Abstract
Cholangiocytes normally express primary cilia, a multisensory organelle that detects signals from the cellular environment. Cilia are significantly reduced in cholangiocarcinoma (CCA) by a mechanism involving overexpression of histone deacetylase 6 (HDAC6). Despite HDAC6 overexpression in CCA, we found no differences in its mRNA level, suggesting a post-transcriptional regulation, possibly involving miRNAs. Here we describe that at least two HDAC6-targeting miRNAs, miR-433 and miR-22, are downregulated in CCA both in vitro and in vivo. Experimental restoration of these miRNAs in CCA cells reduced HDAC6 expression, induced ciliary restoration, and decreased the malignant phenotype. Furthermore, in contrast to the mature forms, levels of precursor forms of these miRNAs were higher in CCA compared to normal cholangiocytes and accumulated in the nuclei, suggesting a defective nuclear export. We assessed the expression of Exportin-5, the protein responsible for transporting miRNA precursors out of the nucleus, and found it to be reduced by 50% in CCA compared to normal cholangiocytes. Experimental overexpression of Exportin-5 in CCA cells restored precursor and mature forms of these miRNAs to normal levels, inducing a decrease in the expression of HDAC6 and a decrease in the malignant phenotype. Conversely, shRNA depletion of Exportin-5 in normal cholangiocytes resulted in increased nuclear retention of precursor miRNAs, decreased mature miRNAs, increased cell proliferation, and shorter cilia. Conclusion: these data suggest that downregulated Exportin-5 impairs the nuclear export of miR-433 and miR-22 precursor forms, causing a decrease in levels of mature miR-433 and miR-22 forms, and leading to overexpression of HDAC6 and ciliary loss in CCA.
Keywords: Cilia, Exportin-5, miR-433, miR-22, bile duct cancer
INTRODUCTION
Cholangiocarcinoma (CCA) is a heterogeneous group of malignancies arising from cholangiocytes, the epithelial cells of the biliary tree. CCA has a poor prognosis and is difficult to diagnose in its early stages due to the absence of clinical manifestations. Over the past two decades, the incidence and mortality rates of CCA have increased worldwide. CCA accounts for 10–20% of all hepatobiliary malignancies and is the second most common primary hepatic cancer (1).
There is currently no effective chemoprevention or treatment for CCA; hence, surgical resection is the only prospect for long-term survival (2). The 5-year survival rate is still as low as 10% and, therefore, early detection or new targeted therapies are urgently needed (3).
The pathogenic pathways involved in the carcinogenesis of this biliary tumor are still unclear and may include alterations of several signaling cascades and molecules, such as Histone deacetylase 6 (HDAC6) (4–7). HDAC6 is a unique member of the HDAC family that is predominately localized in the cytoplasm. One of the main functions of HDAC6 is the deacetylation of a-tubulin, which is a cellular control mechanism for microtubule stability (8, 9).
Cholangiocytes have a single primary cilium, a microtubule-based organelle that extends from the apical membrane and functions as a mechano-, chemo-, and osmosensor (10–13). We recently found that primary cilia expression is decreased in CCA by a mechanism involving HDAC6 protein overexpression (7). It is known that HDAC6 acts as a tubulin deacetylase and the balance between deacetylation and acetylation serves as the molecular basis for stability and function of the ciliary axoneme. We found that the overexpression of HDAC6 is involved in primary cilia loss in CCA. However, the mechanisms of HDAC6 upregulation remain unknown. Despite HDAC6 overexpression, we found that mRNA levels remain unaffected which suggests a potential posttranscriptional regulation, likely microRNAs (miRNAs) (7).
MiRNAs are small non-coding RNAs with an average length of 21 nucleotides (14) that are synthetized as precursor forms in the nucleus. The primary transcript is processed inside the nucleus to the precursor form (Pre-miRNA) by the Drosha-DiGeorge syndrome critical region gene 8 (DGCR8) complex (also known as the Microprocessor complex). Pre-miRNAs are then translocated to the cytoplasm by Exportin-5 and further processed to generate the mature and functional miRNAs. The “seed” sequence (positions 2–7 from the 5′ end of miRNAs) is complementary to the 3′ UTR of mRNA targets. MicroRNA complex interaction with mRNA induces posttranscriptional silencing through mRNA destabilization and/or translational repression (15).
Therefore, in the present work, we tested the hypothesis that decreasing levels of specific miRNAs induce the overexpression of HDAC6 and ciliary loss in CCA.
MATERIALS AND METHODS
Cell lines and culture
The normal human cholangiocytes (H69 and NHC) were maintained as previously described (7). The human cholangiocarcinoma cell lines (HuCCT-1 and KMCH (16, 17)) were cultured in DMEM supplemented with 10% fetal bovine serum (FBS) (Gibco, Grand Island, NY, USA), 100 units/ml penicillin, 100 ug/mI streptomycin, and 100 ug/l insulin at 37°C in a humidified incubator containing 5% CO2.
Tissue samples
The human specimens for immunofluorescences (normal human liver tissue and human cholangiocarcinoma tissue) were obtained from the Mayo Center for Cell Signaling in Gastroenterology Clinical Core. For measuring miR-433 and miR-22 expression by qPCR, tissues were obtained from normal human gallbladder (n=14), and cholangiocarcinoma (CCA) patients (Tumor and matched surrounding tissue, n=19); these samples were provided by the Basque Biobank (http://www.biobancovasco.org; Supplemental Table 1) with approval from the Clinical Research Ethics Committee of Donostia Hospital. Informed consent was obtained from all subjects.
Quantitative reverse PCR for miRNAs and precursor miRNAs expression (qRT-PCR)
Total RNA was performed by lysing cells in TRIzol reagent (Invitrogen, Carlsbad, CA) according to the supplier’s instructions and RNA concentration was determined using a Nanodrop ND-100 Spectrophotometer (Nanodrop). Real-time PCR (qRT-PCR) was applied to quantitate miR-22, miR-433, pri-miR-22, and pri-miR-433 expression in control and CCA cells. For mature miRNA, the TaqMan miRNA Reverse Transcription Kit and TaqMan hsa-miR-22 and hsa-miR-433 primes and TM probes were used (all, Applied Biosystems). Precursor form expression was evaluated using SuperScript III First-Strand System for RT-PCR (Invitrogen, Carlsbad, CA) following the manufacturers’ protocols. Briefly, 5 μg of total RNA was reverse transcribed to cDNA using random hexamer primers. cDNA was stored at −20°C overnight and then used for qPCR. Reverse transcriptase was performed using the Veriti 96 well thermal cycler; q-PCR was performed with TM fluorescent probes using the Light cycler 480 II (Roche). miRNAs amounts were normalized to U6 snRNA and 18S snRNA for mature and precursor forms, respectively (Applied Biosystems). For nuclear pre-miRNA quantifications, nuclear RNA was extracted using the SurePrep Nuclear RNA purification kit (Fisher BioReagents) following the manufacturer’s instructions. cDNA was synthesized using reverse transcriptase. Pre-miRNAs were quantified by real time PCR. U6 was used as an internal control. All reactions were performed in triplicate, and the levels of gene expression were calculated using the 2−ΔΔCt method. Data are given as fold changes of average signals in normal and CCA cholangiocytes.
