Abstract
Rice is a facultative short-day (SD) plant in which flowering is induced under SD conditions or by other environmental factors and internal genetic programs. Overexpression of Histone Deacetylase 701 (HDT701) accelerates flowering in hybrid rice. In this study, mutants defective in HDT701 flowered late under both SD and long-day conditions. Expression levels of florigens Heading date 3a (Hd3a) and Rice Flowering Locus T1 (RFT1), and their immediate upstream floral activator Early heading date 1 (Ehd1), were significantly decreased in the hdt701 mutants, indicating that HDT701 functions upstream of Ehd1 in controlling flowering time. Transcript levels of OsINDETERMINATE SPIKELET 1 (OsIDS1), an upstream repressor of Ehd1, were significantly increased in the mutants while those of OsGI and Hd1 were reduced. Chromatin-immunoprecipitation assays revealed that HDT701 directly binds to the promoter region of OsIDS1. These results suggest that HDT701 induces flowering by suppressing OsIDS1.
Keywords: flowering time, histone deacetylase, OsIDS1, OsGI, Hd1
INTRODUCTION
Flowering is one of the most crucial biological processes in plants because it is a prerequisite for the development of fruits and grains. Transition from the vegetative phase is the first step toward reproductive success. Therefore, producing flowers at the appropriate time is a key factor. Whereas early flowering shortens the vegetative phase to an insufficient period that often leads to reduced yields, deferred flowering may also contribute to yield losses when plants in temperate regions are exposed to characteristically colder temperatures later in the growing season. For rice (Oryza sativa), chilling at the grain ripening stage results in immature grains while high temperatures are associated with heat damage and a reduction in grain quality. Thus, flowering time is highly correlated with total grain yield and quality in rice (Morita et al., 2017; Sun et al., 2014; Zhang et al., 2015).
The timing of floral transition is regulated by many factors, e.g., internal genetic programming, day length, temperature, nutrient availability, and abiotic/biotic stresses (Cho et al., 2017). In Arabidopsis (Arabidopsis thaliana), a long-day (LD) plant, flowering time is accelerated by longer photoperiods. GIGANTEA (GI) merges signals from photoreceptors and a circadian clock to activate CONSTANS (CO), which in turn promotes the expression of Flowering Locus T (FT), a major floral activator that is expressed in the vascular tissues of leaves, all of which lead to the induction of floral transition (Fowler et al.,1999; Park et al., 1999; Samach et al., 2000; Yanovsky and Kay, 2002).
Oryza sativa GIGANTEA (OsGI), Heading date 1 (Hd1), and Heading date 3a (Hd3a) are the rice homologues of GI, CO, and FT, respectively. This core flowering pathway is conserved in many plant species. Although CO enhances flowering in Arabidopsis, Hd1 has a dual function in rice. Whereas Hd1 promotes flowering under short-day (SD) conditions by enhancing the expression of Early heading date 1 (Ehd1) (Zhang et al., 2017), the factor suppresses flowering under LD conditions by inhibiting Ehd1 and Hd3a (Hayama et al., 2003). In addition to this conserved flowering pathway, Flowering Locus C (FLC ) in Arabidopsis and Grain number, plant height, and heading date7 (Ghd7 ) and Early heading date 1 (Ehd1) in rice are unique floral regulators. In these dedicated flowering pathways, FLC and Ghd7 act as major flowering repressors while Ehd1 functions as a floral activator (Cho et al., 2016; Doi et al., 2004; Sun et al., 2014).
Rice is a facultative SD plant. Its heading date is advanced under SD conditions (<13 h of light/day) but retarded under LD conditions (>14 h of light/day) (Cho et al., 2016; Ishikawa et al., 2011; Kim et al., 2013; Lee et al., 2007; Nishida et al., 2002). Rice has two florigens, Hd3a and Rice Flowering Locus T1 (RFT1), that are induced by Ehd1 (Corbesier et al., 2007; Doi et al., 2004; Ryu et al., 2009). Several transcription factors activate or repress the expression of Ehd1, a gene that is a critical convergence point for various flowering signals in rice.
Several genes, including Ghd7 and OsMADS56, preferentially function as suppressors of flowering under LD. However, some constitutive suppressors inhibit flowering regardless of day length. For example, two AP2-like genes, OsINDETERMINATE SPIKELET 1 (OsIDS1) and SUPERNUMERARY BRACT (SNB), repress the expression of Ehd1 and florigens, resulting in delayed flowering under both LD and SD conditions. In this pathway, microRNA172 (miR172 ) degrades transcripts of OsIDS1 and SNB to induce flowering, whereas Oryza sativa Phytochrome B (OsPhyB) enhances the expression of OsIDS1 and SNB by repressing miR172 to inhibit flowering (Lee et al., 2014). OsCOL4, a member of the CONSTANS-like (COL) family in rice, is up-regulated by OsPhyB. The former suppresses flowering under both SD and LD by dampening the transcript levels of Ehd1 and the florigens via upregulation of floral repressors OsIDS1 and SNB (Lee et al., 2010; 2014). Oryza sativa LEAFY COTYLEDON 2 and FUSCA 3-LIKE 1 (OsLFL1) constitutively deters rice flowering by directly attenuating the transcript level of Ehd1 (Peng et al., 2007; 2008). Furthermore, OsLF, which encodes a typical HLH protein, delays flowering regardless of day length by directly repressing Hd1 and OsGI (Zhao et al., 2011).
The histone acetyltransferases (HATs) and histone deacetylases (HDACs) reversibly catalyze acetylation or deacetylation on histone lysine residues for the transcriptional activation and repression, respectively, of target genes. Plant HDACs can be classified into three major families: the RPD3/HDA1 superfamily, the SIR2 family, and the plant-specific HD2 family (Pandey et al., 2002). In Arabidopsis, histone acetylation and deacetylation are involved in various biological processes such as flowering time, leaf development, seed abortion, and abiotic stress responses (Dangl et al., 2001; He et al., 2003; Luo et al., 2012a; 2015; Sridha and Wu, 2006; Ueno et al., 2007; Wu et al., 2000; 2008). The rice genome contains at least 19 HDAC genes (Hu et al., 2009), including at least two HD2 genes -- Histone deacetylase 701 (HDT701) and Histone deacetylase 702 (HDT702) -- based on phylogenic analysis (Fu et al., 2007). HDT702 RNAi plants have smaller-diameter stems and much narrower leaves, implying that this gene has a role in cell division or growth (Hu et al., 2009). HDT701 encodes a histone H4 deacetylase that reduces acetylation levels at the 5th and 16th lysine residues of histone H4. Its overexpression makes rice plants more susceptible to Magnoporthe oryzae and Xanthomonas oryzae pv. oryzae whereas HDT701 RNAi plants are resistant to those pathogens. This suggests that HDT701 functions as a negative regulator in plant innate immunity by modulating histone H4 acetylation of defense-related genes in rice (Ding et al., 2012). Overexpression of HDT701 also leads to late seed germination due to decreased histone H4 acetylation and reduced expression of GA-biosynthetic genes. In addition, HDT701-overexpression transgenic plants display enhanced resistance to salt and osmotic stresses during the seedling stage, thereby denoting the role this gene has in seed germination and responses to abiotic stresses (Zhao et al., 2015). Finally, overexpression of HDT701 accelerates flowering under natural LD conditions by repressing OsGI and Hd1 (Li et al., 2011).
