Abstract
Ulp1 and Ulp2, in the yeast Saccharomyces cerevisiae, are the founding members of deSUMOylating enzymes. These enzymes remove small ubiquitin-like modifier (SUMO) from proteins and are conserved in all eukaryotes. Previous studies have shown that Ulp1 deSUMOylates the bulk of intracellular SUMOylated proteins, whereas Ulp2 is a highly specific enzyme. However, the mechanism for Ulp2's substrate specificity has been insufficiently understood. Here we show that the C-terminal regulatory domain of Ulp2 contains three distinct, yet conserved, motifs that control its in vivo substrate specificity and cell growth. Among them, a SUMO-interacting motif (SIM) was found to coordinate with the domain of Ulp2 that binds to the nucleolar protein Csm1 to ensure maximal deSUMOylation of Ulp2's nucleolar substrates. We found that whereas the Csm1-binding domain of Ulp2 recruits this enzyme to the nucleolus, Ulp2's C-terminal SIM promotes its SUMO protease activity and plays a key role in mediating the in vivo specificity of Ulp2. Thus, the substrate specificity of Ulp2 is controlled by both its subcellular localization and the SUMOylation status of its substrates. These findings illustrate the highly coordinated and dynamic nature of the SUMO pathways in maintaining homeostasis of intracellular SUMOylation.
Keywords: SUMO-interacting motif (SIM), sumoylation, post-translational modification (PTM), protease, substrate specificity, isothermal titration calorimetry (ITC), Cdc14, Net1, Tof2, Ulp1, Ulp2
Introduction
Small ubiquitin-like modifier (SUMO),3 a member of the ubiquitin-like proteins (Ubls), is a highly conserved, ubiquitous and widely utilized post-translational modification (1, 2). SUMO is structurally similar to ubiquitin, despite being mostly different at the primary sequence level (3). Like ubiquitin, the conjugation of SUMO to its substrates also occurs in an ATP-dependent manner that involves a cascade of three enzymes: a heterodimeric SUMO-activating E1 enzyme (Aos1 and Uba2 in yeast), a SUMO-conjugating E2 enzyme (Ubc9 in yeast), and a number of SUMO E3 ligases that impart specificity for the conjugation of SUMO to its substrates (4). Together, these enzymes catalyze the formation of isopeptide bonds between the C terminus of SUMO and lysine side chains on substrate proteins. Although a single E1 and E2 enzyme exists in budding yeast, three mitotic E3 ligases (Siz1, Siz2, and Mms21) are known to exist (5, 6) and have been shown to have overlapping and distinct substrates (7, 8). On the other hand, the removal of SUMO from proteins requires the activity of the SUMO proteases, a family of isopeptidases conserved from yeast to human, which cleave the isopeptide linkages between SUMO and its substrates (2). The first SUMO proteases, Ulp1 and Ulp2, were discovered in the budding yeast Saccharomyces cerevisiae (9, 10); and further investigations led to the discovery of the homolog human SUMO proteases SENP1–SENP3 and SENP5–SENP7 (11–14). Because Ulp1 and Ulp2 are the only two SUMO proteases known to exist in budding yeast, they are expected to deSUMOylate a broad range of SUMOylated targets identified through proteomic studies (7, 15–18). Defining the mechanism by which these enzymes deSUMOylate specific substrates in vivo, will go a long way in understanding their diverse functions.
Prior studies regarding the localization and biochemical activity of the yeast SUMO proteases, Ulp1 and Ulp2, have established their distinct roles in SUMO homeostasis; with Ulp1 being responsible for the maturation of SUMO and Ulp2 playing a larger role in the cleavage of SUMO–SUMO linkages in poly-SUMO chains (9, 19, 20). The maturation of SUMO is essential for cell viability; however, yeast cells expressing a mature form of SUMO, in the ulp1Δ background, are still unable to grow (9). This indicates that Ulp1 plays an essential role in deSUMOylating other cellular proteins. Interestingly, our recent study showed that Ulp1 is not essential in the absence of the main yeast SUMO E3 ligases, Siz1 and Siz2 (21); which perform the bulk of intracellular SUMOylation (7, 8). This genetic finding strongly suggested that the essential function of Ulp1, besides SUMO maturation, is to antagonize the activities of Siz1 and Siz2, and keep the bulk of intracellular SUMOylation levels sufficiently low. Consistent with this idea, loss of Ulp1 in the siz1Δsiz2Δ mutant still resulted in a substantial increase in global SUMOylation even in the absence of Siz1 and Siz2 (21), indicating a major role for Ulp1 in performing global deSUMOylation. When combined with Ulp1's highly potent in vitro activity (9, 19), these findings established a major role for Ulp1 in maintaining global SUMO homeostasis (21).
Unlike Ulp1, Ulp2 is not essential for cell viability, although loss of Ulp2 leads to severe growth defects and interestingly, a rapid accumulation of survivors with aneuploidy (10, 22). The tendency of the ulp2Δ mutant to form survivors has complicated its analysis, although mutations affecting poly-SUMO chain formation appeared to rescue the growth defect of the ulp2Δ mutant (20). Remarkably, Ulp2 was found to be a highly specific enzyme that targets specific protein complexes involved in distinct cellular processes such as ribosomal DNA (rDNA) maintenance, chromosome segregation, and DNA replication (21). With the knowledge of Ulp1 and Ulp2 substrates in hand, a major unsolved question is what dictates the in vivo substrate specificity of Ulp1 and Ulp2? One clue might be related to the distinct subcellular localizations of Ulp1 and Ulp2. Prior studies have shown that Ulp1 and Ulp2 are both localized to the nucleus via a nuclear localization signal; with Ulp1 localizing to the nuclear envelope and Ulp2 localizing throughout the nucleus (19, 23). Moreover, Ulp2 has a C-terminal SUMO-interacting motif (SIM) (10), which is defined by a core consensus of I/V-X-I/V-I/V with an acidic residue usually flanking one side of the consensus (24, 25). Although not yet experimentally confirmed, this putative SIM of Ulp2 could promote it to specifically process poly-SUMO chains (20); however, its role in contributing to Ulp2's in vivo substrate specificity have yet to be investigated.
Disruption of the SUMO pathways in yeast and mammals can lead to genome instability and defects in processes such as rDNA maintenance, chromosome segregation, DNA repair, transcription, and proteostasis (7, 22, 26–31). Prior studies have identified Ulp2 as a key player in rDNA maintenance, and furthermore, Ulp2 was found to localize to the rDNA via its association with the Cohibin complex, Csm1–Lrs4, which connects the nuclear envelope and rDNA (26, 29, 31–34). The Cohibin complex associates with the rDNA through the Tof2 scaffolding protein, which is anchored to the rDNA via its binding to Fob1 at the replication fork barrier of each rDNA repeat (32–34). At each replication fork barrier, the RENT complex (Cdc14–Sir2–Net1) is also anchored by Fob1, and forms a protein network with Cohibin that is important for rDNA stability and silencing (32, 34). This set the stage for our recent study to solve the structure of the Ulp2–Csm1 binding interface, and demonstrate that Ulp2's interaction with the Csm1 subunit, via a newly identified domain in Ulp2's C terminus, was important for rDNA silencing (29). This study demonstrated that Ulp2's Csm1-binding domain targets Ulp2 to the rDNA and is important for SUMO homeostasis and proper silencing at the rDNA (29). In particular, Tof2 becomes poly-SUMOylated in ulp2 mutants, which signals the recruitment of the SUMO-targeted ubiquitin ligase (STUbL) Slx5-Slx8; resulting in the degradation of Tof2 (29, 35, 36). However, the ulp2–781 mutant, which lacks the Csm1-binding domain entirely, does not have an appreciable growth defect, unlike the ulp2Δ mutant (10, 29). Moreover, a relatively modest increase of SUMOylated Net1, Cdc14, and Tof2 is observed in the ulp2–781 mutant, relative to WT cells (29). In contrast, an ∼20-fold accumulation of SUMOylated Net1, Cdc14, and Tof2 was observed in the ulp2Δ mutant (21). These findings indicated that the Csm1-mediated targeting of Ulp2 to the nucleolus is insufficient to account for the exquisite substrate selectivity of Ulp2 toward its nucleolar substrates; despite that eliminating the Csm1–Ulp2 binding is sufficient to cause a defect in rDNA silencing via aberrant degradation of Tof2 (29).
To further understand the substrate specificity of Ulp2, we investigated the role of Ulp2's C-terminal non-catalytic domain, which is largely unstructured, and identified two conserved regions among Ulp2 fungal orthologs, aside from the Csm1-binding domain. We provide evidence supporting that one of these conserved regions contains a SIM between residues 725 and 728 of Ulp2, and further shows that the C-terminal Ulp2 SIM plays an unexpected and major role in contributing to its in vivo substrate specificity toward Ulp2's nucleolar substrates in conjunction with Csm1-mediated Ulp2 recruitment to the nucleolus.
