Abstract
Pemphigus foliaceus (PF) is a rare autoimmune skin disease caused by anti-Dsg1 pathogenic autoantibodies. It is considered as a Th2-mediated disease. Likewise, Th17 cells were recently described in the pathogenesis of the disease but their role is still unclear. We aimed to unravel the eventual implication of the IL23/Th17 pathway in the development of PF. A case-control study was conducted on 115 PF patients and 201 healthy controls using PCR-RFLP and AS-PCR methods. SNPs in IL23R, RORγt, IL17A, IL17F, IL17AR, TNFa, and STAT3 genes were genotyped. mRNA expression of IL23R and RORγt was evaluated using Q-PCR. The frequency of circulating Th17 cells was analyzed by flow cytometry. Genetic associations between IL23R>rs11209026, IL17A>rs3748067, IL17F>rs763780, and TNFa>rs1800629 and the susceptibility to PF were reported. Moreover, we revealed a significant increased frequency of circulating CD4+IL17+ cells as well as higher mRNA levels of RORγt and IL23R in PBMCs of patients. However, no significant increase of RORγt and IL23R mRNA expression was observed in lesional skin biopsies. In spite of the little size of specimens, our results provide converging arguments for the contribution of the IL23/Th17 pathway in the pathogenesis of PF.
1. Introduction
Pemphigus foliaceus (PF) is an autoimmune skin disease with a complex etiology involving genetic and environmental factors [1]. It is mediated by pathogenic auto-antibodies (−Abs) that bind to desmoglein 1 (Dsg1) and lead to acantholysis and consequent blister formation in the subcorneal layer of the epidermis [2-4]. The progression from preclinical phase to clinical active disease is associated with the subclass switching of these auto-Abs from IgG1/IgG2 to IgG4 [5–7]. Therefore, and since Th2 cytokines induce B cells to secrete IgG4, PF was considered as a Th2 disease [8, 9] However, this hypothesis was rapidly contradicted by other researchers who detected Dsg1-responsive Th1 and Th2 cells simultaneously in PF patients' blood [10]. More recently, a significant high frequency of CD4+IL17+ cells in pemphigus patients' PBMCs was reported, particularly in acute onset stage and active chronic stage [11]. Moreover, in PF patients' skin biopsies, while the ratio of IL17+ to CD4+cell count was 1.8%, IL17-positive cells were undetectable in healthy controls' skin [12]. These data that are in favor of a possible implication of Th17 cells in the pathogenesis of PF were based only on the Th17 common marker to characterize this lymphocyte subset whose differentiation and function involve several other specific markers such as the lineage-specific transcription factor RORγt and the IL23 receptor (IL23R).
Indeed, the differentiation of Th17cells from CD4+CD161+ naïve Th cells requires specific cytokines such as TGF-β, IL6, IL1β, IL21, and IL23 [13, 14]. In fact, the synergic action of the different cytokines induces the expression of IL23R, which in turn activates the STAT signaling pathway leading to the expression of the transcription factor RORγt and then the production of Th17-specific cytokines, mainly IL17A and IL17F [15, 16]. Th17 cells play an important role in the clearance of fungal and extracellular bacterial infections by the recruitment of several cell types such as neutrophils, macrophages, and dendritic cells and by the induction of the production of cellular inflammatory mediators, such as TNFα [17–19].
The field of action of Th17 cells is not limited to these infections, and there is increasing evidence for the contribution of these cells in the pathogenesis of various autoimmune diseases (AIDs). In fact, IL23 which is considered as the stabilizing factor for Th17 cell commitment seems to play a key role in the acquisition of the pathogenic character of these cells [20]. IL23 signaling through IL23R can amplify Th17 cell response by inducing proinflammatory cytokines, such as IL1β, TNFα, and IL6, in innate immune cells and by enhancing the activation of STAT3,which is coordinated with RORγt to stabilize them and maintain their function [15]. Moreover, it was reported that the pathogenicity of Th17 cells was associated with the over activation of the IL23/IL23R signaling pathway [16, 21]. Thus, the IL23/Th17 axis is becoming a leading interesting pathway in the field of AIDs such as systemic lupus erythematosus [22] and pemphigus vulgaris [23] and is one of the most promising targets for AID therapy.
In light of these data and to unfold the involvement of the IL23/Th17 pathway in the pathogenesis of Tunisian PF, we aimed to unravel the eventual genetic contribution of IL23/Th17 genes' polymorphisms and functional association of Th17 cells through the expression of their specific markers.
2. Material and Methods
2.1. Subjects
One hundred and fifteen PF patients have been recruited since 2002 at the department of Dermatology in Hedi Chaker University Hospital of Sfax, Tunisia. The diagnosis of PF was confirmed by clinical presentation, histopathology (acantholysis in the upper epidermis either in the granular layer or immediately below with subcorneal bullous formation), direct immunofluorescence (IgG and C3 deposits most often located on the whole epidermis and less frequently predominant in the upper layers of the epidermis), indirect immunofluorescence (IgG Abs directed against the epithelial cell surface), and ELISA test for circulatory anti-Dsg1 Abs that was positive for all patients [24].Two hundred and one healthy controls (HC), who did not suffer from any autoimmune or inflammatory disease, were also recruited (see Table 1).
Table 1.
Characteristics of study populations.
| Features | PF patients | Controls |
|---|---|---|
| Number | 115 | 201 |
| Sex ratio | 1/14 | 1/9 |
| Mean age | 35 (18–84) | 39 (14–73) |
| Origin | Center-southern regions of Tunisia | |
| Anti-Dsg1 | Positive | Negative |
The genetic study is a case-control retrospective study enrolled in the whole population described above. The patient group included 8 men and 107 women, with a mean age of 35 years ranging from 18 years to 84 years. All patients originate from the center and south of Tunisia. The control group included 21 men and 180 women, with a mean age of 39 years ranging from 14 years to 73 years.
Related patients and/or HC were excluded from the genetic study.
The functional study is a perspective one, conducted on subjects from the population described above that were recruited since 2014. Pemphigus disease area index (PDAI) was measured for all retained patients. We enrolled 13 PF patients and 6 HC for mRNA expression analysis on biopsies, 5 PF women patients and 4 women HC for mRNA expression analysis on PBMCs, and 9 women patients in active stage and 5 women HC for flow cytometry analysis.
The patient group was divided into two groups depending on the stage of the disease: active stage and chronic stage. The active stage of PF is defined as de novo development of new blisters on the skin; none of these patients had yet received immunosuppressive therapy. The chronic stage of PF is defined as the expansion and/or the persistence of existing blisters on the skin. Patients in this stage had already received immunosuppressive treatment.
PF patients suffering from other AIDs were excluded. Healthy subjects were selected based on additional criteria; they have not taken any medicines for at least three weeks before the sampling. All HC were negative for anti-Dsg1Abs.
2.1.1. Sample Collection
After obtaining a written informed consent, samples were collected. Our project was approved by the ethical committee of the Habib Bourguiba University Hospital of Sfax (protocol number of the ethical committee, 4/12).
(1) Skin Biopsy. 13 biopsies were taken from 10 patients in the active stage of the disease and from 3 patients in the chronic stage. In addition, 6 control biopsies were taken from healthy subjects undergoing surgeries for a trichilemmal cyst or plastic surgery.
