Abstract
Background: Long non-coding RNAs (LncRNAs) have been reported as key regulators of tumor progression in recent decades. However, the potential molecular mechanisms of breast cancer are still unclear. With the development of sequencing technology, we discovered that LINC00673 is upregulated in tumor tissues. But the main role of LINC00673 in breast cancer has yet to be confirmed. Materials and methods: 35 pairs of breast tumors and normal tissues were selected to real-time quantitative polymerase chain reaction (RT-qPCR) to validate LINC00673 is overexpressed in tumor tissues. We conducted proliferation, colony formation, migration, invasion assays, and EMT-related phenotype to determine the specific role of LINC00673 in breast cancer cell lines (MDA-MB-231, BT-549, and MCF-7) transfected with small interfering RNA. Gene expression profiling was conducted to found LINC00673-associated gene transcriptional changes. Results: We discovered that LINC00673 is significantly upregulated in breast cancer tissues compared to paired adjacent non-tumor tissues by RT-qPCR and highly expressed LINC00673 is positively correlated with lymph node metastasis and clinical stage in the validated cohort. Knocking down LINC00673 inhibited cell proliferation and metastasis, whereas upregulated LINC00673 had the opposite effect. Gene expression profiling results indicated that LINC00673 could influence NCR3LG1(B7-H6) expression in transcriptional level. Western Blot showed us that LINC00673 could regulate epithelial-mesenchymal transition (EMT) and B7-H6 in protein level. Then we demonstrated that knocking down B7-H6 could decrease breast cancer cell proliferation and metastasis. Conclusion: In this study, we identified the role of LINC00673 in inducing proliferation and metastasis of breast cancer cell lines and it might act as an underlying therapeutic target for breast cancer.
Keywords: Breast cancer, LINC00673, proliferation, metastasis, B7-H6
Introduction
For women, breast cancer is one of the most malignancies with an unceasing prevalence of morbidity over the past few years [1,2]. Previous studies estimated that the number of new breast cancers in 2018 was 268,670, with an estimated 41,400 deaths [3]. With the development of medical knowledge, current treatments including surgery, endocrine, or targeted therapies have been made. But the prognosis is not always satisfactory [4-6]. A lot of studies have discovered many oncogenes and tumor suppressor genes, which are acknowledged to be related to the occurrence of breast cancer [7-9]. However, the mechanism of breast cancer progression is complex and much less understood.
Long noncoding RNAs (lncRNAs), which are transcripts longer than 200 nucleotides, are considered to be unable to code proteins [10]. Previous studies have verified that lncRNAs could be identified as biomarkers in tumorigeneses of cancers, such as lncRNA HOTAIR and lncRNA MALAT1 [11-13]. There is growing evidence suggested that dysregulated lncRNAs could induce cancer carcinogenesis via regulating various of the molecular pathway. Sun CC et al found that LINC00511 is upregulated in NSCLC and it induces cell proliferation, metastasis, and apoptosis in multiple NSCLC cell lines through binding to EZH2 and suppressing p57 [14]. LINCXIST is down-regulated in breast cancer and knocking down LINCXIST can inhibit breast cancer cell growth, metastasis via miR-155/CDX1 axis [15]. However, the relationship between breast cancer and lncRNAs still need to further explore.
In our study, 8 pairs of breast cancer tissue samples and adjacent normal tissues were selected to whole transcriptome sequencing in our unpublished article and the results showed that LINC00673 is up-regulated remarkable in breast cancer. As a new lncRNA encoded by chromosome 17q25.1, LINC00673 lacks the potential of coding protein. Many articles have pointed out that LINC00673 influence tumorigenesis in lots of cancers. Mingde Huang et al discovered that sp1 could activate LINC00673 and this lncRNA exerts oncogenic functions by binding LSD1 and EZH2 in gastric cancer [16]. Wei Lu et al showed that LINC00673 is upregulated in non-small cell lung cancer and it can sponge miR-150-5p and modulate ZEB1 indirectly [17]. Ubaidat Abdul-Rahman et al identified LINC00673 as a marker of overall survival (OS) in breast cancer patients [18]. But the underlying relationship between breast cancer and LINC00673 has yet to be further reported.
In our study, 35 paired breast cancer tissue samples and adjacent normal tissues were performed to RT-qPCR assay to validate the dysregulated of LINC00673. Cell culture experiments and western blot were conducted to explore the function of LINC00673 in breast cancer. Our research was designed to test the relationship of LINC00673 expression with the clinical features of breast cancer, to determine the function of LINC00673 in the proliferation and metastasis of breast cancer and to explore the underlying mechanism of LINC00673 in breast cancer.
Materials and methods
Patients and breast tissue samples
35 pairs of tissue samples were gained from patients who underwent surgical resection at Department of Thyroid and Breast Surgery, The First Affiliated Hospital of Wenzhou Medical University. All patient information was collected under the protocols offered by the institutional review board of the Ethics Committee of the First Affiliated Hospital of Wenzhou Medical University. The fresh tissues were snap-frozen in liquid nitrogen immediately and stored at -80°C until further RNA detection.
Cell cultures and growth conditions
MDA-MB-231, BT-549, MDA-MB-468, MCF-7 and MCF-10A cells were used in this study. These cells were obtained from Shanghai Cell Biology, Institute of the Chinese Academy of Sciences (Shanghai, China). MDA-MB-231, MCF-7 were cultured in Dulbecco’s Modified Eagle’s Medium DMEM (Gibco, Grand Island, NY, USA) supplemented with 10% FBS (Gibco, Grand Island, NY, USA). MDA-MB-468 were cultured in L-15 medium (Gibco, Grand Island, NY, USA) supplemented with 10% FBS (Gibco, Grand Island, NY, USA). BT-549 was cultured in RPMI-1640 medium (Gibco, Grand Island, NY, USA) supplemented with 10% FBS (Gibco, Grand Island, NY, USA). MCF-10A cells were cultured in DMEM-F12 (Gibco, Grand Island, NY, USA) supplemented with 100 U/ml of penicillin, 100 μg/ml of streptomycin, 2 mM of L-glutamine, 20 ng/ml of epidermal growth factor and 10% FBS (Gibco, Grand Island, NY, USA). MDA -MB-468 was incubated in a standard cell culture incubator (Thermo, Waltham, MA, USA) at 37°C without CO2. The others were grown in appropriate conditioned medium and maintained at 37°C with 5% CO2.
