Abstract
Synapse maturation is a neural activity–dependent process during brain development, in which active synapses preferentially undergo maturation to establish efficient neural circuits in the brain. Defects in this process are implicated in various neuropsychiatric disorders. We have previously reported that a postsynaptic transmembrane protein, signal regulatory protein-α (SIRPα), plays an important role in activity-dependently directing synapse maturation. In the presence of synaptic activity, the ectodomain of SIRPα is cleaved and released and then acts as a retrograde signal to induce presynaptic maturation. However, how SIRPα detects synaptic activity to promote its ectodomain cleavage and synapse maturation is unknown. Here, we show that activity-dependent tyrosine phosphorylation of SIRPα is critical for SIRPα cleavage and synapse maturation. We found that during synapse maturation and in response to neural activity, SIRPα is highly phosphorylated on its tyrosine residues in the hippocampus, a structure critical for learning and memory. Tyrosine phosphorylation of SIRPα was necessary for SIRPα cleavage and presynaptic maturation, as indicated by the fact that a phosphorylation-deficient SIRPα variant underwent much less cleavage and could not drive presynaptic maturation. However, SIRPα phosphorylation did not affect its synaptic localization. Finally, we show that inhibitors of the Src and JAK kinase family suppress neural activity–dependent SIRPα phosphorylation and cleavage. Together, our results indicate that SIRPα phosphorylation serves as a mechanism for detecting synaptic activity and linking it to the ectodomain cleavage of SIRPα, which in turn drives synapse maturation in an activity-dependent manner.
Keywords: neurodevelopment, synapse, hippocampus, tyrosine, phosphorylation, shedding, immunoglobulin superfamily, neural activity, synaptogenesis
Introduction
The precise development of synapses, the sites of information transfer between neurons in the brain, is critical for proper information processing and brain function. The impaired formation of synapses leads to numerous neurological and psychiatric disorders, such as autism and schizophrenia (1–5). Synapses develop through a series of highly regulated stages: initial synapse differentiation, synapse maturation, and synapse maintenance stages (6–8). These stages of synapse development are orchestrated by a number of bidirectional, trans-synaptic signals that organize and coordinate the development of both pre- and postsynaptic terminals (6–11). Whereas the initial synapse differentiation stage, during which immature synapses are formed, is considered to be synaptic activity-independent, the synapse maturation stage is regulated by synaptic activity (12–15). During synapse maturation, only the active and functional synapses are strengthened and stabilized to form effective synaptic networks in the brain. However, the molecules and mechanisms by which synapses mature in an activity-dependent manner are largely unknown. Defects in synapse maturation have been implicated in various neurological and psychiatric disorders (1–5, 16, 17). Thus, understanding the molecular mechanisms underlying activity-dependent synapse maturation should further our understanding of and provide novel therapeutic strategies for neurological and psychiatric disorders.
To identify the molecular mechanisms by which synapses mature, we have purified synaptogenic molecules from developing mouse brains using a synapse formation assay in cultured neurons (18). One of the synaptogenic molecules we identified was signal regulatory protein-α (SIRPα),2 a member of the Ig superfamily transmembrane proteins. SIRPα is highly expressed in the brain and is concentrated at the postsynaptic terminals of excitatory synapses (19). Using in vitro culture systems and in vivo mouse mutants, we showed that in the hippocampus, the region central to learning, memory, and emotional processing, SIRPα serves as a retrograde, trans-synaptic signal that promotes presynaptic maturation (19). Importantly, synaptic activity is necessary for SIRPα to drive presynaptic maturation. When synapses are active, the ectodomain of SIRPα is cleaved and shed and drives maturation of the presynaptic terminal. Clearly, there are two important questions to address. 1) How does SIRPα detect synaptic activity? 2) How does synaptic activity regulate the cleavage of the SIRPα ectodomain? Addressing these questions will advance our understanding of the mechanisms underlying activity-dependent synapse maturation, a process fundamental to the proper wiring of the brain, and shed light on the pathophysiology of neuropsychiatric disorders caused by impaired synapse maturation.
In nonneural systems, an important way in which SIRPα signaling is transduced is via SIRPα tyrosine phosphorylation. The SIRPα intracellular domain contains immunoreceptor tyrosine-based inhibitory motifs. The tyrosine phosphorylation of these motifs in response to various stimuli, such as growth factors and ligand binding, recruits and subsequently activates the Src homology 2 domain–containing tyrosine phosphatases (SHPs). These phosphatases regulate intracellular signaling pathways, specifically the mitogen-activated protein kinase and the NF-κB pathways, to moderate various cellular functions (20, 21). In cultured cells, the tyrosine phosphorylation of SIRPα regulates cell migration and cell proliferation (22, 23). The expression of WT SIRPα, but not a phosphorylation-deficient SIRPα mutant, positively regulated Chinese hamster ovary cell migration in response to insulin (22) and breast cancer cell proliferation (23). In the immune system, the tyrosine phosphorylation–dependent signaling of SIRPα regulates inflammatory responses, including monocyte adhesion, macrophage migration, phagocytosis, and cytokine release (24–28). SIRPα tyrosine phosphorylation has also been implicated in osteoblast differentiation (29). In the brain, SIRPα is known to be tyrosine-phosphorylated in response to stress, hypothermia, or light exposure (30–33). However, the role of SIRPα tyrosine phosphorylation in the brain is not known.
Here we identify a critical role for the tyrosine phosphorylation of SIRPα in activity-dependent synapse maturation. We show that 1) SIRPα is highly phosphorylated on its tyrosine residues during synapse maturation, 2) neural activity induces tyrosine phosphorylation of SIRPα, 3) SIRPα phosphorylation is critical for its synaptogenic activity, 4) SIRPα phosphorylation is necessary for its ectodomain cleavage, and 5) inhibitors of Src and JAK family tyrosine kinases suppress the activity-dependent tyrosine phosphorylation and cleavage of SIRPα. These results demonstrate a critical role of SIRPα tyrosine phosphorylation in sensing synaptic activity, regulating SIRPα ectodomain cleavage, and, ultimately, driving synapse maturation in an activity-dependent manner. Our results reveal a novel molecular mechanism by which synaptic activity is converted to synaptogenic activity to establish functional synaptic networks.
Results
Characterization of SIRPα mutant constructs and antibodies
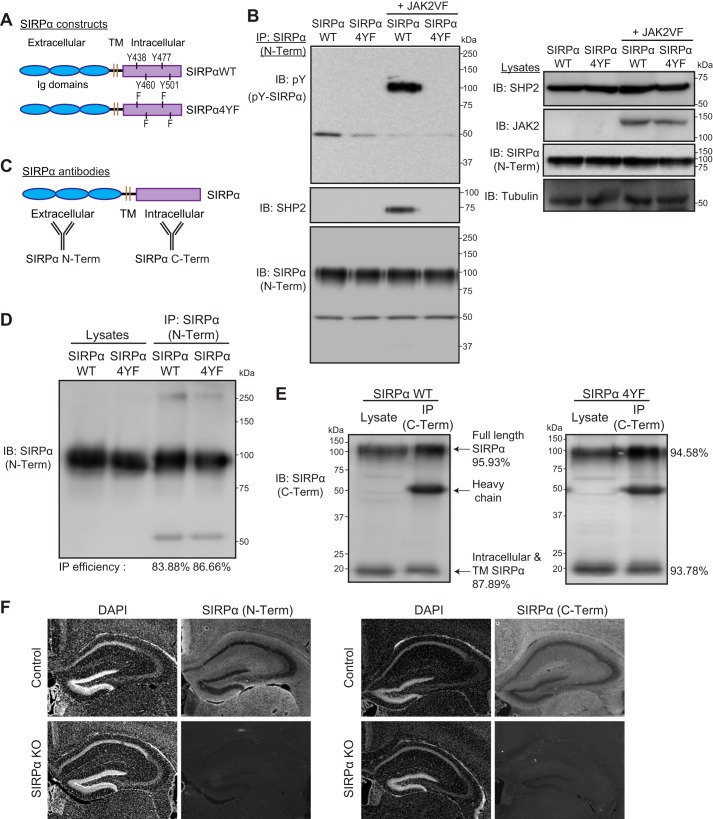
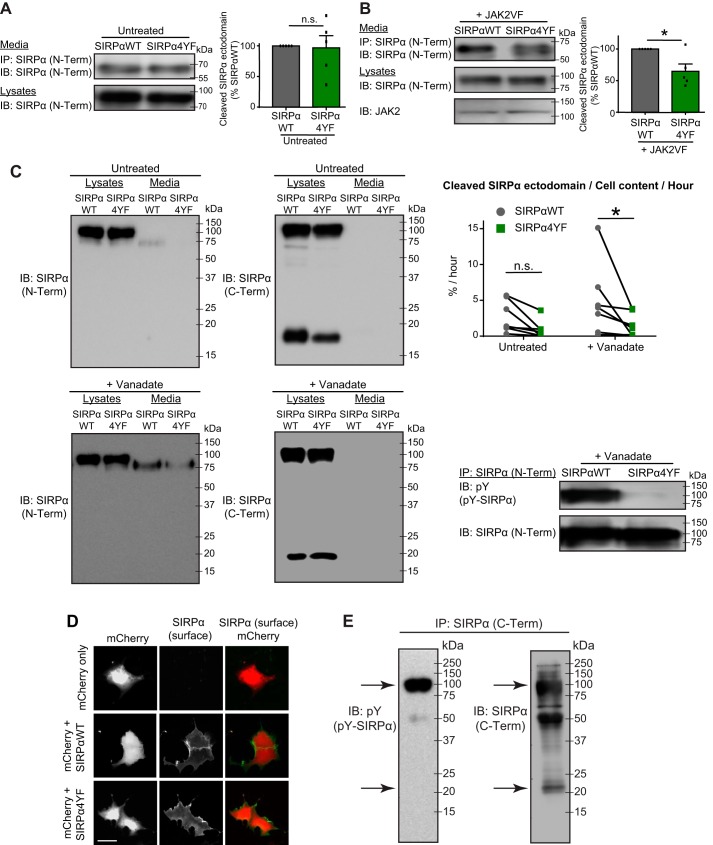
To determine the role of SIRPα tyrosine phosphorylation during synaptic development, we created a tyrosine phosphorylation–deficient SIRPα mutant (SIRPα4YF). In this construct, all four intracellular tyrosine residues in SIRPα were mutated to phenylalanine residues, preventing tyrosine phosphorylation (Fig. 1A). We confirmed that SIRPα4YF is indeed tyrosine phosphorylation–deficient by examining its JAK2-dependent tyrosine phosphorylation (34, 35). In transfected COS cells, SIRPαWT was tyrosine-phosphorylated by an active form of JAK2 (JAK2VF) (36), whereas SIRPα4YF was not (Fig. 1B, top left). We further verified the phosphorylation deficiency of SIRPα4YF by examining its phosphorylation-dependent SHP2 interaction; the adaptor protein SHP2 is known to bind SIRPα following the tyrosine phosphorylation of SIRPα (37–39). We co-transfected COS cells with SIRPα4YF or SIRPαWT, SHP2, with or without JAK2VF. We then immunoprecipitated SIRPα and determined its SHP2 interaction. We found that in the presence of JAK2VF, SHP2 was physically associated with SIRPαWT but not with SIRPα4YF (Fig. 1B, second panel from top left). These results verify that the SIRPα4YF mutant is tyrosine phosphorylation–deficient.
Figure 1.