(2) Blood. A total of 10 ml of blood was taken from patients and HC in sterile endotoxin-free vacutainers with EDTA as anticoagulant for DNA extraction or in heparinized tubes for mRNA expression analysis and cytometry analysis.
2.2. SNP Selection
Tag single-nucleotide polymorphisms (tag SNPs) in the IL23/Th17 axis genes were selected according to their susceptibility to other AIDs and to the genotyping data from the CEU available from the International HapMap project. Selection was undertaken using minor allele frequency (greater than 5%) in Caucasian and sub-Saharan populations.
Four candidate SNPs in the IL23R gene in chromosome 1: rs1884444, rs7517847, rs11209026 and rs10889677, two SNPs in the IL17A gene in chromosome 6: rs3748067 and rs2275913, rs763780 in IL17F in chromosome 6, rs4819554 in the IL17RA gene in chromosome 22, rs9645406 in RORγt within chromosome 1, rs1800629 in the TNFa gene within chromosome 6, and rs744166 in the STAT3 gene within chromosome 17, were selected. All primers and enzymes used in this study are presented in Table 2.
Table 2.
Primary information of genotyped SNPs in Th17/IL23 pathway's genes.
| Gene SNPs |
Base change | MAF | Chromosome regions | Localization | Primers | Enzyme |
|---|---|---|---|---|---|---|
| IL23R rs1884444a |
G/T nonsens |
0.47 | Chr1 1p31.3 |
Exon2 | F:Fam✪ TGCTCTGTTTCCTTCCTTCC R1:CATCCCATTGAATAGTGACC R2:T4CATCCCATTGAATAGTGACA |
— |
| IL23R rs7517847 |
G/T | 0.36 | Intron6 | F:ATTTCTGGATGCCCTTTCCT R:ACATGAATTTGAGGGGCCTA |
BbvI | |
| IL23R rs11209026 |
A/G Arg-gln |
0.022 | Exon8 | F:TTAGACAACAGAGGAGACATTGGA R:CATGTAGTCTAAATCAGAAAACAGAAA |
Hpy188I | |
| IL23R rs10889677 |
A/C | 0.39 | 3′UTR | F:TGCTGGGCCATATGATAAGC R:TCCACCTTCGGGACCTTAAT |
MnII | |
| RORγt rs9645406a |
T/C | Chr1 1q21.3 |
Intron | F1:T4CCTCACAGCAAATCTTTTCTC F2:CCTCACAGCAAATCTTTTCTT R:NED✪AAAAACCGCTGTAGGGATCA |
— | |
| IL17A rs2275913 |
A/G | 0.29 | Chr6 6p12.2 |
5′UTR | F:ATTTCTGCCCTTCCCATTTT R:TTGTGCCTGCTATGAGATGG |
EarI |
| IL17A rs3748067 |
T/C | 0.077 | 3′UTR | R:GGGGCGAAAATGGTTACGAT F:AAGGCCCCTCAGAGATCAAC |
ApoI | |
| IL17F rs763780 |
C/T | 0.093 | Chr6 6p12.2 |
Exon3 | F:TGGGTAAGGAGTGGCATTTC R:CTGCATCAATGCTCAAGGAA |
NlaIII |
| IL17RA rs4819554 |
G/A | 0.233 | Chr22 22q11.1 |
5′UTR | F:TGAAATGTGTAATTCGCTGGC R:TGCTTTCCTTGGCTTTGCTT |
PvuII |
| TNFα rs1800629 |
A/G | 0.090 | Chr6 6p21.33 |
5′UTR | F:AGGCAATAGGTTTTGAGGGGCAT R:TCCTCCCTGCTCCGATTCC |
NcoI |
| STAT3 rs744166 |
C/T | 0.482 | Chr17 17q21.2 |
Intron | F:GCTGGAGTACAAACCCTGAA R:TGGTATTCAGATGGCGGTCA |
AluI |
aGenotyped using the AS-PCR method.
2.3. SNP Genotyping
Genomic DNA was extracted from whole blood samples using a standard proteinase K digestion and phenol/chloroform extraction procedure. Genotyping was performed using the PCR-RLFP method for all SNPs except for rs1884444 in the IL23R gene and rs9645406 in RORγt which were genotyped using the AS-PCR method (see Table 2).
2.3.1. PCR-RFLP
The PCR amplification was carried out in a volume of 25 μl including 1x buffer, 2 mM MgCl2, 0.2–0.4 μmol of each primer (Invitrogen®, CA, USA), 0.12 mM dNTP (Invitrogen, CA, USA),1 U Taq polymerase (Invitrogen, CA, USA), and 50 ηg of DNA template. Enzymatic digestion was performed in a total of 10 μl mixture reaction containing 1x buffer, 0.1x BSA, and 2 U restriction enzyme (Thermo Fisher®, MA, USA). Primers were designed using primer3 software (http://primer3.ut.ee/). Restriction enzymes were selected using the NEBcutter software (http://nc2.neb.com/NEBcutter2/).
2.3.2. AS-PCR
The primers were designed using WASP software (http://bioinfo.biotec.or.th/WASP). Common primer was labeled with 6-FAM fluorescent labels for the rs1884444 and 6-NED fluorescent labels for rs9645406. The mutant primer containing the mutant base was labeled with a poly(A) tail. The PCR amplification was carried out in a volume of 10 μl including 1x buffer, 2.5 pmol of each primer (Invitrogen, CA, USA), 10 mM dNTP (Invitrogen, CA, USA), 1 U AmpliTaq Gold™ DNA Polymerase (Applied Biosystems™, CA, USA), and 1 ηg of DNA template. Amplified products were run on ABI prism DNA sequencer (PerkinElmer®, CT, USA) and the output file was analyzed using GeneScan software analysis.
2.4. Quantitative mRNA Expression of IL23R and RORγt in the Skin and PBMCs by Q-PCR
Peripheral blood mononuclear cells (PBMCs) were isolated by Histopaque-1077 density-gradient method. Total RNA from PBMCs was extracted using One-step RNA reagent (Bio Basic®, Canada) following the manufacturer's instructions and then reversely transcribed using the High-Capacity cDNA Reverse Transcription kit (Applied Biosystems®, CA, USA). The real-time quantitative PCR was performed via the StepOnePlus Real-Time PCR systems (Applied Biosystems, CA, USA). TaqMan analysis was conducted using predesigned and optimized assays from Applied Biosystems: IL23R (ID: Hs 00332759_m1) and RORγt (ID: Hs 01076122_m1). PCR reaction parameters were as follows: 95°C for 10 min, followed by 40 cycles of 95°C for 15 s, 60°C for 1 min. All measurements were performed in triplicate. For the relative quantification, data were analyzed by the comparative 2−ΔΔCt method and normalized to the average of housekeeping gene GAPDH.
2.5. Flow Cytometry Analysis
PBMCs were seeded into 24-well plates at a final concentration of 106 cells/ml, in RPMI1640 complete medium (containing 10% of the fetal bovine serum, 200 g/ml penicillin, and 200 U/ml streptomycin), and were stimulated with 12-myristate 13-acetate (50 ng/ml) and calcium ionomycin (1 μg/ml), in the presence of GolgiStop protein transport inhibitor (BD Biosciences®, CA, USA) at 37°C for four hours in a 5% of CO2 incubator. Cells were subsequently stained for surface markers using PerCP-Cy5.5-labeled anti-human CD4 and PE-labeled anti-human IL17 for intracellular cytokine (BD Bioscience, CA, USA). CD4+T cells were gated, and the percentages of these cells producing IL17 were calculated. The BD FACS Canto II System (BD Biosciences, USA) and Kaluza Analysis 1.5a software (Beckman Coulter, USA) were used for analysis.
