Abstract
The superfamily of heme-copper oxidoreductases (HCOs) include both NO and O2 reductases. Nitric oxide reductases (NORs) are bacterial membrane enzymes that catalyze an intermediate step of denitrification by reducing nitric oxide (NO) to nitrous oxide (N2O). They are structurally similar to heme-copper oxygen reductases (HCOs), which reduce O2 to water. The experimentally observed apparent bimolecular rate constant of NO delivery to the deeply buried catalytic site of NORs was previously reported to approach the diffusion-controlled limit (108–109 M−1s−1). Using the crystal structure of cytochrome-c dependent NOR (cNOR) from Pseudomonas aeruginosa, we employed several protocols of molecular dynamics (MD) simulation, which include flooding simulations of NO molecules, implicit ligand sampling and umbrella sampling simulations, to elucidate how NO in solution accesses the catalytic site of this cNOR. The results show that NO partitions into the membrane, enters the enzyme from the lipid bilayer and diffuses to the catalytic site via a hydrophobic tunnel that is resolved in the crystal structures. This is similar to what has been found for O2 diffusion through the closely related O2 reductases. The apparent second order rate constant approximated using the simulation data is ~5×108 M−1s−1, which is optimized by the dynamics of the amino acid side chains lining in the tunnel. It is concluded that both NO and O2 reductases utilize well defined hydrophobic tunnels to assure that substrate diffusion to the buried catalytic sites is not rate limiting under physiological conditions.
Graphical Abstract
1. Introduction
Nitric oxide (NO), a neutral diatomic molecule, plays many roles in cell signaling and metabolism [1–3] and interacts with numerous heme-containing proteins such as myoglobin, hemoglobin, guanylate cyclase and cytochrome c oxidase, acting as modulating ligand or consumed substrate [4]. NO is a byproduct of denitrification in which nitrate (NO3−) is sequentially reduced to dinitrogen (N2): NO3− → nitrite (NO2−) → NO → nitrous oxide (N2O) → N2 [5–11].
The enzymes that catalyze the reduction of NO during denitrification are members of the hemecopper oxidoreductase (HCO) superfamily, which includes as well the proton-pumping respiratory O2 reductases (e.g. mitochondrial cytochrome c oxidases). The HCO superfamily includes two families of well characterized NO reductases (NORs), the cNORs and qNORs, which utilize cytochrome c and quinol, respectively as the source of electrons. The cNORs and qNORs are integral membrane enzymes that catalyze the following 2NO + 2e− + 2H+ → N2O + H2O reaction.
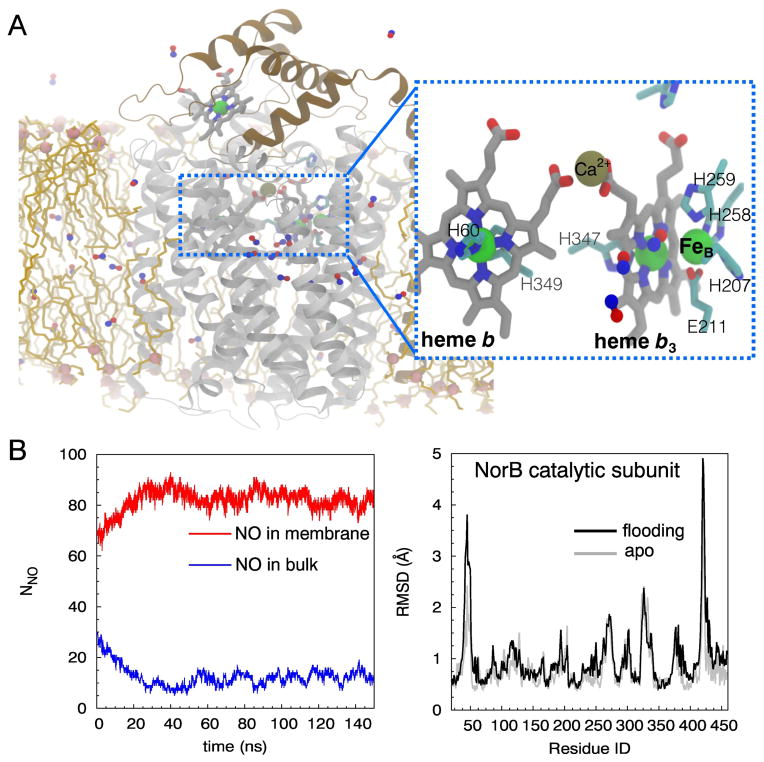
The structures of one cNOR and one qNOR have been obtained from Pseudomonas aeruginosa [12–14] and Geobacillus stearothermophilus [15], respectively. Their structural determination has provided a fundamental basis towards the understanding of their functions [11]. The cNORs are the most extensively studied NORs [14, 16, 17]. They consist of two transmembrane (TM) subunits, NorC with a covalently bound heme c, and NorB, containing the catalytic active site (Fig. 1A). NorB is structurally homologous to Subunit I of the HCO respiratory O2 reductases [18–20] and contain a low-spin heme b and the NO reduction site composed of the high-spin heme b3 and a non-heme Fe ion (FeB). FeB is coordinated to three histidine residues and a glutamate [8, 17]. In the homologous O2 reductases, the non-heme metal is CuB, which is ligated to the equivalent three histidines, one of which is crosslinked to a tyrosine [21, 22]. NORs do not contain the cross-linked tyrosine cofactor, and do not pump protons across the membrane [23, 24].
Figure 1.
Substrate protons diffuse into the catalytic site of cNOR from the periplasm, most probably via hydrated pathways suggested by X-ray crystallography [12], molecular dynamics (MD) simulation [25] and site-directed mutagenesis [26, 27]. Cytochrome c deliver electrons one at a time to heme c in the NorC subunit of cNOR from which they are transferred sequentially to heme b and finally to the heme b3-FeB catalytic active site. The kinetics of diatomic gases NO, CO and O2 to heme b3 has been shown to be second order with rate constants of 107–109 M−1s−1 [23, 27–29]. These rate constants are comparable to those determined for the bovine mitochondrial cytochrome aa3 (A-family O2 reductase) and the Thermus thermophilus cytochrome ba3 (B-family O2 reductase) which are 108 M−1s−1 and 109 M−1s−1 respectively [30–31].
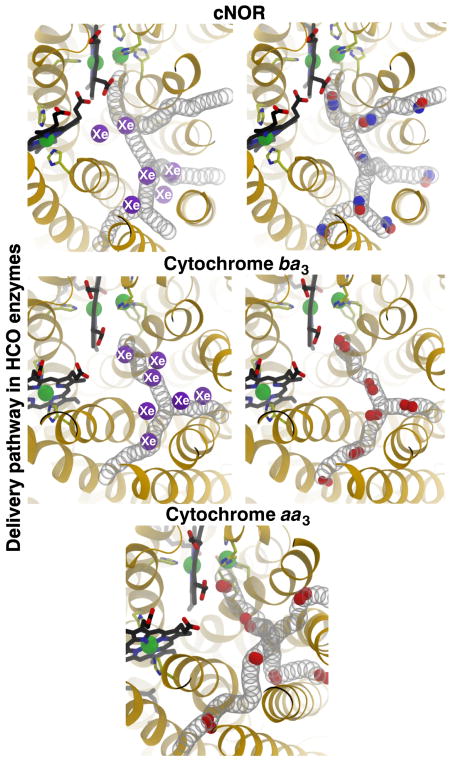
Our previous MD study with cytochrome ba3 demonstrated that O2 diffuses into the catalytic site exclusively via a pathway that is accessible to the hydrophobic membrane core [32]. The pathway corresponds to a pre-formed, static hydrophobic tunnel [33, 34] that has been found to bind Xe atoms [35, 36]. Similar, through more narrow hydrophobic tunnels are observed in the crystal structures of other O2 reductases [37–42] and NORs [12, 15]. Our very recent study with cytochrome aa3 also found that O2 uses a similar pathway to diffuse into the catalytic site but a rate that is ~10 times slower than in cytochrome ba3 [43]
The present study employs “flooding” MD simulation, implicit ligand sampling (ILS) and umbrella sampling (US) techniques to probe NO delivery pathways in cNOR and to describe the diffusion of NO to the catalytic site. The simulations were performed with the cNOR enzyme complex of P. aeruginosa embedded in an ionized-solvated lipid bilayer (Fig. 1A). The use of flooding simulations, in which 100 NO molecules were explicitly added to the simulated system, is to describe the dynamics associated with the NO delivery process. The use of the ILS technique is to identify favorable NO insertion sites within the protein, and the use of the US technique is to examine the partitioning energetics of NO and dynamics of amino acids lining the pathways. It is shown that after partitioning into the lipid bilayer, NO diffuses into the catalytic site of cNOR exclusively via a pathway that is accessible to the membrane and equivalent to the one found in the crystal structures and the previous simulation studies with O2 reductases. Unlike the wide, static tunnel present in cytochrome ba3, rapid NO delivery in cNOR requires the thermal fluctuation of amino acid side chains lining the pathway.
