Abstract
This study aimed to assess the associations of human leukocyte antigen (HLA)-DR and interleukin (IL)-18 gene polymorphisms with hepatitis B virus (HBV).
Clinical data were retrospectively reviewed between December 2006 and December 2015 at Xiangyang Central Hospital. HBV patients were assigned to the high and low viral load groups, respectively, according to HBV copies. HLA-DRB1∗03 polymorphisms and IL-18 polymorphisms were detected by sequence-specific primer-polymerase chain reaction (PCR-SSP) and PCR-ligase detection reaction (PCR-LDR), respectively. T cell subgroups were identified by flow cytometry, and IL-18, IL-12, interferon-γ (IFN-γ), IL-4, and IL-10 expression levels were assessed by ELISA. A total of 630 subjects were included in the analysis.
Compared with healthy controls, the chronic HBV group showed significantly lower IL-18 (P < .001), IL-12 (P < .001), and IFN-γ (P < .001) expression levels, and markedly increased IL-4 (P < .001) and IL-10 (P < .001) amounts. Th2 cytokine expression was high in HLA-DRB1∗03 positive (+) HBV patients, with low Th1 cytokine levels. The ratios of CD4+/CD8+ and Th1/Th2 cells decreased with increasing HBV DNA levels. The chronic HBV group showed a relatively high frequency of -137G in the IL-18 gene, while IL-18 expression was low in homozygous GG genotype individuals.
Polymorphisms in the HLA-DRB1∗03 and IL-18 genes are associated with viral load in HBV. HLA-DRB1 and IL-18 gene polymorphisms are involved in the regulation of the Th1/Th2 balance and expression of relevant cytokines that influence immune responses in HBV.
Keywords: chronic hepatitis B, human leukocyte antigen, interleukin, polymorphism
1. Introduction
Hepatitis B is an inflammatory liver disease caused by hepatitis B virus (HBV) infection.[1,2] The rate of HBV infection in China is high, with approximately 8% of the population affected;[3] meanwhile, HBV infection cannot be completely eliminated during the course of the disease.[4]
Studies have revealed 2 types of T effector cells in humans and mice, namely Th1 and Th2 cells, which secrete different cytokines.[5,6] Th1 cells secrete mainly interleukin-2 (IL-2) and interferon-γ (IFN-γ), and are active in the immune responses against viral infections and intracellular bacterial infections.[7] In contrast, Th2 cells mainly secrete IL-4, IL-5, IL-6, and IL-10, and participate in acute allergic reactions, regulating humoral immune responses.[7] In the chronic stages of HBV infection, Th1 cells are defective and release only low levels of cytokines; therefore, Th2-type cytokines predominate.[8,9] As a result, the cellular immune functions targeting the HBV and helper B cell functions are very poor, and virus elimination is ineffective. IL-18, IL-12, and IFN-γ upregulate Th1 cell immune responses and inhibit Th2 immunity, while IL-4 and IL-10 have the opposite effects.[10–15]
The course of HBV infection is not only influenced by factors associated with the virus, but by the type of immune responses generated in the host.[8,9] The human leukocyte antigen (HLA)-DR and IL-18 genes are involved in immune responses, and play critical roles in the outcomes of HBV-infected patients; polymorphisms in those genes are associated with various disease outcomes.[11,14,16–21]
Recent evidence shows that the HLA-DR genes greatly affect host susceptibility to HBV infection.[17] The effects of the HLA genotype on HBV infection chronicity could be associated with insufficient antiviral immune responses mediated by the critically protective T cells, and the fact that antigen-specific T cell receptors do not recognize antigenic peptides in the context of the HLA molecule.[16–18] Nevertheless, whether the underlying mechanisms are associated with genotype-regulated expression of Th1/Th2 cytokines remains unclear. In addition to reports on HLA-DR, recent studies conducted on the IL-18 gene have shown that IL-18 gene polymorphisms are associated with susceptibility to chronic hepatitis B infection, as well as the severity of liver injury and incidence of liver cancer.[20–22]
Despite emerging studies revealing the roles of HLA-DR and IL-18 gene polymorphisms in HBV infection, the nature of such associations remains unclear. Therefore, this study aimed to assess the effects of immune responses on HBV elimination and to explore the associations of HLA-DRB1∗03 and IL-18 gene polymorphisms with viral load in HBV infection, as well as the underlying mechanisms.
