Abstract
Incidence of pulmonary diseases caused by non-tuberculous mycobacteria (NTM), relatives of Mycobacterium tuberculosis, is increasing at an alarming rate, surpassing tuberculosis in many countries. Current chemotherapies require long treatment times and the clinical outcomes are often disappointing. There is an urgent medical need to discover and develop new, more-efficacious anti-NTM drugs. In this review, we summarize the current status of NTM drug development, and highlight knowledge gaps and scientific obstacles in NTM drug discovery. We propose strategies to reduce biological uncertainties and to begin to populate a NTM drug pipeline with attractive leads and drug candidates.
Keywords: Nontuberculous mycobacteria, pulmonary disease, Mycobacterium avium complex, Mycobacterium abscessus, drug discovery
Introduction
Whereas the incidence of tuberculosis (TB) is decreasing, a new health concern has been raised by non-tuberculous mycobacteria (NTM). Different from their relative: Mycobacterium tuberculosis (the etiologic agent of TB), NTM are opportunistic pathogens, causing mostly TB-like pulmonary diseases largely in immunocompromised patients or patients with pre-existing lung conditions, such as cystic fibrosis (CF), bronchiectasis or chronic obstructive pulmonary disease (COPD). The annual prevalence of NTM pulmonary disease (NTM-PD) varies in different regions, ranging from 0.2/100 000 to 9.8/100 000 [1,2] with an overall alarming growth rate [3,4]. The situation is worse among vulnerable populations. Large-scale epidemiological studies from several countries and regions reported a high prevalence of 3.3–22.6% in CF patients [5], whereas COPD patients treated with inhaled corticosteroid therapy are associated with a 29-fold increased risk of NTM-PD [6]. In developing countries, misdiagnosis of NTM as TB is common, owing to their similar appearance under microscopic examination of sputum smears [7,8]. This is problematic in many ways: NTM incidence is vastly underestimated, it unnecessarily drains resources dedicated to the global fight against TB and it leads to mistreatment of patients because NTM infections do not respond to classic TB drug regimens.
NTM represent >160 species commonly found in soil and water, including municipal and household water supply systems. The species show varying degrees of virulence leading to diverse clinical features. Within this group of bacteria, the Mycobacterium avium complex (MAC: M. avium, Mycobacterium intracellulare and Mycobacterium chimaera) and Mycobacterium abscessus are the most frequently encountered pathogens associated with NTM-PD, accounting for >90% of the total cases reported [5,9]. Until recently, it was thought that the majority of NTM patients were infected with genetically unrelated strains acquired independently through exposure to soil or water [10,11]. However, recent whole-genome analysis of a large global collection of M. abscessus clinical isolates uncovered that most patients were actually infected with genetically clustered strains [12,13]. Although these data are compatible with infection from a common environmental source, studies on transmission events suggest that indirect human-to-human transmission, presumably through fomite spread or infectious aerosols, could have also contributed to the spread of NTM infections [12,13].
Treatment for NTM-PD as recommended by the American Thoracic Society and the British Thoracic Society is largely empirical [14,15]. Despite significant NTM interspecies variability in drug susceptibility, treatment is often lumped together. Therapeutic regimens tailored for specific species are only available for a few commonly encountered pathogens including for instance MAC, M. abscessus, Mycobacterium kansasii and Mycobacterium xenopi. In general, macrolide-based (clarithromycin or azithromycin) multidrug regimens are prescribed. For infection with MAC, the standard regimen includes ethambutol and rifampicin. Whereas, in the case of M. abscessus a macrolide is usually given with parenteral antibiotics, an aminoglycoside and either cefoxitin, imipenem or tigecycline. Standard of care calls for 12 months of negative sputum cultures while on therapy [14], which usually results in 18–24 months of treatment with a minimum of three antibiotics [16,17]. Despite this, treatment outcomes remain poor. Although ~50–88% MAC-PD patients achieve sputum conversion (with 4–12% true relapse rate, as opposed to reinfection) [16,18,19], the cure rate among patients with M. abscessus pulmonary infection is only 25–58% [17,20,21]. Thus, M. abscessus is often referred to as the ‘incurable nightmare’. In addition, the prolonged treatment for NTM-PD not only induces severe adverse events in patients but also creates a high burden to society. It has been estimated that a total of US$815 million was spent in relation to NTM-PD in the USA in 2010 [22].
Clearly, owing to the poor treatment outcomes and lengthy treatment duration accompanied by drug toxicity, there is an urgent medical need to develop more-effective and safe regimens consisting of ideally orally bioavailable drugs with broad-spectrum anti-NTM activities for the treatment of NTM-PD. However, this target product profile is not easy to achieve. In this review, with a focus on NTM-PD caused by MAC or M. abscessus, we summarize the current state of NTM drug discovery and development, present our perspective on the underlying knowledge gaps and challenges in NTM drug discovery and discuss how to focus research efforts to accelerate building a NTM drug pipeline, based on lessons learnt during the past decades in the TB field.
Current status of NTM drug discovery and development
Since the critical shift in the 1990s from anti-TB regimens toward macrolide-based multidrug therapy, not much has been accomplished in the treatment of NTM diseases owing to limited research efforts. Most agents in the current treatment recommendations are derived from clinical practice or in vitro drug susceptibility testing results. The only new antibiotic that has been introduced to the chemotherapy with clinical evidence is tigecycline [23]. Based on literature and information from NIH ClinicalTrials.gov (https://clinicaltrials.gov), we have summarized agents in development for the treatment of NTM infections in Figure 1. Unsurprisingly, in comparison with the TB drug pipeline where >35 chemical entities are in the discovery stage and ~30 interventions are currently in clinical trials (Working Group on New TB Drugs: https://www.newtbdrugs.org), the NTM drug pipeline is nearly empty. Most of the current candidates and leads are derived from repurposing and reformulation of existing antibiotics or ‘cross-testing’ of a few TB active compounds.
Figure 1.

Non-tuberculous mycobacteria (NTM) drug pipeline. Agents currently in discovery or development for the treatment of NTM pulmonary disease are shown. Compounds in the ‘Discovery’ column are from literature. The asterisks (*) indicate repurposed drugs in the discovery stage. A ‘Preclinical’ column is not included because no NTM candidates are currently in preclinical development. Drugs under ‘Phase I–IV’ are from ClinicalTrials.gov (https://clinicaltrials.gov) and more details are shown in Table S1 (see supplementary material online). Phase I and II are combined in one column: the first three trials are in Phase II and the last one is a Phase I/II trial. aGaseous nitric oxide is composed of 0.5% NO and 99.5% nitrogen. Abbreviations: M. abs, Mycobacterium abscessus; MAC, Mycobacterium avium complex; PD, pulmonary disease; CF, cystic fibrosis; FAS-II, type II fatty acid synthase; NDH-2, type II NADH-quinone oxidoreductase.
Clofazimine
Clofazimine is an orally administered drug approved for the treatment of leprosy, currently repurposed as an anti-TB drug. Clinical uses against M. avium infections since the 1990s have demonstrated some efficacy in combination with other drugs [24,25], but its use remained limited owing to lack of demonstrable clinical utility and higher mortality rates reported in a trial among HIV patients where it was added to clarithromycin and ethambutol [26]. Research interest was revived in 2012 after a retrospective review reported that a significantly greater proportion of MAC-PD patients (HIV-negative) treated with clofazimine converted to negative cultures, although relapse still occurred [27]. In vitro, its MIC ranges from 1–4 mg/l against M. avium and is ≤ 1 mg/l against the majority of M. intracellulare isolates [28]. Currently, a Phase II clinical trial of clofazimine is in progress to evaluate its efficacy for the treatment of MAC-PD (ClinicalTrials.gov identifier: NCT02968212; Table S1, see supplementary material online). Clofazimine is also active against M. abscessus and other rapidly growing mycobacteria: a majority of clinical isolates had clofazimine MICs of ≤ 1 mg/l [29,30]. A retrospective study in Korea reviewed the clinical outcomes of 27 refractory M. abscessus lung disease patients after addition of clofazimine for 12 months: 15% of the previously nonresponsive patients achieved sputum culture conversion [31]. Additional safety and efficacy data of clofazimine within a bigger group of patients have recently been documented in another retrospective study by Martiniano et al. [32]. Among a total of 112 patients with NTM infections (MAC, M. abscessus or other species), 80% tolerated treatment with clofazimine for >6 months; and out of 60 patients with pulmonary infections who failed previous treatment 29 (48%) converted to negative cultures within 1 year [32]. These results further imply a contributing effect of clofazimine in the treatment of NTM-PD. In line with clinical observations, in vitro studies have demonstrated additional merits of clofazimine in multidrug regimens for NTM infections: the drug is synergistic with amikacin against M. avium and M. abscessus [29,30] and it significantly prevented regrowth of NTM strains after clarithromycin and amikacin exposure [33].
Linezolid and other new oxazolidinones
Linezolid, the first FDA-approved oxazolidinone, has been shown to exhibit some – but relatively weak – in vitro activity against M. avium and M. abscessus, with MICs ranging from 16 to 64 mg/l [28,34,35]. Substantial toxicity associated with prolonged use of linezolid is a major concern in clinical practice [36]. A Phase IV clinical trial is underway in Bangkok to study the efficacy and tolerability of linezolid in the treatment of NTM diseases (ClinicalTrials.gov identifier: NCT03220074). Recently, two other oxazolidinones were considered based on their potency against NTM. Tedizolid, which has an improved safety profile [37], exhibits an MIC 4- to 16-fold lower than linezolid [38,39]. LCB01-0371, a novel oxazolidinone currently in Phase II clinical development for pulmonary TB, was found to be potent against M. abscessus, with linezolid-like activity in vitro and in vivo [40].
Nitric oxide and aerosolized amikacin
There are two inhaled drugs in clinical development: nitric oxide (NO) and liposomal amikacin for inhalation (LAI). NO is thought to be promising in eliminating NTM infections because of its roles in immune defense and its antimicrobial functions. Two formulations of NO from two companies (AIT Therapeutics and Novoteris) are currently in clinical trials with the objective of evaluating their safety in patients. LAI, a novel formulation of amikacin, seems to be associated with reduced toxicity and improved efficacy in patients with refractory NTM lung disease especially those infected with MAC, as suggested by its Phase II clinical trial results [41]. Its efficacy and safety profiles are being further evaluated in a Phase II trial against M. abscessus and a Phase III trial for recalcitrant MAC lung disease.
Bedaquiline
Bedaquiline, being the only TB drug approved by the FDA during the past 40 years, has low MIC values against a collection of M. avium and M. abscessus clinical isolates [42–45]. However, despite being an excellent growth inhibitor at low doses, it lacks bactericidal activity against NTM in vitro [44,46]. The in vivo efficacy of bedaquiline seems to be limited as well. Although it reduced colony-forming units (CFU) in the lungs by 1 log in an acute M. abscessus mouse model [47], it failed to prevent death of the infected mice in another chronic study [48]. Similarly, bedaquiline was shown to exhibit bacteriostatic activity in a M. avium late-infection mouse model [46]. Preliminary results of bedaquiline as salvage therapy for NTM-PD (infected with either MAC or M. abscessus) treatment suggested that it might have some clinical activity because six out of ten patients had a microbiological response, but its efficacy appears to be relatively moderate as suggested by a low sputum culture conversion rate at 6 months [49]. Furthermore, relapse caused by the emergence of strains resistant to bedaquiline has been reported [50]. Therefore, the clinical utility of bedaquiline remains to be further investigated. Of note, similar to clofazimine which targets type II NADH-quinone oxidoreductase (NDH-2) [51], a key component in the electron transport chain, bedaquiline also targets oxidative phosphorylation via inhibiting ATP synthase [52]. Compared with other TB drugs, such as isoniazid and rifampicin, which lose their potency against most NTM strains, the fact that both oxidative phosphorylation inhibitors exhibit relatively broad-spectrum antimycobacterial activities suggests that targeting this energy-generating pathway could be a useful strategy in anti-NTM drug discovery.
β-lactams in combination with β-lactamase inhibitor avibactam
Owing to the presence of β-lactamases, NTM exhibit high levels of natural resistance to most β-lactams, with the exceptions of cefoxitin and imipenem, which are currently in use for the treatment of M. abscessus infections [53,54]. The recent discovery of the potent inhibitory effect by avibactam against β-lactamases in M. abscessus and M. avium has brought research attentions back to β-lactams. Treatment with avibactam significantly lowered MICs of several β-lactams 4–32-fold in M. abscessus and M. avium [54–56]. Reported combinations are summarized in Table 1.
Table 1.
Synergistic antibiotic combinations in Mycobacterium avium and Mycobacterium abscessusa
| No. | Drug combination | Type of study | No. strains tested | Reported outcomeb | Refs |
|---|---|---|---|---|---|
| (a) M. avium complex | |||||
| 1 | Amikacin + clofazimine | In vitro | 16 | 100% synergy | [29] |
| 2 | Avibactam + ceftazidimec | In vitro | 1 | Synergy | [56] |
| In macrophage | 1 | Synergy | |||
| (b) M. abscessus | |||||
| 1 | Clarithromycin + linezolid | In vitro | 2 | Synergy | [177] |
| 2 | Clarithromycin + tigecycline | In vitro | 20 | 65% synergy for Mycobacterium massiliense | [178] |
| 20 | 25% synergy for M. abscessus | ||||
| In vitro | 31 | 80.6% synergy | [179] | ||
| 3 | Clarithromycin + vancomycin | In vitro | 12 | 100% synergy | [77] |
| 4 | Clarithromycin + moxifloxacin | In vitro | 28 | 39.3% synergy for M. massiliense | [180] |
| 26 | 3.8% synergy and 65.4% antagonism for M. abscessus | ||||
| In macrophage | 15 | 33.3% synergy for M. massiliense | |||
| 15 | 6.6% synergy and 66.7% antagonism for M. abscessus | ||||
| In mice | 6 | 50% synergy for M. massiliense | |||
| 7 | 71.4% antagonism for M. abscessus | ||||
| In vitro | 20 | 85% synergy for M. massiliense | [178] | ||
| 20 | 5% synergy and 45% antagonism for M. abscessus | ||||
| 5 | Azithromycin + moxifloxacin | In vitro | 28 | 35.7% synergy for M. massiliense | [180] |
| 26 | 3.8% synergy and 46.2% antagonism for M. abscessus | ||||
| In macrophage | 15 | 20% synergy for M. massiliense | |||
| 15 | 6.6% synergy and 40.0% antagonism for M. abscessus | ||||
| In mice | 6 | 50% synergy for M. massiliense | |||
| 7 | 71.4% antagonism for M. abscessus | ||||
| 6 | Clarithromycin + linezolid + moxifloxacin/gatifloxacin/levofloxacin | In vitro | 2 | Synergy | [177] |
| 7 | Clarithromycin + ciprofloxacin + rifabutin | In vitro | 2 | Synergy | [177] |
| 8 | Imipenem + clarithromycin | In vitro | 21 | 43% synergy | [181] |
| 9 | Imipenem + levofloxacin | In vitro | 21 | 29% synergy | [181] |
| 10 | Amikacin + clofazimine | In vitro | 40 | 100% synergy | [30] |
| In vitro | 77 | 80.5% synergy | [29] | ||
| 11 | Amikacin + linezolid | In vitro | 32 | 53.1% synergy for M. massiliense | [182] |
| 32 | 37.5% synergy for M. abscessus | ||||
| 12 | Tigecycline + clofazimine | In vitro | 19 | 42% synergy | [183] |
| 13 | Tigecycline + linezolid | In vitro | 32 | 31.3% synergy for M. massiliense | [182] |
| 32 | 21.9% synergy for M. abscessus | ||||
| In fruit fly | 1 | Synergy (dramatically improved survival of infected flies and reduced bacterial population per fly) | [184] | ||
| 14 | Rifampicin + doripenem/biapenem | In vitro | 1 | Synergy | [185] |
| 15 | Avibactam + amoxicillinc | In vitro | 17 | 100% synergy | [54] |
| In macrophage | 1 | synergy | |||
| In zebrafish | 1 | Synergy (increased larva survival and reduced the proportion of embryos with abscesses) | |||
| 16 | Avibactam + ceftarolinec | In vitro | 1 | Synergy (increased kill) | [186] |
| In macrophage | 1 | Synergy (increased kill) | |||
| 17 | Avibactam + imipenem (+ amikacin)c | In vitro | 1 | Synergy (no significant MIC shift, but increased kill) | [187] |
| In macrophage | 1 | Synergy (increased kill) | |||
| In zebrafish | 1 | Synergy (increased larva survival) | |||
| 18 | Avibactam + tebipenem/Ertapenem/panipenemc | In vitro | 29 | 100% synergy | [55] |
| 19 | Avibactam + doripenem/Faropenem/meropenem/biapenemc | In vitro | 29 | 55–75.9% synergy | [55] |
| 20 | Avibactam + cefalotin/Cefuroxime/cefamandole/ceftriaxonec | In vitro | 1 | Synergy | [54] |
| 21 | Clavulanic acid + meropenemc | In vitro | 1 | Synergy | [185] |
Synergy combinations tested before the year 2000 are not included. In vitro synergy is defined as a combination with a fractional inhibitory concentration index (FICI) ≤ 0.5, unless otherwise stated.