Cell transfection
MCF-7, MDA-MB-231, and BT-549 were transfected through Lipofectamine RNAiMAX transfection reagent (Invitrogen). 100,000 breast cancer cells were plated 24 h before transfection. For cell interference, LINC00673 and B7-H6 were synthesized at Shanghai GenePharma Company Limited. The siRNA sequences used in the study are LINC00673 siRNA-1, Forward 5’-GCCUUUGUACUCAGCAAUUTT-3’ and Reverse 5’-AAUUGCUGAGUACAAAGGCTT-3’; LINC00673 siRNA-2, Forward 5’-GGAUACAGAGUGAAUAGUUTT-3’ and Reverse 5’-AACUAUUCACUCUGUAUCCTT-3’; B7-H6 siRNAs target the following sequences: siB7H6-1, Forward 5’-GGAGAUCACCAAGAGGCAUTT-3’ and Reverse 5’-AUGCCUCUUGGUGAUCUCCTT-3’; siB7H6-2, Forward 5’-GGAACACUCGGAUGCAGUUTT-3’ and Reverse 5’-AACUGCAUCCGAGUGUUCCTT-3’; To overexpress LINC00673, vectors which containing the full length of LINC00673 were constructed into CMV-MCS-EGFP-SV40-Neomycin.
RNA extraction and real-time quantitative PCR
According to the manufacturer’s instructions (Invitrogen, USA), total RNA isolated from patient tissue specimens and breast cancer cell lines was extracted using TRIzol reagent. The isolated RNA was measured at 260/280 nm to assess RNA quality and quantity. RNA samples were stored at -80°C. The relative expression of mRNA was calculated using the comparative cycle threshold (CT) (2-ΔΔCT) method with GAPDH as the endogenous control to normalize the data. The primer sequences were used as followed: LINC00673 Forward: 5’-TGCTGATGACACATACACA-3’ and Reverse: 5’-GACAAGGATGAACCATGATAG-3’; B7-H6 Forward: 5’-ACAGTAAATGCCTGATGGACCT-3’ and Reverse: 5’-ATTGGGTATGTGAATGCTGGT-3’; GADPH Forward: 5’-GTCTCCTCTGACTTCAACAGCG-3’ and Reverse: 5’-ACCACCCTGTTGCTGTAGCCAA-3’.
Invasion and migration assay
For cell invasion assays, invasion of cells was measured in Matrigel (BD Bioscience, Franklin Lakes, USA). A total of 40,000 cells transfected with si-NC or si-RNA were transferred into the upper chamber and the lower chamber contained 0.6 mL of medium supplemented with 20% FBS. Then we put the plate into the incubator with 5% CO2 atmosphere. After 24 h, a cotton swab was used to remove the cells that adhered to the upper surface of the well, and fixed it with 4% paraformaldehyde and stained with 0.4% crystal violet solution. Motility assays were similar to invasion assay. Cell migration and invasion were counted in five random fields and images captured under a microscope at a magnification of ×20.
Cell proliferation assay
For colony forming assay, placing breast cancer cells (MDA-MB-231, MCF-7, BT-549) in 6-well plates, maintaining it in RPMI1640 or DMEM supplied with 10% FBS for 14 days and then stained with crystal violet. These images were captured by the camera.
For the CCK-8 assay, breast cancer cells (MDA-MB-231, MCF-7, BT-549) were plated into 96-well plates 24 h and transfected with siRNA. Then incubated it for 24 h, 48 h, 72 h, 96 h. Experiments were performed in triplicate.
Apoptosis detection analysis
Annexin-V-FITC apoptosis detection kit was purchased to detect the proportion of apoptotic cells. Centrifuging cells at 1000 rpm for 5 min three times and resuspending it with PBS. Resuspending cells in 500 μL binding buffer and mixing with 5 μL Annexin V-FITC (Kaijibio, Nanjing, China). The data was analyzed by flow cytometer (Guava easyCyte HT, Millipore, USA).
Western blot analysis
Whole cell lysates were separated using SDS-PAGE on a 10% gel and electrotransferred to PVDF membranes. Then we blocked the blots with 5% non-fat milk for 2 h at room temperature. According to the manufacturer’s protocol, the blots were probed with polyclonal antibody at 4°C overnight. The blots were then incubated with the secondary antibody (Abcam, Cambridge, MA) for 1 h at room temperature. Primary antibodies were as follows: N-cadherin, Vimentin, E-cadherin, B7-H6, human β-actin (all Cell Signaling Technology, Danvers, MA, USA).
Statistical analysis
Statistical significance was established using SPSS 23.0 statistical software package (SPSS, Inc., Chicago, IL, USA). The differences between groups were analyzed using the Student’s t-test and one-way analysis of variance with post hoc contrasts by Student-Newman-Keuls test. Differences were considered to be statistically significant at P < 0.05.
Results
LINC00673 is overexpressed in breast cancer tissues and the relationship between LINC00673 expression and clinical features
The previous study showed that LINC00673 was upregulated in breast cancer compared to normal breast tissue according to the date downloading from the TCGA and the level of LINC00673 was inversely correlated with breast cancer survival across all breast cancer patients [18]. In order to further prove this founding, we detected the expression levels of LINC00673 in 35 pairs of breast cancer samples and paired adjacent non-tumor tissues tissue. As shown in Figure 1A, LINC00673 expression in the breast cancer samples was higher than in adjacent normal tissues (P < 0.05). The relationship between LINC00673 expression and clinicopathologic features was also analyzed. We divided the patients into low-expression group and high-expression group according to the median value in the validate cohort. The high expression of LINC00673 is associated with lymph node metastasis (P = 0.032) and clinical stage (P < 0.01) (Table 1).
Figure 1.
