To the Editor
Mutations in the desmoplakin (DSP) and desmoglein-1 (DSG1) genes have been implicated in patients with the inherited inflammatory skin disease known as severe dermatitis, multiple allergies, and metabolic wasting (SAM) syndrome (MIM#603165, see Tables E1 and E2 in this article’s Online Repository at www.jacionline.org).1,2 The DSP and DSG1 genes encode desmosome components that are critical for the structure of intercellular junctions and maintenance of epithelial barrier integrity. DSP and DSG1 are also key regulators of signaling pathways involved in differentiation, epidermal homeostasis, and carcinogenesis. DSG1 promotes keratinocyte differentiation by inhibiting epidermal growth factor receptor/extracellular signal-regulated kinase signaling through ERBB2-interacting protein (ERBIN), a scaffolding and signaling protein.3 Through characterization of a new syndrome featuring severe allergic dermatitis and DSG1 deficiency, we highlighted the pivotal role of the functional DSG1/ERBIN interaction as an inhibitor of skin inflammation through the nuclear factor κB (NF-κB) signaling pathway.
Patients 1 and 2 (13 and 9 years old, respectively) are 2 unrelated boys born to healthy parents. They were referred for life-long desquamative erythroderma associated with sparse and wooly hair and dysplastic enamel. Both patients had painful palmoplantar keratoderma and dystrophic nails (Fig 1, A and B). Skin manifestations combined recurrent and painful erythrodermic skin flares triggered by infections and episodes of aseptic pustular psoriasiform dermatitis. Patient 1 (but not patient 2) displayed failure to thrive, eosinophilic esophagitis, colitis, and a variety of food allergies (total serum IgE level, 2968 kIU/mL [n < 114]). Cardiac examination of patient 1 revealed an asymptomatic, biventricular, dilated cardiomyopathy. At 9 years of age, patient 2 received a heart transplant because of a severe left-dominant arrhythmogenic cardiomyopathy.
FIG 1.
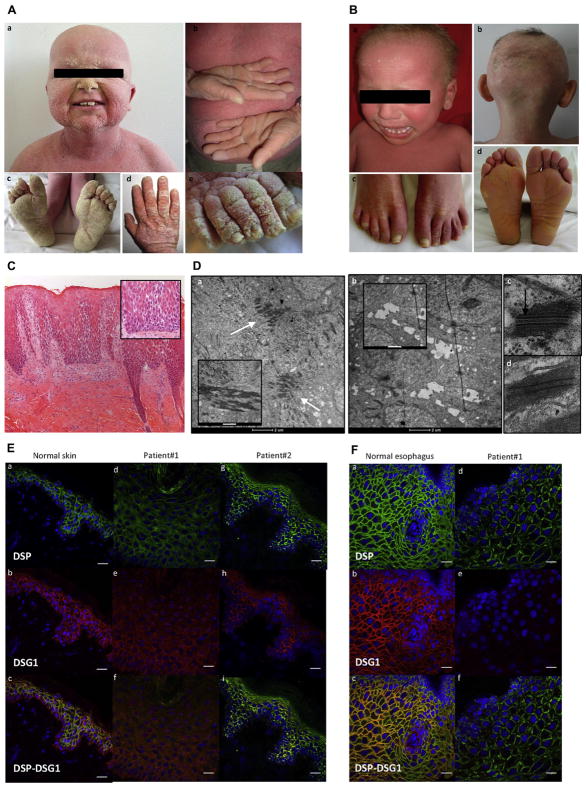
Clinical and histopathologic features of patients 1 and 2. A, Clinical phenotype of patient 1: desquamative erythroderma, thickening of the skin (a–e), sparse hair (a), diffuse palmoplantar keratoderma (PPK; b and c), and thickening of the nail plate (d and e). B, Clinical phenotype of patient 2: desquamative erythroderma (a), sparse hair (a and b), onycholysis (c), and plantar keratoderma (d). C, Skin histology (patient 1) showing epidermal acanthosis, hyperorthokeratosis, extensive acantholysis (inset), and inflammation in the superficial dermis (lymphocytes; ×100 magnification for Fig 1, C, and ×200 magnification for the inset of Fig 1, C). D, Ultrastructural features of a skin biopsy specimen from patient 1 (a, b, and d) and a healthy control subject (c). a, Many desmosomes clustered in the upper epidermis (arrows). Keratin filaments strongly aggregated to the desmosome (inset). b, Complete absence of desmosomes and widened spaces between keratinocytes (cell membrane features filipodium-like processes; inset) in the lower epidermis. d, Patient 1’s desmosome lacks the inner plaque compared with a control subject (arrow, c). Scale bar = 2 μm for Fig 1, D, a and b. Scale bar = 500 nm for the inset of Fig 1, D, a, and 1 μm for the inset of Fig 1, D, b. E, Immunofluorescence on skin sections from a healthy control subject (a–c), patient 1 (d–f), and patient 2 (g–i) showed drastic reduction and mislocalization in staining of DSP and DSG1 in both patients. Scale bar =20 μM. F, Immunofluorescence on esophageal sections from a healthy control subject (a–c) and patient 1 (d–f), showing low levels of DSP staining and absence of DSG1 staining in patient 1. Scale bar = 20 μM.
