Abstract
Genetic determinants of sleep-disordered breathing (SDB), a common set of disorders that contribute to significant cardiovascular and neuropsychiatric morbidity, are not clear. Overnight nocturnal oxygen saturation (SaO2) is a clinically relevant and easily measured indicator of SDB severity but its genetic contribution has never been studied. Our recent study suggests nocturnal SaO2 is heritable. We performed linkage analysis, association analysis and haplotype analysis of average nocturnal oxyhaemoglobin saturation in participants in the Cleveland Family Study (CFS), followed by gene-based association and additional tests in four independent samples. Linkage analysis identified a peak (LOD = 4.29) on chromosome 8p23. Follow-up association analysis identified two haplotypes in angiopoietin-2 (ANGPT2) that significantly contributed to the variation of SaO2 (P = 8 × 10−5) and accounted for a portion of the linkage evidence. Gene-based association analysis replicated the association of ANGPT2 and nocturnal SaO2. A rare missense SNP rs200291021 in ANGPT2 was associated with serum angiopoietin-2 level (P = 1.29 × 10−4), which was associated with SaO2 (P = 0.002). Our study provides the first evidence for the association of ANGPT2, a gene previously implicated in acute lung injury syndromes, with nocturnal SaO2, suggesting that this gene has a broad range of effects on gas exchange, including influencing oxygenation during sleep.
Introduction
Sleep disordered breathing (SDB), affects upwards of 17% and 9% of middle-aged and older men and women, respectively (1). A key consequence of SDB is overnight hypoxaemia, which has been shown to increase risk for numerous adverse health outcomes, including diabetes (2), atrial fibrillation (3), cancer (4) and cognitive decline (5). Development of pharmacological interventions for SDB, and its consequences such as hypoxaemia, has been limited by a lack of understanding of its aetiology. A genetic basis is supported by studies of the chief clinical metric of SDB, the apnea hypopnea index (AHI), which shows evidence of significant familial aggregation (6) and heritability estimates of 0.20 and 0.40 (7). However, studies of the molecular genetics of SDB have been limited to modest-sized association studies of candidate genes and have not yet demonstrated consistent findings (8–15).
Complex traits such as SDB require large samples to discover associations with common frequency variants with modest individual effects. A challenge in elucidating the genetic determinants of SDB is the cost and burden of characterizing overnight sleep-related respiratory phenotypes. Prior studies have focused on the AHI, which requires collection of multiple physiological signals during sleep, and only provides a ‘count’ of the number of respiratory disturbances without quantifying specific clinically important features, such as overnight hypoxaemia level. We observed that average nocturnal oxygen saturation (SaO2) is a highly heritable trait (16) and correlates with AHI. Therefore, we postulated that it may be useful for understanding the genetic basis of nocturnal hypoxaemia associated with SDB. Furthermore, since overnight SaO2 can be reliably and relatively easily measured with a pulse oximeter, it has the potential to be scalable in future large-scale genetic studies. Understanding the genetics of overnight SaO2 furthermore may help elucidate the bases for variation in levels of hypoxaemia across patients with SDB, which may explain differences in susceptibility to many SDB-related morbidities (2–5).
We performed linkage and association analyses of average SaO2 during sleep in the European Americans from Cleveland Family Study (CFS). Family-based cohort studies have multiple advantages compared to population based studies (such as GWAS). For example, family based linkage analysis is robust against allelic heterogeneity (17) and follow-up association analysis in any linkage regions may identify low frequent genetic variants segregating with a trait (18). We also utilized exome chip genotyping from independent community based samples for follow up replication and functional analysis of significant findings.
Results
Linkage analysis
The CFS included 669 European Americans in 139 families who were genotyped using the IBC chip (19). The characteristics of the CFS sample are summarized in Supplementary Material, Table S1. The mean family size is 4.8, with 381 full sibling pairs, 27 half sibling pairs and 502 parent offspring pairs. In this sample, 49.3% have at least mild or more severe sleep apnea (AHI > 5). Median average nocturnal SaO2 was 94 and is differed in men and women (mean = 93.3 vs 94.3, P = 2.5 × 10−4). (IQR: 93–96). Nocturnal SaO2 was strongly associated with AHI (ρ = −0.71) and modestly associated with lung function (percent predicted FVC and FEV1: ρ = 0.34 and 0.43, respectively; Supplementary Material, Table S2).
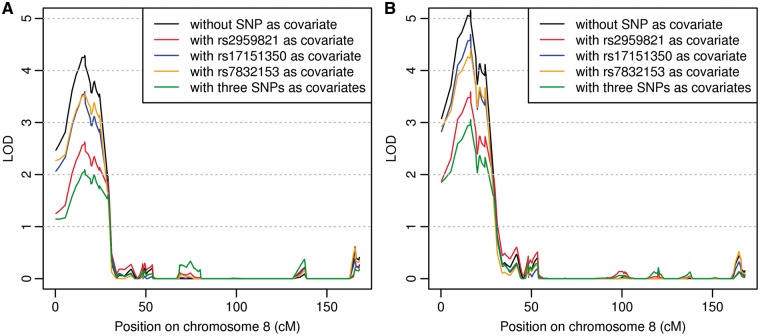
Average nocturnal SaO2 values were highly skewed and therefore transformed using the Box-Cox transformation to fit an approximate normal distribution conditional on covariates (Supplementary Material, Fig. S1). Covariates in the model included sex, age, age2, age × sex, BMI, BMI2, and the first 10 principal components (PCs) calculated from the ancestry informative markers (AIMs) using software FamCC (20) (Materials and Methods), and thus adjusted for trait differences associated with sex, age and BMI and the effect of population structure. The estimated heritability is 35.48% for the transformed average nocturnal SaO2 trait (37.5% in women and 12.83% in men). The most significant linkage peak (LOD = 4.29) was observed on 8p23 (Fig. 1A). A significant peak was observed on 14q21 (LOD = 3.72) and suggestive linkage evidence was observed on chromosomes 1, 3, 4, 5, 7 and 21 (LOD scores > 2). We also conducted linkage analysis without adjusting for BMI and the linkage patterns were very similar to that adjusting for BMI (Supplementary Material, Fig. S2 and Supplementary Material, Table S3). In particular the LOD score on 8p23 was 5.16 (Fig. 1B).
Figure 1.
Linkage analysis of average nocturnal SaO2 traits with and without SNPs as covariates on chromosome 8. (A) Adjusting for sex, age, age2, age × sex, BMI, BMI2, and 10 PCs. (B) Same as (A) but without including BMI and BMI2.
Due to the possibility that the LOD scores may be driven by a few outliers, we winsorized the trait value at four standard deviations (SDs) from the mean and repeated the linkage analysis. The LOD scores were essentially unchanged, including the largest LOD scores on 8p23 (Supplementary Material, Fig. S3 and Supplementary Material, Table S3). Thus, we only report results without eliminating extreme values in the subsequent analysis. Since the CFS families were recently genotyped with the Illumina OmniExpress array, we investigated whether a different set of genetic markers would result in different linkage results. We trimmed the Omni genotyped SNPs on chromosome 8 with minor allele frequencies > 0.2 and linkage disequilibrium (LD) < 0.1, which resulted in 1,719 SNPs for repeating the linkage analysis. Among the 1,719 variants, 29 SNPs overlapped with the original IBC array. The linkage evidence is essentially the same as that from the IBC array but with clearer bimodal peaks (two peaks on 8p23.2 and 8p23.1; Supplementary Material, Fig. S4). We repeated the linkage analysis using another random set of 855 pruned SNPs from the Illumina OmniExpress array (minor allele frequency > 0.2 and LD < 0.03). The two peaks are robust to the choice of SNPs.
