Abstract
The integrative conjugative element (ICE) Tn5253 of Streptococcus pneumoniae, conferring resistance to tetracycline and chloramphenicol, was found integrated at a 83-bp specific target site (attB) located in the rbgA gene of the pneumococcal chromosome. PCR analysis of Tn5253-carrying strains showed evidence of precise excision of Tn5253 from the pneumococcal chromosome with production of (i) circular forms of the ICE in which the ends were joined by a 84-bp sequence (attTn), and (ii) reconstituted chromosomal attB. When integrated into the chromosome, Tn5253 was flanked by attL, identical to attB, and attR, identical to attTn. Circular forms of Tn5253 were present at a concentration of 3.8 × 10-4 copies per chromosome, whereas reconstituted attB sites were at 3.0 × 10-4 copies per chromosome. Deletion of int-xis of Tn5253 abolished production of circular forms (<7.1 × 10-6 copies per chromosome) and was associated to the lack of Tn5253 conjugal transfer suggesting, as expected, that Tn5253 circular form acts as a conjugation intermediate.
Keywords: integrative conjugative element (ICE), circular form, attachment site, conjugative transposon, Tn5253, conjugation, mobile genetic elements
Introduction
Horizontal gene transfer, mediated by MGEs, significantly drives bacterial genome evolution including the acquisition and dissemination of new patterns of antibiotic resistance (Burrus and Waldor, 2004). Functional characterization of MGEs is essential to understand the evolution and spread of antibiotic resistance within a given bacterial species and also among different species (Frost et al., 2005). ICEs, which include CTs, are MGEs that integrate into the bacterial genome and are capable of intracellular transposition to a new genomic location or intercellular transposition to a new genome host upon conjugative transfer (Mullany et al., 2002). ICEs account for up to 25% of the genetic material in a bacterial genome (Paulsen et al., 2003) and are the major promoters of genetic diversity in bacteria (Burrus and Waldor, 2004; Johnson and Grossman, 2015).
The CT Tn916, carrying the tet(M) gene, is the prototype of the Tn916–Tn1545 family of ICEs, and one of the most studied ICEs of gram positive bacteria (Santoro et al., 2014). Tn916 was shown to excise from the bacterial chromosome producing a covalently closed circular form of the element which was called “CI.” Production of CIs of Tn916 was demonstrated to be essential for conjugative transposition of the element (Scott et al., 1988). Recombination processes of ICEs are catalyzed by site specific recombinases (serine or tyrosine) or by DDE transposases (Ambroset et al., 2016). The Tn916 element carries the int and xis genes which code for a tyrosine site specific recombinase and an excisionase, respectively (Lu and Churchward, 1995). Excision and circularization require both Xis and Int, whereas Int alone is sufficient for integration (Storrs et al., 1991). Dosage of Tn916 CIs demonstrated that their number correlates with conjugation frequency and is variable among different strains (Manganelli et al., 1995).
Tn5253 is a 64,528-bp composite ICE of Streptococcus pneumoniae which contains integrated two distinct genetic elements: Tn5251, belonging to the Tn916–Tn1545 family of ICEs, and Ωcat(pC194) which carry tet(M) and cat resistance genes, respectively (Ayoubi et al., 1991; Provvedi et al., 1996; Santoro et al., 2010; Iannelli et al., 2014). Tn5253 contains two pairs of xis/int recombinase genes one of which is part of Tn5251 (Kiliç et al., 1994; Iannelli et al., 2014). Genomic sequence analysis and PCR genotyping studies demonstrated that Tn5253-like elements are very common in multidrug-resistant pneumococcal strains including pandemic isolates (Croucher et al., 2009; Henderson-Begg et al., 2009; Mingoia et al., 2011). A study on 240 different pneumococcal isolates of the multidrug-resistance 23F Spanish strain lineage, carrying the Tn5253-like element ICESpn23FST81, showed that the element is maintained among all derivative strains (Croucher et al., 2011). In this work, we investigated excision and circularization of the composite ICE Tn5253, including the respective contribution of each xis/int recombinase pair to the conjugal transfer of the genetic element.
Materials and Methods
Bacterial Strains, Growth, and Mating Conditions
The bacterial strains used in this study and their relevant properties are described in Table 1. Bacterial growth and plate mating conjugation experiments were performed as reported (Santoro et al., 2010).
Table 1.