The percentage in ‘Reported outcome’ column indicates the percentage of strains tested that showed synergy (or antagonism in some cases). Percentage is not reported when fewer than five strains were tested. Antagonism (a FICI score >2 as defined by Choi et al.) is indicated when the percentage was significantly larger than that of synergy. Almost all combinations reported here were synergistic except for macrolide (clarithromycin or azithromycin) + moxifloxacin which showed a synergistic effect for 30–50% of M. massiliense but an antagonistic effect for 40–70% of M. abscessus strains. ‘M. abscessus’: M. abscessus subsp. abscessus; ‘M. massiliense’: M. abscessus subsp. massiliense.
β-lactam and β-lactamase inhibitor combinations are defined as synergistic if the potency of the β-lactam is significantly improved (e.g., at least fourfold shift in MIC) in the presence of the β-lactamase inhibitor.
Rifabutin
Rifampicin has long been known to be inactive against M. abscessus. Surprisingly, a recent screen of 2700 FDA-approved drugs identified rifabutin, belonging to the same drug class as rifampicin, as active against the bacteria in vitro [57]. It has an MIC of ~2.5 mg/l against a collection of M. abscessus strains and kills 90% of the bacteria at 5 mg/l. A better understanding of the underlying differences of these two drugs (such as differences in intrabacterial metabolism and pharmacokinetic properties) could guide the development of more-potent rifamycins against M. abscessus.
PIPD1 and indole-2-carboxamides
Two recent screens of TB-active hits against M. abscessus resulted in two new leads: PIPD1 and indole-2-carboxamides [58,59]. Coincidently, these two structurally distinct chemical entities both target MmpL3, a transporter crucial for the export of trehalose monomycolates to the periplasmic space and outer membrane of mycobacteria. Disruption of mmpL3 leads to defects in mycolic acid synthesis and thus is fatal for mycobacteria [60]. PIPD1 and indole-2-carboxamides (lead compounds 6 and 12) have excellent activities against M. abscessus with MICs of 0.0625–1 mg/l. They are bactericidal in vitro, reducing CFU 100-fold at 1–2 × MIC. Macrophage assays demonstrated that these compounds could arrest intracellular bacterial growth, but at much higher concentrations. Moreover, treatment of infected zebrafish with PIPD1 at 24 × MIC for 3 days decreased bacterial load by 1 log unit and improved survival of the infected embryos.
Thiacetazone derivatives
Although thiacetazone (TAC) – a former (toxic) TB drug – is inactive against NTM, a few derivatives of TAC synthesized for TB evaluation were found to be effective against M. avium and M. abscessus. SRI-286 and SRI-224 inhibited a panel of M. avium isolates at 2 mg/l or lower, and SRI-286 could reduce the bacterial loads in livers and spleens by 1 log [61]. D6, D15 and D17, second-generation TAC analogs, are active against M. abscessus with MICs ranging from 3.1–12.5 mg/l against the type strains. Similar to TAC in M. tuberculosis, these compounds require cellular activation by the monooxygenase EthA in M. abscessus [62]. These results suggest that modifications of TAC could be an approach to develop new chemical entities active against NTM.
Besides the abovementioned compounds that have been studied extensively for their effect against NTM, TP-271 (a novel fluorocycline antimicrobial related to tetracycline) and some salicylanilide esters and carbamates were also found to exhibit potent activity in vitro against M. abscessus [63,64]. Several TB actives in development, such as SQ109 (a 1,2-ethylenediamine that can target MmpL3 [65,66]), DC-159a (a novel fluoroquinolone), SQ641 (a capuramycin analog), ACH-702 (a new isothiazoloquinolone) and mefloquine (a quinoline used for malaria) are active against some NTM strains [67–71]. In addition, owing to a lack of new active chemical entities, synergy combinations especially with clarithromycin, amikacin, tigecycline or imipenem were explored in vitro, whereas only a few have been studied in vivo (Table 1). Overall, de novo drug discovery efforts have been very limited so far. A few attempts have been made to develop novel therapeutic approaches for NTM disease via repurposing or repositioning of existing antibiotics. Although some candidates exhibit promising activity in vitro, most are at an early stage awaiting in vivo and clinical evaluations. Clearly, more drug discovery efforts are necessary to fill the NTM antibiotic pipeline.
Challenges in NTM drug discovery: from a bacteriology point of view
NTM are naturally resistant to a wide spectrum of antibiotics, including most TB drugs. This poses a major challenge for drug discovery. Hit rates in primary screens for M. abscessus can be lower than 0.1%. Thus, generation of attractive chemical starting points for lead finding presents a bottleneck. The low level of susceptibility of NTM to a wide range of drugs and compounds is attributed to their ‘intrinsic’ drug resistance.
Intrinsic drug resistance
Considering that NTM reside mostly in soil and water, the selection pressure from their antimicrobial-producing neighbors could have driven these bacteria to develop a wide array of resistance mechanisms to allow their survival in hostile environments. Our current understanding of the mechanisms underlying intrinsic drug resistance in NTM has been reviewed [72,73] and is therefore only briefly summarized here. The thick hydrophobic, double-membrane cell envelope of mycobacteria acts as a major permeability barrier. Studies from the 1990s have demonstrated that Mycobacterium chelonae, a species that had not been differentiated from M. abscessus at that time, has a cell envelope that is about 10–20-times less permeable than M. tuberculosis [74]. Morphotypic antibiotic resistance, a phenomenon of varying degrees of drug resistance in M. avium associated with a reversible colony morphology switch (white/red on Congo red containing agar, transparent/opaque), is also attributed to changes in permeability owing to cell-wall modifications [75]. Hence, antibiotics that target the cell envelope are likely to potentiate other drugs with intracellular targets, as implied by the synergistic effect observed between ethambutol and rifampicin in M. avium [76] or vancomycin plus clarithromycin in M. abscessus [77]. Efflux pumps are additional elements that prevent intracellular accumulation of drugs such as fluoroquinolones and macrolides [78]. For drugs that can accumulate inside the bacterial cells, several mechanisms have been identified that render the molecules inactive. Some NTM species harbor polymorphisms in the target gene contributing to natural resistance to the drug, for example amino acid alterations in the arabinosyl transferase EmbB of M. abscessus make ethambutol inactive by preventing drug binding [79]. Upon exposure to the drug, some NTM species induce the expression of certain genes resulting in the modification of the target binding site of the drug. A well characterized example for this strategy is the inducible macrolide resistance in M. abscessus mediated by the erm(41) gene, which encodes a ribosomal methylase. Exposure to clarithromycin or azithromycin increases the expression level of erm(41) dramatically within 24 h. Erm(41) methylates A2058 in the 23S rRNA, leading to reduced binding of macrolides to their target site, thus rendering the drugs inactive [80]. Because such resistance occurs in specific environments (e.g., exposure to antibiotics) and does not involve any genetic alterations, this type of resistance is called adaptive – as opposed to acquired – resistance. Furthermore, NTM possess a large collection of enzymes capable of metabolizing drugs to a less active form. Knockout of these modifying genes such as blaMab (encoding a β-lactamase) and eis2 (encoding a GNAT-acetyltransferase) restored the activity of β-lactams and aminoglycosides in M. abscessus, respectively [81,82]. More examples of these intrinsic resistance mechanisms are shown in Table 2. Interestingly, a conserved transcription factor in mycobacteria: WhiB7, acts as a regulator for many intrinsic drug-resistance mechanisms [83]. In M. abscessus, 128 genes including erm(41) and eis2 have been identified in the WhiB7 regulon (i.e., they are induced via a whiB7-dependent mechanism). Deletion of M. abscessus whiB7 sensitized the bacteria to drugs like clarithromycin, amikacin, erythromycin and tetracycline 2–8-fold [84]. Altogether, the abundant intrinsic resistance mechanisms form an elaborate network leaving behind only a few antibiotics to exhibit inhibitory activity against NTM. A better understanding of these molecular mechanisms might provide insights into overcoming or bypassing some resistance pathways. Development of a tool box to measure – and to understand – compound uptake, metabolism and excretion by the bacteria (a ‘bacterial cell pharmacokinetics platform’) might not only greatly facilitate specific lead finding and optimization projects but also enable rational repositioning programs to improve the potency of poorly active antibiotics targeting pharmacologically validated pathways.
Table 2.
Intrinsic drug resistance mechanisms in Mycobacterium avium and Mycobacterium abscessusa
| Agent | Target | M. avium (genes involved) | M. abscessus (genes involved) | Refs |
|---|---|---|---|---|
| Isoniazid | InhA | Presumably efflux pumps | Presumably efflux pumps | [73] |
| Rifampicin | RNA polymerase | NAb | Inactivation of drug (arrMab) | [188] |
| Ethambutol | Arabinosyl transferase | NAb | Polymorphisms in target gene embB | [79] |
| Pyrazinami de | PanD | Presumably due to active efflux of POAc | Presumably owing to active efflux of POAc | [189,190] |
| Aminoglyc osides | 16S rRNA | NAb | Inactivation of drug [aac(2′), eis2 – whiB7] | [82,84,191] |
| Fluoroquin olones | DNA gyrase | Polymorphisms in target gene gyrA | Polymorphisms in target gene gyrA | [192] |
| β-lactams | Penicillin-binding protein | β-lactamase with mild activity Other unknown reasons |
Inactivation of drug (blaMab) | [81] |
| Thiacetazo ne | FAS-II dehydratased | The target is not essential: redundant dehydratase present | The target is not essential: redundant dehydratase present | [193] |
| BTZ043 | DprE1 | Polymorphisms in target gene dprE1 | Polymorphisms in target gene dprE1 | [194,195] |
| BRD4592 | Tryptophan synthase | NAb | Polymorphisms in target gene trpA | [196] |
| Macrolides | 23S rRNA | Efflux pumps (MAV_1695, MAV_1406) | Modification of drug target [erm(41) – whiB7] | [78,80,84,191] |
Cell envelope, porins and efflux pumps are likely to be involved in intrinsic drug resistance to many antimicrobials and are thus not specified in the table unless roles have been experimentally proven or suggested.
NA: not applicable, the drug is active.
POA: pyrazinoic acid, bioactive metabolite of pyrazinamide.
FAS-II: type II fatty acid synthase.
Acquired drug resistance
Besides being equipped with plentiful intrinsic resistance mechanisms, NTM also have the ability to acquire new resistance through genomic mutations that is inherited by offspring. The prolonged course of treatment has greatly contributed to the emergence of resistant strains, allowing the bacteria to develop mutations in the target or other related genes to confer high-level resistance. As a result, drug efficacy is abolished.
Up to now, there have been limited studies on acquired resistance mechanisms associated with NTM. Acquired resistance to clarithromycin emerged in the early 1990s soon after its introduction for NTM treatment, especially with monotherapy. Mutations at nucleotides 2058 and 2059 in the peptidyl transferase loop of the 23S rRNA (rrl) were found in M. avium and M. abscessus clinical isolates to confer a high level of macrolide resistance (MICs ≥ 256 mg/l) [85,86]. Of note, it has been shown recently in vitro that the chance of acquiring mutations in the 23S rRNA gene is higher in the absence of a functional erm(41) in M. abscessus [87]. Rifampicin is primarily used in the treatment of M. avium infections. Acquired resistance has been documented in clinical isolates and is associated with mutations within the rpoB gene encoding the β-subunit of bacterial RNA polymerase. However, introduction of the mutated rpoB sequence into Mycobacterium smegmatis did not confer resistance to rifampicin, suggesting that there might be other factors contributing to M. avium rifampicin resistance [88]. Acquired resistance to aminoglycosides has been demonstrated in M. abscessus. Mutations at position 1408 of 16S rRNA (rrs) in clinical isolates are associated with aminoglycoside resistance [89]. In addition, an in vitro study by Nessar et al. has shown that mutations at positions 1406, 1409 and 1491 in M. abscessus could also confer a high level of resistance (MICs ≥ 1024 mg/l) [90]. Recently, Alexander et al. reported the emergence of mmpT5 mutations during bedaquiline treatment in all seven patients with M. intracellulare lung infections who relapsed after a positive initial microbiological response [50]. MmpT5 is a transcriptional regulator that represses the expression of the MmpS5–MmpL5 efflux pump. Similar to M. tuberculosis, mutations in M. intracellulare mmpT5 are associated with low-level resistance to bedaquiline (2–8-fold). Two out of seven patients had mutations in the primary target (ATP synthase subunit c), conferring high-level resistance (50-fold).
There is currently limited evidence of lateral gene transfer of drug resistance genes in NTM. However, whole-genome analysis of M. abscessus has revealed a large number of genes in common with two pathogens most frequently isolated from CF patients: Pseudomonas aeruginosa and Bulkholderia cepacia [91], indicating that acquiring drug resistance by horizontal gene exchange is likely to play a part. Although the genetic mechanisms of acquired drug resistance have been extensively studied in M. tuberculosis, not much information is available for NTM with regard to mechanisms, mutation frequency and how fast resistance develops. Recent studies by Ferro and colleagues have exploited a hollow-fiber model to mimic the effect of various drug therapies in the host. Their study demonstrated that M. abscessus could develop genetic resistance against moxifloxacin in 3 days [92], suggesting that acquired drug resistance in NTM can develop very quickly during treatment. Thus, it is of importance to understand the genetic basis for acquired resistance, and more importantly how to optimize regimens to prevent development of resistance.