Family-based association analyses
Below the linkage peak on 8p23, there are seven genes with 329 SNPs genotyped in the IBC array. Using the family-based association test and ASSOC software (21), we found that SNP rs2959821, located in two overlapping genes MCPH1 and ANGPT2, showed the most significant association with SaO2 (empirical P-value < 10−3, Supplementary Material, Table S4). Rs17151350 and rs7832153 showed marginal association evidence (asymptotic P -values < 10−3, Supplementary Material, Table S4). These three SNPs are not in LD (r2 < 0.1). Including all three top associated SNPs in one regression model resulted in virtually no change in the significance level for their association with nocturnal SaO2 (Supplementary Material, Table S5). To examine whether the top three SNPs accounted for the observed linkage evidence, we included the associated SNPs as covariates in the variance component model in linkage analysis. When rs2959821 was included, the maximum LOD score on 8p23 decreased to 2.63 (Table 1 and Fig. 1A), suggesting that the association of rs2959821 accounts for a portion of the observed linkage evidence (22,23). Including the other two SNPs did not result in a significant drop of LOD scores.
Table 1.
Comparison between combined linkage and association models with the baseline linkage model on 8p23
| Modela | SNP covariate | max LOD | Proportion of LOD dropped (%) |
|---|---|---|---|
| BMI adjusted | No SNP (baseline) | 4.287 | |
| rs2959821 | 2.626 | 38.7 | |
| rs17151350 | 3.594 | 16.2 | |
| rs7832153 | 3.570 | 16.7 | |
| 3 SNPs | 2.094 | 51.1 | |
| BMI unadjusted | No SNP (baseline) | 5.160 | |
| rs2959821 | 3.588 | 30.5 | |
| rs17151350 | 4.693 | 9.1 | |
| rs7832153 | 4.417 | 14.4 | |
| 3 SNPs | 3.053 | 40.8 |
aModels all adjusted for sex, age, age2, age × sex, and 10 PCs.
The minor allele of SNP rs2959821 is only present in two siblings from one CFS family. The linkage evidence on 8p23 is consistent for analyses that exclude this family and analyses that condition on rs2959821 (LOD = 2.82 after excluding this family, and LOD = 2.63 with conditional modelling on rs2959821; Supplementary Material, Fig. S5), suggesting that additional variants in this locus may contribute to the association. We therefore performed a haplotype association analysis of 11 SNPs (5 SNPs on each side of rs2959821). 39 haplotypes represented at least three times were analysed using ASSOC software (Supplementary Material, Table S6). We observed 7 haplotype associations (Table 2, P-value < 0.05). The most significantly associated haplotype H1 contains the rs2959821 minor allele and was present three times, including a parent whose haplotypes were inferred. The second most significant haplotype, H2, segregating in five families, has a frequency of 0.0087 and does not carry the rs2959821 minor allele. Since the small asymptotic P-values may be caused by the low frequencies of a SNP or haplotype, we performed simulations to obtain the empirical P-values. The empirical association P-values are 5.2 × 10−4 for H1 alone, and 8.0 × 10−5 for combining H1 and H2 based on 100,000 simulations (Materials and Methods). Furthermore, to assess whether the results could have been influenced by the sample distribution, we tested the association between H1 and the rank normal transformed SaO2 and found that the P-value is similar to the simulated empirical P-value. Subsequent linkage analysis conducted by either conditioning on both H1 and H2 or excluding the 6 families with the H1 or H2 haplotypes did not result in any substantial change in the LOD scores (Supplementary Material, Figs. S5 and S6), suggesting multiple variants under this linkage peak associate with nocturnal SaO2.
Table 2.
The significant haplotype of 11 SNPsa in ANGPT2 associated with nocturnal average SaO2 trait adjusting for sex, age, age2, age × sex, BMI, BMI2, and 10 PCs
| Haplotypea | No. of haplotype counts | Freq | beta | S.E. | P-value |
|---|---|---|---|---|---|
| CGCCAATATGG (H1) | 3 | 0.0019 | −1.169 | 0.176 | 5.2 × 10−4b |
| CGCTAGTATGG (H2) | 14 | 0.0087 | −0.271 | 0.076 | 7.16 × 10−3b |
| CTCCAGTATAG (H3) | 23 | 0.0143 | 0.133 | 0.056 | 0.018 |
| CTCTAGTATGG (H4) | 85 | 0.0529 | −0.067 | 0.030 | 0.028 |
| TTTCGGTATGG (H5) | 4 | 0.0025 | 0.302 | 0.141 | 0.034 |
| TGTCGGTATGG (H7) | 90 | 0.0560 | −0.065 | 0.031 | 0.034 |
| CTCCGGTACGG (H6) | 5 | 0.0031 | −0.269 | 0.135 | 0.046 |
aThe haplotype consists of the following SNPs: rs2922876, rs13269021, rs2922875, rs13255574, rs1988762, rs2959821, rs2442597, rs17077470, rs4478599, rs13250248, rs2959822.
bThe empirical P-values estimated from 100,000 simulations.
Linkage analyses adjusting for AHI and FVC
Because nocturnal hypoxaemia may relate to underlying lung function and/or to SDB, we included the AHI and FVC (percent predicted) as covariates in linkage analysis. With the inclusion of AHI (a common and direct metric of SDB), the linkage peak on 8p23 decreased to 2.54. In contrast, including FVC did not substantively change the linkage peak (Supplementary Material, Fig. S7A). We also performed linkage analysis on AHI and FVC adjusting sex, age, age2, age × sex, BMI, BMI2 and 10 PCs. We observed weak linkage evidence for AHI but not for FVC in that region (Supplementary Material, Fig. S7C). These results suggest that the linkage findings are more likely reflective of genetic variants associated with SDB rather than with forced expiratory lung volume.
Gene-level replication analyses
We performed gene-level rather than variant-level replication analyses due to the low frequency of the two haplotypes associated with nocturnal SaO2 in CFS (<1%) and because of the availability of an exome array or exome sequencing. These analyses used data from four independent community-based cohorts: 1,431 unrelated European Americans from the Atherosclerosis Risk in Communities (ARIC) Study who participated in the Sleep Heart Health Study (24); 701 unrelated European Americans from the Multi-Ethnic Study of Atherosclerosis (MESA) (25); 3,900 European Americans from Framingham Heart Study (FHS) third-generation (26); and 953 Mexican Americans in Starr County Health Study (27) (Supplementary Material, Table S7; Materials and Methods). Given differences in the available phenotype data, these data sets addressed different hypotheses (described below).
The gene-based association analysis of exonic variants in ANGPT2 and nocturnal SaO2 was conducted using software SKAT (Materials and Methods) in MESA and ARIC cohorts. The association evidence between nocturnal SaO2 and ANGPT2 was supported by the results (P-value = 0.029 adjusting for the same age, sex and BMI terms and 10PCs described earlier).
