Abstract
Nidovirus endoribonucleases (NendoUs) include nonstructural protein 15 (nsp15) from coronaviruses and nsp11 from arteriviruses, both of which have been reported to participate in the viral replication process and in the evasion of the host immune system. Results from a previous study of coronaviruses SARS-CoV, HCoV-229E, and MHV nsp15 indicate that it mainly forms a functional hexamer, whereas nsp11 from the arterivirus PRRSV is a dimer. Here, we found that porcine Deltacoronavirus (PDCoV) nsp15 primarily exists as dimers and monomers in vitro. Biological experiments reveal that a PDCoV nsp15 mutant lacking the first 27 amino acids of the N-terminal domain (Asn-1–Asn-27) forms more monomers and displays decreased enzymatic activity, indicating that this region is important for its dimerization. Moreover, multiple sequence alignments and three-dimensional structural analysis indicated that the C-terminal region (His-251–Val-261) of PDCoV nsp15 is 10 amino acids shorter and forms a shorter loop than that formed by the equivalent sequence (Gln-259–Phe-279) of SARS-CoV nsp15. This result may explain why PDCoV nsp15 failed to form hexamers. We speculate that NendoUs may have originated from XendoU endoribonucleases (XendoUs) forming monomers in eukaryotic cells, that NendoU from arterivirus gained the ability to form dimers, and that the coronavirus variants then evolved the capacity to assemble into hexamers. We further propose that PDCoV nsp15 may be an intermediate in this evolutionary process. Our findings provide a theoretical basis for improving our understanding of NendoU evolution and offer useful clues for designing drugs and vaccines against nidoviruses.
Keywords: viral protein, endoribonuclease, protein purification, oligomerization, protein evolution, endoribonuclease, evolution, nidovirus, oligomerization, PDCoV nsp15
Introduction
Nidoviruses are positive-sense, single-stranded, and enveloped RNA viruses with genome sizes ranging from ∼12.7 to 31.7 kb; these viruses include the families of Coronaviridae, Arteriviridae, Roniviridae, and Mesoniviridae (1–3). Nidoviruses are found in a broad range of hosts. Coronaviruses and arteriviruses mainly infect mammals, causing digestive, respiratory, neurological, and immune-mediated diseases, which result in enormous economic losses and threaten public health (4–8). Coronaviruses are currently classified into four genera; these are Alphacoronavirus, Betacoronavirus, Gammacoronavirus, and Deltacoronavirus. Among the coronaviruses, Deltacoronavirus has the smallest genome, ranging in size from ∼25.4 to 26.6 kb (4). In contrast to coronaviruses, arteriviruses possess the smallest genomes of all nidoviruses; their genomes are ∼12.7 to 15.7 kb in size (9, 10).
Nidoviruses contain 5′- and 3′-untranslated regions (UTRs) at the terminal regions of their genomes, and they utilize a discontinuous negative-stranded RNA synthesis model to generate subgenomic mRNA (8, 12). They have similar genomic organizations in which the two largest overlapping open reading frames (ORF1a and ORF1b) are located within the 5′-proximal two-thirds of the genome, and a ribosomal frame-shifting event is involved in the translation process (13, 14). The viral RNA can be translated into 12 or 13 nonstructural proteins (nsps)3 in arteriviruses and 16 nsps in most coronaviruses to exert different biological functions, such as the formation of double-membrane vesicles (DMVs) and replication–transcription complex (RTCs) formation (3, 15, 16). Viral RNA synthesis has been observed in DMVs of SARS-CoV and MHV (15, 17). Among the nsps found in Nidovirales, NendoU is unique and can both excise single- and dsRNAs and specifically recognize uridylates to produce 2′–3′-cyclic phosphodiester products (5, 18). The DMVs are associated with the production of dsRNA intermediates and can sequester them away from host dsRNA sensors to prevent the activation of the host innate immune response (15). A recent study reported that the coronavirus nsp15 cooperates with the viral replication complex to limit the exposure of viral dsRNA to host dsRNA sensors, thereby enabling the evasion of the host cell immune system (15, 19). In addition, the inhibition of IFN-β induction has been verified in PRRSV nsp11, but further investigations need to be conducted because a cytotoxic effect of WT nsp11 resulting in IFN-β inhibition was not excluded (20, 21). Regardless of whether nsp11 contributes to the decline in innate immune functions, its importance in viral replication has been established (22).
Among the Coronaviridae, SARS-CoV nsp15 was first shown to have NendoU activity in vitro (23); similar findings were then obtained for MHV, HCoV-229E, turkey coronavirus (24–27), and PRRSV (5). However, not all nidoviruses possess NendoU activity; some examples are the insect nidovirus Nam Dinh virus and roniviruses, all of which lack the EndoU domain (1, 28). The active sites of three catalytic residues (two histidines and one lysine) in NendoU are highly conserved in its counterpart, XendoU (19, 24), an endoribonuclease found in eukaryotes that processes the intron-encoded box C/D U16 small nucleolar RNA from its host pre-mRNA and produces 2′–3′-cyclic phosphate termini (29). An intriguing finding regarding NendoU and XendoU is the difference in their requirement for the manganese ion. XendoU is a monomer and requires Mn2+ (29, 30). Except for turkey coronavirus nsp15, Mn2+ is generally essential for the catalytic activity of coronavirus nsp15 (25, 31). However, Mn2+ is dispensable and, to some extent, even inhibits the activity of PRRSV nsp11, implying the existence of different enzymatic mechanisms for NendoU in coronaviruses and arteriviruses (5).
The results of previous studies have indicated that nsp15 is important for nidovirus replication. More recent studies have shown that the replacement of the conserved Asp-6408 in HCoV-229E nsp15 with alanine (Ala) completely abolishes its EndoU activity and viral RNA synthesis, thus rendering the virus unrecoverable. A similar result was obtained in an MHV nsp15 mutant in which Ala was substituted for Asp-324 (24, 26). The results reported by Kang et al. (26) indicated that Asp-324 is an important amino acid for the ionic-bond network because the mutant protein is insoluble when MHV nsp15 was expressed in Escherichia coli. Taken together, these results suggest that the failure of nsp15 to fold correctly may play an indirect role in viral replication by affecting the proteolytic processing of the neighboring components by preventing itself from folding correctly, ultimately leading to a nonviable phenotype (32). Furthermore, these findings regarding coronavirus nsp15 demonstrate its critical function in the evasion of the dsRNA sensors in macrophages and suggest that the disruption of the stability of nsp15 in coronaviruses may be helpful for designing new live-attenuated vaccines (19).
In a previous study, nsp15 was found to be active as a hexamer. This finding was confirmed for the nsp15s in several coronaviruses, including those of MHV, SARS-CoV, HCoV-229E, and turkey coronavirus nsp15 (25, 27, 33, 34). Furthermore, the basic biofunctional unit of SARS-CoV nsp15 was confirmed to be a hexamer (33). Multiple sequence alignments and the site-directed mutagenesis experiments using SARS-CoV nsp15 showed that the N-terminal domain was essential for the oligomerization of nsp15 and that destruction of this motif strongly affected the catalytic activity of the enzyme (33). A previous study of the oligomerization's influence on SARS-CoV nsp15 activity showed that a mutant lacking the 28 N-terminal residues and 11 C-terminal residues was assumed a monomeric rather than a hexameric form, and the stabilization of the catalytic loop (residues 234–249) by the supporting loop (residues 276–295) was required to maintain its enzymatic activity. In addition, the supporting loop on the C-terminal domain was supported by the NTD of the adjacent subunit (13). Therefore, the disruption of the hexameric state of nsp15 can indirectly impact the two reactive histidines, namely His-234 and His-249, thus abolishing its enzymatic activity.
In this study, we performed oligomeric and functional analyses of PDCoV nsp15 to elucidate the mechanism underlying the function of NendoU during nidovirus replication and to identify potential drug targets for controlling PDCoV. Our data indicate that PDCoV nsp15 exists as a dimer and monomer rather than as a hexamer in solution and suggest that the dimeric form of PDCoV nsp15 may represent an intermediate in the evolutionary process of nidoviruses. Moreover, our findings further verified the conservatively catalytic sites among NendoU and provided a new hypothesis for the NendoU evolutionary process based on their different oligomeric forms.
Results
Predicted three-dimensional structure of PDCoV nsp15
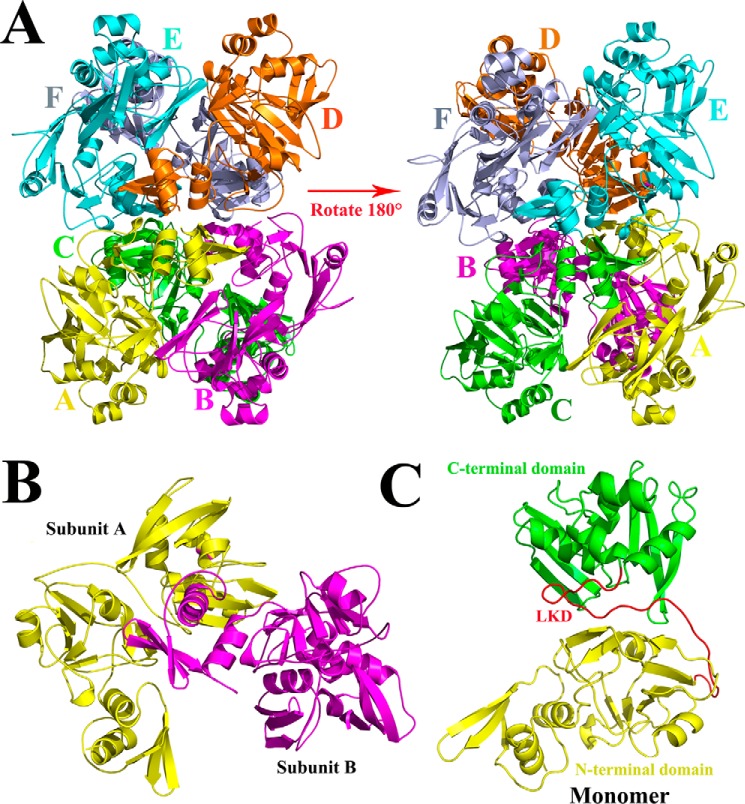
The predicted three-dimensional structure of PDCoV nsp15 is a homohexamer; two monomers interact with each other and form a hexamer with the NTDs facing toward the inside and the C-terminal domains (CTD) facing outward. In addition, the hexamer resembles a triangular scaffold to stand still (Fig. 1A); the basic oligomeric unit is a dimer, and the N-terminal domain is important for the oligomerization (Figs. 1B and 3B). In addition, the monomer consists of three parts: the intact N-terminal domain (Asn-1 to Arg-167); the C-terminal domain (Pro-193 to Gln-327); and a middle linker domain (Tyr-168 to Thr-192) (Fig. 1C).
