Abstract
Chronic inflammation is a key driver of cancer development. Nitrite levels, which are regulated by inducible nitric oxide synthase (iNOS), play a critical role in inflammation. While the anti-oxidant and anti-inflammatory effects of curcumin, a natural product present in the roots of Curcuma longa have been widely studied, the acute pharmacokinetics (PK) and pharmacodynamics (PD) of curcumin in suppressing pro-inflammatory markers and epigenetic modulators remain unclear. In this study, we evaluated the PK and PD of curcumin-induced suppression of lipopolysaccharide (LPS)-mediated inflammation in rat lymphocytes. LPS was administered intravenously either alone or with curcumin to female Sprague-Dawley rats. Plasma samples were analyzed for curcumin concentration and mRNA expression was quantified in lymphocytes. Relative gene expression of several inflammatory and epigenetic modulators was analyzed. To investigate the relationship between curcumin concentration and iNOS, TNF-α, and IL-6 gene expression, PK/PD modeling using Jusko’s indirect response model (IDR) integrating transit compartments (TC) describing the delayed response was conducted. The concentration-time profile of curcumin exhibited a bi-exponential decline, which was well described by a two-compartmental pharmacokinetic model. Importantly our results demonstrate that LPS induced gene expression of pro-inflammatory markers in lymphocytes, with peak expression at approximately 3 h and curcumin suppressed the gene expression in animals administered with LPS. These effects were well captured using the IDR model and an IDR model with the transit compartments. In summary, the PK/PD modeling approach could potentially provide a robust quantitative framework for evaluating the acute anti-inflammatory and epigenetic effects of curcumin in future clinical trials.
Keywords: Curcumin, Inflammation, PKPD modeling, indirect response model, epigenetic modulation
Introduction
Chronic inflammation has been recognized to overlap with the onset of cancer and play an important role in the development of different types of cancers such as colorectal and esophageal cancer and hepatocellular carcinoma (Coussens & Werb, 2002). The enzyme nitric oxide synthase (NOS) is responsible for the production of nitric oxide (NO). NOS is categorized into subfamilies based on the location of expression in the body, such as neuronal tissues (nNOS) and vascular endothelial cells (eNOS), as well as inducible NOS (iNOS). iNOS is commonly expressed in different cells, including macrophages, microglial cells, keratinocytes, hepatocytes, astrocytes, vascular endothelial and epithelial cells have been shown to express iNOS in normal and pathological conditions. It has been widely reported that inflammatory stimuli and infectious diseases trigger the production of NO in micromolar concentrations (Griffith & Stuehr, 1995). iNOS plays an important role in angiogenesis, and collagen deposition. Hemoglobin content, VEGF levels and number of vessels were decreased in implants in iNOS−/− mice compared to WT mice (Cassini-Vieira et al., 2015). The role of iNOS expression in tumors and the adjacent inflammatory microenvironment has been documented as seen with a. strong iNOS expression in prostate tumor cells, whereas normal and hyperplastic prostate tissue exhibit weak or no iNOS expression (Aaltoma, Lipponen, & Kosma, 2001).
Lipopolysaccharide (LPS) is a bacterial toxin which interacts with Toll-like receptor 4 (TLR4) to trigger inflammatory response. LPS-induced systemic inflammation is a classic animal model in studying inflammatory diseases (Juskewitch et al., 2012; Thomas, 1954). Studies of LPS have focused on the roles of macrophages and mediators, e.g., cytokines, released from those cells during the host response. The effects of LPS resulting in an inflammatory cascade demonstrated by early production of pro-inflammatory cytokines have been shown in vitro and in vivo (Terenzi et al., 1995). LPS induces iNOS along with several other pro-inflammatory cytokines, such as tumor necrosis factor-α (TNF-α) and interleukin (IL)-1β (Melillo, Cox, Biragyn, Sheffler, & Varesio, 1994). Quantitative approaches using indirect response models (IDR) and receptor mediated- models have been used to describe the effects of LPS on pro-inflammatory markers (Foteinou, Calvano, Lowry, & Androulakis, 2009; Yang, Calvano, Lowry, & Androulakis, 2011). In one study, a PKPD modeling approach was used to quantify the pharmacodynamics of lung iNOS mRNA expression and plasma NO after LPS-induced acute inflammation in the rats dosed with methylprednisolone (MPL), a well-known anti-inflammatory agent showed induction of iNOS expression post LPS administration. In the animals that received MPL along with LPS, iNOS expression was significantly lowered which was quantitatively described using an IDR (Sukumaran, Lepist, DuBois, Almon, & Jusko, 2012).
Curcumin, a well-studied anti-inflammatory and anti-oxidant dietary phytochemical is a component of the rhizomes of turmeric (Curcuma longa), is used in traditional medicine in India (Ammon & Wahl, 1991). We and others have demonstrated the potential role of curcumin in regulation of epigenetic modifying genes, thus, playing a crucial role in the prevention of several cancers (Khor et al., 2011; Link et al., 2013; Shu et al., 2011). The role of curcumin as an anti-inflammatory agent and its ability to inhibit the expression of pro-inflammatory markers such as Cycoloxygenase-2 (COX-2), iNOS, TNF-α and Interleukin-6 (IL-6) have been well studied (Boyanapalli et al., 2014; Brouet & Ohshima, 1995; Rao, 2007). Curcumin, diacetylcurcumin and diglutarylcurcumin exhibited potent anti-inflammatory properties compared with aspirin in a carrageenan-induced paw edema model in vivo (Jacob, Badyal, Bala, & Toloue, 2013). Curcumin decreases tyrosine phosphorylation in RAW 264.7 murine macrophages by inhibition of ERK 1/2 activation, reducing iNOS enzyme activity, suggesting that curcumin promotes the ubiquitination and degradation of iNOS following LPS stimulation (Ben et al., 2011). The epigenetic modulatory role of curcumin has recently been established, with several studies showing that curcumin can inhibit several DNA methyltransferases (DNMTs) and histone deacetylases (HDACs) and thus reverse the silencing of key genes, such as Neurog1, DLEC1, and Nrf2 (Guo, Shu, Zhang, Su, & Kong, 2015; Khor et al., 2011; Shu et al., 2011). Curcumin elicits anti-inflammatory effects by restoration of the expression of an epigenetically silenced gene called Neprilysin (NEP) which inhibits Akt signaling in Alzheimer’s disease (Deng et al., 2014). In this study, we aim to quantitatively describe the effect of curcumin on LPS-induced inflammation using a PK/PD modeling approach.
