Abstract
Gastric cancer ranks as the third most lethal cancer worldwide. Although many efforts have been made to identify novel markers for early diagnosis and effective drugs for the treatment of gastric cancer, the outcome is still poor due to delayed diagnosis and lack of therapeutic options. MicroRNAs (miRNAs) play crucial roles during tumorigenesis, and several miRNAs were found to be critical for gastric cancer development, offering promise as therapeutic targets. The results of this study indicate that a novel miRNA, miR-1292-5p, is downregulated both in gastric carcinoma in vivo and in gastric cancer cell lines in vitro. In addition, we showed that attenuation of miR-1292-5p inhibited the growth, migration and invasion of the AGS and SGC-7901 gastric cancer cell lines. Importantly, our results demonstrate that the proto-oncogenic protein DEK is a direct target of miR-1292-5p in gastric carcinoma. Our results therefore demonstrate a tumor suppressor role of miR-1292-5p in gastric carcinoma and hint at the diagnostic and therapeutic potential of the miR-1292-5p/DEK pathway in gastric cancer.
Keywords: MicroRNA-1292-5p, gastric carcinoma, DEK, proliferation, migration
Introduction
Gastric cancer is the third most lethal cancer worldwide [1], and its incidence is growing, with over 600000 new cases each year in China alone [2]. Even with the rapid development of healthcare, gastric cancer is still a nightmare since the prognosis is poor due to late diagnosis and lack of therapeutic options [3]. Thus, it is of great importance to explore novel markers for gastric cancer diagnosis and targets for its treatment. A large number of reports has demonstrated that miRNAs are closely related to the progression of gastric cancer, acting in a post-transcriptional manner [4,5]. Similarly, various reports have demonstrated that miRNAs are critical for the proliferation and metastasis of gastric carcinomas [6,7]. In addition, miRNA and transcriptome profiling have revealed dysregulated miRNAs in gastric cancers [8,9]. Moreover, novel and highly effective miRNAs that are crucial for the pathology of gastric carcinomas are being investigated continuously.
MiR-1292-5p was found to be upregulated in the serum of patients with malignant pleural mesothelioma, suggesting a diagnostic value of miR-1292-5p for this neoplasm [10]. In addition, miR-1292-5p was able to differentiate malignant pleural mesothelioma from lung adenocarcinoma, suggesting that miR-1292-5p plays unique roles in tumor progression [10]. Most recently, miR-1292 was found to be overexpressed in cancer cell lines and patient samples of metastatic colorectal carcinoma, and it was found to predict poor outcomes in human colorectal cancer patients, indicating an essential role of the miR-1292 family in cancer migration [11]. Furthermore, the expression of miR1292-5p was also associated with the outcome of bevacizumab treatment in combination with FOLFOX for patients with metastatic colorectal cancer [12]. However, these profiling analyses revealed little about the function of miR-1292-5p in cancer biology. Taken together, limited reports have demonstrated a critical role of miR-1292-5p, but the detailed function of miR-1292-5p in cancers, especially in gastric carcinomas, is completely unknown.
DEK was initially identified as a crucial protein due to the discovery that inappropriate translocation of the dek gene on chromosome 6 generated a dek-can mRNA in acute myeloid leukemia [13-16]. The expression of DEK was subsequently studied systematically, and it was revealed that itsis overexpressed in various types of cancers, including gastric adenocarcinoma [17,18]. It has been indicated that DEK is actively involved in transcriptional regulation [19,20]. Recently, it was found that the overexpression of DEK is positively associated with tumorigenesis and metastasis in multiple cancers [21,22]. In addition, DEK was recently found to be closely associated with DNA repair and cell cycle control [23]. Importantly, miRNA-based regulation of DEK is critical for cancer cell proliferation and migration [24]. Although it has been indicated that DEK is overexpressed in gastric adenocarcinoma [10], the detailed mechanism of how DEK is regulated in gastric cancer and how DEK affects the disease is largely unknown.
Here, we probed the expression pattern of miR-1292-5p and found that its expression is depressed in gastric carcinomas compared to adjoining normal mucosa. We next studied the role of miR-1292-5p in gastric cancer by overexpressing miR-1292-5p in gastric cancer cell lines. In addition, the migration and invasion ability of gastric cancer cells with attenuated miR-1292-5p expression was assessed. Importantly, we explored the direct target of miR-1292-5p in gastric cancer and found that DEK was directly regulated by miR-1292-5p and was responsible for the aberrant proliferation and migration of gastric cells that lose miR-1292-5p expression.
Materials and methods
Human tissue specimens and cell lines
This study complied with the principles of the Declaration of Helsinki, and was approved by the human ethics and research ethics committees of the Second Affiliated Hospital of Xi’an Jiaotong University. This study included human gastric cancer samples and adjacent normal mucosal tissues derived from 10 patients who underwent surgery at the Second Affiliated Hospital of Xi’an Jiaotong University between 2015 and 2016. The human gastric cancer cell lines SGC-7901, MKN45, and AGS, and the human gastric mucosal epithelial cell line GES-1 were purchased from the Cell Resource Center, Shanghai Institute of Biochemistry and Cell Biology, Chinese Academy of Sciences. All cell lines were maintained under the recommended culture conditions according to ATCC (https://www.atcc.org/) and incubated at 37°C in a humidified atmosphere supplied with 5% CO2.
MiRNA isolation and detection
MiRNA isolation and detection was performed as described previously [25]. Total miRNA was extracted from cultured cells or human tissue using the miRcute miRNA isolation kit (Tiangen Biotech, Beijing, China), according to the manufacturer’s instructions. Real-time PCR was performed on a CFX96 Real-Time PCR Detection System (Bio-Rad) using SuperReal PreMix Plus (Tiangen Biotech, Beijing, China), according to the manufacturer’s instructions. The data were normalized against the U6 snRNA. The miRNA sequence of miR-1292-5p used in this study was 5’-UGGGAACGGGUUCCGGCAGACGCUG-3’.