Figure 1.
Predicted three-dimensional structure of PDCoV nsp15. A, from the side face, the predicted three-dimensional structure of PDCoV nsp15 was built in the SWISS MODEL website. It consists of six monomers that are depicted and marked in PyMOL software with yellow, magenta, green, orange, cyan, and light blue for monomers A–F, respectively. B, two monomers interact with each other closely forming into a dimer with subunit A and subunit B. C, monomer of PDCoV nsp15 consists of three parts: the N-terminal domain (Asn-1 to Arg-167) in yellow, the C-terminal domain (Pro-193 to Gln-327) in green, and a middle linker domain (LKD; Tyr-168 to Thr-192) in red.
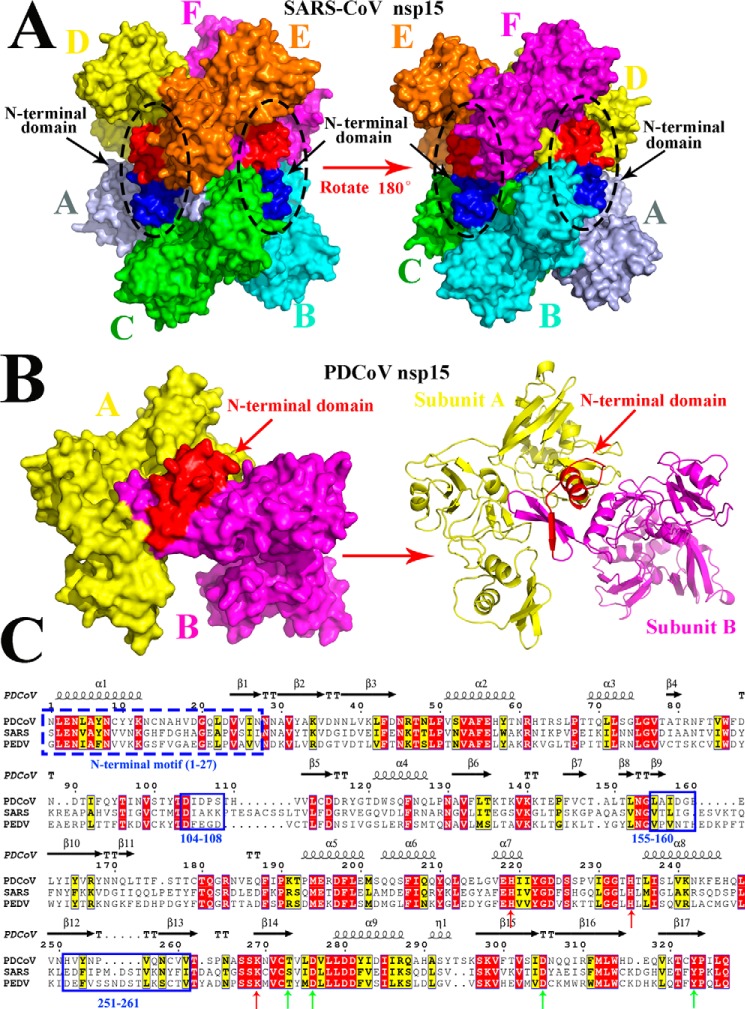
Figure 3.
Predicted residues involved in the dimerization of PDCoV nsp15 determined by sequence and structural alignments. A, structure of SARS-CoV nsp15 (PDB code 2rhb) is a homohexamer with six monomers A (light blue), B (cyan), C (green), D (yellow), E (orange), and F (magenta), and three NTDs are shown in red, and the other three NTDs are shown in blue and are indicated by black arrows. B, predicted three-dimensional structure of PDCoV nsp15 that was built in the SWISS MODEL website. Monomer A (yellow) and monomer B (magenta) form a dimer via the interaction with the N-terminal domain (red), and the right panel is the cartoon formation of PDCoV nsp15 depicted in PyMOL. Monomer A (yellow) and monomer B (magenta) interact with each other through the NTD (red) that are indicated by red arrows. C, amino acid sequence alignment of PDCoV, SARS-CoV, and PDCoV nsp15s. The formal 27 amino acids of these two nsp15s are depicted with a blue dotted line; Asp-104 to Ser-108, Leu-155 to Glu-160, and His-251 to Val-261 residues on PDCoV nsp15 are depicted with the blue solid lines. The conservative sites of three catalytic sites and four binding sites are indicated with red and green arrows, respectively.
Oligomerization of PDCoV nsp15 differs from that of other CoVs nsp15s
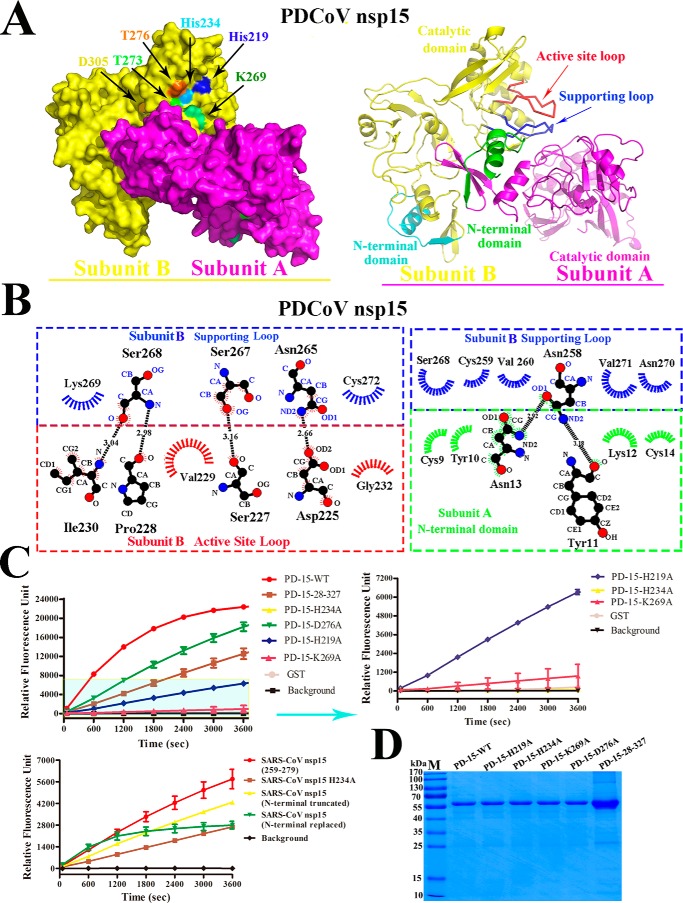
The size-exclusion chromatography results of PDCoV nsp15 show that it is not a hexamer but mainly a monomer and a dimer (Fig. 2, A and B). Thus, PDCoV (Deltacoronavirus), SARS-CoV (Betacoronavirus), and PEDV (Alphacoronavirus), as three representative viruses of the Coronaviridae, were selected among all coronaviruses for nsp15 oligomerization analysis among the whole coronaviruses to compare their different oligomeric stations in vitro. In our work, low production of WT protein was also observed due to its bacterial toxicity, which has been previously reported (20, 33). Previous studies have shown that the key catalytic site histidine does not participate in stabilizing functional oligomerization in coronaviruses and arteriviruses (20, 33). According to the sequence alignment results, the catalytic sites of His-219, His-234, and Lys-269 in PDCoV are highly conserved compared with those in SARS-CoV and PEDV nsp15s. Furthermore, several substrate-binding sites present in PDCoV nsp15 (Thr-273, Asp-276, Asp-305, and Tyr-323) were also conserved at the equivalent sites in SARS-CoV nsp15 (Ser-293, Asp-296, Asp-323, and Tyr-342) (Fig. 3C). According to the conserved catalytic sites, the nsp15 mutants (PDCoV: H219A, H234A, and K269A; PEDV: H241A; and SARS-CoV: H234A) were constructed based on amino acid sequence alignment, and high yields of these proteins were harvested (reached ∼4–8 mg/liter).
