Abstract
The use of T cell receptor (TCR) gene-modified T cells in adoptive cell transfer has had promising clinical success, but often, simple preclinical evaluation does not necessarily accurately predict treatment efficacy or safety. Preclinical studies generally evaluate one or a limited number of type 1 cytokines to assess antigen recognition. However, recent studies have implicated other “typed” T cells in effective anti-tumor/viral immunity, and limited functional evaluations may underestimate cross-reactivity. In this study, we use an altered peptide ligand (APL) model and multi-dimensional flow cytometry to evaluate polyfunctionality of TCR gene-modified T cells. Evaluating six cytokines and the lytic marker CD107a on a per cell basis revealed remarkably diverse polyfunctional phenotypes within a single T cell culture and among peripheral blood lymphocyte (PBL) donors. This polyfunctional assessment identified unexpected phenotypes, including cells producing both type 1 and type 2 cytokines, and highlighted interferon γneg (IFNγneg) antigen-reactive populations overlooked in our previous studies. Additionally, APLs skewed functional phenotypes to be less polyfunctional, which was not necessarily related to changes in TCR-peptide-major histocompatibility complex (pMHC) affinity. A better understanding of gene-modified T cell functional diversity may help identify optimal therapeutic phenotypes, predict clinical responses, anticipate off-target recognition, and improve the design and delivery of TCR gene-modified T cells.
Keywords: T cell, T cell receptor, gene-modified T cells, altered peptide ligand, affinity, polyfunctionality, multi-dimensional flow cytometry, adoptive cell therapy
Spear and colleagues highlight the unexpected and exceptional diversity of polyfunctional phenotypes by T cell receptor gene-modified T cells and demonstrate the impact altered peptide ligand interactions have on skewing polyfunctional populations. These observations hold significant implications in the design and delivery of TCR gene-modified T cells.
Introduction
Treatment of malignancies and viral diseases using T cell receptor (TCR) gene-modified T cells in adoptive cell transfer (ACT) has had moderate but promising clinical success.1 Many of the therapeutic and safety shortcomings of TCR gene-modified T cells have been related to the affinity and specificity of the introduced TCR and preclinical studies failing to predict their in vivo functional capacity. For instance, TCRs with high affinity have recognized low levels of targeted antigen expressed on normal tissue,2 unpredictably cross-reacted against related3 or unrelated antigens.4, 5 Conversely, in diseases with genomic instability, a TCR with the flexibility to recognize altered peptide ligands (APLs) may be advantageous to combat immune escape variants.6 Functional evaluation of TCR gene-modified T cells in pre-clinical studies is generally limited to the production of a single or limited number of type 1 cytokine(s) because many reports suggest they are the best anti-tumor or anti-viral effectors.7, 8, 9 However, more recent studies have identified T cell functional subtypes (type 2, type 17, etc) with anti-tumor potential, acknowledging T cells as multi-functional effectors10, 11, 12 and suggesting that a broader functional evaluation may more appropriately predict the efficacy and safety of TCR gene-modified T cells. The polyfunctional capacity of TCR gene-modified T cells has not yet been well characterized, specifically comparing functional differences between peripheral blood lymphocyte (PBL) donor sources and evaluating how altered or related antigens influence polyfunctional responses.
In this study, we use a well-characterized APL model6, 13, 14, 15, 16 to evaluate the polyfunctional potential of TCR gene-modified T cells and evaluate how polyfunctional phenotypes are skewed by APLs. We have previously shown that HCV1406 TCR is therapeutic in a hepatitis C virus (HCV)-associated hepatocellular carcinoma mouse model15 and is cross-reactive against numerous naturally occurring HCV immune escape epitopes.6 We showed that APL recognition is not solely determined by TCR-peptide-major histocompatibility complex (pMHC) affinity using interferon γ (IFNγ) as a functional readout.16 Here, we use multi-dimensional flow cytometry to measure differential expression of six cytokines (IFNγ, tumor necrosis factor alpha [TNF-α], interleukin-2 [IL-2], IL-4, IL-17A, and IL-22) and the lytic marker CD107a to determine how APLs alter TCR gene-modified T cell polyfunctional responses on a per cell basis. Although other cytokines were initially evaluated, we chose this set of pro-inflammatory markers (limited by immunofluorescence channels) to cast a wide enough net to identify a spectrum of phenotypes believed to be important in anti-viral and anti-tumor immunity. These are described in more detail in the Discussion. Interestingly, there was remarkable diversity of polyfunctional populations in a single T cell culture and among PBL donors despite being engineered with the same TCR. Additionally, a large percentage of IFNγneg T cells were often positive for other functional markers, suggesting that on-target or cross-reactivity can be grossly underestimated when evaluating a single cytokine. Surprisingly, subsets of T cells produced multiple “typed” cytokines, contradicting dogma of a polarized immune response.7 Most strikingly, APLs and lower antigen density skewed functional profiles to be less polyfunctional, producing fewer cytokines simultaneously, and were not necessarily related to TCR-pMHC affinity. Our examination of the crystal structure of TCR and the WT pMHC complex can provide possible explanations for how small changes at the TCR-pMHC interface can induce drastic downstream functional consequences. A broader understanding of the polyfunctional capacity of gene-modified T cells and how recognition of APLs alter their functional behavior can have significant implications into the design, delivery, safety, and efficacy of TCR gene-modified T cells in ACT.
Results
TCR Gene-Modified T Cell Cultures Are Polyfunctional and Individual Functional Phenotypes Vary between Donors
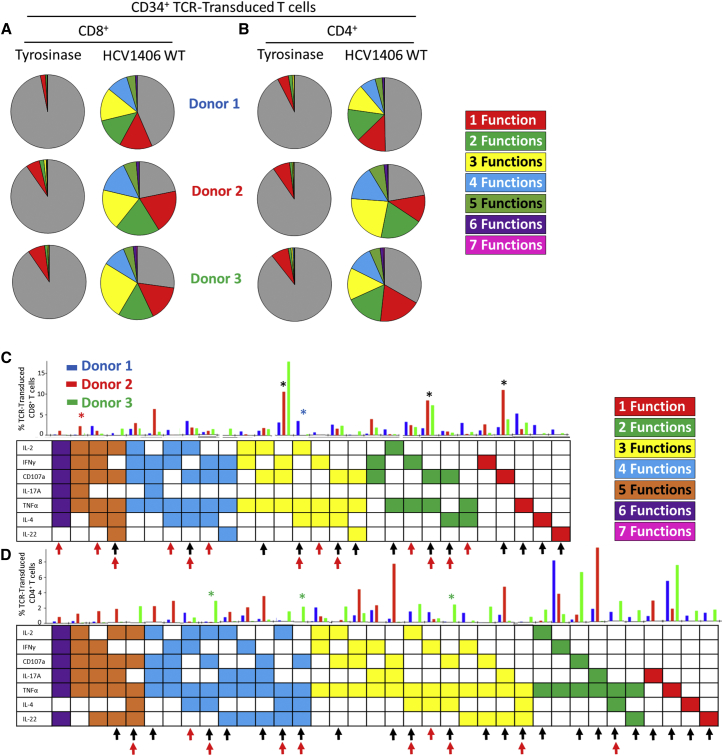
We began characterizing polyfunctionality of TCR gene-modified T cells by stimulating HCV1406 TCR-engineered T cells with the WT HCV NS3:1406-1415 peptide and comparing functional responses among three different healthy PBL donors. We assessed expression of seven functional parameters (IFNγ, TNFα, IL-2, IL-4, IL-17A, IL-22, and CD107a) on a per cell basis, resulting in 128 different possible combinatorial functional phenotypes. We used Boolean gating in FlowJo software to pool phenotypes into categories by the number of functional markers (0–7) identified per cell. Overall, both CD8+ (Figure 1A) and CD4+ (Figure 1B) T cells from all 3 donors were robustly reactive against WT peptide-loaded T2 cells, producing a large range of functional markers. Pestle and SPICE software packages were used to determine the frequency of each of the 128 possible phenotypes for CD8+ and CD4+ TCR-transduced T cells (Figure S1). Figures 1C and 1D portray CD8+ and CD4+ responses, respectively, abridged to display phenotypes with frequencies >1% in at least one donor for simplicity. There are 28 distinct CD8+ functional phenotypes and 36 distinct CD4+ functional phenotypes represented among the three donors evaluated. Some functional phenotypes are shared, whereas others are unique to individual cultures. For example, IFNγ+CD107a+TNFα+ tri-functional, CD107a+TNFα+ bi-functional, and CD107a+ mono-functional phenotypes were robustly present among all three CD8+ subsets (Figure 1C, black asterisks), although their relative magnitudes varied among donors. Conversely, IL-2+TNFα+IL-4+ was unique to donor 1 (blue asterisk) and IL-2+IFNγ+CD107a+IL-17A+TNFα+ was unique to donor 2 (red asterisk). There was even greater diversity within CD4+ functional phenotypes, including the number of phenotypes not shared among donors. For instance, donor 3 exclusively exhibited CD107a+TNFα+IL-4+IL-22+, CD107a+TNFα+IL-4+, and CD107a+TNFα+IL-22+ populations (green asterisks), among others, but lacked IL-17A+TNFα+IL-22+, IL-17A+TNFα+, IFNγ+CD107a+IL-17A+TNFα+, and IL-2+CD107a+IL-17A+TNFα+IL-22+ phenotypes, which were present in donors 1 and/or 2. Additionally, the relative polyfunctionality differed between donors. Donor 2 displayed the greatest number of phenotypes with 4+ functions, whereas donor 1 had a greater proportion of phenotypes limited to 1, 2, or 3 functions. Overall, this type of analysis emphasizes that despite being engineered with the same TCR, inherent differences in individuals’ T cell repertoire and/or physiology can have a drastic impact on the functional phenotypes observed upon antigen encounter.
