Abstract
A defined set of genetic instructions encodes functionality in complex organisms. Delineating these unique genetic signatures is essential to understanding the formation and functionality of specialized tissues. Vision, one of the five central senses of perception, is initiated by the retina and has evolved over time to produce rod and cone photoreceptors that vary in a species-specific manner, and in some cases by geographical region resulting in higher order visual acuity in humans. RNA-sequencing and use of existing and de novo transcriptome assemblies allowed ocular transcriptome mapping from a diverse set of rodent and primate species. Global genomic refinements along with systems-based comparative and co-expression analyses of these transcriptome maps identified gene modules that correlated with specific features of rod versus cone retinal cellular composition. Organization of the ocular transcriptome demonstrated herein defines the molecular basis of photoreceptor architecture and functionality, providing a new paradigm for neurogenetic analyses of the mammalian retina in health and disease.
Introduction
Comparative analysis of genomes can reveal specific pathways that underlie functional (1) and phenotypic variation (2). Transcriptome studies across distinct species have shown a large degree of conservation, even when comparing primates to amniotes (3). Thus, model organisms such as mice (4) and rats (5) have been used to study complex diseases and identify disease-specific genomic loci. Comprehensive genotype to phenotype maps gleaned from such organisms have been essential in identifying evolutionarily conserved pathways affecting human disease (6). A confounding feature of interspecies genetic analysis, however, is that gene expression differences between species can be selectively neutral, and thus have little functional significance (7). Therefore, systems biological approaches must be used in concert to reveal functionally relevant patterns of transcriptional organization and provide novel insights into human genomic functionality. Such approaches have shed light on critical genetic features of neuronal cell types in the human brain (8) and those pathways affected by disease (9).
Neuronal tissue is well suited for this genomic approach as it evolves significantly slower due to a more fine-tuned expression profile compared to other organs (10,11). Conserved genes can highlight the preservation of core functionality whereas divergent genes and gene families can underlie species-specific functions in those tissues (12). Gene expression changes can also reflect alterations in cellular composition and connectivity, which are required for precise functioning in neuronal tissues. An excellent example is the neural retina, composed of diverse cell types that support different ratios of rod and cone photoreceptors that vary in populations with species as well as geographic in primates (13). Recent application of RNA-sequencing (RNA-Seq) technology (14) has begun to uncover complex genetic features of the retina (4,15–17), supporting tissues (18), and specific retinal cell populations (19,20). With dysfunction of the photoreceptor and adjacent supporting cell layers in the retina as the driving force for most retinal degenerative diseases (21) more complete transcriptome analyses of the retina, in both health and disease states is a high priority. A new approach for understanding visual function at the genetic level is essential, because genetic interrogation of human tissue is limited. In addition, the primary rodent models studied, Rattus norvegicus (rat) and Mus musculus (mouse) display a rod-rich retina. Although higher order mammals like humans only have approximately 5% cone photoreceptors in the retina, a central region termed the macula, most specifically the fovea, is rich in cone photoreceptors (22) and is responsible for high-resolution vision. Thus, restricting genetic interrogation to these two rodent models cannot reveal the more complex retinal disease aetiologies afflicting the central cone-rich region of primates (23). Less studied rodents, Arvicanthis niloticus (Nile rat) and Ictidomys tridecemlineatus (ground squirrel), are diurnal cone-dominant rodent species that reflect features of human vision with respect to electroretinogram readings (24), neuronal connections (25), and genomic karyotype (26), but are limited due to lack of comprehensive genomic annotation. Taken together, these four rodent species feature a spectrum of rod versus cone photoreceptor ratios and behavioural patterning that can be used to delineate genetic features that shape the complex human retina.
In this work, RNA-Seq of ocular tissue from Rattus norvegicus, Mus musculus, Arvicanthis niloticus, Ictidomys tridecemlineatus rodent species allowed transcriptome analyses across diverse physiologic retinal architectures that support varying photoreceptor populations. Comprehensive analysis of gene co-expression networks across rodent species revealed enrichment of gene signatures in specific retinal neuroanatomical structures. Ocular tissue from higher order mammals, Macaca fascicularis and Homo sapiens, were also incorporated into the study to elucidate regional specific transcriptome features that support higher order visual function. This approach separated conserved genetic elements essential for ocular function from those divergent features that have shaped higher order primate vision. This work elucidates novel genetic features of the retina and specific photoreceptor subtypes needed to identify pathologic changes affecting visual function and guide future therapies for such retinopathies.
Results
Physiologic consequences of photoreceptor gradation
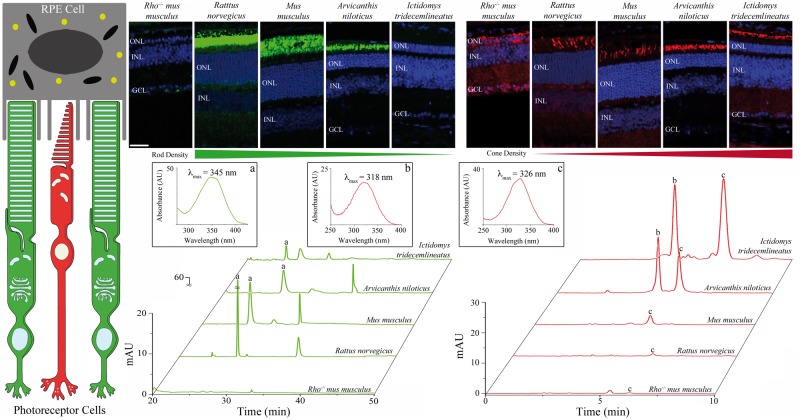
The vertebrate eye evolved over the course of 600 million years to produce the retinal architecture of modern day mammals (27). Visual function is mediated by a complex interplay between the photoreceptor cells and the neighbouring retinal pigmented epithelium (RPE) to drive phototransduction (28) and the retinoid cycle (29) (Fig. 1) . Reliance on the universal visual chromophore, 11-cis-retinal, by both rod and cone photoreceptors indicates that distinct metabolites in the retina must exist to support the difference in the photoreceptor response kinetics (30). Nocturnal species, such as Rattus norvegicus and Mus musculus, rely on scoptic vision and thus exhibit a retina with an abundance of rod photoreceptors, whereas diurnal species, such as Arvicantis niloticus and Ictidomys tridecemlineatus, depend more on photopic vision and thus feature increasing cone dominance in the retina (Fig. 1). Non-polar retinoids extracted and analyzed from eyes revealed that enrichment of particular retinal metabolites was related to the photoreceptor gradation across rodent species (Fig. 1). In nocturnal species, there was enrichment of retinyl-aldehydes, reflecting the high concentration of visual pigment in rod cells, whereas in diurnal species, there was an increase in retinyl-esters, proposed to be preferentially used in vertebrate cone vision (31). These differing retinal metabolic patterns indicate that distinct genetic features exist to mediate vision in unique rod versus cone-dominant retinas. To an extent, mouse models can be used to delineate the genetic drivers of distinct photoreceptor populations (Supplementary Material, Fig. S1). However, such models fail to mimic the retinal architecture and associated pathology seen in humans (23). Understanding the physiologic consequences of photoreceptor gradation relates to the retinal architecture in primates, wherein the peripheral retinal architecture is rod dominant and the central, macular region is cone dominant (22). Therefore, we used RNA-Seq to analyze the genetic signatures unique to varying photoreceptor populations in the physiologic retinal environment across these rodent species.
Figure 1.
Distinct retinal photoreceptor populations account for physiologic differences driving visual function. Phototransduction and regeneration of the visual chromophore, 11-cis-retinal, are mediated by photoreceptor cells and the neighboring retinal-pigmented epithelium (RPE). Rod dominant Rattus norvegicus and Mus musculus retinas (rods stained green against rhodopsin) have distinct structural features compared to cone dominant Arvicanthis niloticus and Ictidomys tridecemlineatus retinas (cones stained red against cone opsin). Spectroscopic characterization of extracted retinoids revealed that rod dominant species are enriched in (A) retinyl-aldehydes whereas cone dominant species display enrichment of (B) 11-cis- and (C) all-trans-retinyl esters. The Rho-/-mus musculus represents a negative control with a retina devoid of photoreceptors as noted by a lack of immunohistochemical staining for rods or cones and no discernable retinal metabolites upon HPLC analysis. Labelled retinal layers are: GCL, ganglion cell layer; INL, inner nuclear layer; ONL, outer nuclear layer. Scale bar is 50 µm.