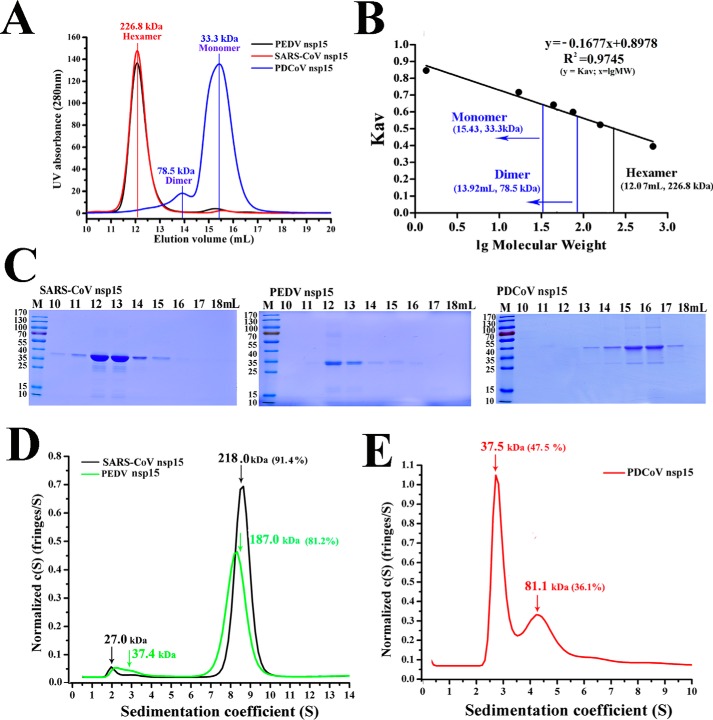
Figure 2.
Oligomerization of PDCoV nsp15 is different from other CoV nsp15s. A, size-exclusion experiment of PDCoV nsp15 (blue), SARS-CoV nsp15 (red), and PEDV nsp15 (black). The calculated molecular masses were determined by fitting to the calibration curve as described in B. B, calculated molecular masses of these nsp15s peaks with the values obtained for known calibration standards (Bio-Rad and GE Healthcare). The calculated molecular mass of nsp15 peaks was determined by fitting to the calibration curve (Kav = volumes of elution (Ves/24)); volumes of elution of 12.07 ml (∼226.8 kDa) in SARS-CoV and PEDV nsp15s (black vertical line) and 13.92 ml (∼78.5 kDa) and 15.43 ml (∼33.3 kDa) in PDCoV nsp15 (blue vertical line) are depicted. C, SDS-PAGE analysis of SARS-CoV nsp15, PEDV nsp15, and PDCoV nsp15. The elution volume is labeled as described in A. Molecular mass markers are shown. D and E, sedimentation velocity analysis of SARS-CoV nsp15 (black), PEDV nsp15 (green), and PDCoV nsp15 (red) with their major peaks of hexamers (∼218.0 and 187.0 kDa), dimers (∼81.1 kDa), and monomers (∼37.5 kDa), respectively. The sedimentation coefficient (s20, w) and the calculated molecular masses are shown in Table 1.
The size-exclusion chromatography results revealed that the oligomeric forms of PDCoV nsp15 were a dimer and a monomer (∼78.5 and 33.3 kDa, respectively). In contrast, both PEDV nsp15 and SARS-CoV nsp15 were hexamers (∼228.6 kDa) (Fig. 2A). The peak elution volumes of PDCoV nsp15 were ∼15.43 and 13.92 ml. PEDV and SARS-CoV nsp15s showed similar peak elution volumes of ∼12.07 ml, which were reflected by the bio-standard protein elution results (Fig. 2B). To further verify the specific distributions of these oligomers, we performed an analytical ultracentrifugation experiment. The hexamer proportions of PEDV and SARS-CoV nsp15s were ∼81.2 and 91.4%, with predicted molecular masses of ∼187.0 and 218.0 kDa, respectively (Fig. 2D). In the case of PDCoV nsp15, dimers and monomers occupied ∼36.1 and 47.5%, with a predicted mass of ∼81.1 and 37.5 kDa, respectively (Fig. 2E). The calculated masses and the sedimentation coefficient (s20, w) data are shown in Table 1. The proteins used in the size-exclusion chromatography experiment were then visualized by SDS-PAGE (Fig. 2C).
Table 1.
Parameters of sedimentation velocity analysis
| Construct | Predicted mass |
AUC |
|||||
|---|---|---|---|---|---|---|---|
| Monomer | Dimer | Hexamer | f/f0 | s20,w | Mass | Percentage | |
| kDa | S | kDa | % | ||||
| SARS-CoV nsp15 (H234A) | 39.2 | 234 | 1.51 | 8.639 | 218.0 | 91.4 | |
| PEDV nsp15 (H241A) | 38.1 | 228 | 1.48 | 8.212 | 187.0 | 81.2 | |
| PDCoV nsp15 (H234A) | 38.2 | 76.4 | 1.40 | 2.88 | 37.5 | 47.5 | |
| 4.824 | 81.1 | 36.1 | |||||
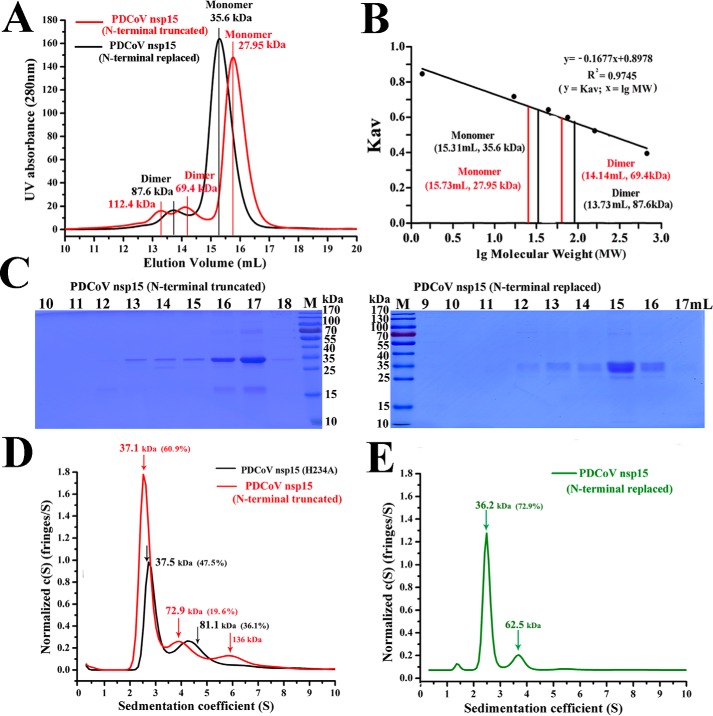
| PDCoV nsp15 (N-terminal truncated) | 35.1 | 71.4 | 1.40 | 2.672 | 37.1 | 60.9 | |
| 4.197 | 72.9 | 19.6 | |||||
| PDCoV nsp15 (N-terminal replaced) | 37.1 | 1.57 | 2.518 | 36.2 | 72.9 | ||
| SARS-CoV nps15Asp-259–Phe-279 | 38.2 | 76.4 | 1.29 | 3.099 | 36.7 | 83.8 | |
| 5.117 | 77.8 | 14.6 | |||||
In conclusion, our result verified that the oligomerization of PDCoV nsp15 was different from that of other CoV nsp15s.
NTD of PDCoV nsp15 is important for its dimerization
The three-dimensional structure of SARS-CV nsp15 (PDB code 2rhb) consists of six homomonomers and forms a compact homohexamer via the interaction of one NTD (1S–27I) with the other CTD (Fig. 3A). The predicted structure of PDCoV nsp15 shows that it is also a homohexamer and that part of the NTD is involved in its higher oligomerization (Figs. 1A and 3B).
To verify that part of the NTD region (1N–27N) participates in the dimerization of PDCoV nsp15, we analyzed a PDCoV nsp15 mutant with its NTD (1N–27N) truncated, called PDCoV nsp15 (N-terminal truncated), and a PDCoV nsp15 NTD (1N–27N) mutant replaced with the SARS-CoV nsp15 NTD (1N-27S), called PDCoV nsp15 (N-terminal replaced) in gel-chromatography experiments. The elution peaks of PDCoV nsp15 (N-terminal truncated) were ∼15.73 and 14.14 ml, with predicted masses of ∼28.0 kDa (monomer) and 69.4 kDa (dimer) (Fig. 4A), respectively, based on the bio-standard protein elution results (Fig. 4B). The main elution volumes of PDCoV nsp15 (N-terminal replaced) were ∼15.31 and 13.73 ml, with predicted masses of ∼35.6 kDa (monomer) and 87.6 kDa (dimer) (Fig. 4A), respectively, which were slightly greater than those of PDCoV nsp15 (H234A) based on the bio-standard protein elution results (Fig. 2B). The AUC results showed that both dimers and monomers were present in PDCoV nsp15 (N-terminal truncated), and they represented ∼19.8% and 61.0% with predicted masses of ∼72.9 and 37.1 kDa, respectively. In addition, few tetramers (∼136.0 kDa) were also observed (Fig. 4D). Compared with those in PDCoV nsp15 (H234A), the percentage of monomers in PDCoV nsp15 (N-terminal truncated) increased by ∼13.5%, and the percentage of dimers decreased by ∼16.3% (Fig. 4D). Additionally, PDCoV nsp15 (N-terminal replaced) had a high proportion of monomers (∼72.9%) with a predicted mass of ∼36.2 kDa and few other oligomers (Fig. 4E). The calculated masses and the sedimentation coefficient (s20, w) data of the AUC are shown in Table 1. The proteins eluted in the size-exclusion chromatography experiment were then visualized by SDS-PAGE (Fig. 4C).
Figure 4.