2.6. Statistical Analysis
A case-control analysis was performed using SHESIS software (http://analysis.bio-x.cn) for each SNP and haplotype. Hardy–Weinberg equilibrium (HWE) was assessed in controls using a χ2 test with one degree of freedom. A threshold P < 0.05 was regarded to indicate deviation from HWE. Odds ratios (OR) and 95% confidence intervals (CI) were calculated for each allele using 2 × 2 contingency tables to estimate the magnitude of association. Binary logistic regression was used to make age and gender adjustment. The linkage disequilibrium (LD) coefficients D' = D/Dmax and r2 values for the pair of the most common alleles at each site were also estimated, and high values of LD were defined as r2 > 0.33 and D' > 0.7. The significance level of P < 0.05 and odds ratios (OR) with 95% confidence intervals (95% CI) was chosen for all sets.
For functional experiments, statistical analyses were carried out using SPSS20.0 software (IBM SPSS® Inc., IL, USA). As the number of samples was less than twenty, the nonparametric Mann–Whitney U test was used to analyze flow cytometry results and mRNA expression of IL23R and RORγt. Spearman correlation rho “r” was used to determine the possible correlation between different results.
3. Results
3.1. Genetic Association of Investigated SNPs with Susceptibility to PF
Genotype frequencies of all SNPs tested in control subjects were consistent with those expected from the Hardy–Weinberg equilibrium (HWE) except for rs1884444 polymorphism in the IL23R gene and rs763780 in IL17F gene (P < 0.001). Minor allele frequency of all the polymorphisms was consistent with that reported in the HapMap database. The genotypic and allelic distributions of the IL23/Th17 gene polymorphisms as well as their associations with the risk to PF are shown in Table 3.
Table 3.
Genotype and allele frequencies of IL23/Th17 pathway's genes polymorphisms in Pemphigus foliaceus patients and matched healthy controls.
| Gene/SNP | Genotype/allele | Case N (%) | Control N (%) | P | OR | 95% CI |
|---|---|---|---|---|---|---|
| IL23R rs11209026 |
AA | 0 | 5 (2.6) | NS | — | — |
| AG | 11 (10.3) | 41 (21.2) | 0.016 | 0.42 | 0.21–0.87 | |
| GG | 96 (89.7) | 147 (76.2) | 0.004 | 2.37 | 1.35–5.53 | |
| A | 11 (5.1) | 51 (13.2) | 0.001 | 0.35 | 0.18–0.69 | |
| G | 203 (94.9) | 335 (86.8) | ||||
|
| ||||||
| IL23R rs1884444 |
GG | 3 (3) | 11 (8.3) | NS | — | — |
| GT | 80 (79.2) | 106 (79.7) | NS | — | — | |
| TT | 18 (17.8) | 16 (12) | NS | — | — | |
| G | 86 (42.6) | 128 (48.1) | NS | — | — | |
| T | 116 (57.4) | 138 (51.9) | ||||
|
| ||||||
| IL23R rs7517847 |
GG | 10 (9.8) | 14 (7.4) | NS | — | — |
| GT | 43 (42.2) | 75 (39.7) | NS | — | — | |
| TT | 49 (48) | 100 (52.9) | NS | — | — | |
| G | 63 (30.9) | 103 (27.2) | NS | — | — | |
| T | 141 (69.1) | 276 (72.8) | ||||
|
| ||||||
| IL23R rs10889677 |
AA | 20 (19.8) | 40 (22.1) | NS | — | — |
| AC | 41 (40.6) | 71 (39.2) | NS | — | — | |
| CC | 40 (39.6) | 70 (38.7) | NS | — | — | |
| A | 81 (40.1) | 151 (41.7) | NS | — | — | |
| C | 121 (59.9) | 211 (58.3) | ||||
|
| ||||||
| IL17A rs2275913 |
AA | 1 (1) | 1 (0.6) | NS | — | — |
| AG | 9 (9.2) | 10 (6.2) | NS | — | — | |
| GG | 88 (89.8) | 150 (93.2) | NS | — | — | |
| A | 11 (5.6) | 12 (3.7) | NS | — | — | |
| G | 185 (94.4) | 310 (96.3) | ||||
|
| ||||||
| IL17A rs3748067 |
TT | 0 | 4 (3.6) | NS | — | — |
| CT | 10 (11.1) | 38 (35) | 9.47e − 4 | 0.23 | 0.11–0.50 | |
| CC | 79 (87.7) | 66 (60.8) | 1.17e − 4 | 5.03 | 2.34–10.78 | |
| T | 10 (5.5) | 46 (21.1) | 8.29e − 6 | 0.218 | 0.10–0.44 | |
| C | 170 (94.4) | 171 (78.8) | ||||
|
| ||||||
| IL17AR rs4819554 |
AA | 78 (74.3) | 137 (71.8) | NS | — | — |
| AG | 24 (22.9) | 48 (25.1) | NS | — | — | |
| GG | 3 (2.9) | 6 (3.1) | NS | — | — | |
| A | 180 (85.7) | 322 (84.3) | NS | — | — | |
| G | 30 (14.3) | 60 (15.7) | ||||
|
| ||||||
| IL17F rs763780 |
CC | 5 (4.7) | 3 (1.6) | NS | — | — |
| CT | 13 (12.3) | 13 (6.7) | NS | — | — | |
| TT | 88 (83) | 177 (91.7) | 0.02 | 0.44 | 0.22–0.91 | |
| C | 23 (10.8) | 19 (4.9) | 0.007 | 2.35 | 1.24–4.42 | |
| T | 189 (89.2) | 367 (95.1) | ||||
|
| ||||||
| TNFa rs1800629 |
AA | 12 (12.8) | 9 (5) | 0.022 | 2.76 | 1.12–6.82 |
| AG | 37 (39.4) | 43 (24) | 0.008 | 2.05 | 1.20–3.51 | |
| GG | 45 (47.9) | 127 (70.9) | 0.0002 | 0.38 | 0.22–0.63 | |
| A | 61 (32.4) | 61 (17) | 4.06e − 5 | 2.33 | 1.55–3.52 | |
| G | 127 (67.6) | 297 (83) | ||||
|
| ||||||
| RORγt rs9645406 |
TT | 0 | 0 | NS | — | — |
| CT | 9 (7.9) | 13 (6.5) | NS | — | — | |
| CC | 106 (92.1) | 188 (93.5) | NS | — | — | |
| T | 11 (4.7) | 13 (3.3) | NS | — | — | |
| C | 219 (95.3) | 389 (96.7) | NS | — | — | |
|
| ||||||
| STAT3 rs744166 |
TT | 22 (23.1) | 29 (18.1) | NS | — | — |
| CT | 47 (49.5) | 77 (48.1) | NS | — | — | |
| CC | 26 (27.4) | 54 (33.8) | NS | — | — | |
| T | 91 (47.9) | 135 (42.2) | NS | — | — | |
| C | 99 (52.1) | 185 (57.8) | NS | — | — | |
Concerning the IL23R gene, the rs11209026 A>G polymorphism seems to be closely associated with the PF's risk. In fact, the homozygous GG genotype was significantly overexpressed in patients (89.7%) compared to HC (76.2%) (P = 0.004, OR = 2.37, 95% CI 1.35–5.53), contrary to the AG genotype which was more expressed in controls (P = 0.01, OR = 0.42, 95% CI 0.21–0.87). Likewise, the decreased expression of the A allele observed in the patient group compared to HC suggests its protective role against PF (P = 0.001, OR = 0.35, 95% CI 0.18–0.69). No association was found either with rs1884444, with rs7517847, or with rs10889677.
