Abstract
MicroRNAs (miRNAs), a class of emerging small non-coding RNAs, serve as vital players in modulating multiple biological processes via the post-transcriptional regulation of gene expression. Dysregulated expression of miRNAs in liver cancer is well documented, and the involvement of miRNAs in liver cancer initiation and progression has also been described. Cancer stem cells (CSCs) are a subset of cells known to be at the root of cancer recurrence and resistance to therapy. In this review, we highlight recent reports indicating that miRNAs participate in the regulation of liver cancer stem cells (LCSCs). The Wnt signaling pathway, TGF-beta signaling pathway, JAK/STAT signaling pathway and epithelial-mesenchymal transition (EMT) are all closely correlated with the miRNA modulation of LCSCs. In addition, several miRNAs have been demonstrated to be involved in the regulation of LCSCs in response to therapy sensitivity. Targeting LCSCs by regulating the expression of these miRNAs represents a potential therapeutic strategy for treating cancer drug resistance, metastasis and recurrence in the near future.
Keywords: MicroRNA (miRNA), cancer stem cell (CSC), liver cancer, EMT, chemo-resistance
Background
Liver cancer is the sixth most common cancer worldwide [1] and the second leading cause of cancer-related mortality, partially due to a deficiency of marked symptoms during early stages [2]. Unfortunately, liver cancer incidence has been increasing, and liver cancer mortality is rising at a much faster pace than that of other types of cancer [3]. The pathogenesis of liver cancer is a remarkably intricate process consisting of a series of linked steps, which are mediated by multiple risk factors, including chronic viral hepatitis, alcohol abuse, aflatoxin, nonalcoholic steatohepatitis and type 2 diabetes [4-8]. Over the past few years, despite great advances in liver cancer prevention along with detection and treatment increasing the possibility of curing the disease at early stages, most patients still eventually relapse and succumb to their disease [9]. Treatment options for liver cancer include surgery, chemotherapy, radiotherapy, embolization therapy, ablation therapy and liver transplantation [10]. However, therapeutic choices for advanced liver cancer are still scarce. The only current effective molecular-targeted drug against advanced liver cancer is the kinase inhibitor sorafenib, which harbors a resistance rate greater than 50% [11]. Therefore, the prognosis of patients with liver cancer remains dismal, with five-year survival rates less than 17% for all stages [12]. Many recent studies have demonstrated that liver cancer stem cells (LCSCs) are one of the major factors contributing to the refractory nature of liver cancer to conventional treatment and are closely associated with liver cancer growth, relapse and metastasis [13]. Thus, successfully seeking and developing potential therapeutic approaches targeting LCSCs holds huge promise for improving the outcomes of patients with liver cancer.
MicroRNAs (miRNAs) are a family of small, endogenous, non-coding RNAs that are approximately 19-25 nucleotides in length. MiRNAs can inversely modulate gene expression via directly inducing messenger RNA (mRNA) degradation or suppressing translation by base pairing with complementary sites in the 3’-untranslated regions (3’-UTRs) of target mRNAs [14,15]. Via this mechanism, miRNAs regulate a number of biological processes, including cellular differentiation, proliferation, migration, and maintenance of LCSCs [16-20]. However, the mechanisms underlying how miRNAs modulate LCSCs remain poorly understood and need to be further elucidated. In this article, we will describe the clinical significance of LCSCs, provide an overview of miRNAs that are dysregulated in LCSCs, summarize the molecular regulatory pathways of selected miRNAs in LCSCs, and finally, discuss the potential utility of miRNAs for treating liver cancer in the future.
Cancer stem cells (CSCs) and LCSCs
Cancer stem cells (CSCs)
Cancer stem cells (CSCs), also known as cancer-initiating cells and tumor-initiating cells, are a rare subset of cancer cells endowed with stem cell properties, including self-renewal and pluripotency [21,22]. The first conclusive evidence of the existence of CSCs came in 1997 from a study by Bonnet et al. [23]. The CD34+/CD38- subpopulation of cells isolated by the authors from leukemia cells could initiate tumors in NOD/SCID mice that were histologically similar to those of the donor. In 2002, cancer stem-like cells were further confirmed in human cortical glial tumors, which contain neural stem-like cells expressing both astroglial and neuronal markers [24].
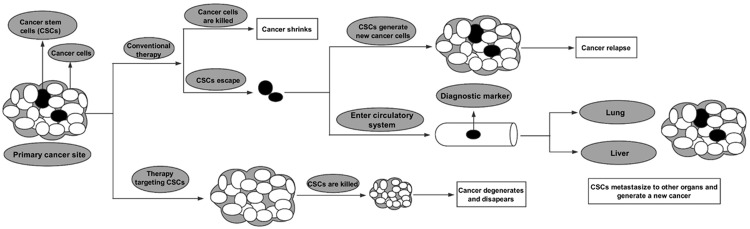
To date, numerous studies have reported that CSCs exist in the context of multiple human cancers, including breast cancer, glioblastoma, prostate cancer, pancreatic cancer and colon cancer [25]. As shown in Figure 1, it is well documented that this specific cancer cell population can survive conventional therapy and is responsible for cancer relapse and metastasis via re-emerging or initiating new cancer.
Figure 1.
Differences between cancer cells and cancer stem cells (CSCs). A cancerous mass includes cancer cells and CSCs. Conventional therapies kill only common cancer cells, while CSCs tend to be resistant to these therapies. After treatment, residual CSCs at the primary cancer site eventually cause cancer re-emergence or relapse. CSCs in the circulatory system can serve as biomarkers and are also responsible for cancer metastasis.
Liver cancer stem cells (LCSCs)
CSCs in liver cancer were first described by Ma et al. [26], wherein the authors identified and isolated a population of CD133+ cells from HCC cell lines and xenograft tumors. It was shown that these cells not only possessed higher levels of proliferation, greater colony-forming efficiency and more potent abilities to form cancer in vivo but also shared stem cell properties, such as the preferential expression of ‘stemness’ genes and the ability to self-renew and differentiate [27]. In addition to CD133, a large number of other LCSC surface markers, including aldehyde dehydrogenase (ALDH), CD13, CD90, CD44, CD24, OV6, Delta-like 1 homolog (DLK1) and epithelial cell-adhesion molecule (EpCAM), have since been discovered [28].
Based on the surface markers shown in Figure 2, LCSCs can be isolated from heterogeneous cancer tissues. In addition, increasing studies have demonstrated that these markers have great potential for use in the diagnosis, therapy and prognosis of liver cancer. Many studies suggest that LCSCs are responsible for the metastasis and relapse of liver cancer. Moreover, it has also been widely accepted that LCSCs account for chemotherapy and radiation resistance in liver cancer. Current therapies against liver cancer successfully kill a majority of cancer cells but may ultimately fail, as they do not eliminate LCSCs, which survive to regenerate new tumors [9]. Therefore, eradication of LCSCs can improve the prognosis of patients with liver cancer. In addition, LCSCs in peripheral blood can serve as diagnostic markers, which are extremely meaningful due to the deficiency of effective measures for early diagnosis of liver cancer. These findings all support the clinical significance of LCSCs.
Figure 2.
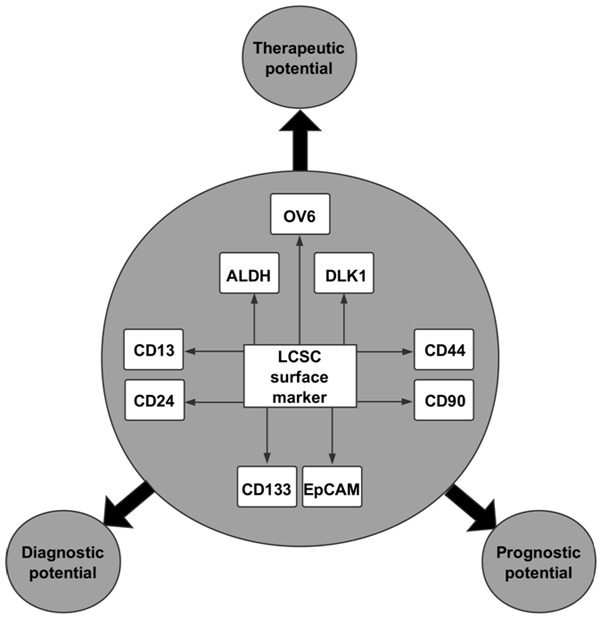
Surface markers of liver CSCs (LCSCs) and their clinical significance. A large number of surface LCSC markers have been discovered. These surface markers include OV6, ALDH, DLK1, CD13, CD24, CD44, CD90, CD133 and EpCAM, which have great potential in the diagnosis, therapy and prognosis of liver cancer.
MiRNAs and LCSCs
MiRNA biogenesis and function
MiRNAs are a class of well-studied non-coding, small, single-stranded RNAs of approximately 22 nucleotides in size. MiRNAs are initially derived from primary transcripts (pri-miRNAs), generally transcribed by RNA polymerase II [29]. Subsequently, pri-miRNAs undergo a two-step processing pathway mediated by two enzymes, Drosha and Dicer, both of which belong to the nuclear ribonuclease III (RNase III) enzyme [30]. Finally, a mature, functional, double-stranded miRNA molecule is yielded. The so-called guide strand, or mature miRNA, is loaded on Argonaute proteins within the RNA-induced silencing complex (RISC) and functions to guide the RISC to complementary sequences in the 3’UTR of specific target mRNAs, resulting in mRNA degradation or translational repression [31-33]. In addition, miRNAs can also activate translation via binding to the 5’UTR of target mRNAs.
MiRNAs function as vital players in regulating a variety of biological processes. Dysregulated expression of miRNAs contributes to multiple human diseases via alterations in proliferation [34], apoptosis [35], cell cycle [36], differentiation [37], angiogenesis [38], autophagy [39], invasion [40] and migration [41]. Increasing findings and data have demonstrated that miRNAs with low expression in cancer often act as tumor suppressors, while miRNAs with high expression usually behave as oncogenes.