For both patients, cutaneous histopathology showed epidermal acantholysis and dermal inflammatory lymphocytic infiltration (Fig 1, C, and see Fig E1 and the Methods section in this article’s Online Repository at www.jacionline.org). Ultrastructural examination revealed large numbers of abnormal clusters of desmosomes in the epidermis (patient 1; Fig 1, D). Histopathology of the explanted heart showed the characteristic fibro-fatty myocardial infiltration of arrhythmogenic dysplasia (see Fig E1).
Two different heterozygous de novo missense mutations were identified by using whole-exome sequencing in exon 14 of the DSP gene: c.A1757C (p.H586P, patient 1) and c.T1828C (p.S610P, patient 2). Substitution of H586 or S610 by a proline is expected to induce a kink in the α-helix of DSP plakin domain that perturbs DSP’s 3-dimensional structure (see Fig E1).
In both patients skin immunohistochemistry showed the following features: (1) low DSP and DSG1 expression in the epidermis (as in patient 1’s primary keratinocytes), (2) irregular and less intense DSP and DSG1 staining at the keratinocyte plasma membrane, and (3) abnormal cytoplasmic accumulation of DSP and DSG1 proteins in keratinocytes (Fig 1, E). Esophageal immunohistochemistry revealed a low level of DSP staining, which was irregular and mottled at the cell border, and the absence of DSG1 expression (patient 1; Fig 1, F). Expression of DSP protein was also low in heart tissue (patient 2; see Fig E1; DSG1 is not expressed in the heart).
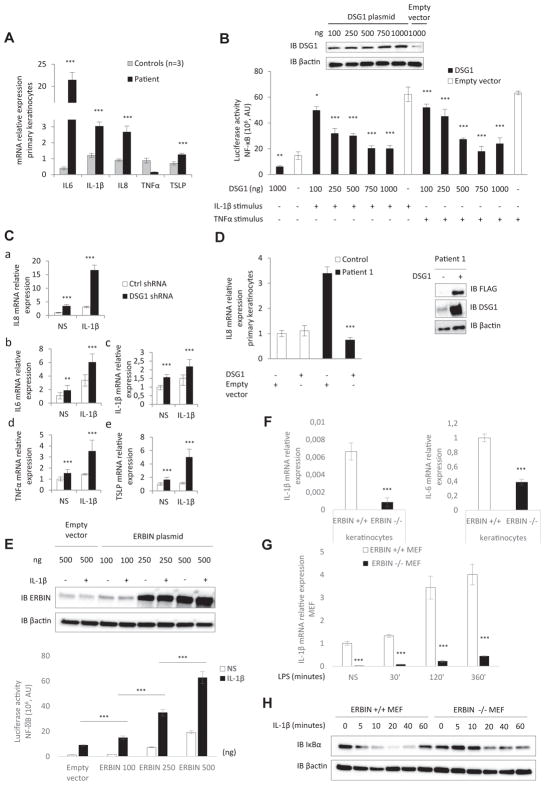
Abnormally high levels of mRNAs encoding proinflammatory cytokines (IL6, IL8, and IL1B), 3 NF-κB target genes, and TSLP were found in patient 1’s keratinocytes (Fig 2, A). Overexpression of IL-6 was confirmed by means of ELISA (see Fig E2 in this article’s Online Repository at www.jacionline.org). In contrast, mRNA levels of TNFA and other pro-TH2 cytokines (IL13, CCL5, and IL4; data not shown) were not increased. Inhibition of the NF-κB signaling pathway by ML120B, which selectively targets the catalytic subunit of inhibitor of NF-κB kinase (IKK) β, restored normal expression of IL8 mRNA by patient 1’s keratinocytes (see Fig E2). In view of the primary DSG1 deficiency reported in patients with SAM syndrome and the very low levels of DSG1 protein expression in our patients, we hypothesized that DSG1 could play a role in the inflammatory phenotype. We demonstrated that DSG1 (but not DSP) inhibits NF-κB reporter activity in a dose-dependent manner after stimulation by IL-1β or TNF-α in HEK293T cells (Fig 2, B, and see Fig E2). Concomitant downregulation of IL6 and IL8 expression was observed on transfection with the DSG1-encoding plasmid (see Fig E2). Silencing of DSG1 by short hairpin RNA (with a mean decrease of DSG1 mRNA of 32%) enhanced transcription of IL6, IL8, IL1B, TNFA, and TSLP genes in control keratinocytes (regardless of stimulation with IL-1β; Fig 2, C, and see Fig E2). Retroviral transduction of wild-type DSG1 into patient 1’s keratinocytes rescued the inflammatory cellular phenotype by restoring IL8 production (Fig 2, D).
