Abstract
This study aimed to explore the role of certain genes and long non-coding RNAs (lncRNAs) in homocysteine (HCY)-induced vascular endothelial injury. HUVECs were treated with HCY, then cell cycle and apoptosis were analyzed by flow cytometry. HUVECs were then sequenced and analyzed using bioinformatics, with a focus on differentially expressed genes/lncRNA (DEGs/DEL), protein-protein interaction (PPI), functional enrichment analyses, and lncRNA-target prediction. Although HCY did not affect the cell cycle, it significantly increased the number of apoptotic cells. In total, 382 DEGs and 147 DELs were identified; DEGs such as CD34, FGF2, and SERPINE1 were the hub nodes in the PPI network, in addition to being the targets of AC005550.3, RP11-415D17.3, and RP1-140K8.5, respectively. Functional enrichment analysis showed that the targets of downregulated AC005550.3 and RP11-415D17.3 were significantly enriched in blood vessel development and those of upregulated RP1-140K8.5 were enriched in fibrinolysis. RT-qPCR showed that the mRNA levels of AC005550.3, RP11-415D17.3, and RP1-140K8.5 were consistent with the results predicted by our bioinformatics analysis. In conclusion, downregulated AC005550.3 and RP11-415D17.3 targeting CD34 and FGF2 and upregulated RP1-140K8.5 targeting SERPINE1 may play an important role in HCY-induced vascular endothelial injury by regulating blood vessel development and fibrinolysis, respectively.
Keywords: Vascular endothelial injury, long non-coding RNA, transcriptome sequence, homocysteine, cardiovascular disease
Introduction
Cardiovascular disease (CVD) is considered the leading cause of disability and mortality, with a high morbidity worldwide [1]. The main risk factors of CAD include advanced age, dyslipidemia, hypertension, diabetes, smoking, and obesity [2]. The existence of cardiovascular risk factors is predicted to cause an additional 23% increase in CVD events and 7.7 million deaths annually from 2010 to 2030 in China [3]. Although an extraordinary effort has been made to control major cardiovascular risk factors, CVD leads to a high burden of disability and mortality [4]. Therefore, it is necessary to ascertain the molecular and pathophysiological mechanisms underlying CVD and search for novel diagnostic and therapeutic targets.
The pathogenesis of CVD is complex, and vascular endothelial injury plays a major role in its development [5]. As the first barrier between blood and tissue, vascular endothelial cells protect vascular smooth muscle and maintain normal organizational structure and function [6]. Under pathological conditions, inflammatory factors can degrade the endothelium and cause increased permeability, leading to increased adherence factors, monocyte adherence, and aggregation, thereby resulting in endothelial dysfunction and major blood vessel atherosclerosis [7]. It has been previously shown that homocysteine (HCY), a sulfhydryl-containing amino acid, is associated with an increased risk of CVD [8]. HCY can promote apoptosis, generate reactive oxide species, and produce endoplasmic reticulum stress in vascular endothelial cells [8]. However, the mechanism underlying vascular endothelial injury in response to HCY remains to be explored.
Long non-coding RNAs (lncRNAs) are non-coding RNA molecules that contain over 200 nucleotides. By mediating target-gene transcription, lncRNAs play important regulatory roles in various diseases [9] such as cancers [10], neurodegeneration [11], autoimmune disorders [12,13], and CVD [14]. Several lncRNAs, including MALAT1 and Tie-1-AS, are involved in the regulation of blood vessel growth and function via endothelial cell proliferation [15]. However, little evidence exists regarding the role of lncRNAs in vascular endothelial injury in response to HCY.
In the present study, human umbilical vein vascular endothelial cells (HUVECs) were treated with HCY to identify the genes and lncRNAs involved in the mechanism underlying the HUVEC response to HCY. HCY-treated and untreated HUVECs were sequenced, and bioinformatics analysis was used to further study the genes and lncRNAs. Our results may offer new insights in understanding the role of lncRNAs in vascular endothelial injury in response to HCY.
Materials and methods
Cell culture
HUVECs were purchased from Lifeline Cell Technology (Walkersville, MD, USA) and were cultured in Endothelial Cell Medium (ECM, Lifeline, Cell Technology) at 37°C with 5% CO2. For the experiment, HUVECs were cultured in ECM containing 2.5 mmol/L HCY at 37°C for 24 h, and the cells cultured only in ECM served as control.
Cell cycle and apoptosis
For cell cycle analysis, HUVECs were fixed overnight with 5 mL 70% ethyl alcohol at 4°C. On the next day, the fixed cells were washed twice with phosphate buffered saline (PBS) and were digested with 50 µg/mL RNase A at 37°C for 30 min. The cells were stained with 5 μL propidium iodide (PI) for 15 min at 4°C in the dark and were then used for flow cytometry (FACSCalibur, BD, Franklin Lakes, NJ, USA). The Annexin V-FITC/PI Apoptosis Detection kit (BD) was used for the detecting apoptosis. The cells were washed once with PBS and were resuspended in 1× Binding Buffer. Subsequently, the cells were stained with 5 μL FITC-Annexin V and 5 μL PI at 25°C for 15 min in the dark. Finally, the cells were analyzed using a flow cytometer (BD).
