Abstract
Background:
Neuropathic pain is one of the common complications after spinal cord injury (SCI), affecting individuals’ quality of life. The molecular mechanism for neuropathic pain after SCI is still unclear. We aimed to discover potential genes and microRNAs (miRNAs) related to neuropathic pain by the bioinformatics method.
Methods:
Microarray data of GSE69901 were obtained from Gene Expression Omnibus (GEO) database. Peripheral blood samples from individuals with or without neuropathic pain after SCI were collected. Twelve samples from individuals with neuropathic pain and 13 samples from individuals without pain as controls were included in the downloaded microarray. Differentially expressed genes (DEGs) between the neuropathic pain group and the control group were detected using the GEO2R online tool. Functional enrichment analysis of DEGs was performed using the DAVID database. Protein-protein interaction network was constructed from the STRING database. MiRNAs targeting these DEGs were obtained from the miRNet database. A merged miRNA-DEG network was constructed and analyzed with Cytoscape software.
Results:
In total, 1134 DEGs were identified between individuals with or without neuropathic pain (case and control), and 454 biological processes were enriched. We identified 4 targeted miRNAs, including mir-204-5p, mir-519d-3p, mir-20b-5p, mir-6838-5p, which may be potential biomarkers for SCI patients.
Conclusion:
Protein modification and regulation of the biological process of the central nervous system may be a risk factor in SCI. Certain genes and miRNAs may be potential biomarkers for the prediction of and potential targets for the prevention and treatment of neuropathic pain after SCI.
Key Words: bioinformatics, biomarkers, neuropathic pain, spinal cord injured
Neuropathic pain is one of the most common symptoms of patients after spinal cord injury (SCI). It is a severe sensory deficit that is experienced and affects about 80% of individuals with SCI.1,2 The condition may lead to lifelong loss of function, affected quality of life, and increased morbidity and mortality.3 It is important to clarify molecular mechanism for treatment of neuropathic pain after SCI.
A number of studies are increasingly focusing on pain after SCI. Multiple mechanisms are raised to explain the neuropathic pain, including peripheral and central sensitization as the major issues. For peripheral sensitization, tissue inflammation may change the chemical environment of the peripheral injured site and cause neuropathic pain. Proinflammatory mediators like leukotriene B4 are contributed to the recruitment of inflammatory cells.4 Expression of the capsaicin-sensitive cation channel transient receptor potential vanilloid type 1 is observed in the dorsal root ganglion neurons after SCI.5,6 Central sensitization may amplify the synaptic transfer from the nociceptor to the spinal cord. The glutamate-activated N-methyl-D-aspartic acid receptor may be related to the process of central sensitization.7,8 Neuroimmune interaction with neurotrophic factor (brain-derived neurotrophic factor), substance P, neurokinin 1 (NK1), dynorphin, and cyclooxygenase 2 are also described as chemical signals associated with the central sensitization process.9–11 However, there have been few studies on microRNAs (miRNAs) involved in neuropathic pain after SCI. Thus, ongoing studies are required to further evaluate potential genes and miRNAs related to neuropathic pain caused by SCI.
In this study, microarray data from the Gene Expression Omnibus (GEO) database, and the differentially expressed genes (DEGs), were identified between individuals with or without neuropathic pain after SCI. miRNAs targeting the DEGs were also included. We aimed to explore new molecular biomarkers or potential therapeutic targets for neuropathic pain after SCI.
METHODS
Microarray Data Search and Selection of Eligible Data Set
We downloaded the microarray data of GSE69901 from the GEO database (www.ncbi.nlm.nih.gov/geo/) with its microarray platform as GPL15207 (Affymetrix Human Gene Expression Array). The gene expression files were uploaded by Yilmaz B and Adigüzel E. In this microarray, 12 samples of peripheral blood were obtained from individuals with neuropathic pain after SCI and 13 samples without pain as controls. Microarray analysis was performed to identify the gene expression pattern.
