Abstract
Synonymous nucleotide variation is increasingly recognized as a factor than can affect protein expression, but the underlying mechanisms are incompletely understood. Here, we investigated whether synonymous changes could affect expression of the potassium voltage-gated channel subfamily H member 2 (KCNH2) gene, encoding the human ether-a-go-go–related gene (hERG) ion channel, which is linked to hereditary cardiac arrhythmia. We examined a previously described synthetic version (hERG-codon modified (CM)) with synonymous substitutions designed to reduce GC content, rare codons, and mRNA secondary structure relative to the native construct (hERG-NT). hERG-CM exhibited lower protein expression than hERG-NT in HEK293T cells. We found that the steady-state abundance of hERG-NT mRNA was greater than hERG-CM because of an enhanced transcription rate and increased mRNA stability for hERG-NT. Translation of hERG-CM was independently reduced, contributing to the overall greater synthesis of hERG-NT channel protein. This was partially offset, however, by a higher aggregation of a newly synthesized hERG-NT channel, resulting in nonfunctional protein. Regional mRNA analyses of chimeras of hERG-NT and hERG-CM revealed that synonymous changes in the 5′ segments of the coding region had the greatest influence on hERG synthesis at both the mRNA and protein levels. Taken together, these results indicate that synonymous nucleotide variations within the coding region, particularly in the 5′ region of the hERG mRNA, can affect both transcription and translation. These findings support the notion that greater attention should be given to the effects of synonymous genetic variation when analyzing hERG DNA sequences in the study of hereditary cardiac disease.
Keywords: genetic polymorphism, genetic disease, potassium channel, protein synthesis, mRNA, transcription, translation, hERG
Introduction
mRNA has previously been viewed as the conveyer of the genetic message, moving information passively from the genetic script, DNA, to the executor, the protein. However, information embedded in the mRNA message outside the codon sequence can alter protein synthesis and function. Codon usage potentially influences translation rate; ribosomes are suspected to move rapidly along common codons, while pausing or slowing at rarer codons and thus influencing co-translational folding, particularly of transmembrane proteins (1–3). mRNA structure can influence the efficiency of translation initiation and plays a role in the speed of translation elongation as well, with highly structured regions causing translation deceleration (4). GC content has also been shown to influence protein quantity, likely due to transcription rates, where a higher GC content in the coding region of some genes leads to higher mRNA quantities (5). These “extra-coding” features of mRNA, when disrupted, are likely to lead to changes in protein synthesis.
The KCNH2 transcript, encoding the human ether-a-go-go–related gene (hERG)3 protein, the α subunit of a potassium ion channel with a critical function in cardiac repolarization, has unique mRNA characteristics. KCNH2 mRNA (termed hERG mRNA) carries a 66% GC content, uses numerous codons with a <10% usage frequency, and has 59 predicted complementary repeats. This gene, when mutated, also causes long QT syndrome, type 2 (LQT2), a cardiac arrhythmia disorder characterized by prolongation of the QT interval on electrocardiogram and with symptoms including palpitation, syncope, and sudden death. Research has focused primarily on the protein hERG and characterization of single nucleotide variants in KCNH2 leading to LQT2, but little attention has been focused on the importance of these interesting hERG mRNA features in protein expression and disease pathogenesis. To interrogate the role of the extra-coding features of hERG mRNA in hERG protein expression, a codon-modified hERG (hERG-CM) construct was created (Fig. S1). (6) This construct decreased the GC content, removed the use of codons under 10% usage frequency, and reduced the number of predicted complementary repeats, all while maintaining a synonymous amino acid sequence. The hERG-CM construct produced less hERG protein than the native hERG construct (hERG-NT) and the hERG-CM protein trafficked to the surface of the cell more efficiently than hERG-NT. In this investigation, we focus on understanding the various ways that these synonymous modifications affect hERG protein production.
Here, we queried multiple steps in hERG protein synthesis as well as protein aggregation and function. We demonstrate that extra-coding features of mRNA influence transcription rate, mRNA stability, translation rate, and protein aggregation. These data highlight the various ways that synonymous change can influence protein production and suggests greater attention be paid to synonymous genetic variation in long QT syndrome pathogenesis.
Results
hERG-CM protein generated from new vector backbone consistently yields decreased protein expression compared with hERG-NT
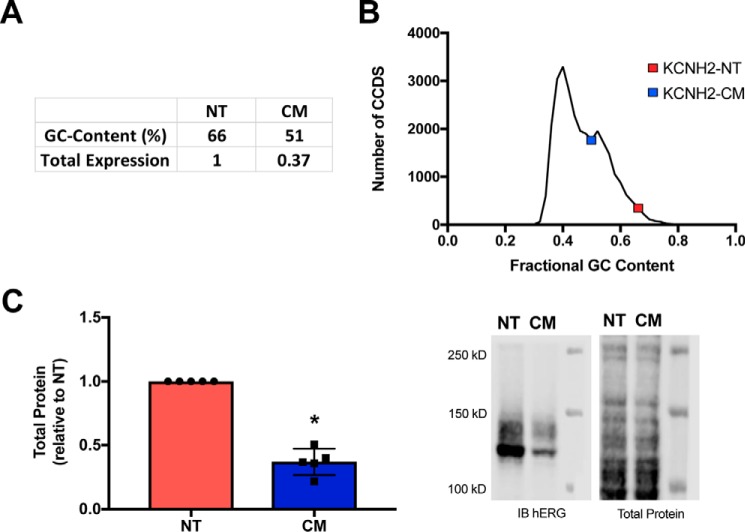
When the hERG-CM was created, we hypothesized that the reduction of GC content and elimination of rare codons would yield higher protein expression than the native hERG (hERG-NT). The primary goal in hERG-CM construction was to reduce GC content to understand the importance of GC content in hERG synthesis (Fig. 1A). As shown in Fig. 1B, the hERG-NT–coding region is in the 97th percentile of GC content among all gene-coding regions (>2 S.D. above the mean). However, as we reported previously (6), hERG-NT protein was expressed at significantly higher levels in human embryonic kidney 293T (HEK293T) cells than the codon-modified version. In this investigation the hERG-NT and hERG-CM constructs were moved into a new backbone without N-terminal tags and were verified for lack of inadvertent variants by sequencing of the entire cDNA, as these features of a vector can influence hERG protein expression (7). To confirm the findings of reduced hERG-CM protein expression in the pcDNA backbone, immunoblots were performed. The immunoblots probing lysate from HEK293T cells expressing hERG-NT exhibited ∼2.7 times more total hERG signal than a corresponding amount of lysate from hERG-CM–expressing cells (Fig. 1C). To determine the mechanism behind this difference of protein expression, we embarked on an analysis of each step in the synthesis of the channel.
Figure 1.
hERG-CM construct exhibits decreased protein expression. A, characteristics of hERG-NT and the codon-modified hERG-CM construct. B, graphical representation of the fractional GC content for the CCDS of all protein-coding genes. Data were obtained from Ensembl using the GrCH37 release of the human genome. C, hERG-NT and hERG-CM expression demonstrated by a graph of immunoblot (IB) data and representative immunoblot from Western blotting analysis of lysate 2 days post-transfection show greater total protein expression for hERG-NT. Data are combined from five independent hERG-NT and hERG-CM transfections (n = 5) performed to generate data used in later figures. Statistically significant difference (p < 0.0001) compared with hERG-NT as determined by a t test analysis is indicated with an asterisk.
hERG-NT mRNA is transcribed at a higher rate, has a longer half-life, and is structurally distinct from hERG-CM mRNA
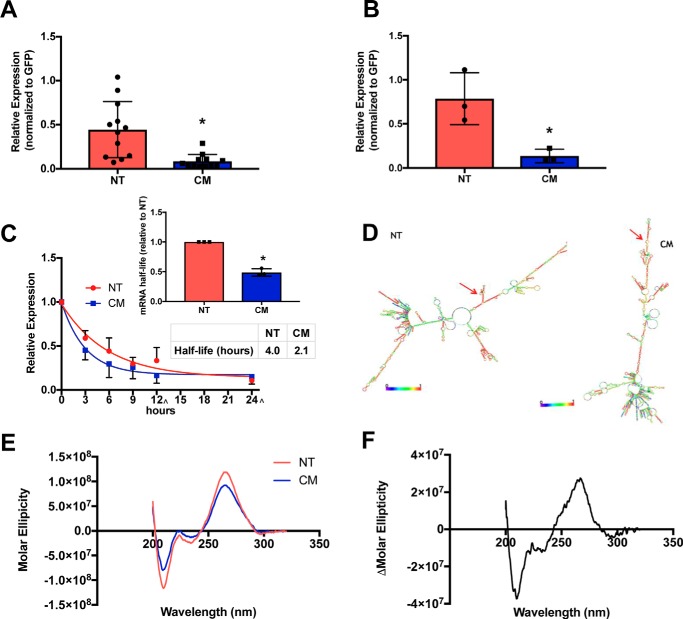
mRNA levels were queried to determine whether differences in transcript abundance could contribute to variation between hERG-NT and hERG-CM protein expression. Quantitative PCR (qPCR) results showed that hERG-NT mRNA levels are 5.3-fold higher than that of hERG-CM mRNA one-day post-transfection (Fig. 2A and Fig. S2). Thus, differential mRNA production and/or mRNA degradation was occurring between NT and CM. To investigate potential differences in transcription rate between hERG-NT and -CM, a pulse experiment was performed using a 30-min 4-thiouridine exposure (5, 8–10). The rate of synthesis of nascent hERG-NT mRNA was at a 5.9-fold greater rate than that of hERG-CM one-day post-transfection (Fig. 2B). These data demonstrate that synonymous modification within the coding region can impact transcription and mRNA expression.
Figure 2.
hERG-NT mRNA is synthesized at a greater rate, has an altered conformation, and a longer half-life than hERG-CM mRNA. A, hERG-NT and -CM mRNA expression levels 24 h post-transfection as determined by quantitative PCR. Four independent transfections were performed (n = 4) with three replicate primer groups per assay. Statistically significant difference (p = 0.0010) was determined by t test and indicated by an asterisk. B, rate of transcription of hERG-NT and hERG-CM determined with a 30-min 4-thiouridine pulse to label nascent mRNA synthesized 24 h post-transfection (n = 3). Statistically significant difference (p = 0.0209) was determined by t test and indicated by asterisk. C, results of one-phase decay analysis for hERG-NT and hERG-CM mRNA half-life determination using actinomycin-D to inhibit transcriptional 1-day post-transfection from three independent transfections (n = 3) with three replicate primer groups per assay. The 12-and 24-h collections were performed at +30 min in the sample 3 collection ([caret]). A bar graph represents the increases in mRNA half-life of hERG-NT compared with hERG-CM for each independent assay (inset). Statistical significance was determined by the difference in half-life values between NT and CM from three independent experiments using a t test (p = 0.0001) (inset) and by comparison of curve fit analysis (regression, p < 0.001). D, minimum free energy structure prediction from RNAFold for hERG-NT and hERG-CM mRNA. Red arrows demonstrate the AUG site. Global differences in the predicted mRNA structure can be noted between hERG-NT and hERG-CM. E, circular dichroism scans of hERG-NT and hERG-CM in in vitro-transcribed mRNA (n = 3). hERG-NT has a higher peak intensity than the hERG-CM peak. F, difference between the hERG-NT and hERG-CM CD spectra across the 320–200-nm wavelength.
