Abstract
Acute respiratory tract infection (ARTI) is the most common causes of outpatient visit and hospital admission for children. The study aimed to report epidemiological data on respiratory viruses in a university-affiliated children's hospital.
The study was a retrospective study conducted in a university affiliated children's hospital from 2016 May to 2017 April. The results of all nasopharyngeal swab and sputum samples sent for the test for respiratory viruses (adenovirus, influenza A, influenza B, and respiratory syncytial virus) were extracted from the electronic healthcare records. Clinical characteristics were compared between groups with positive versus negative results for respiratory viruses. Multivariable regression models were employed by including age, gender, type of sample (swab vs sputum), source (emergency department vs others), and season to explore the independent factors associated with positive results for respiratory viruses.
A total of 34,961 samples were identified during the study period. A total of 3102 (8.9%) samples were positive for adenovirus, 2811 (8.0%) were positive for influenza A, 3460 (9.9%) were positive for influenza B, and 4527 (13.0%) were positive for respiratory syncytial virus. The positive rate of adenovirus was highest in April (50.8%), and lowest in November (3%). The absolute number of positive samples for adenovirus was highest in June (n = 587) and April (n = 544). For the test of influenza A, age was independently associated with positive result. With 1 year increase in age, the odds of positive result increased by 12% (odds ratio [OR]: 1.12; 95% confidence interval [CI]: 1.11–1.13; P < .001). As compared with the autumn, the summer showed significantly lower rate of positive for RSV (OR: 0.49; 95% CI: 0.38–0.62; P < .001), whereas the winter had higher risk of positive result (OR: 3.88; 95% CI: 3.37–4.50; P < .001).
The study reported epidemiological data on the prevalence of respiratory viruses in a large tertiary care children's hospital. Age, gender, type of sample, source, and season were associated with the positive rates for respiratory viruses.
Keywords: acute respiratory tract infection, nasopharyngeal swab, respiratory viruses
1. Introduction
Acute respiratory tract infection (ARTI) is the most common causes of outpatient visit and hospital admission for children.[1] The severity of the ARTI varies substantially depending on the site of infection, the type of viruses, involved organs and comorbidities.[2] For example, viral pneumonia can be life threatening due to uncontrolled systematic inflammatory response, leading to septic shock, acute respiratory distress syndrome (ARDS) and multiple organ failure.[3–5] These later conditions require intensive care unit admission, and are associated with significantly increased risk of death, high medical cost and family distress.[4] It was estimated that there were approximately 1.9 million children died annually from ARTI, and most of them were from developing countries.[6] The type of respiratory viruses is very important for the understanding of the underlying pathogenesis of respiratory infections in children. A recent epidemiological study involving 17 centers and 8 countries showed that rhinovirus/enterovirus (41.5%) is the most prevalent viruses causing children's respiratory infection, followed by influenza (15.8%), adenovirus (9.8%), parainfluenza, and respiratory syncytial virus (RSV) (both 9.7%).[7]
China is a country with the largest population in the world. With the implementation of 2-child policy, the children's hospitals in China are facing great challenge to treat increasing number of children. ARTI is the most common reason for outpatient visit and hospital admission in China,[8] imposing a great challenge for clinicians. Throat swab and sputum samples are usually ordered for children with suspected ARTI and the number is very large.[9,10] However, there is no study reporting epidemiological data on test results of respiratory viruses. The study aimed to report epidemiological data on respiratory viruses in a university-affiliated children's hospital. Furthermore, we investigated distributions of different types of respiratory viruses with seasonal changes. Risk factors for positive results for adenovirus, influenza A, influenza B, and respiratory syncytial virus (RSV) were also investigated.
2. Methods
The study was a retrospective study conducted in a university affiliated children's hospital from 2016 May to 2017 April. The results of all nasopharyngeal swab and sputum samples sent for the test for respiratory viruses (adenovirus, influenza A, influenza B, and respiratory syncytial virus) were extracted from the electronic healthcare records. Samples with missing information on test results were excluded. The study was approved by the ethics committee of Children's Hospital of Zhejiang University School of Medicine (2018-IRB-001).
2.1. Examination of respiratory viruses
Adenovirus was detected using the colloidal gold method (diagnostic kit provided by KaiBiLi Company; Genesis Co.; http://www.hgb.com.cn/En/about.html). Samples were obtained using the throat swab, which was immediately prepared for analysis. If a sample cannot be analyzed immediately, it could be stored in 2°C to 8°C for at most 8 hours. The throat swab should not be touched with saliva. RSV was detected using diagnostic kit (colloidal gold method, KaiBiLi), which was able to identify type A and type B but not to distinguish subtypes. Influenza A and B were detected using nucleoprotein antigen test kit (colloidal gold method, KaiBiLi).
