Abstract
The purpose of our study was to understand the protective effects of reduced expression of dynamin-related protein (Drp1) against amyloid beta (Aβ) induced mitochondrial and synaptic toxicities in Alzheimer’s disease (AD) progression and pathogenesis. Our recent molecular and biochemical studies revealed that impaired mitochondrial dynamics—increased mitochondrial fragmentation and decreased fusion—in neurons from autopsy brains of AD patients and from transgenic AD mice and neurons expressing Aβ, suggesting that Aβ causes mitochondrial fragmentation in AD. Further, our recent co-immunoprecipitation and immunostaining analysis revealed that the mitochondrial fission protein Drp1 interacted with Aβ, and this interaction increased as AD progressed. Based on these findings, we hypothesize that a partial deficiency of Drp1 inhibits Drp1-Aβ interactions and protects Aβ-induced mitochondrial and synaptic toxicities, and maintains mitochondrial dynamics and neuronal function in AD neurons. We crossed Drp1+/− mice with APP transgenic mice (Tg2576 line) and created double mutant (APPXDrp1+/−) mice. Using real-time RT-PCR and immunoblotting analyses, we measured mRNA expressions and protein levels of genes related to the mitochondrial dynamics, mitochondrial biogenesis and synapses from 6-month-old Drp1+/−, APP, APPXDrp1+/− and wild-type (WT) mice. Using biochemical methods, we also studied mitochondrial function and measured soluble Aβ in brain tissues from all lines of mice in our study. Decreased mRNA expressions and protein levels of Drp1 and Fis1 (fission) and CypD (matrix) genes, and increased levels of Mfn1, Mfn2 and Opa1 (fusion), Nrf1, Nrf2, PGC1α, TFAM (biogenesis) and synaptophysin, PSD95, synapsin 1, synaptobrevin 1, neurogranin, GAP43 and synaptopodin (synaptic) were found in 6-month-old APPXDrp1+/− mice relative to APP mice. Mitochondrial functional assays revealed that mitochondrial dysfunction is reduced in APPXDrp1+/− mice relative to APP mice, suggesting that reduced Drp1enhances mitochondrial function in AD neurons. Sandwich ELISA assay revealed that soluble Aβ levels were significantly reduced in APPXDrp1+/− mice relative to APP mice, indicating that reduced Drp1 decreases soluble Aβ production in AD progression. These findings suggest that a partial reduction of Drp1 reduces Aβ production, reduces mitochondrial dysfunction, and maintains mitochondrial dynamics, enhances mitochondrial biogenesis and synaptic activity in APP mice. These findings may have implications for the development of Drp1 based therapeutics for AD patients.
Introduction
Alzheimer’s disease (AD) is a multifactorial, age-related neurodegenerative disease, characterized by multiple cognitive impairments and changes in behaviour and personality (1–9). According to the 2015 World Alzheimer’s Report, 47.5 million people had AD-related dementia worldwide, including 5.4 million Americans, and projected the numbers to rise to 75.6 million by 2030 and to 131.5 million by 2050. Over 9.9 million new cases of AD-related dementia are diagnosed every year worldwide (10). Dementia has a huge economic impact on our society and the estimated total healthcare cost of dementia worldwide in 2015 was $818 billion (10). Currently, there are no drugs or agents available to treat or to prevent fragmented mitochondria in patients with AD.
Several years of intense research have revealed that multiple cellular changes have been implicated in AD pathogenesis, including the loss of synapses, loss of synaptic function, mitochondrial structural and functional abnormalities, inflammatory responses and neuronal loss, in addition to production and accumulation of amyloid beta (Aβ) and hyperphosphorylated tau and neurofibrillary tangles (NFTs) in learning and memory regions of AD brain (3,5,11–22).
Mitochondrial damage and synaptic dysfunction are early events in AD progression and pathogenesis. However, the precise underlying mechanisms of mitochondrial damage and synaptic dysfunction in neuronal damage and cognitive decline in AD are still not well understood. Mitochondrial dysfunction has been identified in AD postmortem brains (17,23–25), AD transgenic mice (15,25–29), and cell lines that express mutant APP and/or cells treated with Aβ (12,13,21,30).
Several lines of evidence suggest that increased free radical production, lipid peroxidation, oxidative DNA damage, oxidative protein damage and decreased ATP production, and cytochrome oxidase activity in brains from AD patients compared to brains from age-matched control subjects (17,25,31,23,24). Further, several groups found that mitochondrial-encoded genes were abnormally expressed in the AD patients and AD transgenic mice, indicating that differentially expressed mitochondrial genes may a compensatory response due to Aβ-induced mitochondrial dysfunction in neurons (32–35) from AD patients and from AD transgenic mice (25,36–38). However, the precise link between Aβ and mitochondria is not known.
Using biochemical, molecular, and electron microscopy studies, and postmortem brain tissues from AD patients and AD transgenic mice, several groups studied the connection between Aβ and mitochondria. They found that Aβ is localized to the mitochondrial membranes and is responsible for generating increased free radicals and initiating mitochondrial dysfunction (15,18,19,27,39). Other groups found presequence protease, ‘Prep Peptidasome’ that degrades the Aβ species in the mitochondria and is capable of degrading mitochondrial Aβ, further supporting the association of Aβ with mitochondria and mitochondrial dysfunction in AD (40). In addition, studies of mitochondrial structure in brain tissue from AD patients and neuronal cells expressing the mutant APP found that Aβ fragments mitochondria and causes structural changes in neurons (12,13,21,41–42).
Recent mitochondrial structural studies revealed that abnormalities in mitochondria are involved in AD (12,13,21,41–43). These abnormalities are caused by an imbalance in highly conserved, GTPase genes that are essential for mitochondrial division and fusion. Fission genes—Drp1, fission 1 (Fis1), mitofusins 1 and 2 (Mfn1, Mfn2), and optic atrophy 1 (Opa1)—regulate, maintain, and remodel mammalian mitochondria (13–14,44–46). In normal mammalian cells, mitochondrial fission and fusion balance equally and maintain mitochondrial dynamics, distribution and mitochondrial function (44–46). However, in aged neurons, in neurons exposed to toxins, and in neurons that express mutant proteins, such as Aβ and phosphorylated tau, an imbalance between fission and fusion leads to abnormalities in mitochondrial structure and function, and to damaged neurons (45,47).
Recently, several groups studied abnormal mitochondrial dynamics in brain tissues from postmortem brains of AD patients (12,13,41), primary neurons from APP transgenic mice (Tg2576 line) and neuroblastoma cells that were treated with the Aβ peptide (21,42,48). They found increased expressions of Drp1 and Fis1 and decreased expressions of Mfn1, Mfn2, and Opa1, indicating abnormal mitochondrial dynamics in AD neurons. These findings suggest that Aβ fragments mitochondria and causes abnormal dynamics, leading to mitochondrial dysfunction and neuronal damage in AD. However, the underlying mechanisms of mitochondrial fragmentation and neuronal damage are not well understood.
To determine whether the interaction of Drp1 and Aβ increases as AD progresses, the Reddy Lab performed co-immunoprecipitation analysis, using a Drp1 antibody, and immunoblotting analysis, using the Aβ-specific antibody 6E10 and protein lysates of the cortical tissues from control subjects and from patients with early, definite, and severe AD (41). They found a 4 kDa Aβ and 110 kDa full-length AβPP in severe AD patients. Using the oligomeric-specific antibody (A11), we also found a band of 50 kDa in these same patients, indicating that Drp1 interacts with both oligomeric and monomeric Aβ in early, definite, and severe AD, and that intensity of 50 kDa band increase with AD progression (17). We found no physical interaction between Drp1 and Aβ in the control subjects. However, the mechanistic link between Drp1 and Aβ in AD neurons is not clear. We proposed that reduced Drp1 protects against Aβ-induced mitochondrial and synaptic toxicities in AD neurons.
In the current study, we sought to determine the protective effects of a partial reduction of Drp1 in transgenic mice that human APP Swedish mutation in the development and progression of the disease process in AD. We crossed Drp1+/− mice and APP mice and generated double mutant (APPXDrp1+/−) mice. Using cortical tissues from 6-month-old APP, Drp1+/−, double mutant (APPXDrp1+/−) and WT mice, we studied 1) mitochondrial structure and activity by measuring mRNA and protein levels of genes related to mitochondrial dynamics, mitochondrial matrix, mitochondrial biogenesis; 2) synaptic activities by measuring mRNA and protein levels of synaptic genes; 3) assessed mitochondrial function by measuring free radical production, lipid peroxidation, mitochondrial ATP and GTPase Drp1 activity and 4) Abeta pathology.
Results
In the current study, our objective was to determine whether a partial reduction of Drp1 protects neurons from Aβ-induced mitochondrial and synaptic toxicities in the progression and pathogenesis of AD. We measured mRNA levels of mitochondrial dynamics, mitochondrial biogenesis and synaptic genes in 6-month-old cortical tissues from Drp1+/−, APP, APPXDrp1+/− mice relative to age-matched WT mice (Table 1). We also compared mRNA data between APP and APPXDrp1+/− mice, in order to understand the protective role of reduced Drp1 in APP transgenic mice for mitochondrial and synaptic toxicities (Table 2).
Table 1.
mRNA fold changes of mitochondrial dynamics, mitochondrial biogenesis and synaptic genes in 6 months old WT, Drp1+/-, APP, APPXDrp1+/- mice
| Genes | mRNA fold changes |
|||
|---|---|---|---|---|
| Drp1+/− | APP | APPXDrp1+/− | ||
| Mitochondrial dynamics genes | Drp1 | −2.3** | 2.3** | 1.1 |
| Fis1 | −2.1** | 1.4* | 1.2 | |
| Mfn1 | 1.5* | −1.5** | −1.2 | |
| Mfn2 | 1.4* | −1.7* | 1.3 | |
| Opa1 | 1.3 | −1.4* | 1.3 | |
| Cyclophilin D | −1.3 | 1.7* | 1.2 | |
| Mitochondrial biogenesis genes | PGC1α | 1.6* | −2.1** | 1.3 |
| Nrf1 | 1.2 | −2.1** | 1.4 | |
| Nrf2 | 1.6** | −1.4* | 1.4 | |
| TFAM | 1.5* | −1.8** | 1.2 | |
| Synaptic genes | Synaptophysin | 1.4 | −1.6** | 1.3 |
| PSD-95 | 2.1** | −1.2 | 1.7** | |
| Synapsin 1 | 1.2 | −1.7* | 1.4* | |
| Synapsin 2 | 1.3 | −1.6* | 1.2 | |
| Synaptobrevin 1 | 1.2 | −1.5* | 1.1 | |
| Synaptopodin | 1.2 | −1.6* | 1.9** | |
| Neurogranin | 1.3 | −1.6* | 1.0 | |
| GAP43 | 1.2 | −1.4* | 1.3 | |
Table 2.
mRNA fold changes of mitochondrial dynamics, mitochondrial biogenesis and synaptic genes in 6 months old APPXDrp1+/− mice relative to APP mice
| Genes | mRNA fold changes | |
|---|---|---|
| APPXDrp1+/− | ||
| Mitochondrial dynamics genes | Drp1 | −2.3** |
| FIS1 | −2.2** | |
| Mfn1 | 1.5* | |
| Mfn2 | 2.0** | |
| OPA1 | 1.4* | |
| Cyclophilin D | −1.5* | |
| Mitochondrial biogenesis genes | PGC1α | 2.1** |
| Nrf1 | 2.6** | |
| Nrf2 | 1.7** | |
| TFAM | 2.1** | |
| Synaptic genes | Synaptophysin | 2.1** |
| PSD-95 | 1.4* | |
| Synapsin 1 | 2.1** | |
| Synapsin 2 | 1.8** | |
| Synaptobrevin 1 | 1.6* | |
| Synaptopodin | 2.5** | |
| Neurogranin | 1.4* | |
| GAP43 | 2.2** | |
Mitochondrial dynamics genes
APP mice versus WT mice
mRNA expression levels significantly increased in Drp1, by 2.3 fold (P = 0.003); and in Fis1, by 1.4 fold (P = 0.04) (Table 1) in APP mice relative to WT mice. In contrast, fusion genes were significantly reduced – Mfn1 by 1.5 fold (P = 0.004), Mfn2 by 1.7 (P = 0.01) and Opa1 by 1.4 fold (P = 0.03). Matrix gene, CypD levels were increased by 1.7 fold (P = 0.02).
