Abstract
The human microbiota interacts with the host immune system in multiple ways to influence the development of diseases, including cancers; however, a detailed understanding of their relationship is unavailable. Accumulating evidence has only revealed an association rather than a causal link between microbial alterations and carcinogenesis. The regulatory loops among the microbiome, human cells and the immune system are far more complicated and require further studies to be revealed. In this review, we discuss the impact of the microbiota on cancer initiation, development and progression in different types of human cells, mainly focusing on the clinical translation from microbiome research to an accurate diagnosis, subtype classification and precision medicine.
Keywords: Microbiota, microbiome, gut, cancer, precision medicine
Introduction
In the early 1990s, the Human Genome Project (HGP), a global scientific research program, was formally launched in the United States with major goals of mapping, sequencing and identifying genes that constitute the human genome. After completing the first human genome sequence by the end of 2003, scientists sought to determine the future of the HGP, since genome-wide sequencing seems insufficient to interpret human diseases in terms of the complex interactions between genes, proteins and cells, as well as environmental factors. In fact, the completion of the HGP, to some extent, is not the end but the beginning of an immense number of scientific explorations that will direct biological studies in the near future [1]. Indeed, in addition to the direct contribution of genomic alterations in diseases, the microenvironment within a tissue and crosstalk among different tissues employ essential mechanisms that may even determine the development of human disease [2]. Among these mechanisms, microbial changes have emerged as a new field to elucidate human disease, particularly cancer.
Historically, the early scientific skepticism, as postulated by Koch 200 years ago, has focused on isolating and featuring a single pathogen in certain diseases [3]. However, these skepticisms were overturned by solid experimental evidence that human diseases are attributed not only to a single pathogenic microorganism but also to a global alteration in the human microbiome [4]. In 2008, a five-year project termed the Human Microbiome Project (HMP) was initiated by the United States National Institutes of Health (NIH) with the goal of identifying and shaping the microorganisms associated with both healthy and ill humans [5]. Phase two of the HMP began in 2014 and is ongoing [6]. Supported by the HMP, many ground-breaking discoveries have substantially expanded our knowledge of microbiome-associated disorders. These findings have prompted researchers to reassess the pathogenesis of human disease and exploit the novel potential of the microbiome for designing novel diagnostics and more precise therapies.
Very recently, the Cell Editorial Team presented a special issue that highlights the complexity of the microbial world, beginning with an introductory remark titled “Living in their world” [7]. As the writer of the remark commented, “They’re just bacteria. They don’t think or plan. They just…exist”; however, “Millions strong, all working together perfectly”, and “If we could even get close, we would have all our problems solved”. Therefore, researchers must move beyond sequencing to learn about our broader ecosystem and to understand these microbes that have quietly resided in human beings for millions of years. In this review, we discuss the latest advances in studies of microbiome-mediated carcinogenesis in different types of human cells, with a particular focus on its possible clinical translation in the future. Here the microbiota is exclusively defined as bacterial microbes unless specified otherwise to maintain a well-defined subject matter.
Microbiome-mediated carcinogenesis in the gut
Gastric cells
Helicobacter pylori (H. pylori, Hp) is a gram-negative microaerophilic bacterium that is commonly found in the stomach. Hp specifically colonizes the gastric mucosal epithelium and induces active chronic gastritis, gastric ulcers and atrophy, which can further induce gastric carcinoma. Although Hp plays a role in the early stage of carcinogenesis [8], and only a small proportion of infected individuals (1%-5%) develop gastric cancer [9,10], Hp remains the strongest well-known risk factor for gastric carcinoma [9]. Among multiple mechanisms of Hp-mediated carcinogenesis, the secreted protein VacA, a vacuolating cytotoxin, is a specific protein linked to gastric malignancy. In addition to inducing vacuolation in gastric epithelial cells, VacA inhibits host immune responses by suppressing IL-2-mediated signaling and T cell activation, thus allowing Hp to evade immune surveillance [11,12]. Another molecule that substantially contributes to carcinogenesis is the cytotoxin-associated gene A (CagA). Transgenic expression of Hp-CagA in mice can lead to gastrointestinal (GI) carcinoma and hematological malignancies in the absence of co-existing gastritis, highlighting the oncogenic role of CagA in the development of Hp-associated neoplasms [13]. Upon Hp infection, CagA is delivered into gastric epithelial cells via the type IV secretory system [14], followed by the successive tyrosine phosphorylation of CagA by SRC and ABL kinases [15-17], leading to the morphological transformation of cells [18,19]. Nonphosphorylated CagA also promotes carcinogenesis by activating several signaling cascades, including the Ras/Erk, NF-κB, E-cadherin/β-catenin and PI3K/Akt signaling pathways [20-23]. Chronic inflammation also contributes to the development of Hp-induced gastric cancer. The release of large amounts of chemokines and cytokines, such as IL-8, IL-1β, TNF-α, IL-6, IL-18, etc. [21,24-28], leads to a marked infiltration of neutrophils, macrophages and bone marrow-derived cells (BMDCs) into the gastric mucosa [21,29,30]. These persistent inflammatory responses further promote gastric mucosal damage and epithelial cell hyper-proliferation, thereby increasing the risk for transformation. Despite its effect on promoting gastric carcinogenesis, Hp infection is inversely correlated with the esophageal adenocarcinoma (EAC) risk, which not only explains the rapid increase in EAC incidence since Hp infection has prominently declined over the past several decades but also highlights a tissue-specific mechanism of carcinogenesis [31,32].
Colorectal cells
Given the large number of microorganisms residing in colorectal (CR) cells, potential infectious agents are, not surprisingly, associated with colorectal cancer (CRC). Barrier dysfunction is one of the etiological factors that lead to CRC. A defective mucus barrier allows bacteria to access the epithelium and increases the permeability of bacterial toxins. Each of these components induces chronic intestinal inflammation by altering the microbial composition and microenvironment, thus contributing to carcinogenesis [33,34]. For example, according to a whole-genome analysis of pairs of CRC/normal tissues, Fusobacterium nucleatum (F. nucleatum) are predominantly enriched in carcinomas compared with their abundance in the adjacent normal mucosa, while Bacteroidetes and Firmicutes are depleted in tumors [35]. Using shotgun sequencing, a similar result was reported in CRC samples, and high abundance of F. nucleatum is associated with lymph node metastasis [36]. Despite the discovery of an infectious cause of CRC, the causal link between F. nucleatum and CRC remains ambiguous. Soon afterwards, the Garrett group utilized mice with a genetic susceptibility for developing intestinal tumors (Apc Min/+) and introduced human isolates of F. nucleatum. F. nucleatum selectively recruited myeloid-derived immune cells and generated a pro-inflammatory microenvironment that potentiated tumor progression [37]. Meanwhile, another group found that FadA, an adhesion molecule encoded by F. nucleatum, promotes inflammatory and pro-oncogenic responses by activating E-cadherin/β-catenin signaling and by increasing other signals, including transcription factors, oncogenes and inflammatory genes, providing further evidence of F. nucleatum-induced CRC [38].
In addition to FadA, Fap2, a surface protein of F. nucleatum, was reported to bind and activate human Ig and ITIM domains (TIGIT), an inhibitory receptor present on all human natural killer (NK) cells and on various T cells; the Fap2-TIGIT interaction is able to protect tumors from immune cell attack by inhibiting NK cell cytotoxicity [39]. Later that year, Fap2 was further proven to be a mediator that facilitated the binding of F. nucleatum to host epithelial Gal-GalNAc residues, revealing a novel homing mechanism for F. nucleatum localization and enrichment [40]. All the findings described above provide mechanistic insights into a potential contribution of F. nucleatum to CRC.
In addition to F. nucleatum, long-term exposure to microbial toxins produced by Escherichia coli (E. coli) [41], Bacteroides fragilis (B. fragilis) [42] or other gram-negative bacteria [43], as well as reactive oxygen and nitrogen species produced by inflammatory myeloid cells, also leads to genomic instability, DNA damage and oncogenic mutations [44,45]. These events allow cells to overcome the tumorigenesis barrier by activating many survival signals, including the p38 MAPK, NF-κB and IL-6/STAT3 axes, ultimately enabling malignant transformation and tumor progression [43]. Collectively, the interplay between the microbiota and inflammation that regulates the development of CRC is complex and requires further in-depth investigations.
Esophageal cells
Like other parts of the human GI and colorectal systems, the esophagus has a distinct and diverse microbiome, which is not well studied [46]. EAC develops from the precursor lesion caused by Barrett’s esophagus (BE), a typical intestinal metaplasia of the distal esophagus [47]. In addition to many identified modifiable risk factors for BE and EAC (cigarette smoking, obesity, etc.) [47], the upper GI microbiome has been regarded as a novel potential co-factor that promotes Barrett’s neoplasia and esophageal cancer [46]. Using biopsy samples of the distal esophagus in 34 patients with BE, reflux esophagitis and normal controls, researchers identified distinct microbiomes by performing a bacterial 16S ribosomal RNA gene survey and found two major clusters, a Streptococcus-predominant cluster I and a gram-negative anaerobe-predominant cluster II, among which, cluster II is primarily linked to reflux esophagitis and BE [48]. Interestingly, another study showed a similar diverse bacterial composition in Japanese patients with a normal esophagus, reflux esophagitis and BE, but no significant differences in the amount of bacteria were observed between groups. The authors attributed this result to the use of a small dataset, younger subjects, and the method for defining species, implying a potential multi-factor effect on bacterial populations [49]. In addition to biopsy samples, mucosal brush samples from the upper GI tract were also used to analyze bacterial communities in a BE cohort. Streptococcus and Prevotella species were highly abundant in the upper GI tract, and the ratios of these two species were associated with two known risk factors for BE. More importantly, this study noted the advantages of brush sampling over tissue biopsies, such as less contamination and a higher yield of bacterial DNA [50].
Unfortunately, the microbiome in patients with EAC is poorly characterized compared to that in patients with BE. An early study was aimed at identifying the leading roles of Treponema denticola (T. denticola), Streptococcus mitis (S. mitis), and Streptococcus anginosus (S. anginosus) in the carcinogenic process of esophageal cancer and found that eradication of these bacteria may decrease the risk of recurrence [51]. In particular, using a large group of patients, Campylobacter concisus (C. concisus) emerged as a new contributor to reflux esophagitis and BE, but not EAC; the presence of C. concisus induces cytokine production coupled with DNA damage-triggered nitrosative and oxidative stress, thus potentiating carcinogenesis [52]. This study highlights a shift in the esophageal microbiota with disease progression, implying that the evolution from a benign condition to cancer might be attributed to the host’s microbiota rather than the changes in the epithelia. However, additional studies are needed to clarify whether the host’s microbiota is a cause or a consequence of disease progression. A modified rat model undergoing surgical esophagojejunostomy was used to study the potential mechanistic links between the microbiota and EAC. Using this model, Toll-like receptors (TLRs) 1-3, 6, 7 and 9 were significantly upregulated in EAC compared to their expression in normal epithelium [53]. High expression of TLR4 correlates with an advanced stage and poor prognosis of EAC [54], indicating that TLR signaling mediated carcinogenesis in patients with EAC. Recently, Peters and his colleagues assessed the association of the oral microbiome with two main types of esophageal cancer, EAC and esophageal squamous cell carcinoma (ESCC). They emphasized the essential influence of the oral microbiota on shaping the esophageal microbiome. For example, Tannerella forsythia (T. forsythia) is associated with a higher EAC risk, and Porphyromonas gingivalis (P. gingivalis) is associated with the ESCC risk. These two species are periodontal pathogens and the greatest contributors to severe periodontitis [55]. In addition, depletion of the commensal genus Neisseria and the species Streptococcus pneumonia is linked to a lower EAC risk [55]. This study provides an insight into the early detection and prevention of highly fatal esophageal cancer, although more convincing data are warranted and many crucial issues remain unaddressed [46].