FIG 2.
Role of DSG1 and ERBIN in NF-κB–mediated epithelial inflammation. A, Relative mRNA expression levels of proinflammatory cytokines in primary keratinocytes (patient 1). B, NF-κB luciferase reporter assay in HEK293T cells transfected with DSG1 vector or with an empty vector and stimulated with IL-1β or TNF-α. AU, Arbitrary units. Inset, Western blot of DSG1 expression in HEK293T cells transfected with DSG1 vector. C, Relative mRNA expression of IL8 (a), IL6 (b), IL1B (c), TNFA (d), and TSLP (e) in control (ctrl) keratinocytes infected with a lentivirus-expressing short hairpin RNA against DSG1 and stimulated or not stimulated (NS) with IL-1β. D, Relative mRNA expression of patient 1’s keratinocytes infected with a retrovirus expressing FLAG DSG1. Inset, Western blot of DSG1 and FLAG expression in patient 1’s keratinocytes after transduction. E, NF-κB luciferase reporter assay in HEK293T cells transfected with ERBIN or empty vector and stimulated with IL-1β. Inset, Western blot of ERBIN expression in HEK293T cells transfected with the ERBIN vector. F, Relative mRNA expression levels of IL1B and IL6 in nonstimulated Erbin+/+ and Erbin−/− keratinocytes. G, Relative mRNA expression levels of IL1B in Erbin+/+ and Erbin−/− MEFs before/after stimulation with LPS. H, Immunoblot assays showing degradation of IκBα proteins in Erbin+/+ and Erbin−/− MEFs induced by IL-1β stimulation. *P < .05, **P < .01, and ***P < .001.
Given that DSG1 might regulate ERBIN’s cellular localization and thus modulate the latter’s ability to block extracellular signal-regulated kinase signaling, we hypothesized that ERBIN can play a role in DSG1-mediated NF-κB inhibition.3 We observed little ERBIN/DSG1 colocalization at the cell membrane and an accumulation of ERBIN in the cytoplasm in a skin biopsy specimen from patient 1 relative to healthy control values (mean proportion of ERBIN/DSG1 colocalization, 40.7% vs 71.2%; P < .001; see Fig E3 in this article’s Online Repository at www.jacionline.org).
Experiments in control keratinocytes highlighted recruitment of ERBIN to the cell membrane and an increase in ERBIN/DSG1 colocalization after stimulation by IL-1β. Interestingly, this recruitment did not occur in patient 1’s keratinocytes (see Fig E3). Contrary to patient 1’s keratinocytes, the level of ERBIN protein increased after stimulation of control keratinocytes by IL-1β (see Fig E3). These results demonstrated that localization of ERBIN at the cell membrane and membrane colocalization of ERBIN/DSG1 in keratinocytes were regulated by inflammatory stimuli (eg, IL-1β).
In a reporter transactivation assay, we found that ERBIN transfection in both nonstimulated and IL-1β–stimulated HEK293T cells led to NF-κB activation in a dose-dependent manner (Fig 2, E). Erbin−/− keratinocytes, derived from Erbin−/− mice, showed abnormally low mRNA expression of the proinflammatory cytokines IL6 and IL1B (Fig 2, F). In addition, abnormally low expression of IL1B mRNA were found in the Erbin−/− mouse embryo fibroblasts (MEFs), regardless of stimulation with LPS (Fig 2, G). Biochemical analysis of NF-κB activation showed that in Erbin−/− MEFs, IκBα degradation was delayed in response to IL-1β (Fig 2, H).