Streptococcus pneumoniae strains.
| Strain | Relevant propertiesa | Origin (Reference) |
|---|---|---|
| D39 | type 2 Avery’s strain | Lanie et al., 2007 |
| FP58 | Conjugation recipient. str-41; SmR derivative of D39 | Iannelli et al., 2004 |
| Rx1 | Unencapsulated derivative of D39 | Pearce et al., 2002 |
| FP10 | Conjugation recipient. ΔcomC, str-41; CmR, SmR; unencapsulated, competence deficient derivative of Rx1 | Santoro et al., 2010 |
| FP11 | Conjugation recipient. ΔcomC, nov-1; CmR, NovR; unencapsulated, competence deficient derivative of Rx1 | Santoro et al., 2010 |
| BM6001 | Tn5253 donor; cat, tet(M); original clinical strain | Dang-Van et al., 1978 |
| DP1322 | Tn5253 donor; cat, tet(M); CmR, TcR; Rx1 derivative transformed with BM6001 DNA | Smith et al., 1981 |
| FR24 | Tn5253; cat, tet(M); CmR, TcR, SmR; transconjugant from mating between DP1322 and FP10 | This study |
| FR51 | Tn5253; cat, tet(M), Δxis-int of Tn5253; CmR, TcR, SmR; KmR; recombinases (orf78-orf79) deletion mutant of FR24 | This study |
| FR82 | Tn5253; cat, tet(M), Δint-xis of Tn5251; CmR, TcR, SmR; SpeR; recombinases (orf21-orf22) deletion mutant of FR24 | This study |
astr-41 indicates a point mutation conferring resistance to streptomycin, while nov-1 indicates a point mutation conferring resistance to novobiocin. Cm, chloramphenicol; Km, kanamycin; Nov, novobiocin; Sm, streptomycin; Tc, tetracycline; Spe, spectinomycin; R, resistance.
Pneumococcal Lysate Preparation
Pneumococcal cultures (1 ml) were harvested in exponential phase (OD590 about 0.2, roughly corresponding to 5 × 108 CFU/ml) and centrifuged at 11,000 ×g for 2 min. Bacterial pellets were resuspended in 30 μl of lysis solution (DOC 0.1%, SDS 0.008%) and incubated at 37°C until clarification (about 10 min). Two hundred and seventy micro liters of TE 1×, pH 8.0 were then added to the lysate.
PCR, Sequencing, and Sequence Analysis
PCR and direct PCR sequencing were carried out following an already described protocol (Iannelli et al., 1998; Santoro et al., 2010) and DNA sequence analysis was obtained with standard softwares. DNA sequence alignments were performed using Clustal Omega1 and Lalign2. Oligonucleotide primers and their characteristics are reported in Table 2.
Table 2.
Oligonucleotide primers.
| Name | Sequence (5′ to 3′) | Notesa | GenBank ID: nucleotides |
|---|---|---|---|
| IF327 | CAA TAT AGC GTG ATG ATT GTA AT | EU351020: 1,103–1,081 | |
| IF328 | AGT GAG AAT CAA ATC AGA GGT T | EU351020: 65,221–65,242 | |
| IF373 | GAT GAT GAT TTG ACA CAA GAA TA | EU351020: 62,969–62,991 | |
| IF521 | ATC AAA CGG ATC CCC AGC TTG TAT TCA TGT CAT CAT CCT TCC T | The first 21 nucleotides are complementary to IF149 | EU351020: 63,439–63,418 |
| IF522 | ATA TTT TAC TGG ATG AAT TGT TTT AGT TTT GGT GTT CGC TTG GTG TTT AG | The first 26 nucleotides are complementary to IF210 | EU351020: 65,260–65,283 |
| IF523 | CGG TGT ATC CAA GAT TTC CAG | EU351020: 65,862–65,842 | |
| IF517 | ATT TCC TTG CGT GAT GTG TGA | EU351020: 16,810–16,830 | |
| IF681 | GTA TCG CTC TTG AAG GGA ATA GTA CAA ATG AAT TTA CTA CTT | The first 19 nucleotides are complementary to IF101 | EU351020: 17,305–17,283 |
| IF682 | GAT CCA CTA GTT CTA GAG CTC CCA AAT AGG AAT GTC AGT | The first 19 nucleotides are complementary to IF100 | EU351020: 18,780–18,799 |
| IF520 | GTA TGG TCG TTG ATG AAG TCT | EU351020: 19,348–19,328 | |
| IF100 | GCT CTA GAA CTA GTG GAT C | AY334020: 1–16 | |
| IF101 | TTC CCT TCA AGA GCG ATA C | AY334020: 890–872 | |
| IF149 | CAA GCT GGG GAT CCG TTT GAT | AY334018: 5–25 | |
| IF210 | CTA AAA CAA TTC ATC CAG TAA AAT AT | AY334019: 880–855 | |
| IF496 | GTT TGG ACA TCA TTC ATT TG | CP000410: 1,043,748–1,043,767 | |
| IF356 | GAC TAG ATA GAG GCA AGC GT | CP000410: 1,043,962–1,043,943 | |
| IF138 | CAG ATC AAG AAA TCA AAC TCC AA | CP000410: 725,024–725,046 | |
| IF139 | CAG CAT CAT CTA CAG AAA CTC | CP000410: 725,194–725,174 | |
aNucleotides complementary to resistance cassettes primers are reported in italics.