APPXDrp1+/− mice versus WT mice
In double mutant mice, relative to WT, increased levels of mRNA were found for fission genes—Drp1 (by 1.1 fold) and Fis1 (by 1.2 fold), but not significant. Fusion genes Mfn2 and Opa1 were increased, but not significant. Mfn 1 was decreased by 1.2 fold, but not significant (Table 1).
Drp1+/− mice versus WT mice
mRNA expression levels significantly decreased in Drp1, by 2.3 fold (P = 0.002); and in Fis1, by 2.1 fold (P = 0.002) (Table 1). However, mRNA expressions of mitochondrial fusion genes were significantly increased – Mfn1 by 1.5 fold (P = 0.04), Mfn2 by 1.4 fold (P = 0.04) and Opa1 by 1.3 fold. mRNA expression of matrix gene CypD was significantly decreased by 1.3 fold, but not significant.
Mitochondrial biogenesis genes
APP mice versus WT mice
Mitochondrial biogenesis genes down-regulated, PGC1α by 2.1 fold (P = 0.003), Nrf1 by 2.1 fold (P = 0.003), Nrf2 by 1.4 fold (P = 0.04) and TFAM by 1.8 fold (P = 0.004) in APP mice relative to WT mice, suggesting that mutant APP and/or Aβ reduces mitochondrial biogenesis activity.
APPXDrp1+/− mice versus WT mice
As shown in Table 1, mitochondrial biogenesis genes (PGC1α by 1.3 fold, Nrf1 by 1.4 fold, Nrf2 by 1.4 fold and TFAM by 1.2 fold) were slightly increased in APPXDrp1+/− mice, but not significant, indicating reduced Drp1 maintains mitochondrial biogenesis in the presence of mutant APP and Aβ in mice.
Drp1+/− mice versus WT mice
mRNA levels of biogenesis genes were increased—PGC1α by 1.6 fold (P = 0.01), Nrf1 by 1.2 fold, Nrf2 by 1.6 fold (P = 0.004) and TFAM by 1.5 fold (P = 0.04) in Drp1+/− mice relative to WT mice indicating that reduced Drp1 increases mitochondrial biogenesis in mice.
Synaptic genes
APP mice versus WT mice
mRNA levels of synaptic genes were significantly decreased—synaptophysin by 1.6 fold (P = 0.003), PSD95 by 1.2 fold, synapsin 1 by 1.7 fold (P = 0.02), synapsin 2 by 1.6 fold (P = 0.02), synaptobrevin by 1.5 fold (P = 0.03), synaptopodin by 1.6 fold (P = 0.01), neurogranin by 1.6 fold (P = 0.02) and GAP43 by 1.4 fold (P = 0.03) (Table 1). These observations suggest that mutant APP and Aβ reduces synaptic activity.
APPXDrp1+/− mice versus WT mice
Synaptic genes were increased (synaptophysin by 1.3 fold, PSD95 by 1.7 fold (P = 0.003), synapsin 1 by 1.4 fold (P = 0.04), synapsin 2 by 1.2 fold, Synaptobrevin by 1.1 fold, synaptopodin by 1.9 fold (P = 0.002) and GAP43 by 1.3 fold in APPXDrp1+/− mice relative to WT mice, indicating that reduced Drp1 enhances synaptic activity in the presence of mutant APP and Aβ (Table 1).
Drp1+/− mice versus WT mice
As shown in Table 1, mRNA levels significantly increased for synaptophysin by 1.4 fold, PSD95 by 2.1 fold (P = 0.001), synapsin 1 by 1.2 fold, synapsin 2 by 1.3 fold, Synaptobrevin by 1.2 fold, neurogranin by 1.3 fold and GAP43 by 1.2 fold in Drp1+/- mice relative to WT, indicating that reduced Drp1 increases synaptic activity.
mRNA differences between APP mice and APPXDrp1+/− mice
To better understand whether reduced Drp1 protects against mutant APP/Aβ-induced mitochondrial and synaptic toxicities, we compared gene expression data between APP mice and APPXDrp1+/- mice. As shown in Table 2, mRNA levels of fission genes were reduced—Drp1 by 2.3 fold (P = 0.004) and Fis1 by 2.2 fold (P = 0.001), fusion genes were increased—Mfn1 by 1.5 fold (P = 0.04), Mfn2 by 2.0 fold (P = 0.001) and Opa1 by 1.4 fold (P = 0.04). CypD levels reduced by 1.5 fold (P = 0.03). These observations suggest that partial reduction of Drp1 increases fusion activity and reduces fission activity in the presence of mutant APP and Aβ in mice.
In double mutant mice relative to APP mice, mitochondrial biogenesis was increased PGC1α by 2.1 fold (P = 0.001), Nrf1 by 2.6 fold (P = 0.003), Nrf2 by 1.7 fold (P = 0.003) and TFAM by 2.1 fold (P = 0.001), indicating that reduced Drp1 enhances mitochondrial biogenesis activity (Table 2).
As shown in Table 2, mRNA levels of synaptic genes were significantly increased—synaptophysin by 2.1 fold (P = 0.001), PSD95 by 1.4 fold (P = 0.04), synapsin 1 by 2.1 fold (P = 0.001), synapsin 2 by 1.8 fold (P = 0.001), synaptobrevin by 1.6 fold (P = 0.02), synaptopodin 2.5 fold (P = 0.001) neurogranin by 1.4 fold (P = 0.04) and GAP43 by 2.2 fold (P = 0.002) (Table 2).
Immunoblotting analysis
To determine the protective effects of a partial reduction of Drp1 on mitochondrial and synaptic proteins, we quantified mitochondrial and synaptic proteins from cortical tissues of 6-month-old Drp1+/−, APP, APPXDrp1+/− and WT mice.
Drp+/− mice versus WT mice
In Drp1+/− mice relative to WT mice, significantly decreased protein levels were found for Drp1 (P = 0.002) and Fis1 (P = 0.01) (Fig. 1A and B) . In contrast, increased levels of mitochondrial fusion proteins, Mfn1, Mfn2 and Opa1 were found in Drp1+/− mice compared to WT mice, but not significant.
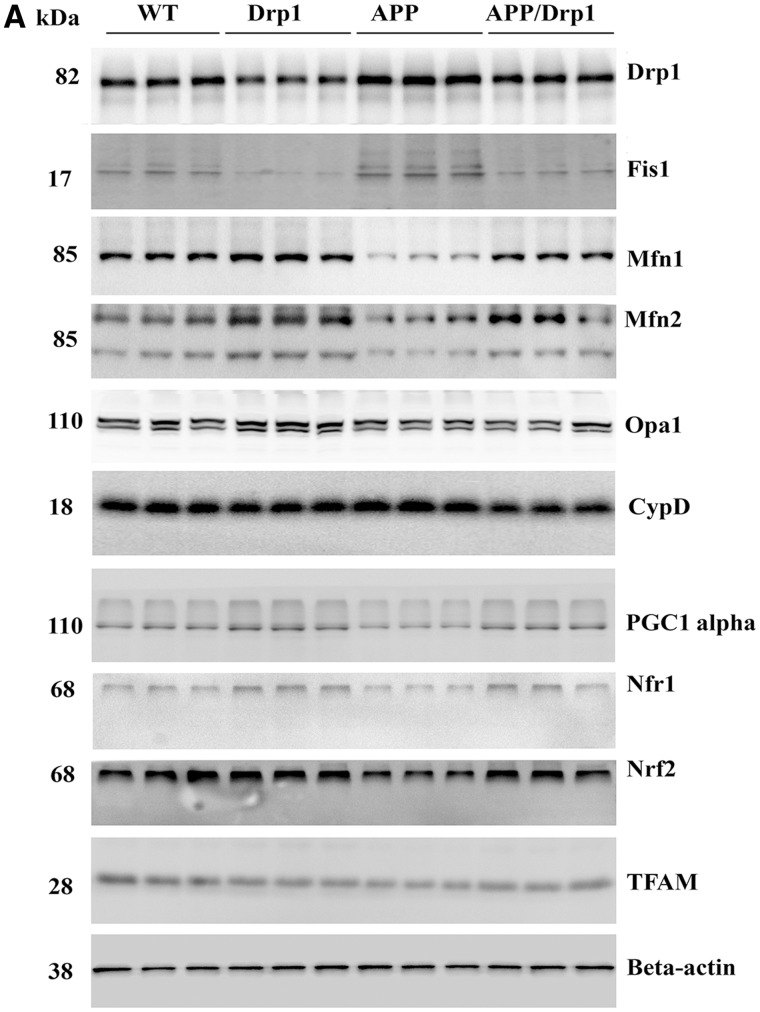
Figure 1.
Immunoblotting analysis of mitochondrial dynamics and biogenesis proteins. (A) Shows representative immunoblotting analysis of 6-month-old WT, Drp1+/−, APP and APPXDrp1+/− mice. (B) Shows quantitative densitometry analysis of mitochondrial dynamics proteins Drp1, Fis1, Mfn1, Mfn2, Opa1 and a matrix protein, CypD. (C) Shows quantitative densitometry analysis of mitochondrial biogenesis proteins PGC1α, Nrf1, Nrf1, and TFAM. The fission proteins Drp1 (P = 0.001) and Fis1 (P = 0.002) and matrix protein CypD (P = 0.01) were significantly increased; and the fusion proteins Mfn1 (P = 0.01), Mfn2 (P = 0.01), and Opa1 (P = 0.03) were significantly decreased in APP mice relative to WT mice, indicating the presence of abnormal mitochondrial dynamics. On the contrary, in APPXDrp1+/− mice relative to APP mice, the fission proteins Drp1 (P = 0.01) and Fis1 (P = 0.03) and matrix protein CypD (P = 0.01) were significantly decreased; and the fusion proteins Mfn1 (P = 0.04), Mfn2 (P = 0.01), and Opa1 (P = 0.04) were significantly increased, indicating that reduced Drp1 protects against mutant APP and Aβ-induced mitochondrial dynamics toxicity. The levels of mitochondrial biogenesis proteins PGC1α (P = 0.01), Nrf1 (P = 0.01), Nrf2 (P = 0.004) and TFAM (P = 0.001) were significantly decreased in APP mice relative to WT mice. In APPXDrp1+/− mice relative APP mice, biogenesis proteins PGC1α (P = 0.01), Nrf1 (P = 0.01), Nrf2 (P = 0.01) and TFAM (P = 0.01).
Mitochondrial biogenesis proteins (PGC1α, P = 0.01; Nrf2, P = 0.01) were increased in Drp1+/− mice relative to WT mice (Fig.1A and C).
Synaptic proteins, synaptophysin and PSD95 levels were increased in Drp1+/− mice relative to WT mice, but not significant (Fig. 2A and B).
Figure 2.