Characterization of SIRPα mutant constructs and antibodies. A, illustration of the structures of SIRPαWT and SIRPα4YF. Four intracellular tyrosine residues in SIRPαWT were replaced with phenylalanine to generate SIRPα4YF. TM, transmembrane domain. B, verification of the tyrosine phosphorylation-deficient mutant of SIRPα (SIRPα4YF). COS cells were transfected with a SIRPα construct (WT or 4YF) and SHP2, together with or without an active form of JAK2 (JAK2VF). Cells were immunoprecipitated for SIRPα protein (with SIRPα N-Term antibody; described below), and the immunoprecipitates were blotted for phosphotyrosine (pY), SHP2, and SIRPα (SIRPα N-Term) (left). SIRPαWT is phosphorylated by JAK2VF and binds SHP2; however, SIRPα4YF is not phosphorylated by JAK2VF and does not bind SHP2. Expression levels of transfected proteins in the lysates are shown in the right panel. C, illustration showing the recognition sites of the anti-SIRPα antibodies used; SIRPα (N-Term) recognizes the SIRPα ectodomain, and SIRPα (C-Term) recognizes the SIRPα intracellular domain. D, immunoprecipitation efficiency of the SIRPα (N-Term) antibody. The same amount of lysates was used for direct blotting (Lysates) and immunoprecipitation (IP) followed by blotting. IP efficiency (%) was calculated by quantifying the ratio of SIRPα (IP) to SIRPα (Lysates). The IP efficiency of the SIRPα (N-Term) antibody in detecting SIRPαWT and SIRPα4YF was 83.88 and 86.66%, respectively. E, immunoprecipitation efficiency of the SIRPα (C-Term) antibody. The C-Term antibody recognizes both the full-length and the intracellular/transmembrane domain (lacking the ectodomain) SIRPα. IP efficiency was calculated as in D. The SIRPα (C-Term) effectively immunoprecipitated SIRPαWT (full-length, 95.93%; intracellular fragment, 87.89%) and SIRPα4YF (full-length, 94.58%; intracellular fragment, 93.78%). F, verification of the specificity of SIRPα antibodies by immunostaining. SIRPα KO brains were stained with either SIRPα (N-Term) or SIRPα (C-Term). SIRPα KO brains showed a lack of staining with both antibodies.
We then characterized the two SIRPα antibodies we used to examine the role of SIRPα tyrosine phosphorylation: an antibody recognizing the extracellular domain of SIRPα (SIRPα N-Term) and an antibody recognizing the intracellular domain of SIRPα (SIRPα C-Term) (Fig. 1C). We have verified that both antibodies 1) similarly recognize SIRPαWT and SIRPα4YF by Western blotting and 2) effectively immunoprecipitate both SIRPαWT and SIRPα4YF (83–96% efficiency; Fig. 1, D and E). The C-Term antibody recognized both the SIRPα full length and intracellular C-terminal fragment, which is produced after ectodomain cleavage, by Western blotting (Fig. 1E). Both antibodies specifically recognize SIRPα by immunostaining, as confirmed by lack of staining in SIRPα knockout brains (Fig. 1F). These results confirm that the antibodies are suitable for examining the role of SIRPα tyrosine phosphorylation by Western blotting, immunoprecipitation, and immunostaining.
SIRPα is highly phosphorylated on tyrosine residues during synapse maturation in the hippocampus in vivo
The synaptic Ig superfamily molecule, SIRPα, is a critical activity-dependent regulator of excitatory synapse maturation in the hippocampus (19). Our aim here was to understand how SIRPα detects synaptic activity and how, in turn, this regulates SIRPα's synaptogenic activity. For this, we focused on tyrosine phosphorylation of SIRPα. SIRPα has four tyrosine residues in its intracellular domain (37). Because tyrosine phosphorylation is a rapid and localized mechanism that moderates the function of a protein (40), we hypothesized that SIRPα is tyrosine-phosphorylated in response to synaptic activity during synapse maturation and that this phosphorylation is necessary for SIRPα's role in driving activity-dependent synapse maturation. To test this idea, we first examined whether SIRPα is tyrosine-phosphorylated during synapse maturation in the mouse hippocampus in vivo.
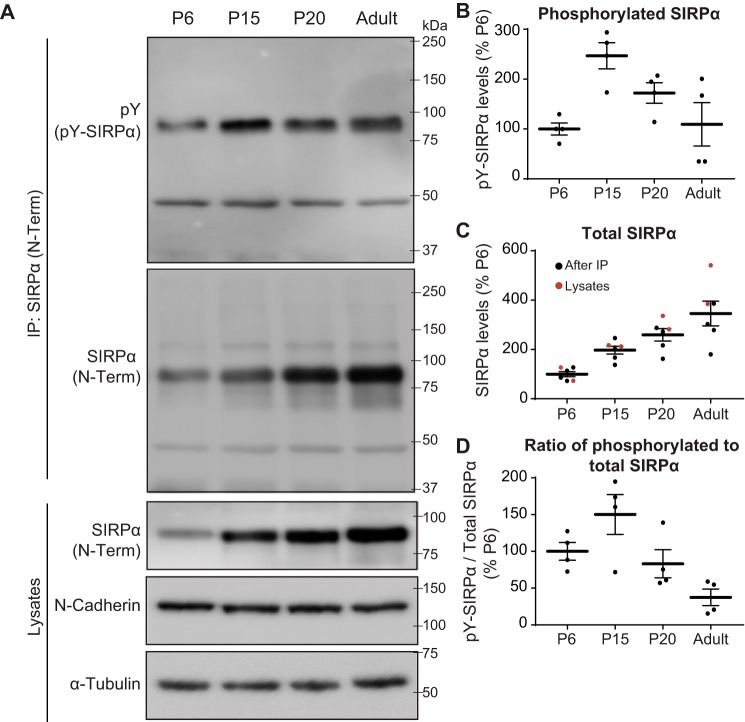
In the mouse hippocampus, initial synapse differentiation occurs during the first two postnatal weeks, between postnatal day 0 (P0) and P14 (19, 41). This differentiation stage is followed by an activity-dependent synapse maturation stage that occurs between P15 and P30 (13, 19). Afterward, synapses are maintained throughout life. Hence, we evaluated tyrosine phosphorylation of SIRPα at P6 (during synapse differentiation), P15 (the start of synapse maturation), and P20 (during synapse maturation) and in adults (during synapse maintenance). We immunoprecipitated SIRPα from hippocampal lysates and assessed the amount of total and tyrosine-phosphorylated SIRPα present (Fig. 2A). The total amount of SIRPα continuously increased from P6 to adulthood (Fig. 2C; the same results were obtained by directly blotting the lysates for SIRPα. Note that the levels of N-cadherin and α-tubulin were similar across all ages tested, suggesting that the lysis buffer is equally capable of solubilizing synaptic proteins). In contrast, the amount of tyrosine-phosphorylated SIRPα robustly increased from P6 to P15, when synapse maturation starts, and then decreased afterward (Fig. 2, A and B). The percentage of phosphorylated SIRPα to total SIRPα also peaked around P15 (Fig. 2D). These results demonstrate that both the absolute amount of tyrosine-phosphorylated SIRPα and the relative percentage of SIRPα that is phosphorylated are highest at the beginning of the synapse maturation stage. This suggests that tyrosine phosphorylation of SIRPα may play important roles in regulating synapse maturation in the hippocampus.
Figure 2.
SIRPα tyrosine phosphorylation peaks during synapse maturation in the mouse hippocampus. A, SIRPα protein was immunoprecipitated (with SIRPα N-Term antibody) from the mouse hippocampal lysates prepared at P6, P15, P20, and adulthood. Immunoprecipitates were subjected to Western blotting using antibodies against phosphotyrosine (pY) and SIRPα (N-Term). Lysates were also directly blotted for SIRPα (N-Term), N-cadherin, and α-tubulin. The levels of N-cadherin and α-tubulin were similar across all ages tested, suggesting that the lysis buffer is equally capable of solubilizing synaptic proteins at different ages. B, quantification of the absolute amount of tyrosine-phosphorylated SIRPα (pY-SIRPα) normalized to that at P6. C, quantification of total SIRPα (black dots, after IP; red dots, lysates) normalized to that at P6. D, quantification of the percentage of tyrosine-phosphorylated SIRPα in total SIRPα (pY-SIRPα/total SIRPα; normalized to P6). Both the absolute amount of phosphorylated SIRPα and the percentage of phosphorylated SIRPα are highest at P15, which corresponds to the beginning of the synapse maturation period in the mouse hippocampus (13, 19). Equal amounts of total protein were used for immunoprecipitation and blotting. n = 4 mice for each age group. Error bars, S.E.
Neural activity induces tyrosine phosphorylation of SIRPα
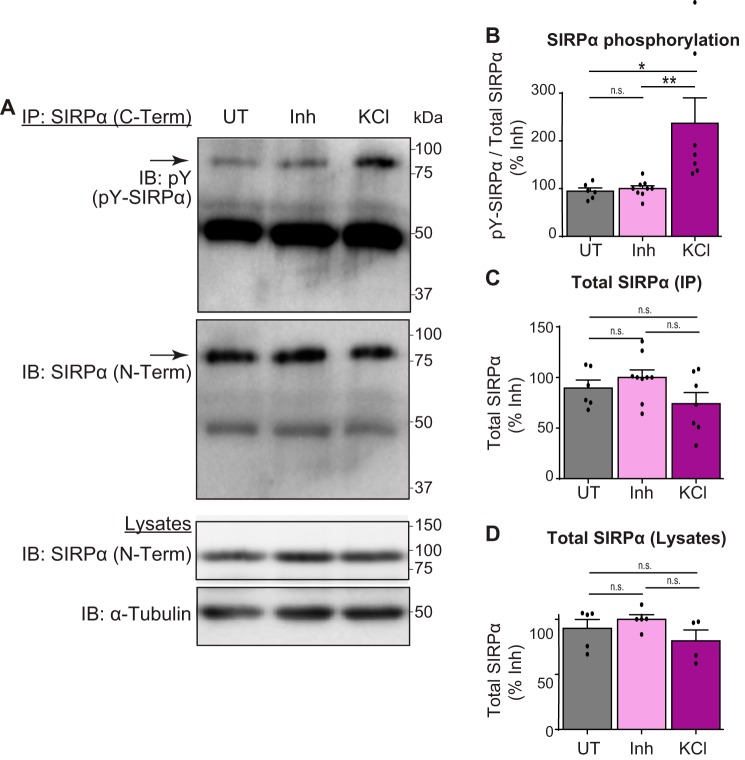
The synapse maturation stage is an activity-dependent stage where active synapses are the ones that undergo maturation (12–15, 19). Because SIRPα phosphorylation peaks during the synapse maturation stage in the hippocampus (Fig. 2), we next asked whether SIRPα phosphorylation is regulated by neural activity. For this, we cultured hippocampal neurons, and at DIV (days in vitro) 12, which is during synapse maturation in cultured hippocampal neurons (19), we treated cultures for 20 min either with a neurotransmitter receptor inhibitor mixture (50 μm picrotoxin, 10 μm CNQX, and 50 μm AP5) to silence all synaptic activity or with 55 mm potassium chloride (KCl) to depolarize neurons. This treatment length was chosen because the KCl treatment for 20 min showed the most robust and consistent response with regard to SIRPα phosphorylation relative to that of 3 min and 1 h (data not shown). After treatment, we immunoprecipitated SIRPα and examined the amount of total and tyrosine-phosphorylated SIRPα. We found that the activation of neurons by KCl treatment induced a more than 2-fold increase in SIRPα phosphorylation relative to untreated and inhibitor-treated cultures (Fig. 3, A and B), indicating that neural activity induces the tyrosine phosphorylation of SIRPα. The total level of SIRPα was not significantly different in all conditions (Fig. 3, C and D). These results support the idea that tyrosine phosphorylation of SIRPα contributes to activity-dependent synapse maturation. The addition of inhibitors showed similar levels of SIRPα phosphorylation as untreated cultures. This may be due to the relatively low level of basal synaptic activity in neuronal cultures in the 20-min period (Fig. 3B).
Figure 3.