PCR Mutagenesis
Isogenic deletion mutant strains were constructed transforming FR24 with linear PCR mutagenic constructs assembled by gene splicing by overlap extension as already described (Pearce et al., 2002; Iannelli and Pozzi, 2004). Deletion of Tn5251 int and xis CDS (orf21 and orf22 of Tn5253) was obtained with a mutagenic construct containing the ami/aad9 spectinomycin resistance cassette flanked at the left by a 496-bp DNA fragment and at the right by a 569-bp fragment corresponding to nucleotides 16,810–17,305 and 18,780–19,348 of Tn5253 (GenBank EU351020), respectively. The primer pair IF100/IF101 was used to amplify the spectinomycin-resistance cassette from plasmid pR412 (Bergé et al., 2002), whereas IF517/IF681 and IF520/IF682 were used to amplify the flanking fragments from FR24.
The xis and int CDSs of Tn5253 (orf78 and orf79) were deleted with a mutagenic construct containing the ami/aphIII kanamycin resistance cassette, flanked at the left by a 471-bp DNA fragment and at the right by a 603-bp corresponding to nucleotides 62,969–63,439 of Tn5253 and 65,260–65,862 of Tn5253, respectively. The primer pair IF149/IF210 was used to amplify the kanamycin-resistance cassette from plasmid pR410 (Bergé et al., 2002), while IF373/IF521 and IF522/IF523 were used to amplify the left and right fragments from FR24. Linear PCR constructs were used directly as donor DNA in transformation experiments. Mutant strains were selected for acquisition of spectinomycin or kanamycin resistance and the correct integration of constructs was confirmed by PCR and sequencing (Iannelli and Pozzi, 2004).
Real-Time PCR Quantification
Real-time PCR experiments were carried out with the KAPA SYBR FAST qPCR kit Master Mix Universal (2X) (Kapa Biosystems) on a LightCycler 1.5 apparatus (Roche). Real-time PCR mixture contained, in a final volume of 20 μl, 1× KAPA SYBR FAST qPCR reaction mix, 5 pmol of each primer and 1 μl of bacterial lysate as starting template. Thermal profile was an initial 3 min denaturation step at 95°C followed by 40 cycles of repeated denaturation (0 s at 95°C), annealing (20 s at 50°C), and polymerisation (10 s at 72°C). The temperature transition rate was 20°C/s in the denaturation and annealing step and 5°C/s in the polymerisation step. The primer pair IF327/IF328 amplified a 411 bp fragment used for CIs quantification, while IF496/IF356 amplified a 215 bp fragment used for free locus quantification, a 171 bp fragment of chromosomal gyrB gene obtained with primers IF138/IF139 was used to standardize results (Table 2). A standard curve for the gyrB gene was built plotting the threshold cycle against the number of chromosome copies using serial dilutions of chromosomal DNA with known concentration. This external standard curve was used to quantify in each sample the number of (i) chromosome copies, (ii) CIs, and (iii) reconstituted attB. Lower limit of detection of the assay was 10 copies/reaction. The quantification was corrected for the primer efficiency. Melting curve analysis was performed to differentiate the amplified products from primer dimers.