DEGs’ Screening
DEGs were screened using the online tool GEO2R/R package limma. In the present study, DEGs between neuropathic pain group and control group were screened and selected by the cut-off point of P-value <0.05 and |logFC|>0.5.
Functional Enrichment Analysis
The selected DEGs were then deposited to the Database for Annotation, Visualization, and Integrated Discovery (DAVID) version 6.8 Beta (https://david-d.ncifcrf.gov/) for further analysis. The DAVID database offers biological function annotation for researchers.12,13 In this study, the DAVID database was applied to investigate GO annotation and KEGG pathways of DEGs. The biological processes and pathways might contribute to the neuropathic pain after SCI. P-value <0.05 was chosen as the threshold.
Protein-Protein Interaction (PPI) Analysis
PPI of DEGs was obtained from Search Tool for the Retrieval of Interacting Genes (STRING, http://string-db.org/).14 The STRING database is an online database resource with comprehensive information of interactions of proteins from prediction or experiments. In our study, confidence score >0.4 of PPI was the selection threshold to construct the PPI network. The list of PPI pairs was downloaded for further analysis.
miRNAs Targeting the DEGs
We uploaded the DEGs to the database, miRNet, (www.mirnet.ca/faces/home.xhtml)15 to obtain the miRNAs targeting the screened DEGs. The miRNet database is an online tool with various kinds of information generated from miRs studies. Researchers can build miRNA-target interaction networks in the help of this database. The generated list of miRNA-DEG pairs was preserved for further analysis.
Construction of miRNA-DEG Network
Both miRNA-DEG pairs and PPI pairs were then merged to be the miRNA-DEG network and visualized in Cytoscape software 3.4.0.16 Module clustering analysis for the network was then performed by the Molecular Complex Detection (MCODE)17 plugin, to detect the potential functional modules in the network. The degree cut-off value to 2 and the node score cut-off to 0.2 were set in the MCODE process.
RESULTS
DEG Screening Between Patients and Controls
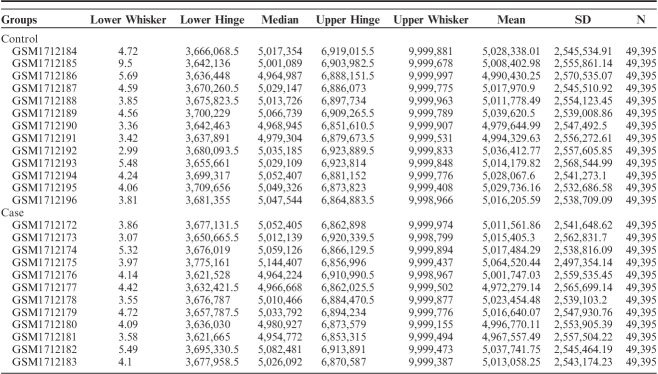
Data normalization and cross-comparability were assessed (Table 1), and then the DEGs analysis was performed. A total of 1134 DEGs were selected by the cut-off point of P-value <0.05 and |logFC|>0.5, which included 489 upregulated and 645 downregulated DEGs. The most significant top 10 upregulated or downregulated genes are shown in Table 2. The upregulated gene with the smallest P-value (P=0.021889) was GLG1 (Golgi glycoprotein 1), and the downregulated gene with the smallest P-value (0.048081) was HPCAL4 (hippocalcin-like 4).
TABLE 1.
Data Normalization and Cross-comparability
TABLE 2.