Immunoblotting analysis of full-length mutant APP, Aβ and synaptic proteins. (A) Represents immunoblotting analysis. (B) Represents quantitative immunoblotting analysis. Synaptophysin (P = 0.001) and PSD95 (P = 0.01) proteins were significantly decreased in APP mice relative to WT mice. On the contrary, synaptophysin (P = 0.002) and PSD95 (P = 0.01) proteins were increased in APPXDrp1+/− mice relative to APP mice. The levels of Full-length APP and Abeta were significantly decreased in APPXDrp1+/− mice relative to APP mice.
APP mice versus WT mice
In APP mice relative to WT mice, Drp1 (P = 0.001) and Fis1 (P = 0.002) proteins were increased (Fig. 1A and B) and fusion proteins Mfn1 (P = 0.01), Mfn2 (P = 0.01) and Opa1 (P = 0.03) were decreased and CypD was increased (P = 0.01). These observations agree with RNA data and suggest that the presence of abnormal mitochondrial dynamics.
Mitochondrial biogenesis proteins (PGC1α, P = 0.01, Nrf1, P = 0.01, Nrf2, P = 0.004 and TFAM, P = 0.001) were decreased in APP mice relative to WT mice (Fig.1A and C).
Synaptic proteins, synaptophysin (P = 0.001) and PSD95 (P = 0.01) were reduced in APP mice relative WT mice (Fig. 2A and B).
APPXDrp1+/− mice versus WT mice
In double mutant (APPXDrp1+/−) mice relative to WT mice, fission protein levels were unchanged for Drp1and Fis1 (Fig. 1A and B). Fusion proteins, Mfn1, Mfn2 and Opa1 were unchanged in APPXDrp1+/− mice compared to WT mice.
The levels of mitochondrial biogenesis proteins, PGC1α, Nrf1, Nrf2, and TFAM, were unchanged in APPXDrp1+/− mice relative to WT (Fig.1A and C).
Synaptic proteins, synaptophysin and PSD95 levels were unchanged in APPXDrp1+/− mice relative WT mice (Fig. 2A and B).
APP mice versus APPXDrp1+/− mice
Protein data were compared between APP mice and APPXDrp1+/− mice, in order to understand whether reduced Drp1 affects mutant APP and Aβ-induced mitochondrial and synaptic proteins. As shown in Fig. 1A and B, significantly reduced levels of fission proteins were found in APPXDrp1+/− mice (Drp1, P = 0.01; Fis1, P = 0.03) relative to APP mice. On the contrary, fusion proteins were increased in APPXDrp1+/− mice (Mfn1, P = 0.04; Mfn2, P = 0.01; Opa1, P = 0.04) relative to APP mice. CypD was decreased (P = 0.01) in APPXDrp1+/− mice relative to APP mice.
Mitochondrial biogenesis proteins were significantly increased in APPXDrp1+/− mice (PGC1α, P = 0.01; Nrf1, P = 0.01; Nrf2, P = 0.01 and TFAM, P = 0.01) and relative to APP mice (Fig. 1A and C), indicating that reduced Drp1 enhances mitochondrial biogenesis.
Synaptic proteins were increased in APPXDrp1+/− mice (synaptophysin, P = 0.002; PSD95, P = 0.01) relative to APP mice (Fig. 2A and B), indicating that reduced Drp1 enhances synaptic activity in APP mice.
Aβ in APP mice and APPXDrp1+/− mice
Full-length APP (P = 0.0001) and Aβ (P = 0.003) levels were significantly decreased in APPXDrp1+/− mice relative to APP mice, indicating that a partial reduction of Drp1 reduces Full-length mutant APP and Aβ.
Immunofluorescence analysis
Using immunofluorescence analysis, localizations and levels of mitochondrial dynamics (Drp1, Fis1—fission; Mfn1, Mfn2 and Opa1- fusion), mitochondrial biogenesis (PGC1α, Nrf1, Nrf2, TFAM) and synaptic proteins (synaptophysin and PSD95) were assessed in cortical sections from WT, Drp1+/−, APP and APPXDrp1+/− mice.
Drp1+/− mice versus WT mice
As shown in Fig. 3A and B, significantly decreased levels of Drp1 (P = 0.01) were found in Drp1+/− mice relative to WT mice, and increased levels of Mfn1, Mfn2 and Opa1were found, but not significant.
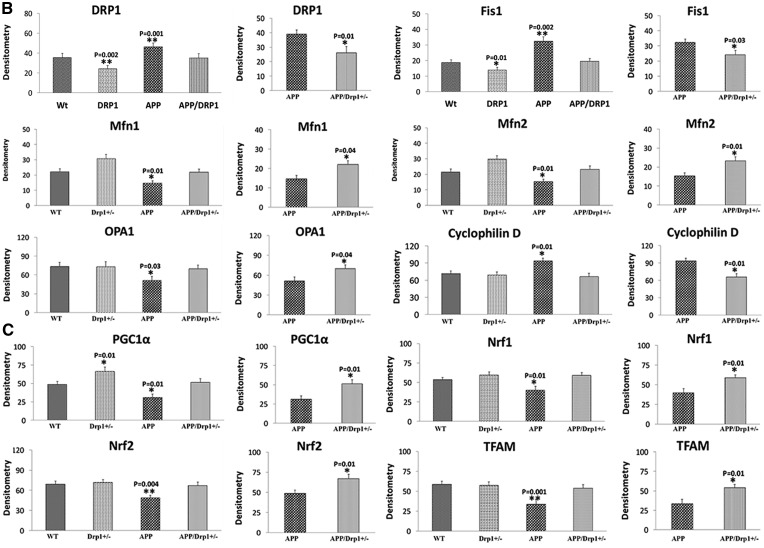
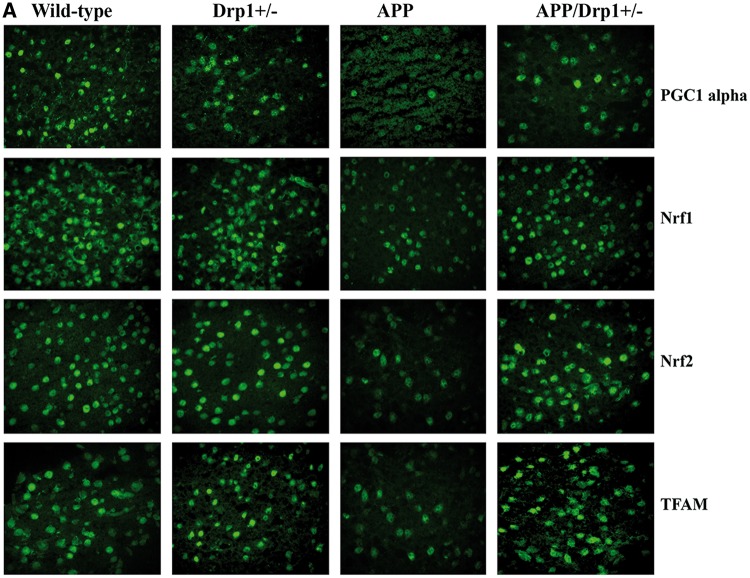
Figure 3.

Immunofluorescence analysis of mitochondrial dynamics proteins. (A) Represents immunofluorescence analysis. (B) Represents quantitative immunofluorescence analysis. The fission proteins Drp1 (P = 0.02) and Fis1 (P = 0.004) were significantly increased; and the fusion proteins Mfn1 (P = 0.04), Mfn2 (P = 0.01), and Opa1 (P = 0.01) were significantly decreased in APP mice relative to WT mice. On the contrary, the fission proteins Drp1 (P = 0.001) and Fis1 (P = 0.03) significantly decreased; and the fusion proteins Mfn1 (P = 0.02), Mfn2 (P = 0.01), and Opa1 (P = 0.02) were significantly increased APPXDrp1+/− mice relative to APP mice.
Mitochondrial biogenesis proteins (PGC1α, Nrf1, Nrf2, and TFAM) were increased in Drp1+/− mice relative to WT mice, but not significant (Fig. 4A and B).
Figure 4.
Immunofluorescence analysis of mitochondrial biogenesis proteins. (A) Represents immunofluorescence analysis. (B) Represents quantitative immunofluorescence analysis. Significantly reduced levels of PGC1α (P = 0.03), Nrf1 (P = 0.01), Nrf2 (P = 0.002) and TFAM (P = 0.04) were found in APP mice relative to WT mice. On the contrary, increased protein levels were found in PGC1α (P = 0.01), Nrf1 (P = 0.01), Nrf2 (P = 0.002) and TFAM (P = 0.02) in APPXDrp1+/− mice relative to APP mice.
Synaptic protein synaptophysin and PSD95 were increased in Drp1+/− mice relative WT mice, but not significant (Fig. 5A and B).
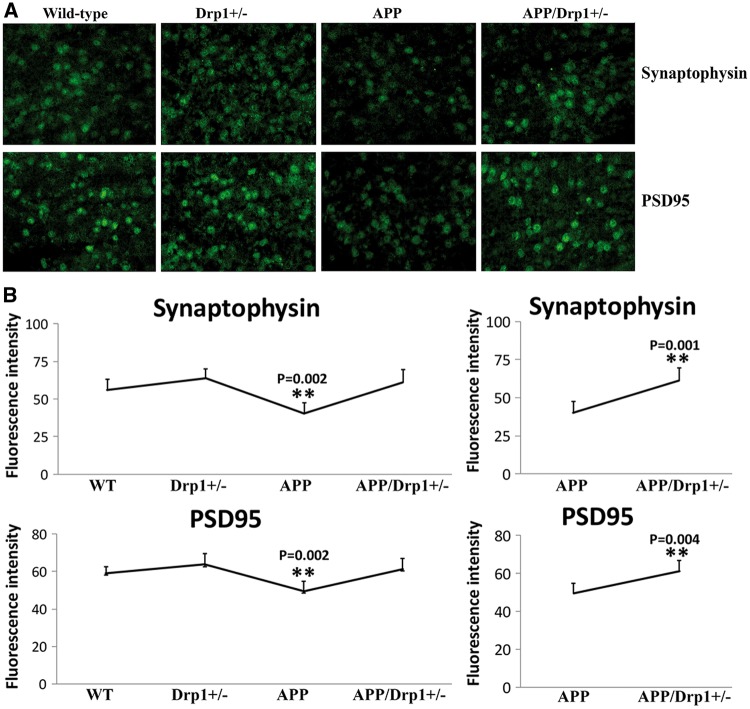
Figure 5.
Immunofluorescence analysis of synaptic proteins. (A) Represents immunofluorescence analysis. (B) Represents quantitative immunofluorescence analysis. Synaptic proteins, synaptophysin (P = 0.002) and PSD95 (P = 0.002) were significantly decreased in APP mice relative WT mice (Figure 5A and 5B). Synaptic proteins synaptophysin (P = 0.001) and PSD95 (P = 0.004) were significantly increased in APPXDrp1+/− mice relative APP mice (Figure 5A and 5B).
APP mice versus WT mice
Mitochondrial fission proteins – Drp1 (P = 0.02) and Fis1 (P = 0.004) were increased and fusion proteins Mfn1 (P = 0.04), Mfn2 (P = 0.01) and Opa1 (P = 0.01) were reduced in APP mice relative to WT mice (Fig. 3A and B).
Mitochondrial biogenesis proteins (PGC1α, P = 0.03; Nrf1, P = 0.01; Nrf2, P = 0.002 and TFAM, P = 0.04) were decreased in APP mice relative to WT mice (Fig. 4A and 4B).
Synaptic protein synaptophysin (P = 0.002) and PSD95 (P = 0.002) were significantly decreased in APP mice relative WT mice (Fig. 5A and 5B).