Results and Discussion
Excision of Tn5253 From the Pneumococcal Chromosome
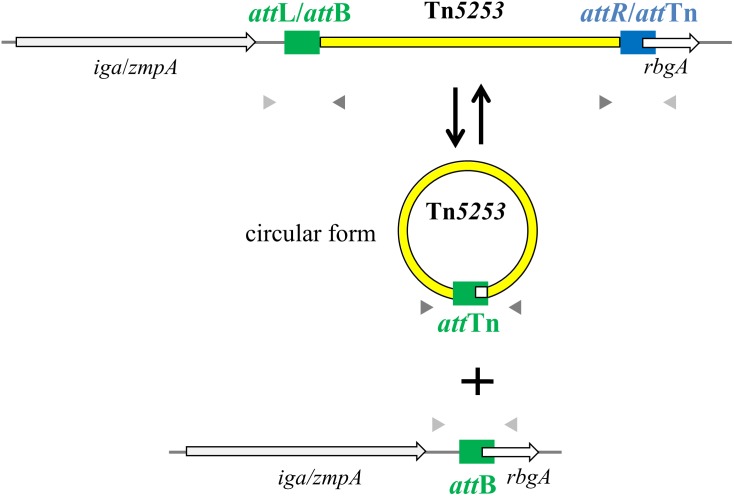
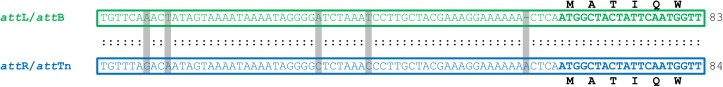
PCR analysis of cell lysates of Tn5253-carrying pneumococcal strains showed evidence of precise excision of Tn5253 from its specific attachment site (attB) in the pneumococcal chromosome. This excision was investigated in liquid cultures of BM6001, the clinical isolate in which Tn5253 was originally found, and four other Tn5253-carrying laboratory strains all deriving from classic type 2 D39 (Table 1). Using divergent primers (IF327, IF328; Table 2) designed on the ends of the element, PCR analysis showed the presence of junctions between the left and right ends of Tn5253 (attTn), whereas with convergent primers (IF496, IF356; Table 2) designed on the regions flanking the insertion site it was possible to show the presence of chromosomes with reconstituted target sites for integration of Tn5253 (attB) (Figure 1). In all Tn5253-carrying pneumococci, DNA sequence analysis of PCR fragments indicated that: (i) attTn was 84 bp in size and was identical to attR, the 84-bp direct repeat present at the right end of the integrated element, whereas (ii) attB was 83 bp in size and was identical to attL, the direct repeat present at the left end of the integrated element (Figure 2). The attR-attTn repeat contained 4 nucleotide changes and 1 insertion compared to attL-attB (Figure 2). These results suggest that in Tn5253-carrying strains, recombination occurs between the two imperfect direct repeats attL and attR leading to precise excision of the element from the chromosome, with production of circular forms of Tn5253 in which the ends are joined by attR/attTn, while the attL/attB repeat remains in the bacterial chromosome (Figure 1).
FIGURE 1.
Tn5253 excises from pneumococcal chromosome producing a circular form and a reconstitution of attB insertion site. In the circular form of Tn5253 the left and right ends are joined by attTn which is identical to attR whereas the reconstituted attB site is identical to attL. att sites are represented as filled rectangles, chromosomal genes as open arrows, Tn5253 as a yellow bar. Arrowheads represent PCR primers used for circular form (dark gray) and reconstituted attB site (light gray) detection.
FIGURE 2.
Sequence alignment of Tn5253 attachment sites in Streptococcus pneumoniae D39 and derivatives. Upon integration into pneumococcal chromosome, Tn5253 is flanked by attL and attR. The element excises from chromosome producing a circular form where the left and right ends are joined by attTn and restoring the attB insertion site. attB is 83-bp long and is identical to attL while attTn is 84-bp long and is identical to attR. attR-attTn contain 4 nucleotide changes and 1 insertion compared to attL-attB. attL-attB contains the first 20 nucleotides of rbgA CDS, whose deduced amino acid sequence is reported, together with the 63 nucleotides upstream of the start codon. Within the sequences, identical nucleotides are indicated by colon, changes are shaded. Amino acids are indicated using one-letter code abbreviations. Lengths of attachment sites are reported on the right.
Quantification of Circular Forms and Reconstituted attB Sites
To obtain a quantitative estimate of Tn5253 excision from the S. pneumoniae chromosome, Real-time PCR was used to quantify concentration of circular forms and reconstituted attB sites in liquid bacterial cultures. Different Tn5253-carrying laboratory strains of D39 ancestry, and BM6001 showed very homogeneous quantitative results (Table 3). In the laboratory strain DP1322 circular forms of Tn5253 were present at a concentration of 5.1 × 10-4 (±2.7 × 10-4) copies per chromosome, whereas reconstituted attB sites were at 2.1 × 10-4 (±3.0 × 10-5) copies per chromosome. These values were comparable to those obtained in the Tn5253-carrying laboratory strains (Table 3). Autonomous plasmid-like replication is common in ICEs and contributes to the stability and maintenance of these elements (Lee et al., 2010; Carraro et al., 2015; Johnson and Grossman, 2015). The hypothesis that also Tn5253 circular forms undergo few cycles of autonomous replication can explain why the copy number of circular forms is higher than the copy number of the reconstituted attB site.