Determination of cell viability and cell cycle status
To determinate the number of actively proliferating viable cells, the Cell Counting Kit-8 (CCK-8; MCE, NJ, USA) was used according to the manufacturer’s instructions. Briefly, 10 μL of the CCK-8 solution were added to each well of a 96-well-plate, incubated for 1 h in the incubator, and the absorbance at 450 nm measured. The Cell Cycle Assay Kit (Abcam, MA, USA) was used to determinate the cell cycle status according to the manufacturer’s instructions.
DEK and miR-1292-5p overexpression
The human miR-1292-5p was cloned into a lentiviral vector. The lentiviruses were generated by the co-transfection of plasmids pGC-LV and pHelper 2.0 using Lipo2000 (Invitrogen, USA). Virions were harvested and the AGS cell line SGC-7901 cells were infected in the presence of 2 µg/µL puromycin (Invitrogen). The ORF sequence of DEK was cloned into the pcDNA3.1 vector and the resulting plasmid was used to transfect AGS and SGC-7901 cells that were infected with viruses using Lipo2000. Cells were harvested 48 hours post-transfection.
DEK RNAi
DEK expression was silenced by transfecting the cells with the targeted siRNA sequences (Sangon Biotech, Shanghai, China) 5’-CGAACCAAAUGUCCUGAAA-3’ and 5’-UUUCAGGACAUUUGGUUCG-3’. Control siRNAs (siC) were generated using 4 base substitutions in the DEK targeting sequence, yielding the sequences 5’-UUCUCCGAACGUGUCACGU-3’ and 5’-ACGUGACACGUUCGGAGAA-3’.
Cell migration and invasion assay
Migration and invasion assays were performed as described previously [26]. Briefly, migration was assessed using a wound-healing assay via scratching of confluent cell monolayers with a pipette tip. Cell invasion was assayed in Biocoat Matrigel invasion chambers (BD Bioscience, San Jose, CA, USA) according to the manufacturer’s protocol. The number of cells was counted by direct visualization under a bright-filed microscope.
Xenografts in nude mice
SGC-7901 cells (1.5 × 106) expressing miR-1292-5p or control miRNA were injected subcutaneously into the flanks of female BALB/c nude mice (aged 6 weeks; 5 mice per group). The tumor size was measured using a caliper every 3 d from the sixth day after injection. The tumor volume was calculated using the formula: volume = (length × width2)/2.
Western blot analysis
Total protein was separated by 10% SDS-PAGE and transferred onto polyvinylidene difluoride membranes (Bio-Rad, Hercules, CA, United States). Primary anti-DEK (ab166624), anti-β-actin (ab8226) and anti-GAPDH (ab8245) antibodies (all from Abcam, USA) were incubated overnight at 4°C, followed by incubation with the secondary antibody Santa Cruz Biotechnology, CA, United States) for 1 h at RT. Protein bands were visualized using the ECL-kit, according to the manufacturer’s instructions. GAPDH or β-actin were used as a loading control.
Immunohistochemistry
The Dako LSAB kit (Dako A/S, Glostrup, Denmark) was applied to perform immunohistochemical analysis of clinical samples. Briefly, 6-μm sections were deparaffinized, rehydrated, and the antigen retrieved. Samples were incubated with a DEK antibody (ab166624; Abcam) at 4°C overnight, followed by incubation with the secondary antibody at room temperature for 1 h and washing for 2 h. The peroxidase reaction was then detected with 3,3’-diaminobenzidine.
Statistical analysis
Student’s t-test was used to analyze cell migration and invasion. All experiments were performed at least three times. Data are displayed as the means ± the standard deviations from at least three independent experiments. RNA expression was examined using the Wilcoxon rank-sum test. Differences with p-values of less than 0.05 were considered statistically significant.
Results
Decrease of miR-1292-5p expression in gastric carcinoma and gastric cancer cell lines
To reveal the role of miR-1292-5p in gastric carcinoma, we first examined the expression levels of miR-1292-5p in clinical gastric carcinoma samples. In the 10 gastric carcinoma tissue samples we collected during surgery, miR-1292-5p was significantly down-regulated compared with the samples of adjacent normal mucosa (Figure 1A). In addition, we investigated the expression levels of miR-1292-5p in three gastric cancer cell lines. As expected, miR-1292-5p expression was decreased in the gastric cancer cell lines AGS, MKN45 and SGC-7901 compared to the normal human gastric cell line GES, further corroborating that miR-1292-5p is down-regulated in gastric carcinoma (Figure 1B).
Figure 1.
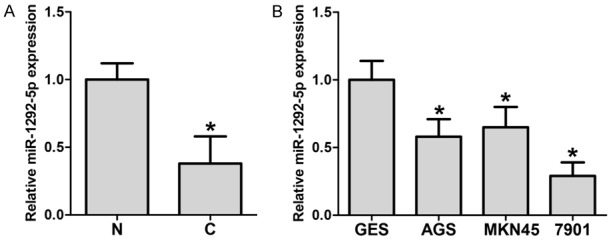
MicroRNA-1292-5p levels in gastric carcinoma, normal tissues and corresponding cell lines. A. qPCR examination of miR-1292-5p levels in gastric carcinoma tissues (C) and adjacent normal mucosa (N). Error bars indicate SD and *indicates P<0.05. B. qPCR examination of miR-1292-5p in human gastric mucosal epithelial cell line GES-1 and gastric cancer cell lines SGC-7901, MKN45 and AGS. Error bars indicate SD and *indicates P<0.05.
