Abstract
Chronic neuroinflammation is a characteristic of Parkinson's disease (PD). Previous investigations have shown that Parkin gene mutations are related to the early-onset recessive form of PD and isolated juvenile-onset PD. Further, Parkin plays important roles in mitochondrial quality control and cytokine-induced cell death. However, whether Parkin regulates other cellular events is still largely unknown. In this study, we performed overexpression and knockout experiments and found that Parkin negatively regulates antiviral immune responses against RNA and DNA viruses. Mechanistically, we show that Parkin interacts with tumor necrosis factor receptor-associated factor 3 (TRAF3) to regulate stability of TRAF3 protein by promoting Lys48-linked ubiquitination. Our findings suggest that Parkin plays a novel role in innate immune signaling by targeting TRAF3 for degradation and maintaining the balance of innate antiviral immunity.
Keywords: innate immunity, cell signaling, Parkin, interferon, ubiquitylation (ubiquitination), TRAF3
Introduction
Parkinson's disease (PD)2 is a neurodegenerative disease caused by genetic mutations or environmental factors. Chronic neuroinflammation is one characteristic of PD pathology (1). Evidence for chronic inflammation and innate immune activation in the incidence of PD comes from the following studies. Postmortem examination detected neuroinflammation in PD patient brains (2), with higher cytokines levels (including interleukin (IL)-1β, transforming growth factor-β, interferon (IFN)-γ, and IL-6) found in cerebrospinal fluid and nigrostriatal regions of PD patients compared with age-matched healthy controls (3). Further, higher plasma concentrations of the proinflammatory cytokine IL-6 were shown to be related to increased incidence of PD development (4), whereas chronic nonsteroidal anti-inflammatory drug regimens decrease the risk of PD (5, 6). However, what leads to this increase in inflammatory cytokines in PD patients is largely unknown.
Previous studies have found that mutations in the Parkin gene (PRKN; also known as PARK2) are a major cause of early-onset recessive forms of PD and isolated juvenile-onset PD (7). The Parkin protein contains one typical RING domain (RING1) and two atypical RING domains (RING0 and RING2) and shows E3 ubiquitin ligase activity in vitro (8). Parkin was found to regulate mitophagy and reactive oxygen species–induced mitochondrial-derived vesicles to maintain mitochondrial homeostasis (9). Similarly, Parkin was also found to enhance linear ubiquitination of NF-κB essential modulator (NEMO), resulting in increased expression of the mitochondrial GTPase, OPA1, and maintenance of mitochondrial integrity (10). Previous studies have shown that PD is caused by neural apoptosis and autophagy (11). Accordingly, Chung et al. (12) showed that Parkin promotes proteasomal degradation of tumor necrosis factor (TNF) receptor–associated factor (TRAF)-2/6 to inhibit inflammation through NF-κB activation and cytokine-induced cell death via c-Jun N-terminal kinase/stress signaling pathways. However, whether Parkin is involved in other cellular events is still largely unknown.
The innate immune system functions as the first line of the host defense, rapidly detecting and eliminating invading pathogens such as viruses (13), bacteria (14, 15), and fungi (16). The innate immune response system comprises various pattern recognition receptors, which recognize different pathogen-associated molecular patterns (17, 18). For virus detection, Toll-like receptors 3/7/8 and retinoic acid–inducible gene I (RIG-I)-like receptors (namely RIG-I and melanoma differentiation–associated protein 5) play important roles in detecting RNA viruses (19, 20), whereas Toll-like receptor 9 and a number of cytoplasmic DNA sensors (such as γ-interferon–inducible protein 16, DNA-dependent activator of IFN-regulatory factor, DEAD-box helicase 41, and cGMP-AMP synthase) function as DNA virus recognition receptors (21). These receptors recruit essential adaptor proteins, including mitochondrial antiviral-signaling protein (MAVS), myeloid differentiation primary response 88, Toll/interleukin-1 receptor (TIR) domain–containing adapter-inducing interferon-β, stimulator of interferon genes (STING), and TRAFs, to activate downstream signaling pathways that contribute to expression of genes encoding type I IFNs and proinflammatory cytokines (such as IL-6 and TNFα) (17, 22). Meanwhile, these innate immune responses must be tightly regulated by host factors to prevent excessive and detrimental inflammation. Thus, greater understanding of the mechanisms by which the host balances the immune response will help treat immune-related diseases.
The incidence of PD increases in an age-dependent manner. Furthermore, the elderly population is at increased susceptibility to infections, which induce inflammatory cytokines (23, 24). Consequently, clarifying the relationship between PD, viral infections, and chronic inflammation will lead to improved understanding of the mechanism underlying PD incidence and treatment. In this study, we investigated the role of Parkin in the antiviral immune response. We found that Parkin negatively regulates the antiviral signaling pathway, and mechanistically, we demonstrate that Parkin modulates stability of TRAF3 protein in an E3 ligase–independent manner. Altogether, our findings suggest that Parkin functions as a negative regulator to control cellular antiviral and inflammatory responses. Moreover, our findings provide novel insight into understanding the molecular mechanisms of PD.
Results
Parkin overexpression suppresses antiviral signaling
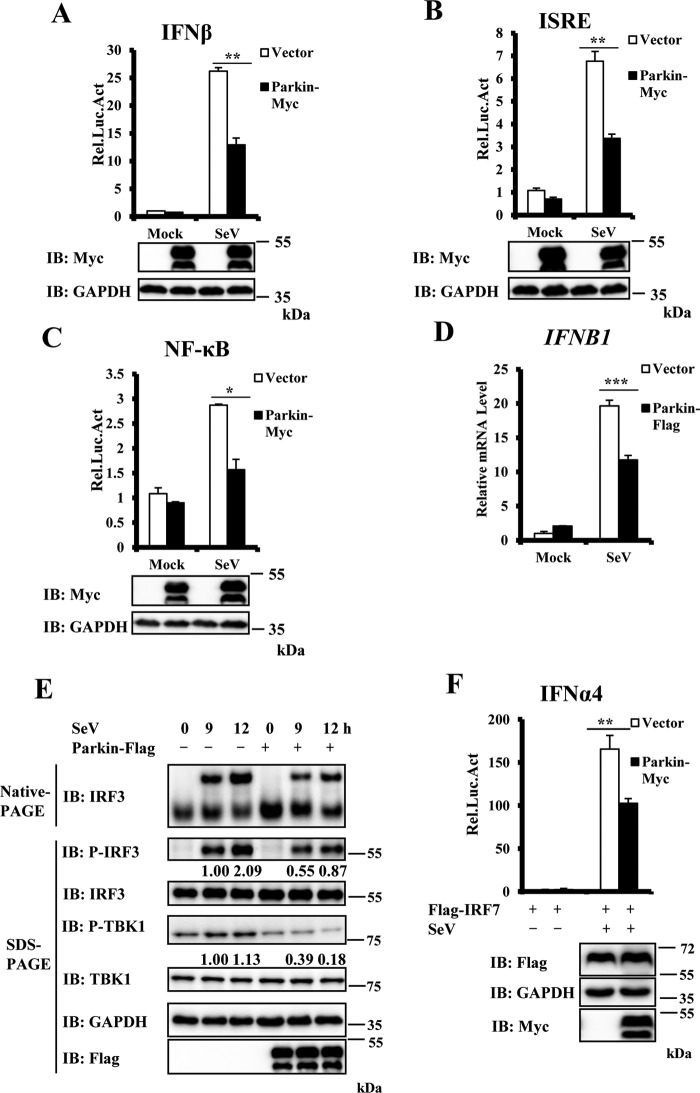
To determine the role of Parkin in regulating antiviral signaling, we first determined whether Parkin overexpression affects activation of the type I IFN promoter (IFN-β) induced by Sendai virus (SeV), a RNA virus of the paramyxoviridae family. As shown in Fig. 1A, Parkin overexpression significantly suppressed IFN-β promoter activation induced by SeV. Because IFN-β activation requires cooperative activation of both IFN regulatory factor 3 (IRF3) and NF-κB transcription factors (17), we next used an IFN-stimulated response element (ISRE) luciferase reporter, which is sufficiently activated by IRF3 activation, and NF-κB luciferase reporter, to test the impact of Parkin on IRF3 and NF-κB activation, respectively. As shown in Fig. 1 (B and C), Parkin overexpression significantly suppressed both ISRE and NF-κB promoter activation induced by SeV infection. To gain further supportive evidence, we performed quantitative real-time PCR (qRT–PCR) assays to measure levels of IFNB1 transcripts in SeV-infected cells with or without Parkin overexpression. As shown in Fig. 1D, Parkin overexpression reduced IFNB1 transcript levels. IRF3 phosphorylation and dimerization are hallmarks of antiviral signaling activation. In agreement with a negative role of Parkin in antiviral signaling, we found that Parkin overexpression significantly decreased levels of IRF3 phosphorylation and dimerization induced by SeV infection (Fig. 1E). TBK1 phosphorylation (at the Ser172 residue) is another important event in the activation of the IFN-β signaling pathway; next we examined whether Parkin overexpression affected TBK1 phosphorylation. As shown in Fig. 1E, levels of TBK1 phosphorylation induced by SeV infection were significantly reduced by Parkin overexpression. Because TBK1 is involved in regulating both IRF3 and IRF7 activation, we also tested whether Parkin overexpression regulated IRF7 activation, as shown in Fig. 1F, Parkin overexpression significantly suppressed IFNα4 promoter activation induced by SeV infection. Taken together, these results indicate that Parkin negatively regulates the antiviral immune response.
Figure 1.
