Abstract
Hirschsprung disease (HSCR) is the most common cause of neonatal intestinal obstruction. It is characterized by the absence of ganglia in the nerve plexuses of the lower gastrointestinal tract. So far, three common disease-susceptibility variants at the RET, SEMA3 and NRG1 loci have been detected through genome-wide association studies (GWAS) in Europeans and Asians to understand its genetic etiologies. Here we present a trans-ethnic meta-analysis of 507 HSCR cases and 1191 controls, combining all published GWAS results on HSCR to fine-map these loci and narrow down the putatively causal variants to 99% credible sets. We also demonstrate that the effects of RET and NRG1 are universal across European and Asian ancestries. In contrast, we detected a European-specific association of a low-frequency variant, rs80227144, in SEMA3 [odds ratio (OR) = 5.2, P = 4.7 × 10−10]. Conditional analyses on the lead SNPs revealed a secondary association signal, corresponding to an Asian-specific, low-frequency missense variant encoding RET p.Asp489Asn (rs9282834, conditional OR = 20.3, conditional P = 4.1 × 10−14). When in trans with the RET intron 1 enhancer risk allele, rs9282834 increases the risk of HSCR from 1.1 to 26.7. Overall, our study provides further insights into the genetic architecture of HSCR and has profound implications for future study designs.
Introduction
Hirschsprung disease (HSCR) is a rare, complex genetic disorder of the enteric nervous system (ENS). It is characterized by the absence of enteric ganglia in the myenteric and submucosal plexuses along a variable length of the hindgut (1). This developmental disorder is attributed to a failure in rostro-caudal migration, proliferation or differentiation of the enteric neural crest cells to the affected portion during embryogenesis, leading to contraction of the bowel and functional colonic obstruction. HSCR exhibits variable phenotypic expressivity and manifests with low, sex-dependent penetrance. Severity of the disease is determined by the extent of aganglionosis and can be classified into short-segment HSCR (S-HSCR; ∼80% of cases), long-segment HSCR (L-HSCR; ∼15%) and total colonic aganglionosis (TCA; ∼5%). Particularly for S-HSCR, gender bias is commonly observed, with males being affected four times more than females. The incidence of the disease (1.4–2.8 in 10 000 newborns) also varies widely across ethnic groups, where Asia has the highest incidence rate reported (1–3).
The mode of inheritance of HSCR varies, from dominant with reduced penetrance or recessive in familial cases to a more complex, non-Mendelian mode of inheritance in the sporadic cases. It has been long recognized as a highly heritable disorder (80–97% for S-HSCR and ∼100% for L-HSCR), although ∼80% of cases are considered sporadic (4). To date, rare variants contributing to HSCR have been found in more than 15 genes, with RET being the most frequently mutated gene (5); however, these rare variants typically have incomplete penetrance and, altogether, only explain a small proportion of the total heritability. Besides, a common variant in intron 1 of RET (rs2435357) also predisposes to HSCR. Despite its smaller effect size, this common, non-coding polymorphism explains 10–20 times more genetic variance than the rare, damaging RET variants (6), implying that common variants might also contribute to a significant proportion of disease risk.
In order to find novel genetic variants associated with HSCR, three genome-wide association studies (GWAS) have been carried out thus far (7–9). In addition to RET, two novel HSCR-associated loci, semaphorin 3C/3D (SEMA3) at 7q21 and neuregulin-1 (NRG1) at 8p12, have been implicated in the GWAS of Europeans and East Asians, respectively. Semaphorin 3C and 3D are secreted glycoproteins involved in axon guidance and plexin/neuropilin-mediated neuronal migration. Likewise, neuregulin-1 also plays an important role in neural crest cell survival and differentiation through binding and interaction with ErbB tyrosine kinase receptors (10). Follow-up mutational scans of these loci revealed similar genetic properties as those reported for RET, whereby both common and rare variants underlies the genetic risk to HSCR (8,11–13). Unlike the RET common variant, the mechanisms by which these novel HSCR-associated alleles influence genetic susceptibility remains largely unknown. Lead SNPs of NRG1 (rs7835688 and rs16879552) and SEMA3 (rs12707682 and rs11766001) loci map to non-coding regions with unclear functional impact and are likely to represent surrogate markers in linkage disequilibrium (LD) with causal variants. To narrow down the potential causal variants for HSCR and to identify novel disease-susceptibility loci, we performed a trans-ethnic meta-analysis by aggregating the association results of all published GWAS on HSCR, totaling 507 HSCR cases and 1191 controls. By exploiting the allelic heterogeneity and difference in LD structure between populations, we aimed to (i) evaluate if the evidence of association is ancestry-specific or shared across ethnicities, and (ii) define credible sets that confidently contain the causal variants for functional interrogation.
Results
To enhance the coverage of genetic variations in the meta-analysis, imputation was initially performed in each study with reference to the 1000 Genomes Project (Phase I Integrated, June 2014 haplotypes). An inverse-variance fixed-effects meta-analysis was carried out across Chinese and Korean studies (Asian ancestry) and the trans-ethnic meta-analysis was performed together with the European GWAS using a Bayesian approach—Meta-Analysis of Trans-ethnic association studies (MANTRA) (Supplementary Material, Figs S2 and S3) (14). We first evaluated the potential of discovering novel HSCR-associated loci across ethnicities by assessing the concordance in the direction of effect between European GWAS and Asian ancestry meta-analysis. We observed an excess in directional concordance (Supplementary Material, Fig. S4) for SNPs showing weak to nominal evidence of association with HSCR, indicating some HSCR-associated loci are likely to be shared between ethnicities.
Localizing variants causal to HSCR in known loci
In the three original two-staged GWAS, RET was the only locus attaining genome-wide significance in the individual discovery stages, without the targeted genotyping in replication cohorts. After the trans-ethnic meta-analysis, all three known loci (RET, NRG1 and SEMA3) showed strong evidence of association with HSCR [log10Bayes factor (BF) >6.1; Table 1]. We further constructed a 99% credible set for each locus, which represent the minimum set of variants accounting for 99% of the posterior probabilities of association in the region (15). SNPs in the credible sets are most likely to be causal based on the statistical evidence from the trans-ethnic meta-analysis.
Table 1.
Trans-ethnic meta-analysis association results for three known HSCR-associated loci
| European GWAS |
Asian-specific meta-analysis |
Trans-ethnic meta-analysis |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 99% credible set |
||||||||||||
| Locus | Lead SNP | Position | Allelea | EAFb | OR (95%CI) | P | EAF | OR (95%CI) | P | LogBF | Number of SNPs | Interval (bp) |
| RET | rs2505998 | 10: 43570925 | A/G | 0.64/0.22 | 4.17 (3.23–5.26) | 1.1 × 10−28 | 0.82/0.48 | 3.57 (2.94–4.35) | 4.44 × 10−43 | 67.25 | 13 | 15 913 |
| NRG1 | rs7005606 | 8: 32401501 | G/T | 0.54/0.42 | 1.64 (1.25–2.15) | 4.0 × 10−4 | 0.36/0.21 | 2.12 (1.70–2.63) | 1.11 × 10−11 | 11.68 | 29 | 31 993 |
| SEMA3C/3D | rs80227144 | 7: 84349842 | A/C | 0.14/0.03 | 5.2 (3.09–8.73) | 4.7 × 10−10 | 0/0 | – | – | 7.99 | 13 | 603 138 |
aEffect allele/other allele.
bEffect allele frequency of cases/controls.
