Abstract
Biochemical, proteomic, and epigenetic studies of chromatin rely on the ability to efficiently isolate native nucleosomes in high yield and purity. However, isolation of native chromatin suitable for many downstream experiments remains a challenging task. This is especially true for the budding yeast Saccharomyces cerevisiae, which continues to serve as an important model organism for the study of chromatin structure and function. Here, we developed a time- and cost-efficient universal protocol for isolation of native chromatin fragments from yeast, insect, and mammalian cells. The resulting protocol preserves histone posttranslational modification in the native chromatin state and is applicable for both parallel multisample spin-column purification and large-scale isolation. This protocol is based on the efficient and stable purification of polynucleosomes and features a combination of optimized cell lysis and purification conditions, three options for chromatin fragmentation, and a novel ion-exchange chromatographic purification strategy. The procedure will aid chromatin researchers interested in isolating native chromatin material for biochemical studies and serve as a mild, acid- and detergent-free sample preparation method for MS analysis.
Keywords: chromatin, chromatin modification, chromatin isolation, yeast, chromatography, chromatin structure, post-translational modification (PTM), acetylation, histone, methylation, nucleosome, purification, mass spectrometry, insect cell, MCF-7, epigenetics, native chromatin fragments, oligonucleosomes
Introduction
Saccharomyces cerevisiae (budding yeast) is a single-celled eukaryote that has proven useful for epigenetic research because of its powerful genetic tractability and extensively annotated genome (1–3). Given that yeast can be grown in large amounts in liquid culture and under a variety of experimentally controlled conditions, this organism is also useful for the biochemical analyses of chromatin, which would greatly complement its genetic and genomic advantages. However, for a number of reasons, the purification of chromatin from yeast has proven particularly challenging (4, 5). There are several features of yeast cell physiology that make quantitative isolation of purified nucleosomes extremely challenging. For example, yeast contain a cell wall that must be removed to access subcellular components, but many common procedures for cell wall removal exacerbate chromatin degradation, particularly proteolytic clipping on the N-terminal histone tails (6). In addition, protecting the integrity of intact chromatin from spontaneous disassembly during subsequent purification steps can be problematic.
Most published procedures targeting direct purification of native yeast chromatin are adapted from protocols previously used for the isolation of mammalian chromatin (5, 7, 8). When applied to yeast, these methods result in relatively low yields, incomplete and/or nonreproducible levels of purity, and/or extensive histone degradation. Metal affinity and micro-ChIP purification methods were previously attempted to overcome the shortcomings of direct yeast chromatin purification (9–11). These methods, however, require overexpression of tagged histone, which can impose a metabolic burden on the cell, and still suffer from low yield and result in material having unnaturally tagged histones.
In this report, we directly address several challenges plaguing various steps of chromatin isolation from yeast and, in doing so, develop a new, flexible scheme for more reproducible, cost- and time-efficient isolation of native yeast, insect, and mammalian nucleosomes. LC-MS/MS analyses of purified nucleosomes from our protocol indicated that the purified chromatin retains relevant post-translational modifications, including important acetylation and methylation marks.
Results and discussion
We found that many existing procedures for chromatin purification are time-consuming, expensive, inconsistent, and, although applicable for higher eukaryotic cells, inadequate for isolating yeast chromatin at sufficient yield and purity for quantitative biochemical or proteomic studies. There are many factors related to yeast biology that contribute to these limitations. Fundamentally, the small, compact genome of yeast cells means that there is simply far less chromatin per cell compared with higher eukaryotes. In addition, yeast nuclei lack lamins and thus are fragile and susceptible to detergents and mechanical degradation (12). Isolation of intact yeast nuclei is known to be a challenging and low-yielding procedure (13). The yeast nuclear membrane provides relatively poor protection for chromatin during the isolation procedure, which significantly contributes to contamination with cationic cytoplasmic impurities. In contrast to more complex eukaryotic chromatin (e.g. mammalian or insect cells), which is largely heterochromatic, the bulk of yeast chromatin is euchromatic (14) and additionally contains no structural H1 histone (15, 16), making yeast chromatin more open and vulnerable to highly abundant yeast proteolytic and nucleolytic enzymes. Finally, yeast core nucleosomes are intrinsically unstable compared with those from higher eukaryotes (14, 17). Together, these factors present significant challenges that additionally impose restrictions on the time and conditions used for chromatin purification.
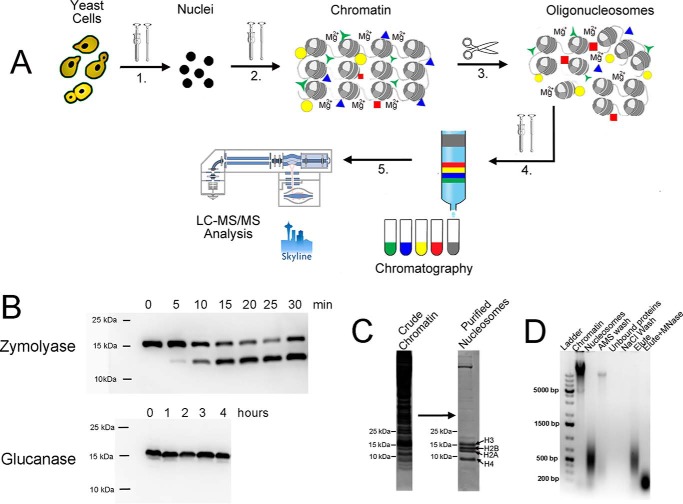
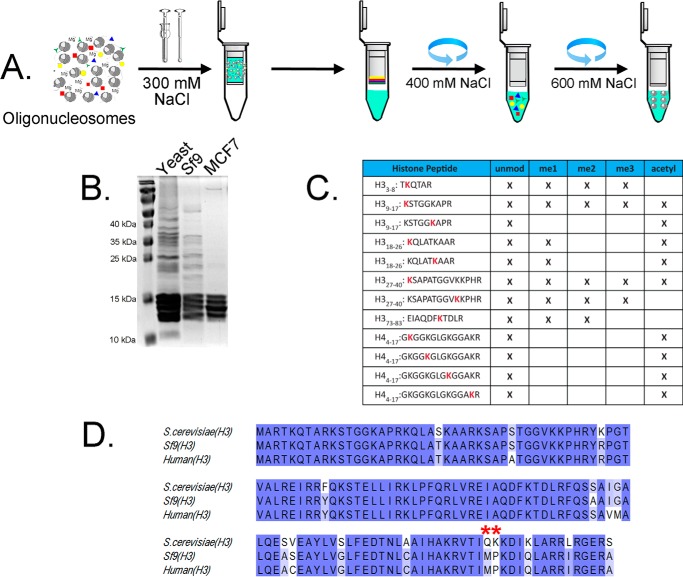
In our efforts to develop a rapid and efficient purification procedure for native chromatin, we considered and solved a number of general and yeast-specific challenges. The result of addressing these concerns was a reproducible protocol for isolating soluble, intact native yeast chromatin (Fig. 1A). Importantly, this purification approach is also applicable to other eukaryotic sources, such as human MCF-7 cells and Sf9 (Spodoptera frugiperda) insect cells (see Fig. 5). The latter are commonly used for heterologous baculovirus-mediated protein overexpression but were not previously considered as an accessible source of chromatin material for in vitro studies. Sf9 cells contain no cell wall, and unlike in yeast, the primary histone sequences resemble mammalian (Fig. 5). These fast-dividing cells can be cultured in suspension using relatively inexpensive serum-free SFX medium at 27 °C in standard shaker-incubators, providing a much higher yield of stable chromatin material. We found no previous reports of chromatin isolation from cultured insect cells but considered that the benefit of such a method would be an inexpensive and readily available source of endogenous chromatin suitable for a range of in vitro studies. The results of each relevant step will be discussed separately below.
Figure 1.
