Abstract
The cornea is the outermost layer of the eye and is a vital component of focusing incoming light on the retina. Central corneal thickness (CCT) is now recognized to have a significant role in ocular health and is a risk factor for various ocular diseases, such as keratoconus and primary open angle glaucoma. Most previous genetic studies utilized European and Asian subjects to identify genetic loci associated with CCT. Minority populations, such as Latinos, may aid in identifying additional loci and improve our understanding of the genetic architecture of CCT. In this study, we conducted a genome-wide association study (GWAS) in Latinos, a traditionally understudied population in genetic research, to further identify loci contributing to CCT. Study participants were genotyped using either the Illumina OmniExpress BeadChip (∼730K markers) or the Illumina Hispanic/SOL BeadChip (∼2.5 million markers). All study participants were 40 years of age and older. We assessed the association between individual single nucleotide polymorphisms (SNPs) and CCT using linear regression, adjusting for age, gender and principal components of genetic ancestry. To expand genomic coverage and to interrogate additional SNPs, we imputed SNPs from the 1000 Genomes Project reference panels. We identified a novel SNP, rs10453441 (P = 6.01E-09), in an intron of WNT7B that is associated with CCT. Furthermore, WNT7B is expressed in the human cornea. We also replicated 11 previously reported loci, including IBTK, RXRA-COL5A1, COL5A1, FOXO1, LRRK1 and ZNF469 (P < 1.25E-3). These findings provide further insight into the genetic architecture of CCT and illustrate that the use of minority groups in GWAS will help identify additional loci.
Introduction
The cornea is the clear, outermost layer of the eye that aids in refracting incoming light onto the lens, which is then further refocused onto the retina. Compared to the lens, the cornea has greater refractive power and thus, is a critical ocular structure to focus light on the retina. Due to the large refractive power, any structural variations of the cornea (i.e. smoothness, curvature, or thickness) may result in adverse effects on vision. Mounting evidence suggests central corneal thickness (CCT) plays a significant factor in ocular health. Thin CCT has been observed in several visual disorders, including brittle cornea syndrome, keratoconus and osteogenesis imperfecta (1–3). CCT is also an important endophenotype for primary open angle glaucoma (POAG). Most notably, thin CCT has been found to be associated with POAG in multiple epidemiologic studies (4–10). Therefore, understanding the factors that influence CCT may help to better predict CCT and potentially, POAG.
Ethnic differences in CCT have been observed in several studies. Generally, Hispanics have slightly higher CCT measurements than Caucasians, while individuals of African descent exhibit the lowest CCT, and CCT from Asian populations representing a wide range of values (7,11–13). While environmental factors may contribute to the observed racial variation, genetic variants specific to certain ethnic populations are more likely to have a major role in CCT determination (14). Moreover, CCT is a highly heritable trait, with heritability ranging from 0.60 to 0.95 (15,16). Taken together, these findings suggest that the primary factors determining CCT are genetic rather than environmental.
Recently, genome-wide association studies (GWAS) have identified numerous genetic loci that are associated with CCT in various ethnic populations. These GWAS, conducted in European, Asian and Latino populations, have identified single nucleotide polymorphisms (SNPs) in or nearby AKAP13 (17,18), COL5A1 (18,19), COL8A2 (19), FAM53B (20), FOXO1 (20), IBTK (17), LRRK1 (17), RXRA-COL5A1 (18,19,21,22) and ZNF469 (18–21) to be associated with CCT. A meta-analysis consisting of a large number of European and Asian individuals identified 16 additional loci associated with CCT, including LPAR1 and ARID5B (23). Furthermore, we reported the first GWAS on CCT in Latinos and replicated previously reported loci, as well as identified novel SNPs (22).
While genetic loci associated with CCT have consistently been identified, these loci have primarily been found in populations of European and Asian descent. We hypothesize performing GWAS in minority populations may identify new loci associated with CCT, as well as elucidate the racial differences of CCT. Latinos, the largest minority group in the United States, are traditionally an understudied population among genetic studies. Furthermore, Latinos exhibit thicker CCT compared to other ethnic groups. Taken together, identifying novel genetic variants associated with CCT in Latinos may elucidate racial differences and further our understanding of the genetic architecture of CCT. Therefore, the purpose of this study is to conduct a larger GWAS using Latino subjects to identify novel genetic loci for CCT. To our knowledge, this is the largest GWAS for CCT conducted in a Latino population.
Results
Study sample
The descriptive statistics of the study sample are presented in Table 1. Overall, a total of 4,515 individuals were used to assess the associations between SNPs and CCT, of which 3,584 unrelated individuals were used as a discovery set and 931 related individuals were used in a replication set. The overall mean age (standard deviation) is 54.8 (10.5), with the discovery set and replication set having mean ages of 54.2 (9.9) and 56.9 (12.5), respectively. The proportion of females in the study sample is 58.8%, with the discovery set composed of 56.5% women and the replication set composed of 68.0% women. Combined, the CCT values are normally distributed with an overall mean of 550.0 (33.4) μm, with the average of the discovery and replicate sets as 550.4 (33.2) μm (range: 443–668) and 548.7 (34.0) μm (range: 451–661), respectively.
Table 1.
Descriptive statistics of the study sample
| Study | Sample Size | Females, % | Age (years), Mean (SD) | CCT (μm), Mean (SD) | CCT (μm) Range |
|---|---|---|---|---|---|
| Discovery Set (stage 1) | 3,584 | 56.5 | 54.2 (9.9) | 550.4 (33.2) | 443–668 |
| Replication Set (stage 2) | 931 | 68.0 | 56.9 (12.5) | 548.7 (34.0) | 451–661 |
| Total | 4,515 | 58.8 | 54.8 (10.5) | 550.0 (33.4) | 443–668 |
Genome-wide association results
The genomic control (GC) inflation factor, λ, was 1.05, after adjusting for age, gender, and principal components of genetic ancestry. A Q-Q plot was generated and is presented in Supplementary Material, Figure S1. Upon visual inspection of the plot, the observed –log10(P values) do not deviate from the null, except at the extreme tail. This plot suggests proper control of population stratification in this Latino sample.
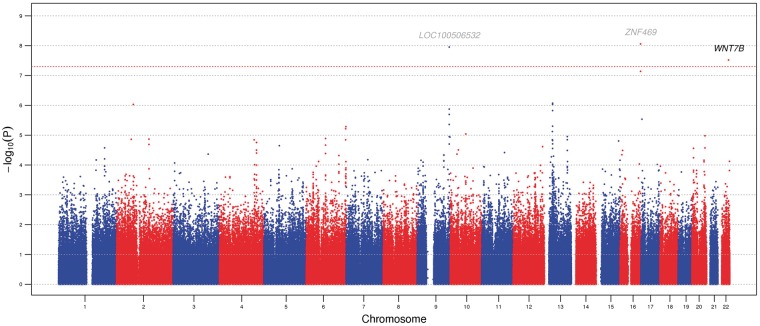
Figure 1 presents a Manhattan plot displaying the discovery set SNP association P values for the 576, 798 SNPs included in this analysis. Table 2 summarizes the SNPs, rs3118515, rs12447690, and rs10453441, that reached genome-wide significance (P < 5 × 10−8) during the discovery set analysis. Among these three SNPs, all have previously been associated with CCT except for rs10453441, which is a novel finding in this study. rs10453441 (P = 3.14 × 10−8, GRCh37/hg19 position 46,363,739, chromosome 22) is located in an intron of the WNT7B gene (wingless-type MMTV integration site family, member 7B). The minor allele G (MAF = 0.455) is associated with a reduction in CCT (β = −4.51). rs3118515 (P = 1.16 × 10−8, GRCh37/hg19 position 137,436,314) resides in the uncharacterized gene LOC100506532, which is situated 86.5 kb 3’ of the RXRA gene (retinoid X receptor, alpha) and 96.5 kb 5’ of the COL5A1 gene (collagen, type V, alpha 1) on chromosome 9. The minor allele of rs3118515, A (MAF = 0.268), is associated with a reduction in CCT (β = −5.15). rs12447690 (P = 9.16 × 10−9, GRCh37/hg19 position 88,298,124) is located 195 kb upstream of the ZNF469 gene (zinc finger protein 469) and 187 kb downstream of the BANP gene (BTG3 associated nuclear protein) on chromosome 16. The minor allele of rs12447690 C (MAF = 0.360) is associated with a reduction in CCT per allele copy (β = −4.86).