Transcriptome sequencing
mRNA extracted from HUVECs was used to construct an mRNA-seq library. Sequencing was performed using the Illumina Genome Analyzer IIx sequencing platform. The raw reads were obtained using the Illumina instrument software and were deposited in the NCBI (National Center for Biotechnology Information) SRA (Sequence Read Archive) database under the accession number of SRP149384 (https://www.ncbi.nlm.nih.gov/sra/?term=SRP149384). Followed by reads and beads with low quality were removed. The clean reads were then mapped to the human reference genome 19 using TopHat2 (v 2.1.0) [16]. The expression values of genes and lncRNAs calculated by counts per million were obtained based on human gene annotation provided by Gencode database [17] using StringTie tool (v 1.2.3) [18].
Bioinformatics analysis of target genes and lncRNAs
DEGs and DELs belonging to HUVECs treated with and without HCY were obtained using the edgeR package in R [19]. The cutoff criteria for DEGs and DELs were set up as follows: |log2 fold change| value > 0.585 and p-value < 0.01. For the functional analysis of DEGs and lncRNA targets, gene ontology (GO) in biological process (BP), cellular component (CC), and molecular function (MF) as well as Kyoto Encyclopedia of Genes and Genomes (KEGG) were checked using the Database for Annotation, Visualization and Integrated Discovery (DAVID, v 6.8) [20]. Protein-protein interaction(PPI) network for DEGs was constructed using the Search Tool for the Retrieval of Interacting Genes online database [21] and was visualized using Cytoscape [22]. In addition, the target genes of lncRNAs (Pearson correlation coefficient > 0.98) among DEGs were screened [23], and the lncRNA-target regulatory network was visualized using Cytoscape. GO in BP terms and KEGG enrichment of lncRNA targets were analyzed using the clusterprofile package [24] in R.
Validation of lncRNAs using real-time quantitative polymerase chain reaction (RT-qPCR)
Total RNA from HUVECs treated with and without HCY was isolated using TRIzol (Invitrogen, Carlsbad, CA, USA) and was reverse transcribed into cDNA using PrimeScript RT Master Mix (Takara, Dalian, China) according to the manufacturer’s instructions. Gene expression was measured using SYBR Green PCR Master (Thermo Fisher Scientific, Waltham, MA, USA) under the following conditions: 50°C for 3 min, 40 cycles at 95°C for 3 min, 95°C for 10 s, and 60°C for 30 s. lncRNA primers are listed in Table 1.
Table 1.
Primer sequence of lncRNAs
| Gene | Primer sequence (5’-3’) |
|---|---|
| GAPDH-hF | TGACAACTTTGGTATCGTGGAAGG |
| GAPDH-hR | AGGCAGGGATGATGTTCTGGAGAG |
| RP1-140K8.5-hF | ACCTTGGCTGAGTCTTGACA |
| RP1-140K8.5-hR | CAATTCCCACCAGCACGAAC |
| AC005550.3-hF | GCATGGATTTTCTTCCGCCTC |
| AC005550.3-hR | TTTCATCACCGTCAGGTTGAGC |
| RP11-415D17.3-hF | TGAGCTGTCATAATCGTGCTT |
| RP11-415D17.3-hR | GCTGGTTAACTGATCTCATCCAC |
| RP11-691H4.4-hF | CCGCCTCAGTTCCCACGGTA |
| RP11-691H4.4-hR | CTTTGTCCGCCTTTATTGTTGGTG |
| CTD-2280E9.1-hF | GTACACCAGCTCAAGATGACT |
| CTD-2280E9.1-hR | TCTTCCTGCCACTTAGAGCAA |
Statistical analysis
Statistical analysis was performed using SPSS 22.0 (SPSS Inc., Chicago, IL, USA). Data were expressed as mean ± SEM and were analyzed by Student’s t-test. A p-value of < 0.05 was considered statistically significant, and P < 0.01 was considered highly significant.
Results
Effect of HCY on cell cycle and apoptosis in HUVECs
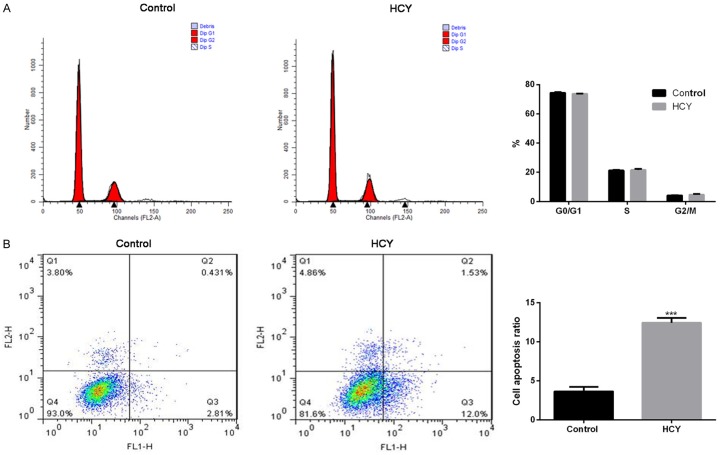
The cell cycle assay showed that HUVECs treated with HCY showed no evident changes with respect to the G0/G1, S, and G2/M phases compared with untreated HUVECs (Figure 1A). However, HCY treatment significantly increased the cell apoptosis ratio compared with untreated HUVECs (12.43% vs 3.65%; P < 0.001, Figure 1B).