Optimized chromatin purification protocol using yeast as an example. A, outline of the purification protocol. 1, yeast cell wall was digested using recombinant glucanase, and spheroplasts were disrupted using a Dounce homogenizer. 2, crude nuclei were treated with 400 mm AMS in the presence of 5 mm magnesium chloride. The suspension was homogenized using the Dounce homogenizer, and the bulk chromatin was isolated by centrifugation. 3, oligonucleosomes were generated from bulk chromatin using MNase, sonication, or an EmulsiFlex C5 homogenizer in the presence of 5 mm magnesium chloride (Fig. 2). 4, oligonucleosomes were extracted from the pellet using NaCl and purified using column chromatography (Fig. 3). 5, purified nucleosomes were isolated and analyzed using LC-MS/MS (Fig. 4). B, immunoblotting reveals H3 degradation (clipping) in the presence of zymolyase, but not glucanase, during cell wall digestion. C, Coomassie-stained polyacrylamide gel of yeast chromatin before and after purification. D, ethidium bromide–stained DNA-agarose gel of yeast chromatin purification procedure.
Figure 5.
Chromatin purification using a spin-column multiparallel approach. A, schematic representation of chromatin purification workflow. Fragmented chromatin material (Fig. 1) is extracted and loaded on anion-exchange spin columns after NaCl adjustment to 300 mm. B, Coomassie-stained protein gel of eluted chromatin from yeast, Sf9, and MSF-7 cells. C, qualitative analysis of histone PTM from Sf9 insect cells. D, protein sequence alignment of primary H3 from yeast, Sf9, and human cell lines. Red asterisks, nucleosome-destabilizing point mutation in native yeast H3 (17) not present in Sf9 cells.
Spheroplasting and chromatin preservation
Yeast contain a thick cell wall that consists of glucans, mannoproteins, and chitin (18). The cell wall must be removed to access nuclei that are then purified away from other subcellular components. Cell wall digestion is conducted in the presence of osmotic stabilizers, such as sorbitol that protects digested yeast cells (spheroplasts) from mechanical stress (19). To increase the efficiency of the lytic enzymes, yeast cells are pretreated with a thiol-containing buffer that eliminates the outer shell of mannoproteins (20, 21). Commercially available lytic enzyme mixtures, such as lyticase or zymolyase, are used to digest the primary cell wall components (22, 23). We found that a commonly used zymolyase-based procedure for yeast cell wall removal led to substantial proteolytic clipping of the N-terminal tail of histone H3, even when a protease-deficient BJ2168 strain was used in combination with a range of inhibitor additives (Fig. 1B). Other reports have documented histone H3 clipping during zymolyase treatment (6). Clipping of histone tails destabilizes native nucleosomes (24, 25) and results in the loss of important post-translational modifications. Small aliquots from zymolyase-containing yeast suspension were used to determine the time course of H3 degradation (Fig. 1B). The time course of cell wall digestion revealed that histone H3 degradation was noticeable after only 5 min of zymolyase treatment and that after 30 min, more than half of the histone H3 tail was clipped (Fig. 1B). The total zymolyase treatment time needed for complete spheroplasting exceeds 30 min and potentially affects other nucleosomal and chromatin-associated protein. Because proteolytic action of zymolyase was responsible in part for the histone H3 degradation (6), and both zymolyase and lyticase commercial blends contain protease components, we turned to purified recombinant 1,3-glucanase for yeast cell wall removal (26). Glucanase cleaves the primary 1,3-β-glucan component of the yeast cell wall and can be overexpressed and purified from Escherichia coli in the active form (27). Recombinantly produced glucanase is an inexpensive alternative to commercial enzymes (lyticase and zymolyase) and is compatible with a broad range of inhibitors used to protect chromatin and histone modifications. Compared with zymolyase, glucanase requires longer incubation times to complete cell wall digestion, but it did not cause detectable H3 degradation even after 4 h (Fig. 1B).
To achieve reproducible and efficient cell wall removal (spheroplasting), we found that yeast cells should be harvested in the mid-log phase, A600 nm (0.6–0.8), when the cells are grown in standard rich liquid medium conditions (YPD). Incomplete cell wall removal impeded enzymatic chromatin fragmentation (Fig. 2A) and downstream chromatographic chromatin purification steps for all of the three fragmentation methods discussed below. Residual undigested cell wall components render chromatin insoluble in the form of disperse 0.3–0.4-μm aggregates that were stable even under high-salt conditions and were not retained during subsequent column purification steps. These observations suggested the existence of very strong and stable interaction between the cell wall debris and chromatin. It is a possibility that, released upon gluconase treatment, reducing sugar ends form a stable Shiff-base covalent interaction with amino-rich chromatin fragments (28). Thus, the ability to use glucanase for longer incubation times to ensure complete cell wall removal, in addition to preventing N-terminal H3-clipping, also promoted more efficient chromatin fragmentation and allowed subsequent purification by column chromatography. To further address proteolysis challenges, we reconsidered a set of protease inhibitors previously used for yeast chromatin isolation. Commonly used phenylmethylsulfonyl fluoride was replaced with the more stable, irreversible serine protease inhibitor aminoethylbenzenesulfonyl fluoride (AEBSF).3 Aprotinin that is deactivated in the presence of reducing agents was replaced with irreversible selective cysteine protease inhibitor E-64. Pepstatin was supplemented to inhibit carboxypeptidases. Finally, the protease-deficient BJ2168 yeast cell line was used for the current work.
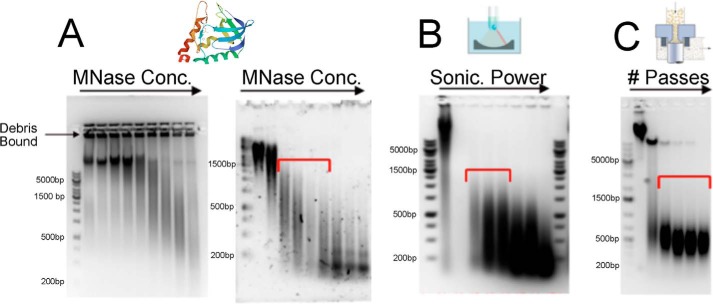
Figure 2.
Optimization and analysis of chromatin fragmentation methods by DNA agarose gel electrophoresis with ethidium bromide staining. MNase (A), sonication (B), and EmulsiFlex C5 homogenization (C) were used to prepare native yeast chromatin fragments. The optimal fragmentation size range is crucial for effective chromatographic purification and is indicated by red brackets. A, chromatin isolated from spheroplasts was digested with a serially increasing concentration of MNase. Left, MNase treatment of digestion-resistant yeast chromatin in the presence of remaining cell wall debris; right, complete digestion of yeast chromatin after the cell wall was completely removed. B, chromatin sonication using a Covaris C5 sonicator after complete removal of cell wall components at various increasing power settings (100–300 W). C, fragmentation of chromatin using an EmulsiFlex C5 homogenizer yields a suitable range of oligonucleosomes for downstream column purification after the second treatment.
Isolation of nuclei and chromatin purification
After spheroplasts were purified from cell wall contaminants, cells were lysed, and nuclei were isolated. The isolation of nuclei is routinely used to eliminate the bulk of cytoplasmic components in a simple centrifugation step or using downstream density gradient purification (29). However, unlike higher eukaryotes, the isolation of intact nuclei from yeast using density gradient sedimentation is a challenging and relatively low-yield process (13, 30). The yeast nucleus lacks lamins, contributing to nuclei fragility and high susceptibility to detergents, osmotic, and mechanical stress during isolation (12). Partial or complete disruption of the nuclear membrane triggers aggregation and contamination of chromatin with cationic (net positively charged) cytoplasmic proteins that complicate downstream purification steps.
Disruption of nuclear membrane integrity also results in a significant loss of chromatin during isolation. Chromatin loss is aggravated by nucleolytic and proteolytic activity, in particular when often-employed detergent-mediated cell lysis disrupts lysosomes. Therefore, we examined preparation conditions that could reduce chromatin losses during all prechromatography steps. In general, we found that chromatin losses and solubility are well-correlated. Hence all prechromatography clean-up conditions were chosen to minimize chromatin solubility and increase stability. Specifically, we used a low ionic strength buffer (31) and gentle mechanical cell lysis using mild osmotic shock (32). Additionally, we found that chromatin losses during cell lysis and downstream clean-up steps could be significantly reduced by incorporating 5 mm magnesium into all buffers, whereas abundant chelating agents, including some that are frequently present in protease inhibitor mixtures, exacerbated chromatin loss. The sensitivity of yeast chromatin condensation state on magnesium concentrations is stipulated by its unique structural properties (e.g. a shorter internucleosomal distance and the absence of structural H1 linker histone) (33–35).