Activation of neurons induces tyrosine phosphorylation of SIRPα. A, hippocampal neurons were cultured and treated at DIV12 with inhibitor mixture (50 μm picrotoxin, 10 μm CNQX, and 50 μm AP5) for 7 h. Cultures were washed with new medium and left untreated (UT), or treated with either inhibitor mixture (Inh) or 55 mm KCl for 20 min. Cultured neurons were lysed, immunoprecipitated for SIRPα (C-Term), and blotted for phosphotyrosine (pY) and SIRPα (N-Term). Lysates were also directly blotted for SIRPα (N-Term) and α-tubulin. B, quantification of the amount of tyrosine-phosphorylated SIRPα (pY-SIRPα/Total SIRPα (IP); normalized to inhibitor group). The amount of phosphorylated SIRPα is significantly increased by KCl-driven neural activation. C, quantification of total SIRPα levels (IP) under each condition (normalized to inhibitor group). D, quantification of total SIRPα levels (Lysates) under each condition (normalized to inhibitor group). Equal amounts of total protein were used for immunoprecipitation and blotting. n = 6 (untreated), n = 9 (inhibitor), or 7 (KCl) dishes from five independent experiments. Data are mean ± S.E. (error bars). n.s., not significant (p > 0.05); *, p < 0.05; **, p < 0.01 by one-way ANOVA followed by Tukey's test.
Tyrosine phosphorylation of SIRPα is necessary for synapse maturation
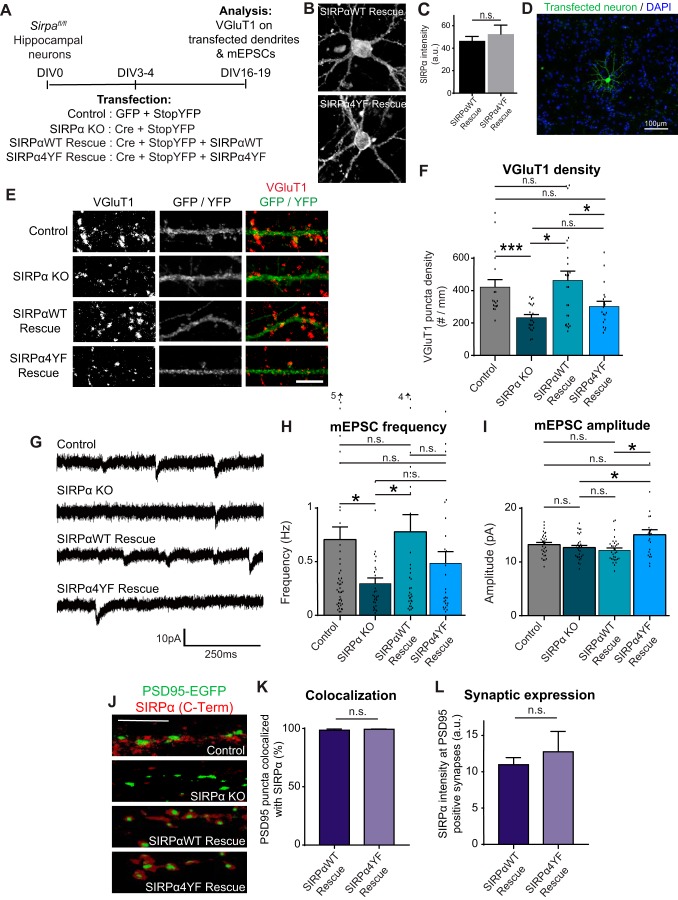
Using the SIRPα4YF mutant (Fig. 1A), we next investigated whether tyrosine phosphorylation of SIRPα is necessary for its synaptogenic activity. For this, we utilized SIRPα-deficient neurons (19). SIRPα-deficient neurons show defects in presynaptic maturation of excitatory (glutamatergic) synapses. We asked whether SIRPα4YF could rescue the impaired presynaptic maturation of SIRPα-deficient neurons. To create the SIRPα-deficient background, we prepared hippocampal cultures from mice bearing the loxP-flanked Sirpa alleles (Sirpafl/fl) (19) and transfected with the Cre expression plasmid (SIRPα KO). For the rescue experiments, we also co-transfected SIRPαWT (SIRPαWT Rescue) or SIRPα4YF (SIRPα4YF Rescue) together with Cre (Fig. 4A). Both SIRPαWT and SIRPα4YF expressed equally in neurons (Fig. 4, B and C). Additionally, to label transfected neurons, we co-transfected StopYFP, which expresses YFP only when Cre is co-expressed (42). This system allows us to temporally control SIRPα inactivation and rescue during synapse development. We transfected neurons sparsely to evaluate the effects of SIRPα KO and rescues in a neuron-specific and cell autonomous manner (Fig. 4D).
Figure 4.
Tyrosine phosphorylation of SIRPα is necessary for its synaptogenic effects. A, experimental scheme to determine the role of SIRPα tyrosine phosphorylation in synapse development. Cultured hippocampal neurons from Sirpafl/fl mice were transfected with GFP and StopYFP plasmids (Control); Cre and StopYFP plasmids (SIRPα KO); Cre, StopYFP, and SIRPαWT plasmids (SIRPαWT Rescue); or Cre, StopYFP and SIRPα4YF plasmids (SIRPα4YF Rescue) at DIV3–4 and stained for VGluT1 or subjected to electrophysiology at DIV16–19. B, representative images of SIRPαWT-rescue and SIRPα4YF-rescue neurons stained with SIRPα C-Term antibody. C, quantification of SIRPα staining intensity in SIRPαWT-rescue and SIRPα4YF-rescue neurons. The SIRPα staining intensity was similar in both rescue conditions. D, image of transfected neuron showing sparse transfection. E, representative images of VGluT1 puncta (red) on transfected dendrites (green). F, graph showing quantification of VGluT1 puncta density (number of puncta/dendrite length). The density of VGluT1 puncta is decreased in SIRPα KO cultures than in control. SIRPαWT rescued these defects, whereas SIRPα4YF did not. G, mEPSCs were recorded at DIV18–19 from cultured hippocampal neurons. H and I, graphs show quantification of mEPSC frequency and amplitude. mEPSC frequency was decreased in SIRPα KO neurons relative to control neurons. SIRPαWT rescued these defects, whereas SIRPα4YF did not. J–L, cultured hippocampal neurons from Sirpafl/fl mice were transfected with PSD95-EGFP plasmid (Control); Cre and PSD95-EGFP plasmids (SIRPα KO); Cre, PSD95-EGFP, and SIRPαWT plasmids (SIRPαWT Rescue); or Cre, PSD95-EGFP, and SIRPα4YF plasmids (SIRPα4YF Rescue) at DIV3 and stained for SIRPα (C-Term) at DIV12. K, percentage of PSD95 puncta that colocalizes with SIRPα. Both SIRPαWT and SIRPα4YF localize to PSD95-positive synapses. L, quantification of SIRPα staining intensity at PSD95-EGFP–positive synapses. Both SIRPαWT and SIRPα4YF express equally at synapses. F, n = 18 (control), 22 (KO), 25 (WT rescue), and 19 (4YF rescue) neurons from three independent cultures. H and I, n (frequency, amplitude) = 50, 31 (control); 30, 30 (KO); 36, 32 (WT rescue); and 26, 23 (4YF rescue) neurons from eight independent cultures. Data are mean ± S.E. (error bars). n.s., not significant (p > 0.05); *, p < 0.05; ***, p < 0.005 by Student's t test or one-way ANOVA followed by Tukey's test. Scale bar, 100 μm (B), 5 μm (E and J).
To evaluate glutamatergic presynaptic maturation, we analyzed the clustering of glutamatergic synaptic vesicles by staining for vesicular glutamate transporter 1 (VGluT1) (19, 41) (Fig. 4E). We quantified the number of VGluT1 puncta on the dendrites of transfected neurons (Fig. 4F). The inactivation of endogenous SIRPα (SIRPα KO) significantly decreased the density of VGluT1 puncta on transfected dendrites relative to control (GFP, instead of Cre, was transfected), consistent with our previous report showing that SIRPα is necessary for glutamatergic presynaptic maturation (19). This decrease was completely rescued by the expression of SIRPαWT. In contrast, SIRPα4YF failed to rescue the decrease in VGluT1 density. These results suggest that tyrosine phosphorylation of SIRPα is necessary for presynaptic maturation.
To confirm the necessity of SIRPα tyrosine-phosphorylation in synapse maturation, we performed electrophysiological experiments. We recorded miniature excitatory postsynaptic currents (mEPSCs) from transfected hippocampal neurons (Fig. 4G). In agreement with our histological data, we found a marked decrease in mEPSC frequency (reflecting the number of presynaptic inputs) in SIRPα KO neurons relative to control neurons. This decrease in mEPSC frequency was completely rescued by the expression of SIRPαWT but not by SIRPα4YF (Fig. 4H). Together, our data demonstrate that SIRPα tyrosine phosphorylation is critical for SIRPα's synaptogenic function in increasing the number of functional presynaptic inputs.
Interestingly, we noticed that the mEPSC amplitude (reflecting postsynaptic response) was increased in SIRPα4YF-rescue neurons (Fig. 4I), whereas there was no difference among control, SIRPα KO, and SIRPαWT-rescue neurons. This suggests that SIRPα phosphorylation may also influence postsynaptic maturation. SIRPα4YF may be interfering with signals that control postsynaptic development.
To rule out the possibility that the inability of SIRPα4YF to drive presynaptic development is due to its inability to localize to synapses, we examined the synaptic localization of SIRPαWT and SIRPα4YF. In neurons, endogenous SIRPα localizes predominantly to excitatory postsynaptic terminals (19). To visualize excitatory postsynaptic terminals in cultured hippocampal neurons, we co-transfected EGFP-tagged postsynaptic density 95 (PSD95-EGFP), which accumulates at excitatory postsynaptic terminals and effectively labels them (43). In control neurons, SIRPα accumulated at PSD95-EGFP–positive synapses. This expression of SIRPα on PSD95-EGFP–positive puncta was diminished in SIRPα KO neurons (Fig. 4J). Importantly, both SIRPαWT and SIRPα4YF localized to PSD95-EGFP–positive puncta (Fig. 4, J and K). Additionally, the intensity of SIRPα staining at synapses was not significantly different between SIRPαWT and SIRPα4YF (Fig. 4L). These results indicate that the tyrosine phosphorylation of SIRPα does not regulate the synaptic targeting of SIRPα.
Tyrosine phosphorylation of SIRPα regulates SIRPα ectodomain cleavage
SIRPα regulates synapse maturation via activity-dependent ectodomain cleavage (19). Because we found that tyrosine phosphorylation of SIRPα is critical for synapse maturation (Fig. 4), we hypothesized that SIRPα tyrosine phosphorylation regulates the cleavage of SIRPα. To test this idea, we transfected COS cells with SIRPαWT only, SIRPα4YF only, SIRPαWT + JAK2VF (to drive SIRPα phosphorylation), or SIRPα4YF + JAK2VF. Two days after transfection, we collected the culture media and immunoprecipitated the secreted SIRPα ectodomain. Without tyrosine kinases, the levels of secreted SIRPα were similar between SIRPαWT and SIRPα4YF (Fig. 5A). In contrast, in the presence of JAK2VF, SIRPαWT had significantly more ectodomain released than SIRPα4YF (Fig. 5B). Similar results were obtained using sodium orthovanadate, which inhibits protein-tyrosine phosphatases and activates tyrosine kinases (44), to drive SIRPα phosphorylation (Fig. 5C). Our quantitative analysis (percentage of cell content released per hour) showed that significantly more ectodomain was released from SIRPαWT than SIRPα4YF following vanadate treatment. Vanadate treatment induced the cleavage of SIRPαWT at the rate of 2.15% per hour (percentage of cell content). In contrast, vanadate induced the cleavage of SIRPα4YF only at the rate of 0.609%. This difference in ectodomain cleavage is not due to lack of surface expression of SIRPα4YF, because both forms of SIRPα were detected on the cell surface when cells were stained for SIRPα without permeabilizing the cell membrane (Fig. 5D). These results indicate that tyrosine phosphorylation of SIRPα plays a critical role in regulating SIRPα ectodomain cleavage in COS cells. Interestingly, after the cleavage of the SIRPα ectodomain, the C-terminal fragment of SIRPα was dephosphorylated (Fig. 5E).