The tumor suppressor role of miR-1292-5p in vitro and in vivo
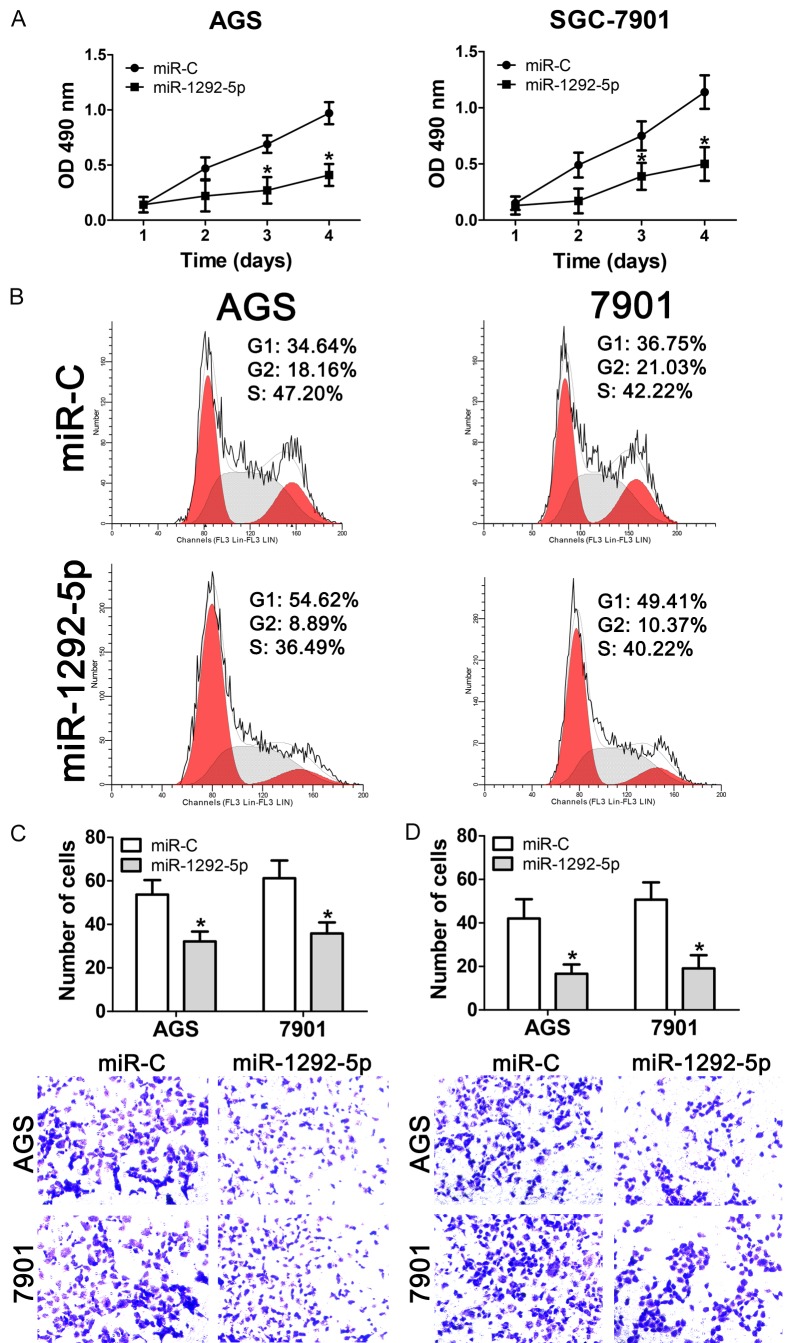
To gain a better understandings of the role that miR-1292-5p plays in gastric carcinoma, we overexpressed miR-1292-5p in gastric cancer cell lines and measured the proliferation of the resulting transgenic cancer cells. Our results showed that overexpression of miR-1292-5p significantly impaired the proliferation of both AGS and SGC-7901 cells compared to overexpression of the control miRNA (Figure 2A). To further understand the differences in cell proliferation, flow cytometry was performed to determine the cell-cycle distribution. Interestingly, the results showed that both miR-1292-5p-overexpressing AGS and SGC-7901 cells showed increased percentages of G1 cells (from 34.64% to 54.62% for AGS cells and from 36.75% to 59.41% of SGC-7901 cells), and decreased subpopulations of G2 cells, suggesting that overexpression of miR-1292-5p resulted in G1 phase arrest (Figure 2B). In addition, we used transwell migration and matrigel invasion assays to evaluate the role miR-1292-5p might play in gastric carcinoma. Our data showed that both the migration and invasion ability of AGS and SGC-7901 cells that overexpressed miR-1292-5p were significantly impaired (Figure 2C and 2D).
Figure 2.
Effect of miR-1292-5p overexpression ongastric cancer cell growth, migration and invasion. A. CCK-8 analysis of cell viability of normal SGC-7901/AGS cells andSGC-7901/AGS cells overexpressing miR-1292-5p. Error bars indicate SD and *indicates P<0.05. B. Cell cycle analysis of normal SGC-7901/AGS cells and SGC-7901/AGS cells overexpressing miR-1292-5p. C. Cell migration analysis of normal SGC-7901/AGS cells and SGC-7901/AGS cells overexpressing miR-1292-5p. D. Cell invasion analysis of normal SGC-7901/AGS cells and SGC-7901/AGS cells overexpressing miR-1292-5p.
Since our results suggested a tumor suppressor role of miR-1292-5p (Figure 2), we next explored the function of miR-1292-5p in gastric cancer growth in vivo. Nude mice were inoculated with the same number of AGS cells overexpressing control miRNA or miR-1292-5p. We found that miR-1292-5p was successfully overexpressed in the tumors that grew in the nude mice (Figure 3A). As expected, overexpression of miR-1292-5p significantly inhibited the tumor growth in vivo, as revealed by measuring the volume and weight of the tumors (Figure 3B-D). Taken together, our data therefore suggest a tumor suppressor role of miR-1292-5p both in vitro and in vivo.
Figure 3.
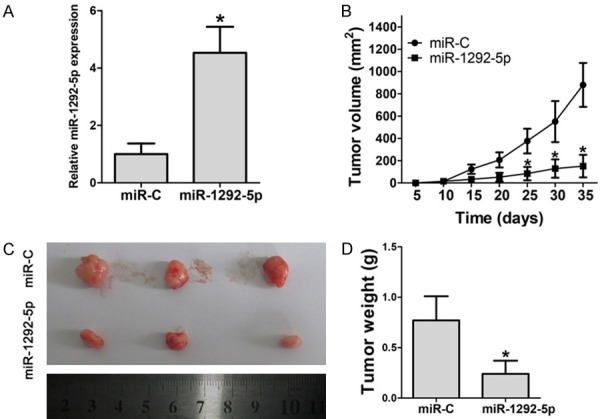
Effect of miR-1292-5p overexpression on tumorigenesis of gastric cancer xenografts in vivo. A. qPCR examination of miR-1292-5p in xenografts comprising normal SGC-7901 cells or SGC-7901 cells overexpressing miR-1292-5p. Error bars indicate SD and *indicates P<0.05. B. Tumor volume of xenografts comprising normal SGC-7901 cells or SGC-7901 cells overexpressing miR-1292-5p. Error bars indicate SD and *indicates P<0.05. C. Tumor size of xenografts comprising normal SGC-7901 cells or SGC-7901 cells overexpressing miR-1292-5p. D. Tumor weight of xenografts comprising normal SGC-7901 cells or SGC-7901 cells overexpressing miR-1292-5p. Error bars indicate SD and *indicates P<0.05.
