Abstract
Somites are a pair of epithelial spheres beside a neural tube and are formed with an accurate periodicity during embryogenesis in vertebrates. It has been known that Hes7 is one of the core clock genes for somitogenesis, and its expression domain is restricted in the presomitic mesoderm (PSM). However, the molecular mechanism of how Hes7 transcription is regulated is not clear. Here, using transgenic mice and luciferase-based reporter assays and in vitro binding assays, we unravel the mechanism by which Hes7 is expressed exclusively in the PSM. We identified a Hes7 essential region residing −1.5 to −1.1 kb from the transcription start site of mouse Hes7, and this region was indispensable for PSM-specific Hes7 expression. We also present detailed analyses of cis-regulatory elements within the Hes7 essential region that directs Hes7 expression in the PSM. Hes7 expression in the PSM was up-regulated through the E-box, T-box, and RBPj-binding element in the Hes7 essential region, presumably through synergistic signaling involving mesogenin1, T-box6 (Tbx6), and Notch. Furthermore, we demonstrate that Tbx18, Ripply2, and Hes7 repress the activation of the Hes7 essential region by the aforementioned transcription factors. Our findings reveal that a unified transcriptional regulatory network involving a Hes7 essential region confers robust PSM-specific Hes7 gene expression.
Keywords: mouse, development, transcription factor, transcription regulation, Notch pathway, Hes7, mesogenin1, somitogenesis, Tbx6
Introduction
Establishment of cellular identities is essential for generating complex patterns during embryogenesis. To establish cellular identities and develop organ/tissue formations properly, gene expressions are spatiotemporally regulated with accuracy in appropriate domains. The lefty1/lefty2 expressions, for example, are restricted to the left side of early developing mouse embryos to direct left–right axis determination (1). Deletion of lefty1 or lefty2 results in left pulmonary isomerism, malpositioning of the cardiac outflow tracts, and other vascular vessels (2) or an expanded primitive streak, formation of excess mesoderm (3), and various situs defects, including left isomerism (4), respectively.
We have identified the Hes7 gene, one of the Hes family transcriptional repressors, which is exclusively expressed in the presomitic mesoderm (PSM)4 and acts as a key molecule for somitogenesis (5–7). Somitogenesis is the process to form somites, which is a pair of epithelial spheres beside a neural tube and appear transiently during embryogenesis, from the anterior PSM (8). It is known that Hes7 expression is restricted in the PSM and is regulated by the Notch, Fgf, and Wnt signaling pathways (9). These signaling pathways regulate various processes during embryogenesis, suggesting that the restricted Hes7 gene expression in the PSM is orchestrated by a combination of transcriptional factors downstream of the Notch, Fgf, and Wnt signaling pathways. However, little is known about the transcriptional regulations that are associated with Hes7 gene expression.
In this study, we describe the presence of a Hes7 essential region, residing from −1.5 to −1.1 kb, from the transcription start site of the mouse Hes7 gene, that directs PSM-specific Hes7 expression. Furthermore, we demonstrate the mechanisms for Hes7 expression in the PSM. Restricted Hes7 expression is controlled through E-box, T-box, and the RBPj-binding element in the Hes7 essential region, presumably activated by a synergistic effect of mesogenin1, Tbx6, and Notch signaling, and repressed by Tbx18, Ripply2, and Hes7. Our study uncovered that the Hes7 essential region directs PSM-restricted expression pattern of Hes7, orchestrated by multiple transcriptional elements.
Results
C region, from −1.5 to −1.1 kb upstream of TSS of mouse Hes7, is sufficient for accurate Hes7 expression in the PSM
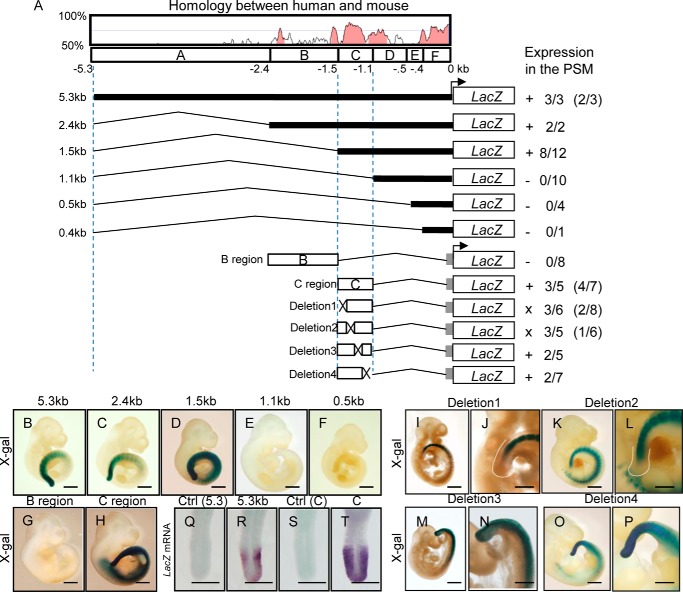
Although Hes7 mRNA is well known to be exclusively expressed in the PSM, the molecular mechanisms that regulate Hes7 expression/repression remain largely unknown. To uncover molecular mechanisms of the unique Hes7 expression in the PSM, we first tried to search for the essential region for PSM-specific Hes7 expression by means of transgenic founder assays. All transgenic embryos (n = 3) carrying the 5.3-kb fragment upstream from the TSS of mouse Hes7, which has been utilized for exogenous Hes7 expression in the PSM (10, 11), followed by a lacZ reporter showed X-gal–positive staining in the PSM at embryonic day (E) 10.5 (Fig. 1B). We next narrowed down the essential region and found that 2.4- and 1.5-kb fragments upstream from TSS were still sufficient for the reporter expression in the PSM (Fig. 1, C and D). However, we could not detect any X-gal–positive staining in the PSM of transgenic mice carrying a 1.1-kb fragment or shorter fragment (Fig. 1, E and F). These results suggest that the 1.5-kb fragment is sufficient for the PSM-specific expression of Hes7 and that the essential region for PSM-restricted expression resides between −1.5 and −1.1 kb upstream of TSS.
Figure 1.
Regulatory elements for Hes7 expression in the PSM. A, upper panel, sequence homology of Hes7 promoter between human and mouse was shown by VISTA. Transcription start site is indicated by the arrow and set to +1. 5.3 kb of Hes7 promoter was divided into six fragments, designated as A–F regions in this study. A, bottom panel, length of each reporter construct is indicated on the left. lacZ was used as a reporter under the control of Hes7 promoter. β-Globin minimal promoter is shown as a gray box. Results of reporter expression in the PSM of transgenic mice are shown on the right. +, positive expression in the PSM; −, negative expression in the PSM; ×, impaired expression in the PSM. Numbers indicated X-gal–positive embryos/genotyping positive embryos. Numbers in parentheses show lacZ mRNA-positive embryos/genotyping-positive embryos. B–F, transgenic mice integrated with 5.3 kb (B), 2.4 kb (C), 1.5 kb (D), 1.1 kb (E), or 0.5 kb (F) of reporter were stained with X-gal at E10.5. G and H, transgenic mice integrated with the B (G) or C (H) region of Hes7 upstream were stained with X-gal at E10.5. I–P, transgenic mice integrated Deletion1 (I and J), Deletion2 (K and J), Deletion3 (M and N), or Deletion4 (O and P) were stained with X-gal at E10.5. J, L, N, and P are magnified photos of I, K, M, and O, respectively. Q–T, transgenic embryos integrated with 5.3 kb (R), Hes7 C region (T), or embryos that did not integrate reporter genes into genome (Q and S) were subjected to in situ hybridization. Scale bars, 1 mm (B–I, K, M, and O), 0.5 mm (J, L, N, and P–T).