Microbiome-mediated tumorigenesis in extra-gut systems
Despite the overwhelming number of studies of microbiome-mediated cancers in the human gut, studies in germ-free animals and human samples have offered solid experimental evidence for distant effects of microbial changes on extra-gut organs.
Lung cells
The lungs were once thought to be a sterile organ in healthy populations until multiple lines of experiments identified distinct microbial communities in the healthy respiratory tract [56] and in diseased lungs, including lungs from patients with chronic obstructive pulmonary disease (COPD) [57-59], asthma [60], idiopathic pulmonary fibrosis [61] and lung cancer [62,63]. Although total bacterial populations within lungs are quite small compared to the gut microbiota [59], an imbalanced lung microbiome may cause respiratory infections and pulmonary inflammation, thus driving lung tumorigenesis [64-66]. An intact commensal bacterial community in the lungs is required for host antitumor defenses in addition to its roles in infection and inflammation. In mice, an antibiotic treatment (Abt) has been shown to enhance lung tumor development [67]. Further mechanistic investigations revealed a defective induction of the γδT17 cell response in the Abt mice, whereas the reintroduction of normal γδT cells or IL-17 was able to restore the impaired immune surveillance. These findings not only highlight the contribution of the lung microbiome in maintaining host immune homeostasis but also a likely tumor-initiating effect of antibiotic exposure. Indeed, recurrent use of antibiotics, particularly penicillin, increases the risk of multiple human cancers, including lung cancer [68].
Interestingly, using a deep sequencing analysis of salivary microbiota from 20 lung cancer patients, researchers identified two potential bacterial biomarkers in saliva, Capnocytophaga and Veillonella, providing a possible noninvasive method for the detection and classification of lung cancer [69]. In contrast, a distinct group of bacterial species were screened and shown to be present at a high abundance in spontaneously collected sputum samples from patients with lung cancer, establishing an alternative noninvasive screening method for lung cancer progression [70].
In addition to the oral-lung axis, communication between the gut and lung microbiota has been recently discovered, illustrating a remote effect of alterations in the microbiota in one compartment on the microbiota in another tissue. This remote effect may be a direct effect caused by aspiration of the GI microbiota [71] or by transmitting immunological information via bacterial metabolites or immune cells [72]. For example, an enrichment of gut-associated bacteria in the lungs is responsible for sepsis and acute respiratory distress syndrome [73]. In turn, the lung microbiota also influences the gut microbiota through the circulation. An instillation of lipopolysaccharide (LPS) into murine lungs leads to changes in the cecal microbiota [74]. Although few studies have examined the contribution of the gut-lung axis to lung tumorigenesis and many technical constraints still exist [75], new findings and improvements are predicted in the near future.
Liver cells
The crosstalk between the liver and gut microbiota was described in 1992 when a novel species of Helicobacter, namely, Helicobacter hepaticus (H. hepaticus), was first identified in mice. H. hepaticus selectively and persistently colonizes the murine hepatic bile canaliculi and results in chronic active hepatitis and hepatocellular neoplasms [76]. Although the inflammatory responses of H. hepaticus infection were later observed in primary human hepatocytes [77], no clear epidemiological report has linked H. hepaticus to human carcinogenesis [4].
Accumulating evidence has identified a strong association between hepatic fibrosis or cirrhosis with hepatocellular carcinoma (HCC) [78]. Intriguingly, the translocation of enteric bacteria is a hallmark of chronic liver disease and subsequently results in severe complications, including advanced cirrhosis [79]. In mice, gut sterilization and genetic TLR4 inactivation suppress hepatic proliferation and fibrogenesis; conversely, prolonged treatment with low-dose LPS, a TLR4 agonist, promotes HCC development by inducing the activation of NF-κB and subsequent production of several inflammatory cytokines and growth factors [80]. Based on these data, the intestinal microbiota and LPS-TLR4 signaling exert profound effects on fibrosis-associated HCC progression; however, the translation of these findings into clinical medicine has yet to be evaluated.
Another connection between the liver and gut microbiota is bile acids (BAs), which are biosynthesized in hepatocytes. As well as being regulated by intestinal bacteria, BAs themselves modulate the gut microbiota composition via the enterohepatic circulation, building a unique liver-bile acid-microbiota axis [81]. Therefore, disruption of BAs is a common etiological factor contributing to liver diseases. In this respect, one elegant study pinpointed a sequential effect of an obesity-induced microbial metabolite on HCC in mice: obesity induces changes in the gut microbiota, subsequently inducing changes in the levels of its metabolite deoxycholic acid (DCA), a secondary bile acid. Elevated DCA levels then elicit the senescence-associated secretory phenotype (SASP) in hepatic stellate cells (HSCs), which in turn release various inflammatory factors; these events ultimately facilitate HCC development. Notably, similar phenotypes were observed in human HSCs from patients with nonalcoholic steatohepatitis, indicating a favorable extension from murine data to human biology [82]. Furthermore, DCA has also been implicated in the development of colon cancer [83] and EAC [84] in the context of obesity, suggesting that an obesity-BA-mediated mechanism is involved in human carcinogenesis.
As evident from the studies above, the TLR4 ligand is a crucial component that promotes fibrosis-associated HCC in mice [80], whereas a TLR4 deficiency fails to protect mice from obesity-associated HCC [82]. This discrepancy is probably due to the distinct mechanistic routes of the gut-liver axis in hepatocarcinogenesis triggered by different pathogenic factors.
Pancreatic cells
Pancreatic cancer remains a major challenge to humans due to its delayed diagnosis, pain, chemoresistance and poor outcome [85]. Hp infection has primarily been implicated in the development of pancreatic cancer; however, the results are variable and paradoxical [86-88]. In contrast, the salivary microbiota has attracted considerable attention as a risk factor, and more importantly, a noninvasive biomarker for distinguishing patients with pancreatic cancer from healthy individuals [89]. High levels of antibodies against the periodontal bacterium P. gingivalis (strain ATCC 53978) have been reported to increase the risk of pancreatic cancer, while elevated levels of antibodies against commensal oral bacteria are inversely correlated with pancreatic cancer risk [90]. Another oral bacteria group, Fusobacterium species, has also been identified in pancreatic abscesses. Although only 8.8% of surgical specimens of pancreatic cancer are colonized by Fusobacterium, the presence of Fusobacterium in pancreatic tumors is associated with a worse prognosis [91]. Recently, a prospective study nested in two large US cohorts was conducted to determine the association between the oral microbiome and subsequent risk of pancreatic cancer. As reported by Ahn and coworkers [90], a link between the periodontal pathogens P. gingivalis and Aggregatibacter actinomycetemcomitans (A. actinomycetemcomitans) and a higher risk of pancreatic cancer is observed, independent of smoking or other potential confounders, supporting the evidence showing that the oral microbiota may play a potential role in the etiology of pancreatic cancer [92]. Although a consistent landscape has been painted in previous studies, many key questions remain unanswered regarding the mechanical crosslinks between pancreatic cancer and poor oral hygiene. For example, do oral pathogens drive or participate in pancreatic carcinogenesis? Do oral bacteria populate distant disease sites? The mechanism by which the oral microbiota determine the termination of its action remains to be determined, since elevated levels of oral P. gingivalis increase the risks of both ESCC and pancreatic cancer, according to the two aforementioned studies [55,92].
Blood cells
Bacterial invasion and colonization were previously linked with leukemia in patients as a consequence of myelosuppressive therapies, which increase the risk of advanced periodontal lesions and abscesses, particularly during pronounced granulocytopenia; these oral pathogens are most likely to seed the blood circulation and hence lead to life-threatening septicemia [93]. Additionally, intestinal toxicity and infections caused by the diminished gut microbial diversity are also very common in immunocompromised patients with leukemia, and prophylactic antibiotics are unable to restore the balance between aerobic and anaerobic bacteria, suggesting a deteriorating progression of secondary infections upon treatment with chemotherapies [94]. Although a close interaction has been identified between the microbiota and host immune system, scientists have only recently begun to bring public attention to the distinct microbiota signature caused by leukemia. The oral microbiota of patients with leukemia is characterized by a lower richness and reduced diversity compared with that of healthy controls. Using high-throughput sequencing, the leukemia status is associated with certain taxa, including the Firmicutes, Bacilli, Lactobacillales, Aerococcaceae and Carnobacteriaceae, as well as the genera Abiotrophia and Granulicatella [95]. Regarding the changes in the structure of the gut microbiota in pediatric patients with acute lymphoblastic leukemia (ALL), the alpha diversity rather than beta diversity is significantly reduced by a short-term treatment with antibiotics [96]. In contrast, adult survivors of pediatric ALL exhibit increased T cell activation and systemic inflammation, which are associated with a reduced microbial diversity and indicate persistent microbial dysbiosis [97]. Research on the link between the microbiome and leukemia remains in its early stages; therefore, unsurprisingly, only association studies between them are ongoing, and the causative mechanisms of these links have rarely been reported and are poorly understood.
Unlike leukemia, lymphoma caused by exposure to certain microbes has been the subject of in-depth investigation. Mouse models housed under specific pathogen-free (SPF) conditions and provided with sterile (SPF-S) food, water and bedding exhibited a longer lifespan and B cell lymphoma latency than mice housed under nonsterile (SPF-N) conditions. The bacterial communities are distinct in mice harboring restricted microbiota (RM) and conventional microbiota (CM): a higher abundance of Lactobacillus johnsonii (L. johnsonii) is observed in RM mice than in CM mice, implying a possible beneficial role. Notably, an enrichment of L. johnsonii in RM mice in oral inoculation experiments revealed its ability to decrease systemic inflammation and micronucleus formation. This effect may be due to i) decreased systemic genotoxicity by the inhibition of basal intestinal inflammation and its systemic sequelae; ii) reduced immune-mediated ROS production and systemic genotoxicity by decreasing NF-κB activation; or iii) a superior colonizing capacity in the intestinal mucosa, and thus outcompeting pathogens and other pro-inflammatory organisms [98]. In addition to the potential favorable role of the gut microbiota in protecting against lymphoma development, mucosal-associated lymphoid tissue (MALT) lymphoma is strongly associated with the presence of H. pylori, because the stomach is the most common site of MALT lymphoma [99]. In a long-term study of H. felis infection in mice, 38% of infected mice had lymphoid follicles and 25% of infected mice developed MALToma-like lesions, whereas these lesions were not observed in noninfected controls [100]. Other microbial pathogens, such as H. helmanii, Campylobacter jejuni (C. jejuni), Borrelia burgdorferi (B. burgdorferi) and Chlamydia psitacci (C. psitacci), have also been identified as contributors to the development of MALT lymphoma in either animal models or humans [101-103], defining a distinct category of infection-associated lymphoid malignant transformation. The mechanisms of microbiota-induced lymphomagenesis have been extensively reviewed [104]; however, a thorough understanding of the exact roles of bacteria in hematological malignancies has yet to be achieved.
Skin cells
As the largest organ of the human body, the skin is another major component of the human microbiome. Many of these microorganisms are harmless or even beneficial to their host, thus protecting humans against the entry of foreign harmful organisms [105]. Therefore, aberrations in the skin microbiome may confer a greater risk for diseases, including cancer. For example, primary cutaneous B-cell lymphoma (PCBCL) has been linked to B. burgdorferi, which was identified when elevated antibody titers against B. burgdorferi were discovered in patients with PCBCL [106]. Based on this finding, B. burgdorferi was also shown to plays a role in the etiology of a significant proportion of PCBCL cases [107,108], although a minimal role of B. burgdorferi was observed in the development or pathogenesis of PCBCL in the United States, highlighting the geographic variations in the clinical manifestations of B. burgdorferi [109]. In addition, colonization of the skin by Staphylococcus aureus (S. aureus) correlates with cutaneous T cell lymphoma (CTCL) [110,111]. Propionibacterium acnes (P. acnes), a human skin-resident commensal bacteria, immediately responds to ultraviolet (UV) radiation, which has been thought to be the major risk factor for most skin cancers [112]. This finding, to some extent, facilitates the diagnosis of radiation risk, although more research is needed to confirm this connection.