NTD of PDCoV nsp15 is important for its dimerization. A, size-exclusion experiment with PDCoV nsp15 (N-terminal truncated) (red) and PDCoV nsp15 (N-terminal replaced) (black). The calculated molecular masses are indicated by a red vertical line for the N-terminal truncated mutant with three peak volumes corresponding to predicted masses of ∼112.4, 69.4, and 27.9 kDa; and the predicted masses of N-terminal replaced mutant were ∼87.6 and 35.6 kDa; they were determined by fitting to the calibration curve as described for B. B, calculated molecular masses of the nsp15 protein peaks with the values obtained for known calibration standards (Bio-Rad and GE Healthcare). The calculated molecular mass of nsp15 peaks was determined by fitting to the calibration curve (Kav = volumes of elution (Ves/24)). C, SDS-PAGE analysis of PDCoV nsp15 (N-terminally truncated) and PDCoV nsp15 (N-terminally replaced). The elution volume is labeled as described for A. Molecular mass markers are shown. D and E, sedimentation velocity analysis of PDCoV nsp15 (N-terminally truncated) (red), PDCoV nsp15 (H234A) (black), and PDCoV nsp15 (N-terminally replaced) (green) with their major peaks is depicted in D and E. The sedimentation coefficient (s20, w) and the calculated masses are shown in Table 1.
The fact that PDCoV nsp15 NTD is important for its dimerization has been validated in PDCoV nsp15 (N-terminal truncated). The reason to explain why PDCoV nsp15 is a dimer and monomer but not a hexamer remains unclear because the oligomeric form of PDCoV nsp15 (N-terminal replaced) remains almost the same as that of PDCoV nsp15 (H234A) in vitro, and it suggests that there should be other regions impeding the hexamerization in PDCoV nsp15. Thus, the following experiments were performed.
Region (251H–261V) of PDCoV nsp15 rendering the PDCoV nsp15 fails to form a hexamer
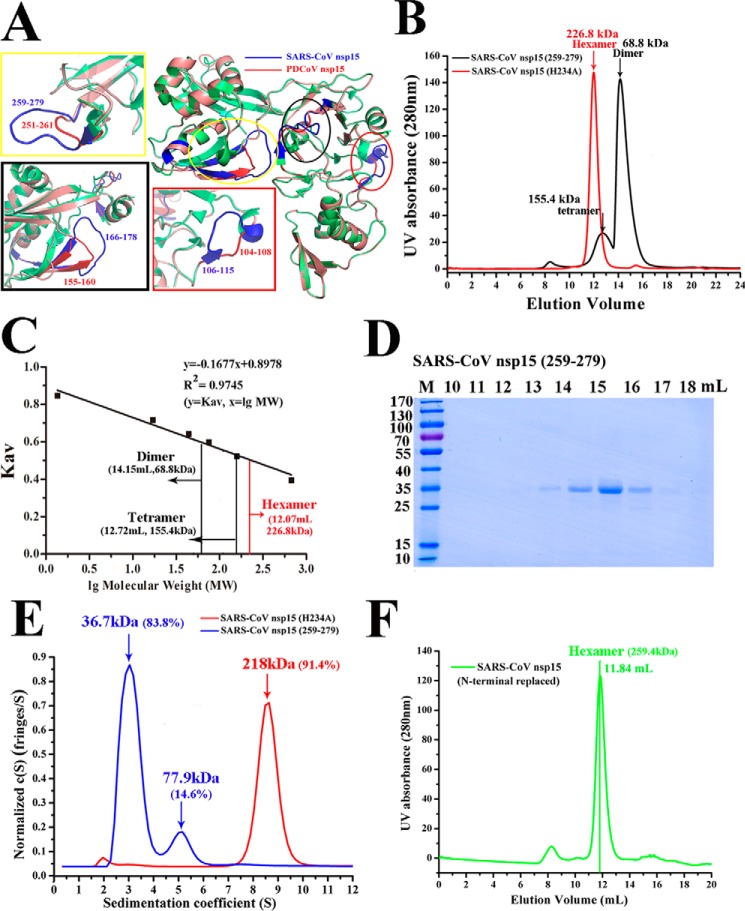
The structural alignment of SARS-CoV and PDCoV nsp15 showed that the Asp-104 to Ser-108, Leu-155 to Glu-160, and His-251 to Val-261 regions of PDCoV nsp15 (Fig. 3C) corresponded to the sequences of the Asp-106 to Ala-115, Thr-166 to Tyr-178, and Gln-259 to Phe-279 regions of SARS-CoV nsp15, respectively (Fig. 5A). In addition, all these three loops formed by SARS-CoV nsp15 are longer than those formed by PDCoV nsp15, especially the loop formed by the region from Gln-259 to Phe-279 (Fig. 5A). Two equivalent-region substitution mutants in SARS-CoV nsp15 were constructed and purified (SARS-CoV nsp15Thr-166–Tyr-178 was not successfully purified). The mutant SARS-CoV nsp15Asp-259–Phe-279 protein was harvested, purified, and used in gel-chromatography experiments. The two peak elution volumes of SARS-CoV nsp15Asp-259–Phe-279 were ∼12.72 ml, corresponding to a tetramer (∼152.8 kDa), and ∼14.15 ml, which was calculated to be a dimer (∼68.8 kDa) (Fig. 5B), as reflected by the bio-standard protein elution results (Fig. 5C). In addition, the peak elution volume of SARS-CoV nsp15 (N-terminal replaced), which was a mutant NTD (1N-27S) of SARS-CoV nsp15 replaced with NTD (1N–27N) of PDCoV nsp15, was 11.84 ml with a predicted mass of ∼259.4 kDa, and it was predicted to be a hexamer (Fig. 5F). The proteins from the size-exclusion chromatography experiment were visualized by SDS-PAGE (Fig. 5D). The AUC experimental results showed that the oligomeric formation completely changed into a mixture of ∼83.8% monomers (∼36.7 kDa), 14.6% dimers (∼77.9 kDa), and some higher-order oligomers in solution (Fig. 5E). The calculated protein mass and sedimentation coefficient (s20, w) data are shown in Table 1.
Figure 5.
Region (His-251–Val-261) of PDCoV nsp15 rendering the PDCoV nsp15 fails to form a hexamer. A, three-dimensional structural alignment of SARS-CoV nsp15 (PDB 2H85) (green) and PDCoV nsp15 (salmon). The predicted residues participating in dimerization are depicted with three ellipses, and they are drawn in rectangles; the residues of Asp-104 to Ser-108, Leu-155 to Glu-160, and His-251 to Val-261 regions of PDCoV nsp15 are highlighted in red and located in red, black, and yellow rectangles, respectively. The corresponding residues of Asp-106 to Ala-115, Thr-166 to Tyr-178, and Gln-259 to Phe-279 regions on SARS-CoV nsp15 are depicted in blue. B, size-exclusion experiment with SARS-CoV nsp15Asp-259–Phe-279 and SARS-CoV nsp15 (H234A). The calculated molecular masses were determined by fitting to the calibration curve as described in C. C, calculated molecular masses of nsp15 protein peaks with the values obtained for known calibration standards (Bio-Rad and GE Healthcare). The calculated molecular mass of nsp15 peaks was determined by fitting to the calibration curve (Kav = volumes of elution (Ves/24)), and the two peak volumes of SARS-CoV nsp15Asp-259–Phe-279 were ∼12.72 and 14.15 ml indicated by black vertical lines with the predicted masses of ∼155.4 and 68.8 kDa, respectively, in B. D, SDS-PAGE analysis of SARS-CoV nsp15Asp-259–Phe-279. E, sedimentation velocity analysis of SARS-CoV nsp15Asp-259–Phe-279 (blue) and SARS-CoV nsp15 (H234A) (red) with their major peaks in E. The sedimentation coefficient (s20, w) and the calculated molecular masses are shown in Table 1. F, size-exclusion experiment result of SARS-CoV nsp15 (N-terminally replaced) (green) with the elution volume of ∼11.84 ml and the calculated molecular mass of ∼259.4 kDa were determined by fitting to the calibration curve as described in C.
In conclusion, this completely oligomeric change found in SARS-CoV nsp15Asp-259–Phe-279 indicated that the region (251H–261V) of PDCoV nsp15 could be a reason to explain why PDCoV nsp15 fails to form a hexamer. In addition, both of the oligomeric forms in SARS-CoV nsp15 (N-terminal replaced) and PDCoV nsp15 (N-terminal replaced) remain the same as those of SARS-CoV nsp15 (H234A) and PDCoV nsp15 (H234A), which is additional evidence to prove that NTD is important for their dimerization but not the hexamerization.
