Abstract
Accumulating studies indicate that long non-coding RNAs (lncRNAs) play important roles in the regulation of diverse biological processes involved in homeostatic control of the vessel wall in health and disease. However, our knowledge of the mechanisms by which lncRNAs control gene expression and cell signaling pathways is still nascent. Furthermore, only a handful of lncRNAs has been functionally evaluated in response to pathophysiological stimuli or in vascular disease states. For example, lncRNAs may regulate endothelial dysfunction by modulating endothelial cell proliferation (e.g. MALAT1, H19) or angiogenesis (e.g. MEG3, MANTIS). LncRNAs have also been implicated in modulating vascular smooth muscle cell (VSMC) phenotypes or vascular remodeling (e.g. ANRIL, SMILR, SENCR, MYOSLID). Finally, emerging studies have implicated lncRNAs in leukocytes activation (e.g. lincRNA-Cox2, linc00305, THRIL), macrophage polarization (e.g. GAS5), and cholesterol metabolism (e.g. LeXis). This review summarizes recent findings on the expression, mechanism, and function of lncRNAs implicated in a range of vascular disease states from mice to human subjects. An improved understanding of lncRNAs in vascular disease may provide new pathophysiological insights and opportunities for the generation of a new class of RNA-based biomarkers and therapeutic targets.
Keywords: lncRNAs, vascular biology, cardiovascular disease, non-coding RNAs
1. Introduction
Impaired vascular remodeling contributes to a wide variety of cardiovascular disease states including atherosclerosis, percutaneous coronary or peripheral interventions, vein graft disease, organ transplantation, among others. Accumulating studies have identified cell- or stage-specific pathophysiological mechanisms in the macro- and microvasculature that may underlie susceptibility to vascular disease. For example, beside its implication in “classical” systemic chronic inflammatory diseases such as rheumatoid arthritis (RA), systemic lupus erythematosus or gout, vascular inflammation contributes also to progression of more complex diseases such as atherosclerosis (1), diabetes (2) or cancer (3). Although the role of inflammation in atherosclerosis has been identified over 150 years ago by Virchow (4), only recently has the “inflammation hypothesis” in atherosclerosis been specifically tested using an anti-inflammatory drug targeting IL-1β (canakinumab), whereby recurrent cardiovascular events were reduced in the canakinumab treatment group independent of changes in lipid levels (5). However, major mechanistic gaps in the understanding of regulatory pathways involved in homeostasis of the vessel wall in response to pathophysiological stimuli remain and contribute to the lack of targeted therapeutics in a range of vascular disease states.
The recent recognition that only about 1.5% of the human genome encodes proteins, has opened new opportunities to better understand regulatory pathways in vascular health and disease (6) (7). Interestingly, a large quantity of the genome is transcribed at some point during development (8). Postnatally, the majority of biologically active RNAs that cannot be translated into proteins are long non-coding RNAs (lncRNAs) measuring more than 200 nucleotides in length and display mRNA-like characteristics such as being 5′-capped, spliced, and polyadenylated. In contrast to microRNAs, which bind to the 3′-UTR of target genes to mediate translational repression thereby altering the biology of diverse disease states (9-12), lncRNAs have emerged as powerful biological regulators by modulating numerous cellular processes, according to their cellular localization, in the nucleus or the cytoplasm (Figure 1). For example, lncRNAs localized in the nucleus can regulate transcription by guiding or sequestering transcription factors (TF) (13), inducing histone modifications, guiding chromatin remodeling complexes to the correct chromosomal locations (14), or acting as enhancer RNAs (15). Other studies demonstrated that lncRNAs may regulate nucleocytoplasmic shuttling of TFs such as nuclear factor of activated T cells (NFAT) (16) or alternative splicing of pre-mRNAs (17). In the cytoplasm, lncRNAs can regulate mRNA stability and control translational events (18), sponge miRNAs (19), and act as a scaffold for proteins complexes (20). Further regulatory functions may include stabilization of ribonucleoprotein (RNP) complexes (21) or protein phosphorylation and activation of signaling pathways (22). In addition, lncRNAs can be circularized by RNA splicing (circular lncRNAs) and act as miRNA sponges (23), or regulate the maturation of ribosomal RNAs (24). Finally, some lncRNAs are released in exosomes or microvesicles, potentially facilitating cell-to-cell communication (25) (26). However, the role of lncRNAs in vascular biology and disease remains poorly understood (27) (28) (29). This review summarizes examples of lncRNAs and their regulatory effects on diverse biological processes important to the macro- and microvasculature in health and disease (Figure 2).
Figure 1. Cellular functions of long non-coding RNAs (lncRNAs).
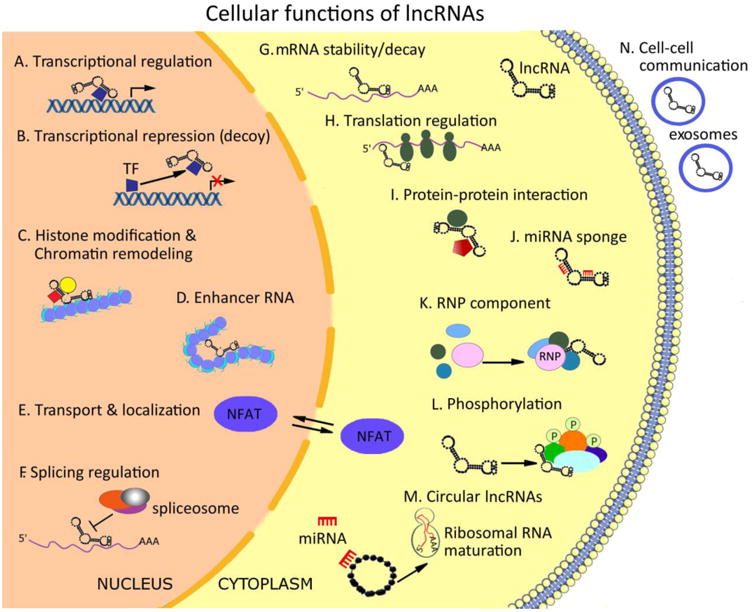
LncRNAs regulate gene expression by multiple mechanisms. Nuclear-localized lncRNA can guide transcription factors (TF) or protein complexes to specific sites in the genome (A) or sequester the TF and repress their function (B). They can induce histone modifications and guide chromatin remodeling complexes to the correct chromosomal locations (C) or induce chromosomal looping to increase association between enhancer and promoter regions (D). LncRNAs can regulate nucleocytoplasmic shuttling (E) of nuclear factor of activated T cells (NFAT) or alternative splicing of pre-mRNAs (F). In the cytoplasm, lncRNAs can regulate mRNA stability (G) and control translational events (H), sponge miRNAs (J) and act as a scaffold for proteins complexes (I). Further regulatory functions may include stabilization of ribonucleoprotein (RNP) complexes (K) or protein phosphorylation and activation of signaling pathways (L); Circular lncRNAs are formed by RNA splicing and were observed to act as miRNA sponges, or regulate the maturation of ribosomal RNAs (M). Finally, some lncRNAs are released in exosomes or microvesicles, potentially facilitating cell-to-cell communication (N).
Figure 2. LncRNAs implicated in vascular disease.
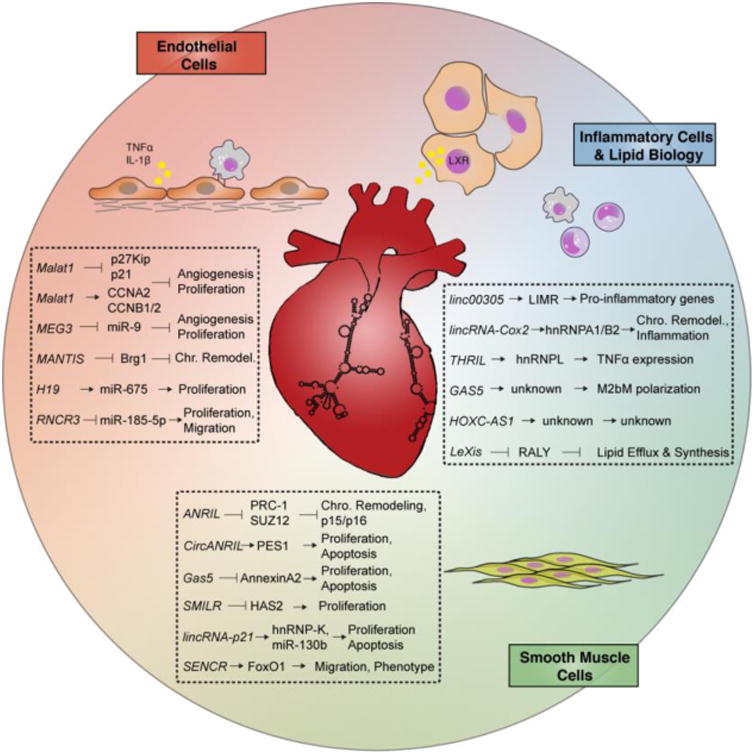
Highlighted lncRNAs involved in endothelial cell biology, vascular smooth muscle cell proliferation, leukocyte inflammation, and lipid metabolism.
2. LncRNAs and endothelial dysfunction
Impaired endothelial function has been linked to a variety of acute and chronic inflammatory disease states. For example, in response to both biochemical (e.g. IL-1β, modified-LDL) and biomechanical (e.g. disturbed blood flow) stimuli, endothelial activation is amongst the earliest processes involved in atherosclerotic lesion initiation (30). Consequently, expression of adhesion molecules (e.g. VCAM-1, E-Selectin) and secretion of chemokines (e.g. MCP-1, fractalkine) facilitates the recruitment of leukocyte subsets into the vessel wall (31). Chronic endothelial dysfunction may lead to loss of endothelial integrity predisposing to vascular inflammation and atherosclerosis (32). Accumulating studies highlight an emerging role for lncRNAs in regulating endothelial dysfunction.
Metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) is a lncRNA highly expressed in endothelial cells in both the macro- and microvasculature. It has been shown to regulate endothelial cells (ECs) inflammation, angiogenesis, and response to oxidative stress (33-35). MALAT1 knockdown decreases ECs proliferation by inhibiting cell cycle progression, decreasing the number of cells in S-phase under basal or hypoxic conditions and after vascular endothelial growth factor (VEGF) stimulation. MALAT1 silencing reduced the S-phase cyclins CCNB1, CCNB2, and CCNA2, while increasing the cell cycle inhibitory genes p21 and p27Kip1 (33). In contrast, MALAT1 overexpression increases the retinal EC proliferation rate (34) and inhibits apoptosis induced by oxygen-glucose deprivation and reoxygenation in human brain microvascular endothelial cells (36). MALAT1 silencing also decreased phosphorylated p38 levels in retinal ECs and the glucose-induced up-regulation of IL-6 and TNFα through activation of SAA3 in ECs (35). In a different study, following oxygen-glucose deprivation and reoxygenation in brain microvascular ECs, lentiviral knockdown of MALAT1 decreased PI3K activities and the activation of Akt phosphorylation, and increased cell apoptosis and caspase 3 activity, suggesting a potential role of MALAT1 in cerebral ischemia/reperfusion (36). Consistent with these in vitro observations, pharmacological inhibition of MALAT1 in mice undergoing hindlimb ischemia reduced blood flow recovery and capillary density, verifying studies that MALAT1 controls EC proliferation and vessel outgrowth in vitro and in vivo (33). MALAT1 also protects the endothelium against ox-LDL-induced dysfunction via upregulating the expression of the miR-22-3p target genes CXCR2 and AKT, hence acting as a miRNA sponge (37). In streptozotocin-induced diabetic rats, intraocular injection of MALAT1 shRNA alleviated vascular leakage induced by hyperglycemia, decreased the number of apoptotic retinal cells, and significantly reduced retinal inflammation (34). Phenotypically, MALAT1-KO mice showed a delayed vessel extension in the retina and a reduction of the vessel density when compared with wild-type littermates while the number of proliferating ECs was significantly reduced (33). While the molecular mechanisms mediating MALAT1's angiogenic effects has not been clarified, a recent study has indicated that MALAT1 may serve as a potential endogenous sponge for miR-26b, regulating EC autophagy and survival (38). Because the related family member miR-26a harbors a very similar seed sequence to miR-26b, a known anti-angiogenic miRNA in diabetic wound healing and post-MI repair (39, 40), future studies will be of interest to assess angiogenic regulation by the MALAT1-miRNA-26a axis. Collectively, these studies highlight an important role for MALAT1 in regulating EC homeostasis, angiogenesis, and vascular inflammation.
