicki J., Neuhauss S. C., Schier A. F., Stemple D. L., Driever W., and Fishman M. C. (1996) Mutations affecting development of zebrafish digestive organs. Development 123, 321–328 [DOI] [PubMed] [Google Scholar]
- 3. Field H. A., Ober E. A., Roeser T., and Stainier D. Y. (2003) Formation of the digestive system in zebrafish: I. liver morphogenesis. Dev. Biol. 253, 279–290 10.1016/S0012-1606(02)00017-9 [DOI] [PubMed] [Google Scholar]
- 4. Janeway C. A. (1989) Approaching the asymptote: evolution and revolution in immunology. Cold Spring Harbor Symp. Quant. Biol. 54, 1–13 10.1101/SQB.1989.054.01.003 [DOI] [PubMed] [Google Scholar]
- 5. Matzinger P. (1994) Tolerance, danger, and the extended family. Annu. Rev. Immunol. 12, 991–1045 10.1146/annurev.iy.12.040194.005015 [DOI] [PubMed] [Google Scholar]
- 6. Kubes P., and Mehal W. Z. (2012) Sterile inflammation in the liver. Gastroenterology 143, 1158–1172 10.1053/j.gastro.2012.09.008 [DOI] [PubMed] [Google Scholar]
- 7. Gao B., Seki E., Brenner D. A., Friedman S., Cohen J. I., Nagy L., Szabo G., and Zakhari S. (2011) Innate immunity in alcoholic liver disease. Am. J. Physiol. Gastroint. Liver Physiol. 300, G516–G525 10.1152/ajpgi.00537.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Schroder K., and Tschopp J. (2010) The inflammasomes. Cell 140, 821–832 10.1016/j.cell.2010.01.040 [DOI] [PubMed] [Google Scholar]
- 9. O'Neill L. A. (2006) Targeting signal transduction as a strategy to treat inflammatory diseases. Nat. Rev. Drug Discov. 5, 549–563 10.1038/nrd2070 [DOI] [PubMed] [Google Scholar]
- 10. He J., Lu H., Zou Q., and Luo L. (2014) Regeneration of liver after extreme hepatocyte loss occurs mainly via biliary transdifferentiation in zebrafish. Gastroenterology 146, 789–800 10.1053/j.gastro.2013.11.045 [DOI] [PubMed] [Google Scholar]
- 11. Choi T.-Y., Ninov N., Stainier D. Y., and Shin D. (2014) Extensive conversion of hepatic biliary epithelial cells to hepatocytes after near total loss of hepatocytes in zebrafish. Gastroenterology 146, 776–788 10.1053/j.gastro.2013.10.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lu H., Ma J., Yang Y., Shi W., and Luo L. (2013) EpCAM is an endoderm-specific Wnt derepressor that licenses hepatic development. Dev. Cell 24, 543–553 10.1016/j.devcel.2013.01.021 [DOI] [PubMed] [Google Scholar]
- 13. Ober E. A., Verkade H., Field H. A., and Stainier D. Y. (2006) Mesodermal Wnt2b signalling positively regulates liver specification. Nature 442, 688–691 10.1038/nature04888 [DOI] [PubMed] [Google Scholar]
- 14. Dong P. D., Munson C. A., Norton W., Crosnier C., Pan X., Gong Z., Neumann C. J., and Stainier D. Y. (2007) Fgf10 regulates hepatopancreatic ductal system patterning and differentiation. Nat. Genet. 39, 397–402 10.1038/ng1961 [DOI] [PubMed] [Google Scholar]
- 15. Mackay J. P., and Crossley M. (1998) Zinc fingers are sticking together. Trends Biochem. Sci. 23, 1–4 10.1016/S0968-0004(97)01168-7 [DOI] [PubMed] [Google Scholar]
- 16. Matthews J. M., and Sunde M. (2002) Zinc fingers: folds for many occasions. IUBMB Life 54, 351–355 10.1080/15216540216035 [DOI] [PubMed] [Google Scholar]