For RET and NRG1, the 99% credible sets contained 13 and 29 non-coding SNPs each, narrowing down the association signals to 16 and 32 kb regions, respectively (Supplementary Material, Tables S3 and S4; Fig. 1, left and middle panels). Both credible set regions comprised promoter and intron 1. We observed little evidence of heterogeneity in allelic effects (posterior probability of heterogeneity <0.5) between the Asian and European samples for the two loci, implying that both loci are robustly associated with HSCR irrespectively of the ancestry.
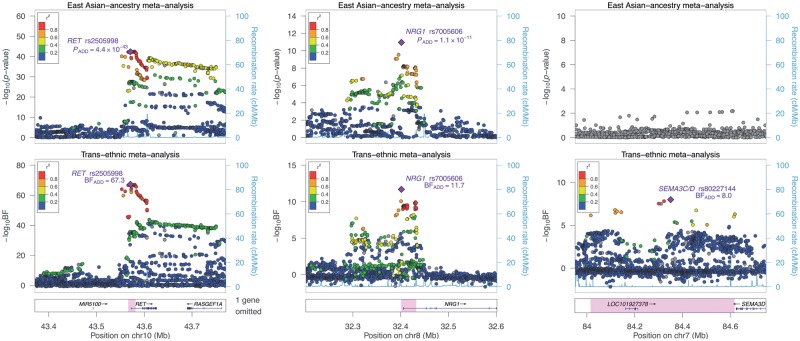
Figure 1.
Regional plots of association for the Asian-specific and trans-ethnic meta-analyses at the three known HSCR-associated loci. Significance of association is reported in P-values for the fixed-effects Asian-specific meta-analysis (upper panel) and in Bayes factors for the trans-ethnic meta-analyses (lower panel). Genomic regions covered by the 99% credible set are highlighted by the purple boxes. The lead SNP of each locus is highlighted by purple diamond. Color indicates the LD (r2) with the lead SNP.
The new lead SNPs—rs2505998 for RET (OR = 3.6, PAsian = 4.4 × 10−43; Table 1) and rs7005606 for NRG1 (OR = 2.1, PAsian = 1.1 × 10−11)—in the Asian-specific meta-analysis also exhibited the strongest association in the trans-ethnic meta-analysis after incorporating the association results from Europeans (RET: rs2505998, log10BF = 67.3; NRG1: rs7005606, log10BF = 11.7). These two lead SNPs are common in both ancestries (minor allele frequency (MAF) >0.2) and are in strong LD with the previously reported lead SNPs, both of which were also included in the 99% credible sets (rs2505998, r2rs2435357 > 0.95; rs7005606, r2rs7835688 > 0.77). Because of the stochastic imputation or genotyping errors as well as statistical fluctuation, effect of SNPs within the same credible set is not easily distinguishable from one another.
To identify the most plausible functional SNP and to better understand the regulatory mechanisms, we annotated the credible set variants against various regulatory databases. Functional annotation revealed that all SNPs in the two credible sets are indeed cis-acting expression quantitative loci (cis-eQTL; Supplementary Material, Tables S3 and S4), affecting NRG1 expression as well as the expression of RET and the immediately downstream CSGALNACT2 gene across multiple tissues in Genotype-Tissue Expression (GTEx) database. In addition, all credible set SNPs showed strong enhancer activities, marked by histone modification signatures of H3K27ac or H3K4me1 known to be enriched in active enhancers. Specifically, the RET lead SNP (rs2505998) overlaps with a CTCF binding site whereas the well-established functional RET enhancer rs2435357 has been shown to be a binding site for the transcriptional co-activator P300.
For SEMA3 loci, 13 intergenic SNPs located downstream of SEMA3D were included in the 99% credible set (Supplementary Material, Table S5; Fig. 1, right panel). Most of the credible set SNPs (9 out of 13), including the lead SNP (rs80227144, MAF = 2.9%), are of low-frequency (MAF = 2.2–4.5%) in the European population but monomorphic in Asians, which plausibly explains why association was detected in neither Asian study. None of these 13 SNPs is in strong LD with the 2 SNPs implicated in the original European GWAS (r2rs11766001 < 0.67 and r2rs12707682 < 0.42). In the European data set, the lead SNP has a strong effect on disease risk (rs80227144, OR = 5.2, PEuropean = 4.7 × 10−10; Table 1) but it is only weakly linked to the reported HSCR-associated SNPs (r2rs11766001 = 0.25 and r2rs12707682 = 0.16). Nevertheless, according to the 1000 Genomes Project, its low frequency risk allele (A) is predominately in phase with the common risk alleles (C) of both rs11766001 (D′ = 0.97) and rs12707682 (D′ = 1). Moderate association was detected for rs80227144 (OR = 4.8, PEuropean = 1.5 × 10−5) when conditioning on both rs11766001 and rs12707682. On the other hand, association of both reported SNPs was completely attenuated once accounting for the effect of rs80227144 (rs11766001, PEuropean = 0.66; rs12707682, PEuropean = 0.08). This implies that the previous finding might reflect an indirect association because of the strong effect of the low frequency rs80227144 variant. Interestingly, the genetic variation of rs80227144 alters a regulatory motif of SOX7 and is reported as a binding site for GATA-6, where both are transcription factors important for cardiac development (16–18). GATA-6 is involved in semaphorin-plexin signaling and, in particular, may regulate the expression of semaphorin 3C in cardiac neural crest cells for cardiovascular morphogenesis (19,20). This putative long-range regulation might represent the missing link in the association of SEMA3C located ∼4Mb upstream of the credible region and the HSCR risk.
Independent association of low frequency Asian-specific RET missense variants
We also attempted to delineate if there are multiple independent association signals by conditioning on the reported lead SNP(s) in each known HSCR locus. A secondary association signal, corresponding to a low-frequency missense variant encoding RET p.Asp489Asn (rs9282834, conditional OR = 20.3, conditional PAsian = 4.1 × 10−14; MAF = 0.02 in Asian controls; Table 2), was discovered from the Asian-specific meta-analysis after adjusting for the effect of the RET enhancer (rs2435357). Further conditioning on rs9282834 in RET or other reported SNPs in NRG1 and SEMA3 did not reveal additional independent association. This protein-altering HSCR-susceptibility allele (rs9282834 A allele) appears to be Asian-specific and is rarely found in non-Asian populations [MAF < 0.001 according to the ExAC database (21)]. Prior to the conditional analysis, the effect of RET p.Asp489Asn was masked as its risk allele (A) is out of phase with the rs2435357 risk allele (T), resulting in an underestimation of the effect sizes of both SNPs if considered individually (Table 2).