Dysregulated miRNAs in LCSCs
Accumulating evidence has shown that miRNAs are involved in controlling self-renewal, proliferation, apoptosis, migration, invasion, differentiation and resistance to therapy in LCSCs. To date, several miRNAs have been reported to modulate LCSCs, such as let-7, the miR-200 family, miR-122, miR-1246, miR-152, miR-181, miR-145, miR-217, miR-500a-3p, and miR-148 (Table 1). These miRNAs exert their roles in LCSCs by directly or indirectly binding to specific target genes that are key molecules in the following pathways: Wnt/beta-catenin signaling, TGF-beta signaling, JAK/STAT signaling, epithelial-mesenchymal transition (EMT) and sensitivity to therapy (Figure 3). Hence, building a therapeutic strategy based on these miRNAs or their relevant pathways can effectively target LCSCs and will be beneficial to rapidly removing the obstacles of metastasis, recurrence and resistance to therapy in liver cancer.
Table 1.
Summary of miRNAs dysregulated in LCSCs
| MiRNA | Up/Down | Surface marker | Target gene | Related pathway | Reference |
|---|---|---|---|---|---|
| let-7b | Down | CD24, CD133 | Frizzled4 | Wnt/beta-catenin pathway | [56] |
| let-7a | Down | Wnt1 | Wnt signaling pathway, EMT | [57] | |
| miR-21 | Up | CD13, EpCAM, CD90, OCT4 | PTEN, RECK, PDCD4 | STAT3 signaling | [85,115,116] |
| miR-25 | Up | PTEN | PTEN/PI3K/AKT pathway | [109] | |
| miR-33a | Down | Lgr5 | ABCB1 | Chemo-resistance to doxorubicin | [105] |
| miR-122 | Down | CD133 | PDK4 | Glycolysis, Smad-independent TGF-beta pathway | [76,110] |
| miR-125b | Down | Smad2, Smad4 | EMT | [73] | |
| miR-130b | Up | CD133 | TP53INP1 | [75] | |
| miR-137 | Down | ANT2 | Chemoresistance to sorafenib | [108] | |
| miR-142-3p | Down | CD133 | CD133 | [111] | |
| miR-145 | Down | OCT4 | [119] | ||
| miR-148a | Down | Smad2, ACVR1, Wnt1 | TGF-beta-Smads signaling pathway, BMPs pathway, EMT | [71,72,102] | |
| miR-148b | Down | NRP1 | [114] | ||
| miR-150 | Down | CD133 | c-Myb | [112] | |
| miR-152 | Down | CD133 | KIT | [113] | |
| miR-155 | Up | CD90, CD133 | TP53INP1, SOCS1 | EMT, STAT3 signaling | [74,86,87] |
| miR-181 | Up | EpCAM | CDX2, GATA6 | Wnt/beta-catenin signaling pathway | [118] |
| miR-200a | Down | CTNNB1 | Wnt/beta-catenin pathway, EMT | [61,99,100] | |
| miR-200b | Down | CD13, CD24, EpCAM, CD133, CD90 | ZEB1 | ZEB1 pathway | [101] |
| miR-205 | Down | CD24 | PLCbeta 1 | [122] | |
| miR-214 | Down | CTNNB1 | Beta-catenin signaling pathway | [62] | |
| miR-217 | Up | DKK1 | Wnt signaling pathway | [59] | |
| miR-429 | Up | EpCAM | RBBP4 | RBBP4/E2F1/OCT4 axis | [120] |
| MiR-452 | Up | SOX7 | Wnt/beta-catenin signaling pathway | [63] | |
| miR-491 | Down | GIT-1 | NF-kappa B pathway, EMT | [103,104] | |
| miR-500a-3p | Up | SOCS2, SOCS4, PTPN11 | JAK3/STAT3 pathway | [84] | |
| miR-589-5p | Down | CD90 | MAP3K8 | [117] | |
| miR-612 | Down | CD133 | AKT2, Sp1 | Wnt/beta-catenin pathway, EMT, Sp1/Nanog signaling pathway | [60,121] |
| miR-1246 | Up | Wnt/beta-catenin pathway | [58] |
Figure 3.
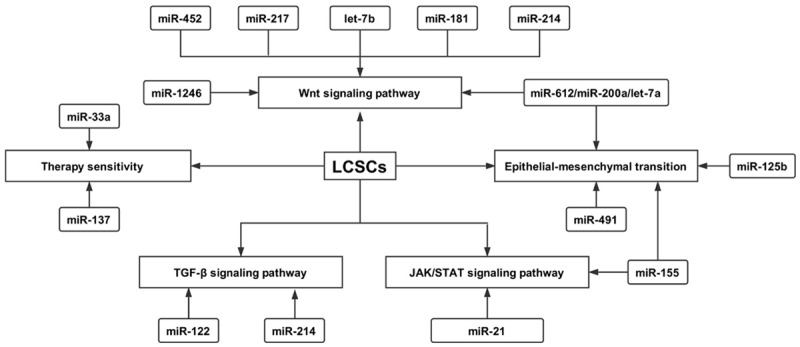
MiRNAs regulate LCSCs via several pathways, including Wnt signaling (miR-1246, miR-452, miR-217, miR-612, miR-214, miR-200a, let-7b), TGF-beta signaling (miR-148a, miR-125b, miR-122, miR-130b, miR-155), JAK/STAT signaling (miR-21, miR-155, miR-500-3p), epithelial-mesenchymal transition (miR-148a, miR-200a, miR-200b) and sensitivity to therapy (miR-33a, miR-25, miR-137).
MiRNAs that regulate LCSCs via the wnt signaling pathway
The Wnt signaling pathway comprises a group of three signaling transduction pathways: the canonical Wnt pathway (Wnt/beta-catenin pathway), the non-canonical planar cell polarity (PCP) pathway and the non-canonical Wnt/calcium pathway [42]. The Wnt signaling pathway’s significant evolutionarily conservation has been well described and is essential for determining cell fates [43].
Dysregulation of the Wnt pathway contributes to several diseases, including cancer [44-46]. The first evidence that Wnt signaling is implicated in cancer came from a study by Nusse et al. in mouse mammary tumor [47], after which the Wnt signaling pathway was subsequently shown to be involved in the occurrence and development of other types of human cancer, including colorectal cancer, lung cancer, prostate cancer and liver cancer [48-51].
A growing body of studies have recently demonstrated that Wnt signaling is deregulated in many kinds of CSCs, including lung [52], prostate [53], colorectal [54] and liver cancers [55]. Interestingly, a large number of studies have demonstrated that miRNAs significantly alter the fraction of LCSCs in liver cancers and effectively regulate several biological behaviors of LCSCs via modulation of the Wnt signaling pathway.
Several lines of evidence underline the pivotal impact of the let-7 family on Wnt signaling in LCSCs. For example, a study by Cai et al. illustrated that upregulation of let-7b not only suppresses the proliferation, invasion and migration of liver cancer cells but also reduces the proportion of LCSCs in liver cancer cells [56]. Subsequent mechanistic analysis indicated that let-7b exerts these anti-cancer effects by inhibiting Wnt/beta-catenin signaling via the downregulation of Frizzled4 [56]. Jin et al. also found that let-7 miRNAs repress self-renewal of LCSCs via the regulation of Wnt signaling [57].
Chai et al. reported that miR-1246 enhances the activation of Wnt/beta-catenin in LCSCs by cooperating with octamer 4, a direct upstream regulator of miR-1246 [58].
Another study suggested that miR-217 markedly increases the fraction of the CSC population by promoting the expression of CSC factors via activation of the Wnt signaling pathway by directly targeting dickkopf-1 (DKK1) in hepatocellular carcinoma [59].
According to Tang et al., miR-612 is capable of inhibiting the stemness of HCC via modulation of the Wnt/beta-catenin signaling pathway [60]. The study identified that overexpression of miR-612 significantly reduces the number and size of tumorspheres and clone formation in soft agar and effectively relieves drug resistance to cisplatin and 5-fluorouracil [60].
Liu et al. also demonstrated that miR-200a is markedly downregulated in F344 rat HCC side population fraction cells using a miRNA microarray [61]. Their subsequent functional investigation confirmed that knockdown of miR-200a confers several CSC-like traits to these cells, which exhibit enhanced spheroid-forming capacity, express LCSCs markers and display enhanced resistance to chemotherapeutic drugs [61]. Finally, they demonstrated that the Wnt/beta-catenin signaling pathway is essential for the impact of miR-200a on these cells.
Xia et al. showed that miR-214 suppresses stem-like traits of HCC as well as invasion and recurrence via regulation of the beta-catenin pathway by targeting the CTNNB1 gene, which encodes the protein beta-catenin [62]. In contrast, knockdown of miR-214 increases EpCAM+ stem-like cells [62].
In addition, a study conducted by Zheng et al. in 2016 demonstrated that miR-452 markedly promotes stem-like cells of HCC both in vitro and in vivo via activation of Wnt/beta-catenin signaling by directly inhibiting SRY-related HMG-box member 7 (Sox7) [63]. Together, these findings underline the key roles of miRNAs on Wnt signaling in LCSCs.
MiRNA regulation of LCSCs through TGF-beta
Transforming growth factor beta (TGF-beta) signaling is another pathway in LCSCs that is often influenced by miRNAs. It contains a series of linked processes, including TGF-beta superfamily ligand binding, receptor recruitment and phosphorylation, small mothers against decapentaplegic (SMAD) phosphorylation, and coSMAD binding, eventually resulting in transcription of DNA [64,65]. The TGF-beta signaling pathway participates in a variety of physiological processes, such as cell growth, differentiation, apoptosis, invasion and migration, and its dysregulation usually contributes to the occurrence and development of multiple diseases, including cancer [66-70]. TGF-beta signaling has recently been implicated in miRNA-mediated regulation of LCSCs.