PDCoV nsp15 is active as a dimer
In arterivirus, the biofunctional unit of the PRRSV NendoU is a dimer. Thus, the unit differs from that of MHV and SARS-CoV NendoUs; both of them function as hexamers (27, 33). The predicted structure of PDCoV nsp15 contains three conserved catalytic sites (His-219, His-234, and Lys-269) and four binding sites (Thr-273, Thr-276, Asp-305, and Tyr-323) that form a pocket for RNA substrate binding (Fig. 6A), which was also reflected by the sequence alignment results (Fig. 3C). Furthermore, the interaction between the two monomers in PDCoV nsp15 was depicted; the NTD on monomer A interacts with the supporting loop on CTD of monomer B. The interactions between the Asp-225, Ser-227, Pro-228, Val-229, Ile-230, and Gly-232 residues on the active loop and the Asn-265, Ser-267, Ser-268, Lys-269, and Cys-272 residues on the supporting loop are depicted. Furthermore, these extensive interactions between the supporting loop and the adjacent monomer NTD were also analyzed and determined in LIGPLOT (35). Interactions between the residues Asn-258, Cys-259, Val-260, Ser-268, Asn-270 and Val271 from the supporting loop and residues Lys-9, Tyr-10, Tyr-11, Lys-12, Asn-13, and Cys-14 of the adjacent monomer NTD are observed (Fig. 6B). To further verify the enzymatic activity of these sites, FRET experiments were performed, and the results confirmed that these positions were the functional sites on PDCoV nsp15. The assay results showed the decreased activity of all PDCoV nsp15 mutants (H219A, H234A K269A, D276A, and the N-terminal truncation). PDCoV nsp15 mutants (T273A, D305A, and Y323A) could not be obtained due to insolubility. Excision by the PDCoV nsp15 mutants (H219A, H234A, and the N-terminal truncated) was dramatically reduced compared with that by WT PDCoV nsp15, and H219A retained approximately one-third of the activity of WT PDCoV nsp15. In addition, the D276A mutant showed a smaller decrease in catalytic activity compared with other mutants. Our experimental results can be explained based on the positions of the predicted catalytic sites within the predicted three-dimensional structure of PDCoV nsp15 (Fig. 6A). The H234A mutant shows the greatest activity decrease among all of these mutants because it lies in the center of the enzymatic groove (Fig. 6A). Therefore, compared with the other two catalytic sites, His-234 may play an important role in the interaction with the RNA substrate. The loss of activity observed in the PDCoV nsp15 (N-terminal truncated) mutant may be due to the dimeric disruption of PDCoV nsp15, which may affect its normal biological function (Fig. 6B). Among the SARS-CoV nsp15 mutants, the nsp15Asp-259–Phe-279 mutant displayed the highest enzymatic activity, and its activity decreased by three-fourths compared with that of WT PDCoV nsp15. The second most active mutant was the N-terminal truncated mutant, followed by the N-terminal replaced mutant, and then the H234A mutant (Fig. 6C). The proteins from the enzymatic assay were then visualized by SDS-PAGE (Fig. 6D).
Figure 6.
NendoU activity of PDCoV nsp15 and SARS-CoV nsp15. A, predicted structure of dimeric PDCoV nsp15. The subunit A (magenta) and subunit B (yellow) are tightly interacted with each other, and the catalytic sites of His-234, His-219, and Lys-269 and binding sites of Thr-273, Asp-276, and Asp-305 are marked with arrows with the cyan, blue, green, light green, orange, and yellow, respectively. The right panel is the cartoon picture of dimeric PDCoV nsp15 with subunit A (magenta) and B (yellow) interactions via the interplay of NTD (green) on subunit A and the supporting loop (Asn-257 to Cys-273) in blue to stabilize the active-site loop (His-219 to His-234) in red on subunit B. The NTD of subunit B is in cyan. B, detailed molecular interactions of the supporting loop (blue) and the active-site loop (red) on subunit B with the NTD (green) on subunit A were determined using LIGPLOT. Carbon, oxygen, and nitrogen atoms are shown as black, red, and blue circles, respectively. Hydrogen bonds are shown with black dashed lines between the donor and acceptor atoms with the bond distance. Hydrophobic interactions are shown by arcs with spokes in blue for the supporting loop, red for the active site loop on subunit A, and green for the NTD on subunit B, which are radiating toward the atoms with which they interact. C, FRET-based enzyme activity experiment. The enzymatic activity of the WT PDCoV nsp15 and mutants (H219A, H234A, K269A, D276A, and N-terminally truncated) is depicted with different colors. The values of the triplicate experiment results are shown. The enzymatic activity of SARS-CoV nsp15 mutants (H234A, N-terminally truncated, N-terminally replaced, and Asp-259–Phe-279) is depicted. The values of the triplicate experiment results are shown. D, SDS-PAGE analysis of PDCoV nsp15 mutants.
Taken together, the enzymatic assay results further verified that the NTD and His-251 to Val-261 regions were important for the dimerization of PDCoV nsp15 and that disruption of its dimerization could decrease its enzymatic activity, which suggests that the biofunctional unit of PDCoV nsp15 may be a dimer in vitro. In addition, the predicted catalytic and binding sites on PDCoV nsp15 could impact its enzymatic activity to different degrees.
Presumption for the evolution of Nidovirales NendoU
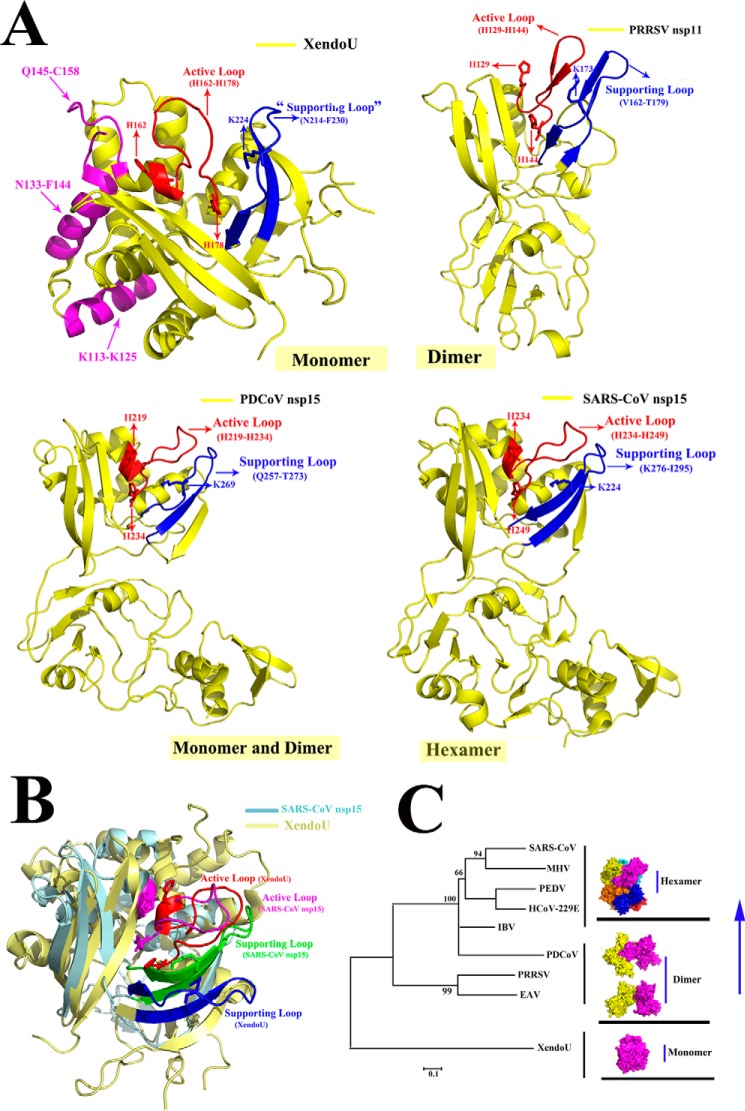
As the counterpart of NendoU, XendoU has been reported to be a monomer (36). According to the three-dimensional structural alignment of XendoU and SARS-CoV nsp15 (Fig. 7B), a similar structural feature on the CTD was found and reflected by the highly conserved catalytic sites of the active loop, which have also been found in other NendoUs, such as PRRSV nsp11 and PDCoV nsp15 (Fig. 7A). We hypothesize that their active loops are stabilized by the supporting loops in PRRSV nsp11, PDCoV, and SARS-CoV nsp15s. However, XendoU, could be active as a monomer via the undergirding support of residues (Lys-113 to Lys-125 and Asn-133 to Phe-144), and the additional upper residues of Gln-145 to Cys-158 could also provide stability to hold up the active loop (His-162 to His-178) (13); these interactions and the predicted supporting loop on XendoU are depicted (Fig. 7A). In PRRSV nsp11, the active loop (His-129 to His-144) and the supporting loop (Val-162 to Thr-179) stand out from the center compared with these features in XendoU; thus, the supporting loop alone was not sufficient for its stabilization, and dimerization was required for the supporting loop to make PRRSV nsp11 functional. In addition, the same condition was found in PDCoV nsp15 with the interaction between the active loop (His-219 to His-234) and the supporting loop (Gln-257 to Thr-273) (Fig. 7A). In addition, the active loop (His-234 to His-249) and the supporting loop (Lys-276 to Ile-295) were also found in SARS-CoV nsp15. However, PDCoV nsp15 was an exception for being composed of dimers and monomers among other nsp15s (Fig. 2A).
Figure 7.
Possible model of the evolutionary process for the nidorivirus NendoU. A, monomeric forms of the three-dimensional structures of XendoU (PDB code 2C1W), PRRSV nsp11 (PDB code 5DA1), PDCoV nsp15 (predicted structure), and SARS-CoV nsp15 (PDB code 2H85) are depicted in yellow, and the catalytic sites of XendoU (His-162, His-178, and Lys-224), PRRSV nsp11 (His-129, His-144, and Lys-173), PDCoV nsp15 (His-219, His-234, and Lys-269), and SARS-CoV nsp15 (His-234, His-249, and Lys-289) are indicated with red arrows. The common active loop and supporting loop on XendoU (His-162 to His-178 and Asn-214 to Phe-230), PRRSV nsp11 (His-129 to His-144 and Val-162 to Thr-179), PDCoV nsp15 (His-219 to His-234 and Gln-257 to Thr-273), and SARS-CoV nsp15 (His-234 to His-249 and Lys-276 to Ile-295) are indicated with red and blue, respectively. Moreover, residues of Lys-113 to Lys-125, Asn-133 to Phe-144, and Gln-145 to Cys-158 on XendoU are shown in magenta. B, three-dimensional structural alignment of SARS-CoV nsp15 (light blue) and XendoU (light yellow) are depicted. The active loop and supporting loop of SARS-CoV nsp15 are shown in magenta and green, respectively. The active loop and supporting loop of XendoU are shown in red and blue, respectively. C, XendoU is a monomer shown in magenta. The dimeric nsp11 is in yellow and magenta, and the hexameric Alpha-, Beta-, and Gammacoronaviruses nsp15s, including six monomers, are shown in different colors. PDCoV nsp15 is an intermediate and depicted as a dimer in yellow and magenta. The phylogenetic tree was analyzed using the distance-based neighbor-joining method in the MEGA package. The different subgenotypes are indicated.