Western blot
Protein fractions were subjected to 4 – 20 % SDS-PAGE and transferred to nitrocellulose membranes. After blocking, blots were incubated overnight at 4°C with HDAC6 (1:1,000, Santa Cruz Biotechnology), Exportin-5 (1:1000, Santa Cruz Biotechology), actin (1:1000, Sigma-Aldrich), or acetylated-a-tubulin (1:1,000, Sigma-Aldrich), washed, and incubated for 1 hour at room temperature with horseradish peroxidase-conjugated (1:5,000, Abcam) corresponding secondary antibody. ECL system was used for protein detection, and the Gel-Pro Analyzer 6.0 software was used for densitometric analysis.
Proliferation assays
Proliferation assays were conducted using the CellTiter 96 AQueous Non-Radioactive Cell Proliferation Assay (MTS; Promega) and/or counting cells using the Cellometer Auto4 (Nexcelom Bioscience) cell counter.
Anchorage-independent growth
Anchorage-independent growth was assessed by growing cells in soft agar. About 25,000 cells suspended in 0.4% agar in culture media were layered over a 1% agar layer in a 6-well plate. Media was added twice a week and pictures were taken after 14 to 21 days of incubation. The number and size of colonies were analyzed using the Gel-Pro software.
Colony formation assay
After 24 h transfection with specific shRNAs, cells were seeded in six-well plates at 300 cells/well and cultured for 5 days. After fixation with methanol for 15 min, the colonies were stained with 0.1% crystal violet in 20% methanol. To quantify, the dye was dissolved with acetic acid solution (10%) and after 15 min, the absorbance was measured at 450nm.
Wound healing assay
Cells were cultured in complete medium at 37 °C in a humidified incubator maintained at an atmosphere of 5% CO2 in 6 well plates until cells were confluent. Cell monolayers were scratched using a 200-μl pipette tip, then rinsed three times with PBS to remove cell debris. Cell migration in the wound area was observed by phase contrast microscopy at 0–8 h and digitally photographed. Wound healing was measured on the images and the migration area was calculated by the area of original wound minus the area of wound during healing using ImageJ software.
Confocal Immunofluorescence (IF) Microscopy
Confocal IF microscopy was performed with a Zeiss LSM 510 confocal microscope using the 63X oil objective as previously described (see Supporting Methods). Briefly, unstained liver sections were deparaffinized and rehydrated, boiled in antigen unmasking solution, and quenched with Image-IT FX signal enhancer (Invitrogen, Grand Island, NY). Normal human cholangiocytes and CCA cells lines were washed with PBS and fixed in ice cold methanol for 5 min at −20°C. Slides were blocked for 1 hour at room temperature and then incubated overnight at 4°C with acetylated α-tubulin (1:500, Sigma-Aldrich), γ-tubulin (1:500, Sigma-Aldrich), and/or HDAC6 (1:100, Abcam) antibodies followed by 1 hour incubation with fluorescent secondary antibodies (1:100). Nuclei were stained with 4′,6-diamino-2-phenylindole (DAPI; Prolong Gold w/DAPI, Invitrogen).
Fluorescence in Situ Hybridization (ISH)
Detection of miR-433 and miR-22 in formalin-fixed paraffin embedded liver tissue was performed as described previously (18). Briefly, tissues were deparaffinized, rehydrated, and subjected to heat-mediated antigen retrieval in 0.01M sodium citrate buffer, pH 6.0. Slides were allowed to cool, washed, and incubated in pre-hybridization solution (3% BSA in 4X SSX) for 20 minutes at 54°C. Tissues were then incubated for 1 hour at 54°C with a fluorescein labeled LNA probe to either miR-433 or miR-22 (Exiqon) diluted to 25 nM in hybridization buffer (10% Dextran Sulfate in 4X SSC). Slides were washed and subjected to a tyramide signal amplification step (Perkin Elmer), then washed again and mounted in Prolong Gold with DAPI (Thermo Fisher Scientific). Slides were analyzed on a Zeiss 710 Confocal Microscope.
Statistical analysis
Data were expressed as the mean ± standard errors (SE). Experiments were repeated at least three times. Statistical analyses were performed using GraphPad Prism 5.0 (GraphPad Software) and conducted by two-tailed Student’s t-tests to compare two groups. A P-value <0.05 was regarded as statistically significant.
RESULTS
The expressions of miR-22 and miR-433, both targeting HDAC6, are down-regulated in CCA cell lines and tissues
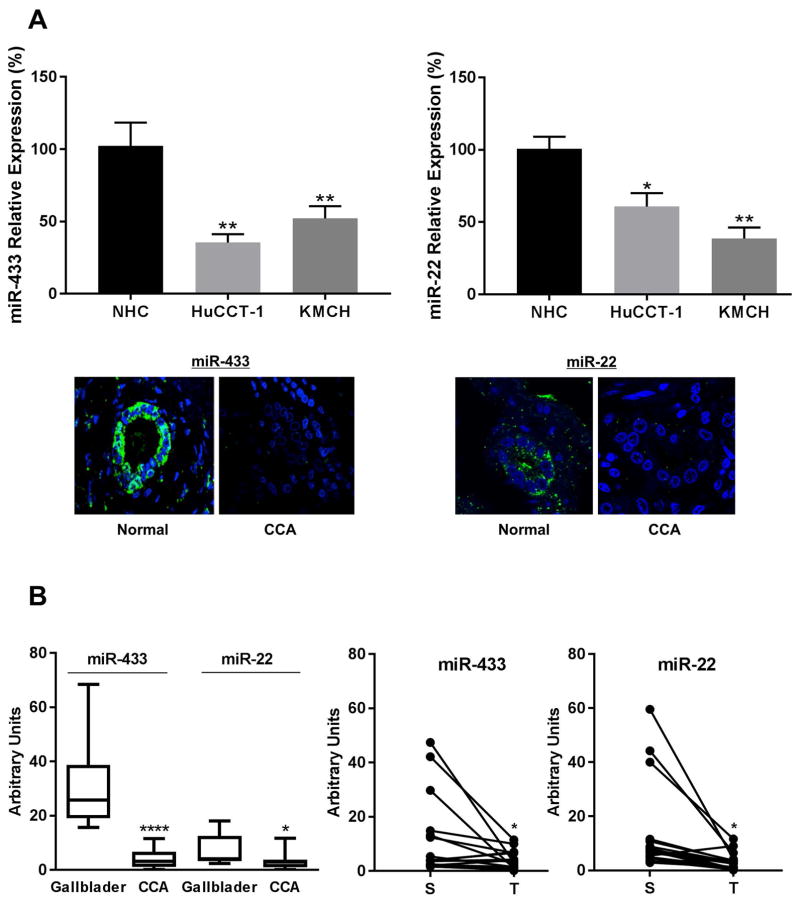
We have previously described that, despite HDAC6 protein overexpression in CCA, HDAC6 mRNA levels were not significantly different between normal and CCA cell lines, suggesting a posttranscriptional regulatory pathway (7). To explore the role of miRNAs in this regulatory mechanism, we assessed in silico the potential miRNAs that could bind to HDAC6 mRNA. Using 3 different target prediction programs, 21 miRNAs were found to have a binding site in the 3′UTR of HDAC6 transcript (Supplemental Table 2). Based on published data, two miRNAs (19, 20), miR-22 and miR-433, were selected and their levels assessed in cells lines and clinical samples by qRT-PCR and in situ hybridization, respectively. MiR-22 and miR-433 were down-regulated in both cholangiocarcinoma cell lines, HuCCT-1 and KMCH, compared to normal cholangiocytes. Moreover, in situ hybridization revealed negative expression of miR-22 and miR-433 in CCA tissue when compared with controls (Figure 1A). Finally, an independent cohort of human CCA samples were analyzed by qPCR and both miRNAs showed a statistically significant decrease when compared to matched surrounding tissue or gallbladder samples used as controls (Figure 1B). To confirm the regulation of HDAC6 by miRNAs, normal cholangiocytes were transfected with scrambled control, anti-miR-433, or miR-433 expression plasmids. As expected, miR-433 reduced and anti-miR-433 increased the levels of HDAC6 protein (Supplemental Figure 1A). The 3′UTR region of HDAC6 was then cloned into a luciferase vector and co-transfected into normal cholangiocyte cells with miR-433 or scrambled control miRs. As shown in Supplemental Figure 1B, miR-433 reduced luciferase activity, consistent with HDAC6 regulation by miR-433.