Figure 1.
HCY promotes HUVEC apoptosis. A. Cell apoptosis ratio with respect to G0/G1, S, and G2/M phases in untreated HUVECs (control group) and HUVECs treated with HCY (HCY group) as analyzed by flow cytometry. B. Cell apoptosis ratio in untreated HUVECs and HUVECs treated with HCY by Annexin V-FITC/PI Apoptosis Detection kit. ***P < 0.001 compared with control. HCY, homocysteine; HUVECs, human umbilical vein vascular endothelial cells.
DEG screening and functional enrichment analyses
According to the sequencing data, 382 DEGs, including 101 upregulated genes and 281 downregulated genes, were identified in HUVECs treated with HCY. GO enrichment analysis showed that DEGs were significantly enriched with respect to extracellular space, cell adhesion, proteinaceous extracellular matrix, platelet activation, response to mechanical stimulus, and regulation of blood pressure (Figure 2A). KEGG enrichment analysis revealed that DEGs belonging to the pathways of complement and coagulation cascades, ECM-receptor interaction, cytokine-cytokine receptor interaction, platelet activation, and renin secretion were significantly enriched (Figure 2A). In addition, PPI among the DEG-encoded proteins (Figure 2B) showed that the top 10 nodes involved in this network were FGF2 (degree = 24), CCL2 (degree = 18), VWF (degree = 16), SERPINE1 (degree = 15), BMP4 (degree = 12), CD34 (degree = 12), PLAU (degree = 11), GNG7 (degree = 10), CD40 (degree = 10), and CDC6 (degree = 9).
Figure 2.
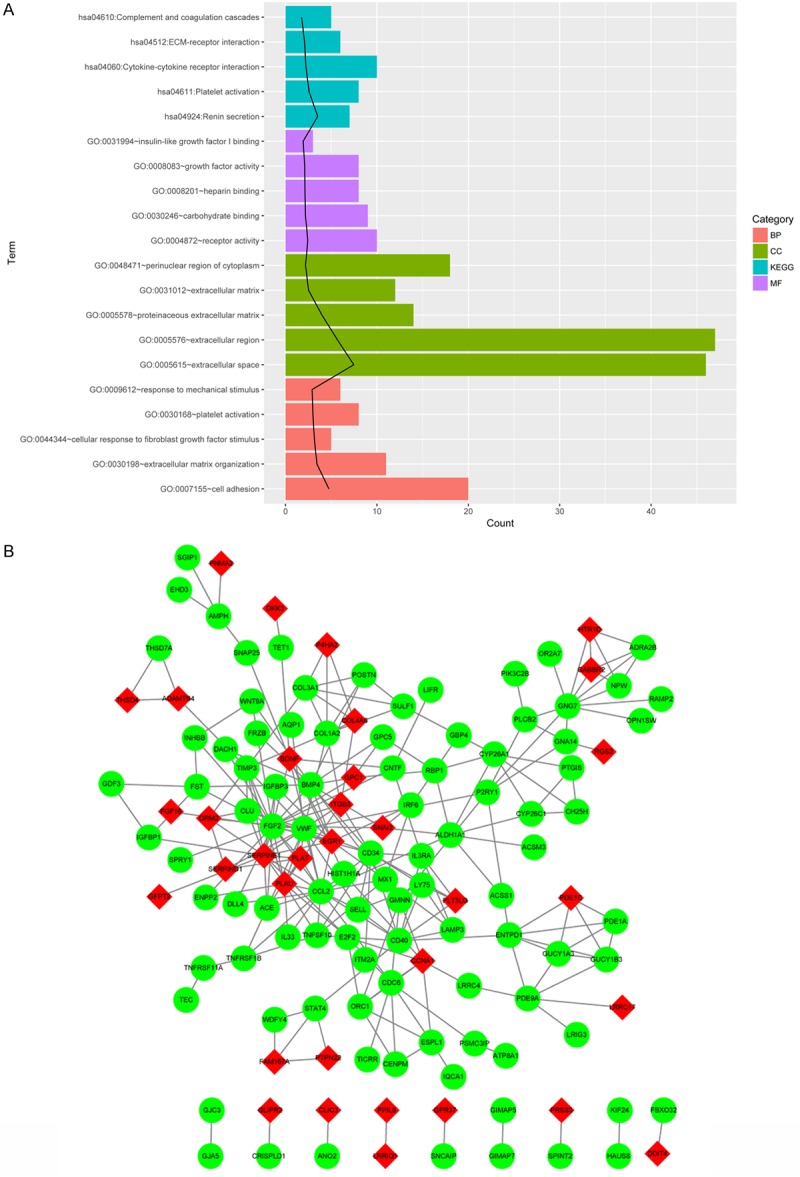
Functional enrichment analyses and PPI network for differentially expressed genes. A. The top five GO in BP, CC, and MF terms as well as KEGG terms for DEGs according to p-values; black line represents -log10 (p-value). B. PPI network for upregulated and downregulated DEGs. Red nodes stand for upregulated DEGs and the green nodes stand for downregulated DEGs. GO, gene ontology; BP, biological process; CC, cellular component; MF, molecular function; KEGG, Kyoto Encyclopedia of Genes and Genomes.