To examine relative mRNA stability, the half-life of the hERG-NT and hERG-CM transcripts was measured. Half-life was determined by quantification of RNA expression at various time points over 24 h after exposure to actinomycin-D. The half-life of hERG-NT mRNA was ∼2-fold greater than that of hERG-CM (4.0 versus 2.1 h, respectively) (Fig. 2C) (p < 0.0001). This result indicates that synonymous modifications can affect mRNA stability.
To continue the investigation of differences in hERG-NT and -CM mRNA characteristics, we sought to test our hypothesis that modifying GC content of the mRNA had changed the structure of the mRNA. mRNA structure predictions were generated using RNAFold (11) that highlight differences between hERG-NT and hERG-CM mRNA (Fig. 2D). hERG-NT has a lower predicted ΔG than CM (−1620.9 kcal/mol versus −1229.9 kcal/mol respectively), indicating more stable structure present in the hERG-NT mRNA. To confirm global differences in mRNA structure, circular dichroism (CD) was utilized. Both hERG-NT and hERG-CM CD profiles demonstrated RNA folded into A-form helices (noted by a positive peak at ∼260 nm and a negative peak at ∼210 nm) (12, 13) and difference in nucleic acid composition can be noted due to a subtle shift in peak maximum amplitude (hERG-NT peak is located at 265.3 nm and hERG-CM peak is at 264.1 nm). The predicted global differences in hERG-NT and hERG-CM mRNA structure (Fig. 2D) were consistent with the mRNA CD results (Fig. 2, E and F), indicating increased base stacking and base interaction in the hERG-NT mRNA (14, 15), shown by an increase in peak amplitude of the hERG-NT CD spectrum when compared with the hERG-CM spectrum. This confirms that synonymous variation in the coding region can affect mRNA structure.
hERG-NT protein is translated at a greater rate than hERG-CM protein
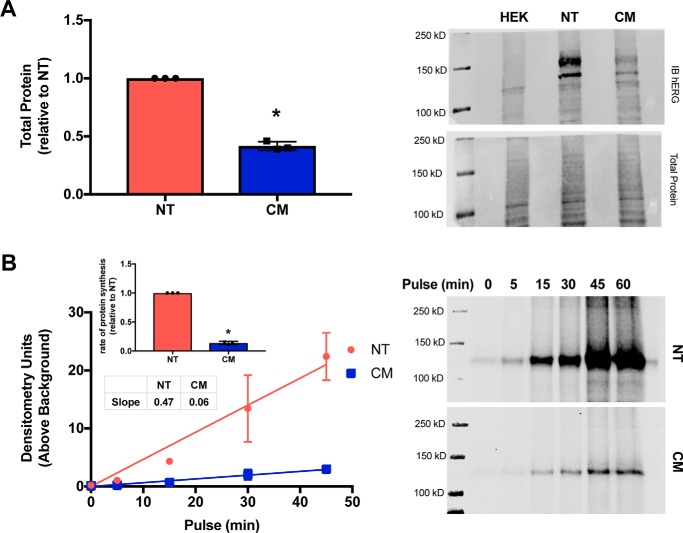
After confirming there were significant differences in hERG-NT and -CM mRNA characteristics, we investigated whether differences in protein expression were solely related to differences in mRNA quantity. To test this, equal amounts of hERG-NT and hERG-CM in vitro transcribed mRNA were transfected into HEK293T cells, and immunoblots were performed to probe protein production. Transfecting an equal amount of mRNA resulted in 2.4 times more hERG-NT than hERG-CM protein production 13 h post-transfection (Fig. 3A). This suggests hERG-NT mRNA is translated more efficiently than hERG-CM. To gain insight into the translation rate for hERG-NT and -CM, pulse experiments utilizing click-chemistry (16, 17) were performed monitoring the rate of nascent hERG synthesis. Cells transfected with the same amount of hERG-NT or -CM plasmid synthesize hERG-NT protein at an ∼7.8-fold greater rate than that of hERG-CM (Fig. 3B). Thus, enhanced translation of hERG-NT over hERG-CM is supported (7.8-fold increased protein synthesis versus 5.9-fold mRNA synthesis). These results, taken together with our prior demonstration that hERG-NT was more robustly produced with a cell-free in vitro translation system (6), indicate that synonymous modification can impact protein synthesis at the translation level.
Figure 3.
hERG-CM translation is reduced. A, graph of immunoblot data and representative immunoblot of hERG-NT and -CM protein expression after transfection with in vitro-transcribed mRNA from three independent transfections (n = 3). These data show increased hERG total protein expression from hERG-NT mRNA when compared with hERG-CM mRNA. Statistically significant difference (p < 0.0001) compared with hERG-NT as determined by a t test analysis is indicated with an asterisk. B, graph and representative immunoblot of an AHA pulse of varied time of hERG-NT and -CM protein production from three independent transfections (n = 3). A bar graph representing the increases in the rate of protein synthesis of hERG-NT compared with hERG-CM for each independent assay (inset). More nascent hERG-NT than hERG-CM protein production was detected 1-day post-transfection. Statistically significant difference (p < 0.0001) compared with hERG-NT as determined by a t test analysis is indicated with an asterisk.
hERG-CM protein trends toward a longer half-life than hERG-NT protein
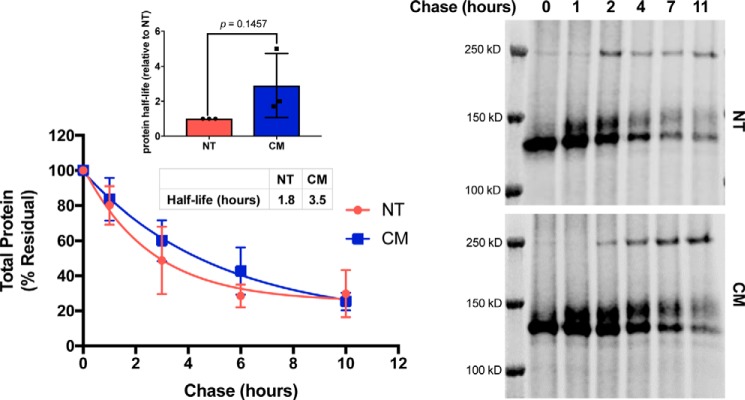
Because protein expression is determined by both production and degradation, we examined the protein stability of hERG-NT and hERG-CM. We measured half-life values for the two variants utilizing a pulse-chase with click-chemistry (16, 17). To decrease the likelihood that the total amount of protein influenced half-life calculation, cells transfected with hERG-CM were transfected with 500 ng of plasmid, whereas cells transfected with hERG-NT were exposed to 125 ng of hERG-NT DNA. This allowed for total protein abundance of hERG-NT and hERG-CM to be equal at the beginning of the pulse-chase experiment. The half-life of hERG-CM was twice that of hERG-NT (3.5 versus 1.8 h, respectively) (Fig. 4) indicating that the hERG-NT channels trended toward being more transient than hERG-CM channels. In addition, two independent assays performed with N-terminally tagged hERG-NT and -CM constructs both demonstrated subtle increases in hERG-CM half-life compared with that of hERG-NT (Fig. S3).
Figure 4.
hERG-CM half-life is increased. Graph and representative immunoblot of AHA pulse-chase to determine hERG-NT and hERG-CM protein half-life. Cells were transfected with 125 ng of hERG-NT or 500 ng of hERG-CM in an attempt to have comparable amounts of nascent protein produced during the 45-min pulse. The amount of residual signal for total protein is displayed. Results of one-phase decay for the hERG-NT and hERG-CM half-life determination are shown from three independent transfections (n = 3). Bar graph represents the increases in the determined half-life of hERG-CM compared with hERG-NT protein for each independent assay (inset). Differences in half-life were assessed by t test analysis. Statistical significance was not reached.
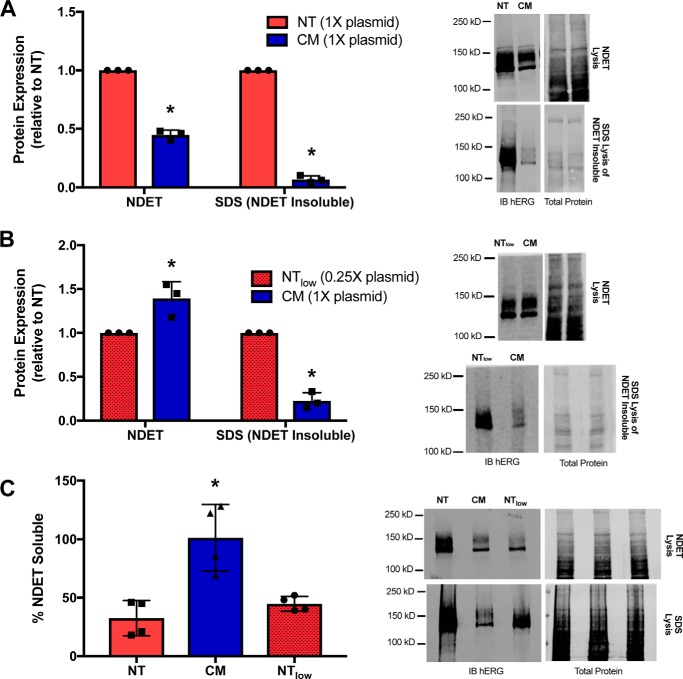
hERG-NT aggregates following translation at a higher rate than hERG-CM
We sought to investigate other factors that could contribute to differences in hERG-NT and hERG-CM protein abundance outside of the hERG protein half-life. Both the mRNA transfection of HEK293T cells and previous polysomal profiling data (6) indicated that hERG-NT mRNA is translated at a greater rate than that of hERG-CM, yet only ∼3-fold more hERG-NT protein than hERG-CM protein is observed. We hypothesized that some hERG-NT protein may potentially aggregate following translation. hERG protein experiences aggregation, ER retention, and subsequent destruction (18–20). These processes are increased due to certain LQT2 pathogenic variants. hERG-NT protein aggregation would lead to under-representation of the true amount of channel protein production as measured by standard SDS-PAGE and immunoblotting techniques. To determine whether the hERG-NT and/or hERG-CM protein was aggregating after lysis with the NDET lysis buffer (NDET (buffer): 1% IGEPAL (CA-630), 0.4% deoxycholic acid, 5 mm EDTA, 25 mm Tris, 150 mm NaCl, pH 7.5), we extracted the pellets obtained from NDET lysis with a more stringent lysis buffer (1.5% SDS) (19). This allowed for quantification of the amount of hERG-NT and hERG-CM protein that was aggregating with NDET lysis. Protein detected in the NDET lysis was deemed “soluble protein,” and protein detected in the SDS buffer extraction of the pellet was deemed “insoluble protein.” hERG-NT was abundantly detected in the insoluble fraction, whereas hERG-CM protein exhibited significantly less signal in the insoluble fraction (Fig. 5A). The protein found in the insoluble fraction is nonfunctional protein that has aggregated prior to full assembly and trafficking to the cell surface (19, 21).