Materials and Methods
Chemicals and reagents
Curcumin (98%), isorhamnetin (>95%, used as internal standard), lipopolysaccharide (LPS), ethanol (99%), diisopropyl ether (98%), ammonium acetate (99%), and formic acid (98%) were purchased from Sigma-Aldrich (St. Louis, MO). Acetonitrile (ACN) and pure water for the LC mobile phase were purchased from Honeywell Burdick & Jackson (Muskegon, MI). De-ionized water was obtained from a Milli-Q system (Millipore, Bedford, MA). Heparin sodium injections (1,000 U/mL) and sodium chloride injections (0.9%) were purchased from Baxter Healthcare Corporation (Deerfield, IL) and Hospira Inc. (Lake Forest, IL), respectively. Ficoll-Paque™ PREMIUM 1.084 was purchased from GE Healthcare (Piscataway, NJ).
Animals
Female Sprague-Dawley rats weighing 250–300 g with a cannulated jugular vein were purchased from Hilltop Laboratories (Scottsdale, PA). The rats were housed in the Laboratory Animal Service facility at Rutgers University and had free access to food and water. The rats were acclimatized for three days, during which they were fed the AIN-76A antioxidant-free diet (Research Diets Inc., New Brunswick, NJ). The experiments were performed in accordance with a protocol approved by the Institutional Animal Care and Use Committee of Rutgers University.
Dosing of animals, plasma collection, and lymphocyte isolation
On the day of the study, the cannula was removed and connected to polyethylene tubes using adapters (Instech Laboratories, PA) for dosing and blood collection. Heparinized saline (50 U/mL) was used to flush the cannulae and tubes. Rats (n=6) were administered Curcumin 40 mg/kg (final injection volume of 0.2 mL) in a vehicle consisting of Cremophor, Tween 80, ethanol and water (1:1:1:7) and LPS 50 µg/kg in PBS. LPS 50 µg/kg was administered (n=6) or vehicle control (n=4) were administered intravenously through the jugular vein. Blood samples (0.3 mL) were collected at 0, 10, 20, and 30 min and 1, 2, 3, 4, 6, 8 12 and 24 hours after the administration of LPS alone or LPS after curcumin, followed by injection of an equal volume of saline. All blood samples were immediately centrifuged at 2,500 rpm at 4°C to obtain plasma samples, which were stored at −80°C until analysis. Due to the limited amount of blood withdrawn from the rats at each time point, the blood cells obtained at the same time point were pooled and re-suspended in a 4× volume of Hank’s balanced salt solution (HBSS, Invitrogen, Grand Island, NY). Lymphocytes were isolated using Ficoll-Paque™ PREMIUM 1.084 according to the manufacturer’s instructions, and the cell pellets were stored at −80°C until further analysis.
Sample preparation and liquid chromatography-mass spectrometry analysis of curcumin in plasma
Aliquots of original or diluted rat plasma samples (100 µL) spiked with isorhamnetin as an internal standard were extracted twice with 200 µL of extraction solvent (diisopropyl ether: ethanol=90:10, v/v), and the layers were separated by centrifugation at 10,000 rpm for 2 min at room temperature. The upper organic layer was transferred to a clean microcentrifuge tube. The combined organic extracts were dried using a vacuum pump, and the dried sample was reconstituted in 200 µL of ACN and water (50:50, v/v). The reconstituted samples were centrifuged at 17,500 rpm for 5 min at 18–20°C, and the supernatant was collected for analysis.
Liquid chromatography-mass spectrometry (LC-MS) analysis was performed using a Finnigan LTQ (Thermo Fisher Scientific Inc., San Jose, CA) consisting of a Surveyor quaternary pump with a build-in degasser, a Surveyor auto-sampler, and a Surveyor PDA detector. Chromatographic separation was achieved using a Zorbax Eclipse XDB C18 column (3.5 µm, 4.6×50 mm, Agilent Technologies, Santa Clara, CA). The aqueous mobile phase A was composed of 0.01% formic acid in water, and the organic mobile phase B was acetonitrile (ACN). The column oven and auto-sampler temperatures were 40°C and 4°C. The mobile phase was 50:50 (A: B) for the first 1 min, followed by 15:85 (A: B) for 5 min. The mobile phase was maintained at 5:95 (A: B) for an additional 2 min and then returned to 50:50 (A: B) for 3 min. The flow rate was 200 µL/min. The volume of the injected sample was 100 µL. The total run time for the present analysis was 11 min. MS/MS system was operated in positive ESI. The analytes were detected in selected ion reaction monitoring (SRM) mode. The ion transition was m/z 369-175 for curcumin and m/z 317-285 for isorhamnetin with normalized collision energy of 30 and 45. The lower and upper limits of quantification were 2 ng/mL and 20 µg/mL. The plasma calibration curve was linear, with r2>0.99.
Measurement of mRNA expression in lymphocytes
Total RNA was extracted from lymphocytes using Picopure RNA isolation kits (Life Technologies) and was reversed transcribed into cDNA. Quantitative PCR was performed using Power SYBR Green mix on an ABI Prism 7900HT system (Applied Biosystems, Foster City, CA). Relative gene expression was calculated using the ΔΔCt method and β-Actin was set as the reference gene for normalization.
Pharmacokinetic (PK) and pharmacodynamic (PD) modeling
The noncompartmental analysis (NCA) was conducted on the time-course of curcumin concentration in plasma. Selected pharmacokinetic (PK) parameters were estimated. The area under the concentration-time curve (AUC) was determined using the linear trapezoidal rule. Clearance (CL) was calculated by dividing the dose by the AUC. The mean residence time (MRT) was obtained by dividing the AUMC by the AUC, where AUMC is the area under the first moment curve. The plasma concentrations were analyzed using the compartmental method in ADAPT 5 (Biomedical Simulations Resource, University of Southern California, Los Angeles, CA) (D'Argenio D, 2009). A two-compartmental model was used to describe the PK profile of curcumin. The differential equations are shown in Eq.1 and Eq.2.
| Eq.1 |
| Eq.2 |
The inter-compartmental clearance between the central (C) and peripheral (P) compartments is represented by CLd; CL is the elimination from the central compartment; and the amount of curcumin in the central and peripheral compartments is represented by X1 and X2 respectively.