Maternally expressed gene 3 (MEG3) is a lncRNA that regulates angiogenesis and diabetes-related microvascular dysfunction (41). MEG3 expression was decreased in retinal ECs upon oxidative stress and high glucose stimulation in vitro and in the retinas of STZ-induced diabetic mice in vivo. MEG3 silencing in vivo exacerbates retinal vessel dysfunction, as observed by increased microvascular leakage, severe capillary degeneration, and inflammation. In retinal ECs MEG3 knockdown increased their proliferation, migration, and tube formation capacity by activating the PI3K/Akt signaling pathway. Mechanistically, MEG3 acts as a miRNA sponge in vascular ECs by negatively regulating miR-9, a key player in angiogenesis and proliferation (42). Alternatively, MEG3 expression is induced in senescent human ECs and MEG3 knockdown rescued the age-induced impairment of angiogenesis. Moreover, in mice undergoing hindlimb ischemia, MEG3 inhibition augmented blood flow recovery (43). Finally, Meg3-KO mice showed increased expression of genes regulated by VEGF. Indeed, the Meg3-null embryos showed increased cortical microvessel density, suggesting the important role of MEG3 in angiogenesis and vascularization (44).
MANTIS (lncRNA n342419) is a lncRNA initially observed at low levels in patients with idiopathic pulmonary arterial hypertension (IPAH) and in a rat PAH disease model. In contrast, it was induced in ECs isolated from human glioblastoma patients as well as in carotid arteries of Macaca fascicularis subjected to atherosclerosis regression diet (45). MANTIS is localized to the nucleus and its expression is controlled by the histone demethylase JARID1B, suggesting a chromatin regulatory function. Functional silencing of MANTIS by oligonucleotide delivery (siRNAs or GapmeRs) or CRISPR/Cas9-mediated deletion inhibited EC migration, angiogenic sprouting, and tube formation in vitro and in vivo in mice injected with matrigel-embedded human umbilical vein endothelial cells (HUVECs). Mechanistically, MANTIS also interacts with Brg1 and regulates SMAD6, COUP-TFII, and SOX18, which are all implicated in angiogenesis modulation.
Polymorphisms in the lncRNA H19 was originally associated with CAD risk in a Chinese population (46). Recent studies have shown that plasma levels of H19 can be an independent predictor for CAD (47). Expression of H19 was significantly increased in the plasma of patients with atherosclerosis compared to healthy volunteers, and was also expressed higher in atherosclerotic plaques of ApoE-/- mice (48). Overexpression of H19 in HUVECs increased their proliferation while decreasing apoptosis by regulating p38-MAPK and NF-κB signaling pathways (48). In contrast, inhibition of H19, a hypoxia-regulated lncRNA, decreased HUVEC growth, inducing their accumulation in G1 phase of the cell cycle (49). Recently, lncRNA H19 was observed to play a role in arterial restenosis, as it is overexpressed in the neointima of balloon-injured arteries (50). As H19 is a host gene for miR-675, gain-of-function studies have revealed that overexpression of H19 increases vascular smooth muscle cell (VSMC) proliferation rate by targeting PTEN in a miR-675-dependent manner (50). Future knockdown studies in atherosclerotic-prone disease models will be important to verify the therapeutic potential of lncRNA H19.
Retinal non-coding RNA3 (RNCR3), also known as LINC00599, is a lncRNA recently implicated to play a role in both atherosclerosis and diabetes mellitus (51-53). Increased levels of RNCR3 were observed in human and mouse aortic atherosclerotic lesions. RNCR3 knockdown in ApoE-/- mice accelerated the development of atherosclerosis, increased LDL plasma levels, and in turn regulated the inflammatory response. RNCR3 knockdown also reduced the proliferation and migration and accelerated apoptosis of ECs and VSMCs in vitro, suggesting that RNCR3 inhibition might impair EC regeneration in injured arteries. Mechanistically, RNCR3 was suggested to function as a competing endogenous RNA (ceRNA) by decreasing the concentration of miR-185-5p, ultimately resulting in the de-repression of Kruppel-like factor 2 (KLF2), a transcriptional factor conferring an endothelial vasoprotective phenotype. This regulatory mechanism was observed by the same group in two different studies on retinal vascular dysfunction in diabetes mellitus (52) (53). However, in all the above studies, an important interpretation issue will still need to be resolved. Because two of the four isoforms of RNCR3 have exonic overlap with microRNA-3078 and microRNA-124a-1, it is not clear if the observed phenotypes after RNCR3 knockdown can be attributed to these miRNAs or to RNCR3 alone. Future studies will need to definitively establish whether the above described phenotypes are indeed independent of miR-3078 and miR-124.
3. LncRNAs and vascular injury
Maladaptive vascular remodeling contributes to a wide range of cardiovascular procedures including percutaneous coronary or peripheral interventions, vein grafts for coronary artery bypass surgery, fistulas for dialysis access, and organ transplantation. In response to mechanical injury often initiated by endothelial denudation or injury, endothelial cells, platelets, and leukocytes release a variety of growth factors (e.g. PDGF-BB, TGF-β1), cytokines (e.g. IL-1, IL-6, and IL-8), chemokines (e.g. MCP-1), metalloproteinases (e.g. MMP-9), and pro-thrombotic mediators (e.g. thrombin) that cooperatively induce the proliferation of vascular smooth muscle cells of the medial layer to form a significant component of the neointima of the damaged vessel wall. Negative medial and adventitial remodeling may result in late lumen loss, restenosis, or complete occlusion of the vessel wall (69) (70) (71). VSMCs also play a prominent role in chronic inflammatory disease states such as atherosclerosis, hypertension, aneurysm formation, and pulmonary artery hypertension. Genetic lineage tracing studies in atherosclerotic models have implicated that VSMCs undergo phenotypic switching to cells that exhibit macrophage-like features with loss of VSMC marker identity (72). Accumulating studies have implicated a growing list of lncRNAs in vascular smooth muscle cell biology, providing potential new levels of functional regulation, mechanistic insights, and targets for therapy in a range of conditions.
Genome-wide association studies (GWAS) have identified that the INK4 locus on chromosome 9p21.3 harbors multiple single nucleotide polymorphisms (SNPs) linked to coronary artery disease (CAD) susceptibility (73) (74) (75), atherosclerosis (76), aortic aneurysm (77), ischemic stroke (75), type II diabetes (78), and specific cancer subtypes (79) (80). Antisense to this locus lies the lncRNA ANRIL (antisense ncRNA in the INK4 locus). The increased risk of SNPs associated with CAD is independent of all known CAD risk factors, suggesting that ANRIL may regulate a different biological pathway relevant for atherosclerosis (76). Two ANRIL transcripts (EU741058 and NR_003529) were found significantly increased in peripheral blood mononuclear cells and human atherosclerotic plaque tissues from CAD patients as compared to healthy subjects, while the most abundant isoform, DQ485454, remained unaffected (81). Consistent with this, loss-of-function studies with siRNAs targeting different exons (exon 1/19) in human SMCs induced different regulatory effects, suggesting that different splicing variants of ANRIL might play distinct roles in cell physiology (82). Mechanistically, Yap et al. showed in chromatin fractions that ANRIL directly binds to CBX7 and SUZ12, components of the polycomb repression complex-1 (PRC-1) and PRC-2, respectively (20, 83). Inhibition of ANRIL disrupts the binding of PRC-1 and PRC-2 at the INK4 locus, increasing the mRNA levels of p15INK4b and p16INK4a, two of the genes encoded by the INK4 locus and limiting cellular life span (20, 83, 84). Adding another layer of complexity, a recent study revealed that ANRIL circularization, resulting from exon skipping events during RNA splicing, activates a different cellular mechanism conferring atheroprotection (24). CircANRIL binds to PES1, an essential 60S-preribosomal assembly factor, impairing pre-rRNA processing and ribosome biogenesis in macrophages and VSMCs. As a consequence, circANRIL induces p53 activation and nucleolar stress, resulting in the inhibition of proliferation and induction of apoptosis. Although ANRIL is an independent risk factor for CAD, future studies will be informative to assess the functionality of ANRIL in relevant disease models.
Reduced levels of lincRNA-p21 were observed in PBMCs and artery tissues of patients with CAD and in the aortic plaques of ApoE–/– mice, as compared to artery tissues of control patients and in C57BL/6 control mice, respectively, thus suggesting a potential role of this lncRNA in disease development (85). Recently, the G-A-A-G haplotype of lincRNA-p21 was found to be associated with a decreased risk of CAD and MI, particularly among premature CAD/MI in the Chinese Han population (86). In vitro studies in VSMCs and mouse mononuclear macrophage cells showed that lincRNA-p21 induces apoptosis and represses cell proliferation (85). Moreover, in the same study in vivo inhibition of lincRNA-p21 resulted in enhanced neointimal hyperplasia in response to carotid artery injury in mice. Mechanistically, lincRNA-p21 expression is regulated by p53, and it physically associates with hnRNP-K to repress hundreds of genes in the p53 pathway (85). Transcriptomic analysis revealed that lincRNA-p21 inhibition deregulated many p53 targets. LincRNA-p21 also binds to mouse double minute 2 (MDM2), an E3 ubiquitin-protein ligase that represses p53 in physiological conditions. The association of lincRNA-p21 with MDM2 de-represses p53, enabling p53 to interact with p300 and to bind to the promoters/enhancers of its target genes, hence participating in a feedback mechanism (85). In addition, lincRNA-p21 promoted cell apoptosis and induced cell cycle progression by acting as an endogenous sponge for miR-130b in vascular endothelial cells (87). LincRNA-p21 has also been implicated in chronic vascular inflammation in patients with rheumatoid arthritis (RA) who expressed lower levels of lincRNA-p21 and increased levels of phosphorylated p65 (RelA), a marker of NF-κB activation. In contrast, patients treated with methotrexate (MTX) had higher levels of lincRNA-p21 (88). Mechanistically, MTX reduced NF-κB activity in TNFα–treated macrophages through a DNA-dependent protein kinase catalytic subunit (DNA PKcs)–dependent mechanism via induction of lincRNA-p21. Finally, lincRNA-p21 can physically bind to RelA mRNA, thus regulating its translation and assembly in the NF-κB complex (88). Taken together, current data highlight that lincRNA-p21 may serve as a potential therapeutic target for vascular injury, atherosclerosis, and potentially other inflammatory diseases such as RA.
Smooth muscle and endothelial cell–enriched migration/differentiation-associated long noncoding RNA (SENCR) is a lncRNA transcribed antisense from the 5′ end of the FLI1 gene and exists as 2 splice variants, localized predominantly in the cytoplasm (89). SENCR knockdown decreased expression of myocardin (MYOCD), a master regulator of numerous smooth muscle contractile genes, whereas several pro-migratory genes were increased. Loss-of-function studies indicated SENCR as an inhibitor of VSMC migration. SENCR-knockdown in human coronary aortic smooth muscle cells (HCASMCs) exhibited reorganization of the actin cytoskeleton with formation of lamellipodia, suggesting a role for SENCR in the regulation of VSMC differentiation and cellular motility (89). In a different study, Zou et al. have reported decreased levels of SENCR in a db/db mouse model and in VSMCs exposed to high glucose, through a mechanism involving FoxO1 regulation (90). However, this latter study requires further clarity since no mouse homologue of SENCR has been identified and the authors did not offer any details on how they identified the mouse transcript of SENCR or the exact transcript sequence used for silencing or overexpression studies, for either human or mouse SMCs employed in the study. Further studies will be needed to verify the existence and characterize the mouse isoform of SENCR in order to interpret the results from the diabetic mouse models. Interestingly, in human subjects with type 2 diabetes the expression of SENCR in the plasma was directly associated with left ventricular (LV) mass to LV end-diastolic volume ratio, a marker of cardiac remodeling (91). This suggests that SENCR may serve as an independent predictor of diastolic function and remodeling in patients with type 2 diabetes. Since the FLI1 gene that overlaps SENCR is a regulator of endothelial development, the expression of SENCR is also markedly regulated during endothelial commitment (65). Although SENCR does not control the pluripotency of pluripotent cells, its overexpression significantly potentiated early mesodermal and endothelial commitment, and induced HUVEC proliferation, migration, and angiogenesis (65). Collectively, these findings suggest SENCR may serve as a master regulator of VSMC and EC differentiation with potential implications in diabetes.