RNA-seq and genomic refinement for global ocular transcriptome mapping
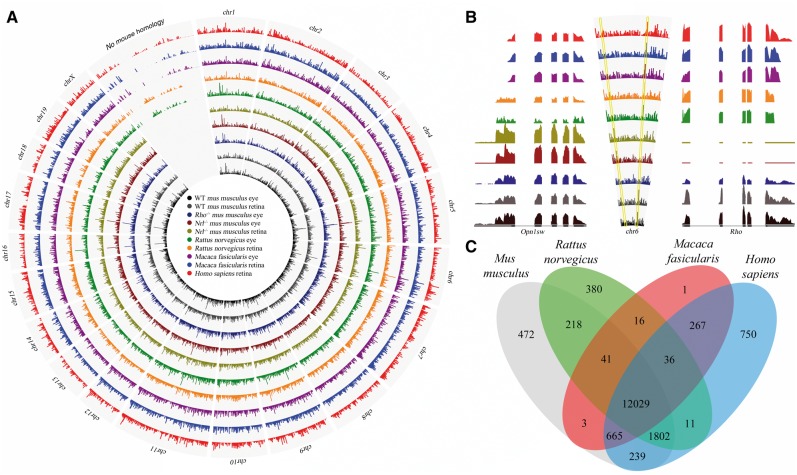
To understand the genetic signatures unique to distinct rod versus cone-dominant photoreceptor populations, we conducted RNA-Seq of rodent species (Rattus norvegicus, Mus musculus, Arvicanthis niloticus, Ictidomys tridecemlineatus) as well as higher order primates (Macaca fascicularis and Homo sapiens). Prepared libraries of poly-adenylated RNA from whole eye and retinal tissue were sequenced with the Illumina platform, yielding ∼1.96 billion reads (Supplementary Materials, Tables S1 and S2). Initially reads from the typical rodent models Rattus norvegicus and Mus musculus, as well as the primate tissue from Macaca fasicularis and Homo sapiens were mapped to their respective genome assemblies from the UCSC genome database (32). These were further refined manually with RefSeq gene models (RNor, GRCm38, Mmul, GRCh37) (33), most notably for Macaca fasicularis. Over 50% of such gene models were missing in the rhesus assembly (34), even essential visual genes such as rod opsin (Rho). Missing homologous genes were identified in each assembly and reads were mapped to nucleotide sequences from the RefSeq gene models. The chromosomal architecture of Mus musculus was then used as a scaffold to visualize coverage of the 16,930 genes detected (≥1 RPKM (35)) across rodent and primate species (Fig. 2A). This global approach provided sensitive assessments of expression levels, highlighting the gradation across species of key genes such as short-wavelength cone opsin, Opn1sw, and rod opsin, Rho (Fig. 2B). The landscape of gene expression across rodent and primate eyes revealed that 71% of reads mapped to 12,029 homologous genes (Fig. 2C), emphasizing that conservation of core visual function across species restricted dramatic transcriptome divergence. Divergent genetic signatures were further investigated to understand possible patterning aspects that have permitted more fine-tuned visual function in primates.
Figure 2.
RNA-Seq transcriptome analyses of ocular tissue from rodent and primate species reveal global genomic profiles. (A) The chromosomal architecture of Mus musculus served as the platform to map genomic coverage of the mapped reads from ocular tissue across various mammalian species investigated by RNA-Seq. (B) Two essential genes related to cone versus rod mediated-vision, Opn1sw and Rho, respectively, reside on chr 6 in Mus musculus and vary in expression level related to rod and cone photoreceptor content across species. (C) Illustrated are homologous genes across species, 12,029 of which are conserved across all four species investigated, as well as those genes that are unique to a particular species.
Divergent expression profiles implicate adaptive genetic signatures in the retina
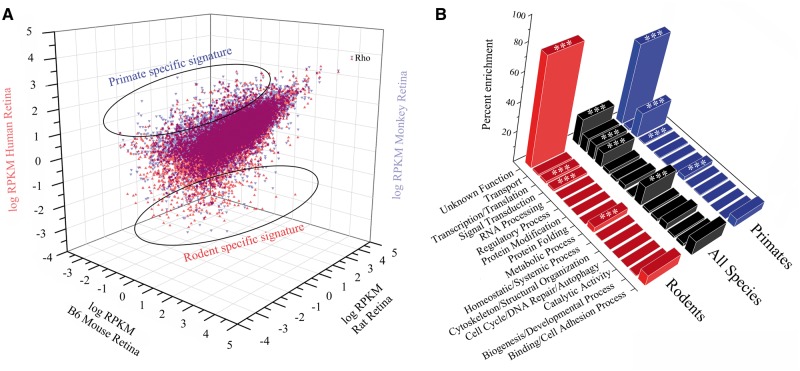
A quantitative map of gene expression across the typical model rodent and primate retinas is illustrated in Fig. 3A, highlighting the genetic signatures unique to a particular species. Breakdown of the categorical enrichment profile of the 2,088 divergent genes between rodents and primates demonstrated species-specific genetic trends (Fig. 3B). Interestingly, both rodents and primates had a large percentage of unique transcripts with no annotated function. The most markedly distinct enrichment profile was seen in transcription/translation genes, with primates featuring enrichment relative to rodents, even when compared to all orthologous transcripts. Further analysis of this transcriptional enrichment pattern provided examples of how the differential expression of genes can underlie unique retinal features driving adaptive aspects of mammalian vision. The Rax family of homeobox genes are essential for vertebrate retinal development (36), wherein our analysis revealed that Rax2, a gene involved in the neural retina development driven by BMP signalling (37), had expression restricted to primates. Most categories of genes, however, failed to show any marked differences in enrichment between rodents and primates. Therefore, to identify those genetic elements unique to a particular photoreceptor composition, more homogeneous photoreceptor populations would be required to distinguish factors unique to rod and cone photoreceptor ratios that shape the unique architecture of the primate retina.
Figure 3.
Genetic signatures and topography of functional enrichment in the rodent and primate retina. (A) Plots of transcript abundance for mapped reads from retinal tissue in rat, mouse, monkey and human revealed rodent and primate specific genetic signatures in the retina. (B) Comparison of topographic maps of these unique signatures evidenced enrichment of gene categories in rodents versus primates compared to homologous genes across all species (denoted by stars).
De novo gene assembly of non-model rodent species
We reasoned that the non-model rodent species Arvicanthis niloticus and Ictidomys tridecemlineatus would provide unique information regarding physiologic retinal environments abundant in cone photoreceptors. De novo assembly was carried out and used as a scaffold for quantitative RNA-seq transcriptome analysis (38). The assembled Arvicanthis niloticus eye transcriptome consisted of 124,439 transcripts totaling 100,656,031 bases, with an N50 value of 1,595 bp for all transcripts. Assembled Ictidomys tridecemlineatus transcriptomes consisted of 165,655 transcripts totaling 103,326,677 bp for the eye and 214,069 transcripts totaling 135,830,374 bp for the retina, with N50 values of 2,205 bp and 1,977 bp, for the eye and retina, respectively (Supplementary Material, Table S2). When mapped back onto the genomic scaffold shown in Fig. 2A, we identified expression of 14,799 genes out of the 16,930 genes detected across species with existing genomic assemblies, representing 87% coverage of the de novo assembled transcripts. These novel ocular transcriptome assemblies allowed us to expand from single gene comparisons to a more systematic approach to identify biologically relevant patterns unique to rod- and cone-dominant retinas.