Cervical cells
Persistent infection with the human papillomavirus (HPV) has been extensively studied and linked to cervical carcinogenesis [113]; however, viral microbes are outside the scope of this review. Less is known about the role of the vaginal or cervical bacterial microbiome in cervical cancer. In healthy women, premenopausal vaginal bacterial communities are dominated by Lactobacillus spp. to maintain a low pH value and constitute the first line of host defense against pathogens [114]. In fact, the vaginal microbiome is far more complicated than previously appreciated, since 282 taxa have been identified in the vagina [115]. In one study, the vaginal microbiome in women with high-grade squamous intraepithelial neoplasia was characterized by higher levels of Sneathia sanguinegens (S. sanguinegens), Anaerococcus tetradius (A. tetradius) and Peptostreptococcus anaerobius (P. anaerobius) and lower levels of Lactobacillus jensenii (L. jensenii) compared to that in women with low-grade squamous intraepithelial neoplasia [116]. Another report further identified a risky microbial pattern using cervical swabs featured by a predominance of Atopobium vaginae (A. vaginae), L. iners and Gardnerella vaginalis (G. vaginalis) and a concomitant paucity of L. crispatus; this pattern is associated with a higher risk of cervical intraepithelial neoplasia (CIN) [117]. Interestingly, species belonging to the same Lactobacillus genus, L. iners and L. crispatus, play opposite roles in cervical carcinogenesis, highlighting that not all Lactobacillus species are protective, as previously recognized. In contrast, a subsequent study failed to show a link between the cervical microbiome diversity and CIN severity, but provided evidence suggesting that a cervical microbiome dominated by L. iners and Lactobacillus is associated with high-grade CIN in women infected with high-risk HPVs [118]. The collection of samples from the cervix is particularly notable. Most studies examine the microbiota from cervical swabs or washes, which, on one hand, are very likely to be contaminated by samples from the vaginal wall and vulva; on the other hand, these samples are quite different than biopsy samples. These limitations surely deserve further investigation.
Breast cells
Little is known about how bacteria potentiate breast carcinogenesis or malignancy, although abundant and distinct microbiota are present in breast cancer tissues [119,120]. For example, significant differences were observed in the microbiome of human breast tissues from women with benign and malignant diseases. Five low-abundant genera, Fusobacterium, Atopobium, Hydrogenophaga, Lactobacillus and Gluconacterobacter, are enriched in malignant breast tissue [119]. An unclassified genus from the Sphingomonadacea family is highly abundant in nipple aspirate fluid (NAF) from healthy control women, whereas increased activity of β-glucuronidase, a pro-carcinogenic enzyme in GI cancer, has been identified in the microbes associated with breast cancer [121].
Similar to other extra-gut organs described above, distant effects of gut dysbiosis by either host immune modulation or metabolic alterations have been observed in breast tumorigenesis and development [122-125]. For instance, levels of systemic estrogens and estrogen metabolites are modulated by Clostridiales and Bacteroides in the intestinal microbiota, thereby influencing the risk of breast cancer [126]. In addition, the percentage and the absolute numbers of certain bacterial groups, namely, Clostridium coccoides (C. coccoides), Faecalibacterium prausnitzii (F. prausnitzii) and Blautia, differ in fecal samples from patients with different clinical stages and histoprognostic grades of breast cancer [127]. In addition to results obtained from patient samples, challenge with enteric bacteria H. hepaticus in Apc Min/+ mice models promotes cancer in the mammary gland [128]. Nevertheless, most current research reports are focusing on associations rather than cause-effect analyses between breast cancer and the microbiome, similar to other types of human cancers.
The impact of the microbiota on tumorigenesis in different types of human cells and the crosstalk between each part are illustrated in Figure 1.
Figure 1.
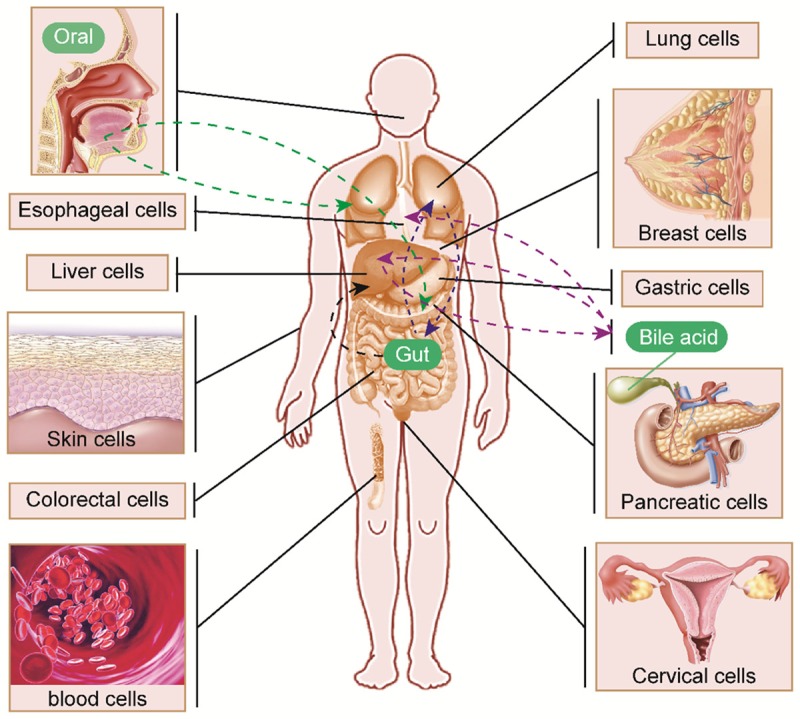
Microbiome-mediated tumorigenesis in different types of human cells. Microbiome alterations in gut and extra-gut systems are associated with cancer initiation and progression. The remote regulation of many axes, including oral-lung/pancreas (green dashed lines), bile acid-esophagus (purple dashed line) and gut-liver axes (black dashed line) as well as dual-directional regulation of the gut-lung (blue dashed lines) and liver-bile acid (purple dashed lines) axes are illustrated.
Translational use of the microbiome in human cancer
An accurate diagnosis and stratification are the initial steps in the treatment of human cancer. Current diagnostic and risk classification methods incorporate clinical features and the combined evaluation of multiple indexes, including endoscopy, pathology, immunology, cytogenetics and molecular biology. However, the early diagnosis of cancer is extremely difficult and remains a substantial challenge for oncologists worldwide. Due to the global alterations in the bacterial communities during the process of carcinogenesis, as reviewed above, these changes may allow researchers to use a noninvasive microbiome analysis as an alternative tool for the diagnosis, risk stratification and screening of the disease. Indeed, several recent enlightening studies have utilized this strategy. For example, gut mucosal microbial communities show distinct alterations across stages of colorectal carcinogenesis [129,130] and in different molecular subtypes of CRC [131]; thus, a profile of the gut or oral microbiome may represent a promising method for predicting CRC [132,133]. Remarkably, gut microbiota profiles have been reported to distinguish pediatric and adolescent patients with ALL upon diagnosis [134]. These findings highlight the potential for developing novel diagnostics based only on stool tests and hopefully the identification of a certain microbiota composition that can be used as a cancer biomarker (Figure 2).
Figure 2.
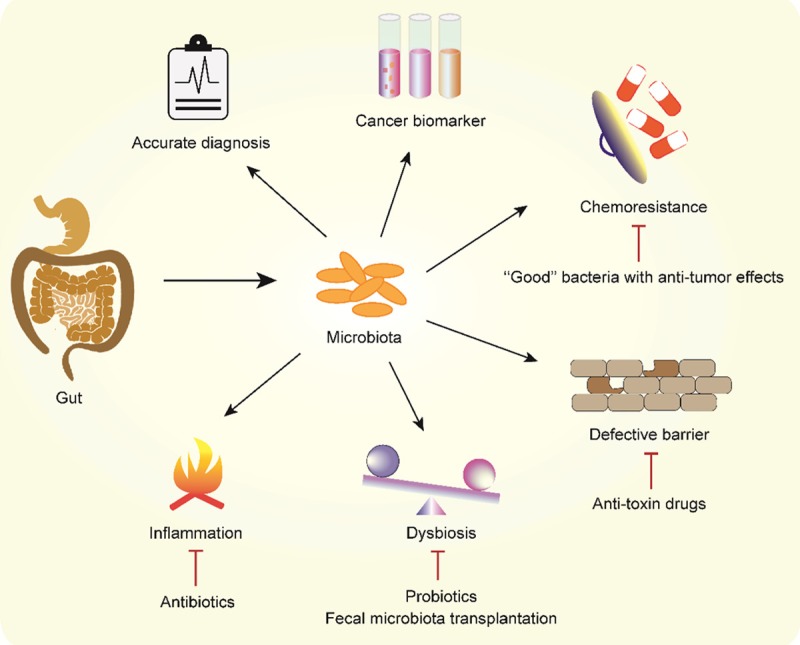
Translational use of the microbiota in human cancer. Global alterations in bacterial communities have the potential to serve as a biomarker in human cancer for an accurate diagnosis, molecular subtype classification and risk stratification. Microbiota-mediated cancer initiation and development are targeted by different mechanisms, including the amelioration of chronic inflammation by antibiotics, the administration of probiotics and fecal microbiota transplantation to rebalance dysbiosis, and the administration of anti-toxin drugs to alleviate defective barrier-induced toxicity. Additional “good” bacteria should be identified and used to prevent chemotherapy resistance.
Due to the important roles of chronic inflammation in cancer initiation and development, the clinical use of antibiotics has become one of the most crucial therapies. One typical example is the use of antibiotics to eradicate Hp infections. During the 1990s, the standard triple therapy included a proton pump inhibitor, clarithromycin and amoxicillin or metronidazole [135]. Because of high resistance to clarithromycin and metronidazole, many modified strategies, such as bismuth quadruple therapy, sequential therapy and hybrid therapy, have emerged [136]. In addition to fighting pathogens using antibiotics, some beneficial living microorganisms themselves have been used as a promising therapy to ameliorate inflammation and rebalance the dysbiosis, based on their effects on modulating host immunity. Most probiotics are derived from bacteria, among which Lactobacillus and Bifidobacterium are the most common species used as probiotics [137]. As expected, their use has extensively proven useful in eradicating certain pathogens [136] or combating different types of cancer [72,138-142]. Nevertheless, some important issues are worth addressing. First, broad-spectrum or persistent antibiotic abuse is one of the most important factors that disrupt commensal microbial homeostasis, thus increasing the susceptibility to certain pathogens, inducing the collapse of the host immune system and even impacting on distal organs [143-145]. In the context of preserving human microbial ecosystems, anti-microbial therapies must be carefully evaluated and reconsidered, and new strategies are urgently needed [145]. Second, despite the effectiveness of probiotics as an anti-tumor therapy, probiotics are not recommended as a single agent for standard treatment, mainly due to their inability to permanently colonize the gut and the lack of profound mechanisms and large-scale studies [146]. In addition to oral administration of probiotics, fecal microbiota transplantation might be the next big innovation in precision medicine [147] (Figure 2).