Figure 1.
A Manhattan plot presenting the –log10(P values) for the 576,798 SNPs included in the discovery set (stage 1) plotted by genomic position The red horizontal dotted line indicates the genome-wide significance level of P = 5.00 × 10−8. Loci previously associated with CCT are shown in grey (LOC100506532 and ZNF469) and the novel locus is shown in black (WNT7B).
Table 2.
Single nucleotide polymorphisms significantly associated (P < 5 × 10−8) with CCT in Latinos
| Discovery |
Replication |
Meta-Analysis |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| SNP | Chr | Position | Gene | A1/A2 | AF1 | β | P | β | P* | P |
| rs3118515 | 9 | 137,436,314 | LOC100506532, RXRA, COL5A1 | A/G | 0.268 | −5.15 | 1.16E-08 | −6.41 | 3.03E-04 | 9.29E-11 |
| rs12447690 | 16 | 88,298,124 | BANP, ZNF469 | C/T | 0.360 | −4.86 | 9.16E-09 | −1.03 | 0.27 | 1.81E-08 |
| rs10453441 | 22 | 46,363,739 | WNT7B | G/A | 0.455 | −4.51 | 3.14E-08 | −2.95 | 0.03 | 6.01E-09 |
Abbreviations: Chr, chromosome; A1/A2, allele 1/allele 2; AF1, allele 1 frequency. β models the expected change in mean CCT per increase of one A1 allele. Gene name is in boldface if the SNP is inside the gene. SNP positions are according to GRCh37/hg19.
One-sided P values computed with regard to the direction of effect found in the discovery set.
For the replication set, we analysed the top SNPs using a set of related individuals and adjusted for the relatedness among individuals using linear mixed-effects models. All three of the SNPs showed similar directionality of the effect estimates for the minor alleles, compared to the discovery set (Table 2). rs10453441 was significant (P = 0.03) in our replicate set. The meta-analysis of the discovery and replication sets strengthened the association for rs10453441, P = 6.01 × 10−9. Therefore, rs10453441 represents a novel genetic locus associated with CCT in Latinos. Furthermore, rs10453441 was associated with CCT in our Indian consanguineous pedigree dataset (240 individuals from 16 pedigrees, P = 5.85 × 10−4) (24), providing another replication for the novel discovery. Similarly, the association result for SNP rs3118515 was also strengthened after the meta-analysis, P = 9.29 × 10−11.
Results from imputed SNPs
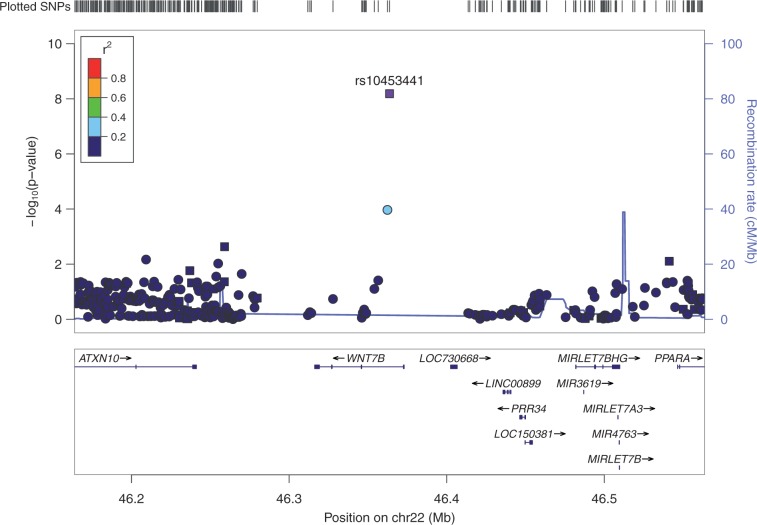
To expand genomic coverage and further identify additional genetic loci associated with CCT, we used the maximum number of unrelated individuals (n = 4,038), including unrelated individuals from the replication set, to analyse imputed SNPs. The GC λ for the imputed dataset was 1.05. Analysis of imputed SNPs did not identify any additional GWAS loci for CCT stronger than directly genotyped SNPs. The SNP rs10453441 was the only significant SNP associated with CCT on chromosome 22 for genotyped and imputed SNPs (Rsq ≥ 0.80). Figure 2 presents the regional SNP association plots for the WNT7B region. Genotyped and imputed SNPs are plotted as squares and circles, respectively.
Figure 2.
Regional SNP association plot for rs10453441 The most significant SNP, using the maximum number of unrelated individuals, is plotted in purple. The lead SNP, rs10453441 (P = 7.15 × 10−9), remained the most significantly associated in the WNT7B gene. Genotyped and imputed SNPs are plotted as squares and circles, respectively. Color-coding of SNPs represents the level of linkage disequilibrium with the most significant SNP, rs10453441. Genes in the region are shown below the SNPs with arrows indicating the strand orientation.
Analysis of previously reported loci for CCT
We evaluated 52 previously reported SNPs that reached genome-wide significance (P < 5 × 10−8) in European and Asian populations to determine whether these associations were consistent in a Latino population. The results are summarized in Table 3. Of the 52 previously reported SNPs, 51 SNPs were either directly genotyped or imputed well and among these SNPs, a majority of SNPs had a P < 0.05 (38/51 = 75%). Furthermore, in our Latino population, all except one SNP exhibited consistent directionality of the effect with these previously reported SNPs. To account for multiple testing among correlated SNPs, we employed simpleM (25,26) to estimate the number of independent tests for the 51 SNPs. This method identified 40 independent tests, resulting in a Bonferroni corrected P value of 0.00125. Using this corrected P value, we replicated 28 SNPs in the following 11 loci, NR3C2, IBTK, C7orf42, LPAR1, RXRA-COL5A1, COL5A1, LCN12-PTGDS, ARHGAP20-POU2AF1, FOXO1, LRRK1, and ZNF469 regions (boldface P values in Table 3). We were, however, unable to replicate previous index SNPs in other regions, including COL8A2, COL4A3, FAM53B, SMAD3, and AKAP13. Intriguingly, a study conducted in individuals of European ancestry was unable to replicate several of these associations (27); however, this later study was restricted to NEIGHBOR and GLAUGEN POAG cases and controls, suggesting that not all CCT loci are POAG related.
Table 3.