APPXDrp1+/− mice versus WT mice
As shown in Fig. 3A and B, mitochondrial fission proteins – Drp1 and Fis1 were increased and fusion proteins Mfn1, Mfn2 and Opa1 were reduced in APPXDrp1+/− mice relative to WT mice, but not significant (Fig. 3A and B).
Mitochondrial biogenesis proteins—PGC1α, Nrf1, Nrf2, and TFAM were increased in APPXDrp1+/− mice relative to WT mice (Fig. 4A and B).
Synaptic protein synaptophysin and PSD95 were increased in APPXDrp1+/− mice relative WT mice, but not significant (Fig. 5A and B).
APP mice versus APPXDrp1+/− mice
Immunofluorescence analysis was compared between APPXDrp1+/− mice and APP for mitochondrial dynamics, biogenesis and synaptic proteins. As shown in Fig. 3A and B, fission proteins Drp1 (P = 0.001) and Fis1 (P = 0.03) reduced and fusion proteins Mfn1 (P = 0.02), Mfn2 (P = 0.01) and Opa1 (P = 0.02) were reduced in APPXDrp1+/− mice relative to APP mice.
Mitochondrial biogenesis proteins PGC1α (P = 0.01), Nrf1 (P = 0.01), Nrf2 (P = 0.002) and TFAM (P = 0.02) were increased in APPXDrp1+/− mice relative to APP mice (Fig. 4A and B).
Synaptic protein synaptophysin (P = 0.001) and PSD95 (P = 0.004) were increased in APPXDrp1+/− mice relative to APP mice (Fig. 5A and B).
Full-length APP and Aβ in APP mice versus APPXDrp1+/− mice
Using 6E10 antibody, we also conducted immunofluorescence analysis in cortical brain sections. Mutant APP and Aβ levels were significantly reduced in APPXDrp1+/− mice relative to APP mice (P = 0.003) (Fig. 6A and B), indicating that a partial reduction of Drp1 reduces mutant APP and Aβ levels in APPXDrp1+/− mice.
Figure 6.

Immunofluorescence analysis of mutant APP and its derivatives in APP mice and APPXDrp1+/− mice. (A) Represents immunofluorescence analysis. (B) Represents quantitative immunofluorescence analysis. Immunoreactivity levels of mutant APP and its derivatives were significantly reduced in APPXDrp1+/− mice relative to APP mice (P = 0.003), indicating that a partial reduction of Drp1 reduces mutant APP and its derivatives in APP mice.
Soluble Aβ level in APP mice versus APPxDrp1+/− mice
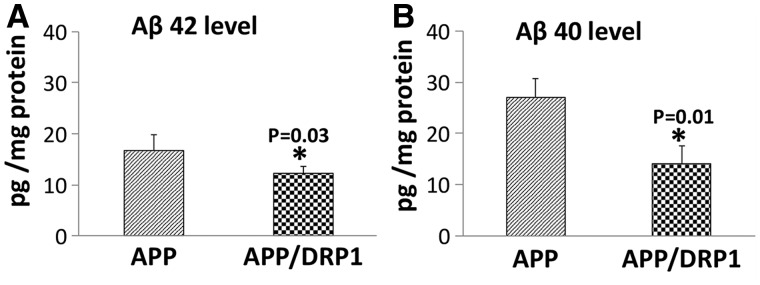
Using the sandwich ELISA method, we measured soluble Aβ42 and Aβ40 levels in 6-moth-old APP and double mutant (APPXDrp1+/−) mice. As shown in Fig. 7A and B, significantly reduced levels of Aβ42 (P = 0.03) and Aβ40 (P = 0.01) in 6-month-old APPXDrp1+/− mice relative to age-matched APP mice. These observations indicate that reduced Drp1 affects Aβ levels APPXDrp1+/− mice.
Figure 7.
Soluble amyloid beta levels in APP and APPXDrp1+/− mice. (A) Represents the levels of soluble Aβ42 and (B) Represents the levels of Aβ40 in APP and APPxDrp1+/− mice. Significantly reduced levels of Aβ42 (P = 0.03) and Aβ40 (P = 0.01) in 6-month-old APPXDrp1+/− mice relative to age-matched APP mice.
Mitochondrial function
Mitochondrial function was assessed in all four lines of 6-month-old mice by measure hydrogen peroxide, lipid peroxidation, cytochrome c oxidase activity, mitochondrial ATP, and GTPase Drp1 activity.
H2O2 production
As shown in Fig. 8A, significantly increased levels of hydrogen peroxide (H2O2) were found in APP mice relative to WT mice (P = 0.01). When the data were compared between APPXDrp1+/− mice with APP mice, H2O2 levels were significantly reduced (P = 0.02), indicating that a partial reduction of Drp1 reduces H2O2 levels in APP mice (Fig. 8A).
Figure 8.
Mitochondrial functional parameters in 6-month-old Drp1+/−, APP, APPXDrp1+/− and WT mice. Mitochondrial function was assessed by measuring: (A) H2O2 production, (B) lipid peroxidation, (C) Cytochrome oxidase activity (D) ATP levels, and (E) GTPase Drp1 activity.
Lipid peroxidation
Similar to hydrogen peroxide, levels of 4-hydroxy-2-nonenol, an indicator of lipid peroxidation were significantly increased in APP mice (P = 0.02) relative to WT mice (Fig. 8B).
Lipid peroxidation levels were significantly reduced in APPXDrp1+/− mice (P = 0.02) relative to APP mice, indicating that reduced Drp1 decreases lipid peroxidation levels in APP mice (Fig. 7B).
Cytochrome oxidase activity
Significantly decreased levels of cytochrome oxidase activity were found in APP mice (P = 0.04) relative to WT mice (Fig. 8C). Cytochrome oxidase activity levels were unchanged in Drp1+/− and APPXDrp1+/− mice relative to WT mice.
Cytochrome oxidase activity levels were increased in APPXDrp1+/− mice relative to APP mice (P = 0.03), indicating that a partial reduction of Drp1 increases cytochrome oxidase activity in APP mice.
ATP production
As shown in Fig. 8D, significantly decreased levels of mitochondrial ATP were found in APP mice relative to WT mice (P = 0.003). Mitochondrial ATP levels were unchanged in Drp1+/− and APPXDrp1+/− mice relative to WT mice.
Significantly increased ATP levels were found in APPXDrp1+/− mice (P = 0.02) relative to APP mice, indicating that reduced Drp1 increases ATP levels in APP mice.
GTPase Drp1 enzymatic activity
Significantly decreased levels of GTPase Drp1 activity were found Drp1+/− mice relative to WT mice (P = 0.01) (Fig. 8E). Interestingly, GTPase Drp1 activity levels were increased in APP mice relative to WT mice (P = 0.03), indicating increased mitochondrial fragmentation in APP mice.
Significantly decreased GTPase Drp1 activity levels were found in APPXDrp1+/− mice (P = 0.04) relative to APP mice (Fig. 8E), indicating that reduced Drp1 decreases GTPase Drp1 activity levels in APP mice.
Discussion
The objective of our study was to understand whether a partial reduction of Drp1 protects against mutant APP/Aβ-induced mitochondrial and synaptic toxicities in the progression of AD. In the current study, we used genetic approach and crossed Drp1 heterozygote knockout (Drp1+/−) mice with APP transgenic mice and created double mutant (APPXDrp1+/−) mice. Our purpose was to understand protective effects of reduced Drp1 in different stages (6, 12 and 20 months old) of disease process in APP transgenic mice. In the current paper, we characterized 6 months old mice. Using cortical tissues from 6-month-old Drp1+/−, APP, APPXDrp1+/− and WT mice, we measured mRNA, protein levels genes related to the mitochondrial dynamics, mitochondrial biogenesis and synapses. We also measured mitochondrial function and soluble Aβ in brain tissues from all lines of mice.
Decreased mRNA expressions and protein levels of fission and matrix genes, and increased levels of mitochondrial fusion, mitochondrial biogenesis and synaptic genes and proteins were found in 6-month-old APPXDrp1+/− mice relative to APP mice. Mitochondrial function was maintained in APPXDrp1+/− mice relative to APP mice. Sandwich ELISA assay revealed that soluble Aβ levels were significantly reduced in APPXDrp1+/− mice relative to APP mice. These findings suggest that a partial reduction of Drp1 reduces Aβ production, reduces mitochondrial dysfunction, and maintains mitochondrial dynamics, enhances mitochondrial biogenesis and synaptic activity in APP mice. These findings may have implications for the development of Drp1 based therapeutics for AD patients.
Aβ-Drp1’s role in mitochondrial fragmentation
Abnormal mitochondrial dynamics (increased fission and decreased fusion) is well documented in AD (12,13,21,41,42,49) and other neurodegenerative diseases (50–62). Further, it is also well documented that abnormal interaction between Aβ and Drp1 elevates the GTPase-Drp1 enzymatic activity and causes excessive fragmentation of mitochondria in mutant neurons. It has been suggested that either reduced levels of either Aβ or Drp1 might inhibit abnormal interaction between Aβ and Drp1, reduce mitochondrial fission activity and mitochondrial fragmentation, ultimately maintain mitochondrial function and synaptic activity in AD neurons.
There are several ways to reduce Drp1 levels, including pharmacological approach using molecular inhibitors such as mitochondrial division inhibitor (Mdivi1) and genetic approach by knocking down of Drp1 allele. In the current study, we used genetic approach, crossed Drp1+/− mice with APP mice and assessed the effects of reduced Drp1 levels on Aβ pathology, mitochondrial and synaptic activities in mice that produces Aβ.
Protective effects of reduced Drp1
Impaired mitochondrial dynamics—increased mRNA and protein levels of fission genes and decreased mRNA and protein levels of fusion genes—were found in 6-month-old APP mice, indicating mutant APP and Aβ causes impaired mitochondrial dynamics early on in disease process of AD. Significantly decreased mRNA and protein levels of mitochondrial biogenesis genes, PGC1α, Nrf1, Nrf2 and TFAM were observed in APP mice, indicating that mutant APP and Aβ reduces biogenesis activity in AD neurons. In addition, mRNA and protein levels were reduced for synaptic genes in APP mice, indicating that mutant APP and Aβ affects synaptic activity. Our study observations agree with earlier postmortem brain studies from AD patients and APP mouse models (21,41–42,63).
On the contrary, our study findings revealed that a partial reduction of Drp1 reduces fission machinery (by reducing Drp1 and Fis1 mRNA and protein levels) and enhances fusion activity (by increasing Mf1, Mfn2 and Opa1mRNA and protein levels) in neurons from WT mice. Further, mitochondrial biogenesis activity is elevated in Drp1+/− mice. In addition, increased levels of mRNA and proteins of synaptic genes in Drp1+/− mice. These observations strongly suggest that reduced Drp1 enhances mitochondrial fusion, biogenesis and synaptic activities in the brains WT mice. Based on these observations, it is concluded that a partial reduction of Drp1 is beneficial to neurons.
Further, we found reduced expression of fission genes and increased levels fusion genes and increased levels of mitochondrial biogenesis and synaptic genes in double mutant – APPXDrp1+/− mice relative to APP mice, indicating that reduced Drp1 is protective against mutant APP and Aβ-induced mitochondrial and synaptic toxicities. These observations agree with our hypothesis that a partial reduction of Drp1 protective against Aβ-induced mitochondrial and synaptic damage in AD neurons. We also expect similar findings in later time points, 12 and 20 months of age in APPXDrp1+/− mice relative to age-matched APP mice. Our study findings may have implications in other neurodegenerative diseases such as Huntington’s, Parkinson’s, ALS and multiple sclerosis, in which increased mitochondrial fragmentation is well documented.
