Abstract
The genetic structure of three contiguous wild chimpanzee communities in West Africa was examined to determine the extent to which the community, the mixed-sex social unit of chimpanzees, represents a closed reproductive unit. An analysis of paternity for 41 offspring resulted in 34 cases of paternity assignment to an adult male belonging to the same community. Among the 14 offspring for which all potential within-community fathers have been tested, one likely case of extra-group paternity (EGP) has been identified, suggesting an incidence of EGP of 7%. This more extensive analysis contradicts a previous genetic study of the Taï chimpanzees that inferred 50% extra-group fathers. We suggest, based on direct comparison of results for 33 individuals at 1 microsatellite locus and direct comparison of paternity assignments for 11 offspring, that the error rate in the previous study was too high to produce accurate genotypes and assignments of paternity and hence caused the false inference of a high rate of EGP. Thus, the community is the primary but not exclusive unit for reproduction in wild chimpanzees, and females do not typically reproduce with outside males. Despite the inferred low level of gene flow from extra-community males, relatedness levels among the community males are not significantly higher than among community females, and the distribution of genetic relationships within the group suggests that, rather than a primarily male-bonded social structure, the group is bonded through relationships between males and females. Kinship may explain cooperative behaviors directed against other communities, but is unlikely to explain the high levels of affiliation and cooperation seen for male within-community interactions.
An understanding of the evolution of primate social systems depends on the accurate description of mating systems and requires a description of the entire repertoire of reproductive strategies used by individuals of both sexes, as well as the outcomes of those strategies. Such data can be provided only by a combination of behavioral observation and molecular genetic analyses, and where such combined approaches have been used, it has frequently been shown that the behavioral dynamics of wild populations are more complex than previously perceived. For example, dominance status does not always correlate in a simple fashion with long-term male reproductive success (1–3), extra-pair paternity has been found to occur at unexpectedly high levels in birds considered monogamous (4), and previously unsuspected or infrequently used reproductive strategies have been described (5–7). In this study, we genotyped all members of three habituated communities of wild West African chimpanzees (Pan troglodytes verus) to examine the paternity of offspring and the relatedness levels of community males and females. A high frequency (>50%) of extra-group paternity (EGP) in chimpanzees has been described (8, 9). That finding was unexpected, as it greatly exceeded reported levels (1%, 13%) of observed extra-group copulations (10, 11), and would seem to represent a hitherto unrecognized opportunity for female mate choice. Such a strategy by females would lessen the significance of male social rank in determining reproductive success, as well as result in a significant amount of gene flow among communities. In fact, Gagneux et al. (9) calculated average relatedness levels among pairs of community males to be low and equivalent to that found among females, contradicting previous work that described average relatedness levels in males as approximating the level of half-siblings (12). Higher male than female relatedness within communities has been suggested to arise from the dispersal pattern in chimpanzees, because in contrast to the usual pattern in primates, male chimpanzees are philopatric, whereas most maturing females transfer out of their natal community at maturity (10, 13). It has been proposed that chimpanzee communities are primarily male-bonded; that is, that inclusive fitness (14) and kin selection theory (15) explain the high rate of occurrence of affiliative and cooperative actions among adult males (16). The role of genetic relatedness in promoting cooperative interaction among chimpanzees is particularly interesting for consideration of human evolution, as it has been suggested that the social organization of the common ancestor of chimpanzees and humans was in all major respects like that of chimpanzees today and was characterized by female dispersal, male philopatry, and in particular the presence of male kin-based associations (17, 18).
Wild chimpanzees (Pan troglodytes) live in multi-male and -female communities (also termed groups) comprised of as few as 10 to as many as 140 adults and dependent offspring (10, 11, 19, 20). Chimpanzee mating has been described as promiscuous and would seem to afford little opportunity for overt female choice, with sexually receptive females mating with different males repeatedly for the 10–12 days of the fertile estrus period and an estimated 800 matings occurring for each conception (10, 21). Most of the matings have been observed to occur within the community, with the socially dominant males attempting to guard the females near the time of ovulation at the end of the maximum genital swelling period. Females often disappear from the community during their sexual swelling period, sometimes together with a male of the community for what is termed a consortship period (21), during which they remain isolated from other community members for up to 3 cycle periods of the females. However, in other situations, no male is known to be absent, and the location and mating behavior of the absent female is unknown. In East African chimpanzees (P. t. schweinfurthii) of the Gombe National Park in Tanzania, 13% of the observed matings occurred with individuals from other communities (11), but lower incidences have been reported from other chimpanzee populations (10).