DEL screening and functional enrichment analyses
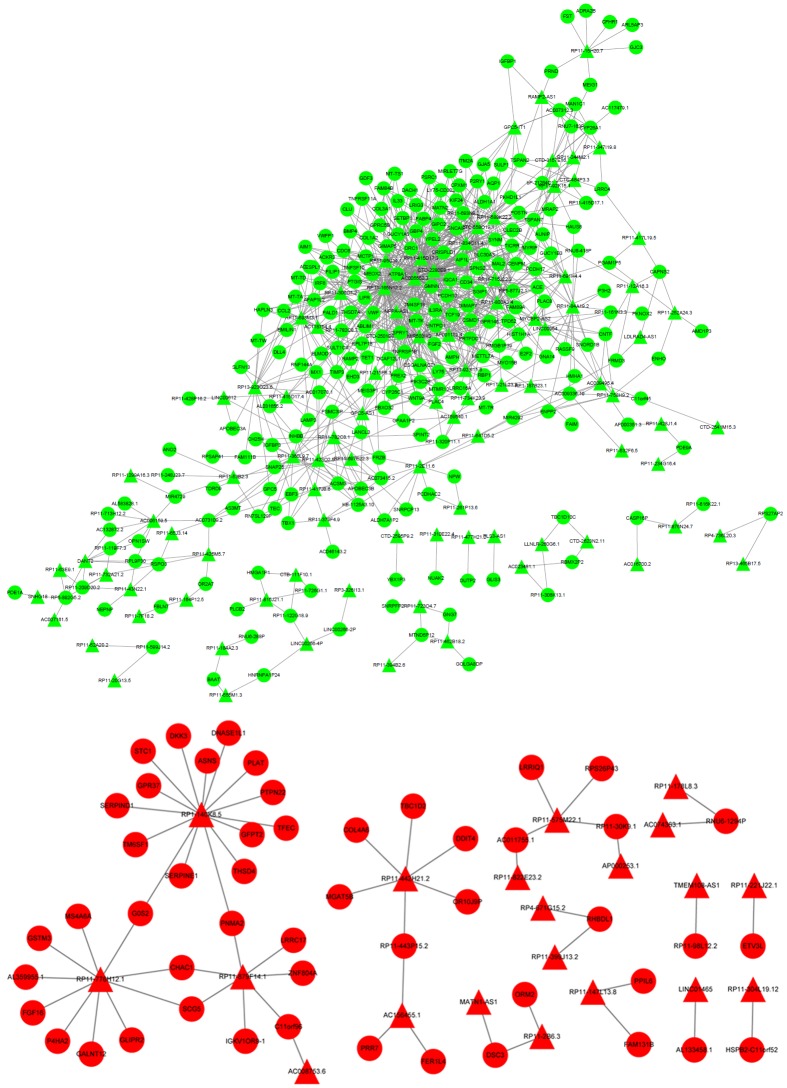
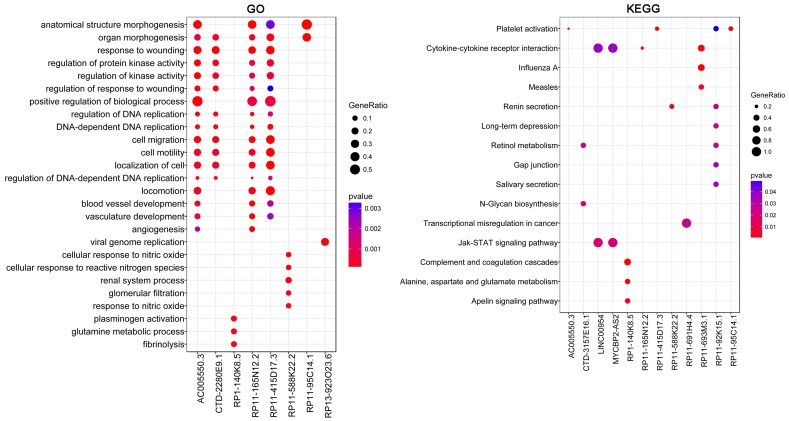
Based on the sequencing data, 147 DELs, including 32 upregulated lncRNAs and 115 downregulated lncRNAs, were identified in HUVECs treated with HCY. Twenty upregulated lncRNAs and 93 downregulated lncRNAs predicted its targets from DEGs, and the lncRNA-target network was constructed with 292 nodes (proteins) and 1097 edges (PPI pairs) (Figure 3). DELs with target genes of over 10 are shown in Table 2, such as RP11-165N12.2 (n = 101), AC005550.3 (n = 81), CTD-2280E9.1 (n = 78), RP11-834C11.4 (n = 47), RP11-415D17.3 (n = 44), RP1-140K8.5 (n = 15), and RP11-776H12.1 (n = 10). To further analyze the lncRNAs involved in regulation, functional enrichment analyses of the lncRNA targets were also performed. GO enrichment analysis showed that the targets for AC005550.3, RP11-165N12.2, and RP11-415D17.3 had similar functional enrichment terms, including anatomical structure morphogenesis, response to wounding, positive regulation of BP, cell motility, and blood vessel development. Targets for CTD-2280E9.1 were significantly enriched in response to wounding, organ morphogenesis, and cell motility, and targets for RP1-140K8.5 were significantly enriched with respect to plasminogen activation, glutamine metabolic process, and fibrinolysis (Figure 4). In addition, KEGG enrichment analysis revealed that targets of AC005550.3 and RP11-415D17.3 were both significantly enriched with regards to platelet activation, and targets for RP1-140K8.5 were significantly enriched with respect to complement and coagulation cascades; alanine, aspartate, and glutamate metabolism; and the apelin signaling pathway (Figure 4).