- 17. Taylor G. A., Carballo E., Lee D. M., Lai W. S., Thompson M. J., Patel D. D., Schenkman D. I., Gilkeson G. S., Broxmeyer H. E., Haynes B. F., and Blackshear P. J. (1996) A pathogenetic role for TNF α in the syndrome of cachexia, arthritis, and autoimmunity resulting from tristetraprolin (TTP) deficiency. Immunity 4, 445–454 10.1016/S1074-7613(00)80411-2 [DOI] [PubMed] [Google Scholar]
- 18. Carballo E., Lai W. S., and Blackshear P. J. (1998) Feedback inhibition of macrophage tumor necrosis factor-α production by tristetraprolin. Science 281, 1001–1005 10.1126/science.281.5379.1001 [DOI] [PubMed] [Google Scholar]
- 19. Lai W. S., Carballo E., Strum J. R., Kennington E. A., Phillips R. S., and Blackshear P. J. (1999) Evidence that tristetraprolin binds to AU-rich elements and promotes the deadenylation and destabilization of tumor necrosis factor α mRNA. Mol. Cell. Biol. 19, 4311–4323 10.1128/MCB.19.6.4311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Carballo E., and Blackshear P. J. (2001) Roles of tumor necrosis factor-α receptor subtypes in the pathogenesis of the tristetraprolin-deficiency syndrome. Blood 98, 2389–2395 10.1182/blood.V98.8.2389 [DOI] [PubMed] [Google Scholar]
- 21. Carrick D. M., Lai W. S., and Blackshear P. J. (2004) The tandem CCCH zinc finger protein tristetraprolin and its relevance to cytokine mRNA turnover and arthritis. Arthritis Res. Ther. 6, 248–264 10.1186/ar1441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Vinuesa C. G., Cook M. C., Angelucci C., Athanasopoulos V., Rui L., Hill K. M., Yu D., Domaschenz H., Whittle B., Lambe T., Roberts I. S., Copley R. R., Bell J. I., Cornall R. J., and Goodnow C. C. (2005) A RING-type ubiquitin ligase family member required to repress follicular helper T cells and autoimmunity. Nature 435, 452–458 10.1038/nature03555 [DOI] [PubMed] [Google Scholar]
- 23. Huang P., Xiao A., Zhou M., Zhu Z., Lin S., and Zhang B. (2011) Heritable gene targeting in zebrafish using customized TALENs. Nat. Biotechnol. 29, 699–700 10.1038/nbt.1939 [DOI] [PubMed] [Google Scholar]
- 24. Cox A. G., and Goessling W. (2015) The lure of zebrafish in liver research: regulation of hepatic growth in development and regeneration. Curr. Opin. Genet. Dev. 32, 153–161 10.1016/j.gde.2015.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Qi F., Song J., Yang H., Gao W., Liu N. A., Zhang B., and Lin S. (2010) Mmp23b promotes liver development and hepatocyte proliferation through the tumor necrosis factor pathway in zebrafish. Hepatology 52, 2158–2166 10.1002/hep.23945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Huang H., Ruan H., Aw M. Y., Hussain A., Guo L., Gao C., Qian F., Leung T., Song H., Kimelman D., Wen Z., and Peng J. (2008) Mypt1-mediated spatial positioning of Bmp2-producing cells is essential for liver organogenesis. Development 135, 3209–3218 10.1242/dev.024406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Li Y., Farooq M., Sheng D., Chandramouli C., Lan T., Mahajan N. K., Kini R. M., Hong Y., Lisowsky T., and Ge R. (2012) Augmenter of liver regeneration (alr) promotes liver outgrowth during zebrafish hepatogenesis. PLoS One 7, e30835 10.1371/journal.pone.0030835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Thomas J. A., Pope C., Wojtacha D., Robson A. J., Gordon-Walker T. T., Hartland S., Ramachandran P., Van Deemter M., Hume D. A., Iredale J. P., and Forbes S. J. (2011) Macrophage therapy for murine liver fibrosis recruits host effector cells improving fibrosis, regeneration, and function. Hepatology 53, 2003–2015 10.1002/hep.24315 [DOI] [PubMed] [Google Scholar]