2.2. Clinical variables
Relevant clinical variables including age, gender, type of sample (throat swab and sputum), source of sample (emergency room vs others), and date of sampling were also collected. The date of sampling was categorized into 4 seasons of spring, summer, autumn, and winter. Age was obtained by the difference between date of birth and the date of sampling. Age was an important factor that might influence the distributions of respiratory virus in Children.[11] Season was also an important factor determining the epidemic waves of ARTI in children.[12] Spring was defined as the period from 2016 May 2, 2016 to May 5, 2016 and from February 3, 2017 to April 9, 2017. Summer was defined from the period from May 5, 2016 to August 7, 2016. Autumn was from August 7, 2016 to November 7, 2016 and winter was from November 7, 2016 to February 3, 2017. Furthermore, samples obtained from emergency room and ward can be quite different, for example, samples from ER were more likely to be due to acute respiratory tract infection, while those from ward were obtained from patients with other medical conditions such as hematological malignancy, and thus source of sample was also included for analysis.
2.3. Statistical analysis
All samples were divided into 2 groups by the results of virus test (positive vs negative). Numeric variables were expressed with mean and standard deviations, and were compared between groups by using t test. Categorical variables were expressed as the number and proportion, and were compared by using Chi-square test or Fisher's exact test as appropriate.[13] The R package CBCgrps was employed for statistical description and bivariate inference.[14] Within each calendar month, the number of positive and negative samples was displayed in a barplot. The percentages of positive samples were reported in the barplot for each respiratory virus. To investigate variables associated with positive results, multivariable Logistic regression model including all potential factors (age, gender, season, source of sample, type of sample) was employed.[15,16] Due to limited number of covariates and the large number of observations, the full model was not reduced (low risk of over fitting). Multivariable Logistic regression model was built upon complete case analysis that observations with missing values on covariates were excluded.[17] Two-tailed P value <.05 was considered as statistical significant. All statistical analyses were performed using R software (version 3.3.2).
3. Results
A total of 34,961 samples were identified during the study period. A total of 3102 (8.9%) samples were positive for adenovirus, 2811 (8.0%) were positive for influenza A, 3460 (9.9%) were positive for influenza B, and 4527 (13.0%) were positive for respiratory syncytial virus. The positive rate of adenovirus was highest in April (50.8%), and lowest in November (3%). The number of sampling was increasing abruptly from November 2017, which was attributable to more outpatient visits for respiratory problem in Autumn and Winter periods (Fig. 1). The absolute number of positive samples for adenovirus was highest in June (n = 587) and April (n = 544). The positive rate of influenza A was highest in December (18.5%), and lowest in July (0) and August (0.2%). The absolute number of positive samples for influenza A was highest in December (n = 1187) and lowest in August (n = 0, Fig. 2). The positive rate of influenza B was highest in April (23.9%), and lowest in December (0.4%) and November (0.6%). The absolute number of positive samples for influenza B was highest in March (n = 1548) and lowest in November (n = 22, Fig. 3). The positive rate of RSV was highest in December (28.5%), and lowest in June (2.9%) and July (4.3%). The absolute number of positive samples for RSV was highest in December (n = 1825) and lowest in May (n = 28, Fig. 4). There were 207 children with combined infection of adenovirus and RSV, 21 children with combined infection of influenza A and RSV, 63 children with combined infection of influenza B and RSV, 9 children with influenza A and adenovirus, 46 children with influenza B and adenovirus and 2 children with combined infection of influenza B, RSV and adenovirus.
Figure 1.
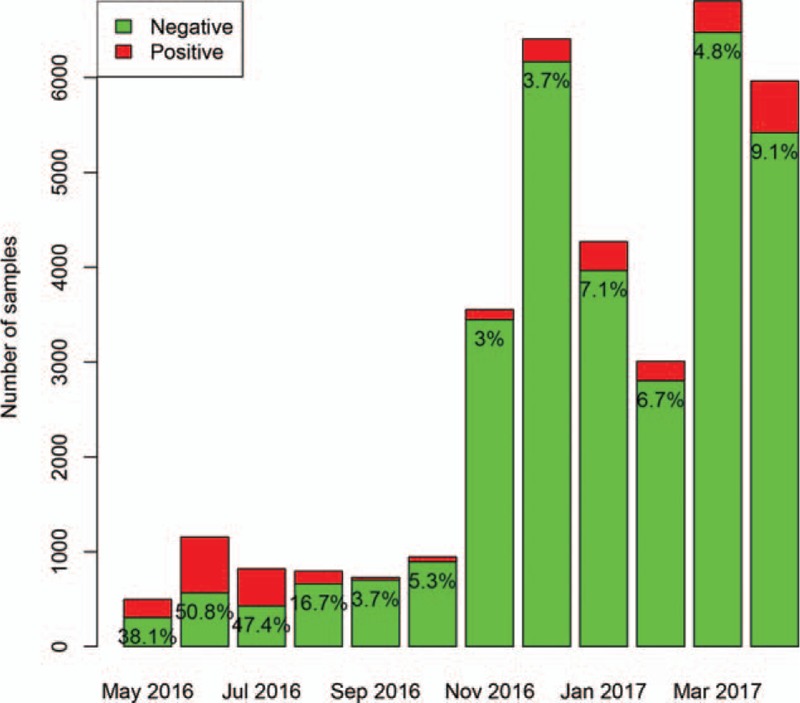
Distribution of positive rate for adenovirus during the study period. The positive rate of adenovirus was highest in April (50.8%), and lowest in November (3%). The absolute number of positive samples for adenovirus was highest in June (n = 587) and April (n = 544).
Figure 2.