2. Methods
2.1. Patient population
The current study retrospectively reviewed data from patients between December 2006 and December 2015 at Xiangyang Central Hospital. The inclusion criteria were: diagnosis of CHB according to published criteria[23] (HBV, >1000 HBV copies/mL; patients with <1000 HBV copies/mL were classified as controls); age>18 years. The exclusion criteria were: other concomitant causes of liver disease or mixed etiologies (autoimmune hepatitis, primary biliary cirrhosis, alcoholic hepatitis, and so on); a family history of HCC; other concomitant malignant neoplasias; a history of autoimmune or inflammatory diseases such as systemic lupus erythematosus, rheumatoid arthritis, or inflammatory bowel disease; or incomplete records. The Ethics Committee of Xiangyang Central Hospital approved the study, and each participant provided a written informed consent.
2.2. Polymerase chain reaction-single specific primer (PCR-SSP)
Patients were genotyped using the PCR-SSP technique. Total DNA was extracted from peripheral blood leukocytes with the QIAamp DNA mini kit (QIAGEN Inc., Hilden, Germany), and stored at –20°C. PCR was carried out on a DNA Thermal Cycler (Gene Amp 9600; Perkin-Elmer Life Sciences, Waltham, MA). After agarose gel electrophoresis, the genotype of each patient was interpreted using the SSP-tool program (Dynal Biotech, Oslo, Norway). The PCR primers are described in Table 1.
Table 1.
The primers and probes used in PCR and LDR-PCR.
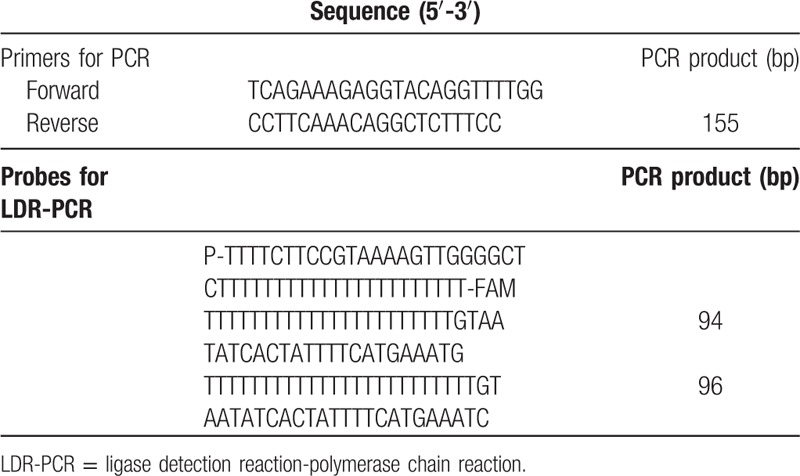
2.3. Ligase detection reaction-polymerase chain reaction (LDR-PCR)
LDR-PCR reactions were carried out according to the manufacturer's instructions (Takara, Otsu, Japan). Briefly, the LDR-PCR procedure was 30 cycles of 30 s at 94oC and 2 minutes at 60oC. Reactions were stopped by adding 0.5 μL of 0.5 mM EDTA. Aliquots of 2.5 μL of the reaction products were mixed with an equal volume of loading buffer (80% formamide, 10 mM EDTA, and 1.2% Blue dextran). The mixture was subjected to polyacrylamide gel electrophoresis (PAGE) under denaturing conditions. The sequences of the LDR-PCR probes are listed in Table 1.
2.4. Sandwich enzyme-linked immunosorbent assay (ELISA)
Sandwich ELISA was performed to measure the plasma concentrations of IL-18, IL-12, IFN-γ, IL-10, and IL-4, with ELISA kits specific to human IL-18, IL-12, IL-10, and IL-4 (Bender Medsystems Inc., Burlingame, CA), and human IFN-γ (R&D Systems, Minneapolis, MN), according to the manufacturers’ instructions.
2.5. Flow cytometry
Whole blood (1 mL) was mixed with 1 mL of either non-activating medium [10% fetal calf serum (FCS)-supplemented RPMI 1640 with 4 μL/mL GolgiStop (BD Biosciences, Franklin Lake, NJ)] or activating medium [non-activating medium with 50 ng/mL phorbol myristate acetate (PMA) and 5 μg/mL ionomycin (BD Biosciences, Franklin Lake, NJ)], and incubated at 37°C in an atmosphere with 5% CO2 for 5 hours. After washing with PBS, the cells were collected by centrifugation and adjusted to 5 × 105 white blood cells per test, and stained with PECy5-labeled anti-human CD4 monoclonal antibody (BD Biosciences, Franklin Lake, NJ). Cell fixation and permeabilization were performed with FACSTM Perm 2 (BD Biosciences), according to the manufacturer's instructions. Intracellular cytokines were stained with FITC-labeled anti-human IFN-γ and PE-labeled anti-human IL-4 monoclonal antibodies (BD Biosciences). IFN-γ- and IL-4-producing CD4+ T cells were analyzed on a FACS Calibur (BD Biosciences). Nonspecific staining with an isotype-matched control monoclonal antibody was <1%.