Lack of bactericidal activity
To make NTM drug discovery more daunting, there is a curious lack of bactericidal activity for most drugs tested against NTM – drugs in the current regimens are either bacteriostatic (tigecycline, imipenem) or exert only weak bactericidal activity at high concentrations (clarithromycin) [93,94]. Bedaquiline, despite a very potent growth-inhibitory effect, exerted only bacteriostatic activity in M. avium – the same concentrations that reduced M. tuberculosis CFU by 5 logs resulted in only 1 log CFU reduction after 14-day exposure in M. avium [46]. Similar observations have been made for a wide spectrum of rapidly growing NTM [95]. Because mycobacteria have a high metabolic rate and divide slowly [96], they are rather unique among bacteria for their ability to adapt to stress before the cells are killed. This could provide a general physiologic basis for the lack of bactericidal activity of drugs. However, the exact reasons behind the lack of bactericidal activity of anti-NTM drugs are not known and should be explored. Insights into why certain drug-induced cell death pathways are not operational in NTM could reveal ways to overcome this phenomenon. Obviously, new anti-NTM agents should be bactericidal (i.e., have sterilizing properties to improve the currently poor treatment outcomes and accelerate cure).
Challenges in NTM drug discovery: from a disease pathology point of view
A low MIC value is often a good starting point for antibacterial drug discovery because it usually predicts eradication of the infection once adequate pharmacokinetic properties have been introduced into the lead compound. However, this general rule does not appear to hold true for NTM pulmonary diseases. Clinical practice has consistently observed a lack of correlation between in vitro MICs and clinical outcomes (i.e., sensitivity for a particular drug as indicated by drug susceptibility testing does not necessarily translate to a positive clinical response) [72]. Why is there such a disconnect? In vitro MIC testing is usually performed with mycobacteria growing exponentially as a suspension under optimal conditions in aerated nutrient-rich broth. These culture conditions are very different from the environments where bacilli reside in the host. TB studies have demonstrated a wide range of lung lesion types, from cellular granuloma mainly composed of macrophages to caseous granuloma with a necrotic core. These lesions are complex and dynamic, giving rise to microenvironments of diverse features and stresses, which in turn drive the tubercle bacilli into distinct physiological and morphological states associated with increased antibiotic tolerance [97]. This increased drug tolerance is also termed ‘phenotypic drug resistance’ to distinguish this phenomenon from drug resistance due to genetic alterations. In TB, several conditions, including intracellular and caseum growth, and quiescent states linked to oxygen or nutrient starvation, have been associated with phenotypic drug resistance of the bacilli [98,99]. Considering similarities between NTM and TB pathology in pulmonary diseases, the same factors are likely to contribute to the persistence of NTM infections despite extensive chemotherapy. Moreover, two features unique to bacterial pathophysiology in NTM-PD – growth in airway mucus and as biofilms – can also lead to physiological adaptations associated with phenotypic drug resistance. Equally important to phenotypic drug resistance is the impact of the local microenvironments on drug penetration, which can lead to subtherapeutic concentrations of antimicrobials at the anatomical sites where bacilli reside, thus affecting treatment efficacy. These factors, altered bacterial physiology associated with phenotypic drug resistance and reduced drug penetration into infection sites are likely to collectively contribute to the observed disconnect between in vitro MICs and clinical outcomes observed for NTM-PD.
Intracellular growth
Similar to M. tuberculosis, NTM can grow and survive extra- as well as intra-cellularly, for instance inside macrophages. In the context of pulmonary infection, NTM invade the mucosa and get phagocytized by macrophages. Whereas some bacilli are killed, those remaining exhibit robust growth within phagocytic vacuoles until autophagy and apoptosis are induced after a few days of infection. M. avium could escape macrophage apoptosis and seize the chance to spread and infect other macrophages [100]. By contrast, M. abscessus, especially the smooth morphotype, could restrict intraphagosomal acidification, induce less apoptosis and block autophagy flux, and thus was able to persist inside macrophages for longer periods of time [101,102]. Regardless of the distinct strategies that can be employed by different NTM species, the selective pressures of macrophages on NTM such as reactive oxygen radicals, NO and low pH as well as carbon source composition (a lipid-rich environment inside macrophages) are likely to invoke mycobacterial adaptations and induce drug tolerance. The Ramakrishnan laboratory has carried out studies on drug tolerance in macrophages [103]. The authors demonstrated that the resistance level of M. tuberculosis and Mycobacterium marinum against isoniazid increased over time inside macrophages: after a 96 h infection period ~49.5% of M. marinum survived isoniazid treatment whereas only 7.6% of bacilli from a 2 h infection period survived. Importantly, it was observed that macrophages harboring drug-tolerant mycobacteria could disperse from the existing granulomas that had shrunk substantially owing to treatment and could disseminate to establish a new site of infection in zebrafish.
Factors contributing to the increased drug tolerance of intracellular mycobacteria are multifold. The tolerance might result from macrophage-induced bacteriostasis because multiple static subpopulations of M. avium have been found in human macrophages after quinolone treatment [104]. The increased resistance could also be attributed to replicating cells with adaptive physiological changes including induction of the master drug-tolerance regulator WhiB7, which has been shown to be strongly induced (14-fold) in M. tuberculosis in macrophages [105]. Upregulated along with WhiB7 are efflux pumps and drug-modifying genes in a WhiB7-dependent or -independent manner. Transcriptome studies on internalized M. tuberculosis suggested that macrophage residence triggered dramatic differential gene expression, among which a large group of efflux pumps (19 out of 55 annotated) were induced [106,107]. Further studies by Adams et al. [103] showed that the increased drug tolerance to isoniazid and rifampicin of M. tuberculosis in macrophages can be reverted by addition of the efflux pump inhibitor reserpine. The same has been observed with M. marinum in macrophages. In addition, two M. tuberculosis transposon mutants defective in Rv1258c – a multidrug efflux pump – were hypersensitive to rifampicin specifically in macrophages, suggesting that the macrophage-induced tolerance in mycobacteria could be mediated by efflux pumps [103]. Besides the abovementioned reasons, drug activity could also be limited intracellularly owing to restricted uptake and accumulation inside macrophages.
Caseum growth and nonreplicating state of persistence
In advanced human TB, most bacteria reside extracellularly in caseum [108]. Owing to lack of vasculatures, tubercle bacilli in the center of caseum are believed to confront oxygen and/or nutrient limitation. Direct measurement with oxygen probes and in situ staining with the hypoxia marker pimonidazole have demonstrated a low oxygen tension in the necrotic and caseous regions of granulomas in patients and animal models [109,110]. M. tuberculosis isolated from lung lesions has also been found to display altered morphology and staining properties similar to bacilli grown in distilled water [111], suggesting that the bacilli are starved in lesions. These lines of direct or indirect evidence have promoted the development of several in vitro models for M. tuberculosis to mimic conditions of oxygen deprivation or nutrient starvation. Studies have shown that, upon hypoxia or nutrient starvation, the bacilli stop growth and enter a nonreplicating state with reduced metabolism [112–115]. Accompanied by such changes is phenotypic drug resistance to most anti-TB drugs. Isoniazid and moxifloxacin for instance are highly active against replicating bacteria; however, they have little or no effect on the viability of nonreplicating nutrient-starved cells [115]. Rifampicin, the remaining active agent against starved bacteria, is active only at a considerably higher concentration [115,116]. The drastic loss of drug potency could be attributed to reduced drug uptake [117] and/or change in drug target essentiality. For instance, absence of cell wall synthesis in nonreplicating starved cells might result in the lack of efficacy of isoniazid [116].
Direct evidence demonstrating drug tolerance of M. tuberculosis present inside caseous necrotic lesions has been provided recently by Sarathy et al. [118]. The authors measured the drug susceptibility of M. tuberculosis bacilli present in ex vivo caseous lesion samples collected from rabbits with active TB. No significant bacterial growth was observed in the caseum homogenate during the first 7 days of incubation, indicating that the bacilli from caseum are largely nongrowing. Intra-caseum M. tuberculosis was highly tolerant to most antibiotics: isoniazid, kanamycin and clofazimine had minimal to no activity. Similar to oxygen- and nutrient-starvation-induced nonreplicating bacteria, rifampicin was active but achieved tenfold killing only at 8 μM, a concentration 100-times higher than the concentration required to kill actively replicating cells. In accordance with the data from this ex vivo caseum model, preclinical efficacy studies that focused on the bacterial population surviving drug treatment also illustrated that lesion compartments that were not sterilized at the end of therapy were mostly necrotic granulomas and caseous foci [119,120].
NTM lung disease shares many traits with TB: pulmonary lesions are broadly classified as nodular bronchiectatic and fibrocavitary [14], highly reminiscent of TB-induced pathology. Advanced lung histopathology of M.-avium-infected mice is characterized by necrotizing granulomas with minimal microvessels and a hypoxic center, strongly resembling human TB lesions [121,122]. Whether M. abscessus and/or M. avium have evolved similar metabolic and physiologic adaptations to M. tuberculosis under hostile conditions remains to be determined. Nevertheless, earlier studies on the non-pathogenic NTM species M. smegmatis showed its capability to maintain long-term viability by entering a nonreplicating state under nutrient or oxygen limitation [123,124]. Reminiscent of M. tuberculosis, nongrowing M. smegmatis is characterized by lower intracellular ATP concentrations, reduced oxygen consumption and extreme tolerance to antibiotic treatment [123]. Interestingly, DosR, the dormancy survival response regulator crucial for hypoxia survival of tubercle bacilli, along with the two histidine kinases, is conserved across NTM species [125]. In a recent M. abscessus transcriptomic study, the entire DosR regulon was strongly upregulated upon NO-induced hypoxia [126], suggesting that there are largely conserved molecular strategies between NTM and M. tuberculosis that persist inside the host. A similar conclusion was drawn by Drapal and colleagues after metabolite profiling of several Mycobacteria species including M. avium under hypoxic conditions [127]. Therefore, it is tempting to hypothesize that, similar to M. tuberculosis, NTM are capable of entering a nonreplicating state and exhibiting phenotypic drug resistance inside lung lesions, and this could be one of the factors contributing to the persistence of NTM infection under prolonged treatment.
In addition to phenotypic drug resistance, the pharmacokinetics of drugs inside lesions also affects the drug efficacy via differential drug access. In a laboratory, in vitro MIC setting, bacteria are exposed to constant drug concentrations present in broth. In patients, however, for successful treatment of NTM pulmonary disease, drugs from the blood vessel need to penetrate the complex lung lesions to reach the bacteria (i.e., they must diffuse from the cellular rim that borders the necrotic center and penetrate the entire caseous region without any assistance) [128]. The Dartois laboratory has carried out several studies on drug penetration into rabbit and human TB granulomas. By visualizing drugs using MALDI MS imaging, the authors observed a heterogeneous distribution of drugs across different lesion compartments. First-line TB drugs with treatment-shortening properties such as pyrazinamide and isoniazid could reach a high accumulation in caseum and maintained therapeutic levels in this compartment throughout the dosing intervals [129]. However, for moxifloxacin, although it accumulates in lesions at relatively high concentrations, it stays predominantly in immune cells and barely diffuses to the acellular caseum [130]. This seems to explain why moxifloxacin, a strong killer of replicating and nonreplicating tubercle bacilli, failed to shorten TB treatment in clinical trials. The correlation between drug distribution into caseous foci and its efficacy in TB is likely to be applicable to NTM-PD as well. Thus, the extent of diffusion into the caseous center by current NTM drugs and new candidates needs to be investigated.
Mucus growth
Different from TB, mucus plays an important part in the development of NTM pulmonary disease, at least in selected patient populations with preexisting chronic lung diseases such as CF or COPD. Owing to the hyper-production of mucus or defective cilia function, sticky mucus cannot be effectively swept out of the lungs. As a result, bacteria residing in mucus remain in the lung, evade the immune system and thrive in the excess of thick stationary mucus adherent to airway surfaces. Excessive mucus in the patient’s airways is likely to trap the bacilli in a unique environment with varied oxygen and nutrient content, as suggested by studies on CF patients infected with P. aeruginosa. By inserting an O2 electrode directly into the right upper lobar bronchi of chronically Pseudomonas-infected CF patients, it was shown that oxygen is depleted in the mucopurulent material obstructing the lobar bronchus possibly owing to restricted oxygen diffusion through thickened mucus, consumption by the CF epithelium or by the bacteria themselves [131]. Nutrient composition in sputum from CF patients also differs with an increased amount of amino acids [132]. Preliminary studies on M. abscessus toward an artificial sputum media that mimic the nutritional composition of CF sputum showed that the organism slowed its growth and induced a ‘low energy’ transcriptional response [126], suggesting that the bacilli indeed undergo a phenotypic switch in the mucus niche. More studies are warranted to further explore how NTM adapt in the mucus and most importantly how mucus growth affects drug susceptibility.
Biofilm growth
Being a common persistence strategy for many microorganisms, biofilms are multicellular structures in which bacterial cells stick to each other and to living or nonliving surfaces. NTM are notorious biofilm producers in nature and human-engineered environments. They are frequently recovered in biofilms from surfaces of water pipes, showerheads and even healthcare equipment, such as heater–cooler units [133,134]. Biofilms of NTM are hard to eliminate by conventional decontamination procedures, thereby raising a serious public health threat. The ability of NTM to form biofilms has been linked to their pathogenicity. This correlation was first speculated by Carter et al. when they observed that a M. avium isolate attenuated in mice happened to form a biofilm less effectively in vitro [135]. Yamazaki et al. further provided evidence by demonstrating that biofilm-deficient transposon mutants of M. avium showed impaired invasion in human bronchiolar epithelial cells and caused limited infection in mice [136]. Importantly, the connection between NTM biofilms and pulmonary disease pathology has been demonstrated recently. M. abscessus has been observed to form biofilms within thickened alveolar walls and airways of CF patients, as well as within the lung cavity in COPD patients [137,138]. One notable attribute of biofilms is their increased tolerance to antimicrobials, and this holds true for NTM biofilms [139–142]. NTM in biofilms are generally ten-times less susceptible to antibiotics than their planktonic counterparts. Even clarithromycin, the cornerstone of NTM treatment therapy, which showed some activities on the initial formation of M. avium biofilms in vitro, is completely inactive in established biofilms [143]. Thus, biofilm growth is likely to contribute to the disconnect between MIC and clinical outcome, and the persistence of NTM infection.