Results
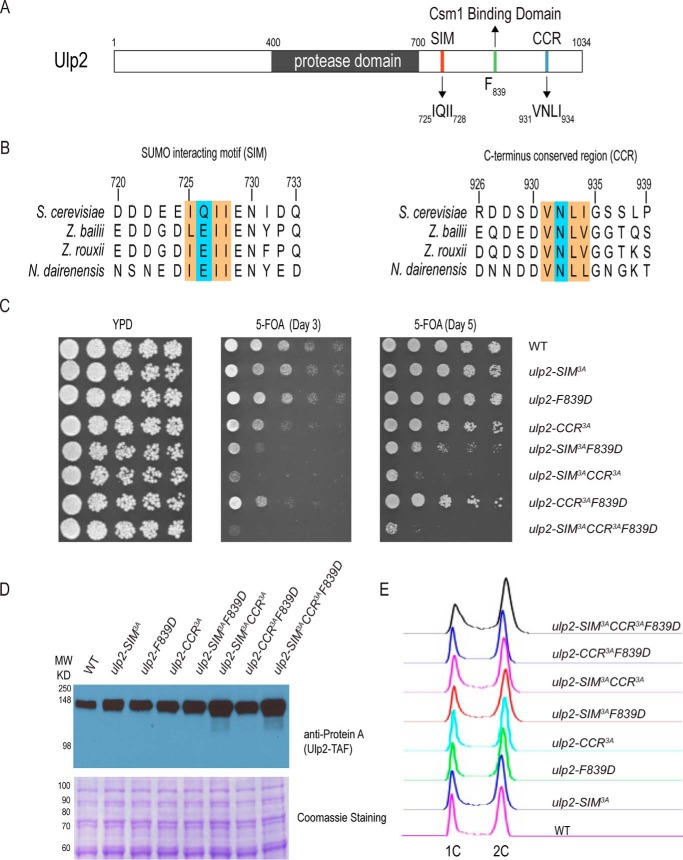
The Ulp2 C terminus contains three conserved regions with distinct and overlapping functions
Our previous study identified a conserved binding interface (residues 821–847) in Ulp2's C terminus that binds to the Csm1 protein to direct Ulp2 to the rDNA region (29). However, the ulp2–781 truncation mutant, in which the binding between Ulp2 and Csm1 is eliminated, does not fully recapitulate the defects of ulp2Δ, suggesting that other parts of Ulp2 are also necessary for its in vivo specificity (29). To address this, sequence alignment of S. cerevisiae Ulp2 was performed across several fungal species (Fig. 1B). This revealed two other conserved regions in the Ulp2 C terminus, besides the Csm1-binding domain, one of which was previously suggested to be a SIM based on its amino acid sequence (23). The other conserved region will be referred to as the C-terminal conserved region, or CCR, as its role remains unknown.
Figure 1.
The Ulp2 C terminus contains three conserved regions with distinct and overlapping functions. A, schematic of the conserved regions of Ulp2. B, sequence alignment of two conserved regions in the C terminus of Ulp2 orthologs; excluding its Csm1-binding domain. C, effect of ulp2 mutations on cell growth in the presence of 5-FOA to acutely remove the complementing WT Ulp2 plasmid. YPD loading control is shown along with growth after 3 and 5 days on 5-FOA plates incubated at 30 °C. D, protein abundance of WT and various mutants of Ulp2 following 5-FOA treatment. Coomassie staining is shown to indicate equal loading. E, flow cytometry of asynchronous yeast cells expressing various forms of the Ulp2 protein following 5-FOA treatment.
Because the ulp2Δ mutant is known to readily accumulate survivors as well as exhibit a severe growth defect (10, 22), any ulp2 mutation that compromises cell growth could potentially accumulate faster growing survivors to obscure the direct effect of such mutation. To address this issue, we introduced specific ulp2 mutations, in a centromeric plasmid under its native promoter, into the ulp2Δ strain that contains a complementing centromeric plasmid with the WT Ulp2 sequence under the control of its native promoter. Upon treatment with 5-fluorooratic acid (5-FOA), to remove the complementing plasmid containing ULP2, the acute effect on growth of these ulp2 mutations could be determined within the first 20–25 generations. To ascertain if Ulp2's C-terminal SIM and CCR are important for its in vivo function, triple alanine (3A) mutants of the SIM and CCR (SIM3A and CCR3A) were generated to substitute the conserved bulky hydrophobic residues within these motifs. Growth analysis revealed that the F839D mutation of Ulp2 does not impair cell growth. Moreover, the SIM3A and CCR3A mutations do not cause a noticeable growth defect on the 5-FOA plate, indicating these mutations have a minimal effect on the overall integrity of Ulp2 (Fig. 1C). Interestingly, double mutations combining SIM3A, and either F839D or CCR3A, result in a significant slower growth phenotype on 5-FOA (Fig. 1C). In contrast, the double mutation combining F839D and CCR3A does not cause an additive growth defect compared with the ulp2–CCR3A single mutant. This slow growth defect was exacerbated further when all three C-terminal-conserved regions were mutated, with very little growth observed on the 5-FOA plate (Fig. 1C). To determine whether these C-terminal mutations affect the overall abundance of Ulp2, whole cell extracts were assayed for the abundance of TAF-tagged Ulp2 using anti-Protein A antibody. As shown in Fig. 1D, none of the C-terminal mutations of Ulp2 reduces the abundance of Ulp2 compared with WT. Instead, the abundance of Ulp2 in the ulp2-SIM3ACCR3A and the ulp2 triple mutant appear to be higher relative to WT, which could be due to an unknown compensatory effect of the mutations. In any case, these findings suggest that these ulp2 mutations unlikely affect the overall integrity of the Ulp2 protein in cells; although they likely affect the mutated regions locally.
We noticed that additional streaking of the ulp2–SIM3ACCR3A and the ulp2 triple mutants, obtained from the 5-FOA plate, rapidly led to faster growing survivors; possibly resembling those generated in the ulp2Δ mutant (22). Because it was reported that truncation of the C terminus of Ulp2 did not appreciably impair cell growth (23), contrary to what we observe here (Fig. 1C), to address these contrasting results, we integrated the same set of ulp2 mutations into the chromosomal locus of Ulp2, and confirmed them via DNA sequencing; a process that allows the cells to grow over 100 generations. We then examined the growth of these chromosomal integrated ulp2 mutants. As shown in Fig. S1A, none of the C-terminal ulp2 mutants exhibit an appreciable growth defect on YPD media, and is in agreement with a previous report (23). These findings suggest that survivor formation may have previously obscured the critical role of Ulp2's C terminus in maintaining cell growth and underscore the importance of observing the growth defects caused by ulp2 C-terminal mutations immediately after the complementing ULP2 allele is removed (Fig. 1C).
Furthermore, flow cytometry was performed to assay for any specific cell cycle arrest that might be present in any of the ulp2 C-terminal mutants that are immediately derived after 5-FOA plating (Fig. 1C). As shown in Fig. 1E, there is no appreciable cell cycle-specific arrest for any of the ulp2 C-terminal mutants, apart from a slight accumulation of S-phase and G2 cells in the ulp2 triple mutant. However, we caution here that this finding could still be affected by the accumulation of unknown survivors at an unknown rate during cell growth. Further study is needed to determine the precise nature of the genome instability defects associated with these ulp2 mutants.
Taken together, these results demonstrate that the C terminus of Ulp2 has distinct and overlapping functions in maintaining cell growth, contrary to previous findings (23). More specifically, the C-terminal SIM of Ulp2 shares redundant roles with either the Csm1-binding domain or the CCR of Ulp2.
The Ulp2 C-terminal SIM shows increasing affinity for longer linear SUMO chains
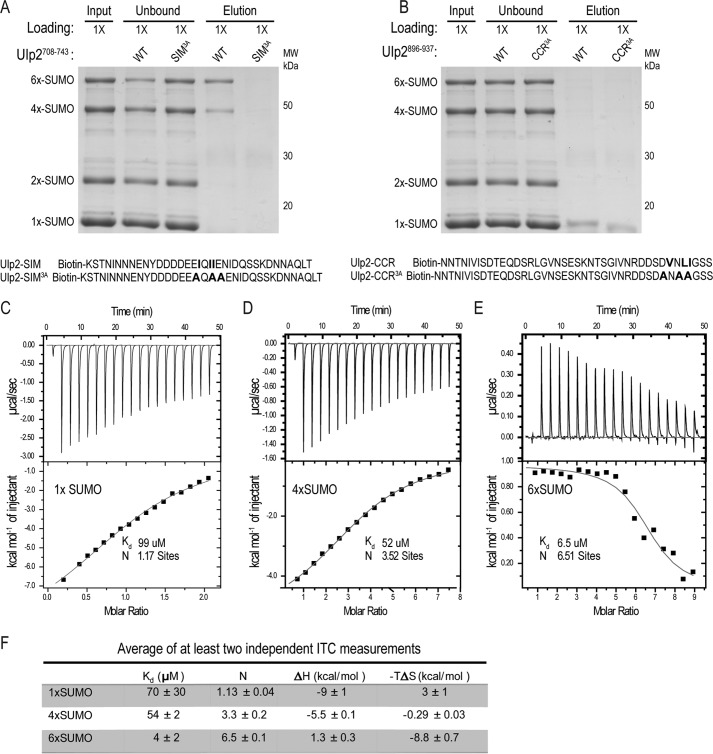
To characterize Ulp2's C-terminal SIM and its CCR further, a binding assay was developed to measure the interaction between a synthetic peptide of each conserved region and varying lengths of linear SUMO chains, which were expressed and purified from bacteria. Synthetic biotinylated peptides of the WT and 3A variants of either the C-terminal SIM or CCR were bound to NeutrAvidin resin and incubated with a mixture of linear poly-SUMO chains containing SUMO, 2×-SUMO, 4×-SUMO, and 6×-SUMO proteins (Fig. 2A). As seen in Fig. 2A, the WT Ulp2 SIM peptide, but not the Ulp2 SIM3A peptide, bound specifically to 4×-SUMO and 6×-SUMO proteins, but not 2×-SUMO or SUMO; confirming that it is a bona fide SUMO-interacting motif with an increasing affinity to longer linear SUMO chains. Furthermore, we replaced Ulp2's C-terminal SIM with the previously described M-IR2 SIM of RanBP2 (Ran-binding protein 2) or the SIM of PIASX (protein inhibitor of activated STAT X) (25, 37). As expected, the growth defect of the ulp2–SIM3ACCR3AF839D on 5-FOA is rescued by the replacement of the SIM3A with either the PIASX SIM (sequence: VDVIDI) or the RanBP2 M-IR2 SIM (sequence: VIIVW); indicating that the slow growth of the triple mutant is due to the loss of a functional SIM (Fig. S1B). In contrast, binding between the CCR peptide and any of the SUMO variants was not detected; suggesting that the CCR does not play a role in the direct binding of SUMO (Fig. 2B). We have not investigated the function of Ulp2's CCR further and chose to focus on characterizing Ulp2's C-terminal SIM in this study.
Figure 2.