The genetic effect of ANGPT2 on nocturnal SaO2 was also observed in Mexican Americans from the Starr County Health Study in whom exome sequencing data were available. Samples with (N = 52) and without (N = 901) ANGPT2 deleterious mutations differed both in average nocturnal SaO2 (median [IQR]: 94.0 [93–96] vs 95.0 [94–96 IQR]; in samples with and without mutations, 2-sided t-test P = 0.040), as well as in average oxyhaemoglobin saturation level with SDB (apneas or hypopneas) events (91.5 [90–92] vs 92.0 [91–93], P = 0.029), despite similar age and BMI (P = 0.184 and 0.331, respectively).
To further investigate the connection between ANGPT2 and SDB traits, we tested the association between serum Ang-2 levels and SDB traits in 105 European Americans from CFS. Ang-2 level was negatively correlated with nocturnal SaO2 (ρ = −0.297, P = 0.002) and positively correlated with AHI (ρ = 0.218, P = 0.025), consistent with an effect of higher Ang-2 levels on more severe SDB (higher AHI) and SDB-related hypoxaemia (lower SaO2). Ang-2 differed in men and women (mean = 2.7 vs 3.3 pg/l, P = 0.028). The correlations decreased after adjusting for sex, age and BMI. In sex-stratified analysis, the correlation between Ang-2 level and nocturnal SaO2 was significant in males (ρ = -0.339, P = 0.028) but not in females (ρ = -0.042, P = 0.745) after adjusting for age and BMI.
We explored the effect of variants in ANGPT2 with Ang-2 serum levels in 3,900 participants from FHS third-generation. Linear regression identified a rare missense SNP, rs200291021 in ANGPT2 (minor allele frequency = 0.0004), that was associated with serum Ang-2 level (β = 2.611; P-value = 1.29 × 10−4). We also examined the three individuals who carry the rare allele. The Ang-2 levels of these three individuals are within 4 standard deviations from the mean, suggesting the association evidence is less likely driven by outliers. Since SaO2 was not available in FHS third generation data and we were not able to test the association between SaO2 and rs20029102 directly. We could not test for association of this variant in CFS because it was not genotyped.
Finally, we explored the associations of methylation of ANGPT2 in purified monocytes from 282 unrelated European Americans from MESA with sleep and methylation data (28). Significant association was observed and presented in Supplementary Material, Table S8.
Discussion
This study provides the first evidence identifying genetic associations for nocturnal SaO2, a clinically relevant and easily measured marker of SDB severity. The findings are based on complementary genetic analyses, including linkage analysis of family data, follow-up association studies, haplotype analysis, and follow-up gene-based association testing in two independent cohorts. Together, these analyses identify: a) a region on chromosome 8 likely harbouring causal variants; and b) two haplotypes in ANGPT2 that segregate within families and partly explain the linkage association. Gene-based tests in independent samples replicated ANGPT2, encoding angiopoietin-2 (Ang-2), an endothelial growth factor known to modulate vascular and inflammatory responses to lung injury (29), as the gene associated with nocturnal SaO2. Sequencing data from a Mexican American sample provided further evidence that deleterious mutations in ANGPT2 associated with both reduced average nocturnal oxyhaemoglobin levels and greater oxyhaemoglobin desaturation occurring with SDB (apneas and hypopneas) events. Associations among ANGPT2 variants and Ang-2 level with nocturnal SaO2 also provide consistent evidence implicating a potential causal association between ANGPT2 and nocturnal SaO2.
There has been growing interest in combining linkage analyses with association studies to enhance the power to detect rare variants that may segregate in families. Using this approach, we observed strong linkage evidence on 8p23 for average nocturnal SaO2. The linkage evidence, not driven by outliers nor was specific for a given set of genetic markers, is consistent with previous study of SDB traits (16). The linkage signal on chromosome 8 persisted in analyses the excluded individual families harbouring rare variants, suggesting that multiple genetic variants in this region contribute to SDB-related traits.
The IBC array includes variants in 7 genes under the peak of 8p23. Family-based association analysis identified that SNP rs2959821 in two overlapping genes (MCPH1 and ANGPT2) is significantly associated with average nocturnal SaO2. When adjusting for rs2959821, we observed a substantial drop of LOD score on 8p23, although the linkage evidence did not disappear, suggesting rs2959821 accounts for a portion of the observed linkage evidence (22,23). Since the minor allele of rs2959821 is rare and only segregates in one family, the asymptotic P-value could be inflated. However, excluding this family resulted in linkage evidence that was almost identical to the evidence obtained from analyses that were conditional on rs2959821. The simulations indicated that two haplotypes in ANGPT2 are significantly associated with the trait (P-value = 8 × 10−5).
The observed linkage evidence on 8p23 is more likely to be driven by multiple rare variants rather than common variants. If the linkage evidence is driven by common variants, we would expect such variants will be detected by large GWAS. Cade et al. conducted a GWAS meta-analysis of 21,055 samples from multi-ethnic groups with dense genotyping data. No variants on 8p23 were associated with average SaO2 at significance level 10−4 (Cade et al., manuscript in preparation). We further tested the trait association of 67 SNPs in ANGPT2 using recently obtained Illumina OmniExpress array. Besides rs2959821, two additional common SNPs (rs13250248 and rs11137037) have P-values < 0.01. However, including three SNPs together as covariates did not reduce the LOD score further, which supports this hypothesis. Follow-up association analyses in this region using whole genome/exome sequencing data are necessary to identify rare causal variants.
Because of the low frequencies of the variant and haplotypes detected in ANGPT2, power to replicate the associations at SNP or haplotype level is low. For example, the power for this variant using the estimated effect size is 0.28 under 0.05 significance level in MESA, and this power can also be overestimated because of the ‘winner’s curse’. In addition, rs2959821 is not available in the exome array data in MESA and ARIC. We have examined the LD between the exonic SNPs in ANGPT2 available in MESA and rs2959821 using the 1000 Genomes Project reference data. None of the exonic SNPs were in LD with rs2959821 (r2 < 0.01). Therefore, we performed gene level replication rather than SNP level replication. Gene-based association analysis using exome array data enriched with functional rare SNPs in independent ARIC and MESA data suggested that functional variants in ANGPT2 contribute to variation in average nocturnal SaO2. This association was further replicated in Mexican Americans from the Starr County Health Study by exome sequencing of ANGPT2 and demonstration that ANGPT2 mutations were associated with lower SaO2 levels.
We were able to examine data from exome arrays and from serum Ang-2 levels to further explore associations. The variants of ANGPT2 in the exome array showed evidence of association with serum Ang-2 levels. Furthermore, lower SaO2 levels were marginally associated with higher serum Ang-2 levels. Interestingly, this latter association was driven by findings in men, which reflects other research suggesting sex-specific differences in SDB (30). This aggregate evidence supports a potential causal role for ANGPT2 and Ang-2 in nocturnal SaO2.
Discovery of an association between ANGPT2 and sleep-related hypoxaemia is of particular interest due to the growing recognition of the role of angiopoietins in modulating lung responses to injury (29,31), including vascular responses to hyperoxia, sepsis and ventilator-induced injury (31,32). Ang-2 is stored in endothelial and pulmonary epithelial cells and is released into tracheal and alveolar epithelial fluid in patients with acute lung injury or those with hydrostatic pulmonary edema (33). In binding to Tie2, an endothelial cell receptor, Ang-2 antagonizes the angiopoietin Ang-1, destabilizing blood vessels and amplifying inflammation. Elevations of Ang-2 are predictive of which critically ill patients develop acute lung injury (34), predict mortality (35) and COPD (36). In the Framingham Heart Study, elevated Ang-2 levels and other markers of vascular remodeling also have been associated with hypertension and diabetes and have been shown to predict incident myocardial infarction (37). Elevated levels of Ang-2 have been implicated in all-cause and cardiovascular mortality (38), mortality associated with kidney disease (39), and heart failure-related morbidity (40). These effects support a central role that Ang-2 plays in stabilizing endothelial barrier function and modulating tissue inflammatory responses, including those that influence pulmonary and bronchial tissue, and vascular remodeling. Our analyses further suggest that higher levels of serum Ang-2 correlate with lower oxygen levels during sleep and higher AHI levels, thus, broadening the potential effects of this molecule to include SDB-related hypoxaemia.