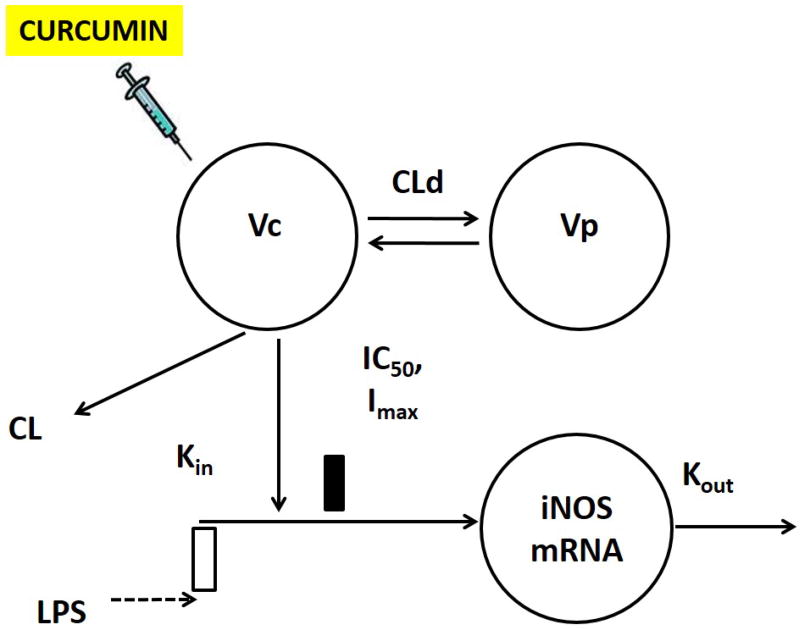
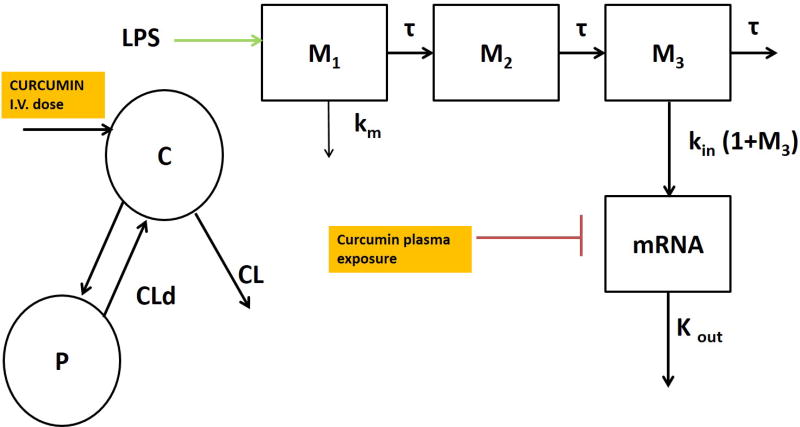
The pro-inflammatory gene expression (iNOS, TNF-α and IL-6) was used as a measure of PD (R) and is described using an indirect response model (Dayneka, Garg, & Jusko, 1993) (Sun & Jusko, 1998). The differential equations shown in Eq. 3 for describe the IDR model without transit compartments (Figure 1). The differential equations in Eq. 4 to 8 describe an indirect response model with transit compartments representing intermediate messengers in the signal transduction system (Figure 4). An indirect response model with transit compartments was used to effectively describe the TNF-α and IL-6 gene expression. The differential equations shown in Eq. 1 and 2 describe the PK while Eq. 4 to 8 explains the PD for iNOS, IL-6 and TNF-α gene expression.
| Eq.3 |
Kin represents the zero-order rate constant for the production of the iNOS mRNA, Kout denotes the first-order rate constant for the degradation of the iNOS mRNA, and R is assumed to be stationary and arbitrarily set as 1 in the initial condition. The stimulation of iNOS mRNA is triggered by (1+Klps), and the duration (T) is fixed for 3H, as observed from the LPS stimulation in all animal groups. The maximum inhibition (Imax) iNOS mRNA by curcumin was fixed at 1 described in Eq.3.
| Eq.4 |
| Eq.5 |
| Eq.6 |
| Eq.7 |
| Eq.8 |
The indirect response model with transit compartments is described by Equations 4–8, where K1 is the rate constant representing the rate of change of LPS, τ represents the mean transit time between compartments, Mn represents the response in the transit compartments and Kin represents the zero-order rate of formation of the iNOS mRNA. Imax is the maximum ability of curcumin to inhibit Kin, and IC50 represents the concentration of curcumin that results in 50% of the maximum inhibition of iNOS. Imax was a fixed parameter (1). The PK/PD modeling was conducted sequentially, where the PK parameters were estimated in ADAPT5 using a generalized least squares method; then, fixing the PK parameters, the gene expression data for LPS alone and in combination with Curcumin were fit to estimate the PD parameters.
Figure 1.
Schematic of the integrated PK-PD model for the curcumin-mediated suppression of LPS-induced iNOS expression. The simple indirect model is defined by Equations 1–3 in the text.
Figure 4.
Schematic of the integrated PKPD model with transit compartments for curcumin-mediated suppression of LPS-induced iNOS, TNF-α, and IL-6 gene expression.
Results
Pharmacokinetics of curcumin
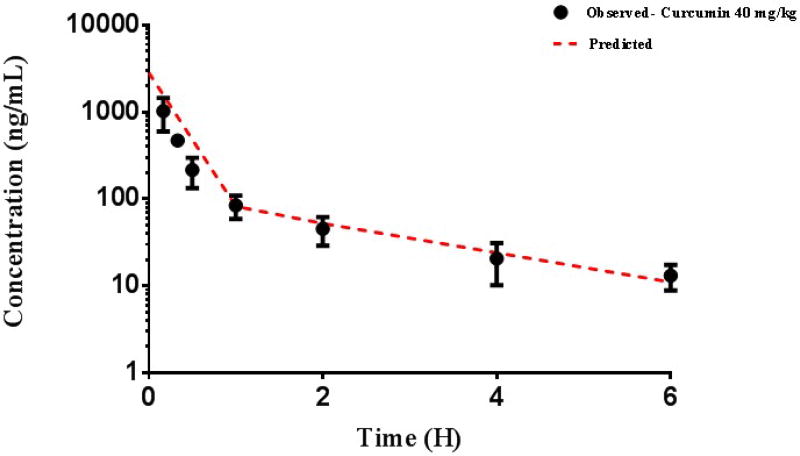
The concentration-time profiles of curcumin in plasma are displayed in Figure 2. The results of noncompartmental analysis are listed in Table 1. The plasma concentration versus time profile showed a bi-exponential decline following i.v administration of 40 mg/kg curcumin which was well-captured by a two compartment pharmacokinetic model. The estimated PK parameters are listed in Table 1. The two-compartment PK parameters for curcumin estimated were used to describe the gene expression data. Briefly, the estimated elimination clearance (CL) was 55.1 L/h/Kg, the distribution clearance was 30.3 L/h/Kg, and the volumes of the central and peripheral compartments were 14.2 and 48.5 L/kg, respectively. The elimination half-life of curcumin was estimated to be 3.36 hours.
