Abstract
Ciliary trafficking defects underlie the pathogenesis of severe human ciliopathies, including Joubert Syndrome (JBTS), Bardet-Biedl Syndrome, and some forms of retinitis pigmentosa (RP). Mutations in the ciliary protein RPGR (retinitis pigmentosa GTPase regulator) are common causes of RP-associated photoreceptor degeneration worldwide. While previous work has suggested that the localization of RPGR to cilia is critical to its functions, the mechanism by which RPGR and its associated cargo are trafficked to the cilia is unclear. Using proteomic and biochemical approaches, we show that RPGR interacts with two JBTS-associated ciliary proteins: PDE6δ (delta subunit of phosphodiesterase; a prenyl-binding protein) and INPP5E (inositol polyphosphate-5-phosphatase 5E). We find that PDE6δ binds selectively to the C-terminus of RPGR and that this interaction is critical for RPGR’s localization to cilia. Furthermore, we show that INPP5E associates with the N-terminus of RPGR and trafficking of INPP5E to cilia is dependent upon the ciliary localization of RPGR. These results implicate prenylation of RPGR as a critical modification for its localization to cilia and, in turn suggest that trafficking of INPP5E to cilia depends upon the interaction of RPGR with PDE6δ. Finally, our results implicate INPP5E, a novel RPGR-interacting protein, in the pathogenesis of RPGR-associated ciliopathies.
Introduction
Cilia are microtubule-based membranous extensions of the apical plasma membrane (1) that serve as critical signalling nodes of diverse developmental processes, including sensory perception (2,3). As such, ciliary dysfunction due to mutations in ciliary proteins or trafficking of ciliary cargo is associated with severe human disorders, called ciliopathies. These include Meckel-Gruber Syndrome, Joubert Syndrome (JBTS), Bardet-Biedl Syndrome, and Usher Syndrome. Although these disorders differ in their age of onset and severity, they share common phenotypic features, including retinal degeneration due to defects in photoreceptor development and maintenance (4–6).
Photoreceptors are polarized sensory neurons with distinct inner segments involved in protein synthesis and trafficking, and photosensory outer segments (OS) (7,8). A photoreceptor OS extends from the apical region of the inner segment in the form of a microtubule based axoneme and contains stacks of membranous discs loaded with proteins involved in light detection. The region between the apical inner segment and the base of the OS is termed the connecting cilium, which is analogous to the transition zone (TZ) of primary cilia (9). The TZ is involved in regulating the composition of the OS. Commensurate with this role, proteins localized to the TZ are involved in regulating the composition of the photoreceptor cilia (6).
RPGR, is a ciliary TZ protein and has been suggested to regulate ciliary trafficking in photoreceptors (10). Mutations in RPGR result in severe photoreceptor degenerative diseases (11–15), the most prevalent of which is X-linked RP, a disorder characterized by progressive vision loss, usually by the third decade of life (16–24). RPGR mutations also account for 15–20% of simplex RP in males, making it a common cause of RP (17).
There are two major isoforms of RPGR: RPGRconst, a 19 exon constitutive transcript encoding a protein of 815 amino acids, and RPGRORF15, encoded by 15 exons (1152 amino acids) (21,22,25). The RPGRORF15 isoform shares exons 1-15 with the RPGRconst isoform; however, it continues into and terminates in intron 15. The terminal exon of this isoform is also termed ORF15. Although RPGRORF15 contains fewer exons, it is longer than RPGRconst because the length of exon ORF15 is greater than the combined length of exons 16-19 of RPGRconst. A schematic representation of these isoforms is depicted in Supplementary Material, Figure 1A and B.
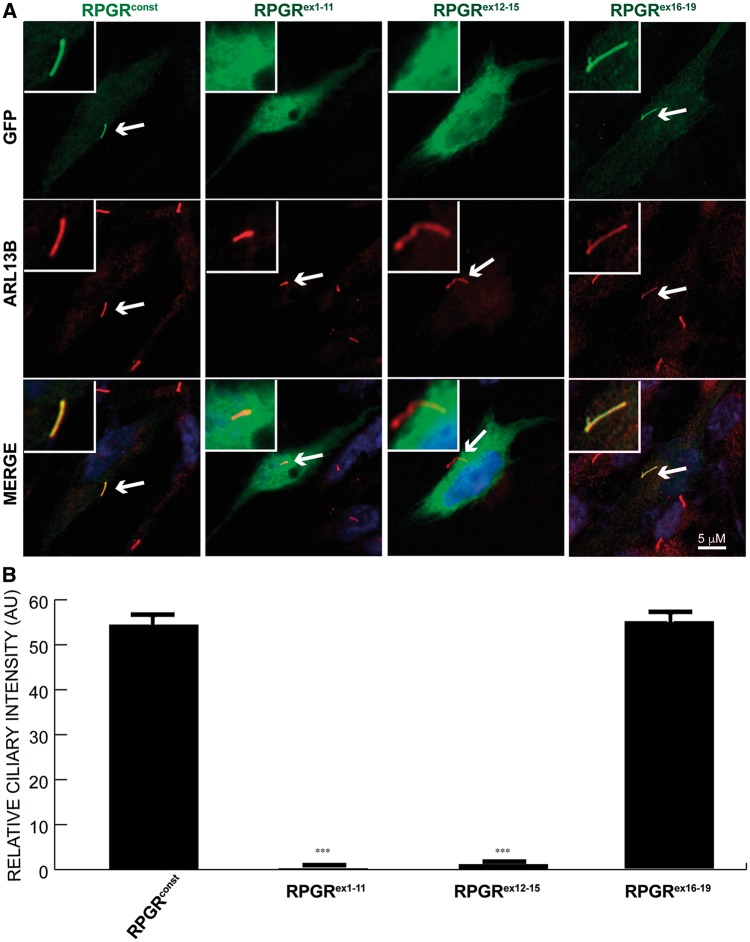
Figure 1.
Localization of RPGR domains to cilia. (A) hTERT-RPE1 cells were transiently transfected with constructs encoding the indicated GFP-fused RPGR proteins. After cilia formation, cells were fixed and stained with anti-GFP (green) and anti-ARL13B (red; cilia marker). Arrows in Merge point to ciliary staining (yellow, if co-localization; red, if no co-localization with ARL13B). Inset depicts enlarged view of ciliary staining of the cells. (B) Quantification of GFP-positive cilia is represented in the lower panel. One-way ANOVA analysis was performed to calculate the statistical significance of the differences in GFP-positive cilia. ***: P < 0.0001. AU: arbitrary units. Number (n) of cells analysed in this experiment: >200.
Both RPGR isoforms share a common N-terminal domain but differ in the C-terminus (26). A region of RPGR’s N-terminal domain, encompassing residues 1-392, shares homology with RCC1 (regulator of chromosome condensation), which is a regulator of GTPase activity (21); consistently, RPGR exhibits guanine exchange factor activity towards small GTPase RAB8A (27). RPGR also interacts with several ciliary and transport proteins, such as PDE6δ, NPHP4, Structural maintenance of Chromosomes (SMC1A), CEP290 (centrosomal protein 290 kDa), RPGR-interacting protein 1 (RPGRIP1), and RPGRIP1-like (RPGRIP1L; FTM) (28–33). Importantly, all RPGR-interacting proteins identified to date are involved in human ciliopathies (34).
Previous studies have revealed that dysfunction of the RPGR-interacting proteins RPGRIP1 and CEP290 result in relatively early and severe photoreceptor degeneration in animal models and in patients. Furthermore, disruption of these proteins leads to mislocalization of RPGR in photoreceptors (31,35,36). Collectively, these results suggest that the localization of RPGR to cilia is critical for optimal ciliary function and for overall photoreceptor health. However, the mechanism whereby RPGR is targeted to cilia and the cargo moieties dependent upon RPGR for their ciliary localization remain unclear. In this study, we present evidence suggesting that the ciliary localization of RPGR is dependent upon its prenylation by PDE6δ and identify INPP5E, a novel RPGR-interacting protein as potential ciliary cargo, which is trafficked by RPGR in photoreceptor cilia.