Growth arrest–specific 5 (GAS5) is a lncRNA that plays important roles in several biological processes including apoptosis, cell proliferation, and differentiation, cell growth arrest (92, 93) and it was recently observed to regulate vascular remodeling in hypertension (94). Silencing of GAS5 accelerated the microvascular dysfunction in a hypertension rat model, as shown by increased capillary leakage and retinal neovascularization. In vitro, GAS5 knockdown regulates VSMC dedifferentiation, accelerating VSMC proliferation and migration, and decreasing the expression of contractile marker proteins including α-smooth muscle actin and calponin by the β-catenin signaling pathway (94). GAS5 knockdown also reversed apoptosis in response to hypoxia stress and partially reversed the H2O2-induced reduction of VSMC and EC viability. Similar results were observed by a different group, where GAS5 regulated the VSMC proliferation and migration through AnnexinA2, a Ca2+-dependent RNA-binding protein (95). GAS5 expression was also decreased in cardiac fibroblasts treated with TGF-β1 and in rat cardiac fibrosis. GAS5 overexpression inhibited the cardiac fibroblast proliferation by decreasing the expression of miR-21 and indirectly regulating one of its targets, PTEN, suggesting the importance of GAS5 in cardiac and vascular remodeling (96). GAS5 was also observed to regulate SMC differentiation from mesenchymal progenitor cells, by modulating the TGF-β/Smad3 signaling pathway (97). Overexpression of GAS5 reduced, while knockdown of GAS5 increased, the expression of SMC contractile markers. Mechanistically, GAS5 binds competitively to the TGF-β effector Smad3 via multiple RNA Smad–binding elements (rSBEs), which prevents Smad3 from binding to the SBE in the promoter regions of TGF-β–responsive genes, resulting in suppression of SMC marker gene transcription and, consequently, inhibition of TGF-β/Smad3-mediated SMC differentiation.
Myocardin-induced Smooth muscle LncRNA, Inducer of Differentiation (MYOSLID) is another lncRNA recently discovered to regulate SMC differentiation in a (MYOCD)/serum response factor (SRF)-responsive manner (22). MYOSLID is a direct transcriptional target of both TGF-β/SMAD and MYOCD/SRF pathways, regulating the HCASMC contractile phenotype. Although MYOSLID does not affect gene expression of MYOCD and SRF transcription factors, its depletion in VSMCs disrupted actin stress fiber formation and blocked nuclear translocation of MYOCD-related transcription factor A (MKL1). Functional studies revealed that MYOSLID promotes VSMC differentiation and inhibits VSMC proliferation. In human samples from patients with end-stage renal disease MYOSLID expression was reduced in failed human arteriovenous fistula samples compared with healthy veins, verifying its implication in vascular disease.
Smooth muscle–induced lncRNA enhanced replication (SMILR) is highly expressed in VSMCs after interleukin-1α and PDGF stimulation. SMILR expression increased in both the cytoplasm and nucleus after stimulation, and was also released in conditioned media. Knockdown of SMILR reduced VSMC proliferation and the expression of the nearby gene HAS2 (98), with no change in the expression of isoforms HAS1 and HAS3, HAS2-AS1 lncRNA, or the ZHX2 gene, indicating the specificity of SMILR silencing for HAS2. SMILR expression is increased in human samples from patients with unstable atherosclerotic plaques and in plasma from patients with high plasma C-reactive protein levels compared to control subjects. Taken together, these findings suggest that SMILR regulates VSMC proliferation with potential implications in vascular injury and atherogenesis, although future studies are required to verify a definitive role in relevant disease models.
4 LncRNAs in inflammatory cells and lipid metabolism
Accumulation of immune cells and low-density lipoproteins (LDL) in the intima occurs during the first stage of plaque formation. Products of oxidative modification of LDL (oxLDL) may activate endothelial cells (ECs) and VSMCs, and native LDL epitopes may instigate adaptive immune responses (109). In response to both biochemical and biomechanical stimuli, EC activation triggers the expression of a number of adhesion molecules, mediating the recruitment of leukocytes to sites of inflammation. Monocytes and T cells may bind to these adhesion molecules and in combination with locally produced chemokines migrate into the arterial intima (110). These mononuclear cells differentiate into macrophages induced by macrophage colony-stimulating factor secreted by ECs and VSMCs (111). Scavenger receptor expression on macrophages enables the uptake of oxLDL particles, inducing the formation of foam cells, and intracellular cholesterol accumulation. Furthermore macrophages and dendritic cells activate T cells to a T helper 1 (TH1) cell response, promoting the production of IFN-γ, TNF-α, and expression of CD40 ligand. Mouse models and plaque analysis in humans revealed that TH1-type cytokines as of IFN-γ, TNF-α, interleukin-12 (IL-12), IL-15, and IL-18 dominate over a few TH2-type cytokines (e.g. IL-4), suggesting that atherosclerosis is a TH1-cell-driven disease (112) (113). The non-coding RNA genome provides opportunities to identify new mediators involved in both innate and adaptive immunity in the multistep disease progression of atherosclerosis.
In order to identify such new mediators, Zhang et al. (114) performed genome wide association studies (GWAS) in a database for atherosclerosis-associated SNPs. Among them, they identified the SNP rs2850711, which lies within the locus of the lncRNA linc00305. linc00305 expression was significantly increased in human atherosclerotic plaques compared to normal artery samples based on RT-qPCR of whole tissue sections. In addition, linc00305 was enriched in PBMCs from patients suffering form atherosclerosis compared to healthy individuals. Moreover, linc00305 was highest expressed in monocyte-like THP-1 cells compared to ECs and VSMCs and in CD14-positive monocytes isolated from cord blood. LPS stimulation induced its expression (114); however, it remains unknown whether linc00305 is implicated in polarization of monocytes. Gain-of-function profiling studies revealed that biological pathways involving inflammation were induced. Because treatment with BAY11-7082, an inhibitor for NF-κB abolished the induction of pro-inflammatory genes, the authors suggest that linc00305 mediates its function in a NF-κB-dependent manner. However, additional studies are required to fully understand whether alternative NF-κB signaling pathways may be involved in response to other pro-inflammatory stimuli. Functionally, THP-1 cells overexpressing linc00305 lead to a phenotypic switch of VSMCs from the contractile to the synthetic phenotype in a co-culture experiment. Mechanistically, biotinylated linc00305 bound to lipocalin-interacting membrane receptor (LIMR) in HeLa cells. LIMR itself was found to bind to the aryl hydrocarbon receptor repressor (AHRR), which is involved in Ahr signaling. Although LIMR and AHRR both increased NF-κB luciferase activity, linc00305 alone had no significant effect on NF-κB activity, but combined with LIMR and AHRR it markedly increased its activity (114). These findings suggest that although linc00305 binding to LIMR is beneficial, it is not required for Ahr-mediated regulation of the NF-κB signaling pathway. Taken together, while these findings suggest a role of linc00305 in the progression of atherosclerosis based on expression data and a SNP associated with atherosclerosis, further investigation is required to address causality and to decipher precisely whether linc00305 observed effects may be due to regulation of overlapping antisense transcripts such as lincRNA01924 and AC100848.
After RNA-Seq profiling of macrophages stimulated with Pam3CSK4, a ligand for TLR2, Carpenter et al. identified the lincRNA-Cox2, which is in close proximity to the Cox2 loci, among the top-induced lncRNAs candidates. TLR7/8 activation by LPS stimulation induced lincRNA-Cox2 expression in both dendritic cells and BMDM in a similar pattern as Ptgs2 (115) (116). However, no regulation of lincRNA-Cox2 could be observed by activation of TLR3 signaling using poly(I:C). In addition, lincRNA-Cox2 expression was shown to be MyD88- and NF-κB–dependent. shRNA-mediated silencing of lincRNA-Cox2 did not affect Cox2 expression, but significantly increased the expression of pro-inflammatory genes such as Irf7 and CCL5 in unstimulated BMDMs, while Pam3CSK4-induced Tlr1 and IL-6 expression was attenuated. Complementary lincRNA-Cox2 gain-of-function experiments decreased expression levels of these genes in macrophages. Taken together, these results demonstrate that lincRNA-Cox2 represses Ccl5, while simultaneously enhancing the expression of TLR-induced IL-6. Mechanistically, lincRNA-Cox2 binds to hnRNP-A/B and hnRNPA2/B1 in cytoplasmic and nuclear compartments as well as affecting IKB-α and SWItch/Sucrose NonFermentable (SWI/SNF) complex stability in the cytosol, suggesting a regulatory role of lincRNA-Cox2 as a co-activator of NF-κB or inducing SWI/SNF-associated chromatin remodeling (117) (118). In sum, these studies identified lincRNA-Cox2 as a critical component of the inflammatory response. However, its causal role in CVD disease is not elucidated and requires further investigation.
In a similar experimental setup as described above (115), Li et al. (119) analyzed the expression of lncRNAs in PMA-activated THP-1 cells using the TLR2 ligand Pam3CSK4 by microarray profiling. They found a panel of 159 differentially expressed lincRNAs (i.e. 1.9-fold up/-down; p-value<0.05). Out of the 159 lncRNA candidates they selected 20 candidates based on their genomic flanking genes within the range of 1 Mb. Those 20 candidates were further validated by RT-qPCR and their expression was compared across different tissues. Subsequently, loss-of-function studies for nine out of the 20 lncRNA candidates revealed that the linc1992, later named as TNFα and hnRNPL related immune-regulatory lincRNA (THRIL) was the lncRNA candidate that most significantly reduced TNFα cytokine expression (119). This approach of stratifying microarray hits based on their proximity to the lncRNA locus assumes the lncRNA acts in a cis and not in trans. Moreover, TNFα expression as a read-out for systematical identification of lncRNAs involved in the activation of innate immune signaling in THP1 macrophages may be overstated. Mechanistically, THRIL was shown to form a complex with hnRNPL and by silencing either of those two components, binding to the TNFα promoter was compromised using ChIP. Those findings suggest that THRIL and hnRNPL form a RNP complex that regulates TNFα transcription by binding to its promoter. Clinically, THRIL expression correlated with the severity of symptoms in patients with Kawasaki disease, an acute inflammatory disease of childhood (119). Future investigations will be of interest to solidify other top hit candidates from those screening platforms and to verify whether those lncRNAs may also have translational value in the context of chronic inflammation such as atherosclerosis or diabetes.
From expression analysis of human atherosclerotic plaques, GAS5 and HOXC-AS1 were found to be differentially expressed compared to healthy controls (120) (121) (122). HOXC-AS1 was found to be expressed lower in carotid atherosclerotic whole tissue sections compared to renal arterial intima tissue using microarray analysis. This lncRNA lies antisense to HOXC6 loci. Both their expression was significantly reduced in THP-1 cells upon oxLDL treatment (121); however, no further results were obtained to show any causal link of HOXC-AS1 and HOX6 expression. The rationale for choosing this particular lncRNA was not clear. While microarray-based methods require less bioinformatics and data processing (which may be an advantage), they typically rely on one transcript or isoform per lncRNA of which annotation may not always be as accurate compared to deep-sequencing-based transcriptomic analysis. In contrast to HOXAC-AS1, GAS5 expression was higher in human atherosclerotic plaques compared to healthy individuals (122). An independent study showed that GAS5 expression levels were reduced in mouse BMDMs polarized to M2bM (LPS+IC stimulation) compared to quiescent macrophages and other subpopulations of M2 and M1. Interestingly overexpression of GAS5 abrogated LPS+IC-induced polarization to M2bM, suggesting that GAS5 plays a role in macrophage polarization (123). However, loss-of-function studies are missing to support this hypothesis. Other studies have showed that silencing of GAS5 blocked oxLDL-induced apoptosis in THP-1 cells, which could be accelerated by overexpressing GAS5 (120).
Disordered lipid metabolism is one of the pathological processes contributing to the onset and progression of atherosclerosis (124). Sallam et al. identified a lncRNA named LeXis that regulates liver X receptor (LXR)-mediated cholesterol synthesis. LeXis promotes cholesterol efflux and inhibition of cholesterol biosynthesis by binding to a heterogeneous ribonucleoprotein named RALY. Mechanistically, LeXis binds to RALY, which affects its ability to interact with DNA, and in turn prevented cholesterol synthesis via transcriptional control of a subset of metabolic genes. As a consequence, total serum cholesterol was reduced in mice ectopically overexpressing LeXis (125). Recently, adenoviral-mediated LeXis overexpression in the liver using a thyroxine-binding globulin promoter, significantly reduced aortic lesion size determined by Oil-red O staining. In line with previous findings, hepatic sterol content and levels of serum cholesterol were significantly lower in these mice (126). Another lncRNA involved in lipid metabolism is LncLSTR, which regulates apoC2 expression through an FXR-mediated pathway, to modulate triglyceride levels in a hyperlipidemia mouse model (127). LncLSTR forms a molecular complex with TDP-43 to regulate expression of Cyp8b1, a key enzyme in the bile acid synthesis (XX add ref). Finally, the expression of the lncRNA Gm16551 suppresses lipogenesis and is induced by SREBP1c in hepatocytes, whereas its expression is reduced in livers of obese mice (128). Collectively, these elegant studies raise the possibility for long-term lncRNA therapy in mice. Future studies that can overexpress lncRNAs in the liver or vessel wall may provide a novel therapeutic approach for regulating vascular inflammation in CVD.