The 10 Most Strongly Upregulated Genes or Downregulated Genes in SCI
Functional Enrichment Analysis
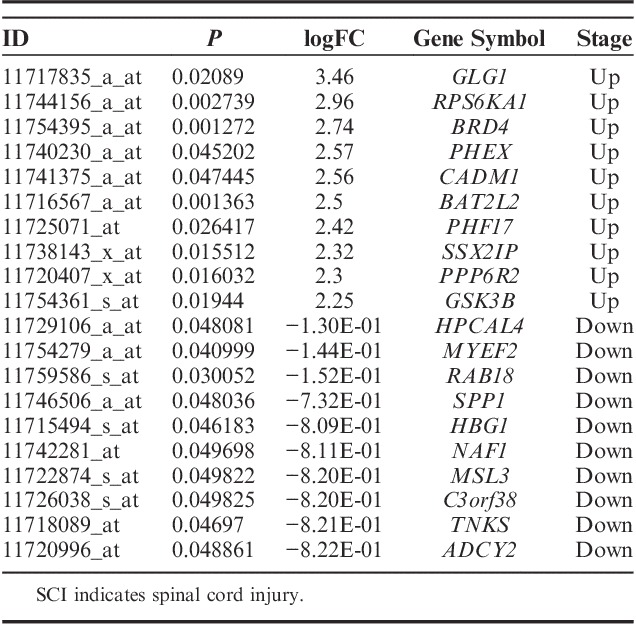
Four hundred eighty-nine GO terms (P<0.05) were significantly enriched by upregulated DEGs, whereas 645 genes (P<0.05) were significantly enriched by downregulated DEGs. Many of these enriched terms were associated with neuron cell development or inflammatory processes involved in protein modification and regulation biological processes in the central nervous system. We recognized significant enrichments of the DEGs (top 20, Table 3), which were classified in 454 GO categories. Most of the categories were about the nervous system, and the extremely significant enrichment GO category was nervous system development with a P-value of 6.31E-07. Other significant categories covered epithelial cell migration and cellular response chemical stimulus processes. To further research the functions of the DEGs, we represented significant enrichments of the DEGs to the KEGG database (top 20 Table 4). We identified 21 significant pathways based on KEGG database analysis. The most significant pathway in our KEGG analysis was dopaminergic synapse with a P-value of 1.88E-05.
TABLE 3.
The Top 20 Enriched GO Terms Among the DEGs in SCI
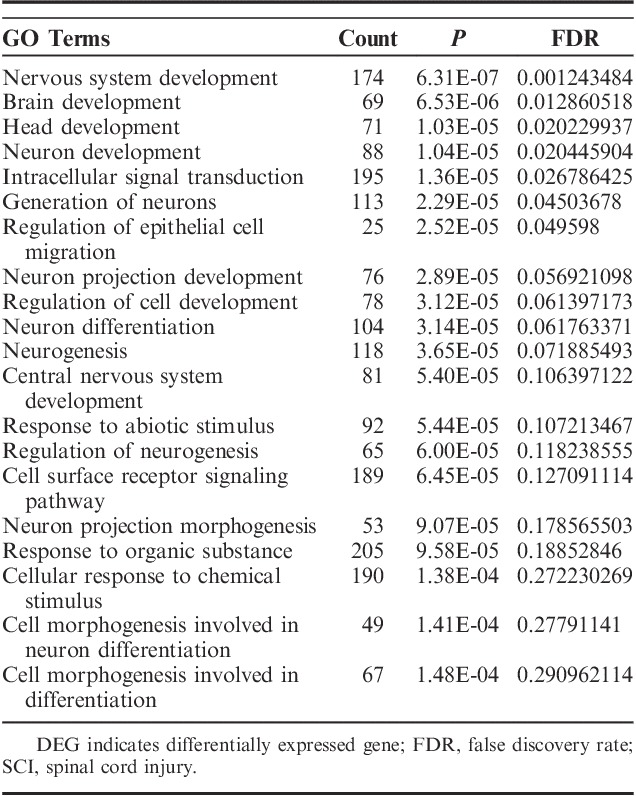
TABLE 4.