Microsatellite genotyping of wild chimpanzees was first used in 1994 to assign fathers to two offspring in a community at Gombe (22). That study also represents an early demonstration of the potential for DNA analysis of noninvasively collected samples such as shed hair from wild animal populations. However, the DNA obtained from such samples is typically degraded and of low concentration, necessitating the use of stringent controls to obtain accurate results (23, 24). Here we present the results of the largest and most rigorously controlled study of wild chimpanzee genetic relationships, encompassing the genotyping of 108 individuals from 3 communities, including the community previously examined (8, 9). Our aims are first, to quantify EGP in these chimpanzees and second, to examine patterns of relatedness within and among communities.
Materials and Methods
Behavioral and Demographic Data.
Three communities of chimpanzees living in the Taï National Park in the Côte d'Ivoire have been the subjects of long-term behavioral observation, and mother–offspring relationships are known (10). These communities are geographically adjacent, with the territory of the Middle community overlapping those of both the North and South communities (25). The number of unhabituated communities in the area in contact with the study communities is estimated at 9. The average numbers of living members in the communities are 38 (North), 12 (Middle), and 63 (South).
DNA Isolation, Quantification, and Microsatellite Genotyping.
Sample material consisted of feces (86 individuals), bones and teeth (13), hairs (8), and chewed fruit wadges (1). Fresh fecal samples weighing ≈5 g were collected from known individuals and placed in 50-ml tubes containing 20 g of silica gel beads. Genomic DNA was extracted from 100 mg of dried feces by using the QIAmp DNA Stool kit (Qiagen, Chatsworth, CA) according to manufacturer's instructions with modifications (24). An ancient DNA facility was used for extraction of DNA from bone by using a modification of an alcohol precipitation protocol (26). Ground bone or tooth root weighing 0.5–1.0 g was incubated overnight at 40°C in 5.0 ml of extraction buffer (0.45 M EDTA, pH 8.0/1% N-lauryl sarcosine/0.4 mg/ml proteinase K). The sample was centrifuged briefly, and the supernatant was extracted twice with phenol:chloroform (1:1) and once with chloroform:isoamylalcohol (24:1). An aliquot was gel-filtered (NAP25 columns, Amersham Pharmacia), eluted into TE (10 mM Tris/1 mM EDTA, pH 8.0), and precipitated with isopropanol [2 vol isopropanol/5/7 vol 4M ammonium acetate/final concentration of 2.5 μg/ml of Dextran Blue (Sigma)/1 mM EDTA pH 8.0]. The pellet was air-dried and resuspended in 50 μl of TE. Single shed hairs were prepared by subjecting the terminal 3-mm root ends to digestion by proteinase K, followed by heat inactivation of the enzyme and direct use of an aliquot in a PCR amplification (27). The wadge sample was collected and extracted in the manner described for feces samples. The nine microsatellite loci used [D2s1329, D9s910, D11s2002, D12s66, D2s136, D5s1470, D7s2204, D7s817, and von Willebrand factor (vWF)] were originally described in humans. Primer sequences and PCR amplification conditions were as described (28). The 5′ ends of the forward primers were fluorescently labeled, amplification products were separated by using capillary electrophoresis (ABI 310), and allele sizes were determined relative to an internal size standard. Several measures were taken to ensure accuracy of genotypes. Sample identification and genotype accuracy was confirmed by molecular sexing, using the X-Y homologous gene amelogenin (29, 30), by verification that all mother–offspring pairs (n = 49) shared an allele at each locus as expected and by use of two or more independently collected samples. Eight of 108 individuals had no known sampled relatives and were genotyped by using DNA from single samples as well. For these now-dead individuals (Ali, Castor, Gitane, Xeres, Joe, Natan, Rafiki, and Totem) the identifications cannot be double-checked, although there is no a priori reason to suspect misidentification. Errors in the genotyping were controlled further by use of a described 5′ nuclease assay to quantify the amount of amplifiable genomic DNA present in the extracts and determine the amount of replication of results necessary to achieve 99% confidence (24). All laboratory investigators were also genotyped. The rate of error stemming from allelic dropout, contamination, and experimental error was estimated (24) by using a subset of the data (1,328 PCRs) at <1%.
Paternity Analysis.
The paternities of 41 offspring were investigated by testing as potential fathers all males, regardless of community affiliation, that were alive and of reproductive age (at least 9 years old; ref. 3) at the time of conception. Paternity assignment was by exclusion, meaning that offspring were required to share one allele at each locus with the mother (maternal genotypes were obtained in all but two cases) and the second allele with the putative father. Assigned fathers could not mismatch at any locus. This approach of assignment by exclusion is the same as that taken in human forensic sciences. Likelihood methods that allow mismatches and choose among multiple unexcluded males were not applied (31). The allele frequencies of both the entire population and of only the pool of potential fathers were used to compute paternity exclusion probabilities (22), and assigned fathers were required to have values in excess of 0.95.