Results
The Carboxyl (C)-terminus of RPGRconst is critical for its ciliary localization
To identify the domains of human RPGR involved in ciliary localization, we generated variants of RPGR harbouring deletions and examined their localization in ciliated cells. As shown in Figure 1A and B, GFP-tagged RPGRconst and GFP-RPGRex16-19 predominantly localized to cilia, as determined by co-localization with ARL13B, a ciliary marker. However, GFP-RPGRex1-11 and GFP-RPGRex12-15 did not localize to cilia, and analysis of additional deletion mutants revealed that the presence of exon 19 of RPGR (RPGRex19) allowed for at least partial localization of RPGR to cilia (Fig. 2A). The reduced ciliary localization of RPGR harbouring shorter C-termini suggests that regions encoded by exons 16-19 contribute together in efficient localization of RPGR to cilia (Fig. 2B).
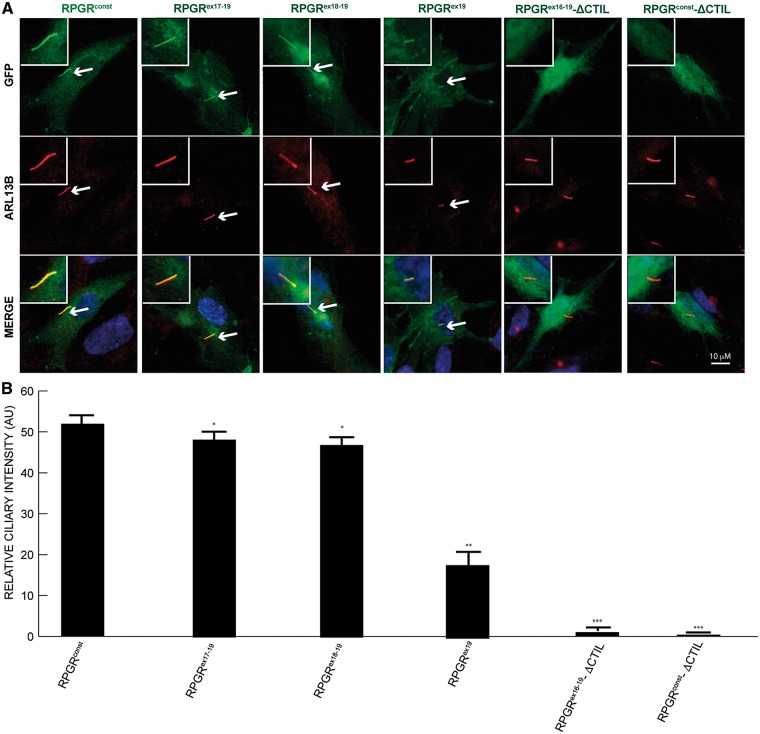
Figure 2.
Localization of deleted variants of RPGR to cilia. (A) The domain of RPGR involved in the localization to cilia was identified by transfecting constructs encoding GFP-fused (green) full length, deleted variants at the C-terminus or without the –CTIL domain of RPGRconst into hTERT-RPE1 cells. After cilia growth, cells were stained with anti-ARL13B antibody (red) or anti-GFP (green) antibody. Inset shows the enlarged image of the cilia-containing region. Arrows in Merge indicate ciliary staining of the proteins that localize to cilia. (B) This panel shows quantification of GFP-positive cilia. *: P < 0.01; **: P < 0.001; ***; P < 0.0001. AU: arbitrary units. Number of cells analysed: n > 200.
Prenylation signal at the C-terminus of RPGRconst is critical for its ciliary localization
The C-terminus of RPGRconst (amino acids 812-815: -CTIL) (37,38) contains a CaaX box, where C is cysteine, A- aliphatic amino acid and X is usually leucine, methionine or phenylalanine (39). The CaaX motif is a predicted prenylation signal, a post-translational modification that regulates membrane association and protein trafficking. We therefore asked whether this putative prenylation site is involved in RPGR’s ciliary localization. To this end, we generated an RPGR isoform harbouring a deletion of the –CTIL motif (RPGR-ΔCTIL) and examined its cellular localization. As shown in Figure 2, deletion of the CTIL motif from RPGRconst or RPGRex16-19 abolished its localization to cilia.
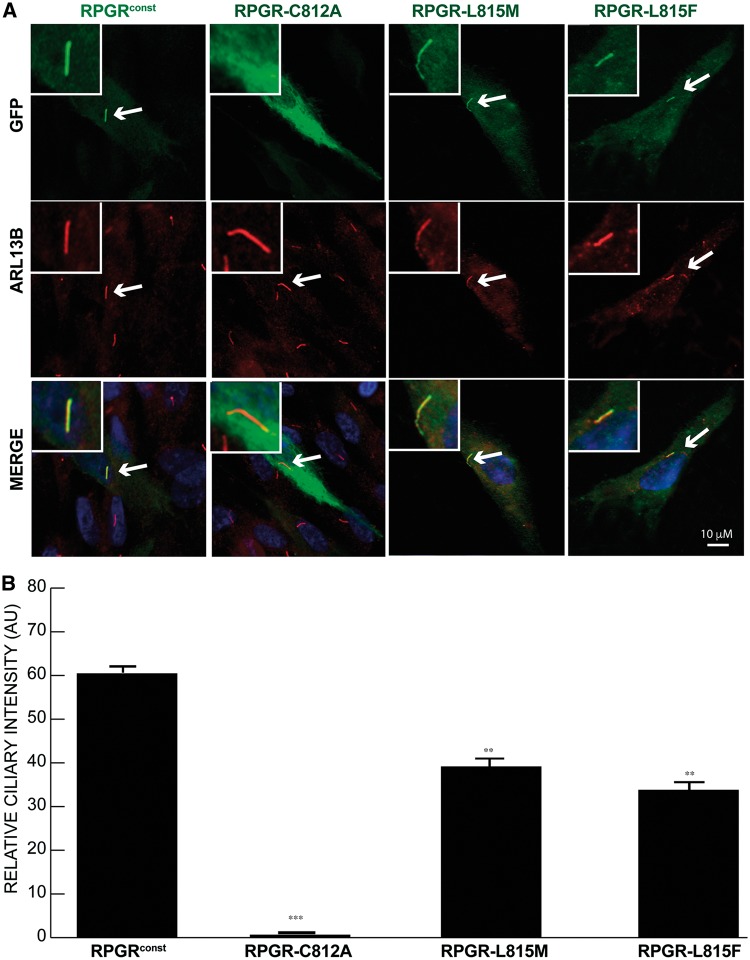
We further assessed the requirement of the conserved C812 residue in its localization to cilia by site-directed mutagenesis of C812A. Whereas WT RPGR localized to cilia, RPGR-C812A was no longer detected in cilia (Fig. 3). Interestingly, previous work has shown that the terminal residue of the –CaaX box determines the type of prenylation that occurs at the site – a leucine or phenylalanine results in geranylgeranylation while a methionine results in farnesylation (40–43). We assessed whether RPGR was geranylgeranylated and whether this modification influenced its ciliary localization by substituting a methionine in place of the leucine 815 (RPGR L815M), such that RPGR became a substrate for farnesylation. As a control, we also generated an L815F variant to promote geranylgeranylation. While the RPGR-L815M or RPGR-L815F variants correctly localized to cilia, they exhibited significantly decreased ciliary signal (Fig. 3). These results suggest that the ciliary localization of RPGR likely depends upon the type of prenylation and the identity of the terminal leucine (L815) residue involved in prenylation.
Figure 3.
Ciliary localization of prenylation-deficient mutants of RPGR. (A) hTERT-RPE1 cells were transiently transfected with constructs encoding GFP-fused wild type or site-directed mutant forms of the –CTIL domain of RPGRconst. After serum starvation to induce cilia formation, cells were stained with anti-GFP (green) and anti-ARL13B (red) antibodies. Arrows indicate cilia in transfected cells. Inset shows the region of cilia growth. (B) shows the quantification of GFP-positive cilia in cells transfected with mutant RPGR with respect to the wild type counterpart. **: P < 0.001; ***; P < 0.0001. AU: arbitrary units. Number of cells analysed: n > 200.
The RPGR C-terminus interacts with PDE6δ
We next investigated the mechanism of localization of RPGR to cilia by identifying its interacting proteins. We performed TAP-mediated isolation of RPGR-containing protein complexes from cells. We first generated stable HEK293T cells expressing RPGRconst protein and purified the RPGRconst-protein complexes followed by MS/MS analysis, as described in the Materials and Methods. Data were collected from three independent experiments. In addition to detecting RPGRconst, we identified several previously reported RPGR-interacting proteins, such as SMC1A, NPHP4, and RPGRIP1L (28,32,33) (Supplementary Material, Table S1).