Comparison with previously reported SNPs associated with CCT
| Previously Reported |
Latinos |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP ID | Chr | Position | Genes Nearby | Effect | Freq | β | Reference(s) | A1/A2 | AF1 | β | P | Imputed | Consistency of Direction |
| Allele | |||||||||||||
| rs3767703 | 1 | 36555758 | COL8A2 | −4.43 | (19) | C/T | 0.93 | −0.03 | 9.81E-01 | NA | |||
| rs7550047 | 1 | 36567343 | COL8A2 | −4.42 | (19) | A/G | 0.91 | 0.88 | 5.00E-01 | Y | NA | ||
| rs96067 | 1 | 36571920 | COL8A2 | −4.80 | (19) | G/A | 0.38 | −1.15 | 1.29E-01 | NA | |||
| rs10189064 | 2 | 219327500 | USP37 | A/G | 0.04 | −0.19 | (23) | A/G | - | - | - | NA | NA |
| rs7606754 | 2 | 228135180 | COL4A3 | A/G | 0.35 | −0.07 | (23) | A/G | 0.41 | −0.79 | 2.91E-01 | Y | Y |
| rs3749260 | 3 | 98250862 | GPR15 | A/C | 0.13 | −0.12 | (23) | A/C | 0.14 | 1.76 | 1.12E-01 | Y | N |
| rs9822953 | 3 | 156472071 | TIPARP | T/C | 0.67 | 0.08 | (23) | T/C | 0.74 | 1.81 | 3.51E-02 | Y | Y |
| rs4894535 | 3 | 171995605 | FNDC3B | T/C | 0.17 | −0.10 | (23) | T/C | 0.25 | −2.16 | 1.09E-02 | Y | |
| rs7620503 | 3 | 177304298 | TBL1XR1-KCNMB2 | T/C | 0.39 | −0.06 | (23) | T/C | 0.44 | −1.18 | 1.16E-01 | Y | Y |
| rs3931397 | 4 | 149079497 | NR3C2 | T/G | 0.07 | −0.12 | (23) | T/G | 0.06 | −6.77 | 1.28E-05 | Y | Y |
| rs1117707 | 5 | 64389665 | CWC27-ADAMTS6 | A/G | 0.7 | −0.11 | (23) | A/G | 0.55 | −0.59 | 4.26E-01 | Y | Y |
| rs1538138 | 6 | 82794594 | IBTK | T | 0.18–0.30 | −4.23 | (17) | T/C | 0.14 | −3.53 | 7.16E-04 | Y | |
| rs11763147 | 7 | 65326821 | VKORC1L1 | A/G | 0.45 | 0.07 | (23) | A/G | 0.45 | 1.62 | 2.85E-02 | Y | |
| rs4718428 | 7 | 66421446 | C7orf42 | G | 0.46–0.74 | −3.18 | (17) | G/T | 0.45 | −2.89 | 1.89E-04 | Y | Y |
| rs1324183 | 9 | 13557491 | 9p23 | A | 0.21–0.27 | −3.37 | (17) | A/C | 0.14 | −2.86 | 1.15E-02 | Y | Y |
| rs1007000 | 9 | 113662681 | LPAR1 | T/C | 0.22 | 0.07 | (23) | T/C | 0.30 | 3.43 | 1.62E-05 | Y | |
| rs1409832 | 9 | 137428425 | RXRA-COL5A1 | −3.95 | (19) | T/G | 0.76 | 4.32 | 7.11E-07 | NA | |||
| rs4842044 | 9 | 137431904 | RXRA-COL5A1 | −4.67 | (19) | C/T | 0.47 | −4.14 | 2.25E-08 | Y | NA | ||
| rs1536478 | 9 | 137432248 | RXRA-COL5A1 | −4.63 | (19) | G/A | 0.47 | −4.17 | 2.03E-08 | Y | NA | ||
| rs3118516 | 9 | 137439792 | RXRA-COL5A1 | A | 0.34 | −0.15 | (21) | A/G | 0.26 | −4.91 | 5.89E-07 | Y* | Y |
| rs3132306 | 9 | 137440212 | RXRA-COL5A1 | T | 0.66 | 0.15 | (21) | T/C | 0.72 | 4.83 | 7.63E-07 | Y* | Y |
| rs1536482 | 9 | 137440528 | RXRA-COL5A1 | G | 0.34 | 0.22 | (18),(21) (A, Freq | G/A | 0.71 | 5.21 | 5.47E-08 | Y* | Y |
| = 0.33, β = −0.15) | |||||||||||||
| rs7044529 | 9 | 137568051 | COL5A1 | (19) | C/T | 0.78 | 3.23 | 2.58E-04 | NA | ||||
| rs11145951 | 9 | 139860264 | LCN12-PTGDS | T/C | 0.49 | 0.09 | (23) | T/C | 0.44 | 3.26 | 1.15E-05 | Y | |
| rs7090871 | 10 | 63830286 | ARID5B | T/C | 0.59 | 0.06 | (23) | T/C | 0.71 | 1.40 | 8.75E-02 | Y | Y |
| rs1006368 | 10 | 126346603 | FAM53B | A/G | (20) | C/T | 0.80 | −2.01 | 2.59E-02 | Y | NA | ||
| rs11245330 | 10 | 126380338 | FAM53B | A/G | (20) | A/G | 0.20 | 2.10 | 1.96E-02 | NA | |||
| rs4938174 | 11 | 110913240 | ARHGAP20-POU2AF1 | A/G | 0.31 | 0.06 | (23) | A/G | 0.20 | 3.34 | 2.97E-04 | Y | |
| rs1564892 | 12 | 104445742 | Near GLT8D2 (5') | A/G | 0.76 | −0.08 | (23) | A/G | 0.74 | −0.37 | 6.66E-01 | Y | Y |
| rs1034200 | 13 | 23228691 | FGF9-FTHL7 | 0.14 | (18) | C/A | 0.75 | −1.69 | 5.06E-02 | NA | |||
| rs2755237 | 13 | 41109429 | FOXO1 | A/C | (20) | A/C | 0.86 | 3.62 | 6.45E-04 | NA | |||
| rs2721051 | 13 | 41110884 | FOXO1 | A/G | (20) | C/T | 0.92 | 4.24 | 1.89E-03 | NA | |||
| rs785422 | 15 | 30173885 | Near TJP1 (5') | T/C | 0.11 | −0.14 | (23) | T/C | 0.07 | −3.24 | 2.66E-02 | Y | |
| rs12913547 | 15 | 67467507 | SMAD3 | T/C | 0.77 | −0.08 | (23) | T/C | 0.71 | −0.44 | 5.87E-01 | Y | |
| rs6496932 | 15 | 85825567 | PDE8A-AKAP13 | 0.13 | (18) | C/A | 0.77 | 1.53 | 8.32E-02 | NA | |||
| rs1828481 | 15 | 85840912 | AKAP13 | C | 0.45–0.56 | 3.12 | (17) | C/A | 0.49 | 1.88 | 1.21E-02 | Y | NA |
| rs7172789 | 15 | 85843517 | AKAP13 | C | 0.45–0.56 | 3.14 | (17) | C/T | 0.48 | 1.72 | 2.14E-02 | Y | |
| rs930847 | 15 | 101558562 | LRRK1 | G | 0.17–0.39 | 3.72 | (17) | G/T | 0.19 | 3.97 | 2.79E-05 | Y | |
| rs4965359 | 15 | 101585336 | LRRK1 | A | 0.40–0.67 | −3.50 | (17) | A/G | 0.54 | −3.77 | 4.32E-07 | Y | Y |
| rs12447690 | 16 | 88298124 | ZNF469 | T/C | 0.17 | (20),(19),(18) (G, | T/C | 0.64 | 4.14 | 1.18E-07 | Y | ||
| β = −0.18), (21) (T, Freq = 0.64, β = 0.16) | |||||||||||||
| rs7500824 | 16 | 88299491 | ZNF469 | A | 0.36 | −0.16 | (21) | A/G | 0.24 | −4.70 | 6.35E-07 | Y | Y |
| rs7405095 | 16 | 88307825 | ZNF469 | A | 0.36 | −0.16 | (21) | A/G | 0.20 | −6.02 | 1.87E-07 | Y* | Y |
| rs7501109 | 16 | 88320862 | ZNF469 | C | 0.64 | 0.16 | (21) | C/G | 0.80 | 4.95 | 2.15E-06 | Y* | Y |
| rs7501402 | 16 | 88320911 | ZNF469 | A | 0.36 | −0.16 | (21) | A/T | 0.36 | −4.38 | 2.94E-07 | Y | Y |
| rs6540223 | 16 | 88321436 | ZNF469 | T | 0.64 | 0.16 | (21) | T/C | 0.80 | 4.96 | 2.22E-06 | Y* | Y |
| rs12448211 | 16 | 88330513 | ZNF469 | A | 0.62 | 0.16 | (21) | A/G | 0.65 | 4.51 | 3.89E-08 | Y | Y |
| rs9938149 | 16 | 88331640 | ZNF469 | A/C | (20),(19),(21) (A, | A/C | 0.79 | 4.56 | 3.62E-07 | Y | |||
| Freq = 0.62, β = 0.16) | |||||||||||||
| rs9922572 | 16 | 88334112 | ZNF469 | A | 0.34 | −0.14 | (21) | A/C | 0.20 | −4.82 | 2.01E-06 | Y | Y |
| rs9925231 | 16 | 88338107 | ZNF469 | −4.79 | (19) | T/C | 0.33 | −4.37 | 4.33E-07 | Y | NA | ||
| rs7204132 | 16 | 88344517 | ZNF469 | −4.95 | (19) | T/G | 0.18 | −4.32 | 4.35E-05 | Y | NA | ||
| rs9927272 | 16 | 88346709 | ZNF469 | −3.95 | (19) | G/A | 0.34 | −2.72 | 5.68E-04 | NA | |||
| rs2323457 | 17 | 14554190 | HS3ST3B1-PMP22 | A/C | 0.29 | −0.07 | (23) | A/C | 0.40 | −0.89 | 2.32E-01 | Y | Y |
Abbreviations: Chr, chromosome; Freq, frequency; A1/A2, allele 1/allele 2. For previously reported SNPs with multiple references, additional information on the allele, frequency, and effect size is given in parentheses. For this Latino population, the frequency for allele 1 is given and is modeled as the effect allele. The program simpleM was used as a multiple testing correction method for correlated SNPs, identifying 40 independent tests and thus, giving a Bonferroni correction P value of 0.05/40 = 0.00125. P values meeting this threshold are shown in bold. The direction of effect was consistent for most SNPs, except rs3749260. SNP positions are according to GRCh37/hg19.