Table 2.
Association results for HSCR-associated SNPs in RET before and after conditional analysis on Asian samples
| Univariate analysis |
Conditional analysis |
||||||
|---|---|---|---|---|---|---|---|
| SNP | Position | Allelea | EAFb | OR (95%CI) | P | OR (95%CI) | P |
| rs2435357 | 10:43582056 | T/C | 0.82/0.48 | 3.54 (2.95–4.24) | 1.36 × 10−42 | 4.01 (3.33–4.84) | 2.98 × 10−48 |
| rs9282834 (D489N) | 10:43606856 | A/G | 0.05/0.03 | 1.80 (1.06–3.04) | 0.029 | 20.3 (9.31–44.4) | 4.09 × 10−14 |
aEffect allele/Other allele.
bEffect allele frequency of cases/controls.
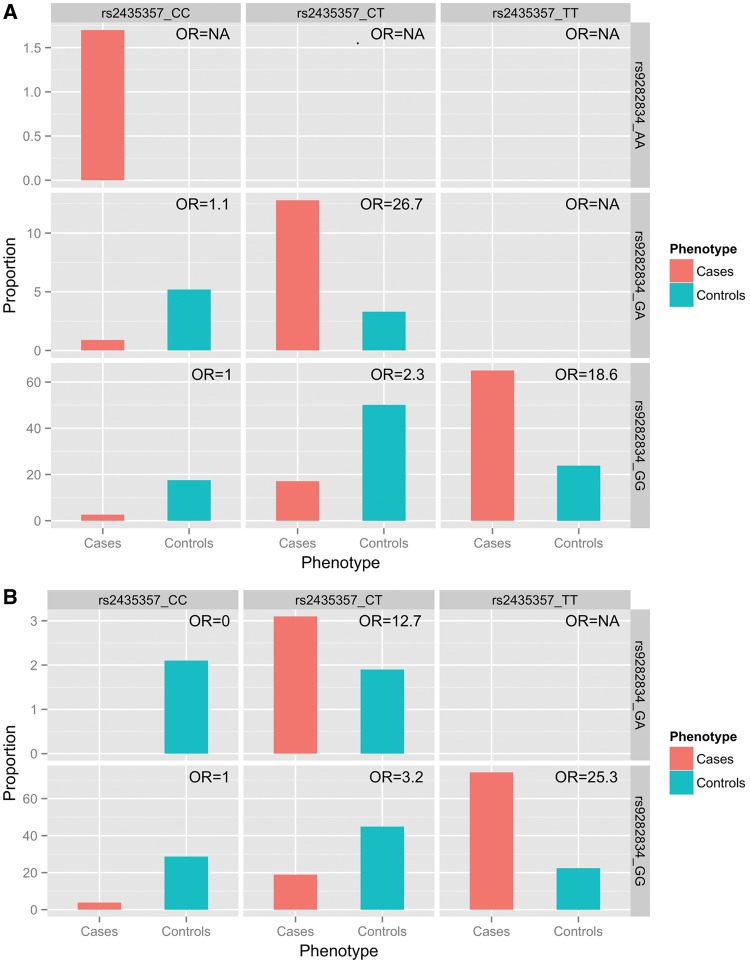
While the OR for HSCR risk is 2.3 to 3.2 for carriers of the rs2435357 risk allele and ∼1.1 for carriers of the rs9282834 risk allele, the risk to HSCR dramatically increased 10-fold (OR = 16.7/26.7 in Chinese/Korean, respectively) in individuals compound heterozygous for both rs9282834 and rs2435357 risk alleles (Fig. 2). Although rs9282834 is predicted to be non-damaging, this missense variant might have a synergistic effect with rs2435357 in gene regulation, possibly by disrupting the functionality of both copies of RET.
Figure 2.
Epistatic effect of the two RET SNPs independently associated with HSCR (rs9282834 and rs2435357) in (A) Korean and (B) Chinese GWAS. Information [proportion and odds ratio (OR)] regarding each genotype combination [rs9282834 (row) × rs2435357 (column)] is represented by cells. In each cell, left bar denotes the proportion of cases with the specified genotype combination among all cases while right bar denotes the same for controls. OR is defined as the odds of being HSCR with the combination relative to the baseline double homozygote carrying neither risk allele (i.e. GG for rs9282834 and CC for rs2435357).
Characterizing the genetic basis of HSCR
Apart from these known loci, no novel genome-wide significant association was detected under the additive model, raising the questions of whether a large proportion of disease risk can still be attributed to common variants and whether an increase in sample size can still discover more novel HSCR-associated loci. To address these questions, we sought to understand the underlying genetic basis of HSCR by evaluating the global contribution of common variants (MAF > 5%) in disease risk using genome-wide complex trait analysis (GCTA) (22). GCTA estimates narrow-sense heritability (ratio of additive genetic variance to total phenotypic variance) on the liability scale by computing the proportion of phenotypic variance captured by all genotyped markers in GWAS simultaneously (23). While the European GWAS is heterogeneous in terms of the genotyping array and the coverage is relatively low for majority of samples, we focused on the two Asian GWAS for the heritability estimation. Using the common SNPs typed in the two Asian GWAS, we estimated that common variants accounted for 0.34 (standard error (s.e.) = 0.11, P = 3.0 × 10−3; Table 3) of variance in risk of HSCR. Compared with the low heritability (∼5.7–6.2%) associated with the three known HSCR-susceptibility variants and the high heritability (>80%) estimated from family studies, the moderate total genetic variance explained implied (i) a non-negligible ‘hidden heritability’ captured by SNPs at other loci on the SNP genotyping arrays and, more importantly, (ii) a significant ‘missing heritability’ unexplained by the additive effects of all typed common variants (24).
Table 3.
Heritability estimates for the two Asian GWAS on HSCR
| Study | Cases | Controls | Heritability Estimate (s.e.) | Pa |
|---|---|---|---|---|
| Chinese | 165 | 427 | 0.330 (0.138) | 5.79 × 10−3 |
| Korean | 121 | 366 | 0.369 (0.208) | 7.11 × 10−3 |
| Combined | 286 | 793 | 0.342 (0.115) | 2.95 × 10−3 |
aP value for likelihood ratio test against null hypothesis of zero heritability.
Hints of the hidden heritability: genes involved in ENS development
To explore the hidden heritability, we first assessed the association of other HSCR candidate genes (n = 109; Supplementary Material, Table S6) involved in ENS development, whereby their association could be hidden under the genome-wide significance threshold because of the lack of power attributed to the inadequate sample size. As shown in the quantile-quantile plot (Supplementary Material, Fig. S5), a significant excess of moderate association (P < 0.001) was observed than expected under the null hypothesis of no association (binomial test P = 6.5 × 10−9), suggesting that some of these candidates might truly associate with risk of HSCR. Among the 109 candidate loci, 2 exhibited moderately strong association with HSCR (log10BF > 4 and/or PAsian < 1 × 10−5) (25), which included JAG1 of the Notch signaling pathway (rs6032942: log10BF = 4.9) (26) and the ENS-expressed transcriptional factor—PHOX2B (rs6826373: OR = 0.6, PAsian = 6.4 × 10−6) (27). While the causal role of PHOX2B rare variants in syndromic HSCR has been well-established, our study further provides further evidence in support of the contribution of PHOX2B common variants to HSCR susceptibility (28).