Interestingly, our analysis also identified PDE6δ and INPP5E in complex with RPGR. Previous studies showed that both PDE6δ and INPP5E interact with each other and that PDE6δ binds to prenylated cargo to regulate its ciliary localization (43–45). Moreover, mutations in PDE6δ and INPP5E are associated with JBTS (46,47). Using yeast two-hybrid analysis and purified proteins, PDE6δ was also identified as an interacting partner of RPGR (29,48,49). We selected PDE6δ and INPP5E for further analysis.
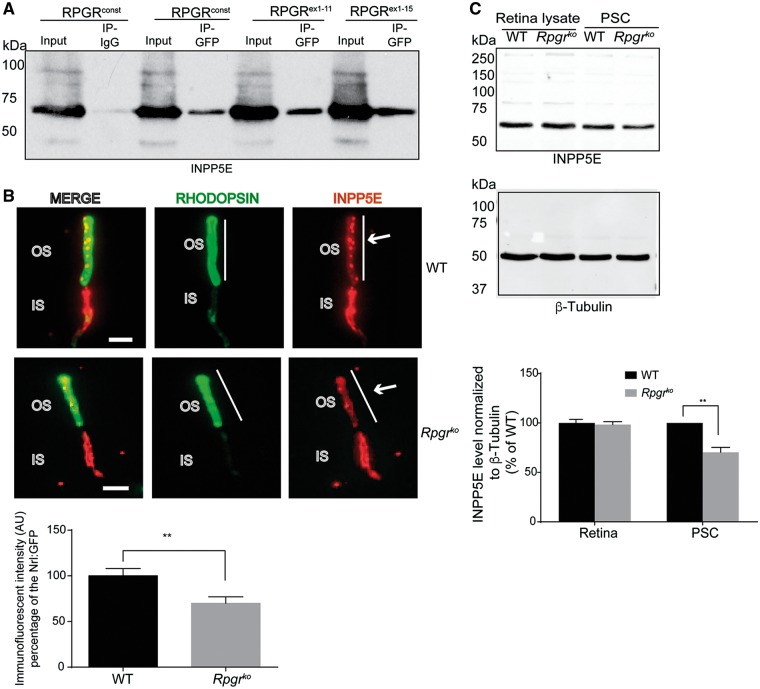
We hypothesized that the interaction of RPGR with PDE6δ is critical for its ciliary localization. We first validated the interaction of RPGR and PDE6δ by co-transfecting GFP-RPGRconst- and MYC-PDE6δ-encoding constructs followed by co-immunoprecipitation (Co-IP) (Supplementary Material, Fig. S2A). As a control, while GFP did not show an association with MYC-tagged PDE6δ (Supplementary Material, Fig. S2B; left panel), the anti-GFP antibody efficiently pulled down GFP from the transfected cells (Supplementary Material, Fig. S2B; right panel). Moreover, we did not detect an interaction of MYC- PDE6δ with RPGRORF15, as indicated by the lack of an anti-MYC immunoreactive band (Supplementary Material, Fig. S2C). To identify the region of RPGRconst that interacts with PDE6δ, we performed co-transfection and Co-IP experiments using constructs encoding deleted variants of RPGRconst. The N-terminal domains of RPGR (RPGRex1-11, RPGRex12-15, and RPGRex1-15) did not exhibit a detectable interaction with PDE6δ (Supplementary Material, Fig. S2C). However, we found that RPGRex16-19 interacted with PDE6δ (Supplementary Material, Fig. S2D). Further analysis revealed that RPGRex19 was sufficient to bind to PDE6δ (Supplementary Material, Fig. S2D).
The RPGR-PDE6δ interaction depends upon the prenylation site in RPGRex19
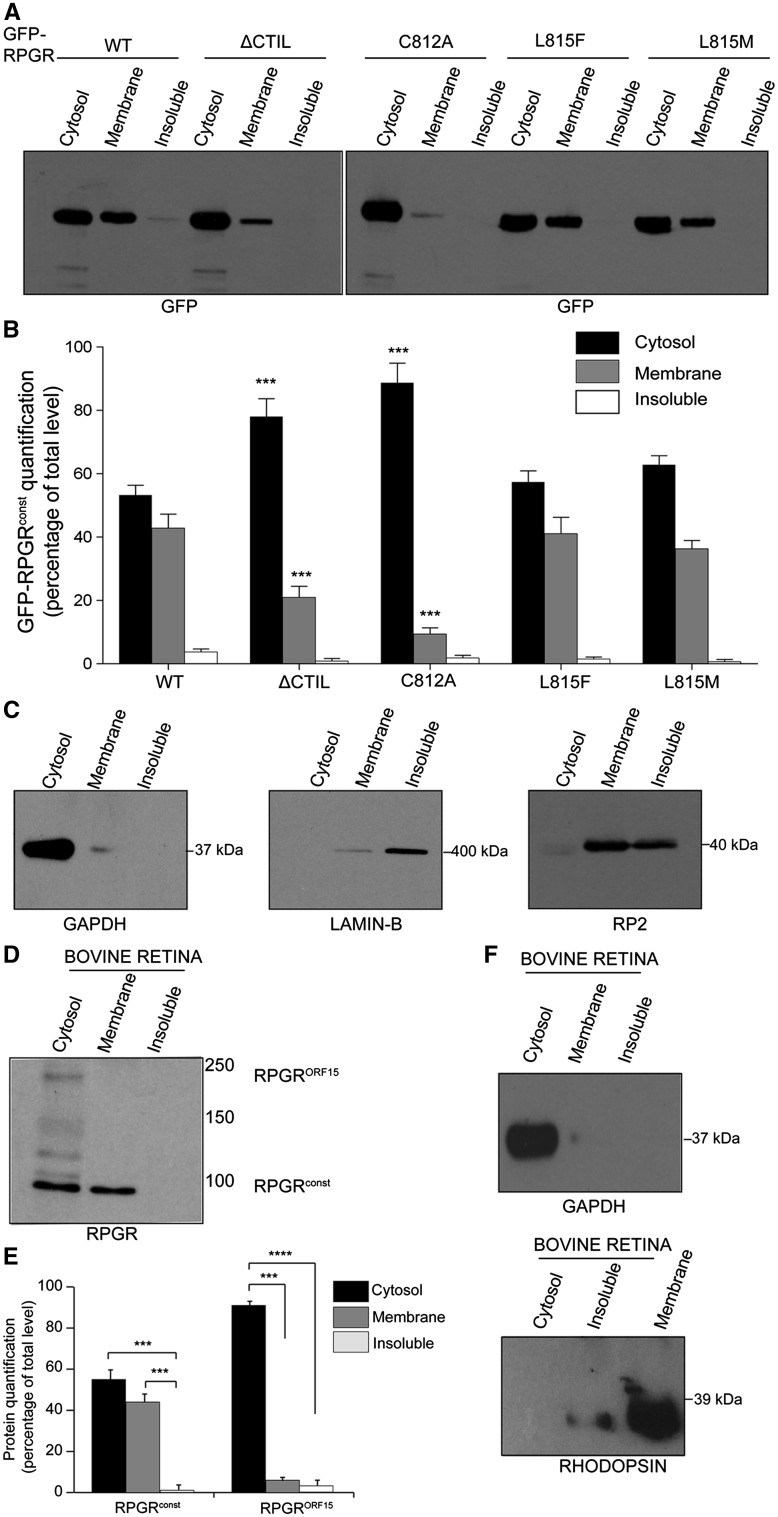
We next tested whether the binding of RPGR to PDE6δ is dependent upon the presence of RPGR’s prenylation site. While the RPGR-C812A variant that failed to localize to cilia did not bind to PDE6δ (Supplementary Material, Fig. S2E), the RPGR-L815M or RPGR-L815F mutants, which both localized to cilia, interact with PDE6δ. We next hypothesized that if the RPGR-PDE6δ interaction is important for RPGR’s ciliary trafficking, then RPGR would associate with membranes in a prenylation-dependent manner. To this end, we fractionated the cells transfected with GFP-RPGRconst or its variants to obtain cytosolic, membrane, and insoluble fractions, followed by immunoblotting using anti-GFP antibody. As shown in Figure 4A and B, GFP-RPGRconst was present in both the membrane and the cytosolic fractions while the RPGR-ΔCTIL variant exhibited an increased abundance in the cytosolic fraction (∼80%) and significantly reduced (∼25%) membrane association. Similarly, the majority of the GFP-RPGR-C812A variant was detected in the cytosol fraction. On the other hand, the GFP-RPGR-L815M and RPGR- L815F variants did not exhibit a detectable difference in their localization as compared to wild type RPGR. We validated the purity of each fraction of transfected cells by assessing the enrichment of cognate marker proteins (Fig. 4C). GAPDH and LaminB markers were used for the cytosolic and insoluble (containing nuclear proteins) fractions, while RP2 (retinitis pigmentosa 2) was enriched in both membrane and insoluble fractions (50,51).