Relatedness Analyses.
Relatedness (R) was estimated for all pairwise combinations of the 108 individuals by using the Queller and Goodnight index (32) and the program RELATEDNESS 5.0 (http://gsoft.smu.edu/GSoft.html). Standard errors for average relatedness values were generated by jackknifing over all loci. Allele frequencies for this analysis were estimated by using 45 individuals of no known relatedness, selected proportionally from the three communities and both sexes. The program KINSHIP 1.3 was used for tests of relatedness by using likelihood ratios for pairs of hypotheses with statistical significance added by use of a simulation routine.
Results
Paternity.
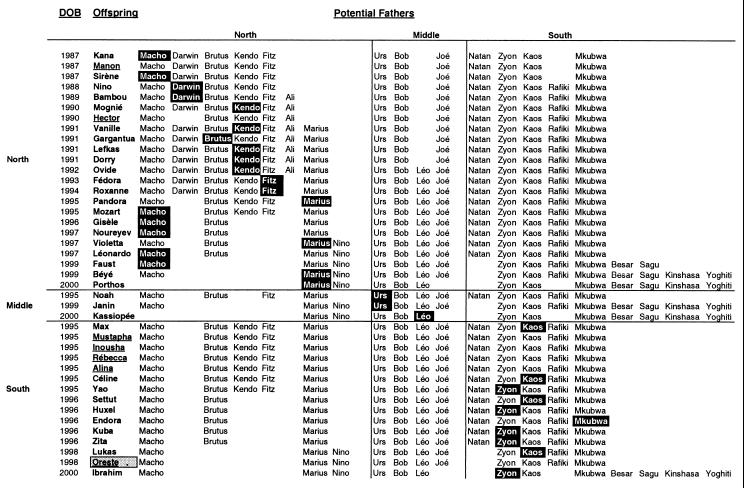
Genotypes at 9 microsatellite loci were generated for 108 individuals, including a total of 21 adult males (Table 3, which is published as supporting material on the PNAS web site, www.pnas.org). Paternity was assigned to 34 of 41 offspring analyzed (Table 1). All assigned fathers had no mismatches, and for no offspring were multiple unexcluded males found. The other potential fathers tested were excluded by mismatches at an average of 3 of the 9 loci compared. There were only two cases in which an excluded male mismatched at only one locus. One of these was the offspring Manon, for which only 7 of 9 loci could be completed, and the second was the offspring Settut, for which no maternal genotype was available. For each of the 34 assignments, the paternity exclusion probability (using allele frequencies from the entire set of potential fathers (was in excess of 0.99. In all cases, the offspring and assigned father belonged to the same community, thus providing no direct evidence for EGP (Fig. 1). Seven offspring could not be assigned to any of the males tested (Table 1; Fig. 1). These unassigned offspring include two individuals from the North community and five from the South community. For the two North offspring, as well as for four of the five South offspring, as a result of deaths and disappearances, not all possible sires from within the community were sampled for this study. However, for one offspring from the South community, Oreste, all males alive in the natal community at the time of conception have been analyzed and excluded. This finding suggests that this offspring represents a case of EGP. Behavioral observations of Oreste's mother, Olivia, were limited but indicate that she was absent some days during the likely time of conception. The program kinship was used to identify all individuals significantly likely to be paternal relatives at the half-sibling level or closer of the seven offspring with unassigned paternity. For six of the offspring, the closest paternal relative was another offspring from the same community. Interestingly, for Oreste, the closest paternal relative was a female (Belle) from the North community (P < 0.05). This finding represents additional evidence that Oreste represents a case of EGP. To summarize, the paternities of a total of 41 offspring were investigated, and 34 highly significant within-community assignments were made, although no assignments could be made for seven offspring. We can thus calculate a maximum frequency of EGP of 7/41 or 17%, but because 6 of those 7 offspring may have been fathered by unsampled within-group males, the actual frequency of EGP may likely be closer to 1/41 or 2.4%. Alternatively, if we restrict ourselves to considering only the 14 offspring for which we have examined all potential within-group fathers (indicated in Table 1), the total of 13 assignments and 1 probable case of EGP produces a frequency of EGP of 1/14 or 7.1%.
Table 1.