Figure 2.
Curcumin pharmacokinetics, as described by a two-compartment model. The dashed line represents the model fit; the circles represent the mean of observed data in rat plasma for a 40 mg/kg dose of curcumin.
Table 1.
Pharmacokinetic (PK) parameters of curcumin in Rat plasma.
| Final Population Pharmacokinetic Parameters | ||
|---|---|---|
| Parameter | Estimate (CV%) | Description |
| CL(L/h/kg) | 55.1 (1.76) | Elimination clearance |
| CLD(L/h/kg) | 30.3 (3.12) | Distribution clearance |
| VC (L/kg) | 14.2 (1.05) | Volume of distribution in the central compartment |
| VP (L/kg) | 48.5 (8.89) | Volume of distribution in the peripheral compartment |
| Non-Compartmental Analysis | ||
| AUC(0–6)(ng/mL*h) | 767 (37.4) | Area under the curve from 0 to 6h |
| AUC(0–∞)(ng/mL*h) | 785 (35.9) | Area under the curve from 0 to infinity |
| AUMC(0–6)(ng/mL*h2) | 962 (26.4) | Area under the first moment curve 0 to 6h |
| AUMC(0–∞)(ng/mL*h2) | 1265 (3.7) | Area under the first moment curve 0 to infinity |
| MRT (h) | 1.75 (33.0) | Mean residence time |
| CL (L/h/kg) | 55.7 (36.5) | Total clearance |
| T1/2(h) | 3.36 (42.9) | Elimination half-life |
Gene expression by qRT-PCR
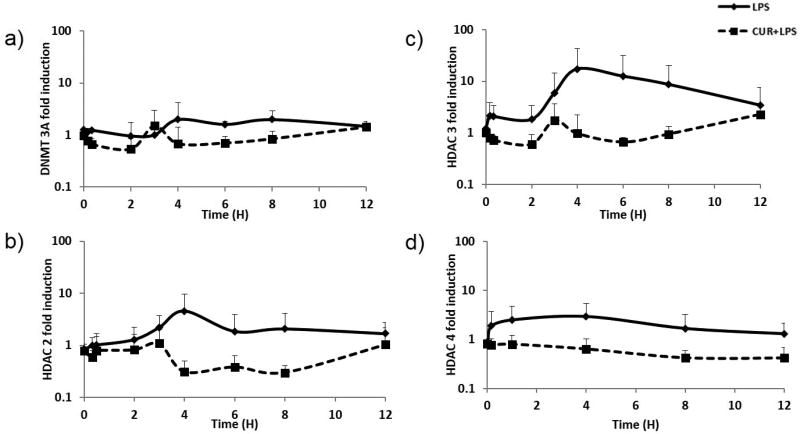
The expression of pro-inflammatory genes such as iNOS, TNF-α and IL-6 increased over time in the LPS-dosed group and reached a maximum at approximately 3 or 4 h. The expression levels of the epigenetic modulatory genes DNMT3A, HDAC2, HDAC3 and HDAC4 (Figure 7) also increased and reached maximum levels at approximately 4 h which were inhibited over time in the animals dosed with curcumin and LPS (Figure 7). A PK/PD modeling approach was used to quantitatively describe the modulation of pro-inflammatory gene expression.
Figure 7.
Changes in the mRNA expression levels of the pro-inflammatory markers a) DNMT3A, b) HDAC2, c) HDAC3, and d) HDAC4 in rat lymphocytes following dosing with LPS alone (50 µg/kg) represented by solid black line or with curcumin (40 mg/kg) and LPS (50 µg/kg) represented by dashed lines. The data presented is the mean of the observed fold change analyzed by qPCR.
Pharmacokinetic-pharmacodynamic correlation
The expression of pro-inflammatory genes such as iNOS, IL-6 and TNF-α was measured by qRT-PCR. Using an IDR with and without transit compartments, the gene expression profile was quantitatively described. The fold changes in mRNA expression levels were calculated for iNOS, TNF-α, and IL-6 mRNA following treatment with LPS alone or curcumin and LPS. iNOS (Figures 3 and 5), TNF-α (Figure 6a), IL-6 (Figure 6b) gene expression was induced by LPS treatment, however, the LPS induced pro-inflammatory gene expression was suppressed in animals administed with curcumin. The estimated PD parameters for iNOS, IL-6 and TNF-α are presented in Tables 2 and 3.
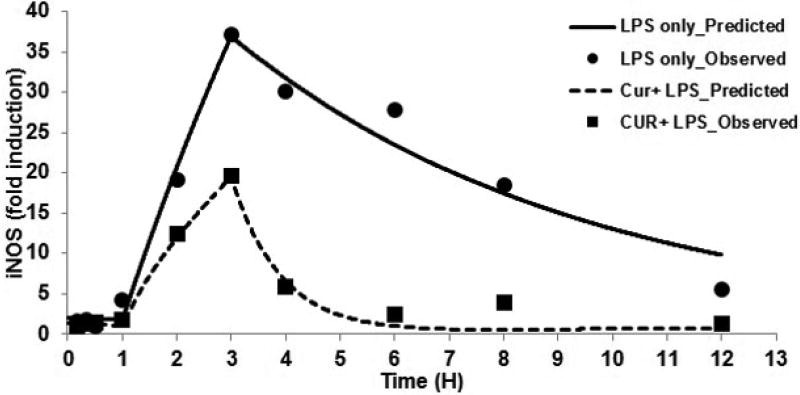
Figure 3.