Figure 4.
Subcellular localization of RPGR mutants. (A–C) hTERT-RPE1 cells were transfected with constructs encoding the indicated RPGR proteins and subjected to subcellular fractionation as described in the Methods section. Equal amounts (50 μg) of proteins were loaded onto SDS-PAGE followed by immunoblotting using indicated antibodies. (B) depicts quantification of the band intensity observed in each lane of (A) relative to the cytosolic fraction. (C) Subcellular fractions were analysed for purity using antibodies against indicated marker proteins. GAPDH: cytosol; LaminB: detergent insoluble; RP2: membrane-associated and detergent insoluble fractions. (D) Bovine retinas were subjected to subcellular fractionation and equal amount of protein was analysed by SDS-PAGE and immunoblotting using anti-RPGR antibody. (E) Quantification of band intensity relative to cytosol fraction was performed as detailed in the Methods section. (F) Immunoblotting using antibodies against marker proteins was performed to assess the purity of the fractions. Rhodopsin was used as a membrane protein marker. *: P < 0.01; **: P < 0.001; ***; P < 0.0001. All experiments were repeated at least three times.
We also tested the enrichment of RPGR in subcellular fractions of mammalian retina. As predicted, fractionation of bovine retina followed by immunoblotting using an anti-RPGR antibody showed that the RPGRconst isoform was enriched in the cytosolic and the membrane fractions (Fig. 4D and E). We validated the purity of the cytosolic and membrane fractions of the retina by validating that these fractions were enriched in GAPDH and rhodopsin antibodies, respectively (Fig. 4F).
PDE6δ is required for the ciliary localization of RPGRconst
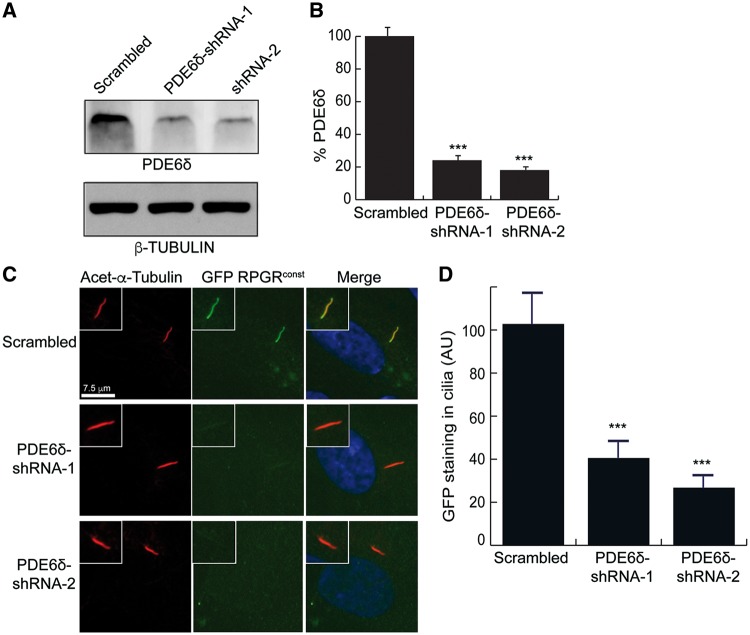
Our results thus far indicated a direct relationship between the localization of RPGRconst to cilia and its interaction with PDE6δ. To directly test the requirement of PDE6δ in mediating the ciliary localization of RPGRconst, we performed shRNA-mediated knockdown of PDE6δ in hTERT-RPE1 cells, followed by assessment of RPGR localization to cilia. Efficient knockdown of PDE6δ was validated by immunoblot analysis of protein extracts transfected with scrambled (negative control) or two specific PDE6δ-shRNAs (shRNA-1 and shRNA-2). Both specific shRNAs exhibited more than 80% knockdown of protein levels of PDE6δ as compared to the scrambled control (Fig. 5A and B). Further analysis revealed that knockdown of PDE6δ significantly perturbed (60–80% reduction) the localization of GFP-RPGRconst to cilia (Fig. 5C and D).
Figure 5.
Role of PDE6δ in RPGR’s localization to cilia. (A) Protein extracts from hTERT-RPE1 cells stably expressing scrambled (negative control) or PDE6δ-specific shRNA-1 and shRNA-2 were analysed by SDS-PAGE and immunoblotting using anti-PDE6D or β-tubulin (loading control) antibodies. (B) Band intensity was quantified relative to that of β-tubulin. The value for scrambled was set as 100%. ***: P < 0.0001. (C) Scrambled or PDE6δ-specific shRNA expressing cells were transiently transfected with GFP-RPGRconst-encoding construct. The cells were then incubated in serum-starved conditions to grow cilia followed by staining with anti-acetylated α-tubulin (Acet- α-Tubulin; ciliary marker; red) or anti-GFP (green) antibodies. (D) GFP-positive ciliary staining in panel C was quantified in n > 100 cells. ***: P < 0.0001. AU: arbitrary units.
The N-terminus of RPGR interacts with INPP5E
As INPP5E was also identified in the MS/MS analysis of RPGR-interacting proteins (Supplementary Material, Table S1) and both INPP5E and RPGR interact with PDE6δ, we hypothesized that RPGR is involved in the ciliary localization of INPP5E. We first investigated whether endogenous PDE6δ and INPP5E can associate with RPGR. Using hTERT-RPE1 cell extracts, we performed co-ip using anti-PDE6δ and anti-INPP5E antibodies followed by immunoblotting using anti-RPGR antibody. As shown in Supplementary Material, Fig. 3a, both INPP5E and PDE6δ can immunoprecipitate RPGR whereas normal immunoglobulin (IgG) do not show any association with RPGR. We next investigated RPGR-INPP5E in further detail. Using co-transfection and Co-IP experiments, we found that GFP-RPGRconst as well as GFP-RPGRex1-11 and GFP-RPGRex1-15 interacted with INPP5E (Fig. 6A). However, the C-terminal domain GFP-RPGRex16-19 did not exhibit an interaction with INPP5E (data not shown).
Figure 6.
RPGR interacts with INPP5E. (A) hTERT-RPE1 cells transfected with constructs encoding GFP-RPGRconst and indicated deleted variants were subjected with IP using anti-GFP antibody followed by immunoblotting using anti-INPP5E antibody. IP using IgG was used as negative control. (B) Dissociated photoreceptors isolated from WT and Rpgrko mouse (2 months old) retinas were stained with anti-rhodopsin (green; photoreceptor OS marker) and anti-INPP5E (red) antibodies. Arrows indicate INPP5E staining in the OS. Lower panel: Images were taken at the same exposure setting using Leica DM5500 microscope. Vertical bar in B represents the region of interest (ROI) marked by rhodopsin staining. The corresponding INPP5E fluorescence intensity was quantified by analysing the grayscale values of the preset ROI using ImageJ. The average intensity of non-OS area was designated as background and was subtracted from the final fluorescence intensity. The experiment was repeated three times and more than 100 photoreceptors were analysed in each experiment. Student t-test was used to calculate statistical significance. **: P < 0.001. IS: inner segment. (C) Total protein extracts (Retinal lysate) or photoreceptor sensory cilium (PSC) isolated from WT and Rpgrko mice were analysed by SDS-PAGE and immunoblotting using anti-INPP5E antibody (upper panel) or anti-β-tubulin antibody (lower panel; loading control). Molecular weight markers are shown in kilo Daltons (kDa). Quantification of INPP5E band intensity normalized to β-tubulin band intensity revealed reduced levels in the Rpgrko PSC as compared to WT PSC. No change in the total levels of INPP5E was observed between the two samples.