*SNPs with suboptimal Rsq (0.62 < Rsq < 0.78).
Biological evidence
The WNT7B gene has been reported to be expressed in the human cornea (28) and the mouse cornea (29). WNT7B was upregulated in the human central cornea and is specific to the mature corneal epithelium (28) and was mainly expressed in the endothelial cell layer in the mouse cornea (29). Genevestigator (see Web Resources), an online database to verify gene expression in different tissue types, also shows that WNT7B is highly expressed in normal human corneas (Supplementary Material, Fig. S2).
Pathway analysis
Table 4 summarizes the top five canonical pathways from IPA and KEGG pathways from WebGestalt through gene-set association testing of directly genotyped SNPs. The most significantly enriched canonical pathway is 14-3-3-mediated signalling (P = 2.9E-03), followed by the IGF-1 signalling (P = 2.9E-03), myc mediated apoptosis signalling (P = 6.45E-03), neuregulin signalling (P = 6.45E-03), and NGF signalling (P = 3.8E-02) pathways, respectively. The top pathway, 14-3-3-mediated signalling, is a family of proteins that are involved in cell cycle, apoptosis, and protein trafficking and the presence of 14-3-3 proteins have been reported in the human cornea (30). Similarly, the Myc mediated apoptosis signalling pathway is related to the cell cycle and may have a role in maintaining the homeostasis of the corneal epithelium. The IGF-1 signalling, neuregulin signalling, and NGF signalling pathways involve growth factors and IGF-1 (31), neuregulin 1 (32), and NGF (33) have been reported to have a biological effect in human corneas. In addition, the top KEGG pathways are metabolic pathways (P = 7.27E-06), ErbB signalling pathway (P = 2.95E-05), tight junction (P = 1.0E-04), pathways in cancer (P = 1.0E-04), and T cell receptor signalling pathway (P = 8.0E-04). The ErbB family consists of receptor tyrosine kinases that regulates various biological responses, including cell proliferation, differentiation, migration, and carcinogenesis, and this family has been shown to play a role in corneal epithelial wound healing (34). Tight junctions are composed of proteins that mediate cell adhesion and serve as diffusion barriers with the tight junction-related proteins occludin, claudin, and ZO-1 expressed in human corneal epithelium (35). And lastly, genes that have been studied in tumour metastasis models were also found to be differentially expressed in the cornea (36). Interestingly, during both wound healing and cancer development, related cells exhibit rapid proliferation and tissue remodelling (36). Taken together, our pathway analysis results from IPA and KEGG yielded comparable pathways with regard to the cornea in that the programs identified pathways relevant to cellular proliferation, migration, and apoptosis, suggesting such pathways may have a role in the integrity and homeostasis of the human cornea.
Table 4.
Top pathways associated with CCT in Latinos
| Pathway | Observed/Total Genes | P | Adjusted P‡ | |
|---|---|---|---|---|
| IPA | ||||
| 14-3-3-mediated signalling | 19/117 | 1.30E-05 | 2.9E-03 | |
| IGF-1 signalling | 17/97 | 1.31E-05 | 2.9E-03 | |
| Myc mediated apoptosis signalling | 12/58 | 4.47E-05 | 6.45E-03 | |
| Neuregulin signalling | 15/88 | 5.81E-05 | 6.45E-03 | |
| NGF signalling | 15/107 | 5.38E-04 | 3.8E-02 | |
| WebGestalt | ||||
| Metabolic pathways | 69/1130 | 4.49E-08 | 7.27E-06 | |
| ErbB signalling pathway | 14/87 | 3.64E-07 | 2.95E-05 | |
| Tight junction | 16/132 | 2.77E-06 | 1.0E-04 | |
| Pathways in cancer | 27/326 | 3.06E-06 | 1.0E-04 | |
| T cell receptor signalling pathway | 13/108 | 2.51E-05 | 8.0E-04 |
‡P-values are Benjamini-Hochberg adjusted. The top five canonical pathways from IPA and KEGG pathways from WebGestalt are displayed.
Discussion
This study is the largest GWAS conducted in Latinos for CCT. We identified rs10453441 in the WNT7B gene on chromosome 22 as a novel SNP for CCT. Furthermore, one previously reported SNP on chromosome 9, rs3118515, and one previously reported SNP on chromosome 16, rs12447690, also reached genome-wide significance. These SNPs were replicated at the 0.05 significance level (one-sided P values being computed with regard to the direction of the effect found in the discovery set) in our Latino replication set, except for rs12447690. The association of rs10453441 with CCT was also replicated in an Indian dataset (P = 5.85 × 10−4) (24). We were also able to replicate 11 previous GWAS loci, such as variants in the RXRA-COL5A1, COL5A1, IBTK, FOXO1, LRRK1, and ZNF469 regions. Though genotype imputation using the 1KGP increased the number of SNPs in our dataset, we did not identify additional GWAS loci for CCT in the imputed dataset. As further support, the WNT7B gene is expressed in the human cornea, which provides biological evidence for its association with CCT.
The novel SNP identified here associated with CCT, rs10453441 is located in the genomic region that includes WNT7B, a member of the WNT gene family, a group of structurally related genes that encode secreted signalling proteins. This SNP has previously been positively associated with axial length and corneal curvature, ocular parameters related to myopia, in a GWAS among Asian and Caucasian populations (29). These investigators further identified a positive association between this SNP and extreme myopia, as well as localized WNT7B expression in the endothelial cell layer of mouse cornea and the downregulation of this gene in extreme myopia. Compared to Miyake et al. (2014), where rs10453441 was associated with an increase in both axial length and corneal curvature, our results identified its association with CCT. CCT is not highly correlated with either axial length or corneal curvature (37), suggesting rs10453441 may influence different ocular phenotypes in different ethnic groups.
As a member of Wnt signalling and one of 19 Wnt proteins, WNT7B was found to be upregulated in the central cornea of humans and the Wnt signalling pathway may have a role in primary limbal stem cell proliferation (28). Similarly, various other components of Wnt signalling have previously been implicated in the cornea. Wnt4 has been detected in the basal and immediate parabasal limbal epithelial cells in both normal foetal and adult human corneas (38). Additionally, during corneal wound healing, Wnt7a was found to promote the proliferation of corneal epithelial cells (39). The results from our study suggest rs10453441 may play an important role in CCT determination, independent of axial length and corneal curvature, and similar to other members of Wnt signalling, WNT7B may have an active biological role in regulating CCT.