Reduced abeta and full-length APP levels in APPXDrp1+/− mice
To understand the effect of reduced Drp1 on mutant APP and soluble Aβ levels in double mutant (APPXDrp1+/−) mice, we performed sandwich ELISA and immunoblotting analysis of cortical/hippocampal tissues from 6-month-old APP mice relative to age-matched APPXDrp1+/− mice. As shown in Fig. 2A and B, both full-length mutant APP and 4-kDa Aβ levels were significantly reduced in double mutant (APPXDrp1+/−) mice, strongly suggest that reduced Drp1 affects mutant APP and its derivatives. As shown in Fig. 7, our sandwich ELISA analysis revealed that significantly reduced levels of soluble Aβ42 and Aβ40 in double mutant (APPXDrp1+/−) mice relative to APP mice, indicating that reduced Drp1 levels decrease Aβ levels. Based on these observations, we strongly conclude that Drp1—and mutant APP/Aβ-induced synaptic (Figs 2A and B, 5A and B) and mitochondrial toxicities (Fig. 8A–E) are reduced in a disease process of AD.
Mitochondrial function
Mitochondria are synthesized at cell soma, travel along axons, dendrites and synapses, supply necessary ATP to multiple synaptic functions at nerve terminals, including neurotransmitter release, neurite outgrowth, synaptic vesicle fusion and synapse formation. Mitochondria constantly undergo fission and fusion, and alter their size and shape while travelling from the cell bodies to nerve terminals. In healthy neurons, fission and fusion are equally balanced and provide ATP to neurons. As extensively documented, in AD neurons, mitochondria are imbalanced – 1) may not be able to travel efficiently from the cell soma to nerve terminals and 2) may not be able to produce and supply sufficient quantities of ATP levels at synapses, ultimately leading to synaptic degeneration. It is possible that the abnormal interaction between Aβ and Drp1, forms complexes of Aβ-Drp1 and 1) blocks axonal transport of mitochondria and 2) causes mitochondrial dysfunction and synaptic damage in AD neurons. Recently, we (42) and others (12,30) reported that defective axonal mitochondrial transport, particularly anterograde moving mitochondria in AD neurons.
As discussed above, reduced Drp1 may inhibit Aβ and Drp1 interaction and increase/maintain normal axonal transport of mitochondria and produce and supply sufficient quantities of ATP in AD neurons, particularly at synapses.
As expected, in APP mice, mitochondrial function was found to be defective (Fig. 8). Our observations agree with others on mutant APP and Aβ-induced defective mitochondrial function (15,26,30,49) (Fig. 8).
On the other hand, mitochondrial function was increased in Drp1+/− mice, indicating that reduced Drp1 boosts and/or maintains mitochondrial function in neurons. As expected, mitochondrial function was increased in double mutant (APPXDrp1+/−) mice relative to APP mice (Fig. 8A–E)—meaning reduced Drp1 exhibited increased mitochondrial ATP, cytochrome oxidase activity and reduced free radicals and oxidative stress. These observations strongly suggest that reduced Drp1 reduces mutant APP and Aβ-induced cellular toxicity and boosts mitochondrial function and may promote neuronal longevity. Our mitochondrial functional data were consistent with gene expression and protein data. In the presence of mutant APP and Aβ, partial reduction of Drp1, reduces free radicals and lipid peroxidation, and increased mitochondrial ATP and cytochrome oxidase activity. Thus, all of our data point to reduced Drp1 protects neurons against Aβ-induced neuronal toxicity.
It is important to note that full-length mutant APP and Aβ levels are significantly decreased in double mutant mice relative to APP mice, further strengthening our hypothesis that mutant APP and Aβ are responsible for mitochondrial and synaptic toxicities in APP mice. In the current study, we found reduced Drp1 decreases mutant APP and Aβ-induced mitochondrial and synaptic toxicities in AD neurons.
It is also important to study 1) axonal transport of mitochondria and 2) the levels of Aβ interaction with Drp1 in APPXDrp1+/− mice relative to APP mice, in order to understand the role of reduced Drp1 in the axonal transport of mitochondria and Aβ-Drp1 interactions in neurons from double mutant mice. Our ongoing studies will provide new information about these questions.
In summary, our current study observations provided protective effects of reduced Drp1 against Abeta-induced mitochondrial and synaptic toxicities in APP mice and provided new evidence to develop Drp1 therapeutic strategies for AD. Ours is the first genetic crossing study to report the beneficial effects of reduced Drp1 in AD.
Methods
Mice and tissue preparation
To study the protective effects of partial deficiency of Drp1, we used Drp1 heterozygote knockout mice (gifted by Hiromi Sesaki, Johns Hopkins University) and APP mice (Tg2576 line) mice were purchased from Taconic Farms, New York and used crossbreeding licensing agreement and used for our experiments.
Amyloid beta precursor protein mice (popularly known as Tg2576 mice) were generated with the mutant human APP gene 695 amino acid isoform and a double mutation (Lys670Asn and Met671Leu) (Swedish mutation) (64). This highly expressed human APP transgenic mouse line exhibits an age-dependent appearance of Aβ plaques, as well as a distribution of the Aβ plaque confined to the cerebral cortex and the hippocampus. The Tg2576 mice show cognitive impairment 6 months of age and older. The human APP transgene is maintained in C57BL6/SJL backgrounds. To determine transgene-positive mice for human APP, genotyping will be performed, (in accordance with TTUHSC Policy on Genotype Tissue Collection) using the DNA prepared from tail biopsy and PCR amplification, as described in Hsiao et al., 1996 (64).
Dynamin-related protein 1 (or Drp1) heterozygote knockout (+/−) mice were generated using genetic a recombination strategy, replaced exons 3–5 of the GTPase domain, of the mouse endogenous Drp1 gene, with a neomycin-resistant gene (65). Homozygous Drp1 (−/−) knockout mice are embryonic-lethal and die by embryonic day 11.5. However, heterozygote Drp1 (+/−) mice are viable, fertile, normal in size, and do not show any phenotypic abnormalities. Heterozygous mice are maintained in a mixed background C57BL6/6-129/SvEv. To determine which mice are homozygous WT (+/+) or heterozygous (+/−) Drp1, we genotyped the mice, using the DNA prepared from tail biopsy and PCR amplification, as described in Wakabayashi et al. 2009 (62).
Double mutant (APPXDrp1+/−) mice were generated by genetic crossing Tg2576 mice with Drp1 +/− mice. We genotyped the Drp1+/−, and human mutant APP, as described in Hsiao et al. 1996 (61) and Wakabayashi et al. 2009 (65).
All the mice were observed daily by a veterinary caretaker and further examined twice a week by laboratory staff, and if animals showed premature signs of neurological deterioration, they were euthanized before experimentation according to the procedure for euthanasia approved by the TTUHSC-IACUC and will not be used in the proposed study.
Quantitative real-time RT-PCR
Using the reagent TriZol (Invitrogen), total RNA was isolated from cortical tissues from APP, Drp+/−, APPXDrp1+/− and WT mice. Using primer express Software (Applied Biosystems, Carlsbad, CA, USA), we designed the oligonucleotide primers for the housekeeping genes β-actin, GAPDH, mitochondrial structural genes, fission genes (Drp1, Fis1), fusion genes (MFN1, MFN2, Opa1), the mitochondrial matrix protein CypD, mitochondrial biogenesis genes PGC1α, Nrf1, Nrf2, TFAM and synaptic genes, synaptophysin, PSD95, synapsins1-2, synaptobrevins1-2, neurogranin, GAP43, and synaptopodin. The primer sequences and amplicon sizes are listed in Table 2. Using SYBR-Green chemistry-based quantitative real-time RT-PCR, we measured the mRNA expression of the above-mentioned genes, as described by Manczak and Reddy (2015) (66).
The mRNA transcript level was normalized against β-actin and the GAPDH at each dilution. The standard curve was the normalized mRNA transcript level, plotted against the log-value of the input cDNA concentration at each dilution. To compare β-actin, GAPDH, and neuroprotective markers, relative quantification was performed according to the CT method (Applied Biosystems). Briefly, the comparative CT method involved averaging triplicate samples, which were taken as the CT values for β-actin, GAPDH, and neuroprotective markers. β-actin normalization was used in the present study because the β-actin CT values were similar for the WT and Drp1+/−, APP and APPXDrp1+/− mice for the mitochondrial dynamics, mitochondrial biogenesis, and the synaptic genes. The ΔCT-value was obtained by subtracting the average β-actin CT value from the average CT-value of the synaptic mitochondrial ETC genes and the mitochondrial structural genes. The ΔCT of WT mice was used as the calibrator. The fold change was calculated according to the formula 2^ − (Δ ΔCT), where ΔΔCT is the difference between ΔCT and the ΔCT calibrator value. To determine the statistical significance of mRNA expression, the CT value differences between WT mice and other lines of mice were used in relation to β-actin normalization. Statistical significance was calculated using one-way ANOVA.
Immunoblotting analysis
To determine whether a partial deficiency of Drp1 alters the protein levels of mitochondrial dynamics, biogenesis and synaptic genes that showed altered mRNA expression, we performed immunoblotting analyses of protein lysates from APP, Drp1+/−, APPXDrp1+/− and WT mice as described in Manczak and Reddy, 2015 (63). Twenty μg protein lysates from cortical/hippocampal tissues of all lines mice were resolved on a 4–12% Nu-PAGE gel (Invitrogen). The resolved proteins were transferred to nylon membranes (Novax Inc., San Diego, CA, USA) and were then incubated for 1 hour at room temperature with a blocking buffer (5% dry milk dissolved in a TBST buffer). The PVDF membranes were incubated overnight with the primary antibodies shown in Table 3. The membranes were washed with a TBST buffer 3 times at 10-minute intervals and were then incubated for 2 hours with appropriate secondary antibodies, followed by 3 additional washes at 10-minute intervals. Mitochondrial and synaptic proteins were detected with chemilumniscence reagents (Pierce Biotechnology, Rockford, IL, USA), and the bands from immunoblots were quantified on a Kodak Scanner (ID Image Analysis Software, Kodak Digital Science, Kennesaw, GA, USA). Briefly, image analysis was used to analyze gel images captured with a Kodak Digital Science CD camera. The lanes were marked to define the positions and specific regions of the bands. An ID fine-band command was used to locate and to scan the bands in each lane and to record the readings.
Table 3.