Gene co-expression network analysis of the rodent eye delineates modules of functionally related genes
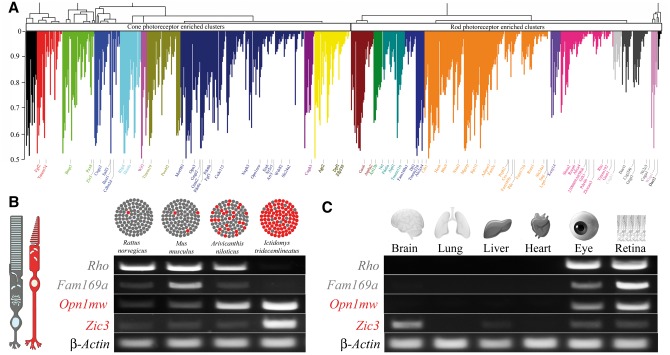
Functional organization of the ocular transcriptome was investigated by applying weighted gene co-expression network analysis to four physiologically diverse rodent species. As shown in Fig. 1, Rattus norvegicus, Mus musculus, Arvicanthis niloticus, and Ictidomys tridecemlineatus exhibited distinct structural patterning of the retina from rod-dominant to cone-dominant, observed on the genomic level as well as in terms of photopigment expression (Supplementary Materials, Fig. S2, Table S3). The co-expression network was constructed based on the Pearson correlation across the rodent species weighted against the pigment composition in the rodent species. By selecting only those that evidenced at least a moderate to strong correlation, we subjected a total of 874 genes to modular analysis. Modules of co-expressed genes were identified by average linkage clustering on the basis of their topological overlap. This identified 21 distinct modules that represent systems-level molecular correlations to cone and rod photoreceptor environmental specificity (Fig 4A). These modules were then interrogated to reveal functional relationships among genes. Analysis of the genes in Fig. 4A revealed that interspersed between known rod and cone photoreceptor genes, such as structural rod disc genes (Supplementary Materials, Table S4) and phototransduction genes (Supplementary Materials, Table S5), are genes with either completely unknown function or unknown relevance to retinal functionality.
Figure 4.
Network analysis of gene expression in the eye across rodent species identifies distinct modules of co-expressed genes relating to photoreceptor function. (A) Genes exhibiting differential expression across Rattus norvegicus, Mus musculus, Arvicanthis niloticus, and Ictidomys tridecemlineatus were used to construct a gene co-expression network based on Pearson correlations weighted against the pigment composition in rodent species. Hierarchical clustering based on topographical overlap identified distinct modules of co-expressed genes, depicted in different colours. Key genes are highlighted and colour-coded according to the module they reside in. The x-axis corresponds to the genes and the y-axis to the co-expression correlation. (B) Module membership identified those genes found in cone- versus rod-enriched clusters and revealed novel genes that could pertain to photoreceptor function. Fam169a exhibited enrichment in rod dominant species similar to the rod pigment Rho, whereas Zic3 exhibited enrichment in cone dominant species similar to the cone pigment Opn1mw. (C) Expression probing of several essential organs revealed the restricted expression of these two genes. Fam169a was enriched in the eye and retina whereas Zic3 was enriched in neuronal tissues such as brain, eye and retina.
Module residence permitted gene function annotation, as genes in modules have a strong likelihood of participating in specific functions represented by their particular modules (Supplementary Material, Fig. S3). For example, Fam169a is a gene of unknown function that resides in a cluster enriched for rod photoreceptor structure and function, and exhibits expression that parallels that of rhodopsin (Rho) across species as well as organ compartments in Mus musculus. Similarly, Zic3, a transcription factor thought to be involved in development, resides in a cluster enriched for cone photoreceptor structure and function, exhibiting expression that parallels that of middle-wavelength cone opsin (Opn1mw), is preferentially expressed in neuronal tissues such as brain and retina (Fig. 4B and C). Modules were also examined for enrichment for known retinal disease-causing genes (RetNet, Supplementary Material, Fig. S3). Notably, Fam169a resides in a module enriched for genes contributing to rod photoreceptor degenerative conditions such as retinitis pigmentosa, Leber congenital amaurosis and Bardet-Biedl syndrome. Therefore, by comparing gene expression profiles across rodent species in discrete modules, we identified distinct networks that influence rod versus cone photoreceptor populations. This analysis permitted identification of novel elements and proposed functional roles based on module membership. The final part of this study was to investigate the genetic signatures of geographically distinct rod and cone photoreceptor patterning that exist in primate retina.
Geographic expression profiles reveal genetic factors driving higher order vision in the primate retina
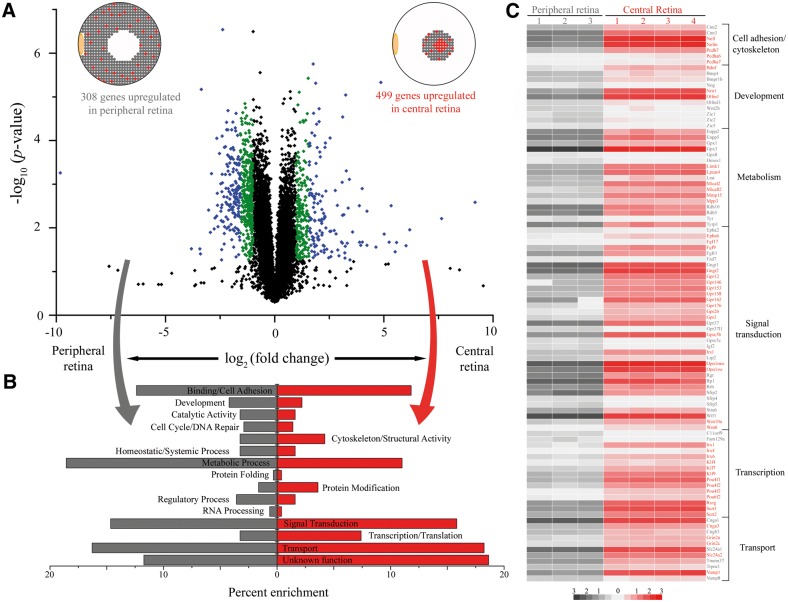
In addition to whole eye and retinal tissue from Macaca fascicularis, three biological replicates of peripheral retina and four biological replicates of central retina tissue were isolated and sequenced for gene expression analysis. Of the 11,060 genes identified in Macaca fascicularis that were expressed at a level of ≥1 RPKM, 807 were differentially expressed by at least 2 fold (P value ≤0.05) between peripheral and central retina samples (Fig. 5A). Similar to the rodent co-expression analysis, known rod and cone photoreceptor genes were found enriched in the peripheral and central retina (Supplementary Materials, Tables S3–S5), respectively, with unannotated genes interspersed as well. Differentially expressed genes were then screened for the enrichment of functional categories to reveal distinct sets of genes that are phenotypically relevant (Fig. 5B). In the peripheral retina, where metabolically active rod photoreceptors are abundant, there was a predictable enrichment of genes involved in cellular metabolism. There was also an enrichment of binding/cell adhesion genes in the peripheral retina, as rods have a strong structural role in maintaining retinal integrity. In the central retina, where there is a specialized cone-rich architecture for high-resolution vision, there was an enrichment of genes related to signal transduction, transcription and transport functional categories. Interrogation of these functional categories revealed overrepresentation of key gene families such as cytoskeletal protocadherin proteins, developmental BMP/WNT signalling proteins, glutathione peroxidase metabolic proteins, orphan G protein-coupled receptors involved in signal transduction, and homeobox and Kruppel-like transcription factors. The differential expression of these gene families highlighted the functional relevance of particular pathways in the maintenance of the specialized primate retina.
Figure 5.
RNA-Seq analysis of central versus peripheral regions of the Macaca fasicularis retina. (A) Volcano plot comparing gene expression in peripheral (left) versus central (right) retinal tissue samples from Macaca fasicularis. Gene expression (log2 scale) across biological replicates plotted against the statistical significance of the difference in expression (P value in a log10 scale) reveal that most transcripts did not show significant changes in expression (black points), whereas those that evidenced a 2-3 fold change in expression are represented by green points, and those with >3 fold change in expression are displayed as blue points. (B) Functional categorization revealed enrichment between the periphery (gray) and central (red) retina. Functional categorization labels on each side represent the region that exhibits enrichment in that category. (C) Heat map of genes differentially expressed from specific functional categories reveals that certain gene families are enriched in particular geographic compartments of the primate retina. The colour scale shows their relative expression profiles (log RPKM).
Discussion
Visual function has been tailored to fit the evolving needs of various species. Rodents such as Rattus norvegicus and Mus musculus have developed rod-dominant retinas for improved scotopic vision for their nocturnal lifestyle. In contrast, diurnal patterning has led Arvicanthis niloticus and Ictidomys tridecemlineatus rodent species to develop cone-dominant retinas that permit improved photopic vision. Higher order primates have further developed a central cone-rich macula that enables high-resolution vision. At the genomic level, Mus musculus and Homo sapiens differ in ∼2.5% of the bases (39) whereas Macaca fascicularis and Homo sapiens differ in only ∼1.2% of bases aligned between species (40). RNA expression level differences seemingly drive the observed phenotypic diversity (41). Thus analytic approaches must expand from single gene comparisons to systems level approaches to reveal the functional relevance of novel genes as drivers of this phenotypic patterning (42). Our work provides a comprehensive analysis of visual adaptation in rod and cone-enriched architectures across species and even specific geographical portions of the primate retina. More importantly, comparative network analyses identified gene modules that correlate with specific neuroanatomical retinal structures. Furthermore, novel genes pertaining to specific photoreceptor cell types in the retina and their relationships to human visual function and associated pathology were elucidated.