Marginal association at VRK2 under recessive model and other candidate genes
Given the dose-dependent nature of the major gene, RET, and considering the fact that non-additive genetic variance is not considered in narrow-sense heritability, we hypothesized that the missing heritability may, in part, be explained by dominance effects. By assuming complete dominance, we therefore performed association tests for all imputed variants by fitting both recessive and dominant models. In addition to RET, which showed the strongest association, we detected, under the recessive model, a novel association at 2p16.1 (rs4672229, recessive log10BF = 6.2 compared with additive log10BF = 4.9; Table 4) mapping to an intronic region of the vaccinia-related kinase 2 gene (VRK2). This lead SNP displayed a heterogeneous effect across ancestries, where the recessive association mainly driven by Asians (OR = 2.5, PAsian = 2.5×10−8 in Asian-specific meta-analysis) was not observed in Europeans (OR = 0.8, PEuropean = 0.24). Taking into account that we tested 3 different models of inheritance, the Asian-specific association at VRK2 did not surpass the stringent Bonferroni correction of multiple testing and, consequently, a follow-up study is required to validate this finding. Although rs4672229 is located in intron 2 of VRK2, it has been reported as an eQTL in tibial nerves that increases the expression of a nearby gene, Fanconi-anemia complementation L polypeptide (FANCL).
Table 4.
Marginal association for VRK2 under recessive model
| Trans-ethnic meta-analysis | European GWAS |
Asian-specific meta-analysis |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Locus | Lead SNP | Position | Genotypea | LogBF | EGFb | OR (95% CI) | P | EGF | OR (95% CI) | P |
| VRK2 | rs4672229 | 2:58276280 | AA/AC + CC | 6.20 | 0.21/0.26 | 0.75 (0.46–1.21) | 0.24 | 0.36/0.19 | 2.48 (1.80–3.41) | 2.52 × 10−8 |
aEffect genotype under recessive model/other genotypes.
bEffect genotype frequency.
Discussion
Here, we present the largest genetic study and the first GWAS meta-analysis of HSCR, aggregating association results of all published GWAS on this disorder. In addition to reconfirming RET as the major gene for HSCR, we fine-mapped the three known loci through trans-ethnic meta-analysis and narrowed down the credible sets of SNPs that might be causal to HSCR. Moreover, we vindicated the important role of NRG1, wherein NRG1 is the only locus other than RET having common variants robustly associated with HSCR across populations. On the other hand, the association of the SEMA3 locus appeared to be European-specific and most likely driven by low-frequency regulatory variants that are monomorphic in Asians. By conditional analysis, we further detected another ancestry-specific association which corresponds to an Asian-specific secondary signal by a low-frequency, missense variant, encoding RET p.Asp489Asn and whose effect is independent of that of the reported common intron 1 RET enhancer.
Like for other complex diseases or traits, trans-ethnic meta-analysis for HSCR did not reveal heterogeneous effects of common variants across populations (29), yet the two low-frequency variants found associated with HSCR are nearly ancestry-specific. This could be explained by the fact that rare variants tend to be more recent and may emerge after the divergence of different populations (30). As a result, increasing sample size involving multiple ancestries by trans-ethnic meta-analysis might not facilitate the discovery of low-frequency variants of larger effect, where their association could be more readily detected by ancestry/population-specific meta-analysis. Indeed, the extent to which trans-ethnic meta-analysis can yield novel associations depends on the underlying genetic architecture of the disease. For HSCR, we estimated that a considerable amount of ‘hidden’ as well as ‘missing heritability’ exists. Proportion of phenotypic variance explained by common SNPs jointly was significantly greater than that of the known disease-susceptibility variants, but did not account for the totality of heritability inferred from family studies. Some of the genetic variance could be simply hidden below the threshold of genome-wide significance, hinting at unraveling more HSCR-associated common variants, for example, of genes involved in ENS development, by further increasing the sample size.
Given our finding here of the two low-frequency HSCR-associated variants, together with reports of causative mutations in known genes and a global burden of copy number variants (CNVs) in HSCR cases, contributions of low-frequency, rare, or even structural variants can be the potential sources of ‘missing heritability’ in HSCR (8,11,31). While our GWAS employed old genotyping platforms, we acknowledge that the limitations on calling CNVs and imputing rare variants made them sub-optimal to evaluate the overall contribution of these types of variations to heritability. Genetic studies using more recent technologies (e.g. the latest SNP chips and/or next generation sequencing) and family-based study designs (32) (e.g. parent-offspring trios) might resolve the mystery of their genetic contribution. Another prevailing hypothesis is that part of the ‘missing heritability’ is attributable to non-additive genetic variance, which includes interaction/epistatic (allelic interaction between loci) and dominance effects (allelic interaction within a locus) (33,34). In fact, the major gene—RET—serves as the best example for both types of non-additive contributions. Firstly, many studies have suggested the genetic interplay between variants in RET and other HSCR-associated loci. For instance, NRG1 common variants have stronger effects on disease risk in the presence of the RET high-risk genotype (7,35). On the other hand, chromosome 21 gene dosage, in which comorbidity of Down syndrome and HSCR is often observed, acts on a genetic background with a smaller contribution of RET intron 1 enhancer (36). Secondly, for the low frequency and common variants, RET appears to follow a recessive model either through homozygosity at the intron 1 enhancer or compound heterozygosity with the missense variant, rs9282834. Thus ‘recessive’ inheritance of gene disrupting alleles at one site (recessive inheritance), or at multiple sites of the same gene (compound heterozygous inheritance) or across multiple genes (oligogenic inheritance) may underlie the non-additive genetic contributions to HSCR. The existence of such non-additive genetic components may lead to confounded estimates of both heritability, the one estimated through family data and the one estimated through genetic variants (37). Only until recently, methods to evaluate the relative importance of these non-additive variations in case–control studies were proposed (34); however, these methods typically require a large sample size for partitioning into variance components or for establishing trans-ethnic genetic correlations and are therefore not feasible in our circumstances given the small sample size. Leveraging power by further increasing sample size would allow us to fully decipher the genetic architecture of HSCR as well as comparing and contrasting the cis- versus trans-haplotype effect of linked variants. Last but not least, we cannot negate the possibility of overestimation of heritability in the original family studies. A revisit of the epidemiology of HSCR, involving more HSCR twins or families, is warranted.