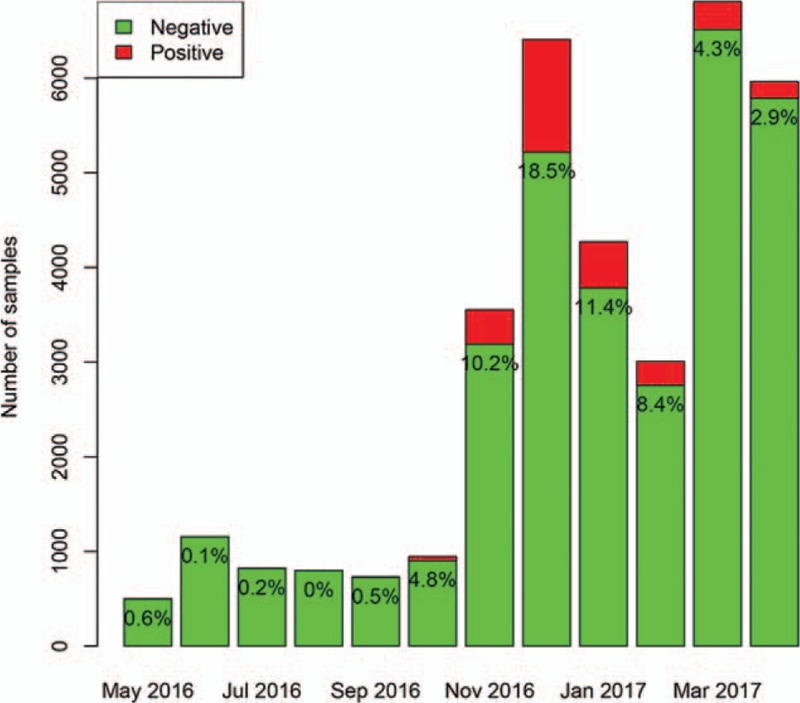
Distribution of positive rate for influenza A during the study period. The positive rate of influenza A was highest in December (18.5%), and lowest in July (0) and August (0.2%). The absolute number of positive samples for influenza A was highest in December (n = 1187) and lowest in August (0).
Figure 3.
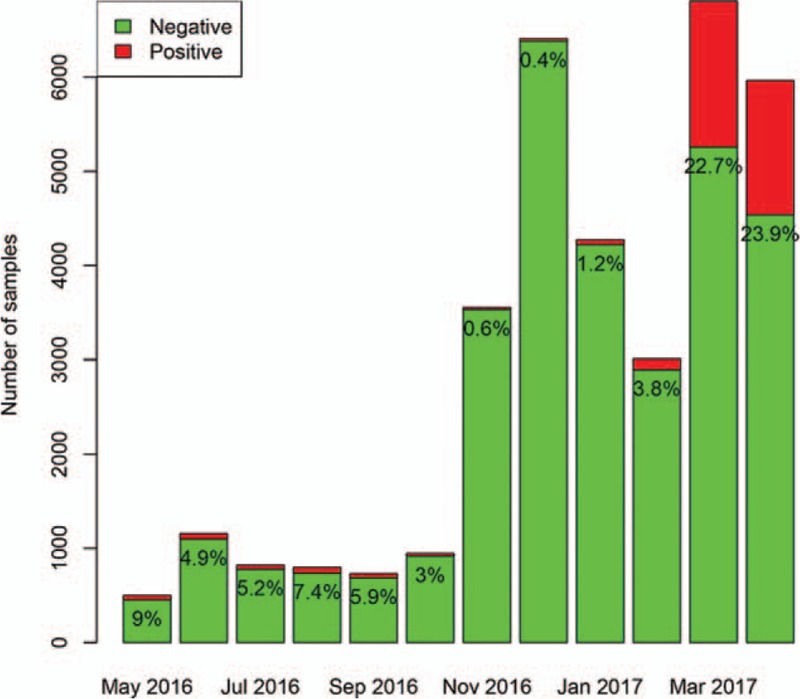
Distribution of positive rate for influenza B during the study period. The positive rate of influenza B was highest in April (23.9%), and lowest in December (0.4%) and November (0.6%). The absolute number of positive samples for influenza B was highest in March (n = 1548) and lowest in November (n = 22).
Figure 4.