Thus, we hypothesize that PDCoV nsp15 is an intermediate during the NendoU evolutionary process of the development of high-order oligomerization to better exert its function via structural analysis (Fig. 7C).
Discussion
The newly emerged PDCoV belongs to the Deltacoronavirus family, whose members can cause similar clinical symptoms to those of PEDV. The highest mortality rate is observed in suckling and weaned pigs with no effective vaccination strategies proposed (4, 6). The large-scale outbreak of PDCoV first emerged in Ohio and Indiana in 2014, and then PDCoV immediately spread across the United States, Canada, China, and Thailand, causing serious economic losses to the pork industry worldwide (37, 38). In addition, few studies on PDCoV nsp15 have been reported; thus, to clarify its biofunction, three-dimensional structure or oligomeric distribution could help us to better understand its exact function in coronaviruses.
NendoU is highly conserved for specifically recognizing the U-pyrimidine and producing 3′–5′-phosphate products (24). In addition, the conserved biofunction of NendoU relies on the conserved catalytic sites (two histidines and one lysine) (Fig. 6A). The stability of the catalytic center in NendoU is the primary requirement to maintain its natural enzymatic capacity, and any degrees of its structural disruption can directly or indirectly destroy its enzymatic activity. The basic biofunctional unit in SARS-CoV nsp15 is a hexamer (33), and it includes a dimer of trimers with the NTDs in the center and the CTDs facing outward, allowing them to possess six active sites to better exert their function in SARS-CoV and MHV nsp15 (27, 31, 39). In addition, for PRRSV nsp11, the dimer is an active unit, and the disruption of its dimerization can markedly impair its endoribonuclease activity (20). Here, we found that both monomers and dimers were present in PDCoV nsp15. In addition, decreased enzymatic activity was observed in the PDCoV nsp15 (N-terminal truncation) mutant (Fig. 6C), which indicates that it functions as a dimer. Furthermore, the SARS-CoV nsp15Asp-259–Phe-279 mutant showed a completely oligomeric change from hexamers to a mixture of monomers and dimers, with the monomer predominating in solution. However, SARS-CoV nsp15 (N-terminal replaced) still remains as a hexamer (Fig. 5F), which needs to be further verified in vivo via reverse genetics system. An alternative hypothesis that the residues required for hexamer formation were lost or mutated during the evolutionary process in PDCoV nsp15 is possible, because the region (His-251–Val-261) is 10 amino acids shorter than the equivalent region (Asp-259–Phe-279) on SARS-CoV nsp15. In conclusion, the disruption of the oligomerization can impair the normal enzymatic activity of NendoU. In addition, further investigations of nsp15s need to be performed to verify whether their different oligomeric forms impact their biofunction or the interaction with the RTCs and other nsps in vivo by using the reverse genetics or replicon systems. In this study, we attempted to screen the PDCoV nsp15 crystal under various conditions. Unfortunately, we were unable to obtain the structure.
As the counterpart of NendoU, XendoU is also an endoribonuclease and was purified from Xenopus laevis oocyte nuclear extracts (29). A previous study revealed that the oligomeric state of XendoU was monomeric, and the three-dimensional structure of XendoU showed that it adopted a different oligomeric pattern to that of NendoU. The support of residues (Lys-113–Lys-125 and Asn-133–Phe-144) undergirding and the additional upper residues (Gln-145–Cys-158) could also provide stability to hold up the active loop (His-165–His-178) with the predicted supporting loop adjacent to its active loop to form into a compact structure, with a globular shape (13). In addition, the highly conserved catalytic sites in XendoU (His-162, His-178, and Lys-224), are also found in PRRSV nsp11, PDCoV, and SARS-CoV nsp15s on the three-dimensional structural level (Fig. 7A), serving as additional evidence of their close relationship at an evolutionary aspect. The conservative catalytic residues may provide a novel target for drug design against nidoviruses in the future.
Regarding the origin of NendoU, we hypothesize that nidoriviruses may acquire this gene from XendoU in eukaryotic cells, and this gene may evolve in the virus to better adapt to the outside environment, a possibility that has already been put forward in previous studies (22, 40). The different oligomeric characteristics of XendoU and NendoU further support this hypothesis. In eukaryotic cells, XendoU exists as a monomer, and in the arterivirus PRRSV nsp11, it exists mainly as a dimer. In PDCoV nsp15, we found it was a mixture of monomers and dimers, and in several Alpha-, Beta-, and Gammacoronavirus nsp15s, the proteins have been identified as hexamers (25, 27, 33, 34). Furthermore, according to the molecular clock analysis, the times to which these four coronaviruses could be traced were ∼2400 BCE, 2800 BCE, 3300 BCE, 3000 BCE, corresponding to Alphacoronavirus, Gammacoronavirus, Betacoronavirus, and Deltacoronavirus, respectively (4). PDCoV is the second oldest among the four genera. This result reflects the possible evolutionary course among all coronavirus NendoUs. PDCoV nsp15 is the intermediate status in the transition process of forming a hexamer from the dimeric form in PRRSV nsp11, and all the NendoUs have the same ancestor, namely XendoU (Fig. 7C). In addition, a relevant study showed that approximately half of the proteins could interact with other copies of themselves and assemble into homomeric complexes, which was often assumed to be a functionally beneficial result of evolutionary selection (11).
Experimental procedures
Three-dimensional structure prediction
The predicted three-dimensional structure of PDCoV nsp15 was accomplished in the SWISS-MODEL website (https://swissmodel.expasy.org/), and then the structural figures were generated with the PyMOL molecular visualization system (Schrödinger).
Gene cloning and plasmid constructs
The genes encoding nsp15s were cloned from the PDCoV strain CHN-HN-2014 (GenBankTM accession number KT336560) and the PEDV FJZZ strain (GenBankTM accession number KC140102.1). A codon-optimized SARS-CoV nsp15 gene (GenBankTM accession number NP_828872.1; gi:29837507) was synthesized by GenScript Corp. (GenScript, Nanjing, China). The PDCoV nsp15 mutants (H234A, N-terminal truncated, N-terminal replaced), the SARS-CoV nsp15 mutants (H234A, N-terminal truncated, and N-terminal replaced), and the PEDV nsp15 mutant (H241A) used in the analytical ultracentrifugation and size-exclusion chromatography experiments were separately cloned into the pET-42b(+) vector with the NdeI and XhoI restriction sites, and a His6 tag was added at the C terminus. The alanine-substituted PDCoV nsp15 mutants (H219A, H234A, K269A, T273A, D276A, D305A, and N-terminal truncated, each with a C-terminal GST tag) were cloned into the pGEX-6p-1 vector by homologous recombination with a C-terminal GST tag for the enzymatic assays. In addition, pET-30a(+) and pET-28a(+) containing the WT PDCoV nsp15 gene with a C-terminal His6 tag were transformed into E. coli DE3 to obtain the WT PDCoV nsp15. For the oligomerization experiment, several important sequences of Phe-145–Ile-164 and His-251–Val-261 on PDCoV nsp15 were replaced by the equivalent sequences of Thr-152–Phe-179 and Asp-259–Phe-279 on SARS-CoV nsp15 to create SARS-CoV nsp15Thr-152–Phe-179 and SARS-CoV nsp15Asp-259–Phe-279, respectively. These constructs, each with a His6 tag at the C terminus, were cloned into pET-42b(+) after the digestion of the plasmid with the NdeI and HindIII restriction enzymes. All of the point mutations and sequence substitution mutations were constructed by the overlapping PCR technique and were confirmed by the GenScript Co.
Protein expression and purification
The recombinant plasmids described above were transformed into the E. coli strain Trans BL21 (DE3) pLysS (Beijing TransGen Biotech, Co., Ltd.). The cells transformed with plasmids encoding proteins with GST tags were cultured at 37 °C in LB medium containing 50 μg/ml ampicillin. LB medium containing 50 μg/ml kanamycin was used for the cells expressing proteins with His6 tags. When the OD600 value of the cell density reached 0.6–0.8, the cells were induced with 0.8 mm isopropyl β-d-1-thiogalactopyranoside. To maximize the protein yield, the cells expressing proteins with GST tags were cultured at 37 °C for an additional 4 h. Protein expression and purification were conducted according to our previously reported procedure (19).
The harvested proteins were further purified on a Superdex 200 gel-filtration column (GE Healthcare) and were washed with buffer B2 (20 mm Tris-HCl and 200 mm NaCl, pH 7.4) when the volume of the filtered protein was less than 2.0 ml in the condensing column for centrifugation at 4 °C. The proteins were flash-frozen in liquid nitrogen and stored at −80 °C until used in biochemical analyses. The concentrations of PDCoV, SARS-CoV, and PEDV nsp15 mutant proteins were determined based on the absorbance values of the samples at 280 nm (A280) using a DS-11 FX+ spectrophotometer combined with a highly sensitive 1-μl UV-visible absorbance spectrophotometer (DeNovix, Inc.).