Last, but not the least, the gut microbiome also ameliorates chemoresistance, which is a major cause of disease relapse and a poor prognosis. Recently, several cutting-edge papers have highlighted the potential of the gut microbiota in conferring drug resistance in cancer. In large cohorts of patients with CRC, F. nucleatum promoted chemoresistance by activating tumor cell autophagy through TLR4- and MYD88-mediated innate immune signaling and specific microRNAs [148], whereas a distinct resistance mechanism was revealed in another study showing that certain bacteria in human pancreatic ductal adenocarcinomas express enzymes capable of metabolizing the cancer chemotherapeutic drug gemcitabine into an inactive form [149]. In contrast, two “good” bacteria, Enterococcus hirae (E. hirae) and Barnesiella intestinihominis (B. intestinihominis), were identified to enhance the anti-tumor effect of cyclophosphamide by increasing the intratumor CD8+ T/Treg ratio and the infiltration of IFN-γ-producing γδT cells [150]. Three recent papers simultaneously published in Science reported that the resident gut bacteria affect patient responses to programmed cell death protein 1 (PD-1)-based immunotherapy, a new FDA-approved treatment. Profiles obtained from patients with lung and kidney cancers indicate that nonresponders have low levels of Akkermansia muciniphila (A. muciniphila); in antibiotic-treated mice, oral supplementation with A. muciniphila restores the efficacy of anti-PD-1 immunotherapy [151]. In two other studies, a greater abundance of “good” bacteria was identified in the guts from patients with melanoma who responded PD-1 blockade, while nonresponders have an imbalance in the gut microbial composition [152,153] (Figure 2).
Once conceived as potential pathogens, the diverse microbial community in the human body has currently been appreciated as a major contributor to the health of the host. As commented, “The next frontier of medicine is to understand the interaction between host factors and the microbiome and to harness this knowledge for therapeutic purpose” [154].
Challenges and prospects
Scientific advances in high-throughput technologies and bioinformatics have promoted a deeper understanding of the human microbiome in healthy and disease states. However, the practical application of this new field is still in its infancy, since the role of the human microbiome in tumorigenesis and its potential for clinical use are only now arousing scientists’ attention. Thus, unsurprisingly, many key questions remain. The main core issue is the complex crosstalk and loops between microbiome alterations and cancer. First, identical human cancers may correlate with diverse forms of dysbiosis, and the same changes in individual bacterial species seem to induce different types of cancers, implying a microbe- and tissue-specific mechanism of tumorigenesis. Second, a specific bacterial pathogen may disrupt the microbial composition, facilitating tumor initiation or progression, which in turn aggravates the bacterial dysbiosis in the host, leading to a “chicken-and-egg” debate. Third, the remote dual-directional regulation of many axes, such as the gut-lung, gut-liver and oral-pancreas axes, forms an inextricably interwoven network in the human body, and thus regulatory loops within the host are far more complicated. More importantly, the precise mechanisms such as which and how bacteria modulate the immune cells remain unaddressed. These issues have essentially impeded the progress of the scientific studies in this field; subsequently, a number of reports have superficially examined the associations rather than elucidating the casual mechanisms underlying the microbiome and tumorigenesis, and longitudinal studies are lacking [155]. In addition, many efforts are currently directed at the gut, which harbors most of our microbes; however, the extra-gut microbiome is poorly understood. The translation of many findings from murine models to humans seems to the most difficult part to accomplish [155]. In particular, the greatest challenge regarding the use of gut microbes in fighting against human cancer is yet to come. For example, fecal microbiota transplantation is very likely to change one individual’s bacterial composition that might predispose them to other heath problems, thus leading to unexpected effects; furthermore, whether a single or several types of bacterial species can help patients against cancer is still uncertain [156]. Despite these limitations and challenges, the modern concept of balancing the microbiome in the whole body intriguingly resembles the conventional philosophy of “harmony” from traditional Chinese medicine in terms of preventing or combating diseases. This novel perspective adds a new layer to the understanding of how microorganisms normally maintain human health and the mechanisms by which the disrupted microbiome supports cancer initiation and progression.
In summary, the HMP is not simply an extension of the HGP, but rather a summation of many inspiring and fundamental questions. Many outcomes of the HMP may provide us with more individualized strategies, ranging from new diagnostic biomarkers of disease to potential clinical applications [157]. As a forgotten organ that was once overlooked in the human body [158], the microbiome has opened a new era in an understanding of the mechanisms underlying cancer and will not be neglected in future studies. We expect that microbiome modulations will become a potentially major translational strategy in future clinical therapies. Strategies targeting the crosstalk among the microbiome, immune system, host metabolism and organ development is most likely a new impetus and the next breakthrough in precision medicine.
Acknowledgements
This work is supported by the Key Project from Institute of Medical Biology, Chinese Academy of Medical Sciences and Peking Union Medical College to H.Z.
Disclosure of conflict of interest
None.
References
- 1.Lander ES. Initial impact of the sequencing of the human genome. Nature. 2011;470:187–97. doi: 10.1038/nature09792. [DOI] [PubMed] [Google Scholar]
- 2.Garg AD, Agostinis P. Cell death and immunity in cancer: from danger signals to mimicry of pathogen defense responses. Immunol Rev. 2017;280:126–48. doi: 10.1111/imr.12574. [DOI] [PubMed] [Google Scholar]
- 3.Koch R. An Address on bacteriological research. Br Med J. 1890;2:380–3. doi: 10.1136/bmj.2.1546.380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schwabe RF, Jobin C. The microbiome and cancer. Nat Rev Cancer. 2013;13:800–12. doi: 10.1038/nrc3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ley RE, Lozupone CA, Hamady M, Knight R, Gordon JI. Worlds within worlds: evolution of the vertebrate gut microbiota. Nat Rev Microbiol. 2008;6:776–88. doi: 10.1038/nrmicro1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Proctor LM. The national institutes of health human microbiome project. Semin Fetal Neonatal Med. 2016;21:368–72. doi: 10.1016/j.siny.2016.05.002. [DOI] [PubMed] [Google Scholar]
- 7.Cell Editorial T. Living in their world. Cell. 2018;172:1137–8. [Google Scholar]
- 8.Brown LM. Helicobacter pylori: epidemiology and routes of transmission. Epidemiol Rev. 2000;22:283–97. doi: 10.1093/oxfordjournals.epirev.a018040. [DOI] [PubMed] [Google Scholar]
- 9.Polk DB, Peek RM Jr. Helicobacter pylori: gastric cancer and beyond. Nat Rev Cancer. 2010;10:403–14. doi: 10.1038/nrc2857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Uemura N, Okamoto S, Yamamoto S, Matsumura N, Yamaguchi S, Yamakido M, Taniyama K, Sasaki N, Schlemper RJ. Helicobacter pylori infection and the development of gastric cancer. N Engl J Med. 2001;345:784–9. doi: 10.1056/NEJMoa001999. [DOI] [PubMed] [Google Scholar]
- 11.Gebert B, Fischer W, Weiss E, Hoffmann R, Haas R. Helicobacter pylori vacuolating cytotoxin inhibits T lymphocyte activation. Science. 2003;301:1099–102. doi: 10.1126/science.1086871. [DOI] [PubMed] [Google Scholar]
- 12.Sundrud MS, Torres VJ, Unutmaz D, Cover TL. Inhibition of primary human T cell proliferation by Helicobacter pylori vacuolating toxin (VacA) is independent of VacA effects on IL-2 secretion. Proc Natl Acad Sci U S A. 2004;101:7727–32. doi: 10.1073/pnas.0401528101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ohnishi N, Yuasa H, Tanaka S, Sawa H, Miura M, Matsui A, Higashi H, Musashi M, Iwabuchi K, Suzuki M, Yamada G, Azuma T, Hatakeyama M. Transgenic expression of helicobacter pylori CagA induces gastrointestinal and hematopoietic neoplasms in mouse. Proc Natl Acad Sci U S A. 2008;105:1003–08. doi: 10.1073/pnas.0711183105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Odenbreit S, Puls J, Sedlmaier B, Gerland E, Fischer W, Haas R. Translocation of helicobacter pylori CagA into gastric epithelial cells by type IV secretion. Science. 2000;287:1497–500. doi: 10.1126/science.287.5457.1497. [DOI] [PubMed] [Google Scholar]
- 15.Selbach M, Moese S, Hauck CR, Meyer TF, Backert S. Src is the kinase of the helicobacter pylori CagA protein in vitro and in vivo. J Biol Chem. 2002;277:6775–8. doi: 10.1074/jbc.C100754200. [DOI] [PubMed] [Google Scholar]
- 16.Stein M, Bagnoli F, Halenbeck R, Rappuoli R, Fantl WJ, Covacci A. c-Src/Lyn kinases activate helicobacter pylori CagA through tyrosine phosphorylation of the EPIYA motifs. Mol Microbiol. 2002;43:971–80. doi: 10.1046/j.1365-2958.2002.02781.x. [DOI] [PubMed] [Google Scholar]
- 17.Tammer I, Brandt S, Hartig R, Konig W, Backert S. Activation of Abl by helicobacter pylori: a novel kinase for CagA and crucial mediator of host cell scattering. Gastroenterology. 2007;132:1309–19. doi: 10.1053/j.gastro.2007.01.050. [DOI] [PubMed] [Google Scholar]
- 18.Higashi H, Tsutsumi R, Muto S, Sugiyama T, Azuma T, Asaka M, Hatakeyama M. SHP-2 tyrosine phosphatase as an intracellular target of helicobacter pylori CagA protein. Science. 2002;295:683–6. doi: 10.1126/science.1067147. [DOI] [PubMed] [Google Scholar]
- 19.Higashi H, Nakaya A, Tsutsumi R, Yokoyama K, Fujii Y, Ishikawa S, Higuchi M, Takahashi A, Kurashima Y, Teishikata Y, Tanaka S, Azuma T, Hatakeyama M. Helicobacter pylori CagA induces Ras-independent morphogenetic response through SHP-2 recruitment and activation. J Biol Chem. 2004;279:17205–16. doi: 10.1074/jbc.M309964200. [DOI] [PubMed] [Google Scholar]
- 20.Mimuro H, Suzuki T, Tanaka J, Asahi M, Haas R, Sasakawa C. Grb2 is a key mediator of helicobacter pylori CagA protein activities. Mol Cell. 2002;10:745–55. doi: 10.1016/s1097-2765(02)00681-0. [DOI] [PubMed] [Google Scholar]