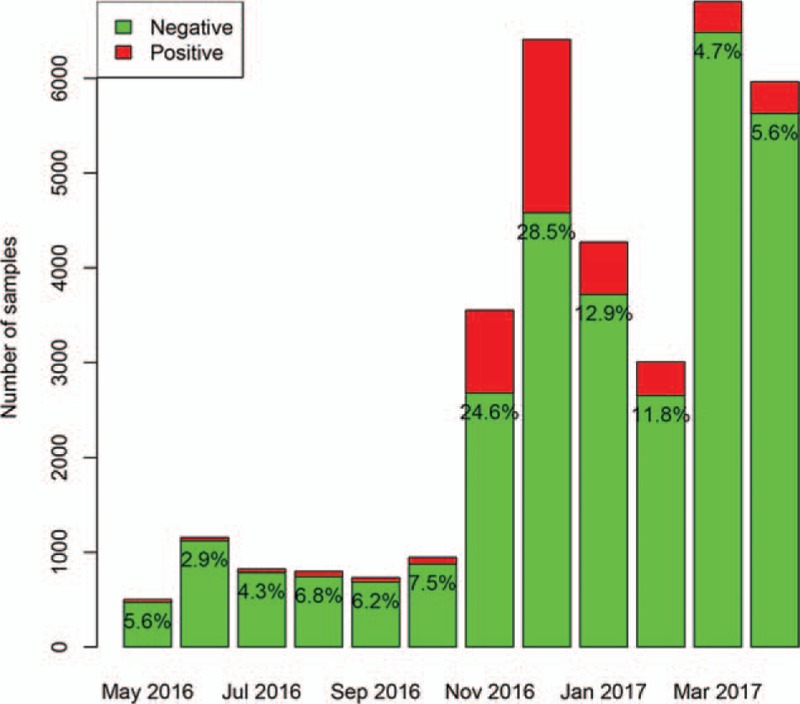
Distribution of positive rate for respiratory syncytial virus (RSV) during the study period. The positive rate of RSV was highest in December (28.5%), and lowest in June (2.9%) and July (4.3%). The absolute number of positive samples for RSV was highest in December (n = 1825) and lowest in May (n = 28).
Samples from boys were more likely to have positive result for adenovirus (0.56 vs 0.54 for positive vs negative; P = .003). Swab samples were more likely to be positive for adenovirus (0.32 vs 0.24; P < .001). However, samples from emergency department were less likely to have positive results for adenovirus (0.01 vs 0.03; P < .001). Most of the samples were ordered during spring (44%) and winter (40%). In summer, the samples were more likely to be positive for adenovirus (0.4 vs 0.04; P < .001, Table 1). In winter, the samples were more likely to be positive for influenza A (0.72 vs 0.37; P < .001). Sputum samples were more likely to be positive for influenza A (0.82 vs 0.75; P < .001, Table 2). However, influenza B was more likely to be positive in Spring (0.89 vs 0.39; P < .001, Table 3). The samples positive for RSV were more likely to be obtained by throat swab (0.26 vs 0.24; P < .001). The positive rate for RSV was the highest in winter (0.72), followed by spring (0.21), autumn (0.05), and summer (0.02). There was no significant difference in the source of sample between groups of positive versus negative for RSV (Table 4).
Table 1.
Comparing baseline characteristics of samples tested negative and positive for adenovirus.
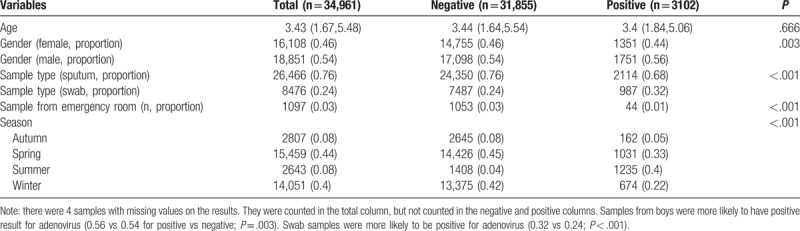
Table 2.
Comparing baseline characteristics of samples tested negative and positive for influenza A virus.
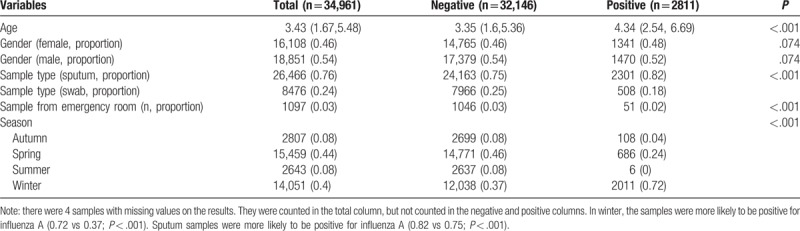
Table 3.
Comparing baseline characteristics of samples tested negative and positive for influenza B virus.
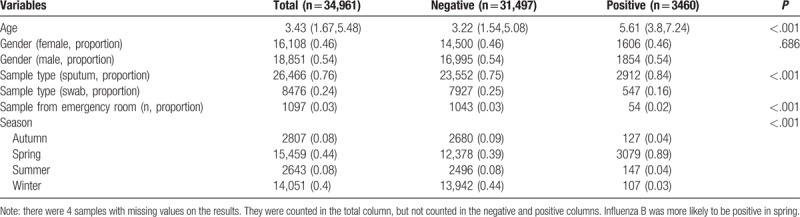
Table 4.
Comparing baseline characteristics of samples tested negative and positive for respiratory syncytial virus.