Figure 1.
Polyfunctional Phenotypes of HCV1406 TCR-Transduced T Cells Are Remarkably Heterogeneous within Individual Cultures and among PBL Donors
HCV1406 TCR-transduced T cells derived from PBL of three healthy donors were co-cultured for 5 hr with T2 cells loaded with 10 μg/mL NS3:1406-1415 or tyrosinase:368-376 peptide. T cells were evaluated for surface lineage markers, CD107a, and intracellular IFNγ, TNFα, IL-2, IL-4, IL-17A, and IL-22. (A and B) Boolean gating seven functional markers for TCR-transduced (A) CD8+ and (B) CD4+ T cells generated functional categories producing no (gray; non-reactive), 1 (red), two (green), 3 (yellow), 4 (blue), 5 (orange), 6 (purple), or 7 (pink) simultaneous functions. Three donors from a representative experiment are shown. (C and D) Of the 128 possible phenotypes of 7-dimensional analysis, frequency of individual populations >1% in at least one donor are shown for (C) CD8+ and (D) CD4+ TCR-transduced T cells. A representative experiment is shown comparing donor 1 (blue bars), donor 2 (red bars), and donor 3 (green bars). Each column represents an individual phenotype, the presence of a functional marker indicated by a shaded box. Black asterisks denote shared phenotypes among more than one donor referenced in the text. Blue, red, or green asterisks represent donor-specific phenotypes referenced in the text. Black arrows indicate antigen-reactive populations lacking IFNγ. Red arrows indicate populations producing both type 1 and type 2 cytokines.
This multi-functional analysis also highlights populations of antigen-reactive cells that could be lost in traditional functional evaluations probing for only IFNγ or other type 1 cytokines. The phenotype matrix in Figures 1C and 1D identifies populations of CD8+ and CD4+ T cells in all three donors lacking IFNγ but performing many other functions (black arrows). Moreover, this type of analysis reveals unexpected phenotypes of individual cells producing both type 1 and type 2 cytokines (combinations of IL-2, IFNγ, and TNFα with IL-4; red arrows), which are generally thought to be mutually exclusive. Together, this type of analysis highlights the remarkable functional diversity of TCR gene-modified T cells within a single culture and among different donors, and acknowledges the gaps present in traditional means of assaying T cell function.
APLs Induce Fewer Simultaneous Functions per T Cell
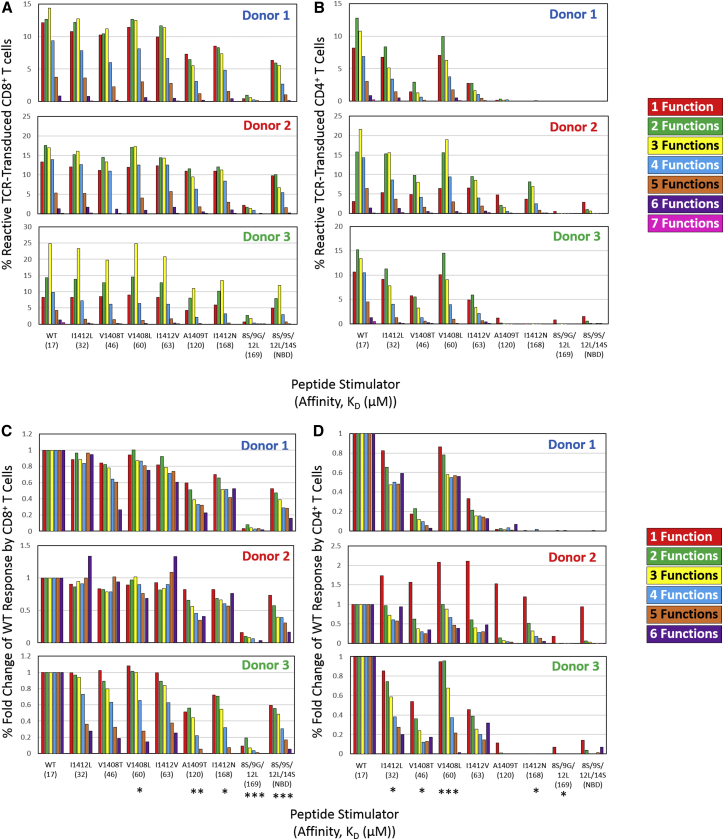
The therapeutic efficacy and safety of TCR gene-modified T cells is often influenced by TCR specificity and cross-reactivity. Given the functional diversity of TCR-engineered T cells against cognate ligand, it is not well defined how APLs influence polyfunctional responses, although seminal APL studies suggest a TCR can have differential signaling and functional output.17, 18, 19 Here, we use multi-dimensional flow cytometry to evaluate if and how APLs impact the polyfunctionality of TCR gene-modified T cell responses relative to WT antigen recognition. Figure 2 displays the percentage of HCV1406 TCR-engineered T cells expressing 1–7 functional markers when stimulated with T2 cells loaded with WT or variant HCV NS3:1406-1415 peptides. Background reactivity against irrelevant tyrosinase:368-376 has been subtracted in each category to show antigen-specific responses (non-background subtractions can be found in Figure S2). Percentages of cells in each functional category are shown for all three donors’ CD8+ (Figure 2A) and CD4+ (Figure 2B) T cell subsets. We have previously reported on equilibrium KD values of each TCR-pMHC interaction.16 These values are provided in Figure 2 for reference, and pMHC ligands are organized from left to right in decreasing TCR-pMHC affinity.
Figure 2.
Altered TCR-pMHC Interactions Induce Fewer Polyfunctional Phenotypes
(A and B) Boolean gating seven functional markers for TCR-transduced (A) CD8+ and (B) CD4+ T cells generated functional categories with same color scheme as Figure 1. Frequencies are background subtracted (irrelevant tyrosinase:368-376 peptide stimulation) to show specific reactivity for each donor. (C and D) Relative fold-change of altered ligand compared to WT for each functional category is shown for (C) CD8+ and (D) CD4+ TCR-transduced T cells. The category of 7 functions was removed from fold change analysis because frequencies were 1%. A linear regression model was used to determine if there was a trend in fold change for each peptide relative to WT. Each model included fold change as the outcome, main effects of the number of functions (continuous), and donor (categorical). p values reported represent the evidence of linear trend between number of functions and fold change (*p < 0.05, **p < 0.01, ***p < 0.0001). Previously reported TCR-pMHC affinity values (KD, μM) are displayed below each peptide ligand for reference.
In agreement with our previous reports comparing IFNγ release, polyfunctional recognition of lower affinity variants A1409T, I1412N, 8S/9G/12L, and 8S/9S/12L/14S was generally CD8-dependent, and 8S/9G/12L was not recognized.6, 16 However, CD4+ T cells of donor 2 appeared to have modest recognition against these mutants, which is accounted for by IFNγneg reactive populations (Figure 2B), described below. Additionally, although changes in total reactivity were not directly related to changes in TCR-pMHC affinity (supporting IFNγ-release studies16), there was an interesting pattern in changes in polyfunctional categories against APLs in both CD4+ and CD8+ T cells. When an APL dampened the overall response, decreases in percent reactive cells was not equal across all functional categories. In these cases, APLs modestly affected percentage of mono- or bi-functional cells, but dramatically reduced cells producing 3, 4, or 5+ functional markers at greater rates. This preferential decrease in higher order polyfunctional populations is consistent across donors and more appropriately displayed as measures of fold-change compared to WT (Figures 2C and 2D). A linear regression model of APL-stimulated polyfunctional phenotypes determined that as the number of simultaneous functions increased, the fold change compared to WT stimulation decreased. However, the magnitude of these differences across APLs was not consistent with their TCR-pMHC affinity, supporting our previous observations.16 Of note, the category of 7 functions was removed from a linear regression analysis because frequencies were <1%. Together, these data suggest that APLs can preferentially induce fewer simultaneous functions, independently of TCR-pMHC affinity.