To confirm the region responsible for PSM-specific expression of Hes7, we next investigated the fragment from −2.4 to −1.5 kb, which is not considered to be an essential region for PSM-specific Hes7 expression, and hereafter it is referred to as the B region; and the fragment from −1.5 to −1.1 kb is hereafter referred to as the C region (Fig. 1A). Transgenic founder assays revealed that the C region drove β-gal protein expression in the PSM, whereas the B region had no β-gal activity in the PSM (Fig. 1, G and H). Next, we narrowed down the essential region in the C region. We constructed four reporter vectors deleting a quarter of fragment C region named Deletion1, -2, -3, or -4 (Fig. 1A and Fig. S1). Deletion1 and -2 showed no X-gal–positive staining at the most posterior end of the PSM (Fig. 1, I–L). However, Deletion3 and -4 resulted in X-gal–positive staining comparable with the WT C region (Fig. 1, M–P). In situ hybridization also demonstrated that the anterior-most regions of the PSM were negative for Deletion1- and -2–driven lacZ mRNA (Fig. S2). We therefore deduce from the above results that the 0.4-kb C region is the essential region, and the distal half of the C region from TSS contains essential transcriptional binding sites for Hes7 expression in the PSM.
Although endogenous Hes7 mRNA was expressed exclusively in the PSM, X-gal–positive staining was present throughout the PSM and newly formed somites, which are derived from the PSM, in transgenic mice carrying 5.3-, 2.4-, 1.5-kb fragments or the C region (Fig. 1, B–D and H). A simple explanation was that the reporter mRNA was transcribed exclusively in PSM cells, whereas its resultant β-gal protein remained in differentiated somite cells due to high protein stability. To assess this possibility, we carried out lacZ mRNA detection by in situ hybridization. The lacZ mRNA driven by a 5.3-kb fragment or the C region was exclusively expressed in the PSM (Fig. 1, R and T), and as expected, the control mice without transgene did not show any signal in the PSM (Fig. 1, Q and S). These results demonstrate that the C region is sufficient for accurate Hes7 expression in the PSM.
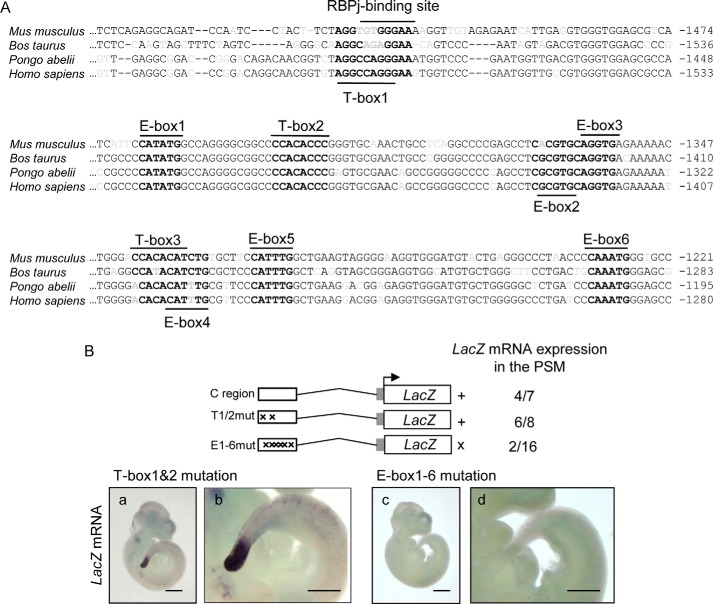
E-boxes in C region are essential to drive Hes7 in the PSM
To address which transcriptional binding elements regulate PSM-specific Hes7 expression, we searched for putative transcriptional binding elements within the C region by in silico analyses. There are several putative regulatory sequences in the C region: three T-boxes (YMACACYY or complementary) (21); six E-boxes (CANNTG) (5); and one RBPj-binding site (YRTGDGAD or complementary) (Fig. 2A and Fig. S1) (37), in particular, E-box1, -3, -5, and -6 and T-box2 were completely conserved among Homo sapiens, Pongo abelii, Bos taurus, and Mus musculus (Fig. 2A). To address whether these putative E-boxes, T-boxes, and the RBPj-binding site in the C region are functional in vivo, we made transgenic mice carrying the mutated E-box1–6/C region or the mutated T-box1 and -2/C region, which also include a mutated RBPj-binding site. In situ hybridization assays using transgenic founder mice revealed that mice with the mutated T-box1 and -2/C region expressed the reporter mRNA in the PSM as well as the mice with WT C region (Fig. 2B, panels a and b). In contrast, 14 of 16 mice with the mutated E-box1–6/C region showed no positive signal, whereas only two embryos showed a dispersed and straggling reporter expression (Fig. 2B, panels c and d, and Fig. S3), indicating that E-boxes in the C region are essential to drive Hes7 in the PSM.
Figure 2.
E-boxes in the C region are necessary for Hes7 expression in the PSM. A, putative T-boxes (YMACACYY or complementary) (21), E-boxes (CANNTG) (5), and the RBPj-binding site (YRTGDGAD or complementary) (37) within the Hes7 C region are shown in bold. One base mismatch is allowed for T-box and RBPj-binding site sequences. Nonconserved sequences among species are shown in gray. Numbers on the right indicate the positions from transcriptional start sites. B, in situ hybridization of transgenic mice embryos carrying lacZ reporter under the control of Tbox1 and -2 mutant (panels a and b) or E-box1–6 mutant (panels c and d) at E10.5 were performed. β-Globin minimal promoter is shown as a gray box. Transcription start site is indicated by the arrow. Results of lacZ expression in the PSM of transgenic mice are shown on the right. +, positive expression in the PSM; ×, impaired expression in the PSM. Numbers on the reporter constructs showed lacZ mRNA-positive embryos/genotyping positive embryos. Panels b and d are magnified photos of panels a and c, respectively. Scale bars, 1 mm (panels a and c), 0.5 mm (panels b and d).
Msgn1, Tbx6, and Notch signaling pathway activate C region in vitro
To investigate the putative elements in the C region in depth, we performed luciferase assays using constructs containing the WT or mutated C region followed by a human β-globin minimal promoter. To perform this, mesogenin1 (Msgn1), Tbx6, and NICD, the intracellular domain of Notch1, were utilized as binding factors for E-box, T-box, and RBPj-binding site, respectively. This is based on previous reports that showed that Msgn1, a bHLH-type transcription factor, is exclusively expressed in the posterior PSM (12), whereas Tbx6, a T-box family of transcriptional factor, and Notch signaling molecules are expressed ubiquitously in mouse PSM (13–15). We confirmed that the Msgn1 expression domain coincided with Hes7 stripes in phase I and II, but not at phase III, whereas Tbx6 mRNA was constantly distributed over the PSM, overlying any phases of the Hes7-transcribed region, besides the anterior-most region of the PSM (Fig. S4). WT C region reporter was activated by all of the transcriptional factors tested (Fig. 3, A–C), whereas the activity of mutated E-box1–6/C or T-box1–3/C region reporter was attenuated by Msgn1 or NICD, respectively, (Fig. 3, A and B), indicating that at least one of the mutated E-boxes is a functional site for Msgn1, whereas the T-box1 is receptive toward Notch signaling molecules. However, the mutated T-box1–3/C region reporter was still responsive to Tbx6, comparable with that of WT C region (Fig. 3C).
Figure 3.