2.6. Statistical analysis
The distribution of demographic and clinical features was evaluated by 1-way ANOVA and the χ2 test for continuous and categorical variables, respectively. Agreement with the Hardy–Weinberg equilibrium (HWE) for each SNP was assessed by the goodness-of-fit χ2 test. Genotype frequencies were compared by the χ2 test and Fisher exact test, as appropriate. A binary logistic regression model was used to obtain the estimated odds ratios (ORs) and 95% confidence intervals (CIs) after adjusting for potential confounding variables. Two-tailed P < .05 was considered statistically significant. All analyses were performed with the SPSS 13.0 software (SPSS Inc., Chicago, IL).
3. Results
3.1. Population characteristics
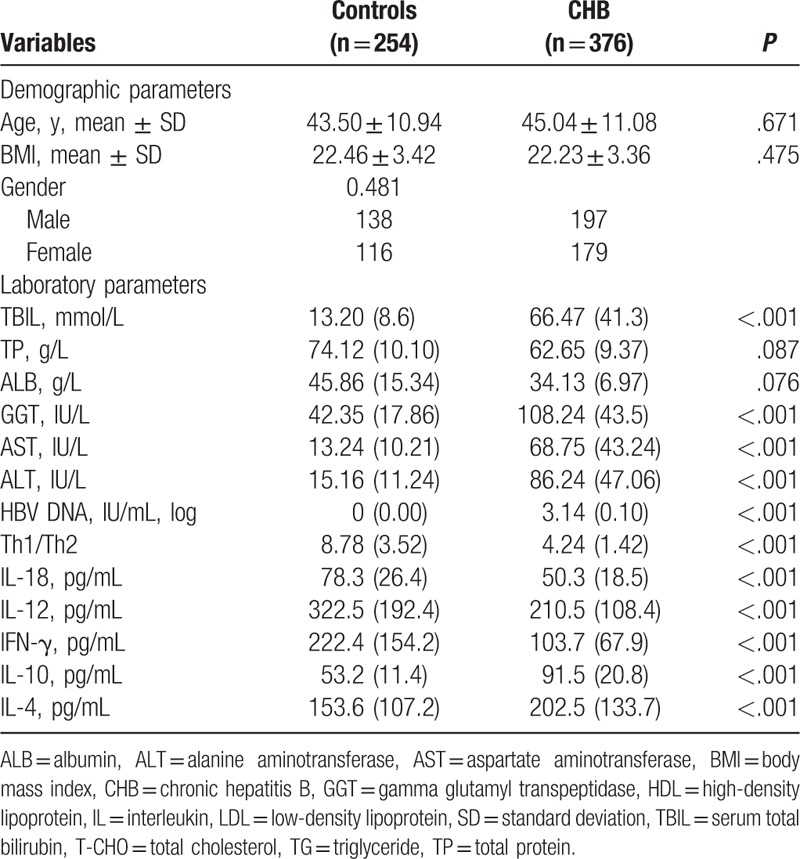
A total of 12 patients were excluded due to incomplete records as well as 1 patient for autoimmune hepatitis. Finally, 376 HBV patients (197 males and 179 females, 45.0 ± 11.1 years) and 254 healthy controls (138 males and 116 females, 43.5 ± 10.9 years) were included in the analysis (Fig. 1). Total bilirubin (TBIL), gamma-glutamyl transferase (GGT), aspartate aminotransferase (AST), alanine aminotransferase (ALT), HBV DNA, IL-10, and IL-4 levels were significantly higher in patients with CHB compared with healthy controls (all P < .001) (Table 2). Th1/Th2 ratios, and IL-18, IL-12, and IFN-γ amounts were significantly lower in patients with CHB than in controls (all P < .001) (Table 2). There were no significant differences in age, BMI, gender, total proteins (TP), and albumin (ALB) between the 2 groups (Table 2).
Figure 1.