APLs Skew Frequency of Individual Polyfunctional Profiles
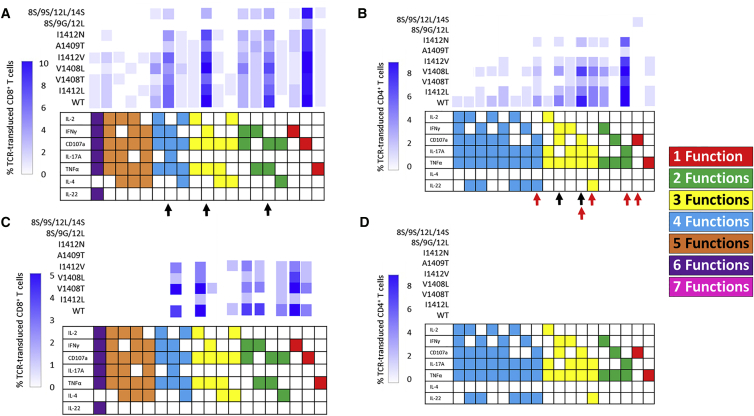
Although APLs impacted the global polyfunctionality (number of functional markers per cell), we wanted to next evaluate the frequency of individual combinatorial phenotypes among APLs and between CD4+ and CD8+ T cells in all three donors. The multi-dimensional data analysis software packages Pestle and SPICE20, 21 were used to generate a heatmap-like representation, called a “cool plot,” displaying frequencies of each of the 128 possible phenotypes across WT and APL stimulations. Trends in changes of ligand-specific reactivity were consistent among donors, and uncondensed cool plots for each can be found in Figure S3. For simplicity, we show a representative cool plot of a single donor, abridged to show only phenotypes above background on the corresponding scale for peptide-stimulated CD8+ (Figure 3A) and CD4+ (Figure 3B) TCR-transduced T cells. Supporting previous observations, APLs did not induce a decrease in the frequency of phenotypes proportional to changes in TCR-pMHC affinity (ordered from bottom to top in decreasing affinity). This is best shown for CD8+ phenotypes, including IFNγ+CD107a+IL-17A+TNFα+, IFNγ+CD107a+TNFα+, and CD107a+TNFa+ (black arrows; Figure 3A). Frequencies of CD107a+ single-positive CD8+ T cells, however, were fairly constant across APLs, suggesting that APLs (and TCR-pMHC affinity) may have the least functional impact on this phenotype. Contrastingly, simultaneous production of 5 and/or 6 functional markers by CD8+ T cells were generally restricted to WT and “moderate affinity” APLs, suggesting that lower affinity may play a role in facilitating higher order polyfunctional phenotypes.
Figure 3.
Changes in pMHC Ligand and Ligand Density Skew Individual Polyfunctional Phenotypes
Software packages Pestle/SPICE were used to generate cool plots based on FlowJo Boolean gating of 7 functional markers. Cool plot is condensed to show relevant populations based on the frequency gradation scale. Data reflect (A) T2-stimulated CD8+ T cells, (B) T2-stimulated CD4+ T cells, (C) HCV+ HepG2-stimulated CD8+ T cells, and (D) HCV+ HepG2-stimulated CD4+ T cells of donor 2 from Figure 2, but uncondensed cool plots for all donors can be found in Figure S3. The presence of a cytokine or CD107a is indicated by a shaded/colored box, and individual phenotypes are listed as columns along the x axis. Phenotypes are also color coordinated to easily show the number of functions similar to Figures 1 and 2. For a given pMHC ligand, elicited phenotypes are read across the x axis. Frequency of cells positive for a given phenotype corresponds to the blue shaded scale. Changes in frequencies of an individual phenotype across various ligand stimulations can be read in the y-direction. TCR-pMHC interactions are ranked from bottom to top by decreasing affinity. Black arrows indicate functional phenotypes with fluctuating frequencies not associated with TCR-pMHC affinity. Red arrows indicate low-affinity APL-reactive populations lacking IFNγ discussed in the text.
Additionally, reactivity patterns by CD4+ T cells supports the inconsistent relationship between TCR-pMHC affinity and T cell function observed in CD8+ T cells. Despite variants I1412L, V1408T, V1408L, and I1412V exhibiting similar TCR-pMHC affinities, there were distinct fluctuations among the frequency of functional phenotypes, including CD107a+IL-17A+TNFα+ and IFNγ+CD107a+TNFα+ (black arrows; Figure 3B). Our previous studies indicated that CD4+ T cells were non-reactive against lower affinity variants A1409T, I1412N, and 8S/9S/12L/14S, which required both TCR-pMHC stabilization as well as lck recruitment by the CD8 co-receptor to facilitate IFNγ release. Here, we unexpectedly found that multiple low-affinity APLs were recognized by CD4+ T cell populations but lacked IFNγ production (red arrows; Figure 3B), suggesting that probing for only a single cytokine can underestimate the functional capacity (and cross-reactivity) of TCR gene-modified T cells. However, there were far less quad-functional CD4+ T cells against APLs compared to mono-, bi-, or tri-functional, suggesting again that APLs may preferentially reduce higher order polyfunctional responses. The general disconnect between changes in phenotypes and TCR-pMHC affinity is consistent among other donor functional profiles (Figure S3). Together, these data indicate that APLs can skew polyfuctional profiles but that changes in TCR-pMHC affinity are not clearly related to changes in certain phenotype frequency. Additionally, APLs can preferentially diminish higher order polyfunctional phenotypes.
Naturally Processed APL by Tumor Cells Induced Fewer Polyfunctional Phenotypes
Peptide-loaded T2 cells are a suitable model for evaluating TCR gene-modified T cell function, but it is also important to evaluate polyfunctionality against physiologically relevant targets, including tumor cells, which we have previously shown present APL at lower densities, directly impacting the magnitude of IFNγ release.16 HepG2 cells, an HLA-A2+ hepatocellular carcinoma cell line, were engineered to express full-length HCV NS3 protein with WT or APL 1406-1415 epitopes. We evaluated the polyfunctional responses of CD8+ (Figure 3C) and CD4+ (Figure 3D) TCR-transduced T cells against these cell lines. Although CD4+ T cells were not able to recognize this low density of antigen consistent with previous reports,16 CD8+ T cells exhibited polyfunctional responses, but were extremely blunted in both number and diversity compared to peptide stimulations. For example, WT stimulation reduced the number of different functional phenotypes from 15 to 8 (Figure 3C). Interestingly, this decrease in ligand density preferentially eliminated higher order polyfunctional phenotypes producing 4, 5, or 6 functional markers. This elimination of higher order phenotypes mimics what is seen in peptide titration experiments, suggesting lower antigen density impacts the diversity and order of polyfunctionality (Figure S4). Although low-affinity APLs were not recognized, the variation in phenotype frequencies against recognized APLs were again not related to changes in TCR-pMHC affinity. Overall, tumor stimulation preferentially reduced higher-order polyfunctional populations and restricted the heterogeneity of responses, suggesting that lower antigen density may result in incomplete T cell activation and impact the quality of a functional response.
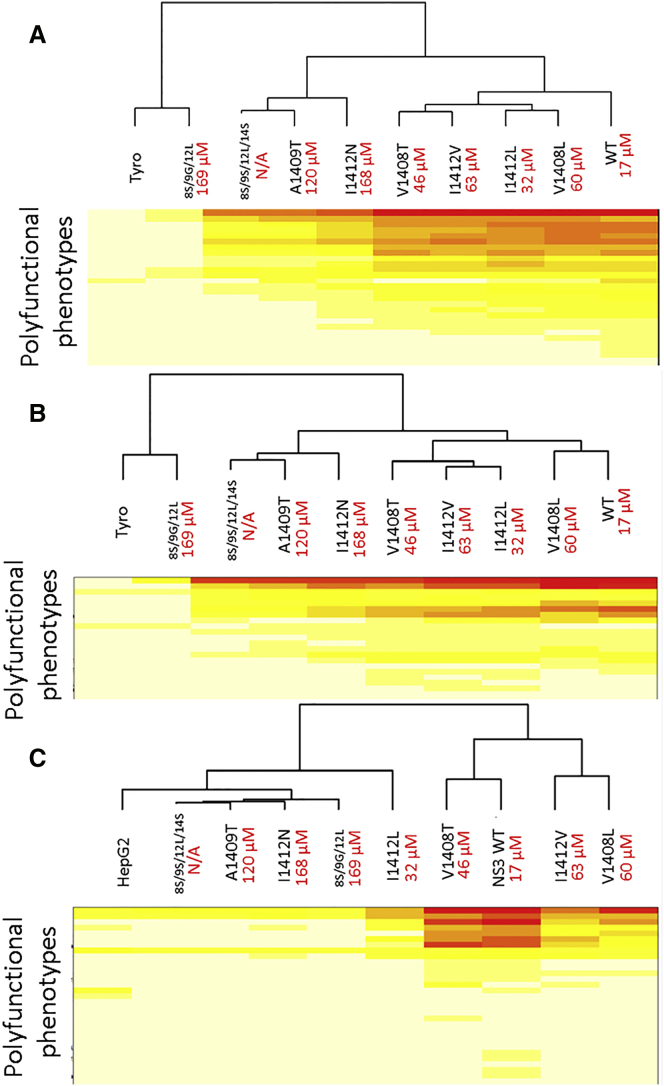
Number and Frequency of Polyfunctional Phenotypes Are Not Dictated by TCR-pMHC Affinity
We have previously argued that TCR-pMHC affinity is not the sole determinant of T cell function.16 Data presented above also argue that changes in polyfunctional responses are also not necessarily dictated by changes in TCR-pMHC affinity either at high or low antigen densities. Hierarchical clustering analysis was used to compare the relatability of polyfunctional responses and TCR-pMHC affinity (Figure 4). Maps are abbreviated to show only minimal functional phenotypes, displaying their relative frequency and diversity of peptide-stimulated CD8+ T cells (Figure 4A), peptide-stimulated CD4+ T cells (Figure 4B), and tumor-stimulated CD8+ T cells (Figure 4C). Complete maps can be found in Figure S5. From these plots, it is evident that functional phenotypes are not necessarily dependent on TCR-pMHC affinity. For example, although overall reactivity by CD4+ T cells is restricted by an affinity threshold somewhere between that of I1412V (63 μM) and A1409T (120 μM), the clustering of functional responses among moderate affinity ligands does not relate to the modest changes in their affinity. Nor does polyfunctionality by CD8+ T cells cluster with affinity. Although responses against I1412N cluster with three other lower affinity variants, 8S/9G/12L with nearly identical TCR-pMHC affinity to I1412N is non-reactive and clusters with an irrelevant peptide stimulation. Additionally, responses among moderate affinity are not necessarily related in a way that TCR-pMHC affinity would predict. Interestingly, the relatedness of pMHC ligands changed upon tumor stimulation of CD8+ T cells (Figure 4C), suggesting that the alterations in TCR-pMHC interactions can have a different impact on functional outcomes at lower densities. Together, these data provide additional evidence and a broader functional overview to our previous observations that TCR-pMHC affinity is not necessarily the sole determinant of T cell function.