For the IL17A gene, a strong association of rs3748067 T>C with the occurrence of PF was revealed. The CC homozygous genotype and the C allele seem to increase the risk of the development of the disease (P = 1.17e − 04, OR = 5.03, 95% CI 2.34–10.78; P = 8.29e − 06, OR = 4.57, 95% CI 2.23–9.36). In addition, the heterozygous genotype CT appears as a PF protection genotype (P = 9.47e − 04, OR = 0.23, 95% CI 0.11–0.50). No significant association was observed for rs2275913.
For IL17RA gene polymorphism, no statistical significant difference in genotype and allele frequencies was observed between patients and controls.
As for the IL17F gene, a significant increase of the rs763780>C allele was observed in the patient group (10.8%) compared to HC (4.9%) (P = 0.007, OR = 2.35, 95% CI 1.24–4.42). On the other hand, the TT genotype was significantly less expressed in patients (89.2%) than in HC (95.1%) (P = 0.02, OR = 0.44, 95% CI 0.22–0.91), suggesting its protective role.
Regarding the TNFa gene, the allelic distribution of rs1800629 A>G in patients' cohort revealed a great significant increase of the A allele (P = 4.06e − 5, OR = 2.33, 95% CI (1.55–3.52)). In addition, while the homozygous AA and heterozygous AG genotypes are associated with the PF's risk (P = 0.02, OR = 2.76, 95% CI 1.12–6.82; P = 0.008, OR = 2.05, 95% CI 1.20–3.51), the GG genotype appeared to be a protector genotype (P = 0.0002, OR = 0.38, 95% CI 0.22–0.63).
No significant association was found with rs9645406 in the RORγt gene or with rs744166 in the STAT3 gene.
All significant associations were conserved after age and gender adjustment using the binary logistic regression.
3.2. Linkage Disequilibrium (LD), Haplotype, and Combination Analysis
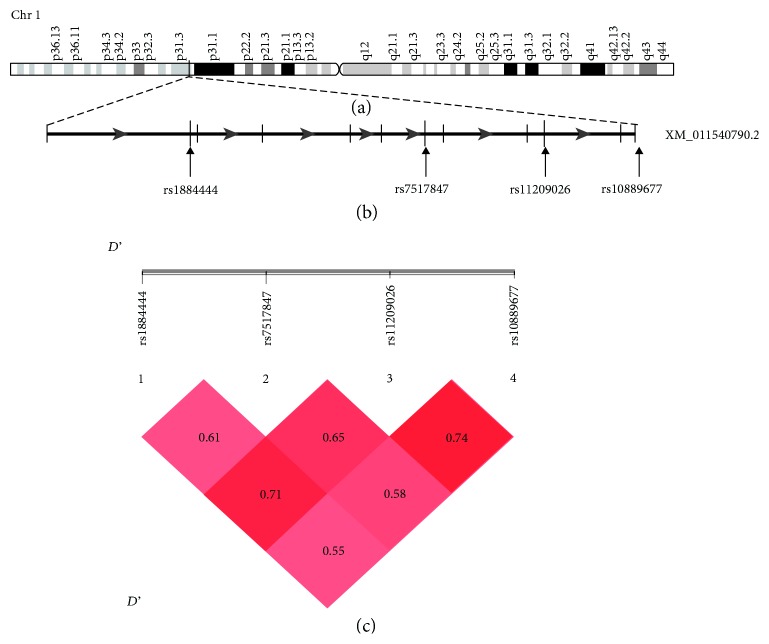
The LD analysis among the patient group was conducted by pairwise comparison of the 4 polymorphisms studied in the IL23R gene within chromosome 1 (see Figure 1).
Figure 1.
Overview and linkage disequilibrium (LD) on chromosome 1q31.1. (a) Overview of chromosome 1. (b) Schematic structure of the IL23R gene. Exons are depicted by black vertical lines. The positions of the four SNPs are shown by the arrows. (c) LD prime charts generated using SHEsis software summarize LD (D') patterns between the 4 SNP. D' > 0.7 is considered as a high value of LD.
An evidence for LD was revealed between rs11209026 and rs10889677 (D' = 0.74) and with rs1884444 (D' = 0.71) suggesting that the presumed haplotype of susceptibility to Tunisian PF is rs1884444>T, rs7517847>T, rs11209026>G, rs10889677>A, which contains the rs11209026>G susceptibility allele. This haplotype was more expressed in patients (23.4%) than in controls (16.3%) and, therefore, appears as a risk haplotype to the disease (P = 1.71e − 04, OR = 2.41, 95% CI 1.51~3.85) (see Table 4).
Table 4.
Haplotypes of genotyped SNPs on chromosomes 1 and 6.
| Chromosome | Haplotype | Case N (%) | Control N (%) | P | OR (95% CI) |
|---|---|---|---|---|---|
| 1 | TTGA | 44 (23.4) | 39 (16.3) | 1.71e − 04 | 2.409 (1.508~3.846) |
| TGGC | 33 (17.5) | 24 (10) | 1.16e − 04 | 2.871 (1.648~5.001) | |
| 6 | AGGT | 42 (0.291) | 30(0.168) | 7.67e − 06 | 3.027 (1.832~5.003) |
| GGAT | 6 (0.041) | 38 (0.213) | 0.002 | 0.276 (0.114~0.664) |
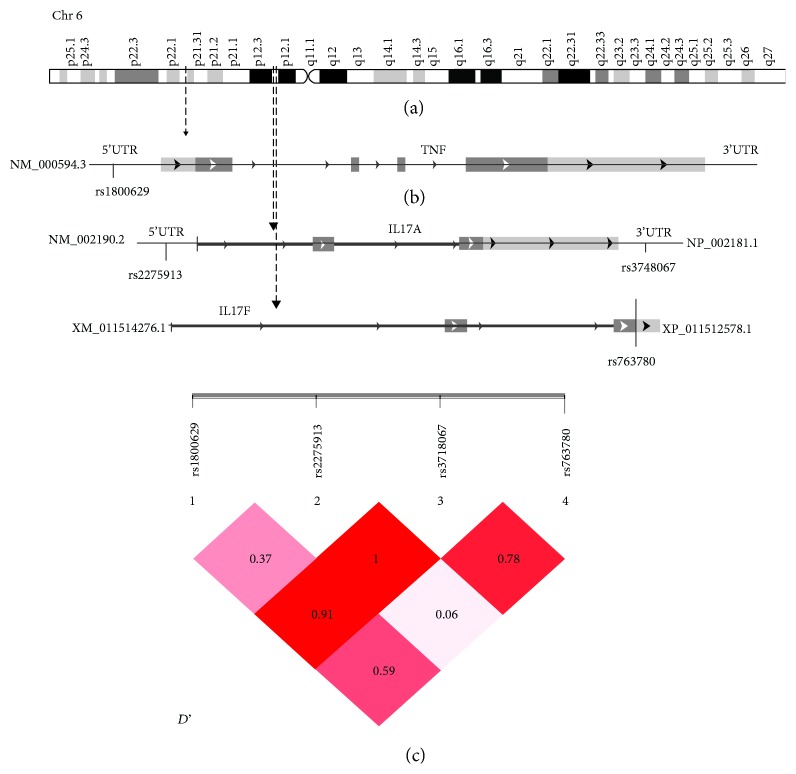
Since IL17A (rs3748067 and rs2275913), IL17F (rs763780), and TNFa (rs1800629) genes are located on nearby regions 6p12.2 and 6p21.33, respectively, LD values may explain the possible effect and interaction between their different alleles (see Figure 2).