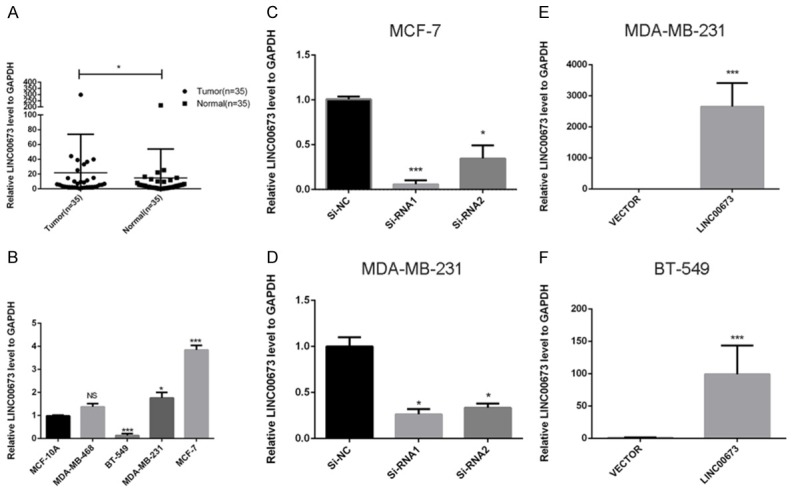
The expression of LINC00673 in breast cancer. A. LINC00673 expression was detected by RT-qPCR in 35 paired breast samples and paired adjacent non-tumor tissues (paired t-test, P < 0.05). B. The relative expression of LINC00673 (compared with the GAPDH gene) was examined via RT-qPCR. Compared to normal breast cell lines MCF-10A, MCF-7 and MDA-MB-231 cell lines have higher LINC00673 expression while BT-549 has lower expression. C and D. The relative expression of LINC00673 (compared with the GAPDH gene) in MCF-7 and MDA-MB-231. Compared with corresponding control group, the expression of LINC00673 in siRNA group was lower. E and F. The relative expression of LINC00673 (compared with the GAPDH gene) in MDA-MB-231 and BT-549. Compared with corresponding Vector group, the expression of LINC00673 in LINC00673 group was higher. *P < 0.05, ***P < 0.001 in comparison with Si-NC or Vector using student’s t-test.
Table 1.
The relationship between LINC00673 and clinicopathologic characteristics in the validated cohort
| Clinicopathologic characteristics | High expression (%) | Low expression (%) | X2 | P |
|---|---|---|---|---|
| Age | 0.122 | 0.726 | ||
| ≤ 60 | 18 (81.8) | 10 (76.9) | ||
| > 60 | 4 (18.2) | 3 (23.1) | ||
| Tumor size | 1.301 | 0.522 | ||
| < 2 cm | 10 (45.5) | 7 (53.8) | ||
| 2-5 cm | 10 (45.5) | 6 (46.2) | ||
| > 5 cm | 2 (9) | 0 (0) | ||
| Lymph node metastasis | 4.609 | 0.032* | ||
| NO | 7 (31.8) | 9 (69.2) | ||
| YES | 15 (68.2) | 4 (30.8) | ||
| Clinical stage | 6.655 | 0.010* | ||
| I-II | 7 (31.8) | 10 (76.9) | ||
| III-IV | 15 (68.2) | 3 (23.1) | ||
| Estrogen receptor | 0.530 | 0.467 | ||
| Negative | 8 (36.3) | 5 (50) | ||
| Positive | 14 (63.6) | 5 (50) | ||
| Progesterone receptor | 0.057 | 0.811 | ||
| Negative | 10 (45.5) | 5 (50) | ||
| Positive | 12 (54.5) | 5 (50) |
P < 0.05.
LINC00673 regulates proliferation and apoptosis of breast cancer cell lines
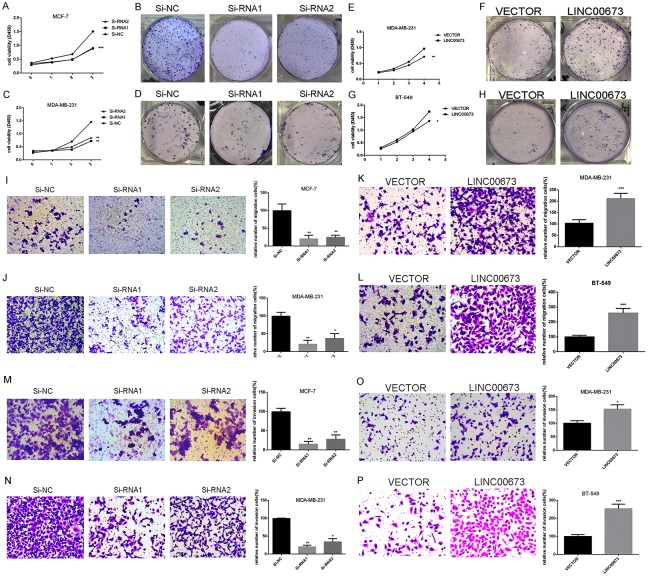
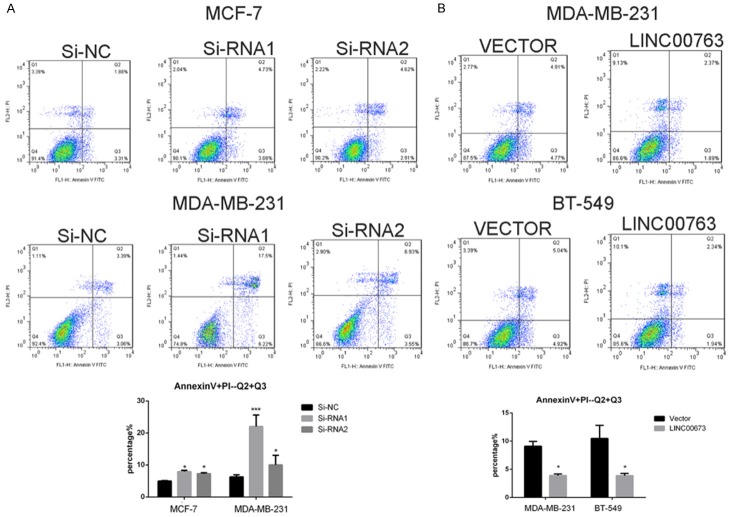
As shown in Figure 1B, LINC00673 expression in MCF-7, MDA-MB-231, and MDA-MB-468 was significantly higher compared with the normal breast cell line MCF-10A while others lower than MCF-10A. Then we selected MCF-7, BT-549, and MDA-MB-231 for the further cell experiment. As confirmed by RT-qPCR, LINC00673 was effectively downregulated in MCF-7 and MDA-MB-231 while LINC00673 was remarkably elevated in BT-549 and MDA-MB-231 (Figure 1C-F). Then, we performed counting kit-8 (CCK8) assay to further determine to check the effects of LINC00673 on cell proliferation. CCK-8 assay results showed that the ability of proliferation was significantly inhibited in MCF-7 and MDA-MB-231 through knocking down LINC00673 expression (transfection with Si-RNA1 or Si-RNA2) compared with control (transfection with si-NC) while upregulated LINC00673 could enhance breast cancer cell lines proliferation (Figure 2A, 2C, 2E, 2G). And further cell colony formation was remarkably inhibited by knocking down LINC00673 compared with control while overexpressed LINC00673 displayed the opposites (Figure 2B, 2D, 2F, 2H). To elucidate the extent by which LINC00673 influences breast tumorigenesis, we conducted apoptosis assays. As shown in Figure 3, we observed that transfected with Si-RNA1 or Si-RNA2 increased the proportion of apoptotic cells in the MCF-7 and MDA-MB-231 while overexpressed LINC00673 decreased the proportion of apoptotic cells in the BT-549 and MDA-MB-231. Taken together, these results indicated that LINC00673 played a significant role in the proliferation and apoptosis capacities of the breast cancer cell lines.