Assigned paternities and associated significance measures
| Offspring | Father | Paternity exclusion probabilities‡ by using allele frequencies from:
|
No. mismatches with next best male | No. unsampled community males | |
|---|---|---|---|---|---|
| Entire population | Potential fathers only | ||||
| North | |||||
| Kana* | Macho | 0.9955 | 0.9971 | 2 | 4 |
| Manon† | — | — | — | 1 | 3 |
| Sirène | Macho | 1.0 | 1.0 | 4 | 3 |
| Nino | Darwin | 1.0 | 0.9999 | 2 | 3 |
| Bambou† | Darwin | 0.9996 | 0.9966 | 2 | 3 |
| Mognié | Kendo | 0.9999 | 0.9996 | 3 | 4 |
| Hector† | — | — | — | 2 | 3 |
| Vanille | Kendo | 1.0 | 1.0 | 3 | 4 |
| Gargantua | Brutus | 1.0 | 1.0 | 5 | 4 |
| Lefkas | Kendo | 0.9999 | 0.9999 | 4 | 4 |
| Dorry | Kendo | 1.0 | 1.0 | 3 | 4 |
| Ovide | Kendo | 0.9999 | 0.9997 | 4 | 4 |
| Fédora | Fitz | 0.9998 | 0.9995 | 2 | 4 |
| Roxanne | Fitz | 0.9996 | 0.9996 | 3 | 4 |
| Pandora | Marius | 0.9997 | 0.9999 | 3 | 2 |
| Mozart | Macho | 0.9998 | 0.9997 | 3 | 0 |
| Gisèle | Macho | 0.9998 | 0.9998 | 3 | 0 |
| Noureyev | Macho | 1.0 | 1.0 | 4 | 0 |
| Violetta | Marius | 0.9999 | 0.9999 | 4 | 0 |
| Léonardo | Macho | 0.9998 | 0.9999 | 3 | 0 |
| Faust | Macho | 1.0 | 1.0 | 3 | 0 |
| Béyé | Marius | 0.9999 | 0.9999 | 3 | 0 |
| Porthos | Marius | 0.9996 | 0.9997 | 4 | 0 |
| Middle | |||||
| Noah | Urs | 0.9996 | 0.9992 | 3 | 0 |
| Janin | Urs | 0.9996 | 0.9993 | 2 | 0 |
| Kassiopée | Léo | 1.0 | 1.0 | 5 | 0 |
| South | |||||
| Max | Kaos | 0.9997 | 1.0 | 2 | 3 |
| Mustapha | — | — | — | 2 | 3 |
| Inousha | — | — | — | 3 | 3 |
| Rébecca | — | — | — | 3 | 3 |
| Alina | — | — | — | 2 | 3 |
| Céline | Kaos | 1.0 | 1.0 | 2 | 3 |
| Yao | Zyon | 1.0 | 1.0 | 4 | 3 |
| Settut* | Kaos | 0.9789 | 0.9979 | 1 | 3 |
| Huxel | Zyon | 1.0 | 1.0 | 5 | 2 |
| Endora | Mkubwa | 1.0 | 1.0 | 3 | 2 |
| Kuba | Zyon | 1.0 | 1.0 | 5 | 2 |
| Zita | Zyon | 0.9999 | 0.9999 | 3 | 2 |
| Lukas | Kaos | 1.0 | 1.0 | 2 | 0 |
| Oreste | — | — | — | 3 | 0 |
| Ibrahim | Zyon | 0.9999 | 1.0 | 4 | 0 |
Genotype of mother unavailable.
Typed at only seven loci.
Values in excess of 0.99995 are presented as 1.0.
Figure 1.
The 41 offspring and the males tested as potential fathers. Offspring and potential fathers are grouped by community affiliation, and all males listed in a horizontal row were tested for a given offspring. Black boxes indicate assigned fathers, and offspring with no assigned fathers are underlined. The EGP offspring Oreste is indicated by the gray box.
This frequency of EGP is significantly lower (P < 0.001) than the >50% incidence of EGP suggested by a previous study of the North community, in which for 7 of 13 offspring, all possible community sires were analyzed and excluded (8, 9). Although samples from some of the individuals analyzed in the previous study are no longer available, the results for several of the offspring can be compared directly (Table 2). The first three offspring are those for which no previous determinations of paternity are made, but because some potential sires were not sampled, no conclusions were drawn. For these cases, we made assignments of paternity to community males (Macho and Darwin) that had been previously excluded. The next four offspring are individuals for which paternity assignments were previously made. We confirmed three of these assignments, and for the fourth made a different assignment while excluding the previously assigned father, Ali, by multiple mismatches. The final set of offspring contains four of the seven previously identified cases of EGP. For three of the offspring, we made assignments to males from within the community that had been excluded in the previous study. In the remaining case of postulated EGP reanalyzed here, that of Hector, we do not identify a father but lack samples from some of the community males and so are unable to make a conclusion about paternity. Thus, as a result of the lack of samples from potential sires, this possible case of EGP remains unprovable.