Figure 1. MicroRNAs targeting HDAC6 are downregulated in CCA.
A, The expression pattern of miR-433 and miR-22 was analyzed in normal cholangiocytes (NHC) and the CCA cell lines HuCCT-1 and KMCH by q-PCR. In situ hybridization for miR-433 and miR-22 in human cholangiocarcinoma tissues. Green, positive signals; blue, counterstained nuclei with DAPI. B, qPCR for miR-433 and miR-22 in a different cohort of CCA human samples (T) compared to matched surrounding tissue (S) and to normal gallbladders as controls. (*p<0.05, **** p<0.001)
Experimental miR-433 and miR-22 upregulations decrease HDAC6 levels, suppress proliferation, colony formation and cellular migration, and induce ciliogenesis in CCA cell lines
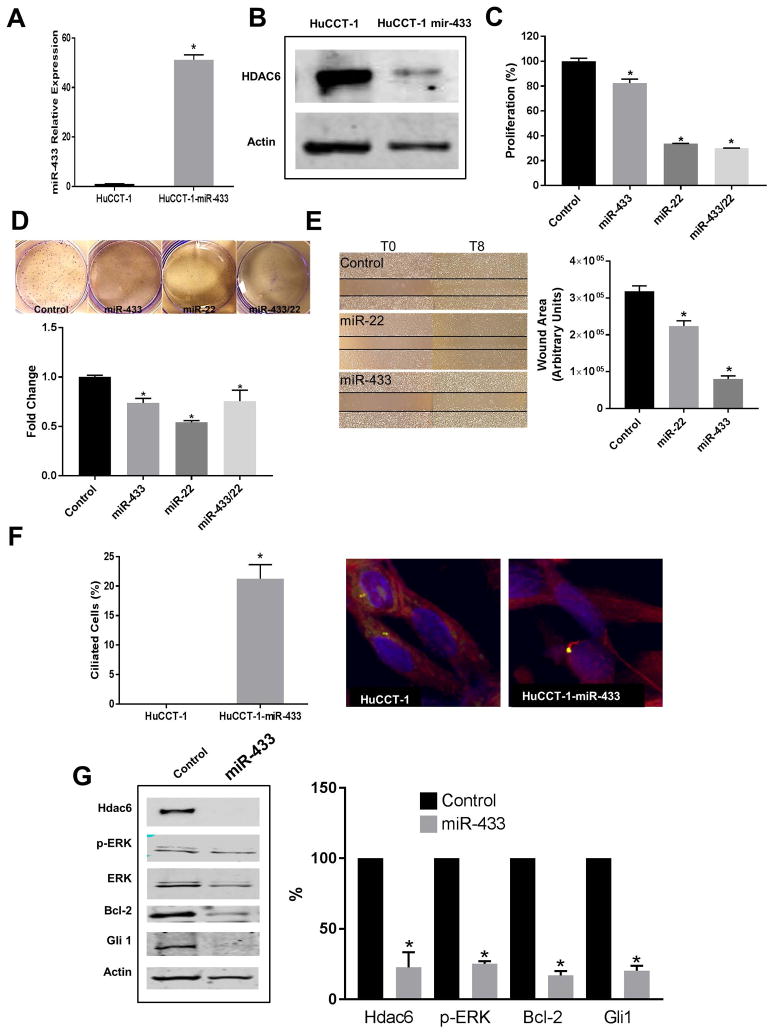
To explore the biologic significance of miR-433 and miR-22 in CCA, we overexpressed miR-433 and/or miR-22 in HuCCT-1 cells. As expected, the overexpression of miR-433 induced downregulation of HDAC6 in transfected cells compared with controls as seen by western blot assays (Figure 2A, B). Furthermore, the overexpression of miR-433 or miR-22 decreased proliferation in this CCA cell line with a slightly greater effect when overexpressed together (Figure 2C). Indeed, overexpression of miR-433 or miR-22 reduces the number of colonies formed by CCA cells, suggesting a decreased malignancy (Figure 2D). Furthermore, these miRNAs also reduced cellular migration (Figure 2E). Importantly, the experimental overexpression of miR-433 correlated with increased expression of primary cilia in CCA cells and decreased signaling in MAPK and Hh pathways, suggesting a less malignant phenotype (Figure 2F and G).
Figure 2. Effect of miR-433 and miR-22 restoration on cholangiocarcinoma cells.