The reason why NTM in biofilms are more prone to escaping attack by antimicrobials remains elusive. Nevertheless, it seems clear that antibiotic tolerance promoted by biofilm growth is adaptive and temporary (i.e., rather than genetic) [139]. A few factors could contribute to the increased tolerance to antibiotics. Similar to other bacterial species, NTM in biofilms are embedded within a matrix composed of extracellular polymeric substances (EPS). Despite interspecies differences, the EPS matrix of NTM has been found to primarily comprise free mycolic acid, glycopeptidolipids, mycolyl-diacylglycerol or lipopeptides [144]. Therefore, it is likely that the waxy nature of the lipid-rich NTM EPS matrix builds a physical barrier that shields antibiotics from penetration. In addition, the presence of extracellular DNA (eDNA) in M. avium and M. abscessus biofilm matrix has also been associated with antibiotic resistance. Disruption of eDNA by DNase treatment led to a significant increase in the susceptibility of NTM biofilm to clarithromycin and moxifloxacin [144]. Besides the physical barrier provided by biofilm architecture, antibiotic resistance could also be attributed to the adaptations that NTM undergo during the biofilm development, as suggested by the transient resistance observed after resuspension of biofilm bacteria [142,145], as well as transcriptional changes observed during M. smegmatis biofilm formation [146].
Although the biofilm structures of NTM have been characterized to some extent, the molecular mechanisms are poorly described. Genes involved in the transition from ‘swimmers’ to ‘stickers’ have not been identified. Interestingly, the fact that exposure to a sub-inhibitory concentration of streptomycin or tetracycline resulted in increased M. avium biofilm formation [139] implies that some of the stress-response-related genes triggered by antibiotic exposure are linked to biofilm formation. Besides the molecular mechanisms, two more knowledge gaps of NTM biofilms remain to be filled. First, biofilms of other bacterial species are usually dynamic and complex, harboring bacterial cells at various growth stages with differential physiological activities in the same biofilm [147]. However, whether NTM biofilms are a dynamic and differentiated community or a relatively more homogenous population remains to be determined. Second, given the nature of NTM infection in patients, especially in CF patients where co-infection with other bacteria such as P. aeruginosa or another NTM species in the respiratory tract is common, mixed-species biofilms appear to be likely. Thus, interspecies interactions in biofilm formation as well as its impact on antibiotic tolerance are important to investigate.
Taken together, it appears that, over the course of evolution, the dual lifestyle of NTM – saprophytic in soil and water vs pathogenic – has equipped these bacteria with a large arsenal of molecular strategies that allows them to adapt swiftly and effectively to a wide spectrum of hostile environments encountered in various habitats. Owing to the complex nature of NTM pulmonary disease, the abovementioned conditions and stresses are not mutually exclusive. For instance, nonreplicating bacilli or biofilm-like structures might be present in thick mucus together with growing organisms. In fact, these factors are likely to collectively contribute to the emergence of NTM persisters. The development of reproducible persister assays and a more comprehensive understanding of molecular mechanisms employed by M. avium and M. abscessus to adjust to various conditions and exert phenotypic drug resistance will assist discovery of leads able to eradicate these physiological forms of NTM and thus increase treatment effectiveness. Additionally, attention should be paid to the assessment of drug penetration pharmacokinetics into macrophages, caseous lesions, biofilms and mucus, because sufficient penetration into the site of infection is crucial for an efficacious treatment.
A road map to new anti-NTM drugs
To date, de novo drug discovery campaigns for novel anti-NTM agents are limited. To facilitate NTM drug discovery, a workflow for compound progression from whole-cell screening to preclinical development (i.e., from primary hit generation to compounds showing tolerability, exposure and efficacy in a mouse model of disease) is proposed in Figure 2. The overall workflow corresponds to the generic compound progression cascade for antibacterial and anti-TB drug discovery that we have described previously [148,149], with a number of biological and pharmacological assays designed specifically to cater for the needs of NTM-PD regimens according to their unique bacteriological and pharmacokinetic requirements. Briefly, to discover new chemical entities active against NTM, a whole-cell-based phenotypic screening should be pursued to generate hits with antimicrobial activity. With the objective to kill two birds with one stone, double hits (i.e., hits active against M. avium and M. abscessus) with reasonable growth-inhibitory and -cidal activity and an acceptable cytotoxicity profile should be identified and further profiled and prioritized for their potency in bacterial persister models, and for their in vitro pharmacokinetic properties in assays that predict lesion penetration. It is important to confirm medium-independent activity of hits as well as activity against a selection of M. avium complex and M. abscessus species and subspecies early in the process. Parallel to hit-to-lead activities, target deconvolution should be carried out to understand drug mechanism-of-action and facilitate medicinal chemistry efforts during lead optimization. Regarding the choice of chemical libraries to be screened for the identification of chemical starting points, collections of TB actives generated over the past two decades should be considered, because they display higher NTM hit rates compared with ‘random’ compound collections [150]. Furthermore, this strategy increases the chance of identifying broad-spectrum antimycobacterials. A few considerations regarding compound profiling and prioritization are elaborated below.
Figure 2.
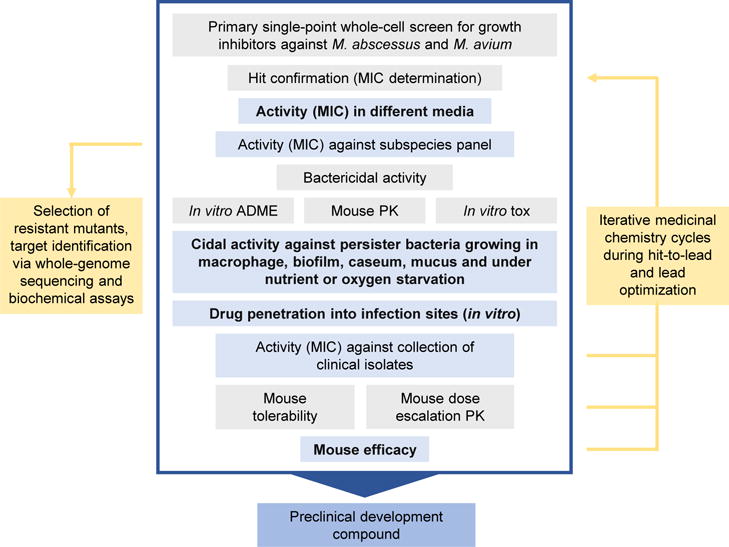
Proposed workflow for non-tuberculous mycobacteria (NTM) drug discovery. Activities and assays from whole-cell library screening to the preclinical development compound are shown. Light blue boxes indicate NTM-specific assays discussed in more detail in the text. Assay and model gaps are in bold. Gray box indicates generic drug discovery activities and assays described elsewhere [148,149]. Abbreviations: PK, pharmacokinetics; tox, cytotoxicity assays.
NTM strains for MIC determinations
Different from the obligate parasite M. tuberculosis, which is genomically rather homogeneous, environmental NTM present a heterogeneous group of species and subspecies. This needs to be considered early in drug discovery projects because different species and subspecies do differ in their drug susceptibility profile. Using surrogates such as the model organism M. smegmatis instead of the actual pathogenic species in drug screening could be counterproductive. Because the most common and the most difficult to cure NTM-PD are caused by members of the M. avium and M. abscessus complexes, respectively, efforts should focus on these two species. For instance, we use the clinical isolates M. avium 11 (belonging to M. avium subsp. hominissuis [151]) and M. abscessus Bamboo (belonging to M. abscessus subsp. abscessus [152]) for primary screens. Immediately after hit confirmation, hits should be tested for activity against reference strains obtained from culture collections representing species and subspecies of M. avium and M. abscessus complexes: M. avium subsp. hominissuis, M. intracellulare and M. chimaera; M. abscessus subsp. abscessus, M. abscessus subsp. bolletii; M. abscessus subsp. massiliense (Figure 2). At a later stage, lead compounds are tested against a large collection of clinical isolates of the two NTM species derived from different regions of the world because geographic variabilities of antibiotic susceptibility have been described [153–156]. Moreover, considering colony phase variation (e.g., smooth vs rough, opaque vs transparent) seen in M. avium and M. abscessus, it is useful to cover all types of colony morphologies in the clinical isolates tested to identify any potential correlation between colony morphology and drug susceptibility.
Media and incubation conditions for MIC determinations
Currently, most in vitro drug testing studies for NTM followed either the Clinical and Laboratory Standards Institute (CLSI) recommendations for NTM susceptibility testing in clinical settings (i.e., cation-adjusted Mueller–Hinton broth and incubation at 30°C) or standard mycobacterial drug discovery culture conditions (i.e., Middlebrook 7H9 broth and incubation at 37°C). We and other researchers have observed that differences in these two assay systems including media composition and presence or absence of detergent (Tween® 80 in 7H9) can affect drug potency and lead to variabilities in the MIC results [54,77,157]. This highlights a need for further investigations to improve the predictive value of current NTM in vitro drug testing assays for clinical outcome. For the time being, primary hits should be tested in at least two different media to provide preliminary evidence of medium-independent activity. Medium dependence of antimycobacterial compounds is not a new phenomenon. During the past ten years of TB drug discovery, we have encountered several cases in which compound activity was dependent on certain components present in the culture media. One well-documented example is a series of pyrimidine-imidazole compounds, which exhibited low MICs in vitro (and adequate pharmacokinetic properties) but had no efficacy in vivo. It was later realized that the activities of these compounds were dependent on glycerol – a major carbon source in TB culture media but apparently not relevant for bacterial growth in mice [158].
Besides media composition, incubation time is another crucial factor that can affect drug susceptibility results for NTM, especially in the case of macrolides when tested in M. abscessus. It was shown by Nash et al. that extended exposure (14 days) to macrolides is required to detect the inducible resistance conferred by erm(41): the MIC on day 3 was ≤ 0.5 mg/l, this value increased to >32 mg/l on day 14 [80]. Therefore, to better evaluate macrolides and their potential synergistic effects with other drugs in M. abscessus, it is recommended to perform 14-day incubation for MIC determinations. Of note, drug stability should be considered when interpreting susceptibility results, particularly when long incubation periods are used. As demonstrated by Rominski et al., severe drug degradation observed for β-lactams (e.g., imipenem and cefoxitin) after 3 days results in misleading MIC values [81].
Persister-specific assays
As elaborated above, the existence of persisters (i.e., bacteria displaying phenotypic drug resistance) in the host might be one of the main factors that hampers the eradication of NTM infection in patients. Hence, conventional MIC alone is not a good predictor of clinical outcome; we need persister-specific assays to identify leads with improved efficacy in vivo. Yet, most persister-specific assays have not been thoroughly characterized, profiled or benchmarked for NTM. Research efforts are warranted to adapt and implement assays developed for either M. tuberculosis or other pathogens such as P. aeruginosa to generate a tool box for NTM drug discovery.
Macrophage infection assays
Macrophage assays have been employed to study the intracellular activity of drugs and chemicals against NTM. A wide variety of macrophages including bone-marrow-derived macrophages, J774 and THP-1 cells are being used [40,58]. Drug treatment usually starts a few hours post-infection followed by incubation for 24–96 h. Drug activity is usually indicated by a reduction in CFU or CFU:macrophage.
Nonreplicating assays
Two in vitro assays, namely the Loebel model (nutrient-starvation model) and the Wayne model (oxygen-depletion model), have been established and widely applied in the TB field to evaluate drug potency against nonreplicating bacilli. In the Loebel model, log-phase cultures are washed and transferred to PBS and nutrient-starved for 14 days before drug treatment [115]. In the Wayne model, hypoxic nonreplicating bacilli are generated under gentle stirring in tightly sealed glass tubes with rubber septa. Oxygen indicator methylene blue is used to monitor oxygen depletion over time in a control tube. Upon reaching a nonreplicating phase, drugs are injected into each tube via a needle through the rubber septa. Because Loebel and Wayne bacilli are nonreplicating, the potency readouts in these systems are concentrations that kill 90% of the bacteria: LCC90 for Loebel cidal concentration and WCC90 for Wayne cidal concentration are the counterparts of minimum bactericidal concentration, or MBC90, for growing bacteria. Characterization of growth and survival kinetics of NTM in the Wayne and Loebel models is required before applying these assays for drug testing. Both models have been established for M. smegmatis [123,124] and should thus be adaptable to M. avium and M. abscessus.
Caseum assay
An ex vivo caseum assay was recently described for M. tuberculosis [118]. Caseum tissue obtained from cavities of TB-infected rabbits is homogenized and dispensed into 96-well plates with drugs. Owing to the nonreplicative nature of bacilli inside caseum, the readout is also the cidal concentration (casMBC90). Currently, this assay might be difficult to develop for NTM because caseum-producing animal models are not well established (see below). Alternatively, the development of artificial, cell-culture-based caseum [129] could be considered.
Biofilm assays
To study NTM biofilms, several laboratory-based biofilm formation models have been developed, among which two systems might be suitable for NTM drug susceptibility testing. The first assay system is known as the Calgary Biofilm Device employed by Bardouniotis et al. to evaluate the cidal activity or the minimal biofilm eradication concentration (MBEC) of biocides on Mycobacterium phlei [159]. The device consists of a 96-peg lid-plate that bacteria can adhere to and a ridged trough into which a standardized inoculum is added. Upon biofilm formation on the pegs, the lid can be transferred to a standard 96-well plate in which the biofilms are challenged by antibiotic treatment. Surviving bacteria in biofilms can be removed from pegs by sonication in media following washing and plated for CFU numeration. This method could be applied to the determination of biofilm MIC and cidal concentrations. The second assay system utilizes a 96-well polyvinyl chloride (PVC) microtiter plate in which NTM such as M. avium and M. smegmatis can adhere and develop biofilm on the PVC surface [141,143]. Following incubation, planktonic bacteria are rinsed away and the remaining adherent bacteria in biofilms with or without antibiotics are stained and quantified by crystal violet. This method is simple and straightforward and enables the determination of concentrations that inhibit biofilm growth. However, because crystal violet is toxic to bacteria, this is usually an end-point measurement and the stained bacteria cannot be used for further studies.
Mucus assay
Kirchner et al. have described the use of synthetic mucus media or artificial sputum medium (ASM) in microtiter plates to test antibiotic efficacy against P. aeruginosa [160]. The ASM contains amino acids, mucin and free extracellular DNA to mimic the thick mucus within the lung of CF patients. P. aeruginosa and Staphylococcus aureus grow as biofilm-like structures (not attached to the surface) in this matrix [161]. This method can be further investigated for NTM (especially for M. abscessus) and exploited as an effective platform to identify antibiotics with potential activity in the lungs of CF patients. The use of patient-derived mucus should be considered to validate assays based on artificial mucus.
Lesion- or infection-site-specific pharmacokinetic assays
NTM reside in many anatomical locations where drugs might not penetrate efficiently owing to differential vascularization and cellular composition. Owing to the complexity of NTM pathology, PK/PD correlations that are made on the basis of drug exposure in plasma are not sufficient to inform drug distribution at the site of infection (inside lesions) in patients. Clinical studies with resected lung lesions have indicated that drugs with better penetration into lesions, especially the necrotic core where most of the persisters locate, tend to play a key part in pathogen sterilization and treatment shortening [130]. To this end, Sarathy et al. developed a medium-throughput caseum-binding assay to predict lesion partitioning of TB drug candidates [129]. In this assay, a surrogate caseum is generated by lysing oleic-acid-induced foamy macrophages followed by protein denaturation to mimic the situation in the necrotic core of granulomas. Based on the negative correlation between ex vivo caseum binding and in vivo passive diffusion into caseum, the authors could predict drug diffusion capacity based on the unbound faction in the surrogate caseum. Combined with drug uptake into macrophages, this assay can be used to predict the partitioning of drugs and newly discovered compounds at the cellular–caseum interface of necrotic lesions [162]. Owing to the similarities in granuloma and cavity structure between TB and NTM, these assays could be applied to predict the lesion penetrance of novel anti-NTM compounds as well.