Perhaps one of the most interesting candidates with non-additive effect is the VRK2 locus, whose variants show a marginal association under a recessive model in the trans-ethnic meta-analysis. VRK2 encodes a serine/threonine protein kinase implicated in neurological function. Variants upstream of VRK2 were reported as one of the 108 loci significantly associated with schizophrenia (38). Nearby variants in LD also showed moderate association with genetic generalized epilepsies (39). In addition, VRK2 together with the downstream gene, FANCL, falls within a common deletion region responsible for the 2p16.1-p15 microdeletion syndrome which is characterized by intellectual disability, microcephaly, and autistic behavior (OMIM:612513) (40,41). Despite multiple lines of evidence supporting a role of VRK2 in central nervous system and neurodevelopmental disorders, its involvement in ENS development remains poorly understood. One possible clue linking VRK2 to HSCR may lie in its interaction with the co-receptor of NRG1, Erb-B2 Receptor Tyrosine Kinase 2 (ErbB2). A recent study of Fernandez et al. (42) showed that, in breast cancer cells, high level of VRK2 inhibited ErbB2 activation of transcription, possibly by blocking phosphorylation of extracellular signal-regulated kinase (ERK) in response to ErbB2 stimulation. On the contrary, knockdown of VRK2 increased the transcriptional response to stimulation. This inverse correlation between VRK2 and ErbB2 expression levels in breast carcinomas suggested a regulatory role of VRK2 in neuregulin signaling, which may represent the missing link underlying the association of VRK2 and HSCR.
With the recent progress of derivation and isolation of ENS progenitor cells, novel therapeutic opportunities, e.g. autologous transplantation of these ENS precursors to repopulate the colon and restore the peristalsis function, might arise as an alternative therapy in the future (43,44); however, the migration and differentiation abilities of these patient-derived precursors are often affected by the genetic background of the patients themselves. It is therefore of utmost importance to apprehend the pathophysiology contributing to, in general, HSCR and, specifically, to each individual in order to develop a patient-specific therapy. Overall, our study not only implicates putative HSCR-associated loci that might help modulate enteric neural crest cell properties, but also provides potential sources of the ‘hidden’ and ‘missing’ heritability. Understanding the genetic architecture of HSCR—the relative contribution of common versus rare variants and additive versus non-additive effects—may have profound implications on future study design.
Materials and Methods
Study subjects and quality controls
The trans-ethnic meta-analysis comprised 507 HSCR cases and 1191 controls from 3 published GWAS on European (8), Chinese (7) and Korean (9) populations. The phenotypic and demographic information for each study is summarized in Supplementary Material, Table S1. On top of the quality controls imposed by the original studies, individuals with low call rate (<90% for Affymetrix 500K or <98% for Illumina 1M arrays) or being an outlier in autosomal heterozygosity (4 SD from the mean) were further removed.
European
Family-based GWAS undertaken by the International HSCR Disease Consortium was carried out on 220 trios (probands together with unaffected parents) of European descent, which included subjects from USA, Italy, The Netherlands, Spain and France (8). Genotyping was performed on either or both of the 250 K arrays (Nsp and Sty) of the Affymetrix GeneChip Human Mapping 500 K array. To convert into case/pseudo-control design, we constructed pseudo-controls from the non-transmitted alleles of parents from each trio. While the resulting pseudo-controls are genetically unrelated to the proband, subsequent association analysis is equivalent to a matched case/control analysis. After quality control, a total of 25 samples genotyped on the Nsp arrays and 23 samples genotyped on the Sty arrays were dropped. The final dataset consisted of 179 cases and 172 controls genotyped on either array—Nsp (130 873 SNPs) or Sty (115 285 SNPs)—as well as 33 cases and 30 controls typed on both arrays (246 158 SNPs) of the 500 K array were used.
Chinese
The Chinese GWAS was originally performed on 181 sporadic cases and 346 controls of matched ancestry (7) using Affymetrix 500K, which assays 493 840 SNPs across the genome. We later included an additional set of 271 controls, of which 181 were genotyped by another array—Affymetrix Genome-Wide Human SNP Array 5.0 (45)—which includes all the SNPs of the original 500K array but differs by inclusion of extra non-polymorphic probes for copy number analysis. More stringent quality controls as described previously were applied on both individual and variant levels (46), resulting in a final set of 320 613 SNPs for 173 cases and 615 controls included in the meta-analysis.
Korean
The original Korean GWAS consisted of 123 sporadic HSCR cases and 432 controls (9). Relatedness in controls was initially modeled and corrected by mixed model association in the original GWAS. To maintain consistency in statistical analysis with the other two studies, these related controls were removed, resulting in 714 024 SNPs of 122 cases and 374 controls for meta-analysis.
Genotype Imputation
For each data set, individual genotypes first underwent pre-phasing using SHAPEIT (v2) to infer the underlying haplotypes (47). Whenever the parental genotypes were available, they were included into the pre-phasing using the SHAPEIT—duohmm module to improve accuracy but these genotypes were later excluded from association analysis. Imputation was then performed using IMPUTE2 with reference to the 1000 Genome Project multi-ancestry reference panel (Phase I Integrated, June 2014 haplotypes) (48,49). The reference panel contains 2184 haplotypes of 1092 samples, with ∼38 million (M) bi-allelic polymorphic but non-singleton markers (∼36.8M SNPs, 1.4 M short bi-allelic insertions/deletions and 14 017 structural variations). Singleton variants—variants appearing only once in the whole data set—were not included in the imputation reference panel as the imputation quality is generally lower and we lack power to detect association for such low frequency variants.
Association Tests
Association analyses of all three imputed data sets were carried out using score tests (option-method score in SNPTEST v2) for three genetic models, (1) additive, (2) recessive and (3) dominant, based on the imputed genotype probabilities after adjustment by the study-specific covariates. For the Chinese GWAS, two principal components were used as covariates to control for population stratification and batch effect simultaneously, whereas, for the European GWAS, two covariates specifying the chip (Nsp only, Sty only and both Nsp and Sty) where genotyping was performed were included. No covariate was used for Korean data set as no population substructure was observed. Variants with low imputation quality (INFO ≤0.6), low population frequency (MAF ≤2%) or violating Hardy–Weinberg equilibrium (P < 10−4) were excluded (Supplementary Material, Table S2). The test statistics, as visualized in a quantile-quantile plot, appeared well calibrated (Supplementary Material, Fig. S1). To further delineate SNPs with independent association, we performed stepwise conditional analyses by SNPTEST using the reported lead SNP for each known HSCR locus as covariate (RET: rs2435357, NRG1: rs7835688 and SEMA3: rs11766001 and rs12707682). While the SEMA3 association is European-specific and one of the reported lead SNP (rs11766001) is monomorphic in East Asians, conditional analysis on SEMA3 was performed only in the European dataset. For RET and NRG1 loci, SNPs with conditional test statistics reaching genome-wide significance (P < 5 × 10−8 in the Asian/European-specific or log10BF > 6.1 in the trans-ethnic meta-analyses) were considered as having independent association and the lead SNPs showing strongest evidence of independent association was later included in the regression model as covariate. The process was repeated until no variant reached genome-wide significance after adjustment.