Size-exclusion chromatography assay
The PDCoV nsp15 mutants (H234A, N-terminal truncated, and N-terminal replaced), the SARS-CoV nsp15 mutants (H234A, N-terminal truncated, N-terminal replaced, and Asp-259–Phe-279), and the PEDV nsp15 mutant (H241A) were purified through a nickel-nitrilotriacetic acid high-affinity filter and then passed through a HiLoad Superdex 200 gel-filtration column (GE Healthcare) in elution buffer (20 mm Tris-HCl, 200 mm NaCl, pH 7.4) at a flow rate of 0.8 ml/min at 4 °C. For the analysis of oligomerization, ∼1 mg of each protein was subjected to chromatography on a Superdex 200 10/300 GL column in the same elution buffer described above at a flow rate of 0.5 ml/min at 4 °C, and the eluted proteins were then assessed by SDS-PAGE analysis. Size-exclusion standards were separated on the column following our previously reported procedure (19). The obtained data were analyzed using Origin 8.0 software. The predicted weight-averaged molar masses were calculated using DNASTAR (version 7.1) software.
Analytical ultracentrifugation assay
To confirm the masses of the individual proteins, sedimentation velocity analysis was conducted in an XL-A model centrifuge (Proteome Lab) at 18 °C and 45,000 rpm in 400-μl double-sector cells. The sedimentation boundary was monitored every 3 min at a wavelength of 280 nm, resulting in 110 scans. The collected data were analyzed using Sedfit software with the model-based distribution of Lamm equation solutions c(s). The data obtained from the size-exclusion chromatography experiments and sedimentation velocity analysis were transformed into curves for analysis in Origin 8.0 software. DNASTAR (version7.1) software was used for protein weight-averaged molar mass prediction.
Enzymatic activity assay
The endoribonuclease activity assay was performed as follows: the WT PDCoV nsp15 and mutant proteins (H219A, H234A, K269A, D276A, and N-terminal truncated) with GST tags at their C-terminal ends and the GST protein as a negative control, each at a concentration of 2 μm, were mixed with 1 μm RNA substrate (5′,6-carboxyfluorescein-dA-rU-dA-dA-6-carboxy-N,N,N′,N′-tetramethylrhodamine-3′, purchased from GenScript Corp.) (32) in the reaction buffer (50 mm KCl, 50 mm HEPES, pH 7.5), 5 mm MnCl2, and 1 mm DTT dissolved in water with 0.1% diethyl pyrocarbonate) and incubated at 25 °C for 30 min. The same protocol was used to assess the endoribonuclease activity of the SARS-CoV nsp15 mutants (H234A, N-terminal truncated, N-terminal replaced, and Asp-259–Phe-279) with the His6 tag. The CFX96 TouchTM Real-Time PCR Detection System (Bio-Rad) was used to quantify the 5′,6-carboxyfluorescein value of each protein for 30 min at wavelengths ranging from 492 to 518 nm every 30 s. Each protein was assayed three times, and the obtained data were analyzed via GraphPad Prism software 5.0 (GraphPad Software Inc., CA). The values obtained in the triplicate measurements are shown. All of the proteins tested in this assay were also analyzed by SDS-PAGE.
Sequence alignment and phylogenetic tree analysis
The genes encoding SARS-CoV (JF29292), PEDV (KU558702), and PDCoV nsp15s (KP757891) were separately cloned from the full-length genomes of these three viruses, and the amino acid prediction results were obtained by analyzing the gene sequences using DNASTAR (version 7.1) software. The amino acid sequences of SARS-CoV, PEDV, and PDCoV nsp15s were prealigned on the website server of Clustal Omega (https://www.ebi.ac.uk/Tools/msa/clustalo/)4 (41), and the resulting file was then uploaded to the ESPript 3 website (http://espript.ibcp.fr)4 (42) to produce the final alignment result. MEGA 6.0 software was used to build the phylogenetic tree to visualize the evolutionary relationships between Nidovirales and Coronaviridae via the distance-based neighbor-joining method. Bootstrap values were calculated from 1000 replicates of the alignment. The different subgenotypes are indicated.
Nucleotide sequence accession numbers
The NCBI accession numbers of the sequences referred in this study are listed as follows: PDCoV (KP757891), SARS-CoV (SARS coronavirus wtic-MB, KF514423), PEDV (AIM47753), HCoV-229E (human coronavirus 229E, AGT21366), transmissible gastroenteritis virus (virulent Purdue, ABG89333), MHV (porcine respiratory and reproductive syndrome virus strain WUH3, ADO33722), equine arteritis virus (NP_705592), simian hemorrhagic fever virus (AHH54245), lactate dehydrogenase-elevating virus (AAA74104), and XendoU (NP_001081040.1).
Author contributions
A. Z. and Y. S. data curation; A. Z. software; A. Z. and Y. S. formal analysis; A. Z. validation; A. Z. visualization; A. Z. writing-original draft; A. Z., Z. S., J. S., and Q. X. project administration; Y. S. and G. P. supervision; Y. S. and G. P. funding acquisition; Y. S. and G. P. writing-review and editing; G. W. methodology; L. F., S. X., Z. F. F., and G. P. resources.
Acknowledgment
We thank research associates at the Center for Protein Research (CPR), Hua Zhong Agricultural University, for technical support.
This work was supported by National Key R&D Plan of China Grant 2018YFD0500100, National Natural Science Foundation of China Grants 31702249 and 31722056, China Postdoctoral Science Foundation Grant 2017M612484, and Natural Science Foundation of Hubei Province of China Grant 2016CFA069. The authors declare that they have no conflicts of interest with the contents of this article.
Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party hosted site.
- nsp
- nonstructural protein
- NendoU
- nidovirus endoribonuclease
- SARS-CoV
- severe acute respiratory syndrome coronavirus
- HCoV
- human coronavirus
- PDCoV
- porcine Deltacoronavirus
- NTD
- N-terminal domain
- DMV
- double-membrane vesicle
- RTC
- replication–transcription complex
- XendoU
- XendoU endoribonuclease
- PDB
- Protein Data Bank
- CTD
- C-terminal domain
- PEDV
- porcine epidemic diarrhea virus
- PRRSV
- porcine respiratory and reproductive syndrome virus
- IFN
- interferon
- MHV
- murine hepatitis virus
- AUC
- analytical ultracentrifugation assay.
References
- 1. Nga P. T., Parquet Mdel C., Lauber C., Parida M., Nabeshima T., Yu F., Thuy N. T., Inoue S., Ito T., Okamoto K., Ichinose A., Snijder E. J., Morita K., and Gorbalenya A. E. (2011) Discovery of the first insect nidovirus, a missing evolutionary link in the emergence of the largest RNA virus genomes. PLoS Pathog. 7, e1002215 10.1371/journal.ppat.1002215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Vasilakis N., Guzman H., Firth C., Forrester N. L., Widen S. G., Wood T. G., Rossi S. L., Ghedin E., Popov V., Blasdell K. R., Walker P. J., and Tesh R. B. (2014) Mesoniviruses are mosquito-specific viruses with extensive geographic distribution and host range. Virol. J. 11, 97 10.1186/1743-422X-11-97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cong Y., Verlhac P., and Reggiori F. (2017) The interaction between nidovirales and autophagy components. Viruses 9, E182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Woo P. C., Lau S. K., Lam C. S., Lau C. C., Tsang A. K., Lau J. H., Bai R., Teng J. L., Tsang C. C., Wang M., Zheng B. J., Chan K. H., and Yuen K. Y. (2012) Discovery of seven novel mammalian and avian coronaviruses in the genus Deltacoronavirus supports bat coronaviruses as the gene source of Alphacoronavirus and Betacoronavirus and avian coronaviruses as the gene source of Gammacoronavirus and Deltacoronavirus. J. Virol. 86, 3995–4008 10.1128/JVI.06540-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Nedialkova D. D., Ulferts R., van den Born E., Lauber C., Gorbalenya A. E., Ziebuhr J., and Snijder E. J. (2009) Biochemical characterization of arterivirus nonstructural protein 11 reveals the nidovirus-wide conservation of a replicative endoribonuclease. J. Virol. 83, 5671–5682 10.1128/JVI.00261-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. McCluskey B. J., Haley C., Rovira A., Main R., Zhang Y., and Barder S. (2016) Retrospective testing and case series study of porcine Deltacoronavirus in U.S. swine herds. Prev. Vet. Med. 123, 185–191 10.1016/j.prevetmed.2015.10.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cao J., and Zhang X. (2012) Comparative in vivo analysis of the Nsp15 endoribonuclease of murine, porcine and severe acute respiratory syndrome coronaviruses. Virus Res. 167, 247–258 10.1016/j.virusres.2012.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Balasuriya U. B., and Carossino M. (2017) Reproductive effects of arteriviruses: equine arteritis virus and porcine reproductive and respiratory syndrome virus infections. Curr. Opin. Virol. 27, 57–70 10.1016/j.coviro.2017.11.005 [DOI] [PubMed] [Google Scholar]
- 9. Snijder E. J., Kikkert M., and Fang Y. (2013) Arterivirus molecular biology and pathogenesis. J. Gen. Virol. 94, 2141–2163 10.1099/vir.0.056341-0 [DOI] [PubMed] [Google Scholar]