2. Materials and Methods
2.1. Construction of the membrane-embedded cNOR model
The simulation system was constructed by embedding the crystal structure of wild-type cNOR from P. aeruginosa resolved at 2.7 Å resolution (PDB 3O0R) [12] into a lipid bilayer (Fig. 1A). The cNOR structure contains the NorB subunit which binds hemes b and b3, a non-heme Fe atom (FeB) and a Ca2+ ion, the NorC subunit which binds a heme c, 28 water molecules belonging to the NorB subunit and 44 water molecules belonging to the NorC subunit. Hydrogen atoms were modeled using PSFGEN of VMD [44]. Histidine residues except H60, H258, H259, H339, H347 and H349 of NorB, and H65 of NorC were in the HSE tautomeric form (Nε of the imidazole ring carrying the proton). H60 and H349 are ligated to the Fe atom of heme b. H211, H258 and H259 are ligated to FeB. H347 is ligated to the Fe atom of heme b3. H339 forms a hydrogen bond with propionate A of heme b3 and has the pKa of 7.2 calculated using PROPKA3 [45]. Since the shift of the pKa from its solution form is small, and as at neutral pH, we will have essentially equal percentages of the protonated and deprotonated forms of the histidine, we opted to use the neutral (uncharged) form of this residues in the simulations. Since the Nδ atom of this residue is in direct contact with the oxygen atoms of the heme, the proton was assigned to the Nδ atom, leaving the other nitrogen atom deprotonated. H65 and M112 of NorC are ligated to the Fe atom of heme c, and two cysteine residues C61 and C65 forming thioether bonds with heme c. In order for NO binding to occur, heme b3 must be unligated, so the oxygen atom bridging the non-heme FeB to the Fe atom of heme b3 was removed. Two other glutamate residues near the catalytic site, E215 and E280, which were found by PROPKA3 to have predicted pKa value of greater than 9, were modeled in the protonated (neutral) form. We note that the side chain of E280 is hydrogen-bonded to that of E211, so these two residues would be both negatively charged, which is energetically unfavorable, if E280 is left in the deprotonated form. The surrounding of E215 is predominantly comprised of nonpolar amino acids, which can explain its predicted high pKa value. This side chain forms a hydrogen bond with a crystal water molecule, which is the only crystal water molecule resolved in this region. These contacts remained during the simulations. The DOWSER program [46] was employed to model additional water molecules in the proteins. Using the maximum energy of −10 kcal/mol as the criteria for water insertion, 32 water molecules were modeled; 14 of these molecules were located in the lower region of NorB (near the cytoplasmic surface) and the rest were located in NorC and its interface with NorB. During the simulations, these modeled water molecules and the X-ray resolved water molecules except those buried near the catalytic site left the protein and exchanged with water molecules in the aqueous solution. No water molecules either entered or left the NO delivery pathway during any of the simulations.
To construct the membrane-embedded cNOR complex, the first principal axis of the protein was aligned with the z axis (membrane normal) using the OPM (Orientations of Proteins in Membranes) database [47, 48]. The protein was inserted into a patch of POPE (1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanoamine) bilayer with the membrane normal. Lipid molecules overlapping the protein were removed, keeping 132 lipids in the periplasmic and cytoplasmic leaflets. The membrane-embedded cNOR complex was then solvated with TIP3P water. Water molecules that were in the membrane, except those from the crystal structure and those added by DOWSER, were removed, keeping 20,637 water molecules. Finally, the bulk aqueous phase was ionized with 0.2 M NaCl by adding 78 Na+ and 79 Cl− ions, maintaining overall neutrality of the entire system, resulting in a fully solvated model of 104,710 atoms. The CHARMM22 force field with φ/ψ corrections [49, 50] was used to describe the protein and heme b cofactors, and the CHARMM36 [51] was used to describe the lipids. Since this study focuses only on the delivery of NO to the catalytic site and substrate NO molecules bind to a fully reduced enzyme, all cofactors were held in the reduced (ferrous) state. The parameters of the heme c cofactor were taken from Autenrieth et al [52]. For FeB, a +2 charge was assigned to the Fe atom and its vdW parameters with ε = 0.067 kcal/mol and Rmin =1.15 Å were from Babu and Lim [53]. The coordinations of the Fe atoms and ligating amino acids were maintained by bonded potentials described in the next section.
2.2. Simulation protocols
MD simulations performed to prepare the systems consisted of the following steps: (1) 0.5-ns melting of lipid tails during which only the lipid tails were allowed to move in order to achieve better packing of lipids around the inserted protein (the rest including the protein were fixed); (2) 0.5-ns simulation with restraints (k = 1 kcal/mol/Å2) applied to heavy atoms of the protein and cofactors (all lipid atoms and water moving) and with harmonic potentials (k = 0.1 kcal/mol/Å2) applied to keep water out of the membrane; (3) 0.5-ns simulations with only backbone atoms of the protein and heavy atoms of the cofactors restrained (k = 1 kcal/mol/Å2); (4) 1-ns simulation with only Cα atoms of the protein and heavy atoms of the cofactors restrained; and (5) 20-ns unrestrained relaxation. Energy minimization (1,000 steps) was performed at the beginning of Steps 1, 2 and 3 using the conjugate gradient algorithm. To maintain the ligation of FeB to H207, H258, H259 and E211 and thereby the structure of the catalytic site, the His-FeB, E211-FeB and heme b3 Fe-FeB connections were described by bonded interactions (k = 200 kcal/mol/Å2 for bonds and k = 50 kcal/mol/rad2 for angles).
All simulations were performed using NAMD2 [54] with a time step of 2 fs and with the periodic boundary condition (PBC). All bonds involving hydrogen atoms were kept rigid using the SHAKE algorithm [55]. To evaluate long-range electrostatic interactions in PBC without truncation, the particle mesh Ewald (PME) method [56] with a grid density of 1/Å3 was used. The cutoff for van der Waals interactions was set at 12 Å. All of simulation steps except the melting of lipid tails (Step 1) were performed in a flexible cell, which allows the membrane-embedded protein model to change its dimensions independently, and were performed under NPT ensemble. The temperature was maintained at 310 K by Langevin dynamics [57] with a damping coefficient γ of 1 ps−1. The Nose-Hoover Langevin piston method [57, 58] with a piston period of 20, fs was used to maintain the pressure at 1 atm.
2.3. Production runs
The production runs consisted of a 100-ns apo simulation performed in the absence of NO molecules (used for ILS analyses and starting flooding simulations) and sets of 150-ns flooding simulations in which NO molecules were explicitly added. The apo simulation was continued from the 20-ns relaxation simulation. The flooding simulations were started from snapshots taken at 0-ns, 50-ns and 100-ns time points of the apo simulation and each simulation set was repeated twice.
2.4. Flooding simulations
To effectively probe NO pathways under a limited timescale of 150 ns, 100 NO molecules, corresponding to the concentration of 0.16 M calculated with respect the volume of the simulation box, were added to an equilibrated membrane-embedded cNOR complex. NO parameter was taken from the previously published works of Cohen et al [59] and Wang et al [60]. At the start of the simulations, 70 NO molecules were initially placed in the membrane, 30 molecules were placed in the aqueous solution, and they were all outside the protein (Fig. 1B). To identify whether NO delivery events took place during the simulations, we identified NO molecules that were within 6 Å of FeB, and then examined the trajectories of those molecules. We first simulated the systems for 50 ns but we soon realized that this timescale is too short to observe even a single NO delivery event. To adequately sample potential delivery pathways, each simulation was extended to 150 ns. Also, in a few of the simulations, we noticed 2–3 water molecules entered the catalytic site from the periplasmic portion of the protein and did not leave, thus compromising the access of NO. To eliminate this problem, only nine flooding simulations trajectories in which the catalytic site remained dehydrated were analyzed, adding up the total of 1,350 ns.
2.5. Implicit ligand sampling (ILS) calculations
In complementary to flooding simulation in which ligand diffusion is explicitly probed, ILS was employed to identify potential regions and pathways for NO insertion including the ones not sufficiently sampled by flooding simulations. ILS calculates ligand-interaction energies (Ei) in any position inside the protein over an ensemble of protein conformations and ligand orientations [59], resulting in a 3D free energy map of NO insertion (ΔGi).
where p0 (in vacuum) = 1 and pi is the probability of inserting a NO molecule at position i.
The method follows the assumption that small, hydrophobic gases such as O2 and NO, which have small molecular volumes, interact weakly with proteins, and therefore do not affect the protein structure and dynamics. This assumption is valid for cNOR because no appreciable conformational changes of the protein by the flooding of NO molecules were observed. This is shown by the similar RMSD profiles of the protein sampled in the apo and flooding simulations (Fig. 1C and Fig. S1). Hence, the 100-ns apo trajectory comprised of 10,000 snapshots of the protein complex was analyzed for NO pathways. NO molecules were sampled in a 55×50×75 Å3 grid with spacing of 1 Å, covering the entire cNOR complex. Ten orientations of NO were sampled in each subgrid, which contained 3×3×3 interaction sites. The solvation free energy of NO (ΔGsol) was used as the reference for calculating the partitioning free energy of NO (ΔGi,sol).
ΔGsol were calculated over a 30×30×30 Å3 ionized water box using ILS and free-energy perturbation (FEP) [61]. Both methods yield ΔGsol values of 1.6 kcal/mol, which is consistent to a previous calculation by Cohen et al [59]. We note that since no electrostatic terms are included in ILS, the atoms effectively carry the partial charges of 0 in ΔG calculations.
2.6. Umbrella sampling simulations
The flooding simulations have identified an NO delivery pathway comprising four entry branches (Branches A1, A2, B and C) (Fig. 3). Energetics characterizations of NO insertion along the delivery pathway can provide insights into functional necessities of these branches. However, this quantity is difficult to be estimated accurately using only the results of the flooding simulations because a few NO entry events were observed (Fig. 2). We therefore employed the umbrella sampling (US) technique [61–63] to calculate 1D free energy profiles of NO partitioning along the delivery pathway. To initiate the starting points for US simulations, sequential sets of steered molecular dynamics (SMD) simulations [64] were performed. To begin SMD, an NO molecule was placed in the final structure of the 20-ns relaxation step at site located 3 Å from the center of the catalytic site defined by the center of FeB and the Fe atom of heme b3. In the first step of SMD, the molecule was pulled towards the intersection of Branches A and B (A/B) defined by the center of residues I65, V66 and F185; the total distance is 13 Å. Following this step, three independent simulations were performed to pull the molecule from 1) A/B towards the entrance of Branches B, 2) the entrance of Branch C, and 3) the intersection of Branches A1 and A2 (A1/A2). The entrance of Branch B is defined by the center of residues V148, L152, A182 and L186 located at the protein-lipid interface, and the total distance is 12 Å. The A1/A2 intersection is defined by the center of residues N62, V66, G105, T108, S143 and I147 and the total distance is 6 Å. Branch C starts at site located 9.5 Å from the catalytic site and its entrance is defined by the center of residues L137, W205 and V205 located ~5 Å away. To completely expel the molecule to the membrane, the pulling distance was extended to 15.5 Å. Once the molecule reached the A1/A2 intersection, two separate simulations were performed: 1) towards the entrance of Branch A1 which is at TM1/TM3 interface (10-Å distance) and 2) towards the entrance of Branch A2 which is at TM3/TM4 interface (8-Å distance). In all of the SMD simulations, NO was pulled at the velocity of 5 Å/ns and k = 20 kcal/mol/Å2, so the total simulation time was 15 ns.