Yeast nuclei separated by centrifugation from cytoplasmic components are prone to aggregation. Nuclei aggregation traps cytoplasmic impurities and complicates conventional purification using density gradients. Additionally, the outer membrane of yeast nuclei forms a continuum with the endoplasmic reticulum (36), which contains a large number of ribosomes, ribosomal proteins, proteases, and nucleases. The removal of such impurities was previously achieved by repeated washings with high NaCl–containing buffers (5). We discovered that repeated washings with high-NaCl buffers resulted in significant chromatin release from the insoluble pellet (Fig. S1) that reduced yield and ultimately provided no significant purification improvement for downstream chromatography. Therefore, to selectively release chromatin-associated impurities without disrupting chromatin itself, a single wash with 400 mm ammonium sulfate in Buffer A containing 5 mm MgCl2 was used. Due to the “salting-out” effect of ammonium sulfate (37), intact chromatin remains insoluble under these conditions and can be easily separated by centrifugation. We found that this single ammonium sulfate treatment significantly improved the yield and purification efficiency during the subsequent chromatography step. Elimination of the multiple NaCl washes with associated mechanical manipulations during repeated resuspension-sedimentation cycles substantially reduced chromatin loss triggered by endogenous proteases and nucleases (Fig. S1) that generate small soluble chromatin fragments.
The yeast lysis and chromatin purification procedure was used successfully for Sf9 and MCF-7 cells, yielding chromatin material suitable for downstream fragmentation.
Chromatin fragmentation
To increase solubility and prepare for the chromatography step, the large-molecular weight chromatin material must be properly fragmented. We determined that chromatin fragments in the 500–1000-bp range were ideal for downstream purification by column chromatography because these polynucleosomes elute later in the salt gradient compared with mononucleosomes, allowing a more efficient separation of chromatin from contaminants. Additionally, larger chromatin fragments are more stable than mononucleosomes due to the formation of higher-order structure.
Due to wide availability of the enzyme, micrococcal nuclease (MNase) digestion is a commonly used method to fragment chromatin. However, MNase digestion can be affected by the degree of chromatin accessibility, which can vary widely. This issue was identified as a major problem in the early stage of our yeast method development. Specifically, we discovered that MNase digestion was inhibited by chromatin insolubility caused by the presence of cell wall debris (Fig. 2A). For this reason, the cell wall must be completely digested so that washed chromatin can be resuspended and digested in MNase digestion buffer (Fig. 2A, right). The primary downside of enzymatic chromatin fragmentation is that the reaction is incubated at 30–37 °C, and the amount of MNase must be optimized for each purification experiment to produce a specific range of chromatin fragments suitable for downstream chromatography (Fig. 3). Unlike MNase digestion that requires incubation of sample at 30–37 °C, we also used two alternative methods, (i) sonication and (ii) high-pressure homogenization, both conducted at 4 °C (Fig. 2B). Mechanical treatment is also believed to be more consistent and unbiased than MNase digestion, but in a practical sense, these mechanical methods are limited by the availability of appropriate equipment. Sonication (Covaris) is more suitable for small-scale (up to 5–10 g of cell pellet) chromatin preparations, whereas an EmulsiFlex C5 homogenizer is ideal for large-scale (up to 100 g of cell pellet) chromatin preparations but also suitable for small cell pellet (3–5 g). MNase-mediated digestion can be used for small- or large-scale preparations and is limited only by the availability and cost of the enzyme supply. An important benefit of using an EmulsiFlex homogenizer is the consistency in producing uniform 500–1000-bp chromatin fragments, which are most suitable for downstream column purification, with no optimization (Fig. 2C). Generally, all three chromatin fragmentation methods, MNase digestion, sonication, and high-pressure homogenization, are applicable, and selection of one over another is largely dictated by equipment availability and purification scale.
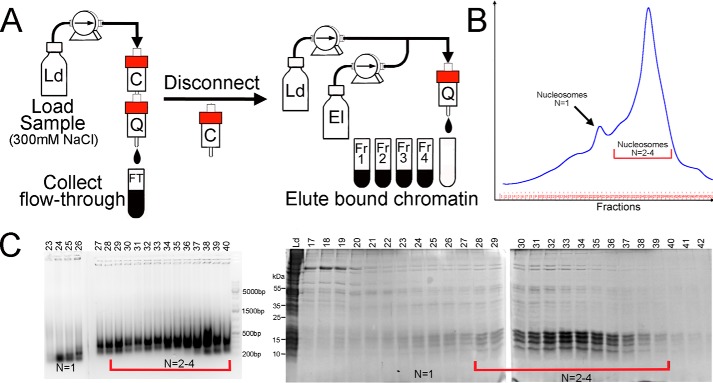
Figure 3.
The details of the column chromatography step used for chromatin purification. A, fragmented and salt-extracted chromatin (Fig. 1) is loaded on the tandem carboxymethyl (C) and quaternary amine (Q) column using loading buffer (Ld). The cation-exchange column traps impurities and is disconnected before Q elution. B, UV (280 nm) elution diagram of the Q anion-exchange column. The column was eluted (El) using a linear salt gradient as described under “Experimental procedures.” C, fractions were analyzed by agarose gel DNA electrophoresis (ethidium bromide staining) (left) and SDS-PAGE (right). Mononucleosomes elute first, followed by oligonucleosomes near the end of the gradient. The highest purity is achieved for larger chromatin fragments, underscoring the importance of controlled and specific chromatin fragmentation. N, the predominant oligomeric form present (e.g. N = 1 = mononucleosome).
After fragmentation, soluble chromatin fragments must be separated from insoluble debris by centrifugation. However, we discovered that in low-ionic strength buffers, even after fragmentation, a significant amount of chromatin remained in the insoluble form. To recover all chromatin, the NaCl concentration must be adjusted to 500 mm immediately after fragmentation. This treatment improves solubility of chromatin fragments and significantly increases chromatin recovery and yield.
Chromatin purification by FPLC column chromatography
To eliminate protein impurities from the above fragmented chromatin preparations, an additional purification is particularly important for native yeast chromatin that is heavily contaminated with passive (cationic DNA-binding proteins) and active (proteases and nucleases) impurities. In mammalian chromatin purifications, repeated high-salt chromatin washes produce relatively clean nucleosomal material (8). Because yeast nucleosomes are unstable and significantly more contaminated, repeated high-salt chromatin washes led to excessive loss of chromatin material (Fig. S1).
To eliminate contaminants from native nucleosome preparations, we used a tandem chromatography setup consisting of CM-FF cation-exchange and Q-HP anion-exchange chromatography columns connected inline. Whereas the CM cation-exchange column is set to capture some cationic contaminants, large polyanionic chromatin fragments (oligonucleosomes) generated by any one of the chromatin fragmentation methods described above associated with the Q-HP column. Selection of anion-exchange chromatographic support to capture oligonucleosomes is based on the intrinsic property and structure of chromatin. Chromatin is a polynucleoprotein complex composed of positively charged histone octamers that bind DNA but neutralize only 50% of DNA's total charge (38, 39). Consequently, the net negative charge on chromatin can be exploited for purification via anion-exchange chromatography. However, many nonhistone chromatin-binding proteins that contaminate the crude chromatin fractions are cationic by nature and can associate with the negatively charged stationary phase. For this reason, we utilized a CM-HP column in front of the Q-HP to capture cationic impurities. Before chromatin elution from the Q-HP column, the CM column was manually disconnected (Fig. 3). This approach allows extensive single-step separation of target chromatin material from associated impurities.
During this step, the chromatin material solubilized in the high-NaCl (600 mm) extraction buffer was immobilized on anion-exchange Q-HP stationary phase (Fig. 3). A linear salt gradient was used to separate impurities from the column (Fig. 3B). We note that longer chromatin fragments elute later compared with mononucleosomes that co-elute with some impurities (Fig. 3, C). Thus, proper chromatin fragmentation that generates oligonucleosomes of ∼500–1000 bp is key to successful separation of chromatin from chromatin-associated impurities using ion-exchange chromatography. After chromatographic purification, the oligonucleosomes can be additionally sonicated or treated with MNase if mononucleosomes are desired.