5. Challenges and Opportunities
Accumulating studies suggest that lncRNAs are important regulators of key biological processes vital for maintaining cellular homeostasis (134) (135) (136). Disease-associated sequence variants exist in the non-coding genome where lncRNAs reside, raising the possibility that lncRNAs may provide genetic links to disease susceptibility that were initially dismissed as “junk” transcripts (137) (134) (134). One example is the ANRIL lncRNA that resides in the chromosome 9p21 CAD susceptibility locus (138). Recent studies provide incipient insights into lncRNA function and their regulatory effects in vascular-related diseases. As detailed above, lncRNAs have been identified as key regulators in various biological processes relevant to vascular homeostasis such as endothelial cells dysfunction, VSMC phenotypes, macrophage differentiation, and lipid metabolism (Figure 2). Several lncRNAs have shown important regulation in the plasma or circulating cells, hence they hold promise as potential biomarkers and therapeutic targets for stage-specific vascular disease (139). However, several challenges exist to the lncRNA field, including their relatively low level of cellular expression as compared to mRNAs. An important challenge here is the standardization of detection methods for reliable reporting. However, as the sensitivity of RNA-seq, microarray technologies, and bioinformatics has gradually increased over recent years, so too has the power to capture lncRNAs even in low-abundant cell types (140)
Although lncRNA regulation has been reported in human plasma and tissue samples and different disease models, their function and mechanism of action are only known for a few lncRNAs (28). The poor conservation between species and the fact that most lncRNAs have various transcript variants challenges the identification of specific biological functions and mechanisms of action (141) and can limit their translational impact in vascular disease. However, often the neighboring genomic locus (the so-called “synteny”) is well-conserved (142), and recent bioinformatics approaches of comparative genomics including use of secondary and tertiary structures can identify lncRNA homologs in different species (143). Future investigation into the precise expression of lncRNA in a cell-type or tissue-type specific manner in human subjects during disease progression and regression will inform lncRNA kinetics and potential use as biomarkers in diagnosis, prognostication, and response to therapies. Moreover, a broader perspective of how lncRNAs interact with RNA, DNA, and proteins to exact functional responses in vascular cells will be important for generating disease-specific networks or interactomes. Technical hurdles to identify such lncRNA interactors using ChiRP, RAP, and RIP pull-down approaches have already made substantial impact in the field (144).
While the translational potential of lncRNAs remains to be elucidated, RNA-based therapeutics have already been approved by the Food and Drug Administration (FDA) and may be successfully implemented for lncRNA regulation in vascular disease. RNA-based silencing strategies include antisense oligonucleotides (ASO), LNA (locked nucleic acid), aptamers or siRNA/shRNA, with great improvements in recent years in terms of stability, tolerability, reduced immunogenicity, and off-target effects (145). One example is an ASO that targets a liver-specific ligand, the liver-specific asialoglycoprotein receptor (ASGPR)) that confers strong efficacy and reasonable safety (145) (146). Another example is Mipomersen, an FDA-approved ASO that targets apolipoprotein B, used for the treatment of homozygous familial hypercholesterolemia (147).
For therapeutic gain-of-function purposes, lncRNAs can be delivered by viral vectors such as lentivirus or by non-viral vectors such as polymeric or lipid nanoparticles. While viral vectors induce immunogenicity (148), there is great anticipation that non-viral vectors with different chemical modifications may be used successfully in clinical trials (149). Important lessons can be learned from mRNA delivery and vaccination studies (149) (150). Challenges for lncRNA delivery remain with respect to their efficiency and tissue specificity. However, these challenges may be overcome using chemical modifications and/or nanoparticles targeted to specific ligands overexpressed by cells in the vessel wall in response to relevant stimuli (151). Finally, new gene editing tools such as CRISPR can be successfully used to manipulate lncRNA expression by both loss- or gain-of function approaches with great specificity and efficiency, at least in vitro (130) (45) (152). Similar delivery issues remain for use of gene editing tools in vivo in the vasculature.
Despite these challenges, accumulating findings from studies using gain- or loss-of function approaches suggest that lncRNAs indeed contribute to vascular dysfunction and their therapeutic regulation can prevent or repair specific pathological processes that lead to maladaptive vascular disease remodeling (126).
6. Conclusions
LncRNAs have been identified as key regulators in biological and pathological processes in a range of vascular disease states. Given the growing massive number of mammalian lncRNAs, the increasing correlation with GWAS hits, and their diverse mechanisms of action in the nucleus, cytosol, or exosomes, increased efforts are desperately needed to define their expression, function, and interactomes. Integrating knowledge of lncRNAs with other non-coding and protein-coding genes will be critical to our understanding of the biological orchestration necessary to finely-tune the vessel wall in health and disease.
Table 1.
List of lncRNAs potentially implicated in endothelial dysfunction and their regulatory mechanisms.
| lncRNA | Target cell type | Regulatory effect | Mechanism | Reference |
|---|---|---|---|---|
| ALT1 | ECs | controls ECs cell cycle and proliferation | targeting ACE2 and Cyclin D1 | (54) |
| ASncmt RNA-2 | ECs | induced in vascular aging and senescence | potentially non-canonical precursor of hsa-miR-4485 and hsa-miR-1973 | (55) |
| cANRIL | VSMCs and PBMCs | atheroprotection; induces vascular cell apoptosis | not investigated | (56) |
| FLJ11812 | ECs | regulates autophagy | binding to miR-4459 and targeting ATG13 | (57) |
| H19 | ECs; VSMCs | increases proliferation and decreases apoptosis; regulated in hypoxia | host gene for miR-675, targeting PTEN; activates p38-MAPK and NF-kB signaling pathways | (46-50) |
| HIF1A-AS2 | ECs | promotes angiogenesis in hypoxia conditions | sponging miR-153-3p | (58) |
| HOTAIR | ECs; PBMCs | decreased in ECs from athero plaques; regulates ECs proliferation and migration | TSLP activates HOTAIR transcription through PI3K/AKT-IRF1 pathway | (59) |
| HOTTIP | ECs | regulates ECs proliferation and migration | Wnt/β-catenin pathway | (60) |
| IGF2-AS | ECs | increased in myocardial microvascular endothelial cells of diabetes rat model; controls angiogenesis | not investigated | (61) |
| LINC00341 | ECs | anti-inflammatory effects | LINC00341 guides EZH-2 to the promoter region of VCAM1 | (15) |
| LINC00305 | ECs | regulates hypoxia-induced apoptosis | sponging of miR-136 | (19) |
| LOC100129973 | ECs | suppression of apoptosis | sponging 4707-5p and miR-4767 | (62) |
| MALAT1 | ECs | controls ECs proliferation and cell cycle; inhibits apoptosis; protects the endothelium against ox-LDL-induced dysfunction; controls vascular homeostasis in diabetic rats and mice undergoing hindlimb ischemia | sponge for miR-22-3p; controls p38 and AKT phosphorylation and signaling pathways | (33-40) |
| MANTIS | ECs | controls ECs migration, angiogenic sprouting, and tube formation | interacts with Brg1 and regulates SMAD6, COUP-TFII, SOX18 | (45) |
| MEG3 | ECs | controls vascularization and angiogenesis, EC proliferation, and senescence | sponge for miR-9; activates the PI3K/AKT signaling pathway | (41-44) |
| MIAT | ECs | Regulates angiogenesis and EC function in diabetes | ceRNA for miR-150-5p | (63) |
| PINC | ECs | EC apoptosis; Kawasaki disease | not investigated | (64) |
| RNCR3 | ECs | atherosclerosis; EC proliferation and migration; due to RNCR3 exonic overlap with miR-3078 and miR-124a, it is unclear if phenotypes are related to RNCR3 or regulation by these microRNAs | ceRNA for miR-185-5p, forming a feedback loop with KLF2; | (51) |
| SENCR | ESCs | CAD; ECs proliferation, migration and angiogenesis | SENCR regulates ESC differentiation into EC | (65) |
| SIRT1-AS | EPCs | EPCs senescence, proliferation, and migration | sponge for mir-22, (relieving miR-22-induced SIRT1 (downregulation) | (66) |
| TGFB2-OT1 | ECs | Regulates autophagy and inflammation | ceRNA for Mir3960, Mir4488 and Mir4459 | (67) |
| TUG1 | ECs | ECs apoptosis, atherosclerosis | potentially a sponge for miR-26a | (68) |
ECs: endothelial cells; ESCs: embryonic stem cells; EPCs: endothelial progenitor cells; PBMCs: peripheral blood mononuclear cells; VSMCs: vascular smooth muscle cells.
Table 2.
List of lncRNAs potentially implicated in vascular injury and their regulatory mechanisms.
| lncRNA | Target cell type | Regulatory effect | Mechanism | Reference |
|---|---|---|---|---|
| ANRIL | HAVSMCs | independent risk factor for CAD; controls VSMC proliferation | binds to CBX7 and SUZ12, components of the PRC-1 and PRC-2 respectively | (20, 76, 81-84) |
| cANRIL | VSMCs; macrophages | atheroprotection; impaires ribosome biogenesis; inhibits proliferation and induces apoptosis | binds to PES1, a 60S-preribosomal assembly factor and induces p53 activation and nucleolar stress | (24) |
| Gas5 | VSMC | regulates vascular remodeling in hypertension; accelerates VSMC proliferation and migration | regulates AnnexinA2; decreases miR-21 and increases one of its targets, PTEN | (92-96) |
| HAS2-AS1 | HASMC | SMC homeostasis | altering the chromatin structure around the HAS2 proximal promoter via O-GlcNAcylation and acetylation | (99) |
| HIF1A-AS2 | VSMC | thoracic aortic aneurysms; controls proliferation and apoptosis | interaction with BRG-1 | (100) |
| HOTAIR | VSMC | downregulated in STAA (sporadic thoracic aortic aneurysm); | regulates extracellular matrix remodelling | (101) |
| HypERLi nc | pericytes | role in idiopathic pulmonary arterial hypertension; heart failure | ER stress regulator | (102) |
| Linc-p21 | VSMC, macrophages and HUVECs | promote apoptosis and repress proliferation | associates with hnRNP-K to repress hundreds of genes in the p53 pathway; feedback mechanism: association with MDM2 to depress p53; endogenous sponge for miR-130b; binds to RelA mRNA regulating NFkB | (85-88) |
| Lnc-Ang362 | VSMCs | VSMC proliferation | host transcript for miR-221 and miR-222, | (103) |
| LnRPT | PASMCs | PASMC proliferation | inhibits the genes Notch3, Jag1, CCNA2 | (104) |
| MEG3 | PASMCs | regulates PASMCs cell cycle, proliferation and migration | regulates p53 pathway | (105) |
| MYOSLI D | CASMCs | promotes VSMC differentiation and inhibits proliferation; actin stress fiber formation | abrogates TGF-β1–induced SMAD2 phosphorylation; modulate nuclear translocation of MKL1 | (22) |
| SENCR | VSMCs | increases proliferation, inhibits migration | decreases FoxO1 and its binding to H3 histone; regulates myocardin | (65, 89-91) |
| SMILR | HSVSMCs | regulates proliferation; decreased in athero plaques | decreases the expression of proximal gene HAS2 | (98), |
| TCONS_34812 | PASMC | proliferation and apoptosis | increase the expression of TF Stox1 | (106) |
| TUG1 | VSMCs | VSMC homeostasis | TUG1 supports the interaction of EZH2 and α-actin, and their co-localization | (107) |
| XR007793 | VSMCs | hypertension, VSMC proliferation and migration | STAT2, LMO2, IRF7 | (108) |
VSMCs: vascular smooth muscle cells; HSVSMCs: human saphenous vein smooth muscle cells; PASMC: pulmonary aortic smooth muscle cells; CASMCs: coronary artery smooth muscle cells; HAVSMCs: human aortic vascular smooth muscle cells
Table 3.