Figure 3.
Figure 2.
US simulations comprised 6 parts, which are 1) Route a: from the catalytic site to A/B (13 Å), 2) Route b: from A/B to Branch B entrance (12 Å), 3) Route c: from A/B to A1/A2 (6 Å), 4) Route d: from 9.5 Å distance of the catalytic site to Branch C entrance (15.5 Å), 5) Route e: from A1/A2 to Branch A1 entrance (10 Å), and 6) Route f: from A1/A2 to Branch A2 entrance (8 Å). Each sampling route was divided into 0.5 Å windows, adding to the total of 136 simulations (Fig. 5). The connection of Routes a, c and e forms Branch A1, that of Routes a, c and f forms Branch A2, and that of Routes a and b forms Branch B. Branch C starts from Route a, ends at 9.5 Å distance and then continues through Route d. In each US simulation which lasted 3 ns, the NO molecule was restrained at the center of a window by harmonic potentials, initially with k = 10 kcal/mol/Å2. If the molecule deviated from the center of the window, the 20 kcal/mol/Å2 force constant was applied and a new simulation was performed. Despite imposing such strong restraining potentials, the histogram of NO molecule sampling in one window partially overlapped with those of its neighbors. The weighted histogram analysis method or WHAM [65] was used to reconstruct the free energy profiles at 0.25 Å bin width, using last 2.5 ns of each US simulation. The WHAM code was kindly implemented by Professor Alan Grossfield at the University of Rochester Medical Center. Effectively, the total simulation time for the US simulations was 600 ns.
Figure 5.
3. Results
3.1. Delivery pathway of NO
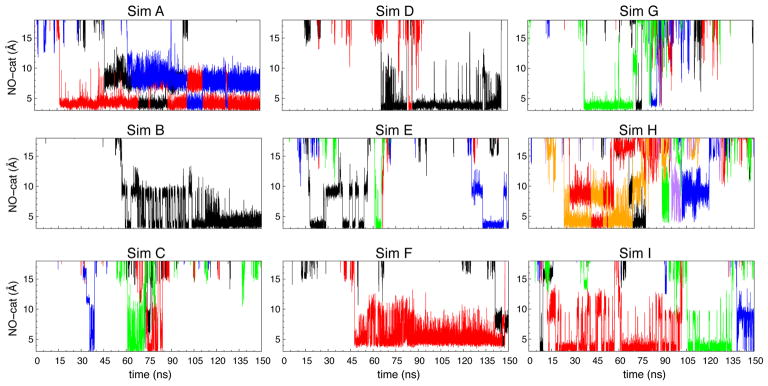
To probe NO delivery pathways, the total of 100 NO molecules were initially placed outside the protein; 70 of them were in the membrane and 30 were in the aqueous solution. During 150-ns simulations, we observed 1–6 entry events of NO to the catalytic site. In one of the nine simulations (Sim I in Fig. 2), the first entry event occurred within the first 10 ns. But, in four of the simulations (Sims B, D, F and G in Fig. 2), it took >30 ns for this event to occur. The duration of an NO molecule residing in the catalytic site or within 6 Å of the center of FeB and the Fe atom of heme b3 ranged from 1–2 ns to >100 ns (Fig. 2).
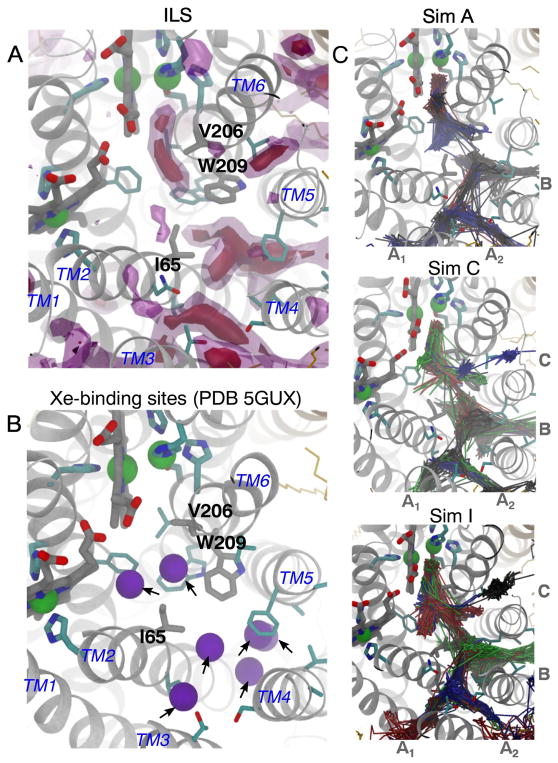
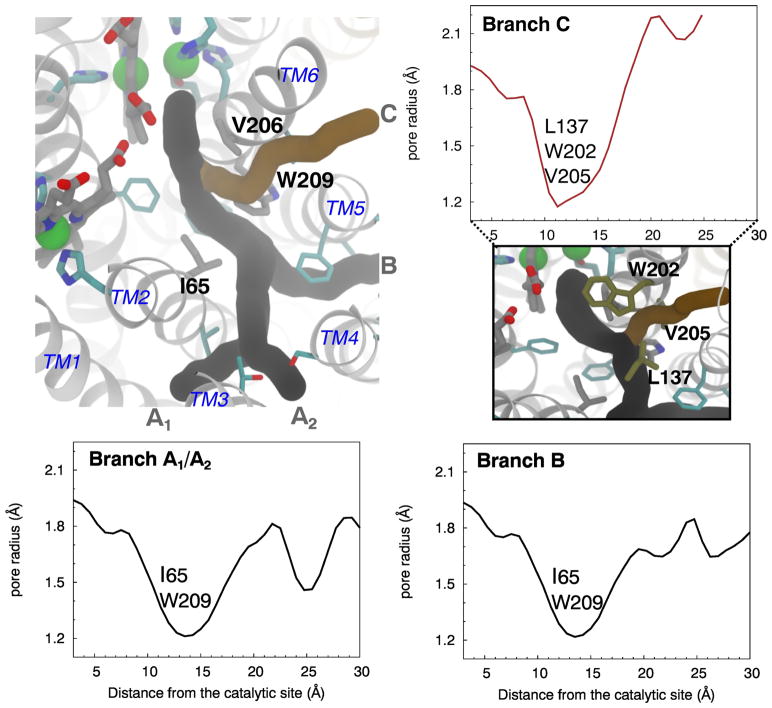
To identify the NO delivery pathways, NO molecules entering the catalytic site were identified and their diffusion dynamics were visually examined. Their trajectories, which are shown in Fig. 3C, indicate that the NO molecules diffused to the catalytic site via a pathway comprised of four entry branches originated from the membrane. Two branches of the pathway, referred to as Branches A1 and A2, intersect at a center of TM2, TM3 and TM4 helices, which is located ~22 Å away from the catalytic site. Branch A1 begins at TM1-TM3 helix-lipid surface, while Branch A2 begins at a TM3-TM4 helix-lipid surface. These branches merge into one pathway, which then merges with the other two branches, referred to as Branches B and C, at site located ~12 Å from the catalytic site. Branch B begins at TM4-TM5 helix-lipid surface, while Branch C begins at TM5-TM6 helix-lipid surface. Branches A1 and B resemble the O2 delivery pathway of cytochrome ba3 characterized by our MD study [32] which coincides with the tunnel inferred from the binding sites for Xe atoms in the crystal structure of cytochrome ba3 [35, 36]
ILS calculations identify favorable NO insertion regions within the protein. The isosurface maps shown in Fig. 3A highlight regions, in which the partitioning ΔG for NO with respect to the aqueous solution (ΔGi,sol) are −2.5 kcal/mol (pink-transparent) and −4 kcal/mol (purple-solid). The observed NO delivery pathway is among the low-energy regions.
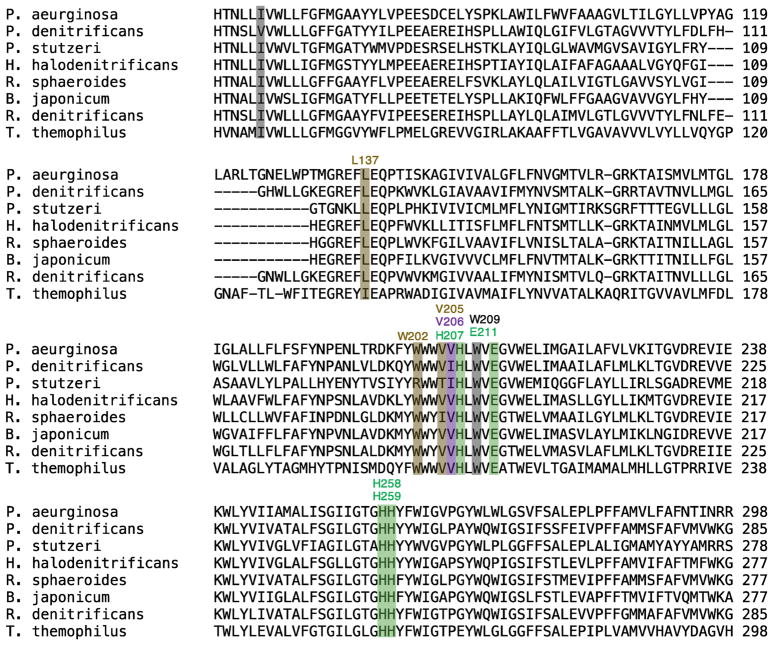
The flooding simulations and ILS calculations identified potential barriers of NO transition along the pathway. One barrier in particular is located ~13 Å from the catalytic site or at the intersection of Branches A1/A2, B and C merge. This barrier is surrounded by residues I65 and W209 and is indicated in Fig. 3A by a discontinuous gap in the ΔGi,sol map. I65 is within TM2 and is replaced by Valine in some of the isolated cNORs (Fig. 8). W209 is within TM6 and is very conserved within cNORs. During the flooding simulations, the transitions of NO through this site, referred to as “the I65/W209 barrier”, rarely occurred and the migrating NO molecules appear to stay very transiently (Fig. 2).