For instance, research conducted by Jiang et al. showed that miR-148a attenuates CSCs-like traits by inhibiting TGF-beta/SMAD2 signaling in several HCC cell lines, including HepG2, Huh7 and MHCC97H [71]. MiR-148a’s effect on LCSCs has been further confirmed by similar results achieved in another study from Li et al. [72]. In this study, investigators found that the subtype characterized by a CSC-like signature displayed much lower expression of miR-148a than other HCC subtypes. By increasing expression of miR-148a, the cancer phenotype was suppressed through direct targeting of the activin A receptor type 1 (ACVR1). ACVR1 is an important receptor in the bone morphogenetic protein (BMP) signaling pathway and is also implicated in the regulation of multiple stem cell markers [72].
Additionally, Zhou et al. reported for the first time that miR-125b overexpression significantly suppressed generation of LCSCs and reduced cancer incidence in a mouse model [73]. They also demonstrated a negative correlation between miR-125b and CSC markers in HCC specimens. Subsequently, using a dual-luciferase reporter assay their research on the molecular mechanism underlying this phenotype confirmed that miR-125b exerts its effect on LCSCs via direct inhibition of SMAD2 and SMAD4 [73]. Most importantly, in a mouse xenograft model, systemic delivery of synthetic miR-125b mimetics significantly decreased surface markers of CSCs and suppressed HCC metastasis, partially owing to the suppressive role of miR-125b in LCSCs.
It was reported by Liu et al. that miR-155 is involved in the promotion of TGF-beta1 in liver cancer stem cell phenotypes [74]. The study demonstrated that upregulation of miR-155 enhances expression of several CSC surface markers, such as CD90 and CD133, and increases the stem-like CSC population among liver cancer cells. Subsequent mechanistic studies have shown that silencing the tumor protein 53-inducible nuclear protein 1 (TP53INP1), at least partially accounted for the effect of miR-155 on LCSCs [74].
Moreover, another study conducted by Ma et al. found that miR-130b facilitates growth and self-renewal of CD133+ liver tumor-initiating cells via targeting TP53INP1 [75]. The authors revealed that antagonizing miR-130b in CD133+ tumor-initiating cells significantly increases sensitivity to chemotherapeutic agents and attenuates tumorigenicity in vivo [75].
Biox L and colleagues, for the first time, demonstrated that reintroduction of miR-122 suppresses stem-like HCC cells, thereby prompting cancer dormancy via a Samd-independent TGF-beta signaling pathway [76]. These findings suggest that upregulation of miR-122 may be a promising approach to weaken liver cancer aggressiveness and decrease cancer recurrence.
MiRNA regulation of LCSCs through JAK/STAT
The JAK-STAT signaling pathway comprises three major components, including cell surface receptors, Janus kinases (JAKs) and two signal transducer and activator of transcription (STAT) proteins. The JAK-STAT pathway plays a crucial role in regulating a wide range of biological processes, including differentiation, proliferation, cell cycle and apoptosis [77-79]. It is well documented that disrupting JAK-STAT signaling can lead to dozens of diseases, such as immune deficiency syndrome and cancer [80,81]. Recent studies have shown that the JAK/STAT pathway serves as a pivotal regulator of miRNA expression in CSCs, including LCSCs, and in return, miRNAs can also modify JAK/STAT signaling in LCSCs [82,83]. For example, Jiang et al. discovered that miR-500a-3p promotes LCSCs characteristics, including enhanced spheroid formation, increased fraction of side population and upregulated expression of cancer stem cell factors [84]. In this study, the authors demonstrated elevated expression of miR-500a-3p in HCC tissues and cells. Further investigation revealed that miR-500a-3p exerts its effects on LCSCs by suppressing several negative regulators of the JAK/STAT signaling pathway, such as suppressor of cytokine signaling 2 (SOCS2), SOCS4 and tyrosine-protein phosphatase non-receptor type 11 (PTPN11), resulting in constitutive activation of STAT3 signaling [84].
Research conducted by Zhang et al. illustrated that both STAT3 and phospho-STAT3 are highly expressed in HCC side populations compared to the primary cell population [85]. They found that upregulation of STAT3 promotes migration and invasion of the stem-like subpopulation. Further, their study revealed that abnormal STAT3 signaling enhances LCSC migration and invasion via a miR-21-dependent mechanism. Activation of STAT3 signaling significantly upregulated expression of miR-21, leading to decreased production of phosphatase and tensin homolog (PTEN), reversion-inducing-cysteine-rich protein with kazal motifs (RECK) and programmed cell death protein 4 (PDCD4), all of which function as anti-metastatic molecules [85].
In addition to its role in LCSCs, the JAK-STAT signaling pathway has been reported to participate in regulation of HCC-associated mesenchymal stem cells (MSCs) according to a study by Yan et al. [86]. They indicated that miR-155 could facilitate the activation of STAT pathway by downregulation of SOCS1, which is a negative regulator of STAT phosphorylation, eventually promoting the expression of matrix metalloproteinases 9 (MMP9). In addition, Liu et al. demonstrated that miR-155 could promote LCSCs acquisition and self-renewal by targeting TP53INP1 [87].
MiRNA regulation of epithelial-mesenchymal transition in LCSCs
The epithelial-mesenchymal transition (EMT) refers to a process by which epithelial cells lose their cell polarity and cell-cell adhesions and gain migratory and invasive properties to become mesenchymal stem cells [88]. EMT can be mediated by activation of transcription factors, such as TWIST [89], Snail [90] and ZEB [91]. In these EMT-activated cells, intercellular junction proteins, such as E-cadherin, are downregulated, while mesenchymal-related proteins, such as N-cadherin, fibronectin and vimentin, are upregulated [92]. EMT plays key roles in a variety of physiological and pathological processes involving neural tube formation, mesoderm formation, wound healing and cancer metastasis [93-97].
Recently, several miRNAs have been confirmed to modulate EMT in LCSCs. The miR-200 family consists of five members, miR-200a, miR-200b, miR-200c, miR-141 and miR-429 [98], some of which have been implicated in EMT in LCSCs. For instance, according to a study conducted by Wang et al., overexpression of miR-200a significantly downregulates expression of N-cadherin, zinc finger E-box-binding homeobox 2 (ZEB2) and vimentin in LCSCs, while upregulating expression of E-cadherin, indicating an inhibitory effect of miR-200a on EMT in LCSCs [99].
Liu et al. also found that downregulating miR-200a induces the EMT phenotype in HCC [61]. Interestingly, another study conducted by Xue et al. also confirmed that miR-200a/b/c-upregulated Vasohibin 2 (VASH2) facilitates malignant transformation of HCC by inducing EMT [100].
In addition, Tsai et al. recently reported that miR-200b could regulate diverse stemness of HCC [101]. In this study, the authors found that the overexpression of miR-200b could decrease the proportion of CD13+ and CD24+ LCSCs whereas increase EpCAM+ LCSCs. All these findings suggest that the miR-200 family can regulate LCSCs via modulation of EMT process.
Several other lines of evidence have also linked miRNAs to EMT in LCSCs. Yan et al. reported a suppressive role for miR-148a on EMT and CSCs-like properties in HCC, consequently resulting in inhibition of liver cancer metastasis [102]. Further analysis demonstrated evidence for a Wnt signaling pathway-dependent mechanism. A study from Zhou et al. showed that miR-491 levels in well-differentiated HCC tissues were higher than those in poorly differentiated HCC tissues [103]. Additionally, they found that miR-491 represses metastasis of HCC via inhibition of EMT and MMP2. Interestingly, miR-491 was also shown to attenuate CSC-like properties in HCC according to a study conducted by Yang et al. [104]. Their subsequent studies revealed that miR-491 exerts its effect on inhibition of CSC-like traits through EMT suppression mediated by the GIT-1/NF-kappa B pathway.
MiRNAs involved in the regulation of LCSC sensitivity to therapy
It is widely acknowledged that LCSCs are responsible for development of chemotherapy or radiotherapy tolerance in liver cancer. Although significant breakthroughs have been made in the management of liver cancer, current therapies against LCSCs remain scarce, accounting for the poor overall and relapse-free survival of patients with liver cancer. Recently, a great deal of research has highlighted the function of miRNAs in modulating LCSC sensitivity to therapy.
An investigation conducted by Hou et al. demonstrated that miR-33a effectively sensitizes Lgr5+ HCC-CSCs to doxorubicin, a recommended agent in the course of liver cancer treatment [105]. They also found that low expression of miR-33a in HCC tissues was correlated with chemo-resistance and poor prognosis. ATP-binding cassette 1 (ABCA1) is a transporter and key regulator of cellular cholesterol and phospholipid homeostasis, and mutations in this gene have been connected to Tangier disease and familial high-density lipoprotein deficiency [106,107]. Mechanistic research revealed that miR-33a regulates drug sensitivity by directly suppressing ABCA1.
Sorafenib, an inhibitor of multiple protein tyrosine kinases, including vascular endothelial growth factor receptor (VEGFR), platelet-derived growth factor receptor (PDGFR) and Raf family kinases, has already been approved for the treatment of advanced primary kidney cancer, liver cancer and thyroid carcinoma. Unfortunately, similar to other chemotherapeutic drugs, tumor develop resistance to sorafenib during the course of treatment. Several recent studies have demonstrated that miRNAs are involved in the modulation of sorafenib resistance in liver cancer. For instance, a study by Lu et al. reported that overexpression of miR-137 in sorafenib resistant Huh7 cells with CSC traits reversed sorafenib resistance, suppressed formation of CSC characteristics, and inhibited migration and invasion in these cells [108]. Further research demonstrated that silencing of adenine nucleotide translocator 2 (ANT2) at least partially accounts for the effect of miR-137 in HCC.