Variants in ANGPT2 have been shown to predict which critically ill patients develop acute lung injury (41,42) and to influence plasma Ang-2 isoform levels (42). To our knowledge, neither Ang-2 nor ANGPT2 has been previously studied in relation to overnight hypoxaemia. The observed association between ANGPT2 and nocturnal SaO2 levels may reflect a causal mechanism by which variants in ANGPT2 influence vascular remodeling and/or inflammation in areas of the brain or airway that influence sleep-related ventilatory control and airway stability. Consistent with this, variations in inflammatory markers and genes have been associated with SDB (43). Alternatively, variants in ANGPT2 may influence nocturnal hypoxaemia through modulation of pulmonary vascular and inflammatory responses to mechanical or oxidative stress resulting from SDB. Studies in acute lung injury have shown that hypoxaemia and ventilator-induced mechanical stresses impact endothelial cell integrity, and that the degree of injury and resulting hypoxaemia correlate with Ang-2 levels (31,44). Oxidant-induced lung injury from exposure to hyperoxia appears to be mediated by Ang-2 (33). Although quantitatively different from acute lung injury, SDB-related stresses related to intermittent hypoxaemia and reoxygenation can result in oxidative stress as well as induce stretch-related injury associated with forced inspiration against a closed glottis. Overnight exposure to these stresses may influence pulmonary vasopermeability and gas exchange during sleep. Related mechanisms have been implicated in explaining associations between SDB and the lung injury found in pulmonary fibrosis (45,46), where obstructive apneas have been implicated in the release of inflammatory proteins (47). Genetic variation in susceptibility to these effects, possibly mediated by Ang-2 and related proteins, may explain the wide variation in level of hypoxaemia and subsequent cardiometabolic morbidity observed with SDB. Our results showing that variants in ANGPT2 are associated with both average nocturnal SaO2 as well as the degree of desaturation with apneas and hypopneas (in Starr County) and serum ANGPT2 levels support causal pathways. Although we did not have a sufficiently large sample to investigate sex-specific genetic associations, the strong associations between Ang-2 level and hypoxaemia in men compared to women is consistent with a large body of research demonstrating that SDB is more severe in men (48), and recent work showing sex differences in common genetic variants for SDB (30). Future studies of larger samples are needed to further investigate the role of genetic variants in mediating sex-specific differences in SDB and its morbidity.
Although prior studies have examined the genetic bases for hypoxaemia occurring in chronic obstructive lung disease, acute lung injury and high altitude, this is the first study to examine the influence of genetic factors on oxyhaemoglobin saturation levels during sleep. Each of these exposures may be considered a ‘stress’ that tests the adequacy of gas exchange in different adverse conditions. To our knowledge, significant associations with ANGPT2 and resting oxyhaemoglobin saturation have not been reported.
The study strengths include the use of complementary analytical strategies, analysis of data from independent samples and analysis of rare genetic variants. A number of sensitivity analyses and simulation studies were conducted to assess for consistency. The use of family data likely improved the power to detect rare variant associations despite the modest sample size. The study limitations include the limited data on serum markers and lack of expression data. In particular, Ang-2 serum levels were available only in a subset of CFS participants, resulting in low power of detecting the association with SaO2. However, we were able to observe the association between a functional variant and Ang-2 serum levels in FHS although without Ang-2 isoform level, which may better associate ANGPT2 variants than serum levels (49). Another limitation is that we could only replicate the association at a gene level instead of individual SNP level. However, replication at an individual SNP level is difficult for rare variants because of the need for very large samples.
In summary, we identified a linkage region on 8p23 that harbour multiple variants associated with average nocturnal oxyhaemoglobin saturation, a trait that correlates strongly with the major SDB phenotype (AHI) but which is measured more easily and provides information on overnight hypoxaemia, which predicts SDB-related morbidities. Complementary analyses provide consistent evidence that variants in ANGPT2 contribute to phenotypic variation in nocturnal SaO2. Future sequencing under the chromosome 8 peak is warranted to identify the causal variants. We identify the potential value of nocturnal SaO2 as a scalable and informative phenotype. Finally, these results also identify the potential importance of Ang-2, a vascular growth factor, in modulating levels of nocturnal hypoxaemia.
Materials and Methods
Samples and genotyping
The Cleveland Family Study (CFS)
The Cleveland Family Study (CFS) is the largest family-based longitudinal study comprised of index cases with laboratory diagnosed sleep apnea, their family members and neighborhood control families since 1990. Four examinations over 16 years included measurements of sleep apnea, anthropometry and other related phenotypes, as detailed previously (13,50). The CFS was genotyped by IBC array, which includes ∼50,000 SNPs in ∼2,000 candidate genes for cardiopulmonary, hematological and sleep-related disorders (19) and 1900 Ancestry Informative Markers (AIMs). 669 European Americans with available phenotypes and genotypes were included in the discovery analyses.
The Atherosclerosis Risk in Community (ARIC) study
The Atherosclerosis Risk in Community (ARIC) study was designed to capture the risk factors for cardiovascular outcomes in a community-based cohort comprised of individuals from Washington County MD, Forsyth County NC, Jackson MS and Minneapolis MN. The baseline examination of 15,792 individuals occurred from 1987 to 1989, with four additional examinations and ongoing follow-up. A European-American sample from the Maryland and Minnesota sites also participated in the baseline Sleep Heart Health Study (1994–1997). ARIC was genotyped using the Illumina HumanExome BeadChip (exome chip), designed to interrogate common and low-frequency coding variants across all genes in the genome. 1,431individuals have available phenotypes and genotypes were included in our supportive analyses.
The Multiethnic Study of Atherosclerosis (MESA)
The Multiethnic Study of Atherosclerosis (MESA) was designed to study the characteristics and risk factors of subclinical cardiovascular disease in multiple ethnic groups in community samples from Baltimore MD, Chicago IL, Los Angeles CA, New York NY, Minneapolis/St. Paul MN and Winston-Salem NC. The first examination of 6,429 individuals occurred in 2000, with four subsequent examinations and ongoing follow-up. 701 European Americans in association with MESA Exam 5 (2010 – 2013) were collected in this study. MESA was also genotyped using the HumanExome BeadChip (exome chip). Among those, 282 individuals have available methylation data obtained from purified monocytes and quantified by Illumina HumanMethylation450 BeadChip (28).