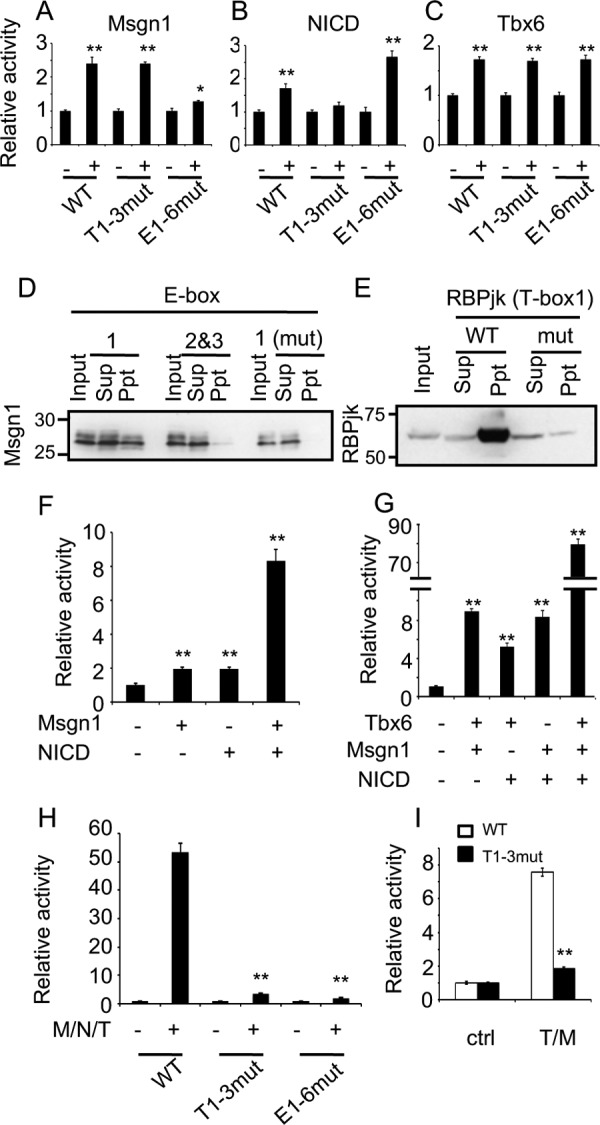
Msgn1, Tbx6, and Notch signaling act on Hes7 expression. A–C, luciferase assays for wildtype (WT), T-box mutant (T1–3mut), or E-box mutant (E1–6mut) Hes7 C region with or without Msgn1 (A), NICD (B), or Tbx6 (C) were performed in NIH3T3 cells. Reporter firefly luciferase activities were normalized by Renilla luciferase activities under SV40 promoter. Data represent the means ± S.E. of three independent samples. D, oligo-DNA pulldown assays against Ebox1 (WT/mut) or E-box2/3 (WT) with Msgn1-Myc were performed. Sup, supernatant; Ppt, Msgn1 was revealed with anti-Myc. E, oligo-DNA pulldown assays against T-box1 (WT/mut) with RBPjk-Myc were performed. RBPjk was revealed with anti-Myc. Sup, supernatant; Ppt, precipitate. F–I, luciferase assays for WT (F and G) or indicated mutant (H and I) Hes7 C region with indicated transcription factors were performed. Reporter firefly luciferase activities were normalized by Renilla luciferase activities under SV40 promoter. Data represent the means ± S.E. of three independent samples. *, p < 0.05; **, p < 0.01 compared with no transcription factor control for each promoter (A–C, F, and G), Msgn1 (M)/NICD (N)/Tbx6 (T) for WT promoter (H) or Tbx/Msgn1 for WT promoter (I) (Student's two-tailed t test).
To further investigate E-boxes, T-boxes, and the RBPj-binding site in the C region in vitro, we performed oligo-DNA pulldown assay and electrophoretic mobility shift assay (EMSA). Pulldown assay revealed that Msgn1 bound to E-box1, but not to E-box2 and -3 (Fig. 3D). Furthermore, we confirmed that Msgn1 binding to the E-box1 was abolished by the mutation of E-box1 (Fig. 3D). EMSA also demonstrated that Msgn1 bound to E-box1, but not to E-box2 and -3 (Fig. S5). Again, we confirmed that Msgn1 binding to E-box1 was abolished by an excess amount of the nonlabeled E-box1 but not by that of the mutant one (Fig. S5). These results raise the possibility that Msgn1 in the PSM activates Hes7 expression via the E-box1 in C region. RBPjk binding was detected by T-box1, in which RBPj-binding site is included, by pulldown assay and EMSA (Fig. 3E and Fig. S6). Moreover, pulldown assay demonstrated that RBPjk binding was dramatically attenuated by the mutated E-box1, and EMSA showed that this binding was attenuated by an excess amount of the nonlabeled E-box1 but not by that of the mutated one (Fig. 3E and Fig. S6). These results raise the possibility that the RBPj-binding site in C region is functional in vivo. Luciferase assays demonstrated that T-box1–3 were not responsive to Tbx6; however, ChIP assay utilizing PSM samples indicated that Tbx6 bound to the C region (Fig. S7A). To investigate whether Tbx6 binds to T-box elements in the C region, we performed oligo-DNA pulldown assay and showed that Tbx6 bound to T-box1 and T-box2 but not to T-box3. In contrast, the Tbx6 binding potential to T-box1 or T-box 2 was eliminated by mutated T-box1 or T-box2, respectively (Fig. S7 and data not shown).
Msgn1, Tbx6, and Notch signaling pathways synergistically activate C region in vitro
As we demonstrated that Msgn1 and NICD increased the luciferase activity of the C region and Msgn1 and RBPjk bound to E-box1 and T-box1, respectively (Fig. 3, A–E), we next investigated whether a combination of Msgn1 and NICD show a coordinated activation. Compared with single Msgn1 or NICD activation, co-expression of Msgn1 and NICD synergistically increased luciferase activity (Fig. 3F). Although, as shown above, T-box1 and -2 were not necessary for the expression of Hes7 in the PSM (Fig. 2B, panels a and b), the ChIP assay, oligo-DNA pulldown assay, and luciferase assay showed that Tbx6 could bind to and activate the C region (Fig. 3C and Fig. S7). Furthermore, because Tbx6 has been known to work synergistically with other transcriptional activators for gene expressions in the PSM (13), we investigated the possibility of Tbx6 working synergistically with Msgn1 or NICD to activate the C region. In contrast to either Msgn1 or NICD alone, the combination of Tbx6 with either Msgn1 or NICD increased the C region-driven luciferase activity (Fig. 3G). Intriguingly, we revealed that Tbx6 together with Msgn1 and NICD accelerated the reporter expression much more than the combinations with Tbx6/Msgn1, Tbx6/NICD, or Msgn1/NICD (Fig. 3G). These results indicate that Tbx6, Msgn1, and Notch signaling activate Hes7 expression coordinately via the C region at least in vitro. To confirm whether their synergistic effect on the C region is due to E-boxes, T-boxes, and the RBPj-binding site, we next performed luciferase assays using the C region with the mutated T-box1–3/or E-box1–6/C region. Synergistic activation by Msgn1/NICD/Tbx6 was almost completely abolished in T-box or E-box mutants (Fig. 3H), although activity in the T-box1–3 mutant was higher than that of the E-box1–6 mutant, supporting the results that embryos with mutated T-box1 and -2 expressed lacZ mRNA, but those with E-box1–6 did not, in vivo (Fig. 2B). We further investigated whether the participation of Tbx6 in the synergistic activation is independent of T-box1–3 elements. To address that, we evaluated the synergistic effect of Tbx6 and Msgn1 to rule out the activation of T-box1 by NICD. Surprisingly, the activation of the mutated T-box1–3/C region by Tbx6 and Msgn1 was almost completely abrogated (Fig. 3I), suggesting that T-box1–3 elements are important for the synergistic effect of Msgn1/NICD/Tbx6 on Hes7 expression, although T-box1–3 elements were not necessary for the single activation by Tbx6 (Fig. 3C). In contrast, the activation of the C region with E-box1–6 mutant by Tbx6 and Msgn1 was completely abolished (Fig. S8), which was similar to the result by Msgn1/NICD/Tbx6 (Fig. 3H), indicating that E-boxes are essential for the synergistic Hes7 expression. Taking these in vitro and in vivo results together (Figs. 2 and 3), we deduce that E-boxes and T-boxes, including the RBPj-binding site, are critical and auxiliary for Hes7 expression, respectively.
Tbx18, Ripply2, and Hes7 repress the activation of C region in vitro
Transgenic mice carrying the mutated T-box1 and -2/C region or Deletion1 expressed lacZ mRNA not only in the PSM but also expressed a dispersed and straggling reporter mRNA in the somites (Fig. 2B, panel b, and Fig. S2), suggesting that the C region, especially T-boxes, has a role in preventing ectopic Hes7 expression. To understand the molecular mechanisms of how Hes7 expression in the anterior-most PSM or in somites is suppressed, we next investigated the repression mechanisms for Hes7. Tbx18 is one of the transcriptional repressors among the T-box family of transcriptional factor genes that is expressed in mouse PSM (13), and as reported previously (16), Tbx18 is expressed in the rostral part of somites and the anterior-most PSM where Hes7 propagation has vanished (Fig. S4, C and C′), demonstrating that Tbx18 and Hes7 expressions are mutually exclusive. We investigated whether Tbx18 binds to T-boxes in the C region by oligo-DNA pulldown assay and EMSA. The pulldown assays demonstrated that Tbx18 bound to WT T-box1 but not to T-box2 or -3 (Fig. 4A and Fig. S9A). Tbx18 binding to T-box1 was diminished by the mutation of T-box1 (Fig. 4A). EMSA also showed that Tbx18 bound to T-box1 but not to T-box2 or -3 (Fig. S9B). Again, we demonstrated that Tbx18 binding to T-box1 was weakened by an excess amount of the nonlabeled T-box1 but not by that of the mutated one (Fig. S9B). Furthermore, luciferase assays revealed that Tbx18 dose-dependently repressed reporter activity induced by Tbx6 and NICD (Fig. 4B), which are expressed at the anterior-most PSM (Fig. 5 and Fig. S4).