The Top 20 Most Significant Pathways Identified in the KEGG Database
Construction of miRNA-DEG Network Analysis
Taking the selected 1134 DEGs into account, we identified 2236 PPI pairs by STRING, as well as 1269 miRNA-DEG pairs by miRNet through the software of Cytoscape, and 2 big pairing pictures were generated. The pictures mentioned above were merged, and an intact tremendous miRNAs-DEG network was generated in Cytoscape. To assess the key functional modules of this network, module clustering based on the miRNA-DEG network mentioned above was then performed by the MCODE plugin of Cytoscape. Three modules were identified and showed DEGs (Fig. 1), such as IL22RA1, IFNA21, IFNA2, NUP50, SSU72, USP42, FZD1, and CSNK1A1, and targeted miRNAs, such as mir-204-5p, mir-519d-3p, mir-20b-5p, and mir-6838-5p. Two outstanding hub DEGs, FZD1 and IL22RA1 (black rhombus, Fig. 1) in these modules also occurred in the GO terms enriched above, and their interacted DEGs, CSNK1A1 and miRNA like mir-204-5p (white rectangle linked with black rhombus, Fig. 2), IFNA21, IFNA2 (white rectangle linked with black rhombus, Fig. 3), associated with inflammatory or neurons development of protein modification and regulation processes. We also observed a key hub miRNA, mir-6838-5p (black rhombus, Fig. 1), that interacted with DEGs, including NUP50, SSU72, and USP42 (white linked with black rectangle, Fig. 4).
FIGURE 1.
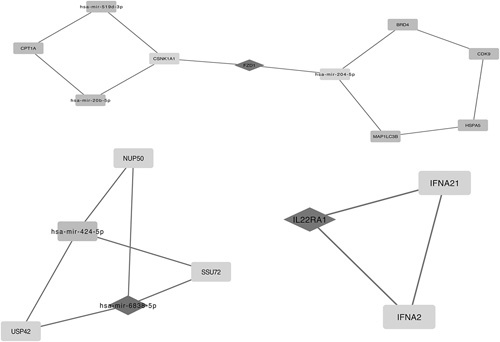
2236 protein-protein interaction pairs by STRING, as well as 1269 microRNAs-differentially expressed gene (miRNA-DEG) pairs by miRNet through the software of Cytoscape, and 2 big pairing pictures were generated. Pictures above were merged, and an intact tremendous miRNAs-DEG network was generated in Cytoscape. Three modules above were identified and showed DEGs, such as IL22RA1, IFNA21, IFNA2, NUP50, SSU72, USP42, FZD1, and CSNK1A1, and targeted miRNAs, such as mir-204-5p, mir-519d-3p, mir-20b-5p, and mir-6838-5p.
FIGURE 2.

An outstanding hub differentially expressed gene (DEG), FZD1 (black rhombus), its interacted DEG such as CSNK1A1, and miR such as mir-204-5p (white linked with black rectangle).
FIGURE 3.
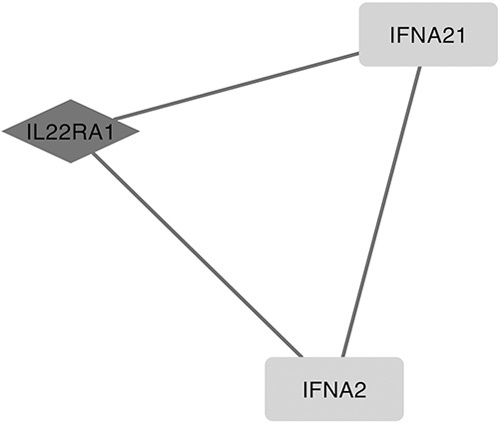
Another outstanding hub differentially expressed gene (DEG), IL22Ra1 (black rhombus), and its interacted DEG such as IFNA21 and IFNA2 (white linked with black rectangle).
FIGURE 4.