Starting from a phenotypic characterization of rodent species with varying rod versus cone photoreceptor ratios and behavioural patterning (Fig. 1), it became evident that key genetic drivers must exist to account for phenotypic diversity in such closely related species. Native and genetically modified mouse models have been employed to approximate various human retinal disease conditions (Supplementary Material, Fig. S1), but this approach often fails for two primary reasons. First, mouse models, or any single rodent model cannot fully approximate the human retina because of the fundamental difference in overall retinal architecture. Second, mouse models often do not recapitulate human pathology and genetic changes lead to unstable retinal patterning. This is exemplified by the primary model system used to study cones, the Nrl-/- mouse, which has an abundance of transmitted cones in an unstable state (17), limiting its relevance. This problem prompted our examination of diverse photoreceptor (rod and cone) architectures to delineate physiologically relevant signatures. The power of this current study lies in the examination of the ocular transcriptome in aggregate across species. Furthermore, in studying whole ocular tissue rather than isolated cell populations we could reduce confounding variables that result from loss in transcript integrity from disruptions of cellular connectivity that occur in preparation of single cell sequencing techniques (43).
Initial analysis of ocular transcriptome reads from species with existing genomes revealed global gene expression profiles, highlighting those essential conserved genes and those with unique signatures that have diverged (Fig. 2). Most importantly, it reinforced the need to use non-model rodent species to better understand physiological consequences of cone enrichment. Examination of genetic signatures between species (Fig. 3) revealed gene families such as GPCR kinases, represented by Grk1 and Grk7, with the former expressed across all species, whereas the latter was restricted to primates and cone dominant rodents, such as Ictidomys tridecemlineatus (44). Restricting genetic analysis to traditional Rattus norvegicus and Mus musculus species would not allow one to understand the specialized cone rich macula of the human retina, prompting the inclusion of non-model cone-dominant rodent species in the genetic analysis. In doing so, this work led to the sequencing and first de novo assemblies of the ocular transcriptomes from Arvicanthis niloticus and Ictidomys tridecemlineatus species. Incorporating these novel genetic assemblies with gene co-expression analyses was essential to reveal their functional significance in the unique cellular composition of the primate retina.
In large datasets as involved in this work, examining distinct modules of gene expression changes across species (45) that are enriched in certain functional categories can highlight phenotypically relevant networks. Co-expression analyses identified genes that corresponded to retinal environments enriched for specific photoreceptor populations (Fig. 4), providing a new methodology for annotating gene function in the retina. Moreover, this enabled the generation of hypotheses related to gene function by examining the module membership. Two such examples are highlighted here. One relates to the cone photoreceptor enrichment of Zic3, a zinc finger transcription factor involved in a variety of developmental processes thought to function in retinal progenitor cell differentiation and proliferation. Zic3 was more highly expressed in cone-dominant rodents and specifically in neuronal tissues across organs (Fig. 4), consistent with previous work (46). The second was Fam169a, a gene of unknown function that was enriched in rod-dominant rodents, but across organs, its expression was restricted to the eye, specifically the retina (Fig. 4). The power of modular analysis is that it can identify genes with a strong likelihood of participating in specific functions represented by each module. In the case of Fam169a, its module is enriched in cell adhesion, metabolism, and transcription related genes as well as in known rod photoreceptor retinopathy genes. This approach can similarly be expanded to other unknown genes to elucidate how they relate to health and ultimately influence disease in the retina. Localization techniques can further elucidate the specific neuroanatomical role of these novel genes.
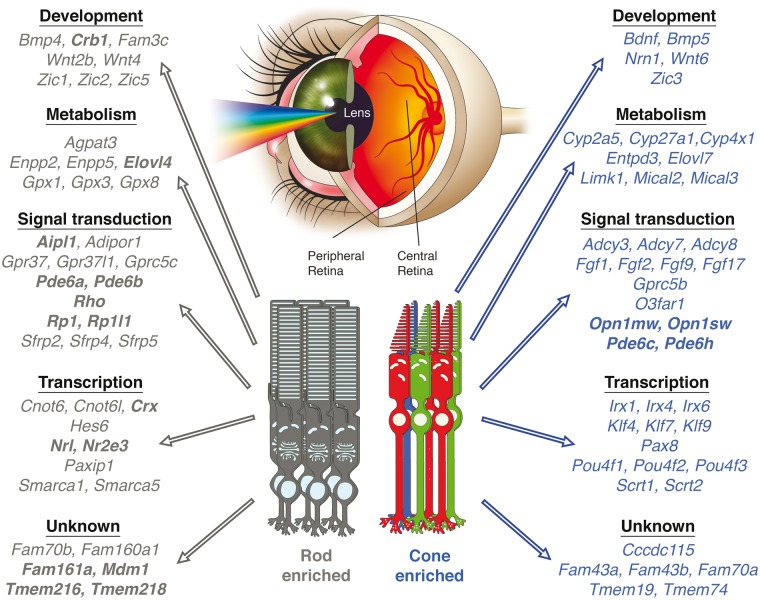
These results were leveraged against data obtained from Macaca fascicularis in which geographical transcriptome dynamics were examined by comparing reads from central versus peripheral retina samples representing cone and rod enriched areas, respectively (Fig. 5). Comparison of gene expression revealed a predominant up-regulation of gene expression in the central region compared to the periphery. This speaks to the increased complexity of the central macular region as it has been shown that increased expression levels can correspond to higher order modelling when proceeding from non-human primate brains to human brains (47). Key gene families related to metabolism, signal transduction and transcription were enriched in a geographic manner in the primate retina consistent with their function in maintaining the specialized architecture in the human retina (Fig. 6). The neural retina consists of post-mitotic photoreceptor cells that rely on the neighbouring RPE cells for a circadian coordinated mechanism of maintenance (48). These highly metabolic cells are subjected to considerable oxidative stress generated by exposure to light and high oxygen tension. Our work reinforces the idea that key metabolic gene products are enriched in particular retinal environments to mediate this homeostatic process. Glutathione-mediated oxidative stress protection by the GPX family of proteins (Gpx1, Gpx3, and Gpx8) seems essential to the peripheral retina, displaying rod-photoreceptor enrichment, and has been implicated to have disrupted expression in age-related retinal phenotypes (4). The differential expression and localization of different metabolic gene products highlights the complex retinal architecture required for normal function and how mutations in gene products can lead to specific retinal phenotypes such as Stargardt disease. Genes linked to this disease, Abca4 and Elovl4, were both found in this study to be enriched in photoreceptor clusters, especially the rod photoreceptor layer and the peripheral retina in Macaca fascicularis. Increased understanding this disease at a molecular level has allowed systems pharmacological approaches to devise new treatment paradigms (49).
Figure 6.
Global gene signatures found unique to rod versus cone photoreceptor enriched retinas. Unified analysis across rodent species and regional differences in the primate retina revealed clustering of specific gene families that highlight the genetics underlying changes in the composition of photoreceptors in the retina. Genes in bolded letters are known retinal disease-causing genes from RetNet.