Figure 4.
Frequency and Number of Polyfunctional Phenotypes Is Not Necessarily Dictated by TCR-pMHC Affinity
Hierarchical clustering analysis of Boolean-gated functional phenotypes and altered pMHC ligands was performed using hierarchical clustering based on the correlation matrix. Data reflect (A) T2-stimulated CD8+ T cells, (B) T2-stimulated CD4+ T cells, and (C) HCV+ HepG2-stimulated CD8+ T cells of donor 2. Relatedness of ligand-stimulated responses is shown in dendograms. Clustering diagrams are shown cut off to positive phenotypes. Complete clustering analyses can be found in Figure S5.
Potential Structural Consequences of Peptide Alterations on T Cell Polyfunction
Because the T cell functional properties induced by the peptide variants did not correlate strongly with TCR binding affinity, we considered other contributors. As we discussed recently, in addition to small changes in affinity, receptor binding kinetics (on/off rates), TCR 2D affinities that occur in the biological setting, and small alterations in peptide-MHC binding affinity, alone or in various combinations, could all influence outcome.16 However, there are conflicting data on the influence of each of these on T cell function.22, 23, 24, 25 One other intriguing possibility is structural consequences in response to peptide modifications because data suggest that T cell signaling can be influenced by architectural changes in how a TCR binds pMHC.26, 27 We therefore examined the recently determined crystal structure of the HCV1406 TCR bound to NS3:1406-1415/HLA-A228 to help ascertain whether gross structural changes could be induced upon peptide modification.
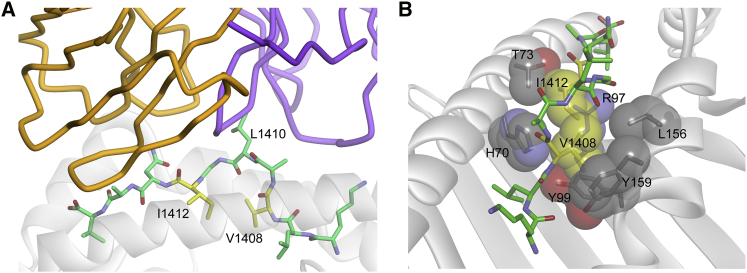
We focused our analysis on two positions where small changes in TCR binding affinity nevertheless resulted in large impacts on polyfunctionality: V1408 and I1412. As discussed, mutations at these two residues had varying impact on the frequency/number of polyfunctional phenotypes and CD8 dependence and exhibited a wide range of TCR-pMHC affinities, making them high-yield positions to focus on. In the published crystal structure, neither the side chains of V1408 nor I1412 interact directly with the TCR.28 Rather, both are directed toward the base of the HLA-A2 peptide binding groove (Figure 5A), packed tightly within the peptide-MHC interface (Figure 5B). Amino acid changes at these positions, even conservative changes, could lead to large alterations in the overall peptide conformation, requiring alterations to TCR binding modes. Replacement of the I1412 with valine, for example, could lead to an unfavorable “cavity” in the interface between the peptide and MHC, necessitating a peptide conformational change to fill it. Overall, we hypothesize that changes at V1408 and I1412 are likely to yield greater structural alterations to the peptide than changes at fully exposed, direct TCR contact residues, such as L1410, potentially altering TCR engagement and signaling. Indeed, this may be why changes to V1408 and I1412 are found in naturally occurring escape variants of the NS3 epitope, as opposed to changes at fully exposed TCR contacting residues such as L1410.
Figure 5.
Key Peptide Variations Occur in MHC-Buried Peptide Residues
(A) Amino acids V1408 and I1412 are not direct TCR contact residues, but instead are buried in the base of the HLA-A2 binding groove. (B) The molecular environments around V1408 and I1412 are tightly packed, such that changes here are likely to alter peptide conformation, potentially altering how the TCR engages the peptide-MHC complex, contributing to alterations in signaling outcomes.
Discussion
Although clinical trials using TCR gene-modified T cells in ACT have had promising results, unpredictable objective responses as well as on- and off-target adverse events call for improvements in therapeutic design and a better understanding of the functional capacity of TCR gene-modified T cells. This includes broadening the way in which we evaluate reactivity and using models to assess how APLs influence functional outcomes. This study provides a model to evaluate polyfunctional responses by TCR gene-modified T cells, compare heterogeneity among PBL donors, and assess the effects of APLs on polyfunctional outcomes.
Traditionally, T cells are often classified by their functional profile (type 1 versus type 2, etc.), which has been related to therapeutic efficacy.8, 9, 29, 30, 31 Specifically, T cells with type 1 responses (IFNγ, TNFα, and IL-2 producing) and lytic behavior are considered to facilitate better effectors in anti-tumor/viral responses.8, 9, 32, 33, 34 In light of these generalizations, preclinical and clinical studies primarily evaluate a limited number of type 1 cytokines to characterize the reactivity of TCR gene-modified T cell cultures.35, 36, 37, 38, 39, 40, 41, 42 However, other emerging T cell subsets have been implicated in facilitating anti-tumor/viral immunity, including type 2 (IL-4 producing) or those secreting IL-17A and IL-22.11, 12, 33, 34 Although there is a greater appreciation that T cells are multi-functional effectors, studies primarily evaluate various type 1 phenotypes43, 44, 45, 46 and have been limited concerning TCR gene-modified T cells. We took a much broader approach than most investigators using multi-dimensional flow cytometry to characterize the polyfunctional capacity of HCV1406 TCR gene-modified T cells against cognate and naturally occurring APLs. Measuring for combinatorial expression of INFγ, TNFα, IL-2, IL-4, IL-17A, IL-22, and CD107a on a per cell basis, we observed a much greater degree of functional phenotype complexity than is typically appreciated in the field. Diversity in phenotypes was present not only within individual T cell cultures, but among normal PBL donors despite all T cells being engineered with the same TCR exhibiting the same specificity. Additionally, the breath of IFNγneg antigen-reactive phenotypes suggests that conventional assays used to evaluate reactivity by IFNγ production alone may grossly underestimate the percentage of reactive cells within a culture and may overgeneralize their functional potential.
Another surprising observation was the presence of populations producing different “typed” cytokines in the same cell. Specifically, populations of T cells in all three donors produced various combinations of IL-4 (type 2), IL-17A (type 17), IL-22 (type 17/22), and type 1 cytokines (IFNγ, TNFα, and IL-2). Generally, type 1 and type 2 cytokine production is thought to be mutually exclusive, driven by antagonizing transcription factors, T-bet and GATA3, respectively, leading to a “polarized” immune response.7 However, our analysis identified unexpected phenotypes, suggesting that the classification of T cells into restricted cytokine-producing populations may be drastically oversimplified. These populations may be playing significant roles in the immune response. Admittedly, the functional markers evaluated here are not all-encompassing, and inclusion of others (granulocyte-macrophage colony-stimulating factor [GM-CSF], IL-10, TGF-β, etc) may help identify other reactive phenotypes and even better evaluate the behavior of TCR gene-modified T cells, despite generating exponentially more complex datasets. However, this breadth of natural diversity among donor and cellular responses should be somewhat expected. The field has largely focused on a reductionist approach, which is useful for digesting large amounts of complex biological systems and for modeling purposes. However, it does not accurately portray the capacity of cellular immune responses. Our novel findings highlight the importance of expanding beyond a reductionist approach. As the ability to gather, analyze, and communicate complex sets of data evolves, the field should be encouraged to modify its approach in evaluating the functional capacity of gene-modified T cells and perhaps other areas in immunomonitoring.