Figure 5.
Tyrosine phosphorylation of SIRPα regulates SIRPα ectodomain cleavage in COS cells. A and B, COS cells were transfected with SIRPαWT or SIRPα4YF (A) or with SIRPαWT + JAK2VF or SIRPα4YF + JAK2VF (B). Two days after transfection, culture medium and cell lysates were collected. Culture medium was subjected to immunoprecipitation for secreted SIRPα protein (with SIRPα N-term antibody) and blotted for SIRPα (N-Term). Cell lysates were subjected to Western blotting for SIRPα (N-Term) and JAK2. Graphs show the quantification of the amount of secreted SIRPα ectodomain. A, when SIRPα was not tyrosine-phosphorylated (without JAK2VF; see Fig. 1B), a similar amount of SIRPα ectodomain was released in the medium from SIRPαWT and SIRPα4YF. B, in the presence of tyrosine kinase (JAK2VF), significantly more SIRPα ectodomain was released in the medium from SIRPαWT compared with SIRPα4YF. C, COS cells were transfected with SIRPαWT or SIRPα4YF. Two days after transfection, cultures were treated with 1 mm sodium orthovanadate for 7–17 h to promote tyrosine phosphorylation of SIRPα (SIRPαWT, but not SIRPα4YF, is tyrosine-phosphorylated (pY-SIRPα; bottom right panel)). Culture medium and cell lysates were collected and subjected to Western blotting; the membranes were first blotted with the SIRPα N-Term antibody, stripped, and then blotted with the SIRPα C-Term antibody. The following SIRPα band intensities were quantified: Media (IB: N-Term, Media), Full-N (N-Term, Lysates), Full-C (C-Term, Lysates, top band), and C-Fragment (C-Term, Lysates, bottom band). The rate of SIRPα ectodomain cleavage was calculated using the formula, Media/((Full-C + C-Fragment) × (Full-N/Full-C))/hours (see “Experimental procedures”). Experiments were done as a pair (WT and 4YF), and the pairs are connected with lines in the graph. Significantly more SIRPα ectodomain was released in the medium from SIRPαWT than SIRPα4YF following vanadate treatment. D, COS cells were transfected with mCherry only, mCherry and SIRPαWT, or mCherry and SIRPα4YF. Cells were fixed and stained for SIRPα (N-Term) without permeabilization to detect surface expression of SIRPα. Both SIRPαWT and SIRPα4YF are expressed on the surface. E, tyrosine phosphorylation of the SIRPα full-length and C-terminal fragment (arrows) after vanadate treatment. n = 5 (A and B) in seven independent experiments (C), reproduced four times (E). Data are mean ± S.E. (error bars). n.s., not significant (p > 0.05); *, p < 0.05, by Student's t test (A and B) or two-way ANOVA (C). Scale bar, 20 μm.
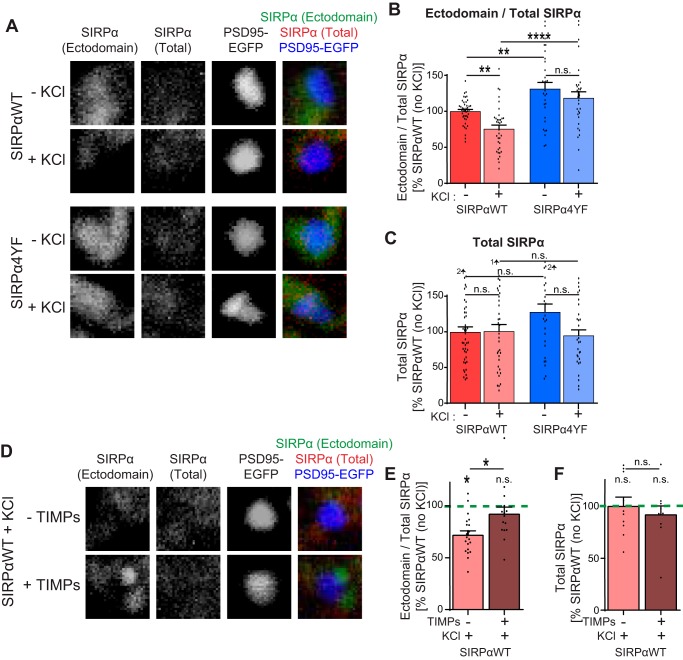
To determine whether SIRPα tyrosine phosphorylation regulates ectodomain cleavage in neurons as well, we prepared hippocampal cultures from Sirpafl/fl mice (19) and transfected the cultures to create the SIRPαWT-rescue or SIRPα4YF-rescue conditions as described above (Fig. 4A). To identify synapses, we co-transfected PSD95-EGFP. We then left cultures untreated, or we treated these cultures with KCl for 2 h at DIV12. After the treatment, we stained the cultures using the SIRPα N-terminal antibody without permeabilization to detect the surface SIRPα ectodomain, followed by the SIRPα C-terminal antibody with permeabilization to evaluate total SIRPα. We imaged these cultures and calculated the ratio of the SIRPα ectodomain to total SIRPα. At the basal level (no KCl conditions), the ectodomain/total SIRPα ratio was significantly smaller in SIRPαWT-rescue neurons relative to SIRPα4YF-rescue neurons, suggesting that the basal level of neuronal activity induced more cleavage of SIRPαWT than SIRPα4YF. In SIRPαWT-rescue neurons, KCl treatment significantly decreased the ectodomain/total SIRPα ratio relative to control, suggesting that the SIRPαWT ectodomain is further cleaved upon neuronal activation. In contrast, in SIRPα4YF neurons, KCl treatment did not significantly change the ectodomain/total SIRPα ratio (Fig. 6, A and B). The total level of SIRPα is not significantly different in all conditions (Fig. 6C). These results suggest that neural activity–dependent SIRPα cleavage requires SIRPα tyrosine phosphorylation.
Figure 6.
Tyrosine phosphorylation of SIRPα regulates activity-driven SIRPα ectodomain cleavage in neurons. A–C, cultured hippocampal neurons from Sirpafl/fl mice were transfected with Cre, PSD95-EGFP, and SIRPαWT plasmids (SIRPαWT) or with Cre, PSD95-EGFP, and SIRPα4YF plasmids (SIRPα4YF) at DIV3 and treated with 55 mm KCl for 2 h at DIV12. Cultures were immunostained with the SIRPα N-Term antibody without permeabilization to detect surface ectodomain-containing SIRPα, followed by the SIRPα C-Term antibody with permeabilization to detect total SIRPα. Ectodomain and total SIRPα staining intensity at synapses (identified by PSD95-EGFP) were quantified. B, ratio of ectodomain/total SIRPα (normalized to SIRPαWT without KCl). KCl-driven neuronal activation decreased the ectodomain/total SIRPα ratio in SIRPαWT-expressing neurons but not in SIRPα4YF-expressing neurons. C, total SIRPα was not significantly different in all groups. D–F, cultured hippocampal neurons expressing SIRPαWT were prepared as described above. At DIV12, cultures were treated with 55 mm KCl or KCl + TIMPs and immunostained as described above. E, ratio of ectodomain/total SIRPα. TIMPs block the KCl-induced decrease in the ectodomain/total SIRPα ratio. F, total SIRPα was not significantly different in all groups. A–C, n = 43 (SIRPαWT, −KCl), 36 (SIRPαWT, +KCl), 30 (SIRPα4YF, −KCl), and 31 (SIRPα4YF, + KCl) neurons from three independent cultures. E, n = 20 (SIRPαWT, +KCl) and 15 (SIRPαWT, +KCl, +TIMPs) neurons. Data are mean ± S.E. (error bars). n.s., not significant (p > 0.05); *, p < 0.05; **, p < 0.01; ****, p < 0.001 by one-way ANOVA followed by Tukey's test.
To ascertain that the change in the ectodomain/total SIRPα ratio is indeed due to SIRPα ectodomain cleavage, we treated SIRPαWT-rescue cultures with the proteinase inhibitors, TIMPs, which inhibit SIRPα ectodomain cleavage (19). The treatment of cultures with TIMPs blocked the KCl-driven decrease in the ectodomain/total SIRPα ratio, suggesting that the change in the ratio is due to SIRPα ectodomain cleavage (Fig. 6, D and E).
Inhibitors of Src and JAK family kinases suppress neural activity–driven SIRPα tyrosine phosphorylation
We next wanted to identify the signaling molecules that regulate the tyrosine phosphorylation of SIRPα in response to neural activity. For this, we focused on two families of tyrosine kinases, the Src and JAK families, because they are highly expressed in hippocampal neurons and can be activated by neural activity (45–47). Additionally, we showed that in COS cells, JAK2 can phosphorylate SIRPα (Fig. 1B) (34, 35).
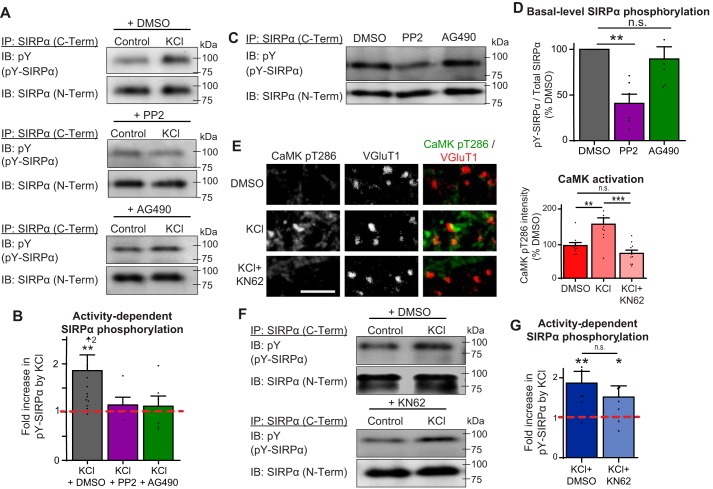
To test whether Src and JAK kinase activity contributes to activity-dependent tyrosine phosphorylation of SIRPα in neurons, we treated hippocampal cultures prepared from WT mice at DIV12 with either 10 μm PP2 (an inhibitor of Src family kinases) (48) or 10 μm AG490 (an inhibitor of JAK family kinases) (46, 49) for 1 h. Control cultures were treated with DMSO only (PP2 and AG490 were dissolved in DMSO). These cultures were then treated with or without 55 mm KCl for 20 min to activate neurons. We then subjected these cultures to immunoprecipitation for SIRPα and immunoblotted for phosphotyrosine and SIRPα. We found that treatment of neurons with either PP2 or AG490 blocked the increase in SIRPα tyrosine phosphorylation in response to neuronal activation (Fig. 7, A and B). These results indicate that the inhibitors, likely through the inhibition of either Src or JAK family kinases, blocked the activity-dependent tyrosine phosphorylation of SIRPα. Interestingly, the treatment of cultures with PP2, but not AG490, diminished the basal level of SIRPα phosphorylation (Fig. 7, C and D), suggesting that Src family kinases, but not JAK kinases, also contribute to the basal level of SIRPα phosphorylation in neurons.
Figure 7.