Figure 5.
hERG-NT protein is more prone to aggregation. Immunoblot analysis of hERG-NT and hERG-CM soluble and aggregated protein production. A, graph and representative immunoblot demonstrating soluble and aggregated (NDET-insoluble) hERG-NT and hERG-CM protein. Results are from three independent transfections (n = 3). Differences in soluble protein expression and aggregated protein between hERG-NT and hERG-CM are statistically significant (p < 0.05) as determined by a t test and indicated with an asterisk. B, graph and representative immunoblot demonstrating the soluble and insoluble protein expression for hERG-NT and hERG-CM. In this assay, the hERG-NT total protein expression was more comparable with hERG-CM expression, which was achieved by decreasing the amount of hERG-NT plasmid to 0.25×. Results are from three independent transfections (n = 3). hERG-CM aggregation is significantly lower than hERG-NT from the same lysis conditions as determined by a t test analysis (p < 0.05) and indicated with an asterisk. C, graph and representative immunoblot analysis from cells transiently transfected with hERG-NT or hERG-CM construct and lysed with NDET or SDS buffer. Graph represents the percentage of total (SDS lysis) hERG protein signal present in the NDET-soluble lysis condition. Results are from three independent transfections (n = 3) with one of the transfections being performed in duplicate. Statistically significant differences compared with hERG-NT determined with an ANOVA analysis followed by the Dunnett's post hoc test are indicated with an asterisk (p < 0.05).
To determine whether the increased propensity of hERG-NT to aggregate was intrinsic to hERG-NT protein expression or solely due to higher levels of protein expression relative to hERG-CM, we decreased the hERG-NT protein expression to mirror hERG-CM protein levels. By reducing the hERG-NT plasmid transfection by 4-fold, the NDET-soluble abundance of hERG-NT and -CM achieved comparable levels (Fig. 5B). Under these conditions hERG-NT protein continued to appear in the NDET-insoluble phase more than hERG-CM. To gain insight on the proportion of hERG-NT or hERG-CM associating with the soluble versus the insoluble fraction, cells were transfected with hERG-NT or hERG-CM plasmid or a reduced amount of hERG-NT plasmid (25% of other transfection) and then directly lysed in either NDET or 1.5% SDS. Lysing with either buffer resulted in similar detection levels of hERG-CM suggesting minimal hERG-CM aggregation into the insoluble fraction, whereas only 33% of the total hERG-NT signal appeared in the soluble fraction. Reducing the level of hERG-NT plasmid still resulted in only 45% of the total hERG-NT signal being detected in the soluble fraction (Fig. 5C). This supports the concept that hERG-NT has a greater propensity to aggregate than hERG-CM. This demonstrates that synonymous modification of the coding sequence can affect the hERG protein product at co-translational or post-translational stages of synthesis.
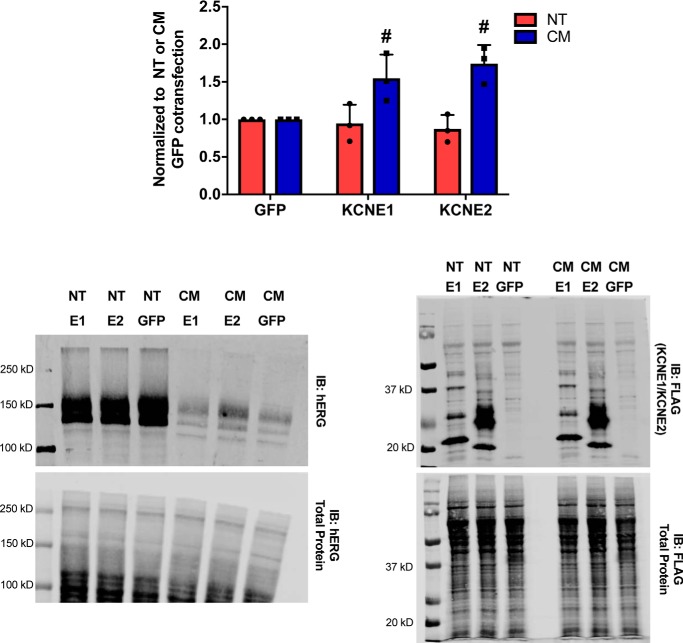
Interestingly, co-expression with FLAG-tagged KCNE1 or KCNE2 augmented hERG-CM expression while having no noticeable effect on hERG-NT (Fig. 6). KCNE2, which has previously been demonstrated to exhibit a greater degree of surface expression than KCNE1 (22), appears to promote the most substantial increase in hERG-CM expression. This result along with the observation that hERG-CM does not have a propensity to aggregate lends evidence that synonymous modification of the hERG protein may affect innate hERG protein characteristics such as aggregation and protein–protein interactions.
Figure 6.
KCNE1 and KCNE2 augments hERG-CM expression. hERG expression is demonstrated by a graph of hERG immunoblot data and representative immunoblot from Western blot analysis of lysate (from NDET lysis) is 2 days post-transfection. Results were obtained from three independent transfections (n = 3) transfecting HEK293T cells in 6-well dishes at ∼60% confluency with 282 ng of hERG construct and 718 ng of N-terminal FLAG-tagged KCNE1, N-terminal FLAG-tagged KCNE2, or GFP. The FLAG-tagged constructs were detected with anti-FLAG M2 antibody (catalogue no. F1804, Sigma) used at 1:1000. hERG-CM expression was significantly augmented when co-expressed with KCNE1 or KCNE2 compared with GFP, p = 0.0146 or 0.0018, respectively, as determined by a two-way ANOVA followed by the Sidak's multiple comparison test and indicated with a number sign (#).
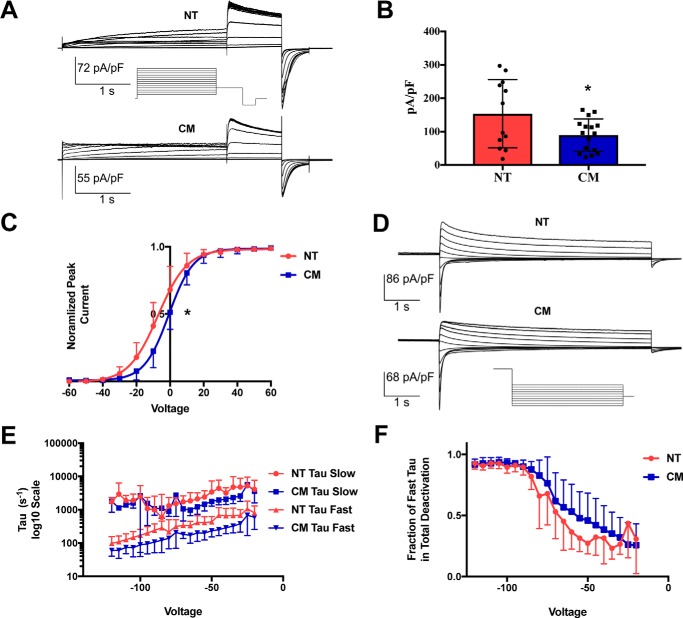
Electrophysiology profiles of hERG-NT and hERG-CM demonstrate subtle differences in IKr parameters
hERG is an essential protein component of the rapidly activating delayed rectifier potassium current (IKr), a current partially responsible for the repolarization of the heart. Because hERG forms an ion channel, we can investigate the function of these two synonymous proteins with the precision inherent in voltage-clamp current measurements. Both hERG-NT- and hERG-CM–expressing cells were subjected to whole-cell patch–clamping to determine channel activation, current density, and deactivation characteristics. Fig. 7A shows representative hERG-NT and hERG-CM current traces during an activation protocol. hERG-CM–expressing cells had significantly lower current density than hERG-NT (p = 0.036) (Fig. 7B), which reflects the lower total protein amounts as expected. Voltage dependence of activation (VDA) (Fig. 7C) showed V½ for hERG-NT at −7.182 ± 0.9453 mV and the V½ for hERG-CM at −0.6763 ± 0.4098 mV. This right shift in hERG-CM voltage dependence of activation was significantly different from the hERG-NT VDA (p < 0.001). Thus, synonymous modification in hERG-CM is associated with a depolarizing shift in its dependence on voltage for channel activation. This could imply differences in the state of the protein folding or differences in the channel interactions.
Figure 7.
hERG-CM exhibits altered electrophysiology properties. Electrophysiology profiles of hERG-NT and hERG-CM activation and deactivation were obtained using whole-cell patch–clamping. A, these images demonstrate representative activation traces for both hERG-NT and hERG-CM. See under “Results” for description of protocol. B, graphical representation of current density, obtained by measuring peak tail current and normalizing to the capacitance of the cell. hERG-NT current density is significantly higher than that of hERG-CM (hERG-NT, mean = 153.70, and hERG-CM, mean = 89.47; results are from 12 cells expressing hERG-NT from three independent transfections, and 16 cells expressing hERG-CM were obtained from 1 transfection). Statistically significant differences (p < 0.05) compared with hERG-NT as determined by a t test analysis are indicated with an asterisk. C, graphical representation of hERG-NT and hERG-CM dependence on voltage for activation. hERG-CM demonstrates a significantly depolarizing shift in voltage dependence of activation (V½ = −0.68 ± 0.41) when compared with the voltage dependence of hERG-NT (V½ = −7.2 ± 0.95); results are from six cells expressing hERG-NT from two independent transfections and 16 hERG-CM cells obtained from one transfection. Significance was determined by comparison of fits after nonlinear regression (p < 0.001) and is represented by an asterisk. D, representative trace shown for hERG-NT and hERG-CM deactivation profile. See protocol under “Results.” E, these graphs represent the slow and fast deactivation constants of hERG-NT and hERG-CM. There is a trend in both the slow and fast τ toward faster deactivation by hERG-CM. These differences were not significant when analyzed using multiple t test (all p values > 0.05). F, graph represents the contribution of fast τ toward total deactivation and demonstrates that hERG-CM trends toward fast τ contributing more to the overall deactivation profile. Results shown in E and F are from nine cells expressing hERG-NT from two independent transfections and five cells expressing hERG-CM from one independent transfection. These differences were not significant when tested using multiple t test (all p values > 0.05).
Another important biophysical characteristic of the IKr current is a slow deactivation rate. Although both hERG-NT and hERG-CM demonstrated typical slow deactivation as seen in the representative traces in Fig. 7D, there were trends toward differences in the time constants. Although deactivation differences were not significant, the fast τ for hERG-CM from −20 to −75 mV trends faster than that of hERG-NT (Fig. 7E). When comparing the relative contribution of the fast τ to overall deactivation (Fig. 7F), there is a trend toward greater fast τ contribution in hERG-CM in the higher voltage range. This, paired with the significant difference in VDA, may suggest slight differences in the folded state of hERG-NT and hERG-CM protein.