Figure 8.
The current results are consistent with a very recent study employing X-ray crystallography and MD simulation [14]. The study resolved a structure of cNOR with 7 Xe atoms bound along the presumed NO delivery pathway (Fig. 3B). Two of the Xe atoms are located between the I65/W209 barrier and the catalytic site. The rest are located towards the entrances of Branches A1/A2 and B. None of the Xe atoms is located in between the side chains of residues I65 and W209 which form the barrier. The same study reported MD simulations of NO diffusion using the structure of the complex of the nitrite reductase (cd1NiR), which generates NO, and cNOR, which consumes NO [14]. The simulations were started by distributing the NO molecules inside cd1NiR, and were observed to exit this enzyme which was complexed to cNOR on the periplasmic side. The NO molecules diffused to the lipid bilayer and then cNOR via a site corresponding to Branch A1 of the NO delivery pathway observed in the present study.
3.2. Structure and accessibility of the pathway
We employed CAVER 3.0 [66] to probe access tunnels from the protein surface to the catalytic site using the trajectory of the last 80 ns of the 100-ns apo simulation. Water and lipid molecules were removed to reduce the size of the system and expedite the calculations. Three of the top five most frequently identified tunnels correspond to Branches A1, A2, B and C, shown in Fig. 4. Branches A1 and A2 were characterized as one tunnel, while Branches B and C were characterized as two separate tunnels.
Figure 4.
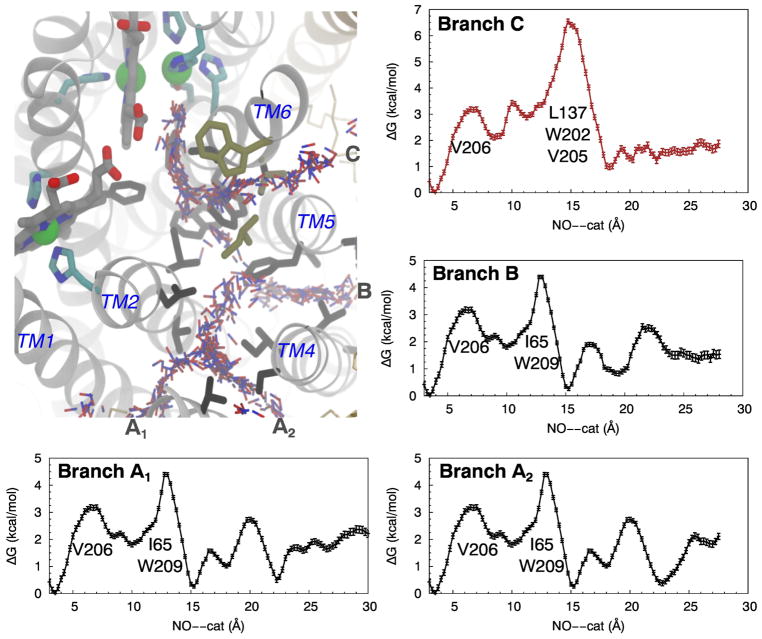
Following the tunnels coinciding to Branches A1, A2 and B from the protein surface leads to the same narrowest point or bottleneck that is ~13 Å from the catalytic site (Fig. 4). This bottleneck is defined by the side chains of I65 and W209, resulting in the minimum pore radius of 1.2 Å which is smaller than the vdW radius of an oxygen atom (1.5 Å). Together with the results from ILS and flooding simulations, this presents a kinetic barrier for NO migration to the catalytic site. There is a second but less important bottleneck (not shown) formed by residues F136 and W214. F136 is in the loop connecting TM3 and TM4 and located on the top of the NO pathway but its phenyl side chain is oriented away from the pathway. W214, located on TM6, is at one turn below W209.
Branch C joins the other branches at a site located ~9 Å from the catalytic site. It has its own bottleneck with the minimum pore radius located at ~11 Å distance (Fig. 4) surrounded by residues E135, L137, W202 and V205. The side chains of L137, W202 and V205 are oriented towards the pathway. E135 is coordinated to a Ca2+ ion that is also coordinated to the propionates of hemes b and b3 [12] and is not directly in the pathway. The equivalence of E135 is found to be important in T. thermophilus cNOR [67], G. stearothermophilus qNOR [15] and R. sphaeroides cytochrome cbb3 O2 reductase [68].
Two hydrophilic tunnels were also found at the periplasmic surface of NorC, connecting to the catalytic site. Because NO delivery through these tunnels was not observed in the flooding simulations and they were not found by ILS to favorably bind NO, they were not considered further in this study.
3.3. Free energy profiles for NO partitioning along the pathway
Free energy calculations performed using the umbrella sampling (US) technique [62] characterized two major NO migration routes within the identified NO delivery pathway (Fig. 5). The free energy profiles show that an NO molecule entering the protein through either Branches A1, A2 or B encounters the highest free energy barrier of ~4 kcal/mol surrounded by residues I65 and W209, 13 Å from the catalytic site. This barrier is at the same location as the bottleneck of these three branches according to the calculated pore radius profiles (Fig. 4). The intersection of these branches, which is located at ~15 Å from the catalytic site, also corresponds to a free energy minimum of NO insertion along the pathway. This free energy minimum and the one within the catalytic site or at ~3-Å distance have about the same height according to the free energy profiles, suggesting that the probabilities of NO insertion are about the same at both sites.
For NO diffusing through Branch C, the highest barrier is at 15 Å distance and is 5.5 kcal/mol, clearly characterizing Branch C as a distinct NO migration route from the other branches. From the transition state theory, the rate constant for NO diffusion, k, is proportional to exp(-ΔGb/RT), where ΔGb corresponds to the height of the free energy barrier. Under this assumption, NO migration through Branch C is presumably 10-fold slower than through Branches A1, A2 and B. We can consider the combined A1–A2-B as “the dominant route” while Branch C is “the alternate route”.
After crossing the main barriers and prior to entering the catalytic site, the NO molecule encounters a smaller barrier of ~1.5 kcal/mol near residue V206 in the single pathway about 6 Å from the catalytic site. In the reverse direction for NO leaving the catalytic site to diffuse to the lipid bilayer, the barrier at V206 is ~3 kcal/mol. Then, going through the dominant route, there is a 2.5 kcal/mol barrier at I65 and W209, or, going through the alternate route, NO encounters a 4 kcal/mol to exiting the protein.
3.4. NO delivery rate
Experimental measurements of the apparent second order rate constant for the association of NO to the enzyme’s catalytic site (kobs) is calculated using the concentration of NO in the aqueous solution [31]. However, according to the flooding simulations performed in the present study, all of NO molecules that reached the catalytic site entered the protein through the lipid bilayer. In simulating the kinetics of the entire delivery process, the equilibration of NO between the aqueous solution and the lipid bilayer is rapid and enters into the consideration simply as a partition constant, resulting in a higher local concentration of NO surrounding the protein within the lipid bilayer. From this point, the diffusion of NO to the catalytic site can be considered in two major steps: 1) Diffusion within the lipid bilayer to the entrance of a pathway; and 2) Diffusion along the pathway. Using the data of the flooding simulations (Table 1), we provide semi-quantitative details of these steps and approximate kobs.
Table 1.
Localization and migration events of NO in cNOR
| Simulation | tcat (ns) | tAB (ns) | tent (ns) | [NO]bulk (mM) | tent--AB (ns) | tcat--AB (ns) |
|---|---|---|---|---|---|---|
| A | 16.7 | 0.4 | 0.11 | 34.1 | 0.2 | 16.3 |
| B | 59.3 | 1.8 | 0.1 | 35.2 | 1.7 | 57.5 |
| C | 35.7 | 5.4 | 5.4 | 37.7 | ~0 | 30.3 |
| D | 65.8 | 1.8 | 1.6 | 36.9 | 0.2 | 64 |
| E | 18 | 10.5 | 2.1 | 38.0 | 8.4 | 7.4 |
| F | 47.6 | 1.8 | 1.2 | 43.9 | 0.6 | 45.8 |
| G | 37.4 | 7.1 | 3.4 | 43.1 | 3.7 | 30.3 |
| H | 23.5 | 9.4 | 1.2 | 36.2 | 8.2 | 14.1 |
| I | 7.4 | 4.7 | 1.2 | 37.1 | 3.5 | 2.7 |
| Average | 34.6 | 4.8 | 1.8 | 38 | 3 | 29.8 |
| Std. err. | 10.6 | 1.9 | 0.9 | 2 | 1.8 | 11.6 |
tcat = time taken to observe the first NO molecule reaching the catalytic site after the start of a simulation. tAB = time taken to observe the first NO molecule reaching the main barrier of the dominant route. tent = time taken to observe the first NO molecule localized at the intersection between Branches A1 and A2 or within the entrance of Branch B. Branches A1/A2 and B form the dominant route. tAB-ent = time taken for NO to diffuse from the entrances to the main barrier of the dominant route. tcat-AB = time taken for NO to diffuse from the main barrier to the catalytic site. Standard errors were calculated based on an 85 % confidence interval, in which lower limits are greater than zero, with the t-score of 1.592. t-score is suitable in this study because the number of simulations (n) is 9, which is less than 30.
From the start of a simulation, it takes 35 ns on average to observe the first NO delivery event (tcat). For the dominant route, it takes ~2 ns for NO to first locate the pathway (tent). The time at which the first NO molecule reaches at the main barrier formed by I65 and W206 (tAB) is 5 ns. This means that on average, an NO molecule spends ~3 ns to transit from the entrance to the main barrier (tAB-ent). It takes on average another 30 ns (tcat-AB) to transit to the catalytic site.