HuCCT-1 cells transfected with miR-433 mimics or scramble were analyzed by qRT-PCR to assess the expression levels of miR-433 (A) and by western blot to assess the proteins levels of HDAC6 (B). C, Proliferation rates were assessed by MTS assays of HuCCT-1 cells transfected with miR-433 and/or miR-22 mimics or scramble (*p<0.05, n=24). D, Colony formation assay (*p<0.05 compared to control, n=3). E, Cellular migration assays (*p<0.01, n=3). F, Percentage of ciliated cells as assessed by confocal immunofluorescence for the ciliary marker acetylated-α-tubulin, in red, and the centrosome marker γ-tubulin in green. Nuclei are stained in blue with DAPI (*p<0.05, n=80). G, Western blot analysis of different proteins involved in CCA cells growth. (*p<0.05, n=3)
Precursor forms of miR-22 and miR-433 are increased in CCA cell lines
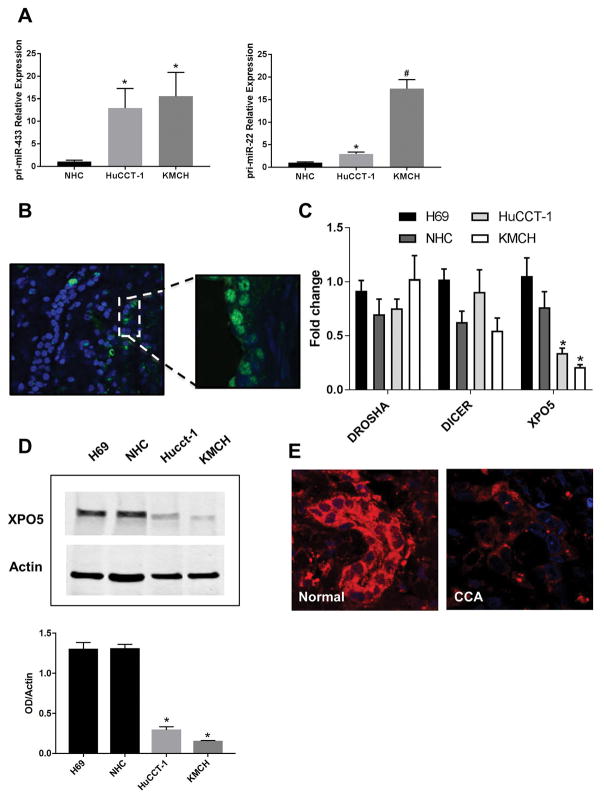
To understand why these miRNAs are affected in cholangiocarcinoma cell lines and human samples, we examined several factors involved in miRNA biogenesis. First, we studied the precursor forms of miR-22 (Pri-miR-22) and miR-433 (Pri-miR-433) by qRT-PCR. We found that, in contrast to the mature forms, pri-miR-22 and pri-miR-433 were upregulated in the CCA cell lines HuCCT-1 and KMCH compared to normal control cholangiocytes (Figure 3A). Interestingly, in situ hybridization for miR-433 showed that the fluorescence signal was concentrated into the nucleus of CCA tissues compared to a more intense and predominantly cytoplasmic signal of normal bile ducts samples (Figures 1B and 3B). Taken together, these findings are suggestive of microRNA precursor forms being trapped in the nucleus.
Figure 3. miRNAs are trapped in the nucleus.
A, qRT-PCR showing increased expression of the precursor forms pri-miR-433 and pri-miR-22 in CCA cell lines compared to a normal control cholangiocyte cell line (*p<0.05, #p<0.01, n=4). B, In situ hybridization showing decreased expression of miR-433 in the cytoplasm and accumulation in the nuclei of cholangiocytes from CCA human liver samples compared to normal control livers. C, mRNA level of genes involved in the biogenesis of miRNAs (*p<0.005, n=6). D, Western blots comparing XPO5 protein expression between normal cholangiocyte cell lines (H69, NHC) and CCA cell lines (HuCCT-1, KMCH) (*p<0.05, n=3). E, Representative Immunofluorescence for XPO5 in CCA human samples compared to normal controls.
Exportin-5 is down regulated both in vitro and in vivo
The mRNA level of genes involved in miRNA biosynthesis, such as Drosha, Dicer and Exportin-5, were compared between normal and CCA cell lines, and only Exportin-5 was found to be significantly down-regulated in cholangiocarcinoma cells compared to normal cholangiocytes (Figure 3C). In addition, to confirm the expression of this miRNA shuttle, we performed western blots in cell lines and immunofluorescences in both cell lines and human samples. As shown in Figure 3D, Exportin-5 is decreased in CCA cell lines compared to normal cholangiocytes. Consistently, immunofluorescences for Exportin-5 showed reduced signal in cholangiocarcinoma cells and human samples compared to normal cholangiocytes and normal liver tissue, respectively (Figure 3E, and Supplemental Fig 2). These results strongly suggest that the primary candidate responsible for impaired nuclear export of miRNA precursor forms is the downregulation of Exportin-5.
Experimental overexpression of Exportin-5 in CCA cells restored miRNAs levels and reduced the malignant phenotype
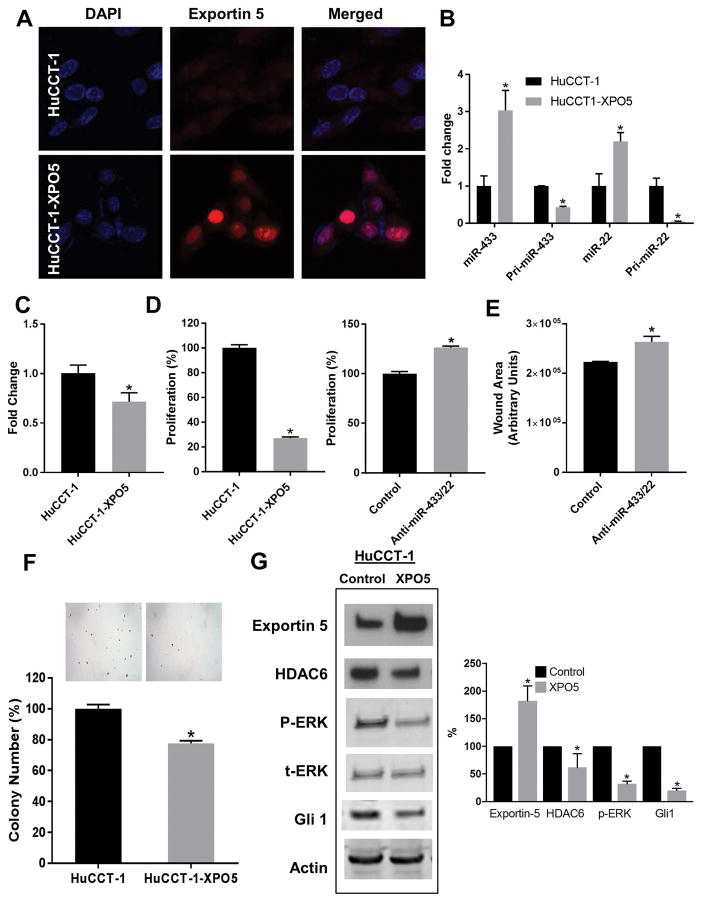
Having established the possible mechanism through which the precursor miRNAs are trapped in the nucleus, we sought to assess the effect of the experimental overexpression of Exportin-5 on tumor cells. The transfection of HuCCT-1 cells with an Exportin-5 expression plasmid demonstrated increased expression at the protein levels by both western blot and immunofluorescence analyses (Figure 4A, 4H), allowing us to test miRNAs expression (mature and precursor miR-22 and miR-433) by qRT-PCR in those transfected cells. Overexpression of Exportin-5 induced a significant upregulation of miR-22 and miR-433, while its precursor forms were significantly downregulated (Figure 4B). To further test the hypothesis, we isolated RNA from nuclear fractions of HuCCT-1 control and Exportin-5 overexpressing cells and found that the nuclear levels of miR-433 precursor forms were reduced upon Exportin-5 overexpression (Figure 4C), suggesting a more efficient export out of the nucleus. Next, we investigated the effect of Exportin-5 on tumor growth activity and found that the overexpression of Exportin-5 decreased cell proliferation rate and reduced ability of colony formation (Figure 4D and 4G). Furthermore, we simultaneously transfected these cells with anti-miR-433 and anti-miR-22 to assess to what extent the beneficial effects of Exportin-5 overexpression were related to miR-433 and miR-22. Increased proliferation and cellular migration suggested that these effects are at least in part mediated by these two miRNAs (Figures 4E, F). Finally, western blot analysis showed that the overexpression of Exportin-5 induced a decrease in HDAC6 and downregulated signaling on Hh and MAPK pathways (Figure 4H). Taken together, these findings suggest that the rescue of Exportin-5 allows the precursor miRNAs to reach the cytoplasm where they are further processed to mature forms that regulate the expression of HDAC6 and the malignant phenotype.