Summary of quantitative real-time RT-PCR oligonucleotide primers used in measuring mRNA expression in mitochondrial dynamics and mitochondrial biogenesis and synaptic genes in 6 months old WT, Drp1+/−, APP and APPXDrp1+/− mice
| Gene | DNA Sequence (5’-3’) | PCR Product Size |
|---|---|---|
| Mitochondrial Dynamics Genes | ||
| Drp1 | Forward Primer ATGCCAGCAAGTCCACAGAA | 86 |
| Reverse Primer TGTTCTCGGGCAGACAGTTT | ||
| Fis1 | Forward Primer CAAAGAGGAACAGCGGGACT | 95 |
| Reverse Primer ACAGCCCTCGCACATACTTT | ||
| MFN1 | Forward Primer GCAGACAGCACATGGAGAGA | 83 |
| Reverse Primer GATCCGATTCCGAGCTTCCG | ||
| MFN2 | Forward Primer TGCACCGCCATATAGAGGAAG | 78 |
| Reverse Primer TCTGCAGTGAACTGGCAATG | ||
| Cyclophilin D | Forward Primer AGATGTCAAATTGGCAGGGGG | 91 |
| Reverse Primer TGCGCTTTTCGGTATAGTGCT | ||
| Opa1 | Forward Primer ACCTTGCCAGTTTAGCTCCC | 82 |
| Reverse Primer TTGGGACCTGCAGTGAAGAA | ||
| Mitochondrial Biogenesis genes | ||
| PGC1α | Forward Primer GCAGTCGCAACATGCTCAAG | 83 |
| Reverse Primer GGGAACCCTTGGGGTCATTT | ||
| Nrf1 | Forward Primer AGAAACGGAAACGGCCTCAT | 96 |
| Reverse Primer CATCCAACGTGGCTCTGAGT | ||
| Nrf2 | Forward Primer ATGGAGCAAGTTTGGCAGGA | 96 |
| Reverse Primer GCTGGGAACAGCGGTAGTAT | ||
| TFAM | Forward Primer TCCACAGAACAGCTACCCAA | 84 |
| Reverse Primer CCACAGGGCTGCAATTTTCC | ||
| Reverse Primer AGACGGTTGTTGATTAGGCGT | ||
| Synaptic genes | ||
| Synaptophysin | Forward Primer CTGCGTTAAAGGGGGCACTA | 81 |
| Reverse Primer ACAGCCACGGTGACAAAGAA | ||
| PSD95 | Forward Primer CTTCATCCTTGCTGGGGGTC | 90 |
| Reverse Primer TTGCGGAGGTCAACACCATT | ||
| Synapsin 1 | Forward Primer TGAGGACATCAGTGTCGGGTAA | 64 |
| Reverse Primer GGCAATCTGCTCAAGCATAGC | ||
| Synapsin 2 | Forward Primer TCCCACTCATTGAGCAGACATACT | 63 |
| Reverse Primer GGGAACGTAGGAAGCGTAAGC | ||
| Synaptobrevin 1 | Forward Primer TGCTGCCAAGCTAAAAAGGAA | 68 |
| Reverse Primer CAGATAGCTCCCAGCATGATCA | ||
| Neurogranin | Forward Primer CTCCAAGCCAGACGACGATA | 83 |
| Reverse Primer AACTCGCCTGGATTTTGGCT | ||
| GAP43 | Forward Primer GCTGCGACCAAAATTCAGGC | 83 |
| Reverse Primer GCTGGTGCATCACCCTTCT | ||
| Synaptopodin | Forward Primer TCCTGCGCCCTGAACCTA | 70 |
| Reverse Primer GACGGGCGACAGAGCATAGA | ||
| Housekeeping Genes | ||
| Beta Actin | Forward Primer AGAAGCTGTGCTATGTTGCTCTA | 91 |
| Reverse Primer TCAGGCAGCTCATAGCTCTTC | ||
| GAPDH | Forward Primer TTCCCGTTCAGCTCTGGG | 59 |
| Reverse Primer CCCTGCATCCACTGGTGC | ||
Immunofluorescence analysis and quantification
Immunofluorescence analysis was performed using midbrain sections from APP, Drp1+/−, APPXDrp1+/− and WT mice as described in Manczak and Reddy (2015) (63). The sections were washed with warm PBS, fixed in freshly prepared 4% paraformaldehyde in PBS for 10 minutes, and then washed with PBS and permeabilized with 0.1% Triton-X100 in PBS. They were blocked with a 1% blocking solution (Invitrogen) for 1 hour at room temperature. All sections were incubated overnight with primary antibodies (see Table 4). After incubation, the sections were washed 3 times with PBS, for 10 minutes each. The sections were incubated with a secondary antibody conjugated with Fluors 488 and 599 (Invitrogen) for 1 hour at room temperature. The sections were washed 3 times with PBS and mounted on slides. Photographs were taken with a multiphoton laser scanning microscope system (ZeissMeta LSM510). To quantify the immunoreactivity of mitochondrial and synaptic antibodies for each treatment, 10–15 photographs were taken at × 40 magnifications, and statistical significance was assessed, using one-way ANOVA for mitochondrial and synaptic and mitochondrial proteins.
Table 5.
Summary of antibody dilutions and conditions used in the immunohistochemistry/immunofluorescence analysis of mitochondrial dynamics, mitochondrial biogenesis and synaptic proteins in 6 months old WT, Drp1+/−. APP and APPXDrp1+/− mice.
| Marker | Primary antibody – species and dilution | Purchased from Company, State | Secondary antibody, dilution, Alexa fluor dye | Purchased from Company, City & State |
|---|---|---|---|---|
| Drp | Rabbit Polyclonal 1:300 | Novus Biological Littleton, CO | Donkey anti-Rabbit IgG Alexa Fluor 488 conjugate | Thermo Fisher Scientific Waltham, MA |
| Fis1 | Rabbit Polyclonal 1:300 | Protein Tech Group, Inc, Chicago, IL | Donkey anti-Rabbit IgG Alexa Fluor 488 conjugate | Thermo Fisher Scientific Waltham, MA |
| Mfn1 | Rabbit Polyclonal 1:300 | Protein Tech Group, Inc, Chicago, IL | Donkey anti-Rabbit IgG Alexa Fluor 488 conjugate | Thermo Fisher Scientific Waltham, MA |
| Mfn2 | Rabbit Polyclonal 1:200 | Protein Tech Group, Inc, Chicago, IL | Donkey anti-Rabbit IgG Alexa Fluor 488 conjugate | Thermo Fisher Scientific Waltham, MA |
| OPA1 | Rabbit Polyclonal 1:500 | Novus Biological Littleton, CO | Donkey anti-Rabbit IgG Alexa Fluor 488 conjugate | Thermo Fisher Scientific Waltham, MA |
| SYN | Rabbit Polyclonal 1:400 | Protein Tech Group, Inc, Chicago, IL | Donkey anti-Rabbit IgG Alexa Fluor 488 conjugate | Thermo Fisher Scientific Waltham, MA |
| PSD95 | Rabbit Polyclonal 1:400 | Protein Tech Group, Inc, Chicago, IL | Donkey anti-Rabbit IgG Alexa Fluor 488 conjugate | Thermo Fisher Scientific Waltham, MA |
| 6E10 | Mouse Monoclonal 1:400 | Biolegend, San Diego, CA | Donkey anti-Mouse IgG Alexa Fluor 594 conjugate | Thermo Fisher Scientific Waltham, MA |
Table 4.
Summary of antibody dilutions and conditions used in the immunoblotting analysis of mitochondrial dynamics, mitochondrial biogenesis and synaptic proteins in 6 months old WT, Drp1+/−, APP, and APPXDrp1 +/− mice.
| Marker | Primary antibody – species and dilution | Purchased from Company, State | Secondary antibody, dilution | Purchased from Company, City & State |
|---|---|---|---|---|
| Drp1 | Rabbit Polyclonal 1:500 | Novus Biological, Littleton, CO | Donkey anti-rabbit HRP 1:10,000 | GE Healthcare Amersham, Piscataway, NJ |
| Fis1 | Rabbit Polyclonal 1:500 | MBL International Corporation Woburn, Ma | Donkey anti-rabbit HRP 1:10,000 | GE Healthcare Amersham, Piscataway, NJ |
| Mfn1 | Rabbit Polyclonal 1:400 | Novus Biological Littleton, CO | Donkey anti-rabbit HRP 1:10,000 | GE Healthcare Amersham, Piscataway, NJ |
| Mfn2 | Rabbit Polyclonal 1:400 | Abcam Cambridge, MA | Donkey anti-rabbit HRP 1:10,000 | GE Healthcare Amersham, Piscataway, NJ |
| Opa1 | Rabbit Polyclonal 1:500 | Novus Biological Littleton, CO | Donkey anti-rabbit HRP 1:10,000 | GE Healthcare Amersham, Piscataway, NJ |
| CypD | Mouse Monoclonal 1:500 | EMD, Calobiochem Chemicals INC, Gibbstown, NJ | Sheep anti-mouse HR1:10,000 | GE Healthcare Amersham, Piscataway, NJ |
| SYN | Rabbit Monoclona 1:400 | Abcam Cambridge, MA | Donkey anti-rabbit HRP 1:10,000 | GE Healthcare Amersham Piscataway, NJ |
| PSD95 | Rabbit Monoclonal 1:300 | Abcam Cambridge, MA | Donkey anti-rabbit HRP 1:10,000 | GE Healthcare Amersham Piscataway, NJ |
| 6E10 | Mouse Monoclonal 1:500 | BioLegen San Diego, CA | Sheep anti-mouse HR 1:10,000 | GE Healthcare Amersham Piscataway, NJ |
| B-actin | Mouse Monoclonal 1:500 | Sigma-Aldrich St Luis, MO | Sheep anti-mouse HR 1:10,000 | GE Healthcare Amersham Piscataway, NJ |
Sandwich ELISA for soluble amyloid beta
Measurement of soluble aβ levels in APP and APPXDrp1+/− mice
The cerebral cortex of each mouse brain was snap-frozen on dry ice at the time of sacrifice and stored at −70°C until a homogenate was prepared. Briefly, protein cortical and hippocampal tissues from all four genotypes of mice were homogenized in a Tris-buffered saline (pH 8.0) containing protease inhibitors (20 mg/ml pepstatin A, aprotinin, phophsoramidon and leupeptin; 0.5 mMphenylmethanesulfonyl fluoride and 1 mM ethyleneglycol-bis(flaminoethyl ether)-NN tetraacetic acid). Samples were sonicated briefly and centrifuged at 10 000 g for 20 min at 4°C. The soluble fraction was used to determine the soluble Aβ by ELISA. For each sample, Aβ1-40 and Aβ1-42 were measured with commercial colorimetric ELISA kits (Biosource International, Camarillo, CA, USA) specific for each species. A 96-well plate reader was used, following the manufacturer's instructions. Each sample was run in duplicate. Protein concentrations of the homogenates were determined following the BSA method, and Aβ was expressed as pg Aβ/mg protein.
Mitochondrial functional assays
H2O2 production. Using an Amplex® Red H2O2 Assay Kit (Molecular Probes, Eugene, OR, USA), the production of H2O2 was measured using cortical tissues from APP, Drp1+/−, APPXDrp1+/− and WT mice as described in Manczak and Reddy, 2015 (66). Briefly, H2O2 production was measured in the mitochondria cortical tissues from all 4 lines of mice. A BCA Protein Assay Kit (Pierce Biotechnology) was used to estimate protein concentration. The reaction mixture contained mitochondrial proteins (μg/μl), Amplex Red reagents (50 μM), horseradish peroxidase (0.1 U/ml), and a reaction buffer (1X). The mixture was incubated at room temperature for 30 minutes, followed by spectrophotometer readings of fluorescence (570 nm). Finally, H2O2 production was determined, using a standard curve equation expressed in nmol/μg mitochondrial protein. Hydrogen peroxide levels were compared between WT mice with APP, Drp+/−, double mutant (APPXDrp1+/−) mice and data were also compared between APP mice versus APPXDrp1+/− mice.
Lipid peroxidation assay
Lipid peroxidates are unstable indicators of oxidative stress in the brain. The final product of lipid peroxidation is 4-hydroxy-2-nonenol (HNE), which was measured in the cell lysates prepared from cortical tissues from APP, Drp1+/−, APPXDrp1+/− and WT mice. We used HNE-His ELISA Kit (Cell BioLabs, Inc., San Diego, CA, USA) as described in Manczak and Reddy, 2015 (63). Briefly, freshly prepared protein as added to a 96-well protein binding plate and incubated overnight at 4°C. It was then washed 3 times with a buffer. After the last wash, the anti-HNE-His antibody was added to the protein in the wells, which was then incubated for 2 hours at room temperature and was washed again three times. Next, the samples were incubated with a secondary antibody conjugated with peroxidase for 2 hours at room temperature, followed by incubation with an enzyme substrate. Optical density was measured (at 450nm) to quantify the level of HNE. Lipid peroxidation levels were compared between WT mice with APP, Drp+/−, double mutant (APPXDrp1+/−) mice and data were also compared between APP versus APPXDrp1+/− mice.