Our study has many strengths, including the use of the largest Latino dataset containing both ophthalmic and genotyped data thus far. We conducted a two stage GWAS, using both population- and family-based study designs to replicate associated loci. Our sample size was able to achieve 80% power to detect variants with MAF 0.455 explaining 1% (like rs10453441) of trait variation (two sided test, αp= 5 × 10−8, calculated by Quanto (40,41)). We imputed and analysed SNPs from the 1KGP to extend genomic coverage to further identify common variants not directly genotyped, using a specialized imputation protocol for Latino populations (42). Both the imputed and genotyped GWAS results are presented in the supplementary data. We also carried out imputation using our own Hispanic/SOL data as the reference panel and analysed these SNPs using linear mixed models, though we did not identify any additional new loci for CCT (data not shown). Gene expression analysis provided biological support for the expression of the WNT7B gene in the cornea. This study, however, also has some limitations. Latinos are historically understudied in genetic research. Despite the largest Latino data for studying ophthalmic phenotypes, our sample size is limited compared to meta-analyses using European and Asian subjects. We emphasize the need to replicate our findings in an independent Latino cohort. Variants of smaller effects may also not be detected. The effect size estimates of GWAS hits may suffer from the winner’s curse (43,44). For example, the bias-corrected beta for rs12447690 in the discovery set was −4.14 with a confidence interval [−2.06, −8.34] calculated using BR-squared software (45) via 1,000 bootstrap re-sampling. Due to the three-way admixture (the ancestral composition of this sample is shown in Supplementary Material, Fig. S3), it is generally more challenging to impute in Latinos (42). The imputation quality of the WNT7B region on chromosome 22 was low, which can mask other possible interesting SNPs, e.g. rs9330813 (P = 5.14 × 10−8) and rs10453458 (P = 1.29 × 10−8) only had imputation scores of Rsq = 0.74 and 0.56, respectively. Chromosome 22 has previously been shown to be difficult to impute for Latinos (42). As reference panels and imputation technologies continue to improve for Latinos, the ability to impute and consequently, identify associations with non-directly genotyped SNPs, may improve as well. We investigated association for the top SNP (rs10453441) in 5 independent European CCT studies, but did not observe a consistent association with this SNP and CCT in these populations (Supplementary Material, Table S1). This suggests that rs10453441 may not be generalized to the European populations. But it is also possible that the association in the European populations was mitigated by the suboptimal imputation quality. Furthermore, we are genotyping our entire cohort using the Illumina HumanExome BeadChip through MAGGS. As our genotyping progresses, we will have more power to uncover additional loci.
In summary, we identified a novel genome-wide significant association between rs10453441 in WNT7B and CCT in the largest GWAS of CCT in Latinos. We also replicated previously reported SNPs identified in other ethnic groups. Furthermore, our pathway analysis identified relevant pathways associated with apoptosis and cell development for CCT. Our results suggest GWAS conducted in minority populations can identify additional loci associated with CCT not previously identified in GWAS or large-scale meta-analyses using European or Asian populations and may aid in furthering our understanding of ethnic differences of CCT, as well as elucidating the genetic architecture of CCT.
Materials and Methods
Ethics statement
The following research was approved by the University of Illinois at Chicago, the University of Southern California Health Sciences Campus, and the Los Angeles Biomedical Research Institute at Harbor-University of California, Los Angeles, (UCLA) institutional review boards. All clinical investigation was conducted according to the principles expressed in the Declaration of Helsinki.
Study sample
We conducted this research using data collected from the Los Angeles Latino Eye Study (LALES), the largest population-based study of visual impairment and ophthalmic diseases conducted among 6,357 Latinos living in six census tracts in the city of La Puente, Los Angeles County, California. All subjects included in this study were 40 years of age and older and written, informed consent was obtained from each study participant.
Measurement of CCT
All study participants underwent a detailed ophthalmologic examination. Three CCT measurements were obtained in each eye using a commercial ultrasonic computation module (A-scan/pachymeter DGH 4000B SBH IOL Computation Module; DGH Tech, Inc., Exton, PA) during examination. The three CCT measurements were averaged to obtain a single value in each eye. The average CCT measurement for the left and right eyes was taken to yield the final CCT value. If CCT measurements were available for only one eye, this value was used as a surrogate for the final CCT measurement.
Genotyping and quality control
We genotyped a total of 4,996 Latinos through LALES and the Mexican American Glaucoma Genetic Study (MAGGS) using either the Illumina OmniExpress BeadChip Kit (730, 525 markers; Illumina, Inc., San Diego, CA; n = 4,278) or the Illumina Hispanic/SOL BeadChip (∼2.5 million markers; Illumina, Inc., San Diego, CA; n = 718). The genotyping for this study was performed at the Genotyping Laboratory of the Institute for Translational Genomics and Population Sciences at the Los Angeles Biomedical Research Institute at Harbor-UCLA. SNPs were called using the software Illumina GenomeStudio (v2011.1; Illumina, Inc.). Study participants whose genotyping call rate was less than 97% were excluded from further analysis. Additionally, study participants with gender inconsistency between reported and genetically inferred gender, missing CCT measurements, CCT outliers, and duplicates were removed. After removal of these study subjects, 4,515 subjects remained for downstream analysis, of which 3,584 unrelated individuals were used as a discovery set (stage 1) and 931 first-degree relatives (416 families) were used as a replication set (stage 2). The program PLINK (v1.90) was used to perform quality control on the genotype data (46). SNPs that overlapped between the genotyping chips were retained for analysis and were further excluded if the minor allele frequency (MAF) < 1%, the call rate < 95%, or the Hardy-Weinberg equilibrium P values < 10−6. After the above quality control parameters, 576,798 SNPs remained for further analysis. SNPs were coded on the forward strand to facilitate the imputation process.
Genotype imputation
To increase genomic coverage, we used Shapeit2 (47), Minimac3 (see Web Resources), and the 1000 Genomes Project (1KGP) reference panels to perform genotype imputation to integrate additional SNPs that were not directly genotyped. Both Minimac3 and the 1KGP Phase 1v3 haplotype reference panels were downloaded from the Center for Statistical Genetics at the University of Michigan (see Web Resources). The 1KGP reference panels used for imputation contain a large number of variants, 39.7 million variants, increasing the ability to interrogate non-genotyped SNPs through the use of linkage disequilibrium (LD). Due to the unique ancestral populations of Latinos, we used the AMR + CEU + YRI reference panels (a combination of Colombian, Mexican, Puerto Rican, CEPH, and Yoruba haplotypes), which we have previously shown to have the highest genotype imputation accuracy for Latinos (42).
We phased our genotypes using Shapeit2 and carried out genotype imputation using Minimac3 on the phased data. Genotypes that were imputed were coded as allelic dosages (estimated counts ranging from 0 to 2). Quality control parameters were applied to filter out low quality imputed SNPs, i.e. Rsq < 0.80 and MAF < 1%. After removal of low quality imputed SNPs, 6,844,888 imputed SNPs remained for further analysis.
Statistical analysis
The program EIGENSOFT was used to infer principal components of genetic ancestry (48). To make inferences of known ancestral populations, we included reference panels of unrelated Northern Europeans (CEU, n = 87) and West Africans (YRI, n = 88) from the 1000 Genomes Project (49), and Native Americans (n = 105) (50). The first four principal components were retained and were included as covariates during linear regression. The genomic control inflation factor (51) was calculated and applied and a quantile-quantile (Q-Q) plot was generated to visually inspect the distribution of test statistics. For the discovery set, we used PLINK to test the association between individual SNPs and CCT using linear regression, adjusting for age, gender, and principal components, assuming an additive genetic effects model (46). For the replication set, the association between SNPs and CCT was assessed using a linear mixed-effects model (Proc Mixed procedure of SAS v9.4; SAS Institute, Cary, NC), adjusting for age, gender, and principal components of genetic ancestry. The compound symmetry covariance structure and the empirical “sandwich” estimator were used. Fixed-effects meta-analysis for the discovery and replication sets was conducted in METAL using inverse-variance weighting (52). Accounting for genotype uncertainty, allelic dosages of imputed SNPs were analysed using the software mach2qtl (see Web Resources). The conventional SNP P value significance threshold of 5 ×10−8 was used to declare genome-wide significant SNPs. We applied the simpleM (25,26,53) method for multiple testing correction to evaluate previously reported loci. The software R (54) and LocusZoom (hg19/1KGP AMR) (55) were used for graphing.