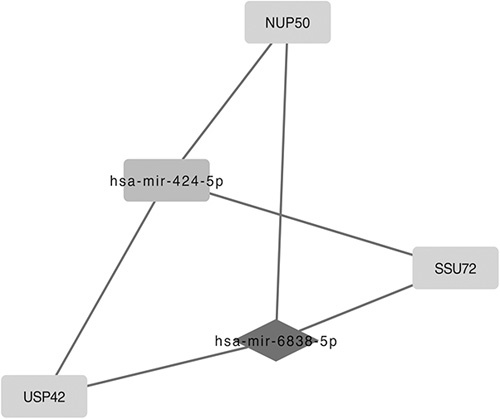
A key hub microRNA, mir-6838-5p (black rhombus) and its interacted differentially expressed genes (DEGs) such as NUP50, SSU72, and USP42 (white linked with black rectangle).
DISCUSSION
Neuropathic pain following SCI is caused by dysfunction of the nervous system or damage of nervous cells.1 Almost one third of people with SCI will experience continuous neuropathic pain. It is difficult to treat and reduces the quality of life for these individuals.18 It is a pity that nonpharmacological approaches may be more useful to neuropathic pain, and the same current management does not achieve satisfactory pain reduction.3 It is necessary to share many more quality data about SCI between different research centers or countries, to better recognize how to treat SCI, as well as more clinical studies or novel management methods to evaluate the effect of treatments.19
As the miRNA-DEG network analysis shows, neuropathic pain after SCI was closely related to the FZD1 gene. Previous studies showed that FZD1 is the transmembrane receptor of Wnt signaling pathways,20,21 Wnt-protein family played an important regulation role in the neural development of the hippocampus in the human central nervous system.22 Wnt proteins were involved in almost all aspects of development in the nervous system.23 Researches had shown that the Wnt signaling was associated with the maintenance of neural stem cells by Muriel et al,24 and the study also found that Wnt signaling pathways regulated new neurons generated from neural stem cells in adult hippocampus. FZD1 was an important receptor for Wnt signaling, highly expressed in the dentate gyrus neural stem cells.24 FZD1 knockdown led to a marked decrease in the differentiation of neural stem cells into neurons, and increased the generation of astrocytes, significantly. Studies had demonstrated that stem cells could be used to treat some common forms of neuropathic pain, including SCI and sciatic nerve injury; this research was confirmed in animal models by Sudhakar et al.25 Moreover, a large number of studies had shown that astrocytes played an important role in the central sensitization of pain, especially for chronic pain.26–28 Thus, we speculated that FDZ1 could regulate the development of neural stem cells, and inhibit the production of astrocytes, relieving the inflammation response triggered by SCI, and alleviate neuropathic pain caused by SCI through Wnt signaling pathways.
As expected, inflammatory factors and mediators, such as interleukins, cyclooxygenase 2, calcitonin gene-related peptide, tumor necrosis factor-α, were involved in the occurrence and development of pain.29–31 In our study, through functional enrichment analysis, we found that IL22RA1 connected with neuropathic pain after SCI. IL22RA1 was a unique receptor of IL-22.32 IL-22 was an IL-10 family cytokine generated by Th17, Th22, and Th1 cells, and IL-22 played an significant role in T cell-mediated inflammatory diseases.33 IL22RA1 and its associated genes through cytoscape analyzing, including IFNA21 and IFNA2, mainly related to inflammation reaction in the GO-BP database. They may be the key genes of initiating the inflammatory response. Through MCODE analysis, we found that mir-6838-5p which may be the core of the module. Mir-6838-5p was associated with genetic RPS6KB1. The studies had shown that RPS6KB1 was closely related to the inflammatory and immune response.34,35 Mir-6838-5p together with its targeted genes, such as NUP50, SSU72, and USP42, were closely associated with protein modification and biosynthesis through GO-BP database analysis. Therefore, we argued that IL22RA1 and mir-6838-5p played a vital role as inflammation reaction mediators in neuropathic pain induced by SCI.