The Ulp2 SIM peptide preferentially binds to longer linear SUMO chains. A, pulldown assay to detect binding between WT and mutant Ulp2 SIM peptide and linear SUMO chains. 1× loading is used for each sample. Alanine substitutions of residues in Ulp2's SIM are indicated in bold. B, pulldown assay to detect binding between WT and mutant Ulp2 CCR peptide and linear SUMO chains. Residues of Ulp2's CCR that were substituted by alanine are indicated in red. C–E, ITC analysis of 1×-, 4×-, and 6×-SUMO binding to the Ulp2 SIM peptide (DDDDEEIQIIENIDQSSKD). Dissociation constant (Kd) and binding stoichiometry (N) are indicated for each titration. F, ITC summary of the average of at least two independent measurements.
To quantitatively assess the binding affinity between the Ulp2 SIM peptide and different lengths of linear SUMO variants, isothermal calorimetry (ITC) was performed. The Ulp2 SIM peptide was injected into ITC cells containing SUMO, 4×-SUMO, or 6×-SUMO, and thermodynamic parameters were recorded (Fig. 2, C–E). The ITC data shows that the Ulp2 SIM peptide only exhibits weak binding to SUMO with a Kd of 99 μm (Fig. 2C). However, the measured binding affinity between the Ulp2 SIM peptide and 4×-SUMO was 2-fold greater than SUMO (Kd 52 μm); and 10-fold greater for 6×-SUMO (Kd 6.5 μm) (Fig. 2, D and E). Moreover, binding between the Ulp2 SIM peptide to SUMO and 4×-SUMO is enthalpy driven (Fig. 2, C, D, and F), whereas in the case of 6×-SUMO the Ulp2 SIM peptide binds with a stronger contribution from entropy. In the absence of structural data, the basis for this entropy contribution is unknown; although this result suggests that poly-SUMO to SIM binding can be modulated via structural rearrangements.
The Ulp2 C-terminal SIM promotes its SUMO protease activity toward linear SUMO chains
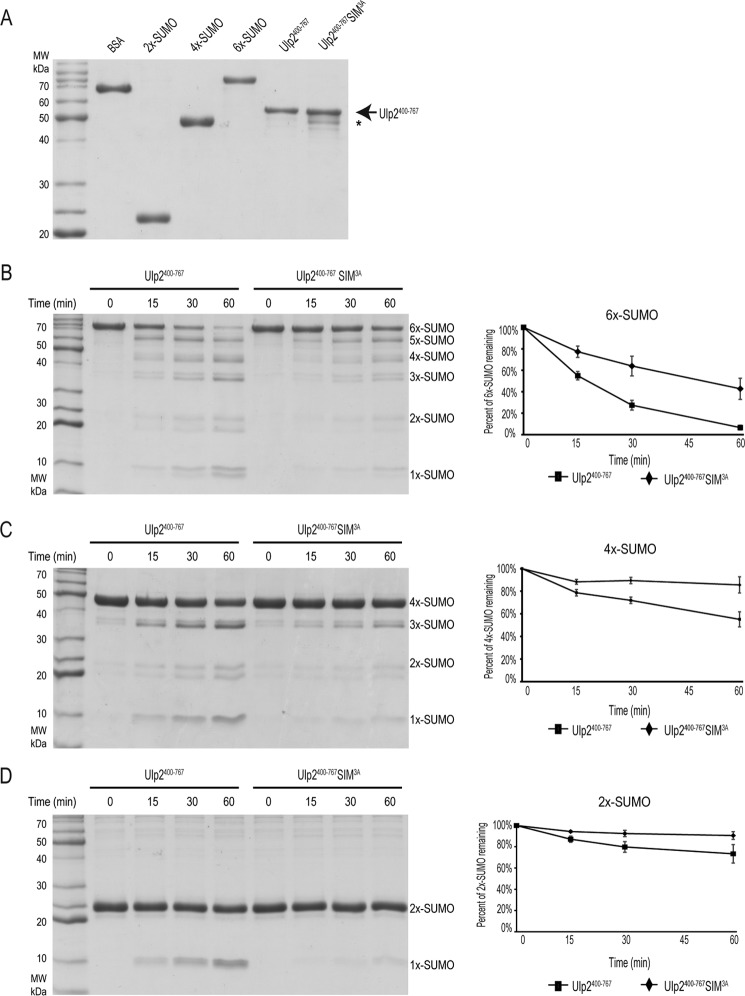
To determine the role that Ulp2's C-terminal SIM plays in its function, we sought to determine its role in regulating in vitro Ulp2 SUMO protease activity. To do so, recombinant 2×-SUMO, 4×-SUMO, and 6×-SUMO were purified from bacteria and used as substrates; whereas a truncated form of recombinant Ulp2 (residues 400–767), containing either the WT or SIM3A variant, was purified from bacteria (Fig. 3A). It is worth noting that the Ulp2 SIM3A mutant protein contains a minor degradation product, but the enzyme used for subsequent biochemical assays were normalized using the non-degraded form of Ulp2 (residues 400–767) (Fig. 3A). Attempts to purify full-length Ulp2 from bacteria were unsuccessful; however, this truncated version of Ulp2 (residues 400–767) contains the catalytic domain as well as the C-terminal extension containing the SIM sequence, allowing us to evaluate the function of the C-terminal SIM in regulating Ulp2's SUMO protease activity.
Figure 3.
The Ulp2 C-terminal SIM promotes Ulp2's SUMO protease activity toward linear SUMO chains. A, Coomassie staining of purified Ulp2400–767 proteins (WT and SIM3A). Degradation products are indicated by an asterisk. B–D, cleavage of 2×-, 4×-, and 6×-SUMO by Ulp2400–767 enzyme (WT and SIM3A). Quantification of Coomassie staining is included to the right of each corresponding reaction.
Ulp2's SUMO protease activity was assessed after incubation with 2×-SUMO, 4×-SUMO, and 6×-SUMO for varying durations. At 15, 30, and 60 min post-incubation; equal amounts of samples were withdrawn and analyzed by SDS-PAGE and Coomassie Blue staining. The intensity of each protein was quantified by densitometry and Ulp2's SUMO protease activity is evaluated according to the percentage of 2×-SUMO, 4×-SUMO, or 6×-SUMO remaining (Fig. 3, B–D). Each experiment was performed in triplicate, and the average of these repeats are shown. The SIM3A mutant significantly reduces Ulp2's SUMO protease activity for all of the linear SUMO substrates (Fig. 3, B–D). However, its effect appears to be more pronounced as the SUMO chain length is increased. Although the WT Ulp2 processes ∼40% more 6×-SUMO compared with the Ulp2 SIM3A (Fig. 3B), WT Ulp2 processes, a relatively reduced, ∼30% more 4×-SUMO and ∼15% more 2×-SUMO than the Ulp2 SIM3A (Fig. 3, C and D). Furthermore, WT Ulp2 processes longer linear SUMO chains more efficiently; as the starting amount of 6×-SUMO remaining after 60 min is ∼5%, whereas there is ∼55 and ∼75% 4×-SUMO and 2×-SUMO remaining, respectively. Thus, Ulp2's C-terminal SIM helps it to preferentially cleave longer SUMO chains, most likely via its increased binding affinity to longer SUMO chains. Our finding here is consistent with a prior study showing that Ulp2 preferentially cleaves longer SUMO chains in vitro (38). However, we have not investigated whether our purified Ulp2 enzyme exhibits a directionality in processing SUMO chains or its detailed mechanism of action; rather, we chose to focus on the role of Ulp2's C-terminal SIM in regulating its cleavage activity here, which was not investigated in prior studies (10, 38).
Ulp2 SIM peptide competitively inhibits the cleavage of linear SUMO chains by Ulp2400–767
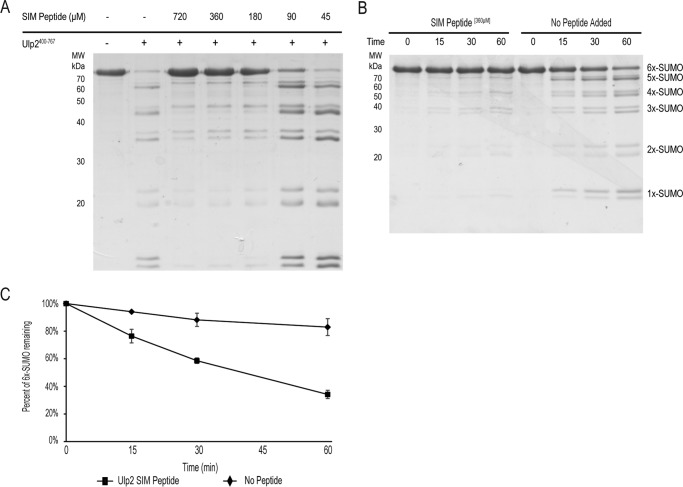
The above finding suggests that the C-terminal SIM of Ulp2 recruits SUMO chains through direct binding, and thus facilitates its SUMO–protease activity. This model predicts that the addition of excess free SIM peptide should competitively inhibit Ulp2's cleavage activity. To test this, different amounts of Ulp2 SIM peptide were added to the Ulp2 cleavage reaction using 6×-SUMO as substrate. As shown in Fig. 4A, free SIM peptide acts to inhibit the cleavage of 6×-SUMO by Ulp2 in a dosage-dependent manner. Moreover, addition of the SIM peptide of Ulp2 causes a significant reduction of cleavage of 6×-SUMO over time (Fig. 4, B and C). Taken together, these findings further support a model in which the C-terminal SIM of Ulp2 acts to recruit SUMOylated targets and facilitate their cleavage by Ulp2's catalytic domain.
Figure 4.
Ulp2400–767 is specifically inhibited by the presence of a SIM peptide. A, decreasing amounts of the WT Ulp2 SIM peptide were added to the Ulp2 cleavage reaction of 6×-SUMO (as performed in Fig. 3B. B, cleavage of 6×-SUMO by Ulp2400–767 in the presence of (360 μm) SIM peptide or no peptide added. C, densitometry quantification of the amount of remaining 6×-SUMO, with error bars representing S.E.