Another avenue of discovery of this work has been elucidating the complex factors that dictate the maintenance of post-mitotic photoreceptor cells. Whereas progress has been made in understanding the transcriptional network involved in rod photoreceptor specification and maintenance, much less is known about how cone photoreceptors are committed and maintained. Understanding retina cell specification, especially those factors that drive cone cell development and survival, is crucial in developing future regenerative therapies for the retina (50). Recent work has shown that a multifunctional BMP, TGF-β and WNT network of genes can drive cone photoreceptor development from human embryonic stem cells (51). Supporting this point, the central primate retina has a reduced expression of Bmp4 compared to the peripheral retina with an increase of inhibitory factors in this multifunctional network such as Wif1 in cone clusters and enhancement of activating factors like Wnt4 and Lrp6 (52) in rod clusters. Even in signal transduction gene products, we see that orphan GPCRs are integrated into this hierarchical network, as Gprc5b mediates neurogenesis via a beta-catenin pathway (53) and is enriched in cone populations. There is an observed cone enrichment pattern of the BMP interacting FGF network (54) (Fgf1, Fgf2, Fgf9, Fgf17) and even a novel downstream gene of Fgf2 such as Ccdc115 (55). Moreover, in the central retina, we noted enrichment of Kruppel-like factors, such as Klf4 that have been implicated in stem cell renewal (56) and are directly influenced by the BMP network (57). Such findings suggest that the central cone-rich macula of primates can have stem cell functionality that allows their life-long survival and maintenance. Taken together these analyses highlight a set of key transcriptional frameworks that are instrumental in the function and maintenance of the adult retina. With technologies emerging that can generate three-dimensional retinal tissue platforms (58), this could shape the future of stem cell based therapeutics as these tissue models are used to restore vision (59).
In summary, this work represents a comprehensive analysis of ocular transcriptome mapping to unveil adaptive elements underlying rod versus cone photoreceptor specialization in mammals. Moreover, the use of gene co-expression relationships in rod and cone photoreceptor-dominant retinas revealed known genes along with novel genetic elements that are crucial for future study, especially in determining changes caused by retinal diseases (Supplementary Material, Fig. S4). An example is the identification of Tmem218, which by our analysis was enriched in a rod photoreceptor cluster and recently identified as a candidate gene for Senior-Løken syndrome (60). Interestingly, we found enrichment of known disease causing retinal genes residing in rod clusters in the co-expression analysis and the primate peripheral retina that likely stem from the reliance on ocular genomes of rod dominant species. The elucidation of transcriptome maps from ocular tissue of cone-dominant mammals in this work could facilitate the identification of novel disease genes related to cone retinopathies. Such associations can enable identification of clinically relevant pathways suitable for therapeutic intervention that have previously been unavailable. With the CRISPR-CAS system used to study the effects of human disease through genetic changes in mice (61), enabling the study of multiple gene mutations (62) and extending to gene editing in primates (63), improved understanding of human disease should result as candidate genes are increasingly identified. This work offers a novel outlook on the structural and functional organization of the mammalian retina, providing a paradigm shift as to how to decipher the genotype to phenotype relationship to better predict outcomes and design therapeutic interventions through a systems-based approach (49).
Materials and Methods
Species information
C57BL/6J mice (Mus musculus) were purchased from the Jackson Laboratory (Bar Harbor, ME, USA). Nrl-deficient mice (64) with a B6 background were kindly provided by Dr. Anand Swaroop (National Eye Institute, Bethesda, MD, USA). Rhodopsin knockout mice (65) with a B6 background were obtained from Dr. Janis Lem (Tufts University, Boston, MA, USA). All mice used were 4 weeks of age. Four-week-old Long-Evans rats (Rattus norvegicus) were purchased from Harlan Laboratories (Madison, WI, USA). Nile rats (Arvicanthis niloticus) at 6 weeks of age, were kindly provided by Dr. Laura Smale (Michigan State University, East Lansing, MI, USA). Wild caught ground squirrels (Ictidomys tridecemlineatus) were purchased from TLS Research (Bloomingdale, IL, USA) and also kindly provided by Dr. Wei Li (National Institutes of Health, Bethesda, MD, USA). Mice, rats, Nile rats, and ground squirrels were all housed in the Case Western Reserve University (CWRU) animal facility where they were maintained on standard chow diets in a 12h light (∼10 lux)/12h dark cycle. Enucleated macaque (Macaca fascicularis) eyes and retinal tissue were obtained from Ricerca Biosciences (Painesville, OH, USA). All animal procedures and experiments were performed in accordance with U.S. animal protection laws and were approved by the CWRU Animal Care Committees and conformed to both the recommendations of the American Veterinary Medical Association Panel on Euthanasia and the Association for Research in Vision and Ophthalmology.
Clinical evaluation of the human patient from whom the retinal tissue was obtained was carried out at the Cleveland Clinic Cole Eye Institute (Cleveland, OH, USA). This research conformed to the tenets of the Declaration of Helsinki.
HPLC analyses of retinoids from the eyes of rat, mouse, nile rat, and ground squirrel species
After dark-adaptation for 3–12 h, animals were euthanized and their eyes were enucleated and flash frozen in liquid nitrogen. To extract retinoids, each eye was transferred to a glass homogenizer in 1 ml of 40 mM NH2OH in a 1:1 solution of PBS:methanol. Each eye was thoroughly homogenized and transferred to a 15 ml Falcon tube, and then kept at room temperature for 20 mins to allow derivatization of retinyl-aldehydes into their retinyl oxime metabolites. Next, 4 ml of hexanes were added to each tube, vigorously shaken to facilitate extraction of retinoids, and then centrifuged at 5000g for 10 min, after which the upper organic phase was collected in a glass test tube. The organic solvent was dried in a rotary SpeedVac, and the residual retinoids were re-dissolved in 250 ml of hexanes, vortexed, and then transferred to standard 2 ml vials with 0.4 ml inserts for HPLC analysis. These analyses were performed with a normal phase column (Agilent Zorbax Sil 5 µm 4.6 x 250 mm) (66). For retinoid separation, a linear gradient of 0.5% ethyl acetate in hexane was applied over 15 min followed by 35 min of 10% ethyl acetate in hexane at a constant flow rate of 1.4 ml/min. Retinyl-esters and retinyl-oxime metabolites were detected at wavelengths of 325 nm and 360 nm, respectively. The column was regenerated by a 10 min equilibration with 0.5% ethyl acetate in hexane after each run.
RNA isolation
Rodent species (Rattus norvegicus, Mus musculus, Arvicanthis niloticus, and Ictidomys tridecemlineatus) were euthanized by cervical dislocation and eyes and retinal samples were dissected out and immediately placed in RNALater stabilization reagent (Qiagen, Valencia, CA, USA). For geographic retinal tissue collection from macaques, each retina was cut into quadrants, and the central region containing the macula was carefully dissected out with scissors to separate it from peripheral retinal tissue before being placed in RNALater. The human retinal sample consisted of a tumour-free hemi-retina that was immediately placed in RNALater from a donor patient requiring enucleation for a large ocular melanoma.
RNA-Seq library preparation. Library preparation for Illumina RNA-Seq (Illumina Inc., San Diego, CA, USA) was carried out as previously described (17). The library preparation for isolated eye and retinal tissue in RNALater, obtained from human and macaque species, was performed similarly to that used for rodent tissues except for the larger scale homogenization required. Each rodent species-specific library preparation was pooled from multiple RNA samples: five mice for Mus musculus, five rats for Rattus norvegicus, five Nile rats for Arvicanthis niloticus, and four ground squirrels for Ictidomys tridecemlineatus. Human and macaque samples were each prepared from one eye. Biological replicates were prepared for both Mus musculus and Macaca fascicularis eye and retina samples.
RNA-seq, read mapping and determination of reads per kilobase per million reads (RPKM)
Libraries of Rattus norvegicus, Mus musculus, and Homo sapiens were sequenced by single-end sequencing, whereas prepared libraries of Arvicanthis niloticus, Ictidomys tridecemlineatus, and Macaca fascicularis were sequenced by paired-end sequencing with the Illumina Genome Analyzer IIx or HiScan SQ. Data for Rattus norvegicus, Mus musculus, Macaca fascicularis and Homo sapiens were processed and aligned with the University of California–Santa Cruz (UCSC; Santa Cruz, CA, USA) rat (rn4), mouse (mm9), rhesus (rheMac3), and human (hg19) genome assemblies and transcript annotations, respectively, using the Genomic Short-read Nucleotide Alignment Program (GSNAP), manual extraction of uniquely-mapped reads, HTseq for raw read counts of genes, and manual calculation of RPKM (35) statistics by gene. Details of sequencing and read statistics are provided in Supplementary Material, Table S1.