In conclusion, FDZ1 could regulate the differentiation of neural stem cells into neurons and induce astrocyte proliferation. An important supporter of neuron cells, astrocytes can generate inflammatory mediators and mediate central sensitization of pain.36,37 Numerous researches suggest that astrocytes have also played a key role in immunoreaction.38–40 As a member of the IL-10 family, IL-22 has an effect in the inflammatory response. Mir-6838-5p could induce an immunoreaction through immunoregulation. Hence, we argue that astrocytes can be activated, releasing inflammatory mediators or immunomodulatory factor when the spinal cord is injured, inducing inflammation or immune response, resulting in central sensitization of acute pain and then producing chronic pain. Downregulation of FDZ1 would inhibit astrocytes activation, inflammatory medium (IL22) release and some genes expression (mir-6838-5p, IL22RA1). Thus, it may stop acute pain to translate into chronic pain. This would be beneficial for the treatment of intractable neuropathic pain induced by SCI.
Nevertheless, the currently popular molecules such as mitogen-activated protein kinase and nuclear factor kappa beta, are all involved in the formation and development of inflammation response pain or neuropathic pain.41–44 But in our study, these genes were not being filtered, it did not mean these signaling molecules or pathways not participation in neuropathic pain after SCI, it maybe not in the core position in the development of developing the pain, or further studies are needed in the future.
CONCLUSION
In conclusion, FZD1, IL22RA1, and mir-6838-5p might become potential biomarkers for the prediction of and new targets for prevention and treatment of neuropathic pain after SCI. However, further studies are necessary for verifying the clinical applications of these findings.
Footnotes
Y.W. and F.Y. contributed equally.
F.Y: data collection and writing up of the first draft of the paper. Y.W: data analysis. W.T: study design and modifying paper.
Supported by the Natural Scientific Foundation of Guangdong Province, 2016A030313255 and the Foundation of Sun Yat-sen University for Young Teachers, 16ykpy36. Both belong to Z.W. The remaining authors declare no conflict of interest.
REFERENCES
- 1.Finnerup NB, Norrbrink C, Trok K, et al. Phenotypes and predictors of pain following traumatic spinal cord injury: a prospective study. J Pain. 2014;15:40–48. [DOI] [PubMed] [Google Scholar]
- 2.Cragg JJ, Noonan VK, Luc N, et al. Neuropathic pain, depression, and cardiovascular disease: a national multicenter study. Neuroepidemiology. 2015;44:130–137. [DOI] [PubMed] [Google Scholar]
- 3.Ellen MH, Tiina R. Management of neuropathic pain associated with spinal cord injury. J Pain Ther. 2015;4:51–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Harry J. Gould III. Complete Freund's adjuvant-induced hyperalgesia: a human perception. Pain. 2000;85:301–303. [DOI] [PubMed] [Google Scholar]
- 5.Ramer LM, van Stolk AP, Inskip JA, et al. Plasticity of TRPV1-expressing sensory neurons mediating autonomic dysreflexia following spinal cord injury. J Front Physiol. 2012;3:257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu Z, Yang Q, Crook R, et al. TRPV1 channels make major contributions to behavioral hypersensitivity and spontaneous activity in nociceptors after spinal cord injury. J Pain. 2013;154:2130–2141. [DOI] [PubMed] [Google Scholar]
- 7.Woolf CJ. Pain: moving from symptom control toward mechanism-specific pharmacologic management. J Ann Intern Med. 2004;140:441–451. [DOI] [PubMed] [Google Scholar]
- 8.Woolf CJ, Thompson SW. The induction and maintenance of central sensitization is dependent on N-methyl-D-aspartic acid receptor activation; implications for the treatment of post-injury pain hypersensitivity states. J Pain. 1991;44:293–299. [DOI] [PubMed] [Google Scholar]