- 29. Sunami Y., Leithäuser F., Gul S., Fiedler K., Güldiken N., Espenlaub S., Holzmann K. H., Hipp N., Sindrilaru A., Luedde T., Baumann B., Wissel S., Kreppel F., Schneider M., Scharffetter-Kochanek K., et al. (2012) Hepatic activation of IKK/NFκB signaling induces liver fibrosis via macrophage-mediated chronic inflammation. Hepatology 56, 1117–1128 10.1002/hep.25711 [DOI] [PubMed] [Google Scholar]
- 30. Shen H., Sheng L., Chen Z., Jiang L., Su H., Yin L., Omary M. B., and Rui L. (2014) Mouse hepatocyte overexpression of NF-κB-inducing kinase (NIK) triggers fatal macrophage-dependent liver injury and fibrosis. Hepatology 60, 2065–2076 10.1002/hep.27348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Barnes P. J., and Karin M. (1997) Nuclear factor-κB: a pivotal transcription factor in chronic inflammatory diseases. N. Engl. J. Med. 336, 1066–1071 10.1056/NEJM199704103361506 [DOI] [PubMed] [Google Scholar]
- 32. Karin M. (2006) NF-κB and cancer: mechanisms and targets. Mol. Carcinog. 45, 355–361 10.1002/mc.20217 [DOI] [PubMed] [Google Scholar]
- 33. Naugler W. E., and Karin M. (2008) NF-κB and cancer-identifying targets and mechanisms. Curr. Opin. Genet. Dev. 18, 19–26 10.1016/j.gde.2008.01.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Koo J. W., Russo S. J., Ferguson D., Nestler E. J., and Duman R. S. (2010) Nuclear factor-κB is a critical mediator of stress-impaired neurogenesis and depressive behavior. Proc. Natl. Acad. Sci. U.S.A. 107, 2669–2674 10.1073/pnas.0910658107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hou Y., Sheng Z., Mao X., Li C., Chen J., Zhang J., Huang H., Ruan H., Luo L., and Li L. (2016) Systemic inoculation of Escherichia coli causes emergency myelopoiesis in zebrafish larval caudal hematopoietic tissue. Sci. Rep. 6, 36853 10.1038/srep36853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zhang Z., Lotti F., Dittmar K., Younis I., Wan L., Kasim M., and Dreyfuss G. (2008) SMN deficiency causes tissue-specific perturbations in the repertoire of snRNAs and widespread defects in splicing. Cell 133, 585–600 10.1016/j.cell.2008.03.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bredesen D. E., Rao R. V., and Mehlen P. (2006) Cell death in the nervous system. Nature 443, 796–802 10.1038/nature05293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Gao G., Guo X., and Goff S. P. (2002) Inhibition of retroviral RNA production by ZAP, a CCCH-type zinc finger protein. Science 297, 1703–1706 10.1126/science.1074276 [DOI] [PubMed] [Google Scholar]
- 39. Dahm K., Nielsen P. J., and Müller A. M. (2001) Transcripts of Fliz1, a nuclear zinc finger protein, are expressed in discrete foci of the murine fetal liver. Genomics 73, 194–202 10.1006/geno.2000.6480 [DOI] [PubMed] [Google Scholar]
- 40. Hwang E. S., and Ho I. C. (2002) Regulation of thymocyte homeostasis by Fliz1. Immunology 106, 464–469 10.1046/j.1365-2567.2002.01455.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hwang E. S., Choi A., and Ho I. C. (2002) Transcriptional regulation of GATA-3 by an intronic regulatory region and fetal liver zinc finger protein 1. J. Immunol. 169, 248–253 10.4049/jimmunol.169.1.248 [DOI] [PubMed] [Google Scholar]