- 21.Brandt S, Kwok T, Hartig R, Konig W, Backert S. NF-kappaB activation and potentiation of proinflammatory responses by the Helicobacter pylori CagA protein. Proc Natl Acad Sci U S A. 2005;102:9300–5. doi: 10.1073/pnas.0409873102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Murata-Kamiya N, Kurashima Y, Teishikata Y, Yamahashi Y, Saito Y, Higashi H, Aburatani H, Akiyama T, Peek RM Jr, Azuma T, Hatakeyama M. Helicobacter pylori CagA interacts with E-cadherin and deregulates the beta-catenin signal that promotes intestinal transdifferentiation in gastric epithelial cells. Oncogene. 2007;26:4617–26. doi: 10.1038/sj.onc.1210251. [DOI] [PubMed] [Google Scholar]
- 23.Suzuki M, Mimuro H, Kiga K, Fukumatsu M, Ishijima N, Morikawa H, Nagai S, Koyasu S, Gilman RH, Kersulyte D, Berg DE, Sasakawa C. Helicobacter pylori CagA phosphorylation-independent function in epithelial proliferation and inflammation. Cell Host Microbe. 2009;5:23–34. doi: 10.1016/j.chom.2008.11.010. [DOI] [PubMed] [Google Scholar]
- 24.Kim SY, Lee YC, Kim HK, Blaser MJ. Helicobacter pylori CagA transfection of gastric epithelial cells induces interleukin-8. Cell Microbiol. 2006;8:97–106. doi: 10.1111/j.1462-5822.2005.00603.x. [DOI] [PubMed] [Google Scholar]
- 25.Tu S, Bhagat G, Cui G, Takaishi S, Kurt-Jones EA, Rickman B, Betz KS, Penz-Oesterreicher M, Bjorkdahl O, Fox JG, Wang TC. Overexpression of interleukin-1beta induces gastric inflammation and cancer and mobilizes myeloid-derived suppressor cells in mice. Cancer Cell. 2008;14:408–19. doi: 10.1016/j.ccr.2008.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.El-Omar EM, Rabkin CS, Gammon MD, Vaughan TL, Risch HA, Schoenberg JB, Stanford JL, Mayne ST, Goedert J, Blot WJ, Fraumeni JF Jr, Chow WH. Increased risk of noncardia gastric cancer associated with proinflammatory cytokine gene polymorphisms. Gastroenterology. 2003;124:1193–201. doi: 10.1016/s0016-5085(03)00157-4. [DOI] [PubMed] [Google Scholar]
- 27.Kanda N, Seno H, Konda Y, Marusawa H, Kanai M, Nakajima T, Kawashima T, Nanakin A, Sawabu T, Uenoyama Y, Sekikawa A, Kawada M, Suzuki K, Kayahara T, Fukui H, Sawada M, Chiba T. STAT3 is constitutively activated and supports cell survival in association with survivin expression in gastric cancer cells. Oncogene. 2004;23:4921–9. doi: 10.1038/sj.onc.1207606. [DOI] [PubMed] [Google Scholar]
- 28.Kanda K, Komekado H, Sawabu T, Ishizu S, Nakanishi Y, Nakatsuji M, Akitake-Kawano R, Ohno M, Hiraoka Y, Kawada M, Kawada K, Sakai Y, Matsumoto K, Kunichika M, Kimura T, Seno H, Nishi E, Chiba T. Nardilysin and ADAM proteases promote gastric cancer cell growth by activating intrinsic cytokine signalling via enhanced ectodomain shedding of TNF-alpha. EMBO Mol Med. 2012;4:396–411. doi: 10.1002/emmm.201200216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oguma K, Oshima H, Aoki M, Uchio R, Naka K, Nakamura S, Hirao A, Saya H, Taketo MM, Oshima M. Activated macrophages promote Wnt signalling through tumour necrosis factoralpha in gastric tumour cells. EMBO J. 2008;27:1671–81. doi: 10.1038/emboj.2008.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Houghton J, Stoicov C, Nomura S, Rogers AB, Carlson J, Li H, Cai X, Fox JG, Goldenring JR, Wang TC. Gastric cancer originating from bone marrow-derived cells. Science. 2004;306:1568–71. doi: 10.1126/science.1099513. [DOI] [PubMed] [Google Scholar]
- 31.Islami F, Kamangar F. Helicobacter pylori and esophageal cancer risk: a meta-analysis. Cancer Prev Res (Phila) 2008;1:329–38. doi: 10.1158/1940-6207.CAPR-08-0109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xie FJ, Zhang YP, Zheng QQ, Jin HC, Wang FL, Chen M, Shao L, Zou DH, Yu XM, Mao WM. Helicobacter pylori infection and esophageal cancer risk: an updated meta-analysis. World J Gastroenterol. 2013;19:6098–107. doi: 10.3748/wjg.v19.i36.6098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hollander D, Vadheim CM, Brettholz E, Petersen GM, Delahunty T, Rotter JI. Increased intestinal permeability in patients with Crohn’s disease and their relatives. A possible etiologic factor. Ann Intern Med. 1986;105:883–5. doi: 10.7326/0003-4819-105-6-883. [DOI] [PubMed] [Google Scholar]
- 34.Bergstrom K, Liu X, Zhao Y, Gao N, Wu Q, Song K, Cui Y, Li Y, McDaniel JM, McGee S, Chen W, Huycke MM, Houchen CW, Zenewicz LA, West CM, Chen H, Braun J, Fu J, Xia L. Defective intestinal mucin-type O-glycosylation causes spontaneous colitis-associated cancer in mice. Gastroenterology. 2016;151:152–64. doi: 10.1053/j.gastro.2016.03.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kostic AD, Gevers D, Pedamallu CS, Michaud M, Duke F, Earl AM, Ojesina AI, Jung J, Bass AJ, Tabernero J, Baselga J, Liu C, Shivdasani RA, Ogino S, Birren BW, Huttenhower C, Garrett WS, Meyerson M. Genomic analysis identifies association of fusobacterium with colorectal carcinoma. Genome Res. 2012;22:292–8. doi: 10.1101/gr.126573.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Castellarin M, Warren RL, Freeman JD, Dreolini L, Krzywinski M, Strauss J, Barnes R, Watson P, Allen-Vercoe E, Moore RA, Holt RA. Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome Res. 2012;22:299–306. doi: 10.1101/gr.126516.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kostic AD, Chun E, Robertson L, Glickman JN, Gallini CA, Michaud M, Clancy TE, Chung DC, Lochhead P, Hold GL, El-Omar EM, Brenner D, Fuchs CS, Meyerson M, Garrett WS. Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host Microbe. 2013;14:207–15. doi: 10.1016/j.chom.2013.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rubinstein MR, Wang X, Liu W, Hao Y, Cai G, Han YW. Fusobacterium nucleatum promotes colorectal carcinogenesis by modulating E-cadherin/beta-catenin signaling via its FadA adhesin. Cell Host Microbe. 2013;14:195–206. doi: 10.1016/j.chom.2013.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gur C, Ibrahim Y, Isaacson B, Yamin R, Abed J, Gamliel M, Enk J, Bar-On Y, Stanietsky-Kaynan N, Coppenhagen-Glazer S, Shussman N, Almogy G, Cuapio A, Hofer E, Mevorach D, Tabib A, Ortenberg R, Markel G, Miklic K, Jonjic S, Brennan CA, Garrett WS, Bachrach G, Mandelboim O. Binding of the Fap2 protein of fusobacterium nucleatum to human inhibitory receptor TIGIT protects tumors from immune cell attack. Immunity. 2015;42:344–55. doi: 10.1016/j.immuni.2015.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Abed J, Emgard JE, Zamir G, Faroja M, Almogy G, Grenov A, Sol A, Naor R, Pikarsky E, Atlan KA, Mellul A, Chaushu S, Manson AL, Earl AM, Ou N, Brennan CA, Garrett WS, Bachrach G. Fap2 mediates fusobacterium nucleatum colorectal adenocarcinoma enrichment by binding to tumor-expressed Gal-GalNAc. Cell Host Microbe. 2016;20:215–25. doi: 10.1016/j.chom.2016.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Arthur JC, Perez-Chanona E, Muhlbauer M, Tomkovich S, Uronis JM, Fan TJ, Campbell BJ, Abujamel T, Dogan B, Rogers AB, Rhodes JM, Stintzi A, Simpson KW, Hansen JJ, Keku TO, Fodor AA, Jobin C. Intestinal inflammation targets cancer-inducing activity of the microbiota. Science. 2012;338:120–3. doi: 10.1126/science.1224820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Boleij A, Hechenbleikner EM, Goodwin AC, Badani R, Stein EM, Lazarev MG, Ellis B, Carroll KC, Albesiano E, Wick EC, Platz EA, Pardoll DM, Sears CL. The Bacteroides fragilis toxin gene is prevalent in the colon mucosa of colorectal cancer patients. Clin Infect Dis. 2015;60:208–15. doi: 10.1093/cid/ciu787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Guidi R, Guerra L, Levi L, Stenerlow B, Fox JG, Josenhans C, Masucci MG, Frisan T. Chronic exposure to the cytolethal distending toxins of Gram-negative bacteria promotes genomic instability and altered DNA damage response. Cell Microbiol. 2013;15:98–113. doi: 10.1111/cmi.12034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Westbrook AM, Wei B, Braun J, Schiestl RH. Intestinal mucosal inflammation leads to systemic genotoxicity in mice. Cancer Res. 2009;69:4827–34. doi: 10.1158/0008-5472.CAN-08-4416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Westbrook AM, Wei B, Braun J, Schiestl RH. Intestinal inflammation induces genotoxicity to extraintestinal tissues and cell types in mice. Int J Cancer. 2011;129:1815–25. doi: 10.1002/ijc.26146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Snider EJ, Freedberg DE, Abrams JA. Potential role of the microbiome in barrett’s esophagus and esophageal adenocarcinoma. Dig Dis Sci. 2016;61:2217–25. doi: 10.1007/s10620-016-4155-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rustgi AK, El-Serag HB. Esophageal carcinoma. N Engl J Med. 2014;371:2499–509. doi: 10.1056/NEJMra1314530. [DOI] [PubMed] [Google Scholar]
- 48.Yang L, Lu X, Nossa CW, Francois F, Peek RM, Pei Z. Inflammation and intestinal metaplasia of the distal esophagus are associated with alterations in the microbiome. Gastroenterology. 2009;137:588–97. doi: 10.1053/j.gastro.2009.04.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu N, Ando T, Ishiguro K, Maeda O, Watanabe O, Funasaka K, Nakamura M, Miyahara R, Ohmiya N, Goto H. Characterization of bacterial biota in the distal esophagus of Japanese patients with reflux esophagitis and Barrett’s esophagus. BMC Infect Dis. 2013;13:130. doi: 10.1186/1471-2334-13-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gall A, Fero J, McCoy C, Claywell BC, Sanchez CA, Blount PL, Li X, Vaughan TL, Matsen FA, Reid BJ, Salama NR. Bacterial composition of the human upper gastrointestinal tract microbiome is dynamic and associated with genomic instability in a barrett’s esophagus cohort. PLoS One. 2015;10:e0129055. doi: 10.1371/journal.pone.0129055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Narikiyo M, Tanabe C, Yamada Y, Igaki H, Tachimori Y, Kato H, Muto M, Montesano R, Sakamoto H, Nakajima Y, Sasaki H. Frequent and preferential infection of treponema denticola, streptococcus mitis, and streptococcus anginosus in esophageal cancers. Cancer Sci. 2004;95:569–74. doi: 10.1111/j.1349-7006.2004.tb02488.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Blackett KL, Siddhi SS, Cleary S, Steed H, Miller MH, Macfarlane S, Macfarlane GT, Dillon JF. Oesophageal bacterial biofilm changes in gastro-oesophageal reflux disease, barrett’s and oesophageal carcinoma: association or causality? Aliment Pharmacol Ther. 2013;37:1084–92. doi: 10.1111/apt.12317. [DOI] [PubMed] [Google Scholar]