Overexpression of Parkin inhibits SeV-induced antiviral signaling. A–C, overexpression of Parkin reduced SeV-induced activation of the IFN-β, ISRE, and NF-κB promoters. HeLa cells were co-transfected with the indicated expression plasmids and luciferase reporter constructs driven by promoter of gene encoding IFN-β (A), ISRE (B), or NF-κB (C). pRSV/lacZ was used as an internal control. 24 h after transfection, the cells were infected with SeV for 24 h and then lysed for luciferase assays (upper panel) and immunoblotting (IB) assays (lower panels). D, Parkin overexpression decreased SeV-induced transcription of the IFNB1 gene. HEK293 cells were transfected with the indicated expression plasmids. 24 h after transfection, the cells were infected with SeV for 6 h, and cell lysates were analyzed by qRT–PCR to examine IFNB1 mRNA levels. E, overexpression of Parkin reduced SeV-induced IRF3 dimerization and IRF3 and TBK1 phosphorylation. HEK293 cells were transfected with the indicated expression plasmids. 24 h after transfection, the cells were left untreated or infected with SeV for the indicated times. The cell lysates were resolved by native gel electrophoresis (upper panel) or SDS–PAGE (lower panels) and analyzed with the indicated antibodies. F, overexpression of Parkin reduced SeV-induced activation of the IFN-α4 promoter. HEK293 cells were co-transfected with the indicated expression plasmids and luciferase reporter construct driven by promoter of gene encoding IFN-α4. Renilla was used as an internal control. 24 h after transfection, the cells were infected with SeV for 24 h and then lysed for luciferase assays (upper panel) and immunoblotting assays (lower panels). The data in A–D and F are from a representative experiment of at least three independent experiments (means ± S.D. of duplicate assays in A–C and F; triplicate experiments in D). Two-tailed Student's t test was used to determine statistical significance. *, p < 0.05:, **, p < 0.01; ***, p < 0.001, versus control groups. The numbers below lanes in E indicate densitometry of the protein presented relative to IRF3 (top) or TBK1 expression in the same lane. Rel. Luc. Act, relative luciferase activity.
Enhanced innate antiviral immune responses in Parkin-deficient embryonic fibroblasts
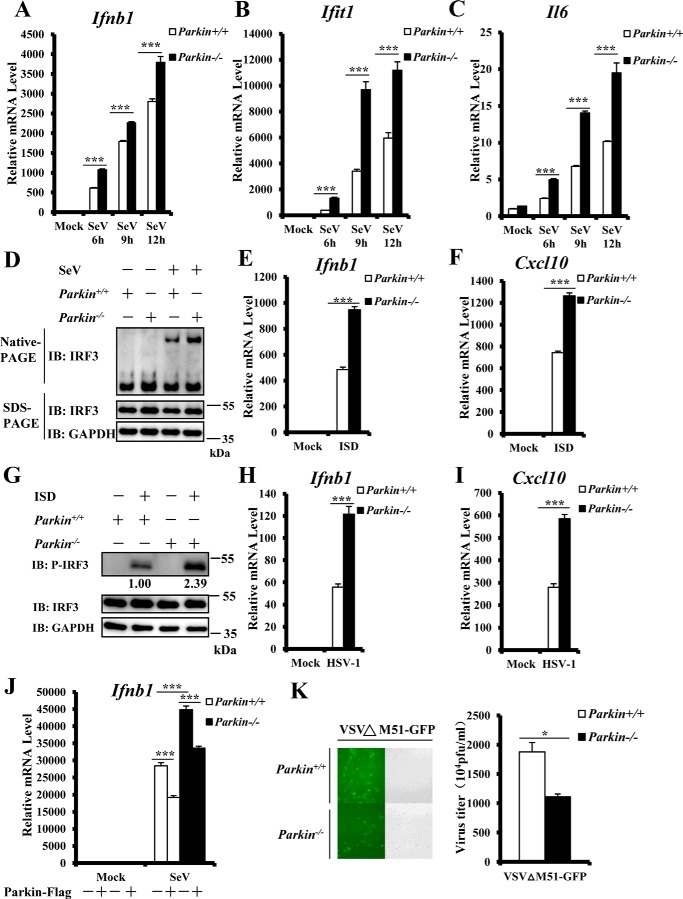
To further investigate the role of Parkin in regulating the innate antiviral immune response, we generated Parkin+/+ and Parkin−/− mouse embryonic fibroblasts (MEFs) and examined the effect of Parkin deficiency on antiviral immunity. qRT–PCR assays showed that after SeV infection, Parkin knockout significantly increased transcript levels of Ifnb1 and IFN-stimulated cytokine genes, such as interferon-induced protein with tetratricopeptide repeats 1 (Ifit1), compared with WT controls (Fig. 2, A and B). Induction of the proinflammatory cytokine, Il-6, by SeV infection was also enhanced in Parkin−/− MEFs (Fig. 2C). Consistently, we observed that Parkin knockout significantly increased IRF3 dimer levels induced by SeV infection (Fig. 2D). To further support this conclusion, we infected Parkin+/+ and Parkin−/− MEFs with another RNA virus, vesicular stomatitis mutant virus (VSVΔM51-GFP), carrying a single amino acid deletion (methionine 51) in the matrix protein. We observed significantly higher transcript levels of Ifnb1, Ifit1, and Il-6 in Parkin−/− MEFs compared with Parkin+/+ MEFs (Fig. S1). To determine whether Parkin is specifically involved in the antiviral response against RNA viruses, we next determined whether Parkin regulates the antiviral response against a DNA virus. First, we transfected Parkin+/+ and Parkin−/− MEFs with interferon stimulatory DNA (ISD), which contains 45-bp dsDNA. Consequently, we observed higher levels of Ifnb1 and CXC motif chemokine ligand 10 (Cxcl10) mRNA in Parkin−/− MEFs compared with control cells (Fig. 2, E and F). Second, we performed Western blotting and found markedly increased IRF3 phosphorylation induced by ISD in Parkin−/− MEFs compared with Parkin+/+ MEFs (Fig. 2G). Third, we infected Parkin+/+ and Parkin−/− MEF cells with the DNA virus, herpes simplex virus 1 (HSV-1) and observed increased Ifnb1 and Cxcl10 mRNA induced in Parkin−/− MEFs compared with Parkin+/+ MEFs (Fig. 2, H and I). Consistently, similar to Ifnβ mRNA, the level of Ifnα4 mRNA induced by infection with SeV or HSV-1 was significantly higher in Parkin−/− cells than control cells (Fig. S2). Altogether, these results indicate that Parkin functions as a negative modulator in innate antiviral immune signaling against both RNA and DNA viruses.
Figure 2.
Enhanced antiviral response in Parkin-deficient MEFs. A–C, Parkin deficiency significantly enhanced SeV-induced transcriptional levels of antiviral genes. Primary Parkin+/+ and Parkin−/− MEFs were infected with SeV for the indicated times. MEFs were then harvested to isolate RNA and measure transcript levels of Ifnb1 (A), Ifit1 (B), and Il6 (C) by qRT–PCR analysis. D, Parkin deficiency increased SeV-induced IRF3 dimerization. Primary Parkin+/+ and Parkin−/− MEFs were left untreated or infected with SeV for 9 h. The cell lysates were separated by native gel electrophoresis (upper panel) or SDS–PAGE (lower panels) and analyzed by immunoblotting (IB) with the indicated antibodies. E and F, ISD-induced transcriptional levels of antiviral genes were increased in Parkin−/− MEFs. Primary Parkin+/+ and Parkin−/− MEFs were transfected with ISD (2 μg/ml) for 3 h, followed by qRT–PCR analysis to measure transcript levels of Ifnb1 (E) and Cxcl10 (F). G, IRF3 phosphorylation induced by ISD was increased in Parkin−/− MEFs. Primary Parkin+/+ and Parkin−/− MEFs were left untreated or transfected with ISD (2 μg/ml) for 3 h. The cell lysates were separated by SDS–PAGE and analyzed by immunoblotting with the indicated antibodies. H and I, Parkin deficiency enhanced transcriptional levels of antiviral genes induced by HSV-1 infection. Primary Parkin+/+ and Parkin−/− MEFs were infected with HSV-1 at a MOI of 5 for 3 h, followed by qRT–PCR analysis to measure transcript levels of Ifnb1 (H) and Cxcl10 (I). J, overexpression of mouse Parkin could reverse the enhancement of Ifnb1 expression induced by SeV infection in Parkin−/− MEFs. Primary Parkin+/+ and Parkin−/− MEFs were first infected with a retrovirus expressing Parkin or an empty vector. After 48 h of infection, the cells were infected with SeV for 6 h, followed by qRT–PCR analysis to measure Ifnb1 transcript levels. K, Parkin deficiency suppressed the amplification of VSVΔM51-GFP. Primary Parkin+/+ and Parkin−/− MEFs were infected with vesicular stomatitis mutant virus (VSVΔM51-GFP) at MOI of 0.01 for 36 h. The cells were imaged by fluorescence microscopy (left panel) or culture supernatants collected to measure viral titer by plaque assay (right panel). The data in A–C, E, F, and H–K are from a representative experiment of at least three independent experiments (means ± S.D. of triplicate experiments in A–C, E, F, and H–J and duplicate experiments in K). Two-tailed Student's t test was used to determine statistical significance. *, p < 0.05; ***, p < 0.001, versus control groups. Numbers below lanes (top) in G indicate densitometry of the protein presented relative to IRF3 expression in the same lane.
To further determine the specific role of Parkin in antiviral signaling, we performed rescue experiments and overexpressed Parkin in Parkin−/− MEF cells. We found that Parkin expression reversed the increase in Ifnb1 expression induced by SeV in Parkin−/− MEFs (Fig. 2J). Consequently, we next investigated the biological function of Parkin in regulating virus amplification. We performed viral titer assays and found a much lower viral titer of VSVΔM51-GFP produced in Parkin−/− MEF cells than Parkin+/+ control cells (Fig. 2K). Taken together, our data provide biological evidence that Parkin regulates virus replication.