The Starr County Health Studies
The Starr County Health Studies have examined Type 2 diabetes and associated risk factors beginning in 1981 and continuing to the present day (27,51,52). As part of the T2D-GENES Consortium, whole exome sequencing was performed on 1616 unrelated samples from the Starr County Health Studies. Sequencing was performed using Agilent SureSelect All Exon Kit v.2 on Illumina HiSeq2000 instruments. Using Picard tools (http://picard.sourceforge.net), sequencing reads were processed and aligned to hg19 reference genome. The GATK HaplotypeCaller was used to call variant sites across 26000 samples that represent the T2D-GENES consortium as well as SIGMA LuCamp, and the Exome Sequencing Project (53). A total of 1,497 unrelated Starr County Health Study samples were successfully sequenced; however 7 failed QC metrics such as excess heterozygosity, excess non-reference alleles, or low GWAS chip concordance. Whole exome sequence data were therefore available for 1490 participants from Starr County Health Studies. Of this sample, 953 individuals have available phenotypes measured between 2010 and 2014. The whole exome sequencing data were annotated using SeattleSeq Annotations (138) (54), and 16 variants mapping to ANGPT2 with functional predictions based on NCBI gene models (including frameshift, missense, splice-donor and near-splice) were identified.
The Framingham Heart Study (FHS)
The Framingham Heart Study (FHS) was a long period cohort study aim to identify the risk factors of cardiovascular diseases in the town of Framingham MA since 1948. Greater phenotypic and genotypic resources were provided on the third generation of 4095 participants. The FHS study third generation samples were genotyped using the HumanExome BeadChip v1.0 (Illumina, Inc., San Diego, CA). The samples were processed at the Illumina Fast Track Services, and the genotyping was called using Illumina GenomeStudio v2011.1 software with the GenTrain 2.0 clustering algorithm as described by Grove et al. (55).
Participants in all samples provided written informed consent through the institutional review boards of each cohort. The Institutional Review Board at Partners HealthCare approved the current analysis.
Phenotyping
Polysomnography
A centralized sleep reading centre with robust quality control measures (56) scored all CFS, ARIC, MESA and Starr County Health Study records. Apneas were scored as nearly complete cessation of breathing for 10 or more seconds. Hypopneas were scored based on a 30% or more decrease in breathing amplitude for 10 or more seconds. The AHI was computed as the average number of apneas and hypopneas associated with a ≥3% desaturation per hour of sleep. The inter-class correlation coefficients for scoring the AHI exceed 0.94. Average nocturnal SaO2 was derived from the recorded SaO2 level during the sleep period, after excluding artefact. It was chosen as the primary phenotype given that it showed strongest linkage evidence in CFS and correlates highly with other SDB related phenotypes (Supplementary Material, Table S2).
In CFS, SDB was measured using either a Type 3 home sleep apnea monitor (SaO2, body position, airflow, respiratory effort and heart rate; Edentrace; Eden Prairie, MN) performed in participants studied prior to 2000 or by 14-channel attended overnight polysomnography (Compumedics E series, Abottsford, AU) after 2000. In ARIC, in-home polysomnography was performed using the 14-channel Compumedics P-Series monitor (Compumedics Ltd., Abbotsville, AU) (24, 56). In MESA, in-home polysomnography was performed using the 15-channel Compumedics Somte monitor (Compumedics Ltd., Abbotsville, AU)(25). Individuals using nightly CPAP, an oral device for sleep apnea, or overnight oxygen were excluded. In the Starr County Health Study, overnight SaO2 was derived from overnight pulse oximetry collected using the Watch-PAT device (Itamar Medical, IS). In addition, average oxyhaemoglobin saturation level with respiratory events identified by the Watch-PAT (57) were available.
Body mass index (BMI) was calculated based on weight and height measured at the same examination as the AHI. Spirometry was performed using a calibrated pneumotach-based spirometer while the participant was in the seated position. The adequacy of the blows was judged using the Epidemiologic Standardization Criteria of: a minimum duration of 6 seconds, back extrapolated volume < 5%, FVC within 5% or 200cc (whichever was less), and assessment by the technician as to the adequacy of the effort. Predicted values of forced expiratory volume in one second (FEV1) and forced vital capacity (FVC) in CFS were obtained from standardized spirometry and calculated using published equations (58).
Angiopoietin-2 (Ang-2) protein levels were assayed in fasting serum samples in 105 CFS participants with a commercial quantitative sandwich enzyme immunoassay that uses a monoclonal antibody specific for human Ang-2 (R&D systems, Minneapolis, MN; Catalog # DANG20). The detectable range y is approximately 234–15000 pg/ml with inter-assay coefficient of variation 2.05% to 4.19%. Selection was based on the availability of samples. In FHS, Serum Ang-2 was assayed using commercial ELISA assay according to the manufacturer’s protocol (R&D Systems, Minneapolis, MN) as previously described (37). The detectable range was between 328 and 21000 pg/ML with inter-assay coefficient of variation was 5.7%.
The methylation level of ANGPT2 in MESA was calculated as the average predicted methylation levels of all probes within that gene. Standard QC and normalization of the methylation data were performed using GenomeStudio and lumi package (59).
Box-cox transformation
Variance component tests require normality of the trait. Average nocturnal SaO2 values were highly skewed and therefore transformed using the Box-Cox transformation to fit an approximate normal distribution conditional on covariates (Supplementary Material, Fig. S1). The Box-Cox transformation is a nonlinear transformation of phenotype y with the general form:
where is the transformed value, and parameters c and g are used to control the scale and shape of the transformed distribution. A maximum likelihood algorithm was implemented to estimate the optimal , such that the residuals of regressed on covariates are normally distributed. We considered two covariate models. The first model contains gender, age, age2, age × sex, BMI and BMI2, and the first 10 principal components calculated from the ancestry informative markers (AIMs) using software FamCC, which is able to calculate PCs for family data (20). The second model does not include: BMI and BMI2. These two models are denoted as ‘BMI adjusted model’ and ‘BMI unadjusted model’, respectively.
Linkage analysis
Variance components linkage analysis was performed on 11,139 trimmed IBC array SNPs with minor allele frequency > 0.2 and linkage disequilibrium (LD) < 0.1 using the software package Merlin (60). In this analysis, the covariance matrix () of the transformed average nocturnal SaO2 residual in a pedigree is decomposed into three variance components: the variance due to the major quantitative trait locus (), the variance due to the random polygenetic effect (), and the variance due to the random environmental effect (),
The elements of the Π matrix are the identical by descent probabilities at the tested quantitative trait locus between two members i and j in a family. is the kinship matrix and is the identity matrix. In particular, the total variance of a trait is . The null hypothesis of no linkage is and the alternative hypothesis is . Merlin uses the likelihood ratio test to test the null hypothesis.
Family-based association analysis
A family based association analysis was performed in the genes under the linkage regions using IBC array SNPs in CFS. We conducted an association analysis using the software ASSOC in the S.A.G.E. package (21), which applied a linear mixed model: , where is the additive genotype value for the ith individual, and is the error term. For family members, follows a multivariate normal distribution . Our null hypothesis is . We reported the likelihood ratio test for testing the null hypothesis from ASSOC.
We performed combined linkage and association analysis by adding the top significant SNP as a covariate in linkage analysis and testing whether the LOD score declined after adding the SNP. We considered a 1 unit LOD score drop as an indication of linkage explained by an SNP (22,23).
We conducted the haplotype association analysis using 5 SNPs in both sides of the index SNP identified by linkage and association analyses. We inferred haplotypes using the software Merlin and performed family-based association analysis for each haplotype present at least three times in the data from CFS. We simulated haplotypes under the same pedigree structure 100,000 times to estimate the empirical significance to account for potential inflation induced by the asymptotic distributions of test statistics for low frequency haplotypes in ASSOC. In each simulation, we first simulated founders’ haplotypes according to estimated haplotype frequencies. For each parent, one of his/her haplotypes was modelled to be randomly transmitted to his/her offspring. We retained the same trait values and performed simulated haplotype association analysis 100,000 times.