Figure 2.
Overview and linkage disequilibrium (LD) on chromosome 6. (a) Overview of chromosome 6. (b) Schematic structure of the IL17A, IL17F, and TNFa genes. The positions of the four SNPs are shown by the arrows (c). LD prime charts generated using SHEsis software summarize LD (D') patterns between the 4 SNPs. D' > 0.7 is considered as a high value of LD.
Thus, an evidence for LD between the SNPs investigated in IL17A, F, and TNFa was observed. Indeed, strong LD between rs3748067 and rs2275913 (D' = 1) and rs3748067 and rs1800629 (D' = 0.91) were found, indicating that the association observed with these SNPs must be interdependent. Additionally, the estimated frequencies of haplotypes were statistically different between PF patients and controls (global χ2 = 23.17, global Pearson's P = 1.17e − 04). In fact, haplotypes AGCT and GGCT which contain the associated risk allele rs3748067>C (P < 0.0001) were associated with an increased risk to PF. In contrast, the GGTT haplotype seems to confer protection against PF (P = 0.001) (see Table 4).
On the other hand, possible combinations between the different SNPs were analyzed and a strong evidence for gene-gene interactions between them was revealed (see Table 5). Thus, three associated combinations that differ statically between patients and controls were observed; TTGAAGCTAC (P = 0.018, OR = 4.49, 95% CI 1.15–17.57), TGGCGGCTAC (P = 0.003, OR = 4.87, 95% CI 1.51–15.74) and TTGAGGCTAT (P = 0.017, OR = 0.017, 95% CI 1.16–8.00).
Table 5.
Combinations of genotyped SNPs in IL23R, IL17A, IL17F, TNFa, RORγt, and STAT3 showing significant differences between PF patients and controls.
| Combinations | Case N (%) | Control N (%) | P | OR (95% CI) |
|---|---|---|---|---|
| TTGAAGCTAC | 7 (5.5) | 3 (2.3) | 0.018 | 4.497 (1.151~17.574) |
| TGGCGGCTAC | 10 (7.9) | 4 (3.1) | 0.003 | 4.877 (1.511~15.742) |
| TTGAGGCTAT | 11 (8.7) | 7 (5.4) | 0.017 | 3.057 (1.167~8.006) |
3.3. Relative mRNA Expression of IL23R and RORγt in the Skin and Blood
In patients' blood, the relative mRNA expression levels of RORγt and IL23R were significantly higher (P < 0.05) compared to the HC (9.64 ± 12.96 versus 0.778 ± 0.382 and 5.01 ± 1.00 versus 0.75 ± 0.22, resp.) as shown in Figure 3. Moreover, a strong positive correlation was revealed between the expression of these two markers (r = 0.943; P < 0.005).
Figure 3.
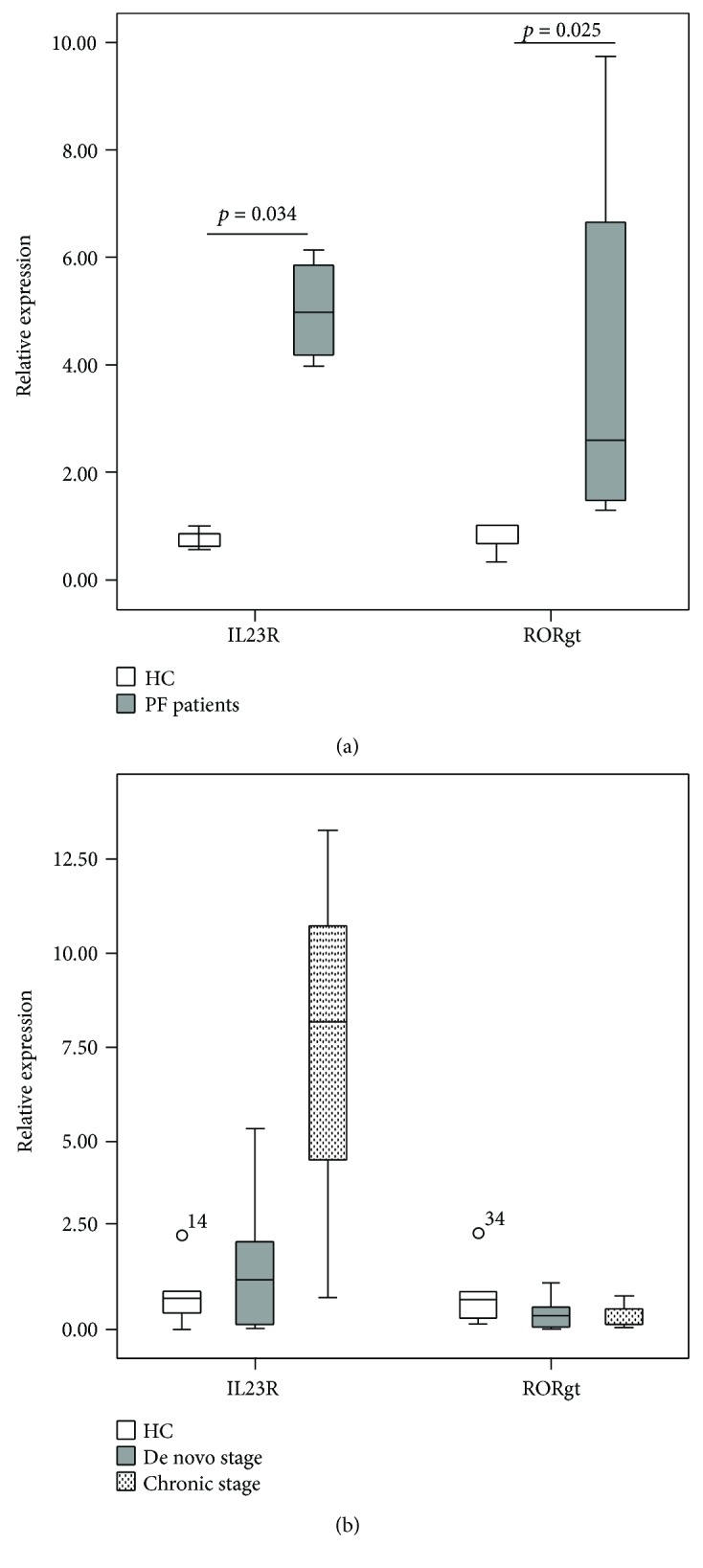
mRNA expression of IL23R and RORγt in peripheral blood (a) and skin biopsies (b). PBMCs were extracted from 5 de novo patients and 4 HC. Biopsies from de novo patients (N = 10), patients in the chronic stage (N = 3), and from HC (N = 5) were analyzed using Q-PCR. The relative expression was estimated using 2−ΔΔCt method and normalized to the average of the GAPDH housekeeping gene. P value was calculated using Mann–Whitney U test.