DEK is a direct target of miR-1292-5p
We used miRNA target prediction software to search for the target genes of miR-1292-5p. Interestingly, among the predicted targets, a complementary region of miR-1292-5p was identified in the 3’-UTR of the oncogene DEK (Figure 4A). Thus, we constructed a wild-type DEK 3’-UTR fragment (3’WT UTR) and amutated DEK 3’-UTR fragment (3’mutant UTR), and inserted them into the Luc-Pair™ Duo-Luciferase vector. Our results showed that miR-1292-5p significantly decreased the luciferase activity compared to control miRNA in both AGS and 7901 cells (Figure 4B). Importantly, miR-1292-5p showed no effect on the mutated 3’-UTR fragment (Figure 4B), suggesting that DEK is a direct target of miR-1292-5p. In addition, both the mRNA and protein levels of DEK were significantly decreased in AGS cells transfected with miR-1292-5p mimics, and both the mRNA level and the protein level of DEK were significantly increased when the cells were transfected with a miR-1292-5p antagonist (Figure 4C and 4D).
Figure 4.
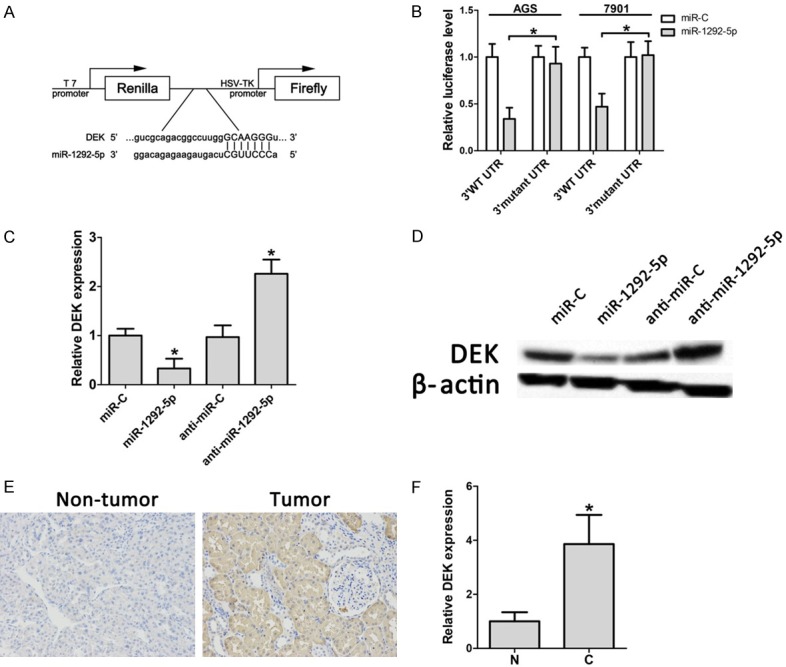
Functional relationship between miR-1292-5p and the 3’UTR of DEK. A. Luciferase reporter plasmids were constructed by the insertion of the full-length NRP1 3’UTR into a region immediately downstream of the luciferase gene. B. The relative luciferase activity of the wild-type (3’WT UTR) and mutated DEK 3’UTR (3’ mutant UTR) in AGS/SGC-7901 cells. C. qPCR examination of DEK in control miRNA-overexpressing SCG-7901 cells, miR-1292-5p-overexpressing SCG-7901 cells, control Antagomir SCG-7901 cells and Antagomir-1292-5p SCG-7901 cells. D. Western bolt analysis of DEK in control miRNA-overexpressing SCG-7901 cells, miR-1292-5p-overexpressing SCG-7901 cells, control Antagomir SCG-7901 cells and Antagomir-1292-5p SCG-7901 cells. E. Representative immunohistochemistry results of staining for DEK in non-tumor and tumor samples. F. Quantification of DEK expression in gastric carcinoma tissues (C) and adjacent normal mucosa (N). Error bars indicate SD and *indicates P<0.05.
Thus, we reasoned that DEK is highly expressed in gastric carcinoma since miR-1292-5p is down-regulated. We next examined the expression of DEK in human gastric cancer samples. Immunohistochemical staining showed that DEK protein levels were dramatically increased in human gastric tumor samples (Figure 4E). In addition, qRT-PCR results showed that mRNA levels of DEK were significantly increased in human gastric cancer samples (Figure 4F). Hence, our results suggested that miR-1292-5p suppresses gastric carcinoma by targeting DEK.
DEK is required for migration and invasion of gastric cancer cells
We next evaluated the expression levels of DEK in human gastric carcinoma by western blot analysis. Our results showed that DEK was significantly upregulated in gastric carcinoma tissues compared to the adjacent normal mucosa (Figure 5A). Additionally, DEK was also upregulated in the gastric cancer cell lines AGS and SGC-7901, suggesting an oncogenic role of DEK in gastric cancer. Accordingly, we designed ansiRNA to knockdown DEK in these gastric cancer cell lines, and our results showed a successful knockdown of DEK in AGS and SGC-7901 cells at both the mRNA and protein level (Figure 5C and 5D). Importantly, both the migration and invasion ability of AGS and SGC-7901 cells that were transfected with the siRNAs targeting DEK were significantly impaired (Figure 5E and 5F), which was identical with the phenotypes that resulted from miR-1292-5p overexpression (Figure 2C and 2D). These data further suggested that miR-1292-5p suppresses the migration and invasion of gastric carcinoma cells by directly targeting DEK.