In this study, we analyzed the role of HDT701 in determining flowering time by analyzing knockout (KO) mutants. Our results demonstrated that this gene controls flowering time in rice mainly by suppressing OsIDS1, which is an upstream suppressor of Ehd1 and florigens.
MATERIALS AND METHODS
Plant materials and growth conditions
Oryza sativa var. japonica cultivars Dongjin and Hwayoung were used to generate the T-DNA tagging lines (Jeon et al., 2000; Jeong et al., 2002). To download the genomic DNA sequences, we accessed the Rice Annotation Project Database (RAP-DB; http://rapdb.dna.affrc.go.jp; Tanaka et al., 2008) and the TIGR Rice Genome Annotation Project Database (http://rice.plantbiology.msu.edu; Ouyang et al., 2007). The hdt701-1 mutant (Line number 1B-05907) was identified from our rice T-DNA insertion sequence database (An et al., 2005a; 2005b; Jeong et al., 2006). Homozygous mutants were confirmed by PCR, using genomic DNA extracted from the leaf blade. The primers for genotyping were TAGCTCCGCCTCCCACCT (F), TGCCCTGGGAGCTGGAATG (R), and AACGCTGATCAATTCCACAG (NGUS1) (Lee et al., 2015). Additional KO alleles of hdt701 were generated in the ‘Nipponbare’ rice background through CRISPR/Cas9 techniques (Miao et al., 2013). The plants were genotyped by sequencing the CRISPR/Cas9 target region using the genomic DNA extracted from leaf blades. Seeds were germinated either on an MS medium or in soil, as previously described (Yi and An, 2013). Plants were cultured naturally in the paddy field or else in controlled growth rooms maintained under LD conditions (14 h light, 28°C/10 h dark, 22°C; humidity approximately 60%) or SD conditions (12 h light, 28°C /12 h dark, 22°C; humidity approximately 70%), as previously described (Cho et al., 2016).
RNA isolation and quantitative real-time PCR analyses
Total RNA was isolated from fully grown uppermost healthy leaves with RNAiso Plus (TaKaRa, Japan; http://www.takarabio.com). RNA samples with 260/280 nm ratios of >1.8 (Nano-Drop 2000; Thermo Scientific, Wilmington, DE, USA; http://www.nanodrop.com) were used. First-strand cDNA synthesis was performed with 2 μg of total RNA plus Moloney murine leukemia virus reverse transcriptase (Promega, USA; http://www.promega.com), RNasin® Ribonuclease Inhibitor (Promega), oligo (dT) 18 primer, and dNTP. Afterward, synthesized cDNAs and SYBR Green I Prime Q-Master mix (GENETBIO, Daejeon, Republic of Korea) were utilized to monitor gene expression via quantitative real-time (qRT)-PCR on a Rotor-Gene Q system (QIAGEN, Germany) (Cho et al., 2016; Ryu et al., 2009). Rice Ubi was used for normalization. All experiments were conducted at least three times and, for each experiment, more than three independent samples were used. To ensure primer specificity, we performed these experiments only when the melting curve displayed a single sharp peak. The ΔΔCT method was applied to calculate changes in relative expression. All primers for quantitative real-time PCR are listed in Supplementary Table S1.
Vector construction and plant transformation
For constructing the CRISPR/Cas9 vector, the rational CRISPR/Cas9 target sequences with protospacer adjacent motifs were screened with the aid of the CRISPRdirect web server (http://crispr.dbcls.jp; Naito et al., 2015) to find potential target sequences with minimal off-target cleavage. A spacer sequence (AAAGATCATTCCAGCTCCCA) was cloned into entry vector pOs-sgRNA for monitoring the expression of sgRNA. The resulting recombinant entry vector, pOs-sgRNA, was further cloned into a destination vector, pH-Ubi-cas9-7, using the GatewayTM system (Miao et al., 2013). For generation of the HA tag transgenic plants, a full-length cDNA of HDT701 was isolated by PCR, using two primers: 5′-AAGCTTTAGCTCCGCCTCCCACCT-3′ and 5′-ACTAGTCTT GGCGGGGTGCTTGGC-3′. Amplified PCR product was digested with restriction enzymes Hind III and SpeI, and inserted into the binary vector pGA3428 under the control of the maize ubiquitin 1 promoter (Kim et al., 2009). The construct was introduced into Agrobacterium tumefaciens LBA4404 by the freeze–thaw method (An et al., 1989). Rice transformation via Agrobacterium-mediated co-cultivation was performed as previously reported (Jeon et al., 2000).
Histochemical assay of GUS activity
The plants were grown for 6 d in MS media under continuous light. After vacuum-infiltration for 30 min, samples were kept overnight at 37°C in a GUS-staining solution containing 100 mM sodium phosphate, 5 mM potassium ferricyanide, 5 mM potassium ferrocyanide, 0.5% Triton X-100, 10 mM EDTA (pH 8.0), 0.1% X-gluc (5-bromo-4-chloro-3-indolyl-β-D-glucuronic acid/cyclohexylammonium salt), 2% DMSO, and 5% methanol (Yoon et al., 2014). Chlorophylls were removed by sequentially incubating the samples in 70% and 95% ethanol at 60°C. The GUS-stained samples were then soaked for 30 to 60 min at room temperature in VISKOL clearing reagent (Phytosys LLC, USA, http://visikol.com/). After resin-sectioning (10 μm thick), GUS activity was visualized with a BX61 microscope (Olympus, Japan).
Chromatin-immunoprecipitation (ChIP) analysis
Transgenic plants expressing HDT701-HA were used for ChIP analysis as previously reported (Yoon et al., 2017). Briefly, 2 g of leaf blade sample were incubated in 3% formaldehyde. After nuclei isolation, chromatins were fragmented to approximately 500- to 1,000-bp lengths by sonication. As an input, 1% of the sample was gathered before pre-clearing. Anti-HA monoclonal antibodies (#2367; Cell Signaling) were used for immunoprecipitation. Data were normalized according to the percent-of-input method (Haring et al., 2007). Tested areas for OsIDS1 were P1; −1,886 ~ −1,766 bp, P2; −1,633 ~ −1,484 bp, P3; −1,139 ~ −1,265 bp, P4; −953 ~ −808 bp, and P5; −252 ~ −143 bp upstream from the translation start codon ATG. Those for SNB were P1; −1,893 ~ −1,766 bp, P2; −1,725 ~ −1,613 bp, P3; −1,517 ~ −1,412 bp, P4; −1,108 ~ −978 bp, P5; −821 ~ −692 bp, and P6; −555 ~ −425 bp upstream from the start codon. The PCR primers for ChIP are listed in Supplementary Table S1. All assays were conducted at least three times, each involving three biological replicates.