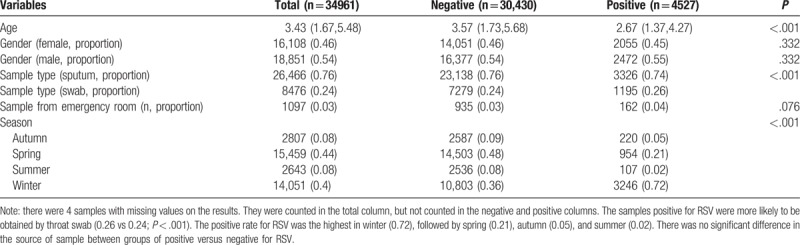
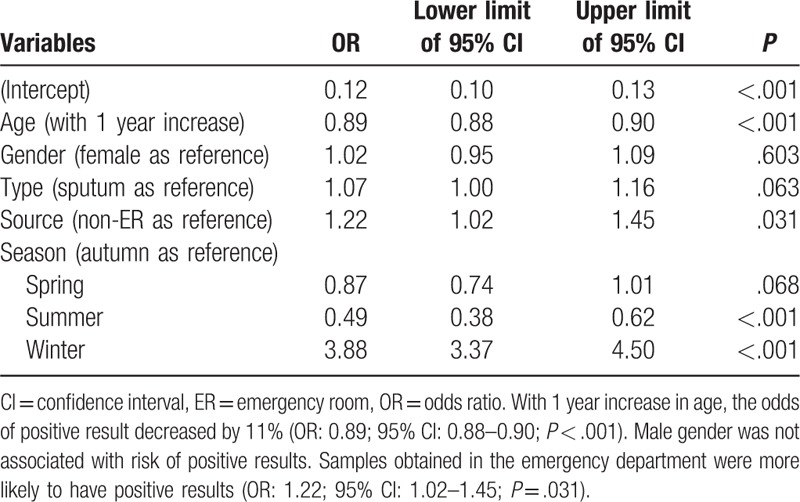
In multivariable logistic regression analysis, age was independently associated with positive result for adenovirus (Table 5). With 1 year increase in age, the odds of positive result increased by 2% (odds ratio [OR]: 1.02; 95% confidence interval [CI]: 1.01–1.04; P = .001). Male gender was associated with higher risk of positive for adenovirus (OR: 1.10; 95% CI: 1.02–1.20; P = .017). The type of sample was not significantly associated with positive result. As compared with the autumn, samples collected in the summer were more likely to be positive for adenovirus (OR: 14.48; 95% CI: 12.17–17.34; P < .001), whereas the winter was associated with lower risk of positive result (OR: 0.81; 95% CI: 0.68–0.97; P = .018). For the test of influenza A (Table 6), age was independently associated with positive result. With 1 year increase in age, the odds of positive result increased by 12% (OR: 1.12; 95% CI: 1.11–1.13; P < .001). Male gender was not associated with risk of positive results. Samples obtained by throat swab was less likely to report positive result for influenza A (OR: 0.68; 95% CI: 0.62–0.76; P < .001). As compared with the autumn, samples obtained in summer were more likely to be positive for influenza A (OR: 0.06; 95% CI: 0.02–0.12; P < .001), whereas the winter was associated with higher risk of positive result (OR: 3.68; 95% CI: 3.03–4.51; P < .001). For the test of influenza B (Table 7), age was independently associated with positive result. With 1 year increase in age, the odds of positive result increased by 25% (OR: 1.25; 95% CI: 1.23–1.27; P < .001). Male gender was not associated with risk of positive results. Samples obtained by throat swab was less likely to report positive result for influenza B (OR: 0.82; 95% CI: 0.74–0.91; P < .001). Samples obtained in the emergency department were less likely to have positive results (OR: 0.39; 95% CI: 0.29–0.52; P < .001). As compared with the autumn, the samples obtained in summer were more likely to be positive for influenza B (OR: 1.3; 95% CI: 1.01–1.67; P = .04), whereas the winter had lower risk of positive result (OR: 0.12; 95% CI: 0.09–0.16; P < .001). For the test of RSV (Table 8), age was independently associated with positive result. With 1 year increase in age, the odds of positive result decreased by 11% (OR: 0.89; 95% CI: 0.88–0.90; P < .001). Male gender was not associated with risk of positive results. Samples obtained in the emergency department were more likely to have positive results (OR: 1.22; 95% CI: 1.02–1.45; P = .031). As compared with the autumn, the samples obtained in summer were less likely to be positive for RSV (OR: 0.49; 95% CI: 0.38–0.62; P < .001), whereas the winter was more likely to have positive result (OR: 3.88; 95% CI: 3.37–4.50; P < .001).
Table 5.
Multivariable logistic regression analysis exploring independent factors associated with positive result for adenovirus.
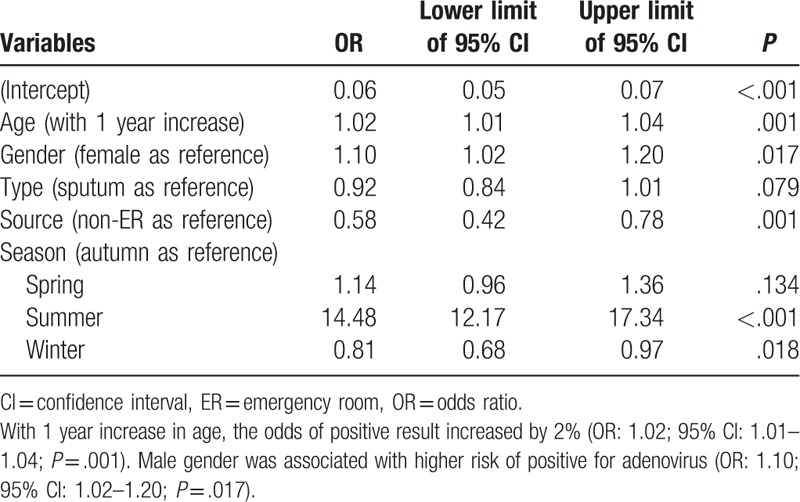
Table 6.