TNF-related apoptosis-inducing ligand (TRAIL), a protein that induces apoptosis through binding to certain death receptors, is widely considered a potential anti-cancer agent. However, cancer cell resistance to TRAIL severely limits its clinical application. Ma et al. reported that knockdown of miR-25 increased the sensitivity of LCSCs to TRAIL-induced apoptosis via targeting the PTEN/PI3K/Akt/Bad signaling pathway [109]. These findings all strongly suggest that regulation of these miRNAs and their relevant target genes represents a potential therapeutic approach against liver cancer drug resistance in the future.
Other miRNAs associated with LCSCs
In addition to those already mentioned, several other miRNAs have been correlated with LCSCs. Recently, Song et al. demonstrated that enhanced glycolysis is closely correlated with CD133+ stem cell characteristics in HCC [110]. In this study, they found that miR-122, a liver-specific miRNA, suppresses LCSC phenotypes via modulation of glycolysis.
MiR-142-3p reportedly possesses an inverse correlation with expression of CD133 [111], and CD133 is a direct target of miR-142-3p. Overexpression of miR-142-3p in HCC cells displayed inhibitory effect on LCSCs, including diminished self-renewal, tumor growth and resistance to chemotherapy.
According to a study from Zhang et al., upregulation of miR-150 leads to cell cycle arrest and apoptosis of CD133+ LCSCs via regulation of c-Myb [112]. In addition, miR-152 suppresses cell proliferation and colony formation of CD133+ LCSCs by targeting KIT [113].
Ma et al. reported that miR-130b facilitates CD133+ LCSC growth and self-renewal [75]. Furthermore, increasing the expression of miR-130b in CD133- LCSCs resulted in enhanced resistance to chemotherapeutic agents and enhanced tumorigenicity in vivo. The impact of miR-130b on LCSCs is, in part, attributed to a reduction of TP53INP1 levels.
Liu et al. recently reported that miR-148b is significantly downregulated in LCSCs, leading to enhanced chemo-sensitivity and decreased metastasis in patients [114]. However, overexpression of miR-148b lead to the opposite outcome. Knockdown of neuropilin-1 (NRP1), mediated by miR-148b, might be part of the mechanism for miR-148b suppression of LCSCs.
Oncogenic miR-21 is one of the most frequently upregulated miRNAs in multiple cancers, including breast cancer, cervical cancer, colon cancer and liver cancer. It was also found to be upregulated in a CSC-like population in HCC, facilitating migration and invasion of these cells [115]. The latter has been further confirmed by similar results achieved in another study conducted by Jiang et al. [116] who reported that miR-589-5p suppresses CD90+ LCSCs by directly targeting mitogen-activated protein kinase kinase kinase 8 (MAP3K8) [117].
Ji et al. found that conserved miR-181 family members are overexpressed in LCSCs with the surface marker EpCAM [118]. Functional studies demonstrated that knockdown of miR-181 resulted in a significant reduction of these LCSCs and tumor initiating ability, whereas upregulation of miR-181 in HCC cells led to an enrichment of EpCAM+ HCC cells, suggesting that targeting miR-181 may completely eradicate HCC [118].
MiR-145 was identified as a tumor suppressor in HCC via inhibition of LCSCs by modulating the downstream target octamer-binding transcription factor 4 (OCT4), which is a stem cell-related gene [119]. Li et al. showed that miR-429, one member of the miR-200 family, facilitates liver tumor-initiating cell properties by targeting the retinoblastoma-binding protein 4 (RBBP4)/E2F1/OCT4 axis [120]. Another investigation conducted by Liu et al. in 2016 revealed that miR-612 markedly suppresses stem cell-like property of HCC via regulation of Sp1/Nanog signaling pathway [121]. Zhou et al. recently reported that dysregulated expression of miR-205 contributes to stemness in HCC [122].
Conclusions
Cancer stem cells have been reported in multiple human cancers and are notorious for their significant contribution to cancer initiation and progression. Compelling studies have demonstrated that cancer stem cells are master regulators of cancer metastasis, recurrence and drug resistance as well as patient prognosis. Most existing therapies against cancer fail to eliminate cancer stem cells despite killing the majority of cancer cells. Unfortunately, therapies targeting CSCs are still rare. Given the clinical significance of CSCs and the lack of effective therapies against CSCs, it is extremely urgent to seek and develop a novel and promising approach to overcome the dilemma in treating CSCs. The thorough discovery of miRNAs inspires researchers and scholars, as miRNAs have been closely associated with CSC properties. A growing number of miRNAs have been identified as key players in regulating a variety of biological behaviors of CSCs. Moreover, miRNA antagonists, mimetics and their delivery technologies have been rapidly developed over the past few years. Therefore, building an miRNA-based therapeutic measure targeting CSCs is theoretically feasible. Currently, it is well established that the expression of miRNA in LCSCs is frequently dysregulated, and the involvement of miRNAs in the modulation of LCSCs has also been well described. Herein, we reviewed recent studies regarding the involvement of miRNAs in LCSCs and elucidated some of the possible mechanisms for the miRNA modulation of LCSC characteristics by summarizing the current knowledge on several signaling pathways. These new findings not only greatly enhance our understanding of the relationship between miRNAs and LCSCs but also inspire us to develop miRNA-based therapeutic strategies against LCSCs. However, there are no drugs based on miRNAs in current clinical practice. Thus, while continuing basic research, we should perform more clinical trials to further assess the effect of these miRNAs in the management of patients with liver cancer. With advances in these arenas, we believe that it is only a matter of time before miRNA-based diagnosis, therapy and prognosis are applied in clinical settings, paving the way for liver cancer eradication.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (81372462, 81572987), the Department of Science and Technology of Zhejiang Province (2014C03012) and the Health and Family Planning Commission of Zhejiang Province (2012KYB045).
Disclosure of conflict of interest
None.
Abbreviations
- LCSC
Liver cancer stem cell
- 3’-UTRs
3’-untranslated regions
- CSC
Cancer stem cell
- ALDH
Aldehyde dehydrogenase
- DLK1
Delta-like 1 homolog
- EpCAM
Epithelial cell-adhesion molecule
- RNase III
Nuclear ribonuclease III
- RISC
RNA-induced silencing complex
- EMT
Epithelial-mesenchymal transition
- DKK1
Dickkopf-1
- Sox7
SRY-related HMG-box member 7
- TGF-beta
Transforming growth factor beta
- SMAD
Small mothers against decapentaplegic
- ACVR1
Activin A receptor type 1
- BMPs
Bone morphogenetic proteins
- TP53INP1
Tumor protein 53-inducible nuclear protein 1
- JAKs
Janus kinases
- STAT
Signal transducer and activator of transcription
- SOCS2
suppressor of cytokine signaling 2
- PTPN11
Tyrosine-protein phosphatase non-receptor type 11
- PTPN
Phosphatase and tensin homolog
- RECK
Reversion-inducing-cysteine-rich protein with kazal motifs
- PDCD4
Programmed cell death protein 4
- MSCs
Mesenchymal stem cells
- MMP9
Matrix metalloproteinases 9
- ZEB2
zinc finger E-box-binding homeobox 2
- VASH2
Vasohibin 2
- ABCA1
ATP-binding cassette 1
- VEGFR
Vascular endothelial growth factor receptor
- PDGFR
Platelet-derived growth factor receptor
- ANT2
Adenine nucleotide translocator 2
- TRAIL
TNF-related apoptosis-inducing ligand
- NRP1
Neuropilin-1
- MAP3K8
Mitogen-activated protein kinase kinase kinase 8
- RBBP4
Retinoblastoma-binding protein 4
References
- 1.Gravitz L. Liver cancer. Nature. 2014;516:S1. doi: 10.1038/516S1a. [DOI] [PubMed] [Google Scholar]
- 2.Chen C, Lou T. Hypoxia inducible factors in hepatocellular carcinoma. Oncotarget. 2017;8:46691–703. doi: 10.18632/oncotarget.17358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Islami F, Miller KD, Siegel RL, Fedewa SA, Ward EM, Jemal A. Disparities in liver cancer occurrence in the United States by race/ethnicity and state. CA Cancer J Clin. 2017;67:273–89. doi: 10.3322/caac.21402. [DOI] [PubMed] [Google Scholar]