- 53.Zaidi AH, Kelly LA, Kreft RE, Barlek M, Omstead AN, Matsui D, Boyd NH, Gazarik KE, Heit MI, Nistico L, Kasi PM, Spirk TL, Byers B, Lloyd EJ, Landreneau RJ, Jobe BA. Associations of microbiota and toll-like receptor signaling pathway in esophageal adenocarcinoma. BMC Cancer. 2016;16:52. doi: 10.1186/s12885-016-2093-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Huhta H, Helminen O, Lehenkari PP, Saarnio J, Karttunen TJ, Kauppila JH. Toll-like receptors 1, 2, 4 and 6 in esophageal epithelium, barrett’s esophagus, dysplasia and adenocarcinoma. Oncotarget. 2016;7:23658–67. doi: 10.18632/oncotarget.8151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Peters BA, Wu J, Pei Z, Yang L, Purdue MP, Freedman ND, Jacobs EJ, Gapstur SM, Hayes RB, Ahn J. Oral microbiome composition reflects prospective risk for esophageal cancers. Cancer Res. 2017;77:6777–87. doi: 10.1158/0008-5472.CAN-17-1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Charlson ES, Bittinger K, Haas AR, Fitzgerald AS, Frank I, Yadav A, Bushman FD, Collman RG. Topographical continuity of bacterial populations in the healthy human respiratory tract. Am J Respir Crit Care Med. 2011;184:957–63. doi: 10.1164/rccm.201104-0655OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Huang YJ, Kim E, Cox MJ, Brodie EL, Brown R, Wiener-Kronish JP, Lynch SV. A persistent and diverse airway microbiota present during chronic obstructive pulmonary disease exacerbations. OMICS. 2010;14:9–59. doi: 10.1089/omi.2009.0100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Erb-Downward JR, Thompson DL, Han MK, Freeman CM, McCloskey L, Schmidt LA, Young VB, Toews GB, Curtis JL, Sundaram B, Martinez FJ, Huffnagle GB. Analysis of the lung microbiome in the “healthy” smoker and in COPD. PLoS One. 2011;6:e16384. doi: 10.1371/journal.pone.0016384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sze MA, Dimitriu PA, Hayashi S, Elliott WM, McDonough JE, Gosselink JV, Cooper J, Sin DD, Mohn WW, Hogg JC. The lung tissue microbiome in chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2012;185:1073–80. doi: 10.1164/rccm.201111-2075OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hilty M, Burke C, Pedro H, Cardenas P, Bush A, Bossley C, Davies J, Ervine A, Poulter L, Pachter L, Moffatt MF, Cookson WO. Disordered microbial communities in asthmatic airways. PLoS One. 2010;5:e8578. doi: 10.1371/journal.pone.0008578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Molyneaux PL, Cox MJ, Willis-Owen SA, Mallia P, Russell KE, Russell AM, Murphy E, Johnston SL, Schwartz DA, Wells AU, Cookson WO, Maher TM, Moffatt MF. The role of bacteria in the pathogenesis and progression of idiopathic pulmonary fibrosis. Am J Respir Crit Care Med. 2014;190:906–13. doi: 10.1164/rccm.201403-0541OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yu G, Gail MH, Consonni D, Carugno M, Humphrys M, Pesatori AC, Caporaso NE, Goedert JJ, Ravel J, Landi MT. Characterizing human lung tissue microbiota and its relationship to epidemiological and clinical features. Genome Biol. 2016;17:163. doi: 10.1186/s13059-016-1021-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lee SH, Sung JY, Yong D, Chun J, Kim SY, Song JH, Chung KS, Kim EY, Jung JY, Kang YA, Kim YS, Kim SK, Chang J, Park MS. Characterization of microbiome in bronchoalveolar lavage fluid of patients with lung cancer comparing with benign mass like lesions. Lung Cancer. 2016;102:89–95. doi: 10.1016/j.lungcan.2016.10.016. [DOI] [PubMed] [Google Scholar]
- 64.de Steenhuijsen Piters WA, Sanders EA, Bogaert D. The role of the local microbial ecosystem in respiratory health and disease. Philos Trans R Soc Lond B Biol Sci. 2015:370. doi: 10.1098/rstb.2014.0294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schreiber H, Nettesheim P, Lijinsky W, Richter CB, Walburg HE Jr. Induction of lung cancer in germfree, specific-pathogen-free, and infected rats by N-nitrosoheptamethyleneimine: enhancement by respiratory infection. J Natl Cancer Inst. 1972;49:1107–14. [PubMed] [Google Scholar]
- 66.Melkamu T, Qian X, Upadhyaya P, O’Sullivan MG, Kassie F. Lipopolysaccharide enhances mouse lung tumorigenesis: a model for inflammation-driven lung cancer. Vet Pathol. 2013;50:895–902. doi: 10.1177/0300985813476061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cheng M, Qian L, Shen G, Bian G, Xu T, Xu W, Shen G, Hu S. Microbiota modulate tumoral immune surveillance in lung through a gammadeltaT17 immune cell-dependent mechanism. Cancer Res. 2014;74:4030–41. doi: 10.1158/0008-5472.CAN-13-2462. [DOI] [PubMed] [Google Scholar]
- 68.Boursi B, Mamtani R, Haynes K, Yang YX. Recurrent antibiotic exposure may promote cancer formation--another step in understanding the role of the human microbiota? Eur J Cancer. 2015;51:2655–64. doi: 10.1016/j.ejca.2015.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yan X, Yang M, Liu J, Gao R, Hu J, Li J, Zhang L, Shi Y, Guo H, Cheng J, Razi M, Pang S, Yu X, Hu S. Discovery and validation of potential bacterial biomarkers for lung cancer. Am J Cancer Res. 2015;5:3111–22. [PMC free article] [PubMed] [Google Scholar]
- 70.Cameron SJS, Lewis KE, Huws SA, Hegarty MJ, Lewis PD, Pachebat JA, Mur LAJ. A pilot study using metagenomic sequencing of the sputum microbiome suggests potential bacterial biomarkers for lung cancer. PLoS One. 2017;12:e0177062. doi: 10.1371/journal.pone.0177062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Budden KF, Gellatly SL, Wood DL, Cooper MA, Morrison M, Hugenholtz P, Hansbro PM. Emerging pathogenic links between microbiota and the gut-lung axis. Nat Rev Microbiol. 2017;15:55–63. doi: 10.1038/nrmicro.2016.142. [DOI] [PubMed] [Google Scholar]
- 72.Bingula R, Filaire M, Radosevic-Robin N, Bey M, Berthon JY, Bernalier-Donadille A, Vasson MP, Filaire E. Desired turbulence? Gut-lung axis, immunity, and lung cancer. J Oncol. 2017;2017:5035371. doi: 10.1155/2017/5035371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Dickson RP, Singer BH, Newstead MW, Falkowski NR, Erb-Downward JR, Standiford TJ, Huffnagle GB. Enrichment of the lung microbiome with gut bacteria in sepsis and the acute respiratory distress syndrome. Nat Microbiol. 2016;1:16113. doi: 10.1038/nmicrobiol.2016.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sze MA, Tsuruta M, Yang SW, Oh Y, Man SF, Hogg JC, Sin DD. Changes in the bacterial microbiota in gut, blood, and lungs following acute LPS instillation into mice lungs. PLoS One. 2014;9:e111228. doi: 10.1371/journal.pone.0111228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Faner R, Sibila O, Agusti A, Bernasconi E, Chalmers JD, Huffnagle GB, Manichanh C, Molyneaux PL, Paredes R, Perez Brocal V, Ponomarenko J, Sethi S, Dorca J, Monso E. The microbiome in respiratory medicine: current challenges and future perspectives. Eur Respir J. 2017:49. doi: 10.1183/13993003.02086-2016. [DOI] [PubMed] [Google Scholar]
- 76.Ward JM, Fox JG, Anver MR, Haines DC, George CV, Collins MJ Jr, Gorelick PL, Nagashima K, Gonda MA, Gilden RV, et al. Chronic active hepatitis and associated liver tumors in mice caused by a persistent bacterial infection with a novel Helicobacter species. J Natl Cancer Inst. 1994;86:1222–7. doi: 10.1093/jnci/86.16.1222. [DOI] [PubMed] [Google Scholar]
- 77.Kleine M, Worbs T, Schrem H, Vondran FW, Kaltenborn A, Klempnauer J, Forster R, Josenhans C, Suerbaum S, Bektas H. Helicobacter hepaticus induces an inflammatory response in primary human hepatocytes. PLoS One. 2014;9:e99713. doi: 10.1371/journal.pone.0099713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Fattovich G, Stroffolini T, Zagni I, Donato F. Hepatocellular carcinoma in cirrhosis: incidence and risk factors. Gastroenterology. 2004;127:S35–50. doi: 10.1053/j.gastro.2004.09.014. [DOI] [PubMed] [Google Scholar]
- 79.Cirera I, Bauer TM, Navasa M, Vila J, Grande L, Taura P, Fuster J, Garcia-Valdecasas JC, Lacy A, Suarez MJ, Rimola A, Rodes J. Bacterial translocation of enteric organisms in patients with cirrhosis. J Hepatol. 2001;34:32–7. doi: 10.1016/s0168-8278(00)00013-1. [DOI] [PubMed] [Google Scholar]
- 80.Dapito DH, Mencin A, Gwak GY, Pradere JP, Jang MK, Mederacke I, Caviglia JM, Khiabanian H, Adeyemi A, Bataller R, Lefkowitch JH, Bower M, Friedman R, Sartor RB, Rabadan R, Schwabe RF. Promotion of hepatocellular carcinoma by the intestinal microbiota and TLR4. Cancer Cell. 2012;21:504–16. doi: 10.1016/j.ccr.2012.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Jia W, Xie G, Jia W. Bile acid-microbiota crosstalk in gastrointestinal inflammation and carcinogenesis. Nat Rev Gastroenterol Hepatol. 2018;15:111–28. doi: 10.1038/nrgastro.2017.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yoshimoto S, Loo TM, Atarashi K, Kanda H, Sato S, Oyadomari S, Iwakura Y, Oshima K, Morita H, Hattori M, Honda K, Ishikawa Y, Hara E, Ohtani N. Obesity-induced gut microbial metabolite promotes liver cancer through senescence secretome. Nature. 2013;499:97–101. doi: 10.1038/nature12347. [DOI] [PubMed] [Google Scholar]
- 83.Bernstein C, Holubec H, Bhattacharyya AK, Nguyen H, Payne CM, Zaitlin B, Bernstein H. Carcinogenicity of deoxycholate, a secondary bile acid. Arch Toxicol. 2011;85:863–71. doi: 10.1007/s00204-011-0648-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Quante M, Bhagat G, Abrams JA, Marache F, Good P, Lee MD, Lee Y, Friedman R, Asfaha S, Dubeykovskaya Z, Mahmood U, Figueiredo JL, Kitajewski J, Shawber C, Lightdale CJ, Rustgi AK, Wang TC. Bile acid and inflammation activate gastric cardia stem cells in a mouse model of barrett-like metaplasia. Cancer Cell. 2012;21:36–51. doi: 10.1016/j.ccr.2011.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Von Hoff DD, Korn R, Mousses S. Pancreatic cancer--could it be that simple? A different context of vulnerability. Cancer Cell. 2009;16:7–8. doi: 10.1016/j.ccr.2009.06.011. [DOI] [PubMed] [Google Scholar]
- 86.Lindkvist B, Johansen D, Borgstrom A, Manjer J. A prospective study of helicobacter pylori in relation to the risk for pancreatic cancer. BMC Cancer. 2008;8:321. doi: 10.1186/1471-2407-8-321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Trikudanathan G, Philip A, Dasanu CA, Baker WL. Association between helicobacter pylori infection and pancreatic cancer. A cumulative meta-analysis. JOP. 2011;12:26–31. [PubMed] [Google Scholar]