Figure 4. Effect of experimental overexpression of Exportin-5 in CCA.
A, The CCA cell line HuCCT-1 was transfected with XPO5 expression vector (HuCCT-1-XPO5) or empty vector (HuCCT-1) and the expression of XPO5 protein analyzed by confocal immunofluorescence. XPO5 is stained in red and nuclei in blue with DAPI. B, The expression levels of the mature forms (miR-433 and miR-22) and the precursor forms (pri-miR-433 and pri-miR-22) were analyzed by qRT-PCR (*p<0.01 n=3). C, qPCR for pre-miR-433 in isolated nuclear fractions (*p<0.05, n=8). D, Proliferation rates were measured by MTS assay (*p<0.001, n=24). E, Cellular migration assay (p<0.05, n=3). F, Anchorage independent growth was assessed by colony number formation in soft agar *(p<0.0001, n=6). G, The effect of XPO5 over-expression on the levels of HDAC6, MAPK and Hh signaling were analyzed by western blots (*p<0.05, n=3).
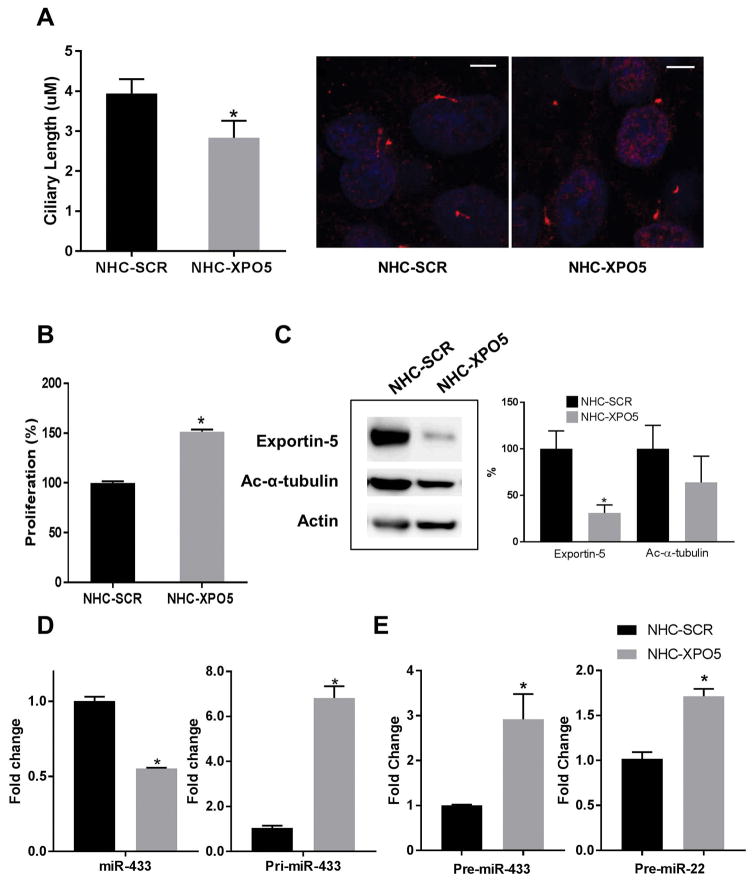
The downregulation of Exportin-5 in normal cells induces shorter cilia and cell proliferation
To further explore the role of Exportin-5 in miRNA dysregulation and cholangiocyte transformation, we downregulated Exportin-5 in normal cholangiocyte cells by shRNA transfection. We found that the downregulation of Exportin-5 induced shorter cilia as well as increased proliferation (Figure 5A, B). Furthermore, Exportin-5 downregulation induced decreased levels of acetylated-a-tubulin, suggesting increased activity of HDAC6 (Figure 5C). Importantly, the downregulation of Exportin-5 induced a decrease in miRNA-433 while increasing the levels of the precursor form (Figure 5D), and, consistent with a nuclear retention of the precursors, the nuclear levels of miRNAs precursor forms were significantly increased upon Exportin-5 knockdown (Figure 5E).
Figure 5. Effect of experimental downregulation of Exportin-5 in normal cholangiocytes.
The normal cholangiocyte cell line NHC was stably transfected with scrambled (NHC-SCR) or specific shRNA targeting XPO5 (NHC-XPO5). A, The effect of XPO5 overexpression on the ciliary phenotype was measured by confocal immunofluorescence microscopy where cilia are stained in red with the ciliary marker acetylated-a-tubulin and nuclei in blue with DAPI (*p<0.05, n=25). B, Proliferation rates were assessed by MTS assay (*p<0.001, n=24). C, Protein levels of XPO5 and acetylated-a-tubulin were assessed by western blots (*p<0.05, n=3). D, Total levels of miR-433 and pri-miR-433 were analyzed by qPCR (p<0.001, n=3). E, Nuclear levels of pre-miR-433 and pre-miR-22 were assessed by qPCR on isolated nuclear fractions (p<0.001, n=3).
DISCUSSION
The key findings reported here relate to the regulation of HDAC6 and ciliary expression by miRNAs and Exportin-5 in cholangiocarcinoma cells. We found that: (i) at least two miRNAs (miR-22 and miR-433) targeting HDAC6 mRNA are down-regulated in CCA; (ii) the experimental overexpression of miR-433 and/or miR-22 in cholangiocarcinoma cells decreases HDAC6 protein levels, partially restores ciliogenesis, and induces reduction in the malignant phenotype (i.e., cell proliferation, anchorage independent growth, cellular migration, and MAPK and Hh signaling pathways); (iii) precursor forms of miR-22 and miR-433 are up-regulated in CCA cell lines; (iv) Exportin-5 is decreased in CCA; and (v) experimental overexpression of Exportin-5 increases miRNAs levels in the cytoplasm and decreases the precursor forms in the nuclear fraction, and induces reduction in malignancy phenotype while the opposite effects were observed when Exportin-5 was downregulated in normal cholangiocyte cells. The results of our study provide the first direct evidence that Exportin-5 is altered in cholangiocarcinoma cells, promoting the accumulation of precursor forms of miR-433 and miR-22 and the subsequent downregulation of mature forms and HDAC6 overexpression and ciliary loss.