Parkin deficiency increases type I IFN signaling in macrophages
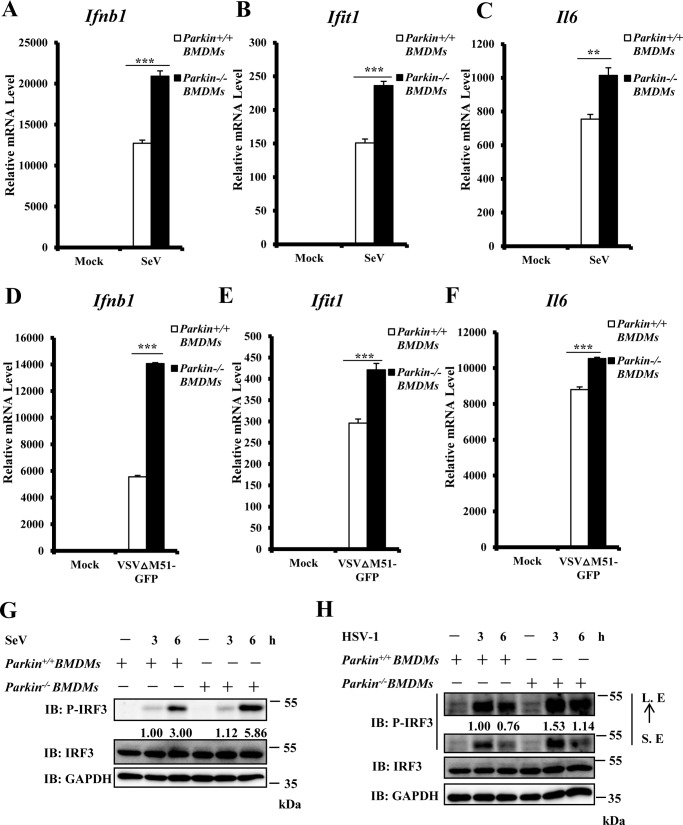
Macrophages play an important role in IFN production induced by viral infections; therefore we next determined whether Parkin plays a negative regulatory role in macrophages. We isolated bone marrow–derived macrophages (BMDMs) from Parkin−/− and Parkin+/+ mice and infected these cells with SeV. As shown in Fig. 3 (A–C), we found increased Ifnb1, Ifit1, and Il6 production induced by SeV infection in Parkin−/− BMDMs compared with Parkin+/+ BMDMs. Corroboratively, we observed similar results with infection by VSVΔM51-GFP (Fig. 3, D–F). In addition, Western blotting showed that Parkin deficiency significantly enhanced IRF3 phosphorylation levels induced by SeV or HSV-1 infection (Fig. 3, G and H). Thus, these data suggest that Parkin suppresses the innate immune response against both RNA and DNA viruses in macrophages.
Figure 3.
Parkin knockout potentiates type I IFN response in macrophage cells. A–C, Parkin deficiency significantly enhanced SeV-induced transcript levels of antiviral genes in BMDMs. Parkin+/+ and Parkin−/− BMDMs were infected with SeV for 6 h and then lysed for qRT–PCR analysis to examine transcript levels of Ifnb1 (A), Ifit1 (B), and Il6 (C). D–F, Parkin deficiency enhanced VSVΔM51-GFP-induced transcript levels of antiviral genes in BMDMs. Parkin+/+ and Parkin−/− BMDMs were infected with VSVΔM51-GFP virus at MOI of 1 for 6 h and then lysed for qRT–PCR analysis to measure transcript levels of Ifnb1 (D), Ifit1 (E), and Il6 (F). G, SeV-induced IRF3 phosphorylation was increased in Parkin−/− BMDMs. Parkin+/+ and Parkin−/− BMDMs were left untreated or infected with SeV for the indicated times. The cell lysates were separated by SDS–PAGE and subjected to immunoblotting (IB) with the indicated antibodies. H, HSV-1–induced IRF3 phosphorylation was enhanced in Parkin−/− BMDMs. Parkin+/+ and Parkin−/− BMDMs were left untreated or infected with HSV-1 at MOI of 5 for the indicated times. The cell lysates were separated by SDS–PAGE, followed by immunoblotting with the indicated antibodies. The data in A–F are from a representative experiment of at least three independent experiments (means ± S.D. of triplicate experiments). Two-tailed Student's t test was used to determine statistical significance. **, p < 0.01; ***, p < 0.001, versus control groups. Numbers below lanes (top) in G and H indicate densitometry of the protein presented relative to IRF3 expression in the same lane. S. E, short exposure; L. E, long exposure.
Identification of TRAF3 as a Parkin-interacting protein
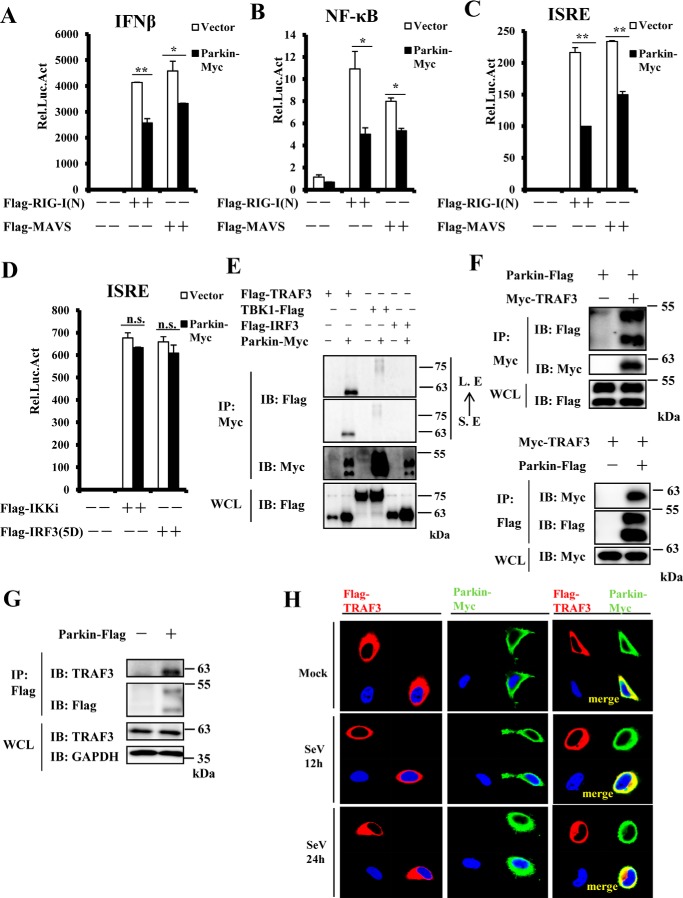
Our results show that Parkin negatively regulates IRF3-mediated and NF-κB pathways of the innate immune response. Interestingly, Parkin is reportedly involved in the NF-κB pathway via TRAF2/6 or NEMO regulation (10, 12). However, it is not known how Parkin is involved in IRF3-mediated signaling. Hence, we next focused on the mechanism by which Parkin regulates the IRF3-mediated pathway. To investigate the epistatic relationship between Parkin and other components of this innate immune pathway, we first determined whether Parkin overexpression affects IFN-β, NF-κB, and ISRE promoter activity in HEK293 cells triggered by overexpression of RIG-I(N) (tandem N-terminal CARD-like domains of RIG-I) or MAVS. Reporter assays showed that Parkin overexpression significantly inhibited activation of IFN-β, NF-κB, and ISRE promoters induced by RIG-I(N) and MAVS (Fig. 4, A–C). In contrast, Parkin failed to suppress ISRE activation induced by IKKi and IRF3–5D, a constitutively active form of IRF3 (25), suggesting that Parkin acts upstream of IKKi to mediate IRF3 signaling (Fig. 4D). We have already shown that Parkin regulates the innate immune response against both RNA and DNA viruses; therefore we subsequently performed co-immunoprecipitation (co-IP) assays to determine which common components of the MAVS/STING-mediated signaling pathways physically associate with Parkin. As shown in Fig. 4E, Parkin strongly interacted with TRAF3 under the same co-IP conditions, but not TBK1 or IRF3. Of note, a very weak interaction between Parkin and TBK1 was detected under longer exposure. To confirm this association, we co-transfected TRAF3 and Parkin plasmids in HEK293 cells, and subsequently performed co-IP experiments. Here, we found that TRAF3 and Parkin reciprocally co-immunoprecipitated in the presence or absence of EDTA (Fig. 4F and Fig. S3). Furthermore, endogenous TRAF3 was co-immunoprecipitated by overexpressed Parkin in HEK293 cells (Fig. 4G). In addition, immunostaining assays showed that Parkin co-localized with TRAF3 in transfected HeLa cells in the presence or absence of SeV infection (Fig. 4H). Collectively, these results indicate that TRAF3 is a potent Parkin target for regulating the antiviral immune response.
Figure 4.