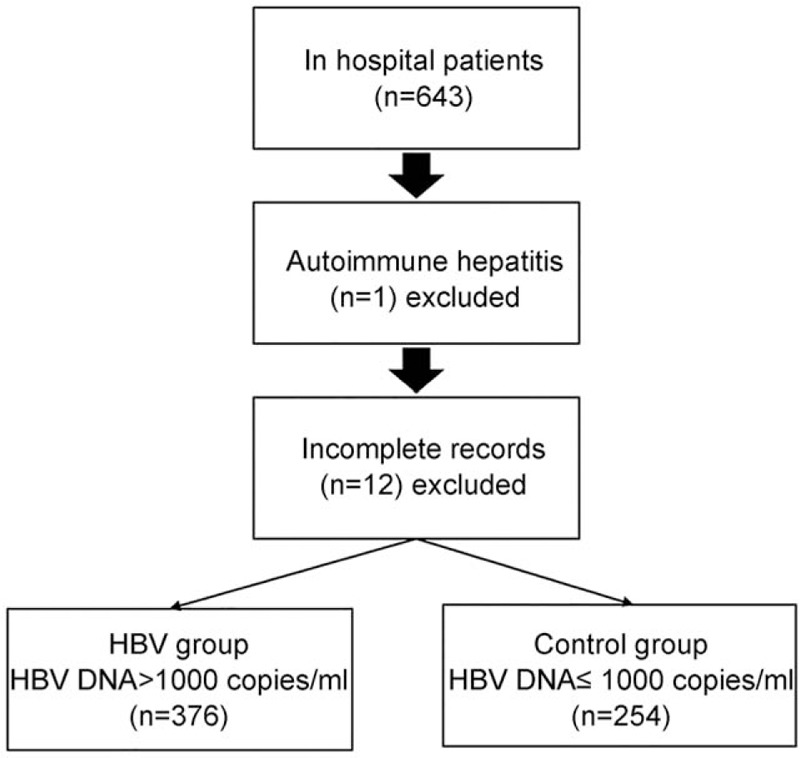
Patient flowchart.
Table 2.
Baseline characteristics of the study population.
3.2. Distribution frequencies of HLA-DRB1∗03 and IL-18 promoter -137G/C gene polymorphisms in patients with CHB
The distribution frequencies of HLA-DRB1 and IL-18 promoter -137 alleles are shown in Table 3. The allele frequency of HLA-DRB1∗03 in the chronic hepatitis B group (58.2%) was higher than that of the control group (42.1%), and there was a significant association between them (OR = 1.92, P = .001). The allele frequency of IL-18 promoter -137G in the chronic hepatitis B group (89.1%) was significantly higher than that of the control group (81.1%), with a significant association between them (OR = 1.90, P = .004). These data indicated that the distribution frequencies of HLA-DRB1∗03 and IL-18 promoter -137G/C gene polymorphisms were significantly associated with CHB.
Table 3.
Distribution frequencies of HLA-DRB1∗03 and IL-18 promoter –137G/C gene polymorphisms in CHB patients and healthy controls.

3.3. Multivariate logistic regression analysis of HLA-DRB1∗03 and IL-18 promoter -137G/C gene polymorphisms in association with CHB
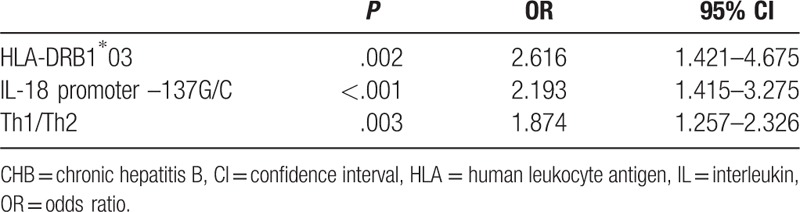
To perform the multivariate analysis of factors associated with CHB, variables with P < .05 in univariate analyses were selected and entered into the logistic regression model. The variables remaining in the equation were HLA-DRB1∗03 and IL-18 promoter –137G/C gene polymorphisms, as well as Th1/Th2 (Table 4). P-values (all P < .003) and 95% CIs listed in Table 4 suggested associations of HLA-DRB1∗03 and IL-18 promoter –137G/C gene polymorphisms with Th1/Th2.
Table 4.
Multivariate logistic regression analysis of HLA-DRB1∗03 and IL-18 promoter –137G/C gene polymorphisms with CHB.
3.4. HLA-DRB1∗03 and IL-18 promoter –137G/C gene polymorphisms are significantly and positively correlated with Th1/Th2
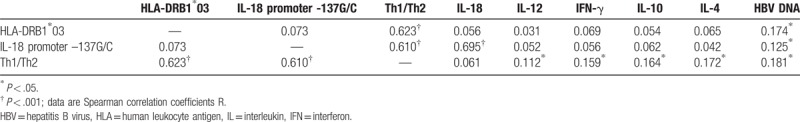
Spearman's correlation analyses showed that HLA-DRB1∗03 and IL-18 promoter –137G/C gene polymorphisms were significantly and positively correlated with Th1/Th2 (r = 0.623, P < .001; r = 0.610, P < .001), meanwhile IL-18 promoter -137G/C gene polymorphisms were significantly and positively correlated with IL-18 expression (r = 0.695, P < .001). No other significant correlations between the analyzed biomarkers were observed (Table 5). These data strongly suggested that HLA-DRB1∗03 and IL-18 promoter -137G/C gene polymorphisms were associated with Th1/Th2, and they were clustered in the factor analysis, suggesting that these polymorphisms could influence the secretion of proinflammatory cytokines.