Chromatin purification using spin-column chromatography
An FPLC-based method is ideal for large-scale purification, but a parallel high-throughput method for small-scale chromatin isolation is frequently preferred for multiple diverse samples. Application of ceramic hydroxyapatite for this purpose has been reported previously (40, 41), but repeated attempts to replicate this published procedure failed to produce significant chromatin material (data not shown). For MS analysis of histones, acid and salt extraction can be used (28), but this method is inappropriate when native chromatin is required. Acid extraction of yeast nuclei yields crude histone material that requires HPLC purification and is not well-suited for high-throughput purification (Fig. S2) (42). Additionally, acid extraction affects labile chromatin posttranslational modifications, such as phosphorylation, whereas mild spin-column purification before MS analysis presents a less harsh high-throughput alternative. We show that application of our protocol for parallel high-throughput native chromatin purification can be accomplished using spin columns prepacked with anion-exchange resin (Fig. 5). After fragmentation, chromatin is loaded onto prepacked anion-exchange spin columns, isocratically washed with 400 mm NaCl-containing buffer, and finally eluted in the presence of inhibitors, yielding concentrated chromatin fragments suitable for downstream analysis.
Analyses of purified nucleosomes by MS/MS
To determine whether the purified intact native nucleosomes retain the endogenous post-translational modifications (PTMs), we performed an extensive LC-MS/MS PTM analysis of native nucleosomes purified using our chromatin purification method. For comparison, we also performed the same PTM analysis on acid-extracted yeast histones from spheroplasts (28), allowing quick preparation of endogenous but denatured histones. The acid-extraction histones were further purified using reversed-phase HPLC (Fig. S2) before subjecting the histone preparation to the LC-MS/MS PTM analysis.
The histone PTM profiling results shown in Fig. 4 quantitatively assessed how the newly designed purification procedure presented here maintains PTM states on native nucleosomes recovered after purification. In the first approach, the bulk chromatin fragmentation by MNase was conducted before chromatographic purification, which was followed by a second MNase treatment after column chromatography to reduce chromatin fragments to mononucleosomes. For all MNase digestion reactions, solutions were prewarmed to 30 °C to sustain sufficient enzymatic activity. In the second approach, an EmulsiFlex homogenizer was used to prepare oligonucleosomes for chromatographic purification, followed by sonication of the column-purified oligonucleosomes to generate mononucleosomes. In the second approach, all steps were performed at 4 °C.
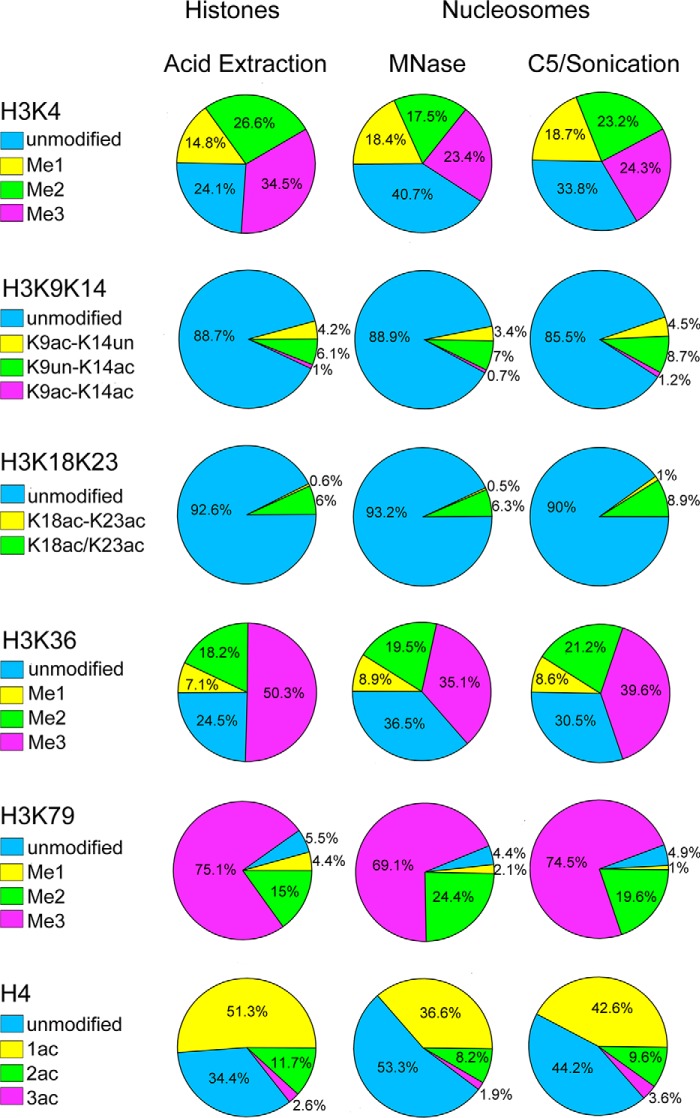
Figure 4.
MS/MS analysis of acid extracted histones and native nucleosomes. Quantitative MS/MS PTM profiling of acid-extracted/HPLC-purified histones was compared with two purification strategies yielding native nucleosomes: MNase-mediated chromatin fragmentation and EmulsiFlex C5 fragmentation followed by sonication to produce mononucleosomes (Fig. 2). Calculation details are presented in Table S1 and Table S2.
The histone profiling quantification revealed that the native chromatin fragment purification methods described in this study display PTM states very similar to those obtained from rapid, acid-extracted histones, which do not maintain native chromatin configuration (Fig. S2). Among the 23 unique peptides quantified, H4 tail mono-, di-, and triacetylations showed a small variation (18–30%) between methods, whereas acetylation states of H3K9, H3K14, H3K18, and H3K23 were essentially identical (Fig. 4). For H3K4 methylation, the native methods showed a 30% reduction of the Me3 state and a 12–30% reduction of the Me2 state with a concomitant increase of unmodified H3K4 peptide and no significant effect on Me1. There was no significant difference for the H3K36Me or H3K36Me2 marks, although the H3K36Me3 state in the acid-extracted histones was ∼30% higher with a simultaneous decrease in unmodified peptide, when compared with the two native methods (Fig. 4). The H3K79Me3 state was similar between methods. Overall, our results demonstrate that the native purification methods yield high-quality chromatin fragments that maintain endogenous PTM states. Yeast chromatin decomposition during purification is caused by intrinsic instability of yeast nucleosomes (33). Such instability affects yield but does not appear to bias the original native PTM landscape, as revealed by proteomic profiling of acid extracted histones and purified native yeast chromatin fragments. The slight differences between acid-extracted histones and histones purified as native fragments could reflect gradual enzymatic demethylation of H3K4Me3 and H3K36Me3 and deacetylation of H4. Consistent with this possibility, the observed losses of PTMs tended to be somewhat greater when MNase was used for chromatin fragmentation, suggesting that the higher temperature used for MNase sped up the reactions. Importantly, we show that purification of native chromatin fragments effectively preserves the endogenous histone PTM landscape on native nucleosomes.
Conclusions
Histone modifications represent a biological “snapshot” that reports important biological information of diverse chromatin states. Here we developed an efficient purification of chromatin fragments that maintains original PTM states for downstream interrogation of the entire chromatinized genome at nucleosome resolution. After native purification described here, intrinsic PTMs become exposed and available for follow-up analysis, such as native ChIP (43), matrix-assisted reader chromatin capture (44), chromatin enzyme characterization, or nucleosome proteoform analysis.
An ideal nucleosome isolation process must be reproducible and time- and cost-effective, yield sufficient amounts of high-quality chromatin material suitable for downstream application, and allow simultaneous parallel isolation from multiple samples. Although chromatin enrichment protocols from mammalian nuclei have been used before (45), in practice, these methods are poorly suited for yeast and suffer from reproducibility issues. Purification of chromatin from yeast has been a challenging undertaking due to a combination of factors stipulated by specific yeast physiology.