iNOS gene expression described by indirect response model; circles and squares represent observed and solid line and dashed lines represent predicted data in LPS alone and Curcumin with LPS group, respectively
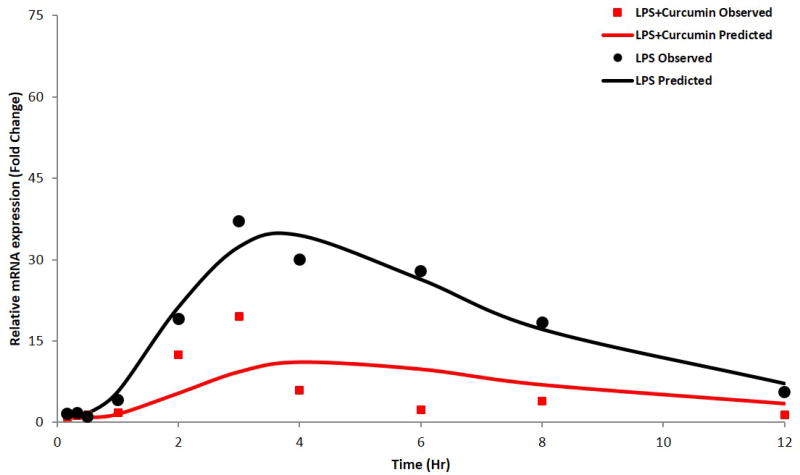
Figure 5.
iNOS gene expression, as described by the indirect response model with transit compartments; the circles and squares represent mean of observed data and the solid lines represent the data predicted by the model for LPS-only and curcumin with LPS groups, respectively.
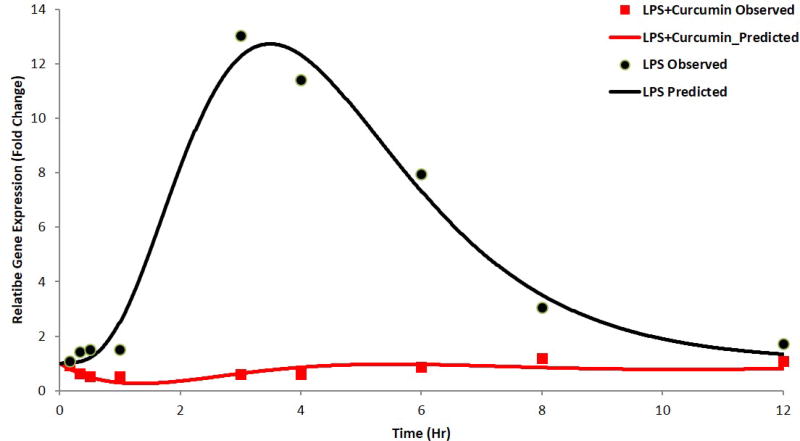
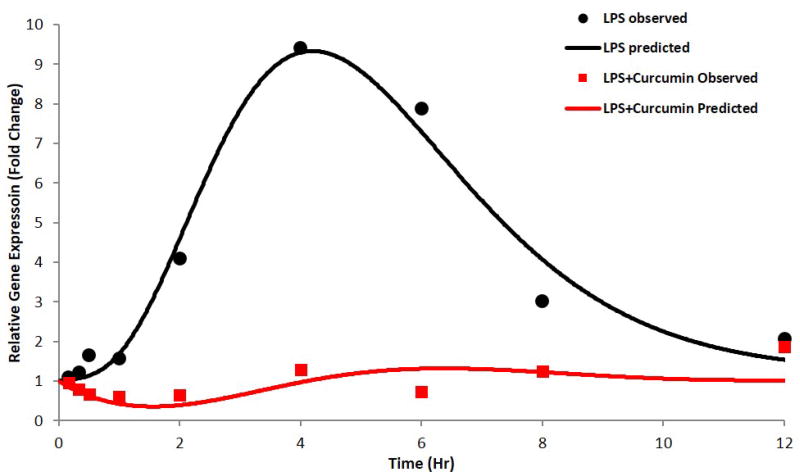
Figure 6.
a) TNF-α gene expression, b) IL-6 gene expression as described by the indirect response model with transit compartments; the circles and squares represent mean of observed data and the solid lines represent the data predicted by the model for LPS-only and curcumin with LPS groups, respectively.
Table 2.
Pharmacodynamic parameters of iNOS expression estimated from the indirect response model and the indirect response model with transit compartments.
| Parameter | Description | Estimates by model | |
|---|---|---|---|
| IDR only | TC + IDR | ||
| Kin (1/h) | Zero-order rate of formation of iNOS mRNA | 1.115 | 0.418 |
| T (h) | Duration of LPS effect | 3 (Fixed) | _ |
| Kout (1/h) | Elimination constant of iNOS mRNA | 1.496 | 0.26 |
| Klps | Stimulation of iNOS mRNA by LPS | 131.8 | 137 |
| IC50 (ng/mL) | 50% inhibition of iNOS by Curcumin | 5.82 | 41.6 |
| τ (h) | Transit time | _ | 0.515 |
| Km (1/h) | Rate constant for LPS elimination | _ | 0.947 |
Table 3.
Pharmacodynamic parameters of TNF-α and IL-6 expression estimated from the indirect response model with transit compartments.