These observations are also intriguing because our data represent the kinds of T cell populations transferred into patients. Similar evaluations of T cell polyfunctionality in the context of individuals’ anti-viral or anti-tumor responses pre- and post-transfer may allow for better treatment correlations, biomarkers for predictive outcomes, or identification of optimal T cell subsets to transfer into patients. For instance, it might be predicted that a T cell producing 3 or 4 cytokines may provide a more effective immune response than a T cell performing 1 or 2 cytokines if there are synergistic or additive effects. Conversely, if the energy requirements of producing many cytokines dampens the overall magnitude of each individual cytokine, a highly polyfunctional T cell may be less advantageous. In that case, it may be more advantageous to have multiple T cells populations with different individual phenotypes.
Another interesting observation was the profound difference in number and frequency of functional phenotypes among PBL donors, despite being engineered with the same TCR. This observation may be important in rationalizing different objective clinical responses between patients transferred with autologous TCR-engineered T cells. For example, if certain functional phenotypes are correlated with better anti-tumor responses, then their relative frequencies in individual patients may predict or dictate therapeutic efficacy. For example, if IL-2+TNFα+IL-4+ T cells elicit the “best” anti-tumor response, then donor 1 (described above) may be a better responder to therapy because this population comprises 5% of its CD8+ T cells but is absent in donors 2 and 3. Further comparison between autologous polyfunctional phenotypes and clinical responses may help identify optimal therapeutic subsets to be prioritized for adoptive transfer.
Additionally, significant variation between PBL donors not only applies to predicting clinical responses in autologous T cell transfer, but is important to consider as the field continues to develop means of generating allogenic universal donors for ACT.47 The choice of donor and its respective functional profile could directly impact therapeutic efficacy and safety. So depending on which functional phenotypes are deemed best effectors, their relative ratios among PBL donors could impact which donor sources are predicted to be the most therapeutic. Conversely, if the diversity or reactivity of PBL sources is too strong, it could lead to a cytokine storm, an adverse event observed in some clinical trials using genetically modified T cells.48, 49 Moreover, although universal agents harboring a large functional diversity of phenotypes may be attractive therapeutics, their inherent diversity would certainly pose regulatory hurdles for Food and Drug Administration (FDA) approval. Such diversity may need to be laboriously characterized or even standardized for use as an off-the-shelf reagent with predictable function. Such characterization of cellular and/or donor diversity as well development of methods to skew or guide cellular phenotypes may help standardize therapeutic subsets, making reagents more predictable and/or approvable by regulatory bodies. Is it important to recognize such barriers when exploring these types of therapeutic agents.
These unresolved questions are beyond the scope of this manuscript, but as technology improves to facilitate isolation, administration, and cellular tracking of individual functional phenotypes in vivo, such models may be helpful in addressing these hypotheses. For instance, TCR-engineered T cells adoptively transferred into tumor-bearing mice could be isolated from tumors or lymphoid compartments, assessed for functional profiles ex vivo, and compared to the tumor regression status of each animal. If T cells isolated from regressing tumors have distinctly different functional phenotypes than those isolated from stable or growing tumors, this may indicate which functional phenotypes are providing better anti-tumor immunity. However, in our hands, adoptively transferred human HCV1406 TCR-engineered T cells do not persist long enough to track and analyze post-transfer in a mouse.6 Development of a fully murinized HCV1406 TCR system or the development of other in vivo TCR/APL systems may help answer these questions. Additionally, if T cells can be sorted into individual polyfunctional phenotypes, tumor-bearing mice could receive distinct functional populations and be evaluated for which functional phenotype(s) are most effective at eradicating tumors and how administered T cells remain or alter their functional phenotypes over time in the host. As technology to perform such complex in vivo evaluations with a large number of functional markers improves, these challenging strategies may help elucidate the importance of certain functional phenotypes and how they can contribute to effective immune responses. A more complete understanding of the behavior of T cells delivered to patients might also enable us to select for or design T cells with an “optimal” polyfunctional profile. This would be an immensely powerful tool to enhance the efficacy and safety of TCR-engineered T cells used in ACT.
The ability to characterize polyfunctional responses by TCR gene-modified T cells is also important for evaluating the functional effects of APL recognition. TCR gene-modified T cell recognition of related and unrelated ligands has led to significant adverse events, including death, in multiple clinical trials.2, 4, 5, 50 However, the ability for a monoclonal therapeutic agent to recognize related ligands has potential benefit in treating cancers and virus-associated disease whose genomic instability leads to immune escape.6 The effects of APLs on the polyfunctional output of TCR gene-modified T cells have not yet been well described. A better understanding will help guide TCR design to maximize their therapeutic efficacy and safety.
Evavold and Allen first described the concept of altered peptide ligands having differential effects on T cell function, showing a single, conservative substitution (E73D) impaired proliferation but not IL-4 production in T cells. Subsequent studies involving additional T cell clones indicated that TCRs have the capacity for differential signaling, leading to a spectrum of functional responses and events.18, 19, 51, 52, 53 Many of the explanations of altered T cell function were initially rationalized by changes in affinity due to small changes in pMHC topology.54 Yet, we show here that changes in TCR-pMHC affinity and APL interactions with similar affinities were not always correlative with polyfunctional responses. Interestingly, APLs reduced polyfunctionality, dampening the number of functions performed per cell.
Others have suggested that altered peptides may induce conformational changes in how the TCR engages altered pMHC, altering TCR-initiated signals, sometimes independent of measurable affinity changes.55, 56, 57 It is also important to realize that despite transcriptional regulation of functional outcomes, the true mechanism behind altered T cell function lies at the origin of the interaction, the interface of TCR-pMHC. Therefore, we cannot fully understand how APL dictates HCV1406 TCR functional outcome without evaluating structurally what happens at this TCR-pMHC interface. We have previously described the crystal structure of HCV1406 TCR with WT HCV NS3:1406-1415/HLA-A2.28 It may be notable that the amino acid variations that show the most dramatic variation in function with the smallest changes in affinity occur at amino acids whose side chains are not direct contact residues, but rather pack between the peptide and the HLA-A2 peptide binding groove. Alterations here would be predicted to have a more substantial impact on peptide conformation, leading to greater variation in the TCR-facing surface than substitutions that occur solely at TCR contact residues. These more substantial structural variations in the peptide-MHC could have consequences for how the receptor engages, potentially altering signaling functions.26, 27
In summary, we have used multi-dimensional flow cytometry to characterize the remarkable polyfunctionality of TCR gene-modified T cells against a spectrum of naturally occurring APLs. Stimulated by cognate ligand, there was significant diversity of functional phenotypes within a single culture and among donor PBL despite being engineered with the same TCR. Additionally, APLs and lower antigen densities skewed functional phenotypes to be less polyfunctional, which were not necessarily related to TCR-pMHC affinity, supporting earlier reports.16 Insight into the structural changes at the TCR-pMHC interface may help rationalize large functional changes despite small alterations in peptide sequence. In light of these observations, we propose that more expansive functional evaluations may help correlate or predict clinical responses of TCR gene-modified T cells and have implications in other areas of immune monitoring. Subsequent in vivo evaluation of T cells with defined multi-cytokine profiles is arduous, but as the technology to do so improves, it may help identify optimal effector phenotypes, anticipate off-target recognition, or even influence the choice and design of “universal” allogeneic donor sources. These strategies will ultimately influence the design and delivery of TCR gene-modified T cells in ACT.
Materials and Methods
Cell Lines and Media
All cell lines were obtained from the American Type Culture Collection (Rockford, MD), unless otherwise noted. All media were obtained from Corning Life Sciences (Corning, NY) unless otherwise noted. HEK293GP, COS (natively HLA-A2−), and HepG2 (natively HLA-A2+) cell lines were maintained in DMEM supplemented with 10% fetal bovine serum (FBS) (Tissue Culture Biologics, Long Beach, CA). HCV+ HepG2 cell lines were generated by transducing HepG2 cells with retroviral vectors containing the entire HCV NS3 gene containing WT or mutant 1406-1415 epitopes, described below, and maintained in DMEM supplemented with 10% FBS. PG13 cells were maintained in Iscove’s DMEM supplemented with 10% FBS. T2 cells were maintained in RPMI 1640 medium supplemented with 10% FBS.
T Cells
Apheresis products of normal, healthy donors were purchased from Key Biologics (Memphis, TN). Peripheral blood mononuclear cells (PBMCs) were purified using Ficoll-Hypaque (Sigma-Aldrich, St. Louis, MO) density gradients as previously described.15 T cells were activated by stimulating PBMCs with 50 ng/mL anti-CD3 mAb (Miltenyi Biotec, Bergisch Gladbach, Germany) in AIM-V medium (Life Technologies, Carlsbad, CA) supplemented with 5% heat-inactivated pooled human AB serum (hAB) (Valley Biomedical, Winchester, VA), 300 IU/mL recombinant human IL-2 (rhIL-2) (Novartis Pharmaceuticals, East Hanover, NJ), and 100 ng/mL recombinant human IL-15 (rhIL-15) (Biological Resources Branch, National Cancer Institute, Bethesda, MD). 3 days following activation, T cells were transduced with an HCV1406 TCR-encoding retroviral vector.