Figure 5.
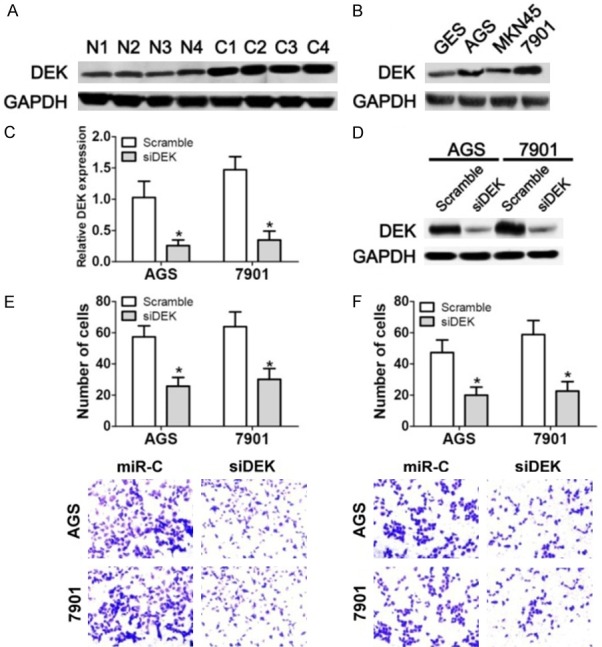
The functional relationship between DEK and the tumor suppressor function of miR-1292-5p. A. Western bolt analysis of DEK expression in gastric carcinoma tissues (C) and adjacent normalmucosa (N). B. Western bolt analysis of DEK expression in human gastric mucosal epithelial cell line GES-1 and gastric cancer cell lines SGC-7901, MKN45 and AGS. C. qPCR examination of DEK expression in control siRNA-and DEK siRNA-transfected SCG-7901/AGS cells. Error bars indicate SD and *indicates P<0.05. D. Western bolt analysis of DEK expression in control siRNA-and DEK siRNA-transfected SCG-7901/AGS cells. E. Cell migration analysis of normal SGC-7901/AGS cells and SGC-7901/AGS cells transfected with DEK siRNA. F. Cell invasion analysis of normal SGC-7901/AGS cells and SGC-7901/AGS cells transfected with DEK siRNA.
MiR-1292-5p suppresses gastric cancer cell growth, migration and invasion by targeting DEK
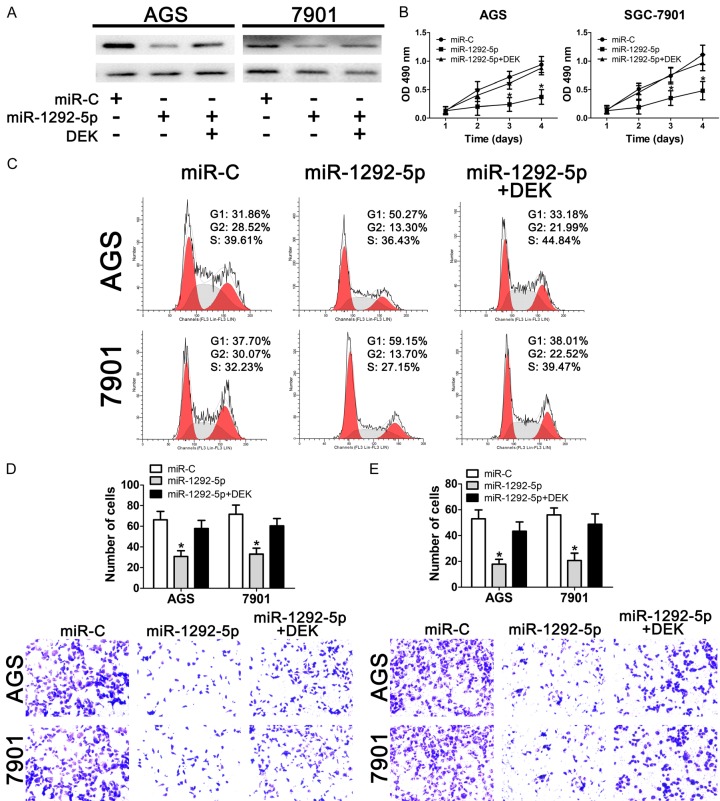
To validate our hypothesis that miR-1292-5p functions by directly targeting DEK, we co-overexpressed DEK together with miR-1292-5p in AGS and SGC-7901 cells. Our results showed that overexpression of miR-1292-5p significantly impaired the expression of DEK while co-overexpression of miR-1292-5p and DEK restored the DEK protein levels in both AGS and SGC-7901 cells (Figure 6A). Importantly, co-overexpression of miR-1292-5p and DEK restored the cell growth of gastric cancer cells (Figure 6B), suggesting that miR-1292-5p functions directly through DEK. In addition, we also examined the cell-cycle distribution of cells with ectopic expression of miR-1292-5p and DEK. Our data showed that the increase of the G1 subpopulation resulting from ectopic miR-1292-5p expression was attenuated by DEK overexpression (Figure 6C). At the same time, our data also showed a slight increase of the S phase subpopulation when gastric cancer cells were co-transfected with miR-1292-5p and DEK (Figure 6C). Finally, our data also revealed that co-overexpression of miR-1292-5p and DEK reversed the impaired migration and invasion capabilities of gastric cancer cells caused by ectopic miR-1292-5p expression. Taken together, the results therefore suggest that miR-1292-5p suppresses the growth, migration and invasion of gastric cancer cells by directly targeting the expression of the onco-protein DEK.
Figure 6.