Figure 4.
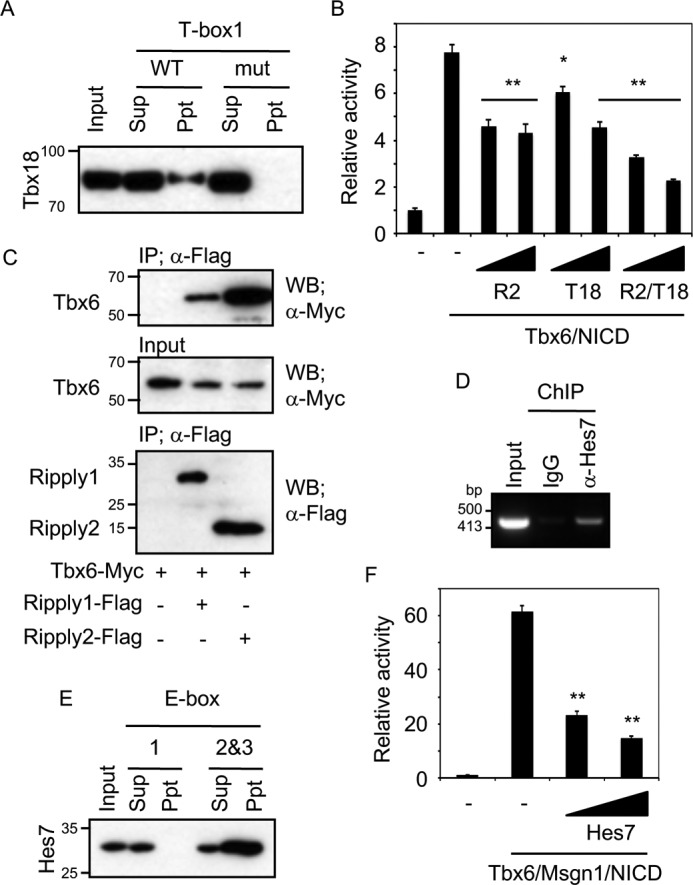
Transcription repressors regulate Hes7 expression. A, oligo-DNA pulldown assays against WT or mutant T-box with Tbx18-FLAG were performed. Tbx18 was revealed with anti-FLAG. B, luciferase assay for Hes7 C region reporter with indicated repressors in NIH3T3 cells. 100 or 200 ng of expression vectors of Ripply2 (R2) and/or Tbx18 (T18) were co-transfected with Tbx6 and NICD. Data represent the means ± S.E. of three independent samples. C, co-immunoprecipitation assays were performed with Ripply1-FLAG or Ripply2-FLAG and Tbx6-Myc in HEK293T cells. Tbx6 and Ripply1/2 were revealed with anti-Myc and anti-FLAG, respectively. D, ChIP assays with anti-Hes7 antibody or normal rabbit IgG (negative control) were performed using mouse PSM. Resultant genomic fragment was amplified using Hes7 C region-specific primer. E, oligo-DNA pulldown assays against E-box1 or E-box2 and -3 with FLAG-Hes7 were performed. Hes7 was revealed with anti-FLAG. F, luciferase assay for Hes7 C region reporter with Hes7 in NIH3T3 cells. 100 or 200 ng of expression vectors of Hes7 were co-transfected with Tbx6, Msgn1, and NICD. *, p < 0.05; **, p < 0.01 compared with Tbx6/NICD without repressors (B) or Tbx6/Msgn1/NICD without Hes7 (F) (Student's two-tailed t test). Sup, supernatant; Ppt, precipitate; WB, Western blotting; IP, immunoprecipitation.
Figure 5.
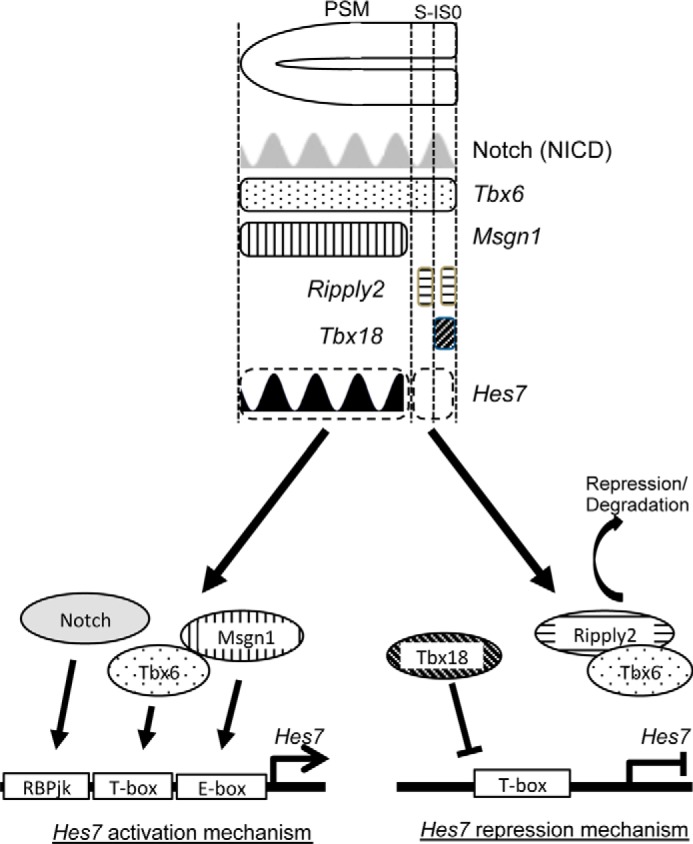
Scheme for restricted Hes7 expression in the PSM. See Fig. S2 for expression domains of Tbx6, Msgn1, Ripply2, and Tbx18. In the PSM (except anterior-most region), constant expressions of Tbx6 and Msgn1 and rhythmic expression of Notch activate Hes7 expression, and at the anterior-most region (S-1 and S0), loss of Msng1 expression and appearance of Ripply2 and Tbx18 expressions suppressed Hes7 expression.
Ripply2 mRNA was strongly expressed, as reported previously (17), in the anterior PSM (prospective somites S0 and S-I) when Hes7 was in phase I (Fig. S4D), whereas in phase III of Hes7, Ripply2 showed two weak stripes at the region where Hes7 is lost (Fig. S4D′). Because Ripply co-repressors have been known to act on the repressor by interacting with Xenopus Tbx6 or zebrafish Tbx24, which are structurally related to mouse Tbx6 (18, 19), we performed co-immunoprecipitation assays in culture cells, HEK293T, to examine whether mouse Ripply co-repressors interact with mouse Tbx6. We revealed that Ripply1/2 form a complex with Tbx6 in vitro (Fig. 4C), especially Ripply2, which had a high affinity to Tbx6. Luciferase assay further uncovered that luciferase activities of the C region induced by Tbx6/NICD were reduced by Ripply2 (Fig. 4B). Moreover, co-expression of Tbx18 and Ripply2 repressed luciferase activity more effectively compared to when either Tbx18 or Ripply2 was expressed, suggesting that Tbx18 and Ripply2 repress the C region independently. These results raise the possibility that Tbx18 and Ripply2 are repressive regulators for Hes7 termination in the anterior-most PSM and somites by binding to T-box1 or by forming a complex with Tbx6 to reduce Tbx6 transcriptional activity, respectively.
Because Hes7 could bind to the E-box to repress transcriptional activity (5), we investigated whether E-boxes in the C region are functional for Hes7. We first demonstrated that Hes7 binding to the C region in vivo was detected by ChIP assay using mouse PSM (Fig. 4D). Furthermore, the oligo-DNA pulldown assay showed that Hes7 could bind to E-box2 and -3 but not to E-box1 (Fig. 4E). Moreover, luciferase assays demonstrated that Hes7 repressed synergistic Msgn1/Tbx6/NICD activation dose-dependently (Fig. 4F). These results suggest that the C region might also be associated with the oscillatory expression of Hes7.