Figure 2.
Dysregulation LINC00673 gene expression in PTC cells inhibiting proliferation and metastasis. A, C. MCF-7 and MDA-MB-231 cell lines transfected with Si-RNA or Si-NC were cultured in 96-well plates for 1-4 days and using CCK-8 measured cell proliferation. Cell proliferation was significantly suppressed in breast cancer cell transfected by Si-RNA. E, G. MDA-MB-231 and BT-549 cell lines transfected with LINC00673 or Vector were cultured in 96-well plates for 1-4 days and using CCK-8 measured cell proliferation. Cell proliferation was significantly enhanced in breast cancer cell transfected by Si-RNA. B, D. MCF-7 and MDA-MB-231 cell lines transfected with Si-RNA or Si-NC were cultured in 6-well plates for 10-14 days. F, H. MDA-MB-231 and BT-549 cell lines transfected with LINC00673 or Vector were cultured in 6-well plates for 10-14 days. I, J. In MCF-7 and MDA-MB-231, transwell migration assays in down-regulation LINC00673 cells and their corresponding control cells. K, L. In MDA-MB-231 and BT-549, transwell migration assays in up-regulation LINC00673 cells and their corresponding control cells. M, N. In MCF-7 and MDA-MB-231, transwell invasion assays in down-regulation LINC00673 cells and their corresponding control cells. O, P. In MDA-MB-231 and BT-549, transwell invasion assays in up-regulation LINC00673 cells and their corresponding control cells. The columns represented the mean number from at least three independent experiments, and the little vertical bars on the top of the columns represent SD. *P < 0.05, **P < 0.01 and ***P < 0.001 in comparison with Si-NC or si-LINC00673 and LINC00673 or Vector using student’s t-test.
Figure 3.
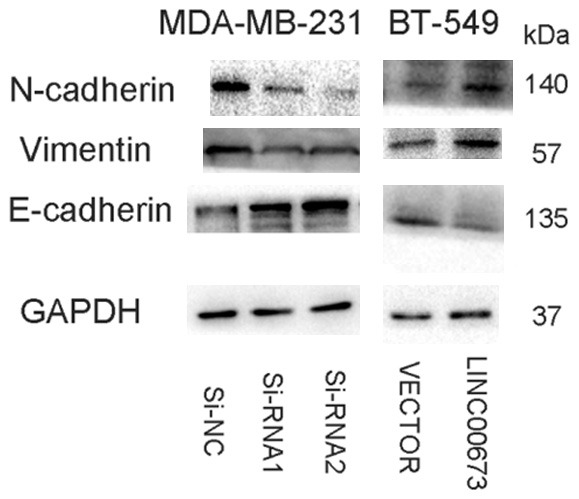
Dysregulation LINC00673 regulates breast cancer cell lines migration and invasion via EMT. A. The influence of LINV00673 expression on the levels of E-cadherin, N-cadherin, and Vimentin in MDA-MB-231 (transfected with si-RNA or si-NC), BT-549 (transfected with Vector or LINC00673) cell lines by western blot.
LINC00673 regulates metastasis of breast cancer cell lines via modulating EMT
Given the obvious relationship between LINC00673 and clinicopathologic features, we continued to probe the function of LINC00673 in the metastasis ability of breast cancer cell lines. Consistent with clinicopathologic features, the migratory and invasive abilities of knocked down LINC00673 cells (transfection with Si-RNA1 or Si-RNA2) were decreased compared with control (transfection with si-NC) (Figure 2I, 2J, 2M, 2N). Further, overexpressed LINC00673 expression raised the metastasis ability of breast cancer cells (Figure 2K, 2L, 2O, 2P). Previous articles pointed that epithelial-mesenchymal transition (EMT) could exert a significant impact in the processes of metastasis [19,20]. Wei Lu et al found that LINC00673 could regulate non-small cell lung cancer cell migration and invasion through EMT [17]. Therefore, we further investigated the potential mechanism by which the LINC00673 contributes to breast cancer metastasis. As shown in Figure 4, down-regulation of LINC00673 had significantly weakened N-cadherin and Vimentin expression and enhanced E-cadherin while overexpressed LINC00673 had opposite results which prompt us that LINC00673 could regulate metastasis by EMT.
Figure 4.
Dysregulation LINC00673 influences breast cancer cell lines apoptosis. A. Annexin V/PI assay was applied to analyze the MCF-7 and MDA-MB-231 cell lines transfected with si-RNA or si-NC. Knocking down LINC00673 could increase apoptotic cell death in breast cancer cells. B. Annexin V/PI assay was applied to analyze the MDA-MB-231 and BT-549 cell lines transfected with LINC00673 or Vector. Overexpressed LINC00673 could decrease apoptotic cell death in breast cancer cells. The Columns represent the mean of death cell numbers from at least three independent experiments. *P < 0.1, ***P < 0.01 in comparison with si-NC using student’s t-test.