Assuming that NO consumption by cNOR is 100% efficient, the rate of NO delivery (kobs) can be approximated from the following model:
E(NO) represents the state at which NO is the intersection of Branches A and B which is before it reaches the main or I65/W209 barrier. E-(NO) represents the state at which NO reaches one of the entrances. “cat” represents the catalytic site of cNOR. The average NO concentration in the aqueous solution is 38 mM. kobs is defined as:
The product, k1[NO], is the number of events per second in which NO diffuses from the aqueous solution to the entrance(s) of the pathway. By using the concentration of NO in the aqueous solution in this expression, the value of k1 is increased to take into account the higher concentration of NO within the lipid bilayer due to equilibrium partitioning. NO is favored by a factor of ~8 in the lipid bilayer (Table 2). Since k1[NO] = 1/(tent), we can determine the value of k1 ~1.5×1010 M−1s−1. k−1 is the rate constant of NO dissociation from the entrance(s) to the membrane. The partitioning free energy profiles show that the chemical potential of NO at the entrances of the pathways is favored by ~1.5 kcal/mol compared to the core of the lipid bilayer (Fig. 5). This means that the probability NO insertion at the entrances of the pathway is about 10-fold higher than in the lipid bilayer, i.e., k1/k−1 = 10. This gives us the value of k−1 ~1.5×109 s− 1. k2 is the rate constant of NO migration from the entrance(s) to the main barrier, and k2 or 1/tABent = 1.4±1×108 s−1. k−2 is the rate constant of NO migration from the main barrier to the entrance(s) and is equal to 1/k2. The free energy profiles (Fig. 5) indicate that the probabilities of NO insertion at the pathway’s entrances and at the intersection of Branches A and B are about the same ([E(NO)]~[E-(NO)]). Hence, k2/k−2 = 1, or k2 = k−2. The rate constant k3 measures the transit of NO from the main barrier to the catalytic site, so k3 = 1/tcat-AB, which is 8±6×107 s−1. Substituting these values into the expression of kobs gives the following: kobs = (5±3×108 M−1s− 1)[NO].
Table 2.
Distributions of NO within bulk aqueous, lipid and protein phases
| Simulation | <N>bulk | <N>lipid | <N>prot | Normalized (Nlipid/Nbulk) |
|---|---|---|---|---|
| A | 11 | 61 | 23 | 9.1 |
| B | 11 | 59 | 24 | 8.8 |
| C | 12 | 61 | 22 | 8.3 |
| D | 12 | 58 | 24 | 7.9 |
| E | 13 | 58 | 23 | 7.3 |
| F | 15 | 56 | 24 | 6.1 |
| G | 14 | 59 | 21 | 6.9 |
| H | 12 | 59 | 24 | 8.1 |
| I | 12 | 60 | 24 | 8.2 |
| Average | 12 | 59 | 23 | 7.9 |
Calculated using the last 125 ns of simulations, Nbulk is the number of NO in the aqueous solution. Nlipid, the number of NO in the lipid phase of the membrane, was calculated by subtracting the number of NO in the membrane (Nmem) and the one within 5 Å of the TM region of the protein (Nprot). NO was considered to be in the membrane when either its oxygen or nitrogen atom resided between the Cα atoms of the palmitate’s ester of both leaflets. Since the length of the simulation box along the z-axis for the aqueous solution was 1.64-time longer than that of the membrane, Nlipid was multiplied by 1.64 in order to determine the partition coefficient or the normalized (Nlipid/Nbulk) ratio.
For the alternate route, the main barrier located at ~15 Å from the catalytic site or near residues W202, L137 and V205, is 1.5 kcal/mol greater than that of the dominant route. This suggests that the rate of NO transition via this route is ~10-fold slower than via the dominant route, so the approximated value of k3 is ~8.4×106 s−1. Because it is the only major barrier between the entrance and the catalytic site,
Hence, the approximated kobs is ~(8×107 M−1s−1)[NO], suggesting that the transition of NO is ~10-fold slower than through the dominant route.
3.5. Dynamics of lining amino acids governing NO diffusion
Structural comparisons of proteins sampled in the apo simulation (absence of NO molecules) and in the flooding simulations (presence of 100 NO molecules) indicate that the delivery of NO in cNOR involves no large scale conformational dynamics of the protein (Fig. 1C and Fig. S1). However, the narrowness of the delivery pathway and the observation of few NO delivery events in the 150-ns flooding simulations suggest that for NO to pass through, only certain orientations of the molecule are allowed and, in addition, the conformational dynamics of its lining amino acids may play a role. Residues V206, I65, W209, L137, V205 and W202, which are very conserved in the cNORs (Fig. 8), constitute major barriers for NO transition along the observed NO delivery pathway. To explore this, minimum pairwise distances (dmin) and dihedral angles of these amino acids were measured and projected as functions of the position of NO within the pathway using the data of the US simulations, in which NO is forced to stay in defined sites, including the bottleneck regions where NO occupancy was rare during the flooding simulations. These results obtained were consistent with those obtained in the flooding simulations (see Supplemental Materials).
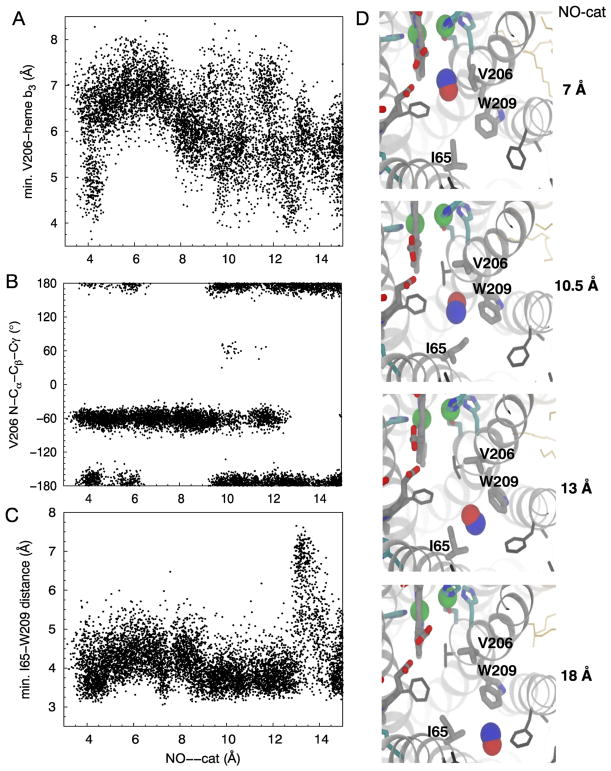
The sidechain of V206 and the methyl group of attached to the D-ring of heme ba3 forms the small 1.5 kcal/mol barrier for an NO molecule to transit from the main barriers or bottlenecks to the catalytic site (Fig. 3–4). When an NO molecule is not localized at this barrier, which is ~6 Å of the catalytic site, dmin between V206 and heme b3 (V206-b3) sample widely from 4 to 8 Å (Fig. 6A). N-Cα-Cβ-Cγ angle or χ1 of V206 is clustered at ±60° and ±180° (Fig. 6B). When NO is localized, V206-b3 becomes clustered at 6–7 Å. χ1 of V206 becomes clustered only at ~-60°, when its side chain rotates out of the pathway to allow an NO molecule to enter the catalytic site (Fig. 6D).
Figure 6.
I65 and W209 constitute the main barrier of the dominant route. I65--W209 dmin and N-Cα-Cβ-Cγ (χ1) and Cα-Cβ-Cγ-Cδ (χ2) angles of I65 and W209 were measured (Fig. 6C and Fig. S3). When an NO molecule is not localized at this site, χ1 of I65 is clustered at ~±180° and −60°, while its χ2 is clustered at ±60° and ±180°. I65-W209 dmin fluctuate around 4 Å (Fig. 6C), and the data of the flooding and apo (unconstrained) simulations show a peak at 4.3 Å (Fig. S5A). The transition of NO across this barrier requires conformational changes of these amino acids (Fig. 6C–D). That is when NO is localized, I65-W209 was found extending to 7 Å (Fig. 6C and Fig. S2). χ1 and χ2 became less populated at ~±180° (Fig. S3). During the US simulations, W209 remained at χ1~−75° and χ2 ~−30°, also predominantly observed in the unconstrained simulations (Fig. S5).
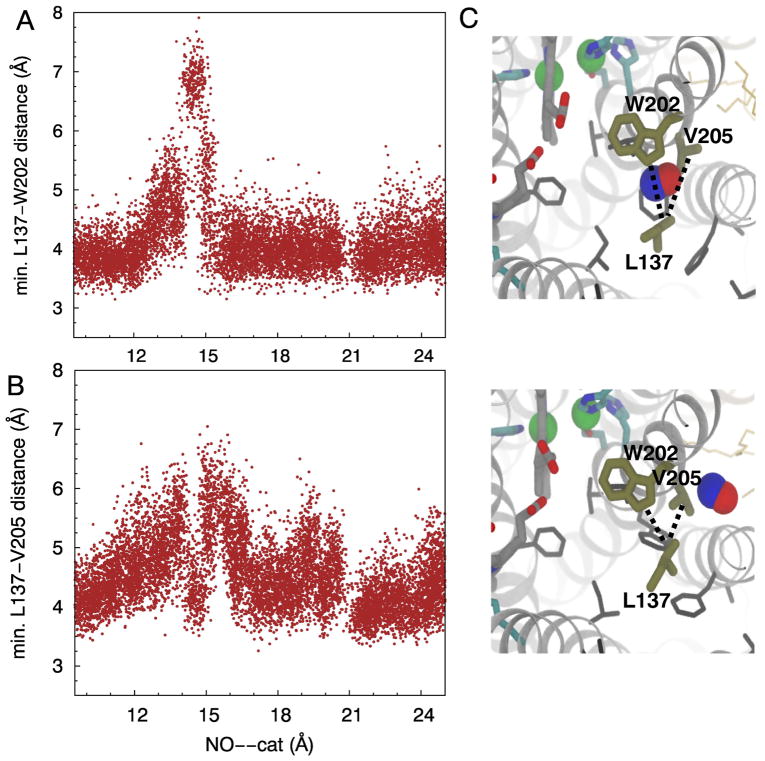
L137, W202 and V205 constitute the main barrier of NO transition through the alternate route, located 15 Å of the catalytic site. L137--V205 and L137--W202 dmin and χ1/χ2 of L137 and W202 were measured. Based on the US and flooding simulation results, L137--V205 and L137--W202 are distributed mainly at 4–4.5 Å (Fig. 7A–B). χ1 and χ2 of L137 are −60° and ±180°, respectively (Fig. S4). The data of the US simulations show that when NO is localized at the barrier site, the side chains of L137 and V205 rotate out of the pathway (Fig. 7C), correlated with the extension of L137--V205 and L137--W202 to 6–7 Å. New clusters of L137 conformations were found to form at χ1 ~180° and χ2 ~60°. The conformations of W202 remained unchanged (Fig. S4).