RESULTS
Identification of late-flowering mutants
A late-flowering mutant line, 1B-05907, was identified by screening T-DNA insertion tagging lines in the paddy field. The T-DNA was inserted in the first intron of HDT701 (Fig. 1A) and the transcript level for that gene was markedly decreased in the mutant (Fig. 1B). That line displayed a phenotype of flowering that was delayed by about two weeks in the field (Fig. 1C). Because flowering time is regulated by multiple pathways, including day length-preferential routes, we studied the mutant phenotypes under controlled SD and LD conditions. When compared with wild type (WT) controls, flowering of hdt701-1 mutant plants was delayed by approximately two weeks under SD and three weeks under LD conditions (Fig. 1D). This demonstrated that HDT701 is a constitutive activator of flowering regardless of day length.
Fig. 1. Schematic diagram of HDT701 structure and comparison of flowering times between WT and hdt701-1 mutants.
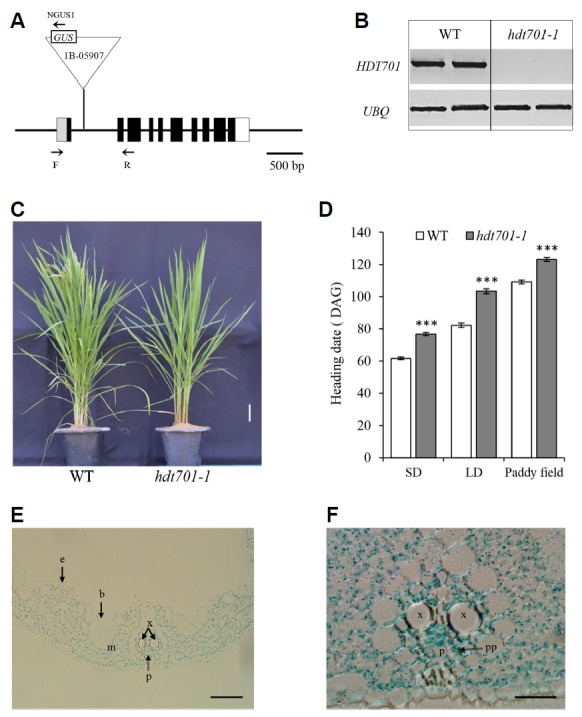
(A) Gene structure of HDT701. Black boxes indicate exons in coding region; lines connecting boxes, introns; gray box, 5′-UTR region; open box, 3′-UTR region. T-DNA is inserted into first intron of HDT701 in Line 1B-05907. Direction of promoterless GUS reporter gene is indicated within T-DNA (triangle). Primers F, R, and NGUS1 (marked with arrows) were used for genotyping. Scale bar, 500 bp. (B) HDT701 transcript levels in WT and hdt701-1 measured by RT-PCR. (C) Phenotypes of hdt701-1 and WT at heading stage under paddy field conditions. Scale bar, 10 cm. (D) Days to heading for WT and hdt701-1 plants under SD, LD, and field conditions. DAG, days after germination. Error bar indicate standard deviation; n = 8. Levels of significant difference are indicated by *** (P < 0.005). (E) GUS-staining of leaf blade cross-section from Line 1B-05907. (F) Close-up of leaf section at vasculature region. b, bulliform cells; e, epidermis; m, mesophyll cells; p, phloem; pp. phloem parenchyma; x, xylem. Scale bars, 50 μm (E) and 20 μm (F).
In the T-DNA tagging line, the GUS coding region was inserted into HDT701 at the same orientation as the tagged gene. GUS analysis of that line showed a positive response, indicating that HDT701 was translationally fused to GUS. We have previously reported that a translational fusion between a tagged gene and GUS can be made even when T-DNA is inserted within an intron (Wei et al., 2017). Analysis of the genomic DNA of the line revealed that only one copy of T-DNA was present in the entire genome, suggesting that GUS expression was likely due to a fusion between HDT701 and GUS. Histochemical GUS analysis of leaf blades showed that GUS signals were ubiquitous in the leaves, including phloem parenchyma cells and mesophyll cells (Fig. 1E). This result is consistent with a previous report that HDT701 is expressed in various organs (Zhao et al., 2015).
To confirm whether the delay in flowering time was indeed due to a mutation in HDT701, we generated additional alleles by the CRISPR/Cas9 method, designing a target site in the 5th exon of HDT701 (Fig. 2A) and obtaining five independent transgenic lines. Sequencing the flanking regions of that site revealed that CRISPR line #4 had deletions in both chromosomes and line #5 had a single-bp insertion, whereas line #1 did not carry any mutation. Further analyses of the two null mutant lines in the next generations (line #4 and #5) showed late flowering when compared to WT controls. While line #1 which has no mutation flowered at the same time as the WT (Figs. 2B and 2C). These experiments confirmed that defects in HDT701 delay flowering.
Fig. 2. Generation of additional hdt701 alleles by CRISPR/Cas9 method.
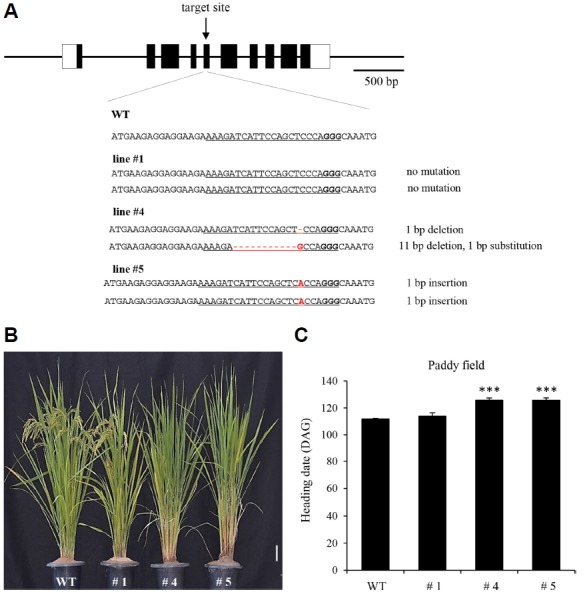
(A) Schematic diagram of HDT701 structure and sequence alignment of sgRNA target region displaying altered bases in mutant lines. The target sequence is underlined. Altered DNA sequences are indicated in red. (B) Phenotypes of hdt701 KO lines at heading stage. Scale bar, 10 cm. (C) Days to heading for WT, and KO Lines #1, 4, and 5 under natural paddy field conditions. Days to heading was scored when first panicle bolted. Error bars indicate standard deviations; n = 10. Levels of significant difference are indicated by ** (P < 0.01) and *** (P < 0.005).
Expression levels for floral regulators
To elucidate the functional roles of HDT701 in controlling flowering time, we monitored expression levels of previously identified genes that play critical roles in that event. We studied the effects of hdt701 mutations under both SD and LD conditions because some regulatory factors function differently depending upon day length. For example, osgi mutants display a significant delay in flowering under SD but only a slight delay under LD, indicating that OsGI controls flowering time preferentially under SD (Lee and An, 2015).