Pathway analysis
To investigate the contribution of combined genetic effects on the underlying biological processes influencing CCT, we performed gene-based and pathway analysis on the maximum number of unrelated individuals using SKAT-O (56), QIAGEN’s Ingenuity Pathway Analysis (IPA) (see Web Resources), and WEB-based GEne SeT AnaLysis Toolkit (WebGestalt) (57). Using all directly genotyped SNPs, we assigned these SNPs to genes according to genomic position based on the hg19 assembly. Moreover, in order to capture proximal regulatory and other functional elements occurring outside of a gene, we extended the gene boundaries to include SNPs ± 10kb of a gene. SNPs occurring in multiple gene boundaries were included in these boundaries separately. Similar to the GWAS model, the gene-set associations were further adjusted for age, gender, and principal components of genetic ancestry. Enrichment of canonical pathways and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways with genes associated with CCT was analysed using IPA and WebGestalt. Canonical pathways and KEGG pathways with a P value < 0.05 after Benjamini-Hochberg multiple testing adjustment (58) were declared significantly enriched. The top five most enriched canonical pathways from IPA and KEGG pathways from WebGestalt are reported.
Supplementary Material
Acknowledgements
The authors would like to thank the study participants from LALES and the staff who aided in data collection and processing.
Conflict of Interest statement. None declared.
Funding
This work was supported in part by National Institutes of Health (NIH; Bethesda, MD, USA) grants R01EY022651 (to XG), U10EY011753 (to RV), P30EY001792 (departmental core grant), and a grant from Midwest Eye-Banks (to XG). The provision of genotyping data was supported in part by the National Center for Advancing Translational Sciences (CTSI) grant UL1TR000124, NIDDK Diabetes Research Center (DRC) Grant DK063491 to the Southern California Diabetes Research Center. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Web Resources
The URLs for downloaded data and programs (date last accessed February 2016):
1000 Genomes Project Phase 1v3 haplotypes, http://csg.sph.umich.edu/abecasis/MACH/download/1000G.2012-03-14.html
EIGENSOFT, https://genetics.med.harvard.edu/reich/Reich_Lab/Software.html
Genevestigator, https://genevestigator.com/
mach2qtl, http://csg.sph.umich.edu/abecasis/MACH/download/
METAL http://csg.sph.umich.edu/abecasis/Metal/download/
Minimac3, http://genome.sph.umich.edu/wiki/Minimac3#Download
PLINK, https://www.cog-genomics.org/plink2
QIAGEN, http://www.ingenuity.com/products/ipa/
simpleM, http://simplem.sourceforge.net
WebGestalt, http://bioinfo.vanderbilt.edu/webgestalt/
Appendix
Tin Aung1,2, Kathryn P. Burdon3, Ching-Yu Cheng1,2, Jamie E. Craig4, Angela J Cree5, Puya Gharahkhani6, Christopher J. Hammond7, Alex W. Hewitt8, René Höhn9, Pirro Hysi7, Adriana I Iglesias Gonzalez10, Jost Jonas11, Anthony Khawaja12,13, Chiea-Cheun Khor14, Caroline CW Klaver15, Francesca Pasutto16, Stuart MacGregor6, David Mackey17, Paul Mitchell18, Aniket Mishra6, Calvin Pang19, Louis R Pasquale20, Henriette Springelkamp15, Gudmar Thorleifsson21, Unnur Thorsteinsdottir21, Cornelia M van Duijn22, Ananth Viswanathan23, Veronique Vitart24, Janey L Wiggs20, Robert Wojciechowski25,26,27, Tien Wong1,2, Terrri L Young28, Tanja Zeller29
1Singapore Eye Research Institute, Singapore National Eye Centre, Singapore; 2Department of Ophthalmology, National University of Singapore and National University Health System, Singapore; 3Menzies Institute for Medical Research, University of Tasmania, Hobart, Tasmania, Australia; 4Department of Ophthalmology, Flinders University, Adelaide, Australia; 5Clinical & Experimental Sciences, Faculty of Medicine, University of Southampton, Southampton, UK; 6Statistical Genetics, Queensland Institute of Medical Research, Brisbane 4029, Australia; 7Department of Twin Research and Genetic Epidemiology, King's College London, St. Thomas’ Hospital, London, UK; 8Centre for Eye Research Australia, University of Melbourne, Royal Victorian Eye and Ear Hospital, Melbourne, Australia; 9Department of Ophthalmology, University Medical Center Mainz, Mainz, Germany; 10Department of Epidemiology, Erasmus University Medical Center, Rotterdam, The Netherlands; 11Department of Ophthalmology, Medical Faculty Mannheim of the Ruprecht-Karls-University of Heidelberg, Mannheim, Germany; 12Department of Public Health and Primary Care, University of Cambridge, Cambridge, UK; 13NIHR Biomedical Research Centre for Ophthalmology, Moorfields Eye Hospital and UCL Institute of Ophthalmology, London, UK; 14Department of Human Genetics, Genome Institute of Singapore, Singapore; 15Department of Epidemiology and Department of Ophthalmology, Erasmus Medical Center, Rotterdam, The Netherlands; 16Institute of Human Genetics, Friedrich-Alexander-Universität Erlangen-Nürnberg (FAU), Erlangen, Germany; 17Centre for Ophthalmology and Visual Science, Lions Eye Institute, University of Western Australia, Perth, Australia; 18Centre for Vision Research, Department of Ophthalmology and Westmead Millennium Institute, University of Sydney, Sydney, Australia; 19Department of Ophthalmology and Visual Sciences, Chinese University of Hong Kong, Hong Kong, China; 20Department of Ophthalmology, Harvard Medical School, Massachusetts Eye and Ear, Boston, MA; 21deCODE Genetics/Amgen, 101 Reykjavik, Iceland; 22Department of Epidemiology, Erasmus Medical Center, Rotterdam, The Netherlands; 23NIHR Biomedical Research Centre, Moorfields Eye Hospital NHS Foundation Trust, University College London, Institute of Ophthalmology, London, UK; 24MRC Human Genetics Unit, IGMM, University of Edinburgh, Edinburgh EH4 2XU, UK; 25Department of Epidemiology, Johns Hopkins Bloomberg School of Public Health, Baltimore, MD, USA; 26Wilmer Eye Institute, Johns Hopkins School of Medicine, Baltimore, MD, USA; 27National Human Genome Research Institute (NIH), Baltimore, MD, USA; 28Department of Ophthalmology and Visual Sciences, School of Medicine and Public Health, University of Wisconsin, Madison, Wisconsin 53705, USA; 29Clinic for General and Interventional Cardiology, University Heart Center Hamburg, Hamburg, Germany.
Supplementary Figure 1. Quantile-Quantile (Q-Q) plot of the genome-wide P values
For the 576, 798 SNPs that passed quality control filters in the discovery set, –log10(P values) is plotted against the corresponding expected –log10(P values). The genomic control inflation factor λ = 1.05.
Supplementary Figure 2. Expression of WNT7B in various tissues from Genevestigator
The boxplot-list displays the WNT7B gene expression in 62 postmortem healthy human anatomical parts, ranked by the level of expression. WNT7B is highly expressed in the human cornea (n = 6).
Supplementary Figure 3. Ancestral composition of our Latino sample.
The first two dimensions from principal component analysis of our Latino subjects are shown along with three ancestral populations. Abbreviations: NA, Native Americans; CEU, CEPH in Utah residents; YRI, Yoruba in Ibadan, Nigeria.
Supplementary Material
Supplementary Material is available at HMG online.