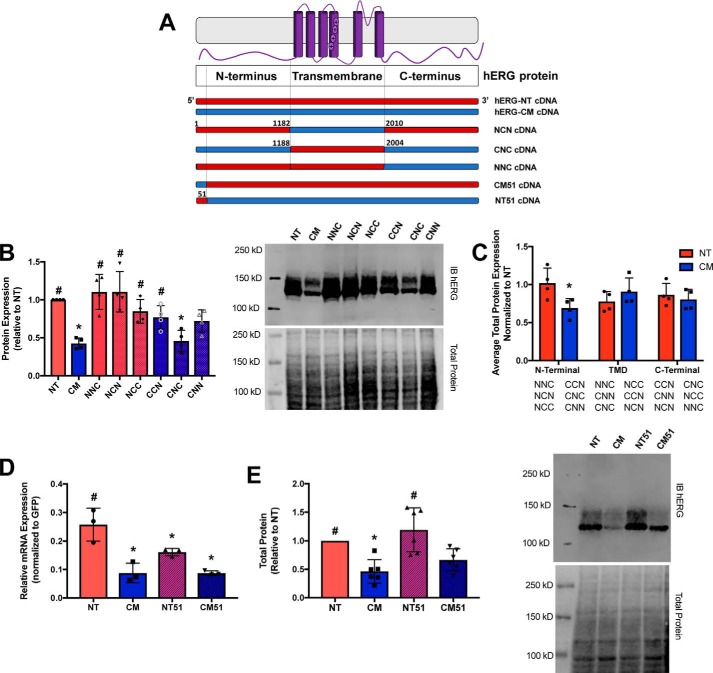
Chimeras of hERG-NT and hERG-CM demonstrate the importance of the 5′-coding region in protein synthesis
To investigate the relative contributions of regional synonymous modification on channel protein synthesis, we utilized a series of chimeras previously described (6) to assess total protein expression (Fig. 8A). hERG-NT and hERG-CM constructs were divided into thirds, and chimeras were generated (N represents hERG-NT, C represents hERG-CM nucleotides). These chimeras were cloned into the pcDNA vector backbone, and protein expression was again interrogated using immunoblots. Expression patterns were consistent with previously reported results (6). To investigate the relative contribution of each region to overall protein expression, the average protein expression was taken for NT and CM at each position. For example, the “first” position is comparing the average protein expression of NNC, NCN, and NCC (chimeras containing the first third of NT) with CNC, CNN, and CCN (chimeras containing the first third of CM). In this analysis, the 5′ third of the mRNA appeared to have the greatest influence on protein translation (Fig. 8, B and C) because the presence of the hERG-NT sequence in the 5′ region yielded higher protein expression regardless of the composition of the remaining two-thirds of the protein.
Figure 8.
Codon modification of the N terminus influences hERG expression. hERG-NT and hERG-CM constructs were divided into thirds, and chimeras were generated (N represents hERG-NT, and C represents hERG-CM nucleotides). A, cartoon representation of the chimeras investigated in this experiment. B, right, representative immunoblot for total protein expression of the chimera. Left, graphical representation of total protein expression for each chimera. Significance was determined through the ANOVA analysis (p < 0.05) from the results of four independent transfections (n = 4). C, expression of each region of the chimera (N-terminal third, transmembrane domain (TMD) third, and C-terminal third) was averaged and compared with the total protein expression of hERG-NT (i.e. 1 on this graph = amount total protein hERG-NT). When NT is in the first third of the chimera, there is a significant increase in protein expression over total protein expression with CM in the first third of the chimera. Statistical significance was determined by t test (p < 0.05). D, mRNA levels of the variants detected 1-day post-transfection are shown. E, right, representative immunoblot of transiently transfected NT51 and CM51 chimeras. Left, graphical representation of total protein expression for each chimera. Total protein expression for each of these chimeras was determined from cells transiently transfected for 2 days. Statistically significant differences (p < 0.05) compared with NT (for B, D, and E) determined with ANOVA followed by the Dunnett's post hoc test are indicated with an asterisk, whereas statistically significant differences (p < 0.05) compared with CM were determined with ANOVA followed by the Dunnett's post hoc test are indicated with a number sign (#).
hERG is a relatively large protein (1159 amino acids from 3480 coding nucleotides), and although we determined that the first third of the construct was most crucial in contributing to protein translation, we were interested in further localizing this region of importance. When evaluating the 5′-coding segment of both hERG-NT and hERG-CM plasmids, we noted local differences between both GC content (71% in hERG-NT and 47% in hERG-CM) and codon usage frequency (many more frequent codons in hERG-NT relative to hERG-CM) in the first 51 nucleotides that exceeded the differences between the hERG-NT and hERG-CM constructs as a whole. To determine whether the effects noted in the previous chimera could be recapitulated with only the first 51 nucleotides, we constructed chimeras made of hERG-NT and hERG-CM in which the first 51 nucleotides of each variant were replaced with the corresponding region from the counterpart (Fig. 8A). In both instances, the first 51 nucleotide fragment tended to promote the channel synthesis phenotype of the construct from which it originated, in both mRNA and protein expression (Fig. 8, D and E). When the first 51 nucleotides of hERG-CM were put into the hERG-NT construct, mRNA expression decreased significantly from the hERG-NT mRNA. When the first 51 nucleotides of hERG-NT were incorporated into the hERG-CM construct, the mRNA expression trended toward an increase relative to hERG-CM mRNA expression. The expression of the hERG-NT and hERG-NT51 proteins was not significantly different, but both had significantly higher protein expression than hERG-CM. This provides evidence to the hypothesis that synonymous modification in localized regions of the cDNA of hERG can influence synthesis both at the mRNA and protein levels and that the 5′-coding region, specifically the first 51 nucleotides, of hERG is essential in maintaining native mRNA and protein expression profiles.
Discussion
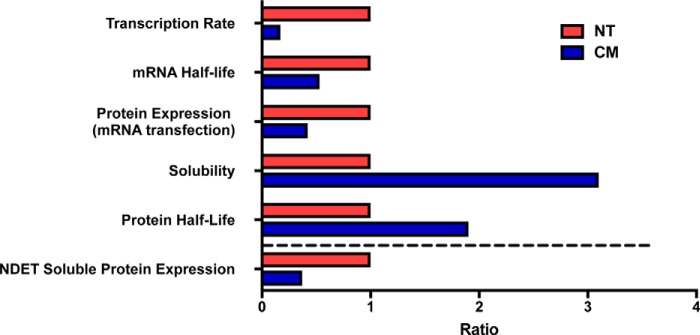
The potassium channel hERG, encoded by KCNH2, is essential for normal cardiac repolarization. Disruption by mutation or other mechanisms results in long QT syndrome type 2, a potentially deadly cardiac arrhythmia disorder. In this study, we sought to investigate whether synonymous modification of KCNH2 could impact hERG protein expression. We had previously demonstrated that hERG-CM had reduced protein expression relative to hERG-NT and that hERG-CM trafficked more efficiently to the cell surface than hERG-NT. In this investigation, we sought to focus on the mechanisms underlying the reduction of hERG-CM protein expression. We studied each aspect of hERG protein synthesis, from transcription to function of the final channel product. We hypothesized that the nucleotide modifications of hERG-CM, while synonymous, could impact each step of synthesis. We found that hERG-NT has a 5.9-fold increase in transcription rate relative to hERG-CM and that the hERG-NT mRNA half-life is approximately double that of the hERG-CM mRNA. Furthermore, hERG-NT protein expression is 2.4-fold greater than that of hERG-CM when evaluated using RNA transfection studies. We observed that hERG-CM trends toward a longer protein half-life, approximately double that of the hERG-NT protein, and that greater than 50% of the hERG-NT protein translated aggregates into nonfunctional protein. These results are summarized in Fig. 9.
Figure 9.
This is a graphical summary of the differences found at each step of synthesis between hERG-NT and hERG-CM (above the dashed line). The summation of these findings results in steady-state NDET-soluble protein expression (represented below the dashed line) of hERG-NT ∼2.7-fold greater than that of hERG-CM detected on immunoblot as shown in Fig. 1.
The most substantial difference in synthesis observed between hERG-NT and hERG-CM was noted at the level of transcription. The hERG-NT construct (66% GC) has 161 GC runs (≥8 nucleotides) that were largely abolished in hERG-CM (51% GC) (6). Accordingly, we hypothesized that reduced hERG-CM transcription may be due to differences in GC content. It has been shown that higher GC content is correlated with increased transcription and mRNA levels (5, 23), which are hypothesized to cause changes in RNA polymerase II (pol II) translocation rates during elongation steps in mRNA transcription. Experimental studies have shown that pol II pauses and backtracks more and for longer periods of time when transcribing AT-rich sequences (24). It has also been hypothesized that codon bias evolved in collaboration with transcription machinery to prevent premature termination and to allow for optimal gene expression (25). Although the mechanism for differences in transcription are not established for hERG-CM and hERG-NT, it is clear that synonymous modification of the codon sequence greatly affects transcription rate.
We also observed that hERG-NT mRNA had a longer half-life than hERG-CM mRNA. Synonymous variation in other mRNA has been shown to alter mRNA stability. A synonymous variant in the dopamine receptor D2 (DRD2) gene resulted in an mRNA that had a shorter half-life than its WT counterpart and was associated with phenotypes such as schizophrenia and alcoholism (26). A synonymous variant in Corneodesmin (CSDN) caused an increase in mRNA half-life and was associated with a psoriasis phenotype (27). Although an exact mechanism defining mRNA stability has not been established, it has been shown that less optimal codon use has been shown to result in mRNA with shorter half-lives (28, 29). There is also evidence that the process of translation itself governs mRNA stability and mRNA decay (30–32). Although rare codons (<10% usage frequency) were eliminated in hERG-CM, the decreased GC content required that some more favored codons were changed to less favored codons. This resulted in an overall reduced codon adaptation index (CAI) of the hERG-CM. CAI is a measure of deviation of codon usage in a gene of interest relative to a reference set of highly expressed genes (33). The CAI of hERG-NT is 0.82, and the CAI of hERG-CM is 0.74 (with most optimized gene being 1). This reduction in overall codon frequency may have caused this observed change in mRNA stability, due to the codons themselves or due to the impact these codons have on translation.
The combined increase in transcription and increase in half-life of hERG-NT mRNA resulted in higher hERG-NT mRNA quantity in preparation for translation. This decrease in hERG-CM mRNA levels is likely not attributed to an inability of hERG-CM plasmid to transfect the cells or a toxic effect induced by hERG-CM transfection as GFP co-transfected with hERG-CM exhibited higher expression levels than when co-transfected with hERG-NT (Fig. S4).