- 4.Fedeles BI, Chawanthayatham S, Croy RG, Wogan GN, Essigmann JM. Early detection of the aflatoxin B1 mutational fingerprint: a diagnostic tool for liver cancer. Mol Cell Oncol. 2017;4:e1329693. doi: 10.1080/23723556.2017.1329693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ringehan M, McKeating JA. Viral hepatitis and liver cancer. Philos Trans R Soc Lond B Biol Sci. 2017:372. doi: 10.1098/rstb.2016.0274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Szabo G, Saha B, Bukong TN. Alcohol and HCV: implications for liver cancer. Adv Exp Med Biol. 2015;815:197–216. doi: 10.1007/978-3-319-09614-8_12. [DOI] [PubMed] [Google Scholar]
- 7.Wang Y, Wang B, Yan S, Shen F, Cao H, Fan J, Zhang R, Gu J. Type 2 diabetes and gender differences in liver cancer by considering different confounding factors: a meta-analysis of cohort studies. Ann Epidemiol. 2016;26:764–72. doi: 10.1016/j.annepidem.2016.09.006. [DOI] [PubMed] [Google Scholar]
- 8.Ning X, Shi Z, Liu X, Zhang A, Han L, Jiang K, Kang C, Zhang Q. DNMT1 and EZH2 mediated methylation silences the microRNA-200b/a/429 gene and promotes tumor progression. Cancer Lett. 2015;359:198–205. doi: 10.1016/j.canlet.2015.01.005. [DOI] [PubMed] [Google Scholar]
- 9.Sun JH, Luo Q, Liu LL, Song GB. Liver cancer stem cell markers: progression and therapeutic implications. World J Gastroenterol. 2016;22:3547–57. doi: 10.3748/wjg.v22.i13.3547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Board PDQATE. PDQ Cancer Information Summaries. Bethesda (MD): National Cancer Institute (US); 2002. Adult Primary Liver Cancer Treatment (PDQ(R)): Patient Version. [Google Scholar]
- 11.Feng F, Jiang Q, Cao S, Cao Y, Li R, Shen L, Zhu H, Wang T, Sun L, Liang E, Sun H, Chai Y, Li X, Liu G, Yang R, Yang Z, Yang Y, Xin S, Li BA. Pregnane X receptor mediates Sorafenib resistance in advanced hepatocellular carcinoma. Biochim Biophys Acta. 2018;1862:1017–30. doi: 10.1016/j.bbagen.2018.01.011. [DOI] [PubMed] [Google Scholar]
- 12.Li HK, Mai RT, Huang HD, Chou CH, Chang YA, Chang YW, You LR, Chen CM, Lee YH. DDX3 represses stemness by epigenetically modulating tumor-suppressive miRNAs in hepatocellular carcinoma. Sci Rep. 2016;6:28637. doi: 10.1038/srep28637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu XL, Xing BC, Han HB, Zhao W, Hu MH, Xu ZL, Li JY, Xie Y, Gu J, Wang Y, Zhang ZQ. The properties of tumor-initiating cells from a hepatocellular carcinoma patient’s primary and recurrent tumor. Carcinogenesis. 2010;31:167–74. doi: 10.1093/carcin/bgp232. [DOI] [PubMed] [Google Scholar]
- 14.Bott A, Erdem N, Lerrer S, Hotz-Wagenblatt A, Breunig C, Abnaof K, Worner A, Wilhelm H, Munstermann E, Ben-Baruch A, Wiemann S. miRNA-1246 induces pro-inflammatory responses in mesenchymal stem/stromal cells by regulating PKA and PP2A. Oncotarget. 2017;8:43897–914. doi: 10.18632/oncotarget.14915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rouleau S, Glouzon JS, Brumwell A, Bisaillon M, Perreault JP. 3’UTR G-quadruplexes regulate miRNA binding. RNA. 2017;23:1172–9. doi: 10.1261/rna.060962.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dong J, Zhu G, Wang TC, Shi FS. Ginsenoside Rg1 promotes neural differentiation of mouse adipose-derived stem cells via the miRNA-124 signaling pathway. J Zhejiang Univ Sci B. 2017;18:445–8. doi: 10.1631/jzus.B1600355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fang L, Zhang HB, Li H, Fu Y, Yang GS. miR-548c-5p inhibits proliferation and migration and promotes apoptosis in CD90(+) HepG2 cells. Radiol Oncol. 2012;46:233–41. doi: 10.2478/v10019-012-0025-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Izarra A, Moscoso I, Canon S, Carreiro C, Fondevila D, Martin-Caballero J, Blanca V, Valiente I, Diez-Juan A, Bernad A. miRNA-1 and miRNA-133a are involved in early commitment of pluripotent stem cells and demonstrate antagonistic roles in the regulation of cardiac differentiation. J Tissue Eng Regen Med. 2017;11:787–99. doi: 10.1002/term.1977. [DOI] [PubMed] [Google Scholar]
- 19.Wang M, Liu C, Su Y, Zhang K, Zhang Y, Chen M, Ge M, Gu L, Lu T, Li N, Yu Z, Meng Q. miRNA-34c inhibits myoblasts proliferation by targeting YY1. Cell Cycle. 2017;16:1661–72. doi: 10.1080/15384101.2017.1281479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhou Q, Sun Q, Zhang Y, Teng F, Sun J. Up-regulation of miRNA-21 expression promotes migration and proliferation of Sca-1+ cardiac stem cells in mice. Med Sci Monit. 2016;22:1724–32. doi: 10.12659/MSM.895753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen W, Zhang X, Chu C, Cheung WL, Ng L, Lam S, Chow A, Lau T, Chen M, Li Y, Nie Y, Wong BC, Pang R. Identification of CD44+ cancer stem cells in human gastric cancer. Hepatogastroenterology. 2013;60:949–54. doi: 10.5754/hge12881. [DOI] [PubMed] [Google Scholar]
- 22.Wang Y, Yu Y, Tsuyada A, Ren X, Wu X, Stubblefield K, Rankin-Gee EK, Wang SE. Transforming growth factor-beta regulates the sphere-initiating stem cell-like feature in breast cancer through miRNA-181 and ATM. Oncogene. 2011;30:1470–80. doi: 10.1038/onc.2010.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bonnet D, Dick JE. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat Med. 1997;3:730–7. doi: 10.1038/nm0797-730. [DOI] [PubMed] [Google Scholar]
- 24.Ignatova TN, Kukekov VG, Laywell ED, Suslov ON, Vrionis FD, Steindler DA. Human cortical glial tumors contain neural stem-like cells expressing astroglial and neuronal markers in vitro. Glia. 2002;39:193–206. doi: 10.1002/glia.10094. [DOI] [PubMed] [Google Scholar]
- 25.Ma S, Lee TK, Zheng BJ, Chan KW, Guan XY. CD133+ HCC cancer stem cells confer chemoresistance by preferential expression of the Akt/PKB survival pathway. Oncogene. 2008;27:1749–58. doi: 10.1038/sj.onc.1210811. [DOI] [PubMed] [Google Scholar]
- 26.Ma S, Chan KW, Hu L, Lee TK, Wo JY, Ng IO, Zheng BJ, Guan XY. Identification and characterization of tumorigenic liver cancer stem/progenitor cells. Gastroenterology. 2007;132:2542–56. doi: 10.1053/j.gastro.2007.04.025. [DOI] [PubMed] [Google Scholar]
- 27.Liu WH, Tao KS, You N, Liu ZC, Zhang HT, Dou KF. Differences in the properties and mirna expression profiles between side populations from hepatic cancer cells and normal liver cells. PLoS One. 2011;6:e23311. doi: 10.1371/journal.pone.0023311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xiao Y, Lin M. The recent advances on liver cancer stem cells: biomarkers, separation, and therapy. Anal Cell Pathol (Amst) 2017;2017:5108653. doi: 10.1155/2017/5108653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zeng Y, Cullen BR. Recognition and cleavage of primary microRNA transcripts. Methods Mol Biol. 2006;342:49–56. doi: 10.1385/1-59745-123-1:49. [DOI] [PubMed] [Google Scholar]
- 30.Kuehbacher A, Urbich C, Dimmeler S. Targeting microRNA expression to regulate angiogenesis. Trends Pharmacol Sci. 2008;29:12–5. doi: 10.1016/j.tips.2007.10.014. [DOI] [PubMed] [Google Scholar]
- 31.John B, Enright AJ, Aravin A, Tuschl T, Sander C, Marks DS. Human MicroRNA targets. PLoS Biol. 2004;2:e363. doi: 10.1371/journal.pbio.0020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–54. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 33.Pasquinelli AE, Reinhart BJ, Slack F, Martindale MQ, Kuroda MI, Maller B, Hayward DC, Ball EE, Degnan B, Muller P, Spring J, Srinivasan A, Fishman M, Finnerty J, Corbo J, Levine M, Leahy P, Davidson E, Ruvkun G. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature. 2000;408:86–9. doi: 10.1038/35040556. [DOI] [PubMed] [Google Scholar]
- 34.Saleh LM, Wang W, Herman SE, Saba NS, Anastas V, Barber E, Corrigan-Cummins M, Farooqui M, Sun C, Sarasua SM, Zhao Z, Abousamra NK, Elbaz O, Abdelghaffar HA, Wiestner A, Calvo KR. Ibrutinib downregulates a subset of miRNA leading to upregulation of tumor suppressors and inhibition of cell proliferation in chronic lymphocytic leukemia. Leukemia. 2017;31:340–9. doi: 10.1038/leu.2016.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang X, Zhang X, Hu S, Zheng M, Zhang J, Zhao J, Zhang X, Yan B, Jia L, Zhao J, Wu K, Yang A, Zhang R. Identification of miRNA-7 by genome-wide analysis as a critical sensitizer for TRAIL-induced apoptosis in glioblastoma cells. Nucleic Acids Res. 2017;45:5930–44. doi: 10.1093/nar/gkx317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhao Y, Ransom JF, Li A, Vedantham V, von Drehle M, Muth AN, Tsuchihashi T, McManus MT, Schwartz RJ, Srivastava D. Dysregulation of cardiogenesis, cardiac conduction, and cell cycle in mice lacking miRNA-1-2. Cell. 2007;129:303–17. doi: 10.1016/j.cell.2007.03.030. [DOI] [PubMed] [Google Scholar]