Gene-based tests
We analysed the MESA and ARIC European American samples using the gene-based test SKAT-O (61), which optimize P-value from the test statistics as a combination of burden test and kernel test (SKAT). SKAT-O is most powerful when functional genetic variants have the same or opposite effect directions. We first performed the analysis in each cohort separately, and then meta-analysed the results using MetaSKAT (62). We used PolyPhen (63–65) scores to weight coding genetic variants using information on sequence and structure to infer function. Two covariate models, BMI adjusted model and BMI unadjusted model, defined in the same way in the linkage and association analyses in CFS were tested in this analysis.
Supplementary Material
Supplementary Material is available at HMG online.
Conflict of Interest statement. None declared.
Funding
The work was supported by the National Institutes of Health, the National Heart, Lung, Blood Institute grant HL113338, R01HL098433, HL46380, the National Human Genome Research Institute grant HG003054, and the Nation Science Foundation grant IIS-1218036. Provision of genotyping services is supported in part by NCATS CTSI grant UL1TR000124 and NIDDK DRC grant DK063491. The Atherosclerosis Risk in Communities (ARIC) Study is conducted and supported by the National Heart, Lung, and Blood Institute in collaboration with the University of North Carolina (N01-HC-55015, N01-HC-55018), Baylor College of Medicine (N01-HC-55016), University of Minnesota (N01-HC-55019), Johns Hopkins University (N01-HC-55020), and University of Mississippi Medical Center (N01-HC-55021). This manuscript was not prepared in collaboration with investigators of the ARIC Study and does not necessarily reflect the opinions or conclusions of the ARIC Study or the NHLBI. Multi-Ethnic Study of Atherosclerosis (MESA) is conducted and supported by the National Heart, Lung, and Blood Institute (NHLBI) in collaboration with MESA investigators. Support for MESA is provided by contracts N01-HC-95159, N01-HC-95160, N01-HC-95161, N01-HC-95162, N01-HC-95163, N01-HC-95164, N01-HC-95165, N01-HC-95166, N01-HC-95167, N01-HC-95168, N01-HC-95169 and CTSA UL1-RR-024156. The MESA epigenomes study is supported by 1R01HL101250. Starr County Health Studies (Starr) is supported in part by grants R01 DK073541, U01 DK085501, R01 AI085014, and R01 HL102830 from the National Institutes of Health, and funds from the University of Texas Health Science Center at Houston. The work was partially supported by the National Heart, Lung and Blood Institute's Framingham Heart Study (Contract No. N01-HC-25195 and HHSN268201500001I) and its contract with Affymetrix, Inc for genotyping services (Contract No. N02-HL-6-4278). A portion of this research utilized the Linux Cluster for Genetic Analysis (LinGA-II) funded by the Robert Dawson Evans Endowment of the Department of Medicine at Boston University School of Medicine and Boston Medical Center. This research was conducted in part using data and resources from the Framingham Heart Study of the National Heart Lung and Blood Institute of the National Institutes of Health and Boston University School of Medicine. The analyses reflect intellectual input and resource development from the Framingham Heart Study investigators participating in the SNP Health Association Resource (SHARe) project.
Supplementary Material
References
- 1. Peppard P.E., Young T., Barnet J.H., Palta M., Hagen E.W., Hla K.M. (2013) Increased prevalence of sleep-disordered breathing in adults. Am. J. Epidemiol., 177, 1006–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Punjabi N.M., Shahar E., Redline S., Gottlieb D.J., Givelber R., Resnick H.E. and Sleep Heart Health Study, I. (2004) Sleep-disordered breathing, glucose intolerance, and insulin resistance: the Sleep Heart Health Study. Am. J. Epidemiol., 160, 521–530. [DOI] [PubMed] [Google Scholar]
- 3. Kanagala R., Murali N.S., Friedman P.A., Ammash N.M., Gersh B.J., Ballman K.V., Shamsuzzaman A.S., Somers V.K. (2003) Obstructive sleep apnea and the recurrence of atrial fibrillation. Circulation, 107, 2589–2594. [DOI] [PubMed] [Google Scholar]
- 4. Nieto F.J., Peppard P.E., Young T., Finn L., Hla K.M., Farre R. (2012) Sleep-disordered breathing and cancer mortality: results from the Wisconsin Sleep Cohort Study. Am. J. Respir. Crit. Care Med., 186, 190–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Yaffe K., Laffan A.M., Harrison S.L., Redline S., Spira A.P., Ensrud K.E., Ancoli-Israel S., Stone K.L. (2011) Sleep-disordered breathing, hypoxia, and risk of mild cognitive impairment and dementia in older women. JAMA, 306, 613–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Redline S., Tosteson T., Tishler P.V., Carskadon M.A., Millman R.P. (1992) Studies in the genetics of obstructive sleep apnea. Familial aggregation of symptoms associated with sleep-related breathing disturbances. Am. Rev. Respir. Dis., 145, 440–444. [DOI] [PubMed] [Google Scholar]
- 7. Patel S.R., Ayas N.T., Malhotra M.R., White D.P., Schernhammer E.S., Speizer F.E., Stampfer M.J., Hu F.B. (2004) A prospective study of sleep duration and mortality risk in women. Sleep, 27, 440–444. [DOI] [PubMed] [Google Scholar]
- 8. Patel S.R., Goodloe R., De G., Kowgier M., Weng J., Buxbaum S.G., Cade B., Fulop T., Gharib S.A., Gottlieb D.J., et al. (2012) Association of genetic loci with sleep apnea in European Americans and African-Americans: the Candidate Gene Association Resource (CARe). PLoS One, 7, e48836.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chatsuriyawong S., Gozal D., Kheirandish-Gozal L., Bhattacharjee R., Khalyfa A.A., Wang Y., Sukhumsirichart W., Khalyfa A. (2013) Polymorphisms in nitric oxide synthase and endothelin genes among children with obstructive sleep apnea. BMC Med. Genomics, 6, 29.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhao Y., Tao L., Nie P., Lu X., Xu X., Chen J., Zhu M. (2013) Association between 5-HT2A receptor polymorphisms and risk of obstructive sleep apnea and hypopnea syndrome: a systematic review and meta-analysis. Gene, 530, 287–294. [DOI] [PubMed] [Google Scholar]
- 11. Qin B., Sun Z., Liang Y., Yang Z., Zhong R. (2014) The association of 5-HT2A, 5-HTT, and LEPR polymorphisms with obstructive sleep apnea syndrome: a systematic review and meta-analysis. PLoS One, 9, e95856.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Xu H., Guan J., Yi H., Yin S. (2014) A systematic review and meta-analysis of the association between serotonergic gene polymorphisms and obstructive sleep apnea syndrome. PLoS One, 9, e86460.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Larkin E.K., Patel S.R., Goodloe R.J., Li Y., Zhu X., Gray-McGuire C., Adams M.D., Redline S. (2010) A candidate gene study of obstructive sleep apnea in European Americans and African Americans. Am. J. Respir. Crit. Care Med., 182, 947–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Foley D.J., Masaki K., White L., Redline S. (2001) Relationship between apolipoprotein E epsilon4 and sleep-disordered breathing at different ages. JAMA, 286, 1447–1448. [DOI] [PubMed] [Google Scholar]