Inhibition of Src and JAK family kinases, but not CaMKs, blocks activity-dependent SIRPα tyrosine phosphorylation in neurons. A and B, hippocampal neurons were cultured to DIV12 and treated with inhibitor mixture (50 μm picrotoxin, 10 μm CNQX, and 50 μm AP5) for 7 h. In the last hour, 10 μm PP2, 10 μm AG490, or DMSO was added to these cultures. Cultures were then washed and treated with either inhibitor mixture or 55 mm KCl, with either PP2, AG490, or DMSO for 20 min. SIRPα was immunoprecipitated (with SIRPα C-Term antibody) from the cell lysate and blotted for phosphotyrosine and SIRPα (N-Term). B, graph shows -fold increase in SIRPα phosphorylation by KCl treatment in the presence of PP2, AG490, or DMSO. KCl treatment significantly increased SIRPα tyrosine phosphorylation in DMSO-treated neurons, but not in the presence of PP2 or AG490. C and D, basal levels of SIRPα tyrosine phosphorylation in the presence of PP2, AG490, or DMSO. Basal-level SIRPα phosphorylation was decreased by PP2 but not by AG490 application. E–G, hippocampal cultures were treated at DIV12, with inhibitor mixture + 5 μm KN62 for 7 h. Cultures were washed and treated with DMSO, DMSO + 55 mm KCl, 5 μm KN62, or KCl + KN62 for 20 min. E, cultures were immunostained for CaMK-pT286 antibody to detect levels of CaMK activation. Treatment of cultures with KN62 effectively suppressed CaMK activation. F and G, SIRPα was immunoprecipitated from the cell lysates and blotted for phosphotyrosine and SIRPα. G, graph shows the amount of tyrosine-phosphorylated SIRPα (normalized to DMSO). KCl-driven tyrosine phosphorylation of SIRPα was not inhibited by KN62. n = 12 (DMSO), 5 (PP2), and 5 (AG490) cultures (B); 6 cultures per group from six independent experiments (D); and 5 (KCl) and 6 (KCl + KN62) cultures from four independent experiments (E and G). Data are mean ± S.E. (error bars). n.s., not significant (p > 0.05); *, p < 0.05; **, p < 0.01; ****, p < 0.001 by Kolmogorov–Smirnov test (B and G) or one-way ANOVA followed by Tukey's test (D and E).
CaMK activity is not involved in SIRPα tyrosine phosphorylation
We have previously shown that the cleavage of SIRPα requires CaMK activity (19). The inhibition of CaMK activity resulted in a substantial decrease in the amount of cleaved SIRPα. Because SIRPα cleavage is regulated by its tyrosine phosphorylation (Figs. 5 and 6), we asked whether CaMK signaling influences tyrosine phosphorylation of SIRPα. To test this, we treated cultured hippocampal neurons at DIV12 with 5 μm KN62 (a CaMK inhibitor) and activated neurons with KCl for 20 min. Treatment of KN62 effectively suppressed CaMK activation at synapses (Fig. 7E). Cultures treated with KN62 still showed a marked increase in the level of SIRPα tyrosine phosphorylation in response to neuronal activation (Fig. 7, F and G), suggesting that CaMK activity is not involved in activity-dependent tyrosine phosphorylation of SIRPα. This implies that SIRPα tyrosine phosphorylation and CaMK signaling regulate SIRPα ectodomain cleavage via distinct pathways.
Inhibitors of Src and JAK family kinases suppress neural activity–driven SIRPα ectodomain cleavage
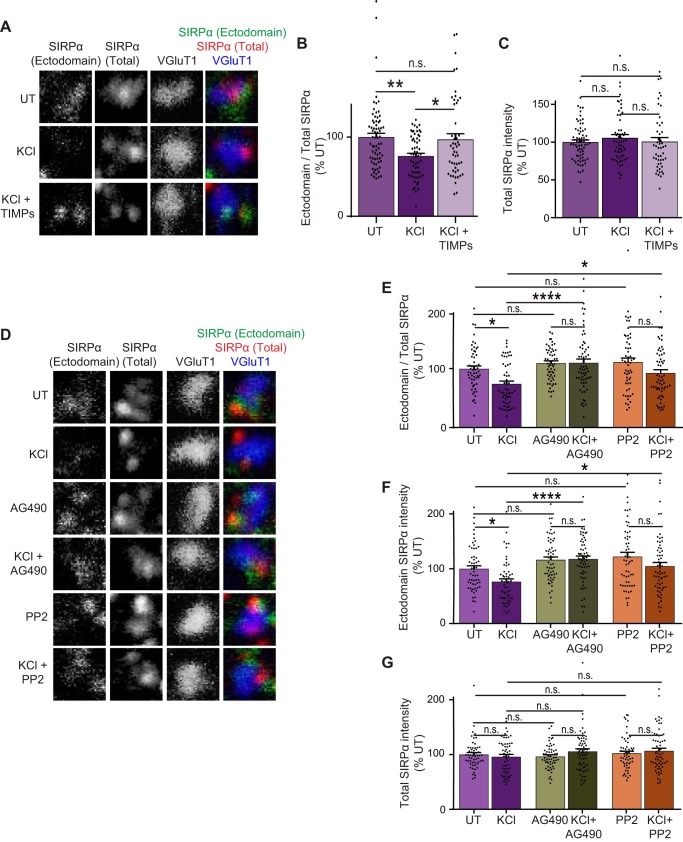
Based on our data that inhibitors of Src and JAK family kinases suppress SIRPα tyrosine phosphorylation in response to neural activity (Fig. 7), we next wanted to determine whether these kinase inhibitors also suppress activity-dependent SIRPα ectodomain cleavage in neurons. We first determined that KCl-dependent neuronal activation affects the SIRPα cleavage by sequential immunostaining (see Fig. 6). After KCl treatment, we stained the neurons with the SIRPα N-terminal antibody without permeabilization (to detect surface “ectodomain-containing SIRPα”) and then with the SIRPα C-terminal antibody (to detect “total SIRPα”) and VGluT1 antibody (to mark synapses) with permeabilization. We quantified the ectodomain/total SIRPα ratio at synapses. We found that KCl treatment resulted in a decrease in this ratio: the intensity of surface ectodomain-containing SIRPα at the synapse significantly decreased following KCl treatment, whereas the level of total SIRPα stayed constant (Fig. 8, A–C). TIMPs, the proteinase inhibitors that inhibit SIRPα ectodomain cleavage, blocked the KCl-driven decrease in the amount of ectodomain SIRPα and the ectodomain/total SIRPα ratio (Fig. 8, A and B). These results support our conclusion that neuronal activation induces the cleavage of SIRPα ectodomain.
Figure 8.
Inhibition of Src and JAK family kinases prevents activity-dependent SIRPα ectodomain cleavage in neurons. A–C, hippocampal neurons were cultured to DIV12 and left untreated or were treated with KCl or KCl + TIMPs for 2 h. The amount of ectodomain and total SIRPα (endogenous) at excitatory synapses (identified by VGluT1 staining) was examined as described in the legend to Fig. 6. B, quantification of the ratio of ectodomain/total SIRPα. KCl-driven neuronal activation decreased the ratio of ectodomain/total SIRPα. This decrease was blocked by TIMPs. C, total levels of SIRPα are not significantly different between all groups. D–G, hippocampal neurons were cultured to DIV12 and treated with either 10 μm PP2, 10 μm AG490, or DMSO, with or without 55 mm KCl for 2 h. The amount of ectodomain and total SIRPα at excitatory synapses was examined as described above. E, quantification of the ratio of ectodomain/total SIRPα. AG490 and PP2 blocked the KCl-induced decrease in the ratio of ectodomain/total SIRPα. F, quantification of ectodomain SIRPα. G, levels of total SIRPα were not changed in all groups. n = 70 (UT), 59 (KCl), and 50 (KCl + TIMPs) (A–C) or 56 (UT), 55 (KCl), 58 (PP2), 62 (KCl + PP2), 58 (AG490), and 62 (KCl + AG490) synapses (D–G) from three independent experiments. Data are mean ± S.E. (error bars). n.s., not significant (p > 0.05); *, p < 0.05; **, p < 0.01; ***, p < 0.005; ****, p < 0.001 by one-way ANOVA followed by Tukey's test.
We next performed these experiments with the addition of the kinase inhibitors, PP2 or AG490, which block activity-dependent phosphorylation of SIRPα (Fig. 8D). We found that PP2 or AG490 treatment inhibited the KCl-driven decrease in the amount of ectodomain SIRPα and the ectodomain/total SIRPα ratio (Fig. 8, E–G). These results indicate that the inhibition of Src or JAK family kinases inhibits the activity-dependent ectodomain cleavage of SIRPα.
Discussion
Selective maturation of active synapses is critical for the establishment of functional neural circuits (1–8, 12–15). Although a few molecules have been identified as regulators of synapse maturation (19, 50–52), the mechanisms through which these molecules detect synaptic activity and signal synapse maturation selectively at active synapses remain unclear. Here, we identified a critical role for the tyrosine phosphorylation of SIRPα in activity-dependent synapse maturation. We showed that 1) tyrosine phosphorylation of SIRPα peaks in vivo during the synapse maturation period; 2) SIRPα phosphorylation is driven by neural activity; 3) in contrast to WT SIRPα, the phosphorylation-deficient SIRPα mutant cannot drive presynaptic maturation; 4) tyrosine phosphorylation of SIRPα controls the ectodomain cleavage of SIRPα, which in turn, signals presynaptic maturation; and 5) inhibitors of Src and JAK family kinases suppress activity-dependent tyrosine phosphorylation and ectodomain cleavage of SIRPα in neurons. Collectively, our results demonstrate that tyrosine phosphorylation of SIRPα serves as a molecular switch that detects neuronal activity and turns on the maturation of active synapses by promoting the cleavage of SIRPα's ectodomain.
Tyrosine phosphorylation of SIRPα as a synaptic activity sensor
SIRPα is an activity-dependent regulator of excitatory presynaptic maturation (19). The overexpression of WT SIRPα in neurons drives presynaptic maturation; however, this only occurs when neuronal activity is present. So, how does SIRPα detect synaptic activity and drive the maturation of active synapses only? Here, we showed that tyrosine phosphorylation of SIRPα is driven by neuronal activity (Fig. 3) and that this phosphorylation is necessary for SIRPα's ability to drive presynaptic maturation during development (Fig. 4). This makes SIRPα tyrosine phosphorylation a key mechanism through which SIRPα senses neuronal activity and converts it to synaptogenic activity. Tyrosine phosphorylation of SIRPα is a temporally and spatially regulated, activity-dependent mechanism that allows SIRPα to function specifically at active synapses during development.
SIRPα tyrosine phosphorylation may be a general mechanism for SIRPα to detect various stimulations, not only during development, but also in adulthood. Light stimulation, hypothermia, and stress can affect SIRPα phosphorylation in the adult brain (30–33). The precise roles of these phosphorylation events are not known, but tyrosine phosphorylation of SIRPα may serve as a sensor of various brain activities even in adults.
Tyrosine phosphorylation of SIRPα as a “cleave me” signal
How does SIRPα tyrosine phosphorylation regulate synapse maturation? Our previous work demonstrated that the ectodomain of SIRPα is cleaved, and this cleavage is essential for presynaptic maturation (18, 19). Here we showed that SIRPα tyrosine phosphorylation regulates this cleavage (Figs. 5 and 6). Thus, we propose that the activity-dependent tyrosine phosphorylation of SIRPα serves as a mechanism to tag SIRPα for cleavage.
How the tyrosine phosphorylation of SIRPα controls the cleavage of its ectodomain is an important next question. Possible mechanisms include the following. 1) SIRPα phosphorylation activates downstream signaling that regulates proteinase expression. One possible downstream signaling molecule is SHP2. Upon phosphorylation, SIRPα recruits SHP2 (Fig. 1B). SHP2 has been implicated in the regulation of MMP-2 and -9 expression (53). 2) SIRPα phosphorylation leads to the recruitment of proteinases. For example, ADAM10, which has an intracellular substrate-binding domain, interacts with the ephrin family of cell adhesion molecules (54, 55). This interaction facilitates ephrin cleavage. Similar mechanisms might exist for SIRPα.
The cleavage of SIRPα is suggested to be regulated by MMP/ADAM10 proteinases (19, 56). At the synapse, there are several other synaptic proteins, including neuroligins, N-cadherin, and nectin, that are known to undergo MMP/ADAM-mediated proteolytic cleavage (51, 57–60). How MMPs and ADAMs recognize these synaptic proteins is unknown, but the phosphorylation of these synaptic proteins may also play a role in tagging them.