Cytochrome oxidase activity
Cytochrome oxidase activity was measured in cortical tissues from all lines of mice. Enzyme activity was assayed spectrophotometrically using a Sigma Kit (Sigma–Aldrich) following manufacturer’s instructions (66). Briefly, 2 μg protein lysate was added to 1.1 ml of a reaction solution containing 50 μl 0.22 mM ferricytochrome c fully reduced by sodium hydrosulphide, Tris–HCl (pH 7.0), and 120 mM potassium chloride. The decrease in absorbance at 550 mM was recorded for 1-min reactions at 10-sec intervals. Cytochrome oxidase activity was measured according to the following formula: mU/mg total mitochondrial protein = (A/min sample – (A/min blank) × 1.1 mg protein × 21.84). The protein concentrations were determined following the BCA method. Cytochrome oxidase activity levels were compared between WT mice with APP, Drp+/−, double mutant (APPXDrp1+/−) mice and data were also compared between APP versus APPXDrp1+/− mice.
ATP levels
ATP levels were measured in mitochondria isolated from cortical tissues of APP, Drp1+/−, APPXDrp1+/− and WT mice and using an ATP determination kit (Molecular Probes) (66). The bioluminescence assay is based on the reaction of ATP with recombinant firefly luciferase and its substract luciferin. Luciferase catalyzes the formation of light from ATP and luciferin. It is the emitted light that is linearly related to the ATP concentration, which is measured with a luminometer. ATP levels were measured from mitochondrial pellets using a standard curve method. ATP levels were compared between WT mice with APP, Drp+/−, double mutant (APPXDrp1+/−) mice and data were also compared between APP versus APPXDrp1+/− mice.
GTPase Drp1 enzymatic activity
Using a calorimetric kit (Novus Biologicals, Littleton, CO, USA), GTPase enzymatic activity was measured in cortical tissues from APP, Drp1+/−, APPXDrp1+/− and WT mice following GTPase assay methods described in Manczak and Reddy, 2015, based on GTP hydrolyzing to GDP and to inorganic Pi (66). GTPase activity was measured, based on the amount of Pi that the GTP produces. By adding the ColorLock Gold (orange) substrate to the Pi generated from GTP, we assessed GTP activity, based on the inorganic complex solution (green). Colorimetric measurements (green) were read in the wavelength range of 650 nm. GTPase activity data were compared between WT mice with APP, Drp+/−, double mutant (APPXDrp1+/−) mice and data were also compared between APP mice versus APPXDrp1+/− mice.
The levels of H2O2 (P = 0.01) (Fig. 8A) and 4-hydroxy-2-nonenol (P = 0.02) (Fig. 8B) were significantly increased and the levels of cytochrome oxidase (P = 0.04) (Fig. 8C) and ATP (P = 0.003) (Fig. 7D) significantly decreased found in APP mice relative to WT mice.
The levels of H2O2 (P = 0.02) (Fig. 8A) and 4-hydroxy-2-nonenol (P = 0.02) (Fig. 8B) were significantly decreased and the levels of cytochrome oxidase (P = 0.03) (Fig. 8C) and ATP (P = 0.02) (Fig. 8D) significantly increased found in APPXDrp1+/− mice relative to APP mice.
The levels of GTPase Drp1 activity were significantly reduced in Drp1+/− mice (P = 0.01) and increased in APP mice (P = 0.03) relative to WT mice. In contrast, significantly decreased levels were found APPXDrp1+/− mice (P = 0.04) relative to APP mice (Fig. 8E).
Acknowledgements
We sincerely thank all the staff at the animal facility for taking care of all lines of mice that are used in the study.
Conflict of Interest statement. None declared.
Funding
Work presented in this article is supported by NIH grants AG042178, AG047812 and the Garrison Family Foundation (to PHR) and GM089853 (to HS).
References
- 1. Selkoe D.J. (2001) Alzheimer's disease: genes, proteins, and therapy. Physiol Rev, 81, 741–766. [DOI] [PubMed] [Google Scholar]
- 2. Reddy P.H., Manczak M., Mao P., Calkins M.J., Reddy A.P., Shirendeb U. (2010) Amyloid-beta and mitochondria in aging and Alzheimer's disease: implications for synaptic damage and cognitive decline. J. Alzheimers Dis., 20 Suppl 2, S499–S512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Mattson M.P. (2004) Pathways towards and away from Alzheimer's disease. Nature, 430, 631–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Reddy P.H., Tripathi R., Troung Q., Tirumala K., Reddy T.P., Anekonda V., Shirendeb U.P., Calkins M.J., Reddy A.P., Mao P., et al. (2012) Abnormal mitochondrial dynamics and synaptic degeneration as early events in Alzheimer's disease: implications to mitochondria-targeted antioxidant therapeutics. Biochim. Biophys. Acta, 1822, 639–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Querfurth H.W., LaFerla F.M. (2010) Alzheimer's disease. N. Engl. J. Med., 362, 329–344. [DOI] [PubMed] [Google Scholar]
- 6. Fang D., Wang Y., Zhang Z., Du H., Yan S., Sun Q., Zhong C., Wu L., Vangavaragu J.R., Yan S., et al. (2015) Increased neuronal PreP activity reduces Aβ accumulation attenuates neuroinflammation and improves mitochondrial and synaptic function in Alzheimer disease's mouse model. Hum. Mol. Genet., 24, 5198–5210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Fang D., Zhang Z., Li H., Yu Q., Douglas J.T., Bratasz A., Kuppusamy P., Yan S.S. (2016) Increased electron paramagnetic resonance signal correlates with mitochondrial dysfunction and oxidative stress in an Alzheimer's disease mouse brain. J. Alzheimers Dis., 51, 571–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Du H., Guo L., Fang F., Chen D., Sosunov A.A., McKhann G.M., Yan Y., Wang C., Zhang H., Molkentin J.D., et al. (2008) Cyclophilin D deficiency attenuates mitochondrial and neuronal perturbation and ameliorates learning and memory in Alzheimer's disease. Nat. Med., 14, 1097–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Jansen W.J., Ossenkoppele R., Knol D.L., Tijms B.M., Scheltens P., Verhey F.R., Visser P.J., Aalten P., Aarsland D., Alcolea D., et al. (2015) Prevalence of cerebral amyloid pathology in persons without dementia: a meta-analysis. JAMA, 313, 1924–1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Alzheimer’s Association(2015) – Facts and Figures, 1-81.
- 11. Swerdlow R.H., Khan S.M. (2004) A “mitochondrial cascade hypothesis” for sporadic Alzheimer's disease. Med. Hypotheses., 63, 8–20. [DOI] [PubMed] [Google Scholar]
- 12. Wang X., Su B., Siedlak S.L., Moreira P.I., Fujioka H., Wang Y., Casadesus G., Zhu X. (2008) Amyloid-beta overproduction causes abnormal mitochondrial dynamics via differential modulation of mitochondrial fission/fusion proteins. Proc. Natl Acad. Sci. U S A, 105, 19318–19323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wang X., Su B., Lee H.G., Li X., Perry G., Smith M.A., Zhu X. (2009) Impaired balance of mitochondrial fission and fusion in Alzheimer's disease. J. Neurosci., 29, 9090–9103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Reddy P.H. (2009) Amyloid beta, mitochondrial structural and functional dynamics in Alzheimer's disease. Exp. Neurol., 218, 286–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Manczak M., Anekonda T.S., Henson E., Park B.S., Quinn J., Reddy P.H. (2006) Mitochondria are a direct site of Abeta accumulation in Alzheimer's disease neurons: implications for free radical generation and oxidative damage in disease progression. Hum. Mol. Genet., 15, 1437–1449. [DOI] [PubMed] [Google Scholar]
- 16. Caspersen C., Wang N., Yao J., Sosunov A., Chen X., Lustbader J.W., Xu H.W., Stern D., McKhann G., Yan S.D. (2005) Mitochondrial Abeta: a potential focal point for neuronal metabolic dysfunction in Alzheimer's disease. Faseb. J., 19, 2040–2041. [DOI] [PubMed] [Google Scholar]
- 17. Devi L., Prabhu B.M., Galati D.F., Avadhani N.G., Anandatheerthavarada H.K. (2006) Accumulation of amyloid precursor protein in the mitochondrial import channels of human Alzheimer's disease brain is associated with mitochondrial dysfunction. J. Neurosci., 26, 9057–9068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hansson Petersen C.A., Alikhani N., Behbahani H., Wiehager B., Pavlov P.F., Alafuzoff I., Leinonen V., Ito A., Winblad B., Glaser E., et al. (2008) The amyloid beta-peptide is imported into mitochondria via the TOM import machinery and localized to mitochondrial cristae. Proc. Natl Acad. Sci. U S A, 105, 13145–13150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yao J., Irwin R.W., Zhao L., Nilsen J., Hamilton R.T., Brinton R.D. (2009) mitochondrial bioenergetic deficit precedes Alzheimer's pathology in female mouse model of Alzheimer's disease. Proc. Natl Acad. Sci. U S A, 106, 14670–14675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kandimalla R.J., Prabhakar S., Binukumar B.K., Wani W.Y., Gupta N., Sharma D.R., Sunkaria A., Grover V.K., Bhardwaj N., Jain K., et al. (2011) Apo-Eepsilon4 allele in conjunction with Abeta42 and tau in CSF: biomarker for Alzheimer's disease. Curr. Alzheimer Res., 8, 187–196. [DOI] [PubMed] [Google Scholar]
- 21. Manczak M., Mao P., Calkins M.J., Cornea A., Reddy A.P., Murphy M.P., Szeto H.H., Park B., Reddy P.H. (2010) Mitochondria-targeted antioxidants protect against amyloid-beta toxicity in Alzheimer's disease neurons. J. Alzheimers Dis., 20 Suppl 2, S609–S631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Butterfield D.A., Drake J., Pocernich C., Castegna A. (2001) Evidence of oxidative damage in Alzheimer's disease brain: central role for amyloid beta-peptide. Trends Mol. Med., 7, 548–554. [DOI] [PubMed] [Google Scholar]
- 23. Maurer I., Zierz S., Moller H.J. (2000) A selective defect of cytochrome c oxidase is present in brain of Alzheimer disease patients. Neurobiol. Aging, 21, 455–462. [DOI] [PubMed] [Google Scholar]
- 24. Smith M.A., Perry G., Richey P.L., Sayre L.M., Anderson V.E., Beal M.F., Kowall N. (1996) Oxidative damage in Alzheimer's. Nature, 382, 120–121. [DOI] [PubMed] [Google Scholar]
- 25. Gibson G.E., Sheu K.F., Blass J.P. (1998) Abnormalities of mitochondrial enzymes in Alzheimer disease. J. Neural. Transm. (Vienna), 105, 855–870. [DOI] [PubMed] [Google Scholar]
- 26. Reddy P.H., McWeeney S., Park B.S., Manczak M., Gutala R.V., Partovi D., Jung Y., Yau V., Searles R., Mori M., et al. (2004) Gene expression profiles of transcripts in amyloid precursor protein transgenic mice: up-regulation of mitochondrial metabolism and apoptotic genes is an early cellular change in Alzheimer's disease. Hum. Mol. Genet., 13, 1225–1240. [DOI] [PubMed] [Google Scholar]
- 27. Lustbader J.W., Cirilli M., Lin C., Xu H.W., Takuma K., Wang N., Caspersen C., Chen X., Pollak S., Chaney M., et al. (2004) ABAD directly links Abeta to mitochondrial toxicity in Alzheimer's disease. Science, 304, 448–452. [DOI] [PubMed] [Google Scholar]