B7-H6 is a potential downstream target of LINC00673
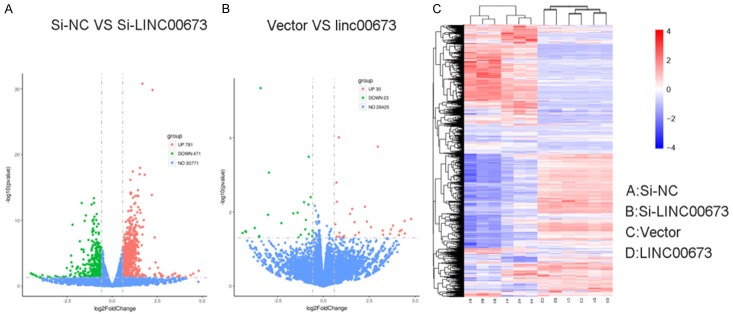
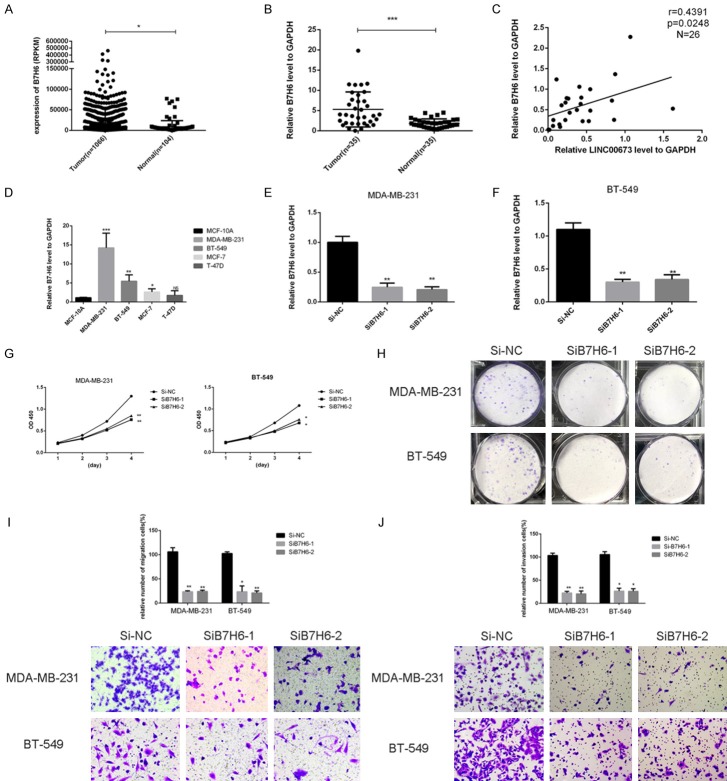
In order to analyze the LINC00673-associated gene transcriptional changes in breast cancer cells, we used an RNA-sequencing analysis. We discovered that B7-H6 was down-regulated after knocking down LINC00673 compared to the control group while B7-H6 was up-regulated in the overexpressed LINC00673 group compared to the control group (Figure 5). As shown in Figure 6, we demonstrated that LINC00673 and B7-H6 had a positive correlation in RNA expression level and protein expression level. Then we continue to experiment to verify whether B7-H6 related to the progression of breast cancer. We found the level of B7-H6 was higher in breast cancer tissue compared to normal breast tissue both in TCGA cohort and validated cohort (Figure 7A, 7B). There was a significantly positive correlation between LINC00673 level and B7-H6 level in breast cancer tissues (Figure 7C). We inferred that B7-H6 may influence proliferation and metastasis in breast cancer cells. To further investigate this hypothesis, we examined the expression of B7-H6 in breast cancer cell lines and found that the B7-H6 is expressed at a higher level in breast cancer cell lines than in the MCF-10A (Figure 7D). To further assess the specific role of B7-H6 in breast cancer cell, we knocked down B7-H6 expression through siRNA. As confirmed by RT-qPCR, B7-H6 expression was effectively downregulated in MDA-MB-231 and BT-549 (Figure 7E, 7F). As shown in Figure 7G-J, knocking down B7-H6 could inhibit the proliferation and metastasis ability of breast cancer cell lines remarkable.
Figure 5.
A. RNA transcriptome sequencing analysis was performed to analyze gene expression profile in MDA-MB-231 cells following linc00673 knock-down. Volcano plot showed all differentially expressed genes. B. RNA transcriptome sequencing analysis was performed to analyze gene expression profile in MDA-MB-231 cells following linc00673 overexpression. Volcano plot showed all differentially expressed genes. C. Heatmap of upregulated and downregulated genes in MDA-MB-231 cells after transfection with Si-NC, LINC00673, Vector or LINC00673.
Figure 6.
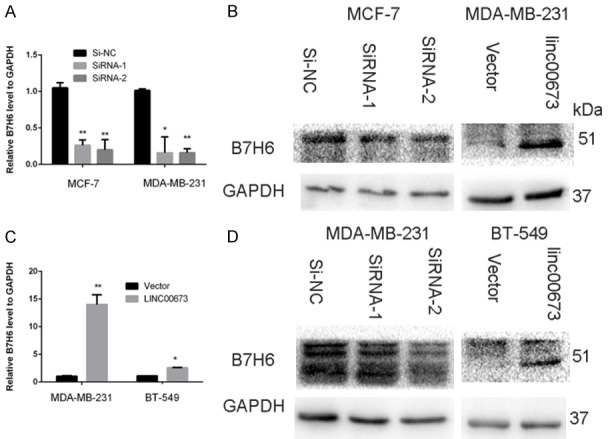
The expression of B7-H6 after regulating LINC00673 in breast cancer cell lines. A and B. RT-qPCR analysis of B7-H6 mRNA level after down-regulated or up-regulated LINC00673. C and D. Western blot analysis of B7-H6 protein level after down-regulated or up-regulated LINC00673.
Figure 7.