Effect of DEK overexpression on the tumor suppressor role of miR-1292-5p. A. Western bolt analysis of DEK expression in control miRNA-overexpressing or miR-1292-5p-overexpressing SCG-7901/AGS cells with/without DEK overexpression. B. CCK-8 analysis of cell viability of normal SGC-7901/AGS cells, SGC-7901/AGS cells overexpressing miR-1292-5p, and SGC-7901/AGS cells co-overexpressing miR-1292-5p and DEK. Error bars indicate SD and *indicates P<0.05. C. Cell cycle analysis of normal SGC-7901/AGS cells, SGC-7901/AGS cells overexpressing miR-1292-5p, and SGC-7901/AGS cells co-overexpressing miR-1292-5p and DEK. Error bars indicate SD and *indicates P<0.05. D. Cell migration analysis of normal SGC-7901/AGS cells, SGC-7901/AGS cells overexpressing miR-1292-5p, and SGC-7901/AGS cells co-overexpressing miR-1292-5p and DEK. Error bars indicate SD and *indicates P<0.05. E. Cell invasion analysis of normal SGC-7901/AGS cells, SGC-7901/AGS cells overexpressing miR-1292-5p, and SGC-7901/AGS cells co-overexpressing miR-1292-5p and DEK. Error bars indicate SD and *indicates P<0.05.
Discussion
MiRNAs have a tremendous diagnostic and therapeutic potential in gastric cancers [27]. Our data outline a scenario in which the decreased expression of a novel miRNA, miR-1292-5p, promotes the development and invasiveness of gastric carcinomas, which suggests a tumor suppressor role of miR-1292-5p that is mediated by the direct targeting of the proto-oncogene DEK. Dysregulated miRNAs are critical for the pathology of gastric cancers, and it is becoming practical to attenuate dysregulated genes using miRNA mimics or antagonists in clinical applications [28-30]. Thus, it is critical to identify cancer-specific miRNAs. Our results show that miR-1292-5p is not only downregulated in the three gastric cancer cell lines AGS, MKN45 and SGC-7901, but also decreased in human gastric carcinoma tissues, suggesting a tumor-suppressor role of miR-1292-5p in gastric cancer. It has been shown that miR-1292-5p levels are elevated in the sera of patients with malignant pleural mesothelioma [10]. Most recently, miR-1292-5p was found to be a potential marker that can predict the outcome of bevacizumab treatment in combination with FOLFOX for metastatic colorectal cancer [12]. However, no targeted studies have been made to examine the mechanistic function of miR-1292-5p. Thus, it is of great importance to examine the expression levels of miR-1292-5p in other types of cancers to gain a complete understanding of the role of miR-1292-5p.
Our data showed a dramatic decrease of cell growth when miR-1292-5p was ectopically expressed in gastric cancer cells, suggesting a remarkable therapeutic potential for gastric cancer treatment. Importantly, our results suggest that the down-regulation of cell growth is due to arrest in G1, the phase of mRNA and protein synthesis. Multiple signaling pathways are involved in regulating G1 phase arrest [31,32], and it would be of great value to further explore the detailed mechanism of how miR-1292-5p regulates the cell cycle using high throughput methods. Our results also revealed a decreased tumorigenicity of SGC-7901 cells that overexpress miR-1292-5p in vivo (Figure 3), further suggesting a therapeutic potential of miR-1292-5p. Moreover, the in vivo results also pointed out the significance of exploring the expression of miR-1292-5p in various types and stages of cancer.
In addition to the inhibitory role of miR-1292-5p in cancer cell growth, our results also revealed that overexpression of miR-1292-5p reduced the migration and invasion of SGC-7901 and AGS cells, suggesting an inhibitory role of miR-1292-5p in cancer cell migration and invasion. In vivo studies on the function of miR-1292-5p in cancer cell migration and invasion should be performed in the future to validate these results. Interestingly, the mouse homolog of miR-1292 exhibited a contrary role in colorectal cancer, whereby it promoted malignant traits and liver metastasis [11]. This may either be due to differences between the cancer types or differences between human and mouse systems. Either way, further validation studies are needed to dissect the exact role of miR-1292-5p in humans.
We also used aluciferase reporter assay to show that miR-1292-5p functions by repressing DEK (Figure 4). Our data revealed that DEK expression was elevated in human gastric carcinomas and gastric cancer cell lines, which was in line with previous findings, in which DEK was found to be highly expressed in human gastric adenocarcinoma and many other types of cancer [18,33]. Furthermore, restoration of DEK expression not only rescued the cell growth inhibition that resulted from miR-1292-5p overexpression, but also attenuated the accompanying G1 phase arrest (Figure 6). Indeed, accumulating evidence seems to indicate that DEK is intimately involved in the control of the cell cycle [34-36], and our results provide further evidence that the onco-protein DEK is involved in cell cycle regulation in the growth of cancer cells. At the same time, our data showed that DEK inhibition through miR-1292-5p overexpression leads to decreased migration and invasion of gastric cancer cells, which was in agreement with previous findings that DEK overexpression is closely related to metastasis in non-small cell lung cancer, colorectal cancer, pancreatic ductal adenocarcinoma, hepatocellular carcinoma and cervical cancer [21,22,24,37,38].
In summary, our results delineate the tumor suppressor role of a novel miRNA, miR-1292-5p, in gastric carcinoma. This study provides experimental evidence that loss of miR-1292-5p expression in gastric cancer results in uncontrolled growth and migration of gastric cancer cells. In addition, our data also demonstrate a direct regulatory interplay between miR-1292-5p and the proto-oncogene DEK. Thus, our data not only demonstrate a novel miRNA target for gastric cancer diagnosis and treatment, but also provide further evidence that DEK is a potential target for gastric cancer treatment.
Acknowledgements
This work is financially surpported by National “12th 5-Year” Plan for Science and Technology Support (No. 2012BAJ8B03) and Shaanxi Province Social Development Science and Technology Breakthrough Project.
Disclosure of conflict of interest
None.