Multiple signaling pathways coordinate to regulate SIRPα tyrosine phosphorylation and cleavage
Src and JAK family kinases are two major tyrosine kinase families in the brain. Furthermore, many members of these kinase families are known to be activated by neuronal activity (45–47). Here we identified that the activity of both Src and JAK family kinases regulates tyrosine phosphorylation of SIRPα during synapse maturation. Interestingly, the inhibition of Src family kinases resulted in a decrease in both basal and activity-dependent SIRPα phosphorylation, whereas the inhibition of JAK family kinases only deterred activity-dependent SIRPα phosphorylation (Fig. 7). These results hint at the possibility that these different kinase groups may coordinate to regulate SIRPα phosphorylation during synapse development, perhaps by phosphorylating different tyrosine residues to send different signals downstream. These data also suggest that Src family kinases may phosphorylate and prime SIRPα to then be phosphorylated by JAK kinases upon stimulation by neuronal activity. Signals from both kinase families need to coincide to drive the activity-dependent tyrosine phosphorylation of SIRPα.
For the ectodomain cleavage of SIRPα, we have previously shown that CaMK signaling is necessary (19). Here we showed that tyrosine phosphorylation of SIRPα is also necessary for SIRPα cleavage (Figs. 5 and 6). However, we found that CaMK signaling is not necessary for tyrosine phosphorylation of SIRPα (Fig. 7). Thus, at least two distinct signaling pathways, the CaMK and SIRPα tyrosine-phosphorylation pathways, are necessary for the cleavage of SIRPα. CaMK signaling may be involved in the direct regulation of proteinase activity in response to neuronal activity and act in parallel with Src and JAK family kinase signaling to regulate SIRPα cleavage.
In summary, our work identified a novel molecular mechanism that governs the function of SIRPα in sensing neural activity and driving the maturation of active synapses. SIRPα-driven hippocampal synaptic maturation is a highly regulated process that requires proper signaling of multiple pathways in response to synaptic activity. A comprehensive understanding of the signals that govern synapse development can further our understanding of and suggest novel therapeutic strategies to prevent and/or treat various neuropsychiatric disorders associated with abnormal synapse maturation.
Experimental procedures
Mouse strains
Sirpafl/fl and Actin-CreER mutant mice were described previously (19). WT mice were ICR/CD-1 (Charles River) (Figs. 3, 7, and 8) or C57/B6 (Jackson) (Fig. 2). All animal care and use was in accordance with institutional guidelines and approved by the institutional animal care and use committee at Boston Children's Hospital.
Primary neuronal cultures, transfection, and treatment
For all neuronal cultures used, hippocampi were dissected from P0-P1 mice and dissociated with 0.5% trypsin (19, 41–43, 52). For immunoprecipitation and Western blotting experiments, 300,000 cells were plated on poly-d-lysine–coated dishes (35 mm). For immunostaining experiments, 75,000–80,000 hippocampal neurons were plated on poly-d-lysine–coated glass coverslips (diameter 12 mm) and grown in culture medium (B27 (Gibco), 2 mm l-glutamine, 100 units/ml penicillin-streptomycin in neurobasal medium (Gibco)). For electrophysiology experiments, 80,000 neurons were plated and grown in culture medium (GS21 (MTI-GlobalStem, Sigma), 2 mm l-glutamine, 100 units/ml penicillin-streptomycin in NeuralQ medium (Sigma)).
For transfection experiments, 1–2 μg of plasmids per coverslip were transfected using the CalPhos transfection kit (Clontech) at DIV3–4 (19, 41–43, 52). Neurons were cultured for 16–19 days and subjected to biochemical, histological, and electrophysiological experiments.
For the treatment with inhibitors described in Figs. 3 and 7, neurons were cultured for 12 days, after which culture medium was replaced by new medium containing inhibitor mixture (50 μm picrotoxin, 10 μm CNQX, and 50 μm AP5) and incubated for 7 h. Cultures were treated with a kinase inhibitor: 10 μm PP2 or 10 μm AG490 (for the last 1 h) or 5 μm KN62 (for 7 h). Because inhibitors were dissolved in DMSO, an equal volume of DMSO was added to control cultures. Medium was then replaced with new culture medium containing inhibitor mixture or 55 mm KCl, with or without the kinase inhibitor for 20 min.
For the treatment with inhibitors described in Fig. 6 and 8, neurons were cultured for 12 days, after which culture medium was replaced by new medium. Cultures were then either untreated or treated with 55 mm KCl with or without inhibitors: 10 μm PP2, 10 μm AG490, or 0.5 μg/ml each of TIMP1 and TIMP2 (for 2 h). Because inhibitors were dissolved in DMSO, an equal volume of DMSO was added to control cultures.
COS cell culture and transfection
COS7 cells (ATCC) were cultured in Dulbecco's modified Eagle's medium (Gibco) supplemented with 10% fetal bovine serum (Corning). Transfections were done via the Lipofectamine 3000 (Thermo Fisher) method or via the calcium phosphate method. Cells were cultured for 1–3 days following transfection and subjected to biochemical and histological experiments. In the sodium orthovanadate experiments, cultures were treated with 1 mm sodium orthovanadate for 7–17 h before biochemical experiments.
DNA constructs
A cDNA encoding the tyrosine phosphorylation–deficient SIRPα (SIRPα4YF) was generated by PCR using the following primer pairs: Set 1, 5′-GCCCACTCGAGTGATCAAGGG-3′ and 5′-TGTGATATCGTTGATGTCATTTG-3′; Set 2, 5′-TCAACGATATCACATTTGCAGACCTGAA-3′ and 5′-TTACCGGTCTCAATGCTTGCAAATTCTGTGTGG-3′; Set 3, 5′-TCAACGATATCACAGAAGCAGACCTGAA-3′ and 5′-TTACCGGTCTCAATGCTTGCTTCTTCTGTGTGG-3′; Set 4, 5′-AGACCGGTAAAGTGCCTAGGCCAGAGGATACCCTCACCTTTGCTGACCTGG-3′ and 5′-CTTCTAGATCACTTCCTCTGGACCTGGACACTAGCAAACTCTGAGAAA-3′; and Set 5, 5′-AGACCGGTAAAGTGCCTAGGCCAGAGGATACCCTCACCGAAGCTGACCTGG-3′ and 5′-CTTCTAGATCACTTCCTCTGGACCTGGACACTAGCTTCCTCTGAGAAA-3′. The amplified fragments were ligated with SIRPαWT cDNA in the pAP-TAG5 vector (18) (digested with XhoI and XbaI). This replaces four intracellular tyrosine amino acids with phenylalanine. All PCR products were verified via sequencing. JAK2VF was generated by site-directed mutagenesis of JAK2 cDNA using the following primer pair: 5′-TTTTGAATTATGGTGTCTGTTTCTGTGGAGAGGAGAACATT-3′ and 5′-AATGTTCTCCTCTCCACAGAAACAGACACCATAATTCAAAA-3′. This substitutes valine 618 with phenylalanine. PSD95-GFP and StopYFP were described previously (42, 43).
Immunoprecipitation and Western blotting
For cultured cells, media and cells were collected separately. Cells were lysed in lysis buffer containing 1% Triton X-100, 50 mm Tris-HCl (pH 8), 150 mm NaCl, 1 mm Na3VO4 with a protease inhibitor mixture tablet (Roche Applied Science). Dissected hippocampi were lysed by homogenization in 10 volumes of lysis buffer/g of tissue. Protein concentrations were measured by protein assays (BCA, Pierce). Media or lysates were incubated with 1 μg of anti-SIRPα (p84; BD Biosciences; BDB552371) or 3 μg of anti-SIRPα C terminus (QED; 2428) antibodies for 2 h at 4 °C. Equal amounts of lysates and equal volume of media from each experimental condition were used for immunoprecipitation. The immune complexes were precipitated with Protein L or Protein A (Pierce). Immunoprecipitates and lysates were subjected to SDS-PAGE. Proteins were transferred to a polyvinylidene difluoride membrane (Bio-Rad), blocked with 3% nonfat milk, and blotted using anti-SIRPα (p84; BD; 1:1000), anti-phosphotyrosine (4G10; Millipore; 05-321X; 1:1000), anti-SHP2 (Santa Cruz Biotechnology, Inc.; SC-7384; 1:1000), anti-α-tubulin (DM1A; Invitrogen; 62204; 1:1000), anti-N-cadherin (BD Biosciences; 610921; 1:1000), and anti-JAK2 (CST; 3230P; 1:1000). The proteins were visualized by chemiluminescence (Bio-Rad) and imaged with ImageQuant LAS4000. Band intensities were quantified with Fiji software. For this, rectangles of equal length and width were drawn around each protein band within the blot and used as the region of interest. The width of the rectangle was drawn to fit the width of the band, whereas the length of the rectangle was drawn to include 3 times the length of the band above and below the protein band of interest. An intensity profile plot was created using ImageJ for each rectangle. The intensity of each peak was measured from this plot, which allows for the background to be subtracted using the intensity of the regions above and below the protein band for each lane. For the quantification of phosphorylated SIRPα to total SIRPα, the blots were probed first for phosphotyrosine (4G10), stripped, and reprobed for SIRPα.
For the calculation of SIRPα ectodomain secretion (Fig. 5C), media and lysates were first blotted with the SIRPα N-Term antibody, stripped, and then blotted with the SIRPα C-Term antibody. The following SIRPα band intensities were quantified (Fig. 5C): Media (IB: N-Term, Media), Full-N (N-Term, Lysates), Full-C (C-Term, Lysates, top band), and C-Fragment (C-Term, Lysates, bottom band). The rate of SIRPα ectodomain cleavage was calculated using the formula, Media/((Full-C + C-Fragment) × (Full-N/Full-C))/hours. (Full-C + C-Fragment), detected by SIRPα C-Term antibody, implies total SIRPα. Because SIRPα in media was detected with SIRPα N-Term antibody, the ratio of Full-N/Full-C was used to normalize between two blots blotted with N-Term and C-Term antibodies.
Immunocytochemistry
Cultured neurons were fixed with 3% paraformaldehyde for 10 min at 37 °C and blocked in 2% BSA, 2% normal goat serum, and 0.1% Triton X-100 for 1 h at room temperature. This was followed by incubation with primary antibodies for 3 h at room temperature or overnight at 4 °C. The cultures were then washed with PBS, and secondary antibodies were applied for 1 h at room temperature. After being washed again with PBS, samples were mounted in glycerol with n-propyl gallate (Sigma). The following antibodies and dilutions were used: anti-VGluT1 (Millipore; AB5905; 1:4000), anti-GFP (Aves Labs; GFP-1020; 1:5000), anti-SIRPα (clone p84; BD; 1:200), anti-CaMKII-pT286 (CST; D21E4; 1:200). For surface staining, a similar protocol as described was used; however, Triton X-100 was omitted to prevent membrane permeabilization.
Image acquisition and analysis
Imaging was done with a confocal microscope (Zeiss LSM 700) using ×40 and ×60 objectives at a resolution of 1024 × 1024 pixels. Images were taken as 16-bit, z-stack images with a 0.5-μm step size. Identical settings for laser power, master gain, digital gain, and offset were used for all acquired images within each experiment.
For VGluT1 density analysis, transfected pyramidal-like neurons were selected for imaging at random. For analysis, images were stacked and merged using Fiji software, and VGluT1 puncta on transfected dendrites after the primary branch point (identified by GFP or YFP expression) were analyzed using MetaMorph (Molecular Devices). The quantification was done blind. Puncta that were within a region 0.3 μm away from the dendritic shaft and spines were included for analysis. The staining intensity of VGluT1 in the dendritic shaft was subtracted as background. This background intensity was similar between conditions. Puncta smaller than 4 pixels (∼0.04 μm2) were excluded from analysis. Dendritic lengths were measured by manual tracing of analyzed dendritic lengths in Fiji.