The expression of B7-H6 and in breast cancer. A. The TCGA cohort contained 1066 breast cancer tissues and 104 normal tissues. Reads Per Kilobases per Million reads (RPKM) was represented for level of B7-H6. The date was managed through the Manne-Whitney U test. B. B7-H6 expression was detected by RT-qPCR in 35 paired breast samples and paired adjacent non-tumor tissues (paired t-test, P < 0.05). C. Analysis of the relationship between LINC00673 expression (ΔCt value) and B7-H6 mRNA level (ΔCt value) in 35 breast cancer tissues. D. The relative expression of B7-H6 (compared with the GAPDH gene) was examined via RT-qPCR. Compared to normal breast cell lines, MDA-MB-231, BT-549, MCF-7 and T-47D cell lines have higher B7-H6 expression. E and F. The relative expression of B7-H6 (compared with the GAPDH gene) in MDA-MB-231 AND BT-549. Compared with corresponding control group, the expression of B7-H6 in siRNA group was lower. G. MDA-MB-231 and BT-549 cell lines transfected with siB7H6 or si-NC were cultured in 96-well plates for 1-4 days, and using CCK-8 measured cell proliferation. Cell proliferation was significantly enhanced in breast cancer cell transfected by Si-RNA. H. MDA-MB-231 and BT-549 cell lines transfected with siB7H6 or Si-NC were cultured in 6-well plates for 10-14 days. I. In MDA-MB-231 and BT-549, transwell migration assays in up-regulation B7-H6 cells and their corresponding control cells. J. In MDA-MB-231 and BT-549, transwell invasion assays in down-regulation B7-H6 cells and their corresponding control cells. *P < 0.05, **P < 0.01 in comparison with Si-NC or Si-B7H6 using student’s t-test.
LINC00673 oncogenic role is partly dependent on repressingB7-H6 expression
To determine whether B7-H6 could influence the LINC00673-related to enhance proliferation and metastasis in breast cancer cells, we continued to conduct the rescue experiments. MDA-MB-231 and BT-549 cells were co-transfected with LINC00673 and siB7H6-1. CCK-8 assay and transwell assays suggested that knocking down B7-H6 could partly rescue the overexpressed LINC00673 mediated cell growth and metastasis decrease in MDA-MB-231 and BT-549 cells (Figure 8). This important evidence showed that B7-H6 was a significant mediator of LINC00673 related proliferation, metastasis.
Figure 8.
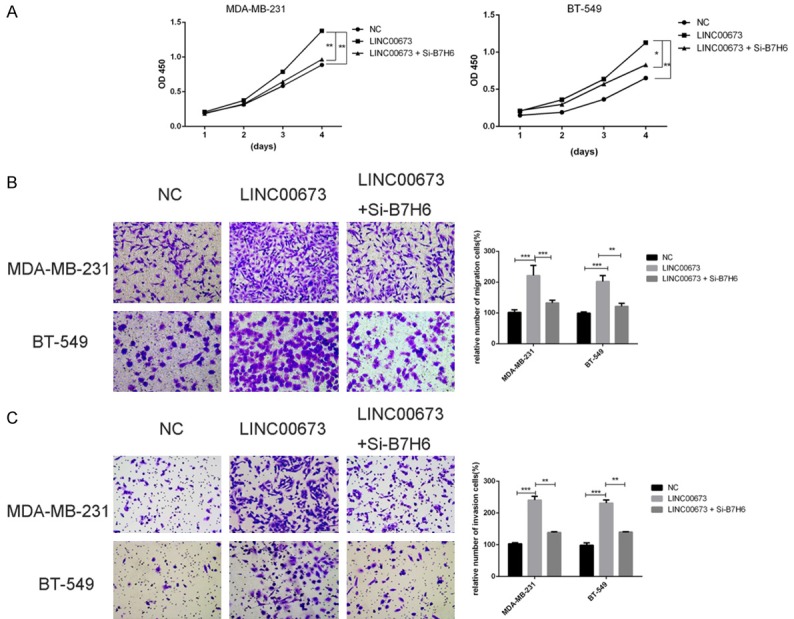
LINC00673 oncogenic role is partly dependent on inducing B7-H6 expression. A. CCK-8 assays were performed to determine the cell viability of MDA-MB-231 and BT-549cells (transfected with NC, LINC00673 or LINC00673+Si-B7-H6). B. Transwell migration assays were performed to determine the cell viability of MDA-MB-231 and BT-549 cells (transfected with NC, LINC00673 or LINC00673+Si-B7-H6). C. Transwell invasion assays were performed to determine the cell viability of MDA-MB-231 and BT-549cells (transfected with NC, LINC00673 or LINC00673+Si-B7-H6). *P < 0.05, **P < 0.01, ***P < 0.001 in comparison with si-NC using student’s t-test.
Discussion
Breast cancer is one of the most widespread malignant tumors with various forms of treatment [3,21]. Despite the advances in medical science, the potential mechanism of pathogenesis is still the major research problem in breast cancer. Emerging evidence indicated that lncRNAs could regulate the expression level of a gene at transcriptional, post-transcriptional and translational regulation levels [22,23]. Previous articles indicated that LncRNAs play a great role in cancer tumorigenesis [24-26]. However, the underlying role of lncRNAs in breast cancer is still unknown.
In case-control studies, LINC00673 is associated with various tumor, including non-small cell lung cancer, gastric cancer, pancreatic cancer, and tongue squamous cell carcinoma [16,27-29]. Ubaidat Abdul-Rahman et al reported that LINC00673 expression was elevated in TCGA cohort and their validated cohort and LINC00673 levels were also inversely correlated with breast cancer survival [18]. However, the potential relationship between LINC00673 and breast cancer has yet to be reported. Our research revealed that the LINC00673 is highly upregulated in breast cancer tissues compared to the adjacent no-tumor tissue in 35 breast cancer patients and high expression of LINC00673 is associated with lymph node metastasis and clinical stage. According to this clinical evidence, we deduced that LINC00673 may influence proliferation and metastasis in breast cancer cells. The cell line experiments sustained the guess that LINC00673 is an oncogene in breast cancer. We discovered that LINC00673 downregulation could remarkably inhibit the ability of cell proliferation and promote apoptotic cell death in vitro while overexpressed LINC00673 had the opposite results. We conducted a loss of function and gain of function assays to demonstrate the role of LINC00673 in the cell metastasis and EMT.
We found B7-H6 might be a potential downstream target of LINC00673 via RNA sequencing. B7-H6 is encoded at 11p15.1 and is approximately 6382 nt in length. Although previous studies have identified B7-H6 is significantly associated with natural killer (NK) cell activation and cytotoxicity [30,31] and tumorigenesis [32], little is known about the biological significance of B7-H6 in breast cancer. After knocking down B7-H6, the proliferation, migration and invasion abilities of breast cancer cell lines were inhibited remarkably. And knocking down B7-H6 could partly rescue the LINC00673 overexpression mediated cell growth and metastasis increase. Based on the previous results presented above, we pointed that the oncogenic role of LINC00673 partly through inducing B7-H6 expression in breast cancer.