Transcriptomes of Arvicanthis niloticus and Ictidomys tridecemlineatus were assembled de novo. The processed reads were assembled using velvet (67), oases, and Trinity (68) with multiple k-mers ranging from 21 to 67 with an increment of 2 whereas reads assembled with ABYSS and trans-ABYSS (69–71) were done with multiple k-mers ranging from 40 to 78 with an increment of 2. The assemblies were combined and the duplicated transcripts were collapsed together to remove redundancy. Transcriptome annotation was done by first searching against the SWISS-PROT database using the BLASTx algorithm (72) and an E value cutoff of 1e-5. Transcripts with the best matches to proteins from viruses and bacteria were considered as potential contaminants and thus were removed from further analysis. The resulting filtered BLASTx results were then used for Gene Ontology (73) analysis with the blast2GO program (74). Protein domains were predicted for all such transcripts by searching the PFAM database (75) with the HMMER program (76). The expression value for each transcript was calculated by aligning the original Illumina reads to the transcriptome using the Bowtie program (77), with all mapping reads reported. The expression value for each transcript was calculated as RPKM (35) using a customized Perl script. Details of these sequencing and read statistics are described in Supplementary Material, Table S2.
The processed and raw fastq files for Mus musculus (accession numbers GSE38359, GSE29752, GSE48974, GSE84927), Rattus norvegicus (GSE84932), Arvicanthis niloticus (GSE84928), Ictidomys tridecemlineatus (GSE84931), Macaca fascicularis (GSE84929), and Homo sapiens (GSE84930) have been deposited in the NCBI GEO database and are available for public use.
Analysis of RNA-seq data
Concordance of R2 ≥ 0.95 has been demonstrated between biological replicates (4) and was noted in this study from biological replicates isolated from pooled or single tissue samples. Orthologous gene relationships across species were extracted with the NCBI biosystems database (78). Genes were then categorized for their biological and functional roles with Gene Ontology using AmiGO software (79). From the total list, an initial set of 4049 genes that displayed differential expression across the rodent species of Rattus norvegicus, Mus musculus, Arvicanthis niloticus, and Ictidomys tridecemlineatus, were considered for gene co-expression network analysis. Co-expression networks were constructed from RNA-Seq data by calculating Pearson correlations across these rodent species. Concordance of gene expression with the Pearson correlation was such that the gene expression pattern was weighted against the pigment composition in rodent species. A threshold (R2 ≥0.5) was set to identify those with moderate to strong correlations. Genes grouped with coherent expression profiles that exhibited topographical overlap were assembled into discreet modules by average linkage hierarchical clustering (80,81), with the gene modules corresponding to branches of a dendrogram. For each module, a heat map of transcript expression was produced with rows corresponding to genes and columns corresponding to the rodent eye sample. These modules were then examined to identify both the global and individual networks to which they corresponded (82).
Immunohistochemistry (IHC)
Histological and IHC procedures were performed as previously described (83). Cross sections of mouse eyecups were incubated with primary antibodies against Rho and Opn1mw. Signals were detected with Cy3- or Alexa488-conjugated secondary antibodies. Nuclear staining was achieved with 4’,6-diamidino-2-phenylindole (DAPI). A Zeiss LSM 510 inverted Laser Scanning Confocal Microscope was used to image the sections and the analysis was accomplished with ImageJ software.
Semi-quantitative real-time PCR
Total RNA was purified by using the RNeasy Mini Kit with on column DNase treatment (Qiagen, Valencia, CA). Selected genes (Supplementary Material, Table S6) were probed with the Qiagen One-step RT-PCR Kit. Twenty-five nanograms of total RNA was used for each 12.5 µl reaction as per the manufacturer’s instructions. Primers used to probe each gene were custom designed to span introns when possible to rule out genomic DNA contamination. To design primers that would amplify from each species, we aligned the human, macaque, mouse and rat sequences for each gene and selected regions with 100% identity. β-Actin primers were designed as a loading control.
Statistical analyses
Experimental results were analyzed by an independent two sample t test. P values of 0.05 or less were considered statistically significant. Data presented graphically in figures are shown as means ± standard deviations.
Supplementary Material
Acknowledgements
We thank Dr. Leslie T. Webster Jr. for helpful comments on this manuscript. We also thank Dr. Akiko Maeda (CWRU) for primate tissue isolation, the laboratories of Dr. Laura Smale (MSU) for Nile rat animals to establish a breeding colony at CWRU and Dr. Wei Li (NIH) for providing ground squirrel animal tissue for retinoid studies. Ms. Simone Edelheit in the CWRU Genomics Core performed the sequencing.
Supplementary Material
Supplementary Material is available at HMG online.
Conflict of interest statement. None declared.
Funding
This work was supported by funding from the National Institutes of Health EY021126 and U01 EY025451, the Arnold and Mabel Beckman Foundation, and the Canadian Institute for Advanced Research (CIFAR). K.P. is the John H. Hord Professor of Pharmacology and Distinguished University Professor.
References
- 1. Zhang G., Cowled C., Shi Z., Huang Z., Bishop-Lilly K.A., Fang X., Wynne J.W., Xiong Z., Baker M.L., Zhao W., et al. (2013) Comparative analysis of bat genomes provides insight into the evolution of flight and immunity. Science, 339, 456–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Keane T.M., Goodstadt L., Danecek P., White M.A., Wong K., Yalcin B., Heger A., Agam A., Slater G., Goodson M., et al. (2011) Mouse genomic variation and its effect on phenotypes and gene regulation. Nature, 477, 289–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Brawand D., Soumillon M., Necsulea A., Julien P., Csardi G., Harrigan P., Weier M., Liechti A., Aximu-Petri A., Kircher M., et al. (2011) The evolution of gene expression levels in mammalian organs. Nature, 478, 343–348. [DOI] [PubMed] [Google Scholar]
- 4. Mustafi D., Maeda T., Kohno H., Nadeau J.H., Palczewski K. (2012) Inflammatory priming predisposes mice to age-related retinal degeneration. J. Clin. Invest., 122, 2989–3001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Atanur S.S., Diaz A.G., Maratou K., Sarkis A., Rotival M., Game L., Tschannen M.R., Kaisaki P.J., Otto G.W., Ma M.C., et al. (2013) Genome sequencing reveals loci under artificial selection that underlie disease phenotypes in the laboratory rat. Cell, 154, 691–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. McGary K.L., Park T.J., Woods J.O., Cha H.J., Wallingford J.B., Marcotte E.M. (2010) Systematic discovery of nonobvious human disease models through orthologous phenotypes. Proc. Natl. Acad. Sci. U S A, 107, 6544–6549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Khaitovich P., Weiss G., Lachmann M., Hellmann I., Enard W., Muetzel B., Wirkner U., Ansorge W., Paabo S. (2004) A neutral model of transcriptome evolution. PLoS Biol., 2, E132.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Oldham M.C., Horvath S., Geschwind D.H. (2006) Conservation and evolution of gene coexpression networks in human and chimpanzee brains. Proc. Natl. Acad. Sci. U S A, 103, 17973–17978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Pramparo T., Libiger O., Jain S., Li H., Youn Y.H., Hirotsune S., Schork N.J., Wynshaw-Boris A. (2011) Global developmental gene expression and pathway analysis of normal brain development and mouse models of human neuronal migration defects. PLoS Genet., 7, e1001331.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Khaitovich P., Enard W., Lachmann M., Paabo S. (2006) Evolution of primate gene expression. Nat. Rev. Genet., 7, 693–702. [DOI] [PubMed] [Google Scholar]
- 11. Khaitovich P., Hellmann I., Enard W., Nowick K., Leinweber M., Franz H., Weiss G., Lachmann M., Paabo S. (2005) Parallel patterns of evolution in the genomes and transcriptomes of humans and chimpanzees. Science, 309, 1850–1854. [DOI] [PubMed] [Google Scholar]