Animal efficacy
Animal models, including many mouse strains, the guinea pig, rhesus macaque, zebrafish and fruit fly, have been investigated for their potential to develop NTM disease. The details of each model have been reviewed by Chan and Bai and Bernut et al. [163,164], and will thus only be briefly mentioned here. Because we are dealing with NTM lung disease, we consider the use of mammalian species as advantageous for in vivo efficacy determinations. Overall, owing to the general low virulence of NTM compared with M. tuberculosis, it is difficult to generate a sustained infection with advanced human-like lung pathology in animals. Although M. avium can cause a relatively high level of infection in the lungs of mice, M. abscessus is cleared rapidly from the lungs of most mouse strains and other animal models including guinea pigs. Only when severely immunocompromised strains of mice such as GKO or SCID mice are used can characteristics of certain lung disease histology (e.g., cellular infiltration, consolidation) be observed, yet non-necrotizing and necrotizing lesions have only been shown in the SCID mouse model [47,165]. A few attempts have been made to evaluate various mouse strains for their adequacy for antimycobacterial testing. However, owing to the differences in disease progression (acute vs chronic infection stage), and disease manifestations (lung vs disseminated infection), in vivo studies in these mouse models have sometimes led to inconsistent results. For instance, bedaquiline treatment reduced CFU by 2 logs in a SCID mouse model but was almost inactive in nude mice [47,48]. As discussed above, the morphological structure and composition of infection sites are likely to impact bacterial pathophysiology and drug penetration. Therefore, robust and practical animal models that present hallmarks of human NTM pathology (e.g., caseous necrosis) are urgently required for improved in vivo assessment of novel anti-NTM compounds.
Design of clinical trials
Owing to the heterogeneity of NTM disease presentation, clinical trials that evaluate the efficacy of new drug candidates or new combinations against NTM-PD are not simple to design [166]. One major challenge lies in the great variabilities among study populations in terms of disease manifestations (e.g., fibro-cavitary vs nodular-bronchiectasis type), pre-existing conditions (nil, CF, COPD, AIDS or other causes of immune-suppression), causative agents (M. avium, M. abscessus or co-infection with other NTM or bacteria) and drug susceptibility profiles. Previous studies have demonstrated that such heterogeneity could result in distinct treatment outcomes [18,167]. Thus, it should be taken into consideration in the clinical trial design whether to target a general population or a particular patient group. If working with a broad patient population, trial results should be stratified by factors such as subspecies and clinical presentations. Another difficulty encountered in NTM clinical trials is the limited choice of endpoints. Very few prospective clinical trials have been conducted for NTM treatment [23,168,169], all of which used sputum culture conversion or clinical improvement at the end of chemotherapy as a measure of treatment success (Table S1, see supplementary material online). In TB studies, primary endpoints such as sputum culture conversion at 2 months, time to sputum culture conversion and serial sputum colony counting, have been introduced in Phase II clinical trials as surrogates of end-of-treatment outcome [170,171]. Unfortunately, none of these markers has been evaluated for NTM pulmonary disease. As a whole, identification of accurate, NTM-specific biomarkers capable of predicting long-term treatment outcome or relapse risk are desired and would greatly accelerate the development of new NTM agents by enabling small and more-reliable proof-of-concept studies. Recently, there has been consensus achieved by international experts from the NTM-NET committee on treatment outcome definitions for NTM-PD [172]. The critical outcome parameters proposed in the statement should be considered in future clinical trial designs to standardize outcome reports for meta-analyses of clinical data.
Concluding remarks
To combat recalcitrant NTM lung diseases with more-effective regimens, we need well-tolerated and preferably orally bioavailable compounds that: (i) are active against at least M. abscessus and M. avium, and ideally against a wider spectrum of mycobacteria including M. tuberculosis; (ii) are bactericidal against growing and ideally against various drug-tolerant persister forms of the bacteria; (iii) show not only adequate standard plasma pharmacokinetic properties but also penetrate the various sites of infection; (iv) eradicate the bacteria in an animal model that presents human-like pathologies. Importantly, drug–drug interactions must be kept to a minimum because NTM treatments are based on multidrug regimens. Furthermore, NTM patients often take additional antibiotic and nonantibiotic medications. Meeting all these requirements seems to be a daunting task at the moment. NTM drug discovery is still in its infancy with many questions of resistance, persistence and pathophysiology remaining to be unveiled, and many assays and models to be developed and validated. Thus, there is an urgent need to reduce the biological uncertainties around NTM-PD by increasing research efforts. Today’s state of NTM drug discovery is reminiscent of the TB situation 20 years ago. Despite a discovery void in the TB field from the 1970s to early 2000s, significant progress has been achieved during the past two decades, owing to a better understanding of the pathogen and disease pathology and – importantly – increased drug discovery efforts. Looking on the bright side, we can benefit from knowledge, bacteriological and pharmacokinetic assays, and from models developed for TB drug discovery. Furthermore, we can make use of chemical matter generated in TB screening campaigns. Libraries of TB actives are available and have been shown to deliver relatively high hit rates for NTM, offering a much-needed impetus to jump-start NTM drug discovery projects, start populating the drug pipeline and discover broad(er)-spectrum antimycobacterials. In addition to de novo drug discovery approaches, repositioning and repurposing efforts must be undertaken (Box 1). We believe that, with increasing research and drug discovery efforts and expanding knowledge on NTM, significant therapeutic advances for NTM diseases will be achieved in the coming years.
Highlights.
Incidence of lung disease caused by non-TB mycobacteria (NTM) is increasing
Current treatments are ineffective
There is an urgent need to establish a drug pipeline
Intrinsic bacterial resistance and persistence present major challenges
Predictive persister assays and animal models need to be developed
Supplementary Material
Box 1. Proposed strategies for populating the non-tuberculous mycobacteria (NTM) drug pipeline.
Repurposing
In the context of drug development, ‘repurposing’ is the application of known drugs to treat new indications (i.e., new diseases). Because de novo drug discovery and development easily takes longer than a decade, the evaluation of existing and clinically used antibiotics is a low-hanging-fruit approach that can dramatically accelerate drug development, and has the potential to bring rapid relief to NTM patients. For example, the rifampicin analog rifabutin was recently shown to be active against Mycobacterium abscessus in vitro through a systematic screen of FDA-approved drugs [57]. Likewise, the systematic exploration of antibiotic combinations could reveal synergistic pairs that could rapidly be tested clinically in patients with very limited therapeutic options such as those with refractory M. abscessus lung disease. In addition, more-targeted synergistic studies based on mechanisms-of-action expected to deliver synergistic effects could yield high hit rates: (i) large-scale combinations of β-lactamase inhibitors with β-lactams; (ii) cell wall targeting drugs with drugs modulating intracellular targets; and (iii) efflux pump inhibitors with other drugs should be further explored, as suggested by several recent studies [55,56,77,78].
Repositioning
We refer to repositioning as the NTM-specific chemical optimization of antibiotic classes that act against pharmacologically validated targets but have been developed for infectious diseases other than NTM. Because these drug classes include members that are FDA-approved, attrition rates are lower and the probability of success is significantly higher than incurred through de novo drug discovery. Examples include the macrolide clarithromycin, the backbone of NTM therapy, but whose efficacy against M. abscessus is affected by inducible resistance conferred by erm(41). Screening of macrolide collections against M. abscessus is a low-risk and high-reward approach to identify analogs that are not affected by the ribosomal modifications. Likewise, the oxazolidinone linezolid is active against NTM but suffers from low potency and toxicity. A few recent studies reported oxazolidinones with improved potency [38,40,150], thus validating the proposed strategy. Finally, although rifampicin is inactive against M. abscessus, other rifamycins appear to exhibit adequate potency [57] (unpublished data).
De novo drug discovery
The third strategy is based on the identification of new chemical entities and targets. Whole-cell screens against mycobacteria suffer from extremely low hit rates. We propose the acceleration of NTM drug discovery by screening compound collections with known activity against Mycobacterium tuberculosis rather than ‘random’ large compound libraries – an approach that has delivered significantly higher hit rates [150]. Furthermore, novel antibacterial discovery concepts such as screening of fragment (MW <300 Da) libraries and membrane targeting molecules should be explored. Although fragment-like compounds are often filtered out in whole-cell screening campaigns, many crucial components of current TB chemotherapies such as isoniazid and pyrazinamide are ‘dirty’ fragments that hit multiple targets and display attractive physicochemical and pharmacokinetic properties, an asset to eradicate the notoriously robust mycobacterial diseases [173]. Targeting the M. tuberculosis membrane has been investigated over the past few years with the attempt to tackle the issues of resistance and persistence. Targeting membrane integrity as opposed to a specific enzyme or pathway also has a lower propensity to enable emergence of genetic resistance. In addition, as opposed to classical pathways targeted by conventional antibiotics, membrane integrity is essential irrespective of the physiological status of the bacilli. Thus, membrane-targeting compounds should retain their activity against slow-growing or nonreplicating persisters. Indeed, several novel compounds targeting the M. tuberculosis membrane do overcome resistance and persistence, with some compounds showing cross-activity against NTM [174–176].
Acknowledgments
We thank all participants of the workshop ‘Advancing Translational Science for Pulmonary NTM Infections’, organized by the National Institute of Allergy and Infectious Diseases and held in Rockville, MD, September 2017, for very stimulating and enlightening presentations and discussions. We also thank the anonymous reviewers for their useful and insightful comments. We are grateful to Pooja Gopal for her comments on the manuscript. We apologize to the many authors whose work could not be discussed and cited owing to space and reference limits. T.D. holds a Toh Chin Chye Visiting Professorship at the Department of Microbiology and Immunology, Yong Loo Lin School of Medicine, National University of Singapore. Research reported in this publication was supported by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health under Award Number R01AI132374 and by the Cystic Fibrosis Foundation under Award Number DICK17XX00 (to T.D.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health or the Cystic Fibrosis Foundation.
Biographies
Thomas Dick
Thomas Dick has 20 years’ experience in mycobacteriology, antibacterial drug discovery and R&D program management, in industry as well as in academia. His research focuses on the discovery of new medicines for the treatment of tuberculosis (TB) and non-tuberculous mycobacterial (NTM) lung disease. Since 2017 he has been Director of Antimicrobial Drug Discovery at the Public Health Research Institute and Associate Professor at the Department of Medicine, New Jersey Medical School, Rutgers University. Dr Dick holds a Toh Chin Chye Visiting Professorship at National University of Singapore where he manages the TB drug discovery project portfolio of the Medical School’s SPRINT TB program which he co-founded in 2014.

Véronique Dartois
Véronique Dartois is a Principal Investigator at the Public Health Research Institute of Rutgers University. Initially trained as a molecular biologist, she has more than 10 years’ experience in the pharmaceutical and biotech industry, acquired through her previous position as Pharmacology Unit Head at the Novartis Institute of Tropical Diseases, and biotech companies in Southern California. Her current research interests include the pharmacokinetics, pharmacodynamics and imaging of antimycobacterial drugs in pulmonary lesions, and the optimization of predictive animal models and in vitro assays to study these questions. Specifically, lesion-specific analysis of drug exposure and bacterial killing help identify and characterize lung granuloma compartments in which organisms are not eliminated.

Mu-Lu Wu
Mu-Lu Wu received her PhD in microbiology from National University of Singapore in 2016, where her research focused on mycobacterial persistence, especially nutrient-starvation-induced phenotypic drug resistance and the cellular differentiation program triggered by various degrees of nutrient starvation. For the past 1.5 years, she has been working as a postdoctoral research fellow at National University of Singapore, leading the NTM drug discovery projects in the laboratory. Her main focus is to utilize existing molecules and explore novel chemicals or strategies to overcome the intrinsic resistance and persistence in NTM.

Dinah B. Aziz
Dinah B. Aziz is a research assistant in the National University of Singapore. She received her degree in Life Sciences from the National University of Singapore in 2015 and is currently a part-time Masters student in the same institution. She has been working in a drug discovery laboratory for the past 3 years. Her current focus is on drug discovery for NTMs, with her wider interests including mechanisms of intrinsic and acquired antibiotic drug resistance.

Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Teaser: From crisis to cures – a review on status, knowledge gaps and major obstacles in NTM drug discovery and development, and how to move forward.
Conflicts of interest
The authors have no conflicts of interest to declare.