List of lncRNAs regulating leukocyte activation and lipid metabolism and their regulatory mechanisms.
| lncRNA | Target cell type | Regulatory effect | Mechanism | Reference |
|---|---|---|---|---|
| GAS5 | BMDM, THP-1 | high expression in human atherosclerotic plaques, macrophage M2bM polarization, apoptosis | NMD pathway, HMBG1, miR-222 | (120-123) |
| HOXC-AS1 | THP-1 | low expression in human atherosclerotic plaques; suppresses Ox-LDL-Induced cholesterol accumulation | unknown | (121) |
| IL7-AS | THP-1, RAW264. 7, A549 | involved in inflammatory response | unknown | (129) |
| LeXis | Hepa1-6 | regulates cholesterol synthesis | binds to RALY | (124, 125) (126) |
| Linc00305 | THP-1 | SNP rs2850711 for atherosclerosis; promotes monocyte activation | binds to LIMR, which activates NF-kB through Ahr signaling | (114) |
| lincRNA-Cox2 | THP-1, dendritic cells, BMDM | mediates immune response; promotes inflammation in macrophages; | co-activator of NF-kB by forming a complex with hnRNPA2/B1 | (115, 116, 118, 130) |
| lncRNA OTTHUMT00000387022 | PBMC, plasma, THP-1 | biomarker for CAD; pro-inflammatory in macrophages | unknown | (131) |
| lincRNA-TNFAIP3 | RAW264. 7, BV2 | likely regulating inflammatory genes | involved in NF-kB/HMGB1 pathway | (132) |
| PACER | U937 | controls COX-2 mRNA transcription and monocyte activation by LPS | chromatin remodeling; regulation of NF-kB through p50 component, binds p300 | (133) |
| THRIL | THP-1 | Kawasaki disease; transcriptional control of TNFa | fomrs complex with hnRNPL | (119). |
| lncLSTR | Primary hepatocytes | regulates apoC2 expression through FXR-mediated pathway. Modulates triglyceride levels in a hyperlipidemia mouse model; | it forms a molecular complex with TDP-43 to regulate expression of Cyp8b1, a key enzyme in the bile acid synthesis' | (127) |
| Gm16551 | Primary hepatocytes | upregulated by SREBP1c in hepatocytes; downregulated in livers of obese mice; suppresses lipogenesis. | unknown | (128) |
BMDMs: bone marrow derived macrophages; PBMCs: peripheral blood mononuclear cells.
Acknowledgments
This work was supported by the National Institutes of Health (HL115141, HL117994, HL134849, and GM115605 to M.W.F.), the Arthur K. Watson Charitable Trust (to M.W.F.), and the Dr. Ralph & Marian Falk Medical Research Trust (to M.W.F.).
Footnotes
Conflicts of Interest: The authors have no conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Manduteanu I, Simionescu M. Inflammation in atherosclerosis: a cause or a result of vascular disorders? J Cell Mol Med. 2012;16:1978–1990. doi: 10.1111/j.1582-4934.2012.01552.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hartge MM, Unger T, Kintscher U. The endothelium and vascular inflammation in diabetes. Diab Vasc Dis Res. 2007;4:84–88. doi: 10.3132/dvdr.2007.025. [DOI] [PubMed] [Google Scholar]
- 3.Coussens LM, Werb Z. Inflammation and cancer. Nature. 2002;420:860–867. doi: 10.1038/nature01322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Virchow R. Cellular pathology. As based upon physiological and pathological histology. Lecture XVI--Atheromatous affection of arteries. 1858. Nutr Rev. 1989;47:23–25. doi: 10.1111/j.1753-4887.1989.tb02747.x. [DOI] [PubMed] [Google Scholar]
- 5.Ridker PM, Everett BM, Thuren T, MacFadyen JG, Chang WH, Ballantyne C, Fonseca F, Nicolau J, Koenig W, Anker SD, et al. Antiinflammatory Therapy with Canakinumab for Atherosclerotic Disease. N Engl J Med. 2017;377:1119–1131. doi: 10.1056/NEJMoa1707914. [DOI] [PubMed] [Google Scholar]
- 6.Okazaki Y, Furuno M, Kasukawa T, Adachi J, Bono H, Kondo S, Nikaido I, Osato N, Saito R, Suzuki H, et al. Analysis of the mouse transcriptome based on functional annotation of 60,770 full-length cDNAs. Nature. 2002;420:563–573. doi: 10.1038/nature01266. [DOI] [PubMed] [Google Scholar]
- 7.Kapranov P, Cheng J, Dike S, Nix DA, Duttagupta R, Willingham AT, Stadler PF, Hertel J, Hackermuller J, Hofacker IL, et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science. 2007;316:1484–1488. doi: 10.1126/science.1138341. [DOI] [PubMed] [Google Scholar]
- 8.Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, Tanzer A, Lagarde J, Lin W, Schlesinger F, et al. Landscape of transcription in human cells. Nature. 2012;489:101–108. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang Y, Sun X, Icli B, Feinberg MW. Emerging Roles for MicroRNAs in Diabetic Microvascular Disease: Novel Targets for Therapy. Endocr Rev. 2017;38:145–168. doi: 10.1210/er.2016-1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Icli B, Feinberg MW. MicroRNAs in dysfunctional adipose tissue: cardiovascular implications. Cardiovasc Res. 2017;113:1024–1034. doi: 10.1093/cvr/cvx098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feinberg MW, Moore KJ. MicroRNA Regulation of Atherosclerosis. Circ Res. 2016;118:703–720. doi: 10.1161/CIRCRESAHA.115.306300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sun X, Belkin N, Feinberg MW. Endothelial microRNAs and atherosclerosis. Curr Atheroscler Rep. 2013;15:372. doi: 10.1007/s11883-013-0372-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kugel JF, Goodrich JA. The regulation of mammalian mRNA transcription by lncRNAs: recent discoveries and current concepts. Epigenomics. 2013;5:95–102. doi: 10.2217/epi.12.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Han P, Chang CP. Long non-coding RNA and chromatin remodeling. RNA Biol. 2015;12:1094–1098. doi: 10.1080/15476286.2015.1063770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang TS, Wang KC, Quon S, Nguyen P, Chang TY, Chen Z, Li YS, Subramaniam S, Shyy J, Chien S. LINC00341 exerts an anti-inflammatory effect on endothelial cells by repressing VCAM1. Physiol Genomics. 2017;49:339–345. doi: 10.1152/physiolgenomics.00132.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Willingham AT, Orth AP, Batalov S, Peters EC, Wen BG, Aza-Blanc P, Hogenesch JB, Schultz PG. A strategy for probing the function of noncoding RNAs finds a repressor of NFAT. Science. 2005;309:1570–1573. doi: 10.1126/science.1115901. [DOI] [PubMed] [Google Scholar]
- 17.Gonzalez I, Munita R, Agirre E, Dittmer TA, Gysling K, Misteli T, Luco RF. A lncRNA regulates alternative splicing via establishment of a splicing-specific chromatin signature. Nat Struct Mol Biol. 2015;22:370–376. doi: 10.1038/nsmb.3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang GQ, Wang Y, Xiong Y, Chen XC, Ma ML, Cai R, Gao Y, Sun YM, Yang GS, Pang WJ. Sirt1 AS lncRNA interacts with its mRNA to inhibit muscle formation by attenuating function of miR-34a. Sci Rep. 2016;6:21865. doi: 10.1038/srep21865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang BY, Jin Z, Zhao Z. Long intergenic noncoding RNA 00305 sponges miR -136 to regulate the hypoxia induced apoptosis of vascular endothelial cells. Biomed Pharmacother. 2017;94:238–243. doi: 10.1016/j.biopha.2017.07.099. [DOI] [PubMed] [Google Scholar]
- 20.Kotake Y, Nakagawa T, Kitagawa K, Suzuki S, Liu N, Kitagawa M, Xiong Y. Long non-coding RNA ANRIL is required for the PRC2 recruitment to and silencing of p15(INK4B) tumor suppressor gene. Oncogene. 2011;30:1956–1962. doi: 10.1038/onc.2010.568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gumireddy K, Li A, Yan J, Setoyama T, Johannes GJ, Orom UA, Tchou J, Liu Q, Zhang L, Speicher DW, et al. Identification of a long non-coding RNA-associated RNP complex regulating metastasis at the translational step. EMBO J. 2013;32:2672–2684. doi: 10.1038/emboj.2013.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhao J, Zhang W, Lin M, Wu W, Jiang P, Tou E, Xue M, Richards A, Jourd'heuil D, Asif A, et al. MYOSLID Is a Novel Serum Response Factor-Dependent Long Noncoding RNA That Amplifies the Vascular Smooth Muscle Differentiation Program. Arterioscler Thromb Vasc Biol. 2016;36:2088–2099. doi: 10.1161/ATVBAHA.116.307879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 24.Holdt LM, Stahringer A, Sass K, Pichler G, Kulak NA, Wilfert W, Kohlmaier A, Herbst A, Northoff BH, Nicolaou A, et al. Circular non-coding RNA ANRIL modulates ribosomal RNA maturation and atherosclerosis in humans. Nat Commun. 2016;7:12429. doi: 10.1038/ncomms12429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ahadi A, Brennan S, Kennedy PJ, Hutvagner G, Tran N. Long non-coding RNAs harboring miRNA seed regions are enriched in prostate cancer exosomes. Sci Rep. 2016;6:24922. doi: 10.1038/srep24922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem. 2012;81:145–166. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sun HJ, Hou B, Wang X, Zhu XX, Li KX, Qiu LY. Endothelial dysfunction and cardiometabolic diseases: Role of long non-coding RNAs. Life Sci. 2016;167:6–11. doi: 10.1016/j.lfs.2016.11.005. [DOI] [PubMed] [Google Scholar]
- 28.Haemmig S, Simion V, Yang D, Deng Y, Feinberg MW. Long noncoding RNAs in cardiovascular disease, diagnosis, and therapy. Curr Opin Cardiol. 2017;32:776–783. doi: 10.1097/HCO.0000000000000454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Freedman JE, Miano JM. Challenges and Opportunities in Linking Long Noncoding RNAs to Cardiovascular, Lung, and Blood Diseases. Arterioscler Thromb Vasc Biol. 2017;37:21–25. doi: 10.1161/ATVBAHA.116.308513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Libby P. Mechanisms of acute coronary syndromes and their implications for therapy. N Engl J Med. 2013;368:2004–2013. doi: 10.1056/NEJMra1216063. [DOI] [PubMed] [Google Scholar]
- 31.Mullick AE, Soldau K, Kiosses WB, Bell TA, 3rd, Tobias PS, Curtiss LK. Increased endothelial expression of Toll-like receptor 2 at sites of disturbed blood flow exacerbates early atherogenic events. J Exp Med. 2008;205:373–383. doi: 10.1084/jem.20071096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Libby P. Inflammation in atherosclerosis. Arterioscler Thromb Vasc Biol. 2012;32:2045–2051. doi: 10.1161/ATVBAHA.108.179705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Michalik KM, You X, Manavski Y, Doddaballapur A, Zornig M, Braun T, John D, Ponomareva Y, Chen W, Uchida S, et al. Long noncoding RNA MALAT1 regulates endothelia l cell function and vessel growth. Circ Res. 2014;114:1389–1397. doi: 10.1161/CIRCRESAHA.114.303265. [DOI] [PubMed] [Google Scholar]