Asian-Specific and Trans-Ethnic Meta-Analyses
Asian-specific meta-analysis
Ethnic-specific meta-analysis was performed for East Asians only in METAL (50) under a fixed-effects model. Association summary statistics (effect size estimates and their standard error) of the Chinese and Korean GWAS were meta-analyzed by the inverse-variance approaches and Cochran's Q statistic was used to estimate the between-study heterogeneity.
Trans-ethnic meta-analysis
We meta-analyzed the association results of all ancestry groups using MANTRA software (14), which takes into account the expected similarity in allelic effects between closely related populations when compared with more diverse ethnic groups. Briefly, the estimated effect sizes, their standard errors and the allele frequencies of each study are used to model the relatedness between studies/populations, assign each population to ‘cluster’ according to the Bayesian partition model, and evaluate the statistical significance. We only considered variants tested in at least two GWAS, except for fine-mapping known loci. Evidence in favor of association was assessed by means of Bayes factor (BF), which represents the probability under the alternative hypothesis that the SNP is associated with the disease divided by the probability under the null hypothesis of no association. The larger the value of BF, the stronger is the evidence of association. An association with a log10BF of 6.1 or above is considered as genome-wide significant (comparable to GWAS significant threshold P < 5 × 10−8 in the frequentist test) (25). A full set of summary statistics of the meta-analysis can be obtained from http://www.surgery.hku.hk/trs/index.php/research-output/#transethnicMeta2016.
Defining Credible Sets for Fine-Mapping Known HSCR Loci
To fine-map the three known HSCR loci, we constructed the 99% credible sets which represent the minimum set of variants accounting for 99% of posterior probabilities in the region. For these loci with prior evidence of association with HSCR, we considered all variants tested for association in any GWAS. Assuming a single causal variant for each locus and that the true causal variant is either genotyped or well imputed, the probability that the 99% credible set contains the causal variant would be ∼0.99. For each locus, we first calculated the posterior probabilities for each variant located ±1 Mb from the top variant (variant showing the lowest P-value in the region), i.e. the corresponding BF divided by the summation of BF over all variants in the region. Variants were then ranked according to their BF and the ranked variants were combined to form the credible set until their cumulative posterior probability attains or exceeds 99%. For visualization purposes, the most distant SNPs within credible sets were defined as boundaries of the set.
Functional Annotation
We annotated the credible set variants for regulatory potentials against RegulomeDB (51) and HaploReg (52) databases. More specifically, we explored evidence of transcription factor binding, DNase hypersensitivity as well as promoter and enhancer activities via histone modification signals. Active promoter is marked by signature of H3K4me3, whereas H3K27ac and H3K4me1 define regions with enhancer activity. In addition, GTEx (53) resources were used to delineate if a variant is an eQTL that affects expression of nearby genes.
Estimation of Variance Explained by Known HSCR SNPS
The heritability contributed by known HSCR susceptibility loci was computed based on population-specific prevalence (0.0001 for Europeans and 0.00028 for Asians) together with allele frequencies and odds ratios (ORs) of the three previously reported HSCR-associated variants (RET rs2435357, NRG1 rs7835688 and SEMA3 rs12707682) based on the So et al. approach (54).
Estimation of Variance Explained by GWAS
To evaluate the narrow-sense heritability attributed to common variants (MAF >5%), we carried out a GCTA analysis on the two Asian population-based GWAS. GCTA estimates heritability by studying the extent to which phenotypic variation can be explained by the genotypic similarity across individuals. The genetic relatedness estimated in terms of the genetic relationship matrix approximates to the coefficient of relationship (two times the kinship coefficient) obtained from pedigrees. For each study, cryptic relatedness was first removed by restricting the genetic relatedness to be <0.025, a coefficient that approximates to fifth-degree relatives or less. A total of 17 Chinese and 9 Korean individuals were removed in this manner. Proportions of variance in disease liability captured by all genotyped SNPs were estimated by a mixed linear model, using a restricted maximum likelihood approach, while assuming the disease prevalence to be 0.0002. For the Chinese GWAS, only samples genotyped by the Affymetrix 500K platform were included to avoid spurious results. Also, two principal components were used as covariates to account for possible confounding effects because of population substructure. GCTA estimates from the two Asian studies were then meta-analyzed under a fixed-effects model to provide a more robust heritability estimate. Evidence for heterogeneity was assessed using Cochran’s Q test.
Supplementary Material
Supplementary Material is available at HMG online.
Supplementary Material
Acknowledgements
We are grateful to the numerous patients, their families and referring physicians that have participated in these studies in our laboratories, and the numerous members of our laboratories for their valuable contributions over many years.
Conflict of Interest statement. None declared.
Funding
This work was supported by Health and Medical Research Fund [grant number 01121796 to C.S.T]; General Research Fund [grant number HKU777612 to P.T.]; Theme-based Research scheme [grant number T12C-714/14-R to P.T.]; Agence Nationale de la Recherche [grant number ANR-10-IAHU-01]; National Research Foundation of Korea funded by the Ministry of Education, Science and Technology [grant number 2010-0007857 and 2009-0093822]; National Institutes of Health [grant numbers HD28088 (Merit Award), R37HD28088]; Italian Ministry of Health (‘Cinque per mille’ and Ricerca Corrente to the Gaslini Institute); the Institute of Health Carlos III (ISCIII), Spanish Ministry of Economy and Competitiveness, Spain [grant number PI13/01560]; Andalusian Regional Ministry of Innovation, Science and Enterprise [grant number CTS-7447]; ZonMW [grant number TOP-subsidie 40-00812-98-10042 to R.M.W.H]; and the Maag Lever Darm stichting [grant number WO09-62 to R.M.W.H].