The Ulp2 C-terminal SIM facilitates the processing of branched SUMO chains
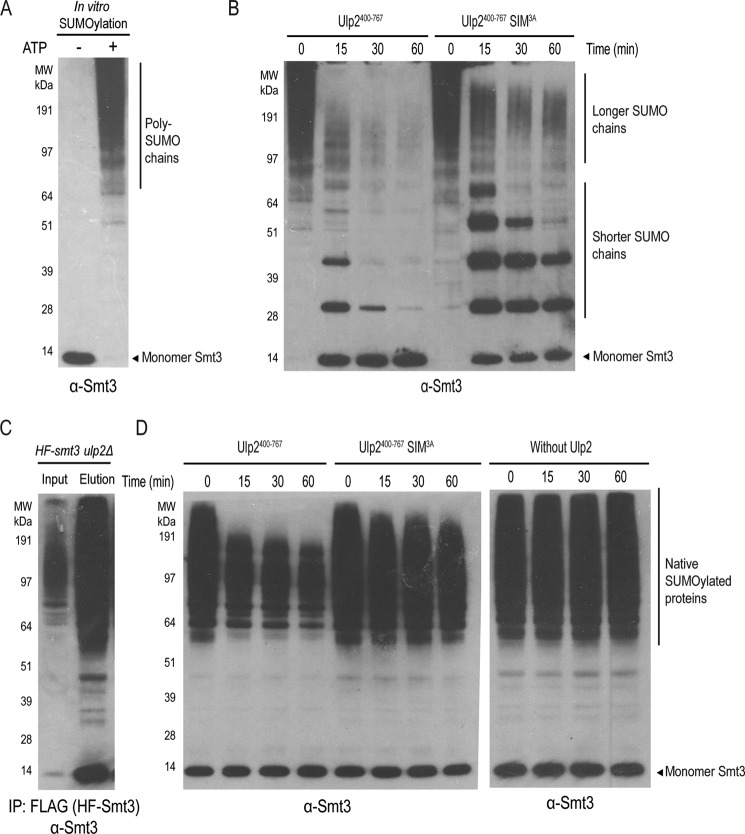
The majority of poly-SUMO chains in yeast cells are known to be branched as opposed to linear (20). To assess the role of Ulp2's C-terminal SIM in its ability to cleave branched poly-SUMO chains, poly-SUMO chains were synthesized through an in vitro SUMOylation reaction (Fig. 5A). Previous studies have also shown that these poly-SUMO chains are formed through branched lysine SUMO–SUMO linkages (20), and thus were used as a substrate to determine the SUMO isopeptidase activity of Ulp2 containing either WT SIM or the SIM3A mutant. Similar to its effect on linear SUMO chains, the Ulp2 SIM3A mutant is severely compromised in the cleavage of shorter SUMO chains, which are still detected 60 min post-incubation; whereas they are not detected when incubated with WT Ulp2 (Fig. 5B). It is difficult to assess the effect of the Ulp2 SIM3A mutant on the cleavage of longer SUMO chains, as we also cannot exclude the possibility that Ulp2's catalytic domain likely possesses additional recognition for SUMO itself. This demonstrates that the Ulp2 C-terminal SIM increases its protease activity for both linear and branched poly-SUMO chains, irrespective of the peptide linkage between SUMO involved, and is particularly important for the efficient cleavage of shorter poly-SUMO chains.
Figure 5.
The C-terminal SIM of Ulp2 facilitates its cleavage of in vitro synthesized poly-SUMO chains as well as native sumoylated proteins. A, in vitro sumoylation to generate poly-SUMO chains. B, cleavage kinetics of in vitro synthesized poly-SUMO chains by Ulp2400–767 enzyme (WT and SIM3A). C, purification of sumoylated proteins from the ulp2Δ mutant using anti-FLAG affinity resins. D, cleavage kinetics of native sumoylated proteins by Ulp2400–767 enzyme (WT and SIM3A).
The Ulp2 C-terminal SIM promotes the desumoylation of endogenous sumoylated proteins by Ulp2
Considering that Ulp2's C-terminal SIM is needed for efficient cleavage of shorter poly-SUMO chains, we sought to evaluate its role further in the cleavage of endogenous SUMOylated proteins. For this purpose, we chose to purify total SUMOylated proteins from an ulp2Δ mutant that expresses His6–3×FLAG–Smt3 (HF-Smt3) (7). This mutant allowed us to purify total SUMOylated proteins using anti-FLAG affinity resins, which were eluted under native conditions using an excess of 3×FLAG peptide (Fig. 5C). One caveat of this purified sample is that the relative amount of poly-SUMO chains versus mono-SUMOylated proteins is difficult to determine; largely because there are hundreds of SUMOylated proteins, along with a few identified poly-SUMOylated Ulp2 substrates (21). Moreover, multiple subunits of the same protein complex are often SUMOylated and each SUMOylated protein could have multiple lysines being modified by SUMO (39). Despite these caveats, recombinant Ulp2 WT and the Ulp2 SIM3A mutant proteins were incubated with the purified total SUMOylated proteins, and as seen in Fig. 5D, little to none of the endogenous SUMOylated proteins were processed by the Ulp2 SIM3A mutant, yet WT Ulp2 was able to cleave the majority of the SUMOylated proteins. Unlike the processing of in vitro synthesized poly-SUMO chains (Fig. 5B), treatment by WT or SIM3A mutant of Ulp2 here did not lead to an accumulation of shorter SUMO chains, suggesting that poly-SUMO chains likely exist at a level below our detection limit. Therefore, most of the SUMO signals of the endogenous SUMOylated proteins are likely attributed to either mono-SUMOylated protein species or proteins with shorter SUMO chains, which were observably cleaved by the WT Ulp2 but not the Ulp2 SIM3A mutant (Fig. 5D). This finding indirectly suggests that WT Ulp2 is able to process proteins with shorter or mono-SUMOylated moieties; and requires its C-terminal SIM to do so. Considering that SUMOylation can occur on multiple lysines on a given protein, or on multiple proteins of the same protein complex, it is conceivable that multiple SUMOs on a single protein–protein complex could make it a better substrate for Ulp2. In any event, the apparent difference in the cleavage of endogenous SUMOylated proteins by WT Ulp2 and Ulp2 SIM3A revealed a critical role for Ulp2's C-terminal SIM in facilitating its SUMO-cleavage activity from native SUMOylated substrates.
Both the Ulp2 C-terminal SIM- and Csm1-binding domains contribute to its in vivo substrate specificity toward its nucleolar substrates
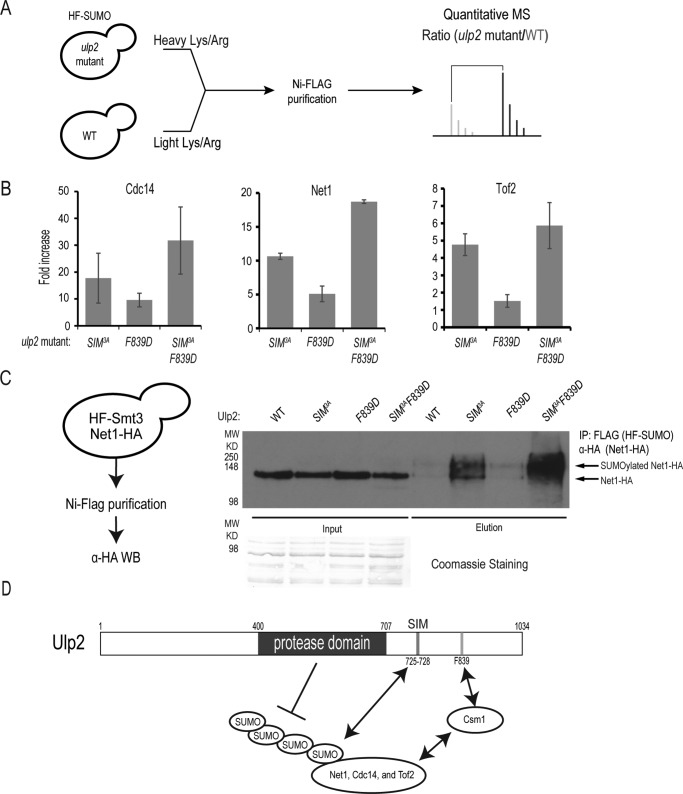
To determine whether Ulp2's C-terminal SIM plays any role in the processing of in vivo Ulp2 substrates, a quantitative proteomic assay was performed to evaluate the SUMOylation status of three known Ulp2 targets in the nucleolus: Net1, Cdc14, and Tof2 (Fig. 6A). In each case, we performed stable isotope labeling of WT and ulp2 mutant cells, and purified total SUMOylated proteins for quantitative mass spectrometry (MS) analysis as previously described (7, 29). Using our quantitative SUMO proteomics assay, the relative abundance of SUMOylated Ulp2 targets, Cdc14, Net1, and Tof2, were determined (Fig. 6B). The F839D mutant of Ulp2 was previously shown to have a partial loss of Ulp2 function (29), which is observed here as the relative abundance of Cdc14 and Net1 are moderately increased relative to WT by 10- and 5-fold, respectively (Fig. 6B and Table S1). The abundance of SUMOylated Tof2 is not appreciably changed in the ulp2–F839D mutant; and this can be attributed to our earlier observation that the overall abundance of Tof2 is reduced in this ulp2–F839D mutant, whereas the abundance of Net1 and Cdc14 are largely unchanged (Fig. 6B) (29). Interestingly, the ulp2–SIM3A mutant substantially increases the amount of SUMOylated Cdc14, Net1, and Tof2 to a much higher level, 5–18–fold, than the effect of the ulp2–F839D mutation (Fig. 6B and Table S1), indicating that the Ulp2's C-terminal SIM has a stronger role than its Csm1-binding domain for its substrate selectivity in vivo. Furthermore, the double mutant, ulp2–SIM3AF839D, exhibits an over 20-fold accumulation of SUMOylated Net1 and Cdc14, which is comparable with the effect of ulp2Δ on these proteins (21). To address the possibility that these effects are simply due to a lower abundance of Ulp2, Western blots were performed to assess the abundance of Ulp2 in these strains. As seen in Fig. S2A, we did not see a change in the abundance of Ulp2 in these strains. Furthermore, flow cytometry of these strains also indicated that the asynchronous cell cycle profiles were no different from WT (Fig. S2B), thus the changes in SUMOylated Net1 and Cdc14 are unlikely caused by any cell cycle perturbation. Taken together, the ulp2–SIM3AF839D mutant recapitulates the effect of the ulp2Δ on the SUMOylation of Cdc14, Net1, and Tof2, indicating that the C-terminal SIM and Csm1-binding domain of Ulp2 are both necessary and sufficient for Ulp2's specificity toward these proteins.