DNA methylation is not correlated with the mechanism for the PSM-specific expression of Hes7
Finally, we sought to investigate whether epigenetic regulations, especially DNA methylation, of the C region take part in the repression of Hes7 ectopic expression. To that end, we examined the DNA methylation status of the C region by bisulfite sequencing. However, no CpG sites within the C region were methylated in all the tissues tested, including PSM, head, and caudal trunk from E10.5 embryo (Fig. S10). Experimental procedures for this bisulfite sequence are provided in the supporting Experimental procedures. This result suggests that the regulation by DNA methylation is not correlated with the mechanism for the PSM-specific expression of Hes7.
Discussion
Spatiotemporal Hes7 expression pattern is very unique, whereby it is restricted to the PSM and propagates (oscillates) from the posterior-end to the anterior-end of the PSM with 2-h periodicity in mice. In this study, we identified a narrow region within −1.5 to −1.1 kb from TSS in mouse Hes7, referred to as the C region in this study, that directs the specific expression of reporter gene in the PSM during mouse embryogenesis. At the molecular level, we found E-boxes, T-boxes, and RBPj-binding sites in the C region and further demonstrated that these elements are crucial for the restricted expression of the reporter gene in the PSM. Furthermore, this study raises the possibility that the Notch signaling pathway, the transcriptional activators, Msgn1 and Tbx6, and the transcriptional repressors, Tbx18, Ripply1/2, and Hes7, participate as novel factors for the C region's activation/repression.
Our study using transgenic founder assays revealed that this C region, which is highly conserved among several species, is sufficient to direct expression of the reporter gene in the PSM specifically during mouse embryogenesis (Figs 1 and 2). Our current study further dissected the molecular mechanisms for PSM-specific Hes7 expression. In situ hybridization for lacZ mRNA (Fig. 2B) showed that only two of 16 embryos with mutated E-box demonstrated positive, but obscure, signals. In addition, luciferase assays (Fig. 3H) demonstrated that the mutated E-box was completely unresponsive to the activation by Tbx6/Msng1/NICD. These findings suggest that the mutated E-box has no potential to activate gene expression in the PSM. In contrast to the mutated E-box, the mutated T-box had produced lacZ signals in six of eight embryos (Fig. 2B) and had increased the luciferase activity by Tbx6/Msng1/NICD (Fig. 3H), although the activity was much lower than that of WT C region. We therefore conclude that E-boxes and T-boxes, including the RBPj-binding site, are critical and auxiliary, respectively. Moreover, X-gal staining and in situ hybridization for Deletion1 or -2 (Fig. 1, I–L, and Fig. S2) suggest that E-box1–3 and T-box1/2 are functional, because these results showed no/weak signals at the posterior-end PSM. The reason no/weak signals were restricted at the posterior-end PSM could be that the gene expression driven by the C region without any of these elements is very weak and under detectable levels by X-gal staining and in situ hybridization at the posterior-end PSM; however, after a while, reporter mRNA and protein slowly accumulated to reach detectable levels.
It has been reported that the somite formation does not take place when Msgn1 or Tbx6 is knocked out in mice (12, 20, 21). In addition to Msgn1 and Tbx6, somite formation does not occur properly when Hes7 is deficient (6). Moreover, Msgn1 or components of the Notch pathway knockout mice express less Hes7 in the PSM (10, 22, 23). These reports strongly support our findings that Msgn1, Tbx6, and Notch pathway are upstream of Hes7. Furthermore, we raise the possibility that the combination of Msgn1, Notch signaling, and Tbx6 induces Hes7 expression via the C region in the mouse PSM. Interestingly, synergistic transcriptional activation by Tbx6 with other transcriptional factors during somitogenesis has been reported; for example, Tbx6 cooperates with Wnt signaling for Dll1 and Msgn1 induction (24, 25) and with Notch signaling for Mesp2 induction in mouse (26). In Xenopus, Tbx6 activates bowline, a Xenopus Ripply homologue, in synergy with bHLH transcription factors, Thylacine1 and E47 (27). Moreover, we also raise the possibility that T-box1–3 elements are essential to form the optimal three-dimensional structure of the C region with Tbx6/Msng1/NICD for the synergistic activation. Although further investigations will be required to address whether and how Tbx6/Msng1/NICD form a complex with the C region for the complete Hes7 activation, our finding nonetheless had shed light on a new role of Tbx6 in somitogenesis.
Our findings in this study also raised the possibility that T-box elements function to repress Hes7 expression in the anterior-most PSM (S-I and S0) and somites by binding with Tbx18. This dual function of T-boxes could be one of the mechanisms for the termination and inhibition of Hes7 expression at the anterior-most PSM and somites, respectively. Another key factor for Hes7 repression is Ripply1/2. Ripply1/2 have already been shown to repress Tbx6 expression by two different ways. One is the conversion of Xenopus and zebrafish Tbx6 from transcriptional activator to suppressor by binding with Groucho/TLE co-repressors (18, 19). The other is the elimination of Tbx6 protein by unknown mechanisms (28). In this study, we demonstrated that mouse Ripply1/2 could bind with mouse Tbx6 and repress the expression of Hes7 in vitro (Fig. 4, B and C). Our data suggest that, at the anterior-most PSM in mouse, Ripply suppresses Hes7 expression through recruitment to T-box elements with Tbx6/Groucho/TLE co-repressors and/or by eliminating Tbx6 protein (Fig. 5). However, because Hes7 expression patterns are normal even in Ripply1 and -2 double knockout embryos (28), the Hes7 termination mechanism by Ripply1/2 might be ancillary and that by Tbx18 is primary. It is noteworthy that an interesting paper has reported that Mesp2, which expresses S-I, suppresses Notch signaling via destabilizing Mastermind-like 1, a coactivator of Notch signaling (29). The suppression of Notch signaling by Mesp2 at the anterior-most PSM shown in that report could be another potential mechanism for the termination of Hes7 expression. Taking their findings and our current analyses together, we establish that to spatiotemporally express and terminate Hes7 expression at the PSM, a web of transcriptional mechanisms is required during somitogenesis and that the Hes7 suppressor element would be important for a proper maintenance of the functional Hes7 domain as well as activator elements.
González et al. (30) reported that Hes7 expression is controlled by Tbx6 and Wnt signaling. They have identified an essential 400-bp region (−1.4 to −1.0 kbp from TSS) for proper Hes7 expression, which is almost identical to our Hes7 essential region (−1.5 to −1.1 kbp from TSS). Furthermore, they have also found that the activity of the Hes7 promoter in mouse PSM requires Tbx6-binding sites within this 400-bp region. These finding support our current study, although their distal T-box corresponds to T-box2 in our study, and the proximal one, which we missed as T-box, overlaps with E-box3 identified in our study. Intriguingly, they have mentioned that downstream molecules of the Wnt pathway activate the Hes7 promoter cooperatively with Tbx6 in cell culture and are necessary for its proper expression in the mouse PSM. More interestingly, they have shown that the expression of Msgn1, one of the Wnt target genes and the activator for the Hes7 essential region revealed in our study, is activated in embryos treated with LiCl, the inhibitor of GSK3β, in which Wnt signaling is activated. Taken together, their study strongly supports our results that Msgn1 is associated with the activation of Hes7 expression with Tbx6.
Our current findings demonstrated that the C region, including the specific elements of the C region, is sufficient for the restricted reporter mRNA expression in the PSM, and the C region is activated by a synergistic effect of Msgn1, Tbx6, and Notch signaling and repressed by Tbx18, Ripply2, and Hes7 in cell culture. However, we have not addressed whether the C region and the regulatory factors are indispensable for the endogenous Hes7 expression in the PSM. In addition, we cannot rule out the possibility that other existing shadow or cryptic enhancers such as long range (1 Mb or more) enhancer (31) and enhancer residing in introns (32) or in 3′ downstream (33) may also be essential to Hes7 expression. To uncover whether the C region, and which elements in the C region, are indispensable for the PSM-specific endogenous Hes7 expression, further in vivo transcriptional analyses of the endogenous Hes7 promoter deleting the whole C region or knocking-in the C region with mutated elements will be required. As mentioned above, previous reports suggest that Msgn1 and Notch pathway are upstream of Hes7; however, detailed analyses of endogenous Hes7 expression in gene-modified mice in which the expression levels of Msgn1, Tbx6, components of Notch signaling, Tbx18, or Ripply2 are altered will be required.