In spite of the insignificant difference in RORγt and IL23R expression levels in skin biopsies between the patient and control groups (1.09 ± 1.74 versus 0.95 ± 0.96 and 2.99 ± 3.89 versus 1.18 ± 0.90, resp.), the IL23R mRNA was slightly more expressed in specimens from patients in the chronic stage than those de novo (7.427 ± 6.25 versus 1.661 ± 1.734) (see Figure 3).
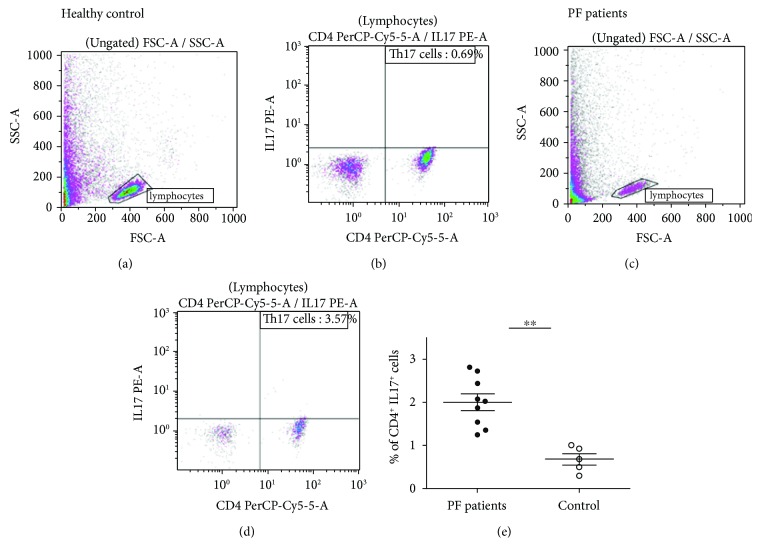
3.4. Increased Frequency of CD4+IL17+ Circulatory Cells in Patients' Blood
After stimulation, the frequency of CD4+IL17+cells was significantly higher in patients compared to controls as shown in Figure 4 (P = 0.001). In patients, the percentage of Th17 cells varied from 1.36 to 3.57% of CD4+cells; however, in the control group, the percentage does not exceed 1% (see Figure 4).
Figure 4.
Frequency of IL17+ cells in the peripheral blood mononuclear cells of PF patients and healthy controls. Th17 cells identified using specific antibodies CD4-PerCP-Cy5 and IL17A-PE. Representative flow cytometry analysis of Th17 cells in healthy controls (a, b) and in PF patients (c, d). (e) Percentage of Th17 cells in control and patients' PBMCs.
4. Discussion
The IL23/Th17 pathway has taken a lot of interest recently, as it is pivotal both in tissue immunosurveillance and autoimmunity mechanisms. Based on genetic and functional studies, strong evidence for its implication in the pathogenesis of several autoimmune diseases, such as systemic lupus erythematosus [22] and rheumatoid arthritis [23], has emerged. Bearing in mind that substitutions of one base pair (SNPs) may have functional importance, we investigated, in the present data, an eventual functional contribution of the most important IL23/Th17 axis markers in the pathogenesis of PF.
Concerning the four Tag SNPs genotyped in the IL23R gene, only rs11209026>G was found to be strongly associated with PF susceptibility (P = 0.001, OR = 2.81). Moreover, this SNP in strong LD with the others generates the susceptibility haplotype rs1884444>T-rs7517847>T-rs11209026>G-rs10889677>A. Lesional skin biopsies from PF patients showed a slight increase of IL23R mRNA expression as compared to those from HC. Interestingly, specimens of patients in the chronic stage showed a higher mRNA level than those of de novo patients. Furthermore, this relative expression was more significantly pronounced in PF's PBMCs compared to HC ones (P = 0.03). The critical role of the IL23/IL23R signaling pathway was proved in favoring terminal differentiation, maintenance, and pathogenicity of Th17 cells [16]. In addition, strong data demonstrate that the lack of the proinflammatory cytokine IL23 in mice makes them resistant to experimental models of arthritis and multiple sclerosis (MS) [25, 26]. Thus, overexpression of this cytokine was reported in several AIDs such as psoriasis [27] and PV [28] but not in PF. The genetic association of rs11209026 was widely described in the context of AID. This variant is located between the transmembrane domain and putative JAK2 binding site in the cytoplasmic portion of IL23R protein and is extremely conserved across different species [29]. Consequently, a change in the highly conserved Arg to Gln might have functional consequences on the IL23R transduction pathway. Several functional data have proved that the protective effect of A allele against autoimmune disease is exerted by the impairment of IL23-induced Th17 cell functions without interfering with Th17 differentiation nor affecting the IL17 production [21, 30]. Unfortunately, as all our patients express the GG genotype, the analysis of the expression in patients with GG genotypes versus others with AG genotypes was not possible.
On the other hand, we found significant associations with genotyped SNPs in the 6p12.2 region in chromosome 6: rs3748067>C inIL17A and rs763780>C in IL17F. While few studies have explored the association of rs3748067 in IL17A with the cancer risk but not with AIDs [31, 32], rs763780 in IL17F was associated with many AIDs such rheumatoid arthritis in Tunisian population [33].
Little amount of RORγt, the specific transcription factor of Th17 cells, was observed in PF patients' skin biopsies, but the difference when compared to HC was not statistically significant. PBMCs from PF patients showed a significantly higher level of RORγt mRNA, when compared to specimens from HC. A possible explanation for the lack of correlation may reside in the sample in which mRNA was measured and/or in the little size of the skin sample group. Thus, Th17 cells have low-frequency lesional tissue. So, it is possible that RORγt mRNA is expressed at an undetectable low level in skin biopsies as compared to PBMC. Besides, cellular infiltration in PF includes essentially eosinophils and a little amount of CD4+ cells as compared to other autoimmune intraepidermal bullous diseases, such as that in PV [24]. In fact, Arakawa et al. revealed the presence of IL17+ cells in PF biopsies with a ratio count to CD4+ cell of 1.8%, but the difference compared to HC was not statistically significant. On the contrary, a higher frequency of IL17+ cells reaching a level of 5.2% was reported in skin biopsies of PV patients, in which cellular infiltration is more important [12]. As was expected, correlation analysis showed a positive correlation between the expression of RORγt and IL23R, as the expression of this receptor is necessary for the induction of RORγt expression. Several data showed that cytokines, including TGF-β, IL6, IL1, and IL21, are important in the differentiation of Th17 cells and have proved the necessity of IL23 for the maintenance and the proliferation of established Th17 cells.