- 88.Ertz-Archambault N, Keim P, Von Hoff D. Microbiome and pancreatic cancer: a comprehensive topic review of literature. World J Gastroenterol. 2017;23:1899–908. doi: 10.3748/wjg.v23.i10.1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Farrell JJ, Zhang L, Zhou H, Chia D, Elashoff D, Akin D, Paster BJ, Joshipura K, Wong DT. Variations of oral microbiota are associated with pancreatic diseases including pancreatic cancer. Gut. 2012;61:582–8. doi: 10.1136/gutjnl-2011-300784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Michaud DS, Izard J, Wilhelm-Benartzi CS, You DH, Grote VA, Tjonneland A, Dahm CC, Overvad K, Jenab M, Fedirko V, Boutron-Ruault MC, Clavel-Chapelon F, Racine A, Kaaks R, Boeing H, Foerster J, Trichopoulou A, Lagiou P, Trichopoulos D, Sacerdote C, Sieri S, Palli D, Tumino R, Panico S, Siersema PD, Peeters PH, Lund E, Barricarte A, Huerta JM, Molina-Montes E, Dorronsoro M, Quiros JR, Duell EJ, Ye W, Sund M, Lindkvist B, Johansen D, Khaw KT, Wareham N, Travis RC, Vineis P, Bueno-de-Mesquita HB, Riboli E. Plasma antibodies to oral bacteria and risk of pancreatic cancer in a large European prospective cohort study. Gut. 2013;62:1764–70. doi: 10.1136/gutjnl-2012-303006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Mitsuhashi K, Nosho K, Sukawa Y, Matsunaga Y, Ito M, Kurihara H, Kanno S, Igarashi H, Naito T, Adachi Y, Tachibana M, Tanuma T, Maguchi H, Shinohara T, Hasegawa T, Imamura M, Kimura Y, Hirata K, Maruyama R, Suzuki H, Imai K, Yamamoto H, Shinomura Y. Association of fusobacterium species in pancreatic cancer tissues with molecular features and prognosis. Oncotarget. 2015;6:7209–20. doi: 10.18632/oncotarget.3109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Fan X, Alekseyenko AV, Wu J, Peters BA, Jacobs EJ, Gapstur SM, Purdue MP, Abnet CC, Stolzenberg-Solomon R, Miller G, Ravel J, Hayes RB, Ahn J. Human oral microbiome and prospective risk for pancreatic cancer: a population-based nested case-control study. Gut. 2018;67:120–7. doi: 10.1136/gutjnl-2016-312580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Slots J, Rams TE. New views on periodontal microbiota in special patient categories. J Clin Periodontol. 1991;18:411–20. doi: 10.1111/j.1600-051x.1991.tb02309.x. [DOI] [PubMed] [Google Scholar]
- 94.van Vliet MJ, Tissing WJ, Dun CA, Meessen NE, Kamps WA, de Bont ES, Harmsen HJ. Chemotherapy treatment in pediatric patients with acute myeloid leukemia receiving antimicrobial prophylaxis leads to a relative increase of colonization with potentially pathogenic bacteria in the gut. Clin Infect Dis. 2009;49:262–70. doi: 10.1086/599346. [DOI] [PubMed] [Google Scholar]
- 95.Wang Y, Xue J, Zhou X, You M, Du Q, Yang X, He J, Zou J, Cheng L, Li M, Li Y, Zhu Y, Li J, Shi W, Xu X. Oral microbiota distinguishes acute lymphoblastic leukemia pediatric hosts from healthy populations. PLoS One. 2014;9:e102116. doi: 10.1371/journal.pone.0102116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Bai L, Zhou P, Li D, Ju X. Changes in the gastrointestinal microbiota of children with acute lymphoblastic leukaemia and its association with antibiotics in the short term. J Med Microbiol. 2017 doi: 10.1099/jmm.0.000568. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 97.Chua LL, Rajasuriar R, Azanan MS, Abdullah NK, Tang MS, Lee SC, Woo YL, Lim YA, Ariffin H, Loke P. Reduced microbial diversity in adult survivors of childhood acute lymphoblastic leukemia and microbial associations with increased immune activation. Microbiome. 2017;5:35. doi: 10.1186/s40168-017-0250-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Yamamoto ML, Maier I, Dang AT, Berry D, Liu J, Ruegger PM, Yang JI, Soto PA, Presley LL, Reliene R, Westbrook AM, Wei B, Loy A, Chang C, Braun J, Borneman J, Schiestl RH. Intestinal bacteria modify lymphoma incidence and latency by affecting systemic inflammatory state, oxidative stress, and leukocyte genotoxicity. Cancer Res. 2013;73:4222–32. doi: 10.1158/0008-5472.CAN-13-0022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Isaacson PG, Du MQ. MALT lymphoma: from morphology to molecules. Nat Rev Cancer. 2004;4:644–53. doi: 10.1038/nrc1409. [DOI] [PubMed] [Google Scholar]
- 100.Enno A, O’Rourke JL, Howlett CR, Jack A, Dixon MF, Lee A. MALToma-like lesions in the murine gastric mucosa after long-term infection with Helicobacter felis. A mouse model of Helicobacter pylori-induced gastric lymphoma. Am J Pathol. 1995;147:217–22. [PMC free article] [PubMed] [Google Scholar]
- 101.O’Rourke JL, Dixon MF, Jack A, Enno A, Lee A. Gastric B-cell mucosa-associated lymphoid tissue (MALT) lymphoma in an animal model of ‘Helicobacter heilmannii’ infection. J Pathol. 2004;203:896–903. doi: 10.1002/path.1593. [DOI] [PubMed] [Google Scholar]
- 102.Suarez F, Lortholary O, Hermine O, Lecuit M. Infection-associated lymphomas derived from marginal zone B cells: a model of antigen-driven lymphoproliferation. Blood. 2006;107:3034–44. doi: 10.1182/blood-2005-09-3679. [DOI] [PubMed] [Google Scholar]
- 103.Suzuki A, Kobayashi M, Matsuda K, Matsumoto T, Kawakubo M, Kumazawa S, Koide N, Miyagawa S, Ota H. Induction of high endothelial venule-like vessels expressing GlcNAc6ST-1-mediated L-selectin ligand carbohydrate and mucosal addressin cell adhesion molecule 1 (MAdCAM-1) in a mouse model of “Candidatus Helicobacter heilmannii”-induced gastritis and gastric mucosa-associated lymphoid tissue (MALT) lymphoma. Helicobacter. 2010;15:538–48. doi: 10.1111/j.1523-5378.2010.00801.x. [DOI] [PubMed] [Google Scholar]
- 104.Yamamoto ML, Schiestl RH. Intestinal microbiome and lymphoma development. Cancer J. 2014;20:190–4. doi: 10.1097/PPO.0000000000000047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Grice EA, Segre JA. The skin microbiome. Nat Rev Microbiol. 2011;9:244–53. doi: 10.1038/nrmicro2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Garbe C, Stein H, Dienemann D, Orfanos CE. Borrelia burgdorferi-associated cutaneous B cell lymphoma: clinical and immunohistologic characterization of four cases. J Am Acad Dermatol. 1991;24:584–90. doi: 10.1016/0190-9622(91)70088-j. [DOI] [PubMed] [Google Scholar]
- 107.Cerroni L, Zochling N, Putz B, Kerl H. Infection by Borrelia burgdorferi and cutaneous B-cell lymphoma. J Cutan Pathol. 1997;24:457–61. doi: 10.1111/j.1600-0560.1997.tb01318.x. [DOI] [PubMed] [Google Scholar]
- 108.Goodlad JR, Davidson MM, Hollowood K, Ling C, MacKenzie C, Christie I, Batstone PJ, Ho-Yen DO. Primary cutaneous B-cell lymphoma and Borrelia burgdorferi infection in patients from the Highlands of Scotland. Am J Surg Pathol. 2000;24:1279–85. doi: 10.1097/00000478-200009000-00012. [DOI] [PubMed] [Google Scholar]
- 109.Wood GS, Kamath NV, Guitart J, Heald P, Kohler S, Smoller BR, Cerroni L. Absence of Borrelia burgdorferi DNA in cutaneous B-cell lymphomas from the United States. J Cutan Pathol. 2001;28:502–7. doi: 10.1034/j.1600-0560.2001.281002.x. [DOI] [PubMed] [Google Scholar]
- 110.Jackow CM, Cather JC, Hearne V, Asano AT, Musser JM, Duvic M. Association of erythrodermic cutaneous T-cell lymphoma, superantigenpositive Staphylococcus aureus, and oligoclonal T-cell receptor V beta gene expansion. Blood. 1997;89:32–40. [PubMed] [Google Scholar]
- 111.Nguyen V, Huggins RH, Lertsburapa T, Bauer K, Rademaker A, Gerami P, Guitart J. Cutaneous T-cell lymphoma and Staphylococcus aureus colonization. J Am Acad Dermatol. 2008;59:949–52. doi: 10.1016/j.jaad.2008.08.030. [DOI] [PubMed] [Google Scholar]
- 112.Wang Y, Zhu W, Shu M, Jiang Y, Gallo RL, Liu YT, Huang CM. The response of human skin commensal bacteria as a reflection of UV radiation: UV-B decreases porphyrin production. PLoS One. 2012;7:e47798. doi: 10.1371/journal.pone.0047798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Walboomers JM, Jacobs MV, Manos MM, Bosch FX, Kummer JA, Shah KV, Snijders PJ, Peto J, Meijer CJ, Munoz N. Human papillomavirus is a necessary cause of invasive cervical cancer worldwide. J Pathol. 1999;189:12–9. doi: 10.1002/(SICI)1096-9896(199909)189:1<12::AID-PATH431>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 114.Boskey ER, Cone RA, Whaley KJ, Moench TR. Origins of vaginal acidity: high D/L lactate ratio is consistent with bacteria being the primary source. Hum Reprod. 2001;16:1809–13. doi: 10.1093/humrep/16.9.1809. [DOI] [PubMed] [Google Scholar]
- 115.Ravel J, Gajer P, Abdo Z, Schneider GM, Koenig SS, McCulle SL, Karlebach S, Gorle R, Russell J, Tacket CO, Brotman RM, Davis CC, Ault K, Peralta L, Forney LJ. Vaginal microbiome of reproductive-age women. Proc Natl Acad Sci U S A. 2011;108(Suppl 1):4680–7. doi: 10.1073/pnas.1002611107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Mitra A, MacIntyre DA, Lee YS, Smith A, Marchesi JR, Lehne B, Bhatia R, Lyons D, Paraskevaidis E, Li JV, Holmes E, Nicholson JK, Bennett PR, Kyrgiou M. Cervical intraepithelial neoplasia disease progression is associated with increased vaginal microbiome diversity. Sci Rep. 2015;5:16865. doi: 10.1038/srep16865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Oh HY, Kim BS, Seo SS, Kong JS, Lee JK, Park SY, Hong KM, Kim HK, Kim MK. The association of uterine cervical microbiota with an increased risk for cervical intraepithelial neoplasia in Korea. Clin Microbiol Infect. 2015;21:674, e1–9. doi: 10.1016/j.cmi.2015.02.026. [DOI] [PubMed] [Google Scholar]
- 118.Piyathilake CJ, Ollberding NJ, Kumar R, Macaluso M, Alvarez RD, Morrow CD. Cervical microbiota associated with higher grade cervical intraepithelial neoplasia in women infected with high-risk human papillomaviruses. Cancer Prev Res (Phila) 2016;9:357–66. doi: 10.1158/1940-6207.CAPR-15-0350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Hieken TJ, Chen J, Hoskin TL, Walther-Antonio M, Johnson S, Ramaker S, Xiao J, Radisky DC, Knutson KL, Kalari KR, Yao JZ, Baddour LM, Chia N, Degnim AC. The microbiome of aseptically collected human breast tissue in benign and malignant disease. Sci Rep. 2016;6:30751. doi: 10.1038/srep30751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Urbaniak C, Gloor GB, Brackstone M, Scott L, Tangney M, Reid G. The microbiota of breast tissue and its association with breast cancer. Appl Environ Microbiol. 2016;82:5039–48. doi: 10.1128/AEM.01235-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Chan AA, Bashir M, Rivas MN, Duvall K, Sieling PA, Pieber TR, Vaishampayan PA, Love SM, Lee DJ. Characterization of the microbiome of nipple aspirate fluid of breast cancer survivors. Sci Rep. 2016;6:28061. doi: 10.1038/srep28061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Shapira I, Sultan K, Lee A, Taioli E. Evolving concepts: how diet and the intestinal microbiome act as modulators of breast malignancy. ISRN Oncol. 2013;2013:693920. doi: 10.1155/2013/693920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Goedert JJ, Jones G, Hua X, Xu X, Yu G, Flores R, Falk RT, Gail MH, Shi J, Ravel J, Feigelson HS. Investigation of the association between the fecal microbiota and breast cancer in postmenopausal women: a population-based casecontrol pilot study. J Natl Cancer Inst. 2015:107. doi: 10.1093/jnci/djv147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Lakritz JR, Poutahidis T, Mirabal S, Varian BJ, Levkovich T, Ibrahim YM, Ward JM, Teng EC, Fisher B, Parry N, Lesage S, Alberg N, Gourishetti S, Fox JG, Ge Z, Erdman SE. Gut bacteria require neutrophils to promote mammary tumorigenesis. Oncotarget. 2015;6:9387–96. doi: 10.18632/oncotarget.3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Gunter MJ, Xie X, Xue X, Kabat GC, Rohan TE, Wassertheil-Smoller S, Ho GY, Wylie-Rosett J, Greco T, Yu H, Beasley J, Strickler HD. Breast cancer risk in metabolically healthy but overweight postmenopausal women. Cancer Res. 2015;75:270–4. doi: 10.1158/0008-5472.CAN-14-2317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Fuhrman BJ, Feigelson HS, Flores R, Gail MH, Xu X, Ravel J, Goedert JJ. Associations of the fecal microbiome with urinary estrogens and estrogen metabolites in postmenopausal women. J Clin Endocrinol Metab. 2014;99:4632–40. doi: 10.1210/jc.2014-2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Luu TH, Michel C, Bard JM, Dravet F, Nazih H, Bobin-Dubigeon C. Intestinal proportion of blautia sp. is associated with clinical stage and histoprognostic grade in patients with earlystage breast cancer. Nutr Cancer. 2017;69:267–75. doi: 10.1080/01635581.2017.1263750. [DOI] [PubMed] [Google Scholar]