- 37.Kassambara A, Jourdan M, Bruyer A, Robert N, Pantesco V, Elemento O, Klein B, Moreaux J. Global miRNA expression analysis identifies novel key regulators of plasma cell differentiation and malignant plasma cell. Nucleic Acids Res. 2017;45:5639–52. doi: 10.1093/nar/gkx327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pencheva N, Tran H, Buss C, Huh D, Drobnjak M, Busam K, Tavazoie SF. Convergent multi-miRNA targeting of ApoE drives LRP1/LRP8-dependent melanoma metastasis and angiogenesis. Cell. 2012;151:1068–82. doi: 10.1016/j.cell.2012.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kovaleva V, Mora R, Park YJ, Plass C, Chiramel AI, Bartenschlager R, Dohner H, Stilgenbauer S, Pscherer A, Lichter P, Seiffert M. miRNA-130a targets ATG2B and DICER1 to inhibit autophagy and trigger killing of chronic lymphocytic leukemia cells. Cancer Res. 2012;72:1763–72. doi: 10.1158/0008-5472.CAN-11-3671. [DOI] [PubMed] [Google Scholar]
- 40.Xu L, Hou Y, Tu G, Chen Y, Du YE, Zhang H, Wen S, Tang X, Yin J, Lang L, Sun K, Yang G, Tang X, Liu M. Nuclear Drosha enhances cell invasion via an EGFR-ERK1/2-MMP7 signaling pathway induced by dysregulated miRNA-622/197 and their targets LAMC2 and CD82 in gastric cancer. Cell Death Dis. 2017;8:e2642. doi: 10.1038/cddis.2017.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Staton AA, Knaut H, Giraldez AJ. miRNA regulation of Sdf1 chemokine signaling provides genetic robustness to germ cell migration. Nat Genet. 2011;43:204–11. doi: 10.1038/ng.758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kahn M. Can we safely target the WNT pathway? Nat Rev Drug Discov. 2014;13:513–32. doi: 10.1038/nrd4233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Klingenberg M, Matsuda A, Diederichs S, Patel T. Non-coding RNA in hepatocellular carcinoma: mechanisms, biomarkers and therapeutic targets. J Hepatol. 2017;67:603–18. doi: 10.1016/j.jhep.2017.04.009. [DOI] [PubMed] [Google Scholar]
- 44.Deng F, Peng L, Li Z, Tan G, Liang E, Chen S, Zhao X, Zhi F. YAP triggers the Wnt/beta-catenin signalling pathway and promotes enterocyte self-renewal, regeneration and tumorigenesis after DSS-induced injury. Cell Death Dis. 2018;9:153. doi: 10.1038/s41419-017-0244-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li M, Han Y, Zhou H, Li X, Lin C, Zhang E, Chi X, Hu J, Xu H. Transmembrane protein 170B is a novel breast tumorigenesis suppressor gene that inhibits the Wnt/beta-catenin pathway. Cell Death Dis. 2018;9:91. doi: 10.1038/s41419-017-0128-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhao M, Xu P, Liu Z, Zhen Y, Chen Y, Liu Y, Fu Q, Deng X, Liang Z, Li Y, Lin X, Fang W. Dual roles of miR-374a by modulated c-Jun respectively targets CCND1-inducing PI3K/AKT signal and PTEN-suppressing Wnt/beta-catenin signaling in non-small-cell lung cancer. Cell Death Dis. 2018;9:78. doi: 10.1038/s41419-017-0103-7. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 47.Nusse R, Theunissen H, Wagenaar E, Rijsewijk F, Gennissen A, Otte A, Schuuring E, van Ooyen A. The Wnt-1 (int-1) oncogene promoter and its mechanism of activation by insertion of proviral DNA of the mouse mammary tumor virus. Mol Cell Biol. 1990;10:4170–9. doi: 10.1128/mcb.10.8.4170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bordonaro M, Lazarova DL. Hypothesis: cell signalling influences age-related risk of colorectal cancer. J Cell Mol Med. 2015;19:74–81. doi: 10.1111/jcmm.12366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cheng AS, Lau SS, Chen Y, Kondo Y, Li MS, Feng H, Ching AK, Cheung KF, Wong HK, Tong JH, Jin H, Choy KW, Yu J, To KF, Wong N, Huang TH, Sung JJ. EZH2-mediated concordant repression of Wnt antagonists promotes beta-catenin-dependent hepatocarcinogenesis. Cancer Res. 2011;71:4028–39. doi: 10.1158/0008-5472.CAN-10-3342. [DOI] [PubMed] [Google Scholar]
- 50.Guan H, Liu C, Fang F, Huang Y, Tao T, Ling Z, You Z, Han X, Chen S, Xu B, Chen M. MicroRNA-744 promotes prostate cancer progression through aberrantly activating Wnt/beta-catenin signaling. Oncotarget. 2017;8:14693–707. doi: 10.18632/oncotarget.14711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Peng Y, Cao J, Yao XY, Wang JX, Zhong MZ, Gan PP, Li JH. TUSC3 induces autophagy in human non-small cell lung cancer cells through Wnt/beta-catenin signaling. Oncotarget. 2017;8:52960–74. doi: 10.18632/oncotarget.17674. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 52.Hu X, Liu T, Wu X, Chen T, Luo Z. Downregulation of DNMT3A by miR-708-5p inhibits lung cancer stem cell-like phenotypes through repressing Wnt/beta-catenin signaling. Clin Cancer Res. 2018;24:1748–60. doi: 10.1158/1078-0432.CCR-17-1169. [DOI] [PubMed] [Google Scholar]
- 53.Zhang K, Guo Y, Wang X, Zhao H, Ji Z, Cheng C, Li L, Fang Y, Xu D, Zhu HH, Gao WQ. WNT/beta-catenin directs self-renewal symmetric cell division of hTERT(high) prostate cancer stem cells. Cancer Res. 2017;77:2534–47. doi: 10.1158/0008-5472.CAN-16-1887. [DOI] [PubMed] [Google Scholar]
- 54.Catalano V, Dentice M, Ambrosio R, Luongo C, Carollo R, Benfante A, Todaro M, Stassi G, Salvatore D. Activated thyroid hormone promotes differentiation and chemotherapeutic sensitization of colorectal cancer stem cells by regulating wnt and BMP4 signaling. Cancer Res. 2016;76:1237–44. doi: 10.1158/0008-5472.CAN-15-1542. [DOI] [PubMed] [Google Scholar]
- 55.Wang Y, He L, Du Y, Zhu P, Huang G, Luo J, Yan X, Ye B, Li C, Xia P, Zhang G, Tian Y, Chen R, Fan Z. The long noncoding RNA lncTCF7 promotes self-renewal of human liver cancer stem cells through activation of Wnt signaling. Cell Stem Cell. 2015;16:413–25. doi: 10.1016/j.stem.2015.03.003. [DOI] [PubMed] [Google Scholar]
- 56.Cai H, Chen Y, Yang X, Ma S, Wang Q, Zhang Y, Niu X, Ding G, Yuan Y. Let7b modulates the Wnt/beta-catenin pathway in liver cancer cells via downregulated Frizzled4. Tumour Biol. 2017;39:1010428317716076. doi: 10.1177/1010428317716076. [DOI] [PubMed] [Google Scholar]
- 57.Jin B, Wang W, Meng XX, Du G, Li J, Zhang SZ, Zhou BH, Fu ZH. Let-7 inhibits self-renewal of hepatocellular cancer stem-like cells through regulating the epithelial-mesenchymal transition and the Wnt signaling pathway. BMC Cancer. 2016;16:863. doi: 10.1186/s12885-016-2904-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chai S, Ng KY, Tong M, Lau EY, Lee TK, Chan KW, Yuan YF, Cheung TT, Cheung ST, Wang XQ, Wong N, Lo CM, Man K, Guan XY. Octamer 4/microRNA-1246 signaling axis drives Wnt/beta-catenin activation in liver cancer stem cells. Hepatology. 2016;64:2062–76. doi: 10.1002/hep.28821. [DOI] [PubMed] [Google Scholar]
- 59.Jiang C, Yu M, Xie X, Huang G, Peng Y, Ren D, Lin M, Liu B, Liu M, Wang W, Kuang M. miR-217 targeting DKK1 promotes cancer stem cell properties via activation of the Wnt signaling pathway in hepatocellular carcinoma. Oncol Rep. 2017;38:2351–9. doi: 10.3892/or.2017.5924. [DOI] [PubMed] [Google Scholar]
- 60.Tang J, Tao ZH, Wen D, Wan JL, Liu DL, Zhang S, Cui JF, Sun HC, Wang L, Zhou J, Fan J, Wu WZ. MiR-612 suppresses the stemness of liver cancer via Wnt/beta-catenin signaling. Biochem Biophys Res Commun. 2014;447:210–5. doi: 10.1016/j.bbrc.2014.03.135. [DOI] [PubMed] [Google Scholar]
- 61.Liu J, Ruan B, You N, Huang Q, Liu W, Dang Z, Xu W, Zhou T, Ji R, Cao Y, Li X, Wang D, Tao K, Dou K. Downregulation of miR-200a induces EMT phenotypes and CSC-like signatures through targeting the beta-catenin pathway in hepatic oval cells. PLoS One. 2013;8:e79409. doi: 10.1371/journal.pone.0079409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Xia H, Ooi LL, Hui KM. MiR-214 targets beta-catenin pathway to suppress invasion, stem-like traits and recurrence of human hepatocellular carcinoma. PLoS One. 2012;7:e44206. doi: 10.1371/journal.pone.0044206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zheng Z, Liu J, Yang Z, Wu L, Xie H, Jiang C, Lin B, Chen T, Xing C, Liu Z, Song P, Yin S, Zheng S, Zhou L. MicroRNA-452 promotes stem-like cells of hepatocellular carcinoma by inhibiting Sox7 involving Wnt/beta-catenin signaling pathway. Oncotarget. 2016;7:28000–12. doi: 10.18632/oncotarget.8584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Du X, Pan Z, Li Q, Liu H, Li Q. SMAD4 feedback regulates the canonical TGF-beta signaling pathway to control granulosa cell apoptosis. Cell Death Dis. 2018;9:151. doi: 10.1038/s41419-017-0205-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jung B, Staudacher JJ, Beauchamp D. Transforming growth factor beta superfamily signaling in development of colorectal cancer. Gastroenterology. 2017;152:36–52. doi: 10.1053/j.gastro.2016.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Funaki S, Shintani Y, Kawamura T, Kanzaki R, Minami M, Okumura M. Chemotherapy enhances programmed cell death 1/ligand 1 expression via TGF-beta induced epithelial mesenchymal transition in non-small cell lung cancer. Oncol Rep. 2017;38:2277–84. doi: 10.3892/or.2017.5894. [DOI] [PubMed] [Google Scholar]