Table 3.
Real-time PCR quantification of Tn5253 circular form and reconstituted attBa.
| Strain | Circular forms | Reconstituted attB sites | Conjugation frequencyb |
|---|---|---|---|
| BM6001 | 1.8 × 10-4 (±1.5 × 10-5) | 2.4 × 10-4 (±3.6 × 10-5) | 3.4 × 10-7 |
| DP1322 | 5.1 × 10-4 (±2.7 × 10-4) | 2.1 × 10-4 (±3.0 × 10-5) | 1.6 × 10-4 |
| FR24 | 5.4 × 10-4 (±3.1 × 10-4) | 3.7 × 10-4 (±3.3 × 10-5) | 2.0 × 10-4 |
| FR51 | none (≤7.1 × 10-6) | none (≤7.1 × 10-6) | none (<9.9 × 10-8) |
aConcentration was expressed as copies per chromosome. bFrequency refers to mating experiments where S. pneumoniae FP11 was the recipient.
Site-Specific Integration of Tn5253 Into the rbgA Gene
In S. pneumoniae D39 and in its derivatives used as conjugation recipients (Table 1) DNA sequence analysis showed that attB of Tn5253 was 83 bp in size, and was always located within the rbgA gene (nucleotides 1,043,779 to 1,043,861, GenBank CP000410) (Figure 1). The ribosomal biogenesis GTPase A encoded by rbgA is a conserved, essential bacterial protein involved in the 50S ribosome subunit assembly (Uicker et al., 2006). The attB site contained the first 20 nucleotides of the rbgA CDS together with the 63 nucleotides upstream of the start codon (Figures 1, 2). The junction fragments of Tn5253 with the bacterial chromosome were investigated in a total of 12 transconjugants obtained in independent matings in which Tn5253 was transferred by conjugation from 2 pneumococcal donors (BM6001 and DP1322; Table 1) to 3 pneumococcal recipients (FP58, FP10, FP11; Table 1). Left and right junction fragments were amplified by PCR (using primer pairs IF496/IF327 and IF328/IF356) and sequenced in all transconjugants. DNA sequence analysis showed that in all cases Tn5253 integration occurred at the same site within rbgA, between the two direct repeats: (i) attL, corresponding to the attB of the recipient and (ii) attR, corresponding to the attTn of the circular forms of Tn5253 (Figure 2). These results indicated that attR, one of the two repeats flanking Tn5253 in the donor chromosome, was always transferred by conjugation to the recipients. Since the integrated form of Tn5253 was invariably flanked by attB at the left end (attL) and attTn at the right end (attR), we hypothesize a polarization in the DNA integration process. Site specific integration of MGEs often occurs at one end of essential and highly conserved genes, such as the 3′ end of tRNA genes and the 3′ or 5′ end of genes coding for ribosomal proteins (Ambroset et al., 2016). Also for Tn5253 integration occurs at the 5′ end of an essential gene, with target site duplication allowing restoration of an intact CDS. The use of essential and conserved genes as target sites guarantees the presence and conservation of attB in bacterial genomes favoring the spread of ICEs such as Tn5253, which can overpass the border of a single species and thus favor the dissemination of multiple antibiotic resistance genes. In fact, in other bacterial species such as Streptococcus pyogenes and Streptococcus mitis, Tn5253-like elements are found integrated at the 5′ end of rbgA orthologous genes (Mingoia et al., 2014; Petrosyan et al., 2016).