- 34.Liu JY, Yao J, Li XM, Song YC, Wang XQ, Li YJ, Yan B, Jiang Q. Pathogenic role of lncRNA-MALAT1 in endothelial cell dysfunction in diabetes mellitus. Cell Death Dis. 2014;5:e1506. doi: 10.1038/cddis.2014.466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Puthanveetil P, Chen S, Feng B, Gautam A, Chakrabarti S. Long non-coding RNA MALAT1 regulates hyperglycaemia induced inflammatory process in the endothelial cells. J Cell Mol Med. 2015;19:1418–1425. doi: 10.1111/jcmm.12576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xin JW, Jiang YG. Long noncoding RNA MALAT1 inhibits apoptosis induced by oxygen-glucose deprivation and reoxygenation in human brain microvascular endothelial cells. Exp Ther Med. 2017;13:1225–1234. doi: 10.3892/etm.2017.4095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tang Y, Jin X, Xiang Y, Chen Y, Shen CX, Zhang YC, Li YG. The lncRNA MALAT1 protects the endothelium against ox-LDL-induced dysfunction via upregulating the expression of the miR-22-3p target genes CXCR2 and AKT. FEBS Lett. 2015;589:3189–3196. doi: 10.1016/j.febslet.2015.08.046. [DOI] [PubMed] [Google Scholar]
- 38.Li Z, Li J, Tang N. Long noncoding RNA Malat1 is a potent autophagy inducer protecting brain microvascular endothelial cells against oxygen-glucose deprivation/reoxygenation-induced injury by sponging miR-26b and upregulating ULK2 expression. Neuroscience. 2017;354:1–10. doi: 10.1016/j.neuroscience.2017.04.017. [DOI] [PubMed] [Google Scholar]
- 39.Icli B, Nabzdyk CS, Lujan-Hernandez J, Cahill M, Auster ME, Wara AK, Sun X, Ozdemir D, Giatsidis G, Orgill DP, et al. Regulation of impaired angiogenesis in diabetic dermal wound healing by microRNA-26a. J Mol Cell Cardiol. 2016;91:151–159. doi: 10.1016/j.yjmcc.2016.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Icli B, Wara AK, Moslehi J, Sun X, Plovie E, Cahill M, Marchini JF, Schissler A, Padera RF, Shi J, et al. MicroRNA-26a regulates pathological and physiological angiogenesis by targeting BMP/SMAD1 signaling. Circ Res. 2013;113:1231–1241. doi: 10.1161/CIRCRESAHA.113.301780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Qiu GZ, Tian W, Fu HT, Li CP, Liu B. Long noncoding RNA-MEG3 is involved in diabetes mellitus-related microvascular dysfunction. Biochem Biophys Res Commun. 2016;471:135–141. doi: 10.1016/j.bbrc.2016.01.164. [DOI] [PubMed] [Google Scholar]
- 42.He C, Yang W, Yang J, Ding J, Li S, Wu H, Zhou F, Jiang Y, Teng L. Long Noncoding RNA MEG3 Negatively Regulates Proliferation and Angiogenesis in Vascular Endothelial Cells. DNA Cell Biol. 2017;36:475–481. doi: 10.1089/dna.2017.3682. [DOI] [PubMed] [Google Scholar]
- 43.Boon RA, Hofmann P, Michalik KM, Lozano-Vidal N, Berghauser D, Fischer A, Knau A, Jae N, Schurmann C, Dimmeler S. Long Noncoding RNA Meg3 Controls Endothelial Cell Aging and Function: Implications for Regenerative Angiogenesis. J Am Coll Cardiol. 2016;68:2589–2591. doi: 10.1016/j.jacc.2016.09.949. [DOI] [PubMed] [Google Scholar]
- 44.Gordon FE, Nutt CL, Cheunsuchon P, Nakayama Y, Provencher KA, Rice KA, Zhou Y, Zhang X, Klibanski A. Increased expression of angiogenic genes in the brains of mouse meg3-null embryos. Endocrinology. 2010;151:2443–2452. doi: 10.1210/en.2009-1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Leisegang MS, Fork C, Josipovic I, Richter F, Preussner J, Hu J, Miller MJ, Epah JN, Hofmann P, Gunther S, et al. Long Noncoding RNA MANTIS Facilitates Endothelial Angiogenic Function. Circulation. 2017 doi: 10.1161/CIRCULATIONAHA.116.026991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gao W, Zhu M, Wang H, Zhao S, Zhao D, Yang Y, Wang ZM, Wang F, Yang ZJ, Lu X, et al. Association of polymorphisms in long non-coding RNA H19 with coronary artery disease risk in a Chinese population. Mutat Res. 2015;772:15–22. doi: 10.1016/j.mrfmmm.2014.12.009. [DOI] [PubMed] [Google Scholar]
- 47.Zhang Z, Gao W, Long QQ, Zhang J, Li YF, Liu DC, Yan JJ, Yang ZJ, Wang LS. Increased plasma levels of lncRNA H19 and LIPCAR are associated with increased risk of coronary artery disease in a Chinese population. Sci Rep. 2017;7:7491. doi: 10.1038/s41598-017-07611-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pan JX. LncRNA H19 promotes atherosclerosis by regulating MAPK and NF-kB signaling pathway. Eur Rev Med Pharmacol Sci. 2017;21:322–328. [PubMed] [Google Scholar]
- 49.Voellenkle C, Garcia-Manteiga JM, Pedrotti S, Perfetti A, De Toma I, Da Silva D, Maimone B, Greco S, Fasanaro P, Creo P, et al. Implication of Long noncoding RNAs in the endothelial cell response to hypoxia revealed by RNA-sequencing. Sci Rep. 2016;6:24141. doi: 10.1038/srep24141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lv J, Wang L, Zhang J, Lin R, Sun W, Wu H, Xin S. Long noncoding RNA H19-derived miR-675 aggravates restenosis by targeting PTEN. Biochem Biophys Res Commun. 2017 doi: 10.1016/j.bbrc.2017.01.011. [DOI] [PubMed] [Google Scholar]
- 51.Shan K, Jiang Q, Wang XQ, Wang YN, Yang H, Yao MD, Liu C, Li XM, Yao J, Liu B, et al. Role of long non-coding RNA-RNCR3 in atherosclerosis-related vascular dysfunction. Cell Death Dis. 2016;7:e2248. doi: 10.1038/cddis.2016.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu C, Li CP, Wang JJ, Shan K, Liu X, Yan B. RNCR3 knockdown inhibits diabetes mellitus-induced retinal reactive gliosis. Biochem Biophys Res Commun. 2016;479:198–203. doi: 10.1016/j.bbrc.2016.09.032. [DOI] [PubMed] [Google Scholar]
- 53.Shan K, Li CP, Liu C, Liu X, Yan B. RNCR3: A regulator of diabetes mellitus-related retinal microvascular dysfunction. Biochem Biophys Res Commun. 2017;482:777–783. doi: 10.1016/j.bbrc.2016.11.110. [DOI] [PubMed] [Google Scholar]
- 54.Li W, Wang R, Ma JY, Wang M, Cui J, Wu WB, Liu RM, Zhang CX, Wang SM. A Human Long Non-Coding RNA ALT1 Controls the Cell Cycle of Vascular Endothelial Cells Via ACE2 and Cyclin D1 Pathway. Cell Physiol Biochem. 2017;43:1152–1167. doi: 10.1159/000481756. [DOI] [PubMed] [Google Scholar]
- 55.Bianchessi V, Badi I, Bertolotti M, Nigro P, D'Alessandra Y, Capogrossi MC, Zanobini M, Pompilio G, Raucci A, Lauri A. The mitochondrial lncRNA ASncmtRNA-2 is induced in aging and replicative senescence in Endothelial Cells. J Mol Cell Cardiol. 2015;81:62–70. doi: 10.1016/j.yjmcc.2015.01.012. [DOI] [PubMed] [Google Scholar]
- 56.Song CL, Wang JP, Xue X, Liu N, Zhang XH, Zhao Z, Liu JG, Zhang CP, Piao ZH, Liu Y, et al. Effect of Circular ANRIL on the Inflammatory Response of Vascular Endothelial Cells in a Rat Model of Coronary Atherosclerosis. Cell Physiol Biochem. 2017;42:1202–1212. doi: 10.1159/000478918. [DOI] [PubMed] [Google Scholar]
- 57.Ge D, Han L, Huang S, Peng N, Wang P, Jiang Z, Zhao J, Su L, Zhang S, Zhang Y, et al. Identification of a novel MTOR activator and discovery of a competing endogenous RNA regulating autophagy in vascular endothelial cells. Autophagy. 2014;10:957–971. doi: 10.4161/auto.28363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li L, Wang M, Mei Z, Cao W, Yang Y, Wang Y, Wen A. lncRNAs HIF1A-AS2 facilitates the up-regulation of HIF-1alpha by sponging to miR-153-3p, whereby promoting angiogenesis in HUVECs in hypoxia. Biomed Pharmacother. 2017;96:165–172. doi: 10.1016/j.biopha.2017.09.113. [DOI] [PubMed] [Google Scholar]
- 59.Peng Y, Meng K, Jiang L, Zhong Y, Yang Y, Lan Y, Zeng Q, Cheng L. Thymic stromal lymphopoietin-induced HOTAIR activation promotes endothelial cell proliferation and migration in atherosclerosis. Biosci Rep. 2017;37 doi: 10.1042/BSR20170351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Liao B, Chen R, Lin F, Mai A, Chen J, Li H, Xu Z, Dong S. Long noncoding RNA HOTTIP promotes endothelial cell proliferation and migration via activation of the Wnt/beta-catenin pathway. J Cell Biochem. 2017 doi: 10.1002/jcb.26448. [DOI] [PubMed] [Google Scholar]
- 61.Zhao Z, Liu B, Li B, Song C, Diao H, Guo Z, Li Z, Zhang J. Inhibition of long noncoding RNA IGF2AS promotes angiogenesis in type 2 diabetes. Biomed Pharmacother. 2017;92:445–450. doi: 10.1016/j.biopha.2017.05.039. [DOI] [PubMed] [Google Scholar]
- 62.Lu W, Huang SY, Su L, Zhao BX, Miao JY. Long Noncoding RNA LOC100129973 Suppresses Apoptosis by Targeting miR-4707-5p and miR-4767 in Vascular Endothelial Cells. Sci Rep. 2016;6:21620. doi: 10.1038/srep21620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yan B, Yao J, Liu JY, Li XM, Wang XQ, Li YJ, Tao ZF, Song YC, Chen Q, Jiang Q. lncRNA-MIAT regulates microvascular dysfunction by functioning as a competing endogenous RNA. Circ Res. 2015;116:1143–1156. doi: 10.1161/CIRCRESAHA.116.305510. [DOI] [PubMed] [Google Scholar]
- 64.Jiang C, Fang X, Jiang Y, Shen F, Hu Z, Li X, Huang X. TNF-alpha induces vascular endothelial cells apoptosis through overexpressing pregnancy induced noncoding RNA in Kawasaki disease model. Int J Biochem Cell Biol. 2016;72:118–124. doi: 10.1016/j.biocel.2016.01.011. [DOI] [PubMed] [Google Scholar]
- 65.Boulberdaa M, Scott E, Ballantyne M, Garcia R, Descamps B, Angelini GD, Brittan M, Hunter A, McBride M, McClure J, et al. A Role for the Long Noncoding RNA SENCR in Commitment and Function of Endothelial Cells. Mol Ther. 2016;24:978–990. doi: 10.1038/mt.2016.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ming GF, Wu K, Hu K, Chen Y, Xiao J. NAMPT regulates senescence, proliferation, and migration of endothelial progenitor cells through the SIRT1 AS lncRNA/miR-22/SIRT1 pathway. Biochem Biophys Res Commun. 2016;478:1382–1388. doi: 10.1016/j.bbrc.2016.08.133. [DOI] [PubMed] [Google Scholar]
- 67.Huang S, Lu W, Ge D, Meng N, Li Y, Su L, Zhang S, Zhang Y, Zhao B, Miao J. A new microRNA signal pathway regulated by long noncoding RNA TGFB2-OT1 in autophagy and inflammation of vascular endothelial cells. Autophagy. 2015;11:2172–2183. doi: 10.1080/15548627.2015.1106663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chen C, Cheng G, Yang X, Li C, Shi R, Zhao N. Tanshinol suppresses endothelial cells apoptosis in mice with atherosclerosis via lncRNA TUG1 up-regulating the expression of miR-26a. Am J Transl Res. 2016;8:2981–2991. [PMC free article] [PubMed] [Google Scholar]
- 69.Gomez D, Swiatlowska P, Owens GK. Epigenetic Control of Smooth Muscle Cell Identity and Lineage Memory. Arterioscler Thromb Vasc Biol. 2015;35:2508–2516. doi: 10.1161/ATVBAHA.115.305044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Curcio A, Torella D, Indolfi C. Mechanisms of smooth muscle cell proliferation and endothelial regeneration after vascular injury and stenting: approach to therapy. Circ J. 2011;75:1287–1296. doi: 10.1253/circj.cj-11-0366. [DOI] [PubMed] [Google Scholar]
- 71.Owens GK, Kumar MS, Wamhoff BR. Molecular regulation of vascular smooth muscle cell differentiation in development and disease. Physiol Rev. 2004;84:767–801. doi: 10.1152/physrev.00041.2003. [DOI] [PubMed] [Google Scholar]
- 72.Bennett MR, Sinha S, Owens GK. Vascular Smooth Muscle Cells in Atherosclerosis. Circ Res. 2016;118:692–702. doi: 10.1161/CIRCRESAHA.115.306361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Samani NJ, Erdmann J, Hall AS, Hengstenberg C, Mangino M, Mayer B, Dixon RJ, Meitinger T, Braund P, Wichmann HE, et al. Genomewide association analysis of coronary artery disease. N Engl J Med. 2007;357:443–453. doi: 10.1056/NEJMoa072366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.de los Campos G, Gianola D, Allison DB. Predicting genetic predisposition in humans: the promise of whole-genome markers. Nat Rev Genet. 2010;11:880–886. doi: 10.1038/nrg2898. [DOI] [PubMed] [Google Scholar]