- 10. Guo Z., Chen X. X., Li R., Qiao S., and Zhang G. (2018) The prevalent status and genetic diversity of porcine reproductive and respiratory syndrome virus in China: a molecular epidemiological perspective. Virol. J. 15, 2 10.1186/s12985-017-0910-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bergendahl L. T., and Marsh J. A. (2017) Functional determinants of protein assembly into homomeric complexes. Sci. Rep. 7, 4932 10.1038/s41598-017-05084-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Di H., McIntyre A. A., and Brinton M. A. (2018) New insights about the regulation of Nidovirus subgenomic mRNA synthesis. Virology 517, 38–43 10.1016/j.virol.2018.01.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Joseph J. S., Saikatendu K. S., Subramanian V., Neuman B. W., Buchmeier M. J., Stevens R. C., and Kuhn P. (2007) Crystal structure of a monomeric form of severe acute respiratory syndrome coronavirus endonuclease Nsp15 suggests a role for hexamerization as an allosteric switch. J. Virol. 81, 6700–6708 10.1128/JVI.02817-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Brierley I., Boursnell M. E., Binns M. M., Bilimoria B., Blok V. C., Brown T. D., and Inglis S. C. (1987) An efficient ribosomal frame-shifting signal in the polymerase-encoding region of the coronavirus IBV. EMBO J. 6, 3779–3785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Knoops K., Kikkert M., Worm S. H., Zevenhoven-Dobbe J. C., van der Meer Y., Koster A. J., Mommaas A. M., and Snijder E. J. (2008) SARS-coronavirus replication is supported by a reticulovesicular network of modified endoplasmic reticulum. PLoS Biol. 6, e226 10.1371/journal.pbio.0060226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chen Q., Fang L., Wang D., Wang S., Li P., Li M., Luo R., Chen H., and Xiao S. (2012) Induction of autophagy enhances porcine reproductive and respiratory syndrome virus replication. Virus Res. 163, 650–655 10.1016/j.virusres.2011.11.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gosert R., Kanjanahaluethai A., Egger D., Bienz K., and Baker S. C. (2002) RNA replication of mouse hepatitis virus takes place at double-membrane vesicles. J. Virol. 76, 3697–3708 10.1128/JVI.76.8.3697-3708.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bhardwaj K., Sun J., Holzenburg A., Guarino L. A., and Kao C. C. (2006) RNA recognition and cleavage by the SARS coronavirus endoribonuclease. J. Mol. Biol. 361, 243–256 10.1016/j.jmb.2006.06.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Deng X., Hackbart M., Mettelman R. C., O'Brien A., Mielech A. M., Yi G., Kao C. C., and Baker S. C. (2017) Coronavirus nonstructural protein 15 mediates evasion of dsRNA sensors and limits apoptosis in macrophages. Proc. Natl. Acad. Sci. U.S.A. 114, E4251–E4260 10.1073/pnas.1618310114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Shi Y., Li Y., Lei Y., Ye G., Shen Z., Sun L., Luo R., Wang D., Fu Z. F., Xiao S., and Peng G. (2016) A dimerization-dependent mechanism drives the endoribonuclease function of porcine reproductive and respiratory syndrome virus nsp11. J. Virol. 90, 4579–4592 10.1128/JVI.03065-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Shi X., Wang L., Li X., Zhang G., Guo J., Zhao D., Chai S., and Deng R. (2011) Endoribonuclease activities of porcine reproductive and respiratory syndrome virus nsp11 was essential for nsp11 to inhibit IFN-β induction. Mol. Immunol. 48, 1568–1572 10.1016/j.molimm.2011.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Posthuma C. C., Nedialkova D. D., Zevenhoven-Dobbe J. C., Blokhuis J. H., Gorbalenya A. E., and Snijder E. J. (2006) Site-directed mutagenesis of the Nidovirus replicative endoribonuclease NendoU exerts pleiotropic effects on the arterivirus life cycle. J. Virol. 80, 1653–1661 10.1128/JVI.80.4.1653-1661.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bhardwaj K., Guarino L., and Kao C. C. (2004) The severe acute respiratory syndrome coronavirus Nsp15 protein is an endoribonuclease that prefers manganese as a cofactor. J. Virol. 78, 12218–12224 10.1128/JVI.78.22.12218-12224.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ivanov K. A., Hertzig T., Rozanov M., Bayer S., Thiel V., Gorbalenya A. E., and Ziebuhr J. (2004) Major genetic marker of nidoviruses encodes a replicative endoribonuclease. Proc. Natl. Acad. Sci. U.S.A. 101, 12694–12699 10.1073/pnas.0403127101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Cao J., Wu C. C., and Lin T. L. (2008) Turkey coronavirus non-structure protein Nsp15–an endoribonuclease. Intervirology 51, 342–351 10.1159/000175837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kang H., Bhardwaj K., Li Y., Palaninathan S., Sacchettini J., Guarino L., Leibowitz J. L., and Kao C. C. (2007) Biochemical and genetic analyses of murine hepatitis virus Nsp15 endoribonuclease. J. Virol. 81, 13587–13597 10.1128/JVI.00547-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Xu X., Zhai Y., Sun F., Lou Z., Su D., Xu Y., Zhang R., Joachimiak A., Zhang X. C., Bartlam M., and Rao Z. (2006) New antiviral target revealed by the hexameric structure of mouse hepatitis virus nonstructural protein Nsp15. J. Virol. 80, 7909–7917 10.1128/JVI.00525-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lauber C., Ziebuhr J., Junglen S., Drosten C., Zirkel F., Nga P. T., Morita K., Snijder E. J., and Gorbalenya A. E. (2012) Mesoniviridae: a proposed new family in the order Nidovirales formed by a single species of mosquito-borne viruses. Arch. Virol. 157, 1623–1628 10.1007/s00705-012-1295-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Laneve P., Altieri F., Fiori M. E., Scaloni A., Bozzoni I., and Caffarelli E. (2003) Purification, cloning, and characterization of XendoU, a novel endoribonuclease involved in processing of intron-encoded small nucleolar RNAs in Xenopus laevis. J. Biol. Chem. 278, 13026–13032 10.1074/jbc.M211937200 [DOI] [PubMed] [Google Scholar]
- 30. Gioia U., Laneve P., Dlakic M., Arceci M., Bozzoni I., and Caffarelli E. (2005) Functional characterization of XendoU, the endoribonuclease involved in small nucleolar RNA biosynthesis. J. Biol. Chem. 280, 18996–19002 10.1074/jbc.M501160200 [DOI] [PubMed] [Google Scholar]
- 31. Ricagno S., Egloff M. P., Ulferts R., Coutard B., Nurizzo D., Campanacci V., Cambillau C., Ziebuhr J., and Canard B. (2006) Crystal structure and mechanistic determinants of SARS coronavirus nonstructural protein 15 define an endoribonuclease family. Proc. Natl. Acad. Sci. U.S.A. 103, 11892–11897 10.1073/pnas.0601708103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ziebuhr J., Snijder E. J., and Gorbalenya A. E. (2000) Virus-encoded proteinases and proteolytic processing in the Nidovirales. J. Gen. Virol. 81, 853–879 10.1099/0022-1317-81-4-853 [DOI] [PubMed] [Google Scholar]
- 33. Guarino L. A., Bhardwaj K., Dong W., Sun J., Holzenburg A., and Kao C. (2005) Mutational analysis of the SARS virus Nsp15 endoribonuclease: identification of residues affecting hexamer formation. J. Mol. Biol. 353, 1106–1117 10.1016/j.jmb.2005.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Huo T., and Liu X. (2015) Crystallization and preliminary X-ray crystallographic analysis of a nonstructural protein 15 mutant from human coronavirus 229E. Acta Crystallogr. F Struct. Biol. Commun. 71, 1156–1160 10.1107/S2053230X15007359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Laskowski R. A., and Swindells M. B. (2011) LigPlot+: multiple ligand–protein interaction diagrams for drug discovery. J. Chem. Inf. Model. 51, 2778–2786 10.1021/ci200227u [DOI] [PubMed] [Google Scholar]
- 36. Renzi F., Caffarelli E., Laneve P., Bozzoni I., Brunori M., and Vallone B. (2006) The structure of the endoribonuclease XendoU: from small nucleolar RNA processing to severe acute respiratory syndrome coronavirus replication. Proc. Natl. Acad. Sci. U.S.A. 103, 12365–12370 10.1073/pnas.0602426103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ma Y., Zhang Y., Liang X., Lou F., Oglesbee M., Krakowka S., and Li J. (2015) Origin, evolution, and virulence of porcine Deltacoronavirus in the United States. MBio 6, e00064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lorsirigool A., Saeng-Chuto K., Temeeyasen G., Madapong A., Tripipat T., Wegner M., Tuntituvanont A., Intrakamhaeng M., and Nilubol D. (2016) The first detection and full-length genome sequence of porcine deltacoronavirus isolated in Lao PDR. Arch. Virol. 161, 2909–2911 10.1007/s00705-016-2983-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Deng X., and Baker S. C. (2018) An “Old” protein with a new story: coronavirus endoribonuclease is important for evading host antiviral defenses. Virology 517, 157–163 10.1016/j.virol.2017.12.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Orr H. A. (2009) Fitness and its role in evolutionary genetics. Nat. Rev. Genet. 10, 531–539 10.1038/nrg2603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Li W., Cowley A., Uludag M., Gur T., McWilliam H., Squizzato S., Park Y. M., Buso N., and Lopez R. (2015) The EMBL-EBI bioinformatics web and programmatic tools framework. Nucleic Acids Res. 43, W58–W584 10.1093/nar/gkv279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Robert X., and Gouet P. (2014) Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 42, 320–324 10.1093/nar/gku316 [DOI] [PMC free article] [PubMed] [Google Scholar]