- 9.Neumann S, Doubell TP, Leslie T, et al. Inflammatory pain hypersensitivity mediated by phenotypic switch in myelinated primary sensory neurons. J Nature. 1996;384:360–364. [DOI] [PubMed] [Google Scholar]
- 10.Mannion RJ, Costigan M, Decosterd I, et al. Neurotrophins: peripherally and centrally acting modulators of tactile stimulus-induced inflammatory pain hypersensitivity. J Proc Natl Acad Sci USA. 1999;96:9385–9390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ji RR, Befort K, Brenner GJ, et al. ERK-MAP kinase activation in superficial spinal cord neurons induces prodynorphin and NK-1 upregulation and contributes to persistent inflammatory pain hypersensitivity. J Neurosci. 2002;22:478–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. J Nat Protoc. 2009;4:44–57. [DOI] [PubMed] [Google Scholar]
- 13.Huang DW, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. J Nucleic Acids Res. 2009;37:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Szklarczyk D, Franceschini A, Kuhn M, et al. The STRING database in 2011: functional interaction networks of proteins, globally integrated and scored. J Nucleic Acids Res. 2011;3:D561–D568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fan Y, Siklenka K, Arora SK, et al. miRNet-dissecting miRNA-target interactions and functional associations through network-based visual analysis. J Nucleic Acids Res. 2016;44:W135–W141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Saito R, Smoot ME, Ono K, et al. A travel guide to cytoscape plugins. J Nat Methods. 2012;9:1069–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang J, Zhong J, Chen G, et al. ClusterViz: a cytoscape APP for cluster analysis of biological network. IEEE/ACM Trans Comput Biol Bioinform. 2015;12:815–822. [DOI] [PubMed] [Google Scholar]
- 18.Woller SA, Hook MA. Opioid administration following spinal cord injury: implications for pain and locomotor recovery. J Exp Neurol. 2013;247:328–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fin BS, Vanessa KN. Standardization of data for clinical use and research in spinal cord injury. J Brain Sci. 2016;12:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yua SB, Laura MY, Chua YX, et al. E2F1 effects on osteoblast differentiation and mineralization are mediated through up-regulation of frizzled-1. J Bone. 2013;56:234–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Flahaut M, Meier R, Coulon A, et al. The Wnt receptor FZD1 mediates chemoresistance in neuroblastoma through activation of the Wnt/beta-catenin pathway. J Oncogene. 2009;11:2245–2256. [DOI] [PubMed] [Google Scholar]
- 22.Theologos M, Michaelidis D, Chichung L. Wnt signaling and neural stem cells: caught in the Wnt web. J Cell Tissue Res. 2008;331:193–210. [DOI] [PubMed] [Google Scholar]
- 23.Jensen M, Brockie PJ, Maricq AV. Wnt signaling regulates experience-dependent synaptic plasticity in the adult nervous system. J Cell Cycle. 2012;15:2585–2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Muriel DM, Gabriela AA, Manuel VG, et al. Frizzled-1 receptor regulates adult hippocampal neurogenesis. J Molecular Brain. 2016;9:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sudhakar V, Matthew W, Daniel J, et al. Potential role of stem cells for neuropathic pain disorders. J Neurosurg Focus. 2013;35:E11. [DOI] [PubMed] [Google Scholar]