- 75.Gschwendtner A, Bevan S, Cole JW, Plourde A, Matarin M, Ross-Adams H, Meitinger T, Wichmann E, Mitchell BD, Furie K, et al. Sequence variants on chromosome 9p21.3 confer risk for atherosclerotic stroke. Ann Neurol. 2009;65:531–539. doi: 10.1002/ana.21590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Jarinova O, Stewart AF, Roberts R, Wells G, Lau P, Naing T, Buerki C, McLean BW, Cook RC, Parker JS, et al. Functional analysis of the chromosome 9p21.3 coronary artery disease risk locus. Arterioscler Thromb Vasc Biol. 2009;29:1671–1677. doi: 10.1161/ATVBAHA.109.189522. [DOI] [PubMed] [Google Scholar]
- 77.Helgadottir A, Thorleifsson G, Magnusson KP, Gretarsdottir S, Steinthorsdottir V, Manolescu A, Jones GT, Rinkel GJ, Blankensteijn JD, Ronkainen A, et al. The same sequence variant on 9p21 associates with myocardial infarction, abdominal aortic aneurysm and intracranial aneurysm. Nat Genet. 2008;40:217–224. doi: 10.1038/ng.72. [DOI] [PubMed] [Google Scholar]
- 78.Zeggini E, Weedon MN, Lindgren CM, Frayling TM, Elliott KS, Lango H, Timpson NJ, Perry JR, Rayner NW, Freathy RM, et al. Replication of genome-wide association signals in UK samples reveals risk loci for type 2 diabetes. Science. 2007;316:1336–1341. doi: 10.1126/science.1142364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bishop DT, Demenais F, Iles MM, Harland M, Taylor JC, Corda E, Randerson-Moor J, Aitken JF, Avril MF, Azizi E, et al. Genome-wide association study identifies three loci associated with melanoma risk. Nat Genet. 2009;41:920–925. doi: 10.1038/ng.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Shete S, Hosking FJ, Robertson LB, Dobbins SE, Sanson M, Malmer B, Simon M, Marie Y, Boisselier B, Delattre JY, et al. Genome-wide association study identifies five susceptibility loci for glioma. Nat Genet. 2009;41:899–904. doi: 10.1038/ng.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Holdt LM, Beutner F, Scholz M, Gielen S, Gabel G, Bergert H, Schuler G, Thiery J, Teupser D. ANRIL expression is associated with atherosclerosis risk at chromosome 9p21. Arterioscler Thromb Vasc Biol. 2010;30:620–627. doi: 10.1161/ATVBAHA.109.196832. [DOI] [PubMed] [Google Scholar]
- 82.Congrains A, Kamide K, Katsuya T, Yasuda O, Oguro R, Yamamoto K, Ohishi M, Rakugi H. CVD-associated non-coding RNA, ANRIL, modulates expression of atherogenic pathways in VSMC. Biochem Biophys Res Commun. 2012;419:612–616. doi: 10.1016/j.bbrc.2012.02.050. [DOI] [PubMed] [Google Scholar]
- 83.Yap KL, Li S, Munoz-Cabello AM, Raguz S, Zeng L, Mujtaba S, Gil J, Walsh MJ, Zhou MM. Molecular interplay of the noncoding RNA ANRIL and methylated histone H3 lysine 27 by polycomb CBX7 in transcriptional silencing of INK4a. Mol Cell. 2010;38:662–674. doi: 10.1016/j.molcel.2010.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wan G, Mathur R, Hu X, Liu Y, Zhang X, Peng G, Lu X. Long non-coding RNA ANRIL (CDKN2B-AS) is induced by the ATM-E2F1 signaling pathway. Cell Signal. 2013;25:1086–1095. doi: 10.1016/j.cellsig.2013.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wu G, Cai J, Han Y, Chen J, Huang ZP, Chen C, Cai Y, Huang H, Yang Y, Liu Y, et al. LincRNA-p21 regulates neointima formation, vascular smooth muscle cell proliferation, apoptosis, and atherosclerosis by enhancing p53 activity. Circulation. 2014;130:1452–1465. doi: 10.1161/CIRCULATIONAHA.114.011675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Tang SS, Cheng J, Cai MY, Yang XL, Liu XG, Zheng BY, Xiong XD. Association of lincRNA-p21 Haplotype with Coronary Artery Disease in a Chinese Han Population. Dis Markers. 2016;2016:9109743. doi: 10.1155/2016/9109743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.He C, Ding JW, Li S, Wu H, Jiang YR, Yang W, Teng L, Yang J. The Role of Long Intergenic Noncoding RNA p21 in Vascular Endothelial Cells. DNA Cell Biol. 2015;34:677–683. doi: 10.1089/dna.2015.2966. [DOI] [PubMed] [Google Scholar]
- 88.Spurlock CF, 3rd, Tossberg JT, Matlock BK, Olsen NJ, Aune TM. Methotrexate inhibits NF-kappaB activity via long intergenic (noncoding) RNA-p21 induction. Arthrit is Rheumatol. 2014;66:2947–2957. doi: 10.1002/art.38805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Bell RD, Long X, Lin M, Bergmann JH, Nanda V, Cowan SL, Zhou Q, Han Y, Spector DL, Zheng D, et al. Identification and initial functional characterization of a human vascular cell-enriched long noncoding RNA. Arterioscler Thromb Vasc Biol. 2014;34:1249–1259. doi: 10.1161/ATVBAHA.114.303240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zou ZQ, Xu J, Li L, Han YS. Down-regulation of SENCR promotes smooth muscle cells proliferation and migration in db/db mice through up-regulation of FoxO1 and TRPC6. Biomed Pharmacother. 2015;74:35–41. doi: 10.1016/j.biopha.2015.06.009. [DOI] [PubMed] [Google Scholar]
- 91.de Gonzalo-Calvo D, Kenneweg F, Bang C, Toro R, van der Meer RW, Rijzewijk LJ, Smit JW, Lamb HJ, Llorente-Cortes V, Thum T. Circulating long-non coding RNAs as biomarkers of left ventricular diastolic function and remodelling in patients with well-controlled type 2 diabetes. Sci Rep. 2016;6:37354. doi: 10.1038/srep37354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Williams GT, Mourtada-Maarabouni M, Farzaneh F. A critical role for non-coding RNA GAS5 in growth arrest and rapamycin inhibition in human T-lymphocytes. Biochem Soc Trans. 2011;39:482–486. doi: 10.1042/BST0390482. [DOI] [PubMed] [Google Scholar]
- 93.Kino T, Hurt DE, Ichijo T, Nader N, Chrousos GP. Noncoding RNA gas5 is a growth arrest- and starvation-associated repressor of the glucocorticoid receptor. Sci Signal. 2010;3:ra8. doi: 10.1126/scisignal.2000568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wang YN, Shan K, Yao MD, Yao J, Wang JJ, Li X, Liu B, Zhang YY, Ji Y, Jiang Q, et al. Long Noncoding RNA-GAS5: A Novel Regulator of Hypertension-Induced Vascular Remodeling. Hypertension. 2016;68:736–748. doi: 10.1161/HYPERTENSIONAHA.116.07259. [DOI] [PubMed] [Google Scholar]
- 95.Li L, Li X, The E, Wang LJ, Yuan TY, Wang SY, Feng J, Wang J, Liu Y, Wu YH, et al. Low expression of lncRNA-GAS5 is implicated in human primary varicose great saphenous veins. PLoS One. 2015;10:e0120550. doi: 10.1371/journal.pone.0120550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Tao H, Zhang JG, Qin RH, Dai C, Shi P, Yang JJ, Deng ZY, Shi KH. LncRNA GAS5 controls cardiac fibroblast activation and fibrosis by targeting miR-21 via PTEN/MMP-2 signaling pathway. Toxicology. 2017;386:11–18. doi: 10.1016/j.tox.2017.05.007. [DOI] [PubMed] [Google Scholar]
- 97.Tang R, Zhang G, Wang YC, Mei X, Chen SY. The long non-coding RNA GAS5 regulates transforming growth factor beta (TGF-beta)-induced smooth muscle cell differentiation via RNA Smad-binding elements. J Biol Chem. 2017;292:14270–14278. doi: 10.1074/jbc.M117.790030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ballantyne MD, Pinel K, Dakin R, Vesey AT, Diver L, Mackenzie R, Garcia R, Welsh P, Sattar N, Hamilton G, et al. Smooth Muscle Enriched Long Noncoding RNA (SMILR) Regulates Cell Proliferation. Circulation. 2016;133:2050–2065. doi: 10.1161/CIRCULATIONAHA.115.021019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Vigetti D, Deleonibus S, Moretto P, Bowen T, Fischer JW, Grandoch M, Oberhuber A, Love DC, Hanover JA, Cinquetti R, et al. Natural antisense transcript for hyaluronan synthase 2 (HAS2-AS1) induces transcription of HAS2 via protein O-GlcNAcylation. J Biol Chem. 2014;289:28816–28826. doi: 10.1074/jbc.M114.597401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wang S, Zhang X, Yuan Y, Tan M, Zhang L, Xue X, Yan Y, Han L, Xu Z. BRG1 expression is increased in thoracic aortic aneurysms and regulates proliferation and apoptosis of vascular smooth muscle cells through the long non-coding RNA HIF1A-AS1 in vitro. Eur J Cardiothorac Surg. 2015;47:439–446. doi: 10.1093/ejcts/ezu215. [DOI] [PubMed] [Google Scholar]
- 101.Guo X, Chang Q, Pei H, Sun X, Qian X, Tian C, Lin H. Long Non-coding RNA-mRNA Correlation Analysis Reveals the Potential Role of HOTAIR in Pathogenesis of Sporadic Thoracic Aortic Aneurysm. Eur J Vasc Endovasc Surg. 2017;54:303–314. doi: 10.1016/j.ejvs.2017.06.010. [DOI] [PubMed] [Google Scholar]
- 102.Bischoff FC, Werner A, John D, Boeckel JN, Melissari MT, Grote P, Glaser SF, Demolli S, Uchida S, Michalik KM, et al. Identification and Functional Characterization of Hypoxia-Induced Endoplasmic Reticulum Stress Regulating lncRNA (HypERlnc) in Pericytes. Circ Res. 2017;121:368–375. doi: 10.1161/CIRCRESAHA.116.310531. [DOI] [PubMed] [Google Scholar]
- 103.Leung A, Trac C, Jin W, Lanting L, Akbany A, Saetrom P, Schones DE, Natarajan R. Novel long noncoding RNAs are regulated by angiotensin II in vascular smooth muscle cells. Circ Res. 2013;113:266–278. doi: 10.1161/CIRCRESAHA.112.300849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Chen J, Guo J, Cui X, Dai Y, Tang Z, Qu J, Raj JU, Hu Q, Gou D. Long Non-Coding RNA LnRPT is Regulated by PDGF-BB and Modulates Proliferation of Pulmonary Artery Smooth Muscle Cells. Am J Respir Cell Mol Biol. 2017 doi: 10.1165/rcmb.2017-0111OC. [DOI] [PubMed] [Google Scholar]
- 105.Sun Z, Nie X, Sun S, Dong S, Yuan C, Li Y, Xiao B, Jie D, Liu Y. Long Non-Coding RNA MEG3 Downregulation Triggers Human Pulmonary Artery Smooth Muscle Cell Proliferation and Migration via the p53 Signaling Pathway. Cell Physiol Biochem. 2017;42:2569–2581. doi: 10.1159/000480218. [DOI] [PubMed] [Google Scholar]
- 106.Liu Y, Sun Z, Zhu J, Xiao B, Dong J, Li X. LncRNA-TCONS00034812 in cell proliferation and apoptosis of pulmonary artery smooth muscle cells and its mechanism. J Cell Physiol. 2017 doi: 10.1002/jcp.26279. [DOI] [PubMed] [Google Scholar]
- 107.Chen R, Kong P, Zhang F, Shu YN, Nie X, Dong LH, Lin YL, Xie XL, Zhao LL, Zhang XJ, et al. EZH2-mediated alpha-actin methylation needs lncRNA TUG1, and promotes the cortex cytoskeleton formation in VSMCs. Gene. 2017;616:52–57. doi: 10.1016/j.gene.2017.03.028. [DOI] [PubMed] [Google Scholar]
- 108.Yao QP, Xe ZW, Wang KX, Zhang P, Han Y, Qi YX, Jiang ZL. Profiles of long noncoding RNAs in hypertensive rats: long noncoding RNA XR007793 regulates cyclic strain-induced proliferation and migration of vascular smooth muscle cells. J Hypertens. 2017;35:1195–1203. doi: 10.1097/HJH.0000000000001304. [DOI] [PubMed] [Google Scholar]