Finally, although we demonstrated that (i) C region has multiple E-boxes, (ii) the bHLH-type activator Msgn1 and the repressor Hes7 occupy E-box1 and E-box2 and -3, respectively (Figs. 3D and 4E), and (iii) Hes7 can repress Msgn1/Tbx6/NICD-dependent activation (Fig. 4D). Although we were unable to demonstrate the oscillation by the 5.3-kb fragment (Fig. 1R), we were unable to evince that the C region is enough for Hes7 oscillation, in that we could not detect the oscillatory lacZ mRNA expression pattern driven by the C region (Fig. 1T). As it has been known that multiple enhancers act on gene expression to ensure robustness (34), multiple additional elements, such as E/N-boxes near the transcription start site as reported previously (35), might be needed for Hes7 to achieve and maintain oscillatory expression. Further analysis therefore will be required to find a minimum set of oscillatory elements for establishment of Hes7 oscillatory expression.
Experimental procedures
Animals
CD1 mice used in this study were purchased from SLC (Japan). Our experiments with mice have been approved by The Animal Care Committee of Nara Institute of Science and Technology (NAIST). These experiments were conducted in accordance with guidelines that were established by the Science Council of Japan.
Reporter constructs and transgenic mice
Hes7 upstream region was cloned by conventional molecular biological methods. Upstream fragment was PCR-amplified and inserted into pBluescriptII (Stratagene) with lacZ gene and SV40 poly(A) signal. Human β-globin minimal promoter, being synthesized as a double-stranded oligonucleotide, was inserted into a reporter vector for the enhancer assay. Sequences for human β-globin minimal promoter were as follows: Fw, TCC CGG GCT GGG CAT AAA AGT CAG GGC AGA GCC ATC TAT TGC TTA CAT TTG CTT C, and Rv, GAA GCA AAT GTA AGC AAT AGA TGG CTC TGC CCT GAC TTT TAT GCC CAG CCC GGG A. To make transgenic mice, the constructs were linearized and injected into fertilized eggs from CD1 mice by the animal facility of NAIST.
X-gal staining and in situ hybridization
Transgenic mice were dissected and analyzed at embryonic day 10.5 (E10.5). For X-gal staining, embryos were fixed in 0.5% glutaraldehyde with 2 mm MgCl2 at 4 °C for 30 min. Then, these embryos were soaked in color solution (1 mg/ml 5-bromo-4-chloro-3-indolyl-β-d-galactoside, 5 mm potassium ferricyanide, 5 mm potassium ferrocyanide, 2 mm MgCl2, Nonidet P-40, 0.01% sodium deoxycholate) at 37 °C overnight. Whole-mount in situ hybridization of mouse embryos was performed as described previously (5).
Luciferase assay
Hes7 C region followed by human β-globin minimal promoter were inserted into pGL3-Basic vector (Promega). Transcription factors were cloned by PCR with cDNA from mouse PSM. 5′-UTR and the coding region of each gene were inserted into pcDNA3 (Invitrogen), including FLAG, HA, or Myc tag at the 3′-end. 3 × 104 NIH3T3 cells were plated in each well of a 24-well plate and were cultured in 10% fetal bovine serum/Dulbecco's modified Eagle's medium at 5% CO2. After 24 h, cells were co-transfected with 300 ng of Hes7 reporter and 200 ng of expression vector of transcription factors using TransIT LT1 (Mirus). Transfected cells were lysed after 24 h of culture, and reporter activity was measured using a Dual-Luciferase assay system (Promega) and analyzed by ARBO (PerkinElmer Life Sciences). Firefly luciferase activity of the reporter was normalized by the activity of Renilla luciferase under control of SV40 promoter.
Mutagenesis
Site-directed mutagenesis was performed as described previously (36). Sequences were substituted as follows: T-box1, AGG TGT GGG AA to AGG TtT taa Ac; T-box2, CCA CAC CC to CgA tAt CC; T-box3, CCA CAC AT to CgA tAt AT; E-box1, CAT ATG to gtT Aac; E-box2, CAC GTG to gtC Gac; E-box3, CAG GTG to gtG Gac; E-box4, CAT CTG to gtT Cac; E-box5, CAT TTG to gtT Tac; and E-box6, CAA ATG to gtA Aac. Small letters indicate mutated nucleotides.
In silico promoter analysis
A homology between human Hes7 upstream and mouse Hes7 upstream was analyzed by VISTA (37, 38). To predict T-boxes, E-boxes, or RBPj-binding sites in the C region, YMACACYY or complementary, CANNTG, or YRTGDGAD or complementary were referred to as T-box, E-box, or RBPj-binding site, respectively (5, 21, 39). Homology search among C regions of H. sapiens, P. abelii, B. taurus, and M. musculus was performed by ClustalW version 2.1.
Oligo-DNA pulldown assay
COS7 cells were seeded in 10-cm dish (4 × 105 cells/dish), transfected with 15 μg of expression vectors, and cultured for 48 h. Cells were lysed in binding buffer (10 mm Tris, pH 8.0, 150 mm NaCl, 1 mm MgCl2, 0.5% Nonidet P-40, 5% glycerol) with protease inhibitor mixture (Nacalai Tesque, Japan). After removing debris by centrifugation, 30 μl of 50% slurry streptavidin-Sepharose beads (GE Healthcare) and 200 pmol of double-stranded oligonucleotide conjugated with biotin at each 5′-end were added to the cell lysates. The mixture was incubated at 4 °C for 30 min with rotation. Sepharose beads were washed with the binding buffer three times. Then, the resultant pulldown samples were separated by SDS-PAGE on precast 5–20% polyacrylamide gels (Nacalai Tesque, Japan). Proteins were then transferred to PVDF membranes using a wet electroblotting apparatus. The membranes were blocked using 5% skim milk in TBS with 0.1% Tween (TBS-T) for 1 h and incubated with anti-Myc (monoclonal, PL14, MBL, Japan) overnight, followed by incubation with anti-mouse IgG conjugated with horseradish peroxidase (GE Healthcare). Signals were visualized by the enhanced chemiluminescence detection system according to the manufacturer's instruction (Nacalai Tesque, Japan). Oligo-DNA sequences are as follows: E-box1 FW, 5′-AAA GTC ATT CCA TAT GGC CAG GGG CG-3′, and E-box1 RV, 5′-CGC CCC TGG CCA TAT GGA ATG ACT TT-3′; E-box1 mut FW, 5′-AAA GTC ATT Cgc TAg GGC CAG GGG CG-3′, and E-box1 mut RV, 5′-CGC CCC TGG CCc TAg cGA ATG ACT TT-3′; E-box2 and -3 FW, 5′-CCC CGA GCC TCA CGT GCA GGT GAG AAA AAC TC-3′, and E-box2 and -3 RV, 5′-GAG TTT TTC TCA CCT GCA CGT GAG GCT CGG GG-3′; T-box1 FW, 5′-ACT TCT AGG TGT GGG AAA AGG TTG TAG-3′, and T-box1 RV, 5′-CTA CAA CCT TTT CCC ACA CCT AGA AGT-3′; T-box1 mut FW, 5′-ACT TCT AGG TtT taa AcA AGG TTG TAG-3′, and T-box1 mut RV, 5′-CTA CAA CCT TgT tta AaA CCT AGA AGT-3′. Small letters indicate mutated nucleotides.