References
- 1.Prevots DR, Marras TK. Epidemiology of human pulmonary infection with nontuberculous mycobacteria: a review. Clin Chest Med. 2015;36:13–34. doi: 10.1016/j.ccm.2014.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stout JE, et al. Update on pulmonary disease due to non-tuberculous mycobacteria. Int J Infect Dis. 2016;45:123–134. doi: 10.1016/j.ijid.2016.03.006. [DOI] [PubMed] [Google Scholar]
- 3.Prevots DR, et al. Nontuberculous mycobacterial lung disease prevalence at four integrated health care delivery systems. Am J Respir Crit Care Med. 2010;182:970–976. doi: 10.1164/rccm.201002-0310OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Adjemian J, et al. Prevalence of nontuberculous mycobacterial lung disease in U.S. Medicare beneficiaries. Am J Respir Crit Care Med. 2012;185:881–886. doi: 10.1164/rccm.201111-2016OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Park IK, Olivier KN. Nontuberculous mycobacteria in cystic fibrosis and non-cystic fibrosis bronchiectasis. Semin Respir Crit Care Med. 2015;36:217–224. doi: 10.1055/s-0035-1546751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Andrejak C, et al. Chronic respiratory disease, inhaled corticosteroids and risk of non-tuberculous mycobacteriosis. Thorax. 2013;68:256–262. doi: 10.1136/thoraxjnl-2012-201772. [DOI] [PubMed] [Google Scholar]
- 7.Raju RM, et al. Leveraging advances in tuberculosis diagnosis and treatment to address nontuberculous mycobacterial disease. Emerg Infect Dis. 2016;22:365–369. doi: 10.3201/eid2203.151643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shahraki AH, et al. “Multidrug-resistant tuberculosis” may be nontuberculous mycobacteria. Eur J Intern Med. 2015;26:279–284. doi: 10.1016/j.ejim.2015.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.van Ingen J, et al. Poor adherence to management guidelines in nontuberculous mycobacterial pulmonary diseases. Eur Respir J. 2017;49:1601855. doi: 10.1183/13993003.01855-2016. [DOI] [PubMed] [Google Scholar]
- 10.Bange FC, et al. Lack of transmission of Mycobacterium abscessus among patients with cystic fibrosis attending a single clinic. Clin Infect Dis. 2001;32:1648–1650. doi: 10.1086/320525. [DOI] [PubMed] [Google Scholar]
- 11.Jonsson BE, et al. Molecular epidemiology of Mycobacterium abscessus, with focus on cystic fibrosis. J Clin Microbiol. 2007;45:1497–1504. doi: 10.1128/JCM.02592-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bryant JM, et al. Whole-genome sequencing to identify transmission of Mycobacterium abscessus between patients with cystic fibrosis: a retrospective cohort study. Lancet. 2013;381:1551–1560. doi: 10.1016/S0140-6736(13)60632-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bryant JM, et al. Emergence and spread of a human-transmissible multidrug-resistant nontuberculous mycobacterium. Science. 2016;354:751–757. doi: 10.1126/science.aaf8156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Griffith DE, et al. An official ATS/IDSA statement: diagnosis, treatment, and prevention of nontuberculous mycobacterial diseases. Am J Respir Crit Care Med. 2007;175:367–416. doi: 10.1164/rccm.200604-571ST. [DOI] [PubMed] [Google Scholar]
- 15.Haworth CS, et al. British Thoracic Society guidelines for the management of non-tuberculous mycobacterial pulmonary disease (NTM-PD) Thorax. 2017;72:ii1–ii64. doi: 10.1136/thoraxjnl-2017-210927. [DOI] [PubMed] [Google Scholar]
- 16.Wallace RJ, Jr, et al. Macrolide/azalide therapy for nodular/bronchiectatic Mycobacterium avium complex lung disease. Chest. 2014;146:276–282. doi: 10.1378/chest.13-2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jeon K, et al. Antibiotic treatment of Mycobacterium abscessus lung disease: a retrospective analysis of 65 patients. Am J Respir Crit Care Med. 2009;180:896–902. doi: 10.1164/rccm.200905-0704OC. [DOI] [PubMed] [Google Scholar]
- 18.Zweijpfenning S, et al. Treatment and outcome of non-tuberculous mycobacterial pulmonary disease in a predominantly fibro-cavitary disease cohort. Respir Med. 2017;131:220–224. doi: 10.1016/j.rmed.2017.08.031. [DOI] [PubMed] [Google Scholar]
- 19.Koh WJ, et al. Outcomes of Mycobacterium avium complex lung disease based on clinical phenotype. Eur Respir J. 2017;50:1602503. doi: 10.1183/13993003.02503-2016. [DOI] [PubMed] [Google Scholar]
- 20.Jarand J, et al. Clinical and microbiologic outcomes in patients receiving treatment for Mycobacterium abscessus pulmonary disease. Clin Infect Dis. 2011;52:565–571. doi: 10.1093/cid/ciq237. [DOI] [PubMed] [Google Scholar]
- 21.Koh WJ, et al. Mycobacterial characteristics and treatment outcomes in Mycobacterium abscessus lung disease. Clin Infect Dis. 2017;64:309–316. doi: 10.1093/cid/ciw724. [DOI] [PubMed] [Google Scholar]
- 22.Strollo SE, et al. The burden of pulmonary nontuberculous mycobacterial disease in the United States. Ann Am Thorac Soc. 2015;12:1458–1464. doi: 10.1513/AnnalsATS.201503-173OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wallace RJ, Jr, et al. Clinical experience in 52 patients with tigecycline-containing regimens for salvage treatment of Mycobacterium abscessus and Mycobacterium chelonae infections. J Antimicrob Chemother. 2014;69:1945–1953. doi: 10.1093/jac/dku062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Roussel G, Igual J. Clarithromycin with minocycline and clofazimine for Mycobacterium avium intracellulare complex lung disease in patients without the acquired immune deficiency syndrome. Int J Tuberc Lung Dis. 1998;2:462–470. [PubMed] [Google Scholar]
- 25.Field SK, Cowie RL. Treatment of Mycobacterium avium-intracellulare complex lung disease with a macrolide, ethambutol, and clofazimine. Chest. 2003;124:1482–1486. doi: 10.1378/chest.124.4.1482. [DOI] [PubMed] [Google Scholar]
- 26.Chaisson RE, et al. Clarithromycin and ethambutol with or without clofazimine for the treatment of bacteremic Mycobacterium avium complex disease in patients with HIV infection. AIDS. 1997;11:311–317. doi: 10.1097/00002030-199703110-00008. [DOI] [PubMed] [Google Scholar]
- 27.Jarand J, et al. Long-term follow-up of mycobacterium avium complex lung disease in patients treated with regimens including clofazimine and/or rifampin. Chest. 2016;149:1285–1293. doi: 10.1378/chest.15-0543. [DOI] [PubMed] [Google Scholar]
- 28.Huang CC, et al. In vitro activity of aminoglycosides, clofazimine, D-cycloserine and dapsone against 83 Mycobacterium avium complex clinical isolates. J Microbiol Immunol Infect. 2017 doi: 10.1016/j.jmii.2017.05.001. [DOI] [PubMed] [Google Scholar]
- 29.van Ingen J, et al. In vitro synergy between clofazimine and amikacin in treatment of nontuberculous mycobacterial disease. Antimicrob Agents Chemother. 2012;56:6324–6327. doi: 10.1128/AAC.01505-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shen GH, et al. High efficacy of clofazimine and its synergistic effect with amikacin against rapidly growing mycobacteria. Int J Antimicrob Agents. 2010;35:400–404. doi: 10.1016/j.ijantimicag.2009.12.008. [DOI] [PubMed] [Google Scholar]
- 31.Yang B, et al. Clofazimine-containing regimen for the treatment of Mycobacterium abscessus lung disease. Antimicrob Agents Chemother. 2017;61:e02052–16. doi: 10.1128/AAC.02052-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Martiniano SL, et al. Safety and effectiveness of clofazimine for primary and refractory nontuberculous mycobacterial infection. Chest. 2017;152:800–809. doi: 10.1016/j.chest.2017.04.175. [DOI] [PubMed] [Google Scholar]
- 33.Ferro BE, et al. Clofazimine prevents the regrowth of Mycobacterium abscessus and Mycobacterium avium type strains exposed to amikacin and clarithromycin. Antimicrob Agents Chemother. 2016;60:1097–1105. doi: 10.1128/AAC.02615-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wallace RJ, Jr, et al. Activities of linezolid against rapidly growing mycobacteria. Antimicrob Agents Chemother. 2001;45:764–767. doi: 10.1128/AAC.45.3.764-767.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brown-Elliott BA, et al. In vitro activity of linezolid against slowly growing nontuberculous mycobacteria. Antimicrob Agents Chemother. 2003;47:1736–1738. doi: 10.1128/AAC.47.5.1736-1738.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wasserman S, et al. Linezolid in the treatment of drug-resistant tuberculosis: the challenge of its narrow therapeutic index. Expert Rev Anti Infect Ther. 2016;14:901–915. doi: 10.1080/14787210.2016.1225498. [DOI] [PubMed] [Google Scholar]
- 37.Yuste JR, et al. Prolonged use of tedizolid in a pulmonary non-tuberculous mycobacterial infection after linezolid-induced toxicity. J Antimicrob Chemother. 2017;72:625–628. doi: 10.1093/jac/dkw484. [DOI] [PubMed] [Google Scholar]
- 38.Brown-Elliott BA, Wallace RJ., Jr In vitro susceptibility testing of tedizolid against nontuberculous mycobacteria. J Clin Microbiol. 2017;55:1747–1754. doi: 10.1128/JCM.00274-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vera-Cabrera L, et al. In vitro activities of the novel oxazolidinones DA-7867 and DA-7157 against rapidly and slowly growing mycobacteria. Antimicrob Agents Chemother. 2006;50:4027–4029. doi: 10.1128/AAC.00763-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim TS, et al. Activity of LCB01-0371, a novel oxazolidinone, against Mycobacterium abscessus. Antimicrob Agents Chemother. 2017;61:e02752–16. doi: 10.1128/AAC.02752-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Olivier KN, et al. Randomized trial of liposomal amikacin for inhalation in nontuberculous mycobacterial lung disease. Am J Respir Crit Care Med. 2017;195:814–823. doi: 10.1164/rccm.201604-0700OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huitric E, et al. In vitro antimycobacterial spectrum of a diarylquinoline ATP synthase inhibitor. Antimicrob Agents Chemother. 2007;51:4202–4204. doi: 10.1128/AAC.00181-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pang Y, et al. In vitro activity of bedaquiline against nontuberculous mycobacteria in China. Antimicrob Agents Chemother. 2017;61:e02627–16. doi: 10.1128/AAC.02627-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dupont C, et al. Bedaquiline inhibits the ATP synthase in Mycobacterium abscessus and is effective in infected zebrafish. Antimicrob Agents Chemother. 2017;61:e01225–17. doi: 10.1128/AAC.01225-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brown-Elliott BA, et al. In vitro susceptibility testing of bedaquiline against Mycobacterium avium complex. Antimicrob Agents Chemother. 2017;61:e01798–16. doi: 10.1128/AAC.01798-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lounis N, et al. ATP synthase inhibition of Mycobacterium avium is not bactericidal. Antimicrob Agents Chemother. 2009;53:4927–4929. doi: 10.1128/AAC.00689-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Obregon-Henao A, et al. Susceptibility of Mycobacterium abscessus to antimycobacterial drugs in preclinical models. Antimicrob Agents Chemother. 2015;59:6904–6912. doi: 10.1128/AAC.00459-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lerat I, et al. In vivo evaluation of antibiotic activity against Mycobacterium abscessus. J Infect Dis. 2014;209:905–912. doi: 10.1093/infdis/jit614. [DOI] [PubMed] [Google Scholar]
- 49.Philley JV, et al. Preliminary results of bedaquiline as salvage therapy for patients with nontuberculous mycobacterial lung disease. Chest. 2015;148:499–506. doi: 10.1378/chest.14-2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Alexander DC, et al. Emergence of mmpT5 variants during bedaquiline treatment of Mycobacterium intracellulare lung disease. J Clin Microbiol. 2017;55:574–584. doi: 10.1128/JCM.02087-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yano T, et al. Reduction of clofazimine by mycobacterial type 2 NADH:quinone oxidoreductase: a pathway for the generation of bactericidal levels of reactive oxygen species. J Biol Chem. 2011;286:10276–10287. doi: 10.1074/jbc.M110.200501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Andries K, et al. A diarylquinoline drug active on the ATP synthase of Mycobacterium tuberculosis. Science. 2005;307:223–227. doi: 10.1126/science.1106753. [DOI] [PubMed] [Google Scholar]
- 53.Lavollay M, et al. In vitro activity of cefoxitin and imipenem against Mycobacterium abscessus complex. Clin Microbiol Infect. 2014;20:O297–300. doi: 10.1111/1469-0691.12405. [DOI] [PubMed] [Google Scholar]
- 54.Dubee V, et al. β-Lactamase inhibition by avibactam in Mycobacterium abscessus. J Antimicrob Chemother. 2015;70:1051–1058. doi: 10.1093/jac/dku510. [DOI] [PubMed] [Google Scholar]
- 55.Kaushik A, et al. Combinations of avibactam and carbapenems exhibit enhanced potencies against drug-resistant Mycobacterium abscessus. Future Microbiol. 2017;12:473–480. doi: 10.2217/fmb-2016-0234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Deshpande D, et al. The discovery of ceftazidime/avibactam as an anti-Mycobacterium avium agent. J Antimicrob Chemother. 2017;72:i36–i42. doi: 10.1093/jac/dkx306. [DOI] [PubMed] [Google Scholar]
- 57.Aziz DB, et al. Rifabutin is active against Mycobacterium abscessus complex. Antimicrob Agents Chemother. 2017;61:e00155–17. doi: 10.1128/AAC.00155-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dupont C, et al. A new piperidinol derivative targeting mycolic acid transport in Mycobacterium abscessus. Mol Microbiol. 2016;101:515–529. doi: 10.1111/mmi.13406. [DOI] [PubMed] [Google Scholar]
- 59.Kozikowski AP, et al. Targeting mycolic acid transport by indole-2-carboxamides for the treatment of Mycobacterium abscessus infections. J Med Chem. 2017;60:5876–5888. doi: 10.1021/acs.jmedchem.7b00582. [DOI] [PubMed] [Google Scholar]
- 60.Li W, et al. Therapeutic potential of the Mycobacterium tuberculosis mycolic acid transporter, MmpL3. Antimicrob Agents Chemother. 2016;60:5198–5207. doi: 10.1128/AAC.00826-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bermudez LE, et al. Thiosemicarbazole (thiacetazone-like) compound with activity against Mycobacterium avium in mice. Antimicrob Agents Chemother. 2003;47:2685–2687. doi: 10.1128/AAC.47.8.2685-2687.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Halloum I, et al. Resistance to thiacetazone derivatives active against Mycobacterium abscessus involves mutations in the MmpL5 transcriptional repressor MAB_4384. Antimicrob Agents Chemother. 2017;61:e02509–16. doi: 10.1128/AAC.02509-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cynamon M, et al. In vitro activity of TP-271 against Mycobacterium abscessus, Mycobacterium fortuitum, and Nocardia species. Antimicrob Agents Chemother. 2012;56:3986–3988. doi: 10.1128/AAC.00743-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Baranyai Z, et al. Combating highly resistant emerging pathogen Mycobacterium abscessus and Mycobacterium tuberculosis with novel salicylanilide esters and carbamates. Eur J Med Chem. 2015;101:692–704. doi: 10.1016/j.ejmech.2015.07.001. [DOI] [PubMed] [Google Scholar]