An earlier study showed that overexpression of HDT701 represses the expression of OsGI and Hd1, and induces flowering under natural LD conditions (Li et al., 2011). To verify that expression of these genes was also affected in our KO mutants, we performed qRT-PCR experiments with plants grown under controlled LD conditions. Expression was examined at 49 days after germination (DAG) because florigens and most upstream regulatory genes are active at that time when plants are grown under LD (Lee et al., 2016). Leaves were sampled nine times (2- to 4-h intervals) the day to observe any diurnal patterns. We first analyzed HDT701 and confirmed that the gene was completely silent throughout the 24-h period in the hdt701-1 mutant (Fig. 3A and Supplementary Fig. S1A). In the WT, the gene was expressed at higher levels in the dark but at reduced levels under illuminated conditions. This diurnal pattern of expression is similar to that previously reported (Li et al., 2011). Expression of Hd3a and RFT1 was significantly lower in the leaves from mutant plants, indicating that the delay in flowering was due to reduced expression of the florigens (Figs. 3B and 3C; Supplementary Figs. S1B and S1C). Ehd1, an immediate upstream regulator of those genes, was also significantly affected by the mutation (Fig. 3D). Activities of OsGI and Hd1 were slightly decreased in the hdt701 mutant, especially during the dark period (Figs. 3E and 3F; Supplementary Figs. S1E and S1F). We had not expected to make these observations because overexpression of hdt701-1 in ‘YS63′ hybrid rice also reduces the expression of OsGI and Hd1 (Li et al., 2011). If these genes were the main regulatory elements contributing to the flowering phenotype in the hdt701 mutant, then the KO mutants should have flowered early because OsGI functions upstream of Hd1, a floral repressor under LD conditions. Therefore, the OsGI-Hd1 pathway does not seem to be the main downstream route from hdt701-1 to the florigens. Because the hdt701 mutants flowered late under both SD and LD, the HDT701 target gene is likely a constitutive repressor that functions upstream of Ehd1. We previously determined that two AP2 family genes, OsIDS1 and SNB, are constitutive flowering repressors (Lee et al., 2014). Here, expression levels of the former were significantly increased in the mutant (Figs. 3G and Supplementary S1G) while those of the latter were not affected by the mutations (Figs. 3H and Supplementary Fig. S1H). These results suggested that OsIDS1 is downstream of HDT701. Expression levels of other constitutive repressors, i.e., OsLFL1, OsLF, and OsPhyB, were not altered in the mutant (Figs. 3J–3L; Supplementary Figs. S1J-S1L).
Fig. 3. Diurnal expression patterns of floral regulators in leaf blades of WT and hdt701-1 plants at 49 DAG under LD.
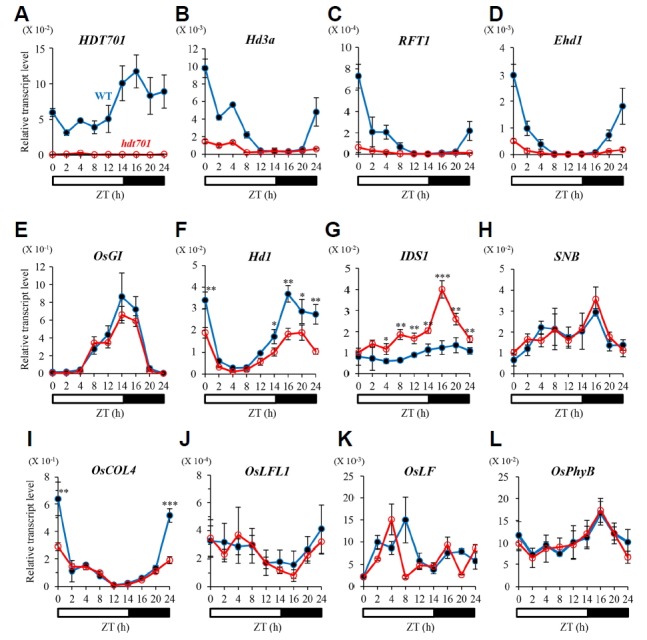
Quantitative RT-PCR analyses of HDT701 (A), Hd3a (B), RFT1 (C), Ehd1 (D), OsGI (E), Hd1 (F), OsIDS1 (G), SNB (H), OsCOL4 (I), OsLFL1 (J), OsLF (K), and OsPhyB (L). Closed circles, WT; open circles, hdt701-1. y-axis, relative transcript level of each gene compared with that of rice Ubi. Error bars indicate standard deviations; n = 4 (technical replicates). Levels of significant difference are indicated by * (P < 0.05), ** (P < 0.01), and *** (P < 0.005).
Because flowering by hdt701 mutants was also delayed under SD, we measured the expression levels of regulatory genes from plants grown under SD conditions. Mature leaf blades were sampled at 28 DAG, when the florigens started to be expressed in SD-grown plants. As we had observed from the LD-grown plants, the mutants expressed no detectable levels of HDT701 transcript (Figs. 4A and Supplementary Fig. S2A). Expression of the florigens and Ehd1 was significantly lower in the mutant leaves than in the WT leaves (Figs. 4B–4D; Supplementary Figs. S2B–S2D). Transcript levels of OsGI and Hd1 were also reduced in the mutants, as noted from LD-grown plants, and especially so under SD (Figs. 4E and 4F; Supplementary Figs. S2E and S2F). Because both OsGI and Hd1 function as positive regulatory elements under SD conditions, their decreased expression should have caused late flowering, consistent with the mutant phenotype. Transcript levels of OsIDS1 were reduced at all nine sampling times, as observed under LD conditions (Figs. 4G; Supplementary Fig. S2G). These results suggested that OsIDS1 is an important regulator that functions downstream of HDT701. Expression was not altered for the other constitutive repressors -- SNB1, OsCOL4, OsLFL1, OsLF, and OsPhyB -- in mutant plants grown under SD (Figs. 4H–4L; Supplementary Figs. S2H–S2L). Although expression of Os-COL4 was reduced at ZT 0 and 24 h under LD (Fig. 3I), it may not be related to the regulatory pathway mediated by HDT701 because the gene is a constitutive floral repressor and the hdt701 mutants flowered late under both SD and LD.
Fig. 4. Diurnal expression patterns of floral regulators in leaf blades of WT and hdt701-1 plants at 28 DAG under SD.
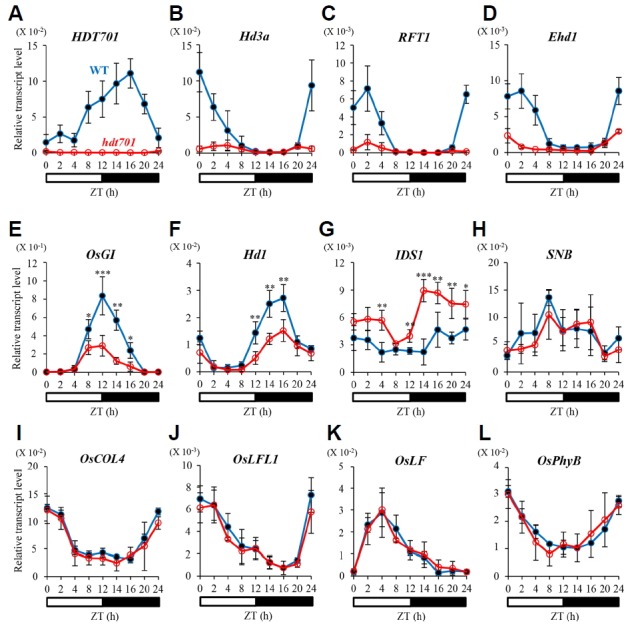
Quantitative RT-PCR analyses of HDT701 (A), Hd3a (B), RFT1 (C), Ehd1 (D), OsGI (E), Hd1 (F), OsIDS1 (G), SNB (H), OsCOL4 (I), OsLFL1 (J), OsLF (K), and OsPhyB (L). Closed circles, WT; open circles, hdt701-1. y-axis, relative transcript level of each gene compared with that of rice Ubi. Error bars indicate standard deviations; n = 4 (technical replicates). Levels of significant difference are indicated by * (P < 0.05), ** (P < 0.01), and *** (P <0.005).