Recombinase Genes Involved in Tn5253 Excision
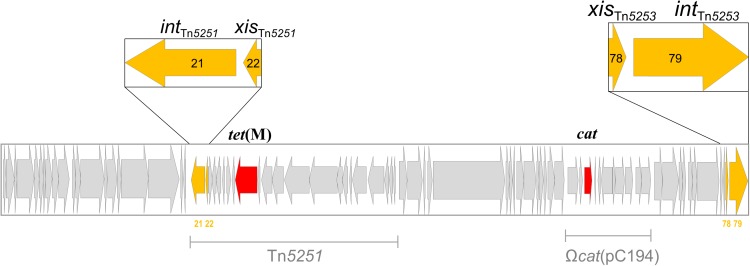
Two sets of xis/int recombinase genes are present in the sequence of Tn5253, one set (orf78/orf79) is at the right end of the element,while the other (orf21/orf22) belongs to Tn5251 (Figure 3). Excisionase Xis and tyrosine integrase Int are known to work in synergy, for this reason we decided to construct mutants where the xis and int genes were both deleted. For each set of xis/int recombinase genes, we constructed an isogenic deletion mutant in the Tn5253-carrying strain FR24. In FR51 a 1,820-bp DNA fragment (position 63,440–65,259, GenBank No. EU351020) encompassing orf78/orf79 CDSs was deleted and replaced with the 876-bp ami/aphIII cassette. In FR82 a 1,474-bp DNA fragment (position 17,306–18,779, GenBank No. EU351020) encompassing orf21/orf22 CDS was deleted and replaced with the 894-bp ami/aad9 cassette (Table 2). The deletion of xis/int of Tn5251 abolished the production of circular forms and the conjugal transfer of Tn5251, but did not affect the frequencies of Tn5253 circular forms, of attB site reconstitution, and of Tn5253 conjugal transfer (data not shown). Deletion of xis/int of Tn5253 in FR51 abolished the circular forms generation and the reconstitution of attB site (<7.1 × 10-6 copies per chromosome for both genetic structures, Table 3). The absence of circular forms in FR51 was associated to the lack of Tn5253 conjugal transfer suggesting, that the circular form of Tn5253 acts as a conjugation intermediate as proposed for other characterized ICEs including Tn916. Data obtained using xis/int deletion mutants showed that the two recombinase pairs act independently and do not complement each other. This finding suggests that the association between the two elements is physical but not functional.
FIGURE 3.
The composite integrative conjugative element (ICE) Tn5253 contains two distinct genetic elements integrated: (i) Tn5251 which carries tet(M) and is able to transfer by conjugation autonomously; (ii) Ωcat(pC194) which contains cat, is not conjugative, but is capable of intracellular transposition. Two sets of xis/int recombinase genes are carried by Tn5253, one set (orf78/orf79) is at the right end of the element, while the other (orf21/orf22) belongs to Tn5251. ORFs and their transcription direction are indicated as arrows, sequences corresponding to Tn5251 and Ωcat(pC194) are indicated by solid bars.
Conclusion
In this work we have shown that: (i) Tn5253 is capable of precise excision from the chromosome, producing circular forms of the element, and leaving chromosomes with reconstituted attB sites; (ii) in the circular forms, the two ends of Tn5253 are joined by attTn, an 84-bp DNA fragment identical to the attR junction fragment flanking the element in its integrated form; (iii) attR is always transferred to the recipient strain during conjugation; (iv) production of Tn5253 circular forms and their conjugal transfer were abolished when xis/int of Tn5253 were deleted. Even if the importance of ICEs in shaping bacterial genomes is widely recognized and nucleotide sequences of ICEs are increasingly available, a functional characterization is available only for a few of these genetic elements. This work on Tn5253 contributes to elucidating the transfer functions of one of the prototypes of ICEs of gram positive bacteria.
Author Contributions
FS, FI, and GP designed the experiments. FS and AR performed the experimental work. All authors analyzed and interpreted the data. FI, FS, and GP wrote the paper.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are grateful to Marc Prudhomme and Jean-Pierre Claverys for kindly providing us plasmids pr410 and pr412.
Abbreviations
- CI
circular intermediate
- CDS
coding sequence
- CT
conjugative transposon
- HGT
horizontal gene transfer
- ICE
integrative conjugative element
- MGE
mobile genetic element
Funding. The research leading to these results has received funding from the European Union Seventh Framework Programme (FP7/2007-2013) under grant agreement no. 241446 (project ANTIRESDEV).