References
- 1.Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359–386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 2.Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 3.Oliveira C, Pinheiro H, Figueiredo J, Seruca R, Carneiro F. Familial gastric cancer: genetic susceptibility, pathology, and implications for management. Lancet Oncol. 2015;16:e60–70. doi: 10.1016/S1470-2045(14)71016-2. [DOI] [PubMed] [Google Scholar]
- 4.Liu W, Ma R, Yuan Y. Post-transcriptional regulation of genes related to biological behaviors of gastric cancer by long noncoding RNAs and MicroRNAs. J Cancer. 2017;8:4141–4154. doi: 10.7150/jca.22076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hao NB, He YF, Li XQ, Wang K, Wang RL. The role of miRNA and lncRNA in gastric cancer. Oncotarget. 2017;8:81572–81582. doi: 10.18632/oncotarget.19197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tao Y, Yang S, Wu Y, Fang X, Wang Y, Song Y, Han T. MicroRNA-216a inhibits the metastasis of gastric cancer cells by targeting JAK2/STAT3-mediated EMT process. Oncotarget. 2017;8:88870–88881. doi: 10.18632/oncotarget.21488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Safaralizadeh R, Ajami N, Nemati M, Hosseinpourfeizi M, Azimzadeh Isfanjani A, Moaddab SY. Disregulation of miR-216a and miR-217 in gastric cancer and their clinical significance. J Gastrointest Cancer. 2017 doi: 10.1007/s12029-017-0019-6. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 8.Sierzega M, Kaczor M, Kolodziejczyk P, Kulig J, Sanak M, Richter P. Evaluation of serum microRNA biomarkers for gastric cancer based on blood and tissue pools profiling: the importance of miR-21 and miR-331. Br J Cancer. 2017;117:266–273. doi: 10.1038/bjc.2017.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Meng Q, Xiang L, Fu J, Chu X, Wang C, Yan B. Transcriptome profiling reveals miR-9-3p as a novel tumor suppressor in gastric cancer. Oncotarget. 2017;8:37321–37331. doi: 10.18632/oncotarget.16310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gayosso-Gomez LV, Zarraga-Granados G, Paredes-Garcia P, Falfan-Valencia R, Vazquez-Manriquez ME, Martinez-Barrera LM, Castillo-Gonzalez P, Rumbo-Nava U, Guevara-Gutierrez R, Rivera-Bravo B, Ramirez-Venegas A, Sansores R, Negrete-Garcia MC, Ortiz-Quintero B. Identification of circulating miRNAs profiles that distinguish malignant pleural mesothelioma from lung adenocarcinoma. EXCLI J. 2014;13:740–750. [PMC free article] [PubMed] [Google Scholar]
- 11.Torres S, Garcia-Palmero I, Bartolome RA, Fernandez-Acenero MJ, Molina E, Calvino E, Segura MF, Casal JI. Combined miRNA profiling and proteomics demonstrates that different miRNAs target a common set of proteins to promote colorectal cancer metastasis. J Pathol. 2017;242:39–51. doi: 10.1002/path.4874. [DOI] [PubMed] [Google Scholar]
- 12.Kiss I, Mlcochova J, Souckova K, Fabian P, Poprach A, Halamkova J, Svoboda M, Vyzula R, Slaby O. MicroRNAs as outcome predictors in patients with metastatic colorectal cancer treated with bevacizumab in combination with FOLFOX. Oncol Lett. 2017;14:743–750. doi: 10.3892/ol.2017.6255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Soekarman D, von Lindern M, Daenen S, de Jong B, Fonatsch C, Heinze B, Bartram C, Hagemeijer A, Grosveld G. The translocation (6;9) (p23;q34) shows consistent rearrangement of two genes and defines a myeloproliferative disorder with specific clinical features. Blood. 1992;79:2990–2997. [PubMed] [Google Scholar]
- 14.von Lindern M, Fornerod M, van Baal S, Jaegle M, de Wit T, Buijs A, Grosveld G. The translocation (6;9), associated with a specific subtype of acute myeloid leukemia, results in the fusion of two genes, dek and can, and the expression of a chimeric, leukemia-specific dek-can mRNA. Mol Cell Biol. 1992;12:1687–1697. doi: 10.1128/mcb.12.4.1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.von Lindern M, Breems D, van Baal S, Adriaansen H, Grosveld G. Characterization of the translocation breakpoint sequences of two DEK-CAN fusion genes present in t(6;9) acute myeloid leukemia and a SET-CAN fusion gene found in a case of acute undifferentiated leukemia. Genes Chromosomes Cancer. 1992;5:227–234. doi: 10.1002/gcc.2870050309. [DOI] [PubMed] [Google Scholar]
- 16.Soekarman D, von Lindern M, van der Plas DC, Selleri L, Bartram CR, Martiat P, Culligan D, Padua RA, Hasper-Voogt KP, Hagemeijer A, et al. Dek-can rearrangement in translocation (6;9)(p23;q34) Leukemia. 1992;6:489–494. [PubMed] [Google Scholar]
- 17.Carro MS, Spiga FM, Quarto M, Di Ninni V, Volorio S, Alcalay M, Muller H. DEK expression is controlled by E2F and deregulated in diverse tumor types. Cell Cycle. 2006;5:1202–1207. doi: 10.4161/cc.5.11.2801. [DOI] [PubMed] [Google Scholar]
- 18.Piao J, Shang Y, Liu S, Piao Y, Cui X, Li Y, Lin Z. High expression of DEK predicts poor prognosis of gastric adenocarcinoma. Diagn Pathol. 2014;9:67. doi: 10.1186/1746-1596-9-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hu HG, Scholten I, Gruss C, Knippers R. The distribution of the DEK protein in mammalian chromatin. Biochem Biophys Res Commun. 2007;358:1008–1014. doi: 10.1016/j.bbrc.2007.05.019. [DOI] [PubMed] [Google Scholar]