- 67.Jiang J, Zhang Y, Peng K, Wang Q, Hong X, Li H, Fan G, Zhang Z, Gong T, Sun X. Combined delivery of a TGF-beta inhibitor and an adenoviral vector expressing interleukin-12 potentiates cancer immunotherapy. Acta Biomater. 2017;61:114–23. doi: 10.1016/j.actbio.2017.05.009. [DOI] [PubMed] [Google Scholar]
- 68.Lu Y, Ma J, Li Y, Huang J, Zhang S, Yin Z, Ren J, Huang K, Wu G, Yang K, Xu S. CDP138 silencing inhibits TGF-beta/Smad signaling to impair radioresistance and metastasis via GDF15 in lung cancer. Cell Death Dis. 2017;8:e3036. doi: 10.1038/cddis.2017.434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sui H, Zhao J, Zhou L, Wen H, Deng W, Li C, Ji Q, Liu X, Feng Y, Chai N, Zhang Q, Cai J, Li Q. Tanshinone IIA inhibits beta-catenin/VEGF-mediated angiogenesis by targeting TGF-beta1 in normoxic and HIF-1alpha in hypoxic microenvironments in human colorectal cancer. Cancer Lett. 2017;403:86–97. doi: 10.1016/j.canlet.2017.05.013. [DOI] [PubMed] [Google Scholar]
- 70.Yu C, Zhou JJ, Fan HY. Studying the functions of TGF-beta signaling in the ovary. Methods Mol Biol. 2016;1344:301–11. doi: 10.1007/978-1-4939-2966-5_19. [DOI] [PubMed] [Google Scholar]
- 71.Jiang F, Mu J, Wang X, Ye X, Si L, Ning S, Li Z, Li Y. The repressive effect of miR-148a on TGF beta-SMADs signal pathway is involved in the glabridin-induced inhibition of the cancer stem cells-like properties in hepatocellular carcinoma cells. PLoS One. 2014;9:e96698. doi: 10.1371/journal.pone.0096698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Li L, Liu Y, Guo Y, Liu B, Zhao Y, Li P, Song F, Zheng H, Yu J, Song T, Niu R, Li Q, Wang XW, Zhang W, Chen K. Regulatory MiR-148a-ACVR1/BMP circuit defines a cancer stem cell-like aggressive subtype of hepatocellular carcinoma. Hepatology. 2015;61:574–84. doi: 10.1002/hep.27543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhou JN, Zeng Q, Wang HY, Zhang B, Li ST, Nan X, Cao N, Fu CJ, Yan XL, Jia YL, Wang JX, Zhao AH, Li ZW, Li YH, Xie XY, Zhang XM, Dong Y, Xu YC, He LJ, Yue W, Pei XT. MicroRNA-125b attenuates epithelial-mesenchymal transitions and targets stem-like liver cancer cells through small mothers against decapentaplegic 2 and 4. Hepatology. 2015;62:801–15. doi: 10.1002/hep.27887. [DOI] [PubMed] [Google Scholar]
- 74.Liu F, Kong X, Lv L, Gao J. TGF-beta1 acts through miR-155 to down-regulate TP53INP1 in promoting epithelial-mesenchymal transition and cancer stem cell phenotypes. Cancer Lett. 2015;359:288–98. doi: 10.1016/j.canlet.2015.01.030. [DOI] [PubMed] [Google Scholar]
- 75.Ma S, Tang KH, Chan YP, Lee TK, Kwan PS, Castilho A, Ng I, Man K, Wong N, To KF, Zheng BJ, Lai PB, Lo CM, Chan KW, Guan XY. miR-130b Promotes CD133(+) liver tumor-initiating cell growth and self-renewal via tumor protein 53-induced nuclear protein 1. Cell Stem Cell. 2010;7:694–707. doi: 10.1016/j.stem.2010.11.010. [DOI] [PubMed] [Google Scholar]
- 76.Boix L, Lopez-Oliva JM, Rhodes AC, Bruix J. Restoring miR122 in human stem-like hepatocarcinoma cells, prompts tumor dormancy through Smad-independent TGF-beta pathway. Oncotarget. 2016;7:71309–29. doi: 10.18632/oncotarget.11885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mary Photini S, Chaiwangyen W, Weber M, Al-Kawlani B, Favaro RR, Jeschke U, Schleussner E, Morales-Prieto DM, Markert UR. PIM kinases 1, 2 and 3 in intracellular LIF signaling, proliferation and apoptosis in trophoblastic cells. Exp Cell Res. 2017;359:275–83. doi: 10.1016/j.yexcr.2017.07.019. [DOI] [PubMed] [Google Scholar]
- 78.Tsai YC, Sun YH. Long-range effect of upd, a ligand for Jak/STAT pathway, on cell cycle in Drosophila eye development. Genesis. 2004;39:141–53. doi: 10.1002/gene.20035. [DOI] [PubMed] [Google Scholar]
- 79.Zhang YL, Zhou Z, Han WW, Zhang LL, Song WS, Huang JH, Liu S. Oleanolic acid inhibiting the differentiation of neural stem cells into astrocyte by down-regulating JAK/STAT signaling pathway. Am J Chin Med. 2016;44:103–17. doi: 10.1142/S0192415X16500075. [DOI] [PubMed] [Google Scholar]
- 80.Mi C, Ma J, Wang KS, Wang Z, Li MY, Li JB, Li X, Piao LX, Xu GH, Jin X. Amorfrutin A inhibits TNF-alpha induced JAK/STAT signaling, cell survival and proliferation of human cancer cells. Immunopharmacol Immunotoxicol. 2017:338–347. doi: 10.1080/08923973.2017.1371187. [DOI] [PubMed] [Google Scholar]
- 81.Minegishi Y, Karasuyama H. Defects in Jak-STAT-mediated cytokine signals cause hyper-IgE syndrome: lessons from a primary immunodeficiency. Int Immunol. 2009;21:105–12. doi: 10.1093/intimm/dxn134. [DOI] [PubMed] [Google Scholar]
- 82.Huang JS, Yao CJ, Chuang SE, Yeh CT, Lee LM, Chen RM, Chao WJ, Whang-Peng J, Lai GM. Honokiol inhibits sphere formation and xenograft growth of oral cancer side population cells accompanied with JAK/STAT signaling pathway suppression and apoptosis induction. BMC Cancer. 2016;16:245. doi: 10.1186/s12885-016-2265-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yang MY, Lee HT, Chen CM, Shen CC, Ma HI. Celecoxib suppresses the phosphorylation of STAT3 protein and can enhance the radiosensitivity of medulloblastoma-derived cancer stem-like cells. Int J Mol Sci. 2014;15:11013–29. doi: 10.3390/ijms150611013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhang Y, Wei C, Guo CC, Bi RX, Xie J, Guan DH, Yang CH, Jiang YH. Prognostic value of microRNAs in hepatocellular carcinoma: a meta-analysis. Oncotarget. 2017;8:107237–57. doi: 10.18632/oncotarget.20883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zhang N, Duan WD, Leng JJ, Zhou L, Wang X, Xu YZ, Wang XD, Zhang AQ, Dong JH. STAT3 regulates the migration and invasion of a stemlike subpopulation through microRNA21 and multiple targets in hepatocellular carcinoma. Oncol Rep. 2015;33:1493–8. doi: 10.3892/or.2015.3710. [DOI] [PubMed] [Google Scholar]
- 86.Yan XL, Jia YL, Chen L, Zeng Q, Zhou JN, Fu CJ, Chen HX, Yuan HF, Li ZW, Shi L, Xu YC, Wang JX, Zhang XM, He LJ, Zhai C, Yue W, Pei XT. Hepatocellular carcinoma-associated mesenchymal stem cells promote hepatocarcinoma progression: role of the S100A4-miR155-SOCS1-MMP9 axis. Hepatology. 2013;57:2274–86. doi: 10.1002/hep.26257. [DOI] [PubMed] [Google Scholar]
- 87.Liu F, Kong X, Lv L, Gao J. MiR-155 targets TP53INP1 to regulate liver cancer stem cell acquisition and self-renewal. FEBS Lett. 2015;589:500–506. doi: 10.1016/j.febslet.2015.01.009. [DOI] [PubMed] [Google Scholar]
- 88.Cicchini C, Amicone L, Alonzi T, Marchetti A, Mancone C, Tripodi M. Molecular mechanisms controlling the phenotype and the EMT/MET dynamics of hepatocyte. Liver Int. 2015;35:302–10. doi: 10.1111/liv.12577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Jung HY, Yang J. Unraveling the TWIST between EMT and cancer stemness. Cell Stem Cell. 2015;16:1–2. doi: 10.1016/j.stem.2014.12.005. [DOI] [PubMed] [Google Scholar]
- 90.Lander R, Nordin K, LaBonne C. The F-box protein Ppa is a common regulator of core EMT factors Twist, Snail, Slug, and Sip1. J Cell Biol. 2011;194:17–25. doi: 10.1083/jcb.201012085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Krebs AM, Mitschke J, Lasierra Losada M, Schmalhofer O, Boerries M, Busch H, Boettcher M, Mougiakakos D, Reichardt W, Bronsert P, Brunton VG, Pilarsky C, Winkler TH, Brabletz S, Stemmler MP, Brabletz T. The EMT-activator Zeb1 is a key factor for cell plasticity and promotes metastasis in pancreatic cancer. Nat Cell Biol. 2017;19:518–29. doi: 10.1038/ncb3513. [DOI] [PubMed] [Google Scholar]