Multivariable logistic regression analysis exploring independent factors associated with positive result for influenza A.
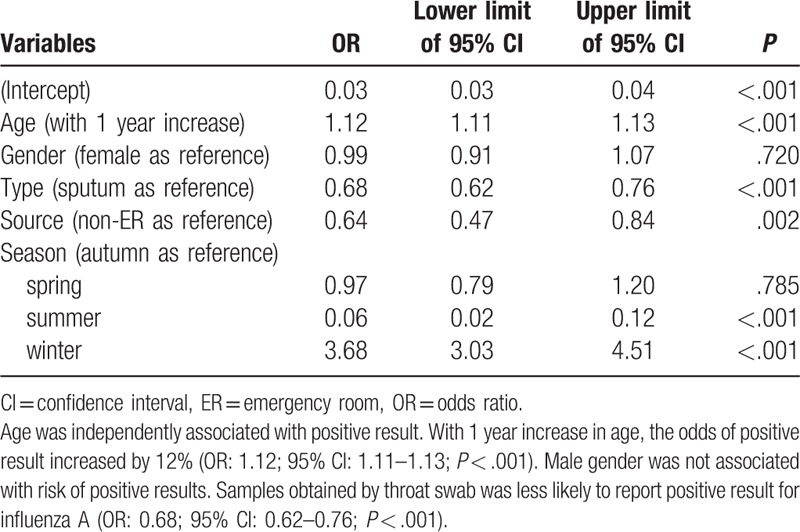
Table 7.
Multivariable logistic regression analysis exploring independent factors associated with positive result for influenza B.
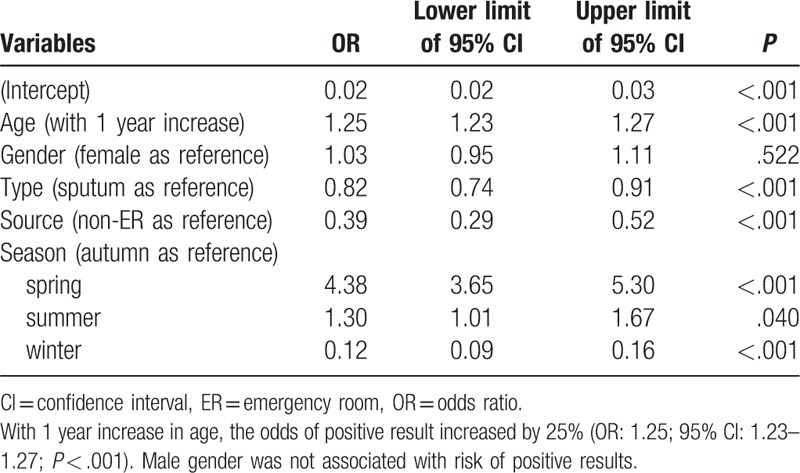
Table 8.
Multivariable logistic regression analysis exploring independent factors associated with positive result for respiratory syncytial virus.
4. Discussion
The study showed that among samples tested for respiratory virus, RSV had the highest positive rate (13.0%), followed by influenza B (9.9%), adenovirus (8.9%), and influenza A (8.0%). The positive rates for the 4 respiratory viruses changed in distinct seasonal patterns. Adenovirus was most likely to be detected in summer, followed by spring, winter and autumn. Influenza A was most likely to be detected in winter, followed by spring, autumn, and summer. Influenza B was most likely to be detected in spring, followed by autumn, summer, and winter. RSV was most likely to be detected in winter, followed by spring, autumn, and summer. The multivariable analysis confirmed these results. The seasonal pattern of these common ARIT viruses can help to develop annual vaccination policy for children. Vaccination can effectively reduce the incidence and severity of respiratory infection in children older than 6 months.[18]
Taylor et al conducted an epidemiological study involving 17 centers in 8 countries, which showed very similar prevalence for influenza (15.8%), adenovirus (9.8%), and RSV (9.7%). While the prevalence of RSV in our study was higher than Taylor's study (13.0% vs 9.7%), the prevalence of influenza was lower than that study (8%–9% vs 15.8%). Consistent with Taylor's study, the rate of influenza virus increased with age, while the RSV declined with age. However, while our study showed increased positivity of adenovirus with age (OR: 1.02; 95% CI: 1.01–1.04; P = .001), Taylor's study showed declining prevalence of adenovirus with age.[7] The difference might be attributable to different methodological designs for the 2 studies. While Taylor's study was a population-based study that all eligible healthy children receiving H1N1 vaccine were enrolled, our study was based on a tertiary care hospital. Furthermore, Taylor's study analyzed their data in patient level, and each case was confirmed to have the diagnosis of virus infection. However, our study was based on swab or sputum samples, some positive samples may not represent a clinical infection and 1 patient can have several samples.