Figure 6.
Ulp2's C-terminal SIM and Csm1-binding domain play a redundant role in controlling the desumoylation of the RENT complex in vivo. A, schematic of the quantitative MS assay used to determine sumoylation levels in WT and various ulp2 mutants. B, average abundance ratios of sumoylated Net1, Tof2, and Cdc14 in various ulp2 mutants, including ulp2–SIM3A, ulp2–F839D, and ulp2–SIM3AF839D mutants. Further details of the MS results are summarized in Table S1 and Tables S3–S5. C, maximum desumoylation of Net1 requires Ulp2's C-terminal SIM and its Csm1-binding domain. Total sumoylated proteins in various ulp2 mutants were purified and probed by anti-HA antibody to detect sumoylated Net1–HA; schematic for the immunoprecipitation (IP) is shown. D, mechanism of Ulp2 substrate specificity toward its nucleolar substrates Net1, Tof2, and Cdc14. Ulp2's Csm1-binding domain and SUMO-interacting motif (SIM) are both needed for maximal desumoylation of the RENT complex and Tof2 in the nucleolus.
Finally, to determine whether mutating Ulp2's C-terminal SIM or Csm1-binding domain causes a defect in the processing of poly-SUMOylated Ulp2 substrates; we chose to purify total sumoylated proteins from Net1–3xHA and HF-Smt3–tagged cells carrying the ulp2–SIM3A, F839D, or the SIM3AF839D double mutant (Fig. 6C). Following enrichment for SUMOylated proteins using Ni-NTA and anti-FLAG affinity columns (29), immunoblots for Net1-HA revealed that the ulp2–SIM3A and SIM3AF839D mutations drastically increased the amount of poly-SUMOylated Net1, whereas the effect of ulp2–F839D mutation alone on Net1 SUMOylation is modest; which is in agreement with the quantitative MS result (Fig. 6B). Together, these findings show that both the Ulp2 C-terminal SIM and Csm1-binding domain are involved in the deSUMOylation of Ulp2's nucleolar substrates; with the Ulp2 C-terminal SIM playing a greater role compared with its binding to Csm1.
Discussion
The findings here indicate that Ulp2's C-terminal SIM and its Csm1-binding domain both promote deSUMOylation of its nucleolar substrates; including the RENT complex and Tof2 (Fig. 6D). We have previously shown that Ulp2 binds to Csm1, which recruits Ulp2 to the nucleolus where it is poised to deSUMOylate Tof2 and likely the RENT complex (29). Here we demonstrated that the C-terminal SIM of Ulp2 plays an unexpected and major role in controlling Ulp2's in vivo specificity, specifically its ability to deSUMOylate the RENT complex and Tof2 in the nucleolus.
Removal of the C terminus of Ulp2 was previously shown to cause minimal growth defect (23), which differs from our finding here (Fig. 1C). However, a major difference lies in the approaches used to analyze the growth of the ulp2 C-terminal mutants. Because the ulp2Δ mutant was shown to accumulate faster growing survivors via the formation of aneuploidy (22), we reasoned that mutations to the C terminus of Ulp2 could also generate survivors. Thus, we chose a plasmid shuffling approach to observe the growth of ulp2 mutants within the first 25 generations after the complementing ULP2 plasmid is removed (Fig. 1C). Our approach allowed us to identify the specific regions of the Ulp2 C terminus that are needed to maintain cell growth (Fig. 1C). Interestingly, and consistent with a prior study (23), when the same ulp2 mutations were integrated into the chromosomal Ulp2 locus, which involved about 100 generations of cell division, the growth defect of ulp2 mutant was no longer detectable (Fig. S1A). Because formation of survivors, like all other genetically unstable mutants, is directly correlated with the number of cell divisions, the relatively larger number of cell divisions needed to generate chromosomally integrated ulp2 mutants has likely masked their growth defects. However, plasmid shuffling allowed us to identify the specific regions in the Ulp2 C terminus that play a major role in maintaining cell growth; as relatively fewer cell divisions are needed before analysis, thereby reducing the chance of survivor formation (Fig. 1C). At present, the nature or rate of survivor formation in these ulp2 mutants, particularly the ulp2–sim3ACCR3A double mutant, is unknown. It should be interesting to determine whether aneuploidy of a specific chromosome will accumulate in this ulp2 mutant, much like what was previously reported for the ulp2Δ mutant (22).
The SUMO–SIM interaction generally has a relatively modest affinity (25). Consistent with this notion, the Ulp2 C-terminal SIM also exhibits a modest affinity to monomeric SUMO but a stronger affinity to longer SUMO chains (Fig. 2), which allows Ulp2 to cleave poly-SUMO chains more efficiently (Figs. 3 and 4). Although these in vitro results strongly implicate the function of Ulp2's C-terminal SIM in SUMO-binding, the in vivo data suggests a surprisingly strong requirement for the SIM in the Ulp2-dependent deSUMOylation of its nucleolar substrates (RENT complex and Tof2) (Fig. 6D); raising the possibility that poly-SUMO chains or multisite SUMO modifications, on these nucleolar proteins, may act to recruit Ulp2 toward their own cleavage. It is worth noting that deletion of Ulp2 in the 4R-smt3 mutant, which lacks the major poly-SUMOylation sites in the N terminus of Smt3 (21), still resulted in substantial accumulation of SUMOylated Net1, Tof2, and Cdc14 (21). Thus, poly-SUMO chains are unlikely to be the sole determining factor for the recruitment of Ulp2. Instead, SUMO attached to multiple lysines on the same protein, or different proteins in the same protein complex, could also recruit Ulp2 for their deSUMOylation. In support of this idea, multiple lysines of Net1 and Tof2 have been found to be SUMOylated, although MS identification of SUMO modification sites of these proteins could still be incomplete (40). Moreover, Net1, Cdc14, and Tof2 have been shown to form a large protein complex (32); suggesting a mechanism by which cells would continuously SUMOylate Net1, Cdc14, and Tof2 to a high level, and thus, necessitate the presence of Ulp2 to keep sumoylation of these nucleolar proteins low in WT cells. One potential concern is whether the ulp2-SIM3AF839D mutant could form survivors, and if so, whether its effect on SUMOylation of Ulp2's nucleolar substrates could be altered. We consider this unlikely for several reasons. First, to detect SUMOylated proteins in yeast cells (Fig. 6), we used the HF-Smt3 strain in which the HF tag at the N terminus of Smt3 appears to compromise poly-SUMO formation and thus suppress the growth defect of the ulp2Δ mutant (21). Thus, it is unlikely that any ulp2 mutants containing HF-Smt3 would accumulate survivors. Second, the ulp2–SIM3A single mutant, even without HF-Smt3, exhibits little to no growth defect (Fig. 1C), yet a significant amount of SUMOylated Net1 and Cdc14 accumulates in this mutant (Fig. 6). Third, although the ulp2–SIM3ACCR3A mutant is prone to generate survivors in the absence of HF-Smt3 (Fig. 1C), this mutation likely affects SUMOylation of other Ulp2 substrates that are outside of the nucleolus. Further studies are needed to investigate the defect of this mutant, including its tendency to accumulate survivors.
Given the ability of Ulp2 to bind SUMOylated proteins via its C-terminal SIM, one may ask why Ulp2 specifically deSUMOylates relatively fewer substrates compared with Ulp1 (21). Prior studies have shown that Ulp1's protease domain directly binds to SUMO with nanomolar affinity (Kd = 12.9 nm) (41). In contrast, binding between SUMO and the catalytic domain of Ulp2, or its human ortholog Senp7, has not been reported (42), or detected via our own pulldown assay.4 We suggest that the modest affinity between Ulp2's C-terminal SIM and SUMO may ensure that only those substrates with multiple SUMO modifications, either via poly-SUMO chains or multiple lysines on the substrates, would provide a strong enough signal to recruit Ulp2 for their destruction. Besides the RENT complex, several other nucleolar SUMOylated proteins have been identified, including Rpa135, Rpa190, and others (21); yet deletion of Ulp2 does not appreciably alter SUMOylation of these proteins. One possible explanation is that these proteins are not SUMOylated to a high enough level to be recognized by Ulp2. In contrast, the RENT complex and Tof2 are likely continuously SUMOylated, although the mechanism behind their SUMOylation is presently unknown.
Ulp1 is unable to deSUMOylate the nucleolar substrates of Ulp2; despite its stronger binding to SUMO and its potent activity toward SUMOylated proteins (9). This is likely due to the distinct localization of Ulp1 at the nuclear periphery (19, 43), and its noted absence in the nucleolus (44). Indeed, the removal of the N-terminal domain of Ulp1 mislocalizes it throughout the cell (19, 43), and also results in the aberrant deSUMOylation of the nucleolar Ulp2 substrates in an Ulp2-independent manner (21). We therefore speculate that the potent activity of Ulp1 is restricted by its localization to the nuclear periphery, despite the fact that Ulp1 still performs the majority of deSUMOylation in cells. In doing so, Ulp1 keeps most of the intracellular SUMOylation relatively low, so that they are not recognized by Ulp2, even though Ulp2 appears throughout the nucleoplasm. On the other hand, Ulp2 is localized to the nucleolus via its Csm1-binding domain, whereas its C-terminal SIM further directs it toward poly-SUMO and/or multi-SUMO to selectively deSUMOylate proteins that would otherwise be SUMOylated to a high level, such as the RENT complex and Tof2 (Fig. 6).