References
- Ambroset C., Coluzzi C., Guédon G., Devignes M. D., Loux V., Lacroix T., et al. (2016). New insights into the classification and integration specificity of Streptococcus integrative conjugative elements through extensive genome exploration. Front. Microbiol. 6:1483. 10.3389/fmicb.2015.01483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayoubi P., Kilic A. O., Vijayakumar M. N. (1991). Tn5253, the pneumococcal omega (cat tet) BM6001 element, is a composite structure of two conjugative transposons, Tn5251 and Tn5252. J. Bacteriol. 173 1617–1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergé M., Moscoso M., Prudhomme M., Martin B., Claverys J.-P. (2002). Uptake of transforming DNA in Gram-positive bacteria: a view from Streptococcus pneumoniae. Mol. Microbiol. 45 411–421. 10.1046/j.1365-2958.2002.03013.x [DOI] [PubMed] [Google Scholar]
- Burrus V., Waldor M. K. (2004). Shaping bacterial genomes with integrative and conjugative elements. Res. Microbiol. 155 376–386. 10.1016/j.resmic.2004.01.012 [DOI] [PubMed] [Google Scholar]
- Carraro N., Poulin D., Burrus V. (2015). Replication and active partition of integrative and conjugative elements (ICEs) of the SXT/R391 family: the line between ICEs and conjugative plasmids is getting thinner. PLoS Genet. 11:e1005298. 10.1371/journal.pgen.1005298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croucher N. J., Harris S. R., Fraser C., Quail M. A., Burton J., van der Linden M., et al. (2011). Rapid pneumococcal evolution in response to clinical interventions. Science 331 430–434. 10.1126/science.1198545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croucher N. J., Walker D., Romero P., Lennard N., Paterson G. K., Bason N. C., et al. (2009). Role of conjugative elements in the evolution of the multidrug-resistant pandemic clone Streptococcus pneumoniae Spain23F ST81. J. Bacteriol. 191 1480–1489. 10.1128/JB.01343-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang-Van A., Tiraby G., Acar J. F., Shaw W. V., Bouanchaud D. H. (1978). Chloramphenicol resistance in Streptococcus pneumoniae: enzymatic acetylation and possible plasmid linkage. Antimicrob. Agents Chemother. 13 577–583. 10.1128/AAC.13.4.577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frost L. S., Leplae R., Summers A. O., Toussaint A. (2005). Mobile genetic elements: the agents of open source evolution. Nat. Rev. Microbiol. 3 722–732. 10.1038/nrmicro1235 [DOI] [PubMed] [Google Scholar]
- Henderson-Begg S. K., Roberts A. P., Hall L. M. C. (2009). Diversity of putative Tn5253-like elements in Streptococcus pneumoniae. Int. J. Antimicrob. Agents 33 364–367. 10.1016/j.ijantimicag.2008.10.002 [DOI] [PubMed] [Google Scholar]
- Iannelli F., Chiavolini D., Ricci S., Oggioni M. R., Pozzi G. (2004). Pneumococcal surface protein C contributes to sepsis caused by Streptococcus pneumoniae in mice. Infect. Immun. 72 3077–3080. 10.1128/IAI.72.5.3077-3080.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iannelli F., Giunti L., Pozzi G. (1998). Direct sequencing of long polymerase chain reaction fragments. Mol. Biotechnol. 10 183–185. 10.1007/BF02760864 [DOI] [PubMed] [Google Scholar]
- Iannelli F., Pozzi G. (2004). Method for introducing specific and unmarked mutations into the chromosome of _Streptococcus pneumoniae_. Mol. Biotechnol. 26 81–86. 10.1385/MB:26:1:81 [DOI] [PubMed] [Google Scholar]
- Iannelli F., Santoro F., Oggioni M. R., Pozzi G. (2014). Nucleotide sequence analysis of integrative conjugative element Tn5253 of Streptococcus pneumoniae. Antimicrob. Agents Chemother. 58 1235–1239. 10.1128/AAC.01764-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson C. M., Grossman A. D. (2015). Integrative and conjugative elements (ICEs): what they do and how they work. Annu. Rev. Genet 49 577–601. 10.1146/annurev-genet-112414-055018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiliç A. O., Vijayakumar M. N., al-Khaldi S. F. (1994). Identification and nucleotide sequence analysis of a transfer-related region in the streptococcal conjugative transposon Tn5252. J. Bacteriol 176 5145–5150. 10.1128/jb.176.16.5145-5150.1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanie J. A., Ng W.-L., Kazmierczak K. M., Andrzejewski T. M., Davidsen T. M., Wayne K. J., et al. (2007). Genome sequence of Avery’s virulent serotype 2 strain D39 of Streptococcus pneumoniae and comparison with that of unencapsulated laboratory strain R6. J. Bacteriol. 