Not only were there differences in mRNA levels, we also noted differences in the rate of translation, with hERG-NT being translated faster than hERG-CM, even when normalizing for mRNA abundance. We hypothesize that changed mRNA structure and an overall decrease of codon frequency in hERG-CM resulted in a changed pattern of ribosomal movement across the transcript, causing a decrease in translation efficiency. This agrees with previous data from Sroubek et al. (6), which demonstrated a decreased ribosomal occupancy on the hERG-CM transcript determined by polysomal profiling and a decreased in vitro protein production rate.
With the increase in mRNA combined with a higher translation rate for hERG-NT, we would expect to observe significantly higher differential protein expression between hERG-NT and hERG-CM. However, after translation, it was noted that the hERG-CM protein trended toward a longer half-life than hERG-NT and that more than half of the hERG-NT protein produced was partitioned into an insoluble fraction of the cell lysate rather than becoming functional hERG protein. Unlike hERG-NT, the majority of nascent hERG-CM remains in the soluble fraction where it undergoes complex glycosylation and trafficking to the cell surface (6, 34). Native hERG protein has a propensity toward aggregation and misfolding that is exacerbated by nonsynonymous pathogenic variants, resulting in LQT2 (6, 35–40). There have not been previous variants that demonstrate a decrease in protein aggregation, but the observations here support the hypothesis that a changed ribosomal movement pattern across the hERG-CM transcript results in a slightly altered final folded conformation that promotes decreased ER aggregation and improved trafficking (6), potentially through improved contact with other proteins, as seen in the increased protein expression of hERG-CM after interaction with KCNE1 and KCNE2. The differential association between hERG-NT and hERG-CM is also interesting; hERG-CM has improved expression with both KCNE1 and KCNE2, but hERG-NT has been shown to preferentially interact with KCNE1. KCNE2 seemed to interact with the hERG channel present at the cell surface (22). These protein–protein interactions will need to be further investigated but suggest differences in hERG-NT and hERG-CM protein structure. The combination of these observed differences at the transcriptional, translational, and post-translational states resulted in the ∼2.7-fold increase of hERG-NT protein expression over the hERG-CM protein noted in Fig. 1.
Subtle differences in the electrophysiology profiles of the hERG-NT and hERG-CM channels also contribute to the evidence that there may be differences in the folding of the two proteins. Previous electrophysiology comparison of the hERG-CM and hERG-NT channels only investigated the activation of the channel (6). Current density was lower for hERG-CM, consistent with our findings. Lower current density for hERG-CM was not unexpected, due to decreased protein expression for hERG-CM. Interestingly, the difference in current density is much less than the difference in protein expression between hERG-NT and hERG-CM (a 1.7-fold difference in current density versus a 2.7-fold difference in protein expression). This can be explained through the previous observation that hERG-CM traffics more efficiently to the surface of the cell (6). Thus, even though there is less protein produced, there is a higher proportion of functional channels presenting to the surface. Also, in the study by Sroubek et al. (6), the hERG-CM VDA was slightly hyperpolarized relative to hERG-NT. This is likely due to the presence of the N-terminal Myc epitope tag on the hERG constructs, which has been shown to cause hyperpolarization of the hERG VDA (7), but it still supports the observation that there are slight differences in the hERG-NT and hERG-CM folded structures, resulting in differences in the voltage dependence of activation. In this investigation, deactivation of the hERG-CM and hERG-NT channels was queried and demonstrated slight differences in the speed of deactivation that supports a slightly altered structure between hERG-NT and hERG-CM channels.
Taken together, the data presented in this study indicate that synonymous modification in the coding region can impact each level of synthesis: transcription, translation, mRNA, protein half-life, and protein folding. It also suggests that the mRNA characteristics such as GC content and codon usage observed in KCNH2 play an important role in regulation and expression of the native hERG protein.
Previous studies have focused on individual aspects of synthesis when studying how synonymous changes at the DNA level can alter the protein product and have effects on the protein function. There are numerous proposed mechanisms through which these changes occur centered around changes in GC content, mRNA structure, codon usage bias, and mRNA stability (2, 4, 5). Most hypotheses center on the effect of these synonymous changes affecting the process of either transcription, through changes in pol II processing, or translation through ribosomal pausing or stalling. One mechanism that would change protein production rates, as well as mRNA t½, is that of no-go decay (41–44), in which an event such as stable mRNA structure or rare codons cause a ribosomal stall, leading to ribosomal pile-up. The ribosome is removed from the transcript, and the transcript is degraded. Although we cannot yet determine which of these mechanisms contribute the most to changes in the hERG protein expression due to extreme synonymous modification, we were able to use chimeras to narrow down the region of importance. Using chimeras focusing on the proximal third and then the proximal 51 bp, we were able to show that the 5′ region of the coding sequence of hERG-NT is able to confer mRNA and protein expression comparable with hERG-NT. The initial coding bp have been hypothesized to play a role in translation efficiency, although the exact mechanism through which this occurs is unknown, and most research has focused on prokaryotic systems (45). Evidence suggests that excessive mRNA structure in the initial nucleotides of a coding sequence has been detrimental to translation (46). Others have hypothesized that rare codon usage bias (47) or enhanced secondary structure (48) in the initial codons of a coding sequence could augment translation by reducing the likelihood of ribosome jamming immediately downstream of the initiation site. Here, we demonstrate in a eukaryotic system that the first 51 nucleotides of the coding sequence of KCNH2 play an important role in hERG protein production. We hypothesize that the first 51 bp of hERG-NT is able to confer its phenotype in a method primarily mediated through GC content, which increases transcription and mRNA stability, but more investigation is needed to confirm this hypothesis.
Limitations of this study must be noted. This study does not take into account tRNA concentrations in cardiomyocytes, so some codons we have determined as “rare” may not have a limited tRNA pool in cardiomyocytes. In addition, this study uses only cDNA constructs, and it thus does not take splicing into account as another mechanism that could be affected by synonymous modification. Also, the hERG-NT and hERG-CM constructs share identical Kozak sequences and upstream and downstream untranslated regions (UTRs), allowing a focus to be placed on the role of synonymous modification in the coding sequence. However, both the 5′ and 3′ UTR may have significant regulatory roles in mRNA regulation and protein expression. How untranslated regions of hERG mRNA can affect these synonymous coding region variations is an important area of future investigation.
The results in this study demonstrate that synonymous modification can affect hERG protein production and function. This suggests there may be a role of synonymous nucleotide variation in long QT syndrome type 2 pathogenesis. Genetic variation responsible for LQTS is generally determined by sequencing of the candidate gene exons and reporting only missense or insertion/deletion mutations. Approximately 25% of patients diagnosed with LQTS do not exhibit such genetic DNA changes (49). Furthermore, there is considerable variation in penetrance of LQTS that is not entirely understood. Presently, synonymous variants are not reported unless they are predicted to alter mRNA splicing. Our study, along with mounting evidence from others, increasingly lends support that synonymous changes at the DNA level can alter protein expression. Such variation has the potential to modify severity or penetrance of genetic disease or even be responsible for a disease phenotype. Moric-Janiszewska and Glowacka (50) have provided evidence that some patients with no identifiable genetic variant responsible for their clinical LQTS have been reported to have significantly less KCNH2 mRNA than the control population. This could be due to decreased mRNA transcription or half-life. Whether synonymous variation existed in hERG is not known, but it could play a role in the expression changes noted. A better understanding of the impact of individual synonymous variants in KCNH2 could result in more precise genetic analyses of disease and may reduce the fraction of patients who test “negative” for a genetic etiology of their LQTS (49).
Here, we demonstrate multiple ways in which synonymous hERG variation can alter transcription, mRNA half-life, translation, protein stability, and protein function. Thus, past and future synonymous variants identified in clinical conditions do not merit automatic designation as “benign” and should be considered carefully.
Experimental procedures
Molecular biology
hERG-NT (GenBankTM accession number: NM_000238.3) or hERG-CM cDNA was cloned into pcDNA 3.1+ (Invitrogen, ThermoFisher Scientific, Waltham, MA) using BamHI (5′) and XbaI (3′) restriction enzymes (New England Biolabs, Ipswich, MA). As described previously (7), the constructs contain a Kozak sequence (GCCACC) immediately preceding the start codon. The super-folder GFP (sfGFP) plasmid used in the qPCR experiments was generated from a plasmid obtained through GenBankTM accession number U55762. The sfGFP-pcDNA 3.1 plasmid was generated by digesting out the gene using BamHI/XbaI and inserting the sfGFP into the pcDNA 3.1 vector. The hERG-NT51 and hERG-CM51 plasmids were created by ATUM bio (Newark, CA). The NCC, NCN, NNC, CCN, CNN, and CNC hERG constructs originally generated by Sroubek et al. (6) were moved into pcDNA 3.1+, and inadvertent mutations V198E/P202L (7, 51, 52) were removed. Sanger sequencing services from Genewiz (South Plainfield, NJ) were used to confirm the validity of the constructs. Alignment of the two nucleotide sequences was performed using Geneious Software (version 9.1.7) (53). Alignment of the hERG-NT and hERG-CM amino acid sequences was performed using Clustal Omega Web Services (54–56).
Cell culture
HEK293T cells were maintained in Roswell Park Memorial Institute 1640 medium (RPMI) (Hyclone, GE Healthcare) supplemented with 10% fetal bovine serum (Hyclone) and 10,000 IU penicillin/streptomycin (Hyclone). Cells were cultured in a humidified incubator at 5% CO2 and 37 °C. Transfection grade plasmid was generated using either the PureYieldTM plasmid miniprep system (catalogue no. A1223, Promega, Madison, WI) or the HiSpeed plasmid midi kit (catalogue no. 12643, Qiagen, Hilden, Germany). A nanodrop was used to determine plasmid concentration. Standard plasmid transfections were done using FuGENE 6 (catalogue no. E2692, Promega) (10 μl per 60-mm dish, 4 μl per well of a 6-well dish, or 2 μl per well of a 12-well dish) in a 1:8 DNA (μg)/FuGENE 6 (μl) ratio. The transfection solution was replaced with cell growth media the day following the transfection, unless the end point of the assay was 1 day or less following the transfection.