Traditionally, nasopharyngeal washes or swabs were used for the diagnosis of respiratory viral infection. Sputum is secreted from lower respiratory tract and is seldom used for viral testing. In our study, sputum showed higher positive rate for the detection of respiratory virus than swab. For example, adenovirus was more likely to be detected in swab than that in sputum (11.6% vs 8.0%; P < .001). This is inconsistent with the results obtained in a study conducted in adult outpatient, which showed that respiratory viruses were detected more frequently in sputum than that in swab.[19] Another study also showed that while bacteria were more frequently detected in sputum, virus was more frequently detected in swabs.[20] However, there are also other studies showing consistent results with our study. Branche et al showed that the sputum samples were able to increase the diagnostic yield of nasopharyngeal swab, which might be to higher viral loads in sputum than that in swabs.[21] Similar finding that sputum samples can increase diagnostic yield were replicated in other studies.[22,23] Collectively, current evidences are inconsistent with regard to the diagnostic yield of sputum versus swab and further well controlled studies are required to settle this debate.
Seasonal variations of different types of viruses have also been reported in other studies. Saraya et al[24] showed that influenza viruses were more frequently detected in spring and winter, and RSV was more common in autumn. The variation pattern of influenza viruses was generally consistent. However, our study showed that RSV was more common in winter and spring, which was different from that reported in Saraya's study. Their study was conducted in adult patients with asthma, which might be responsible for the difference. In another study conducted from the year 2002 to 2014 involving 5102 samples, RSV was most frequently detected from December to March, influenza viruses from November to March, and HRV from December to June.[25] Yang et al's[11] study found 2-peak patterns across age groups, one in winter and the other in spring/summer. The study was conducted in subtropical area, which was consistent with our study.
Several limitations must be acknowledged in the present study. First, the study was based on clinical samples and some relevant clinical characteristics were not included. For example, the severity of illness of the children, the diagnosis (upper respiratory tract infection vs pneumonia), and clinical outcomes (outpatient visit, hospital or ICU admission) can provide further insights into infection caused by different pathogens. Second, the study was retrospective in design, with inherent limitations such as selection bias and data missing.[26,27] However, the study was based on electronic healthcare records (EHR), which benefits from large number of samples and could be considered as a kind of big data.[28–30] With the development of computer technology, the big data analytics have found its way into all walks of life.[31,32] In our study, the utilization of EHR can provide more insights into the epidemiology of viral respiratory tract infection in children. Third, the indications for the test of respiratory viruses in sputum and/or swab samples were not clearly defined in the study. Because the study was not prospectively designed and the ordering of sample test was largely determined by the treating physician. However, in our clinical practice, there were protocols for ordering tests for respiratory viruses and included patients were suspected to have ARTI. Finally, there are other minor respiratory viruses such as human bocavirus and rhinovirus that were not included in this analysis. Since they are also important pathogens for causing ARTI in children, we will plan future studies by incorporating these viruses.
In conclusion, the study reported epidemiological data on the prevalence of respiratory viruses in a large tertiary care children's hospital. The study showed that among samples tested for respiratory virus, RSV had the highest positive rate (13.0%), followed by influenza B (9.9%), adenovirus (8.9%), and influenza A (8.0%). The positive rates of the 4 respiratory viruses showed distinct seasonal patterns. Such a seasonal pattern may help to develop a specific vaccination program, which was shown to be effective in reducing the incidence and severity of respiratory infection in children.[18]
Author contributions
Conceptualization: Sheng Ye.
Data curation: Sheng Ye.
Formal analysis: Sheng Ye.
Investigation: Tianlin Wang.
Methodology: Sheng Ye, Tianlin Wang.
Supervision: Tianlin Wang.
Validation: Tianlin Wang.
Visualization: Tianlin Wang.
Writing – review & editing: Sheng Ye, Tianlin Wang.
Footnotes
Abbreviations: ARDS = acute respiratory distress syndrome, ARTI = acute respiratory tract infection, ER = emergency room, HER = electronic healthcare record, RSV = respiratory syncytial virus.
Sheng Ye received grants from the Zhejiang Medical and Health Science and Technology Plan project (2017 B119), the Zhejiang Medical and Health Science and Technology Plan project (2012 KYB119) and the Natural Science Foundation of Zhejiang province (LY12H19006). The authors have no conflicts of interest to disclose.