For the TNFa gene, we found a strong association with rs1800629>A to the PF risk, similar to the result found by Torzecka et al. on Polish population [34]. However, other studies in Indian and Egyptian population estimated no significant genotype and/or allelic frequency difference between PF patients and HC [35, 36]. The investigation of the relation between this polymorphism and secreted levels of TNFα showed inconsistent results. Thus, some studies revealed that A allele is associated with a higher level of TNFα compared to the common G allele [37, 38], whereas others failed to demonstrate any functional change related to allele change [39, 40]. Previously, we reported the association of an STR within the TNFa gene with the PF risk [41]. As the TNFa locus is located in the HLA area near the DR and DQ regions, several data suggest that the TNFa association with PF could be explained by its linkage disequilibrium with DRB1 locus. However, after multivariant analysis since multiple regression analysis, we showed that, in our population, TNF and HLA DR susceptibility alleles were independently associated with PF [41].
5. Conclusion
In this case-control study, a high frequency of circulating Th17 cells was revealed in patients' blood compared to healthy controls. This increased level was confirmed by the mRNA expression analysis of IL23R and RORγt, specific markers of these cells. Moreover, we reported for the first time, to our knowledge, the genetic contribution of the IL23/Th17 pathway gene's variants in PF pathogenesis. Thus, these results could provide an argument for the contribution of the IL23/Th17 pathway in the pathogenesis of PF. However, much deeper investigation on larger patient groups should be done to support these findings.
Acknowledgments
The authors thank Ms Fekria Abdennadher and Ms Emna Hentati, proficient in the English language, for proofreading this article. This study was supported by the Ministry of High Education and Scientific Research of Tunisia, the Habib Bourguiba University hospital of Sfax, and the “Comité de Recherche de I'hôpital Habib Bourguiba, Sfax, Tunisia”.
Data Availability
The data used to support the findings of this study are available from the corresponding author upon request.
Conflicts of Interest
The authors declare that there is no conflict of interest regarding the publication of this paper.
References
- 1.Kasperkiewicz M., Ellebrecht C. T., Takahashi H., et al. Pemphigus. Nature Reviews Disease Primers. 2017;3:p. 17026. doi: 10.1038/nrdp.2017.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Roscoe J. T., Diaz L., Sampaio S. A. P., et al. Brazilian pemphigus foliaceus autoantibodies are pathogenic to BALB/c mice by passive transfer. Journal of Investigative Dermatology. 1985;85(6):538–541. doi: 10.1111/1523-1747.ep12277362. [DOI] [PubMed] [Google Scholar]
- 3.Beutner E. H., Prigenzi L. S., Hale W., Leme C. D. A., Bier O. G. Immunofluorescent studies of autoantibodies to intercellular areas of epithelia in Brazilian pemphigus foliaceus. Proceedings of the Society for Experimental Biology and Medicine. 1968;127(1):81–86. doi: 10.3181/00379727-127-32626. [DOI] [PubMed] [Google Scholar]
- 4.Kowalczyk A. P., Anderson J. E., Borgwardt J. E., Hashimoto T., Stanley J. R., Green K. J. Pemphigus sera recognize conformationally sensitive epitopes in the amino-terminal region of desmoglein-1. The Journal of Investigative Dermatology. 1995;105(2):147–152. doi: 10.1111/1523-1747.ep12316680. [DOI] [PubMed] [Google Scholar]
- 5.Hacker M. K., Janson M., Fairley J. A., Lin M.-S. Isotypes and antigenic profiles of pemphigus foliaceus and pemphigus vulgaris autoantibodies. Clinical Immunology. 2002;105(1):64–74. doi: 10.1006/clim.2002.5259. [DOI] [PubMed] [Google Scholar]
- 6.Warren S. J. P., Arteaga L. A., Diaz L. A., et al. The role of subclass switching in the pathogenesis of endemic pemphigus foliaceus. Journal of Investigative Dermatology. 2003;120(1):1–5. doi: 10.1046/j.1523-1747.2003.12017.x. [DOI] [PubMed] [Google Scholar]
- 7.Futei Y., Amagai M., Ishii K., Kuroda-Kinoshita K., Ohya K., Nishikawa T. Predominant IgG4 subclass in autoantibodies of pemphigus vulgaris and foliaceus. Journal of Dermatological Science. 2001;26(1):55–61. doi: 10.1016/S0923-1811(00)00158-4. [DOI] [PubMed] [Google Scholar]
- 8.Armitage R. J., Macduff B. M., Spriggs M. K., Fanslow W. C. Human B cell proliferation and Ig secretion induced by recombinant CD40 ligand are modulated by soluble cytokines. The Journal of Immunology. 1993;150(9):3671–3680. [PubMed] [Google Scholar]
- 9.Lin M. S., Fu C. L., Aoki V., et al. Desmoglein-1-specific T lymphocytes from patients with endemic pemphigus foliaceus (fogo selvagem) Journal of Clinical Investigation. 2000;105(2):207–213. doi: 10.1172/JCI8075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gebhard K. L., Veldman C. M., Wassmuth R., Schultz E., Schuler G., Hertl M. Ex vivo analysis of desmoglein 1-responsive T-helper (th) 1 and Th2 cells in patients with pemphigus foliaceus and healthy individuals. Experimental Dermatology. 2005;14(8):586–592. doi: 10.1111/j.0906-6705.2005.00329.x. [DOI] [PubMed] [Google Scholar]
- 11.Xu R. C., Zhu H. Q., Li W. P., et al. The imbalance of Th17 and regulatory T cells in pemphigus patients. European Journal of Dermatology. 2013;23(6):795–802. doi: 10.1684/ejd.2013.2177. [DOI] [PubMed] [Google Scholar]
- 12.Arakawa M., Dainichi T., Yasumoto S., Hashimoto T. Lesional Th17 cells in pemphigus vulgaris and pemphigus foliaceus. Journal of Dermatological Science. 2009;53(3):228–231. doi: 10.1016/j.jdermsci.2008.09.008. [DOI] [PubMed] [Google Scholar]
- 13.Cosmi L., de Palma R., Santarlasci V., et al. Human interleukin 17-producing cells originate from a CD161+CD4+ T cell precursor. The Journal of Experimental Medicine. 2008;205(8):1903–1916. doi: 10.1084/jem.20080397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Annunziato F., Cosmi L., Liotta F., Maggi E., Romagnani S. Defining the human T helper 17 cell phenotype. Trends in Immunology. 2012;33(10):505–512. doi: 10.1016/j.it.2012.05.004. [DOI] [PubMed] [Google Scholar]
- 15.Maddur M. S., Miossec P., Kaveri S. V., Bayry J. Th17 cells: Biology, pathogenesis of autoimmune and inflammatory diseases, and therapeutic strategies. American Journal of Pathology. 2012;181(1):8–18. doi: 10.1016/j.ajpath.2012.03.044. [DOI] [PubMed] [Google Scholar]