Western blotting
Cells were lysed 2 days post-transfection in NDET buffer (1% IGEPAL (CA-630), 0.4% deoxycholic acid, 5 mm EDTA, 25 mm Tris, 150 mm NaCl, pH 7.5) supplemented with complete protease inhibitor (Roche Diagnostics, Basel, Switzerland) or in SDS lysis buffer (1.5% SDS, 20 mm Tris-HCl) where noted. The 48-h time point for protein expression was determined by examining protein expression 24, 48, and 72 h post-transfection. The highest protein signal for both hERG-NT and hERG-CM protein was found at 48 h post-transfection, but the difference in expression levels was consistent over the 72-h period (data not shown). Following lysis, the cell lysate was cleared by centrifugation at 21,000 × g. Protein concentration was determined using the PierceTM detergent compatible Bradford assay reagent (catalogue no. 23246, ThermoFisher Scientific). 20–50 μg of lysate combined with 4× SDS-PAGE sample loading buffer with DTT was heated at 37 °C for at least 30 min and subsequently loaded and separated in a 7.5% SDS-polyacrylamide gel. The protein was transferred to 0.22-μm nitrocellulose membranes (catalogue no. 1620097 (Bio-Rad)), and REVERT stain (catalogue no. 92611015, Li-Cor, Lincoln, NE) was used to normalize for protein loaded. The membrane was then blocked with a solution of 50% Odyssey® blocking buffer (TBS) (catalogue no. 927–50000, Li-Cor) and 50% TBS-T (TBS containing 0.5% Tween 20). Primary hERG antibody (catalogue no. SC-20130, Santa Cruz Biotechnology, Inc., Santa Cruz, CA) was prepared in blocking solution at 1:500 or 1:1000 and then incubated with the membrane overnight at 4 °C. The following day, the membrane was washed with TBST and incubated with IRDye® 800CW donkey anti-rabbit secondary antibody (catalogue no. 925-32213, Li-Cor) in blocking buffer at 1: 10,000 at room temperature and subsequently washed with TBST. Images were obtained using an Odyssey FC or CLX imaging system (Li-Cor) or a GE Typhoon imager (GE Healthcare).
Determination of GC content of all coding regions of human genes
To determine GC content for all coding regions of human genes, first the human coding genome (only coding regions of human genes) was downloaded from the Ensembl database (version GrCH37). Then BEDTools nuc (57) was used to calculate the per gene GC content of the consensus coding sequence (CCDS) for each gene of the genomes. The GC content per CCDS, highlighting hERG-NT and hERG-CM, was graphed in GraphPad Prism.
Quantification of mRNA level
RNA for qPCR analysis was extracted from cells plated in 12- or 6-well plates ∼24 h post-transfection with either hERG-NT or hERG-CM and transfected in a 1:3 ratio (GFP: Gene of interest). The 24-h time point was used because both hERG-NT and hERG-CM mRNA levels from the transient transfection were low at the 48-h mark, which coincided with other data demonstrating significant decrease in hERG protein expression after 48 h post-transfection. RNA was isolated using the RNeasy kit (catalogue no. 74104, Qiagen), and cell lysate was homogenized using the QiaShredder (catalogue no. 79654, Qiagen), and the optional on-column DNase treatment was performed. RNA was converted to cDNA using the SuperScript VILO cDNA synthesis kit (catalogue no. 11754050, ThermoFisher Scientific). 2 μg of total RNA was converted to cDNA. cDNA was diluted 1:200, and qPCR was performed using unique primers directed at specific gene targets created by Integrated DNA Technologies (IDT, Coralville, IA), hERG-NT, hERG-CM, actin, and GFP. Because of the absence of conserved stretches of base pairs between the two constructs long enough to generate real-time PCR primers capable of annealing to both variants, we opted to utilize multiple pairs of primers targeting the corresponding sites on the mRNA of the two constructs. See Table S1 for primer sequences. qPCR was performed using 4.5 μl of 1:200 diluted cDNA (or 1:20, or 1:2000 in the primer efficiency calculations), 5 μl of SYBR Green (catalogue no. 4367659, Applied Biosystems, Foster City, CA), and 0.5 μl of 5 μm primer. qPCR was run on the StepOnePlus system (Applied Biosystems) in a 96-well plate (Applied Biosystems). The reaction cycle was as follows: 98 °C for 10 min then 95 °C for 15 s, 60 °C for 1-min run for 40 cycles, followed by a melt curve. All samples were performed in triplicate. Slope for primer efficiency is calculated across three dilutions of template. Primer efficiency (PE) was then calculated using the following formula: PE = 10−[slope]. Primer efficiency is then used to calculate mRNA quantity using: PE−Ct. Graph Pad prism was used for statistical analysis.
Rate of nascent mRNA production
To determine the rate of nascent mRNA hERG-NT and hERG-CM production, the protocol from Newman et al. (5) was modified. 60-mm dishes of HEK cells were transfected 1:3 (GFP: gene of interest) with either hERG-NT or -CM. 24 h post-transfection, cells were exposed to 4-thiouridine (catalogue no. T4509-25MG, Sigma) for 30 min. Total RNA was then harvested, and RNA was isolated using RNeasy mini kit (catalogue no. 74104, Qiagen); RNA was then labeled with EZ-Link HPDP-Biotin (catalogue no. 21341, ThermoFisher Scientific), and RNA was purified with Zymo RNA Clean and Concentrator-25 (catalogue no. R1017, Zymo, Irvine, CA). Biotin-labeled RNA was then isolated with streptavidin magnetic beads (catalogue no. S1420, New England Biolabs) and eluted in 5% 2-mercaptoethanol. Isolated RNA was then cleaned up using Zymo RNA Clean and Concentrator-5 kit (catalogue no. R1015, Zymo). 1 μg of total RNA was then converted to cDNA using the SuperScript VILO cDNA synthesis kit (catalogue no. 11754050, ThermoFisher Scientific), and the qPCR protocol as described above was followed.
mRNA structure prediction
RNAfold (http://rna.tbi.univie.ac.at/cgi-bin/RNAWebSuite/RNAfold.cgi)4 was used to generate hERG-NT and hERG-CM mRNA structure and free energy predictions. This software predicts structure using minimum free energy calculations based on the mRNA sequence (11, 58).
Circular dichroism spectroscopy of in vitro transcribed mRNA
mRNA for CD was generated using the T7 mMessage mMachine kit (catalogue no. AM1344, Ambion, Austin, TX) and purified using the MegaClear Transcription Clean Up kit (catalogue no. AM1908, ThermoFisher Scientific). pcDNA plasmid constructs for hERG-NT and hERG-CM were digested using DraIII-HF (New England Biolabs), and vectors were purified using the Monarch PCR purification kit (catalogue no. T1030S, New England Biolabs). 1 μg of digested plasmid was used per transcription reaction. RNA was then diluted to 50 ng/μl in 1 m NaCl and 10 mm potassium phosphate. mRNA was folded by heating to 95 °C and then cooled to 37 °C for 15 min. CD spectra of refolded hERG-NT and hERG-CM samples were acquired with a JASCO J-815 spectrophotometer at room temperature (∼23 °C) or at 70 °C, where specified, in a cell with a 0.1-cm path length, a data pitch of 0.1 nm, scanning speed of 100 nm/min, response time of 1 s, and a bandwidth of 1 nm. Three separate mRNA samples from each construct were tested. Each sample was run six times to obtain the CD spectra. The results represent the average of 18 samples. The results are expressed in molar ellipticity [θ] as function of the wavelength λ [θ] = 100 × θ/(CI), where C is the molar concentration of the RNA species, and I is path length. Data from the JASCO is given in ellipticity. Graphical representation created using GraphPad Prism.
mRNA half-life determination
To determine mRNA half-life, the qPCR protocol described above was followed with the following modifications. Actinomycin-D (catalogue no. 11805017, ThermoFisher Scientific) was added to 6 wells of the 12-well plate at a concentration of 7.5 μg/ml. RNA samples were then isolated from the cells at 0, 3, 6, or 12 h (or 12.5-sample 2), 24 h (or 24.5-sample 2) post-actinomycin-D addition. cDNA was generated with 1/8 total isolated RNA rather than 2 μg to account for reduction in total RNA in the cells at the later time points. Primer targets used to evaluate half-life were NT 1, 2, 4; CM 1, 2, 3; and 18S rRNA (for housekeeping control) and c-MYC (for positive actinomycin-D control). One-phase nonlinear regression on GraphPad Prism (La Jolla, CA) was performed to determine half-life.
mRNA transfection
hERG containing pcDNA 3.1+ plasmid was linearized with DRAIII (New England Biolabs) and purified with a PCR cleanup kit (catalogue no. T1030S, New England Biolabs). The HiScribeTM T7 ARCA mRNA kit (with tailing) (catalogue no. E2060S, New England Biolabs) was used to generate polyadenylated mRNA. mRNA was purified either by LiCl precipitation or by using the RNeasy mini kit (catalogue no. 74104, Qiagen) and resuspended in water. mRNA concentration was determined with a nanodrop spectrophotometer. HEK293T cells at ∼80% confluency were transfected with mRNA using the Mirus TransIT transfection reagents (catalogue no. MIR 2225, Mirus Bio LLC, Madison). 2 μl of TransIT-mRNA reagent and 2 μl of mRNA Boost Reagent were complexed with 3 μg of mRNA to transfect the cells of each well of a 12-well dish. Cells were lysed 13 h post-transfection.
Protein half-life determination
Approximately 3.5 × 105 HEK293T cells were plated in a 6-well dish. The following day, cells were transfected with plasmid using FuGENE 6 in a manner as described above. 1-Day post-transfection, cells were starved of methionine by changing the media to methionine-free RPMI 1640 medium (Gibco) containing 10% dialyzed FBS (Sigma) with penicillin/streptomycin. 1 h later, l-azidohomoalanine (AHA) (Kerafast, Boston) was added into the wells (200 μm concentration in solution) for 45 min. Standard growth media supplemented with 1 mm l-methionine (Acros, Bridgewater, NJ) replaced the “pulse” media. Cells lysates collected in 500 μl of NDET buffer supplemented with protease inhibitor were transferred to the −20 °C freezer at the time points indicated on the figure legend. After thawing, the lysates were centrifuged at 18,000–21,000 × g. Supernatants were transferred to new microcentrifuge tubes, and typically 1 μg of hERG C20 antibody (catalogue no. sc-15968, Santa Cruz Biotechnology, Inc.) was added to each sample. After rocking for at least 2 h at room temperature 50 μl of resuspended SureBeadsTM Protein G Magnetic Beads (catalogue no. 1614023, Bio-Rad) were added to each tube, and the samples were incubated at 4 °C overnight with constant agitation. The following day, the beads were washed with 950 μl of chilled PBS (catalogue no. 10010-023, Gibco, Grand Island, NY). 35 μl of PBS was then added to each tube, and a “click-it” reaction was performed in each tube using a click-it buffer reagent kit (ThermoFisher Scientific) to conjugate IRDye 800CW alkyne (∼10 μm in solution, Li-Cor) onto the azido moiety-containing analogue of methionine. Following a 1-h incubation, the beads were washed with 950 μl of prechilled PBS, resuspended in SDS-PAGE loading buffer supplemented with DTT, and then heated at 37 °C for 30 min, and the samples were loaded into and separated on 7.5% SDS-polyacrylamide gels. The gels were directly scanned using the 800 channel of the Odyssey FC, and the obtained images were analyzed with the Image Studio Lite Version 5.2 program (Li-Cor) to determine the densitometry values. Subtle increases often detected in protein detection from the 0- to 1-h time points suggested residual methionine in the cells may not be immediately outcompeted by the excess unlabeled methionine; thus, the 1-h chase time point of both hERG-NT and hERG-CM was used to define 100% total protein. GraphPad Prism was then used to fit the data to a one-phase decay analysis.