Retroviral Vectors
Retroviral vectors encoding HCV NS3 antigen or HCV1406 TCR were used to transduce HepG2 cells or T cells, respectively. Target genes were encoded in a modified SAMEN retroviral vector, as previously described.6 The WT HCV NS3 or sequences containing mutant 1406-1415 epitopes were linked to a GFP reporter gene by a self-cleaving P2A sequence. The retroviral vector encoding HCV1406 TCR contained the TCR α chain linked to the TCR β chain and a truncated CD34 marker gene by P2A or T2A self-cleaving sequences, respectively. Generation of stable producer cells lines expressing these retroviral vectors and the collection of retrovirus have been previously described.6
Retroviral Transduction
HepG2 cells were seeded in a 24-well tissue culture plate to yield 70%–80% confluency. 2 mL of 0.45 μm-filtered respective retroviral supernatants were applied to each well and incubated for 48 hr at 37°C in 5% CO2. HCV-GFP+ positive cells were sorted for high and uniform expression of GFP using a FACSAria IIIu instrument (BD Biosciences, San Jose, CA). Activated T cells were transduced to express the HCV1406 TCR by spinoculation as previously described.6, 15, 58, 59, 60, 61 T cells were then sorted for TCR-transduced cells by positive selection using anti-CD34 mAb-coated immunomagnetic beads (Miltenyi Biotec) and used in functional assays.
Peptides
All peptides used in functional assays were purchased from Synthetic Biomolecules (San Diego, CA) at 95% purity. Peptide sequences are as follows: HCV NS3:1406-1415 WT (KLVALGINAV); and NS3 mutants A1409T (KLVTLGINAV), I1412L (KLVALGLNAV), I1412V (KLVALGVNAV), I1412N (KLVAALGNNAV), V1408S/A1409G/I1412L (KLSGLGLNAV), V1408T (KLTALGINAV), V1408S/A1409S/I1412L/A1414S (KLSSLGLNSV), V1408L (KLLALGINAV) tyrosinase:368-376 (TMDGTMSQV) was used as a negative control.
Antibodies
All fluorochrome-conjugated antibodies were purchased from BioLegend (San Diego, CA). These included anti-CD3-APC/Cy7, anti-CD4-PE/Cy7, anti-CD8-FITC, anti-CD34-Alexa Fluor (AF)700, anti-CD107a-Brilliant Violet (BV)510, anti-IFNγ-BV421, anti-TNFα-BV711, anti-IL-2-PerCP/Cy5.5, anti-IL-4-AF647, anti-IL-17A-BV570, and anti-IL-22-PE.
Polyfunctional T Cell Lysis and Multi-intracellular Cytokine Assay
3 × 105 stimulator cells (peptide-loaded T2 cells or HCV+ HepG2 cells) and 3 × 105 HCV1406 TCR-transduced PBL-derived T cells were mixed in a 1:1 ratio in 96-well U-bottom tissue culture plates in 200 μL complete medium. 2.5 μg/mL anti-CD107a mAb, 5.0 ng/mL brefeldin-A, and 2.0 nM monensin (all BioLegend) were added at the beginning of the co-culture. Co-cultures were incubated at 37°C for 5 hr, and cells were stained for cell surface antigens for 20 min at room temperature (RT). Subsequently, cells were incubated in Fixation Buffer (BioLegend) for 20 min, washed 3 times in Permeabilization and Wash Buffer (BioLegend), and stained for intracellular cytokines for 20 min at RT. Cells were washed, resuspended in Cell Staining Buffer (BioLegend), and analyzed by flow cytometry. Samples were acquired using a LSRFortessa flow cytometer (BD Biosciences). Staining profiles were gated and analyzed using FlowJoX software (Tree Star, Ashland, OR). Lymphocyte populations were discerned by forward scatter (FSC) versus side scatter (SSC) comparison, and events were gated on CD4+CD8− or CD4−CD8+ populations. CD4+ or CD8+ T cells were then gated on CD34+ expression to define our TCR-transduced T cell population. These CD4+CD8−CD34+ or CD4−CD8+CD34+ were subsequently used as starting points for Boolean gating on functional parameters CD107a, IFNγ, TNFα, IL-2, IL-4, IL-17A, and IL-22. Identification of functional populations was performed using software packages Pestle and SPICE (https://exon.niaid.nih.gov/spice).
Statistical Analysis
A linear regression model was used to determine if there was a trend in fold change for each variant HCV peptide relative to WT. Each model included fold change as the outcome, and the main effects of the number of functions (continuous) and donor (categorical). p values reported represent the evidence of linear trend between number of functions and fold change. Agglomerative hierarchical clustering analysis was also performed to assess the similarity of functional phenotypes across altered pMHC ligand stimulation. Frequencies of the 128 possible combinations of 7 functional parameters generated by Boolean Gating in FlowJoX was formed into a 127 × k matrix, where k = number of different stimulation conditions to be compared after removing the matrix row corresponding to the frequency of cells with zero positive parameters (128 – 1). The correlation matrix of the 127 patterns was calculated, which was used to perform agglomerative hierarchical clustering.62
Ethics Statement
All recombinant DNA and retroviral transduction work was done under approved Loyola University of Chicago Institutional Biosafety Committee protocols. Human materials used were either established, de-identified tumor cell lines or PBMCs purchased from commercial sources and not considered Human Subjects Research. Therefore, no Institutional Review Board approval was required.
Author Contributions
Conceptualization, T.T.S., B.M.B, and M.I.N.; Methodology, T.T.S., B.M.B, and M.I.N.; Investigation, T.T.S., Y.W., T.W.S., E.G.-M., L.M.H, B.M.B., and M.I.N.; Resources, P.E.S., B.M.B, and M.I.N.; Writing – Original Draft, T.T.S., B.M.B, and M.I.N.; Writing – Review and Editing, T.T.S., B.M.B, and M.I.N.; Visualization, T.T.S. and P.E.S.; Funding Acquisition, T.T.S., B.M.B, and M.I.N.
Acknowledgments
Funding for these studies was provided by the National Cancer Institute: P01 CA154779 (M.I.N.), R01 CA102280 (M.I.N.), R01 CA104947 (M.I.N.), R01 CA90873 (M.I.N.), R21 CA153789 (M.I.N.), and F30 CA180731 (T.T.S.); and the National Institute of General Medical Sciences: R35 GM118166 (B.M.B.).
Footnotes
Supplemental Information includes five figures and can be found with this article online at https://doi.org/10.1016/j.ymthe.2018.01.015.
Supplemental Information
References
- 1.Spear T.T., Nagato K., Nishimura M.I. Strategies to genetically engineer T cells for cancer immunotherapy. Cancer Immunol. Immunother. 2016;65:631–649. doi: 10.1007/s00262-016-1842-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Parkhurst M.R., Yang J.C., Langan R.C., Dudley M.E., Nathan D.A., Feldman S.A., Davis J.L., Morgan R.A., Merino M.J., Sherry R.M. T cells targeting carcinoembryonic antigen can mediate regression of metastatic colorectal cancer but induce severe transient colitis. Mol. Ther. 2011;19:620–626. doi: 10.1038/mt.2010.272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Morgan R.A., Chinnasamy N., Abate-Daga D., Gros A., Robbins P.F., Zheng Z., Dudley M.E., Feldman S.A., Yang J.C., Sherry R.M. Cancer regression and neurological toxicity following anti-MAGE-A3 TCR gene therapy. J. Immunother. 2013;36:133–151. doi: 10.1097/CJI.0b013e3182829903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cameron B.J., Gerry A.B., Dukes J., Harper J.V., Kannan V., Bianchi F.C., Grand F., Brewer J.E., Gupta M., Plesa G. Identification of a Titin-derived HLA-A1-presented peptide as a cross-reactive target for engineered MAGE A3-directed T cells. Sci. Transl. Med. 2013;5:197ra103. doi: 10.1126/scitranslmed.3006034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Linette G.P., Stadtmauer E.A., Maus M.V., Rapoport A.P., Levine B.L., Emery L., Litzky L., Bagg A., Carreno B.M., Cimino P.J. Cardiovascular toxicity and titin cross-reactivity of affinity-enhanced T cells in myeloma and melanoma. Blood. 2013;122:863–871. doi: 10.1182/blood-2013-03-490565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Spear T.T., Riley T.P., Lyons G.E., Callender G.G., Roszkowski J.J., Wang Y., Simms P.E., Scurti G.M., Foley K.C., Murray D.C. Hepatitis C virus-cross-reactive TCR gene-modified T cells: a model for immunotherapy against diseases with genomic instability. J. Leukoc. Biol. 2016;100:545–557. doi: 10.1189/jlb.2A1215-561R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Broere F., Apasov S.G., Sitkovsky M.V., van Edn W. T cell subsets and T cell-mediated immunity. In: Nijkamp F.P., Parnham M.J., editors. Principes of Immunopharmacology. Springer; 2011. pp. 15–27. [Google Scholar]