Unlike previously published procedures, our protocol can be completed in 1 day and does not require large quantities of expensive protease inhibitors. The method yields native yeast chromatin fragments containing 83 μg of intact yeast octamers or 300 μg by DNA from a 1-liter culture of yeast cells. We tested several chromatin fragmentation methods and show for the first time the utility of an Avestin EmulsiFlex C5 homogenizer for chromatin fragmentation and its unique ability to consistently yield chromatin fragments suitable for ChIP-quantitative PCR (41). Our chromatographic approach allows size-based separation of fragmented chromatin with concomitant purification from associated proteins, from free DNA, and, importantly, from nonchromatin histones. The chromatographic method is more time-efficient compared with previously used density-gradient ultracentrifugation methods that are low-yielding, take hours to complete, require specialized equipment, and are not suitable for particularly unstable yeast chromatin. When chromatin separation by size is not required, commercially available spin columns prepacked with anion-exchange resins can be used for quick parallel purification of multiple chromatin samples. This method proved to be more scalable, cost-effective, and reproducible than a previously published ceramic hydroxyapatite–based chromatin purification approach (41) that failed when applied to yeast or insect chromatin. Our purification methodology avoids an acidic extraction environment that is detrimental to labile PTMs and, when applied to fungi, produces a histone preparation that contains carbohydrates that readily cross-link amine-rich histone proteins (28).
Although the methods revealed in the current study were primarily developed to address the issues presented in yeast, we also used this method in other systems (Fig. 5). It resulted in an unprecedented time- and cost-efficient preparation of high-purity chromatin material from several types of eukaryotic cells. Whereas yeast may serve as a great epigenetic model, it is nevertheless distinct from higher eukaryotes in several key aspects (42). A number of challenges we faced and resolved during isolation and purification spurred us to search for an alternative, more accessible yet readily scalable source of stable chromatin material for in vitro studies. We demonstrate that cultured insect cells (Sf9) harbor these desirable features and exhibit high sequence homology to mammalian systems (Fig. 5). Because of the ease of manipulation and culturing that does not require any specialized equipment or expensive serum-based medium, high homology to mammalian cells, chromatin stability, and accessibility without enzymatic cell treatment, we believe this largely untapped insect model could serve the needs of a broad scientific community interested in studying chromatin biology and epigenetics.
Experimental procedures
All buffers were prepared from components provided by Sigma-Aldrich and Fisher. pH of the buffers was adjusted at room temperature with consideration of temperature effect on thermodynamic properties of primary buffering agent.
Chromatin isolation buffers
Chromatin isolation buffers were as follows: Buffer 1 (sensitizing buffer), 50 mm Tris, pH 9.0, 10 mm DTT (reducing agent and inhibitors were added before use); Buffer 2 (glucanase digestion buffer), 50 mm HEPES, pH 7.8, 1 m sorbitol, 5 mm MgCl2, 5 mm DTT (reducing agent and inhibitors were added before use); Buffer 3 (spheroplast wash buffer), 25 mm MES, pH 6.0, 1 m sorbitol, 5 mm MgCl2 (inhibitors were added before use); Buffer 4 (lysis buffer), 20 mm MES, pH 6.0, 5 mm MgCl2, 8% PVP-40, 0.02% Triton X-100, inhibitors (all inhibitors and Triton X-100 were added fresh before use); Buffer M (MNase digestion buffer), 50 mm HEPES, pH 7.5, 10% sucrose, 10% glycerol, 3 mm CaCl2, 1 mm MgCl2, 1 mm DTT (reducing agent and inhibitors should be added fresh before use); Buffer Q (2× MNase quench buffer), 50 mm MES, pH 5.0, 1 m NaCl, 10 mm EDTA, 10 mm EGTA, and all inhibitor additives.
Chromatography buffers
Chromatography buffers were as follows: Buffer A (low-salt FPLC buffer), 25 mm MES, pH 6.0, 10% sucrose, 10% glycerol; Buffer B (high-salt FPLC buffer), 25 mm MES, pH 6.0, 750 mm NaCl, 1 mm EDTA, 0.1 mm EGTA, 10% sucrose, 10% glycerol; Buffer C (cryoprotection/storage buffer), 20 mm HEPES, pH 7.2, 10 mm KCl, 1 mm EDTA, 0.1 mm EGTA, 0.5 mm spermine, 1 mm tris(2-carboxyethyl)phosphine, 50% glycerol.
Protease inhibitors
10 mm (1000×) pepstatin in methanol, 100 mm (100×) AEBSF in DMSO, 10 mm (1000×) E-64 in DI water were from Gold-Bio and prepared using instructions in the literature.
Concentrated stock solutions were prepared in 50-ml conical tubes as follows. 5 m NaCl was prepared using 14.6 g of NaCl dissolved in warm DI water. 4 m ammonium sulfate (AMS) buffer was prepared using 26.5 g of solid AMS dissolved in warm water. pH was adjusted to 7.0 using dilute NaOH. EGTA, 100 mm stock, was prepared using 1.95 g of EGTA solid dissolved in up to 50 ml of 150 mm NaOH. pH 8.0 was adjusted using concentrated 10 n NaOH. EDTA was prepared using 1.5 g of EDTA solid dissolved in up to 50 ml of 150 mm NaOH. pH 8.0 was adjusted using concentrated 10 n NaOH. Chromatography FPLC columns were from GE Healthcare. Anion-exchange Midi spin columns were from Epoch Life Sciences. Zymolyase was obtained from Zymo Research. MNase (45 kilounits) was obtained from Worthington. PVP-40 was from Sigma.
Recombinant glucanase expression and purification
Prof. Craig Peterson (University of Massachusetts) generously donated bacterial plasmid containing the glucanase gene (26). The plasmid was transformed into E. coli Bl21(DE3) that was cultured in in 2xYT medium to A(600 nm) ∼ 0.4–0.6. Isopropyl 1-thio-β-d-galactopyranoside (0.1 g/liter) was added to induce protein expression, and cells were harvested after 4 h by centrifugation. Collected cells were osmotically disrupted to release periplasmic proteins, and the lysate was extensively dialyzed in 5 liters of 50 mm sodium acetate buffer, pH 5.0, 1 mm β-mercaptoethanol, 1 mm EDTA, overnight. Precipitated proteins were removed by centrifugation, and the clarified lysate was loaded onto an SP-FF anion-exchange column pre-equilibrated with dialysis buffer. Protein was eluted using a 120-ml linear gradient in the same buffer, using 300 mm NaCl as the eluting agent (Fig. S3 shows SDS-PAGE of eluted glucanase). The eluted single peak was concentrated to 10 ml, and flash-frozen fractions were stored at −80 °C.
Cell growth and medium conditions
One liter of starter culture of S. cerevisiae (strain BJ2168) was grown in YPD medium using an shaker-incubator at 30 °C (200–225 rpm) until A600 nm of 0.6–0.8 in the presence of kanamycin sulfate. Nine liters of YPD medium were inoculated and cultured to A600 nm of 0.6–0.8 (∼5 × 107 cells/ml for BJ2168) in 12–14 h (averaged doubling time of 120 min). Cells were harvested by centrifugation (5000 × g, 10 min) and washed (resuspended) in 200 ml of freshly prepared Buffer 1 at room temperature. After 10 min, cells were transferred to four 50-ml conical tubes and pelleted by centrifugation, yielding on average 3.2 g of wet cell pellet from 1 liter of culture (7.5 g/tube). Pelleted cells should not be frozen at this point and should be used for the next step.
Sf9 cells were cultured in SFX serum-free medium using spinner flasks at 27 °C. Cell concentration was maintained at 2 × 106 cells/ml. Cells were harvested by centrifugation (5000 × g, 10 min) and immediately used for purification, avoiding freezing.
MCF-7 cells were cultured in high-glucose Dulbecco's modified Eagle's medium supplemented with 10% FBS in 150-mm dishes. Cells were trypsinized at 90% confluence, harvested by centrifugation (5000 × g, 10 min), and immediately used for purification, avoiding freezing.