189 38–51. 10.1128/JB.01148-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C. A., Babic A., Grossman A. D. (2010). Autonomous plasmid-like replication of a conjugative transposon. Mol. Microbiol. 75 268–279. 10.1111/j.1365-2958.2009.06985.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu F., Churchward G. (1995). Tn916 target DNA sequences bind the C-terminal domain of integrase protein with different affinities that correlate with transposon insertion frequency. J. Bacteriol. 177 1938–1946. 10.1128/jb.177.8.1938-1946.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manganelli R., Romano L., Ricci S., Zazzi M., Pozzi G. (1995). Dosage of Tn916 circular intermediates in Enterococcus faecalis. Plasmid 34 48–57. 10.1006/plas.1995.1032 [DOI] [PubMed] [Google Scholar]
- Mingoia M., Morici E., Morroni G., Giovanetti E., Del Grosso M., Pantosti A., et al. (2014). Tn5253 family integrative and conjugative elements carrying mef(I) and catQ determinants in Streptococcus pneumoniae and Streptococcus pyogenes. Antimicrob. Agents Chemother. 58 5886–5893. 10.1128/AAC.03638-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mingoia M., Tili E., Manso E., Varaldo P. E., Montanari M. P. (2011). Heterogeneity of Tn5253-like composite elements in clinical Streptococcus pneumoniae isolates. Antimicrob. Agents Chemother 55 1453–1459. 10.1128/AAC.01087-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullany P., Roberts A. P., Wang H. (2002). Mechanism of integration and excision in conjugative transposons. Cell. Mol. Life Sci. 59 2017–2022. 10.1007/s000180200001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulsen I. T., Banerjei L., Myers G. S. A., Nelson K. E., Seshadri R., Read T. D., et al. (2003). Role of mobile DNA in the evolution of vancomycin-resistant Enterococcus faecalis. Science 299 2071–2074. 10.1126/science.1080613 [DOI] [PubMed] [Google Scholar]
- Pearce B. J., Iannelli F., Pozzi G. (2002). Construction of new unencapsulated (rough) strains of Streptococcus pneumoniae. Res. Microbiol. 153 243–247. 10.1016/S0923-2508(02)01312-8 [DOI] [PubMed] [Google Scholar]
- Petrosyan V., Holder M., Ajami N. J., Petrosino J. F., Sahasrabhojane P., Thompson E. J., et al. (2016). Complete genome sequence of Streptococcus mitis strain SVGS_061 isolated from a neutropenic patient with viridans group streptococcal shock syndrome. Genome Announc 4:e00259-16. 10.1128/genomeA.00259-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Provvedi R., Manganelli R., Pozzi G. (1996). Characterization of conjugative transposon Tn5251 of Streptococcus pneumoniae. FEMS Microbiol. Lett. 135 231–236. 10.1111/j.1574-6968.1996.tb07994.x [DOI] [PubMed] [Google Scholar]
- Santoro F., Oggioni M. R., Pozzi G., Iannelli F. (2010). Nucleotide sequence and functional analysis of the tet(M)-carrying conjugative transposon Tn5251 of Streptococcus pneumoniae. FEMS Microbiol. Lett. 308 150–158. 10.1111/j.1574-6968.2010.02002.x [DOI] [PubMed] [Google Scholar]
- Santoro F., Vianna M. E., Roberts A. P. (2014). Variation on a theme; an overview of the Tn916/Tn1545 family of mobile genetic elements in the oral and nasopharyngeal streptococci. Front. Microbiol. 5:535. 10.3389/fmicb.2014.00535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott J. R., Kirchman P. A., Caparon M. G. (1988). An intermediate in transposition of the conjugative transposon Tn916. Proc. Natl. Acad. Sci. U.S.A. 85 4809–4813. 10.1073/pnas.85.13.4809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith M. D., Hazum S., Guild W. R. (1981). Homology among tet determinants in conjugative elements of streptococci. J. Bacteriol. 148 232–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storrs M. J., Poyart-Salmeron C., Trieu-Cuot P., Courvalin P. (1991). Conjugative transposition of Tn916 requires the excisive and integrative activities of the transposon-encoded integrase. J. Bacteriol. 173 4347–4352. 10.1128/jb.173.14.4347-4352.1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uicker W. C., Schaefer L., Britton R. A. (2006). The essential GTPase RbgA (YlqF) is required for 50S ribosome assembly in Bacillus subtilis. Mol. Microbiol. 59 528–540. 10.1111/j.1365-2958.2005.04948.x [DOI] [PubMed] [Google Scholar]