HDT701 directly regulates the expression of OsIDS1
HDT701 is an active histone H4 deacetylase that suppresses expression of target genes via histone deacetylation (Ding et al., 2012; Li et al., 2011; Zhao et al., 2015). To study how HDT701 might directly regulate OsIDS1 expression, we performed ChIP assays using transgenic plants that express HA-tagged HDT701 as well as transgenics expressing HA alone as a negative control. Four areas (P1, P2, P3, and P4) in the OsIDS1 promoter region and one area (P5) in the 5′-untranslated region (UTR) were selected for the binding assay (Fig. 5A). Results from the experiments with anti-HA antibodies showed that P4 was preferentially enriched in the chromatins expressing the HDT701-HA fusion protein when compared with the chromatins from transgenic plants expressing HA tag alone (Fig. 5B). However, chromatin enrichment in P1, P2, P3, and P5 was similar between the two types of transgenic plants.
Fig. 5. Chromatin-immunoprecipitation (ChIP) assays of OsIDS1 chromatin and SNB chromatin.
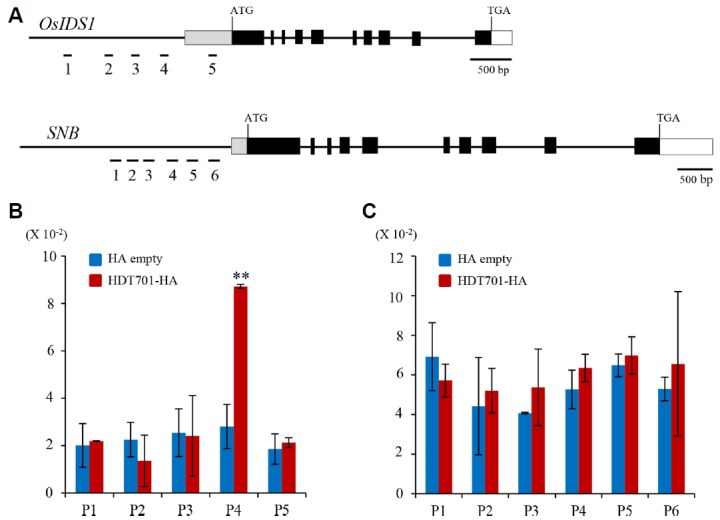
(A) Genomic structures of OsIDS1 and SNB. Tested areas are numbered. (B) ChIP assay of HDT701 enrichment on OsIDS1 chromatin, using HDT701-HA-tagged transgenic plants. Transgenic plants expressing HA tag alone served as control. Leaf blades were harvested at 30 DAG under SD. Percent-of-input method was used for normalization. (C) ChIP assay of HDT701-HA enrichment on SNB chromatin, as described in (B). Levels of significant difference are indicated by ** (P < 0.01). Error bars show standard deviations, n = 3. The entire experiment was conducted two times.
As a negative control, we performed ChIP assays of SNB chromatins because this gene encodes a protein that is highly homologous to OsIDS1. Six areas in the SNB promoter region were selected for the analysis using plants expressing HDT701-HA or HA tag alone (Fig. 5A). The chromatin enrichment experiments with HA antibodies demonstrated that all six areas were selected equally in the HDT701-HA and HA plants (Fig. 5C). This implied that the promoter region of OsIDS1 is a potential target of HDT701.
Regulatory genes that function upstream of HDT701
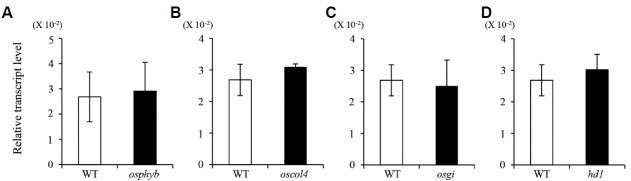
To identify the regulatory genes that function upstream of HDT701, we elucidated its expression patterns in various flowering-time mutants. Transcript levels of HDT701 were not changed in mutants defective in OsPhyB and OsCOL4, two positive regulators of OsIDS1 (Figs. 6A and 6B, Supplementary Figs. S3A and S3B). Likewise, expression was not altered in the hd1 and osgi mutants (Figs. 6C and 6D; Supplementary Figs. S3C and S3D).
Fig. 6. Expression levels of HDT701 in osphyb (A), oscol4 (B), osgi (C), and hd1 (D).
Total RNAs were prepared from leaf blades at 42 DAG under LD. Error bars display standard deviations; n = 4 (technical replicates).
DISCUSSION
We investigated the role of HDT701 in controlling flowering time using KO mutants generated by T-DNA insertions and CRISPR/Cas9. The mutant plants flowered later than the WT due to reductions in the expression levels of Hd3a, RFT1, and Ehd1. This indicated that HDT701 is a floral activator that functions upstream of Ehd1. Our result is consistent with other observations of HDT701-overexpression plants, which flower early because of induced expression of the three genes (Li et al., 2011). The previous experiments were conducted under natural LD conditions (Li et al., 2011). In our current study, we observed that the gene is a constitutive repressor of flowering under both LD and SD. Because HDT701 encodes histone 4 deacetylase, deacetylation of floral repressors would enhance florigen expression. Several histone deacetylation (HDA) genes also control flowering time in Arabidopsis (He et al., 2003). Constitutive delayed-flowering phenotypes of mutants defective in HDA5 and HDA6 under both LD and SD conditions imply that histone deacetylation accelerates flowering time in Arabidopsis, similar to that observed in our study (c.f., Luo et al., 2015; Wu et al., 2008).
Histochemical staining of hdt701-1 transgenic plants showed that HDT701 is expressed not only in mesophyll cells but also in phloem parenchyma cells, indicating that the gene has multiple functions. In addition to its role in controlling flowering time, this gene is involved in plant innate immunity, GA biosynthesis, and abiotic stress responses (Ding et al., 2012; Zhao et al., 2015). Florigens as well as upstream regulatory genes such as Ehd1 and Ghd7 are preferentially expressed in phloem parenchyma cells, whereas other regulatory genes such as OsCOL4, Hd1, OsGI, and OsPhyB are strongly expressed in mesophyll cells (Komiya et al., 2008; Lee et al., 2010; Saito et al., 2012; Tamaki et al., 2007; Xue et al., 2008). Therefore, these findings suggest that HDT701 may function in multiple pathways to influence flowering time.