Identification of TRAF3 as a Parkin-interacting protein. A–C, overexpression of Parkin reduced RIG-I(N)- and MAVS-mediated activation of the IFN-β, NF-κB, and ISRE promoters. HEK293 cells were co-transfected with the indicated expression plasmids and luciferase reporter constructs driven by the promoter of gene encoding IFN-β (A), NF-κB (B), or ISRE (C). Renilla was used as an internal control. 24 h after transfection, the cells were lysed for luciferase assays. D, Parkin overexpression failed to suppress activation of the ISRE promoter induced by IKKi and IRF3–5D. HEK293 cells were co-transfected with the indicated expression plasmids and a luciferase reporter construct driven by the promoter of the gene encoding ISRE, with Renilla used as an internal control. 24 h after transfection, the cells were lysed for luciferase assays. E and F, Parkin interacted with TRAF3. HEK293 cells were transfected with the indicated expression plasmids. 24 h after transfection, the cell lysates were immunoprecipitated with anti-Myc beads or anti-Flag beads and analyzed by immunoblotting (IB) with the indicated antibodies. Bottom panel of E or F, respectively, expression of exogenous proteins in WCL. G, endogenous TRAF3 interacted with exogenous Parkin in HEK293 cells. HEK293 cells were transfected with empty vector or Parkin Flag-tagged plasmids. 24 h after transfection, lysates were immunoprecipitated with anti-Flag beads, followed by immunoblotting with the indicated antibodies. Bottom panel, expression of endogenous proteins in WCL. H, Parkin was co-localized with TRAF3 in transfected HeLa cells in the presence or absence of SeV infection. HeLa cells were transfected with the indicated expression plasmids. 24 h after transfection, the cells were untreated or infected with SeV for the indicated times, then fixed, and stained with DAPI, anti-Flag, and anti-Myc antibodies. The cells were observed by confocal microscopy. The data shown in A–D are from a representative experiment of at least three independent experiments (means ± S.D. of duplicate assays). Two-tailed Student's t test was used to determine statistical significance. *, p < 0.05; **, p < 0.01; n.s., not significant, versus control groups. S. E, short exposure; L. E, long exposure; Rel. Luc. Act, relative luciferase activity.
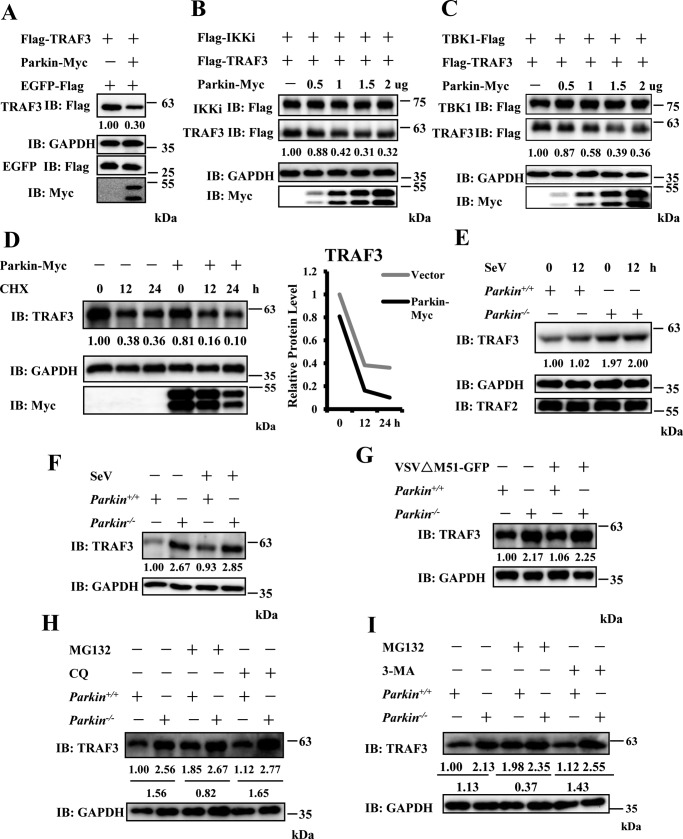
Parkin influences TRAF3 stability
Previous evidence has shown that Parkin functions as an E3 ligase to modulate protein turnover in a proteasomally or mitochondrial autophagy (mitophagy)-dependent manner (26). First, we determined whether Parkin regulates TRAF3 stability, because Parkin interacts with TRAF3. As shown in Fig. 5A, overexpression of Parkin reduced the abundance of ectopic TRAF3 protein. A previous study demonstrated that IKKi plays a role in TRAF3 protein turnover (27). Consistently, we found that with co-transfection of IKKi in 293T cells, Parkin overexpression enhanced TRAF3 degradation in a dose-dependent manner (Fig. 5B). We observed similar results with co-transfection of TBK1, Parkin and TRAF3 (Fig. 5C). In addition, we found that Parkin overexpression shortened the half-life of endogenous TRAF3 in HEK293 cells (Fig. 5D). To further confirm these results, we examined the impact of Parkin deficiency on TRAF3 protein turnover in the presence or absence of SeV infection. As shown in Fig. 5E, TRAF3 protein was more stable in primary Parkin−/− MEFs than control cells. Similar results were observed in Parkin+/+ and Parkin−/− immortalized MEFs, with or without SeV or VSVΔM51-GFP infection (Fig. 5, F and G). Next, we investigated how Parkin mediates TRAF3 turnover. To determine which pathway (proteasome or mitophagy) is involved in Parkin-mediated TRAF3 turnover, we pretreated Parkin+/+and Parkin−/− immortalized MEFs with a proteasome inhibitor (MG132) and autophagy inhibitor (chloroquine (CQ)) and performed Western blotting assays. Our results show that MG132, but not CQ treatment, significantly inhibited the decrease in TRAF3 turnover in WT cells (Fig. 5H). Similarly, we found that another autophagy inhibitor, 3-methyladenine (3-MA), could inhibit vesicular stomatitis mutant virus–induced autophagy, but failed to affect the stability of TRAF3 (Fig. 5I and Fig. S4). These data suggest that Parkin regulates TRAF3 turnover through the proteasomal pathway.
Figure 5.
Parkin affects TRAF3 stability. A, overexpression of Parkin reduced the abundance of exogenous TRAF3 protein. HEK293 cells were transfected with Flag–TRAF3 and EGFP–Flag together with empty vector or Parkin–Myc plasmids. EGFP–Flag was used to determine transfection efficiency. 24 h after transfection, the cell lysates were analyzed by immunoblotting with the indicated antibodies. B and C, Parkin overexpression enhanced TRAF3 degradation in a dose-dependent manner with co-expression of IKKi or TBK1. HEK293 cells were co-transfected with TRAF3 and IKKi (B) or TBK1 (C) together with empty vector or increased dose of Parkin–Myc plasmids. 24 h after transfection, the cell lysates were analyzed by immunoblotting with the indicated antibodies. D, overexpression of Parkin shortened the half-life of endogenous TRAF3 in HEK293 cells. HEK293 cells were transfected with the indicated expression plasmids. 24 h after transfection, the cells were treated with CHX (final concentration, 50 μg/ml) for the indicated times. The cell lysates were analyzed by immunoblotting with the indicated antibodies. Densitometry analysis to quantify TRAF3 expression is shown in the right panel. E, Parkin deficiency enhanced TRAF3 protein stability in primary Parkin−/− MEFs. Primary Parkin+/+ and Parkin−/− MEFs were infected with SeV for 12 h. The cell lysates were analyzed by immunoblotting with the indicated antibodies. F and G, TRAF3 protein was more stable in Parkin−/− immortalized MEFs. Immortalized Parkin+/+ and Parkin−/− MEFs were infected with SeV (F) or VSVΔM51-GFP at MOI of 0.5 (G) for 6 h. The cell lysates were analyzed by immunoblotting with the indicated antibodies. H, the treatment of MG132, but not CQ, affected the stability of TRAF3 protein in Parkin+/+ immortalized MEFs. Immortalized Parkin+/+ and Parkin−/− MEFs were left untreated or treated with MG132 (final concentration, 25 μm) or CQ (final concentration, 25 μg/ml) for 10 h. The cell lysates were analyzed by immunoblotting with the indicated antibodies. I, autophagy inhibitor 3-MA failed to affect the stability of TRAF3 protein in Parkin+/+ immortalized MEFs. Immortalized Parkin+/+ and Parkin−/− MEFs were left untreated or treated with MG132 (final concentration, 25 μm) or 3-MA (final concentration, 5 mm) for 9 h. The cell lysates were analyzed by immunoblotting with the indicated antibodies. Numbers below lanes (top) indicate densitometry of the protein presented relative to GAPDH expression in the same lane.