We previously reported that HDAC6 is overexpressed in cholangiocarcinoma which induces the loss of primary cilia and promotes cholangiocyte malignancy. The loss of primary cilia dysregulates tumorigenic signaling pathways like MAPK and Hh (7). Likewise, overexpression of HDAC6 has been reported in different types of tumors, such as breast cancer, melanoma, and pancreatic cancer (21–23). Liu et al, found that HDAC6 was up-regulated in both melanoma cells and tissues compared to normal skin cells and tissues (22). Indeed, HDAC6 knockdown in melanoma cells decreased cell growth and colonization. These findings are consistent with our results; however, the levels of HDAC6 mRNA are higher in melanoma cell lines than in normal skin cells. In contrast, we found unaffected levels of HDAC6 mRNA despite protein overexpression, suggesting a potential posttranscriptional regulation.
The study of the role of microRNAs and its dysregulation in the initiation and progression of cancer has become a very important subject. MicroRNAs regulate gene expression by binding to complementary target mRNAs and promoting their decay or inhibiting their translation. According to the role of their target genes, its aberrant expression functions as tumor suppressors or oncogenes (24).
There are several miRNAs dysregulated in CCA (15, 25). Although the mechanisms by which different miRNAs modify CCA cells phenotype and tumor cells in general are well studied, the reasons for miRNAs alterations remain less known. Epigenetic alterations, DNA copy anomalies, transcriptional deregulations, and mutations might have roles in miRNAs deregulation (26). For instance, the treatment of HuCCT-1 cells with histone deacetylase inhibitor in combination with the DNA-demethylating agent 5-aza-29-deoxycytidine induces upregulation of miR-376c. Based on this, Iwaki et al suggested that miR-376c expression is suppressed by HDAC modification and DNA methylation (27). He et al., reported that Arsenic resistance protein 2 (Ars2), a protein involved in miRNAs biogenesis, is overexpressed in biliary cancer. Depletion of Ars2 induces decreased expression of miR-21 and consequent reduction of cell proliferation and invasion rates (28). Also, the regulatory region of miR-21 is inducible by interleukin-6 (IL-6)/signal transducer and activator of transcription 3 (STAT3) and can be post transcriptionally regulated by the transforming growth factor beta (TGF-β) and bone morphogenetic protein (BMP) signaling (29). The transcription factor c-Myc regulates around 10–15% of the human genes and it has been identified that c-Myc binds to the regulatory region of the miR-17-92 cluster (30). Therefore, an increased expression of c-Myc might cause the stimulation of the miRNAs in the cluster (31, 32).
As we described here, another mechanism involved in the dysregulation of miRNAs may be a defective nuclear/cytoplasmic transport via the downregulation of Exportin-5. In agreement with the described mechanism, Melo et al showed that mutations in the Exportin-5 gene is another mechanism to explain deregulated miRNA expressions (33, 34). Mutated XPO5 is mainly accumulated in the nucleus of colon-rectal cancer cell lines (HCT-15 and DLD-1) and leads to pre-miRNA being trapped in the nucleus. Our study also found that miRNA precursors are trapped in the nucleus likely as a consequence of XPO5 downregulation in CCA cell lines. Whether XPO5 mutations, as suggested by TCGA database, epigenetic modifications, or other regulatory mechanisms induce the downregulation of Exportin-5 in CCA cells remains to be elucidated.
Interestingly, it has been recently reported that there are decreased levels of both mRNA and protein XPO5 in hepatocellular carcinoma tissues compared to healthy control tissues in concordance with our findings in this small number of cholangiocarcinoma human samples (35). Additionally, a homozygous single nucleotide polymorphism site on the XPO5 gene in hepatocellular carcinoma patients showed shorter overall survival compared to both wild type and heterozygous SNP genes (36). Although there is no data about miRNAs amounts in this study, based on other papers and in our own experiments, it is reasonable to assume that levels of mature miRNAs would decrease, inducing a more severe tumor phenotype. In our studies, we showed that recovering mature miRNAs by rescuing the expression of Exportin-5 have an anti-proliferative effect and a reduced cell anchorage-independent growth.
Interestingly, our findings suggest that Exportin-5 may not be the only nuclear export mechanism available for miRNAs, at least in tumor cells. Even though the downregulation of miRNAs is a common feature of tumors, which may be explained by a downregulation of Exportin-5, there are other miRNAs that are overexpressed. Indeed, consistent with our observations, mature miRNAs are markedly decreased in the cytoplasm and the precursor forms are accumulated in the nucleus of the colorectal cancer cell line HCT116 with XPO5 knockout; however, a significant number of microRNAs are still produced in those cells (37). On the other hand, consistent with our data, overexpression of XPO5 in HeLa cells led to an increase in mature miRNAs (38). Interestingly, decreased expression of some miRNAs was described, suggesting that the biogenesis of miRNAs is a process that is individually and intricately regulated. Taken together, these results suggest that an alternative pathway to mediate nuclear translocation of pre-miRNAs may exist. In fact, miR-320, a non-canonical pre-miRNA, is exported by XPO1 and this shuttle involves exporting pri-miRNAs in C. elegans and Drosophila as well as mature miRNAs in mammals (39).
In summary, we found that mature forms of miR-22 and miR-433 are significantly decreased in CCA cell lines and human samples, while their precursor forms are trapped in the nucleus. Exportin-5 downregulation may be the underlying mechanism for this deregulation. We previously speculated that the loss of primary cilia by the overexpression of HDAC6 in CCA induces disengagement between the environment and the cell interior and induces the derepression of tumorigenic pathways, and thereby proposed that the restoration of primary cilia would be a potential approach to decrease cholangiocarcinoma progression. Here, we have further defined the mechanisms of ciliary loss by the dysregulation of miRNAs and identified new potential targets such as miR-22, miR-433, and Exportin-5 to reduce CCA cell growth (Supplemental Fig 3).
Supplementary Material
Supplemental Figure 1. A, Western blotting was performed to determine endogenous HDAC6 protein in HuCCT-1 cells that were transfected with pTarget, pTarget-miR-433, or anti-miR-433. β-Actin served as an internal control. B, Luciferase reporter assays were performed to verify the binding of miR-433 in the 3′-UTR of HDAC6. Reporter vector pMIR-HDAC6 and miR-433 or scrambled negative control [Scr] were co-transfected into HuCCT1 cells.
Supplemental Figure 2. Confocal immunofluorescence analysis of Exportin-5 expression in different CCA cells (HuCCT-1, KMCH, TFK7) compared to normal cholangiocytes (NHC).
Supplemental Figure 3. Working model. A, the downregulation of Exportin-5 in CCA induces the accumulation of the precursor forms of miR-22 and miR-433 in the nucleus. Therefore, the levels of mature miR-22 and miR-433 in the cytoplasm are significantly decreased and the translational repression of target messenger RNAs like Hdac6 is lost, inducing an increase in HDAC6 protein levels. B, the loss of primary cilia by the overexpression of HDAC6 in CCA induces the disengagement between the environment and the cell interior and induces the derepression of tumorigenic pathways like MAPK and Hh. Therefore, we propose that the restoration of primary cilia by targeting HDAC6, miR-22, miR-433, and/or Exportin-5 would be a potential approach to reduce CCA growth.
Supplemental Table 1. Cholangiocarcinoma samples cohort description.
Supplemental Table 2. MicroRNAs predicted to target HDAC6: expression in hepatobiliary cancers and described biological functions.