- 28. Li F., Calingasan N.Y., Yu F., Mauck W.M., Toidze M., Almeida C.G., Takahashi R.H., Carlson G.A., Flint Beal M., Lin M.T., et al. (2004) Increased plaque burden in brains of APP mutant MnSOD heterozygous knockout mice. J. Neurochem., 89, 1308–1312. [DOI] [PubMed] [Google Scholar]
- 29. Eckert A., Hauptmann S., Scherping I., Rhein V., Muller-Spahn F., Gotz J., Muller W.E. (2008) Soluble beta-amyloid leads to mitochondrial defects in amyloid precursor protein and tau transgenic mice. Neurodegener. Dis., 5, 157–159. [DOI] [PubMed] [Google Scholar]
- 30. Du H., Guo L., Yan S., Sosunov A.A., McKhann G.M., Yan S.S. (2010) Early deficits in synaptic mitochondria in an Alzheimer's disease mouse model. Proc. Natl Acad. Sci. U S A, 107, 18670–18675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Parker W.D., Jr., Filley C.M., Parks J.K. (1990) Cytochrome oxidase deficiency in Alzheimer's disease. Neurology, 40, 1302–1303. [DOI] [PubMed] [Google Scholar]
- 32. Chandrasekaran K., Giordano T., Brady D.R., Stoll J., Martin L.J., Rapoport S.I. (1994) Impairment in mitochondrial cytochrome oxidase gene expression in Alzheimer disease. Brain Res. Mol. Brain Res., 24, 336–340. [DOI] [PubMed] [Google Scholar]
- 33. Chandrasekaran K., Hatanpaa K., Rapoport S.I., Brady D.R. (1997) Decreased expression of nuclear and mitochondrial DNA-encoded genes of oxidative phosphorylation in association neocortex in Alzheimer disease. Brain Res. Mol. Brain Res., 44, 99–104. [DOI] [PubMed] [Google Scholar]
- 34. Simonian N.A., Hyman B.T. (1994) Functional alterations in Alzheimer's disease: selective loss of mitochondrial-encoded cytochrome oxidase mRNA in the hippocampal formation. J Neuropathol. Exp. Neurol., 53, 508–512. [DOI] [PubMed] [Google Scholar]
- 35. Manczak M., Park B.S., Jung Y., Reddy P.H. (2004) Differential expression of oxidative phosphorylation genes in patients with Alzheimer's disease: implications for early mitochondrial dysfunction and oxidative damage. Neuromolecular Med., 5, 147–162. [DOI] [PubMed] [Google Scholar]
- 36. Johnson L.V., Leitner W.P., Rivest A.J., Staples M.K., Radeke M.J., Anderson D.H. (2002) The Alzheimer's A beta -peptide is deposited at sites of complement activation in pathologic deposits associated with aging and age-related macular degeneration. Proc. Natl Acad. Sci. U S A, 99, 11830–11835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Moreira P.I., Carvalho C., Zhu X., Smith M.A., Perry G. (2010) Mitochondrial dysfunction is a trigger of Alzheimer's disease pathophysiology. Biochim. Biophys. Acta, 1802, 2–10. [DOI] [PubMed] [Google Scholar]
- 38. Selwood S.P., Parvathy S., Cordell B., Ryan H.S., Oshidari F., Vincent V., Yesavage J., Lazzeroni L.C., Murphy G.M., Jr. (2009) Gene expression profile of the PDAPP mouse model for Alzheimer's disease with and without Apolipoprotein E. Neurobiol. Aging, 30, 574–590. [DOI] [PubMed] [Google Scholar]
- 39. Crouch P.J., Blake R., Duce J.A., Ciccotosto G.D., Li Q.X., Barnham K.J., Curtain C.C., Cherny R.A., Cappai R., Dyrks T., et al. (2005) Copper-dependent inhibition of human cytochrome c oxidase by a dimeric conformer of amyloid-beta1-42. J. Neurosci., 25, 672–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Falkevall A., Alikhani N., Bhushan S., Pavlov P.F., Busch K., Johnson K.A., Eneqvist T., Tjernberg L., Ankarcrona M., Glaser E. (2006) Degradation of the amyloid beta-protein by the novel mitochondrial peptidasome, PreP. J. Biol. Chem., 281, 29096–29104. [DOI] [PubMed] [Google Scholar]
- 41. Manczak M., Calkins M.J., Reddy P.H. (2011) impaired mitochondrial dynamics and abnormal interaction of amyloid beta with mitochondrial protein Drp1 in neurons from patients with Alzheimer's disease: implications for neuronal damage. Hum. Mol. Genet., 20, 2495–2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Calkins M.J., Manczak M., Mao P., Shirendeb U., Reddy P.H. (2011) Impaired mitochondrial biogenesis, defective axonal transport of mitochondria, abnormal mitochondrial dynamics and synaptic degeneration in a mouse model of Alzheimer's disease. Hum. Mol. Genet., 20, 4515–4529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Silva D.F., Selfridge J.E., Lu J., Cardoso S.M., Swerdlow R.H. (2013) Mitochondrial abnormalities in Alzheimer's disease: possible targets for therapeutic intervention. Adv. Pharmacol., 64, 83–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Reddy P.H., Reddy T.P., Manczak M., Calkins M.J., Shirendeb U., Mao P. (2011) Dynamin-related protein 1 and mitochondrial fragmentation in neurodegenerative diseases. Brain Res. Rev., 67, 103–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kandimalla R., Reddy P.H. (2016) Multiple faces of dynamin-related protein 1 and its role in Alzheimer's disease pathogenesis. Biochim. Biophys. Acta, 1862, 814–828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kageyama Y., Zhang Z., Sesaki H. (2011) Mitochondrial division: molecular machinery and physiological functions. Curr. Opin. Cell Biol., 23, 427–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Chen H., Chan D.C. (2009) Mitochondrial dynamics–fusion, fission, movement, and mitophagy–in neurodegenerative diseases. Hum. Mol. Genet., 18, R169–R176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Barsoum M.J., Yuan H., Gerencser A.A., Liot G., Kushnareva Y., Graber S., Kovacs I., Lee W.D., Waggoner J., Cui J., et al. (2006) Nitric oxide-induced mitochondrial fission is regulated by dynamin-related GTPases in neurons. EMBO J., 25, 3900–3911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Manczak M., Reddy P.H. (2012) Abnormal interaction between the mitochondrial fission protein Drp1 and hyperphosphorylated tau in Alzheimer's disease neurons: implications for mitochondrial dysfunction and neuronal damage. Hum. Mol. Genet., 21, 2538–2547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Shirendeb U., Reddy A.P., Manczak M., Calkins M.J., Mao P., Tagle D.A., Reddy P.H. (2011) Abnormal mitochondrial dynamics, mitochondrial loss and mutant huntingtin oligomers in Huntington's disease: implications for selective neuronal damage. Hum. Mol. Genet., 20, 1438–1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Shirendeb U.P., Calkins M.J., Manczak M., Anekonda V., Dufour B., McBride J.L., Mao P., Reddy P.H. (2012) Mutant huntingtin's interaction with mitochondrial protein Drp1 impairs mitochondrial biogenesis and causes defective axonal transport and synaptic degeneration in Huntington's disease. Hum. Mol. Genet., 21, 406–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Song W., Chen J., Petrilli A., Liot G., Klinglmayr E., Zhou Y., Poquiz P., Tjong J., Pouladi M.A., Hayden M.R., et al. (2013) Mutant huntingtin binds the mitochondrial fission GTPase dynamin-related protein-1 and increases its enzymatic activity. Nat. Med., 17, 377–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Kim J., Moody J.P., Edgerly C.K., Bordiuk O.L., Cormier K., Smith K., Beal M.F., Ferrante R.J. (2010) Mitochondrial loss, dysfunction and altered dynamics in Huntington's disease. Hum. Mol. Genet., 19, 3919–3935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Costa V., Giacomello M., Hudec R., Lopreiato R., Ermak G., Lim D., Malorni W., Davies K.J., Carafoli E., Scorrano L. (2010) Mitochondrial fission and cristae disruption increase the response of cell models of Huntington's disease to apoptotic stimuli. EMBO Mol. Med., 2, 490–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Song W., Song Y., Kincaid B., Bossy B., Bossy-Wetzel E. (2013) Mutant SOD1G93A triggers mitochondrial fragmentation in spinal cord motor neurons: neuroprotection by SIRT3 and PGC-1alpha. Neurobiol. Dis., 51, 72–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Magrane J., Sahawneh M.A., Przedborski S., Estevez A.G., Manfredi G. (2012) Mitochondrial dynamics and bioenergetic dysfunction is associated with synaptic alterations in mutant SOD1 motor neurons. J. Neurosci., 32, 229–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Magrane J., Cortez C., Gan W.B., Manfredi G. (2014) Abnormal mitochondrial transport and morphology are common pathological denominators in SOD1 and TDP43 ALS mouse models. Hum. Mol. Genet., 23, 1413–1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Wang X., Winter D., Ashrafi G., Schlehe J., Wong Y.L., Selkoe D., Rice S., Steen J., LaVoie M.J., Schwarz T.L. (2011) PINK1 and Parkin target Miro for phosphorylation and degradation to arrest mitochondrial motility. Cell, 147, 893–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Wang X., Yan M.H., Fujioka H., Liu J., Wilson-Delfosse A., Chen S.G., Perry G., Casadesus G., Zhu X. (2011) LRRK2 regulates mitochondrial dynamics and function through direct interaction with DLP1. Hum. Mol. Genet., 21, 1931–1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Binukumar B.K., Bal A., Kandimalla R.J., Gill K.D. (2010) Nigrostriatal neuronal death following chronic dichlorvos exposure: crosstalk between mitochondrial impairments, alpha synuclein aggregation, oxidative damage and behavioral changes. Mol. Brain, 3, 35–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Kamp F., Exner N., Lutz A.K., Wender N., Hegermann J., Brunner B., Nuscher B., Bartels T., Giese A., Beyer K., et al. (2010) Inhibition of mitochondrial fusion by alpha-synuclein is rescued by PINK1, Parkin and DJ-1. EMBO J., 29, 3571–3589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Xie H., Guan J., Borrelli L.A., Xu J., Serrano-Pozo A., Bacskai B.J. (2012) Mitochondrial alterations near amyloid plaques in an Alzheimer's disease mouse model. J. Neurosci., 33, 17042–17051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Cai Q., Tammineni P. (2016) Alterations in mitochondrial quality control in Alzheimer's disease. Front Cell Neurosci., 10, 24–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Hsiao K., Chapman P., Nilsen S., Eckman C., Harigaya Y., Younkin S., Yang F., Cole G. (1996) Correlative memory deficits, Abeta elevation, and amyloid plaques in transgenic mice. Science, 274, 99–102. [DOI] [PubMed] [Google Scholar]
- 65. Wakabayashi J., Zhang Z., Wakabayashi N., Tamura Y., Fukaya M., Kensler T.W., Iijima M., Sesaki H. (2009) The dynamin-related GTPase Drp1 is required for embryonic and brain development in mice. J. Cell Biol., 186, 805–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Manczak M., Reddy P.H. (2015) Mitochondrial division inhibitor 1 protects against mutant huntingtin-induced abnormal mitochondrial dynamics and neuronal damage in Huntington's disease. Hum. Mol. Genet., 24, 7308–7325. [DOI] [PMC free article] [PubMed] [Google Scholar]