In a previous study, Li et al. (2011) proposed that HDT701 induces flowering by suppressing OsGI and Hd1 under LD; this was based on observations that overexpression of the former caused a reduction in expression for the latter two. However, the decline in expression of OsGI in the HDT701-overexpression plants should have resulted in delayed flowering because OsGI is a flowering enhancer. That research group also reported that transcript levels of OsGI and Hd1 were not altered under SD conditions. We found here that transcript levels of the two upstream regulatory genes were reduced in hdt701 KO mutants regardless of day length. This discrepancy might have been due to the cultivar used for generating the transgenic plants. Alternatively, overexpression of the gene may have caused side effects by forming unusual protein complexes.
We identified OsIDS1 as being downstream of HDT701 because expression of the former was significantly enhanced under both SD and LD in the hdt701 mutant. Direct interaction of HDT701 on OsIDS1 chromatins was indicated by our ChIP assay. OsIDS1 is a member of the AP2 family, which is involved in various processes (Lee et al., 2014). For example, six Arabidopsis members in this family delay flowering and are suppressed by miR172 (Lee et al., 2014). Similarly, increasing expression of Zea mays GROSSY 15, an AP2 member, delays flowering (Zhu and Helliwell, 2011). We previously reported that rice AP2 members OsIDS1 and SNB act as negative regulatory elements in flowering, and their transcripts are targeted by miR172 (Lee et al., 2014). Although SNB is closely related to OsIDS1, its transcript levels were not affected in hdt701 mutants. This suggests that HDT701 specifically selects OsIDS1 chromatin even though the chromatin-remodeling factor appears to target multiple genes.
In hdt701 mutants, the mRNA levels of OsGI were constitutively down-regulated. That gene plays a positive role in enhancing florigen expression and flowering induction under both LD and SD, although the effect is more severe under SD (Lee et al., 2015). Therefore, the delayed flowering phenotype of the mutant could be explained by lower expression of OsGI. However, that reduction in expression was not very significant under LD, although the delay in flowering by hdt701 mutants was equally significant under both LD and SD.
Transcript levels of Hd1 were also significantly diminished regardless of day length. Because OsGI positively controls the expression of Hd1 (Hayama et al., 2003), the decrease in expression for Hd1 could have resulted from the down-regulation of OsGI in the mutants. Although Hd1 advances flowering under SD, the regulatory element inhibits flowering under LD. Therefore, the reduction in Hd1 expression in the hdt701 mutant under LD would accelerate flowering rather than suppress that process. Therefore, we conclude that the delay in flowering by the mutants under LD was not due to an alteration of the OsGI and Hd1 pathway. It is probable that HDT701-OsIDS1-Ehd1 is the major pathway under LD. However, under SD, both the HDT701-OsGI-Hd1-Ehd1 and HDT701-OsIDS1-Ehd1 pathways appear to modulate florigen expression (Fig. 7).
Fig. 7.
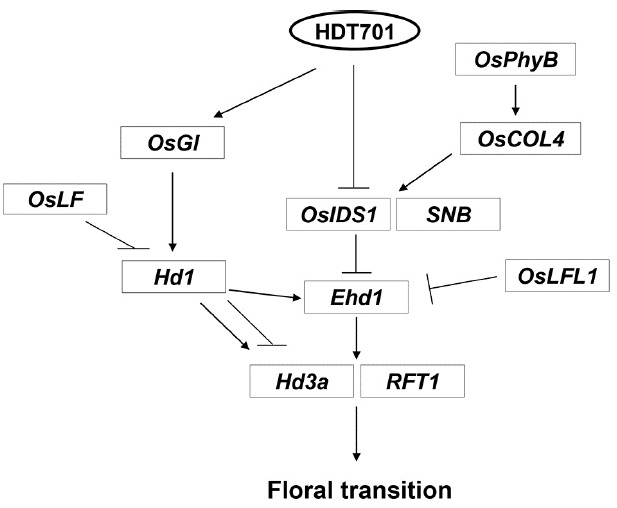
Model for regulatory pathway governed by HDT701 for control of flowering time.
Supplementary data
ACKNOWLEDGMENTS
We thank Kyungsook An for generating the T-DNA transgenic lines and managing the seed stocks, and Priscilla Licht for editing the English composition of the article. This work was supported in part by grants from the Next-Generation BioGreen 21 Program (Plant Molecular Breeding Center, No. PJ013210), Rural Development Administration, Republic of Korea.
Footnotes
Note: Supplementary information is available on the Molecules and Cells website (www.molcells.org).
REFERENCES
- An G., Ebert P.R., Mitra A., Ha S.B. Plant Molecular Biology Manual. A3. Dordrecht: Kluwer Academic Publisher; 1989. Binary vectors; pp. 1–19. [Google Scholar]
- An G., Jeong D.H., Jung K.H., Lee S. Reverse genetic approaches for functional genomics of rice. Plant Mol Biol. 2005a;59:111–123. doi: 10.1007/s11103-004-4037-y. [DOI] [PubMed] [Google Scholar]
- An G., Lee S., Kim S.H., Kim S.R. Molecular genetics using T-DNA in rice. Plant Cell Physiol. 2005b;46:14–22. doi: 10.1093/pcp/pci502. [DOI] [PubMed] [Google Scholar]
- Cho L.H., Yoon J., Pasriga R., An G. Homodimerization of Ehd1 is required to induce flowering in rice. Plant Physiol. 2016;170:2159–2171. doi: 10.1104/pp.15.01723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho L.H., Yoon J., An G. The control of flowering time by environmental factors. Plant J. 2017;90:708–719. doi: 10.1111/tpj.13461. [DOI] [PubMed] [Google Scholar]
- Corbesier L., Vincent C., Jang S., Fornara F., Fan Q., Searle I., Giakountis A., Farrona S., Gissot L., Turnbull C., Coupland G. FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science. 2007;316:1030–1033. doi: 10.1126/science.1141752. [DOI] [PubMed] [Google Scholar]
- Dangl M., Brosch G., Haas H., Loidl P., Lusser A. Comparative analysis of HD2 type histone deacetylases in higher plants. Planta. 2001;213:280–285. doi: 10.1007/s004250000506. [DOI] [PubMed] [Google Scholar]
- Ding B., del Bellizzi M.R., Ning Y., Meyers B.C., Wang G.L. HDT701, a histone H4 deacetylase, negatively regulates plant innate immunity by modulating histone H4 acetylation of defense-related genes in rice. Plant Cell. 2012;24:3783–3794. doi: 10.1105/tpc.112.101972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doi K., Izawa T., Fuse T., Yamanouchi U., Kubo T., Shimatani Z., Yano M., Yoshimura A. Ehd1, a B-type response regulator in rice, confers short-day promotion of flowering and controls FT-Iike gene expression independently of Hd1. Genes Dev. 2004;18:926–936. doi: 10.1101/gad.1189604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler S., Lee K., Onouchi H., Samach A., Richardson K., Coupland G., Putterill J. GIGANTEA: a circadian clock-controlled gene that regulates photoperiodic flowering in Arabidopsis and encodes a protein with several possible membrane-spanning domains. EMBO J. 1999;18:4679–4688. doi: 10.1093/emboj/18.17.4679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu W., Wu K., Duan J. Sequence and expression analysis of histone deacetylases in rice. Biochem Biophys Res Commun. 2007;356:843–850. doi: 10.1016/j.bbrc.2007.03.010. [DOI] [PubMed] [Google Scholar]