References
- 1. Burkitt Wright E.M., Porter L.F., Spencer H.L., Clayton-Smith J., Au L., Munier F.L., Smithson S., Suri M., Rohrbach M., Manson F.D., et al. (2013) Brittle cornea syndrome: recognition, molecular diagnosis and management. Orphanet. J. Rare Dis., 8, 68.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Romero-Jimenez M., Santodomingo-Rubido J., Wolffsohn J.S. (2010) Keratoconus: a review. Cont. Lens Anterior Eye, 33, 157–166. quiz 205. [DOI] [PubMed] [Google Scholar]
- 3. Dimasi D.P., Chen J.Y., Hewitt A.W., Klebe S., Davey R., Stirling J., Thompson E., Forbes R., Tan T.Y., Savarirayan R., et al. (2010) Novel quantitative trait loci for central corneal thickness identified by candidate gene analysis of osteogenesis imperfecta genes. Hum. Genet., 127, 33–44. [DOI] [PubMed] [Google Scholar]
- 4. Leske M.C., Wu S.Y., Hennis A., Honkanen R., Nemesure B., Group B.E.S. (2008) Risk factors for incident open-angle glaucoma: the Barbados Eye Studies. Ophthalmology, 115, 85–93. [DOI] [PubMed] [Google Scholar]
- 5. Jiang X., Varma R., Wu S., Torres M., Azen S.P., Francis B.A., Chopra V., Nguyen B.B., Los Angeles Latino Eye Study G. (2012) Baseline risk factors that predict the development of open-angle glaucoma in a population: the Los Angeles Latino Eye Study. Ophthalmology, 119, 2245–2253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. European Glaucoma Prevention Study G., Miglior S., Pfeiffer N., Torri V., Zeyen T., Cunha-Vaz J., Adamsons I. (2007) Predictive factors for open-angle glaucoma among patients with ocular hypertension in the European Glaucoma Prevention Study. Ophthalmology, 114, 3–9. [DOI] [PubMed] [Google Scholar]
- 7. Aghaian E., Choe J.E., Lin S., Stamper R.L. (2004) Central corneal thickness of Caucasians, Chinese, Hispanics, Filipinos, African Americans, and Japanese in a glaucoma clinic. Ophthalmology, 111, 2211–2219. [DOI] [PubMed] [Google Scholar]
- 8. Gordon M.O., Beiser J.A., Brandt J.D., Heuer D.K., Higginbotham E.J., Johnson C.A., Keltner J.L., Miller J.P., Parrish R.K., 2nd, Wilson M.R., et al. (2002) The Ocular Hypertension Treatment Study: baseline factors that predict the onset of primary open-angle glaucoma. Arch. Ophthalmol., 120, 714–720. discussion 829-730. [DOI] [PubMed] [Google Scholar]
- 9. Varma R., Ying-Lai M., Francis B.A., Nguyen B.B., Deneen J., Wilson M.R., Azen S.P., Los Angeles Latino Eye Study G. (2004) Prevalence of open-angle glaucoma and ocular hypertension in Latinos: the Los Angeles Latino Eye Study. Ophthalmology, 111, 1439–1448. [DOI] [PubMed] [Google Scholar]
- 10. Leske M.C., Heijl A., Hyman L., Bengtsson B., Dong L., Yang Z., Group E. (2007) Predictors of long-term progression in the early manifest glaucoma trial. Ophthalmology, 114, 1965–1972. [DOI] [PubMed] [Google Scholar]
- 11. Nemesure B., Wu S.Y., Hennis A., Leske M.C., Barbados Eye Study G. (2003) Corneal thickness and intraocular pressure in the Barbados eye studies. Arch. Ophthalmol., 121, 240–244. [DOI] [PubMed] [Google Scholar]
- 12. Hahn S., Azen S., Ying-Lai M., Varma R., Los Angeles Latino Eye Study G. (2003) Central corneal thickness in Latinos. Invest. Ophthalmol. Vis. Sci., 44, 1508–1512. [DOI] [PubMed] [Google Scholar]
- 13. Wolfs R.C., Klaver C.C., Vingerling J.R., Grobbee D.E., Hofman A., de Jong P.T. (1997) Distribution of central corneal thickness and its association with intraocular pressure: The Rotterdam Study. Am. J. Ophthalmol., 123, 767–772. [DOI] [PubMed] [Google Scholar]
- 14. Dimasi D.P., Burdon K.P., Craig J.E. (2010) The genetics of central corneal thickness. Br. J. Ophthalmol., 94, 971–976. [DOI] [PubMed] [Google Scholar]
- 15. Alsbirk P.H. (1978) Corneal thickness. II. Environmental and genetic factors. Acta. Ophthalmol. (Copenh.), 56, 105–113. [DOI] [PubMed] [Google Scholar]
- 16. Toh T., Liew S.H., MacKinnon J.R., Hewitt A.W., Poulsen J.L., Spector T.D., Gilbert C.E., Craig J.E., Hammond C.J., Mackey D.A. (2005) Central corneal thickness is highly heritable: the twin eye studies. Invest. Ophthalmol. Vis. Sci., 46, 3718–3722. [DOI] [PubMed] [Google Scholar]
- 17. Cornes B.K., Khor C.C., Nongpiur M.E., Xu L., Tay W.T., Zheng Y., Lavanya R., Li Y., Wu R., Sim X., et al. (2012) Identification of four novel variants that influence central corneal thickness in multi-ethnic Asian populations. Hum. Mol. Genet., 21, 437–445. [DOI] [PubMed] [Google Scholar]
- 18. Vitart V., Bencic G., Hayward C., Skunca Herman J., Huffman J., Campbell S., Bucan K., Navarro P., Gunjaca G., Marin J., et al. (2010) New loci associated with central cornea thickness include COL5A1, AKAP13 and AVGR8. Hum. Mol. Genet., 19, 4304–4311. [DOI] [PubMed] [Google Scholar]
- 19. Vithana E.N., Aung T., Khor C.C., Cornes B.K., Tay W.T., Sim X., Lavanya R., Wu R., Zheng Y., Hibberd M.L., et al. (2011) Collagen-related genes influence the glaucoma risk factor, central corneal thickness. Hum. Mol. Genet., 20, 649–658. [DOI] [PubMed] [Google Scholar]
- 20. Lu Y., Dimasi D.P., Hysi P.G., Hewitt A.W., Burdon K.P., Toh T., Ruddle J.B., Li Y.J., Mitchell P., Healey P.R., et al. (2010) Common genetic variants near the Brittle Cornea Syndrome locus ZNF469 influence the blinding disease risk factor central corneal thickness. PLoS. Genet., 6, e1000947.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hoehn R., Zeller T., Verhoeven V.J., Grus F., Adler M., Wolfs R.C., Uitterlinden A.G., Castagne R., Schillert A., Klaver C.C., et al. (2012) Population-based meta-analysis in Caucasians confirms association with COL5A1 and ZNF469 but not COL8A2 with central corneal thickness. Hum. Genet., 131, 1783–1793. [DOI] [PubMed] [Google Scholar]
- 22. Gao X., Gauderman W.J., Liu Y., Marjoram P., Torres M., Haritunians T., Kuo J.Z., Chen Y.D., Allingham R.R., Hauser M.A., et al. (2013) A genome-wide association study of central corneal thickness in Latinos. Invest. Ophthalmol. Vis. Sci., 54, 2435–2443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lu Y., Vitart V., Burdon K.P., Khor C.C., Bykhovskaya Y., Mirshahi A., Hewitt A.W., Koehn D., Hysi P.G., Ramdas W.D., et al. (2013) Genome-wide association analyses identify multiple loci associated with central corneal thickness and keratoconus. Nat. Genet., 45, 155–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Fan B.J., Chen X., Sondhi N., Sharmila PFm., Soumittra N., Sripriya S., Sacikala S., Asokan R., Friedman D.S., Pasquale L.R., et al. Family-based Genome-wide Association Study of South Indian Pedigrees Supports WNT7B as a Central Corneal Thickness Locus (Submitted). [DOI] [PMC free article] [PubMed]
- 25. Gao X., Starmer J., Martin E.R. (2008) A multiple testing correction method for genetic association studies using correlated single nucleotide polymorphisms. Genet. Epidemiol., 32, 361–369. [DOI] [PubMed] [Google Scholar]