References
- 1. Amiel J., Sproat-Emison E., Garcia-Barcelo M., Lantieri F., Burzynski G., Borrego S., Pelet A., Arnold S., Miao X., Griseri P., et al. (2008) Hirschsprung disease, associated syndromes and genetics: a review. J. Med. Genet., 45, 1–14. [DOI] [PubMed] [Google Scholar]
- 2. Tam P.K., Garcia-Barcelo M. (2009) Genetic basis of Hirschsprung's disease. Pediatr. Surg. Int., 25, 543–558. [DOI] [PubMed] [Google Scholar]
- 3. Chakravarti A., McCallion A.S., Lyonnet S. (2014) In Beaudet A.L., Vogelstein B., Kinzler K.W., Antonarakis S.E., Ballabio A., Gibson K.M., Mitchell G. (eds), The Online Metabolic and Molecular Bases of Inherited Disease. The McGraw-Hill Companies, Inc, New York, NY. [Google Scholar]
- 4. Badner J.A., Sieber W.K., Garver K.L., Chakravarti A. (1990) A genetic study of Hirschsprung disease. Am. J. Hum. Genet., 46, 568–580. [PMC free article] [PubMed] [Google Scholar]
- 5. Alves M.M., Sribudiani Y., Brouwer R.W., Amiel J., Antinolo G., Borrego S., Ceccherini I., Chakravarti A., Fernandez R.M., Garcia-Barcelo M.M., et al. (2013) Contribution of rare and common variants determine complex diseases-Hirschsprung disease as a model. Dev. Biol, 382, 320–329. [DOI] [PubMed] [Google Scholar]
- 6. Emison E.S., McCallion A.S., Kashuk C.S., Bush R.T., Grice E., Lin S., Portnoy M.E., Cutler D.J., Green E.D., Chakravarti A. (2005) A common sex-dependent mutation in a RET enhancer underlies Hirschsprung disease risk. Nature, 434, 857–863. [DOI] [PubMed] [Google Scholar]
- 7. Garcia-Barcelo M.M., Tang C.S., Ngan E.S., Lui V.C., Chen Y., So M.T., Leon T.Y., Miao X.P., Shum C.K., Liu F.Q., et al. (2009) Genome-wide association study identifies NRG1 as a susceptibility locus for Hirschsprung's disease. Proc. Natl. Acad. Sci. U. S. A., 106, 2694–2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Jiang Q., Arnold S., Heanue T., Kilambi K.P., Doan B., Kapoor A., Ling A.Y., Sosa M.X., Guy M., Jiang Q., et al. (2015) Functional loss of semaphorin 3C and/or semaphorin 3D and their epistatic interaction with ret are critical to Hirschsprung disease liability. Am J Hum. Genet., 96, 581–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kim J.H., Cheong H.S., Sul J.H., Seo J.M., Kim D.Y., Oh J.T., Park K.W., Kim H.Y., Jung S.M., Jung K., et al. (2014) A genome-wide association study identifies potential susceptibility loci for Hirschsprung disease. PLoS One, 9, e110292.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Paratore C., Goerich D.E., Suter U., Wegner M., Sommer L. (2001) Survival and glial fate acquisition of neural crest cells are regulated by an interplay between the transcription factor Sox10 and extrinsic combinatorial signaling. Development, 128, 3949–3961. [DOI] [PubMed] [Google Scholar]
- 11. Tang C.S., Ngan E.S., Tang W.K., So M.T., Cheng G., Miao X.P., Leon T.Y., Leung B.M., Hui K.J., Lui V.H., et al. (2012) Mutations in the NRG1 gene are associated with Hirschsprung disease. Hum. Genet., 131, 67–76. [DOI] [PubMed] [Google Scholar]
- 12. Luzon-Toro B., Fernandez R.M., Torroglosa A., de Agustin J.C., Mendez-Vidal C., Segura D.I., Antinolo G., Borrego S. (2013) Mutational spectrum of semaphorin 3A and semaphorin 3D genes in Spanish Hirschsprung patients. PLoS One, 8, e54800.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Luzon-Toro B., Torroglosa A., Nunez-Torres R., Enguix-Riego M.V., Fernandez R.M., de Agustin J.C., Antinolo G., Borrego S. (2012) Comprehensive analysis of NRG1 common and rare variants in Hirschsprung patients. PLoS One, 7, e36524.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Morris A.P. (2011) Transethnic meta-analysis of genomewide association studies. Genet. Epidemiol., 35, 809–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wellcome Trust Case Control Consortium, Maller J.B., McVean G., Byrnes J., Vukcevic D., Palin K., Su Z., Howson J.M., Auton A., Myers S., et al. (2012) Bayesian refinement of association signals for 14 loci in 3 common diseases. Nat. Genet., 44, 1294–1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kamachi Y., Kondoh H. (2013) Sox proteins: regulators of cell fate specification and differentiation. Development, 140, 4129–4144. [DOI] [PubMed] [Google Scholar]
- 17. Hermkens D.M., van Impel A., Urasaki A., Bussmann J., Duckers H.J., Schulte-Merker S. (2015) Sox7 controls arterial specification in conjunction with hey2 and efnb2 function. Development, 142, 1695–1704. [DOI] [PubMed] [Google Scholar]
- 18. Verzi M.P., Shin H., He H.H., Sulahian R., Meyer C.A., Montgomery R.K., Fleet J.C., Brown M., Liu X.S., Shivdasani R.A. (2010) Differentiation-specific histone modifications reveal dynamic chromatin interactions and partners for the intestinal transcription factor CDX2. Dev. Cell, 19, 713–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lepore J.J., Mericko P.A., Cheng L., Lu M.M., Morrisey E.E., Parmacek M.S. (2006) GATA-6 regulates semaphorin 3C and is required in cardiac neural crest for cardiovascular morphogenesis. J. Clin. Invest., 116, 929–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kodo K., Nishizawa T., Furutani M., Arai S., Yamamura E., Joo K., Takahashi T., Matsuoka R., Yamagishi H. (2009) GATA6 mutations cause human cardiac outflow tract defects by disrupting semaphorin-plexin signaling. Proc. Natl. Acad. Sci. U. S. A., 106, 13933–13938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lek M., Karczewski K., Minikel E., Samocha K., Banks E., Fennell T., O'Donnell-Luria A., Ware J., Hill A., Cummings B., et al. (2015) Analysis of protein-coding genetic variation in 60,706 humans. bioRxiv, in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yang J., Lee S.H., Goddard M.E., Visscher P.M. (2011) GCTA: a tool for genome-wide complex trait analysis. Am. J. Hum. Genet., 88, 76–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lee S.H., Wray N.R., Goddard M.E., Visscher P.M. (2011) Estimating missing heritability for disease from genome-wide association studies. Am. J. Hum. Genet., 88, 294–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Manolio T.A., Collins F.S., Cox N.J., Goldstein D.B., Hindorff L.A., Hunter D.J., McCarthy M.I., Ramos E.M., Cardon L.R., Chakravarti A., et al. (2009) Finding the missing heritability of complex diseases. Nature, 461, 747–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wang X., Chua H.X., Chen P., Ong R.T., Sim X., Zhang W., Takeuchi F., Liu X., Khor C.C., Tay W.T., et al. (2013) Comparing methods for performing trans-ethnic meta- analysis of genome-wide association studies. Hum. Mol. Genet., 22, 2303–2311. [DOI] [PubMed] [Google Scholar]
- 26. Okamura Y., Saga Y. (2008) Notch signaling is required for the maintenance of enteric neural crest progenitors. Development, 135, 3555–3565. [DOI] [PubMed] [Google Scholar]