In spite of our interesting discovery, there are still some limitations in the study. On the one hand, the quantity of the validated cohort should be expanded to provide more convincing results. On the other hand, we need to conduct more examination in vivo experiments to better validate the oncogene effects of LINC00673.
Conclusion
In the summary, LINC00673 is up-regulated in breast cancer and the abnormal expression exerts oncogene function role in breast cancer cell lines. The findings of our study provide a potential new marker and a target for gene therapy in breast cancer.
Acknowledgements
This study was funded by National Natural Science Foundation of China (NO 81572291) and Natural Science Foundation of Zhejiang province (LY17H160053, LGF18H160031, GF18H160071, and LGF18H160032) and the Medical and Health Technology Projects of Zhejiang province (NO 2017187475) and the Science and Technology Project of Wenzhou (Y20170030). Written informed consent was obtained from each individual participant.
Disclosure of conflict of interest
None.
References
- 1.Chen L, Huang Z, Yao G, Lyu X, Li J, Hu X, Cai Y, Li W, Li X, Ye C. The expression of CXCL13 and its relation to unfavorable clinical characteristics in young breast cancer. J Transl Med. 2015;13:168. doi: 10.1186/s12967-015-0521-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hong W, Dong E. The past, present and future of breast cancer research in China. Cancer Lett. 2014;351:1–5. doi: 10.1016/j.canlet.2014.04.007. [DOI] [PubMed] [Google Scholar]
- 3.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68:7–30. doi: 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 4.Sørlie T, Tibshirani R, Parker J, Hastie T, Marron JS, Nobel A, Deng S, Johnsen H, Pesich R, Geisler S, Demeter J, Perou CM, Lønning PE, Brown PO, Børresen-Dale AL, Botstein D. Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc Natl Acad Sci U S A. 2003;100:8418–8423. doi: 10.1073/pnas.0932692100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Perou CM, Sørlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA, Fluge O, Pergamenschikov A, Williams C, Zhu SX, Lønning PE, Børresen-Dale AL, Brown PO, Botstein D. Molecular portraits of human breast tumours. Nature. 2000;406:747–52. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 6.Hu G, Chong RA, Yang Q, Wei Y, Blanco MA, Li F, Reiss M, Au JL, Haffty BG, Kang Y. MTDH activation by 8q22 genomic gain promotes chemoresistance and metastasis of poor-prognosis breast cancer. Cancer Cell. 2009;15:9–20. doi: 10.1016/j.ccr.2008.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bhandari A, Xia E, Zhou Y, Guan Y, Xiang J, Kong L, Wang Y, Yang F, Wang O, Zhang X. ITGA7 functions as a tumor suppressor and regulates migration and invasion in breast cancer. Cancer Manag Res. 2018;10:969–976. doi: 10.2147/CMAR.S160379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu G, Hu X, Zhou G. Long non-coding RNA OR3A4 promotes proliferation and migration in breast cancer. Biomed Pharmacother. 2017;96:426–433. doi: 10.1016/j.biopha.2017.10.011. [DOI] [PubMed] [Google Scholar]
- 9.Liu Y, Su C, Shan Y, Yang S, Ma G. Targeting Notch1 inhibits invasion and angiogenesis of human breast cancer cells via inhibition Nuclear Factor-κB signaling. Am J Transl Res. 2016;8:2681–92. [PMC free article] [PubMed] [Google Scholar]
- 10.Cerk S, Schwarzenbacher D, Adiprasito JB, Stotz M, Hutterer GC, Gerger A, Ling H, Calin GA, Pichler M. Current status of long non-coding rnas in human breast cancer. Int J Mol Sci. 2016;17 doi: 10.3390/ijms17091485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yan TH, Lu SW, Huang YQ, Que GB, Chen JH, Chen YP, Zhang HB, Liang XL, Jiang JH. Upregulation of the long noncoding RNA HOTAIR predicts recurrence in stage Ta/T1 bladder cancer. Tumour Biol. 2014;35:10249–10257. doi: 10.1007/s13277-014-2344-8. [DOI] [PubMed] [Google Scholar]
- 12.Vaiopoulos AG, Kostakis ID, Koutsilieris M, Papavassiliou AG. Colorectal cancer stem cells. Stem Cells. 2012;30:363–371. doi: 10.1002/stem.1031. [DOI] [PubMed] [Google Scholar]
- 13.Wu DC, Wang SSW, Liu CJ, Wuputra K, Kato K, Lee YL, Lin YC, Tsai MH, Ku CC, Lin WH, Wang SW, Kishikawa S, Noguchi M, Wu CC, Chen YT, Chai CY, Lin CS, Kuo KK, Yang YH, Miyoshi H, Nakamura Y, Saito S, Nagata K, Lin CS, Yokoyama KK. Reprogramming antagonizes the oncogenicity of HOXA13-Long noncoding RNA HOTTIP axis in gastric cancer cells. Stem Cells. 2017;35:2115–2128. doi: 10.1002/stem.2674. [DOI] [PubMed] [Google Scholar]
- 14.Sun CC, Li SJ, Li G, Hua RX, Zhou XH, Li DJ. Long intergenic noncoding RNA 00511 acts as an oncogene in non-small-cell lung cancer by binding to EZH2 and suppressing p57. Mol Ther Nucleic Acids. 2016;5:e385. doi: 10.1038/mtna.2016.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zheng R, Lin S, Guan L, Yuan H, Liu K, Liu C, Ye W, Liao Y, Jia J, Zhang R. Long non-coding RNA XIST inhibited breast cancer cell growth, migration, and invasion via miR-155/CDX1 axis. Biochem Biophys Res Commun. 2018;498:1002–1008. doi: 10.1016/j.bbrc.2018.03.104. [DOI] [PubMed] [Google Scholar]