Quantification of nascent protein production
The protein half-life determination protocol described above was followed with various pulse lengths used, which are indicated on the figure legend. The no-chase step was performed, i.e. the cells were lysed immediately after the introduction of AHA at the indicated times. The 60-min time point was excluded from the linear regression analysis for the calculation of the slope representing the rate of protein synthesis as it was outside the linear range of detection under the conditions used.
Electrophysiology
For all electrophysiology experiments, HEK293T cells were transiently transfected with either hERG-NT or hERG-CM using the transfection protocol described above. Cells were plated on sterile glass coverslips 24 h after transfection and subsequently placed into an acrylic/polystyrene perfusion chamber (Warner Instruments, Hamden, CT). Chambers were mounted on an inverted microscope equipped with fluorescence optics and patch pipette micromanipulators. All recordings were done at room temperature (∼22 °C) and at 48–72 h post-transfection. Cells were bathed in extracellular solution (150 mm NaCl, 1.8 mm CaCl2, 4 mm KCl, 1 mm MgCl2, 5 mm glucose, and 10 mm HEPES buffer, pH 7.4). Intracellular solution was used in the micropipette (126 mm KCl, 4 mm Mg-ATP, 2 mm MgSO4, 5 mm EGTA, 0.5 mm CaCl2, and 25 mm HEPES buffer, pH 7.2). Patch-clamp pipettes with a tip resistance of ∼2–3 megohms were used to obtain the whole-cell configuration (59). A MultiClamp 700B amplifier (Molecular Devices, LLC, Sunnyvale, CA) was used, and patch-clamp protocols were performed through the pCLAMP10 acquisition and analysis software (Molecular Devices, Sunnyvale, CA). The pipette offset potential in these solutions was zeroed just prior to seal formation. Whole-cell capacitance (generally 10–30 picofarads) was compensated electronically. Whole-cell series resistance was compensated to 85–90% using amplifier circuitry so that the voltage errors for currents of 2 nA were always less than 6 mV. A holding potential of −80 mV was used for patching and whole-cell configurations, and figure insets illustrate the voltage command protocol. The data filtering (8-pole Bessel) at 1.4 kHz and sampling at 5 kHz were done for both the activation and deactivation protocols. The channel activation protocol entails the membrane stepped to various depolarizing levels (−60 to 60 mV) for 1.5 s, then repolarized to −40 mV for 0.5 s, and finally hyperpolarized to −120 mV for 0.5 s. A voltage-dependent activation curve was generated by measuring the peak outward current at the −40 mV repolarizing step and plotted against the preceding activation voltage. The voltage-dependent activation (VDA) data were fit to a Boltzmann function: I = 1/(1 + exp((V½ − V)/k)), where I is the relative tail current amplitude; V is the applied membrane voltage; V½ is the voltage at half-maximal activation, and k is the slope factor. To determine the deactivation rate, a pulse depolarizing the membrane to +20 mV was applied for 1.7 s, followed by voltage steps from −20 to −120 mV for 4.9 s. Deactivation of the current during the final voltage steps was fitted with a double-exponential function (Itail = A0 + AMPF·expt/τF + AMPS·expt/τS, where A0 is the initial amplitude; AMPF and AMPS are the relative amplitudes of the fast and slow components, and τF (fast Tau) and τS (slow Tau) are the time constants (60). All data were analyzed with CLAMPFIT software (Molecular Devices), and statistics were performed using GraphPad Prism software.
Data analysis
Unless otherwise noted, at least three independent experiments were carried out for each assay. Error bars represent standard deviation (S.D.) of the displayed mean value. Statistical significance is defined as p values < 0.05 using the analysis described in the figure legends. Analysis include t tests, one- or two-way ANOVAs, and comparison of fits of nonlinear regression analysis. Dunnett or Sidak multiple comparison tests were performed as described in the figure legends.
Author contributions
A. C. B. and M. L. O. B. data curation; A. C. B., M. L. O. B., and T. V. M. formal analysis; A. C. B. and M. L. O. B. validation; A. C. B. and M. L. O. B. investigation; A. C. B. and M. L. O. B. visualization; A. C. B., M. L. O. B., and T. V. M. methodology; A. C. B. and M. L. O. B. writing-original draft; A. C. B., M. L. O. B., and T. V. M. writing-review and editing; M. L. O. B. and T. V. M. funding acquisition; T. V. M. conceptualization; T. V. M. supervision; T. V. M. project administration.
Supplementary Material
This work was supported by National Institutes of Health Grants R01HL118437 and F30HL126283 from NHLBI. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Figs. S1–S4 and Table S1.
Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party hosted site.
- hERG
- human ether-a-go-go-related gene
- LQT2
- long QT syndrome, type 2
- hERG-NT
- native hERG construct
- hERG-CM
- codon-modified hERG construct
- HEK293T
- human embryonic kidney 293T
- sfGFP
- super-folder GFP
- qPCR
- quantitative PCR
- ER
- endoplasmic reticulum
- CCDS
- consensus coding sequence
- VDA
- voltage dependence of activation
- IKr
- rapidly activating delayed rectifier potassium current
- τF
- fast τ
- τS
- slow τ
- ANOVA
- analysis of variance
- AHA
- l-azidohomoalanine
- pol
- polymerase
- CAI
- codon adaptation index
- LQTS
- long QT syndrome.
References
- 1. Tsai C. J., Sauna Z. E., Kimchi-Sarfaty C., Ambudkar S. V., Gottesman M. M., and Nussinov R. (2008) Synonymous mutations and ribosome stalling can lead to altered folding pathways and distinct minima. J. Mol. Biol. 383, 281–291 10.1016/j.jmb.2008.08.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kimchi-Sarfaty C., Oh J. M., Kim I. W., Sauna Z. E., Calcagno A. M., Ambudkar S. V., and Gottesman M. M. (2007) A “silent” polymorphism in the MDR1 gene changes substrate specificity. Science 315, 525–528 10.1126/science.1135308 [DOI] [PubMed] [Google Scholar]
- 3. Purvis I. J., Bettany A. J., Santiago T. C., Coggins J. R., Duncan K., Eason R., and Brown A. J. (1987) The efficiency of folding of some proteins is increased by controlled rates of translation in vivo. A hypothesis. J. Mol. Biol. 193, 413–417 10.1016/0022-2836(87)90230-0 [DOI] [PubMed] [Google Scholar]
- 4. Bartoszewski R. A., Jablonsky M., Bartoszewska S., Stevenson L., Dai Q., Kappes J., Collawn J. F., and Bebok Z. (2010) A synonymous single nucleotide polymorphism in ΔF508 CFTR alters the secondary structure of the mRNA and the expression of the mutant protein. J. Biol. Chem. 285, 28741–28748 10.1074/jbc.M110.154575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Newman Z. R., Young J. M., Ingolia N. T., and Barton G. M. (2016) Differences in codon bias and GC content contribute to the balanced expression of TLR7 and TLR9. Proc. Natl. Acad. Sci. U.S.A. 113, E1362–E1371 10.1073/pnas.1518976113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sroubek J., Krishnan Y., and McDonald T. V. (2013) Sequence and structure-specific elements of HERG mRNA determine channel synthesis and trafficking efficiency. FASEB J. 27, 3039–3053 10.1096/fj.12-227009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Osterbur Badhey M. L., Bertalovitz A. C., and McDonald T. V. (2017) Express with caution: epitope tags and cDNA variants effects on hERG channel trafficking, half-life and function. J. Cardiovasc. Electrophysiol. 28, 1070–1082 10.1111/jce.13259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dolken L., Ruzsics Z., Rädle B., Friedel C. C., Zimmer R., Mages J., Hoffmann R., Dickinson P., Forster T., Ghazal P., and Koszinowski U. H. (2008) High-resolution gene expression profiling for simultaneous kinetic parameter analysis of RNA synthesis and decay. RNA 14, 1959–1972 10.1261/rna.1136108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cleary M. D., Meiering C. D., Jan E., Guymon R., and Boothroyd J. C. (2005) Biosynthetic labeling of RNA with uracil phosphoribosyltransferase allows cell-specific microarray analysis of mRNA synthesis and decay. Nat. Biotechnol. 23, 232–237 10.1038/nbt1061 [DOI] [PubMed] [Google Scholar]
- 10. Miller M. R., Robinson K. J., Cleary M. D., and Doe C. Q. (2009) TU-tagging: cell type-specific RNA isolation from intact complex tissues. Nat. Methods 6, 439–441 10.1038/nmeth.1329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lorenz R., Bernhart S. H., Höner Zu Siederdissen C., Tafer H., Flamm C., Stadler P. F., and Hofacker I. L. (2011) ViennaRNA Package 2.0. Algorithms Mol. Biol. 6, 26 10.1186/1748-7188-6-26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ivanov V. I., Minchenkova L. E., Schyolkina A. K., and Poletayev A. I. (1973) Different conformations of double-stranded nucleic acid in solution as revealed by circular dichroism. Biopolymers 12, 89–110 10.1002/bip.1973.360120109 [DOI] [PubMed] [Google Scholar]
- 13. Williams A. L. Jr., Cheong C., Tinoco I. Jr., and Clark L. B. (1986) Vacuum ultraviolet circular dichroism as an indicator of helical handedness in nucleic acids. Nucleic Acids Res. 14, 6649–6659 10.1093/nar/14.16.6649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hashizume H., and Imahori K. (1967) Circular dichroism and conformation of natural and synthetic polynucleotides. J. Biochem. 61, 738–749 10.1093/oxfordjournals.jbchem.a128608 [DOI] [PubMed] [Google Scholar]
- 15. Johnson W. C. Jr, Itzkowitz M. S., and Tinoco I. Jr. (1972) Circular dichroism of polynucleotides: dimers as a function of conformation. Biopolymers 11, 225–234 10.1002/bip.1972.360110117 [DOI] [PubMed] [Google Scholar]
- 16. Kolb H. C., Finn M. G., and Sharpless K. B. (2001) Click chemistry: diverse chemical function from a few good reactions. Angew. Chem. Int. Ed. Engl. 40, 2004–2021 10.1002/1521-3773(20010601)40:11%3C2004::AID-ANIE2004%3E3.0.CO%3B2-5 [DOI] [PubMed] [Google Scholar]
- 17. Wang S., Wang Q., Wang Y., Liu L., Weng X., Li G., Zhang X., and Zhou X. (2008) Novel anthraquinone derivatives: synthesis via click chemistry approach and their induction of apoptosis in BGC gastric cancer cells via reactive oxygen species (ROS)-dependent mitochondrial pathway. Bioorg. Med. Chem. Lett. 18, 6505–6508 10.1016/j.bmcl.2008.10.047 [DOI] [PubMed] [Google Scholar]