- 42. Luedde T., Beraza N., Kotsikoris V., van Loo G., Nenci A., De Vos R., Roskams T., Trautwein C., and Pasparakis M. (2007) Deletion of NEMO/IKKγ in liver parenchymal cells causes steatohepatitis and hepatocellular carcinoma. Cancer Cell 11, 119–132 10.1016/j.ccr.2006.12.016 [DOI] [PubMed] [Google Scholar]
- 43. Luedde T., Heinrichsdorff J., de Lorenzi R., De Vos R., Roskams T., and Pasparakis M. (2008) IKK1 and IKK2 cooperate to maintain bile duct integrity in the liver. Proc. Natl. Acad. Sci. U.S.A. 105, 9733–9738 10.1073/pnas.0800198105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Bettermann K., Vucur M., Haybaeck J., Koppe C., Janssen J., Heymann F., Weber A., Weiskirchen R., Liedtke C., Gassler N., Müller M., de Vos R., Wolf M. J., Boege Y., Seleznik G. M., et al. (2010) TAK1 suppresses a NEMO-dependent but NF-κB-independent pathway to liver cancer. Cancer Cell 17, 481–496 10.1016/j.ccr.2010.03.021 [DOI] [PubMed] [Google Scholar]
- 45. Inokuchi S., Aoyama T., Miura K., Osterreicher C. H., Kodama Y., Miyai K., Akira S., Brenner D. A., and Seki E. (2010) Disruption of TAK1 in hepatocytes causes hepatic injury, inflammation, fibrosis, and carcinogenesis. Proc. Natl. Acad. Sci. U.S.A. 107, 844–849 10.1073/pnas.0909781107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Liang J., Wang J., Azfer A., Song W., Tromp G., Kolattukudy P. E., and Fu M. (2008) A novel CCCH-zinc finger protein family regulates proinflammatory activation of macrophages. J. Biol. Chem. 283, 6337–6346 10.1074/jbc.M707861200 [DOI] [PubMed] [Google Scholar]
- 47. Kimmel C. B., Ballard W. W., Kimmel S. R., Ullmann B., and Schilling T. F. (1995) Stages of embryonic development of the zebrafish. Dev. Dyn. 203, 253–310 10.1002/aja.1002030302 [DOI] [PubMed] [Google Scholar]
- 48. Doyle E. L., Booher N. J., Standage D. S., Voytas D. F., Brendel V. P., Vandyk J. K., and Bogdanove A. J. (2012) TAL effector–nucleotide targeter (TALE-NT) 2.0: tools for TAL effector design and target prediction. Nucleic Acids Res. 40, W117–W122 10.1093/nar/gks608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Bedell V. M., Wang Y., Campbell J. M., Poshusta T. L., Starker C. G., Krug R. G. 2nd, Tan W., Penheiter S. G., Ma A. C., Leung A. Y., Fahrenkrug S. C., Carlson D. F., Voytas D. F., Clark K. J., Essner J. J., et al. (2012) In vivo genome editing using a high-efficiency TALEN system. Nature 491, 114–118 10.1038/nature11537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Grabher C., Joly J. S., and Wittbrodt J. (2004) Highly efficient zebrafish transgenesis mediated by the meganuclease I-SceI. Methods Cell Biol. 77, 381–401 10.1016/S0091-679X(04)77021-1 [DOI] [PubMed] [Google Scholar]
- 51. Zheng L., Baumann U., and Reymond J. L. (2004) An efficient one-step site-directed and site-saturation mutagenesis protocol. Nucleic Acids Res. 32, e115 10.1093/nar/gnh110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Mudumana S. P., Wan H., Singh M., Korzh V., and Gong Z. (2004) Expression analyses of zebrafish transferrin, ifabp, and elastaseB mRNAs as differentiation markers for the three major endodermal organs: Liver, intestine, and exocrine pancreas. Dev. Dyn. 230, 165–173 10.1002/dvdy.20032 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.