| Parameter | Description | Estimates by TC+IDR model | |
|---|---|---|---|
| TNF-α | IL-6 | ||
| Kin (1/h) | Zero-order rate of formation of TNF-α mRNA | 1.49 | 1.14 |
| Kout (1/h) | Elimination constant of TNF-α mRNA | 1.39 | 0.941 |
| Klps | Stimulation of TNF-α mRNA by LPS | 30.4 | 33.7 |
| IC50 (ng/mL) | 50% inhibition of TNF-α by Curcumin | 4.58 | 7.37 |
| τ (h) | Transit time | 0.727 | 0.941 |
| Km (1/h) | Rate constant for LPS elimination | 0.581 | 0.946 |
Discussion
Curcumin is an anti-inflammatory agent with potential applications in the prevention of inflammation because inflammation is a precursor to several diseases, such as cancers and neurological disorders (Boyanapalli & Tony Kong, 2015; Deguchi, 2015). The low oral bioavailability of curcumin is one challenge that must be overcome for the drug to reach adequate circulating levels and induce a pharmacodynamic (PD) response (Gutierres et al., 2015). Several studies have attempted to improve the bioavailability of curcumin by adding compounds such as piperine to the formulation (Jager et al., 2014; Klickovic et al., 2014; Shoba et al., 1998). The ability of curcumin to inhibit DNA hypermethylation and to reactivate the expression of key genes such as DLEC1 and NEP, that are responsible for preventing cancers and neurological disorders establishes it as a potential epigenetic modulator (Deng et al., 2014; Guo et al., 2015; Rodriguez-Paredes & Esteller, 2011). Role of epigenetic modulators in inflammation is an interesting area of research with the link between them not fully understood yet. Activated IL-6 signaling is known to responsible for induction of DNMT 1 and 3B in ulcerative colitis and oral cancers respectively (Chen, Chen, & Lin, 2014; Li et al., 2012). HDAC inhibitors which are another potential class of epigenetic modulators have been reported in several diseases and are kown to inhibit TNF-α expression, thereby could play an important role in the regulation of inflammatory disorders (Chung, Lee, Wang, & Yao, 2003; Nishida et al., 2004). Studies have shown that Trichostatin-A a well-known HDAC inhibitor reduced joint swelling due to hyperacetylation of p21 promoter that was accompanied by reduced TNF-α expression (Chung et al., 2003). Our results showed that LPS induced HDAC and DNMT gene expression which was inhibited by co-administration with curcumin. The same effects were also seen in the pro-inflammatory cytokines. Although several studies have reported the role of epigenetic modulators like DNMTs and HDACs on pro-inflammatory genes, the link to the epigenetic modulation and inflammatory gene expression and the subsequent post transcriptional events remains to be clearly understood (Wang et al., 2018). Recent study showed that sulforaphane (SFN) a HDAC inhibitor that was shown to inhibit HDACs and induce Nrf2, has inhibited HDAC gene expression levels upon induction with LPS thereby paving way to study the epigenetic modulation of LPS induced innate immune responses (Qu et al., 2015). In this study, the plasma concentraion of curcumin and pro-inflammatory as well as epigenetic regulatory gene expression was measured in lymphocytes. The concentration-response of curcumin in lowering acute inflammation following LPS administration was quantitatively described using PK/PD modeling. However, keeping in context the lack of physiological relevance in using the transit compartments in PK/PD moedling to quantitatively describe the epigenetic modulations, the model was not used to describe the epigenetic modulatory gene expression.
Curcumin disposition in plasma exhibited a bi-exponential decline, as described by a two-compartment model with linear clearance (Figure 2). The PK results indicated high clearance and an increased distribution volume, suggesting extensive tissue distribution of curcumin. iNOS and TNF-α mRNA expression increased upon LPS dosing after an initial delay of 1 h, with maximum expression at 3 h. The administration of both curcumin and LPS reduced the expression of iNOS mRNA, which exhibited a peak at 3 h and was considerably lower than the levels elicited by LPS administration alone. Treatment with Curcumin and LPS also reduced the expression of TNF-α mRNA. A simple IDR with and without transit compartments was used to describe the induction of iNOS mRNA expression; the iNOS gene expression was described successfully in the absence of the transit compartments (Figures 3). However, the delay and duration of the effects of LPS were fixed in this model based on the observed gene expression data from all animals following LPS treatment.
Considering the signal transduction process in the activation of inflammatory molecules and the time-dependent nature of these processes(Foteinou et al., 2009), an IDR model with transit compartments was used to describe the delay in iNOS mRNA expression. The transit compartments serve as messenger molecules (e.g. kinases in TLR4 signals) essential for the transcriptional activation and known to be involved in LPS induced mRNA expression of pro-inflammatory genes (Figures 4, 5 and 6). The delay in the PD response was successfully captured by both modeling approaches; with the transit compartments in the IDRmodel describing the delay which could be physiologically explained by the LPS triggered signal transduction processes. In LPS alone and LPS plus curcumin groups, this model underestimated iNOS mRNA expression, with the model failing to capture the maximum effects of LPS and curcumin on iNOS expression likely due to the simplified nature of the model where the transit compartments serve to capture the delay rather than estimating the different parameters of the genes which are part of the downstream signaling event of the pro-inflammatory pathway. IDR modeling with transit compartments were carried out as an iterative process, with various numbers of compartments tested. The number of transit compartments selected for the modeling was based on the goodness of fit with three transit compartments effectively describing the observed data. Similar results were obtained when the inhibition of LPS induced TNF-α and IL-6 expression was modeled using the indirect response model with transit compartments (Figure 6). However, the effect of curcumin was not very well described at later time points. This lack of flexibility in effectively modeling gene expression using transit compartments was improved by estimating the initial conditions of the transit compartments, which were close to zero (data not shown). Although the PKPD model described the pro-inflammatory gene expression governing acute inflammation, there exist certain limitations that limit the direct translation of the model in predicting chronic inflammatory processes. To improve the predictability of such processes, it may be important to take into account any post-transcriptional or translational changes that play an important role in chronic inflammation.
Hence, the present IDR model with transit compartments could potentially be used as an initial framework to describe the effects of LPS and curcumin in the regulation of iNOS and TNF-α transcription. This model was able to describe the observed data but did not provide sufficient flexibility to capture the dynamics of iNOS and TNF-α expression following curcumin and LPS treatment since there was only one dose of curcumin tested. However, this model could be further enhanced by measuring the expression of downstream genes in the TLR4 signaling pathway at multiple time points, thereby building a more robust model that may explain the intermediary steps involved in the transcription of downstream genes in the LPS and TLR4 signaling cascade, as previously described (Foteinou et al., 2009).
Our results demonstrate that the gene expression of pro-inflammatory cytokines (iNOS, TNF-α and IL-6) and epigenetic modulators (DNMT 3A, HDAC2, 3 and 4) were inhibited in animals dosed with 40 mg/kg of curcumin compared with animals administered with LPS alone. The dose used in this study (40 mg/kg) corresponds to a dose of approximately 440 mg for a 70 kg human, calculated based on “Guidance for Industry: Estimating the Maximum Safe Starting Dose in Initial Clinical Trials for Therapeutics in Adult Healthy” published by the US FDA in 2005. So far there are limited clinical studies conducted on curcumin with I.V. administration, but a dose of 120 mg/m2 (~200mg dose, multiplied by population average body surface area) appeared to be safe (Storka et al., 2015). On the other hand, as evidenced from clinical trials and dose escalation studies, oral administration of 12,000 mg curcumin is well-tolerated in humans (Lao et al., 2006). By changing the formulation to improve the bioavailability of curcumin, a maximum plasma concentration of approximately 300 ng/mL was observed after a non-toxic oral dose of 210 mg curcumin in human (Kanai et al., 2012). This concentration is much higher than the IC50 estimates of curcumin anti-inflammatory effect in our current model. Therefore, it is likely that a similar inhibitory response of inflammatory genes may be achievable in humans.
In conclusion, the current results are indicative of the effect of curcumin in reducing LPS induced pro-inflammatory gene expression and in modulating the expression of epigenetic regulatory genes. Furthermore, the description of iNOS and TNF-α expression by the PD model suggests that these two genes may be good potential PD markers for future trials studying anti-inflammatory effects of curcumin.