Table 5.
Spearman correlation analysis of the assessed biomarkers (n = 376).
3.5. CD4+/CD8+ and Th1/Th2 ratios decrease with increasing HBV DNA replication
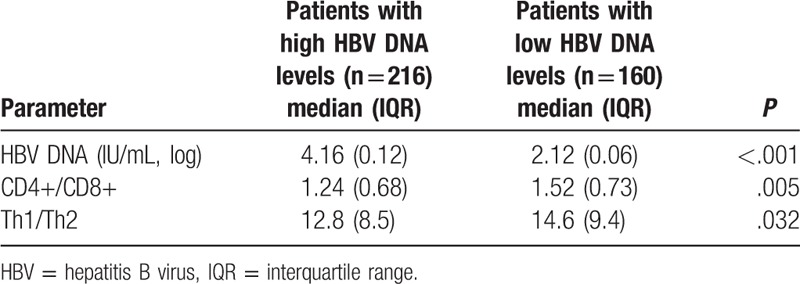
The patients were further sub-divided into 2 groups according to HBV DNA levels, including the high (n = 216) and low (n = 160) groups. As shown in Table 6, CD4+/CD8+ ratios were elevated in patients with low HBV DNA levels compared with those with high HBV DNA amounts (P = .005); the same trend was observed for Th1/Th2 ratios.
Table 6.
CD4+/CD8+ and Th1/Th2 ratios in subgroups with low- and high HBV DNA levels.
4. Discussion
The present study showed that polymorphisms in the HLA-DRB1∗03 and IL-18 genes are associated with viral load in HBV. These polymorphisms can influence the secretion of proinflammatory cytokines, probably affecting the course of the disease.
Indeed, HBV elimination by the human body is mainly dependent on viral activity, the host immune response, and genetic diversity.[24] Non-specific immune cells and cytokines play important roles in HBV elimination by regulating the Th1/Th2 balance and the type of immune response.[25–28] It is believed that the genetic background of patients with HBV infection is associated with the polymorphisms of cytokine genes involved in the immune response to HBV infection.[29] Previous studies have demonstrated the existence of single nucleotide polymorphisms at position –137 of the IL-18 gene promoter.[19,30–33] Furthermore, the HLA complex is highly polymorphic and considered the most complex genetic system identified so far, and participates in immune responses. Indeed, HLA plays a very important role in antigen recognition and presentation, immune response, and immune regulation.[16] Thursz et al[18] investigated HLA-II gene polymorphisms in patients infected with HBV and found that the frequency of HLA-DRB1∗1302 is significantly higher in self-restricted HBV infections than in chronic HBV infections. These observations suggest that the HLA-DRB1∗1302 genotype could be associated with a greater ability to eliminate the virus after infection. In the present study, the HLA-DRB1∗03 polymorphism was associated with chronic HBV infection, corroborating previous studies.[17,18]
As shown above, chronic hepatitis B was associated with polymorphisms at position –137 in the IL-18 gene promoter. IL-18 is a unique cytokine that enhances innate immunity and the Th1 cell immune response.[34] During HBV infection, the HBx protein induces IL-18 expression in the liver, and is associated with liver damage induced by enhanced FasL expression.[35] IL-18 also increases IFN-γ release from peripheral blood mononuclear cells in chronic hepatitis B patients, and improves the clearance of infected cells. The present study showed that plasma IL-18 levels in patients with chronic hepatitis B and healthy controls were significantly lower in individuals carrying the GG genotype compared with those harboring the C allele. Furthermore, IL-18 levels in patients with chronic hepatitis B were also lower than in healthy controls, which further confirmed that the position –137 of the IL-18 gene is a functional domain, and that the –137 G/C polymorphism may affect the expression of the IL-18 protein. These findings provide additional evidence for the influence of host genetic diversity on the immune response to HBV infection.[36]
Cytokine profiles produced by T cells largely determine the immune response; stimulation by antigen exposure causes T-helper lymphocytes to differentiate into 2 distinct phenotypes, Th1 and Th2.[37] Acute HBV infection mainly stimulates the cellular immune response, while non-specific cytokines involved in the humoral immune response play important roles in chronic HBV infection.