- 15. Gottlieb D.J., DeStefano A.L., Foley D.J., Mignot E., Redline S., Givelber R.J., Young T. (2004) APOE epsilon4 is associated with obstructive sleep apnea/hypopnea: the Sleep Heart Health Study. Neurology, 63, 664–668. [DOI] [PubMed] [Google Scholar]
- 16. Liang J., Cade B.E., Wang H., Chen H., Gleason K.J., Larkin E.K., Saxena R., Lin X., Redline S., Zhu X. (2016) Comparison of heritability estimation and linkage analysis for multiple traits using principal component analyses. Genet. Epidemiol., 40, 222–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zhu X., Feng T., Li Y., Lu Q., Elston R.C. (2010) Detecting rare variants for complex traits using family and unrelated data. Genet. Epidemiol., 34, 171–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bowden D.W., An S.S., Palmer N.D., Brown W.M., Norris J.M., Haffner S.M., Hawkins G.A., Guo X., Rotter J.I., Chen Y.D., et al. (2010) Molecular basis of a linkage peak: exome sequencing and family-based analysis identify a rare genetic variant in the ADIPOQ gene in the IRAS Family Study. Hum. Mol. Genet., 19, 4112–4120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Keating B.J., Tischfield S., Murray S.S., Bhangale T., Price T.S., Glessner J.T., Galver L., Barrett J.C., Grant S.F., Farlow D.N., et al. (2008) Concept, design and implementation of a cardiovascular gene-centric 50 k SNP array for large-scale genomic association studies. PloS One, 3, e3583.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhu X., Li S., Cooper R.S., Elston R.C. (2008) A unified association analysis approach for family and unrelated samples correcting for stratification. Am. J. Hum. Genet., 82, 352–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Elston R.C., Gray-McGuire C. (2004) A review of the ‘Statistical Analysis for Genetic Epidemiology’ (S.A.G.E.) software package. Hum. Genomics, 1, 456–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Martin L.J., Ding L., Zhang X., Kissebah A.H., Olivier M., Benson D.W. (2014) A novel method, the Variant Impact On Linkage Effect Test (VIOLET), leads to improved identification of causal variants in linkage regions. Eur. J. Hum. Genet., 22, 243–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Almasy L., Blangero J. (2004) Exploring positional candidate genes: linkage conditional on measured genotype. Behav. Genet., 34, 173–177. [DOI] [PubMed] [Google Scholar]
- 24. Lind B.K., Goodwin J.L., Hill J.G., Ali T., Redline S., Quan S.F. (2003) Recruitment of healthy adults into a study of overnight sleep monitoring in the home: experience of the Sleep Heart Health Study. Sleep Breath, 7, 13–24. [DOI] [PubMed] [Google Scholar]
- 25. Chen X., Wang R., Zee P., Lutsey P.L., Javaheri S., Alcantara C., Jackson C.L., Williams M.A., Redline S. (2015) Racial/Ethnic Differences in Sleep Disturbances: The Multi-Ethnic Study of Atherosclerosis (MESA). Sleep, 38, 877–888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Splansky G.L., Corey D., Yang Q., Atwood L.D., Cupples L.A., Benjamin E.J., D'Agostino R.B., Sr., Fox C.S., Larson M.G., Murabito J.M., et al. (2007) The Third Generation Cohort of the National Heart, Lung, and Blood Institute's Framingham Heart Study: design, recruitment, and initial examination. Am. J. Epidemiol., 165, 1328–1335. [DOI] [PubMed] [Google Scholar]
- 27. Hallman D.M., Huber J.C., Jr., Gonzalez V.H., Klein B.E., Klein R., Hanis C.L. (2005) Familial aggregation of severity of diabetic retinopathy in Mexican Americans from Starr County, Texas. Diabetes Care, 28, 1163–1168. [DOI] [PubMed] [Google Scholar]
- 28. Liu Y., Ding J., Reynolds L.M., Lohman K., Register T.C., De La Fuente A., Howard T.D., Hawkins G.A., Cui W., Morris J., et al. (2013) Methylomics of gene expression in human monocytes. Hum. Mol. Genet., 22, 5065–5074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. van der Heijden M., van Nieuw Amerongen G.P., Chedamni S., van Hinsbergh V.W., Johan Groeneveld A.B. (2009) The angiopoietin-Tie2 system as a therapeutic target in sepsis and acute lung injury. Expert Opin. Ther. Targets, 13, 39–53. [DOI] [PubMed] [Google Scholar]
- 30. Cade B.E., Chen H., Stilp A.M., Gleason K.J., Sofer T., Ancoli-Israel S., Arens R., Bell G.I., Below J.E., Bjonnes A.C., et al. (2016) Genetic associations with obstructive sleep apnea traits in Hispanic/Latino Americans. Am. J. Respir. Crit. Care Med., 194, 886–897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Gallagher D.C., Parikh S.M., Balonov K., Miller A., Gautam S., Talmor D., Sukhatme V.P. (2008) Circulating angiopoietin 2 correlates with mortality in a surgical population with acute lung injury/adult respiratory distress syndrome. Shock, 29, 656–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. van der Heijden M., van Nieuw Amerongen G.P., Koolwijk P., van Hinsbergh V.W., Groeneveld A.B. (2008) Angiopoietin-2, permeability oedema, occurrence and severity of ALI/ARDS in septic and non-septic critically ill patients. Thorax, 63, 903–909. [DOI] [PubMed] [Google Scholar]
- 33. Bhandari V., Choo-Wing R., Lee C.G., Zhu Z., Nedrelow J.H., Chupp G.L., Zhang X., Matthay M.A., Ware L.B., Homer R.J., et al. (2006) Hyperoxia causes angiopoietin 2-mediated acute lung injury and necrotic cell death. Nat. Med., 12, 1286–1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Agrawal A., Matthay M.A., Kangelaris K.N., Stein J., Chu J.C., Imp B.M., Cortez A., Abbott J., Liu K.D., Calfee C.S. (2013) Plasma angiopoietin-2 predicts the onset of acute lung injury in critically ill patients. Am. J. Respir. Crit. Care Med., 187, 736–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Tseliou E., Bakakos P., Kostikas K., Hillas G., Mantzouranis K., Emmanouil P., Simoes D., Alchanatis M., Papiris S., Loukides S. (2012) Increased levels of angiopoietins 1 and 2 in sputum supernatant in severe refractory asthma. Allergy, 67, 396–402. [DOI] [PubMed] [Google Scholar]
- 36. Bessa V., Loukides S., Hillas G., Delimpoura V., Simoes D., Kontogianni K., Papiris S., Kostikas K., Alchanatis M., Bakakos P. (2012) Levels of angiopoietins 1 and 2 in induced sputum supernatant in patients with COPD. Cytokine, 58, 455–460. [DOI] [PubMed] [Google Scholar]
- 37. Lieb W., Zachariah J.P., Xanthakis V., Safa R., Chen M.H., Sullivan L.M., Larson M.G., Smith H.M., Yang Q., Mitchell G.F., et al. (2010) Clinical and genetic correlates of circulating angiopoietin-2 and soluble Tie-2 in the community. Circ. Cardiovasc. Genet., 3, 300–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lorbeer R., Baumeister S.E., Dorr M., Nauck M., Grotevendt A., Volzke H., Vasan R.S., Wallaschofski H., Lieb W. (2013) Circulating angiopoietin-2, its soluble receptor Tie-2, and mortality in the general population. Eur. J. Heart Fail., 15, 1327–1334. [DOI] [PubMed] [Google Scholar]