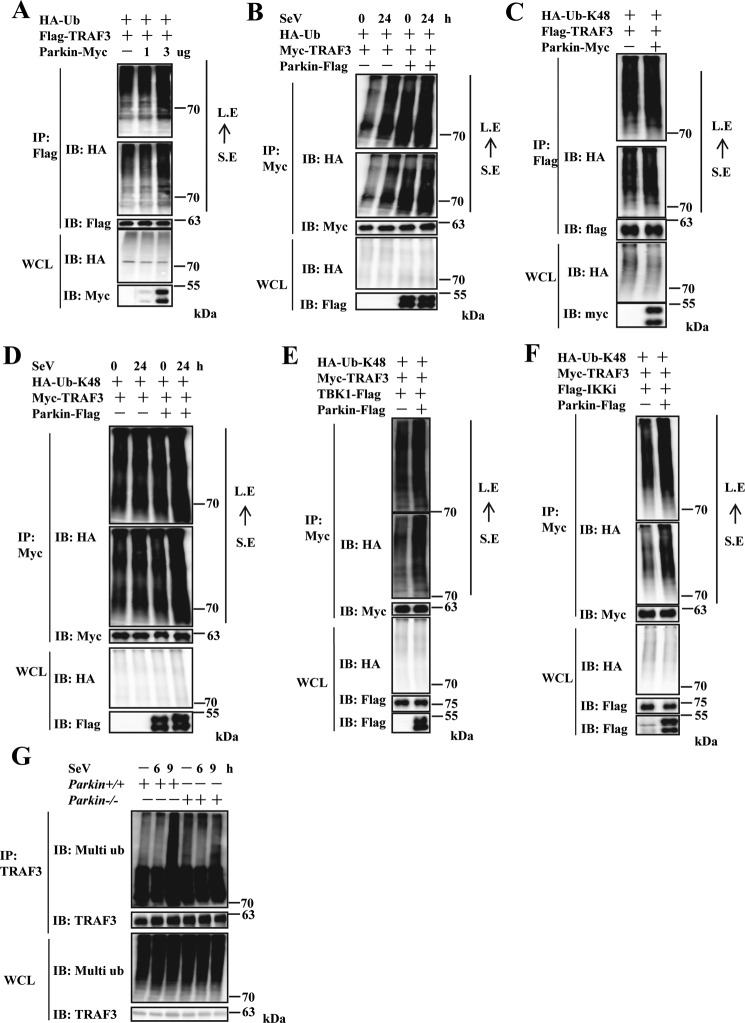
Parkin modulates Lys48-linkage ubiquitination of TRAF3
Previous studies have shown that Parkin has E3 ubiquitin ligase activity in vitro and modulates ubiquitination of many cytosolic and outer mitochondrial membrane proteins (such as mitofusin Mfn1 (28), Mfn2 (29, 30), and mitochondrial rho GTPase 1 (Miro1) (31, 32)) to regulate protein turnover. Thus, we determined whether Parkin regulates TRAF3 turnover through TRAF3 ubiquitination. First, we performed in vivo ubiquitination assays in HEK293 cells. As shown in Fig. 6A, TRAF3 was polyubiquitinated in cells co-transfected with a vector expressing WT ubiquitin. Further, enhanced TRAF3 ubiquitination was observed with Parkin co-expression in a dose-dependent manner. Next, we determined whether viral infection had an effect on Parkin-mediated ubiquitination of TRAF3. As shown in Fig. 6B, Parkin overexpression significantly increased TRAF3 ubiquitination levels in the presence or absence of SeV infection. Of note, TRAF3 ubiquitination levels increased after SeV infection. Taken together, these results suggest that Parkin regulates TRAF3 ubiquitination. Because Lys48-linkage ubiquitination of proteins is predominantly involved in protein turnover, we used an ubiquitin mutant (in which only one lysine (at position 48) is available for ubiquitination) to determine whether Parkin modulates Lys48-linkage ubiquitination of TRAF3. As shown in Fig. 6C, Parkin significantly increased Lys48-linked ubiquitination of TRAF3. Similar results were observed with SeV infection and TBK1 or IKKi co-expression (Fig. 6, D–F). Consistent with the overexpression experiments, endogenous TRAF3 ubiquitination levels were markedly reduced in Parkin−/− MEFs compared with WT MEFs at 9 h postinfection with SeV (Fig. 6G). Collectively, these results demonstrate that Parkin regulates Lys48-linkage ubiquitination of TRAF3.
Figure 6.
Parkin increases Lys48-linked ubiquitination of TRAF3. A, overexpression of Parkin enhanced exogenous TRAF3 polyubiquitination in a dose-dependent manner. HEK293 cells were transfected with Flag–TRAF3 and HA–Ub together with empty vector or increased dose of Parkin–Myc plasmids. 24 h after transfection, the cells were treated with MG132 (final concentration, 25 μm) for 4 h. The cell lysates were immunoprecipitated with anti-Flag beads and analyzed by immunoblotting (IB) with the indicated antibodies. Bottom panel, expression of exogenous proteins in WCL. B, overexpression of Parkin increased exogenous TRAF3 polyubiquitination in the presence of SeV infection. HEK293 cells were transfected with Myc-TRAF3 and HA–Ub together with empty vector or Parkin–Flag plasmids. 24 h after transfection, the cells were infected with SeV for 24 h and then treated with MG132 (final concentration, 25 μm) for 4 h. The cell lysates were immunoprecipitated with anti-Myc beads and analyzed by immunoblotting with the indicated antibodies. Bottom panel, expression of exogenous proteins in WCL. C, Parkin overexpression increased Lys48-linked ubiquitination of exogenous TRAF3. HEK293 cells were transfected with Flag–TRAF3 and HA–Ub–Lys48 together with empty vector or Parkin–Myc plasmids. 24 h after transfection, the cells were treated with MG132 (final concentration, 25 μm) for 4 h. The cell lysates were immunoprecipitated with anti-Flag beads and analyzed by immunoblotting with the indicated antibodies. Bottom panel, expression of exogenous proteins in WCL. D, Parkin overexpression enhanced Lys48-linked ubiquitination of exogenous TRAF3 in the presence of SeV infection. HEK293 cells were transfected with Myc-TRAF3 and HA–Ub–Lys48 together with empty vector or Parkin–Flag plasmids. 24 h after transfection, the cells were infected with SeV for 24 h and then treated with MG132 (final concentration, 25 μm) for 4 h. The cell lysates were immunoprecipitated with anti-Myc beads and analyzed by immunoblotting with the indicated antibodies. Bottom panel, expression of exogenous proteins in WCL. E and F, Parkin overexpression increased Lys48-linked ubiquitination of exogenous TRAF3 with co-expression of IKKi or TBK1. HEK293 cells were transfected with Myc-TRAF3, HA–Ub–Lys48, and TBK1–Flag (E) or Flag–IKKi (F) together with empty vector or Parkin–Flag plasmids. 24 h after transfection, the cells were treated with MG132 (final concentration, 25 μm) for 4 h. The cell lysates were immunoprecipitated with anti-Myc beads and analyzed by immunoblotting with the indicated antibodies. Bottom panel of A, B, C, D, E, or F, respectively, expression of exogenous proteins in WCL. G, Parkin deficiency significantly reduced endogenous TRAF3 ubiquitination induced by SeV infection in Parkin−/− immortalized MEFs. Immortalized Parkin+/+ and Parkin−/− MEFs were left untreated or infected with SeV for the indicated times, then treated with MG132 (final concentration, 25 μm) for 4 h. The cell lysates were immunoprecipitated with anti-TRAF3 antibody and analyzed by immunoblotting with the indicated antibodies. Bottom panel, expression of endogenous proteins in WCL. S. E, short exposure; L. E, long exposure.
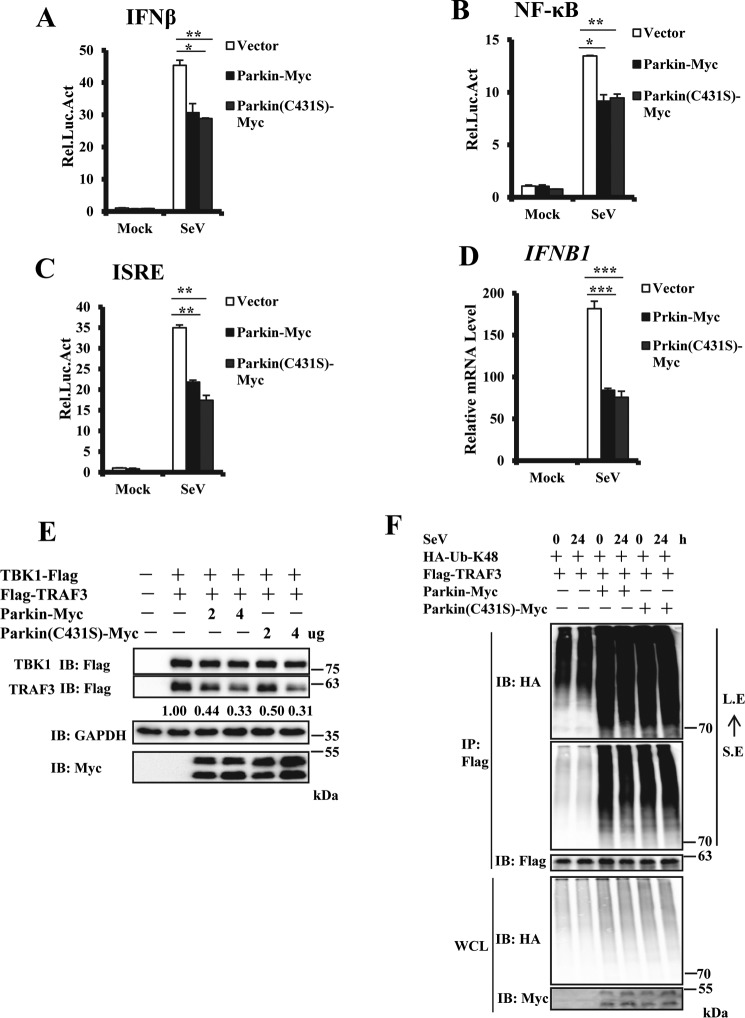
Parkin attenuates TRAF3 function in an E3 ligase–independent manner
Because Parkin is an E3 ubiquitin ligase, we determined whether enzymatic activity of Parkin is required for regulating TRAF3 ubiquitination. First, we generated a catalytically inactive mutant of Parkin (Parkin C431S) and performed reporter assays. As shown in Fig. 7 (A–C), overexpression of WT Parkin or catalytically inactive mutant C431S suppressed IFN-β, NF-κB, and ISRE promoter activation induced by SeV infection. Second, qRT–PCR assays showed that overexpression of both WT and C431S mutant forms of Parkin significantly reduced IFNB1 transcript levels induced by SeV infection (Fig. 7D). Third, Western blotting showed that overexpression of Parkin C431S mutant significantly reduced TRAF3 protein levels, similar to WT Parkin (Fig. 7E). Fourth, in vivo ubiquitination assays found no significant distinction in TRAF3 ubiquitination levels modulated by WT and mutant C431S of Parkin (Fig. 7F). Taken together, these data suggest that Parkin inhibits TRAF3 function in an E3 ligase–independent manner.
Figure 7.