- Haring M., Offermann S., Danker T., Horst I., Peterhansel C., Stam M. Chromatin immunoprecipitation: optimization, quantitative analysis and data normalization. Plant Methods. 2007;3:1–16. doi: 10.1186/1746-4811-3-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayama R., Yokoi S., Tamaki S., Yano M., Shimamoto K. Adaptation of photoperiodic control pathways produces short-day flowering in rice. Nature. 2003;422:719–722. doi: 10.1038/nature01549. [DOI] [PubMed] [Google Scholar]
- He Y., Michaels S.D., Amasino R.M. Regulation of flowering time by histone acetylation in Arabidopsis. Science. 2003;302:1751–1754. doi: 10.1126/science.1091109. [DOI] [PubMed] [Google Scholar]
- Hu Y., Qin F., Huang L., Sun Q., Li C., Zhao Y., Zhou D.X. Rice histone deacetylase genes display specific expression patterns and developmental functions. Biochem Biophys Res Commun. 2009;388:266–271. doi: 10.1016/j.bbrc.2009.07.162. [DOI] [PubMed] [Google Scholar]
- Ishikawa R., Aoki M., Kurotani K., Yokoi S., Shinomura T., Takano M., Shimamoto K. Phytochrome B regulates Heading date 1(Hd1)-mediated expression of rice florigen Hd3a and critical day length in rice. Mol Genet Genom. 2011;285:461–470. doi: 10.1007/s00438-011-0621-4. [DOI] [PubMed] [Google Scholar]
- Jeon J.S., Lee S., Jung K.H., Jun S.H., Jeong D.H., Lee J., Kim C., Jang S., Lee S., Yang K., et al. T-DNA insertional mutagenesis for functional genomics in rice. Plant J. 2000;22:561–570. doi: 10.1046/j.1365-313x.2000.00767.x. [DOI] [PubMed] [Google Scholar]
- Jeong D.H., An S., Kang H.G., Moon S., Han J.J., Park S., Lee H.S., An K., An G. T-DNA insertional mutagenesis for activation tagging in rice. Plant Physiol. 2002;130:1636–1644. doi: 10.1104/pp.014357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong D.H., An S., Park S., Kang H.G., Park G.G., Kim S.R., Sim J., Kim Y.O., Kim M.K., Kim S.R., et al. Generation of a flanking sequence-tag database for activation-tagging lines in japonica rice. Plant J. 2006;45:123–132. doi: 10.1111/j.1365-313X.2005.02610.x. [DOI] [PubMed] [Google Scholar]
- Kim S.R., Lee D.Y., Yang J.I., Moon S., An G. Cloning vectors for rice. J Plant Biol. 2009;52:73–78. [Google Scholar]
- Kim S.L., Choi M., Jung K.H., An G. Analysis of the early-flowering mechanisms and generation of T-DNA tagging lines in Kitaake, a model rice cultivar. J Exp Bot. 2013;64:4169–4182. doi: 10.1093/jxb/ert226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komiya R., Ikegami A., Tamaki S., Yokoi S., Shimamoto K. Hd3a and RFT1 are essential for flowering in rice. Development. 2008;135:767–774. doi: 10.1242/dev.008631. [DOI] [PubMed] [Google Scholar]
- Lee Y.S., An G. OsGI controls flowering time by modulating rhythmic flowering time regulators preferentially under short day in rice. J Plant Biol. 2015;58:137–145. [Google Scholar]
- Lee Y.S., Jeong D.H., Lee D.Y., Yi J., Ryu C.H., Kim S.L., Jeong H.J., Choi S.C., Jin P., Yang J., et al. OsCOL4 is a constitutive flowering repressor upstream of Ehd1 and downstream of OsphyB. Plant J. 2010;63:18–30. doi: 10.1111/j.1365-313X.2010.04226.x. [DOI] [PubMed] [Google Scholar]
- Lee Y.S., Lee D.Y., Cho L.H., An G. Rice miR172 induces flowering by suppressing OsIDS1 and SNB, two AP2 genes that negatively regulate expression of Ehd1 and florigens. Rice. 2014;7:31. doi: 10.1186/s12284-014-0031-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y.S., Yi J., Jung K.H., An G. Comparison of rice flowering-time genes under paddy conditions. J Plant Biol. 2016;59:238–246. [Google Scholar]
- Li C., Huang L., Xu C., Zhao Y., Zhou D.X. Altered levels of histone deacetylase OsHDT1 affect differential gene expression patterns in hybrid rice. PLoS One. 2011;6:e21789. doi: 10.1371/journal.pone.0021789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo M., Wang Y.Y., Liu X., Yang S., Lu Q., Cui Y., Wu K. HD2C interacts with HDA6 and is involved in ABA and salt stress response in Arabidopsis. J Exp Bot. 2012;63:3297–3306. doi: 10.1093/jxb/ers059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo M., Tai R., Yu C.W., Yang S., Chen C.Y., Lin W.D., Schmidt W., Wu K. Regulation of flowering time by the histone deacetylase HDA5 in Arabidopsis. Plant J. 2015;82:925–936. doi: 10.1111/tpj.12868. [DOI] [PubMed] [Google Scholar]
- Miao J., Guo D., Zhang J., Huang Q., Qin G., Zhang X., Wan J., Gu H., Qu L.J. Targeted mutagenesis in rice using the CRISPR-Cas system. Cell Res. 2013;23:1233–1236. doi: 10.1038/cr.2013.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita S., Wada H., Matsue Y. Countermeasures for heat damage in rice grain quality under climate change. Plant Prod Sci. 2017;19:1–11. [Google Scholar]
- Naito Y., Hino K., Bono H., Ui-Tei K. CRISPRdirect: software for designing CRISPR/Cas guide RNA with reduced off-target sites. Bioinformatics. 2015;31:1120–1123. doi: 10.1093/bioinformatics/btu743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishida H., Inoue H., Okumoto Y., Tanisakao T. A novel gene ef1-h conferring an extremely long basic vegetative growth period in rice. Crop Sci. 2002;42:348–354. doi: 10.1007/s00122-009-1078-2. [DOI] [PubMed] [Google Scholar]
- Ouyang S., Zhu W., Hamilton J., Lin H., Campbell M., Childs K., Thibaud-Nissen F., Malek R.L., Lee Y., Zheng L., et al. The TIGR rice genome annotation resource: improvements and new features. Nucleic Acids Res. 2007;35:D883–D887. doi: 10.1093/nar/gkl976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey R., Müller A., Napoli C.A., Selinger D.A., Pikaard C.S., Richards E.J., Bender J., Mount D.W., Jorgensen R.A. Analysis of histone acetyltransferase and histone deacetylase families of Arabidopsis thaliana suggests functional diversification of chromatin modification among multicellular eukaryotes. Nucleic Acids Res. 2002;30:5036–5055. doi: 10.1093/nar/gkf660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park D.H., Somers D.E., Kim Y.S., Choy Y.H., Lim H.K., Soh M.S., Kim H.J., Kay S.A., Nam H.G. Control of circadian rhythms and photoperiodic flowering by the Arabidopsis GIGANTEA gene. Science. 1999;285:1579–1582. doi: 10.1126/science.285.5433.1579. [DOI] [PubMed] [Google Scholar]