Here we report on 2 unrelated patients in whom severe dermatitis and loss of epithelial barrier integrity were related to 2 different heterozygous mutations in DSP. Both patients presented with a combination of SAM syndrome, ectodermal dysplasia (combination of hair, nail, and tooth anomalies), and arrhythmogenic cardiomyopathy (hereby named SAMEC syndrome). Because of the major risk of severe cardiac involvement in patients carrying a DSP mutation, a thorough cardiac assessment should always be carried out in patients presenting with the SAM syndrome phenotype unless DSP mutations have been ruled out.
Our results demonstrate that DSG1 inhibits skin inflammation by inhibiting the NF-κB signaling pathway. Various observations support that DSG1 deficiency is linked to inflammation. First, DSG1 deficiency has been reported in patients with atopic dermatitis and Netherton syndrome,4,5 which share chronic inflammatory dermatitis and allergic manifestations with SAMEC. Second, 2 siblings presented with Netherton syndrome and normal DSG1 epidermal staining, whereas no skin inflammation was observed.6 Third, DSG1 deficiency is probably responsible for the mucosal barrier impairment and inflammation observed in patients with eosinophilic esophagitis.7
We suggest that DSG1 normally inhibits the NF-κB signaling pathway, at least in part by retaining ERBIN at the cell membrane. Involvement of ERBIN in other inflammatory diseases has been shown. In the context of inflammatory bowel disease, ERBIN seems to specifically inhibit the NF-κB signaling pathway through nucleotide-binding oligomerization domain 2.8 In addition, 5 related patients presenting with allergic inflammatory manifestations similar to SAMEC syndrome (eczema, eosinophilic esophagitis, and an increased serum IgE level) were shown to carry a heterozygous mutation in the gene coding for ERBIN.9 Thus ERBIN mislocalization in patients with SAMEC syndrome might explain our patients’ allergic manifestations.
Through characterization of SAMEC syndrome, we showed that DSG1 acts as a novel and hitherto unexpected inhibitor of epithelial inflammation by inhibiting the NF-κB signaling pathway, probably through functional DSG1/ERBIN interaction. The lack of an epithelial barrier protein, here DSG1, appears to be a crucial link between loss of epithelial barrier integrity and immunologic dysregulation.
METHODS
Molecular genetics
DNA was extracted from peripheral blood lymphocytes by using the Nucleon BACC3 DNA extraction kit (GE Healthcare, Piscataway, NJ), according to the manufacturer’s instructions. Samples of genomic DNA (1 μg) from patient 1 and his parents underwent whole-exome sequencing. The exons were captured with an in-solution enrichment technique (Sure-Select Human All Exon Kits Version 3; Agilent, Massy, France) based on a biotinylated oligonucleotide probe library (Human All Exon v3 50 Mb; Agilent). Each genomic DNA fragment was sequenced by using the paired-end strategy and an average read length of 75 bases (Illumina HISEQ Sequencer; Illumina, San Diego, Calif). Image analysis and base calling were performed with the Illumina Real-Time Analysis pipeline (version 1.9, Illumina) by using its default parameters. Sequences were aligned with the Human Genome Project reference sequence (hg19 assembly), and single nucleotide polymorphisms were called on the basis of allele calls and read depth by using the Consensus Assessment of Sequence and Variation pipeline (version 1.8; Illumina).
Genetic variations were annotated by using an in-house pipeline (Integra-Gen, Evry, France), and the results for each sample were made available online for analysis with ERIS (http://eris.integragen.com/). The DSP variant was confirmed by using Sanger sequencing with specific primers for exon 14 of the DSP gene. For patient 2 and his relatives, DSP’s 24 exons were amplified by using PCR with specific primers. The PCR products were sequenced by using the Sanger method and a BigDye Terminator Cycle Sequencing Ready Reaction Kit (version 3.1; Applied Biosystems, Foster City, Calif) and then analyzed with SeqScape software (version 3.0; Applied Biosystems). Whole-exome sequencing was then performed for patient 2.
Primary keratinocyte culture
Primary human keratinocytes were obtained from 4-mm punch biopsy specimens from patient 1 and 3 healthy control subjects. Keratinocytes were cultured on a feeder layer of lethally irradiated mouse 3T3-J2 fibroblasts, as described previously.E1,E2 Perhaps because of intrinsic factors linked to desmosomal disease, several tentative keratinocyte cultures for patient 2 failed. Similarly, keratinocytes from patient 1’s biopsy specimen were obtained after several culture assays.
Cell lines
HEK293T cells were purchased from ATCC (Teddington, Middlesex, United Kingdom).