- 39. David S., John S.G., Jefferies H.J., Sigrist M.K., Kumpers P., Kielstein J.T., Haller H., McIntyre C.W. (2012) Angiopoietin-2 levels predict mortality in CKD patients. Nephrol. Dial. Transplant, 27, 1867–1872. [DOI] [PubMed] [Google Scholar]
- 40. Poss J., Ukena C., Kindermann I., Ehrlich P., Fuernau G., Ewen S., Mahfoud F., Kriechbaum S., Bohm M., Link A. (2014) Angiopoietin-2 and outcome in patients with acute decompensated heart failure. Clin. Res. Cardiol., 4:380–387. [DOI] [PubMed] [Google Scholar]
- 41. Su L., Zhai R., Sheu C.C., Gallagher D.C., Gong M.N., Tejera P., Thompson B.T., Christiani D.C. (2009) Genetic variants in the angiopoietin-2 gene are associated with increased risk of ARDS. Intensive Care Med., 35, 1024–1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Meyer N.J., Li M., Feng R., Bradfield J., Gallop R., Bellamy S., Fuchs B.D., Lanken P.N., Albelda S.M., Rushefski M., et al. (2011) ANGPT2 genetic variant is associated with trauma-associated acute lung injury and altered plasma angiopoietin-2 isoform ratio. Am. J. Respir. Crit. Care Med., 183, 1344–1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Larkin E.K., Patel S.R., Zhu X., Tracy R.P., Jenny N.S., Reiner A.P., Walston J., Redline S. (2010) Study of the relationship between the interleukin-6 gene and obstructive sleep apnea. Clin. Transl Sci., 3, 337–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Parikh S.M., Mammoto T., Schultz A., Yuan H.T., Christiani D., Karumanchi S.A., Sukhatme V.P. (2006) Excess circulating angiopoietin-2 may contribute to pulmonary vascular leak in sepsis in humans. PLoS Med., 3, e46.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lederer D.J., Jelic S., Bhattacharya J., Basner R.C. (2012) Is obstructive sleep apnea a cause of idiopathic pulmonary fibrosis?. Arch. Pathol. Lab. Med., 136, 470. author reply 470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Leslie K.O. (2012) Idiopathic pulmonary fibrosis may be a disease of recurrent, tractional injury to the periphery of the aging lung: a unifying hypothesis regarding etiology and pathogenesis. Arch. Pathol. Lab. Med., 136, 591–600. [DOI] [PubMed] [Google Scholar]
- 47. Lederer D.J., Jelic S., Basner R.C., Ishizaka A., Bhattacharya J. (2009) Circulating KL-6, a biomarker of lung injury, in obstructive sleep apnoea. Eur. Respir. J., 33, 793–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. O'Connor C., Thornley K.S., Hanly P.J. (2000) Gender differences in the polysomnographic features of obstructive sleep apnea. Am. J. Respir. Crit. Care Med., 161, 1465–1472. [DOI] [PubMed] [Google Scholar]
- 49. McDonald M.L., Cho M.H., Sorheim I.C., Lutz S.M., Castaldi P.J., Lomas D.A., Coxson H.O., Edwards L.D., MacNee W., Vestbo J., et al. (2014) Common genetic variants associated with resting oxygenation in chronic obstructive pulmonary disease. Am. J. Respir. Cell Mol. Biol., 51, 678–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Redline S., Schluchter M.D., Larkin E.K., Tishler P.V. (2003) Predictors of longitudinal change in sleep-disordered breathing in a nonclinic population. Sleep, 26, 703–709. [DOI] [PubMed] [Google Scholar]
- 51. Hanis C.L., Ferrell R.E., Barton S.A., Aguilar L., Garza-Ibarra A., Tulloch B.R., Garcia C.A., Schull W.J. (1983) Diabetes among Mexican Americans in Starr County, Texas. Am. J. Epidemiol., 118, 659–672. [DOI] [PubMed] [Google Scholar]
- 52. Brown S.A., Garcia A.A., Kouzekanani K., Hanis C.L. (2002) Culturally competent diabetes self-management education for Mexican Americans: the Starr County border health initiative. Diabetes Care, 25, 259–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. DePristo M.A., Banks E., Poplin R., Garimella K.V., Maguire J.R., Hartl C., Philippakis A.A., del Angel G., Rivas M.A., Hanna M., et al. (2011) A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet., 43, 491–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ng S.B., Turner E.H., Robertson P.D., Flygare S.D., Bigham A.W., Lee C., Shaffer T., Wong M., Bhattacharjee A., Eichler E.E., et al. (2009) Targeted capture and massively parallel sequencing of 12 human exomes. Nature, 461, 272–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Grove M.L., Yu B., Cochran B.J., Haritunians T., Bis J.C., Taylor K.D., Hansen M., Borecki I.B., Cupples L.A., Fornage M., et al. (2013) Best practices and joint calling of the HumanExome BeadChip: the CHARGE Consortium. PLoS One, 8, e68095.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Redline S., Sanders M.H., Lind B.K., Quan S.F., Iber C., Gottlieb D.J., Bonekat W.H., Rapoport D.M., Smith P.L., Kiley J.P. (1998) Methods for obtaining and analyzing unattended polysomnography data for a multicenter study. Sleep Heart Health Research Group. Sleep, 21, 759–767. [PubMed] [Google Scholar]
- 57. White D.P. (2008) Monitoring peripheral arterial tone (PAT) to diagnose sleep apnea in the home. J. Clin. Sleep Med., 4, 73.. [PMC free article] [PubMed] [Google Scholar]
- 58. Hankinson J.L., Odencrantz J.R., Fedan K.B. (1999) Spirometric reference values from a sample of the general U.S. population. Am. J. Respir. Crit. Care Med., 159, 179–187. [DOI] [PubMed] [Google Scholar]
- 59. Du P., Kibbe W.A., Lin S.M. (2008) lumi: a pipeline for processing Illumina microarray. Bioinformatics, 24, 1547–1548. [DOI] [PubMed] [Google Scholar]
- 60. Abecasis G.R., Cherny S.S., Cookson W.O., Cardon L.R. (2002) Merlin–rapid analysis of dense genetic maps using sparse gene flow trees. Nat. Genet., 30, 97–101. [DOI] [PubMed] [Google Scholar]
- 61. Lee S., Wu M.C., Lin X. (2012) Optimal tests for rare variant effects in sequencing association studies. Biostatistics, 13, 762–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Lee S., Teslovich T.M., Boehnke M., Lin X. (2013) General framework for meta-analysis of rare variants in sequencing association studies. Am. J. Hum. Genet., 93, 42–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Sunyaev S.R., Lathe W.C., 3rd, Ramensky V.E., Bork P. (2000) SNP frequencies in human genes an excess of rare alleles and differing modes of selection. Trends Genet., 16, 335–337. [DOI] [PubMed] [Google Scholar]
- 64. Sunyaev S., Ramensky V., Koch I., Lathe W., 3rd, Kondrashov A.S., Bork P. (2001) Prediction of deleterious human alleles. Hum. Mol. Genet., 10, 591–597. [DOI] [PubMed] [Google Scholar]
- 65. Ramensky V., Bork P., Sunyaev S. (2002) Human non-synonymous SNPs: server and survey. Nucleic Acids Res., 30, 3894–3900. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.