ChIP assay
Mouse embryos (more than 120 embryos) were dissected at E10.5, and PSMs were collected in ice-cold PBS with protease inhibitor mixture. PSMs were dispersed by 0.05 w/v % trypsin/EDTA treatment for 3 min. Cells were fixed with 1% formaldehyde in PBS for 10 min at room temperature, and the fixation was stopped by adding 0.1 amount of 1.5 m glycine. After a brief centrifugation, cells were lysed with 200 ml of SDS lysis buffer (50 mm Tris-HCl, pH 8.0, 10 mm EDTA, 1% SDS). Cell lysates were sonicated genome using a bioruptor (Cosmo Bio) with power set at high and a 30-s on and 60-s off interval. After centrifugation to remove the insoluble fraction, supernatants were diluted with nine times the amount of dilution buffer (50 mm Tris-HCl, pH 8.0, 167 mm NaCl, 1.1% Triton X-100, 0.11% sodium deoxycholate), including protease inhibitor mixture. For pre-cleaning, samples were added 20 μl of 50% slurry protein A-Sepharose (Nacalai Tesque, Japan) and incubated more than 4 h at 4 °C, followed by centrifugation to remove protein A-Sepharose. Tbx6 antibody (29) or Hes7 antiserum (35) were added to the pre-cleaned sample, respectively, and were incubated overnight at 4 °C. The sample was added with 20 μl of 50% slurry protein A-Sepharose and incubated for more than 3 h at 4 °C. Resultant immunoprecipitated samples were washed with a series of buffers (RIPA buffer 1: 50 mm Tris-HCl, pH 8.0, 150 mm NaCl, 1 mm EDTA, 1% Triton X-100, 0.1% SDS, 0.1% sodium deoxycholate; RIPA buffer 2: 50 mm Tris-HCl, pH 8.0, 500 mm NaCl, 1 mm EDTA, 1% Triton X-100, 0.1% SDS, 0.1% sodium deoxycholate; LiCl buffer: 10 mm Tris-HCl, pH 8.0, 0.25 m LiCl, 1 mm EDTA, 0.5% Nonidet P-40, 0.5% sodium deoxycholate; TE buffer: 10 mm Tris-HCl, pH 8.0, 1 mm EDTA, two times). For decross-linking and elution, elution buffer (10 mm Tris-HCl, pH 8.0, 300 mm NaCl, 5 mm EDTA, 0.5% SDS) was added to samples and incubated at 65 °C for 4 h. After decross-linking, DNA were purified and subjected to PCR amplification. The sequences of the forward and reverse primers are as follows: Hes7 C ChIP FW: 5′-ATG TGA ACT TCT CAG AGG CAG ATC CAA TCC-3′, and Hes7 C ChIP RV: 5′-CCT TCC CAG AGG CCC TCC ACA TCC TG-3′.
Immunoprecipitation
HEK293T cells were seeded at 4 × 105 cells in a 10-cm dish, cultured for 24 h, and transfected with 5 μg of each expression vectors or empty pcDNA3 vector. After 48 h of culture, cells were lysed in ice-cold TNE buffer (20 mm Tris-HCl, pH 8.0, 1 mm EDTA, pH 8.0, 1% Nonidet P-40, 150 mm NaCl) with protease inhibitor mixture. After removing debris by centrifugation, supernatants were incubated with 15 μl of 50% slurry anti-FLAG M2-agarose (Sigma) overnight at 4 °C with rotation. The next day, agarose beads were washed three times with ice-cold TNE buffer, and resultant immunoprecipitated samples were separated by SDS-PAGE on precast 5–20% polyacrylamide gels (Nacalai Tesque, Japan). Proteins were then transferred to PVDF membranes using a wet electroblotting apparatus. The membranes were blocked using 5% skim milk in TBS-T for 1 h and incubated with anti-Myc or anti-FLAG (monoclonal, FLA-1, MBL, Japan) overnight, followed by incubation with anti-mouse IgG conjugated with horseradish peroxidase. Signals were visualized by the enhanced chemiluminescence detection system according to the manufacturer's instruction.
Author contributions
S. H., Y. N., T. M., and Y. B. data curation; S. H. formal analysis; S. H. and Y. N. investigation; S. H. and Y. N. writing-original draft; Y. N., T. M., and Y. B. conceptualization; Y. N. and Y. B. supervision; Y. N. and Y. B. project administration; Y. N. and Y. B. writing-review and editing; K. K. resources; Y. N. and Y. B. funding acquisition.
Supplementary Material
Acknowledgments
We thank Drs. Y. Yasuhiko and M. Saito for providing anti-Tbx6 antibody and for the generation of transgenic mice, respectively. We also thank R. Ahmed and Dr. F. D. Khaidizar for providing critical comments on the manuscript.
This work was supported by Japan Society for the Promotion of Science KAKENHI Grant JP24659086 (to Y. N.) and by Ministry of Education, Culture, Sports, Science and Technology (MEXT) KAKENHI Grant JP17H05768 (to Y. B.) and in part by The Uehara Memorial Foundation and Senri Life Science Foundation. The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1–S10 and supporting Experimental procedures.
- PSM
- presomitic mesoderm
- TSS
- transcription start site
- FW
- forward
- RV
- reverse
- bHLH
- basic helix-loop-helix
- PVDF
- polyvinylidene difluoride.
References
- 1. Saijoh Y., Adachi H., Mochida K., Ohishi S., Hirao A., and Hamada H. (1999) Distinct transcriptional regulatory mechanisms underlie left-right asymmetric expression of lefty-1 and lefty-2. Genes Dev. 13, 259–269 10.1101/gad.13.3.259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Meno C., Shimono A., Saijoh Y., Yashiro K., Mochida K., Ohishi S., Noji S., Kondoh H., and Hamada H. (1998) Lefty-1 is required for left-right determination as a regulator of lefty-2 and nodal. Cell 94, 287–297 10.1016/S0092-8674(00)81472-5 [DOI] [PubMed] [Google Scholar]
- 3. Meno C., Gritsman K., Ohishi S., Ohfuji Y., Heckscher E., Mochida K., Shimono A., Kondoh H., Talbot W. S., Robertson E. J., Schier A. F., and Hamada H. (1999) Mouse lefty2 and zebrafish antivin are feedback inhibitors of nodal signaling during vertebrate gastrulation. Mol. Cell 4, 287–298 10.1016/S1097-2765(00)80331-7 [DOI] [PubMed] [Google Scholar]
- 4. Meno C., Takeuchi J., Sakuma R., Koshiba-Takeuchi K., Ohishi S., Saijoh Y., Miyazaki J., ten Dijke P., Ogura T., and Hamada H. (2001) Diffusion of nodal signaling activity in the absence of the feedback inhibitor lefty2. Dev. Cell 1, 127–138 10.1016/S1534-5807(01)00006-5 [DOI] [PubMed] [Google Scholar]
- 5. Bessho Y., Miyoshi G., Sakata R., and Kageyama R. (2001) Hes7: a bHLH-type repressor gene regulated by Notch and expressed in the presomitic mesoderm. Genes Cells 6, 175–185 10.1046/j.1365-2443.2001.00409.x [DOI] [PubMed] [Google Scholar]
- 6. Bessho Y., Sakata R., Komatsu S., Shiota K., Yamada S., and Kageyama R. (2001) Dynamic expression and essential functions of Hes7 in somite segmentation. Genes Dev. 15, 2642–2647 10.1101/gad.930601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kageyama R., Ohtsuka T., and Kobayashi T. (2007) The Hes gene family: repressors and oscillators that orchestrate embryogenesis. Development 134, 1243–1251 10.1242/dev.000786 [DOI] [PubMed] [Google Scholar]
- 8. Dequéant M. L., and Pourquié O. (2008) Segmental patterning of the vertebrate embryonic axis. Nat. Rev. Genet. 9, 370–382 10.1038/nrg2320 [DOI] [PubMed] [Google Scholar]
- 9. Pourquié O. (2011) Vertebrate segmentation: from cyclic gene networks to scoliosis. Cell 145, 650–663 10.1016/j.cell.2011.05.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Niwa Y., Masamizu Y., Liu T., Nakayama R., Deng C. X., and Kageyama R. (2007) The initiation and propagation of Hes7 oscillation are cooperatively regulated by Fgf and notch signaling in the somite segmentation clock. Dev. Cell 13, 298–304 10.1016/j.devcel.2007.07.013 [DOI] [PubMed] [Google Scholar]
- 11. Hayashi S., Shimoda T., Nakajima M., Tsukada Y., Sakumura Y., Dale J. K., Maroto M., Kohno K., Matsui T., and Bessho Y. (2009) Sprouty4, an FGF inhibitor, displays cyclic gene expression under the control of the notch segmentation clock in the mouse PSM. PLoS One 4, e5603 10.1371/journal.pone.0005603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Yoon J. K., Moon R. T., and Wold B. (2000) The bHLH class protein pMesogenin1 can specify paraxial mesoderm phenotypes. Dev. Biol. 222, 376–391 10.1006/dbio.2000.9717 [DOI] [PubMed] [Google Scholar]