Cell disruption and chromatin isolation
Yeast cell pellet (30 g) in 50-ml conical tubes were resuspended in DI water (room temperature) containing 1 mm DTT and centrifuged to eliminate residual components of Buffer 1. Washed cells were resuspended in 30 ml/tube of freshly prepared glucanase digestion Buffer 2 supplemented with the following inhibitor additives: protease inhibitors (10 μm pepstatin, 1 mm AEBSF, 10 μm E-64) and deacetylase inhibitors (10 mm sodium butyrate, 5 mm nicotinamide, 5 μm trichostatin A). Purified recombinant glucanase was added (1 mg/1 g of wet cell pellet, 1.5 × 1010 cells/g for BJ2168 strain), and the suspension was incubated at 37 °C with gentle rocking for 1 h. Digestion was monitored and verified by measuring OD reduction at 600 nm (46) of a small (10-μl) aliquot diluted with 1 ml of lysis Buffer 4. Residual A600 nm <20% indicated sufficient spheroplasting. To completely eliminate the remaining cell wall debris components, digested spheroplasts were centrifuged in a swinging bucket centrifuge at 4000 × g (10 min, 4 °C) through 15 ml of Buffer 3 supplemented with 5% (w/v) PVP containing all previously used inhibitors. Spheroplasts were finally resuspended in 15 ml of ice-cold Buffer 3 supplemented with inhibitors, transferred into a Dounce homogenizer (pestle A), and mixed with 30 ml of ice-cold lysis Buffer 4 containing all inhibitors. Cells were allowed to swell on ice for 5 min and lysed using a Dounce homogenizer (20 strokes pestle A, 5 strokes pestle B). Lysis completeness was verified by microscope, and repeated Dounce homogenization (pestle B) was used until nearly all visible cells were disrupted. Crude nuclei were centrifuged at 10,000 × g for 10 min. The pellet was resuspended using a Dounce homogenizer in 50 ml of Buffer A supplemented with 5 mm MgCl2 and inhibitors followed by centrifugation (10 min, 5000 × g). The process was repeated until supernatant remained clear. Finally, the pellet was resuspended in 45 ml of Buffer A supplemented with 5 mm MgCl2 and all inhibitors and then quickly added to 5 ml of 4 m ammonium sulfate (10×) in a 50-ml conical tube. The conical tube was gently inverted 4–5 times, followed immediately by high-speed centrifugation (25,000 × g, 15 min). Pelleted chromatin was resuspended in 25 ml of Buffer A or 50 ml of Buffer M (supplemented with inhibitors) and used for downstream fragmentation.
Cell disruption and chromatin isolation for Sf9 and MCF-7 cells
One g of Sf9 cells and 0.3 g of MCF-7 cells were lysed using a Dounce homogenizer (20 strokes, pestle A) in 10 and 20 ml of ice-cold lysis Buffer 4, respectively, supplemented with all inhibitors. Lysis completeness was verified by microscope. Nuclei were centrifuged at 5000 × g for 10 min. The pellet was washed in 10 ml of Buffer A supplemented with 5 mm MgCl2 and inhibitors followed by centrifugation (10 min, 5000 × g). The process was repeated until supernatant remained clear. Finally, the pellet was resuspended in 13.5 ml of Buffer A supplemented with 5 mm MgCl2 and all inhibitors and then quickly added to 1.5 ml of 4 m (10×) ammonium sulfate in a 15-ml conical tube. The conical tube was gently inverted 4–5 times, followed by high-speed centrifugation (25,000 × g, 30 min). Pelleted chromatin was resuspended in 15 ml of Buffer A for downstream EmulsiFlex fragmentation.
Chromatin fragmentation and extraction
Yeast chromatin was fragmented using any one of three different methods, as indicated in Fig. 2: enzymatic (MNase), sonication, or high-pressure homogenization (EmulsiFlex C5). Depending on the purification scale and equipment availability, these methods can generate equivalent material for further purification by column chromatography. Only EmulsiFlex C5 homogenization was used for insect and MCF-7 cell chromatin.
MNase digestion of chromatin
For MNase digestion, 600 μl of resuspended chromatin from the previous step were equally aliquoted in 12 PCR tubes. MNase was diluted to contain 1 unit/μl using fresh Buffer M, and 12 2-fold serial dilutions were prepared. Five μl of each enzyme dilution were added to each tube containing 50 μl of chromatin prewarmed to 30 °C for 5 min in a thermal cycler. Digestion continued for 5 min, and reactions were stopped by the addition of 55 μl of ice-cold 2× Buffer Q. Reactions were phenol/chloroform-extracted; DNA was precipitated with ethanol/sodium acetate, followed by analysis of fragments by 1% agarose gel electrophoresis (Fig. 2A). The concentration of MNase yielding 500–1000-bp DNA fragments is considered optimal and was used for the remaining chromatin. After quenching the MNase reactions, the chromatin solution was incubated on ice for 15 min and clarified by centrifugation. Chromatin in the form of oligonucleosomes remained in the supernatant, and they were used immediately for downstream chromatography purification.
Chromatin fragmentation by sonication
Aliquots (800 μl) of resuspended chromatin from the previous step were transferred into a sonication vial compatible with the Covaris S220 sonicator. Treatment conditions producing oligonucleosomes were optimized to yield chromatin fragments of 500–1000 bp (Fig. 2B). After sonication, all aliquots were recombined, and NaCl concentration was adjusted to 500 mm using 5 m NaCl. The solution was clarified by centrifugation (15 min, 45,000 × g), after which the NaCl concentration of the supernatant was reduced to 300 mm using fresh Buffer A.
Chromatin fragmentation by EmulsiFlex C5 (high-pressure) homogenization
Resuspended chromatin was passed three times through an EmulsiFlex C5 homogenizer prechilled to 4 °C with a temperature-controlled water-circulating bath. No optimization was required, as fragmentation consistently produced chromatin fragments of 500–1000 bp and larger (Fig. 2C). Similarly, chromatin from Sf9 and MCF-7 cells was fragmented using an EmulsiFlex C5 homogenizer without prior optimization. After fragmentation, the concentration of NaCl was adjusted to 500 mm using concentrated 5 m NaCl stock. The solution was clarified by centrifugation (15 min, 45,000 × g), after which the NaCl concentration was adjusted to 300 mm using fresh Buffer A.
Chromatographic FPLC purification of chromatin
A 5-ml Q-HP anion-exchange column (GE Healthcare; trimethylamine, Sepharose, particle size 30 μm) was used to capture chromatin fragments produced by MNase digestion, sonication, or homogenization from yeast or Sf9 cells. A 5-ml CM-FF column was connected inline and in front of the Q-HP to remove remaining cationic impurities during loading. Salt-extracted chromatin was loaded onto the twin column at a rate of 0.7 ml/min and extensively washed with 50% Buffer B. Bound proteins were eluted using a 50–100%, 250-ml linear NaCl gradient against Buffer B.
Chromatographic spin-column purification of chromatin
Anion-exchange midi columns (Epoch Life Sciences, Inc.) were used for postfragmentation purification of chromatin from Sf9, MCF-7, and yeast cells (Fig. 5). Midi spin columns (Epoch Life Science) packed with anion-exchange resin were equilibrated with Buffer A, and NaCl-extracted chromatin was gravity-loaded (20 ml/column for MCF-7 and Sf9 cells, 50 ml/column for yeast cells). Flow-through was discarded, and each column was gravity-washed with 3 × 10 ml of Buffer A supplemented with 400 mm NaCl and inhibitors. Chromatin fragments were gravity-eluted using 5 ml of Buffer B supplemented with inhibitors. Columns were additionally centrifuged at 5000 × g for 5 min to collect the matrix-retained elution buffers.
Quantification of purified chromatin fragments
A small aliquot (270 μl) from each fraction after purification was ethanol-precipitated (70% final) at −80 °C for 1 h, centrifuged, and analyzed using agarose gel electrophoresis (Fig. 3). Separately, 300 μl from each fraction were mixed with acetone and methanol to precipitate proteins (47). After centrifugation, proteins were analyzed using PAGE (Fig. 3). Selected fractions were pulled together and concentrated using a stirred Millipore Amicon concentrator with a 30-kDa PES ultrafiltration membrane. Total combined yield by DNA was evaluated spectrophotometrically by absorbance at 260 nm (41). Total histone concentration was determined using a BCA assay (Bio-Rad). Nucleosome concentration was estimated based on the total protein concentration using an octamer molecular mass of 108,000 Da. On average (calculated from two separate chromatin purifications), the final concentrated yeast nucleosomal fractions contained 0.75 mg of protein (octamers) component per 2.8 mg of total DNA from 30 g of yeast. Purification of chromatin fragments from Sf9 cells on average yields 0.3 mg (by octamers) and 0.8 mg (by DNA) from 1 g of cells. Chromatin yield from MCF-7 cells was 200 μg (DNA) and 90 μg of histones per 107 cells.