- 12. Merkin J., Russell C., Chen P., Burge C.B. (2012) Evolutionary dynamics of gene and isoform regulation in Mammalian tissues. Science, 338, 1593–1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Masland R.H. (2012) The neuronal organization of the retina. Neuron, 76, 266–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wang Z., Gerstein M., Snyder M. (2009) RNA-Seq: a revolutionary tool for transcriptomics. Nat. Rev. Genet., 10, 57–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Farkas M.H., Grant G.R., White J.A., Sousa M.E., Consugar M.B., Pierce E.A. (2013) Transcriptome analyses of the human retina identify unprecedented transcript diversity and 3.5 Mb of novel transcribed sequence via significant alternative splicing and novel genes. BMC Genomics, 14, 486.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mustafi D., Kevany B.M., Bai X., Maeda T., Sears J.E., Khalil A.M., Palczewski K. (2013) Evolutionarily conserved long intergenic non-coding RNAs in the eye. Hum. Mol. Genet., 22, 2992–3002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Mustafi D., Kevany B.M., Genoud C., Okano K., Cideciyan A.V., Sumaroka A., Roman A.J., Jacobson S.G., Engel A., Adams M.D., et al. (2011) Defective photoreceptor phagocytosis in a mouse model of enhanced S-cone syndrome causes progressive retinal degeneration. Faseb J., 25, 3157–3176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Li M., Jia C., Kazmierkiewicz K.L., Bowman A.S., Tian L., Liu Y., Gupta N.A., Gudiseva H.V., Yee S.S., Kim M., et al. (2014) Comprehensive analysis of gene expression in human retina and supporting tissues. Hum. Mol. Genet., 23, 4001–4014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Siegert S., Cabuy E., Scherf B.G., Kohler H., Panda S., Le Y.Z., Fehling H.J., Gaidatzis D., Stadler M.B., Roska B. (2012) Transcriptional code and disease map for adult retinal cell types. Nat. Neurosci., 15, 487–495. [DOI] [PubMed] [Google Scholar]
- 20. Macosko E.Z., Basu A., Satija R., Nemesh J., Shekhar K., Goldman M., Tirosh I., Bialas A.R., Kamitaki N., Martersteck E.M., et al. (2015) Highly Parallel Genome-wide Expression Profiling of Individual Cells Using Nanoliter Droplets. Cell, 161, 1202–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wright A.F., Chakarova C.F., Abd El-Aziz M.M., Bhattacharya S.S. (2010) Photoreceptor degeneration: genetic and mechanistic dissection of a complex trait. Nat. Rev. Genet., 11, 273–284. [DOI] [PubMed] [Google Scholar]
- 22. Curcio C.A., Sloan K.R., Kalina R.E., Hendrickson A.E. (1990) Human photoreceptor topography. J. Comp. Neurol., 292, 497–523., [DOI] [PubMed] [Google Scholar]
- 23. Veleri S., Lazar C.H., Chang B., Sieving P.A., Banin E., Swaroop A. (2015) Biology and therapy of inherited retinal degenerative disease: insights from mouse models. Dis. Model. Mech., 8, 109–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gilmour G.S., Gaillard F., Watson J., Kuny S., Mema S.C., Bonfield S., Stell W.K., Sauve Y. (2008) The electroretinogram (ERG) of a diurnal cone-rich laboratory rodent, the Nile grass rat (Arvicanthis niloticus). Vision Res., 48, 2723–2731. [DOI] [PubMed] [Google Scholar]
- 25. Gaillard F., Bonfield S., Gilmour G.S., Kuny S., Mema S.C., Martin B.T., Smale L., Crowder N., Stell W.K., Sauve Y. (2008) Retinal anatomy and visual performance in a diurnal cone-rich laboratory rodent, the Nile grass rat (Arvicanthis niloticus). J. Comp. Neurol., 510, 525–538. [DOI] [PubMed] [Google Scholar]
- 26. Stanyon R., Stone G., Garcia M., Froenicke L. (2003) Reciprocal chromosome painting shows that squirrels, unlike murid rodents, have a highly conserved genome organization. Genomics, 82, 245–249. [DOI] [PubMed] [Google Scholar]
- 27. Lamb T.D., Collin S.P., Pugh E.N., Jr. (2007) Evolution of the vertebrate eye: opsins, photoreceptors, retina and eye cup. Nat. Rev. Neurosci., 8, 960–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Burns M.E., Arshavsky V.Y. (2005) Beyond counting photons: trials and trends in vertebrate visual transduction. Neuron, 48, 387–401. [DOI] [PubMed] [Google Scholar]
- 29. Wald G. (1968) The molecular basis of visual excitation. Nature, 219, 800–807. [DOI] [PubMed] [Google Scholar]
- 30. Mustafi D., Engel A.H., Palczewski K. (2009) Structure of cone photoreceptors. Prog. Retin. Eye Res., 28, 289–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Babino D., Perkins B.D., Kindermann A., Oberhauser V., von Lintig J. (2015) The role of 11-cis-retinyl esters in vertebrate cone vision. Faseb J., 29, 216–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Rosenbloom K.R., Armstrong J., Barber G.P., Casper J., Clawson H., Diekhans M., Dreszer T.R., Fujita P.A., Guruvadoo L., Haeussler M., et al. (2015) The UCSC Genome Browser database: 2015 update. Nucleic Acids Res., 43, D670–D681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Pruitt K.D., Brown G.R., Hiatt S.M., Thibaud-Nissen F., Astashyn A., Ermolaeva O., Farrell C.M., Hart J., Landrum M.J., McGarvey K.M., et al. (2014) RefSeq: an update on mammalian reference sequences. Nucleic Acids Res., 42, D756–D763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zhang X., Goodsell J., Norgren R.B., Jr. (2012) Limitations of the rhesus macaque draft genome assembly and annotation. BMC Genomics, 13, 206.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Mortazavi A., Williams B.A., McCue K., Schaeffer L., Wold B. (2008) Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods, 5, 621–628. [DOI] [PubMed] [Google Scholar]
- 36. Irie S., Sanuki R., Muranishi Y., Kato K., Chaya T., Furukawa T. (2015) Rax Homeoprotein Regulates Photoreceptor Cell Maturation and Survival in Association with Crx in the Postnatal Mouse Retina. Mol. Cell. Biol., 35, 2583–2596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Pandit T., Jidigam V.K., Patthey C., Gunhaga L. (2015) Neural retina identity is specified by lens-derived BMP signals. Development, 142, 1850–1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hornett E.A., Wheat C.W. (2012) Quantitative RNA-Seq analysis in non-model species: assessing transcriptome assemblies as a scaffold and the utility of evolutionary divergent genomic reference species. BMC Genomics, 13, 361.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Mural R.J., Adams M.D., Myers E.W., Smith H.O., Miklos G.L., Wides R., Halpern A., Li P.W., Sutton G.G., Nadeau J., et al. (2002) A comparison of whole-genome shotgun-derived mouse chromosome 16 and the human genome. Science, 296, 1661–1671. [DOI] [PubMed] [Google Scholar]
- 40. Chimpanzee S., Analysis C. (2005) Initial sequence of the chimpanzee genome and comparison with the human genome. Nature, 437, 69–87. [DOI] [PubMed] [Google Scholar]
- 41. Lin S., Lin Y., Nery J.R., Urich M.A., Breschi A., Davis C.A., Dobin A., Zaleski C., Beer M.A., Chapman W.C., et al. (2014) Comparison of the transcriptional landscapes between human and mouse tissues. Proc. Natl. Acad. Sci. U S A, 111, 17224–17229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Chen S., Krinsky B.H., Long M. (2013) New genes as drivers of phenotypic evolution. Nat. Rev. Genet., 14, 645–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ilicic T., Kim J.K., Kolodziejczyk A.A., Bagger F.O., McCarthy D.J., Marioni J.C., Teichmann S.A. (2016) Classification of low quality cells from single-cell RNA-seq data. Genome Biol., 17, 29.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Weiss E.R., Ducceschi M.H., Horner T.J., Li A., Craft C.M., Osawa S. (2001) Species-specific differences in expression of G-protein-coupled receptor kinase (GRK) 7 and GRK1 in mammalian cone photoreceptor cells: implications for cone cell phototransduction. J. Neurosci., 21, 9175–9184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ihmels J., Bergmann S., Barkai N. (2004) Defining transcription modules using large-scale gene expression data. Bioinformatics, 20, 1993–2003. [DOI] [PubMed] [Google Scholar]
- 46. Nagai T., Aruga J., Takada S., Gunther T., Sporle R., Schughart K., Mikoshiba K. (1997) The expression of the mouse Zic1, Zic2, and Zic3 gene suggests an essential role for Zic genes in body pattern formation. Dev. Biol., 182, 299–313. [DOI] [PubMed] [Google Scholar]