[38–42] In addition, the mechanisms involved in HBV infection chronicity are associated with Th1/Th2 cell imbalance. The present study showed that compared with healthy individuals, HLA-DR expression and T cell proliferative capacity were significantly lower in patients with chronic HBV. Furthermore, the results showed that IL-18, IL-12, and IFN-γ expression levels were significantly higher in HLA-DRB1∗03-negative patients with chronic hepatitis B compared with HLA-DRB1∗03-positive patients, while IL-4 and IL-10 amounts were significantly lower. These observations suggest that the Th1 response in HLA-DRB1∗03-positive patients with chronic hepatitis B is suppressed, allowing the Th2 response to dominate, which could initiate and induce progression to chronic HBV infection. Accordingly, in the present study, the Th1 cell response was relatively high in patients not carrying the HLA-DRB1∗03 allele, suggesting that the HLA-DRB1∗03 allele could affect the response of CD4+ T cells. This study also showed that CD4+ T cells in hepatitis B patients were significantly less abundant than in controls, while CD8+ T cells were significantly more abundant. These effects of HLA-DRB1∗03 probably affect HBV chronicity. The decreased production of CD4+ T cells could result in insufficient antigen-specific secretion, causing incomplete elimination of the virus and affecting the viability of CD8+ T cells.[39] CD8+ T cells are potent immune effector cells; increased CD8+ T cell counts are associated with improved capability of virus elimination.[43] Chronic hepatitis B patients exhibit various degrees of cellular immune response dysfunction; the high viral load caused by increased viral replication can induce immune tolerance, affect the functions of CD4+ and CD8+ T cells, and result in an inadequate immune response.[43,44] Since the effects of CD4+ T cells are HLA-II restricted, it can be speculated that the mechanism underlying this effect involves the inability of antigen-specific T cell receptors to recognize antigen peptides in the context of the HLA molecule, which affects immune recognition, response, and tolerance to the HBV antigen. Nevertheless, additional studies are necessary to test this hypothesis.
The present study was not without limitations. The number of subjects was relatively small and from a single institution, possibly introducing some biases. What's more, only a limited panel of inflammatory markers and polymorphisms were assessed. Additional studies are necessary to determine the factors leading to HBV chronic infection. Finally, subgroup analysis was performed between low and high HBV DNA load groups only for CD4+/CD8+ and Th1/Th2 ratios in this study. Further subgroup analyses should be performed in future studies for other parameters assessed in this work, especially HLA and IL-18 gene polymorphisms.
5. Conclusion
In summary, the dynamic changes in IL-18 and Th1/Th2 cytokine expression, as well as in the CD4+/CD8+ ratio suggest alterations in the immune function of patients with HBV chronic infection. It can be hypothesized that such indexes can be used in assessing and monitoring the immune status of patients with HBV infection, which could provide more effective treatments for hepatitis B patients.
Author contributions
Conceptualization: Hua Jiang, Fengsheng Cao, Hong Cao.
Data curation: Hua Jiang, Hong Cao, Qun Rao.
Formal analysis: Hua Jiang, Fengsheng Cao, Ying Yang.
Funding acquisition: Ying Yang.
Investigation: Hua Jiang, Hong Cao, Ying Yang.
Methodology: Hua Jiang, Fengsheng Cao, Qun Rao.
Project administration: Ying Yang.
Resources: Hong Cao.
Software: Qun Rao.
Visualization: Qun Rao.
Writing – original draft: Hua Jiang, Hong Cao.
Writing – review & editing: Fengsheng Cao, Qun Rao, Ying Yang.
Footnotes
Abbreviations: CHB = chronic hepatitis B, HBV = hepatitis B virus, HIV-1 = human immunodeficiency virus 1, IFN-γ = interferon γ, IL = interleukin, LDR-PCR = ligase detection reaction-polymerase chain reaction, PCR-SSP = sequence-specific primed-polymerase chain reaction, SNP = single nucleotide polymorphism, TNF-α = tumor necrosis factor α.
HJ and HC contributed equally to this work.
This work was supported by the Hubei Natural Science Foundation of China (2012FFA071) and Xiangyang Science Foundation of China (2012[40]10).
The authors declare no conflicts of interest.
The author(s) of this work have nothing to disclose.