Figure 3.
LncRNA-target network. The red circle represents the upregulated DEGs, the green circle represents the downregulated DEGs, the red triangle represents upregulated lncRNAs, and the green triangle represents downregulated lncRNAs. lncRNA, long non-coding RNA; DEGs, differentially expressed genes.
Table 2.
LncRNA and the number of its targets
| lncRNA (down) | Number | lncRNA (up) | Number |
|---|---|---|---|
| RP11-165N12.2 | 101 | RP1-140K8.5 | 15 |
| AC005550.3 | 81 | RP11-776H12.1 | 10 |
| CTD-2280E9.1 | 78 | ||
| RP11-834C11.4 | 47 | ||
| RP11-415D17.3 | 44 | ||
| MIR503HG | 40 | ||
| CTC-558O19.1 | 35 | ||
| RP11-95C14.1 | 34 | ||
| RP11-306O1.2 | 33 | ||
| RP11-800A3.4 | 31 | ||
| RP11-588K22.2 | 28 | ||
| AP001189.4 | 23 | ||
| NPPA-AS1 | 23 | ||
| RP11-923I11.6 | 23 | ||
| RP11-715J22.3 | 22 | ||
| RP11-693M3.1 | 21 | ||
| GPC5-AS1 | 17 | ||
| RP11-360L9.7 | 17 | ||
| RP13-923O23.6 | 16 | ||
| AC118754.4 | 13 | ||
| RP11-691H4.4 | 13 | ||
| RP11-84A19.2 | 13 | ||
| CTD-3157E16.1 | 12 | ||
| RP11-782C8.1 | 12 | ||
| RP11-423O2.5 | 11 | ||
| LINC00954 | 10 | ||
| MYCBP2-AS2 | 10 | ||
| RP11-92K15.1 | 10 |
Figure 4.
Functional enrichment analyses for differentially expressed lncRNAs. P-value represents significance of terms; GeneRatio represents the ratio of the number of lncRNA targets in this GO/KEGG term to the number of lncRNA targets in all GO/KEGG term. GO, gene ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; lncRNA, long non-coding RNA.
Validation of DELs
According to the bioinformatics results, changes in the expression of several DELs in HUVECs were validated using RT-qPCR. The results showed that following HUVEC treatment with HCY, the mRNA levels of AC005550.3 and RP11-415D17.3 decreased significantly, whereas those of RP1-140K8.5 increased compared with those in untreated HUVECs (Figure 5).
Figure 5.
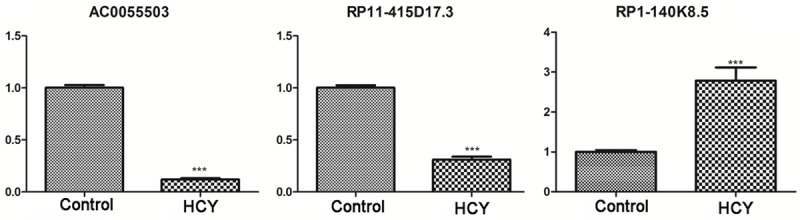
Validation of differentially expressed lncRNAs using real-time quantitative polymerase chain reaction. ***P < 0.001 compared with control. lncRNA, long non-coding RNA.
Discussion
HCY has been demonstrated to be an independent risk factor for CAD [25]. The present study found that HCY significantly promoted HUVEC apoptosis but had no effect on the cell cycle. To find the putative lncRNAs involved in HCY-related CAD, the present bioinformatics study included HUVECs treated with and without HCY. Following HCY treatment, our results revealed 101 upregulated and 281 downregulated genes in addition to 32 upregulated and 115 downregulated lncRNAs. CD34, FGF2, and SERPINE1 were the hub nodes in the PPI network, and AC005550.3, RP11-415D17.3, and RP1-140K8.5 were found to play a critical role in the regulation of these DEGs. Furthermore, the downregulation of AC005550.3 and RP11-415D17.3 as well as upregulation of RP1-140K8.5 were detectable by RT-qPCR, which was consistent with the results predicted by our bioinformatics analysis.
Both AC005550.3 and RP11-415D17.3 were found to be downregulated in HCY-treated HUVECs. These two lncRNAs had similar targets such as CD34 and FGF2 in addition to similar functional enrichment pathways. We also found that CD34 and FGF2, which are associated with blood vessel development, were downregulated in HCY-treated HUVECs compared with those in untreated HUVECs. CD34, a highly glycosylated transmembrane glycoprotein, usually expresses in hematopoietic stem and progenitor cells as well as in vascular endothelial cells [26]. CD34 plays an important role in cell adhesion and participates in hematopoietic stem cell transport as well as inflammatory response [27]. In particular, previous studies have shown that CD34 plays an antiadhesive role during lumen formation in blood vessel development [28,29], and CD34 expression in endothelial cells is closely associated with active angiogenesis [26]. This indicates that CD34 may be beneficial to endothelial repair and vascular reconstruction. Along these lines, we detected downregulated CD34 in HCY-treated HUVECs, which may lead to vascular endothelial injury.