- 13. Wardle F. C., and Papaioannou V. E. (2008) Teasing out T-box targets in early mesoderm. Curr. Opin. Genet. Dev. 18, 418–425 10.1016/j.gde.2008.07.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chapman D. L., Agulnik I., Hancock S., Silver L. M., and Papaioannou V. E. (1996) Tbx6, a mouse T-box gene implicated in paraxial mesoderm formation at gastrulation. Dev. Biol. 180, 534–542 10.1006/dbio.1996.0326 [DOI] [PubMed] [Google Scholar]
- 15. Barrantes I. B., Elia A. J., Wünsch K., Hrabe de Angelis M. H., Mak T. W., Rossant J., Conlon R. A., Gossler A., and de la Pompa J. L. (1999) Interaction between Notch signalling and Lunatic fringe during somite boundary formation in the mouse. Curr. Biol. 9, 470–480 10.1016/S0960-9822(99)80212-7 [DOI] [PubMed] [Google Scholar]
- 16. Kraus F., Haenig B., and Kispert A. (2001) Cloning and expression analysis of the mouse T-box gene Tbx18. Mech. Dev. 100, 83–86 10.1016/S0925-4773(00)00494-9 [DOI] [PubMed] [Google Scholar]
- 17. Biris K. K., Dunty W. C. Jr, and Yamaguchi T. P. (2007) Mouse Ripply2 is downstream of Wnt3a and is dynamically expressed during somitogenesis. Dev. Dyn. 236, 3167–3172 10.1002/dvdy.21342 [DOI] [PubMed] [Google Scholar]
- 18. Kawamura A., Koshida S., and Takada S. (2008) Activator-to-repressor conversion of T-box transcription factors by the Ripply family of Groucho/TLE-associated mediators. Mol. Cell. Biol. 28, 3236–3244 10.1128/MCB.01754-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kondow A., Hitachi K., Okabayashi K., Hayashi N., and Asashima M. (2007) Bowline mediates association of the transcriptional co-repressor XGrg-4 with Tbx6 during somitogenesis in Xenopus. Biochem. Biophys. Res. Commun. 359, 959–964 10.1016/j.bbrc.2007.05.211 [DOI] [PubMed] [Google Scholar]
- 20. Chapman D. L., and Papaioannou V. E. (1998) Three neural tubes in mouse embryos with mutations in the T-box gene Tbx6. Nature 391, 695–697 10.1038/35624 [DOI] [PubMed] [Google Scholar]
- 21. Nikaido M., Kawakami A., Sawada A., Furutani-Seiki M., Takeda H., and Araki K. (2002) Tbx24, encoding a T-box protein, is mutated in the zebrafish somite-segmentation mutant fused somites. Nat. Genet. 31, 195–199 10.1038/ng899 [DOI] [PubMed] [Google Scholar]
- 22. Ferjentsik Z., Hayashi S., Dale J. K., Bessho Y., Herreman A., De Strooper B., del Monte G., de la Pompa J. L., and Maroto M. (2009) Notch is a critical component of the mouse somitogenesis oscillator and is essential for the formation of the somites. PLoS Genet. 5, e1000662 10.1371/journal.pgen.1000662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chalamalasetty R. B., Dunty W. C. Jr, Biris K. K., Ajima R., Iacovino M., Beisaw A., Feigenbaum L., Chapman D. L., Yoon J. K., Kyba M., and Yamaguchi T. P. (2011) The Wnt3a/β-catenin target gene Mesogenin1 controls the segmentation clock by activating a Notch signalling program. Nat. Commun. 2, 390 10.1038/ncomms1381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hofmann M., Schuster-Gossler K., Watabe-Rudolph M., Aulehla A., Herrmann B. G., and Gossler A. (2004) WNT signaling, in synergy with T/TBX6, controls Notch signaling by regulating Dll1 expression in the presomitic mesoderm of mouse embryos. Genes Dev. 18, 2712–2717 10.1101/gad.1248604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wittler L., Shin E. H., Grote P., Kispert A., Beckers A., Gossler A., Werber M., and Herrmann B. G. (2007) Expression of Msgn1 in the presomitic mesoderm is controlled by synergism of WNT signalling and Tbx6. EMBO Rep. 8, 784–789 10.1038/sj.embor.7401030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Yasuhiko Y., Kitajima S., Takahashi Y., Oginuma M., Kagiwada H., Kanno J., and Saga Y. (2008) Functional importance of evolutionally conserved Tbx6 binding sites in the presomitic mesoderm-specific enhancer of Mesp2. Development 135, 3511–3519 10.1242/dev.027144 [DOI] [PubMed] [Google Scholar]
- 27. Hitachi K., Kondow A., Danno H., Inui M., Uchiyama H., and Asashima M. (2008) Tbx6, Thylacine1, and E47 synergistically activate bowline expression in Xenopus somitogenesis. Dev. Biol. 313, 816–828 10.1016/j.ydbio.2007.10.015 [DOI] [PubMed] [Google Scholar]
- 28. Takahashi J., Ohbayashi A., Oginuma M., Saito D., Mochizuki A., Saga Y., and Takada S. (2010) Analysis of Ripply1/2-deficient mouse embryos reveals a mechanism underlying the rostro-caudal patterning within a somite. Dev. Biol. 342, 134–145 10.1016/j.ydbio.2010.03.015 [DOI] [PubMed] [Google Scholar]
- 29. Sasaki N., Kiso M., Kitagawa M., and Saga Y. (2011) The repression of Notch signaling occurs via the destabilization of mastermind-like 1 by Mesp2 and is essential for somitogenesis. Development 138, 55–64 10.1242/dev.055533 [DOI] [PubMed] [Google Scholar]
- 30. González A., Manosalva I., Liu T., and Kageyama R. (2013) Control of Hes7 expression by Tbx6, the Wnt pathway and the chemical Gsk3 inhibitor LiCl in the mouse segmentation clock. PLoS One 8, e53323 10.1371/journal.pone.0053323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kleinjan D. A., and van Heyningen V. (2005) Long-range control of gene expression: emerging mechanisms and disruption in disease. Am. J. Hum. Genet. 76, 8–32 10.1086/426833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Garnett A. T., Han T. M., Gilchrist M. J., Smith J. C., Eisen M. B., Wardle F. C., and Amacher S. L. (2009) Identification of direct T-box target genes in the developing zebrafish mesoderm. Development 136, 749–760 10.1242/dev.024703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lieven O., Knobloch J., and Rüther U. (2010) The regulation of Dkk1 expression during embryonic development. Dev. Biol. 340, 256–268 10.1016/j.ydbio.2010.01.037 [DOI] [PubMed] [Google Scholar]
- 34. Frankel N., Davis G. K., Vargas D., Wang S., Payre F., and Stern D. L. (2010) Phenotypic robustness conferred by apparently redundant transcriptional enhancers. Nature 466, 490–493 10.1038/nature09158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bessho Y., Hirata H., Masamizu Y., and Kageyama R. (2003) Periodic repression by the bHLH factor Hes7 is an essential mechanism for the somite segmentation clock. Genes Dev. 17, 1451–1456 10.1101/gad.1092303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sawano A., and Miyawaki A. (2000) Directed evolution of green fluorescent protein by a new versatile PCR strategy for site-directed and semi-random mutagenesis. Nucleic Acids Res. 28, E78 10.1093/nar/28.16.e78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Mayor C., Brudno M., Schwartz J. R., Poliakov A., Rubin E. M., Frazer K. A., Pachter L. S., and Dubchak I. (2000) VISTA: visualizing global DNA sequence alignments of arbitrary length. Bioinformatics 16, 1046–1047 10.1093/bioinformatics/16.11.1046 [DOI] [PubMed] [Google Scholar]
- 38. Frazer K. A., Pachter L., Poliakov A., Rubin E. M., and Dubchak I. (2004) VISTA: computational tools for comparative genomics. Nucleic Acids Res. 32, W273–W279 10.1093/nar/gkh458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Yasuhiko Y., Haraguchi S., Kitajima S., Takahashi Y., Kanno J., and Saga Y. (2006) Tbx6-mediated Notch signaling controls somite-specific Mesp2 expression. Proc. Natl. Acad. Sci. U.S.A. 103, 3651–3656 10.1073/pnas.0508238103 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.