RPGR regulates the ciliary localization of INPP5E
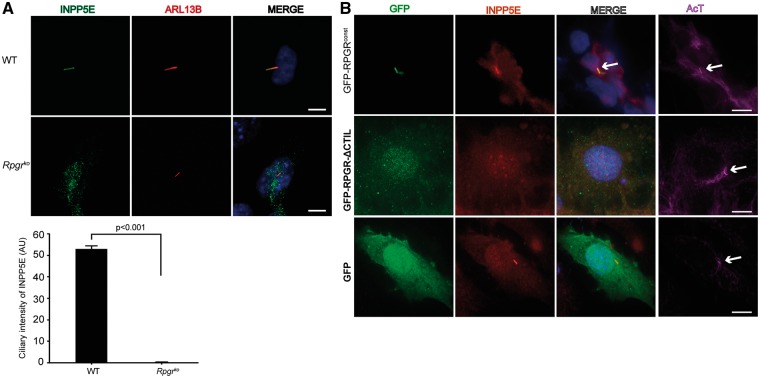
To test if RPGR is involved in the trafficking of INPP5E, we examined the localization of INPP5E in dissociated photoreceptors of wild type and Rpgrko mice. Consistent with previous studies (52), we detected a predominant localization of INPP5E to the inner segment (IS) of photoreceptors (Fig. 6B). However, we also detected an OS localization of INPP5E in dissociated photoreceptors, as determined by co-staining with rhodopsin. Such discrepancy may have arisen because the previous study was performed using mouse retinal tissue sections whereas we used dissociated photoreceptors. Further analysis showed that the OS content of INPP5E was significantly reduced in the Rpgrko photoreceptors as compared to their wild type counterparts. To corroborate these results, we quantified the amount of INPP5E in purified photoreceptor sensory cilia (PSC) from wild type and Rpgrko mouse retinas. As shown in Figure 6C, diminished levels of INPP5E were detected in the Rpgrko PSC as compared to wild type PSC. Similar results were obtained using mouse embryonic fibroblasts (MEFs) derived from wild type and Rpgrko mice (Fig. 7A).
Figure 7.
RPGR regulates ciliary localization of INPP5E. (A) WT and Rpgrko MEFs were serum-starved to grow cilia and stained with anti-ARL13B (red; ciliary marker) and anti-INPP5E antibody (green). Arrow in Merge shows co-localization of INPP5E in the WT MEFs. Ciliary intensity of INPP5E was calculated using Image J and represented in arbitrary units (AU). (B) The Rpgrko MEFs were transiently transfected with constructs encoding GFP, GFP-RPGRconst or GFP-RPGR-ΔCTIL followed by staining with anti-GFP (green), anti-INPP5E (red) or anti-acetylated α-tubulin (AcT; cilia marker; purple) antibodies. Nuclei were stained with Hoechst (blue). Arrows in the AcT panel depict the cilia growth in all cells.
While our results showed that RPGR is involved in the ciliary localization of INPP5E, it was unclear whether RPGR’s localization to cilia influenced the ciliary localization of INPP5E. To test this, we investigated the ability of RPGR-ΔCTIL, which cannot localize to cilia, to rescue the ciliary localization of INPP5E in Rpgrko MEFs. As a positive control, we examined the potential of wild type RPGR to mediate ciliary localization of INPP5E in Rpgrko MEFs. As shown in Figure 7B, cells expressing GFP-RPGRconst, but not GFP-RPGR-ΔCTIL, exhibited proper ciliary localization of INPP5E. Overall, these results suggest that RPGR traffics INPP5E to cilia via the PDE6δ-mediated targeting of RPGR itself to the cilia. If this were true, then PDE6δ-INPP5E association should be altered in the absence of RPGR. To test this hypothesis, we assessed the interaction of INPP5E and PDE6δ in Rpgrko MEFs. As shown in Supplementary Material, Figure 3B, reduced PDE6δ is observed in MYC-INPP5E immunoprecipitated cell extracts from Rpgrko MEFs as compared to that in the wild type MEFs.
Discussion
Ciliary protein trafficking is essential for the development and maintenance of photoreceptors (34,53,54). Although several major players involved in the genesis of photoreceptor sensory cilium are now being identified, it is still unclear how such a specialized cilium is maintained during physiologic circumstances and undergoes degeneration during disease. Examination of RPGR provides a unique opportunity to answer these questions, as RPGR mutations are common causes of RP and loss of RPGR alters ciliary trafficking and maintenance but spares ciliogenesis (10). In this study, we provide a mechanism of RPGR’s localization to cilia and further demonstrate its involvement in the ciliary localization of INPP5E, a protein involved in ciliary stability and photoreceptor maintenance.
Although both RPGR and INPP5E are prenylated, the C-terminus of RPGR binds PDE6δ whereas N-terminus of RPGR interacts with INPP5E. This finding indicates that RPGR has the capability to bind to both the proteins simultaneously. Mutations in both INPP5E and PDE6δ are associated with severe retinal degeneration. A Pde6δko mouse model exhibits relatively earlier onset photoreceptor dysfunction (at 2 months age) (45) as compared to the Rpgrko mice, which show dysfunction around 6 months of age. The Inpp5e-/- mice, on the other hand, die soon after birth and exhibit defects in eye development (55). These studies suggest that both PDE6δ and INPP5E, in addition to participating in RPGR-mediated ciliary trafficking, are involved in additional functions. Moreover, based on the observation that the PDE6δ-INPP5E complex is only partially perturbed in the absence of RPGR and a relatively milder reduction in INPP5E levels in the OS of the Rpgrko mice, we suggest that additional proteins may be involved in the ciliary targeting of INPP5E.
Based on the findings reported here, we propose that prenylation of RPGR promotes its binding to PDE6δ and localization to cilia. As PDE6δ shuttles prenylated cargo and assists in its release from the membrane, we propose that RPGRconst provides a docking site for the INPP5E at the cilia. INPP5E is likely released from the PDE6δ-RPGRconst complex by the action of ARL13B or ARL3 or a hitherto unknown small GTPases. As RPGR can also act as a local regulator of small GTPases at the cilia, it is possible that RPGR recruits small GTPases for efficient delivery of INPP5E. Further studies are needed to test the validity of this model.
It was previously reported that PDE6δ interacts with N-terminus of RPGRconst (29). However, we found that the C-terminus of RPGRconst predominantly binds to PDE6δ. This discrepancy could be due to differences in experimental platform; earlier experiments were performed in yeast two-hybrid system or using bacterially purified proteins in vitro (29). However, we have utilized a physiological platform in a eukaryotic system. Nonetheless, it is possible that the N-terminal region of RPGR may present a weaker binding site for PDE6δ or assist in the optimal binding to the C-terminus of RPGRconst by stabilizing the conformation of the complex. While this study was being prepared for consideration, two reports were published showing that both N- and C-termini of RPGRconst can interact with PDE6δ (38). Although these studies corroborate our observations, they do not provide the physiological relevance of this association. It is possible that in endogenous conditions, the binding of the N-terminus of RPGR to PDE6δ is perturbed due to the binding of INPP5E. Further studies are needed to test this scenario. Our investigation shows that the RPGR-PDE6δ interaction is critical for the localization of RPGRconst to cilia. As predicted, the RPGRORF15 isoform, which lacks the C-terminal prenylation signal, does not interact with PDE6δ. However, RPGRORF15 isoform localizes to cilia in mouse retina (32). Such differences could be because of an as yet unknown cilia-targeting signal in this isoform or the differences in the cell type investigated in this study. We propose that isoform-specific differences in the mechanisms of localization of RPGR to cilia may underlie the heterogenic ciliary dysfunction observed in RPGR-associated disease (24,56).
Our data suggest a potential crosstalk between RPGR and PDE6δ in regulating the ciliary localization of INPP5E. As these proteins are involved in severe human pathologies that share retinal degeneration as a common feature, our studies provide critical insights into the pathogenesis of retinal degeneration associated with distinct ciliopathies.
Materials and Methods
Animals
The Rpgrko mice were described earlier (12). Studies using animals were approved by the Institutional Animal Care and Use Committee of UMASS Medical School. Since Rpgr is an X-linked gene, only hemizygous Rpgrko males were used in this study. As controls, we used C57BL6/J mice, which were procured from Jackson Labs.