Sample preparation for MS/MS analysis
Yeast nucleosomes were denatured in the presence of 4 m deionized urea added from 8 m freshly prepared stock and sonicated using a Covaris sonicator (300 W, 25%, 10 min, 25 °C) to fragment released DNA. 5 μg of total protein were used for MS sample preparation. Urea concentration was adjusted to 1 m using DI water, and 1 m triethylammonium bicarbonate solution (Sigma), pH 8.5, was added to a final concentration of 100 mm. 10 μl of propionic anhydride were mixed with 990 μl of DI water (10× solution) and added to the samples to a final concentration of 7 mm. Propionylation proceeded at room temperature for 2 min, and excessive propionic anhydride was quenched using aqueous 80 mm (10×) hydroxylamine solution (20 min, room temperature). pH was verified by a litmus paper test using 0.5 μl of sample and adjusted if needed to pH 8.5 using 1 m triethylamine bicarbonate solution. Samples were trypsinized for 6 h at 37 °C, and peptides were derivatized at their N termini with phenylisocyanate (48). Derivatized histone extracts were then desalted using in-house C18 stage tips. Peptides were eluted with 30 μl of acetonitrile (0.1% (v/v) formic acid). Solvent was removed under vacuum using a SpeedVac and resuspended in 30 μl of DI water (0.1% TFA).
MS/MS data collection and analysis
Derivatized peptides (0.3 μg) were injected onto a Dionex Ultimate 3000 nanoflow HPLC with a Waters nanoAcquity UPLC C18 column coupled to a Thermo Fisher Q-Exactive mass spectrometer at 700 nl/min. Mobile phase consisted of water + 0.1% formic acid (A) and acetonitrile + 0.1% formic acid (B). Histone peptides were resolved with a two-step linear gradient of 2–25% mobile phase B over 60 min, followed by 25–40% mobile phase B over 15 min. Data were acquired using data-independent acquisition (DIA) mode. The mass spectrometer was operated with an MS1 scan at resolution = 35,000, automatic gain control target = 1 × 106, and scan range = 390–910 m/z, followed by a DIA scan with a loop count of 10. DIA settings were as follows: window size = 10 m/z, resolution = 17,500, automatic gain control target = 1 × 106, DIA maximum fill time = AUTO, and normalized collision energy = 30. For each cycle, one full MS1 scan was followed by 10 MS2 scans using an isolation window size of 10 m/z.
Author contributions
V. I. K., C. A. F., and J. M. D. conceptualization; V. I. K. resources; V. I. K. data curation; V. I. K., S. A. H., C. A. F., and J. M. D. formal analysis; V. I. K. and S. A. H. validation; V. I. K., S. A. H., and C. A. F. investigation; V. I. K. visualization; V. I. K. methodology; V. I. K. writing-original draft; V. I. K., S. A. H., C. A. F., and J. M. D. writing-review and editing; C. A. F. and J. M. D. supervision; C. A. F. and J. M. D. funding acquisition.
Supplementary Material
This work was supported by NIGMS, National Institutes of Health Grants GM056890 (to C. A. F.) and GM059785 (to J. M. D). J. M. D. is a consultant for Bio-Techne and FORGE Life Science and is cofounder of Galilei BioSciences. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Tables S1 and S2 and Figs. S1–S3.
- AEBSF
- aminoethylbenzenesulfonyl fluoride
- DI
- deionized
- MNase
- micrococcal nuclease
- PTM
- post-translational modification
- AMS
- ammonium sulfate
- DIA
- data-independent acquisition
- H3
- histone H3
- Me or Me1
- monomethylation
- Me2
- dimethylation
- Me3
- trimethylation.
References
- 1. Grunstein M., and Gasser S. M. (2013) Epigenetics in Saccharomyces cerevisiae. Cold Spring Harb. Perspect. Biol. 5, a017491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Krebs J. E. (2007) Moving marks: dynamic histone modifications in yeast. Mol. Biosyst. 3, 590–597 10.1039/b703923a [DOI] [PubMed] [Google Scholar]
- 3. Fuchs S. M., and Quasem I. (2014) Budding yeast as a model to study epigenetics. Drug Discov. Today Dis. Models 12, 1–6 10.1016/j.ddmod.2014.04.004 [DOI] [Google Scholar]
- 4. Lowary P. T., and Widom J. (1989) Higher-order structure of Saccharomyces cerevisiae chromatin. Proc. Natl. Acad. Sci. U.S.A. 86, 8266–8270 10.1073/pnas.86.21.8266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lacoste N., Bhat W., and Côté J. (2017) Purification of yeast native reagents for the analysis of chromatin function-I: nucleosomes for reconstitution and manipulation of histone marks. Methods Mol. Biol. 1528, 39–51 10.1007/978-1-4939-6630-1_3 [DOI] [PubMed] [Google Scholar]
- 6. Rodríguez-Peña J. M., Díez-Muñiz S., Bermejo C., Nombela C., and Arroyo J. (2013) Activation of the yeast cell wall integrity MAPK pathway by zymolyase depends on protease and glucanase activities and requires the mucin-like protein Hkr1 but not Msb2. FEBS Lett. 587, 3675–3680 10.1016/j.febslet.2013.09.030 [DOI] [PubMed] [Google Scholar]
- 7. Sidoli S., and Garcia B. A. (2017) Characterization of individual histone posttranslational modifications and their combinatorial patterns by mass spectrometry-based proteomics strategies. Methods Mol. Biol. 1528, 121–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Côté J., Utley R. T., and Workman J. L. (1995) Basic analysis of transcription factor binding to nucleosomes. Methods Mol. Genet. 6, 108–128 10.1016/S1067-2389(06)80009-9 [DOI] [PubMed] [Google Scholar]
- 9. Lambert J.-P., Mitchell L., Rudner A., Baetz K., and Figeys D. (2009) A novel proteomics approach for the discovery of chromatin-associated protein networks. Mol. Cell Proteomics 8, 870–882 10.1074/mcp.M800447-MCP200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lorch Y., and Kornberg R. D. (1994) Isolation of the yeast histone octamer. Proc. Natl. Acad. Sci. U.S.A. 91, 11032–11034 10.1073/pnas.91.23.11032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lacoste N., Bhat W., and Côté J. (2017) Purification of yeast native reagents for the analysis of chromatin function-II: multiprotein complexes and biochemical assays. Methods Mol. Biol. 1528, 53–67 10.1007/978-1-4939-6630-1_4 [DOI] [PubMed] [Google Scholar]
- 12. Taddei A., and Gasser S. M. (2012) Structure and function in the budding yeast nucleus. Genetics 192, 107–129 10.1534/genetics.112.140608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Aris J. P., and Blobel G. (1991) Isolation of yeast nuclei. Methods Enzymol. 194, 735–749 10.1016/0076-6879(91)94056-I [DOI] [PubMed] [Google Scholar]
- 14. Truong D. M., and Boeke J. D. (2017) Resetting the yeast epigenome with human nucleosomes. Cell 171, 1508–1519.e13 10.1016/j.cell.2017.10.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Rodríguez-Campos A., Shimamura A., and Worcel A. (1989) Assembly and properties of chromatin containing histone H1. J. Mol. Biol. 209, 135–150 10.1016/0022-2836(89)90177-0 [DOI] [PubMed] [Google Scholar]
- 16. Bustin M., Catez F., and Lim J.-H. (2005) The dynamics of histone H1 function in chromatin. Mol. Cell 17, 617–620 10.1016/j.molcel.2005.02.019 [DOI] [PubMed] [Google Scholar]
- 17. Leung A., Cheema M., González-Romero R., Eirin-Lopez J. M., Ausió J., and Nelson C. J. (2016) Unique yeast histone sequences influence octamer and nucleosome stability. FEBS Lett. 590, 2629–2638 10.1002/1873-3468.12266 [DOI] [PubMed] [Google Scholar]
- 18. Klis F. M., Boorsma A., and De Groot P. W. J. (2006) Cell wall construction in Saccharomyces cerevisiae. Yeast 23, 185–202 10.1002/yea.1349 [DOI] [PubMed] [Google Scholar]
- 19. Yu M. Y., and Chang S. T. (1987) Effects of osmotic stabilizers on the activities of mycolytic enzymes used in fungal protoplast liberation. MIRCEN J. 3, 161–167 10.1007/BF00933616 [DOI] [Google Scholar]