Figure 7.
4. Discussion
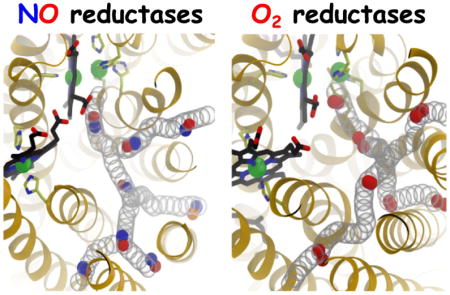
In both cNOR and O2 reductases, NO and O2 diffuse to the catalytic site through similar hydrophobic tunnels that are accessible from the membrane (Fig. 9). The gas-diffusion tunnels have a different number of branches, depending on the enzyme, but in all cases the entrances are within the hydrophobic lipid bilayer and they merge into a single pathway leading to the catalytic site. Not only are these enzymes structurally homologous, but the properties of their diatomic substrates are very similar. O2 and NO are both 4–8 fold more soluble in lipids than in the aqueous solution [70–73] and have similar diffusion constants of ~4.5×10−5 cm2/s in an aqueous buffer solution [70]. They also diffuse similarly in proteins as demonstrated in small proteins, such as myoglobin, in which their diffusion rates from the aqueous solution to the distal pocket were found to be the same [74]. These correlations suggest that the results from the studies of NORs and O2 reductases can be compared.
Figure 9.
In the O2 reductases, time-resolved absorption spectroscopic experiments shows that O2 and NO react to the Fe atom of the catalytic-site heme at the rate constants of 1×109 M−1s−1 in cytochrome ba3 and 1×108 M−1s−1 in bovine cytochrome aa3 [30, 31, 75, 76]. In the present study, the apparent second order rate constant for NO delivery to the catalytic site of the P. aeruginosa cNOR (kobs/[NO]) approximated using the data of the flooding simulations is 5±3×108 M−1s−1, which is similar to the kobs/[NO] value determined experimentally in Paracoccus denitrificans cNOR [23, 27–29]. Hence, both experimental as well as computational studies indicate that second order rate constants of substrate (O2 or NO) diffusion to the catalytic site of cNOR are ~3-fold slower than for cytochrome ba3.
The reason for the slower diffusion of the diatomic gas molecules in cNOR compared to cytochrome ba3 is that the delivery pathway in cNOR is partially occluded (Fig. 9) and the migration of these molecules relies on thermal motions of the lining residues. The crystal structures of cytochrome ba3 with Xe within the hydrophobic tunnels [35, 36] indicate no appreciable conformational changes of lining residues required to accommodate the binding of Xe. The minimum pore radius of the pathway of only ~1.5 Å essentially provide unimpeded access of an O2 or NO molecule to the catalytic site. This is not the case for the delivery pathway for NO in cNOR, of which the minimum pore radii are as narrow as 1.2 Å. The Xe-bound structure of cNOR [14] shows seven Xe atoms along the dominant NO delivery pathway but none at the bottleneck formed by residues I65 and W209 (Fig. 3B). The present study shows that the transition of NO through this bottleneck involves conformational fluctuations of I65 and/or W209 (Fig. 6C–D), which is apparently not the case for O2 and presumably NO to pass the equivalent residues in cytochrome ba3, I78 and I235.
On the other hand, the putative O2 delivery pathway in cytochrome aa3 is also structurally constricted [35, 39, 40] similar to that in cNOR (Fig 9). Residue I65 of cNOR is equivalent to F67 and F108 of the bovine and R. sphaeroides enzymes respectively. W209 of cNOR is equivalent to E242bov and E286Rb, which are critical residues in the D proton delivery channel in these enzymes, but is absent in cytochrome ba3 [77] and all the NORs [12, 15] Nearby tryptophan (W126bov or W172Rb) and phenylalanine (F238bov or F282Rb) residues also can restrict O2 migration in the three strucurally resolved cytochrome aa3 species from bovine, R. sphaeroides and P. denitricans [35, 43, 75]. W172Rb forms a hydrogen bond to the propionate D of heme a3 and is replaced by tyrosine (Y133) in cytochrome ba3. F282Rb is equivalent to W202 of cNOR, which is one of the residues that restricts NO diffusion via the alternate route (Fig. 7), and is replaced by T231 in cytochrome ba3. Diffusion of O2 to the catalytic site of the cytochrome aa3 species through these tunnels is considerably slower than in cytochrome ba3 as shown both experimentally [30, 31] and computationally [43, 78]
Although the diatomic substrate NO migrates more slowly in cNOR than does O2 in cytochrome ba3 the more meaningful comparison is with the limiting catalytic rate for NO reduction (kcat), which is 10−2 to 10−1s−1 [79, 80]. This means that the enzyme can effectively function at maximum velocity even when the NO concentration of 100 nM, the upper threshold of toxicity [23] Rapid NO delivery is pertinent to pathogenic bacteria, such as P. aeruginosa and Neisseria meningitidis, which encode NORs that function to detoxify NO and combat defenses against infection [81, 82].
Supplementary Material
Highlights.
Denitrifying NO and respiratory O2 reductases are in the heme/copper superfamily
Hydrophobic tunnels guide NO/O2 to the active site buried within the protein
The hydrophobic tunnels have entrances facing the membrane lipid bilayer
The rate of NO through the cNOR tunnel depends on amino acid side chain orientation
Computed optimal rate constant for NO to reach the cNOR active site is 5×108 M−1s−1
Acknowledgments
The authors acknowledge funding supports from the National Institutes of Health (NIH P41-GM104601, U01-GM111251 and U54-GM087519 to E.T., and R01-HL016101 to R.B.G.) and the Office of Naval Research (ONR N00014-16-1-2535 to E.T.). P.M. gratefully acknowledges NIH support as a trainee of the Molecular Biophysics Training Program (T32-GM008276) during his graduate study. Computational resources were provided by XSEDE (XSEDE MCA06N060) and Blue Waters (ACI-1440026).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kerwin JF, Jr, Lancaster JR, Jr, Feldman PL. Nitric oxide: A new paradigm for second messengers. J Med Chem. 1995;38:4343–4362. doi: 10.1021/jm00022a001. [DOI] [PubMed] [Google Scholar]
- 2.Ignarro LJ. Nitric oxide as a unique signaling molecule in the vascular system: a historical overview. J Physiol Pharmacol. 2002;53:503–14. [PubMed] [Google Scholar]
- 3.Evgenov OV, Pacher P, Schmidt PM, Haskó G, Schmidt HH, Stasch JP. NO - independent stimulators and activators of soluble guanylate cyclase: discovery and therapeutic potential. Nat Rev Drug Disc. 2006;5:755–768. doi: 10.1038/nrd2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cooper CE. Nitric oxide and iron proteins. Biochim Biophys Acta. 1999;1411:290–309. doi: 10.1016/s0005-2728(99)00021-3. [DOI] [PubMed] [Google Scholar]
- 5.Wasser IM, de Vries S, Moenne-Loccoz P, Schröder I, Karlin KD. Nitric oxide in biological denitrification: Fe/Cu metalloenzyme and metal complex NO2 redox chemistry. Chem Rev. 2002;102:1201–1234. doi: 10.1021/cr0006627. [DOI] [PubMed] [Google Scholar]
- 6.Canfield DE, Glazer AN, Falkowski PG. The evolution and future of earth’s nitrogen cycle. Science. 2010;330:192–196. doi: 10.1126/science.1186120. [DOI] [PubMed] [Google Scholar]
- 7.Zumft W. Cell biology and molecular basis of denitrification. Microbiol Mol Biol Rev. 1997;61:533–616. doi: 10.1128/mmbr.61.4.533-616.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Watmough NJ, Butland G, Chessman MR, Moir JW, Richardson DJ, Spiro S. Nitric oxide in bacteria: synthesis and consumption. Biochim Biophys Acta – Bioener. 1999;1411:456–474. doi: 10.1016/s0005-2728(99)00032-8. [DOI] [PubMed] [Google Scholar]
- 9.Einsle O, Kroneck PM. Structural basis of denitrification. Biol Chem. 2004;385:875–883. doi: 10.1515/BC.2004.115. [DOI] [PubMed] [Google Scholar]
- 10.Zumft W. Nitric oxide reductases of prokaryotes with emphasis on the respiratory, hemecopper oxidase type. J Inorg Biochem. 2005;99:194–215. doi: 10.1016/j.jinorgbio.2004.09.024. [DOI] [PubMed] [Google Scholar]
- 11.Moenne-Loccoz P, Fee JA. Catalyzing NO to N2O in the nitrogen cycle. Science. 2010;330:1632–1633. doi: 10.1126/science.1200247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hino T, Matsumoto Y, Nagano S, Sugimoto H, Fukumori Y, Murata T, Iwata S, Shiro Y. Structural basis of biological N2O generation by bacterial nitric oxide reductase. Nature. 2010;330:1666–1670. doi: 10.1126/science.1195591. [DOI] [PubMed] [Google Scholar]
- 13.Sato N, Ishi S, Sugimoto H, Hino T, Fukumori Y, Sako Y, Shiro Y, Tosha T. Structures of reduced and ligand-bound nitric oxide reductase provide insights into functional differences in respiratory enzymes. Proteins: Struct, Func, Bioinf. 2013;82:1258–1271. doi: 10.1002/prot.24492. [DOI] [PubMed] [Google Scholar]