Antibodies and plasmids
Anti-GFP, anti-INPP5E, anti-ARL13B and anti-PDE6δ were purchased from Abcam (Cambridge, MA, USA), Proteintech Group (Chicago, IL), and Thermo Scientific (Grand Island, NY), respectively. The anti-RPGR antibody was described previously (32,37). Alexa 488-conjugated, and Alexa 546-conjugated secondary antibodies were purchased from Invitrogen (Carlsbad, CA). Constructs encoding N-terminal GFP tagged full-length and deleted variants of human RPGRconst were generated using site-directed mutagenesis and cloned into pEGFC1 (Invitrogen). A detailed description of the constructs used in this study is presented in Supplementary Material, Figure S1C. For tandem affinity purification (TAP), RPGR was cloned into pNTAP-B vector (Agilent Technologies, La Jolla, CA).
Cell culture, immunofluorescence analysis and shRNA-knockdown
hTERT-RPE1 cells and HEK293 cells (American Type Culture Collection; ATCC) were maintained in DMEM/F12 (Invitrogen) supplemented with 10% Fetal Bovine Serum and penicillin/streptomycin. These cells have been recently authenticated and tested for contamination. For immunofluorescence, the cells were seeded on glass cover slips in 12-well plates and grown to 95% confluency. Transient transfections were performed using Lipofectamine 2000 (Invitrogen). After 30 h, cells were serum starved to induce cilia formation, fixed for 15 min with 4% paraformaldehyde in PBS and washed three times with PBS. Samples were blocked with 5% Normal Goat Serum in PBS containing 0.5% TritonX100 followed by incubation with primary antibody overnight. After washing with PBS, Alexa 488-conjugated or Alexa 546-conjugated secondary antibodies were added and further incubated for 1 h. After washing, nuclei were stained with Hoechst dye and cells were imaged using a Leica microscope (DM5500).
For PDE6δ knockdown experiments, the shRNA constructs were selected from human lentiviral short hairpin RNA (shRNA) libraries from Open Biosystems/GE Dharmacon (Lafayette, CO) in the pLKO.1 vector purchased from the University of Massachusetts Medical School RNAi Core Facility. The sequences of oligos are: shRNA-1: 5’< AAAGTCTCAC TCTG GATGTGC > 3’; shRNA-2: 5’< AACCATTCTTCTAGGCATTGC > 3’. Lentivirus was generated by co-transfection of lentiviral vectors encoding the PDE6δ silencing sequence or scrambled sequence with plasmids coding for the lentiviral envelope (pMD2G) and packaging proteins (psPAX2) into 293T cells using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA). RPE cells were transduced with lentiviral supernatants according to the manufacturer’s instructions. After 24 h, 10 μg/ml puromycin was added to the culture medium to select stable knockdown cells. Knockdown efficiency was monitored by immunoblotting for PDE6δ. Al experiments were performed three independent times.
Tandem affinity purification (TAP) and mass spectrometry analysis
HEK293 cells were transfected with pNTAP-B vector alone or pNTAPB-RPGRconst. Stable cell lines were selected with Geneticin (500 μg/mL). TAP was performed according to manufacturer’s instructions (Agilent Technologies). Mass spectrometry of the protein complexes was performed by in-gel tryptic digestion and LC-MS/MS using an LTQ Orbitrap XL hybrid mass spectrometer, as previously described (10). Peptide identification was performed via the MASCOT search engine (Matrix Science, London, UK; version 2.4.0) to search the Swiss-Prot database. Data were subsequently filtered through SCAFFOLD viewer (version Scaffold_4.3.4, proteome Software Inc., Portland, OR). Greater than 95% confidence of probability by the Peptide Prophet algorithm was accepted. Data from three independent experiments were used to calculate the SAINT (Significance Analysis of Interactome) score, which utilizes spectral counts from experimental and control experiments (57). A score of 0.5 or higher (maximum score of 1) was considered significant.
Immunoprecipitation (IP)
Plasmids encoding GFP tagged RPGRconst and MYC tagged PDE6δ were co-transfected into hTERT-RPE1 cells. Cells were lysed 48 h post-transfection in IP lysis buffer containing 25mM Tris, 150mM NaCl, 1mM EDTA, 1% NP-40, 5% glycerol (pH 7.4) with Complete protease inhibitors (Roche) and the resultant lysates were spun at 13000xg for 15 min at 4°C. Anti-GFP antibody was conjugated to AminoLink Plus Coupling Resin according to manufacturer’s guidelines. Briefly, 10 μg of GFP antibody was conjugated with AminoLink Plus Coupling Resin in the presence of sodium cyanoborohydride solution for 90 min at room temperature. Unbound antibody was washed and quenched and conjugated beads were further washed for six times. For IP, GFP antibody conjugated resin was added to the lysate and incubated on a nutator at 4°C for overnight. Immunoprecipitates were washed three times with IP lysis buffer. The samples were eluted with glycine buffer (pH 2.8), neutralized by the addition of 1 M Tris (pH 9.5) and analysed by SDS-PAGE.
For identification of the RPGR- PDE6δ interaction in bovine retina, the tissue was first lysed in IP lysis buffer and the immunoprecipitation was performed overnight by adding 5 μg of anti-RPGR antibody or normal IgG, as previously described (32).
Retinal dissociation and PSC preparation
Dissociation of WT and Rpgrko retinas was performed, essentially as described previously (58). In brief, retinas were chopped into 1-2mm square pieces followed by incubation in 0.25% trypsin at 37°C for 20 min. The reaction was stopped by exchanging the buffer with DMEM containing 10% FBS. Dissociated photoreceptors were obtained by pipetting gently 50 times and letting the solution settle and collecting the supernatant. The cells were fixed with 10% formalin for 10 min and processed for immunostaining. PSC from mouse retinas was prepared as described previously (10). All analyses were performed at least three times with independent samples.
Subcellular fractionation
For detection of membrane association of RPGR, pEGFPC1 plasmids encoding RPGRconst or different domains were transfected into cells. After 48 h, cells were lysed in 20mM Tris-HCl, pH 7.4 by three cycles of freeze/thaw and passed five times through a syringe with 27G needle. Cell lysates were incubated on ice for 20 min and centrifuged at 13000g for 20 min. The supernatant was transferred to a fresh eppendorf tube and was designated as cytosolic fraction. The pellet was gently washed in Tris buffer and was resuspended in NP40 buffer (50 mM Tris, pH 7.4, 150mM NaCl, and 1% NP-40 and 0.1% CHAPS). After incubating for 30 min on ice, the samples were centrifuged at 18000g for 20 min. The supernatant was collected and designated as the detergent-soluble fraction. The pellet was carefully washed in NP40 buffer, resuspended in RIPA (RadioImmunoPrecipitation Assay) buffer (50 mM Tris pH 7.5, 150 mM NaCl, 0.1% SDS, 0.5% sodium deoxycholate, 1% NP40, and Complete protease inhibitor cocktail) and lysed by sonication. Subsequently, the lysate was incubated on ice for 20 min and centrifuged at 18000xg for 20 min. The supernatant was collected and designated as the detergent insoluble fraction. Bovine retinas were fractionated using similar procedures. All fractions were analysed by immunoblotting.
For the quantification of bands, we used ImageJ image processing software (National Institutes of Health). The intensity of the bands was measured in the linear range of detection. The percentage of each band intensity was calculated using the following equation:
The percentage of each fraction was compared with the corresponding control (wild type) fraction using the student t-test. P < 0.05 is considered statistically significant.
Statistical analysis
The statistical analyses were performed done by Student’s t-tests (2-group data, compared with wild type) or one-way ANOVA (3 or more-group data, compared with wild type), with P < 0.05 being considered as statistically significant. The results were presented as mean ± SEM (standard error of mean).
Supplementary Material
Supplementary Material is available at HMG online.
Supplementary Material
Acknowledgements
We thank Dr. John Leszyk, Associate Director of UMMS Proteomics Core facility for mass spectrometry; Dr. Hyungwon Choi for SAINT analysis of MS/MS data and; Drs. Gregory Pazour, George Witman, Raju Rajala and Visvanathan Ramamurthy for constructive discussions about this work and; Vishesh Khanna for critical reading of the manuscript.
Conflict of Interest statement. None declared.