- 20.Sawatsubashi S, Murata T, Lim J, Fujiki R, Ito S, Suzuki E, Tanabe M, Zhao Y, Kimura S, Fujiyama S, Ueda T, Umetsu D, Ito T, Takeyama K, Kato S. A histone chaperone, DEK, transcriptionally coactivates a nuclear receptor. Genes Dev. 2010;24:159–170. doi: 10.1101/gad.1857410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu X, Zou L, Yao Q, Zhang Y, Gan L, Tang L. Silencing DEK downregulates cervical cancer tumorigenesis and metastasis via the DEK/p-Ser9-GSK-3beta/p-Tyr216-GSK-3beta/beta-catenin axis. Oncol Rep. 2017;38:1035–1042. doi: 10.3892/or.2017.5721. [DOI] [PubMed] [Google Scholar]
- 22.Yu L, Huang X, Zhang W, Zhao H, Wu G, Lv F, Shi L, Teng Y. Critical role of DEK and its regulation in tumorigenesis and metastasis of hepatocellular carcinoma. Oncotarget. 2016;7:26844–26855. doi: 10.18632/oncotarget.8565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Smith EA, Gole B, Willis NA, Soria R, Starnes LM, Krumpelbeck EF, Jegga AG, Ali AM, Guo H, Meetei AR, Andreassen PR, Kappes F, Vinnedge LM, Daniel JA, Scully R, Wiesmuller L, Wells SI. DEK is required for homologous recombination repair of DNA breaks. Sci Rep. 2017;7:44662. doi: 10.1038/srep44662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu X, Wu G, Wu Z, Yao X, Li G. MiR-200a suppresses the proliferation and metastasis in pancreatic ductal adenocarcinoma through downregulation of DEK gene. Transl Oncol. 2016;9:25–31. doi: 10.1016/j.tranon.2015.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Peng Y, Liu YM, Li LC, Wang LL, Wu XL. MicroRNA-338 inhibits growth, invasion and metastasis of gastric cancer by targeting NRP1 expression. PLoS One. 2014;9:e94422. doi: 10.1371/journal.pone.0094422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yoon JH, Choi WS, Kim O, Choi BJ, Nam SW, Lee JY, Park WS. Gastrokine 1 inhibits gastric cancer cell migration and invasion by downregulating RhoA expression. Gastric Cancer. 2017;20:274–285. doi: 10.1007/s10120-016-0617-1. [DOI] [PubMed] [Google Scholar]
- 27.Yang W, Ma J, Zhou W, Cao B, Zhou X, Yang Z, Zhang H, Zhao Q, Fan D, Hong L. Molecular mechanisms and theranostic potential of miRNAs in drug resistance of gastric cancer. Expert Opin Ther Targets. 2017;21:1063–1075. doi: 10.1080/14728222.2017.1389900. [DOI] [PubMed] [Google Scholar]
- 28.Detassis S, Grasso M, Del Vescovo V, Denti MA. microRNAs make the call in cancer personalized medicine. Front Cell Dev Biol. 2017;5:86. doi: 10.3389/fcell.2017.00086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rodriguez-Aguayo C, Monroig PDC, Redis RS, Bayraktar E, Almeida MI, Ivan C, Fuentes-Mattei E, Rashed MH, Chavez-Reyes A, Ozpolat B, Mitra R, Sood AK, Calin GA, Lopez-Berestein G. Regulation of hnRNPA1 by microRNAs controls the miR-18a-K-RAS axis in chemotherapy-resistant ovarian cancer. Cell Discov. 2017;3:17029. doi: 10.1038/celldisc.2017.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pulito C, Mori F, Sacconi A, Goeman F, Ferraiuolo M, Pasanisi P, Campagnoli C, Berrino F, Fanciulli M, Ford RJ, Levrero M, Pediconi N, Ciuffreda L, Milella M, Steinberg GR, Cioce M, Muti P, Strano S, Blandino G. Metformin-induced ablation of microRNA 21-5p releases Sestrin-1 and CAB39L antitumoral activities. Cell Discov. 2017;3:17022. doi: 10.1038/celldisc.2017.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Donjerkovic D, Scott DW. Regulation of the G1 phase of the mammalian cell cycle. Cell Res. 2000;10:1–16. doi: 10.1038/sj.cr.7290031. [DOI] [PubMed] [Google Scholar]
- 32.Lalande M. A reversible arrest point in the late G1 phase of the mammalian cell cycle. Exp Cell Res. 1990;186:332–339. doi: 10.1016/0014-4827(90)90313-y. [DOI] [PubMed] [Google Scholar]
- 33.Sanden C, Gullberg U. The DEK oncoprotein and its emerging roles in gene regulation. Leukemia. 2015;29:1632–1636. doi: 10.1038/leu.2015.72. [DOI] [PubMed] [Google Scholar]
- 34.Takahashi M, Seki N, Ozaki T, Kato M, Kuno T, Nakagawa T, Watanabe K, Miyazaki K, Ohira M, Hayashi S, Hosoda M, Tokita H, Mizuguchi H, Hayakawa T, Todo S, Nakagawara A. Identification of the p33(ING1)-regulated genes that include cyclin B1 and proto-oncogene DEK by using cDNA microarray in a mouse mammary epithelial cell line NMuMG. Cancer Res. 2002;62:2203–2209. [PubMed] [Google Scholar]
- 35.Kavanaugh GM, Wise-Draper TM, Morreale RJ, Morrison MA, Gole B, Schwemberger S, Tichy ED, Lu L, Babcock GF, Wells JM, Drissi R, Bissler JJ, Stambrook PJ, Andreassen PR, Wiesmuller L, Wells SI. The human DEK oncogene regulates DNA damage response signaling and repair. Nucleic Acids Res. 2011;39:7465–7476. doi: 10.1093/nar/gkr454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Matrka MC, Hennigan RF, Kappes F, DeLay ML, Lambert PF, Aronow BJ, Wells SI. DEK over-expression promotes mitotic defects and micronucleus formation. Cell Cycle. 2015;14:3939–3953. doi: 10.1080/15384101.2015.1044177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang J, Sun L, Yang M, Luo W, Gao Y, Liu Z, Qiu X, Wang E. DEK depletion negatively regulates Rho/ROCK/MLC pathway in non-small cell lung cancer. J Histochem Cytochem. 2013;61:510–521. doi: 10.1369/0022155413488120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Martinez-Useros J, Rodriguez-Remirez M, Borrero-Palacios A, Moreno I, Cebrian A, Gomez del Pulgar T, del Puerto-Nevado L, Vega-Bravo R, Puime-Otin A, Perez N, Zazo S, Senin C, Fernandez-Acenero MJ, Soengas MS, Rojo F, Garcia-Foncillas J. DEK is a potential marker for aggressive phenotype and irinotecan-based therapy response in metastatic colorectal cancer. BMC Cancer. 2014;14:965. doi: 10.1186/1471-2407-14-965. [DOI] [PMC free article] [PubMed] [Google Scholar]