Besides the RENT complex and Tof2 in the nucleolus, Ulp2 also specifically deSUMOylates several inner kinetochore proteins, as well as specific subunits of the MCM complex (21). We do not know whether these SUMOylated kinetochore and MCM subunits are found on the centromeres and origins of DNA replication of all chromosomes, or exclusively those on chromosome 12, which reside in the nucleolus. However, mutations that eliminate Csm1–Ulp2 binding do not appreciably alter SUMOylation of either the inner kinetochore or MCM subunits, unlike the deletion of Ulp2, and argue against the latter idea (29). Analogous to Ulp2's mechanism for deSUMOylating its nucleolar substrates, we speculate that the same paradigm could apply for other Ulp2 substrates. In other words, Ulp2 may utilize additional protein–protein interactions to gain access to other substrates, in addition to its C-terminal SIM, to target those proteins that become hyper-SUMOylated. In support of this idea, the CCR of Ulp2 appears to be needed for optimal cell growth (Fig. 1C), especially when the Ulp2 C-terminal SIM and Csm1-binding domain are also mutated. Thus, the CCR of Ulp2 may have an unknown but important role in mediating other Ulp2 functions in cells, and further studies are needed to fully understand how Ulp2 achieves all of its substrate specificity.
Extensive studies of the deubiquitination enzymes have also shown that these enzymes possess distinct ubiquitin-binding properties; a critical aspect in their ability to process ubiquitinated substrates (45). The findings here support the general concept that SUMO-mediated interaction plays a major role in the specificity of deSUMOylating enzymes. Although Ulp1 utilizes its stronger interaction with SUMO for its potent activity as well as its distinct localization to keep its activity under control; Ulp2 employs two modes of substrate recognition via a specific adaptor protein such as Csm1 and a relatively modest SUMO–SIM interaction to selectively target its hyper-SUMOylated substrates, as a way to antagonize the actions of the SUMOylation enzymes. Thus, our findings have provided new insights into the mechanism of SUMO protease substrate recognition; however, future studies are needed to fully characterize how Ulp2 selectively desumoylates its other substrates, as well as the mechanism that underlies hyper-SUMOylation of specific Ulp2 substrates in unperturbed cells.
Experimental procedures
Standard yeast genetic methods were used to construct the yeast strains used in this study. The genotypes of all strains and plasmids used in this work can be found in Table S2. ULP2 was deleted by polymerase chain reaction (PCR)-based methods using natMX4 to confer resistance to nourseothricin. All deletions were confirmed by PCR to ensure both the presence of the deletion and the removal of the WT allele. All mutations were integrated in yeast chromosomal loci, and were confirmed by DNA sequencing.
The 5-FOA sensitivity assay was performed as follows: eight different ULP2 alleles were carried on pRS315 plasmids that were transformed into HZY3658; carrying ULP2 on a pRS316 plasmid. These eight transformants were grown in 5 ml of Complete Synthetic Medium lacking leucine (0.69 g/liter of CSM-Leu drop out mixture, 7 g/liter of yeast nitrogen base, and 20 g/liters of glucose) until reaching an A600 of ∼2.0. Cell density of the eight cultures were normalized according to their A600 to ensure equal plating. Five 1:10 serial dilutions were then performed for each transformant in a sterile 96-well plate using sterile-deionized water as a diluent. 4 μl of each well was spotted onto either YPD (1.0% yeast extract, 2.0% peptone, 2% d-glucose, and 2.4% agar) or 5-FOA (0.67% yeast nitrogen base, 0.77 g/liters of CSM–Ura drop-out mixture, 20 mg/liter of uracil 0.1% 5-FOA plates, and 2.4% agar). Both plates were incubated at 30 °C, between 2 and 5 days, and representative images were acquired using a Bio-Rad ChemiDocTM MP imaging system.
To measure the growth of chromosomally integrated ulp2 mutants, a plating assay was performed as follows: eight different yeast strains (Table S2: HZY621, HZY001–HZY006, and HZY035) carrying ULP2 or C-terminal mutations of ulp2 were grown in YPD overnight until saturation ∼A600 3.0. Cells were then normalized to an A600 of 2.0 and 1:10 serial dilutions were performed in a sterile 96-well plate using sterile-deionized water. 4 μl of each well was spotted onto YPD plates and allowed to grow for 3 days at 30 °C, and representative images were acquired using a Bio-Rad ChemiDocTM MP imaging system.
To evaluate the relative abundance of Ulp2, whole cell extracts were generated using a base–acid lysis approach. Specifically, the eight strains used in the 5-FOA sensitivity assay were taken from 5-FOA plates and grown in 50 ml of YPD to an A600 of 0.5. Cells were then centrifuged at 2,000 × g for 5 min and the supernatants were decanted. Then to each cell pellet the following was added: 250 μl of glass beads, 200 μl of 1 m NaOH, 500 μl of H2O, 100 μl of 1 m phosphate buffer, pH 8.0, and 200 μl of 10% SDS. The cells were then lysed at 4 °C by vortexing for 5 min, after which 200 μl of 1 m HCl was added to the sample to neutralize pH. Samples were then heated for 10 min at 65 °C and centrifuged at 15,000 × g. Bradford reagent was used to normalize protein concentrations, and equal amounts of whole cell extract were loaded and run through a 10% SDS-PAGE gel to be either visualized by Coomassie staining or Protein A-Western blotting. The relative abundance of Ulp2 was assayed by Western blotting using α-Protein A primary antibody (Sigma P3775) and α-rabbit HRP secondary antibody (Millipore Sigma).
Samples for flow cytometry were prepared as follows: yeast cells were grown to A600 of 0.5 and 300 μl of yeast cell culture was mixed with 700 μl of pure ethanol for fixation. Samples were then incubated overnight at 37 °C in 1 ml of buffer (50 mm sodium citrate, pH 7.0, 250 μg/ml of RNase A, and 1 mg/ml of proteinase K). The following day, samples were resuspended by sonication in 1 ml of buffer containing (50 mm sodium citrate, pH 7.0, and 1 μm Sytox Green). Samples were incubated 30 min in the dark before being analyzed by flow cytometry. All flow cytometry was performed on a BD LSR II flow cytometer.
Mature Smt3 (SUMO) was cloned into the pET21B plasmid, whereas the other Smt3 constructs containing Smt3 monomer, Smt3 dimer (pKDC297), Smt3 tetramer (pKDC298), and Smt3 hexamer (pKDC296), were a generous gift from Dr. Kevin D. Corbett (University of California, San Diego) and contain an N-terminal His6–tobacco etch virus cleavable tag. These SUMO constructs were expressed in Escherichia coli RosettaTM-2(DE3)pLysS (Novagen) cells, and were grown in 4 liters of LB (Luria Broth) media containing 100 μg/ml of ampicillin. Constructs were induced overnight when cells reached an A600 of 0.6 at 18 °C with 0.2 mm isopropyl β-d-1-thiogalactopyranoside. The Smt3 constructs were lysed in phosphate-buffered saline (PBS) with 10% glycerol and 14 mm β-mercaptoethanol (β-ME), and purified using a Ni-NTA column (Qiagen). Constructs were further purified by FPLC gel filtration (Superdex 200 10/300 GL) using ÄKTA pure FPLC system, and fractions containing the Smt3 proteins were pooled and stored at −80 °C.
WT Ulp2400–767 and SIM3A–Ulp2400–767 were cloned into a 2BT LIC (ligation independent cloning) plasmid containing a His6 tag. They were then expressed in E. coli RosettaTM-2(DE3)pLysS cells as described for the Smt3 constructs. WT Ulp2400–767 and SIM3A–Ulp2400–767 were then lysed in PBS with 10% glycerol and 14 mm β-ME and purified via FPLC using Ni-NTA in PBS followed by cation exchange (HiTrap SP), gel filtration (Superdex 200 10/300 GL), and anion exchange (monoQ 5/50 GL) via ÄKTA pure FPLC system. Fractions containing WT Ulp2400–767 and SIM3A–Ulp2400–767 were pooled and stored at −80 °C. Expression and purification of Aos1-Uba2 (E1), Ubc9 (E2), and Siz1 (E3, residues 167–465) enzymes were described previously (40).
For the endogenous purification of sumoylated proteins, 2 liters of HF-SMT3 ulp2Δ NET1-HA (HZY3725) were grown in YPD to an A600 of 1.5. Cells were harvested and washed with 10 ml of PBS/Nonidet P-40 (0.2% Nonidet P-40) containing 10 mm iodoacetamide and 10 mm N-ethylmaleimide. Cells were then resuspended in 2.5 ml of PBS/Nonidet P-40 with 10 mm iodoacetamide, 10 mm N-ethylmaleimide, and protease inhibitor mixture (2 mm phenylmethylsulfonyl fluoride, 200 μm benzamidine, 0.5 μg/ml of leupeptin, 1 μg/ml of pepstatin A). The resuspended cells were frozen dropwise in liquid nitrogen. Cells were pulverized using a SPEX SamplePrep 6875D freezer/mill and then thawed in ¼ volume of PBS/Nonidet P-40 with protease inhibitor mixture. Cell lysate was cleared by ultracentrifugation and incubated with 100 μl of packed anti-FLAG-M2 resin (Sigma) for 2 h at 4 °C. Following incubation, the resin was washed four times with 2 ml of PBS/Nonidet P-40 containing protease inhibitor mixture. Proteins were eluted in 1000 μl of PBS containing 10% glycerol, 12 mm β-ME, 0.2 mg/ml of 3x-FLAG peptide and protease inhibitor mixture.