Funding
This work is supported by grants from National Eye Institute (EY022372); University of Massachusetts Center for Clinical and Translational Sciences (UMCCTS); Foundation Fighting Blindness and UMASS Cell Biology Confocal Core and Electron Microscopy Core (Award # S10RR027897).
References
- 1. Pedersen L.B., Veland I.R., Schroder J.M., Christensen S.T. (2008) Assembly of primary cilia. Dev. Dyn., 237, 1993–2006. [DOI] [PubMed] [Google Scholar]
- 2. Singla V., Reiter J.F. (2006) The primary cilium as the cell's antenna: signaling at a sensory organelle. Science, 313, 629–633. [DOI] [PubMed] [Google Scholar]
- 3. Pazour G.J., Witman G.B. (2003) The vertebrate primary cilium is a sensory organelle. Curr. Opin. Cell. Biol., 15, 105–110. [DOI] [PubMed] [Google Scholar]
- 4. Adams N.A., Awadein A., Toma H.S. (2007) The retinal ciliopathies. Ophthalmic Genet., 28, 113–125. [DOI] [PubMed] [Google Scholar]
- 5. Badano J.L., Mitsuma N., Beales P.L., Katsanis N. (2006) The ciliopathies: an emerging class of human genetic disorders. Annu. Rev. Genomics Hum. Genet., 7, 125–148. [DOI] [PubMed] [Google Scholar]
- 6. Khanna H. (2015) Photoreceptor Sensory Cilium: Traversing the Ciliary Gate. Cells, 4, 674–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wang J., Deretic D. (2014) Molecular complexes that direct rhodopsin transport to primary cilia. Prog. Retin. Eye Res., 38, 1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kandel E.R. (2013) Principles of neural science. McGraw-Hill, New York. [Google Scholar]
- 9. Besharse J.C., Bok D. (2011) The retina and its disorders. Academic Press, Amsterdam; Boston. [Google Scholar]
- 10. Rao K.N., Li L., Anand M., Khanna H. (2015) Ablation of retinal ciliopathy protein RPGR results in altered photoreceptor ciliary composition. Sci. Rep., 5, 11137.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhang Q., Acland G.M., Wu W.X., Johnson J.L., Pearce-Kelling S., Tulloch B., Vervoort R., Wright A.F., Aguirre G.D. (2002) Different RPGR exon ORF15 mutations in Canids provide insights into photoreceptor cell degeneration. Hum. Mol. Genet., 11, 993–1003. [DOI] [PubMed] [Google Scholar]
- 12. Hong D.H., Pawlyk B.S., Shang J., Sandberg M.A., Berson E.L., Li T. (2000) A retinitis pigmentosa GTPase regulator (RPGR)-deficient mouse model for X-linked retinitis pigmentosa (RP3). Proc. Natl Acad. Sci. U S A, 97, 3649–3654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Huang W.C., Wright A.F., Roman A.J., Cideciyan A.V., Manson F.D., Gewaily D.Y., Schwartz S.B., Sadigh S., Limberis M.P., Bell P., et al. (2012) RPGR-associated retinal degeneration in human X-linked RP and a murine model. Invest. Ophthalmol. Vis. Sci., 53, 5594–5608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Thompson D.A., Khan N.W., Othman M.I., Chang B., Jia L., Grahek G., Wu Z., Hiriyanna S., Nellissery J., Li T., et al. (2012) Rd9 is a naturally occurring mouse model of a common form of retinitis pigmentosa caused by mutations in RPGR-ORF15. PLoS One, 7, e35865.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ghosh A.K., Murga-Zamalloa C.A., Chan L., Hitchcock P.F., Swaroop A., Khanna H. (2010) Human retinopathy-associated ciliary protein retinitis pigmentosa GTPase regulator mediates cilia-dependent vertebrate development. Hum. Mol. Genet., 19, 90–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bird A.C. (1975) X-linked retinitis pigmentosa. Br. J. Ophthalmol., 59, 177–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Churchill J.D., Bowne S.J., Sullivan L.S., Lewis R.A., Wheaton D.K., Birch D.G., Branham K.E., Heckenlively J.R., Daiger S.P. (2013) Mutations in the X-linked retinitis pigmentosa genes RPGR and RP2 found in 8.5% of families with a provisional diagnosis of autosomal dominant retinitis pigmentosa. Invest. Ophthalmol. Vis. Sci., 54, 1411–1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Fishman G.A. (1978) Retinitis pigmentosa. Genetic percentages. Arch. Ophthalmol., 96, 822–826. [DOI] [PubMed] [Google Scholar]
- 19. Fishman G.A., Farber M.D., Derlacki D.J. (1988) X-linked retinitis pigmentosa. Profile of clinical findings. Arch. Ophthalmol., 106, 369–375. [DOI] [PubMed] [Google Scholar]
- 20. Heckenlively J.R., Yoser S.L., Friedman L.H., Oversier J.J. (1988) Clinical findings and common symptoms in retinitis pigmentosa. Am. J. Ophthalmol., 105, 504–511. [DOI] [PubMed] [Google Scholar]
- 21. Meindl A., Dry K., Herrmann K., Manson F., Ciccodicola A., Edgar A., Carvalho M.R., Achatz H., Hellebrand H., Lennon A., et al. (1996) A gene (RPGR) with homology to the RCC1 guanine nucleotide exchange factor is mutated in X-linked retinitis pigmentosa (RP3). Nat. Genet., 13, 35–42. [DOI] [PubMed] [Google Scholar]
- 22. Roepman R., van Duijnhoven G., Rosenberg T., Pinckers A.J., Bleeker-Wagemakers L.M., Bergen A.A., Post J., Beck A., Reinhardt R., Ropers H.H., et al. (1996) Positional cloning of the gene for X-linked retinitis pigmentosa 3: homology with the guanine-nucleotide-exchange factor RCC1. Hum. Mol. Genet., 5, 1035–1041. [DOI] [PubMed] [Google Scholar]
- 23. Sharon D., Bruns G.A., McGee T.L., Sandberg M.A., Berson E.L., Dryja T.P. (2000) X-linked retinitis pigmentosa: mutation spectrum of the RPGR and RP2 genes and correlation with visual function. Invest. Ophthalmol. Vis. Sci., 41, 2712–2721. [PubMed] [Google Scholar]
- 24. Sharon D., Sandberg M.A., Rabe V.W., Stillberger M., Dryja T.P., Berson E.L. (2003) RP2 and RPGR mutations and clinical correlations in patients with X-linked retinitis pigmentosa. Am. J. Hum. Genet., 73, 1131–1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Vervoort R., Lennon A., Bird A.C., Tulloch B., Axton R., Miano M.G., Meindl A., Meitinger T., Ciccodicola A., Wright A.F. (2000) Mutational hot spot within a new RPGR exon in X-linked retinitis pigmentosa. Nat. Genet., 25, 462–466. [DOI] [PubMed] [Google Scholar]
- 26. He S., Parapuram S.K., Hurd T.W., Behnam B., Margolis B., Swaroop A., Khanna H. (2008) Retinitis pigmentosa GTPase regulator (RPGR) protein isoforms in mammalian retina: Insights into X-linked retinitis pigmentosa and associated ciliopathies. Vision Res., 48, 366–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Murga-Zamalloa C.A., Atkins S.J., Peranen J., Swaroop A., Khanna H. (2010) Interaction of retinitis pigmentosa GTPase regulator (RPGR) with RAB8A GTPase: implications for cilia dysfunction and photoreceptor degeneration. Hum. Mol. Genet., 19, 3591–3598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Khanna H., Davis E.E., Murga-Zamalloa C.A., Estrada-Cuzcano A., Lopez I., den Hollander A.I., Zonneveld M.N., Othman M.I., Waseem N., Chakarova C.F., et al. (2009) A common allele in RPGRIP1L is a modifier of retinal degeneration in ciliopathies. Nat. Genet., 41, 739–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Linari M., Ueffing M., Manson F., Wright A., Meitinger T., Becker J. (1999) The retinitis pigmentosa GTPase regulator, RPGR, interacts with the delta subunit of rod cyclic GMP phosphodiesterase. Proc. Natl Acad. Sci. U S Aa, 96, 1315–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Roepman R., Bernoud-Hubac N., Schick D.E., Maugeri A., Berger W., Ropers H.H., Cremers F.P., Ferreira P.A. (2000) The retinitis pigmentosa GTPase regulator (RPGR) interacts with novel transport-like proteins in the outer segments of rod photoreceptors. Hum. Mol. Genet., 9, 2095–2105. [DOI] [PubMed] [Google Scholar]