- 14.Terasaka E, Yamada K, Wang P, Hosokawa K, Yamagiwa R, Matsumoto K, Ishii S, Mori T, Yagi K, Sawai H, Arai H, Sugimoto H, Sugita Y, Shiro Y, Tosha T. Dynamics of nitric oxide controlled by protein complex in bacterial system. Proc Natl Acad Sci USA. 2017;114:9888–9893. doi: 10.1073/pnas.1621301114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Matsumoto Y, Tosha T, Pisliakov AV, Hino T, Sugimoto H, Nagano S, Sugita Y, Shiro Y. Crystal structure of quinol-dependent nitric oxide reductase from Geobacillus stearothermophilus. Nat Struct Biol. 2012;19:238–245. doi: 10.1038/nsmb.2213. [DOI] [PubMed] [Google Scholar]
- 16.Shiro Y, Sugimoto H, Tosha T, Nagano S, Hino T. Structural basis for nitrous oxide generation by bacterial nitric oxide reductases. Phil Trans R Soc Lond B. 2012;367:1195–1203. doi: 10.1098/rstb.2011.0310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hino T, Nagano S, Sugimoto H, Tosha T, Shiro Y. Molecular structure and function of bacterial nitric oxide reductase. Biochim Biophys Acta – Bioener. 2012;1817:680–687. doi: 10.1016/j.bbabio.2011.09.021. [DOI] [PubMed] [Google Scholar]
- 18.Wikstrom M. Active site intermediates in the reactions of O2 by cytochrome oxidase, and their derivatives. Biochim Biophys Acta – Bioener. 2012;1817:468–475. doi: 10.1016/j.bbabio.2011.10.010. [DOI] [PubMed] [Google Scholar]
- 19.Wikstrom M, Sharma V, Kaila VR, Hosler JP, Hummer G. New perspective on proton pumping in cellular respiration. Chem Rev. 2015;115:2196–2221. doi: 10.1021/cr500448t. [DOI] [PubMed] [Google Scholar]
- 20.Yoshikawa S, Shimada A. Reaction mechanism of cytochrome c oxidase. Chem Rev. 2015;115:1936–1989. doi: 10.1021/cr500266a. [DOI] [PubMed] [Google Scholar]
- 21.Pereira M, Santana M, Teixeira M. A novel scenario for the evolution of haem-copper oxygen reductases. Biochim Biophys Acta – Bioener. 2001;1505:185–208. doi: 10.1016/s0005-2728(01)00169-4. [DOI] [PubMed] [Google Scholar]
- 22.Sousa FL, Alves RJ, Ribeiro MA, Pereira-Leal JB, Teixeira M, Pereira MM. The superfamily of heme-copper oxygen reductases: Types and evolutionary considerations. Biochim Biophys Acta – Bioener. 2012;1817:629–637. doi: 10.1016/j.bbabio.2011.09.020. [DOI] [PubMed] [Google Scholar]
- 23.Hendriks JH, Jasaitis A, Saraste M, Verkhovsky MI. Proton and electron pathways in the bacterial nitric oxide reductase. Biochemistry. 2002;41:2331–2340. doi: 10.1021/bi0121050. [DOI] [PubMed] [Google Scholar]
- 24.Reimann J, Flock U, Lepp H, Honigmann A, Adelroth P. A pathway for protons in nitric oxide reductase from Paracoccus denitrificans. Biochim Biophys Acta. 2007;1767:362–373. doi: 10.1016/j.bbabio.2007.03.006. [DOI] [PubMed] [Google Scholar]
- 25.Pisliakov AV, Hino T, Shiro Y, Sugita Y. Molecular dynamics simulations reveal proton transfer pathways in cytochrome c-dependent nitric oxide reductase. PLoS Comput Biol. 2012;8:e1002674. doi: 10.1371/journal.pcbi.1002674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Flock U, Thorndycroft FH, Matorin AD, Richardson DJ, Watmough NJ, Adelroth P. Defining the proton entry point in the bacterial respiratory nitric oxide reductase. J Biol Chem. 2006;283:3839–3845. doi: 10.1074/jbc.M704615200. [DOI] [PubMed] [Google Scholar]
- 27.ter Beek J, Kruse N, Reimann J, Lachmann P, Adelroth P. The nitric-oxidereductase from Paracoccus denitrificans uses a single specific proton pathway. J Biol Chem. 2013;288:30626–30635. doi: 10.1074/jbc.M113.497347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hendriks JH, Prior L, Baker AR, Thomson AJ, Saraste M, Watmough NJ. Reaction of carbon monoxide with the reduced active site of bacterial nitric oxide reductase. Biochemistry. 2001;40:13361–13369. doi: 10.1021/bi011428t. [DOI] [PubMed] [Google Scholar]
- 29.Flock U, Watmough NJ, Adelroth P. Electron/proton coupling in bacterial nitric oxide reductase during reduction of oxygen. Biochemistry. 2005;44:10711–10719. doi: 10.1021/bi050524h. [DOI] [PubMed] [Google Scholar]
- 30.Szundi I, Funatogawa C, Fee JA, Soulimane T, Einarsdottir O. CO impedes superfast O2 binding in ba3 cytochrome oxidase from Thermus thermophilus. Proc Natl Acad Sci USA. 2010;107:21010–21015. doi: 10.1073/pnas.1008603107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Einarsdottir O, Funatogawa C, Soulimane T, Szundi I. Kinetic studies of the reactions of O2 and NO with reduced Thermus thermophilus ba3 and bovine aa3 using photolabile carriers. Biochim Biophys Acta – Bioener. 2012;1817:672–679. doi: 10.1016/j.bbabio.2011.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mahinthichaichan P, Gennis R, Tajkhorshid E. All the O2 consumed by Thermus thermophilus cytochrome ba3 is delivered to the active site through a long, open hydrophobic tunnel with entrances within the lipid bilayer. Biochemistry. 2016;55(8):1265–1278. doi: 10.1021/acs.biochem.5b01255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Soulimane T, Buse G, Bourenkov GP, Bartunik HD, Huber R, Than ME. Structure and mechanism of the aberrant ba3-cytochrome c oxidase from Thermus thermophilus. EMBO J. 2000;19:1766–1776. doi: 10.1093/emboj/19.8.1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tiefenbrunn T, Liu W, Chen Y, Katritch V, Stout CD, Fee JA, Cherezov V. High resolution structure of the ba3 cytochrome c oxidase from Thermus thermophilus in a lipidic environment. PLoS One. 2011;6:e22348. doi: 10.1371/journal.pone.0022348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Luna VM, Chen Y, Fee JA, Stout CD. Crystallographic studies of Xe and Kr binding within the large internal cavity of cytochrome ba3 from Thermus thermophilus: Structural analysis and role of oxygen transport channels in the Heme-Cu oxidases. Biochemistry. 2008;47:4657–4665. doi: 10.1021/bi800045y. [DOI] [PubMed] [Google Scholar]
- 36.Luna VM, Fee JA, Deniz AA, Stout CD. Mobility of Xe atoms within the oxygen diffusion channel of cytochrome ba3 oxidase. Biochemistry. 2012;51:4669–4676. doi: 10.1021/bi3003988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Iwata S, Ostermeier C, Ludwig B, Michel H. Structure at 2.8 Å resolution of cytochrome c oxidase from Paracoccus denitrificans. Nature. 1995;376:660–669. doi: 10.1038/376660a0. [DOI] [PubMed] [Google Scholar]
- 38.Tsukihara T, Aoyama H, Yamashita E, Tomizaki T, Yamaguchi H, Shinnzawha-Itoh K, Nakashima R, Yaono R, Yoshikawa S. Structures of metal sites of oxidized bovine heart cytochrome c oxidase at 2.8 Å. Science. 1995;269:1069–1074. doi: 10.1126/science.7652554. [DOI] [PubMed] [Google Scholar]
- 39.Svensson-Ek M, Abramson J, Larsson G, Tornroth S, Brzezinski P, Iwata S. The X-ray crystal structures of wild-type and EQ(I-286) mutant cytochrome c oxidases from Rhodobacter sphaeroides. J Mol Biol. 2002;321(2):329–339. doi: 10.1016/s0022-2836(02)00619-8. [DOI] [PubMed] [Google Scholar]
- 40.Shinzawa-Itoh K, Aoyama H, Muramoto K, Terada H, Kurauchi T, Tadehara Y, Yamasaki A, Sugimura T, Kurono S, Tsujimoto K, Mizushima T, Yamashita E, Tsukihara T, Yoshikawa S. Structures and physiological roles of 13 integral lipids of bovine heart cytochrome c oxidase. EMBO J. 2007;26(6):1713–1725. doi: 10.1038/sj.emboj.7601618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Buschmann S, Warkentin E, Xie H, Langer JD, Ermler U, Michel H. structure of cbb3 cytochrome oxidase provides insights into proton pumping. Science. 2010;329:327–330. doi: 10.1126/science.1187303. [DOI] [PubMed] [Google Scholar]
- 42.Lyons JA, Aragao D, Slattery O, Pisliakov AV, Soulimane T, Caffrey M. Structural insights into electron transfer in caa3-type cytochrome oxidase. Nature. 2012;487:514–518. doi: 10.1038/nature11182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mahinthichaichan P, Gennis RB, Tajkhorshid E. Cytochrome aa3 oxygen reductase utilizes the tunnel observed in the crystal structures to deliver O2 for catalysis. Biochemistry. 2018;57:2150–2161. doi: 10.1021/acs.biochem.7b01194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Humphrey W, Dalke A, Schulten K. VMD – Visual Molecular Dynamics. J Mol Graphics. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 45.Olsson MH, Sondergaard CR, Rostkowski M, Jensen JH. Propka3: consistent treatment of internal and surface residues in empirical pKa predictions. J Chem Theory Comput. 2011;7:525–537. doi: 10.1021/ct100578z. [DOI] [PubMed] [Google Scholar]
- 46.Zhang L, Hermans J. Hydrophilicity of cavities in proteins. Proteins: Struct, Func, Gen. 1996;24:433–438. doi: 10.1002/(SICI)1097-0134(199604)24:4<433::AID-PROT3>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 47.Lomize MA, Lomize AL, Pogozheva LD, Mosberg HI. Opm: Orientations of proteins in membranes database. Bioinformatics. 2006;22:623–625. doi: 10.1093/bioinformatics/btk023. [DOI] [PubMed] [Google Scholar]