Fibroblast growth factor 2 (FGF2), another target of AC005550.3 and RP11-415D17.3, is involved in diverse BPs, including tumor growth, wound healing, and nervous system development [30]. It also participates in mitogenic and angiogenic activities and can promote cell proliferation and differentiation along with inducing angiogenesis and cell migration [31]. It has been previously shown that vascular endothelial cell apoptosis is related to reduced FGF2 levels [32], which is consistent with our present results. In addition, angiogenesis is associated with endothelial cell dysfunction, and angiogenesis or vascular remodeling is beneficial in the treatment of CVD [33]. Several clinical studies have further demonstrated that FGF2 can improve myocardial perfusion and function and symptoms of patients with CVD [34-36]. All these results indicate that AC005550.3 and RP11-415D17.3 may play a role in blood vessel development by targeting CD34 and FGF2, thereby regulating HCY-treated vascular endothelial injury.
We additionally found that RP1-140K8.5 is upregulated in HCY-treated HUVECs. SERPINE1, a target of RP1-140K8.5, was upregulated in HCY-treated HUVECs and is associated with plasminogen activation and fibrinolysis. Fibrinolysis abnormality has been shown to be associated with vascular endothelial injury [37], and vascular endothelial injury may be involved in thrombosis and atherosclerosis [38]. It has also been shown that impaired fibrinolytic activity can be caused by elevated levels of plasminogen activator inhibitor type 1 (PAI-1) [39]. PAI-1, encoded by SERPINE1, is a member of the serine proteinase inhibitor superfamily [40]. It is a major inhibitor of fibrinolysis, which it achieves by inhibiting urokinase (uPA) and tissue-type plasminogen activator (tPA) [40]. PAI-1 deficiency can induce bleeding disorders, and high levels of PAI-1 are related to thrombophilia [41]. In particular, PAI-1 is associated with endothelial dysfunction [42]. Thus, elevated PAI-1 levels are closely linked to CVD, including reinfarction, coronary heart disease, acute ischemic stroke, and atherosclerosis [43,44]. In addition, tPA administration can be used for treating acute ischemic stroke because it can inhibit PAI-1 action [45]. This indicates that RP1-140K8.5 may be involved in fibrinolysis because it targets SERPINE1, thereby regulating HCY-treated vascular endothelial injury.
Our study revealed that downregulated AC005550.3 and RP11-415D17.3 targeting CD34 and FGF2 and upregulated RP1-140K8.5 targeting SERPINE1 may be associated with HCY-induced vascular endothelial injury by regulating blood vessel development and fibrinolysis, respectively. Further investigation of the functions of these lncRNAs in CVD is necessary.
Acknowledgements
This work was supported by Shanghai Municipal Commission of Health and Family Planning (Project number: 2015ZB0502).
Disclosure of conflict of interest
None.
References
- 1.Pagidipati NJ, Gaziano TA. Estimating deaths from cardiovascular disease: a review of global methodologies of mortality measurement. Circulation. 2013;127:749–56. doi: 10.1161/CIRCULATIONAHA.112.128413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.American Diabetes Association. 8. Cardiovascular disease and risk management. Diabetes Car. 2016;39(Suppl 1):S60–71. doi: 10.2337/dc16-S011. [DOI] [PubMed] [Google Scholar]
- 3.Moran A, Gu D, Zhao D, Coxson P, Wang YC, Chen CS, Liu J, Cheng J, Bibbins-Domingo K, Shen YM. Future cardiovascular disease in China. Circ Cardiovasc Qual Outcomes. 2010;3:243–252. doi: 10.1161/CIRCOUTCOMES.109.910711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Perk J, De Backer G, Gohlke H, Graham I, Reiner Ž, Verschuren WM, Albus C, Benlian P, Boysen G, Cifkova R, Deaton C, Ebrahim S, Fisher M, Germanò G, Hobbs R, Hoes A, Karadeniz S, Mezzani A, Prescott E, Ryden L, Scherer M, Syvanne M, Scholte Op Reimer WJ, Vrints C, Wood D, Zamorano JL, Zannad F Comitato per Linee Guida Pratiche (CPG) dell’ESC. [European Guidelines on cardiovascular disease prevention in clinical practice (version 2012). The Fifth Joint Task Force of the European Society of Cardiology and other societies on cardiovascular disease prevention in clinical practice (constituted by representatives of nine societies and by invited experts)] G Ital Cardiol (Rome) 2013;14:328–92. doi: 10.1714/1264.13964. [DOI] [PubMed] [Google Scholar]
- 5.Siti HN, Kamisah Y, Kamsiah J. The role of oxidative stress, antioxidants and vascular inflammation in cardiovascular disease (a review) Vascul Pharmacol. 2015;71:40–56. doi: 10.1016/j.vph.2015.03.005. [DOI] [PubMed] [Google Scholar]