- 27. Heanue T.A., Pachnis V. (2006) Expression profiling the developing mammalian enteric nervous system identifies marker and candidate Hirschsprung disease genes. Proc. Natl. Acad. Sci. U. S. A., 103, 6919–6924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Garcia-Barcelo M., Sham M.H., Lui V.C., Chen B.L., Ott J., Tam P.K. (2003) Association study of PHOX2B as a candidate gene for Hirschsprung's disease. Gut, 52, 563–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Marigorta U.M., Navarro A. (2013) High trans-ethnic replicability of GWAS results implies common causal variants. PLoS Genet., 9, e1003566.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. The 1000 Genomes Project Consortium, Auton A., Brooks L.D., Durbin R.M., Garrison E.P., Kang H.M., Korbel J.O., Marchini J.L., McCarthy S., McVean G.A., et al. (2015) A global reference for human genetic variation. Nature, 526, 68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Tang C.S., Cheng G., So M.T., Yip B.H., Miao X.P., Wong E.H., Ngan E.S., Lui V.C., Song Y.Q., Chan D., et al. (2012) Genome-wide copy number analysis uncovers a new HSCR gene: NRG3. PLoS Genet., 8, e1002687.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Luzon-Toro B., Gui H., Ruiz-Ferrer M., Sze-Man Tang C., Fernandez R.M., Sham P.C., Torroglosa A., Kwong-Hang Tam P., Espino-Paisan L., Cherny S.S., et al. (2015) Exome sequencing reveals a high genetic heterogeneity on familial Hirschsprung disease. Sci. Rep., 5, 16473.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Chen X., Kuja-Halkola R., Rahman I., Arpegard J., Viktorin A., Karlsson R., Hagg S., Svensson P., Pedersen N.L., Magnusson P.K. (2015) Dominant genetic variation and missing heritability for human complex traits: insights from twin versus genome-wide common SNP models. Am. J. Hum. Genet., 97, 708–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Brown B.C., Asian Genetic Epidemiology Network Type 2 Diabetes Consortium. Ye C.J., Price A.L., Zaitlen N. (2016) Transethnic Genetic-Correlation Estimates from Summary Statistics. Am. J. Hum. Genet., 99, 76-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Gui H., Tang W.K., So M.T., Proitsi P., Sham P.C., Tam P.K., Ngan E.S., Cherny S.S., Garcia-Barcelo M.M. (2013) RET and NRG1 interplay in Hirschsprung disease. Hum. Genet., 132, 591–600. [DOI] [PubMed] [Google Scholar]
- 36. Arnold S., Pelet A., Amiel J., Borrego S., Hofstra R., Tam P., Ceccherini I., Lyonnet S., Sherman S., Chakravarti A. (2009) Interaction between a chromosome 10 RET enhancer and chromosome 21 in the Down syndrome-Hirschsprung disease association. Hum. Mutat., 30, 771–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Genin E., Clerget-Darpoux F. (2015) The missing heritability paradigm: a dramatic resurgence of the GIGO syndrome in genetics. Hum. Hered., 79, 1–4. [DOI] [PubMed] [Google Scholar]
- 38. Schizophrenia Working Group of the Psychiatric Genomics Consortium (2014) Biological insights from 108 schizophrenia-associated genetic loci. Nature, 511, 421–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. EPICURE Consortium, EMINet Consortium, Steffens M., Leu C., Ruppert A.K., Zara F., Striano P., Robbiano A., Capovilla G., Tinuper P., et al. (2012) Genome-wide association analysis of genetic generalized epilepsies implicates susceptibility loci at 1q43, 2p16.1, 2q22.3 and 17q21.32. Hum. Mol. Genet., 21, 5359–5372. [DOI] [PubMed] [Google Scholar]
- 40. Rajcan-Separovic E., Harvard C., Liu X., McGillivray B., Hall J.G., Qiao Y., Hurlburt J., Hildebrand J., Mickelson E.C., Holden J.J., et al. (2007) Clinical and molecular cytogenetic characterisation of a newly recognised micro deletion syndrome involving 2p15-16.1. J. Med. Genet., 44, 269–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. de Leeuw N., Pfundt R., Koolen D.A., Neefs I., Scheltinga I., Mieloo H., Sistermans E.A., Nillesen W., Smeets D.F., de Vries B.B., et al. (2008) A newly recognised microdeletion syndrome involving 2p15p16.1: narrowing down the critical region by adding another patient detected by genome wide tiling path array comparative genomic hybridisation analysis. J. Med. Genet., 45, 122–124. [DOI] [PubMed] [Google Scholar]
- 42. Fernandez I.F., Blanco S., Lozano J., Lazo P.A. (2010) VRK2 inhibits mitogen-activated protein kinase signaling and inversely correlates with ErbB2 in human breast cancer. Mol. Cell. Biol., 30, 4687–4697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hotta R., Stamp L.A., Foong J.P., McConnell S.N., Bergner A.J., Anderson R.B., Enomoto H., Newgreen D.F., Obermayr F., Furness J.B., et al. (2013) Transplanted progenitors generate functional enteric neurons in the postnatal colon. J. Clin. Invest., 123, 1182–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Fattahi F., Steinbeck J.A., Kriks S., Tchieu J., Zimmer B., Kishinevsky S., Zeltner N., Mica Y., El-Nachef W., Zhao H., et al. (2016) Deriving human ENS lineages for cell therapy and drug discovery in Hirschsprung disease. Nature 531, 105-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Garcia-Barcelo M.M., Yeung M.Y., Miao X.P., Tang C.S., Cheng G., So M.T., Ngan E.S., Lui V.C., Chen Y., Liu X.L., et al. (2010) Genome-wide association study identifies a susceptibility locus for biliary atresia on 10q24.2. Hum. Mol. Genet., 19, 2917–2925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Tang C.S., Tang W.K., So M.T., Miao X.P., Leung B.M., Yip B.H., Leon T.Y., Ngan E.S., Lui V.C., Chen Y., et al. (2011) Fine mapping of the NRG1 Hirschsprung's disease locus. PLoS One, 6, e16181.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. O'Connell J., Gurdasani D., Delaneau O., Pirastu N., Ulivi S., Cocca M., Traglia M., Huang J., Huffman J.E., Rudan I., et al. (2014) A general approach for haplotype phasing across the full spectrum of relatedness. PLoS Genet., 10, e1004234.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Howie B., Fuchsberger C., Stephens M., Marchini J., Abecasis G.R. (2012) Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nat. Genet., 44, 955–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Marchini J., Howie B., Myers S., McVean G., Donnelly P. (2007) A new multipoint method for genome-wide association studies by imputation of genotypes. Nat. Genet., 39, 906–913. [DOI] [PubMed] [Google Scholar]
- 50. Willer C.J., Li Y., Abecasis G.R. (2010) METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics, 26, 2190–2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Boyle A.P., Hong E.L., Hariharan M., Cheng Y., Schaub M.A., Kasowski M., Karczewski K.J., Park J., Hitz B.C., Weng S., et al. (2012) Annotation of functional variation in personal genomes using RegulomeDB. Genome Res., 22, 1790–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Ward L.D., Kellis M. (2016) HaploReg v4: systematic mining of putative causal variants, cell types, regulators and target genes for human complex traits and disease. Nucleic. Acids. Res., 44, D877–D881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. GTEx Consortium. (2013) The Genotype-Tissue Expression (GTEx) project. Nat. Genet., 45, 580–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. So H.C., Gui A.H., Cherny S.S., Sham P.C. (2011) Evaluating the heritability explained by known susceptibility variants: a survey of ten complex diseases. Genet. Epidemiol., 35, 310–317. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.