Acknowledgments
Financial Support: This work was supported by National Institutes of Health Grant R01CA183764 (to S.A.G.), The Randy Shaver Cancer Research and Community Fund Award (to S.A.G.), The Hormel Foundation, Grant P30DK084567 and R01DK24031 (to N.F.L.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. The miRNA qPCR study in human samples were supported by Spanish Ministry of Economy and Competitiveness [J.M.B. (FIS PI15/01132 and Miguel Servet Program CON14/00129) and M.J.P. (FIS PI14/00399) cofinanced by “Fondo Europeo de Desarrollo Regional” (FEDER)], and BIOEF (Basque Foundation for Innovation and Health Research: EiTB Maratoia BIO15/CA/016/BD to J.M.B).
Abbreviations
- CCA
cholangiocarcinoma
- HDAC6
histone-deacetylase-6
- miRNA
microRNA
BIBLIOGRAPHY
- 1.Bergquist A, von Seth E. Epidemiology of cholangiocarcinoma. Best Pract Res Clin Gastroenterol. 2015;29:221–232. doi: 10.1016/j.bpg.2015.02.003. [DOI] [PubMed] [Google Scholar]
- 2.Maemura K, Natsugoe S, Takao S. Molecular mechanism of cholangiocarcinoma carcinogenesis. J Hepatobiliary Pancreat Sci. 2014;21:754–760. doi: 10.1002/jhbp.126. [DOI] [PubMed] [Google Scholar]
- 3.Williams TM, Majithia L, Wang SJ, Thomas CR., Jr Defining the role of adjuvant therapy: cholangiocarcinoma and gall bladder cancer. Semin Radiat Oncol. 2014;24:94–104. doi: 10.1016/j.semradonc.2014.01.001. [DOI] [PubMed] [Google Scholar]
- 4.Jiao Y, Pawlik TM, Anders RA, Selaru FM, Streppel MM, Lucas DJ, Niknafs N, et al. Exome sequencing identifies frequent inactivating mutations in BAP1, ARID1A and PBRM1 in intrahepatic cholangiocarcinomas. Nat Genet. 2013;45:1470–1473. doi: 10.1038/ng.2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Voss JS, Holtegaard LM, Kerr SE, Fritcher EG, Roberts LR, Gores GJ, Zhang J, et al. Molecular profiling of cholangiocarcinoma shows potential for targeted therapy treatment decisions. Hum Pathol. 2013;44:1216–1222. doi: 10.1016/j.humpath.2012.11.006. [DOI] [PubMed] [Google Scholar]
- 6.Borger DR, Tanabe KK, Fan KC, Lopez HU, Fantin VR, Straley KS, Schenkein DP, et al. Frequent mutation of isocitrate dehydrogenase (IDH)1 and IDH2 in cholangiocarcinoma identified through broad-based tumor genotyping. Oncologist. 2012;17:72–79. doi: 10.1634/theoncologist.2011-0386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gradilone SA, Radtke BN, Bogert PS, Huang BQ, Gajdos GB, LaRusso NF. HDAC6 inhibition restores ciliary expression and decreases tumor growth. Cancer research. 2013;73:2259–2270. doi: 10.1158/0008-5472.CAN-12-2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang PH, Zhang L, Zhang YJ, Zhang J, Xu WF. HDAC6: physiological function and its selective inhibitors for cancer treatment. Drug Discov Ther. 2013;7:233–242. doi: 10.5582/ddt.2013.v7.6.233. [DOI] [PubMed] [Google Scholar]
- 9.Seidel C, Schnekenburger M, Dicato M, Diederich M. Histone deacetylase 6 in health and disease. Epigenomics. 2015;7:103–118. doi: 10.2217/epi.14.69. [DOI] [PubMed] [Google Scholar]
- 10.Gradilone SA, Masyuk AI, Splinter PL, Banales JM, Huang BQ, Tietz PS, Masyuk TV, et al. Cholangiocyte cilia express TRPV4 and detect changes in luminal tonicity inducing bicarbonate secretion. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:19138–19143. doi: 10.1073/pnas.0705964104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Masyuk AI, Masyuk TV, Splinter PL, Huang BQ, Stroope AJ, LaRusso NF. Cholangiocyte cilia detect changes in luminal fluid flow and transmit them into intracellular Ca2+ and cAMP signaling. Gastroenterology. 2006;131:911–920. doi: 10.1053/j.gastro.2006.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Masyuk AI, Gradilone SA, Banales JM, Huang BQ, Masyuk TV, Lee SO, Splinter PL, et al. Cholangiocyte primary cilia are chemosensory organelles that detect biliary nucleotides via P2Y12 purinergic receptors. American journal of physiology Gastrointestinal and liver physiology. 2008;295:G725–734. doi: 10.1152/ajpgi.90265.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Masyuk AI, Huang BQ, Ward CJ, Gradilone SA, Banales JM, Masyuk TV, Radtke B, et al. Biliary exosomes influence cholangiocyte regulatory mechanisms and proliferation through interaction with primary cilia. American journal of physiology Gastrointestinal and liver physiology. 2010;299:G990–999. doi: 10.1152/ajpgi.00093.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 15.Pisarello MJ, Loarca L, Ivanics T, Morton L, LaRusso N. MicroRNAs in the Cholangiopathies: Pathogenesis, Diagnosis, and Treatment. J Clin Med. 2015;4:1688–1712. doi: 10.3390/jcm4091688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miyagiwa M, Ichida T, Tokiwa T, Sato J, Sasaki H. A new human cholangiocellular carcinoma cell line (HuCC-T1) producing carbohydrate antigen 19/9 in serum-free medium. In vitro cellular & developmental biology: journal of the Tissue Culture Association. 1989;25:503–510. doi: 10.1007/BF02623562. [DOI] [PubMed] [Google Scholar]
- 17.Murakami T, Yano H, Maruiwa M, Sugihara S, Kojiro M. Establishment and characterization of a human combined hepatocholangiocarcinoma cell line and its heterologous transplantation in nude mice. Hepatology. 1987;7:551–556. doi: 10.1002/hep.1840070322. [DOI] [PubMed] [Google Scholar]
- 18.de Planell-Saguer M, Rodicio MC, Mourelatos Z. Rapid in situ codetection of noncoding RNAs and proteins in cells and formalin-fixed paraffin-embedded tissue sections without protease treatment. Nat Protoc. 2010;5:1061–1073. doi: 10.1038/nprot.2010.62. [DOI] [PubMed] [Google Scholar]
- 19.Simon D, Laloo B, Barillot M, Barnetche T, Blanchard C, Rooryck C, Marche M, et al. A mutation in the 3′-UTR of the HDAC6 gene abolishing the post-transcriptional regulation mediated by hsa-miR-433 is linked to a new form of dominant X-linked chondrodysplasia. Human molecular genetics. 2010;19:2015–2027. doi: 10.1093/hmg/ddq083. [DOI] [PubMed] [Google Scholar]