- 20. Klis F. M., de Jong M., Brul S., and de Groot P. W. J. (2007) Extraction of cell surface-associated proteins from living yeast cells. Yeast 24, 253–258 10.1002/yea.1476 [DOI] [PubMed] [Google Scholar]
- 21. Valentin E., Herrero E., Pastor F. I. J., and Sentandreu R. (1984) Solubilization and analysis of mannoprotein molecules from the cell wall of Saccharomyces cerevisiae. J. Gen Microbiol. 130, 1419–1428 10.1099/00221287-130-6-1419 [DOI] [Google Scholar]
- 22. Scott J. H., and Schekman R. (1980) Lyticase: endoglucanase and protease activities that act together in yeast cell lysis. J. Bacteriol. 142, 414–423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Knorr D., Shetty K. J., and Kinsella J. E. (1979) Enzymatic lysis of yeast cell walls. Biotechnol. Bioeng. 21, 2011–2021 10.1002/bit.260211109 [DOI] [Google Scholar]
- 24. Nurse N. P., Jimenez-Useche I., Smith I. T., and Yuan C. (2013) Clipping of flexible tails of histones H3 and H4 affects the structure and dynamics of the nucleosome. Biophys. J. 104, 1081–1088 10.1016/j.bpj.2013.01.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Iwasaki W., Miya Y., Horikoshi N., Osakabe A., Taguchi H., Tachiwana H., Shibata T., Kagawa W., and Kurumizaka H. (2013) Contribution of histone N-terminal tails to the structure and stability of nucleosomes. FEBS Open Bio. 3, 363–369 10.1016/j.fob.2013.08.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Shen S. H., Chrétien P., Bastien L., and Slilaty S. N. (1991) Primary sequence of the glucanase gene from Oerskovia xanthineolytica: expression and purification of the enzyme from Escherichia coli. J. Biol. Chem. 266, 1058–1063 [PubMed] [Google Scholar]
- 27. Palumbo J. D., Sullivan R. F., and Kobayashi D. Y. (2003) Molecular characterization and expression in Escherichia coli of three β-1,3-glucanase genes from Lysobacter enzymogenes strain N4-7. J. Bacteriol. 185, 4362–4370 10.1128/JB.185.15.4362-4370.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Shechter D., Dormann H. L., Allis C. D., and Hake S. B. (2007) Extraction, purification and analysis of histones. Nat. Protoc. 2, 1445–1457 10.1038/nprot.2007.202 [DOI] [PubMed] [Google Scholar]
- 29. Zinser E., and Daum G. (1995) Isolation and biochemical characterization of organelles from the yeast, Saccharomyces cerevisiae. Yeast 11, 493–536 10.1002/yea.320110602 [DOI] [PubMed] [Google Scholar]
- 30. von Holt C., Brandt W. F., Greyling H. J., Lindsey G. G., Retief J. D., Rodrigues J. D., Schwager S., and Sewell B. T. (1989) Isolation and characterization of histones. Methods Enzymol. 170, 431–523 10.1016/0076-6879(89)70061-6 [DOI] [PubMed] [Google Scholar]
- 31. Guo X. W., and Cole R. D. (1989) Chromatin aggregation changes substantially as pH varies within the physiological range. J. Biol. Chem. 264, 11653–11657 [PubMed] [Google Scholar]
- 32. Kiseleva E., Allen T. D., Rutherford S. A., Murray S., Morozova K., Gardiner F., Goldberg M. W., and Drummond S. P. (2007) A protocol for isolation and visualization of yeast nuclei by scanning electron microscopy (SEM). Nat. Protoc. 2, 1943–1953 10.1038/nprot.2007.251 [DOI] [PubMed] [Google Scholar]
- 33. White C. L., Suto R. K., and Luger K. (2001) Structure of the yeast nucleosome core particle reveals fundamental changes in internucleosome interactions. EMBO J. 20, 5207–5218 10.1093/emboj/20.18.5207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Allahverdi A., Chen Q., Korolev N., and Nordenskiöld L. (2015) Chromatin compaction under mixed salt conditions: opposite effects of sodium and potassium ions on nucleosome array folding. Sci. Rep. 5, 8512 10.1038/srep08512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lee K. P., Baxter H. J., Guillemette J. G., Lawford H. G., and Lewis P. N. (1982) Structural studies on yeast nucleosomes. Can. J. Biochem. 60, 379–388 10.1139/o82-045 [DOI] [PubMed] [Google Scholar]
- 36. Preuss D., Mulholland J., Kaiser C. A., Orlean P., Albright C., Rose M. D., Robbins P. W., and Botstein D. (1991) Structure of the yeast endoplasmic reticulum: localization of ER proteins using immunofluorescence and immunoelectron microscopy. Yeast 7, 891–911 10.1002/yea.320070902 [DOI] [PubMed] [Google Scholar]
- 37. Wingfield P. (1998) Protein precipitation using ammonium sulfate. Curr. Protoc. Protein Sci. 13, A.3F.1–A.3F.8 10.1002/0471140864.psa03fs13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Fan Y., Korolev N., Lyubartsev A. P., and Nordenskiöld L. (2013) An advanced coarse-grained nucleosome core particle model for computer simulations of nucleosome-nucleosome interactions under varying ionic conditions. PLoS One 8, e54228–16 10.1371/journal.pone.0054228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Korolev N., Allahverdi A., Lyubartsev A. P., and Nordenskiöld L. (2012) The polyelectrolyte properties of chromatin. Soft Matter 8, 9322–9333 10.1039/c2sm25662b [DOI] [Google Scholar]
- 40. Simon R. H., and Felsenfeld G. (1979) A new procedure for purifying histone pairs H2A+H2B and H3+H4 from chromatin using hydroxylapatite. Nucleic Acids Res. 6, 689–696 10.1093/nar/6.2.689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Brand M., Rampalli S., Chaturvedi C.-P., and Dilworth F. J. (2008) Analysis of epigenetic modifications of chromatin at specific gene loci by native chromatin immunoprecipitation of nucleosomes isolated using hydroxyapatite chromatography. Nat. Protoc. 3, 398–409 10.1038/nprot.2008.8 [DOI] [PubMed] [Google Scholar]
- 42. Garcia B. A., Hake S. B., Diaz R. L., Kauer M., Morris S. A., Recht J., Shabanowitz J., Mishra N., Strahl B. D., Allis C. D., and Hunt D. F. (2007) Organismal differences in post-translational modifications in histones H3 and H4. J. Biol. Chem. 282, 7641–7655 10.1074/jbc.M607900200 [DOI] [PubMed] [Google Scholar]
- 43. O'Neill L. P., and Turner B. M. (2003) Immunoprecipitation of native chromatin: NChIP. Methods 31, 76–82 10.1016/S1046-2023(03)00090-2 [DOI] [PubMed] [Google Scholar]
- 44. Su Z., and Denu J. M. (2001) MARCC (matrix-assisted reader chromatin capture): an antibody-free method to enrich and analyze combinatorial nucleosome modifications. Curr. Protoc. Mol. Biol. 6, 21.32.1–21.32.21 10.1002/0471142727.mb2132s111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lacoste N., Utley R. T., Hunter J. M., Poirier G. G., and Côte J. (2002) Disruptor of telomeric silencing-1 is a chromatin-specific histone H3 methyltransferase. J. Biol. Chem. 277, 30421–30424 10.1074/jbc.C200366200 [DOI] [PubMed] [Google Scholar]
- 46. Ovalle R., Lim S. T., Holder B., Jue C. K., Moore C. W., and Lipke P. N. (1998) A spheroplast rate assay for determination of cell wall integrity in yeast. Yeast 14, 1159–1166 10.1002/(SICI)1097-0061(19980930)14:13%3C1159::AID-YEA317%3E3.0.CO%3B2-3 [DOI] [PubMed] [Google Scholar]
- 47. Fic E., Kedracka-Krok S., Jankowska U., Pirog A., and Dziedzicka-Wasylewska M. (2010) Comparison of protein precipitation methods for various rat brain structures prior to proteomic analysis. Electrophoresis 31, 3573–3579 10.1002/elps.201000197 [DOI] [PubMed] [Google Scholar]
- 48. Wang D., Fang S., and Wohlhueter R. M. (2009) N-terminal derivatization of peptides with isothiocyanate analogues promoting Edman-type cleavage and enhancing sensitivity in electrospray ionization tandem mass spectrometry analysis. Anal. Chem. 81, 1893–1900 10.1021/ac8021136 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.