- 26. Gao X. (2011) Multiple testing corrections for imputed SNPs. Genet. Epidemiol., 35, 154–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ulmer M., Li J., Yaspan B.L., Ozel A.B., Richards J.E., Moroi S.E., Hawthorne F., Budenz D.L., Friedman D.S., Gaasterland D., et al. (2012) Genome-wide analysis of central corneal thickness in primary open-angle glaucoma cases in the NEIGHBOR and GLAUGEN consortia. Invest. Ophthalmol. Vis. Sci., 53, 4468–4474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Nakatsu M.N., Ding Z., Ng M.Y., Truong T.T., Yu F., Deng S.X. (2011) Wnt/beta-catenin signaling regulates proliferation of human cornea epithelial stem/progenitor cells. Invest. Ophthalmol. Vis. Sci., 52, 4734–4741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Miyake M., Yamashiro K., Tabara Y., Suda K., Morooka S., Nakanishi H., Khor C.C., Chen P., Qiao F., Nakata I., et al. (2015) Identification of myopia-associated WNT7B polymorphisms provides insights into the mechanism underlying the development of myopia. Nat. Commun., 6, 6689.. [DOI] [PubMed] [Google Scholar]
- 30. Shankardas J., Senchyna M., Dimitrijevich S.D. (2008) Presence and distribution of 14-3-3 proteins in human ocular surface tissues. Mol. Vis., 14, 2604–2615. [PMC free article] [PubMed] [Google Scholar]
- 31. Lee H.K., Lee J.H., Kim M., Kariya Y., Miyazaki K., Kim E.K. (2006) Insulin-like growth factor-1 induces migration and expression of laminin-5 in cultured human corneal epithelial cells. Invest. Ophthalmol. Vis. Sci., 47, 873–882. [DOI] [PubMed] [Google Scholar]
- 32. Brown D.J., Lin B., Holguin B. (2004) Expression of neuregulin 1, a member of the epidermal growth factor family, is expressed as multiple splice variants in the adult human cornea. Invest. Ophthalmol. Vis. Sci., 45, 3021–3029. [DOI] [PubMed] [Google Scholar]
- 33. Lambiase A., Manni L., Bonini S., Rama P., Micera A., Aloe L. (2000) Nerve growth factor promotes corneal healing: structural, biochemical, and molecular analyses of rat and human corneas. Invest. Ophthalmol. Vis. Sci., 41, 1063–1069. [PubMed] [Google Scholar]
- 34. Xu K.P., Riggs A., Ding Y., Yu F.S. (2004) Role of ErbB2 in Corneal Epithelial Wound Healing. Invest. Ophthalmol. Vis. Sci., 45, 4277–4283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ban Y., Dota A., Cooper L.J., Fullwood N.J., Nakamura T., Tsuzuki M., Mochida C., Kinoshita S. (2003) Tight junction-related protein expression and distribution in human corneal epithelium. Exp. Eye Res., 76, 663–669. [DOI] [PubMed] [Google Scholar]
- 36. Diehn J.J., Diehn M., Marmor M.F., Brown P.O. (2005) Differential gene expression in anatomical compartments of the human eye. Genome Biol., 6, R74.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Chen M.J., Liu Y.T., Tsai C.C., Chen Y.C., Chou C.K., Lee S.M. (2009) Relationship between central corneal thickness, refractive error, corneal curvature, anterior chamber depth and axial length. J. Chin. Med. Assoc., 72, 133–137. [DOI] [PubMed] [Google Scholar]
- 38. Figueira E.C., Di Girolamo N., Coroneo M.T., Wakefield D. (2007) The phenotype of limbal epithelial stem cells. Invest. Ophthalmol. Vis. Sci., 48, 144–156. [DOI] [PubMed] [Google Scholar]
- 39. Lyu J., Joo C.K. (2005) Wnt-7a up-regulates matrix metalloproteinase-12 expression and promotes cell proliferation in corneal epithelial cells during wound healing. J. Biol. Chem., 280, 21653–21660. [DOI] [PubMed] [Google Scholar]
- 40. Gauderman W.J. (2002) Sample size requirements for matched case-control studies of gene-environment interaction. Stat. Med., 21, 35–50. [DOI] [PubMed] [Google Scholar]
- 41. Gauderman W.J. (2002) Sample size requirements for association studies of gene-gene interaction. Am. J. Epidemiol., 155, 478–484. [DOI] [PubMed] [Google Scholar]
- 42. Gao X., Haritunians T., Marjoram P., McKean-Cowdin R., Torres M., Taylor K.D., Rotter J.I., Gauderman W.J., Varma R. (2012) Genotype Imputation for Latinos Using the HapMap and 1000 Genomes Project Reference Panels. Front. Genet., 3, 117.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Xiao R., Boehnke M. (2009) Quantifying and correcting for the winner's curse in genetic association studies. Genet. Epidemiol., 33, 453–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Xiao R., Boehnke M. (2011) Quantifying and correcting for the winner's curse in quantitative-trait association studies. Genet. Epidemiol., 35, 133–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Sun L., Dimitromanolakis A., Faye L.L., Paterson A.D., Waggott D., Group D.E.R., Bull S.B. (2011) BR-squared: a practical solution to the winner's curse in genome-wide scans. Hum. Genet., 129, 545–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Chang C.C., Chow C.C., Tellier L.C., Vattikuti S., Purcell S.M., Lee J.J. (2015) Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience, 4, 7.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Delaneau O., Marchini J., Genomes Project C., Genomes Project C. (2014) Integrating sequence and array data to create an improved 1000 Genomes Project haplotype reference panel. Nat. Commun., 5, 3934.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Patterson N., Price A.L., Reich D. (2006) Population structure and eigenanalysis. PLoS. Genet., 2, e190.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Genomes Project C., Abecasis G.R., Auton A., Brooks L.D., DePristo M.A., Durbin R.M., Handsaker R.E., Kang H.M., Marth G.T., McVean G.A. (2012) An integrated map of genetic variation from 1,092 human genomes. Nature, 491, 56–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Mao X., Bigham A.W., Mei R., Gutierrez G., Weiss K.M., Brutsaert T.D., Leon-Velarde F., Moore L.G., Vargas E., McKeigue P.M., et al. (2007) A genomewide admixture mapping panel for Hispanic/Latino populations. Am. J. Hum. Genet., 80, 1171–1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Devlin B., Roeder K. (1999) Genomic control for association studies. Biometrics, 55, 997–1004. [DOI] [PubMed] [Google Scholar]
- 52. Willer C.J., Li Y., Abecasis G.R. (2010) METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics, 26, 2190–2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Gao X., Becker L.C., Becker D.M., Starmer J.D., Province M.A. (2010) Avoiding the high Bonferroni penalty in genome-wide association studies. Genet. Epidemiol., 34, 100–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. R Development Core Team. (2009), In R Foundation for Statistical Computing, Vienna, Austria.
- 55. Pruim R.J., Welch R.P., Sanna S., Teslovich T.M., Chines P.S., Gliedt T.P., Boehnke M., Abecasis G.R., Willer C.J. (2010) LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics, 26, 2336–2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Lee S., Emond M.J., Bamshad M.J., Barnes K.C., Rieder M.J., Nickerson D.A., Team N.G.E.S.P.E.L.P., Christiani D.C., Wurfel M.M., Lin X. (2012) Optimal unified approach for rare-variant association testing with application to small-sample case-control whole-exome sequencing studies. Am. J. Hum. Genet., 91, 224–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Wang J., Duncan D., Shi Z., Zhang B. (2013) WEB-based GEne SeT AnaLysis Toolkit (WebGestalt): update 2013. Nucleic Acids Res., 41, W77–W83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Benjamini Y., Hochberg Y. (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Series B, 57, 289–300. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.