Acknowledgments
We thank all the members of Kong lab for their suggestions and helpful discussion in the preparation of this manuscript. This work was supported in part by institutional funds and R01AT007065 from the National Center for Complementary and Alternative Medicines (NCCAM) and the Office of Dietary Supplements (ODS).
This work is supported in part by R01-AT007065 awarded to Dr. Ah-Ng Tony Kong from National Center for Complementary and Alternative Medicine (NCCAM), Office of Dietary Supplements (ODS). The authors thank all the members in Dr. Ah-Ng Tony Kong’s lab for their helpful discussion of this work.
Footnotes
Conflict of Interest
The authors declare that there is no conflict of interest associated with this work.
References
- Aaltoma SH, Lipponen PK, Kosma VM. Inducible nitric oxide synthase (iNOS) expression and its prognostic value in prostate cancer. Anticancer Res. 2001;21(4B):3101–3106. [PubMed] [Google Scholar]
- Ammon HP, Wahl MA. Pharmacology of Curcuma longa. Planta Med. 1991;57(1):1–7. doi: 10.1055/s-2006-960004. [DOI] [PubMed] [Google Scholar]
- Ben P, Liu J, Lu C, Xu Y, Xin Y, Fu J, Yin Z. Curcumin promotes degradation of inducible nitric oxide synthase and suppresses its enzyme activity in RAW 264.7 cells. Int Immunopharmacol. 2011;11(2):179–186. doi: 10.1016/j.intimp.2010.11.013. [DOI] [PubMed] [Google Scholar]
- Boyanapalli SS, Paredes-Gonzalez X, Fuentes F, Zhang C, Guo Y, Pung D, Kong AN. Nrf2 knockout attenuates the anti-inflammatory effects of phenethyl isothiocyanate and curcumin. Chem Res Toxicol. 2014;27(12):2036–2043. doi: 10.1021/tx500234h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyanapalli SS, Tony Kong AN. "Curcumin, the King of Spices": Epigenetic Regulatory Mechanisms in the Prevention of Cancer, Neurological, and Inflammatory Diseases. Curr Pharmacol Rep. 2015;1(2):129–139. doi: 10.1007/s40495-015-0018-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouet I, Ohshima H. Curcumin, an anti-tumour promoter and anti-inflammatory agent, inhibits induction of nitric oxide synthase in activated macrophages. Biochem Biophys Res Commun. 1995;206(2):533–540. doi: 10.1006/bbrc.1995.1076. [DOI] [PubMed] [Google Scholar]
- Cassini-Vieira P, Araujo FA, da Costa Dias FL, Russo RC, Andrade SP, Teixeira MM, Barcelos LS. iNOS Activity Modulates Inflammation, Angiogenesis, and Tissue Fibrosis in Polyether-Polyurethane Synthetic Implants. Mediators Inflamm. 2015;2015:138461. doi: 10.1155/2015/138461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen WC, Chen MF, Lin PY. Significance of DNMT3b in oral cancer. PLoS One. 2014;9(3):e89956. doi: 10.1371/journal.pone.0089956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung YL, Lee MY, Wang AJ, Yao LF. A therapeutic strategy uses histone deacetylase inhibitors to modulate the expression of genes involved in the pathogenesis of rheumatoid arthritis. Mol Ther. 2003;8(5):707–717. doi: 10.1016/s1525-0016(03)00235-1. [DOI] [PubMed] [Google Scholar]
- Coussens LM, Werb Z. Inflammation and cancer. Nature. 2002;420(6917):860–867. doi: 10.1038/nature01322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Argenio D SA, Wang X. ADAPT 5 user's guide: pharmacokinetic/pharmacodynamic systems analysis software 2009 [Google Scholar]
- Dayneka NL, Garg V, Jusko WJ. Comparison of four basic models of indirect pharmacodynamic responses. J Pharmacokinet Biopharm. 1993;21(4):457–478. doi: 10.1007/BF01061691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deguchi A. Curcumin targets in inflammation and cancer. Endocr Metab Immune Disord Drug Targets. 2015;15(2):88–96. doi: 10.2174/1871530315666150316120458. [DOI] [PubMed] [Google Scholar]
- Deng Y, Lu X, Wang L, Li T, Ding Y, Cao H, Yu G. Curcumin inhibits the AKT/NF-kappaB signaling via CpG demethylation of the promoter and restoration of NEP in the N2a cell line. AAPS J. 2014;16(4):649–657. doi: 10.1208/s12248-014-9605-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foteinou PT, Calvano SE, Lowry SF, Androulakis IP. Modeling endotoxin-induced systemic inflammation using an indirect response approach. Math Biosci. 2009;217(1):27–42. doi: 10.1016/j.mbs.2008.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffith OW, Stuehr DJ. Nitric oxide synthases: properties and catalytic mechanism. Annu Rev Physiol. 1995;57:707–736. doi: 10.1146/annurev.ph.57.030195.003423. [DOI] [PubMed] [Google Scholar]
- Guo Y, Shu L, Zhang C, Su ZY, Kong AN. Curcumin inhibits anchorage-independent growth of HT29 human colon cancer cells by targeting epigenetic restoration of the tumor suppressor gene DLEC1. Biochem Pharmacol. 2015;94(2):69–78. doi: 10.1016/j.bcp.2015.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierres VO, Campos ML, Arcaro CA, Assis RP, Baldan-Cimatti HM, Peccinini RG, Brunetti IL. Curcumin Pharmacokinetic and Pharmacodynamic Evidences in Streptozotocin-Diabetic Rats Support the Antidiabetic Activity to Be via Metabolite(s) Evid Based Complement Alternat Med. 2015;2015:678218. doi: 10.1155/2015/678218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob JN, Badyal DK, Bala S, Toloue M. Evaluation of the in vivo anti-inflammatory and analgesic and in vitro anti-cancer activities of curcumin and its derivatives. Nat Prod Commun. 2013;8(3):359–362. [PubMed] [Google Scholar]