Isolation and culture of MEFs and epidermal keratinocytes from Erbin-null mice
Primary MEFs were isolated from 14.5-day-old Erbin+/+ and Erbin−/− FVB mouse embryos. After dissection, fetuses were transferred in sterile PBS, livers and heads were removed and discarded, and the remaining part of each fetus was teased into fine pieces. Tissues were broken up into a suspension by means of vigorous pipetting, and after a short sedimentation, the remaining cell suspension was grown in Dulbecco modified Eagle medium supplemented with 10% FCS and antibiotics.
Primary mouse epidermal keratinocytes were isolated from 19.5-day-old Erbin+/+ and Erbin−/− BALB/c mouse embryos. After dissection, fetuses were transferred in sterile PBS; heads, paws, and tails were removed and discarded; and back skins were removed and treated with Dispase (Gibco/Invitrogen, Carlsbad, Calif). The epidermis was separated from the dermis with forceps and incubated with a trypsin/versene mixture. The epidermis was cut into small pieces, and after pipetting vigorously up and down, the basal layer keratinocytes were separated out from the epidermis. The cell suspension was filtered, washed with medium, and grown in CnT-57 medium (CELLnTEC, Bern, Switzerland) containing a low concentration of bovine pituitary extract.
RNA extraction, RT-PCR, and real-time PCR
Total RNA was isolated from HEK293T cells, MEFs, and cultured keratinocytes from patient 1 and different control subjects by using the RNeasy Plus Mini Kit (Qiagen GmbH, Hilden, Germany), according to the manufacturer’s instructions. RNA samples were reverse transcribed into cDNA by using the High Capacity cDNA Reverse Transcriptase Kit (Applied Biosystems). Real-time PCR was carried out with the Fast SYBR Green PCR Master Mix (Applied Biosystems) on an ABI Prism 7000 (PE Applied Biosystems). Quantitative RT-PCR primers were designed by using the sequences available in Ensembl and spanned an intron-exon boundary.
Amounts of the various mRNAs were normalized against the amount of ACTB RNA in each sample (measured by using quantitative RT-PCR). Results were analyzed with DataAssist (version 3.01; Applied Biosystems), which uses the comparative cycle threshold (ddCt) method. All experiments were performed in triplicates.
All data are expressed as the means ±SDs. Values were calculated by using an unpaired t test as follows.
Immunoblotting analysis
HEK293T cells were transiently transfected with increasing doses (100–1000 ng) of DSG1 plasmid (plasmid #55029; Addgene, Cambridge, Mass) or 250 ng of DSP plasmid (plasmid #32227; Addgene) and/or ERBIN plasmid (pEF1-Erbin-Myc-HisA construct) together with 0.2 μg of Igκ-Luc (see above). Cells were then lysed in RIPA buffer (150 mmol/L NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS, and 50 mmol/LTris-HCl [pH 8]) with protease inhibitors (Roche Diagnostics GmbH, Mannheim, Germany). Western blotting was performed by using mouse anti-DSP I/II antibody (diluted 1:1000, sc-390975; Santa Cruz Biotechnology, Heidelberg, Germany), rabbit anti-DSG1 antibody (diluted 1:1000, sc-20114; Santa Cruz Biotechnology), and rabbit anti-ERBIN antibody (diluted 1:10000, a gift from Lionel Fontao, Laboratory of Dermatology, University Hospitals and Medical School of Geneva, Geneva, Switzerland). Bound antibodies were visualized with horseradish peroxidase–conjugated antibodies against rabbit or mouse IgG (Santa Cruz Biotechnology) and an Enhanced Chemiluminescence kit (SuperSignal West Dura Extended Duration Substrate; Thermo Scientific, Rockford, Ill). Western blotting to assess degradation of IκBα proteins in MEFs was performed with a mouse anti-IκBα antibody (diluted 1:1000, L35A5; Cell Signaling Technology, Danvers, Mass). MEFs were stimulated with 20 ng/mL mouse IL-1β (R&D Systems, Minneapolis, Minn).
Inhibition of IKK-2 by ML120B
Keratinocytes from a healthy control subject and patient 1 were seeded into 12-well plates (100,000 cells/well). Twenty-four hours later, keratinocytes were preincubated with 20 μmol/L ML120B at 37°C for 1 hour and then stimulated with 10 ng/mL IL-1β. Twenty-four hours after stimulation, cells were pelleted for RNA extraction. ML120B was sent as a gift by Emmanuel Laplantine (Institut Pasteur, Paris, France).