- 128.Rao VP, Poutahidis T, Ge Z, Nambiar PR, Boussahmain C, Wang YY, Horwitz BH, Fox JG, Erdman SE. Innate immune inflammatory response against enteric bacteria helicobacter hepaticus induces mammary adenocarcinoma in mice. Cancer Res. 2006;66:7395–400. doi: 10.1158/0008-5472.CAN-06-0558. [DOI] [PubMed] [Google Scholar]
- 129.Nakatsu G, Li X, Zhou H, Sheng J, Wong SH, Wu WK, Ng SC, Tsoi H, Dong Y, Zhang N, He Y, Kang Q, Cao L, Wang K, Zhang J, Liang Q, Yu J, Sung JJ. Gut mucosal microbiome across stages of colorectal carcinogenesis. Nat Commun. 2015;6:8727. doi: 10.1038/ncomms9727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Peters BA, Dominianni C, Shapiro JA, Church TR, Wu J, Miller G, Yuen E, Freiman H, Lustbader I, Salik J, Friedlander C, Hayes RB, Ahn J. The gut microbiota in conventional and serrated precursors of colorectal cancer. Microbiome. 2016;4:69. doi: 10.1186/s40168-016-0218-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Purcell RV, Visnovska M, Biggs PJ, Schmeier S, Frizelle FA. Distinct gut microbiome patterns associate with consensus molecular subtypes of colorectal cancer. Sci Rep. 2017;7:11590. doi: 10.1038/s41598-017-11237-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Ai L, Tian H, Chen Z, Chen H, Xu J, Fang JY. Systematic evaluation of supervised classifiers for fecal microbiota-based prediction of colorectal cancer. Oncotarget. 2017;8:9546–56. doi: 10.18632/oncotarget.14488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Flemer B, Warren RD, Barrett MP, Cisek K, Das A, Jeffery IB, Hurley E, O’Riordain M, Shanahan F, O’Toole PW. The oral microbiota in colorectal cancer is distinctive and predictive. Gut. 2018;67:1454–1463. doi: 10.1136/gutjnl-2017-314814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Rajagopala SV, Yooseph S, Harkins DM, Moncera KJ, Zabokrtsky KB, Torralba MG, Tovchigrechko A, Highlander SK, Pieper R, Sender L, Nelson KE. Gastrointestinal microbial populations can distinguish pediatric and adolescent acute lymphoblastic leukemia (ALL) at the time of disease diagnosis. BMC Genomics. 2016;17:635. doi: 10.1186/s12864-016-2965-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Malfertheiner P, Megraud F, O’Morain C, Hungin AP, Jones R, Axon A, Graham DY, Tytgat G European Helicobacter Pylori Study Group (EHPSG) Current concepts in the management of helicobacter pylori infection--the maastricht 2-2000 consensus report. Aliment Pharmacol Ther. 2002;16:167–80. doi: 10.1046/j.1365-2036.2002.01169.x. [DOI] [PubMed] [Google Scholar]
- 136.Goderska K, Agudo Pena S, Alarcon T. Helicobacter pylori treatment: antibiotics or probiotics. Appl Microbiol Biotechnol. 2018;102:1–7. doi: 10.1007/s00253-017-8535-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Fuller R. Probiotics in human medicine. Gut. 1991;32:439–42. doi: 10.1136/gut.32.4.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Wollowski I, Rechkemmer G, Pool-Zobel BL. Protective role of probiotics and prebiotics in colon cancer. Am J Clin Nutr. 2001;73:451S–5S. doi: 10.1093/ajcn/73.2.451s. [DOI] [PubMed] [Google Scholar]
- 139.de Moreno de LeBlanc A, Matar C, Perdigon G. The application of probiotics in cancer. Br J Nutr. 2007;98(Suppl 1):S105–10. doi: 10.1017/S0007114507839602. [DOI] [PubMed] [Google Scholar]
- 140.Zhu Y, Michelle Luo T, Jobin C, Young HA. Gut microbiota and probiotics in colon tumorigenesis. Cancer Lett. 2011;309:119–27. doi: 10.1016/j.canlet.2011.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Feyisetan O, Tracey C, Hellawell GO. Probiotics, dendritic cells and bladder cancer. BJU Int. 2012;109:1594–7. doi: 10.1111/j.1464-410X.2011.10749.x. [DOI] [PubMed] [Google Scholar]
- 142.Gui QF, Lu HF, Zhang CX, Xu ZR, Yang YH. Well-balanced commensal microbiota contributes to anti-cancer response in a lung cancer mouse model. Genet Mol Res. 2015;14:5642–51. doi: 10.4238/2015.May.25.16. [DOI] [PubMed] [Google Scholar]
- 143.Hill DA, Artis D. Intestinal bacteria and the regulation of immune cell homeostasis. Annu Rev Immunol. 2010;28:623–67. doi: 10.1146/annurev-immunol-030409-101330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Buffie CG, Pamer EG. Microbiota-mediated colonization resistance against intestinal pathogens. Nat Rev Immunol. 2013;13:790–801. doi: 10.1038/nri3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Modi SR, Collins JJ, Relman DA. Antibiotics and the gut microbiota. J Clin Invest. 2014;124:4212–8. doi: 10.1172/JCI72333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Yu LX, Schwabe RF. The gut microbiome and liver cancer: mechanisms and clinical translation. Nat Rev Gastroenterol Hepatol. 2017;14:527–39. doi: 10.1038/nrgastro.2017.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Bashan A, Gibson TE, Friedman J, Carey VJ, Weiss ST, Hohmann EL, Liu YY. Universality of human microbial dynamics. Nature. 2016;534:259–62. doi: 10.1038/nature18301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Yu T, Guo F, Yu Y, Sun T, Ma D, Han J, Qian Y, Kryczek I, Sun D, Nagarsheth N, Chen Y, Chen H, Hong J, Zou W, Fang JY. Fusobacterium nucleatum promotes chemoresistance to colorectal cancer by modulating autophagy. Cell. 2017;170:548–563. e16. doi: 10.1016/j.cell.2017.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Geller LT, Barzily-Rokni M, Danino T, Jonas OH, Shental N, Nejman D, Gavert N, Zwang Y, Cooper ZA, Shee K, Thaiss CA, Reuben A, Livny J, Avraham R, Frederick DT, Ligorio M, Chatman K, Johnston SE, Mosher CM, Brandis A, Fuks G, Gurbatri C, Gopalakrishnan V, Kim M, Hurd MW, Katz M, Fleming J, Maitra A, Smith DA, Skalak M, Bu J, Michaud M, Trauger SA, Barshack I, Golan T, Sandbank J, Flaherty KT, Mandinova A, Garrett WS, Thayer SP, Ferrone CR, Huttenhower C, Bhatia SN, Gevers D, Wargo JA, Golub TR, Straussman R. Potential role of intratumor bacteria in mediating tumor resistance to the chemotherapeutic drug gemcitabine. Science. 2017;357:1156–60. doi: 10.1126/science.aah5043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Daillere R, Vetizou M, Waldschmitt N, Yamazaki T, Isnard C, Poirier-Colame V, Duong CPM, Flament C, Lepage P, Roberti MP, Routy B, Jacquelot N, Apetoh L, Becharef S, Rusakiewicz S, Langella P, Sokol H, Kroemer G, Enot D, Roux A, Eggermont A, Tartour E, Johannes L, Woerther PL, Chachaty E, Soria JC, Golden E, Formenti S, Plebanski M, Madondo M, Rosenstiel P, Raoult D, Cattoir V, Boneca IG, Chamaillard M, Zitvogel L. Enterococcus hirae and barnesiella intestinihominis facilitate cyclophosphamide-induced therapeutic immunomodulatory effects. Immunity. 2016;45:931–43. doi: 10.1016/j.immuni.2016.09.009. [DOI] [PubMed] [Google Scholar]
- 151.Routy B, Le Chatelier E, Derosa L, Duong CPM, Alou MT, Daillere R, Fluckiger A, Messaoudene M, Rauber C, Roberti MP, Fidelle M, Flament C, Poirier-Colame V, Opolon P, Klein C, Iribarren K, Mondragon L, Jacquelot N, Qu B, Ferrere G, Clemenson C, Mezquita L, Masip JR, Naltet C, Brosseau S, Kaderbhai C, Richard C, Rizvi H, Levenez F, Galleron N, Quinquis B, Pons N, Ryffel B, Minard-Colin V, Gonin P, Soria JC, Deutsch E, Loriot Y, Ghiringhelli F, Zalcman G, Goldwasser F, Escudier B, Hellmann MD, Eggermont A, Raoult D, Albiges L, Kroemer G, Zitvogel L. Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science. 2018;359:91–7. doi: 10.1126/science.aan3706. [DOI] [PubMed] [Google Scholar]
- 152.Gopalakrishnan V, Spencer CN, Nezi L, Reuben A, Andrews MC, Karpinets TV, Prieto PA, Vicente D, Hoffman K, Wei SC, Cogdill AP, Zhao L, Hudgens CW, Hutchinson DS, Manzo T, Petaccia de Macedo M, Cotechini T, Kumar T, Chen WS, Reddy SM, Szczepaniak Sloane R, Galloway-Pena J, Jiang H, Chen PL, Shpall EJ, Rezvani K, Alousi AM, Chemaly RF, Shelburne S, Vence LM, Okhuysen PC, Jensen VB, Swennes AG, McAllister F, Marcelo Riquelme Sanchez E, Zhang Y, Le Chatelier E, Zitvogel L, Pons N, Austin-Breneman JL, Haydu LE, Burton EM, Gardner JM, Sirmans E, Hu J, Lazar AJ, Tsujikawa T, Diab A, Tawbi H, Glitza IC, Hwu WJ, Patel SP, Woodman SE, Amaria RN, Davies MA, Gershenwald JE, Hwu P, Lee JE, Zhang J, Coussens LM, Cooper ZA, Futreal PA, Daniel CR, Ajami NJ, Petrosino JF, Tetzlaff MT, Sharma P, Allison JP, Jenq RR, Wargo JA. Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Science. 2018;359:97–103. doi: 10.1126/science.aan4236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Matson V, Fessler J, Bao R, Chongsuwat T, Zha Y, Alegre ML, Luke JJ, Gajewski TF. The commensal microbiome is associated with anti-PD-1 efficacy in metastatic melanoma patients. Science. 2018;359:104–8. doi: 10.1126/science.aao3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Ray K. Gut microbiota: colorectal cancer-driven by inflammation and gut bacteria? Nat Rev Gastroenterol Hepatol. 2012;9:558. doi: 10.1038/nrgastro.2012.172. [DOI] [PubMed] [Google Scholar]
- 155.Schmidt TSB, Raes J, Bork P. The human gut microbiome: from association to modulation. Cell. 2018;172:1198–215. doi: 10.1016/j.cell.2018.02.044. [DOI] [PubMed] [Google Scholar]
- 156.Guglielmi G. How gut microbes are joining the fight against cancer. Nature. 2018;557:482–4. doi: 10.1038/d41586-018-05208-8. [DOI] [PubMed] [Google Scholar]
- 157.Turnbaugh PJ, Ley RE, Hamady M, Fraser-Liggett CM, Knight R, Gordon JI. The human microbiome project. Nature. 2007;449:804–10. doi: 10.1038/nature06244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.O’Hara AM, Shanahan F. The gut flora as a forgotten organ. EMBO Rep. 2006;7:688–93. doi: 10.1038/sj.embor.7400731. [DOI] [PMC free article] [PubMed] [Google Scholar]