References
- [1].Song JE, Kim DY. Diagnosis of hepatitis B. Ann Transl Med 2016;4:338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Trepo C, Chan HL, Lok A. Hepatitis B virus infection. Lancet 2014;384:2053–63. [DOI] [PubMed] [Google Scholar]
- [3].Yan YP, Su HX, Ji ZH, et al. Epidemiology of hepatitis B virus infection in China: current status and challenges. J Clin Transl Hepatol 2014;2:15–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Liang TJ. Hepatitis B: the virus and disease. Hepatology 2009;49:S13–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Hsieh CS, Macatonia SE, Tripp CS, et al. Development of TH1 CD4+ T cells through IL-12 produced by Listeria-induced macrophages. Science 1993;260:547–9. [DOI] [PubMed] [Google Scholar]
- [6].Mosmann TR, Sad S. The expanding universe of T-cell subsets: Th1, Th2 and more. Immunol Today 1996;17:138–46. [DOI] [PubMed] [Google Scholar]
- [7].Jankovic D, Trinchieri G. IL-10 or not IL-10: that is the question. Nat Immunol 2007;8:1281–3. [DOI] [PubMed] [Google Scholar]
- [8].Jiang Y, Ma Z, Xin G, et al. Th1 and Th2 immune response in chronic hepatitis B patients during a long-term treatment with adefovir dipivoxil. Mediators Inflamm 2010;2010:143026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Saxena R, Kaur J. Th1/Th2 cytokines and their genotypes as predictors of hepatitis B virus related hepatocellular carcinoma. World J Hepatol 2015;7:1572–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Bossu P, Del Giudice E, Ciaramella A, et al. IL-18 and IL-18 receptors in the development of autoimmunity. Eur Cytokine Netw 2000;11:515–6. [PubMed] [Google Scholar]
- [11].Kimura K, Kakimi K, Wieland S, et al. Interleukin-18 inhibits hepatitis B virus replication in the livers of transgenic mice. J Virol 2002;76:10702–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Leung BP, McInnes IB, Esfandiari E, et al. Combined effects of IL-12 and IL-18 on the induction of collagen-induced arthritis. J Immunol 2000;164:6495–502. [DOI] [PubMed] [Google Scholar]
- [13].Yoshimoto T, Takeda K, Tanaka T, et al. IL-12 up-regulates IL-18 receptor expression on T cells, Th1 cells, and B cells: synergism with IL-18 for IFN-gamma production. J Immunol 1998;161:3400–7. [PubMed] [Google Scholar]
- [14].Nakanishi K, Yoshimoto T, Tsutsui H, et al. Interleukin-18 is a unique cytokine that stimulates both Th1 and Th2 responses depending on its cytokine milieu. Cytokine Growth Factor Rev 2001;12:53–72. [DOI] [PubMed] [Google Scholar]
- [15].Szkaradkiewicz A, Jopek A, Wysocki J. Effects of IL-12 and IL-18 on HBcAg-specific cytokine production by CD4 T lymphocytes of children with chronic hepatitis B infection. Antiviral Res 2005;66:23–7. [DOI] [PubMed] [Google Scholar]
- [16].Mellins E, Arp B, Singh D, et al. Point mutations define positions in HLA-DR3 molecules that affect antigen presentation. Proc Natl Acad Sci U S A 1990;87:4785–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Ramezani A, Hasanjani Roshan MR, Kalantar E, et al. Association of human leukocyte antigen polymorphism with outcomes of hepatitis B virus infection. J Gastroenterol Hepatol 2008;23:1716–21. [DOI] [PubMed] [Google Scholar]
- [18].Thursz MR, Kwiatkowski D, Allsopp CE, et al. Association between an MHC class II allele and clearance of hepatitis B virus in the Gambia. N Engl J Med 1995;332:1065–9. [DOI] [PubMed] [Google Scholar]
- [19].Arimitsu J, Hirano T, Higa S, et al. IL-18 gene polymorphisms affect IL-18 production capability by monocytes. Biochem Biophys Res Commun 2006;342:1413–6. [DOI] [PubMed] [Google Scholar]
- [20].Bao J, Lu Y, Deng Y, et al. Association between IL-18 polymorphisms, serum levels, and HBV-related hepatocellular carcinoma in a Chinese population: a retrospective case-control study. Cancer Cell Int 2015;15:72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Ferreira Sda C, Chacha SG, Souza FF, et al. IL-18, TNF, and IFN-gamma alleles and genotypes are associated with susceptibility to chronic hepatitis B infection and severity of liver injury. J Med Virol 2015;87:1689–96. [DOI] [PubMed] [Google Scholar]
- [22].Karra VK, Gumma PK, Chowdhury SJ, et al. IL-18 polymorphisms in hepatitis B virus related liver disease. Cytokine 2015;73:277–82. [DOI] [PubMed] [Google Scholar]