Pulldown assay for detecting binding between the Ulp2 SIM peptide and linear SUMO chains was performed using biotinylated peptides purchased from EZbio: UIp2708–743SIM (Biotin-KSTNINNNENYDDDDEEIQIIENIDQSSKDNNAQLT), UIp2708–743SIM3A (Biotin-KSTNINNNENYDDDDEEAQAAENIDQSSKDNNAQLT), UIp2896–937CCR (Biotin-NNTNIVISDTEQDSRLGVNSESKNTSGIVNRDDSDVNLIGSS), and UIp2896–937CCR3A (Biotin-NNTNIVISDTEQDSRLGVNSESKNTSGIVNRDDSDANAAGSS). 20 μg of each linear SUMO construct, 1×, 2×, 4×, and 6×, was mixed and incubated with 10 μl of NeutrAvidin-agarose resin (preincubated with the biotinylated peptide) in 100 μl of PBS buffer for 2 h at 4 °C. Beads were washed 3 times with 200 μl of PBS, and eluted by boiling at 100 °C in 100 μl of 2% SDS. 5% of the input, flow-through, and elution were loaded and run through a 15% SDS-PAGE gel and visualized by Coomassie staining.
In vitro SUMOylation for generating poly-SUMO chains was carried out in 500 μl of reaction buffer containing 20 mm HEPES, pH 7.5, 0.15 m NaCl, 1 mm ATP, and 2 mm MgCl2. Working concentrations of E1, E2, E3, and Smt3 were: 0.4 μm Aos1–Uba2, 2 μm Ubc9, 0.4 mm Siz1 (residues 167–465), and 5 μm Smt3. Reactions were incubated for 1 h at 30 °C, and 100 mm EDTA was used to quench and stop the reaction.
In vitro assays for measuring the activity of UIp2400–767 WT or SIM3A toward linear SUMO chains was performed as follows: 100 μg of each respective construct, dimer, tetramer, or hexamer, to 50 μl of PBS containing 1 mm DTT and 1 μg of UIp2400–767 WT or SIM3A at room temperature. 10-μl aliquots of the reaction were then collected for SDS-PAGE analysis at 0, 15, 30, and 60 min after the initial addition of each construct. 2 μg of SUMO from each fraction was loaded to a 15% SDS-PAGE gel and visualized by Coomassie staining and quantified by densitometry using the ImageJ processing and analysis software.
In vitro assays for measuring the activity of UIp2400–767 WT or SIM3A toward in vitro poly-SUMO chains were performed as described above but with the following changes: 75 μg of in vitro synthesized poly-SUMO chains were used as a substrate, and 0.75 μg of either WT Ulp2400–767 or SIM3A–Ulp2400–767 was used in a total reaction volume of 50 μl. 30 ng of SUMO for each time point was analyzed by Western blotting using a 4–12% gradient gel (Invitrogen); blots were performed using an α-Smt3 (rabbit polyclonal antibody made via Covance, Inc.) and an α-rabbit HRP antibody (Millipore Sigma).
In vitro cleavage activity of UIp2400–767 WT or SIM3A toward endogenous sumoylated proteins was measured as described above with the following changes: 100 μl of endogenous sumoylated protein was used as a substrate (purification described above), and 0.75 μg of either WT Ulp2400–767 or SIM3A–Ulp2400–767 was used. 20-μl aliquots for each time point were collected, and 5 μl was loaded onto a 4–12% gradient gel (Invitrogen). Western blotting was performed using α-Smt3 and α-rabbit HRP. In vitro cleavage inhibition assays of Ulp2 were performed with the same parameters as the in vitro cleavage assay, except for the addition of SIM peptide at the indicated final concentrations.
ITC experiments were carried out at the Sanford Burnham Prebys Medical Discovery Institute Protein Analysis Core Facility. The SUMO proteins used for ITC were dialyzed in PBS, and two replicates of ITC was performed using an ITC200 calorimeter from Microcal (Northampton, MA) at 23 °C. 2.0-μl aliquots of solution containing 2 mm Ulp2SIM peptide (DDDDEEIQIIENIDQSSKD, GenScript) were injected into the cell containing 0.04 to 0.2 mm of the SUMO constructs in PBS. Nineteen injections of 2 μl were performed. ITC data were analyzed using Origin software provided by Microcal.
The preparation of samples for quantitative MS analysis to measure changes in SUMOylated protein abundance between two strains was performed as previously described in Ref. 7, but with the following changes. FLAG elution was performed using 500 μl of elution buffer containing: 8 m urea, 40 mm sodium carbonate in 50 mm phosphate buffer, pH 8, and incubated for 5 min at room temperature. The elution was then neutralized with 18 μl of 1 m HCl and then bound to 75 μl of packed Ni-NTA-agarose beads (Qiagen). The beads were washed once with 1 ml of 8 m urea in 50 mm phosphate buffer, pH 8, followed by 2 ml of ammonium bicarbonate containing 0.04% Nonidet P-40. Samples were then incubated with 500 μl of ammonium bicarbonate with 0.5 μg of trypsin (Promega; sequencing grade) for 12 h. Samples were then dried and fractionated on a HILIC column, before being analyzed by LC-MS/MS (Thermo Scientific Orbitrap Fusion Lumos mass spectrometer; acquired via NIH S10 OD023498) (7).
SILAC abundance ratios of Cdc14, Net1, and Tof2 were determined by first applying several filters to the data set: 1) a peptide probability score of greater than 0.8, 2) a parental ion mass accuracy filter of ±10 ppm, and 3) a minimum spectral intensity cutoff of 1.0 × 104. Then the top five most abundant spectra, assigned to each protein, was used to determine the average abundance ratio for each protein along with standard deviations of each protein ratio. In the case of Cdc14 only four peptides met this requirement in the ulp2-SIM3AF839D versus WT experiment. The complete list of SUMOylated proteins identified in each MS experiment are provided in Tables S3–S5, which include the number of peptides identified for each protein and the median abundance ratio of each sumoylated protein. To evaluate SUMOylated Net1–HA in various ulp2 mutants, total SUMOylated proteins from the indicated strains were purified using the method described previously (29), and then analyzed by α-HA (Sigma 3F10) antibody to detect Net1–3HA.
Author contributions
C. P. d. A., R. T. S., C. R. C., and H. Z. conceptualization; C. P. d. A., R. T. S., C. R. C., and H. Z. data curation; C. P. d. A., R. T. S., C. R. C., W.-T. Y., and H.Z. investigation; C.P.d.A., R. T. S., C. R. C., W.-T. Y., and H. Z. methodology; C. P. d. A., R. T. S., C. R. C., and H. Z. writing-original draft; R. T. S., C. R. C., W.-T. Y., and H. Z. formal analysis; R. T. S. and H. Z. validation; R. T. S., C. R. C., and H. Z. writing-review and editing; H. Z. resources; H. Z. supervision; H. Z. funding acquisition; H. Z. project administration.
Supplementary Material
Acknowledgments
We thank Dr. Kevin Corbett for the generous gift of plasmids expressing linear SUMO chains and Andrey Bobkov (Sanford Burnham Prebys Medical Discovery Institute Protein Analysis Core Facility) for performing the ITC titrations. We also thank members of the Zhou lab for discussions and assistance in the experiments described in this work.
This work was supported by National Institutes of Health NIGMS Grant R01 GM116897 and the Ludwig Institute for Cancer Research (to H. Z.) and National Institutes of Health NCI Grant NCI T32 CA009523 (to R. T. S.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Figs. S1 and S2 and Tables S1–S5.
A lack of binding between Ulp2's catalytic domain and SUMO was demonstrated in the Zhou lab (Zhou, H., and Albuquerque, C. P., unpublished results).
- SUMO
- small ubiquitin-like modifier
- Ubl
- ubiquitin-like protein
- HA
- hemagglutinin
- HRP
- horseradish peroxidase
- LC-MS/MS
- liquid chromatography-tandem mass spectrometry
- MCM
- mini-chromosome maintenance complex
- NTA
- nitrilotriacetic acid
- RENT
- regulator of nucleolar silencing and telophase
- rDNA
- ribosomal DNA
- SIM
- SUMO-interacting motif
- CCR
- C-terminal conserved region
- 5-FOA
- 5-fluorooratic acid
- ITC
- isothermal calorimetry.