- 109.Libby P, Lichtman AH, Hansson GK. Immune effector mechanisms implicated in atherosclerosis: from mice to humans. Immunity. 2013;38:1092–1104. doi: 10.1016/j.immuni.2013.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Galkina E, Ley K. Immune and inflammatory mechanisms of atherosclerosis (*) Annu Rev Immunol. 2009:27165–197. doi: 10.1146/annurev.immunol.021908.132620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Libby P, Nahrendorf M, Swirski FK. Monocyte heterogeneity in cardiovascular disease. Semin Immunopathol. 2013;35:553–562. doi: 10.1007/s00281-013-0387-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Stemme S, Faber B, Holm J, Wiklund O, Witztum JL, Hansson GK. T lymphocytes from human atherosclerotic plaques recognize oxidized low density lipoprotein. Proc Natl Acad Sci U S A. 1995;92:3893–3897. doi: 10.1073/pnas.92.9.3893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Hansson GK, Libby P. The immune response in atherosclerosis: a double-edged sword. Nat Rev Immunol. 2006;6:508–519. doi: 10.1038/nri1882. [DOI] [PubMed] [Google Scholar]
- 114.Zhang DD, Wang WT, Xong J, Xe XM, Cui SS, Zhao ZG, Li MJ, Zhang ZQ, Hao DL, Zhao X, et al. Long noncoding RNA LINC00305 promotes inflammation by activating the AHRR-NF-kappaB pathway in human monocytes. Sci Rep. 2017;7:46204. doi: 10.1038/srep46204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Carpenter S, Aiello D, Atianand MK, Ricci EP, Gandhi P, Hall LL, Byron M, Monks B, Henry-Bezy M, Lawrence JB, et al. A long noncoding RNA mediates both activation and repression of immune response genes. Science. 2013:341789–792. doi: 10.1126/science.1240925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009:458223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Covarrubias S, Robinson EK, Shapleigh B, Vollmers A, Katzman S, Hanley N, Fong N, McManus MT, Carpenter S. CRISPR/Cas-based screening of long non-coding RNAs (lncRNAs) in macrophages with an NF-κB reporter. Journal of Biological Chemistry. 2017;292:20911–20920. doi: 10.1074/jbc.M117.799155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Hu G, Gong AY, Wang Y, Ma S, Chen X, Chen J, Su CJ, Shibata A, Strauss-Soukup JK, Drescher KM, et al. LincRNA-Cox2 Promotes Late Inflammatory Gene Transcription in Macrophages through Modulating SWI/SNF-Mediated Chromatin Remodeling. J Immunol. 2016:1962799–2808. doi: 10.4049/jimmunol.1502146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Li Z, Chao TC, Chang KY, Lin N, Patil VS, Shimizu C, Head SR, Burns JC, Rana TM. The long noncoding RNA THRIL regulates TNFalpha expression through its interaction with hnRNPL. Proc Natl Acad Sci U S A. 2014;111:1002–1007. doi: 10.1073/pnas.1313768111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Chen L, Yang W, Guo Y, Chen W, Zheng P, Zeng J, Tong W. Exosomal lncRNA GAS5 regulates the apoptosis of macrophages and vascular endothelial cells in atherosclerosis. PLoS One. 2017;12:e0185406. doi: 10.1371/journal.pone.0185406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Huang C, Hu YW, Zhao JJ, Ma X, Zhang Y, Guo FX, Kang CM, Lu JB, Xiu JC, Sha YH, et al. Long Noncoding RNA HOXC-AS1 Suppresses Ox-LDL-Induced Cholesterol Accumulation Through Promoting HOXC6 Expression in THP-1 Macrophages. DNA Cell Biol. 2016;35:722–729. doi: 10.1089/dna.2016.3422. [DOI] [PubMed] [Google Scholar]
- 122.Chen L, Yao H, Hui JY, Ding SH, Fan YL, Pan YH, Chen KH, Wan JQ, Jiang JY. Global transcriptomic study of atherosclerosis development in rats. Gene. 2016;592:43–48. doi: 10.1016/j.gene.2016.07.023. [DOI] [PubMed] [Google Scholar]
- 123.Ito I, Asai A, Suzuki S, Kobayashi M, Suzuki F. M2b macrophage polarization accompanied with reduction of long noncoding RNA GAS5. Biochem Biophys Res Commun. 2017;493:170–175. doi: 10.1016/j.bbrc.2017.09.053. [DOI] [PubMed] [Google Scholar]
- 124.Libby P, Ridker PM, Hansson GK. Progress and challenges in translating the biology of atherosclerosis. Nature. 2011;473:317–325. doi: 10.1038/nature10146. [DOI] [PubMed] [Google Scholar]
- 125.Sallam T, Jones MC, Gilliland T, Zhang L, Wu X, Eskin A, Sandhu J, Casero D, Vallim TQ, Hong C, et al. Feedback modulation of cholesterol metabolism by the lipid-responsive non-coding RNA LeXis. Nature. 2016;534:124–128. doi: 10.1038/nature17674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Tontonoz P, Wu X, Jones M, Zhang Z, Salisbury D, Sallam T. Long Noncoding RNA Facilitated Gene Therapy Reduces Atherosclerosis in a Murine Model of Familial Hypercholesterolemia. Circulation. 2017;136:776–778. doi: 10.1161/CIRCULATIONAHA.117.029002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Li P, Ruan X, Yang L, Kiesewetter K, Zhao Y, Luo H, Chen Y, Gucek M, Zhu J, Cao H. A liver-enriched long non-coding RNA, lncLSTR, regulates systemic lipid metabolism in mice. Cell Metab. 2015;21:455–467. doi: 10.1016/j.cmet.2015.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Yang L, Li P, Yang W, Ruan X, Kiesewetter K, Zhu J, Cao H. Integrative Transcriptome Analyses of Metabolic Responses in Mice Define Pivotal LncRNA Metabolic Regulators. Cell Metab. 2016;24:627–639. doi: 10.1016/j.cmet.2016.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Roux BT, Heward JA, Donnelly LE, Jones SW, Lindsay MA. Catalog of Differentially Expressed Long Non-Coding RNA following Activation of Human and Mouse Innate Immune Response. Front Immunol. 2017;8:1038. doi: 10.3389/fimmu.2017.01038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Covarrubias S, Robinson EK, Shapleigh B, Vollmers A, Katzman S, Hanley N, Fong N, McManus MT, Carpenter S. CRISPR/Cas9-based screening of long noncoding RNAs (lncRNAs) in macrophages with an NF-kappa B reporter. J Biol Chem. 2017 doi: 10.1074/jbc.M117.799155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Cai Y, Yang Y, Chen X, Wu G, Zhang X, Liu Y, Yu J, Wang X, Fu J, Li C, et al. Circulating ‘lncRNA OTTHUMT00000387022’ from monocytes as a novel biomarker for coronary artery disease. Cardiovasc Res. 2016;112:714–724. doi: 10.1093/cvr/cvw022. [DOI] [PubMed] [Google Scholar]
- 132.Ma S, Ming Z, Gong AY, Wang Y, Chen X, Hu G, Zhou R, Shibata A, Swanson PC, Chen XM. A long noncoding RNA, lincRNA-Tnfaip3, acts as a coregulator of NF-kappaB to modulate inflammatory gene transcription in mouse macrophages. FASEB J. 2017;31:1215–1225. doi: 10.1096/fj.201601056R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Krawczyk M, Emerson BM. p50-associated COX-2 extragenic RNA (PACER) activates COX-2 gene expression by occluding repressive NF-kappaB complexes. Elife. 2014;3:e01776. doi: 10.7554/eLife.01776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Nguyen Q, Carninci P. Expression Specificity of Disease-Associated lncRNAs: Toward Personalized Medicine. Curr Top Microbiol Immunol. 2016;394:237–258. doi: 10.1007/82_2015_464. [DOI] [PubMed] [Google Scholar]
- 135.Batista PJ, Chang HY. Long noncoding RNAs: cellular address codes in development and disease. Cell. 2013;152:1298–1307. doi: 10.1016/j.cell.2013.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Schmitz SU, Grote P, Herrmann BG. Mechanisms of long noncoding RNA function in development and disease. Cell Mol Life Sci. 2016;73:2491–2509. doi: 10.1007/s00018-016-2174-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Kumar V, Westra HJ, Karjalainen J, Zhernakova DV, Esko T, Hrdlickova B, Almeida R, Zhernakova A, Reinmaa E, Vosa U, et al. Human disease-associated genetic variation impacts large intergenic non-coding RNA expression. PLoS Genet. 2013;9:e1003201. doi: 10.1371/journal.pgen.1003201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Congrains A, Kamide K, Oguro R, Yasuda O, Miyata K, Yamamoto E, Kawai T, Kusunoki H, Yamamoto H, Takeya Y, et al. Genetic variants at the 9p21 locus contribute to atherosclerosis through modulation of ANRIL and CDKN2A/B. Atherosclerosis. 2012;220:449–455. doi: 10.1016/j.atherosclerosis.2011.11.017. [DOI] [PubMed] [Google Scholar]
- 139.Shi T, Gao G, Cao Y. Long Noncoding RNAs as Novel Biomarkers Have a Promising Future in Cancer Diagnostics. Dis Markers. 2016;2016:9085195. doi: 10.1155/2016/9085195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Xue C, Zhang X, Zhang H, Ferguson JF, Wang Y, Hinkle CC, Li M, Reilly MP. De novo RNA sequence assembly during in vivo inflammatory stress reveals hundreds of unannotated lincRNAs in human blood CD14(+) monocytes and in adipose tissue. Physiol Genomics. 2017;49:287–305. doi: 10.1152/physiolgenomics.00001.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Uchida S, Dimmeler S. Long noncoding RNAs in cardiovascular diseases. Circ Res. 2015;116:737–750. doi: 10.1161/CIRCRESAHA.116.302521. [DOI] [PubMed] [Google Scholar]
- 142.Ulitsky I. Evolution to the rescue: using comparative genomics to understand long non-coding RNAs. Nat Rev Genet. 2016;17:601–614. doi: 10.1038/nrg.2016.85. [DOI] [PubMed] [Google Scholar]
- 143.Ventola GM, Noviello TM, D'Aniello S, Spagnuolo A, Ceccarelli M, Cerulo L. Identification of long non-coding transcripts with feature selection: a comparative study. BMC Bioinformatics. 2017;18:187. doi: 10.1186/s12859-017-1594-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Haemmig S, Feinberg MW. Targeting LncRNAs in Cardiovascular Disease: Options and Expeditions. Circ Res. 2017;120:620–623. doi: 10.1161/CIRCRESAHA.116.310152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Khvorova A, Watts JK. The chemical evolution of oligonucleotide therapies of clinical utility. Nat Biotechnol. 2017;35:238–248. doi: 10.1038/nbt.3765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Feinberg MW. No small task: therapeutic targeting of Lp(a) for cardiovascular disease. Lancet. 2016;388:2211–2212. doi: 10.1016/S0140-6736(16)31329-0. [DOI] [PubMed] [Google Scholar]
- 147.Bell DA, Hooper AJ, Watts GF, Burnett JR. Mipomersen and other therapies for the treatment of severe familial hypercholesterolemia. Vasc Health Risk Manag. 2012;8:651–659. doi: 10.2147/VHRM.S28581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Nayak S, Herzog RW. Progress and prospects: immune responses to viral vectors. Gene Ther. 2010;17:295–304. doi: 10.1038/gt.2009.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Kanasty R, Dorkin JR, Vegas A, Anderson D. Delivery materials for siRNA therapeutics. Nat Mater. 2013;12:967–977. doi: 10.1038/nmat3765. [DOI] [PubMed] [Google Scholar]
- 150.Reichmuth AM, Oberli MA, Jeklenec A, Langer R, Blankschtein D. mRNA vaccine delivery using lipid nanoparticles. Ther Deliv. 2016;7:319–334. doi: 10.4155/tde-2016-0006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Aagaard L, Rossi JJ. RNAi therapeutics: principles, prospects and challenges. Adv Drug Deliv Rev. 2007;59:75–86. doi: 10.1016/j.addr.2007.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Elling R, Chan J, Fitzgerald KA. Emerging role of long noncoding RNAs as regulators of innate immune cell development and inflammatory gene expression. Eur J Immunol. 2016;46:504–512. doi: 10.1002/eji.201444558. [DOI] [PMC free article] [PubMed] [Google Scholar]