- [23].Lok AS, Heathcote EJ, Hoofnagle JH. Management of hepatitis B: 2000–summary of a workshop. Gastroenterology 2001;120:1828–53. [DOI] [PubMed] [Google Scholar]
- [24].Thursz M. Genetic susceptibility in chronic viral hepatitis. Antiviral Res 2001;52:113–6. [DOI] [PubMed] [Google Scholar]
- [25].Maddrey WC. Hepatitis B: an important public health issue. J Med Virol 2000;61:362–6. [DOI] [PubMed] [Google Scholar]
- [26].Maruyama T, McLachlan A, Iino S, et al. The serology of chronic hepatitis B infection revisited. J Clin Invest 1993;91:2586–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Chu CJ, Lok AS. Clinical significance of hepatitis B virus genotypes. Hepatology 2002;35:1274–6. [DOI] [PubMed] [Google Scholar]
- [28].Frodsham AJ. Host genetics and the outcome of hepatitis B viral infection. Transpl Immunol 2005;14:183–6. [DOI] [PubMed] [Google Scholar]
- [29].Ben-Ari Z, Mor E, Papo O, et al. Cytokine gene polymorphisms in patients infected with hepatitis B virus. Am J Gastroenterol 2003;98:144–50. [DOI] [PubMed] [Google Scholar]
- [30].Giedraitis V, He B, Huang WX, et al. Cloning and mutation analysis of the human IL-18 promoter: a possible role of polymorphisms in expression regulation. J Neuroimmunol 2001;112:146–52. [DOI] [PubMed] [Google Scholar]
- [31].An P, Thio CL, Kirk GD, et al. Regulatory polymorphisms in the interleukin-18 promoter are associated with hepatitis C virus clearance. J Infect Dis 2008;198:1159–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Cheong JY, Cho SW, Oh B, et al. Association of interleukin-18 gene polymorphisms with hepatitis B virus clearance. Dig Dis Sci 2010;55:1113–9. [DOI] [PubMed] [Google Scholar]
- [33].Zhang PA, Wu JM, Li Y, et al. Association of polymorphisms of interleukin-18 gene promoter region with chronic hepatitis B in Chinese Han population. World J Gastroenterol 2005;11:1594–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Dinarello CA. Interleukin-18. Methods 1999;19:121–32. [DOI] [PubMed] [Google Scholar]
- [35].Lee MO, Choi YH, Shin EC, et al. Hepatitis B virus X protein induced expression of interleukin 18 (IL-18): a potential mechanism for liver injury caused by hepatitis B virus (HBV) infection. J Hepatol 2002;37:380–6. [DOI] [PubMed] [Google Scholar]
- [36].Sun Y, Chen HY, Wang F, et al. Effect of IL-18 on peripheral blood monocytes from chronic hepatitis B patients. Zhonghua Gan Zang Bing Za Zhi 2003;11:470–3. [PubMed] [Google Scholar]
- [37].Chang J, Block TM, Guo JT. The innate immune response to hepatitis B virus infection: implications for pathogenesis and therapy. Antiviral Res 2012;96:405–13. [DOI] [PubMed] [Google Scholar]
- [38].Ait-Goughoulte M, Lucifora J, Zoulim F, et al. Innate antiviral immune responses to hepatitis B virus. Viruses 2010;2:1394–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Szkaradkiewicz A, Jopek A, Wysocki J, et al. HBcAg-specific cytokine production by CD4 T lymphocytes of children with acute and chronic hepatitis B. Virus Res 2003;97:127–33. [DOI] [PubMed] [Google Scholar]
- [40].Chen CH, Lin CL, Kao CH. Association between chronic hepatitis B virus infection and risk of osteoporosis: a nationwide population-based study. Medicine (Baltimore) 2015;94:e2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Liu Y, Xie L, Zhao J, et al. Association between catalase gene polymorphisms and risk of chronic hepatitis B, hepatitis B virus-related liver cirrhosis and hepatocellular carcinoma in Guangxi population: a case-control study. Medicine (Baltimore) 2015;94:e702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Cho JH, Yoon YD, Jang HM, et al. Immunologic monitoring of T-lymphocyte subsets and Hla-Dr-positive monocytes in kidney transplant recipients: a prospective, observational cohort study. Medicine (Baltimore) 2015;94:e1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Thimme R, Wieland S, Steiger C, et al. CD8(+) T cells mediate viral clearance and disease pathogenesis during acute hepatitis B virus infection. J Virol 2003;77:68–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Schmidt J, Blum HE, Thimme R. T-cell responses in hepatitis B and C virus infection: similarities and differences. Emerg Microbes Infect 2013;2:e15. [DOI] [PMC free article] [PubMed] [Google Scholar]