References
- 1. Gareau J. R., and Lima C. D. (2010) The SUMO pathway: emerging mechanisms that shape specificity, conjugation and recognition. Nat. Rev. Mol. Cell Biol. 11, 861–871 10.1038/nrm3011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hickey C. M., Wilson N. R., and Hochstrasser M. (2012) Function and regulation of SUMO proteases. Nat. Rev. Mol. Cell Biol. 13, 755–766 10.1038/nrm3478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bayer P., Arndt A., Metzger S., Mahajan R., Melchior F., Jaenicke R., and Becker J. (1998) Structure determination of the small ubiquitin-related modifier SUMO-1. J. Mol. Biol. 280, 275–286 10.1006/jmbi.1998.1839 [DOI] [PubMed] [Google Scholar]
- 4. Johnson E. S. (2004) Protein modification by SUMO. Annu. Rev. Biochem. 73, 355–382 10.1146/annurev.biochem.73.011303.074118 [DOI] [PubMed] [Google Scholar]
- 5. Johnson E. S., and Gupta A. A. (2001) An E3-like factor that promotes SUMO conjugation to the yeast septins. Cell 106, 735–744 10.1016/S0092-8674(01)00491-3 [DOI] [PubMed] [Google Scholar]
- 6. Zhao X., and Blobel G. (2005) A SUMO ligase is part of a nuclear multiprotein complex that affects DNA repair and chromosomal organization. Proc. Natl. Acad. Sci. U.S.A. 102, 4777–4782 10.1073/pnas.0500537102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Albuquerque C. P., Wang G., Lee N. S., Kolodner R. D., Putnam C. D., and Zhou H. (2013) Distinct SUMO ligases cooperate with Esc2 and Slx5 to suppress duplication-mediated genome rearrangements. PLoS Genet. 9, e1003670 10.1371/journal.pgen.1003670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Reindle A., Belichenko I., Bylebyl G. R., Chen X. L., Gandhi N., and Johnson E. S. (2006) Multiple domains in Siz SUMO ligases contribute to substrate selectivity. J. Cell Sci. 119, 4749–4757 10.1242/jcs.03243 [DOI] [PubMed] [Google Scholar]
- 9. Li S. J., and Hochstrasser M. (1999) A new protease required for cell-cycle progression in yeast. Nature 398, 246–251 10.1038/18457 [DOI] [PubMed] [Google Scholar]
- 10. Li S. J., and Hochstrasser M. (2000) The yeast ULP2 (SMT4) gene encodes a novel protease specific for the ubiquitin-like Smt3 protein. Mol. Cell. Biol. 20, 2367–2377 10.1128/MCB.20.7.2367-2377.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gong L., Millas S., Maul G. G., and Yeh E. T. (2000) Differential regulation of sentrinized proteins by a novel sentrin-specific protease. J. Biol. Chem. 275, 3355–3359 10.1074/jbc.275.5.3355 [DOI] [PubMed] [Google Scholar]
- 12. Gong L., and Yeh E. T. (2006) Characterization of a family of nucleolar SUMO-specific proteases with preference for SUMO-2 or SUMO-3. J. Biol. Chem. 281, 15869–15877 10.1074/jbc.M511658200 [DOI] [PubMed] [Google Scholar]
- 13. Hang J., and Dasso M. (2002) Association of the human SUMO-1 protease SENP2 with the nuclear pore. J. Biol. Chem. 277, 19961–19966 10.1074/jbc.M201799200 [DOI] [PubMed] [Google Scholar]
- 14. Mukhopadhyay D., and Dasso M. (2007) Modification in reverse: the SUMO proteases. Trends Biochem. Sci. 32, 286–295 10.1016/j.tibs.2007.05.002 [DOI] [PubMed] [Google Scholar]
- 15. Denison C., Rudner A. D., Gerber S. A., Bakalarski C. E., Moazed D., and Gygi S. P. (2005) A proteomic strategy for gaining insights into protein sumoylation in yeast. Mol. Cell. Proteomics 4, 246–254 [DOI] [PubMed] [Google Scholar]
- 16. Panse V. G., Hardeland U., Werner T., Kuster B., and Hurt E. (2004) A proteome-wide approach identifies sumoylated substrate proteins in yeast. J. Biol. Chem. 279, 41346–41351 10.1074/jbc.M407950200 [DOI] [PubMed] [Google Scholar]
- 17. Zhou W., Ryan J. J., and Zhou H. (2004) Global analyses of sumoylated proteins in Saccharomyces cerevisiae: induction of protein sumoylation by cellular stresses. J. Biol. Chem. 279, 32262–32268 10.1074/jbc.M404173200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wohlschlegel J. A., Johnson E. S., Reed S. I., and Yates J. R. 3rd. (2004) Global analysis of protein sumoylation in Saccharomyces cerevisiae. J. Biol. Chem. 279, 45662–45668 10.1074/jbc.M409203200 [DOI] [PubMed] [Google Scholar]
- 19. Li S. J., and Hochstrasser M. (2003) The Ulp1 SUMO isopeptidase: distinct domains required for viability, nuclear envelope localization, and substrate specificity. J. Cell Biol. 160, 1069–1081 10.1083/jcb.200212052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bylebyl G. R., Belichenko I., and Johnson E. S. (2003) The SUMO isopeptidase Ulp2 prevents accumulation of SUMO chains in yeast. J. Biol. Chem. 278, 44113–44120 10.1074/jbc.M308357200 [DOI] [PubMed] [Google Scholar]
- 21. de Albuquerque C. P., Liang J., Gaut N. J., and Zhou H. (2016) Molecular circuitry of the SUMO (small ubiquitin-like modifier) pathway in controlling sumoylation homeostasis and suppressing genome rearrangements. J. Biol. Chem. 291, 8825–8835 10.1074/jbc.M116.716399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ryu H. Y., Wilson N. R., Mehta S., Hwang S. S., and Hochstrasser M. (2016) Loss of the SUMO protease Ulp2 triggers a specific multichromosome aneuploidy. Genes Dev. 30, 1881–1894 10.1101/gad.282194.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kroetz M. B., Su D., and Hochstrasser M. (2009) Essential role of nuclear localization for yeast Ulp2 SUMO protease function. Mol. Biol. Cell 20, 2196–2206 10.1091/mbc.e08-10-1090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hannich J. T., Lewis A., Kroetz M. B., Li S. J., Heide H., Emili A., and Hochstrasser M. (2005) Defining the SUMO-modified proteome by multiple approaches in Saccharomyces cerevisiae. J. Biol. Chem. 280, 4102–4110 10.1074/jbc.M413209200 [DOI] [PubMed] [Google Scholar]
- 25. Song J., Durrin L. K., Wilkinson T. A., Krontiris T. G., and Chen Y. (2004) Identification of a SUMO-binding motif that recognizes SUMO-modified proteins. Proc. Natl. Acad. Sci. U.S.A. 101, 14373–14378 10.1073/pnas.0403498101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Srikumar T., Lewicki M. C., and Raught B. (2013) A global S. cerevisiae small ubiquitin-related modifier (SUMO) system interactome. Mol. Systems Biol. 9, 668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lee M. T., Bakir A. A., Nguyen K. N., and Bachant J. (2011) The SUMO isopeptidase Ulp2p is required to prevent recombination-induced chromosome segregation lethality following DNA replication stress. PLoS Genet. 7, e1001355 10.1371/journal.pgen.1001355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Rosonina E., Duncan S. M., and Manley J. L. (2010) SUMO functions in constitutive transcription and during activation of inducible genes in yeast. Genes Dev. 24, 1242–1252 10.1101/gad.1917910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Liang J., Singh N., Carlson C. R., Albuquerque C. P., Corbett K. D., and Zhou H. (2017) Recruitment of a SUMO isopeptidase to rDNA stabilizes silencing complexes by opposing SUMO targeted ubiquitin ligase activity. Genes Dev. 31, 802–815 10.1101/gad.296145.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Cremona C. A., Sarangi P., Yang Y., Hang L. E., Rahman S., and Zhao X. (2012) Extensive DNA damage-induced sumoylation contributes to replication and repair and acts in addition to the mec1 checkpoint. Mol. Cell 45, 422–432 10.1016/j.molcel.2011.11.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Gillies J., Hickey C. M., Su D., Wu Z., Peng J., and Hochstrasser M. (2016) SUMO pathway modulation of regulatory protein binding at the ribosomal DNA locus in Saccharomyces cerevisiae. Genetics 202, 1377–1394 10.1534/genetics.116.187252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Huang J., Brito I. L., Villen J., Gygi S. P., Amon A., and Moazed D. (2006) Inhibition of homologous recombination by a cohesin-associated clamp complex recruited to the rDNA recombination enhancer. Genes Dev. 20, 2887–2901 10.1101/gad.1472706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Corbett K. D., Yip C. K., Ee L. S., Walz T., Amon A., and Harrison S. C. (2010) The monopolin complex crosslinks kinetochore components to regulate chromosome-microtubule attachments. Cell 142, 556–567 10.1016/j.cell.2010.07.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Corbett K. D., and Harrison S. C. (2012) Molecular architecture of the yeast monopolin complex. Cell Rep. 1, 583–589 10.1016/j.celrep.2012.05.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Prudden J., Pebernard S., Raffa G., Slavin D. A., Perry J. J., Tainer J. A., McGowan C. H., and Boddy M. N. (2007) SUMO-targeted ubiquitin ligases in genome stability. EMBO J. 26, 4089–4101 10.1038/sj.emboj.7601838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sun H., Leverson J. D., and Hunter T. (2007) Conserved function of RNF4 family proteins in eukaryotes: targeting a ubiquitin ligase to SUMOylated proteins. EMBO J. 26, 4102–4112 10.1038/sj.emboj.7601839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Namanja A. T., Li Y. J., Su Y., Wong S., Lu J., Colson L. T., Wu C., Li S. S., and Chen Y. (2012) Insights into high affinity small ubiquitin-like modifier (SUMO) recognition by SUMO-interacting motifs (SIMs) revealed by a combination of NMR and peptide array analysis. J. Biol. Chem. 287, 3231–3240 10.1074/jbc.M111.293118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Eckhoff J., and Dohmen R. J. (2015) In vitro studies reveal a sequential mode of chain processing by the yeast SUMO (small ubiquitin-related modifier)-specific protease Ulp2. J. Biol. Chem. 290, 12268–12281 10.1074/jbc.M114.622217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Johnson E. S., and Blobel G. (1999) Cell cycle-regulated attachment of the ubiquitin-related protein SUMO to the yeast septins. J. Cell Biol. 147, 981–994 10.1083/jcb.147.5.981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Albuquerque C. P., Yeung E., Ma S., Fu T., Corbett K. D., and Zhou H. (2015) A chemical and enzymatic approach to study site-specific sumoylation. PloS One 10, e0143810 10.1371/journal.pone.0143810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Elmore Z. C., Donaher M., Matson B. C., Murphy H., Westerbeck J. W., and Kerscher O. (2011) Sumo-dependent substrate targeting of the SUMO protease Ulp1. BMC Biol. 9, 74 10.1186/1741-7007-9-74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lima C. D., and Reverter D. (2008) Structure of the human SENP7 catalytic domain and poly-SUMO deconjugation activities for SENP6 and SENP7. J. Biol. Chem. 283, 32045–32055 10.1074/jbc.M805655200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Panse V. G., Küster B., Gerstberger T., and Hurt E. (2003) Unconventional tethering of Ulp1 to the transport channel of the nuclear pore complex by karyopherins. Nat. Cell Biol. 5, 21–27 10.1038/ncb893 [DOI] [PubMed] [Google Scholar]
- 44. Zhao X., Wu C. Y., and Blobel G. (2004) Mlp-dependent anchorage and stabilization of a desumoylating enzyme is required to prevent clonal lethality. J. Cell Biol. 167, 605–611 10.1083/jcb.200405168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Mevissen T. E. T., and Komander D. (2017) Mechanisms of deubiquitinase specificity and regulation. Annu. Rev. Biochem. 86, 159–192 10.1146/annurev-biochem-061516-044916 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.