- 65.Tahlan K, et al. SQ109 targets MmpL3, a membrane transporter of trehalose monomycolate involved in mycolic acid donation to the cell wall core of Mycobacterium tuberculosis. Antimicrob Agents Chemother. 2012;56:1797–1809. doi: 10.1128/AAC.05708-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Xu Z, et al. MmpL3 is the flippase for mycolic acids in mycobacteria. Proc Natl Acad Sci U S A. 2017;114:7993–7998. doi: 10.1073/pnas.1700062114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bermudez LE, et al. Mefloquine is active in vitro and in vivo against Mycobacterium avium complex. Antimicrob Agents Chemother. 1999;43:1870–1874. doi: 10.1128/aac.43.8.1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sacksteder KA, et al. Discovery and development of SQ109: a new antitubercular drug with a novel mechanism of action. Future Microbiol. 2012;7:823–837. doi: 10.2217/fmb.12.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Disratthakit A, Doi N. In vitro activities of DC-159a, a novel fluoroquinolone, against Mycobacterium species. Antimicrob Agents Chemother. 2010;54:2684–2686. doi: 10.1128/AAC.01545-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Dubuisson T, et al. In vitro antimicrobial activities of capuramycin analogues against non-tuberculous mycobacteria. J Antimicrob Chemother. 2010;65:2590–2597. doi: 10.1093/jac/dkq372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Molina-Torres CA, et al. In vitro activity of a new isothiazoloquinolone, ACH-702, against Mycobacterium tuberculosis and other mycobacteria. Antimicrob Agents Chemother. 2010;54:2188–2190. doi: 10.1128/AAC.01603-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.van Ingen J, et al. Resistance mechanisms and drug susceptibility testing of nontuberculous mycobacteria. Drug Resist Updat. 2012;15:149–161. doi: 10.1016/j.drup.2012.04.001. [DOI] [PubMed] [Google Scholar]
- 73.Nessar R, et al. Mycobacterium abscessus: a new antibiotic nightmare. J Antimicrob Chemother. 2012;67:810–818. doi: 10.1093/jac/dkr578. [DOI] [PubMed] [Google Scholar]
- 74.Jarlier V, Nikaido H. Mycobacterial cell wall: structure and role in natural resistance to antibiotics. FEMS Microbiol Lett. 1994;123:11–18. doi: 10.1111/j.1574-6968.1994.tb07194.x. [DOI] [PubMed] [Google Scholar]
- 75.Philalay JS, et al. Genes required for intrinsic multidrug resistance in Mycobacterium avium. Antimicrob Agents Chemother. 2004;48:3412–3418. doi: 10.1128/AAC.48.9.3412-3418.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zimmer BL, et al. In vitro synergistic activity of ethambutol, isoniazid, kanamycin, rifampin, and streptomycin against Mycobacterium avium-intracellulare complex. Antimicrob Agents Chemother. 1982;22:148–150. doi: 10.1128/aac.22.1.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mukherjee D, et al. Vancomycin and clarithromycin show synergy against Mycobacterium abscessus in vitro. Antimicrob Agents Chemother. 2017;61:e01298–17. doi: 10.1128/AAC.01298-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Machado D, et al. Boosting effect of 2-phenylquinoline efflux inhibitors in combination with macrolides against Mycobacterium smegmatis and Mycobacterium avium. ACS Infect Dis. 2015;1:593–603. doi: 10.1021/acsinfecdis.5b00052. [DOI] [PubMed] [Google Scholar]
- 79.Alcaide F, et al. Role of embB in natural and acquired resistance to ethambutol in mycobacteria. Antimicrob Agents Chemother. 1997;41:2270–2273. doi: 10.1128/aac.41.10.2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Nash KA, et al. A novel gene, erm(41), confers inducible macrolide resistance to clinical isolates of Mycobacterium abscessus but is absent from Mycobacterium chelonae. Antimicrob Agents Chemother. 2009;53:1367–1376. doi: 10.1128/AAC.01275-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Rominski A, et al. Effect of β-lactamase production and β-lactam instability on MIC testing results for Mycobacterium abscessus. J Antimicrob Chemother. 2017;72:3070–3078. doi: 10.1093/jac/dkx284. [DOI] [PubMed] [Google Scholar]
- 82.Rominski A, et al. Elucidation of Mycobacterium abscessus aminoglycoside and capreomycin resistance by targeted deletion of three putative resistance genes. J Antimicrob Chemother. 2017;72:2191–2200. doi: 10.1093/jac/dkx125. [DOI] [PubMed] [Google Scholar]
- 83.Ramon-Garcia S, et al. WhiB7, an Fe-S-dependent transcription factor that activates species-specific repertoires of drug resistance determinants in actinobacteria. J Biol Chem. 2013;288:34514–34528. doi: 10.1074/jbc.M113.516385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hurst-Hess K, et al. Mycobacterium abscessus WhiB7 regulates a species-specific repertoire of genes to confer extreme antibiotic resistance. Antimicrob Agents Chemother. 2017;61:e01347–17. doi: 10.1128/AAC.01347-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Meier A, et al. Molecular mechanisms of clarithromycin resistance in Mycobacterium avium: observation of multiple 23S rDNA mutations in a clonal population. J Infect Dis. 1996;174:354–360. doi: 10.1093/infdis/174.2.354. [DOI] [PubMed] [Google Scholar]
- 86.Wallace RJ, Jr, et al. Genetic basis for clarithromycin resistance among isolates of Mycobacterium chelonae and Mycobacterium abscessus. Antimicrob Agents Chemother. 1996;40:1676–1681. doi: 10.1128/aac.40.7.1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Mougari F, et al. Selection of resistance to clarithromycin in Mycobacterium abscessus subspecies. Antimicrob Agents Chemother. 2017;61:e00943–16. doi: 10.1128/AAC.00943-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Obata S, et al. Association of rpoB mutations with rifampicin resistance in Mycobacterium avium. Int J Antimicrob Agents. 2006;27:32–39. doi: 10.1016/j.ijantimicag.2005.09.015. [DOI] [PubMed] [Google Scholar]
- 89.Prammananan T, et al. A single 16S ribosomal RNA substitution is responsible for resistance to amikacin and other 2-deoxystreptamine aminoglycosides in Mycobacterium abscessus and Mycobacterium chelonae. J Infect Dis. 1998;177:1573–1581. doi: 10.1086/515328. [DOI] [PubMed] [Google Scholar]
- 90.Nessar R, et al. Genetic analysis of new 16S rRNA mutations conferring aminoglycoside resistance in Mycobacterium abscessus. J Antimicrob Chemother. 2011;66:1719–1724. doi: 10.1093/jac/dkr209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Sapriel G, et al. Genome-wide mosaicism within Mycobacterium abscessus: evolutionary and epidemiological implications. BMC Genomics. 2016;17:118. doi: 10.1186/s12864-016-2448-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ferro BE, et al. Moxifloxacin’s limited efficacy in the hollow-fiber model of Mycobacterium abscessus disease. Antimicrob Agents Chemother. 2016;60:3779–3785. doi: 10.1128/AAC.02821-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Maurer FP, et al. Lack of antimicrobial bactericidal activity in Mycobacterium abscessus. Antimicrob Agents Chemother. 2014;58:3828–3836. doi: 10.1128/AAC.02448-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Heifets LB, et al. Clarithromycin minimal inhibitory and bactericidal concentrations against Mycobacterium avium. 1992;145:856–858. doi: 10.1164/ajrccm/145.4_Pt_1.856. [DOI] [PubMed] [Google Scholar]
- 95.Aguilar-Ayala DA, et al. In vitro activity of bedaquiline against rapidly growing nontuberculous mycobacteria. J Med Microbiol. 2017;66:1140–1143. doi: 10.1099/jmm.0.000537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Falkinham JO., 3rd The biology of environmental mycobacteria. Environ Microbiol Rep. 2009;1:477–487. doi: 10.1111/j.1758-2229.2009.00054.x. [DOI] [PubMed] [Google Scholar]
- 97.Lenaerts A, et al. Heterogeneity in tuberculosis pathology, microenvironments and therapeutic responses. Immunol Rev. 2015;264:288–307. doi: 10.1111/imr.12252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Sacchettini JC, et al. Drugs versus bugs: in pursuit of the persistent predator Mycobacterium tuberculosis. Nat Rev Microbiol. 2008;6:41–52. doi: 10.1038/nrmicro1816. [DOI] [PubMed] [Google Scholar]
- 99.Guinn KM, Rubin EJ. Tuberculosis: just the FAQs. MBio. 2017;8:e01910–17. doi: 10.1128/mBio.01910-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Early J, et al. Mycobacterium avium uses apoptotic macrophages as tools for spreading. Microb Pathog. 2011;50:132–139. doi: 10.1016/j.micpath.2010.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Roux AL, et al. The distinct fate of smooth and rough Mycobacterium abscessus variants inside macrophages. Open Biol. 2016;6:pii160185. doi: 10.1098/rsob.160185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kim SW, et al. Clinical Mycobacterium abscessus strain inhibits autophagy flux and promotes its growth in murine macrophages. Pathog Dis. 2017;75 doi: 10.1093/femspd/ftx107. [DOI] [PubMed] [Google Scholar]
- 103.Adams KN, et al. Drug tolerance in replicating mycobacteria mediated by a macrophage-induced efflux mechanism. Cell. 2011;145:39–53. doi: 10.1016/j.cell.2011.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Bermudez LE, et al. Isolation of two subpopulations of Mycobacterium avium within human macrophages. FEMS Microbiol Lett. 1999;178:19–26. doi: 10.1111/j.1574-6968.1999.tb13754.x. [DOI] [PubMed] [Google Scholar]
- 105.Larsson C, et al. Gene expression of Mycobacterium tuberculosis putative transcription factors whiB1-7 in redox environments. PLoS One. 2012;7:e37516. doi: 10.1371/journal.pone.0037516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Schnappinger D, et al. Transcriptional adaptation of Mycobacterium tuberculosis within macrophages: insights into the phagosomal environment. J Exp Med. 2003;198:693–704. doi: 10.1084/jem.20030846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Szumowski JD, et al. Antimicrobial efflux pumps and Mycobacterium tuberculosis drug tolerance: evolutionary considerations. Curr Top Microbiol Immunol. 2013;374:81–108. doi: 10.1007/82_2012_300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Eum SY, et al. Neutrophils are the predominant infected phagocytic cells in the airways of patients with active pulmonary TB. Chest. 2010;137:122–128. doi: 10.1378/chest.09-0903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Haapanen JH, et al. Studies on the gaseous content of tuberculous cavities. Am Rev Respir Dis. 1959;80:1–5. doi: 10.1164/arrd.1959.80.1P1.1. [DOI] [PubMed] [Google Scholar]
- 110.Via LE, et al. Tuberculous granulomas are hypoxic in guinea pigs, rabbits, and nonhuman primates. Infect Immun. 2008;76:2333–2340. doi: 10.1128/IAI.01515-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Nyka W. Studies on the effect of starvation on mycobacteria. Infect Immun. 1974;9:843–850. doi: 10.1128/iai.9.5.843-850.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Wayne LG, Hayes LG. An in vitro model for sequential study of shiftdown of Mycobacterium tuberculosis through two stages of nonreplicating persistence. Infect Immun. 1996;64:2062–2069. doi: 10.1128/iai.64.6.2062-2069.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Betts JC, et al. Evaluation of a nutrient starvation model of Mycobacterium tuberculosis persistence by gene and protein expression profiling. Mol Microbiol. 2002;43:717–731. doi: 10.1046/j.1365-2958.2002.02779.x. [DOI] [PubMed] [Google Scholar]
- 114.Rao SP, et al. The protonmotive force is required for maintaining ATP homeostasis and viability of hypoxic, nonreplicating Mycobacterium tuberculosis. Proc Natl Acad Sci U S A. 2008;105:11945–11950. doi: 10.1073/pnas.0711697105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Gengenbacher M, et al. Nutrient-starved, non-replicating Mycobacterium tuberculosis requires respiration, ATP synthase and isocitrate lyase for maintenance of ATP homeostasis and viability. Microbiology. 2010;156:81–87. doi: 10.1099/mic.0.033084-0. [DOI] [PubMed] [Google Scholar]
- 116.Xie Z, et al. Differential antibiotic susceptibilities of starved Mycobacterium tuberculosis isolates. Antimicrob Agents Chemother. 2005;49:4778–4780. doi: 10.1128/AAC.49.11.4778-4780.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Sarathy J, et al. Reduced drug uptake in phenotypically resistant nutrient-starved nonreplicating Mycobacterium tuberculosis. Antimicrob Agents Chemother. 2013;57:1648–1653. doi: 10.1128/AAC.02202-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Sarathy JP, et al. Extreme drug tolerance of Mycobacterium tuberculosis in caseum. Antimicrob Agents Chemother. 2017;62:e02266–17. doi: 10.1128/AAC.02266-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Hoff DR, et al. Location of intra- and extracellular M. tuberculosis populations in lungs of mice and guinea pigs during disease progression and after drug treatment. PLoS One. 2011;6:e17550. doi: 10.1371/journal.pone.0017550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Lenaerts AJ, et al. Location of persisting mycobacteria in a guinea pig model of tuberculosis revealed by r207910. Antimicrob Agents Chemother. 2007;51:3338–3345. doi: 10.1128/AAC.00276-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Aly S, et al. Interferon-gamma-dependent mechanisms of mycobacteria-induced pulmonary immunopathology: the role of angiostasis and CXCR3-targeted chemokines for granuloma necrosis. J Pathol. 2007;212:295–305. doi: 10.1002/path.2185. [DOI] [PubMed] [Google Scholar]
- 122.Aly S, et al. Oxygen status of lung granulomas in Mycobacterium tuberculosis-infected mice. J Pathol. 2006;210:298–305. doi: 10.1002/path.2055. [DOI] [PubMed] [Google Scholar]
- 123.Wu ML, et al. Mild nutrient starvation triggers the development of a small-cell survival morphotype in mycobacteria. Front Microbiol. 2016;7:947. doi: 10.3389/fmicb.2016.00947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Dick T, et al. Oxygen depletion induced dormancy in Mycobacterium smegmatis. FEMS Microbiol Lett. 1998;163:159–164. doi: 10.1111/j.1574-6968.1998.tb13040.x. [DOI] [PubMed] [Google Scholar]
- 125.Gerasimova A, et al. Comparative genomics of the dormancy regulons in mycobacteria. J Bacteriol. 2011;193:3446–3452. doi: 10.1128/JB.00179-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Miranda-CasoLuengo AA, et al. Functional characterization of the Mycobacterium abscessus genome coupled with condition specific transcriptomics reveals conserved molecular strategies for host adaptation and persistence. BMC Genomics. 2016;17:553. doi: 10.1186/s12864-016-2868-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Drapal M, et al. Metabolite analysis of Mycobacterium species under aerobic and hypoxic conditions reveals common metabolic traits. Microbiology. 2016;162:1456–1467. doi: 10.1099/mic.0.000325. [DOI] [PubMed] [Google Scholar]
- 128.Dartois V. The path of anti-tuberculosis drugs: from blood to lesions to mycobacterial cells. Nat Rev Microbiol. 2014;12:159–167. doi: 10.1038/nrmicro3200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Sarathy JP, et al. Prediction of drug penetration in tuberculosis lesions. ACS Infect Dis. 2016;2:552–563. doi: 10.1021/acsinfecdis.6b00051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Prideaux B, et al. The association between sterilizing activity and drug distribution into tuberculosis lesions. Nat Med. 2015;21:1223–1227. doi: 10.1038/nm.3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Worlitzsch D, et al. Effects of reduced mucus oxygen concentration in airway Pseudomonas infections of cystic fibrosis patients. J Clin Invest. 2002;109:317–325. doi: 10.1172/JCI13870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Thomas SR, et al. Increased sputum amino acid concentrations and auxotrophy of Pseudomonas aeruginosa in severe cystic fibrosis lung disease. Thorax. 2000;55:795–797. doi: 10.1136/thorax.55.9.795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Walker J, et al. Microbiological problems and biofilms associated with Mycobacterium chimaera in heater-cooler units used for cardiopulmonary bypass. J Hosp Infect. 2017;96:209–220. doi: 10.1016/j.jhin.2017.04.014. [DOI] [PubMed] [Google Scholar]