References
- [1].Monto AS. Epidemiology of viral respiratory infections. Am J Med 2002;112(suppl 6A):4S–12S. [DOI] [PubMed] [Google Scholar]
- [2].Pavia AT. Viral infections of the lower respiratory tract: old viruses, new viruses, and the role of diagnosis. Clin Infect Dis 2011;52(suppl 4):S284–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Zhang Z, Smischney NJ, Zhang H, et al. AME evidence series 001—The Society for Translational Medicine: clinical practice guidelines for diagnosis and early identification of sepsis in the hospital. J Thorac Dis 2016;8:2654–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Zhang Z, Ni H. Prediction model for critically ill patients with acute respiratory distress syndrome. PLoS One 2015;10:e0120641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Zhang Z, Ni H, Qian Z. Effectiveness of treatment based on PiCCO parameters in critically ill patients with septic shock and/or acute respiratory distress syndrome: a randomized controlled trial. Intensive Care Med 2015;41:444–51. [DOI] [PubMed] [Google Scholar]
- [6].Williams BG, Gouws E, Boschi-Pinto C, et al. Estimates of world-wide distribution of child deaths from acute respiratory infections. Lancet Infect Dis 2002;2:25–32. [DOI] [PubMed] [Google Scholar]
- [7].Taylor S, Lopez P, Weckx L, et al. Respiratory viruses and influenza-like illness: epidemiology and outcomes in children aged 6 months to 10 years in a multi-country population sample. J Infect 2017;74:29–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Huang Y, Hua J, Wang D, et al. Risk factors of respiratory syncytial virus infection among pediatric influenza-like illness and severe acute respiratory infections in Suzhou. China J Med Virol 2017;375:1545. [DOI] [PubMed] [Google Scholar]
- [9].Hara M, Takao S, Shimazu Y. Comparison of throat swab and nasopharyngeal aspirate specimens for rapid detection of adenovirus. Diagn Microbiol Infect Dis 2015;82:135–6. [DOI] [PubMed] [Google Scholar]
- [10].Heikkinen T. Comparative study of nasopharyngeal aspirate and nasal swab specimens for detection of influenza. BMJ 2001;322:138–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Yang L, Chan KH, Suen LKP, et al. Age-specific epidemic waves of influenza and respiratory syncytial virus in a subtropical city. Sci Rep 2015;5:10390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Zhang XL, Ji W, Ji ZH, et al. Epidemiological study on respiratory syncytial virus and its bronchopneumonia among children in Suzhou. Zhonghua Yu Fang Yi Xue Za Zhi 2007;41:371–4. [PubMed] [Google Scholar]
- [13].Zhang Z. Univariate description and bivariate statistical inference: the first step delving into data. Ann Transl Med 2016;4:91–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Zhang Z, Gayle AA, Wang J, et al. Comparing baseline characteristics between groups: an introduction to the CBCgrps package. Ann Transl Med 2017;5:484–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Tolles J, Meurer WJ. Logistic regression: relating patient characteristics to outcomes. JAMA 2016;316:533–4. [DOI] [PubMed] [Google Scholar]
- [16].Zhang Z. Model building strategy for logistic regression: purposeful selection. Ann Transl Med 2016;4:111–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Zhang Z. Missing values in big data research: some basic skills. Ann Transl Med 2015;3:323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Flannery B, Chung JR, Belongia EA, et al. Interim estimates of 2017–2018 seasonal influenza vaccine effectiveness—United States, February 2018. MMWR Morb Mortal Wkly Rep 2018;67:180–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Jeong JH, Kim KH, Jeong SH, et al. Comparison of sputum and nasopharyngeal swabs for detection of respiratory viruses. J Med Virol 2014;86:2122–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Yoshii Y, Shimizu K, Morozumi M, et al. Detection of pathogens by real-time PCR in adult patients with acute exacerbation of bronchial asthma. BMC Pulm Med 2017;17:150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Branche AR, Walsh EE, Formica MA, et al. Detection of respiratory viruses in sputum from adults by use of automated multiplex PCR. J Clin Microbiol 2014;52:3590–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Falsey AR, Formica MA, Walsh EE. Yield of sputum for viral detection by reverse transcriptase PCR in adults hospitalized with respiratory illness. J Clin Microbiol 2012;50:21–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Memish ZA, Al-Tawfiq JA, Makhdoom HQ, et al. Respiratory tract samples, viral load, and genome fraction yield in patients with Middle East respiratory syndrome. J Infect Dis 2014;210:1590–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Saraya T, Kimura H, Kurai D, et al. The molecular epidemiology of respiratory viruses associated with asthma attacks: a single-center observational study in Japan. Medicine (Baltimore) 2017;96:e8204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Çiçek C, Arslan A, Karakuş HS, et al. Prevalence and seasonal distribution of respiratory viruses in patients with acute respiratory tract infections, 2002–2014. Mikrobiyol Bul 2015;49:188–200. [DOI] [PubMed] [Google Scholar]
- [26].Zhang Z. Missing data imputation: focusing on single imputation. Ann Transl Med 2016;4:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Gopalakrishna G, Mustafa RA, Davenport C, et al. Applying Grading of Recommendations Assessment, Development and Evaluation (GRADE) to diagnostic tests was challenging but doable. J Clin Epidemiol 2014;67:760–8. [DOI] [PubMed] [Google Scholar]
- [28].Zhang Z. Big data and clinical research: focusing on the area of critical care medicine in mainland China. Quant Imaging Med Surg 2014;4:426–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Butte AJ. Big data opens a window onto wellness. Nat Biotechnol 2017;35:720–1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Zhang Z. Big data and clinical research: perspective from a clinician. J Thorac Dis 2014;6:1659–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Jensen PB, Jensen LJ, Brunak S. Mining electronic health records: towards better research applications and clinical care. Nat Rev Genet 2012;13:395–405. [DOI] [PubMed] [Google Scholar]
- [32].Wu P-Y, Cheng C-W, Kaddi C, et al. Advanced big data analytics for -omic data and electronic health records: toward precision medicine. IEEE Trans Biomed Eng 2016;64:263–73. [DOI] [PMC free article] [PubMed] [Google Scholar]