- 20.Huang S, Wang S, Bian C, Yang Z, Zhou H, Zeng Y, Li H, et al. Upregulation of miR-22 promotes osteogenic differentiation and inhibits adipogenic differentiation of human adipose tissue-derived mesenchymal stem cells by repressing HDAC6 protein expression. Stem cells and development. 2012;21:2531–2540. doi: 10.1089/scd.2012.0014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Seo J, Min SK, Park HR, Kim DH, Kwon MJ, Kim LS, Ju YS. Expression of Histone Deacetylases HDAC1, HDAC2, HDAC3, and HDAC6 in Invasive Ductal Carcinomas of the Breast. Journal of breast cancer. 2014;17:323–331. doi: 10.4048/jbc.2014.17.4.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu J, Gu J, Feng Z, Yang Y, Zhu N, Lu W, Qi F. Both HDAC5 and HDAC6 are required for the proliferation and metastasis of melanoma cells. Journal of translational medicine. 2016;14:7. doi: 10.1186/s12967-015-0753-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li D, Sun X, Zhang L, Yan B, Xie S, Liu R, Liu M, et al. Histone deacetylase 6 and cytoplasmic linker protein 170 function together to regulate the motility of pancreatic cancer cells. Protein & cell. 2014;5:214–223. doi: 10.1007/s13238-013-0010-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Acunzo M, Romano G, Wernicke D, Croce CM. MicroRNA and cancer--a brief overview. Adv Biol Regul. 2015;57:1–9. doi: 10.1016/j.jbior.2014.09.013. [DOI] [PubMed] [Google Scholar]
- 25.O’Hara SP, Gradilone SA, Masyuk TV, Tabibian JH, LaRusso NF. MicroRNAs in Cholangiopathies. Curr Pathobiol Rep. 2014;2:133–142. doi: 10.1007/s40139-014-0048-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang N, Xia S, Chen K, Xiang X, Zhu A. Genetic alteration regulated by microRNAs in biliary tract cancers. Crit Rev Oncol Hematol. 2015;96:262–273. doi: 10.1016/j.critrevonc.2015.05.015. [DOI] [PubMed] [Google Scholar]
- 27.Iwaki J, Kikuchi K, Mizuguchi Y, Kawahigashi Y, Yoshida H, Uchida E, Takizawa T. MiR-376c down-regulation accelerates EGF-dependent migration by targeting GRB2 in the HuCCT1 human intrahepatic cholangiocarcinoma cell line. PLoS One. 2013;8:e69496. doi: 10.1371/journal.pone.0069496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.He Q, Cai L, Shuai L, Li D, Wang C, Liu Y, Li X, et al. Ars2 is overexpressed in human cholangiocarcinomas and its depletion increases PTEN and PDCD4 by decreasing microRNA-21. Mol Carcinog. 2013;52:286–296. doi: 10.1002/mc.21859. [DOI] [PubMed] [Google Scholar]
- 29.Lu L, Byrnes K, Han C, Wang Y, Wu T. miR-21 targets 15-PGDH and promotes cholangiocarcinoma growth. Mol Cancer Res. 2014;12:890–900. doi: 10.1158/1541-7786.MCR-13-0419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fernandez PC, Frank SR, Wang L, Schroeder M, Liu S, Greene J, Cocito A, et al. Genomic targets of the human c-Myc protein. Genes Dev. 2003;17:1115–1129. doi: 10.1101/gad.1067003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.O’Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT. c-Myc-regulated microRNAs modulate E2F1 expression. Nature. 2005;435:839–843. doi: 10.1038/nature03677. [DOI] [PubMed] [Google Scholar]
- 32.Saito Y, Suzuki H, Matsuura M, Sato A, Kasai Y, Yamada K, Saito H, et al. MicroRNAs in Hepatobiliary and Pancreatic Cancers. Front Genet. 2011;2:66. doi: 10.3389/fgene.2011.00066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Melo SA, Moutinho C, Ropero S, Calin GA, Rossi S, Spizzo R, Fernandez AF, et al. A genetic defect in exportin-5 traps precursor microRNAs in the nucleus of cancer cells. Cancer Cell. 2010;18:303–315. doi: 10.1016/j.ccr.2010.09.007. [DOI] [PubMed] [Google Scholar]
- 34.Melo SA, Esteller M. A precursor microRNA in a cancer cell nucleus: get me out of here! Cell Cycle. 2011;10:922–925. doi: 10.4161/cc.10.6.15119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li Y, Wang X, He B, Cai H, Gao Y. Downregulation and tumor-suppressive role of XPO5 in hepatocellular carcinoma. Mol Cell Biochem. 2016;415:197–205. doi: 10.1007/s11010-016-2692-3. [DOI] [PubMed] [Google Scholar]
- 36.Liu S, An J, Lin J, Liu Y, Bao L, Zhang W, Zhao JJ. Single nucleotide polymorphisms of microRNA processing machinery genes and outcome of hepatocellular carcinoma. PLoS One. 2014;9:e92791. doi: 10.1371/journal.pone.0092791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim YK, Kim B, Kim VN. Re-evaluation of the roles of DROSHA, Export in 5, and DICER in microRNA biogenesis. Proc Natl Acad Sci U S A. 2016;113:E1881–1889. doi: 10.1073/pnas.1602532113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Volk N, Shomron N. Versatility of MicroRNA biogenesis. PLoS One. 2011;6:e19391. doi: 10.1371/journal.pone.0019391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xie M, Li M, Vilborg A, Lee N, Shu MD, Yartseva V, Sestan N, et al. Mammalian 5′-capped microRNA precursors that generate a single microRNA. Cell. 2013;155:1568–1580. doi: 10.1016/j.cell.2013.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1. A, Western blotting was performed to determine endogenous HDAC6 protein in HuCCT-1 cells that were transfected with pTarget, pTarget-miR-433, or anti-miR-433. β-Actin served as an internal control. B, Luciferase reporter assays were performed to verify the binding of miR-433 in the 3′-UTR of HDAC6. Reporter vector pMIR-HDAC6 and miR-433 or scrambled negative control [Scr] were co-transfected into HuCCT1 cells.
Supplemental Figure 2. Confocal immunofluorescence analysis of Exportin-5 expression in different CCA cells (HuCCT-1, KMCH, TFK7) compared to normal cholangiocytes (NHC).
Supplemental Figure 3. Working model. A, the downregulation of Exportin-5 in CCA induces the accumulation of the precursor forms of miR-22 and miR-433 in the nucleus. Therefore, the levels of mature miR-22 and miR-433 in the cytoplasm are significantly decreased and the translational repression of target messenger RNAs like Hdac6 is lost, inducing an increase in HDAC6 protein levels. B, the loss of primary cilia by the overexpression of HDAC6 in CCA induces the disengagement between the environment and the cell interior and induces the derepression of tumorigenic pathways like MAPK and Hh. Therefore, we propose that the restoration of primary cilia by targeting HDAC6, miR-22, miR-433, and/or Exportin-5 would be a potential approach to reduce CCA growth.
Supplemental Table 1. Cholangiocarcinoma samples cohort description.
Supplemental Table 2. MicroRNAs predicted to target HDAC6: expression in hepatobiliary cancers and described biological functions.