- 92.Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. J Clin Invest. 2009;119:1420–8. doi: 10.1172/JCI39104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Luo W, Tan P, Rodriguez M, He L, Tan K, Zeng L, Siwko S, Liu M. Leucine-rich repeat-containing G protein-coupled receptor 4 (Lgr4) is necessary for prostate cancer metastasis via epithelial-mesenchymal transition. J Biol Chem. 2017;292:15525–37. doi: 10.1074/jbc.M116.771931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ray HJ, Niswander LA. Grainyhead-like 2 downstream targets act to suppress epithelial-to-mesenchymal transition during neural tube closure. Development. 2016;143:1192–204. doi: 10.1242/dev.129825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Richter A, Valdimarsdottir L, Hrafnkelsdottir HE, Runarsson JF, Omarsdottir AR, Ward-van Oostwaard D, Mummery C, Valdimarsdottir G. BMP4 promotes EMT and mesodermal commitment in human embryonic stem cells via SLUG and MSX2. Stem Cells. 2014;32:636–48. doi: 10.1002/stem.1592. [DOI] [PubMed] [Google Scholar]
- 96.Yun Y, Gao R, Yue H, Guo L, Li G, Sang N. Sulfate aerosols promote lung cancer metastasis by epigenetically regulating the epithelial-to-mesenchymal transition (EMT) Environ Sci Technol. 2017;51:11401–11. doi: 10.1021/acs.est.7b02857. [DOI] [PubMed] [Google Scholar]
- 97.Shen J, Zhou S, Shi L, Liu X, Lin H, Yu H, Xiaoliang , Tang J, Yu T, Cai X. DUSP1 inhibits cell proliferation, metastasis and invasion and angiogenesis in gallbladder cancer. Oncotarget. 2017;8:12133–44. doi: 10.18632/oncotarget.14815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Korpal M, Lee ES, Hu G, Kang Y. The miR-200 family inhibits epithelial-mesenchymal transition and cancer cell migration by direct targeting of E-cadherin transcriptional repressors ZEB1 and ZEB2. J Biol Chem. 2008;283:14910–4. doi: 10.1074/jbc.C800074200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Wang J, Yang X, Ruan B, Dai B, Gao Y, Duan J, Qu S, Tao K, Dou K, Li H. Overexpression of miR-200a suppresses epithelial-mesenchymal transition of liver cancer stem cells. Tumour Biol. 2015;36:2447–56. doi: 10.1007/s13277-014-2856-2. [DOI] [PubMed] [Google Scholar]
- 100.Xue X, Zhang Y, Zhi Q, Tu M, Xu Y, Sun J, Wei J, Lu Z, Miao Y, Gao W. MiR200-upregulated Vasohibin 2 promotes the malignant transformation of tumors by inducing epithelial-mesenchymal transition in hepatocellular carcinoma. Cell Commun Signal. 2014;12:62. doi: 10.1186/s12964-014-0062-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Tsai SC, Lin CC, Shih TC, Tseng RJ, Yu MC, Lin YJ, Hsieh SY. The miR-200b-ZEB1 circuit regulates diverse stemness of human hepatocellular carcinoma. Mol Carcinog. 2017;56:2035–47. doi: 10.1002/mc.22657. [DOI] [PubMed] [Google Scholar]
- 102.Yan H, Dong X, Zhong X, Ye J, Zhou Y, Yang X, Shen J, Zhang J. Inhibitions of epithelial to mesenchymal transition and cancer stem cells-like properties are involved in miR-148a-mediated anti-metastasis of hepatocellular carcinoma. Mol Carcinog. 2014;53:960–9. doi: 10.1002/mc.22064. [DOI] [PubMed] [Google Scholar]
- 103.Zhou Y, Li Y, Ye J, Jiang R, Yan H, Yang X, Liu Q, Zhang J. MicroRNA-491 is involved in metastasis of hepatocellular carcinoma by inhibitions of matrix metalloproteinase and epithelial to mesenchymal transition. Liver Int. 2013;33:1271–80. doi: 10.1111/liv.12190. [DOI] [PubMed] [Google Scholar]
- 104.Yang X, Ye J, Yan H, Tang Z, Shen J, Zhang J, Yang L. MiR-491 attenuates cancer stem cells-like properties of hepatocellular carcinoma by inhibition of GIT-1/NF-kappaB-mediated EMT. Tumour Biol. 2016;37:201–9. doi: 10.1007/s13277-015-3687-5. [DOI] [PubMed] [Google Scholar]
- 105.Hou H, Kang Y, Li Y, Zeng Y, Ding G, Shang J. miR-33a expression sensitizes Lgr5+ HCC-CSCs to doxorubicin via ABCA1. Neoplasma. 2017;64:81–91. doi: 10.4149/neo_2017_110. [DOI] [PubMed] [Google Scholar]
- 106.Oram JF, Vaughan AM. ABCA1-mediated transport of cellular cholesterol and phospholipids to HDL apolipoproteins. Curr Opin Lipidol. 2000;11:253–60. doi: 10.1097/00041433-200006000-00005. [DOI] [PubMed] [Google Scholar]
- 107.Ordovas JM. ABC1: the gene for Tangier disease and beyond. Nutr Rev. 2000;58:76–9. doi: 10.1111/j.1753-4887.2000.tb01843.x. [DOI] [PubMed] [Google Scholar]
- 108.Lu AQ, Lv B, Qiu F, Wang XY, Cao XH. Upregulation of miR-137 reverses sorafenib resistance and cancer-initiating cell phenotypes by degrading ANT2 in hepatocellular carcinoma. Oncol Rep. 2017;37:2071–8. doi: 10.3892/or.2017.5498. [DOI] [PubMed] [Google Scholar]
- 109.Ma S, Feng X, Jiang J, Shi S, Xie H, Zhou L, Zheng S. Knockdown of miR-25 increases the sensitivity of liver cancer stem cells to TRAIL-induced apoptosis via PTEN/PI3K/Akt/Bad signaling pathway. Hepatology. 2016;49:2600–10. doi: 10.3892/ijo.2016.3751. [DOI] [PubMed] [Google Scholar]
- 110.Song K, Kwon H, Han C, Zhang J, Dash S, Lim K, Wu T. Active glycolytic metabolism in CD133(+) hepatocellular cancer stem cells: regulation by MIR-122. Oncotarget. 2015;6:40822–35. doi: 10.18632/oncotarget.5812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Chai S, Tong M, Ng KY, Kwan PS, Chan YP, Fung TM, Lee TK, Wong N, Xie D, Yuan YF, Guan XY, Ma S. Regulatory role of miR-142-3p on the functional hepatic cancer stem cell marker CD133. Oncotarget. 2014;5:5725–35. doi: 10.18632/oncotarget.2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Zhang J, Luo N, Luo Y, Peng Z, Zhang T, Li S. microRNA-150 inhibits human CD133-positive liver cancer stem cells through negative regulation of the transcription factor c-Myb. Int J Oncol. 2012;40:747–56. doi: 10.3892/ijo.2011.1242. [DOI] [PubMed] [Google Scholar]
- 113.Huang H, Hu M, Li P, Lu C, Li M. Mir-152 inhibits cell proliferation and colony formation of CD133(+) liver cancer stem cells by targeting KIT. Tumour Biol. 2015;36:921–8. doi: 10.1007/s13277-014-2719-x. [DOI] [PubMed] [Google Scholar]
- 114.Liu Q, Xu Y, Wei S, Gao W, Chen L, Zhou T, Wang Z, Ying M, Zheng Q. miRNA-148b suppresses hepatic cancer stem cell by targeting neuropilin-1. Biosci Rep. 2015;35:e00229. doi: 10.1042/BSR20150084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Zhou L, Yang ZX, Song WJ, Li QJ, Yang F, Wang DS, Zhang N, Dou KF. MicroRNA-21 regulates the migration and invasion of a stem-like population in hepatocellular carcinoma. Int J Oncol. 2013;43:661–9. doi: 10.3892/ijo.2013.1965. [DOI] [PubMed] [Google Scholar]
- 116.Jiang J, Yang P, Guo Z, Yang R, Yang H, Yang F, Li L, Xiang B. Overexpression of microRNA-21 strengthens stem cell-like characteristics in a hepatocellular carcinoma cell line. World J Surg Oncol. 2016;14:278. doi: 10.1186/s12957-016-1028-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Zhang X, Jiang P, Shuai L, Chen K, Li Z, Zhang Y, Jiang Y, Li X. miR-589-5p inhibits MAP3K8 and suppresses CD90+ cancer stem cells in hepatocellular carcinoma. J Exp Clin Cancer Res. 2016;35:176. doi: 10.1186/s13046-016-0452-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Ji J, Yamashita T, Budhu A, Forgues M, Jia HL, Li C, Deng C, Wauthier E, Reid LM, Ye QH, Qin LX, Yang W, Wang HY, Tang ZY, Croce CM, Wang XW. Identification of microRNA-181 by genome-wide screening as a critical player in EpCAM-positive hepatic cancer stem cells. Hepatology. 2009;50:472–80. doi: 10.1002/hep.22989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Jia Y, Liu H, Zhuang Q, Xu S, Yang Z, Li J, Lou J, Zhang W. Tumorigenicity of cancer stem-like cells derived from hepatocarcinoma is regulated by microRNA-145. Oncol Rep. 2012;27:1865–72. doi: 10.3892/or.2012.1701. [DOI] [PubMed] [Google Scholar]
- 120.Li L, Tang J, Zhang B, Yang W, LiuGao M, Wang R, Tan Y, Fan J, Chang Y, Fu J, Jiang F, Chen C, Yang Y, Gu J, Wu D, Guo L, Cao D, Li H, Cao G, Wu M, Zhang MQ, Chen L, Wang H. Epigenetic modification of MiR-429 promotes liver tumour-initiating cell properties by targeting Rb binding protein 4. Gut. 2015;64:156–67. doi: 10.1136/gutjnl-2013-305715. [DOI] [PubMed] [Google Scholar]
- 121.Liu Y, Liu DL, Dong LL, Wen D, Shi DM, Zhou J, Fan J, Wu WZ. miR-612 suppresses stem cell-like property of hepatocellular carcinoma cells by modulating Sp1/Nanog signaling. Cell Death Dis. 2016;7:e2377. doi: 10.1038/cddis.2016.282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Zhao J, Xu G, Qian YW, Li YW. Down-regulation of miR-205 promotes stemness of hepatocellular carcinoma cells by targeting PLCbeta1 and increasing CD24 expression. Neoplasma. 2015;62:567–73. doi: 10.4149/neo_2015_068. [DOI] [PubMed] [Google Scholar]