- 6.Tang X, Luo YX, Chen HZ, Liu DP. Mitochondria, endothelial cell function, and vascular diseases. Front Physiol. 2014;5 doi: 10.3389/fphys.2014.00175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mangge H, Becker K, Fuchs D, Gostner JM. Antioxidants, inflammation and cardiovascular disease. World J Cardiol. 2014;6:462–77. doi: 10.4330/wjc.v6.i6.462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ganguly P, Alam SF. Role of homocysteine in the development of cardiovascular disease. Nutr J. 2015;14:6. doi: 10.1186/1475-2891-14-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shi X, Sun M, Liu H, Yao Y, Song Y. Long non-coding RNAs: a new frontier in the study of human diseases. Cancer Lett. 2013;339:159–66. doi: 10.1016/j.canlet.2013.06.013. [DOI] [PubMed] [Google Scholar]
- 10.Spizzo R, Almeida MI, Colombatti A, Calin GA. Long non-coding RNAs and cancer: a new frontier of translational research? Oncogene. 2012;31:4577–87. doi: 10.1038/onc.2011.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Johnson R. Long non-coding RNAs in Huntington’s disease neurodegeneration. Neurobiol Dis. 2012;46:245–54. doi: 10.1016/j.nbd.2011.12.006. [DOI] [PubMed] [Google Scholar]
- 12.Heward JA, Lindsay MA. Long non-coding RNAs in the regulation of the immune response. Trends Immunol. 2014;35:408–19. doi: 10.1016/j.it.2014.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu GC, Pan HF, Leng RX, Wang DG, Li XP, Li XM, Ye DQ. Emerging role of long noncoding RNAs in autoimmune diseases. Autoimmun Rev. 2015;14:798–805. doi: 10.1016/j.autrev.2015.05.004. [DOI] [PubMed] [Google Scholar]
- 14.Congrains A, Kamide K, Oguro R, Yasuda O, Miyata K, Yamamoto E, Kawai T, Kusunoki H, Yamamoto H, Takeya Y, Yamamoto K, Onishi M, Sugimoto K, Katsuya T, Awata N, Ikebe K, Gondo Y, Oike Y, Ohishi M, Rakugi H. Genetic variants at the 9p21 locus contribute to atherosclerosis through modulation of ANRIL and CDKN2A/B. Atherosclerosis. 2012;220:449–55. doi: 10.1016/j.atherosclerosis.2011.11.017. [DOI] [PubMed] [Google Scholar]
- 15.Uchida S, Dimmeler S. Long noncoding RNAs in cardiovascular diseases. Circ Res. 2015;116:737–50. doi: 10.1161/CIRCRESAHA.116.302521. [DOI] [PubMed] [Google Scholar]
- 16.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–11. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harrow J, Frankish A, Gonzalez JM, Tapanari E, Diekhans M, Kokocinski F, Aken BL, Barrell D, Zadissa A, Searle S, Barnes I, Bignell A, Boychenko V, Hunt T, Kay M, Mukherjee G, Rajan J, Despacio-Reyes G, Saunders G, Steward C, Harte R, Lin M, Howald C, Tanzer A, Derrien T, Chrast J, Walters N, Balasubramanian S, Pei B, Tress M, Rodriguez JM, Ezkurdia I, van Baren J, Brent M, Haussler D, Kellis M, Valencia A, Reymond A, Gerstein M, Guigó R, Hubbard TJ. GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res. 2012;22:1760–74. doi: 10.1101/gr.135350.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pertea M, Pertea GM, Antonescu CM, Chang TC, Mendell JT, Salzberg SL. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat Biotechnol. 2015;33:290–5. doi: 10.1038/nbt.3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Robinson MD, Oshlack A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010;11:R25. doi: 10.1186/gb-2010-11-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 21.Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos A, Tsafou KP, Kuhn M, Bork P, Jensen LJ, von Mering C. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2014;43:D447–52. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pearson K. Note on regression and inheritance in the case of two parents. Proceedings of the Royal Society of London. 1895;58:240–2. [Google Scholar]
- 24.Yu G, Wang LG, Han Y, He QY. ClusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16:284–7. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wald DS, Law M, Morris JK. Homocysteine and cardiovascular disease: evidence on causality from a meta-analysis. BMJ. 2002;325:1202. doi: 10.1136/bmj.325.7374.1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Siemerink MJ, Klaassen I, Vogels IM, Griffioen AW, Van Noorden CJ, Schlingemann RO. CD34 marks angiogenic tip cells in human vascular endothelial cell cultures. Angiogenesis. 2012;15:151–63. doi: 10.1007/s10456-011-9251-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Krause DS, Fackler MJ, Civin CI, May WS. CD34: structure, biology, and clinical utility. Blood. 1996;87:1–13. [PubMed] [Google Scholar]
- 28.Nielsen JS, McNagny KM. CD34 is a key regulator of hematopoietic stem cell trafficking to bone marrow and mast cell progenitor trafficking in the periphery. Microcirculation. 2009;16:487–96. doi: 10.1080/10739680902941737. [DOI] [PubMed] [Google Scholar]