Luciferase NF-κB reporter assays
HEK293T cells were seeded into 24-well plates and transfected in triplicates by using jetPRIM reagent (Polyplus Transfection, New York, NY) with increasing doses (100–1000 ng) of DSG1 plasmid (mCherry–desmoglein-1–C-18, a gift from Michael Davidson; plasmid #55029; Addgene) or 250 ng of DSP plasmid (1136-Desmoplakin–GFP, a gift from Kathleen Green (Addgene plasmid #32227) and/or increasing doses (100–500 ng) of ERBIN plasmid (pEF1-Erbin-Myc-HisA construct, a gift from Masaki Inagaki, Division of Biochemistry, Aichi Cancer Center Research Institute, Chikusa-ku, Nagoya, Aichi, Japan), together with 0.2 μg of a plasmid carrying the firefly luciferase gene under the control of the NF-κB promoter (Igκ-Luc; a gift from Gilles Courtois, Biosciences and Biotechnology Institute of Grenoble, Grenoble, France). Sixteen hours after transfection, cells were stimulated with 10 ng/mL IL-1β or 20 ng/mLTNF-α. Eight hours after stimulation, luciferase activity was determined by using a dual luciferase assay kit (Promega, Madison, Wis).
Retroviral vector production
pRetro-DSG1 was a gift from Kathleen Green (Northwestern University, Chicago, Ill). For virus preparation, pRetro-DSG1 or blank vector was cotransfected with jetPRIME reagent (Polyplus Transfection) and packaging vectors pGag/Pol and pVSVG into HEK293T cells. Infectious retroviruses were harvested at 24, 48, and 72 hours after transfection and filtered through 0.8-μm-pore cellulose acetate filters. Recombinant retroviruses were concentrated by means of ultracentrifugation (2 hours at 20,000g) and resuspended in Hank balanced salt solution. Viral aliquots were frozen and stored.
Lentiviral and retroviral transductions
Primary keratinocytes from a healthy control subject were seeded into 12-well plates. Twelve hours later, keratinocytes were infected with lentivirus containing (or not) DSG1 short hairpin RNA (sc-35224-V; Santa Cruz Biotechnology). Twenty-four hours after infection, keratinocytes were stimulated or not with 10 ng/mL human IL-1β (R&D Systems). Twenty-four hours after stimulation, cells were pelleted for RNA extraction. Expression of DSG1 was assessed by using quantitative RT-PCR. Primary keratinocytes from patient 1 and a healthy control subject were seeded into 12-well plates (80,000 cells/well). Twelve hours later, keratinocytes (20% confluent) were infected with retrovirus containing (or not) the DSG1 construct. Twenty-four hours after infection, keratinocytes were stimulated with 10 ng/mL IL-1β (R&D Systems). Lastly, 24 hours after stimulation, cells were pelleted for RNA extraction. DSG1 expression was assessed by using quantitative RT-PCR.
Reagents
Anti-FLAG antibody (diluted 1:10 000; Sigma-Aldrich, St Louis, Mo) was used as a reagent.
Light microscopy
Skin and heart biopsy specimens were fixed in 10% formalin, embedded in paraffin, and processed by using standard procedures. Three-micrometer-thick sections were stained with hematoxylin and eosin reagent and examined under LEICA DFC280 light microscopy (Leica, Buffalo Grove, Ill) at different magnifications. Images were acquired with Leica Application Suite Software.
Electron microscopy
Skin biopsy specimen was immersed in 2.5% glutaraldehyde fixative in 0.1 mol/L cacodylate buffer at pH 7.4 for 3 to 5 hours at 4°C, washed thoroughly in cacodylate buffer overnight at 4°C, and then postfixed in 1% osmium tetroxide for 1 hour at room temperature. The skin biopsy slices were then dehydrated in graded ethanol and impregnated with epoxy resin. After selection of suitable areas, semithin sections were stained with 1% toluidine blue and examined under a light microscope. Ultrathin sections were prepared and stained with uranyl acetate and lead citrate for electron microscopy (Tecnai T12; FEI, Hillsboro, Ore).