For ectodomain/total SIRPα analysis, transfected pyramidal-like neurons (Fig. 6) or fields of neuronal cultures (Fig. 8) were selected for imaging at random. For analysis, images were stacked using Fiji software, and regions of interest (ROI) were selected as follows. 1) For analysis in Fig. 6, PSD95-EGFP–positive puncta were selected as the ROI, and 2) for analysis in Fig. 8, SIRPα (C-Term)–positive puncta with >10% overlap with VGluT1 puncta were selected as the ROI. The average intensity of SIRPα (C-Term) and SIRPα (N-Term) staining within the ROI was measured. 5–10 ROIs were selected at random per transfected neuron (Fig. 6) or per field of image (Fig. 8).
Electrophysiology
mEPSCs were recorded via whole-cell patch-clamp recordings of cultured hippocampal neurons as described previously (19, 41). Neurons were placed in HEPES-buffered saline (119 mm NaCl, 5 mm KCl, 2 mm CaCl2, 2 mm MgCl2, 30 mm glucose, 10 mm HEPES) supplemented with 1 μm tetrodotoxin and 50 μm picrotoxin. Recordings were made with glass pipettes (resistance 4–6 megaohms) filled with internal solution (100 mm gluconic acid, 0.2 mm EGTA, 5 mm MgCl2, 2 mm ATP, 0.3 mm GTP, and 40 mm HEPES). Recordings were made with a Multiclamp 700 amplifier (Molecular Devices), and data were collected with Clampex version 10.3. (Molecular Devices). mEPSCs were analyzed using Minianalysis version 6.0 (Synaptosoft). All recordings and mEPSC analyses were done blind.
Statistical analysis
Statistical analyses were performed using GraphPad Prism software. Statistical tests used were the Kolmogorov–Smirnov test, Student's t test, and one-way ANOVA or two-way ANOVA, as indicated in the figure legends. In the case of one-way ANOVA, post hoc analysis was done with Tukey's test. In the case of two-way ANOVA, post hoc analysis was done with Sidak's test. All data are mean ± S.E. In all figures, significance is indicated as follows: n.s., p > 0.05; *, p < 0.05; **, p < 0.01; ***, p < 0.005; ****, p < 0.001. Sample sizes were similar to those reported in previous publications (19, 41–43, 52). Post hoc statistical calculation of sample sizes for imaging and electrophysiology was also done to ensure that sample sizes had sufficient power for subsequent statistical analyses (at least 80% power at the 0.05 level of significance for each set of experiments).
Author contributions
S. N.-C. and H. U. conceptualization; S. N.-C., E. M. J.-V., and H. U. data curation; S. N.-C., E. M. J.-V., and H. U. formal analysis; S. N.-C. and E. M. J.-V. investigation; S. N.-C., E. M. J.-V., and H. U. methodology; S. N.-C. and H. U. writing-original draft; S. N.-C., E. M. J.-V., and H. U. writing-review and editing; H. U. supervision; H. U. funding acquisition; H. U. project administration.
Acknowledgment
We thank Masahiro Yasuda for help with molecular cloning.
Note added in proof
Fig. 1F contained an inadvertently duplicated image in the version of this article that was published as a Paper in Press on June 18, 2018. This error has now been corrected and does not affect the results or conclusions of this work.
This work was supported by National Institutes of Health Grants R01-NS092578 and R01-MH111647 (to H. U.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- SIRP
- signal regulatory protein
- JAK
- Janus kinase
- CaMK
- calcium/calmodulin protein kinase
- P
- postnatal day
- DIV
- days in vitro
- CNQX
- 6-cyano-7-nitroquinoxaline-2,3-dione
- AP5
- (2R)-amino-5-phosphonopentanoate
- VGluT
- vesicular glutamate transporter
- mEPSC
- miniature excitatory postsynaptic current
- SHP
- Src homology 2 domain-containing phosphatase
- PSD95
- postsynaptic density 95
- ADAM
- A disintegrin and metalloproteinase domain–containing protein
- MMP
- matrix metalloproteinase
- CD47
- cluster of differentiation 47
- NFκB
- nuclear factor κB
- KO
- knockout
- EGFP
- enhanced green fluorescent protein
- TIMP
- tissue inhibitor of metalloproteinases
- ANOVA
- analysis of variance
- C-Term and N-Term
- C-terminal and N-terminal, respectively
- DAPI
- 4′,6-diamidino-2-phenylindole
- IB
- immunoblotting
- YFP
- yellow fluorescent protein
- IP
- immunoprecipitation.
References
- 1. Kasai H., Fukuda M., Watanabe S., Hayashi-Takagi A., and Noguchi J. (2010) Structural dynamics of dendritic spines in memory and cognition. Trends Neurosci. 33, 121–129 10.1016/j.tins.2010.01.001 [DOI] [PubMed] [Google Scholar]
- 2. Santini E., Huynh T. N., MacAskill A. F., Carter A. G., Pierre P., Ruggero D., Kaphzan H., and Klann E. (2013) Exaggerated translation causes synaptic and behavioural aberrations associated with autism. Nature 493, 411–415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Tang G., Gudsnuk K., Kuo S. H., Cotrina M. L., Rosoklija G., Sosunov A., Sonders M. S., Kanter E., Castagna C., Yamamoto A., Yue Z., Arancio O., Peterson B. S., Champagne F., Dwork A. J., et al. (2014) Loss of mTOR-dependent macroautophagy causes autistic-like synaptic pruning deficits. Neuron 83, 1131–1143 10.1016/j.neuron.2014.07.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Penzes P., Buonanno A., Passafaro M., Sala C., and Sweet R. A. (2013) Developmental vulnerability of synapses and circuits associated with neuropsychiatric disorders. J. Neurochem. 126, 165–182 10.1111/jnc.12261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sekar A., Bialas A. R., de Rivera H., Davis A., Hammond T. R., Kamitaki N., Tooley K., Presumey J., Baum M., Van Doren V., Genovese G., Rose S. A., Handsaker R. E., Schizophrenia Working Group of the Psychiatric Genomics Consortium, Daly M. J., et al. (2016) Schizophrenia risk from complex variation of complement component 4. Nature 530, 177–183 10.1038/nature16549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Waites C. L., Craig A. M., and Garner C. C. (2005) Mechanisms of vertebrate synaptogenesis. Annu. Rev. Neurosci. 28, 251–274 10.1146/annurev.neuro.27.070203.144336 [DOI] [PubMed] [Google Scholar]
- 7. Fox M. A., and Umemori H. (2006) Seeking long-term relationship: axon and target communicate to organize synaptic differentiation. J. Neurochem. 97, 1215–1231 10.1111/j.1471-4159.2006.03834.x [DOI] [PubMed] [Google Scholar]
- 8. Shen K., and Scheiffele P. (2010) Genetics and cell biology of building specific synaptic connectivity. Annu. Rev. Neurosci. 33, 473–507 10.1146/annurev.neuro.051508.135302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Siddiqui T. J., and Craig A. M. (2011) Synaptic organizing complexes. Curr. Opin. Neurobiol. 21, 132–143 10.1016/j.conb.2010.08.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chia P. H., Li P., and Shen K. (2013) Cellular and molecular mechanisms underlying presynapse formation. J. Cell Biol. 203, 11–22 10.1083/jcb.201307020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Johnson-Venkatesh E. M., and Umemori H. (2010) Secreted factors as synaptic organizers. Eur. J. Neurosci. 32, 181–190 10.1111/j.1460-9568.2010.07338.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Katz L. C., and Shatz C. J. (1996) Synaptic activity and the construction of cortical circuits. Science 274, 1133–1138 10.1126/science.274.5290.1133 [DOI] [PubMed] [Google Scholar]
- 13. Yasuda M., Johnson-Venkatesh E. M., Zhang H., Parent J. M., Sutton M. A., and Umemori H. (2011) Multiple forms of activity-dependent competition refine hippocampal circuits in vivo. Neuron 70, 1128–1142 10.1016/j.neuron.2011.04.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hashimoto K., and Kano M. (2013) Synapse elimination in the developing cerebellum. Cell. Mol. Life Sci. 70, 4667–4680 10.1007/s00018-013-1405-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sanes J. R., and Lichtman J. W. (1999) Development of the vertebrate neuromuscular junction. Annu. Rev. Neurosci. 22, 389–442 10.1146/annurev.neuro.22.1.389 [DOI] [PubMed] [Google Scholar]
- 16. Lipska B. K., Halim N. D., Segal P. N., and Weinberger D. R. (2002) Effects of reversible inactivation of the neonatal ventral hippocampus on behavior in the adult rat. J. Neurosci. 22, 2835–2842 10.1523/JNEUROSCI.22-07-02835.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Meyer F., and Louilot A. (2012) Early prefrontal functional blockade in rats results in schizophrenia-related anomalies in behavior and dopamine. Neuropsychopharmacology 37, 2233–2243 10.1038/npp.2012.74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Umemori H., and Sanes J. R. (2008) Signal regulatory proteins (SIRPS) are secreted presynaptic organizing molecules. J. Biol. Chem. 283, 34053–34061 10.1074/jbc.M805729200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Toth A. B., Terauchi A., Zhang L. Y., Johnson-Venkatesh E. M., Larsen D. J., Sutton M. A., and Umemori H. (2013) Synapse maturation by activity-dependent ectodomain shedding of SIRPα. Nat. Neurosci. 16, 1417–1425 10.1038/nn.3516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Takada T., Matozaki T., Takeda H., Fukunaga K., Noguchi T., Fujioka Y., Okazaki I., Tsuda M., Yamao T., Ochi F., and Kasuga M. (1998) Roles of the complex formation of SHPS-1 with SHP-2 in insulin-stimulated mitogen-activated protein kinase activation. J. Biol. Chem. 273, 9234–9242 10.1074/jbc.273.15.9234 [DOI] [PubMed] [Google Scholar]
- 21. Neznanov N., Neznanova L., Kondratov R. V., Burdelya L., Kandel E. S., O'Rourke D. M., Ullrich A., and Gudkov A. V. (2003) Dominant negative form of signal-regulatory protein-α (SIRPα/SHPS-1) inhibits tumor necrosis factor-mediated apoptosis by activation of NF-κB. J. Biol. Chem. 278, 3809–3815 10.1074/jbc.M210698200 [DOI] [PubMed] [Google Scholar]
- 22. Motegi S., Okazawa H., Ohnishi H., Sato R., Kaneko Y., Kobayashi H., Tomizawa K., Ito T., Honma N., Bühring H.-J., Ishikawa O., and Matozaki T. (2003) Role of the CD47-SHPS-1 system in regulation of cell migration. EMBO J. 22, 2634–2644 10.1093/emboj/cdg278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Galbaugh T., Feeney Y. B., and Clevenger C. V. (2010) Prolactin receptor-integrin cross-talk mediated by SIRPα in breast cancer cells. Mol. Cancer Res. 8, 1413–1424 10.1158/1541-7786.MCR-10-0130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Smith R. E., Patel V., Seatter S. D., Deehan M. R., Brown M. H., Brooke G. P., Goodridge H. S., Howard C. J., Rigley K. P., Harnett W., and Harnett M. M. (2003) A novel MyD-1 (SIRP-1α) signaling pathway that inhibits LPS-induced TNFα production by monocytes. Blood 102 2532–2540 [DOI] [PubMed] [Google Scholar]