- 29.Strilić B, Kučera T, Eglinger J, Hughes MR, McNagny KM, Tsukita S, Dejana E, Ferrara N, Lammert E. The molecular basis of vascular lumen formation in the developing mouse aorta. Dev Cell. 2009;17:505–15. doi: 10.1016/j.devcel.2009.08.011. [DOI] [PubMed] [Google Scholar]
- 30.Baird A, Esch F, Mormede P, Ueno N, Ling N, Bohlen P, Ying SY, Wehrenberg WB, Guillemin R. Molecular characterization of fibroblast growth factor: distribution and biological activities in various tissues. Recent Prog Horm Res. 1986;42:143–205. doi: 10.1016/b978-0-12-571142-5.50008-2. [DOI] [PubMed] [Google Scholar]
- 31.Okada-Ban M, Thiery JP, Jouanneau J. Fibroblast growth factor-2. Int J Biochem Cell Biol. 2000;32:263–7. doi: 10.1016/s1357-2725(99)00133-8. [DOI] [PubMed] [Google Scholar]
- 32.Chen CH, Jiang T, Yang JH, Jiang W, Lu J, Marathe GK, Pownall HJ, Ballantyne CM, McIntyre TM, Henry PD, Yang CY. Low-density lipoprotein in hypercholesterolemic human plasma induces vascular endothelial cell apoptosis by inhibiting fibroblast growth factor 2 transcription. Circulation. 2003;107:2102–8. doi: 10.1161/01.CIR.0000065220.70220.F7. [DOI] [PubMed] [Google Scholar]
- 33.Kolluru GK, Bir SC, Kevil CG. Endothelial dysfunction and diabetes: effects on angiogenesis, vascular remodeling, and wound healing. Int J Vasc Med. 2012;2012:918267. doi: 10.1155/2012/918267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Simons M, Annex BH, Laham RJ, Kleiman N, Henry T, Dauerman H, Udelson JE, Gervino EV, Pike M, Whitehouse M, Moon T, Chronos NA. Pharmacological treatment of coronary artery disease with recombinant fibroblast growth factor-2. Circulation. 2002;105:788–93. doi: 10.1161/hc0802.104407. [DOI] [PubMed] [Google Scholar]
- 35.Post MJ, Laham R, Sellke FW, Simons M. Therapeutic angiogenesis in cardiology using protein formulations. Cardiovasc Res. 2001;49:522–31. doi: 10.1016/s0008-6363(00)00216-9. [DOI] [PubMed] [Google Scholar]
- 36.Laham RJ, Chronos NA, Pike M, Leimbach ME, Udelson JE, Pearlman JD, Pettigrew RI, Whitehouse M, Yoshizawa C, Simons M. Intracoronary basic fibroblast growth factor (FGF-2) in patients with severe ischemic heart disease: results of a phase I open-label dose escalation study. J Am Coll Cardiol. 2000;36:2132–9. doi: 10.1016/s0735-1097(00)00988-8. [DOI] [PubMed] [Google Scholar]
- 37.Ueno H, Hirasawa H, Oda S, Shiga H, Nakanishi K, Matsuda K. Coagulation/fibrinolysis abnormality and vascular endothelial damage in the pathogenesis of thrombocytopenic multiple organ failure. Crit Care Med. 2002;30:2242–8. doi: 10.1097/00003246-200210000-00011. [DOI] [PubMed] [Google Scholar]
- 38.Gimbrone MA, García-Cardeña G. Endothelial cell dysfunction and the pathobiology of atherosclerosis. Circ Res. 2016;118:620–36. doi: 10.1161/CIRCRESAHA.115.306301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chapin JC, Hajjar KA. Fibrinolysis and the control of blood coagulation. Blood Rev. 2015;29:17–24. doi: 10.1016/j.blre.2014.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yasar Yildiz S, Kuru P, Toksoy Oner E, Agirbasli M. Functional stability of plasminogen activator inhibitor-1. ScientificWorldJournal. 2014;2014 doi: 10.1155/2014/858293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fay WP, Parker AC, Condrey LR, Shapiro AD. Human plasminogen activator inhibitor-1 (PAI-1) deficiency: characterization of a large kindred with a null mutation in the PAI-1 gene. Blood. 1997;90:204–08. [PubMed] [Google Scholar]
- 42.Widlansky ME, Gokce N, Keaney JF, Vita JA. The clinical implications of endothelial dysfunction. J Am Coll Cardiol. 2003;42:1149–60. doi: 10.1016/s0735-1097(03)00994-x. [DOI] [PubMed] [Google Scholar]
- 43.Song C, Burgess S, Eicher JD, O’Donnell CJ, Johnson AD. Causal effect of plasminogen activator inhibitor type 1 on coronary heart disease. J Am Heart Assoc. 2017;6:e004918. doi: 10.1161/JAHA.116.004918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kohler HP, Grant PJ. Plasminogen-activator inhibitor type 1 and coronary artery disease. N Engl J Med. 2000;342:1792–801. doi: 10.1056/NEJM200006153422406. [DOI] [PubMed] [Google Scholar]
- 45.Thögersen AM, Jansson JH, Boman K, Nilsson TK, Weinehall L, Huhtasaari F, Hallmans G. High plasminogen activator inhibitor and tissue plasminogen activator levels in plasma precede a first acute myocardial infarction in both men and women: evidence for the fibrinolytic system as an independent primary risk factor. Circulation. 1998;98:2241–7. doi: 10.1161/01.cir.98.21.2241. [DOI] [PubMed] [Google Scholar]