Parkin inhibits TRAF3 function in E3 ligase–independent manner. A–C, overexpression of Parkin (WT) or Parkin (C431S) showed similar inhibition in SeV-induced activation of the IFN-β, NF-κB and ISRE promoters. HEK293 cells were co-transfected with the indicated expression plasmids and luciferase reporter constructs driven by promoter of gene encoding IFN-β (A), NF-κB (B), or ISRE (C). pRSV/LacZ was used as an internal control. 24 h after transfection, the cells were infected with SeV for 24 h and then lysed for luciferase assays. D, overexpression of Parkin (WT) or Parkin (C431S) showed no different inhibition in SeV-induced transcription of the IFNB1 gene. HEK293 cells were transfected with the indicated expression plasmids. 24 h after transfection, the cells were infected with SeV for 6 h, and cell lysates were analyzed by qRT–PCR to examine IFNB1 transcript levels. E, overexpression of Parkin (WT) or Parkin (C431S) showed similar enhancement of TRAF3 degradation in a dose-dependent manner with co-expression of TBK1. HEK293 cells were transfected with the indicated expression plasmids. The cell lysates were analyzed by immunoblotting with the indicated antibodies. F, overexpression of Parkin (WT) or Parkin (C431S) showed similar increased Lys48-linked ubiquitination of exogenous TRAF3. HEK293 cells were transfected with Flag–TRAF3 and HA–Ub–Lys48 together with Parkin–Myc or Parkin (C431S)–Myc plasmids. 24 h after transfection, the cells were infected with SeV for 24 h and treated with MG132 (final concentration, 25 μm) for 4 h. The cell lysates were immunoprecipitated with anti-Flag beads and analyzed by immunoblotting with the indicated antibodies. Bottom panel, expression of exogenous proteins in WCL. The data in A–D are from a representative experiment of at least three independent experiments (means ± S.D. of duplicate assays in A-C; triplicate assays in D). Two-tailed Student's t test was used to determine statistical significance. *, p < 0.05; **, p < 0.01; ***, p < 0.001, versus control groups. S. E, short exposure; L. E, long exposure; Rel. Luc. Act, relative luciferase activity.
Discussion
Parkinson's disease is a neurodegenerative disorder, with chronic neuroinflammation being one characteristic of PD pathology (1). Previous investigations have shown that Parkin gene mutations are related to the early-onset recessive form of PD and isolated juvenile-onset PD (7). It is well-documented that Parkin plays a key role in modulating degradation of damaged mitochondria via selective autophagy (mitophagy) (33). However, whether Parkin is involved in other cellular events is still largely unknown. Because the frequency of PD rapidly increases over the age of 60 years and with elderly people at enhanced susceptibility to infection, in this study we investigated whether Parkin is involved in regulation of antiviral signaling. We observed that in addition to modulating NF-κB signaling induced by viral infection, Parkin also suppressed IRF3-mediated antiviral signaling. Mechanistically, we show that Parkin inhibits IRF3 signaling via ubiquitination-mediated degradation of the adaptor protein, TRAF3.
Previous studies have shown that Parkin modulates neuroprotection through activation of NF-κB signaling, in which Parkin is recruited to the linear ubiquitin assembly complex (LUBAC) to enhance linear ubiquitination of IKKγ/NEMO, the regulatory subunit of the IκB kinase complex (10). Parkin was also found to regulate degradation-independent ubiquitination of IKKγ and TRAF2 to active NF-κB signaling (34). Nonetheless, the role of Parkin in classical NF-κB signaling is controversial. A recent study demonstrated that Parkin inhibits NF-κB signaling by increasing proteasomal degradation of TRAF2/6 and thereby reducing inflammation and TNFα/IL-1β–induced cell death (12). Here, we show that Parkin suppressed NF-κB signaling induced by DNA and RNA viral infection. Compared with the very strong interaction between Parkin and TRAF3, we detected the weak interaction between Parkin and TRAF6 and much weaker interaction between Parkin and TRAF2 at the same co-IP condition (Fig. S5). Thus, further investigation is needed to determine how Parkin regulates NF-κB signaling induced by viral infection.
Parkin was recently reported to modulate MAVS-mediated signaling of innate immunity. Under hepatitis B virus infection, Parkin interacts with MAVS, recruits LUBAC to mitochondria and accumulates unanchored linear polyubiquitin chains on MAVS to affect MAVS-mediated signaling (35). However, in our study, we failed to detect the interaction between Parkin and MAVS (Fig. S6). Our results indicated that Parkin modulated Lys48-linkage ubiquitination of TRAF3 to subsequently reduce TRAF3 stability. Consequently, this results in suppression of TRAF3-mediated signaling during the antiviral immune response against RNA and DNA viruses.
Previous studies showed that Parkin functions as an E3 ligase to regulate a variety of cellular events, such as mitophagy, inflammation, and TNFα-induced cell death (36, 37). In this study, our results suggested that Parkin played an important role in antiviral signaling by regulating TRAF3 Lys48-linked ubiquitination in an E3 ligase–independent manner; this raised the question about how Parkin modulated TRAF3 protein stability. We propose several possible models. First, Parkin enhances the interaction of Parkin with other E3 ligases, such as Triad3A (38), which targets TRAF3 to degrade. Second, the presence of Parkin affects TRAF3 ubiquitination by antagonizing activities of DUBs, such as OTUD7B and USP25, which targets TRAF3 for deubiquitination (39, 40). Third, Parkin affects LUBAC's function, which was reported to play an important role in innate immunity. Finally, the interaction of Parkin with TRAF3 maybe promotes TRAF3 autoubiquitination by enhancing the TRAF3 E3 ligase activity. To understand the exact mechanism of Parkin regulation on TRAF3, further investigation is needed in future. Taken together, our results provide new insight into understanding the novel function of Parkin in innate antiviral immunity and the mechanism by which Parkin regulates chronic inflammation in PD.
Experimental procedures
Ethics statements
All animal studies were performed in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the Ministry of Science and Technology of the People's Republic of China. The protocols for animal studies were approved by the Committee on the Ethics of Animal Experiments of the Institute of Zoology, Chinese Academy of Sciences (Beijing, China; approval number IOZ15001).
Cell culture and animals
HEK293, HeLa, and Vero cells were obtained from the Shanghai Cell Bank of Chinese Academy of Sciences (Shanghai, China). The cells were maintained in DMEM supplemented with 10% fetal bovine serum, 1% penicillin, and 1% streptomycin. Parkin+/+ and Parkin−/− MEFs were generated from 13.5-day embryos of Parkin+/+ and Parkin−/− mice (41) (The Jackson Laboratory). MEFs were maintained in complete DMEM containing 1 mm sodium pyruvate, 10 μm l-glutamine, 10 μm β-mercaptoethanol, and 1% nonessential amino acids. BMDMs were prepared as described previously (42). Briefly, bone marrow cells were harvested from mouse femurs and tibiae and cultured for 5 days in complete DMEM containing conditioned medium from L929 cell culture before collection for experiments. Parkin+/+ and Parkin−/− MEF cells were immortalized using SV40 large T antigen and kindly provided by Dr. Tie-shan Tang (Institute of Zoology, Chinese Academy of Sciences, Beijing, China).
Plasmids
Mammalian expression plasmids encoding Myc-tagged Parkin, TRAF3, HA-tagged ubiquitin, ubiquitin-Lys48, and Flag-tagged RIG-I(N), MAVS, TBK1, IKKi, Parkin, TRAF2, TRAF6, IRF3, IRF3–5D, and IRF7 were constructed by standard molecular biology methods. Parkin mutant was generated by PCR using Pfu DNA polymerase. IFN-β-, NF-κB-, and ISRE-luciferase reporter plasmids have been described previously (43).
Transfection and luciferase reporter analysis
HEK293 cells were seeded in 24-well plates and transfected the following day by the standard calcium phosphate transfection method. pRSV/lacZ or Renilla reporter plasmid and firefly luciferase reporter plasmids were co-transfected with the indicated expression plasmids. An empty control plasmid was used in the same experiment to ensure that the same amount of total DNA was transfected. The cells were lysed to measure luciferase activity. Firefly luciferase activity was normalized to lacZ or Renilla activity. All reporter assays were repeated at least three times.
Antibodies
Rabbit anti-phospho-IRF3 and anti-IRF3 antibodies were from Cell Signaling Technology. Rabbit anti-TRAF3 and anti-TRAF2 were from Santa Cruz Biotechnology. Mouse anti-glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and mouse anti-HA antibodies were from Sungene Biotechnology. Rabbit anti-Flag antibody was from Sigma. Mouse anti-Flag, rabbit anti-HA, rabbit anti-Myc, mouse anti-Myc, and mouse multiubiquitin antibodies were from MBL. Rabbit anti-LC3 antibody was from NOVUS.
Viruses and viral plaque assay
SeV, VSVΔM51-GFP, and HSV-1 have been described previously (42, 43). For viral plaque assay, confluent Vero cells were infected with diluted VSVΔM51-GFP virus for 1 h. Culture medium containing 2% methylcellulose was overlaid and incubated for ∼36 h. The cells were then fixed for 15 min with methanol and stained with 1% crystal violet to identify plaques. Plaques were counted to quantitate viral titer as plaque-forming units/ml.
Co-immunoprecipitation, immunoblotting analysis, and native gel electrophoresis
These experiments were performed using the following method (43). For overexpression and semiendogenous co-IP, the cells were lysed in 0.5% Triton X-100 lysis buffer (20 mm Tris-Cl, pH 7.5, 150 mm NaCl, 0.5% Triton X-100, 10% glycerol, and 1 mm EDTA). The cell lysates were incubated with anti-Flag beads (Sigma) or anti-Myc beads (Biotool) for 4–6 h at 4 °C. Immune complexes were washed three times with lysis buffer and analyzed by immunoblotting. Loading amount of IP samples, and epitope tag protein was 20% of whole-cell lysate (WCL) epitope tag protein. Loading amount of WCL samples, and epitope tag protein was 2% of WCL epitope tag protein.