- 25. Liu S. Q., Alkema P. K., Tieché C., Tefft B. J., Liu D. Z., Li Y. C., Sumpio B. E., Caprini J. A., and Paniagua M. (2005) Negative regulation of monocyte adhesion to arterial elastic laminae by signal regulatory protein α and Src homology 2 domain-containing protein-tyrosine phosphatase-1. J. Biol. Chem. 280, 39294–39301 10.1074/jbc.M503866200 [DOI] [PubMed] [Google Scholar]
- 26. Bian Z., Shi L., Guo Y.-L., Lv Z., Tang C., Niu S., Tremblay A., Venkataramani M., Culpepper C., Li L., Zhou Z., Mansour A., Zhang Y., Gewirtz A., Kidder K., et al. (2016) CD47-Sirpα interaction and IL-10 constrain inflammation-induced macrophage phagocytosis of healthy self-cells. Proc. Natl. Acad. Sci. U.S.A. 113, E5434–E5443 10.1073/pnas.1521069113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Alblas J., Honing H., de Lavalette C. R., Brown M. H., Dijkstra C. D., and van den Berg T. K. (2005) Signal regulatory protein α ligation induces macrophage nitric oxide production through JAK/STAT- and phosphatidylinositol 3-kinase/Rac1/NAPDH oxidase/H2O2-dependent pathways. Mol. Cell. Biol. 25 7181–7192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Barclay A. N., and Van den Berg T. K. (2014) The interaction between signal regulatory protein α (SIRPα) and CD47: structure, function and therapeutic target. Annu. Rev. Immunol. 32, 25–50 [DOI] [PubMed] [Google Scholar]
- 29. Holm C. K., Engman S., Sulniute R., Matozaki T., Oldenborg P.-A., and Lundberg P. (2016) Lack of SIRPα phosphorylation and concomitantly reduced SHP-2-PI3K-Akt2 signaling decrease osteoblast differentiation. Biochem. Biophys. Res. Commun. 478, 268–273 10.1016/j.bbrc.2016.07.048 [DOI] [PubMed] [Google Scholar]
- 30. Nakahata Y., Okumura N., Shima T., Okada M., and Nagai K. (2000) Light-induced tyrosine phosphorylation of BIT in the rat suprachiasmatic nucleus. J. Neurochem. 74, 2436–2444 [DOI] [PubMed] [Google Scholar]
- 31. Hamada J., Okumura N., Inagaki M., Taniguchi H., Nakahata Y., Sano S., and Nagai K. (2004) Tyrosine phosphorylation of BIT on photic stimulation in the rat retina. FEBS Lett. 557, 204–208 10.1016/S0014-5793(03)01493-5 [DOI] [PubMed] [Google Scholar]
- 32. Ohnishi H., Murata T., Kusakari S., Hayashi Y., Takao K., Maruyama T., Ago Y., Koda K., Jin F.-J., Okawa K., Oldenborg P.-A., Okazawa H., Murata Y., Furuya N., Matsuda T., et al. (2010) Stress-evoked tyrosine phosphorylation of signal regulatory protein regulates behavioral immobility in the forced swim test. J. Neurosci. 30, 10472–10483 10.1523/JNEUROSCI.0257-10.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Maruyama T., Kusakari S., Sato-Hashimoto M., Hayashi Y., Kotani T., Murata Y., Okazawa H., Oldenborg P. A., Kishi S., Matozaki T., and Ohnishi H. (2012) Hypothermia-induced tyrosine phosphorylation of SIRPα in the brain. J. Neurochem. 121, 891–902 10.1111/j.1471-4159.2012.07748.x [DOI] [PubMed] [Google Scholar]
- 34. Stofega M. R., Wang H., Ullrich A., and Carter-Su C. (1998) Growth hormone regulation of SIRP and SHP-2 tyrosyl phosphorylation and association. J. Biol. Chem. 273, 7112–7117 10.1074/jbc.273.12.7112 [DOI] [PubMed] [Google Scholar]
- 35. Stofega M. R., Argetsinger L. S., Wang H., Ullrich A., and Carter-Su C. (2000) Negative regulation of growth hormone receptor/JAK2 signaling by signal regulatory protein α. J. Biol. Chem. 275, 28222–28229 [DOI] [PubMed] [Google Scholar]
- 36. Kralovics R., Passamonti F., Buser A. S., Teo S. S., Tiedt R., Passweg J. R., Tichelli A., Cazzola M., and Skoda R. C. (2005) A gain-of-function mutation of JAK2 in myeloproliferative disorders. N. Engl. J. Med. 352, 1779–1790 10.1056/NEJMoa051113 [DOI] [PubMed] [Google Scholar]
- 37. Kharitonenkov A., Chen Z., Sures I., Wang H., Schilling J., and Ullrich A. (1997) A family of proteins that inhibit signalling through tyrosine kinase receptors. Nature 386, 181–186 10.1038/386181a0 [DOI] [PubMed] [Google Scholar]
- 38. Fujioka Y., Matozaki T., Noguchi T., Iwamatsu A., Yamao T., Takahashi N., Tsuda M., Takada T., and Kasuga M. (1996) A novel membrane glycoprotein, SHPS-1, that binds the SH2-domain-containing protein tyrosine phosphatase SHP-2 in response to mitogens and cell adhesion. Mol. Cell. Biol. 16, 6887–6899 10.1128/MCB.16.12.6887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Timms J. F., Carlberg K., Gu H., Chen H., Kamatkar S., Nadler M. J. S., Rohrschneider L. R., and Neel B. G. (1998) Identification of major binding proteins and substrates for the SH2-containing protein tyrosine phosphatase SHP-1 in macrophages. Mol. Cell. Biol. 18, 3838–3850 10.1128/MCB.18.7.3838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Browning M. D., Huganir R., and Greengard P. (1985) Protein phosphorylation and neuronal function. J. Neurochem. 45, 11–23 10.1111/j.1471-4159.1985.tb05468.x [DOI] [PubMed] [Google Scholar]
- 41. Terauchi A., Johnson-Venkatesh E. M., Toth A. B., Javed D., Sutton M. A., and Umemori H. (2010) Distinct FGFs promote differentiation of excitatory and inhibitory synapses. Nature 465, 783–787 10.1038/nature09041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Dabrowski A., Terauchi A., Strong C., and Umemori H. (2015) Distinct sets of FGF receptors sculpt excitatory and inhibitory synaptogenesis. Development 142, 1818–1830 10.1242/dev.115568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Terauchi A., Timmons K. M., Kikuma K., Pechmann Y., Kneussel M., and Umemori H. (2015) Selective synaptic targeting of the excitatory and inhibitory presynaptic organizers FGF22 and FGF7. J. Cell Sci. 128, 281–292 10.1242/jcs.158337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Umemori H., Hayashi T., Inoue T., Nakanishi S., Mikoshiba K., and Yamamoto T. (1999) Involvement of protein tyrosine phosphatases in activation of the trimeric G protein Gq/11. Oncogene 18, 7399–7402 10.1038/sj.onc.1203152 [DOI] [PubMed] [Google Scholar]
- 45. Lu Y. M., Roder J. C., Davidow J., and Salter M. W. (1998) Src activation in the induction of long-term potentiation in CA1 hippocampal neurons. Science 279, 1363–1367 10.1126/science.279.5355.1363 [DOI] [PubMed] [Google Scholar]
- 46. Nicolas C. S., Peineau S., Amici M., Csaba Z., Fafouri A., Javalet C., Collett V. J., Hildebrandt L., Seaton G., Choi S. L., Sim S. E., Bradley C., Lee K., Zhuo M., Kaang B. K., et al. (2012) The JAK/STAT pathway is involved in synaptic plasticity. Neuron 73, 374–390 10.1016/j.neuron.2011.11.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Niimura M., Moussa R., Bissoon N., Ikeda-Douglas C., Milgram N. W., and Gurd J. W. (2005) Changes in phosphorylation of the NMDA receptor in the rat hippocampus induced by status epilepticus. J. Neurochem. 92, 1377–1385 10.1111/j.1471-4159.2005.02977.x [DOI] [PubMed] [Google Scholar]
- 48. Bain J., Plater L., Elliott M., Shpiro N., Hastie C. J., McLauchlan H., Klevernic I., Arthur J. S., Alessi D. R., and Cohen P. (2007) The selectivity of protein kinase inhibitors: a further update. Biochem. J. 408, 297–315 10.1042/BJ20070797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Chiba T., Yamada M., Sasabe J., Terashita K., Shimoda M., Matsuoka M., and Aiso S. (2009) Amyloid-β causes memory impairment by disturbing the JAK2/STAT3 axis in hippocampal neurons. Mol. Psychiatry 14, 206–222 10.1038/mp.2008.105 [DOI] [PubMed] [Google Scholar]
- 50. Bemben M. A., Shipman S. L., Hirai T., Herring B. E., Li Y., Badger J. D. 2nd, Nicoll R. A., Diamond J. S., and Roche K. W. (2014) CaMKII phosphorylation of neuroligin-1 regulates excitatory synapses. Nat. Neurosci. 17, 56–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Nagappan-Chettiar S., Johnson-Venkatesh E. M., and Umemori H. (2017) Activity-dependent proteolytic cleavage of cell adhesion molecules regulates excitatory synaptic development and function. Neurosci. Res. 116, 60–69 10.1016/j.neures.2016.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Terauchi A., Johnson-Venkatesh E. M., Bullock B., Lehtinen M. K., and Umemori H. (2016) Retrograde fibroblast growth factor 22 (FGF22) signaling regulates insulin-like growth factor 2 (IGF2) expression for activity-dependent synapse stabilization in the mammalian brain. eLife 5, e12151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Zhao H., and Agazie Y. M. (2015) Inhibition of SHP2 in basal-like and triple-negative breast cells induces basal-to-luminal transition, hormone dependency, and sensitivity to anti-hormone treatment. BMC Cancer 15, 109 10.1186/s12885-015-1131-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Janes P. W., Saha N., Barton W. A., Kolev M. V., Wimmer-Kleikamp S. H., Nievergall E., Blobel C. P., Himanen J. P., Lackmann M., and Nikolov D. B. (2005) Adam meets Eph: an ADAM substrate recognition module acts as a molecular switch for ephrin cleavage in trans. Cell 123, 291–304 10.1016/j.cell.2005.08.014 [DOI] [PubMed] [Google Scholar]
- 55. Atapattu L., Saha N., Llerena C., Vail M. E., Scott A. M., Nikolov D. B., Lackmann M., and Janes P. W. (2012) Antibodies binding the ADAM10 substrate recognition domain inhibit Eph function. J. Cell Sci. 125, 6084–6093 10.1242/jcs.112631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Londino J. D., Gulick D., Isenberg J. S., and Mallampalli R. K. (2015) Cleavage of signal regulatory protein α (SIRPα) enhances inflammatory signaling. J. Biol. Chem. 290, 31113–31125 10.1074/jbc.M115.682914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Peixoto R. T., Kunz P. A., Kwon H., Mabb A. M., Sabatini B. L., Philpot B. D., and Ehlers M. D. (2012) Transsynaptic signaling by activity-dependent cleavage of Neuroligin-1. Neuron 76, 396–409 10.1016/j.neuron.2012.07.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Suzuki K., Hayashi Y., Nakahara S., Kumazaki H., Prox J., Horiuchi K., Zeng M., Tanimura S., Nishiyama Y., Osawa S., Sehara-Fujisawa A., Saftig P., Yokoshima S., Fukuyama T., Matsuki N., et al. (2012) Activity-dependent proteolytic cleavage of Neuroligin-1. Neuron 76, 410–422 10.1016/j.neuron.2012.10.003 [DOI] [PubMed] [Google Scholar]
- 59. Kim J., Lilliehook C., Dudak A., Prox J., Saftig P., Federoff H. J., and Lim S. T. (2010) Activity-dependent α-cleavage of nectin-1 is mediated by a disintegrin and metalloprotease 10 (ADAM10). J. Biol. Chem. 285, 22919–22926 10.1074/jbc.M110.126649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Reiss K., Maretzky T., Ludwig A., Tousseyn T., de Strooper B., Hartmann D., and Saftig P. (2005) ADAM10 cleavage of N-cadherin and regulation of cell-cell adhesion and β-catenin nuclear signaling. EMBO J. 24, 742–752 10.1038/sj.emboj.7600548 [DOI] [PMC free article] [PubMed] [Google Scholar]