- Jager R, Lowery RP, Calvanese AV, Joy JM, Purpura M, Wilson JM. Comparative absorption of curcumin formulations. Nutr J. 2014;13:11. doi: 10.1186/1475-2891-13-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juskewitch JE, Knudsen BE, Platt JL, Nath KA, Knutson KL, Brunn GJ, Grande JP. LPS-induced murine systemic inflammation is driven by parenchymal cell activation and exclusively predicted by early MCP-1 plasma levels. Am J Pathol. 2012;180(1):32–40. doi: 10.1016/j.ajpath.2011.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanai M, Imaizumi A, Otsuka Y, Sasaki H, Hashiguchi M, Tsujiko K, Chiba T. Dose-escalation and pharmacokinetic study of nanoparticle curcumin, a potential anticancer agent with improved bioavailability, in healthy human volunteers. Cancer Chemother Pharmacol. 2012;69(1):65–70. doi: 10.1007/s00280-011-1673-1. [DOI] [PubMed] [Google Scholar]
- Khor TO, Huang Y, Wu TY, Shu L, Lee J, Kong AN. Pharmacodynamics of curcumin as DNA hypomethylation agent in restoring the expression of Nrf2 via promoter CpGs demethylation. Biochem Pharmacol. 2011;82(9):1073–1078. doi: 10.1016/j.bcp.2011.07.065. [DOI] [PubMed] [Google Scholar]
- Klickovic U, Doberer D, Gouya G, Aschauer S, Weisshaar S, Storka A, Wolzt M. Human pharmacokinetics of high dose oral curcumin and its effect on heme oxygenase-1 expression in healthy male subjects. Biomed Res Int. 2014;2014:458592. doi: 10.1155/2014/458592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lao CD, Ruffin MTt, Normolle D, Heath DD, Murray SI, Bailey JM, Brenner DE. Dose escalation of a curcuminoid formulation. BMC Complement Altern Med. 2006;6:10. doi: 10.1186/1472-6882-6-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Deuring J, Peppelenbosch MP, Kuipers EJ, de Haar C, van der Woude CJ. IL-6-induced DNMT1 activity mediates SOCS3 promoter hypermethylation in ulcerative colitis-related colorectal cancer. Carcinogenesis. 2012;33(10):1889–1896. doi: 10.1093/carcin/bgs214. [DOI] [PubMed] [Google Scholar]
- Link A, Balaguer F, Shen Y, Lozano JJ, Leung HC, Boland CR, Goel A. Curcumin modulates DNA methylation in colorectal cancer cells. PLoS One. 2013;8(2):e57709. doi: 10.1371/journal.pone.0057709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melillo G, Cox GW, Biragyn A, Sheffler LA, Varesio L. Regulation of nitric-oxide synthase mRNA expression by interferon-gamma and picolinic acid. J Biol Chem. 1994;269(11):8128–8133. [PubMed] [Google Scholar]
- Nishida K, Komiyama T, Miyazawa S, Shen ZN, Furumatsu T, Doi H, Asahara H. Histone deacetylase inhibitor suppression of autoantibody-mediated arthritis in mice via regulation of p16INK4a and p21(WAF1/Cip1) expression. Arthritis Rheum. 2004;50(10):3365–3376. doi: 10.1002/art.20709. [DOI] [PubMed] [Google Scholar]
- Qu X, Proll M, Neuhoff C, Zhang R, Cinar MU, Hossain MM, Uddin MJ. Sulforaphane epigenetically regulates innate immune responses of porcine monocyte-derived dendritic cells induced with lipopolysaccharide. PLoS One. 2015;10(3):e0121574. doi: 10.1371/journal.pone.0121574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao CV. Regulation of COX and LOX by curcumin. Adv Exp Med Biol. 2007;595:213–226. doi: 10.1007/978-0-387-46401-5_9. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Paredes M, Esteller M. Cancer epigenetics reaches mainstream oncology. Nat Med. 2011;17(3):330–339. doi: 10.1038/nm.2305. [DOI] [PubMed] [Google Scholar]
- Shoba G, Joy D, Joseph T, Majeed M, Rajendran R, Srinivas PS. Influence of piperine on the pharmacokinetics of curcumin in animals and human volunteers. Planta Med. 1998;64(4):353–356. doi: 10.1055/s-2006-957450. [DOI] [PubMed] [Google Scholar]
- Shu L, Khor TO, Lee JH, Boyanapalli SS, Huang Y, Wu TY, Kong AN. Epigenetic CpG demethylation of the promoter and reactivation of the expression of Neurog1 by curcumin in prostate LNCaP cells. AAPS J. 2011;13(4):606–614. doi: 10.1208/s12248-011-9300-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storka A, Vcelar B, Klickovic U, Gouya G, Weisshaar S, Aschauer S, Wolzt M. Safety, tolerability and pharmacokinetics of liposomal curcumin in healthy humans. Int J Clin Pharmacol Ther. 2015;53(1):54–65. doi: 10.5414/CP202076. [DOI] [PubMed] [Google Scholar]
- Sukumaran S, Lepist EI, DuBois DC, Almon RR, Jusko WJ. Pharmacokinetic/pharmacodynamic modeling of methylprednisolone effects on iNOS mRNA expression and nitric oxide during LPS-induced inflammation in rats. Pharm Res. 2012;29(8):2060–2069. doi: 10.1007/s11095-012-0733-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun YN, Jusko WJ. Transit compartments versus gamma distribution function to model signal transduction processes in pharmacodynamics. J Pharm Sci. 1998;87(6):732–737. doi: 10.1021/js970414z. [DOI] [PubMed] [Google Scholar]
- Terenzi F, Diaz-Guerra MJ, Casado M, Hortelano S, Leoni S, Bosca L. Bacterial lipopeptides induce nitric oxide synthase and promote apoptosis through nitric oxide-independent pathways in rat macrophages. J Biol Chem. 1995;270(11):6017–6021. doi: 10.1074/jbc.270.11.6017. [DOI] [PubMed] [Google Scholar]
- Thomas L. The physiological disturbances produced by endotoxins. Annu Rev Physiol. 1954;16:467–490. doi: 10.1146/annurev.ph.16.030154.002343. [DOI] [PubMed] [Google Scholar]
- Wang J, Hodes GE, Zhang H, Zhang S, Zhao W, Golden SA, Pasinetti GM. Epigenetic modulation of inflammation and synaptic plasticity promotes resilience against stress in mice. Nat Commun. 2018;9(1):477. doi: 10.1038/s41467-017-02794-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q, Calvano SE, Lowry SF, Androulakis IP. A dual negative regulation model of Toll-like receptor 4 signaling for endotoxin preconditioning in human endotoxemia. Math Biosci. 2011;232(2):151–163. doi: 10.1016/j.mbs.2011.05.005. [DOI] [PubMed] [Google Scholar]