- 47. Caceres M., Lachuer J., Zapala M.A., Redmond J.C., Kudo L., Geschwind D.H., Lockhart D.J., Preuss T.M., Barlow C. (2003) Elevated gene expression levels distinguish human from non-human primate brains. Proc. Natl. Acad. Sci. U S A, 100, 13030–13035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Mustafi D., Kevany B.M., Genoud C., Bai X., Palczewski K. (2013) Photoreceptor phagocytosis is mediated by phosphoinositide signaling. Faseb J., 27, 4585–4595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Chen Y., Palczewska G., Mustafi D., Golczak M., Dong Z., Sawada O., Maeda T., Maeda A., Palczewski K. (2013) Systems pharmacology identifies drug targets for Stargardt disease-associated retinal degeneration. J. Clin. Invest., 123, 5119–5134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Brzezinski J.A., Reh T.A. (2015) Photoreceptor cell fate specification in vertebrates. Development, 142, 3263–3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zhou S., Flamier A., Abdouh M., Tetreault N., Barabino A., Wadhwa S., Bernier G. (2015) Differentiation of human embryonic stem cells into cone photoreceptors through simultaneous inhibition of BMP, TGFbeta and Wnt signaling. Development, 142, 3294–3306. [DOI] [PubMed] [Google Scholar]
- 52. Hunter D.D., Zhang M., Ferguson J.W., Koch M., Brunken W.J. (2004) The extracellular matrix component WIF-1 is expressed during, and can modulate, retinal development. Mol. Cell. Neurosci., 27, 477–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Kurabayashi N., Nguyen M.D., Sanada K. (2013) The G protein-coupled receptor GPRC5B contributes to neurogenesis in the developing mouse neocortex. Development, 140, 4335–4346. [DOI] [PubMed] [Google Scholar]
- 54. Greber B. (2011) When BMP meets FGF. Cell Stem Cell, 9, 91–92. [DOI] [PubMed] [Google Scholar]
- 55. Pellicano F., Inglis-Broadgate S.L., Pante G., Ansorge W., Iwata T. (2006) Expression of coiled-coil protein 1, a novel gene downstream of FGF2, in the developing brain. Gene Expr. Patterns, 6, 285–293. [DOI] [PubMed] [Google Scholar]
- 56. Zhang P., Andrianakos R., Yang Y., Liu C., Lu W. (2010) Kruppel-like factor 4 (Klf4) prevents embryonic stem (ES) cell differentiation by regulating Nanog gene expression. J. Biol. Chem., 285, 9180–9189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Morikawa M., Koinuma D., Mizutani A., Kawasaki N., Holmborn K., Sundqvist A., Tsutsumi S., Watabe T., Aburatani H., Heldin C.H., et al. (2016) BMP Sustains Embryonic Stem Cell Self-Renewal through Distinct Functions of Different Kruppel-like Factors. Stem Cell Reports, 6, 64–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Zhong X., Gutierrez C., Xue T., Hampton C., Vergara M.N., Cao L.H., Peters A., Park T.S., Zambidis E.T., Meyer J.S., et al. (2014) Generation of three-dimensional retinal tissue with functional photoreceptors from human iPSCs. Nat. Commun., 5, 4047.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Shirai H., Mandai M., Matsushita K., Kuwahara A., Yonemura S., Nakano T., Assawachananont J., Kimura T., Saito K., Terasaki H., et al. (2016) Transplantation of human embryonic stem cell-derived retinal tissue in two primate models of retinal degeneration. Proc. Natl. Acad. Sci. U S A, 113, E81–E90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Vogel P., Gelfman C.M., Issa T., Payne B.J., Hansen G.M., Read R.W., Jones C., Pitcher M.R., Ding Z.M., DaCosta C.M., et al. (2015) Nephronophthisis and retinal degeneration in tmem218-/- mice: a novel mouse model for Senior-Loken syndrome? Vet. Pathol., 52, 580–595. [DOI] [PubMed] [Google Scholar]
- 61. Li D., Qiu Z., Shao Y., Chen Y., Guan Y., Liu M., Li Y., Gao N., Wang L., Lu X., et al. (2013) Heritable gene targeting in the mouse and rat using a CRISPR-Cas system. Nat. Biotechnol., 31, 681–683. [DOI] [PubMed] [Google Scholar]
- 62. Wang H., Yang H., Shivalila C.S., Dawlaty M.M., Cheng A.W., Zhang F., Jaenisch R. (2013) One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell, 153, 910–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Niu Y., Shen B., Cui Y., Chen Y., Wang J., Wang L., Kang Y., Zhao X., Si W., Li W., et al. (2014) Generation of gene-modified cynomolgus monkey via Cas9/RNA-mediated gene targeting in one-cell embryos. Cell, 156, 836–843. [DOI] [PubMed] [Google Scholar]
- 64. Mears A.J., Kondo M., Swain P.K., Takada Y., Bush R.A., Saunders T.L., Sieving P.A., Swaroop A. (2001) Nrl is required for rod photoreceptor development. Nat. Genet., 29, 447–452. [DOI] [PubMed] [Google Scholar]
- 65. Lem J., Krasnoperova N.V., Calvert P.D., Kosaras B., Cameron D.A., Nicolo M., Makino C.L., Sidman R.L. (1999) Morphological, physiological, and biochemical changes in rhodopsin knockout mice. Proc. Natl. Acad. Sci. U S A, 96, 736–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Golczak M., Bereta G., Maeda A., Palczewski K. (2010) Molecular biology and analytical chemistry methods used to probe the retinoid cycle. Methods Mol. Biol., 652, 229–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Zerbino D.R., Birney E. (2008) Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res., 18, 821–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Grabherr M.G., Haas B.J., Yassour M., Levin J.Z., Thompson D.A., Amit I., Adiconis X., Fan L., Raychowdhury R., Zeng Q., et al. (2011) Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol., 29, 644–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Birol I., Jackman S.D., Nielsen C.B., Qian J.Q., Varhol R., Stazyk G., Morin R.D., Zhao Y., Hirst M., Schein J.E., et al. (2009) De novo transcriptome assembly with ABySS. Bioinformatics, 25, 2872–2877. [DOI] [PubMed] [Google Scholar]
- 70. Robertson G., Schein J., Chiu R., Corbett R., Field M., Jackman S.D., Mungall K., Lee S., Okada H.M., Qian J.Q., et al. (2010) De novo assembly and analysis of RNA-seq data. Nat. Methods, 7, 909–912. [DOI] [PubMed] [Google Scholar]
- 71. Simpson J.T., Wong K., Jackman S.D., Schein J.E., Jones S.J., Birol I. (2009) ABySS: a parallel assembler for short read sequence data. Genome Res., 19, 1117–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Altschul S.F., Madden T.L., Schaffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res., 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., et al. (2000) Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet., 25, 25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Gotz S., Garcia-Gomez J.M., Terol J., Williams T.D., Nagaraj S.H., Nueda M.J., Robles M., Talon M., Dopazo J., Conesa A. (2008) High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res., 36, 3420–3435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Finn R.D., Mistry J., Tate J., Coggill P., Heger A., Pollington J.E., Gavin O.L., Gunasekaran P., Ceric G., Forslund K., et al. (2010) The Pfam protein families database. Nucleic Acids Res., 38, D211–D222. 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Eddy S.R. (1998) Profile hidden Markov models. Bioinformatics, 14, 755–763. [DOI] [PubMed] [Google Scholar]
- 77. Langmead B., Trapnell C., Pop M., Salzberg S.L. (2009) Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol., 10, R25.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Geer L.Y., Marchler-Bauer A., Geer R.C., Han L., He J., He S., Liu C., Shi W., Bryant S.H. (2010) The NCBI BioSystems database. Nucleic Acids Res., 38, D492–D496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Gene Ontology C. (2015) Gene Ontology Consortium: going forward. Nucleic Acids Res., 43, D1049–D1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Ravasz E., Somera A.L., Mongru D.A., Oltvai Z.N., Barabasi A.L. (2002) Hierarchical organization of modularity in metabolic networks. Science, 297, 1551–1555. [DOI] [PubMed] [Google Scholar]
- 81. Yip A.M., Horvath S. (2007) Gene network interconnectedness and the generalized topological overlap measure. BMC Bioinformatics, 8, 22.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Barabasi A.L., Oltvai Z.N. (2004) Network biology: understanding the cell's functional organization. Nat. Rev. Genet., 5, 101–113. [DOI] [PubMed] [Google Scholar]
- 83. Maeda A., Maeda T., Imanishi Y., Kuksa V., Alekseev A., Bronson J.D., Zhang H., Zhu L., Sun W., Saperstein D.A., et al. (2005) Role of photoreceptor-specific retinol dehydrogenase in the retinoid cycle in vivo. J. Biol. Chem., 280, 18822–18832. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.