- 16.McGeachy M. J., Chen Y., Tato C. M., et al. The interleukin 23 receptor is essential for the terminal differentiation of interleukin 17–producing effector T helper cells in vivo. Nature Immunology. 2009;10(3):314–324. doi: 10.1038/ni.1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Park H., Li Z., Yang X. O., et al. A distinct lineage of CD4 T cells regulates tissue inflammation by producing interleukin 17. Nature Immunology. 2005;6(11):1133–1141. doi: 10.1038/ni1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Essakalli M., Brick C., Bennani N., Benseffaj N., Ouadghiri S., Atouf O. Le lymphocyte TH17 dernier-né de la famille des lymphocytes T CD4+ Pathologie Biologie. 2010;58(6):437–443. doi: 10.1016/j.patbio.2009.01.001. [DOI] [PubMed] [Google Scholar]
- 19.Jovanovic D. V., di Battista J. A., Martel-Pelletier J., et al. IL-17 stimulates the production and expression of proinflammatory cytokines, IL-beta and TNF-alpha, by human macrophages. The Journal of Immunology. 1998;160(7):3513–3521. [PubMed] [Google Scholar]
- 20.Croxford A. L., Mair F., Becher B. IL-23: one cytokine in control of autoimmunity. European Journal of Immunology. 2012;42(9):2263–2273. doi: 10.1002/eji.201242598. [DOI] [PubMed] [Google Scholar]
- 21.Di Meglio P., Di Cesare A., Laggner U., et al. The IL23R R381Q gene variant protects against immune-mediated diseases by impairing IL-23-induced Th17 effector response in humans. PLoS One. 2011;6(2, article e17160) doi: 10.1371/journal.pone.0017160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Puwipirom H., Hirankarn N., Sodsai P., Avihingsanon Y., Wongpiyabovorn J., Palaga T. Increased interleukin-23 receptor(+) T cells in peripheral blood mononuclear cells of patients with systemic lupus erythematosus. Arthritis Research & Therapy. 2010;12(6):p. R215. doi: 10.1186/ar3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shen H., Goodall J. C., Hill Gaston J. S. Frequency and phenotype of peripheral blood Th17 cells in ankylosing spondylitis and rheumatoid arthritis. Arthritis and Rheumatism. 2009;60(6):1647–1656. doi: 10.1002/art.24568. [DOI] [PubMed] [Google Scholar]
- 24.Joly P., Litrowski N. Pemphigus group (vulgaris, vegetans, foliaceus, herpetiformis, brasiliensis) Clinics in Dermatology. 2011;29(4):432–436. doi: 10.1016/j.clindermatol.2011.01.013. [DOI] [PubMed] [Google Scholar]
- 25.Cua D. J., Sherlock J., Chen Y., et al. Interleukin-23 rather than interleukin-12 is the critical cytokine for autoimmune inflammation of the brain. Nature. 2003;421(6924):744–748. doi: 10.1038/nature01355. [DOI] [PubMed] [Google Scholar]
- 26.Murphy C. A., Langrish C. L., Chen Y., et al. Divergent pro- and antiinflammatory roles for IL-23 and IL-12 in joint autoimmune inflammation. The Journal of Experimental Medicine. 2003;198(12):1951–1957. doi: 10.1084/jem.20030896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee E., Trepicchio W. L., Oestreicher J. L., et al. Increased expression of interleukin 23 p19 and p40 in lesional skin of patients with psoriasis vulgaris. The Journal of Experimental Medicine. 2004;199(1):125–130. doi: 10.1084/jem.20030451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xue J., Su W., Chen Z., Ke Y., Du X., Zhou Q. Overexpression of interleukin-23 and interleukin-17 in the lesion of pemphigus vulgaris: a preliminary study. Mediators of Inflammation. 2014;2014:5. doi: 10.1155/2014/463928.463928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Abdollahi E., Tavasolian F., Momtazi-Borojeni A. A., Samadi M., Rafatpanah H. Protective role of R381Q (rs11209026) polymorphism inIL-23Rgene in immune-mediated diseases: A comprehensive review. Journal of Immunotoxicology. 2016;13(3):286–300. doi: 10.3109/1547691X.2015.1115448. [DOI] [PubMed] [Google Scholar]
- 30.Pidasheva S., Trifari S., Phillips A., et al. Functional studies on the IBD susceptibility gene IL23R implicate reduced receptor function in the protective genetic variant R381Q. PLoS One. 2011;6(10, article e25038) doi: 10.1371/journal.pone.0025038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang S., Li C., Li X., et al. Relationship of IL-17A and IL-17F genetic variations to cervical cancer risk: a meta-analysis. Biomarkers in Medicine. 2017;11(5):459–471. doi: 10.2217/bmm-2016-0315. [DOI] [PubMed] [Google Scholar]
- 32.Gao Y.-W., Xu M., Xu Y., Li D., Zhou S. Effect of three common IL-17 single nucleotide polymorphisms on the risk of developing gastric cancer. Oncology Letters. 2015;9(3):1398–1402. doi: 10.3892/ol.2014.2827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marwa O. S., Kalthoum T., Wajih K., Kamel H. Association of IL17A and IL17F genes with rheumatoid arthritis disease and the impact of genetic polymorphisms on response to treatment. Immunology Letters. 2017;183:24–36. doi: 10.1016/j.imlet.2017.01.013. [DOI] [PubMed] [Google Scholar]
- 34.Torzecka J. D., Narbutt J., Sysa-jedrzejowska A., et al. Tumour necrosis factor-α polymorphism as one of the complex inherited factors in pemphigus. Mediators of Inflammation. 2003;12(5):307. doi: 10.1080/09629350310001619735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mosaad Y. M., Fathy H., Fawzy Z., El-Saied M. A. Tumor necrosis factor-α -308 G>A and interleukin-6 -174 G>C promoter polymorphisms and pemphigus. Human Immunology. 2012;73(5):560–565. doi: 10.1016/j.humimm.2012.02.001. [DOI] [PubMed] [Google Scholar]
- 36.Dar S. A., Akhter N., Haque S., et al. Tumor necrosis factor (TNF)-α -308G/a (rs1800629) polymorphism distribution in North India and its association with pemphigus: case-control study and meta-analysis. Autoimmunity. 2016;49(3):179–187. doi: 10.3109/08916934.2015.1134512. [DOI] [PubMed] [Google Scholar]
- 37.Bidwell J., Keen L., Gallagher G., et al. Cytokine gene polymorphism in human disease: on-line databases. Genes and Immunity. 1999;1(1):3–19. doi: 10.1038/sj.gene.6363645. [DOI] [PubMed] [Google Scholar]
- 38.Juszczynski P., Kalinka E., Bienvenu J., et al. Human leukocyte antigens class II and tumor necrosis factor genetic polymorphisms are independent predictors of non-Hodgkin lymphoma outcome. Blood. 2002;100(8):3037–3040. doi: 10.1182/blood-2002-02-0654. [DOI] [PubMed] [Google Scholar]
- 39.Pociot F., Wilson A. G., Nerup J., Duff G. W. No independent association between a tumor necrosis factor-α promotor region polymorphism and insulin-dependent diabetes mellitus. European Journal of Immunology. 1993;23(11):3050–3053. doi: 10.1002/eji.1830231148. [DOI] [PubMed] [Google Scholar]
- 40.Brinkman B., Zuijdeest D., Kaijzel E., Breedveld F., Verweij C. Relevance of the tumor necrosis factor alpha (TNF alpha) -308 promoter polymorphism in TNF alpha gene regulation. Journal of Inflammation. 1995;46(1):32–41. [PubMed] [Google Scholar]
- 41.Abida O., Mahfoudh N., Kammoun A., et al. Polymorphisms of HLA microsatellite marker in Tunisian pemphigus foliaceus. Human Immunology. 2013;74(1):104–109. doi: 10.1016/j.humimm.2012.10.013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data used to support the findings of this study are available from the corresponding author upon request.