- 18. Zhou T., Ko E. A., Gu W., Lim I., Bang H., and Ko J. H. (2012) Non-silent story on synonymous sites in voltage-gated ion channel genes. PLoS One 7, e48541 10.1371/journal.pone.0048541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Keller S. H., Platoshyn O., and Yuan J. X. (2005) Long QT syndrome-associated I593R mutation in HERG potassium channel activates ER stress pathways. Cell Biochem. Biophys. 43, 365–377 10.1385/CBB:43:3:365 [DOI] [PubMed] [Google Scholar]
- 20. Gong Q., Keeney D. R., Molinari M., and Zhou Z. (2005) Degradation of trafficking-defective long QT syndrome type II mutant channels by the ubiquitin-proteasome pathway. J. Biol. Chem. 280, 19419–19425 10.1074/jbc.M502327200 [DOI] [PubMed] [Google Scholar]
- 21. Manganas L. N., Akhtar S., Antonucci D. E., Campomanes C. R., Dolly J. O., and Trimmer J. S. (2001) Episodic ataxia type-1 mutations in the Kv1.1 potassium channel display distinct folding and intracellular trafficking properties. J. Biol. Chem. 276, 49427–49434 10.1074/jbc.M109325200 [DOI] [PubMed] [Google Scholar]
- 22. Um S. Y., and McDonald T. V. (2007) Differential association between HERG and KCNE1 or KCNE2. PLoS One 2, e933 10.1371/journal.pone.0000933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kudla G., Lipinski L., Caffin F., Helwak A., and Zylicz M. (2006) High guanine and cytosine content increases mRNA levels in mammalian cells. PLoS Biol. 4, e180 10.1371/journal.pbio.0040180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zamft B., Bintu L., Ishibashi T., and Bustamante C. (2012) Nascent RNA structure modulates the transcriptional dynamics of RNA polymerases. Proc. Natl. Acad. Sci. U.S.A. 109, 8948–8953 10.1073/pnas.1205063109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zhou Z., Dang Y., Zhou M., Yuan H., and Liu Y. (2018) Codon usage biases co-evolve with transcription termination machinery to suppress premature cleavage and polyadenylation. eLife 7, e33569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Duan J., Wainwright M. S., Comeron J. M., Saitou N., Sanders A. R., Gelernter J., and Gejman P. V. (2003) Synonymous mutations in the human dopamine receptor D2 (DRD2) affect mRNA stability and synthesis of the receptor. Hum. Mol. Genet. 12, 205–216 10.1093/hmg/ddg055 [DOI] [PubMed] [Google Scholar]
- 27. Capon F., Allen M. H., Ameen M., Burden A. D., Tillman D., Barker J. N., and Trembath R. C. (2004) A synonymous SNP of the corneodesmosin gene leads to increased mRNA stability and demonstrates association with psoriasis across diverse ethnic groups. Hum. Mol. Genet. 13, 2361–2368 10.1093/hmg/ddh273 [DOI] [PubMed] [Google Scholar]
- 28. Radhakrishnan A., Chen Y. H., Martin S., Alhusaini N., Green R., and Coller J. (2016) The DEAD-Box protein Dhh1p couples mRNA decay and translation by monitoring codon optimality. Cell 167, 122–132.e9 10.1016/j.cell.2016.08.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Presnyak V., Alhusaini N., Chen Y. H., Martin S., Morris N., Kline N., Olson S., Weinberg D., Baker K. E., Graveley B. R., and Coller J. (2015) Codon optimality is a major determinant of mRNA stability. Cell 160, 1111–1124 10.1016/j.cell.2015.02.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Huch S., and Nissan T. (2014) Interrelations between translation and general mRNA degradation in yeast. Wiley Interdiscip. Rev. RNA 5, 747–763 10.1002/wrna.1244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Roy B., and Jacobson A. (2013) The intimate relationships of mRNA decay and translation. Trends Genet. 29, 691–699 10.1016/j.tig.2013.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Radhakrishnan A., and Green R. (2016) Connections underlying translation and mRNA stability. J. Mol. Biol. 428, 3558–3564 10.1016/j.jmb.2016.05.025 [DOI] [PubMed] [Google Scholar]
- 33. Sharp P. M., and Li W. H. (1987) The rate of synonymous substitution in enterobacterial genes is inversely related to codon usage bias. Mol. Biol. Evol. 4, 222–230 zpmid3328816 [DOI] [PubMed] [Google Scholar]
- 34. Lamothe S. M., Hulbert M., Guo J., Li W., Yang T., and Zhang S. (2017) Glycosylation stabilizes hERG channels on the plasma membrane by decreasing proteolytic susceptibility. FASEB J. 2018, fj201700832R 10.1096/fj.201700832R [DOI] [PubMed] [Google Scholar]
- 35. Thomas D., Kiehn J., Katus H. A., and Karle C. A. (2003) Defective protein trafficking in hERG-associated hereditary long QT syndrome (LQT2): molecular mechanisms and restoration of intracellular protein processing. Cardiovasc. Res. 60, 235–241 10.1016/j.cardiores.2003.08.002 [DOI] [PubMed] [Google Scholar]
- 36. Foo B., Williamson B., Young J. C., Lukacs G., and Shrier A. (2016) hERG quality control and the long QT syndrome. J. Physiol. 594, 2469–2481 10.1113/JP270531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Young J. C. (2014) The role of the cytosolic HSP70 chaperone system in diseases caused by misfolding and aberrant trafficking of ion channels. Dis. Model Mech. 7, 319–329 10.1242/dmm.014001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Smith J. L., Reloj A. R., Nataraj P. S., Bartos D. C., Schroder E. A., Moss A. J., Ohno S., Horie M., Anderson C. L., January C. T., and Delisle B. P. (2013) Pharmacological correction of long QT-linked mutations in KCNH2 (hERG) increases the trafficking of Kv11.1 channels stored in the transitional endoplasmic reticulum. Am. J. Physiol. Cell Physiol. 305, C919–C930 10.1152/ajpcell.00406.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Walker V. E., Atanasiu R., Lam H., and Shrier A. (2007) Co-chaperone FKBP38 promotes HERG trafficking. J. Biol. Chem. 282, 23509–23516 10.1074/jbc.M701006200 [DOI] [PubMed] [Google Scholar]
- 40. Walker V. E., Wong M. J., Atanasiu R., Hantouche C., Young J. C., and Shrier A. (2010) Hsp40 chaperones promote degradation of the HERG potassium channel. J. Biol. Chem. 285, 3319–3329 10.1074/jbc.M109.024000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Doma M. K., and Parker R. (2006) Endonucleolytic cleavage of eukaryotic mRNAs with stalls in translation elongation. Nature 440, 561–564 10.1038/nature04530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Isken O., and Maquat L. E. (2007) Quality control of eukaryotic mRNA: safeguarding cells from abnormal mRNA function. Genes Dev. 21, 1833–1856 10.1101/gad.1566807 [DOI] [PubMed] [Google Scholar]
- 43. Simms C. L., Thomas E. N., and Zaher H. S. (2017) Ribosome-based quality control of mRNA and nascent peptides. Wiley Interdiscip. Rev. RNA 8, 10.1002/wrna.1366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Simms C. L., Yan L. L., and Zaher H. S. (2017) Ribosome collision is critical for quality control during no-go decay. Mol. Cell 68, 361–373 10.1016/j.molcel.2017.08.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Tuller T., and Zur H. (2015) Multiple roles of the coding sequence 5′ end in gene expression regulation. Nucleic Acids Res. 43, 13–28 10.1093/nar/gku1313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Bentele K., Saffert P., Rauscher R., Ignatova Z., and Blüthgen N. (2013) Efficient translation initiation dictates codon usage at gene start. Mol. Syst. Biol. 9, 675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Tuller T., Carmi A., Vestsigian K., Navon S., Dorfan Y., Zaborske J., Pan T., Dahan O., Furman I., and Pilpel Y. (2010) An evolutionarily conserved mechanism for controlling the efficiency of protein translation. Cell 141, 344–354 10.1016/j.cell.2010.03.031 [DOI] [PubMed] [Google Scholar]
- 48. Tuller T., Waldman Y. Y., Kupiec M., and Ruppin E. (2010) Translation efficiency is determined by both codon bias and folding energy. Proc. Natl. Acad. Sci. U.S.A. 107, 3645–3650 10.1073/pnas.0909910107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Tester D. J., and Ackerman M. J. (2014) Genetics of long QT syndrome. Methodist DeBakey Cardiovasc. J. 10, 29–33 10.14797/mdcj-10-1-29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Moric-Janiszewska E., and Gl̸owacka M. (2012) Molecular diagnostics of families with long-QT syndrome. Cardiol. J. 19, 159–167 10.5603/CJ.2012.0028 [DOI] [PubMed] [Google Scholar]
- 51. Trudeau M. C., Warmke J. W., Ganetzky B., and Robertson G. A. (1995) HERG, a human inward rectifier in the voltage-gated potassium channel family. Science 269, 92–95 10.1126/science.7604285 [DOI] [PubMed] [Google Scholar]
- 52. Trudeau M. C., Warmke J. W., Ganetzky B., and Robertson G. A. (1996) HERG sequence correction. Science 272, 1087 [PubMed] [Google Scholar]
- 53. Kearse M., Moir R., Wilson A., Stones-Havas S., Cheung M., Sturrock S., Buxton S., Cooper A., Markowitz S., Duran C., Thierer T., Ashton B., Meintjes P., and Drummond A. (2012) Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28, 1647–1649 10.1093/bioinformatics/bts199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Goujon M., McWilliam H., Li W., Valentin F., Squizzato S., Paern J., and Lopez R. (2010) A new bioinformatics analysis tools framework at EMBL-EBI. Nucleic Acids Res. 38, W695–W699 10.1093/nar/gkq313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. McWilliam H., Li W., Uludag M., Squizzato S., Park Y. M., Buso N., Cowley A. P., and Lopez R. (2013) Analysis tool web services from the EMBL-EBI. Nucleic Acids Res. 41, W597–W600 10.1093/nar/gkt376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Sievers F., Wilm A., Dineen D., Gibson T. J., Karplus K., Li W., Lopez R., McWilliam H., Remmert M., Söding J., Thompson J. D., and Higgins D. G. (2011) Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7, 539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Quinlan A. R. (2014) BEDTools: The Swiss-Army tool for genome feature analysis. Curr. Protoc. Bioinformatics 47, 11.12.11–34 10.1002/0471250953.bi1112s47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Mathews D. H. (2004) Using an RNA secondary structure partition function to determine confidence in base pairs predicted by free energy minimization. RNA 10, 1178–1190 10.1261/rna.7650904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Hamill O. P., Marty A., Neher E., Sakmann B., and Sigworth F. J. (1981) Improved patch-clamp techniques for high-resolution current recording from cells and cell-free membrane patches. Pflugers Arch. 391, 85–100 10.1007/BF00656997 [DOI] [PubMed] [Google Scholar]
- 60. Sanguinetti M. C., Jiang C., Curran M. E., and Keating M. T. (1995) A mechanistic link between an inherited and an acquired cardiac arrhythmia: HERG encodes the IKr potassium channel. Cell 81, 299–307 10.1016/0092-8674(95)90340-2 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.