- Peng L.T., Shi Z.Y., Li L., Shen G.Z., Zhang J.L. Ectopic expression of OsLFL1 in rice represses Ehd1 by binding on its promoter. Biochem Biophys Res Commun. 2007;360:251–256. doi: 10.1016/j.bbrc.2007.06.041. [DOI] [PubMed] [Google Scholar]
- Peng L.T., Shi Z.Y., Li L., Shen G.Z., Zhang J.L. Overexpression of transcription factor OsLFL1 delays flowering time in Oryza sativa. Plant Physiol. 2008;165:876–885. doi: 10.1016/j.jplph.2007.07.010. [DOI] [PubMed] [Google Scholar]
- Ryu C.H., Lee S., Cho L.H., Kim S.L., Lee Y.S., Choi S.C., Jeong H.J., Yi J., Park S.J., Han C.D., et al. OsMADS50 and OsMADS56 function antagonistically in regulating long day (LD) -dependent flowering in rice. Plant Cell Environ. 2009;32:1412–1427. doi: 10.1111/j.1365-3040.2009.02008.x. [DOI] [PubMed] [Google Scholar]
- Saito H., Ogiso-Tanaka E., Okumoto Y., Yoshitake Y., Izumi H., Yokoo T., Matsubara K., Hori K., Yano M., Inoue H., et al. Ef7 encodes an ELF3-likeprotein and promotes rice flowering by negatively regulating the floral repressor gene Ghd7 under both short- and long-day conditions. Plant Cell Physiol. 2012;53:717–728. doi: 10.1093/pcp/pcs029. [DOI] [PubMed] [Google Scholar]
- Samach A., Onouchi H., Gold S.E., Ditta G.S., Schwarz-Sommer Z., Yanofsky M.F., Coupland G. Distinct roles of CONSTANS target genes in reproductive development of Arabidopsis. Science . 2000;288:1613–1616. doi: 10.1126/science.288.5471.1613. [DOI] [PubMed] [Google Scholar]
- Sridha S., Wu K. Identification of AtHD2C as a novel regulator of abscisic acid responses in Arabidopsis. Plant J. 2006;46:124–133. doi: 10.1111/j.1365-313X.2006.02678.x. [DOI] [PubMed] [Google Scholar]
- Sun C., Chen D., Fang J., Wang P., Deng X., Chu C. Understanding the genetic and epigenetic architecture in complex network of rice flowering pathways. Protein Cell. 2014;5:889–898. doi: 10.1007/s13238-014-0068-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamaki S., Matsuo S., Wong H.L., Yokoi S., Shimamoto K. Hd3a protein is a mobile flowering signal in rice. Science. 2007;316:1033–1036. doi: 10.1126/science.1141753. [DOI] [PubMed] [Google Scholar]
- Tanaka T., Antonio B.A., Kikuchi S., Matsumoto T., Nagamura Y., Numa H., Sakai H., Wu J., Itoh T., Sasaki T., et al. The rice annotation project database (RAP-DB): 2008 update. Nucleic Acids Res. 2008;36:D1028–D1033. doi: 10.1093/nar/gkm978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueno Y., Ishikawa T., Watanabe K., Terakura S., Iwakawa H., Okada K., Machida C., Machida Y. Histone deacetylases and ASYMMETRIC LEAVES2 are involved in the establishment of polarity in leaves of Arabidopsis. Plant Cell. 2007;19:445–457. doi: 10.1105/tpc.106.042325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei J., Wu Y., Cho L.H., Yoon J., Choi H., Yoon H., Jin P., Yi J., Lee Y.S., Jeong H.J., et al. Identification of root-preferential transcription factors in rice by analyzing GUS expression patterns of T-DNA tagging lines. J Plant Biol. 2017;60:268–277. [Google Scholar]
- Wu K., Tian L., Malik K., Brown D., Miki B. Functional analysis of HD2 histone deacetylase homologues in Arabidopsis thaliana. Plant J. 2000;22:19–27. doi: 10.1046/j.1365-313x.2000.00711.x. [DOI] [PubMed] [Google Scholar]
- Wu K., Zhang L., Zhou C., Yu C.W., Chaikam V. HDA6 is required for jasmonate response, senescence and flowering in Arabidopsis. J Exp Bot. 2008;59:225–234. doi: 10.1093/jxb/erm300. [DOI] [PubMed] [Google Scholar]
- Xue W., Xing Y., Weng X., Zhao Y., Tang W., Wang L., Zhou H., Yu S., Xu C., Li X., et al. Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice. Nat Genet. 2008;40:761–767. doi: 10.1038/ng.143. [DOI] [PubMed] [Google Scholar]
- Yanovsky M.J., Kay S.A. Molecular basis of seasonal time measurement in Arabidopsis. Nature. 2002;419:308–312. doi: 10.1038/nature00996. [DOI] [PubMed] [Google Scholar]
- Yoon J., Cho L.H., Kim S.L., Choi H., Koh H.J., An G. The BEL1-type homeobox gene SH5 induces seed shattering by enhancing abscission-zone development and inhibiting lignin biosynthesis. Plant J. 2014;79:717–728. doi: 10.1111/tpj.12581. [DOI] [PubMed] [Google Scholar]
- Yoon J., Cho L.H., Antt H.W., Koh H.J., An G. KNOX protein OSH15 induces grain shattering by repressing lignin biosynthesis genes. Plant Physiol. 2017;174:312–325. doi: 10.1104/pp.17.00298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi J., An G. Utilization of T-DNA tagging lines in rice. J Plant Biol. 2013;56:85–90. [Google Scholar]
- Zhang J., Zhou X., Yan W., Zhang Z., Lu L., Han Z., Zhao H., Liu H., Song P., Hu Y., et al. Combinations of the Ghd7, Ghd8 and Hd1 genes largely define the ecogeographical adaptation and yield potential of cultivated rice. New Phytol. 2015:1056–1066. doi: 10.1111/nph.13538. [DOI] [PubMed] [Google Scholar]
- Zhang Z., Hu W., Shen G., Liu H., Hu Y., Zhou X., Liu T., Xing Y. Alternative functions of Hd1 in repressing or promoting heading are determined by Ghd7 status under long-day conditions. Sci Rep. 2017;7:5388. doi: 10.1038/s41598-017-05873-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X.L., Shi Z.Y., Peng L.T., Shen G.Z., Zhang J.L. An atypical HLH protein OsLF in rice regulates flowering time and interacts with OsPIL13 and OsPIL15. N Biotechnol. 2011;28:788–797. doi: 10.1016/j.nbt.2011.04.006. [DOI] [PubMed] [Google Scholar]
- Zhao J., Zhang J., Zhang W., Wu K., Zheng F., Tian L., Liu X., Duan J. Expression and functional analysis of the plant-specific histone deacetylase HDT701 in rice. Front Plant Sci. 2015;5:764. doi: 10.3389/fpls.2014.00764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Q.H., Helliwell C.A. Regulation of flowering time and floral patterning by miR172. J Exp Bot. 2011;62:487–495. doi: 10.1093/jxb/erq295. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.