- 16.Huang M, Hou J, Wang Y, Xie M, Wei C, Nie F, Wang Z, Sun M. Long noncoding RNA LINC00673 is activated by SP1 and exerts oncogenic properties by interacting with LSD1 and EZH2 in gastric cancer. Mol Ther. 2017;25:1014–1026. doi: 10.1016/j.ymthe.2017.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 17.Lu W, Zhang H, Niu Y, Wu Y, Sun W, Li H, Kong J, Ding K, Shen HM, Wu H, Xia D, Wu Y. Long non-coding RNA linc00673 regulated non-small cell lung cancer proliferation, migration, invasion and epithelial mesenchymal transition by sponging miR-150-5p. Mol Cancer. 2017;16:118. doi: 10.1186/s12943-017-0685-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Abdul-Rahman U, Gyorffy B, Adams BD. linc00673 (ERRLR01) is a prognostic indicator of overall survival in breast cancer. Transcription. 2018;9:17–29. doi: 10.1080/21541264.2017.1329684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Adam L, Zhong M, Choi W, Qi W, Nicoloso M, Arora A, Calin G, Wang H, Siefker-Radtke A, McConkey D, Bar-Eli M, Dinney C. miR-200 expression regulates epithelial-to-mesenchymal transition in bladder cancer cells and reverses resistance to epidermal growth factor receptor therapy. Clin Cancer Res. 2009;15:5060–5072. doi: 10.1158/1078-0432.CCR-08-2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hung CM, Kuo DH, Chou CH, Su YC, Ho CT, Way TD. Osthole suppresses hepatocyte growth factor (HGF)-induced epithelial-mesenchymal transition via repression of the c-Met/Akt/mTOR pathway in human breast cancer cells. J Agric Food Chem. 2011;59:9683–9690. doi: 10.1021/jf2021489. [DOI] [PubMed] [Google Scholar]
- 21.Ogretmen B, Hannun YA. Biologically active sphingolipids in cancer pathogenesis and treatment. Nat Rev Cancer. 2004;4:604–616. doi: 10.1038/nrc1411. [DOI] [PubMed] [Google Scholar]
- 22.Batista PJ, Chang HY. Long noncoding RNAs: cellular address codes in development and disease. Cell. 2013;152:1298–1307. doi: 10.1016/j.cell.2013.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Latos PA, Pauler FM, Koerner MV, Senergin HB, Hudson QJ, Stocsits RR, Allhoff W, Stricker SH, Klement RM, Warczok KE, Aumayr K, Pasierbek P, Barlow DP. Airn transcriptional overlap, but not its lncRNA products, induces imprinted Igf2r silencing. Science. 2012;338:1469–1472. doi: 10.1126/science.1228110. [DOI] [PubMed] [Google Scholar]
- 24.Dai W, Tian Y, Jiang B, Chen W. Down-regulation of long non-coding RNA AFAP1-AS1 inhibits tumor growth, promotes apoptosis and decreases metastasis in thyroid cancer. Biomed Pharmacother. 2018;99:191–197. doi: 10.1016/j.biopha.2017.12.105. [DOI] [PubMed] [Google Scholar]
- 25.Yan K, Tian J, Shi W, Xia H, Zhu Y. LncRNA SNHG6 is associated with poor prognosis of gastric cancer and promotes cell proliferation and EMT through epigenetically silencing p27 and sponging miR-101-3p. Cell Physiol Biochem. 2017;42:999–1012. doi: 10.1159/000478682. [DOI] [PubMed] [Google Scholar]
- 26.Qi D, Li J, Que B, Su J, Li M, Zhang C, Yang M, Zhou G, Ji W. Long non-coding RNA DBCCR1-003 regulate the expression of DBCCR1 via DNMT1 in bladder cancer. Cancer Cell Int. 2016;16:81. doi: 10.1186/s12935-016-0356-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ma C. Long intergenic noncoding RNA 00673 promotes non-small-cell lung cancer metastasis by binding with EZH2 and causing epigenetic silencing of HOXA5. Oncotarget. 2017;8:32696–32705. doi: 10.18632/oncotarget.16158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zheng J, Huang X, Tan W, Yu D, Du Z, Chang J, Wei L, Han Y, Wang C, Che X, Zhou Y, Miao X, Jiang G, Yu X, Yang X, Cao G, Zuo C, Li Z, Wang C, Cheung ST, Jia Y, Zheng X, Shen H, Wu C, Lin D. Pancreatic cancer risk variant in LINC00673 creates a miR-1231 binding site and interferes with PTPN11 degradation. Nat Genet. 2016;48:747–757. doi: 10.1038/ng.3568. [DOI] [PubMed] [Google Scholar]
- 29.Yu J, Liu Y, Gong Z, Zhang S, Guo C, Li X, Tang Y, Yang L, He Y, Wei F, Wang Y, Liao Q, Zhang W, Li X, Li Y, Li G, Xiong W, Zeng Z. Overexpression long non-coding RNA LINC00673 is associated with poor prognosis and promotes invasion and metastasis in tongue squamous cell carcinoma. Oncotarget. 2017;8:16621–16632. doi: 10.18632/oncotarget.14200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu F, Wang J, Ke X. Knockdown of B7-H6 inhibits tumor progression and enhances chemosensitivity in B-cell non-Hodgkin lymphoma. Int J Oncol. 2016;48:1561–1570. doi: 10.3892/ijo.2016.3393. [DOI] [PubMed] [Google Scholar]
- 31.Trabanelli S, Chevalier MF, Martinez-Usatorre A, Gomez-Cadena A, Salome B, Lecciso M, Salvestrini V, Verdeil G, Racle J, Papayannidis C, Morita H, Pizzitola I, Grandclement C, Bohner P, Bruni E, Girotra M, Pallavi R, Falvo P, Leibundgut EO, Baerlocher GM, Carlo-Stella C, Taurino D, Santoro A, Spinelli O, Rambaldi A, Giarin E, Basso G, Tresoldi C, Ciceri F, Gfeller D, Akdis CA, Mazzarella L, Minucci S, Pelicci PG, Marcenaro E, McKenzie ANJ, Vanhecke D, Coukos G, Mavilio D, Curti A, Derre L, Jandus C. Tumour-derived PGD2 and NKp30-B7H6 engagement drives an immunosuppressive ILC2-MDSC axis. Nat Commun. 2017;8:593. doi: 10.1038/s41467-017-00678-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Che F, Xie X, Wang L, Su Q, Jia F, Ye Y, Zang L, Wang J, Li H, Quan Y, You C, Yin J, Wang Z, Li G, Du Y, Wang L. B7-H6 expression is induced by lipopolysaccharide and facilitates cancer invasion and metastasis in human gliomas. Int Immunopharmacol. 2018;59:318–327. doi: 10.1016/j.intimp.2018.03.020. [DOI] [PubMed] [Google Scholar]