- 31. Chang B., Khanna H., Hawes N., Jimeno D., He S., Lillo C., Parapuram S.K., Cheng H., Scott A., Hurd R.E., et al. (2006) In-frame deletion in a novel centrosomal/ciliary protein CEP290/NPHP6 perturbs its interaction with RPGR and results in early-onset retinal degeneration in the rd16 mouse. Hum. Mol. Genet., 15, 1847–1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Khanna H., Hurd T.W., Lillo C., Shu X., Parapuram S.K., He S., Akimoto M., Wright A.F., Margolis B., Williams D.S., et al. (2005) RPGR-ORF15, which is mutated in retinitis pigmentosa, associates with SMC1, SMC3, and microtubule transport proteins. J. Biol. Chem., 280, 33580–33587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Murga-Zamalloa C.A., Desai N.J., Hildebrandt F., Khanna H. (2010) Interaction of ciliary disease protein retinitis pigmentosa GTPase regulator with nephronophthisis-associated proteins in mammalian retinas. Mol. Vis., 16, 1373–1381. [PMC free article] [PubMed] [Google Scholar]
- 34. Anand M., Khanna H. (2012) Ciliary transition zone (TZ) proteins RPGR and CEP290: role in photoreceptor cilia and degenerative diseases. Expert Opin. Ther. Targets, 16, 541–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Da Costa R., Glaus E., Tiwari A., Kloeckener-Gruissem B., Berger W., Neidhardt J. (2015) Localizing the RPGR protein along the cilium: a new method to determine efficacies to treat RPGR mutations. Gene Ther., 22, 413–420. [DOI] [PubMed] [Google Scholar]
- 36. Zhao Y., Hong D.H., Pawlyk B., Yue G., Adamian M., Grynberg M., Godzik A., Li T. (2003) The retinitis pigmentosa GTPase regulator (RPGR)- interacting protein: subserving RPGR function and participating in disk morphogenesis. Proc. Natl Acad. Sci. U S Aa, 100, 3965–3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Yan D., Swain P.K., Breuer D., Tucker R.M., Wu W., Fujita R., Rehemtulla A., Burke D., Swaroop A. (1998) Biochemical characterization and subcellular localization of the mouse retinitis pigmentosa GTPase regulator (mRpgr). J. Biol. Chem., 273, 19656–19663. [DOI] [PubMed] [Google Scholar]
- 38. Lee J.J., Seo S. (2015) PDE6D binds to the C-terminus of RPGR in a prenylation-dependent manner. EMBO Rep., 16, 1581–1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Glomset J.A., Farnsworth C.C. (1994) Role of protein modification reactions in programming interactions between ras-related GTPases and cell membranes. Annu. Rev. Cell Biol., 10, 181–205. [DOI] [PubMed] [Google Scholar]
- 40. Reiss Y., Goldstein J.L., Seabra M.C., Casey P.J., Brown M.S. (1990) Inhibition of purified p21ras farnesyl:protein transferase by Cys-AAX tetrapeptides. Cell, 62, 81–88. [DOI] [PubMed] [Google Scholar]
- 41. Zhang F.L., Casey P.J. (1996) Protein prenylation: molecular mechanisms and functional consequences. Annu. Rev. Biochem., 65, 241–269. [DOI] [PubMed] [Google Scholar]
- 42. Zhang F.L., Kirschmeier P., Carr D., James L., Bond R.W., Wang L., Patton R., Windsor W.T., Syto R., Zhang R., et al. (1997) Characterization of Ha-ras, N-ras, Ki-Ras4A, and Ki-Ras4B as in vitro substrates for farnesyl protein transferase and geranylgeranyl protein transferase type I. J. Biol. Chem., 272, 10232–10239. [DOI] [PubMed] [Google Scholar]
- 43. Baehr W. (2014) Membrane protein transport in photoreceptors: the function of PDEdelta: the Proctor lecture. Invest Ophthalmol. Vis. Sci., 55, 8653–8666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Humbert M.C., Weihbrecht K., Searby C.C., Li Y., Pope R.M., Sheffield V.C., Seo S. (2012) ARL13B, PDE6D, and CEP164 form a functional network for INPP5E ciliary targeting. Proc. Natl Acad. Sci. U S Aa, 109, 19691–19696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Zhang H., Li S., Doan T., Rieke F., Detwiler P.B., Frederick J.M., Baehr W. (2007) Deletion of PrBP/delta impedes transport of GRK1 and PDE6 catalytic subunits to photoreceptor outer segments. Proc. Natl Acad. Sci. U S Aa, 104, 8857–8862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Thomas S., Wright K.J., Le Corre S., Micalizzi A., Romani M., Abhyankar A., Saada J., Perrault I., Amiel J., Litzler J., et al. (2014) A homozygous PDE6D mutation in Joubert syndrome impairs targeting of farnesylated INPP5E protein to the primary cilium. Hum. Mutat., 35, 137–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Bielas S.L., Silhavy J.L., Brancati F., Kisseleva M.V., Al-Gazali L., Sztriha L., Bayoumi R.A., Zaki M.S., Abdel-Aleem A., Rosti R.O., et al. (2009) Mutations in INPP5E, encoding inositol polyphosphate-5-phosphatase E, link phosphatidyl inositol signaling to the ciliopathies. Nat. Genet., 41, 1032–1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Watzlich D., Vetter I., Gotthardt K., Miertzschke M., Chen Y.X., Wittinghofer A., Ismail S. (2013) The interplay between RPGR, PDEdelta and Arl2/3 regulate the ciliary targeting of farnesylated cargo. EMBO Rep., 14, 465–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Fansa E.K., O'Reilly N.J., Ismail S., Wittinghofer A. (2015) The N- and C-terminal ends of RPGR can bind to PDE6δ. EMBO Rep, 16, 1583–1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Chapple J.P., Hardcastle A.J., Grayson C., Willison K.R., Cheetham M.E. (2002) Delineation of the plasma membrane targeting domain of the X-linked retinitis pigmentosa protein RP2. Invest. Ophthalmol. Vis. Sci., 43, 2015–2020. [PubMed] [Google Scholar]
- 51. Yoon J.H., Qiu J., Cai S., Chen Y., Cheetham M.E., Shen B., Pfeifer G.P. (2006) The retinitis pigmentosa-mutated RP2 protein exhibits exonuclease activity and translocates to the nucleus in response to DNA damage. Exp. Cell Res., 312, 1323–1334. [DOI] [PubMed] [Google Scholar]
- 52. Hanke-Gogokhia C., Wu Z., Gerstner C.D., Frederick J.M., Zhang H., Baehr W. (2016) Arf-like Protein 3 (ARL3) Regulates Protein Trafficking and Ciliogenesis in Mouse Photoreceptors. J. Biol. Chem., 291, 7142–7155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Young R.W. (1967) The renewal of photoreceptor cell outer segments. J. Cell Biol., 33, 61–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Young R.W. (1968) Passage of newly formed protein through the connecting cilium of retina rods in the frog. J. Ultrastruct. Res., 23, 462–473. [DOI] [PubMed] [Google Scholar]
- 55. Jacoby M., Cox J.J., Gayral S., Hampshire D.J., Ayub M., Blockmans M., Pernot E., Kisseleva M.V., Compere P., Schiffmann S.N., et al. (2009) INPP5E mutations cause primary cilium signaling defects, ciliary instability and ciliopathies in human and mouse. Nat. Genet., 41, 1027–1031. [DOI] [PubMed] [Google Scholar]
- 56. Rao K.N., Li L., Zhang W., Brush R.S., Rajala R.V.S., Khanna H. (2016) Loss of human disease protein retinitis pigmentosa GTPase regulator (RPGR) differentially affects rod or cone-enriched retina. Hum Mol Genet, 25, 1345–1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Choi H., Larsen B., Lin Z.Y., Breitkreutz A., Mellacheruvu D., Fermin D., Qin Z.S., Tyers M., Gingras A.C., Nesvizhskii A.I. (2011) SAINT: probabilistic scoring of affinity purification-mass spectrometry data. Nat. Methods, 8, 70–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Li L., Anand M., Rao K.N., Khanna H. (2015) Cilia in photoreceptors. Methods Cell Biol., 127, 75–92. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.