- 8.Mosmann T.R., Coffman R.L. TH1 and TH2 cells: different patterns of lymphokine secretion lead to different functional properties. Annu. Rev. Immunol. 1989;7:145–173. doi: 10.1146/annurev.iy.07.040189.001045. [DOI] [PubMed] [Google Scholar]
- 9.Tada T., Takemori T., Okumura K., Nonaka M., Tokuhisa T. Two distinct types of helper T cells involved in the secondary antibody response: independent and synergistic effects of Ia- and Ia+ helper T cells. J. Exp. Med. 1978;147:446–458. doi: 10.1084/jem.147.2.446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Burel J.G., Apte S.H., Groves P.L., McCarthy J.S., Doolan D.L. Polyfunctional and IFN-γ monofunctional human CD4+ T cell populations are molecularly distinct. JCI Insight. 2017;2:e87499. doi: 10.1172/jci.insight.87499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Foster R.G., Golden-Mason L., Rutebemberwa A., Rosen H.R. Interleukin (IL)-17/IL-22-producing T cells enriched within the liver of patients with chronic hepatitis C viral (HCV) infection. Dig. Dis. Sci. 2012;57:381–389. doi: 10.1007/s10620-011-1997-z. [DOI] [PubMed] [Google Scholar]
- 12.Lu Y., Hong S., Li H., Park J., Hong B., Wang L., Zheng Y., Liu Z., Xu J., He J. Th9 cells promote antitumor immune responses in vivo. J. Clin. Invest. 2012;122:4160–4171. doi: 10.1172/JCI65459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Callender G.G., Rosen H.R., Roszkowski J.J., Lyons G.E., Li M., Moore T., Brasic N., McKee M.D., Nishimura M.I. Identification of a hepatitis C virus-reactive T cell receptor that does not require CD8 for target cell recognition. Hepatology. 2006;43:973–981. doi: 10.1002/hep.21157. [DOI] [PubMed] [Google Scholar]
- 14.Rosen H.R., Hinrichs D.J., Leistikow R.L., Callender G., Wertheimer A.M., Nishimura M.I., Lewinsohn D.M. Cutting edge: identification of hepatitis C virus-specific CD8+ T cells restricted by donor HLA alleles following liver transplantation. J. Immunol. 2004;173:5355–5359. doi: 10.4049/jimmunol.173.9.5355. [DOI] [PubMed] [Google Scholar]
- 15.Spear T.T., Callender G.G., Roszkowski J.J., Moxley K.M., Simms P.E., Foley K.C., Murray D.C., Scurti G.M., Li M., Thomas J.T. TCR gene-modified T cells can efficiently treat established hepatitis C-associated hepatocellular carcinoma tumors. Cancer Immunol. Immunother. 2016;65:293–304. doi: 10.1007/s00262-016-1800-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Spear T.T., Wang Y., Foley K.C., Murray D.C., Scurti G.M., Simms P.E., Garrett-Mayer E., Hellman L.M., Baker B.M., Nishimura M.I. Critical biological parameters modulate affinity as a determinant of function in T-cell receptor gene-modified T-cells. Cancer Immunol. Immunother. 2017;66:1411–1424. doi: 10.1007/s00262-017-2032-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Evavold B.D., Allen P.M. Separation of IL-4 production from Th cell proliferation by an altered T cell receptor ligand. Science. 1991;252:1308–1310. doi: 10.1126/science.1833816. [DOI] [PubMed] [Google Scholar]
- 18.Evavold B.D., Sloan-Lancaster J., Hsu B.L., Allen P.M. Separation of T helper 1 clone cytolysis from proliferation and lymphokine production using analog peptides. J. Immunol. 1993;150:3131–3140. [PubMed] [Google Scholar]
- 19.Evavold B.D., Williams S.G., Hsu B.L., Buus S., Allen P.M. Complete dissection of the Hb(64-76) determinant using T helper 1, T helper 2 clones, and T cell hybridomas. J. Immunol. 1992;148:347–353. [PubMed] [Google Scholar]
- 20.Spear T.T., Nishimura M.I., Simms P.E. Comparative exploration of multidimensional flow cytometry software: a model approach evaluating T cell polyfunctional behavior. J. Leukoc. Biol. 2017;102:551–561. doi: 10.1189/jlb.6A0417-140R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Roederer M., Nozzi J.L., Nason M.C. SPICE: exploration and analysis of post-cytometric complex multivariate datasets. Cytometry A. 2011;79:167–174. doi: 10.1002/cyto.a.21015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chervin A.S., Stone J.D., Holler P.D., Bai A., Chen J., Eisen H.N., Kranz D.M. The impact of TCR-binding properties and antigen presentation format on T cell responsiveness. J. Immunol. 2009;183:1166–1178. doi: 10.4049/jimmunol.0900054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Martinez R.J., Evavold B.D. Lower affinity T cells are critical components and active participants of the immune response. Front. Immunol. 2015;6:468. doi: 10.3389/fimmu.2015.00468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stone J.D., Kranz D.M. Role of T cell receptor affinity in the efficacy and specificity of adoptive T cell therapies. Front. Immunol. 2013;4:244. doi: 10.3389/fimmu.2013.00244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhu C., Jiang N., Huang J., Zarnitsyna V.I., Evavold B.D. Insights from in situ analysis of TCR-pMHC recognition: response of an interaction network. Immunol. Rev. 2013;251:49–64. doi: 10.1111/imr.12016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Adams J.J., Narayanan S., Liu B., Birnbaum M.E., Kruse A.C., Bowerman N.A., Chen W., Levin A.M., Connolly J.M., Zhu C. T cell receptor signaling is limited by docking geometry to peptide-major histocompatibility complex. Immunity. 2011;35:681–693. doi: 10.1016/j.immuni.2011.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gras S., Chadderton J., Del Campo C.M., Farenc C., Wiede F., Josephs T.M., Sng X.Y.X., Mirams M., Watson K.A., Tiganis T. Reversed T cell receptor docking on a major histocompatibility class I complex limits involvement in the immune response. Immunity. 2016;45:749–760. doi: 10.1016/j.immuni.2016.09.007. [DOI] [PubMed] [Google Scholar]
- 28.Wang Y., Singh N.K., Spear T.T., Hellman L.M., Piepenbrink K.H., McMahan R.H., Rosen H.R., Vander Kooi C.W., Nishimura M.I., Baker B.M. How an alloreactive T-cell receptor achieves peptide and MHC specificity. Proc. Natl. Acad. Sci. USA. 2017;114:E4792–E4801. doi: 10.1073/pnas.1700459114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Harrington L.E., Hatton R.D., Mangan P.R., Turner H., Murphy T.L., Murphy K.M., Weaver C.T. Interleukin 17-producing CD4+ effector T cells develop via a lineage distinct from the T helper type 1 and 2 lineages. Nat. Immunol. 2005;6:1123–1132. doi: 10.1038/ni1254. [DOI] [PubMed] [Google Scholar]
- 30.Jäger A., Dardalhon V., Sobel R.A., Bettelli E., Kuchroo V.K. Th1, Th17, and Th9 effector cells induce experimental autoimmune encephalomyelitis with different pathological phenotypes. J. Immunol. 2009;183:7169–7177. doi: 10.4049/jimmunol.0901906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li H., Rostami A. IL-9: basic biology, signaling pathways in CD4+ T cells and implications for autoimmunity. J. Neuroimmune Pharmacol. 2010;5:198–209. doi: 10.1007/s11481-009-9186-y. [DOI] [PubMed] [Google Scholar]
- 32.Kang W., Li Y., Zhuang Y., Zhao K., Huang D., Sun Y. Dynamic analysis of Th1/Th2 cytokine concentration during antiretroviral therapy of HIV-1/HCV co-infected patients. BMC Infect. Dis. 2012;12:102. doi: 10.1186/1471-2334-12-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Price P., Keane N.M., Lee S., Lim A.F., McKinnon E.J., French M.A. A T2 cytokine environment may not limit T1 responses in human immunodeficiency virus patients with a favourable response to antiretroviral therapy. Immunology. 2006;119:74–82. doi: 10.1111/j.1365-2567.2006.02407.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Villacres M.C., Literat O., Degiacomo M., Du W., La Rosa C., Diamond D.J., Kovacs A. Reduced type 1 and type 2 cytokines in antiviral memory T helper function among women coinfected with HIV and HCV. J. Clin. Immunol. 2005;25:134–141. doi: 10.1007/s10875-005-2819-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bestard O., Lucia M., Crespo E., Van Liempt B., Palacio D., Melilli E., Torras J., Llaudó I., Cerezo G., Taco O. Pretransplant immediately early-1-specific T cell responses provide protection for CMV infection after kidney transplantation. Am. J. Transplant. 2013;13:1793–1805. doi: 10.1111/ajt.12256. [DOI] [PubMed] [Google Scholar]