Immunohistochemical analysis of skin and esophageal biopsy specimens
Immunohistochemical reactions were performed on 4-μm-thick frozen tissue sections by using rabbit anti-DSG1 antibody (diluted 1:50, sc-20114; Santa Cruz Biotechnology), mouse anti-DSP I/II antibody (diluted 1:50, sc-390975; Santa Cruz Biotechnology), or sheep anti-ERBIN antibody (diluted 1:20, AF-7866; R&D Systems). Secondary antibodies were anti-rabbit Alexa Fluor 546, anti-mouse Alexa Fluor 488, and anti-sheep Alexa Fluor 488 (Life Technologies, Grand Island, NY) diluted 1:500 in 1% normal goat serum and incubated for 1 hour at 37°C. Sections were washed with once with PBS. Coverslips were mounted with In Situ Mounting Medium with DAPI (Duo-link; Olink Biosciences, Uppsala, Sweden). Images were acquired and processed with an LSM700 microscope (Zeiss, Jena, Germany) and Zen Software (Zeiss). Data were quantified with ImageJ software (https://imagej.nih.gov/ij/). Quantifications of the ERBIN/DSG1 colocalization have been made by using a surface-based colocalization measurement on Fiji (with immunofluorescence and confocal microscopy). Data corresponded to the average of the quantification of 5 images per skin (n = 5). Data are quoted as means ± SDs. ***P <.001. All scale bars = 20 μm.
Immunohistochemistry of heart biopsy specimens
Immunohistochemistry was performed on formalin-fixed, paraffin-embedded sections. After the deparaffination steps, reactions were carried out according to the automatized system Bond (A. Menarini Diagnosis system, Firenze, Italy). Staining was performed with the Menarini refine detection kit. Anti-desmoplakin 1+2 antibody (clone 65146; Progen, Heidelberg, Germany) was used.
Study approval
Clinical investigations were conducted according to Declaration of Helsinki principles and approved by the local ethics committee. Written informed consent was provided for a picture of the 3 patients appearing in the article. All patients, parents, and control subjects provided informed consent for genetic studies.
Statistical analysis
Results were expressed as the means ± SDs. Statistical significance was determined by using unpaired 2-sample t tests (equal variance). All data were normally distributed, and variance was similar in groups that were compared in statistical tests. The threshold for statistical significance was set at a P value of less than .05.
Supplementary Material
Acknowledgments
We thank the families involved in this study and the following physicians for their collaboration: Jean-Jacques Grob (Marseille, France), Véronique Fressart (Paris, France), Estelle Gandjbakhch (Paris, France), and Lionel Fontao (Geneva, Switzerland).
Supported by grants from the French Society of Dermatology (SFD), Laboratoires Expanscience, and the French National Research Agency (ANR) under the “Investments for the Future” program (reference: ANR-10-IAHU-01). Expanscience Laboratories helped to fund the molecular and immunologic analyses. J.-P.B.’s laboratory is funded by La Ligue Nationale Contre le Cancer (Label Ligue) and SIRIC (INCa-DGOS-Inserm 6038). J.-P.B. is a scholar of Institut Universitaire de France.
Footnotes
Disclosure of potential conflict of interest: J.-L. Casanova reports personal fees from Genentech, Sanofi, Novartis, Pfizer, Bioaster, Regeneron, BiogenIdec, and Merck outside the submitted work. The rest of the authors declare that they have no relevant conflicts of interest.
References
- 1.Samuelov L, Sarig O, Harmon RM, Rapaport D, Ishida-Yamamoto A, Isakov O, et al. Desmoglein 1 deficiency results in severe dermatitis, multiple allergies and metabolic wasting. Nat Genet. 2013;45:1244–8. doi: 10.1038/ng.2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McAleer MA, Pohler E, Smith FJD, Wilson NJ, Cole C, MacGowan S, et al. Severe dermatitis, multiple allergies, and metabolic wasting syndrome caused by a novel mutation in the N-terminal plakin domain of desmoplakin. J Allergy Clin Immunol. 2015;136:1268–76. doi: 10.1016/j.jaci.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Harmon RM, Simpson CL, Johnson JL, Koetsier JL, Dubash AD, Najor NA, et al. Desmoglein-1/Erbin interaction suppresses ERK activation to support epidermal differentiation. J Clin Invest. 2013;123:1556–70. doi: 10.1172/JCI65220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Broccardo CJ, Mahaffey S, Schwarz J, Wruck L, David G, Schlievert PM, et al. Comparative proteomic profiling of patients with atopic dermatitis based on history of eczema herpeticum infection and Staphylococcus aureus colonization. J Allergy Clin Immunol. 2011;127:186–93. e1–11. doi: 10.1016/j.jaci.2010.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fortugno P, Furio L, Teson M, Berretti M, El Hachem M, Zambruno G, et al. The 420K LEKTI variant alters LEKTI proteolytic activation and results in protease deregulation: implications for atopic dermatitis. Hum Mol Genet. 2012;21:4187–200. doi: 10.1093/hmg/dds243. [DOI] [PubMed] [Google Scholar]