Cell immunofluorescence
For microscopy images, HeLa cells were grown on gelatin-coated glass coverslips and transfected or infected as described. After washing with PBS, the cells were fixed with 4% paraformaldehyde for 10 min, permeabilized, and blocked with 0.2% Triton X-100 in PBS containing 5% BSA for 30 min at room temperature. The cells were then incubated with primary antibody, followed by secondary antibody. The cells were washed with PBST (PBS with 0.2% Tween 20) between each step. The coverslips were mounted, and the cells were imaged on a LSM 510 META analyzer (Zeiss, Oberkochen, Germany).
In vivo ubiquitination assay
In vivo ubiquitination assays have been described previously (44). Briefly, HEK293 cells were transfected with the indicated plasmids. 24 h post-transfection or after being infected for 24 h by SeV after transfection, the cells were treated with MG132 (final concentration, 25 μm) for 4 h. The cells were lysed in 100 μl of lysis buffer A (150 mm NaCl, 50 mm Tris-HCl, pH 7.4, 10% glycerol, 0.5% Nonidet P-40, 1% SDS, 1 mm EDTA, and 10 mm N-ethylmaleimide), diluted with 1 ml of buffer B (150 mm NaCl, 50 mm Tris-HCl, pH 7.4, 10% glycerol, 1 mm EDTA, and 0.5% Nonidet P-40), and sonicated briefly. After incubating with anti-Flag beads or anti-Myc beads for 4 h, the beads were extensively washed three times for 1 h with lysis buffer containing 0.1% SDS and 500 mm NaCl. The samples were then subjected to Western blotting analysis with the indicated antibodies. For endogenous ubiquitination assay, the cells were infected with SeV for the indicated time and treated with MG132 (final concentration, 25 μm) for 4 h. The cells were lysed in 100 μl of lysis buffer A, diluted with 1 ml of buffer B, and sonicated briefly. The cell lysates were incubated overnight at 4 °C with anti-TRAF3 antibody and then incubated for 2–3 h with protein A/G-agarose beads. Immune complexes were extensively washed, followed by immunoblotting with the indicated antibodies.
qRT–PCR
Total RNA was extracted using TRIzol reagent (Invitrogen). cDNA was synthesized using the SuperScript III first-strand cDNA synthesis kit (Invitrogen). Real-time PCR was performed in triplicate using SYBR Green Master Mix (Thermo Fisher Scientific) on a Light Cycler 480® (Roche). Relative mRNA levels were normalized to GAPDH or actin mRNA levels in each sample. The 2−ΔΔCt method was used to calculate relative expression changes. The data are shown as mRNA abundance relative to control groups. The primers used were as follows (5′–3′): hGAPDH-S, ATGACATCAAGAAGGTGGTG; hGAPDH-AS, CATACCAGGAAATGAGCTTG; hIFNB1-S, AGGACAGGATGAACTTTGAC; hIFNB1-AS, TGATAGACATTAGCCAGGAG; mActin-S, TCCAGCCTTCCTTCTTGGGT; mActin-AS, GCACTGTGTTGGCATAGAGGT; mCxcl10-S, GAATCCGGAATCTAAGACCATCAA; mCxcl10-AS, GTGCGTGGCTTCACTCCAGT; mIfnb1-S, ATGGTGGTCCGAGCAGAGAT; mIfnb1-AS, CCACCACTCATTCTGAGGCA; mIl-6-S, TCCATCCAGTTGCCTTCTTG; mIl-6-AS,GGTCTGTTGGGAGTGGTATC; mIfit1-S, CTGAGATGTCACTTCACATGGAA; mIfit1-AS, GTGCATCCCCAATGGGTTCT; mIfnα4-S, AGCCTGTGTGATGCAGGAACC; and mIfnα4-AS, CAGCAAGTTGGTTGAGGAAGAG.
Statistical analyses
The results of all statistical analyses are shown as means ± S.D. Significant differences between values under different experimental conditions were performed using two-tailed Student's t test. For all tests, p values < 0.05 were considered statistically significant.
Author contributions
D. X. resources; D. X. data curation; D. X. software; D. X. formal analysis; D. X. validation; D. X., H. G., and E. L. investigation; D. X. visualization; D. X., H. G., and E. L. methodology; D. X. and Q. S. writing-original draft; Q. S. conceptualization; Q. S. supervision; Q. S. writing-review and editing.
Supplementary Material
This work was supported by Strategic Priority Research Program of Chinese Academy of Sciences Grant XDB13000000 and National Natural Science Foundation of China Grants 31570916, 31370882, and 31371450. The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Fig. S1–S6.
- PD
- Parkinson's disease
- IL
- interleukin
- IFN
- interferon
- NEMO
- NF-κB essential modulator
- TNF
- tumor necrosis factor
- TRAF
- TNF receptor–associated factor
- RIG
- retinoic acid–inducible gene
- MAVS
- mitochondrial antiviral-signaling protein
- STING
- stimulator of interferon genes
- SeV
- Sendai virus
- IRF
- IFN regulatory factor
- ISRE
- IFN-stimulated response element
- qRT–PCR
- quantitative real-time PCR
- MEF
- mouse embryonic fibroblast
- ISD
- interferon stimulatory DNA
- HSV
- herpes simplex virus
- BMDM
- bone marrow–derived macrophage
- IP
- immunoprecipitation
- CQ
- chloroquine
- 3-MA
- 3-methyladenine
- LUBAC
- linear ubiquitin assembly complex
- IKK
- inhibitor of κB kinase
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase
- Ub
- ubiquitin
- WCL
- whole-cell lysate
- MOI
- multiplicity of infection.
References
- 1. Lee J. K., Tran T., and Tansey M. G. (2009) Neuroinflammation in Parkinson's disease. J. Neuroimmune Pharmacol. 4, 419–429 10.1007/s11481-009-9176-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Whitton P. S. (2007) Inflammation as a causative factor in the aetiology of Parkinson's disease. Br. J. Pharmacol. 150, 963–976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kannarkat G. T., Boss J. M., and Tansey M. G. (2013) The role of innate and adaptive immunity in Parkinson's disease. J. Parkinsons Dis. 3, 493–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chen H., O'Reilly E. J., Schwarzschild M. A., and Ascherio A. (2008) Peripheral inflammatory biomarkers and risk of Parkinson's disease. Am. J. Epidemiol. 167, 90–95 [DOI] [PubMed] [Google Scholar]
- 5. Chen H., Jacobs E., Schwarzschild M. A., McCullough M. L., Calle E. E., Thun M. J., and Ascherio A. (2005) Nonsteroidal antiinflammatory drug use and the risk for Parkinson's disease. Ann. Neurol. 58, 963–967 10.1002/ana.20682 [DOI] [PubMed] [Google Scholar]
- 6. Chen H., Zhang S. M., Hernán M. A., Schwarzschild M. A., Willett W. C., Colditz G. A., Speizer F. E., and Ascherio A. (2003) Nonsteroidal anti-inflammatory drugs and the risk of Parkinson disease. Arch. Neurol. Chicago 60, 1059–1064 10.1001/archneur.60.8.1059 [DOI] [PubMed] [Google Scholar]
- 7. Lücking C. B., Dürr A., Bonifati V., Vaughan J., De Michele G., Gasser T., Harhangi B. S., Meco G., Denèfle P., Wood N. W., Agid Y., Brice A., French Parkinson's Disease Genetics Study Group, and European Consortium on Genetic Susceptibility in Parkinson's Disease (2000) Association between early-onset Parkinson's disease and mutations in the parkin gene. New Engl. J. Med. 342, 1560–1567 10.1056/NEJM200005253422103 [DOI] [PubMed] [Google Scholar]
- 8. Shimura H., Hattori N., Kubo S., Mizuno Y., Asakawa S., Minoshima S., Shimizu N., Iwai K., Chiba T., Tanaka K., and Suzuki T. (2000) Familial Parkinson disease gene product, parkin, is a ubiquitin-protein ligase. Nat. Genet. 25, 302–305 10.1038/77060 [DOI] [PubMed] [Google Scholar]
- 9. Sai Y., Zou Z., Peng K., and Dong Z. (2012) The Parkinson's disease-related genes act in mitochondrial homeostasis. Neurosci. Biobehav. Rev. 36, 2034–2043 10.1016/j.neubiorev.2012.06.007 [DOI] [PubMed] [Google Scholar]
- 10. Müller-Rischart A. K., Pilsl A., Beaudette P., Patra M., Hadian K., Funke M., Peis R., Deinlein A., Schweimer C., Kuhn P. H., Lichtenthaler S. F., Motori E., Hrelia S., Wurst W., Trümbach D., et al. (2013) The E3 ligase parkin maintains mitochondrial integrity by increasing linear ubiquitination of NEMO. Mol. Cell 49, 908–921 10.1016/j.molcel.2013.01.036 [DOI] [PubMed] [Google Scholar]
- 11. Anglade P., Vyas S., Javoy-Agid F., Herrero M. T., Michel P. P., Marquez J., Mouatt-Prigent A., Ruberg M., Hirsch E. C., and Agid Y. (1997) Apoptosis and autophagy in nigral neurons of patients with Parkinson's disease. Histol. Histopathol. 12, 25–31 [PubMed] [Google Scholar]
- 12. Chung J. Y., Park H. R., Lee S. J., Lee S. H., Kim J. S., Jung Y. S., Hwang S. H., Ha N. C., Seol W. G., Lee J., and Park B. J. (2013) Elevated TRAF2/6 expression in Parkinson's disease is caused by the loss of Parkin E3 ligase activity. Lab. Invest. 93, 663–676 10.1038/labinvest.2013.60 [DOI] [PubMed] [Google Scholar]