- 48.Lomize MA, Pogozheva ID, Joo H, Mosberg HI, Lomize AL. OPM database and ppm web server: resources for positioning of proteins in membranes. Nucleic Acids Res. 2012;40:D370–376. doi: 10.1093/nar/gkr703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.MacKerell AD, Jr, Bashford D, Bellott M, Dunbrack RL, Jr, Evanseck JD, Field MJ, Fischer S, Gao J, Guo H, Ha S, Joseph D, Kuchnir L, Kuczera K, Lau FTK, Mattos C, Michnick S, Ngo T, Nguyen DT, Prodhom B, Reiher IWE, Roux B, Schlenkrich M, Smith J, Stote R, Straub J, Watanabe M, Wiorkiewicz-Kuczera J, Yin D, Karplus M. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J Phys Chem B. 1998;102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 50.MacKerell AD, Jr, Feig M, Brooks CL., III Extending the treatment of backbone energetics in protein force fields: Limitations of gas-phase quantum mechanics in reproducing protein conformational distributions in molecular dynamics simulations. J Comp Chem. 2004;25(11):1400–1415. doi: 10.1002/jcc.20065. [DOI] [PubMed] [Google Scholar]
- 51.Klauda JB, Venable RM, Freites JA, O’Connor JW, Tobias DJ, Mondragon-Ramirez C, Vorobyov I, MacKerell AD, Jr, Pastor RW. Update of the CHARMM all-atom additive force field for lipids: Validation on six lipid types. J Phys Chem B. 2010;114(23):7830–7843. doi: 10.1021/jp101759q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Autenrieth F, Tajkhorshid E, Baudry J, Luthey-Schulten Z. Classical force field parameters for the heme prosthetic group of cytochrome c. J Comp Chem. 2004;25:1613–1622. doi: 10.1002/jcc.20079. [DOI] [PubMed] [Google Scholar]
- 53.Babu CS, Lim C. Empirical force fields for biological active divalent metal cations in water. J Phys Chem B. 2006;110:691–699. doi: 10.1021/jp054177x. [DOI] [PubMed] [Google Scholar]
- 54.Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kale L, Schulten K. Scalable molecular dynamics with NAMD. J Comp Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ryckaert JP, Ciccotti G, Berendsen HJC. Numerical integration of the Cartesian equations of motion of a system with constraints: Molecular dynamics of n-alkanes. J Comp Phys. 1977;23:327–341. [Google Scholar]
- 56.Darden T, York D, Pedersen LG. Particle mesh Ewald: An N•log(N) method for Ewald sums in large systems. J Chem Phys. 1993;98:10089–10092. [Google Scholar]
- 57.Martyna GJ, Tobias DJ, Klein ML. Constant pressure molecular dynamics algorithms. J Chem Phys. 1994;101(5):4177–4189. [Google Scholar]
- 58.Feller SE, Zhang Y, Pastor RW, Brooks BR. Constant pressure molecular dynamics simulation: The Langevin piston method. J Chem Phys. 1995;103(11):4613–4621. [Google Scholar]
- 59.Cohen J, Arkhipov A, Braun R, Schulten K. Imaging the migration pathways for O2, CO, NO, and Xe inside myoglobin. Biophys J. 2006;91:1844–1857. doi: 10.1529/biophysj.106.085746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang Y, Tajkhorshid E. Nitric oxide conduction by the brain aquaporin AQP4. Proteins: Struct, Func, Bioinf. 2010;78:661–670. doi: 10.1002/prot.22595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Frenkel D, Smit B. Understanding Molecular Simulation From Algorithms to Applications. Academic Press; California: 2002. [Google Scholar]
- 62.Torrie GM, Valleau JP. Nonphysical sampling distributions in Monte Carlo free-energy estimation: Umbrella sampling. J Comp Phys. 1977;23:187–199. [Google Scholar]
- 63.Roux B. The calculation of the potential of mean force using computer simulations. Comput Phys Commun. 1995;91:275–282. [Google Scholar]
- 64.Izrailev S, Stepaniants S, Isralewitz B, Kosztin D, Lu H, Molnar F, Wriggers W, Schulten K. Steered molecular dynamics. In: Deuflhard P, Hermans J, Leimkuhler B, Mark AE, Reich S, Skeel RD, editors. Computational Molecular Dynamics: Challenges, Methods, Ideas, Vol. 4 of Lecture Notes in Computational Science and Engineering. Springer-Verlag; Berlin: 1998. pp. 39–65. [Google Scholar]
- 65.Kumar S, Bouzida D, Swendsen RH, Kollman PA, Rosenberg JM. The weighted histogram analysis method for free-energy calculations on biomolecules. I. The method. J Comp Chem. 1992;13:1011–1021. [Google Scholar]
- 66.Chovancova E, Pavelka A, Benes P, Strnad O, Brezovsky J, Kozlikova B, Gora A, Sustr V, Klvana M, Medek P, Biedermannova L, Sochor J, Damborsky J. CAVER 3.0: A tool for the analysis of transport pathways in dynamic protein structures. PLoS Comput Biol. 2012;8(10):e1002708. doi: 10.1371/journal.pcbi.1002708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Schurig-Briccio LA, Venkatakrishnan P, Hemp J, Bricio C, Berenguer J, Gennis RB. Characterization of the nitric oxide reductase from Thermus thermophilus. Proc Natl Acad Sci USA. 2013;110:12613–12618. doi: 10.1073/pnas.1301731110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ouyang H, Han H, Roh JH, Hemp J, Hosler JP, Gennis RB. The functional importance of a pair of conserved glutamic acid residues and of Ca2+ binding in the cbb3 type oxygen reductases from Rhodobacter sphaeroides and Vibrio cholerae. Biochemistry. 2012;51:7290–7296. doi: 10.1021/bi3006847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.The UniProt Consortium. UniProt: the universal protein knowledgebase. Nucleic Acids Res. 2017;45:D158–169. doi: 10.1093/nar/gkw1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Moller M, Botti H, Batthyany C, Rubbo H, Radi R, Denicola A. Direct measurement of nitric oxide and oxygen partitioning into liposomes and low density lipoprotein. J Biol Chem. 2005;280:8850–8854. doi: 10.1074/jbc.M413699200. [DOI] [PubMed] [Google Scholar]
- 71.Shiva S, Brookes PS, Patel RP, Anderson PG, Darley-Usmar VM. Nitric oxide partitioning into mitochondrial membranes and the control of respiration at cytochrome c oxidase. Proc Natl Acad Sci USA. 2001;98:7212–7217. doi: 10.1073/pnas.131128898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Marsh D, Dzikovski BG, Livshits VA. Oxygen profiles in membranes. Biophys J. 2006;90:L49–L51. doi: 10.1529/biophysj.106.081042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Al-Abdul-Wahid MS, Evanics F, Prosser RS. Dioxygen transmembrane distributions and partitioning thermodynamics in lipid bilayers and micelles. Biochemistry. 2011;50:3975–3983. doi: 10.1021/bi200168n. [DOI] [PubMed] [Google Scholar]
- 74.Quillin ML, Li T, Olson JS, Phillips GN, Jr, Dou Y, Ikeda-Saito M, Regan R, Carlson M, Gibson QH, Li H, Elber R. Structural and functional effects of apolar mutations of the distal valine in myoglobin. J Mol Biol. 1995;245:416–436. doi: 10.1006/jmbi.1994.0034. [DOI] [PubMed] [Google Scholar]
- 75.McDonald W, Funatogawa C, Li Y, Szundi I, Fee YCJA, Stout CD, Einarsdottir O. Ligand access to the active site in Thermus thermophilus ba3 and bovine heart aa3 cytochrome oxidases. Biochemistry. 2013;52:640–652. doi: 10.1021/bi301358a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.McDonald W, Funatogawa C, Li Y, Chen Y, Szundi I, Fee JA, Stout CD, Einarsdottir O. Conserved glycine 232 in the ligand channel of ba3 cytochrome oxidase of Thermus thermophilus. Biochemistry. 2014;53:4467–4475. doi: 10.1021/bi500289h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chang HY, Hemp J, Chen Y, Fee JA, Gennis RB. The cytochrome ba3 oxygen reductase from Thermus thermophilus uses a single input channel for proton delivery to the active site and for proton pumping. Proc Natl Acad Sci USA. 2009;106:16169–16173. doi: 10.1073/pnas.0905264106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Oliveira AS, Damas JM, Baptista A, Soares CM. Exploring O2 diffusion in A-type cytochrome c oxidases: molecular dynamics simulations uncover two alternative channels towards the binuclear site. PLoS Comput Biol. 2014;10:e1004010. doi: 10.1371/journal.pcbi.1004010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Forte E, Urbani A, Saraste M, Sarti P, Brunori M, Giuffre A. The cytochrome cbb3 from Pseudomonas stutzeri displays nitric oxide reductase activity. Eur J Biochem. 2001;268:6486–6490. doi: 10.1046/j.0014-2956.2001.02597.x. [DOI] [PubMed] [Google Scholar]
- 80.Giuffre A, Strubauer G, Sarti P, Brunori M, Zumft WG, Buse G, Soulimane T. The heme-copper oxidases of Thermus thermophilus catalyze the reduction of nitric oxide: evolutionary implications. Proc Natl Acad Sci USA. 1999;96:14718–14723. doi: 10.1073/pnas.96.26.14718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kolpen M, Kuhl M, Bjarnsholt T, Moser C, Ronne Hansen C, Liengaard L, Kharazmi A, Pressler T, Hoiby N, Oestrup Jensen P. Nitrous oxide production in sputum from cystic fibrosis patients with chronic Pseudomonas aeruginosa lung infection. PLoS One. 2014;9:e84353. doi: 10.1371/journal.pone.0084353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Stevanin TM, Moir JWB, Read RC. Nitric oxide detoxification systems enhance survival of Neisseria meningitidis in human macrophages and in nasopharyngeal mucosa. Infection and Immunity. 2005;73:3322–3329. doi: 10.1128/IAI.73.6.3322-3329.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.