- 134.Revetta RP, et al. Changes in bacterial composition of biofilm in a metropolitan drinking water distribution system. J Appl Microbiol. 2016;121:294–305. doi: 10.1111/jam.13150. [DOI] [PubMed] [Google Scholar]
- 135.Carter G, et al. Characterization of biofilm formation by clinical isolates of Mycobacterium avium. J Med Microbiol. 2003;52:747–752. doi: 10.1099/jmm.0.05224-0. [DOI] [PubMed] [Google Scholar]
- 136.Yamazaki Y, et al. The ability to form biofilm influences Mycobacterium avium invasion and translocation of bronchial epithelial cells. Cell Microbiol. 2006;8:806–814. doi: 10.1111/j.1462-5822.2005.00667.x. [DOI] [PubMed] [Google Scholar]
- 137.Brodlie M, et al. Bile acid aspiration in people with cystic fibrosis before and after lung transplantation. Eur Respir J. 2015;46:1820–1823. doi: 10.1183/13993003.00891-2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Fennelly KP, et al. Biofilm formation by Mycobacterium abscessus in a lung cavity. Am J Respir Crit Care Med. 2016;193:692–693. doi: 10.1164/rccm.201508-1586IM. [DOI] [PubMed] [Google Scholar]
- 139.McNabe M, et al. Mycobacterium avium ssp. hominissuis biofilm is composed of distinct phenotypes and influenced by the presence of antimicrobials. Clin Microbiol Infect. 2011;17:697–703. doi: 10.1111/j.1469-0691.2010.03307.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Greendyke R, Byrd TF. Differential antibiotic susceptibility of Mycobacterium abscessus variants in biofilms and macrophages compared to that of planktonic bacteria. Antimicrob Agents Chemother. 2008;52:2019–2026. doi: 10.1128/AAC.00986-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Teng R, Dick T. Isoniazid resistance of exponentially growing Mycobacterium smegmatis biofilm culture. FEMS Microbiol Lett. 2003;227:171–174. doi: 10.1016/S0378-1097(03)00584-6. [DOI] [PubMed] [Google Scholar]
- 142.Falkinham JO., 3rd Growth in catheter biofilms and antibiotic resistance of Mycobacterium avium. J Med Microbiol. 2007;56:250–254. doi: 10.1099/jmm.0.46935-0. [DOI] [PubMed] [Google Scholar]
- 143.Carter G, et al. A subinhibitory concentration of clarithromycin inhibits Mycobacterium avium biofilm formation. Antimicrob Agents Chemother. 2004;48:4907–4910. doi: 10.1128/AAC.48.12.4907-4910.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Rose SJ, et al. Mycobacterium avium possesses extracellular DNA that contributes to biofilm formation, structural integrity, and tolerance to antibiotics. PLoS One. 2015;10:e0128772. doi: 10.1371/journal.pone.0128772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Steed KA, Falkinham JO., 3rd Effect of growth in biofilms on chlorine susceptibility of Mycobacterium avium and Mycobacterium intracellulare. Appl Environ Microbiol. 2006;72:4007–4011. doi: 10.1128/AEM.02573-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Ojha A, Hatfull GF. The role of iron in Mycobacterium smegmatis biofilm formation: the exochelin siderophore is essential in limiting iron conditions for biofilm formation but not for planktonic growth. Mol Microbiol. 2007;66:468–483. doi: 10.1111/j.1365-2958.2007.05935.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Werner E, et al. Stratified growth in Pseudomonas aeruginosa biofilms. Appl Environ Microbiol. 2004;70:6188–6196. doi: 10.1128/AEM.70.10.6188-6196.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Dick T, et al. TB drug discovery from target identification to proof of concept studies. In: Kaufmann SHE, Rubin E, editors. Handbook of Tuberculosis. Wiley-VCH; 2008. pp. 143–163. [Google Scholar]
- 149.Dartois V, et al. Tuberculosis drug discovery: issues, gaps and the way forward. In: Selzer PM, editor. Antiparasitic and Antibacterial Drug Discovery. Wiley-VCH; 2009. pp. 415–440. [Google Scholar]
- 150.Low JL, et al. Screening of TB actives for activity against nontuberculous mycobacteria delivers high hit rates. Front Microbiol. 2017;8:1539. doi: 10.3389/fmicb.2017.01539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Yee M, et al. Draft genome sequence of Mycobacterium avium 11. Genome Announc. 2017;5:e00766–17. doi: 10.1128/genomeA.00766-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Yee M, et al. Draft genome sequence of Mycobacterium abscessus bamboo. Genome Announc. 2017;5:e00388–17. doi: 10.1128/genomeA.00388-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Cowman S, et al. The antimicrobial susceptibility of non-tuberculous mycobacteria. J Infect. 2016;72:324–331. doi: 10.1016/j.jinf.2015.12.007. [DOI] [PubMed] [Google Scholar]
- 154.Nie W, et al. Species identification of Mycobacterium abscessus subsp. abscessus and Mycobacterium abscessus subsp. bolletii using rpoB and hsp65, and susceptibility testing to eight antibiotics. Int J Infect Dis. 2014;25:170–174. doi: 10.1016/j.ijid.2014.02.014. [DOI] [PubMed] [Google Scholar]
- 155.Hatakeyama S, et al. Antimicrobial susceptibility testing of rapidly growing mycobacteria isolated in Japan. BMC Infect Dis. 2017;17:197. doi: 10.1186/s12879-017-2298-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.van Ingen J, et al. In vitro drug susceptibility of 2275 clinical non-tuberculous Mycobacterium isolates of 49 species in The Netherlands. Int J Antimicrob Agents. 2010;35:169–173. doi: 10.1016/j.ijantimicag.2009.09.023. [DOI] [PubMed] [Google Scholar]
- 157.Truffot-Pernot C, et al. Effect of pH on the in vitro potency of clarithromycin against Mycobacterium avium complex. Antimicrob Agents Chemother. 1991;35:1677–1678. doi: 10.1128/aac.35.8.1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Pethe K, et al. A chemical genetic screen in Mycobacterium tuberculosis identifies carbon-source-dependent growth inhibitors devoid of in vivo efficacy. Nat Commun. 2010;1:57. doi: 10.1038/ncomms1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Bardouniotis E, et al. Characterization of biofilm growth and biocide susceptibility testing of Mycobacterium phlei using the MBEC assay system. FEMS Microbiol Lett. 2001;203:263–267. doi: 10.1111/j.1574-6968.2001.tb10851.x. [DOI] [PubMed] [Google Scholar]
- 160.Kirchner S, et al. Use of artificial sputum medium to test antibiotic efficacy against Pseudomonas aeruginosa in conditions more relevant to the cystic fibrosis lung. J Vis Exp. 2012;2012:e3857. doi: 10.3791/3857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Haley CL, et al. Characterization of biofilm-like structures formed by Pseudomonas aeruginosa in a synthetic mucus medium. BMC Microbiol. 2012;12:181. doi: 10.1186/1471-2180-12-181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Zimmerman M, et al. Ethambutol partitioning in tuberculous pulmonary lesions explains its clinical efficacy. Antimicrob Agents Chemother. 2017;61:e00924–17. doi: 10.1128/AAC.00924-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Chan ED, Bai X. Animal models of non-tuberculous mycobacterial infections. Mycobact Dis. 2016;6:216. [Google Scholar]
- 164.Bernut A, et al. The diverse cellular and animal models to decipher the physiopathological traits of Mycobacterium abscessus infection. Front Cell Infect Microbiol. 2017;7:100. doi: 10.3389/fcimb.2017.00100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Ordway D, et al. Animal model of Mycobacterium abscessus lung infection. J Leukoc Biol. 2008;83:1502–1511. doi: 10.1189/jlb.1007696. [DOI] [PubMed] [Google Scholar]
- 166.Daley CL, Glassroth J. Treatment of pulmonary nontuberculous mycobacterial infections: many questions remain. Ann Am Thorac Soc. 2014;11:96–97. doi: 10.1513/AnnalsATS.201311-399ED. [DOI] [PubMed] [Google Scholar]
- 167.Koh WJ, et al. Clinical significance of differentiation of Mycobacterium massiliense from Mycobacterium abscessus. Am J Respir Crit Care Med. 2011;183:405–410. doi: 10.1164/rccm.201003-0395OC. [DOI] [PubMed] [Google Scholar]
- 168.Research Committee of the British Thoracic Socierty. First randomised trial of treatments for pulmonary disease caused by M avium intracellulare, M malmoense, and M xenopi in HIV negative patients: rifampicin, ethambutol and isoniazid versus rifampicin and ethambutol. Thorax. 2001;56:167–172. doi: 10.1136/thorax.56.3.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Jenkins PA, et al. Clarithromycin vs ciprofloxacin as adjuncts to rifampicin and ethambutol in treating opportunist mycobacterial lung diseases and an assessment of Mycobacterium vaccae immunotherapy. Thorax. 2008;63:627–634. doi: 10.1136/thx.2007.087999. [DOI] [PubMed] [Google Scholar]
- 170.Lienhardt C, Davies G. Methodological issues in the design of clinical trials for the treatment of multidrug-resistant tuberculosis: challenges and opportunities. Int J Tuberc Lung Dis. 2010;14:528–537. [PubMed] [Google Scholar]
- 171.Wallis RS, et al. Tuberculosis biomarkers discovery: developments, needs, and challenges. Lancet Infect Dis. 2013;13:362–372. doi: 10.1016/S1473-3099(13)70034-3. [DOI] [PubMed] [Google Scholar]
- 172.van Ingen J, et al. Treatment outcome definitions in nontuberculous mycobacterial pulmonary disease – an NTM-NET consensus statement. Eur Respir J. 2018;51:1800170. doi: 10.1183/13993003.00170-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Gopal P, Dick T. Reactive dirty fragments: implications for tuberculosis drug discovery. Curr Opin Microbiol. 2014;21C:7–12. doi: 10.1016/j.mib.2014.06.015. [DOI] [PubMed] [Google Scholar]
- 174.Mukherjee D, et al. Membrane-targeting AM-0016 kills mycobacterial persisters and shows low propensity for resistance development. Future Microbiol. 2016;11:643–650. doi: 10.2217/fmb-2015-0015. [DOI] [PubMed] [Google Scholar]
- 175.Moreira W, et al. Boromycin kills mycobacterial persisters without detectable resistance. Front Microbiol. 2016;7:199. doi: 10.3389/fmicb.2016.00199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Yang T, et al. Amphiphilic indole derivatives as antimycobacterial agents: structure–activity relationships and membrane targeting properties. J Med Chem. 2017;60:2745–2763. doi: 10.1021/acs.jmedchem.6b01530. [DOI] [PubMed] [Google Scholar]
- 177.Cremades R, et al. Mycobacterium abscessus from respiratory isolates: activities of drug combinations. J Infect Chemother. 2009;15:46–48. doi: 10.1007/s10156-008-0651-y. [DOI] [PubMed] [Google Scholar]
- 178.Zhang Z, et al. In vitro activity of clarithromycin in combination with other antimicrobial agents against Mycobacterium abscessus and Mycobacterium massiliense. Int J Antimicrob Agents. 2017;49:383–386. doi: 10.1016/j.ijantimicag.2016.12.003. [DOI] [PubMed] [Google Scholar]
- 179.Huang CW, et al. Synergistic activities of tigecycline with clarithromycin or amikacin against rapidly growing mycobacteria in Taiwan. Int J Antimicrob Agents. 2013;41:218–223. doi: 10.1016/j.ijantimicag.2012.10.021. [DOI] [PubMed] [Google Scholar]
- 180.Choi GE, et al. Activities of moxifloxacin in combination with macrolides against clinical isolates of Mycobacterium abscessus and Mycobacterium massiliense. Antimicrob Agents Chemother. 2012;56:3549–3555. doi: 10.1128/AAC.00685-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Miyasaka T, et al. In vitro efficacy of imipenem in combination with six antimicrobial agents against Mycobacterium abscessus. Int J Antimicrob Agents. 2007;30:255–258. doi: 10.1016/j.ijantimicag.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 182.Zhang Z, et al. In vitro activity between linezolid and other antimicrobial agents against Mycobacterium abscessus complex. Diagn Microbiol Infect Dis. 2018;90:31–34. doi: 10.1016/j.diagmicrobio.2017.09.013. [DOI] [PubMed] [Google Scholar]
- 183.Singh S, et al. In vitro evaluation of a new drug combination against clinical isolates belonging to the Mycobacterium abscessus complex. Clin Microbiol Infect. 2014;20:O1124–7. doi: 10.1111/1469-0691.12780. [DOI] [PubMed] [Google Scholar]
- 184.Oh CT, et al. Novel drug combination for Mycobacterium abscessus disease therapy identified in a Drosophila infection model. J Antimicrob Chemother. 2014;69:1599–1607. doi: 10.1093/jac/dku024. [DOI] [PubMed] [Google Scholar]
- 185.Kaushik A, et al. Carbapenems and rifampin exhibit synergy against Mycobacterium tuberculosis and Mycobacterium abscessus. Antimicrob Agents Chemother. 2015;59:6561–6567. doi: 10.1128/AAC.01158-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Lefebvre AL, et al. Bactericidal and intracellular activity of β-lactams against Mycobacterium abscessus. J Antimicrob Chemother. 2016;71:1556–1563. doi: 10.1093/jac/dkw022. [DOI] [PubMed] [Google Scholar]
- 187.Lefebvre AL, et al. Inhibition of the β-lactamase BlaMab by avibactam improves the in vitro and in vivo efficacy of imipenem against Mycobacterium abscessus. Antimicrob Agents Chemother. 2017;61:e02240–16. doi: 10.1128/AAC.02440-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Rominski A, et al. Intrinsic rifamycin resistance of Mycobacterium abscessus is mediated by ADP-ribosyltransferase MAB_0591. J Antimicrob Chemother. 2017;72:376–384. doi: 10.1093/jac/dkw466. [DOI] [PubMed] [Google Scholar]
- 189.Zhang Y, et al. Role of acid pH and deficient efflux of pyrazinoic acid in unique susceptibility of Mycobacterium tuberculosis to pyrazinamide. J Bacteriol. 1999;181:2044–2049. doi: 10.1128/jb.181.7.2044-2049.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Sun Z, et al. The pncA gene from naturally pyrazinamide-resistant Mycobacterium avium encodes pyrazinamidase and confers pyrazinamide susceptibility to resistant M. tuberculosis complex organisms. Microbiology. 1997;143:3367–3373. doi: 10.1099/00221287-143-10-3367. [DOI] [PubMed] [Google Scholar]
- 191.Pryjma M, et al. Antagonism between front-line antibiotics clarithromycin and amikacin in the treatment of Mycobacterium abscessus infections is mediated by the whiB7 Gene. Antimicrob Agents Chemother. 2017;61:e01353–17. doi: 10.1128/AAC.01353-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Guillemin I, et al. Correlation between quinolone susceptibility patterns and sequences in the A and B subunits of DNA gyrase in mycobacteria. Antimicrob Agents Chemother. 1998;42:2084–2088. doi: 10.1128/aac.42.8.2084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Carrere-Kremer S, et al. A new dehydratase conferring innate resistance to thiacetazone and intra-amoebal survival of Mycobacterium smegmatis. Mol Microbiol. 2015;96:1085–1102. doi: 10.1111/mmi.12992. [DOI] [PubMed] [Google Scholar]
- 194.Makarov V, et al. Benzothiazinones kill Mycobacterium tuberculosis by blocking arabinan synthesis. Science. 2009;324:801–804. doi: 10.1126/science.1171583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Incandela ML, et al. DprE1, a new taxonomic marker in mycobacteria. FEMS Microbiol Lett. 2013;348:66–73. doi: 10.1111/1574-6968.12246. [DOI] [PubMed] [Google Scholar]
- 196.Wellington S, et al. A small-molecule allosteric inhibitor of Mycobacterium tuberculosis tryptophan synthase. Nat Chem Biol. 2017;13:943–950. doi: 10.1038/nchembio.2420. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