- 36.Kirkwood J.M., Lee S., Moschos S.J., Albertini M.R., Michalak J.C., Sander C., Whiteside T., Butterfield L.H., Weiner L. Immunogenicity and antitumor effects of vaccination with peptide vaccine+/-granulocyte-monocyte colony-stimulating factor and/or IFN-alpha2b in advanced metastatic melanoma: Eastern Cooperative Oncology Group Phase II Trial E1696. Clin. Cancer Res. 2009;15:1443–1451. doi: 10.1158/1078-0432.CCR-08-1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee K.H., Wang E., Nielsen M.B., Wunderlich J., Migueles S., Connors M., Steinberg S.M., Rosenberg S.A., Marincola F.M. Increased vaccine-specific T cell frequency after peptide-based vaccination correlates with increased susceptibility to in vitro stimulation but does not lead to tumor regression. J. Immunol. 1999;163:6292–6300. [PubMed] [Google Scholar]
- 38.Reynolds S.R., Oratz R., Shapiro R.L., Hao P., Yun Z., Fotino M., Vukmanović S., Bystryn J.C. Stimulation of CD8+ T cell responses to MAGE-3 and Melan A/MART-1 by immunization to a polyvalent melanoma vaccine. Int. J. Cancer. 1997;72:972–976. doi: 10.1002/(sici)1097-0215(19970917)72:6<972::aid-ijc9>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 39.Salgaller M.L., Lodge P.A., McLean J.G., Tjoa B.A., Loftus D.J., Ragde H., Kenny G.M., Rogers M., Boynton A.L., Murphy G.P. Report of immune monitoring of prostate cancer patients undergoing T-cell therapy using dendritic cells pulsed with HLA-A2-specific peptides from prostate-specific membrane antigen (PSMA) Prostate. 1998;35:144–151. doi: 10.1002/(sici)1097-0045(19980501)35:2<144::aid-pros8>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- 40.Scheibenbogen C., Schmittel A., Keilholz U., Allgäuer T., Hofmann U., Max R., Thiel E., Schadendorf D. Phase 2 trial of vaccination with tyrosinase peptides and granulocyte-macrophage colony-stimulating factor in patients with metastatic melanoma. J. Immunother. 2000;23:275–281. doi: 10.1097/00002371-200003000-00012. [DOI] [PubMed] [Google Scholar]
- 41.Schreiber S., Kämpgen E., Wagner E., Pirkhammer D., Trcka J., Korschan H., Lindemann A., Dorffner R., Kittler H., Kasteliz F. Immunotherapy of metastatic malignant melanoma by a vaccine consisting of autologous interleukin 2-transfected cancer cells: outcome of a phase I study. Hum. Gene Ther. 1999;10:983–993. doi: 10.1089/10430349950018382. [DOI] [PubMed] [Google Scholar]
- 42.Tey S.K., Kennedy G.A., Cromer D., Davenport M.P., Walker S., Jones L.I., Crough T., Durrant S.T., Morton J.A., Butler J.P. Clinical assessment of anti-viral CD8+ T cell immune monitoring using QuantiFERON-CMV® assay to identify high risk allogeneic hematopoietic stem cell transplant patients with CMV infection complications. PLoS One. 2013;8:e74744. doi: 10.1371/journal.pone.0074744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Betts M.R., Nason M.C., West S.M., De Rosa S.C., Migueles S.A., Abraham J., Lederman M.M., Benito J.M., Goepfert P.A., Connors M. HIV nonprogressors preferentially maintain highly functional HIV-specific CD8+ T cells. Blood. 2006;107:4781–4789. doi: 10.1182/blood-2005-12-4818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Franzese O., Palermo B., Di Donna C., Sperduti I., Ferraresi V., Stabile H., Gismondi A., Santoni A., Nisticò P. Polyfunctional Melan-A-specific tumor-reactive CD8(+) T cells elicited by dacarbazine treatment before peptide-vaccination depends on AKT activation sustained by ICOS. OncoImmunology. 2016;5:e1114203. doi: 10.1080/2162402X.2015.1114203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kannanganat S., Ibegbu C., Chennareddi L., Robinson H.L., Amara R.R. Multiple-cytokine-producing antiviral CD4 T cells are functionally superior to single-cytokine-producing cells. J. Virol. 2007;81:8468–8476. doi: 10.1128/JVI.00228-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lepone L., Rappocciolo G., Knowlton E., Jais M., Piazza P., Jenkins F.J., Rinaldo C.R. Monofunctional and polyfunctional CD8+ T cell responses to human herpesvirus 8 lytic and latency proteins. Clin. Vaccine Immunol. 2010;17:1507–1516. doi: 10.1128/CVI.00189-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schmitt T.M., Stromnes I.M., Chapuis A.G., Greenberg P.D. New strategies in engineering T-cell receptor gene-modified T cells to more effectively target malignancies. Clin. Cancer Res. 2015;21:5191–5197. doi: 10.1158/1078-0432.CCR-15-0860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee D.W., Gardner R., Porter D.L., Louis C.U., Ahmed N., Jensen M., Grupp S.A., Mackall C.L. Current concepts in the diagnosis and management of cytokine release syndrome. Blood. 2014;124:188–195. doi: 10.1182/blood-2014-05-552729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.van den Berg J.H., Gomez-Eerland R., van de Wiel B., Hulshoff L., van den Broek D., Bins A., Tan H.L., Harper J.V., Hassan N.J., Jakobsen B.K. Case report of a fatal serious adverse event upon administration of T cells transduced with a MART-1-specific T-cell receptor. Mol. Ther. 2015;23:1541–1550. doi: 10.1038/mt.2015.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Morgan R.A., Yang J.C., Kitano M., Dudley M.E., Laurencot C.M., Rosenberg S.A. Case report of a serious adverse event following the administration of T cells transduced with a chimeric antigen receptor recognizing ERBB2. Mol. Ther. 2010;18:843–851. doi: 10.1038/mt.2010.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Arstila T.P., Casrouge A., Baron V., Even J., Kanellopoulos J., Kourilsky P. A direct estimate of the human alphabeta T cell receptor diversity. Science. 1999;286:958–961. doi: 10.1126/science.286.5441.958. [DOI] [PubMed] [Google Scholar]
- 52.Kersh G.J., Allen P.M. Essential flexibility in the T-cell recognition of antigen. Nature. 1996;380:495–498. doi: 10.1038/380495a0. [DOI] [PubMed] [Google Scholar]
- 53.Sloan-Lancaster J., Allen P.M. Altered peptide ligand-induced partial T cell activation: molecular mechanisms and role in T cell biology. Annu. Rev. Immunol. 1996;14:1–27. doi: 10.1146/annurev.immunol.14.1.1. [DOI] [PubMed] [Google Scholar]
- 54.Persaud S.P., Donermeyer D.L., Weber K.S., Kranz D.M., Allen P.M. High-affinity T cell receptor differentiates cognate peptide-MHC and altered peptide ligands with distinct kinetics and thermodynamics. Mol. Immunol. 2010;47:1793–1801. doi: 10.1016/j.molimm.2010.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Baker B.M., Gagnon S.J., Biddison W.E., Wiley D.C. Conversion of a T cell antagonist into an agonist by repairing a defect in the TCR/peptide/MHC interface: implications for TCR signaling. Immunity. 2000;13:475–484. doi: 10.1016/s1074-7613(00)00047-9. [DOI] [PubMed] [Google Scholar]
- 56.Ding Y.H., Baker B.M., Garboczi D.N., Biddison W.E., Wiley D.C. Four A6-TCR/peptide/HLA-A2 structures that generate very different T cell signals are nearly identical. Immunity. 1999;11:45–56. doi: 10.1016/s1074-7613(00)80080-1. [DOI] [PubMed] [Google Scholar]
- 57.Garboczi D.N., Ghosh P., Utz U., Fan Q.R., Biddison W.E., Wiley D.C. Structure of the complex between human T-cell receptor, viral peptide and HLA-A2. Nature. 1996;384:134–141. doi: 10.1038/384134a0. [DOI] [PubMed] [Google Scholar]
- 58.Clay T.M., Custer M.C., Sachs J., Hwu P., Rosenberg S.A., Nishimura M.I. Efficient transfer of a tumor antigen-reactive TCR to human peripheral blood lymphocytes confers anti-tumor reactivity. J. Immunol. 1999;163:507–513. [PubMed] [Google Scholar]
- 59.Lyons G.E., Moore T., Brasic N., Li M., Roszkowski J.J., Nishimura M.I. Influence of human CD8 on antigen recognition by T-cell receptor-transduced cells. Cancer Res. 2006;66:11455–11461. doi: 10.1158/0008-5472.CAN-06-2379. [DOI] [PubMed] [Google Scholar]
- 60.Norell H., Zhang Y., McCracken J., Martins da Palma T., Lesher A., Liu Y., Roszkowski J.J., Temple A., Callender G.G., Clay T. CD34-based enrichment of genetically engineered human T cells for clinical use results in dramatically enhanced tumor targeting. Cancer Immunol. Immunother. 2010;59:851–862. doi: 10.1007/s00262-009-0810-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Roszkowski J.J., Lyons G.E., Kast W.M., Yee C., Van Besien K., Nishimura M.I. Simultaneous generation of CD8+ and CD4+ melanoma-reactive T cells by retroviral-mediated transfer of a single T-cell receptor. Cancer Res. 2005;65:1570–1576. doi: 10.1158/0008-5472.CAN-04-2076. [DOI] [PubMed] [Google Scholar]
- 62.Garrett-Mayer E. Overview of standard clustering approaches for gene microarray data analysis. In: Allison D.B., Page G.P., Beasley T.M., Edwards J.W., editors. DNA Microarrays and Related Genomics Techniques: Design, Analysis, and Interpretation of Experiments. Chapman & Hall/CRC; 2006. pp. 131–159. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.