- 6.Guerra L, Fortugno P, Pedicelli C, Mazzanti C, Proto V, Zambruno G, et al. Ichthyosis linearis circumflexa as the only clinical manifestation of Netherton syndrome. Acta Derm Venereol. 2015;95:720–4. doi: 10.2340/00015555-2075. [DOI] [PubMed] [Google Scholar]
- 7.Sherrill JD, Kc K, Wu D, Djukic Z, Caldwell JM, Stucke EM, et al. Desmoglein-1 regulates esophageal epithelial barrier function and immune responses in eosinophilic esophagitis. Mucosal Immunol. 2014;7:718–29. doi: 10.1038/mi.2013.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McDonald C, Chen FF, Ollendorff V, Ogura Y, Marchetto S, Lécine P, et al. A role for Erbin in the regulation of Nod2-dependent NF-kappaB signaling. J Biol Chem. 2005;280:40301–9. doi: 10.1074/jbc.M508538200. [DOI] [PubMed] [Google Scholar]
- 9.Lyons JJ, Liu Y, Ma CA, Yu X, O’Connell MP, Lawrence MG, et al. ERBIN deficiency links STAT3 and TGF-β pathway defects with atopy in humans. J Exp Med. 2017;214:669–80. doi: 10.1016/j.jaci.2018.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
REFERENCES
- E1.Green H, Rheinwald JG, Sun TT. Properties of an epithelial cell type in culture: the epidermal keratinocyte and its dependence on products of the fibroblast. Prog Clin Biol Res. 1977;17:493–500. [PubMed] [Google Scholar]
- E2.Rheinwald JG, Green H. Serial cultivation of strains of human epidermal keratinocytes: the formation of keratinizing colonies from single cells. Cell. 1975;6:331–43. doi: 10.1016/s0092-8674(75)80001-8. [DOI] [PubMed] [Google Scholar]
- E3.Choi H-J, Weis WI. Crystal structure of a rigid four-spectrin-repeat fragment of the human desmoplakin plakin domain. J Mol Biol. 2011;409:800–12. doi: 10.1016/j.jmb.2011.04.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- E4.Samuelov L, Sarig O, Harmon RM, Rapaport D, Ishida-Yamamoto A, Isakov O, et al. Desmoglein 1 deficiency results in severe dermatitis, multiple allergies and metabolic wasting. Nat Genet. 2013;45:1244–8. doi: 10.1038/ng.2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- E5.Has C, Jakob T, He Y, Kiritsi D, Hausser I, Bruckner-Tuderman L. Loss of desmoglein 1 associated with palmoplantar keratoderma, dermatitis and multiple allergies. Br J Dermatol. 2015;172:257–61. doi: 10.1111/bjd.13247. [DOI] [PubMed] [Google Scholar]
- E6.Schlipf NA, Vahlquist A, Teigen N, Virtanen M, Dragomir A, Fismen S, et al. Whole-exome sequencing identifies novel autosomal recessive DSG1 mutations associated with mild SAM syndrome. Br J Dermatol. 2016;174:444–8. doi: 10.1111/bjd.14079. [DOI] [PubMed] [Google Scholar]
- E7.Cheng R, Yan M, Ni C, Zhang J, Li M, Yao Z. Report of Chinese family with severe dermatitis, multiple allergies and metabolic wasting syndrome caused by novel homozygous desmoglein-1 gene mutation. J Dermatol. 2016;43:1201–4. doi: 10.1111/1346-8138.13431. [DOI] [PubMed] [Google Scholar]
- E8.Dănescu S, Leppert J, Cosgarea R, Zurac S, Pop S, Baican A, et al. Compound heterozygosity for dominant and recessive DSG1 mutations in a patient with atypical SAM syndrome (severe dermatitis, multiple allergies, metabolic wasting) J Eur Acad Dermatol Venereol. 2017;31:e144–6. doi: 10.1111/jdv.13967. [DOI] [PubMed] [Google Scholar]
- E9.McAleer MA, Pohler E, Smith FJD, Wilson NJ, Cole C, MacGowan S, et al. Severe dermatitis, multiple allergies, and metabolic wasting syndrome caused by a novel mutation in the N-terminal plakin domain of desmoplakin. J Allergy Clin Immunol. 2015;136:1268–76. doi: 10.1016/j.jaci.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- E10.Boyden LM, Kam CY, Hernández-Martín A, Zhou J, Craiglow BG, Sidbury R, et al. Dominant de novo DSP mutations cause erythrokeratodermia-cardiomyopathy syndrome. Hum Mol Genet. 2016;25:348–57. doi: 10.1093/hmg/ddv481. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.