- 26.Koyanagi S, Kusunose N, Taniguchi M, et al. Glucocorticoid regulation of ATP release from spinal astrocytes underlies diurnal exacerbation of neuropathic mechanical allodynia. J Nat Commun. 2016;7:131102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lu Y, Jiang BC, Cao DL, et al. TRAF6 up-regulation in spinal astrocytes maintains neuropathic pain by integrating TNF-α and IL-1βsignaling. J Pain. 2014;155:2618–2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ikeda H, Kiritoshi T, Murase K. Contribution of microglia and astrocytes to the central sensitization, inflammatory and neuropathic pain in the juvenile rat. J Mol Pain. 2012;15:43–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Van DB, Potier E, van DM, et al. Reduced tonicity stimulates an inflammatory response in nucleus pulposus tissue that can be limited by a COX-2-specific inhibitor. J Orthop Res. 2015;33:1724–1731. [DOI] [PubMed] [Google Scholar]
- 30.Zychowska M, Rojewska E, Makuch W, et al. Participation of pro- and anti-nociceptive interleukins in botulinum toxin A-induced analgesia in a rat model of neuropathic pain. Eur J Pharmacol. 2016;15:377–388. [DOI] [PubMed] [Google Scholar]
- 31.Hirsch S, Birklein F. Pain-relieving effect of CGRP antagonism on inflammatory pain. J Schmerz. 2014;28:532–535. [DOI] [PubMed] [Google Scholar]
- 32.Lim C, Savan R. The role of the IL-22/IL-22R1 axis in cancer. J Cytokine Growth Factor Rev. 2014;25:257–271. [DOI] [PubMed] [Google Scholar]
- 33.Lerman G, Sharon M, Leibowitz-Amit R, et al. The crosstalk between IL-22 signaling and miR-197 in human keratinocytes. PLoS One. 2014;10:e107467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Catalán V, Gómez-AJ, Rodríguez A, et al. Expression of S6K1 in human visceral adipose tissue is unregulated in obesity and related to insulin resistance and inflammation. J Acta Diabetol. 2015;52:257–266. [DOI] [PubMed] [Google Scholar]
- 35.Zhao XF, Zhao MY, Chai L, et al. Amplified RPS6KB1 and CDC2 genes are potential biomarkers for aggressive HIV+/EBV+ diffuse large B-cell lymphomas. Int J Clin Exp Pathol. 2013;6:148–154. [PMC free article] [PubMed] [Google Scholar]
- 36.Ji RR, Berta T, Nedergaard M. Glia and pain: Is chronic pain a gliopathy? J Pain. 2013;154(suppl 1):S10–S28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chiang CY, Sessle BJ, Dostrovsky JO. Role of astrocytes in pain. J Neurochem Res. 2012;37:2419–2431. [DOI] [PubMed] [Google Scholar]
- 38.Alvarez MI, Rivas L, Lacruz C, et al. Astroglial cell subtypes in the cerebella of normal adults, elderly adults, and patients with Alzheimer’s disease: a histological and immunohistochemical comparison. J Glia. 2015;63:287–312. [DOI] [PubMed] [Google Scholar]
- 39.Hwang IK, Yoo KY, Kim DW, et al. Changes in the expression of mitochondrial peroxiredoxin and thioredoxin in neurons and glia and their protective effects in experimental cerebral ischemic damage. J Free Radic Biol Med. 2010;48:1242–1251. [DOI] [PubMed] [Google Scholar]
- 40.Park OK, Yoo KY, Lee CH, et al. Arylalkylamine N-acetyltransferase (AANAT) is expressed in astrocytes and melatonin treatment maintains AANAT in the gerbil hippocampus induced by transient cerebral ischemia. J Neurol Sci. 2010;294:7–17. [DOI] [PubMed] [Google Scholar]
- 41.Luo C, Zhang YL, Luo W, et al. Differential effects of general anesthetics on anxiety-like behavior in formalin-induced pain: involvement of ERK activation in the anterior cingulate cortex. J Psychopharmacology. 2015;232:4433–4444. [DOI] [PubMed] [Google Scholar]
- 42.Sanna MD, Ghelardini C, Galeotti N. Blockade of the spinal BDNF-activated JNK pathway prevents the development of antiretroviral-induced neuropathic pain. Neuropharmacology. 2016;105:543–552. [DOI] [PubMed] [Google Scholar]
- 43.Dominguez E, Rivat C, Pommier B, et al. JAK/STAT3 pathway is activated in spinal cord microglia after peripheral nerve injury and contributes to neuropathic pain development in rat. J Neurochem. 2008;107:50–60. [DOI] [PubMed] [Google Scholar]
- 44.Arora V, Kuhad A, Tiwari V, et al. Curcumin ameliorates reserpine-induced pain-depression dyad: behavioural, biochemical, neurochemical and molecular evidences. Psychoneuroendocrinology. 2011;36:1570–1581. [DOI] [PubMed] [Google Scholar]