- 13. Thompson M. R., Kaminski J. J., Kurt-Jones E. A., and Fitzgerald K. A. (2011) Pattern recognition receptors and the innate immune response to viral infection. Viruses 3, 920–940 10.3390/v3060920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Abdullah Z., and Knolle P. A. (2014) Scaling of immune responses against intracellular bacterial infection. EMBO J. 33, 2283–2294 10.15252/embj.201489055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Koppe U., Suttorp N., and Opitz B. (2012) Recognition of Streptococcus pneumoniae by the innate immune system. Cell Microbiol. 14, 460–466 10.1111/j.1462-5822.2011.01746.x [DOI] [PubMed] [Google Scholar]
- 16. Cottier F., and Pavelka N. (2012) Complexity and dynamics of host-fungal interactions. Immunol. Res. 53, 127–135 10.1007/s12026-012-8265-y [DOI] [PubMed] [Google Scholar]
- 17. Akira S., Uematsu S., and Takeuchi O. (2006) Pathogen recognition and innate immunity. Cell 124, 783–801 10.1016/j.cell.2006.02.015 [DOI] [PubMed] [Google Scholar]
- 18. Kumagai Y., and Akira S. (2010) Identification and functions of pattern-recognition receptors. J. Allergy Clin. Immun. 125, 985–992 10.1016/j.jaci.2010.01.058 [DOI] [PubMed] [Google Scholar]
- 19. Baum A., and García-Sastre A. (2010) Induction of type I interferon by RNA viruses: cellular receptors and their substrates. Amino Acids 38, 1283–1299 10.1007/s00726-009-0374-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jensen S., and Thomsen A. R. (2012) Sensing of RNA viruses: a review of innate immune receptors involved in recognizing RNA virus invasion. J. Virol. 86, 2900–2910 10.1128/JVI.05738-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Paludan S. R., and Bowie A. G. (2013) Immune sensing of DNA. Immunity 38, 870–880 10.1016/j.immuni.2013.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sun L., Liu S., and Chen Z. J. (2010) SnapShot: pathways of antiviral innate immunity. Cell 140, 436 10.1016/j.cell.2010.01.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tansey M. G., and Goldberg M. S. (2010) Neuroinflammation in Parkinson's disease: its role in neuronal death and implications for therapeutic intervention. Neurobiol. Dis. 37, 510–518 10.1016/j.nbd.2009.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. McGeer E. G., Klegeris A., and McGeer P. L. (2005) Inflammation, the complement system and the diseases of aging. Neurobiol. Aging 26, 94–97 10.1016/j.neurobiolaging.2005.08.008 [DOI] [PubMed] [Google Scholar]
- 25. Servant M. J., ten Oever B., LePage C., Conti L., Gessani S., Julkunen I., Lin R., and Hiscott J. (2001) Identification of distinct signaling pathways leading to the phosphorylation of interferon regulatory factor 3. J. Biol. Chem. 276, 355–363 10.1074/jbc.M007790200 [DOI] [PubMed] [Google Scholar]
- 26. Yoshii S. R., Kishi C., Ishihara N., and Mizushima N. (2011) Parkin mediates proteasome-dependent protein degradation and rupture of the outer mitochondrial membrane. J. Biol. Chem. 286, 19630–19640 10.1074/jbc.M110.209338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Paz S., Vilasco M., Werden S. J., Arguello M., Joseph-Pillai D., Zhao T., Nguyen T. L., Sun Q., Meurs E. F., Lin R., and Hiscott J. (2011) A functional C-terminal TRAF3-binding site in MAVS participates in positive and negative regulation of the IFN antiviral response. Cell Res. 21, 895–910 10.1038/cr.2011.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Glauser L., Sonnay S., Stafa K., and Moore D. J. (2011) Parkin promotes the ubiquitination and degradation of the mitochondrial fusion factor mitofusin 1. J. Neurochem. 118, 636–645 10.1111/j.1471-4159.2011.07318.x [DOI] [PubMed] [Google Scholar]
- 29. Gegg M. E., Cooper J. M., Chau K. Y., Rojo M., Schapira A. H., and Taanman J. W. (2010) Mitofusin 1 and mitofusin 2 are ubiquitinated in a PINK1/parkin-dependent manner upon induction of mitophagy. Hum. Mol. Genet. 19, 4861–4870 10.1093/hmg/ddq419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Tanaka A., Cleland M. M., Xu S., Narendra D. P., Suen D. F., Karbowski M., and Youle R. J. (2010) Proteasome and p97 mediate mitophagy and degradation of mitofusins induced by Parkin. J. Cell Biol. 191, 1367–1380 10.1083/jcb.201007013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Birsa N., Norkett R., Wauer T., Mevissen T. E., Wu H. C., Foltynie T., Bhatia K., Hirst W. D., Komander D., Plun-Favreau H., and Kittler J. T. (2014) Lysine 27 ubiquitination of the mitochondrial transport protein Miro is dependent on serine 65 of the Parkin ubiquitin ligase. J. Biol. Chem. 289, 14569–14582 10.1074/jbc.M114.563031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kazlauskaite A., Kelly V., Johnson C., Baillie C., Hastie C. J., Peggie M., Macartney T., Woodroof H. I., Alessi D. R., Pedrioli P. G., and Muqit M. M. (2014) Phosphorylation of Parkin at serine 65 is essential for activation: elaboration of a Miro1 substrate-based assay of Parkin E3 ligase activity. Open Biol. 4, 130213 10.1098/rsob.130213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Narendra D., Tanaka A., Suen D. F., and Youle R. J. (2008) Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J. Cell Biol. 183, 795–803 10.1083/jcb.200809125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Henn I. H., Bouman L., Schlehe J. S., Schlierf A., Schramm J. E., Wegener E., Nakaso K., Culmsee C., Berninger B., Krappmann D., Tatzelt J., and Winklhofer K. F. (2007) Parkin mediates neuroprotection through activation of IκB kinase/nuclear factor-κB signaling. J. Neurosci. 27, 1868–1878 10.1523/JNEUROSCI.5537-06.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Khan M., Syed G. H., Kim S. J., and Siddiqui A. (2016) Hepatitis B virus-induced Parkin-dependent recruitment of linear ubiquitin assembly complex (LUBAC) to mitochondria and attenuation of innate immunity. PLoS Pathog. 12, e1005693 10.1371/journal.ppat.1005693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Narendra D., Tanaka A., Suen D. F., and Youle R. J. (2009) Parkin-induced mitophagy in the pathogenesis of Parkinson disease. Autophagy 5, 706–708 10.4161/auto.5.5.8505 [DOI] [PubMed] [Google Scholar]
- 37. Dzamko N., Geczy C. L., and Halliday G. M. (2015) Inflammation is genetically implicated in Parkinson's disease. Neuroscience 302, 89–102 10.1016/j.neuroscience.2014.10.028 [DOI] [PubMed] [Google Scholar]
- 38. Nakhaei P., Mesplede T., Solis M., Sun Q., Zhao T., Yang L., Chuang T. H., Ware C. F., Lin R., and Hiscott J. (2009) The E3 ubiquitin ligase Triad3A negatively regulates the RIG-I/MAVS signaling pathway by targeting TRAF3 for degradation. PLoS Pathog. 5, e1000650 10.1371/journal.ppat.1000650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hu H., Brittain G. C., Chang J. H., Puebla-Osorio N., Jin J., Zal A., Xiao Y., Cheng X., Chang M., Fu Y. X., Zal T., Zhu C., and Sun S. C. (2013) OTUD7B controls non-canonical NF-κB activation through deubiquitination of TRAF3. Nature 494, 371–374 10.1038/nature11831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lin D., Zhang M., Zhang M. X., Ren Y., Jin J., Zhao Q., Pan Z., Wu M., Shu H. B., Dong C., and Zhong B. (2015) Induction of USP25 by viral infection promotes innate antiviral responses by mediating the stabilization of TRAF3 and TRAF6. Proc. Natl. Acad. Sci. U.S.A. 112, 11324–11329 10.1073/pnas.1509968112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Goldberg M. S., Fleming S. M., Palacino J. J., Cepeda C., Lam H. A., Bhatnagar A., Meloni E. G., Wu N., Ackerson L. C., Klapstein G. J., Gajendiran M., Roth B. L., Chesselet M. F., Maidment N. T., Levine M. S., et al. (2003) Parkin-deficient mice exhibit nigrostriatal deficits but not loss of dopaminergic neurons. J. Biol. Chem. 278, 43628–43635 10.1074/jbc.M308947200 [DOI] [PubMed] [Google Scholar]
- 42. Li Z., Liu G., Sun L., Teng Y., Guo X., Jia J., Sha J., Yang X., Chen D., and Sun Q. (2015) PPM1A regulates antiviral signaling by antagonizing TBK1-mediated STING phosphorylation and aggregation. PLoS Pathog. 11, e1004783 10.1371/journal.ppat.1004783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Zhao Y., Sun X., Nie X., Sun L., Tang T. S., Chen D., and Sun Q. (2012) COX5B regulates MAVS-mediated antiviral signaling through interaction with ATG5 and repressing ROS production. PLoS Pathog. 8, e1003086 10.1371/journal.ppat.1003086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Lin W., Zhang J., Lin H., Li Z., Sun X., Xin D., Yang M., Sun L., Li L., Wang H., Chen D., and Sun Q. (2016) Syndecan-4 negatively regulates antiviral signalling by mediating RIG-I deubiquitination via CYLD. Nat. Commun. 7, 11848 10.1038/ncomms11848 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.