Table 2.
Comparison of paternity assignments of North community offspring
| Individual | Birth year | Paternity assignment
|
|
|---|---|---|---|
| Previous study* | Current study | ||
| Kana | 1987 | All males excluded† | Macho |
| Sirène | 1987 | All males excluded† | Macho |
| Nino | 1988 | All males excluded† | Darwin |
| Vanille | 1991 | Ali | Kendo |
| Gargantua | 1991 | Brutus | Brutus |
| Dorry | 1991 | Kendo | Kendo |
| Fedora | 1993 | Fitz | Fitz |
| Mognié | 1990 | EGP | Kendo |
| Hector | 1990 | EGP | All males excluded† |
| Lefkas | 1991 | EGP | Kendo |
| Pandora | 1995 | EGP | Marius |
Although the current study relied primarily on a newly selected set of highly variable tri- and tetranucleotide repeat microsatellite markers (28), one marker (vWF) used in the previous study was included for comparison. This locus was retyped in 33 individuals examined previously and, in accordance with recommended practice for genotyping from noninvasive samples (23), all heterozygous genotypes were scored only after each allele had been observed twice from separate PCRs, and similarly each homozygous genotype was considered final only after it had been obtained from 7 or more independent PCRs. Of the 66 alleles scored at vWF, different results were obtained for 10 alleles as compared with the previous study. Because our study used more extensive replication, the multiple checks for accuracy listed in the Materials and Methods section, and fluorescent labeling for automated determination of allele sizes, we suggest that the previous study contained errors in ≈15% of the genotypes at this single locus. The incorrect alleles were distributed among 9 individuals; thus, 27% of the individuals examined contained inaccuracies in the genotypes at vWF. Only 2 of the 10 errors found could plausibly be the result of allelic dropout. Similarly, examination of the published genotypes (9) reveals four instances at various loci of mother–offspring mismatches, of which only two could possibly be the result of allelic dropout. Hence, other sources of error must have significantly influenced the previous study.
Relatedness.
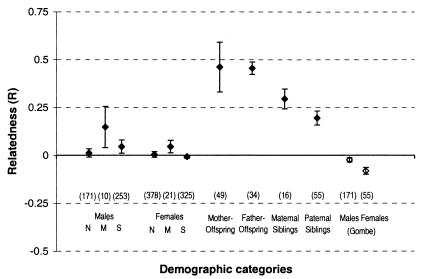
We calculated relatedness (R) values for subsets of community members (Fig. 2) and found first-order relatives (mother–offspring, father–offspring) had mean R values close to the expected value of 0.5 (0.46 for both). Maternal sibling pairs had an average R of 0.29, and paternal sibling pairs an average R of 0.20, both similar to the theoretical expectation of 0.25. Comparison of average R values between males and females within each community (North, 0.012 vs. 0.004; Middle 0.147 vs. 0.045; South 0.045 vs. −0.003), using the R difference test (RELATEDNESS 5.0) in each case, showed no significant difference (North, Middle, and South P > 0.85, 0.65, and 0.57, respectively), suggesting that on average males within a community are not more related than are the females. Another way of analyzing the data is to generate an array of pairwise relatedness values for each within-community set of males and females and examine this array to determine the proportion of comparisons for which the observed average R value was significantly better than an R of 0, the expected value for nonrelatives. In all cases, the proportion of comparisons for which the hypothesis R = 0 was rejected was low (P < 0.05; males North 12%, Middle 10%, South 10%; females North 5%, Middle 14%, South 0), implying that the majority of the comparisons within a community is between unrelated individuals. Finally, both to test the hypothesis that on average males but not females within a community are related at the level of half-siblings (10) and because the ability in primates to detect kin is low for more distant relationships (34, 35), the proportion of comparisons for which R = 0.195 (the average for known paternal half-siblings) was significantly better than the average observed R was also calculated. Consistent with the previous result, only a small proportion of comparisons were compatible with the hypothesis of R = 0.195 (P < 0.05; males North 5%, Middle 10%, South 10%; females North 6%, Middle 14%, South 4%).
Figure 2.
Average relatedness values (R) and standard errors for pairs of individuals in various demographic categories. Individuals were classified according to the results of the genetic analysis. N, M, and S denote the North, Middle, and South communities, respectively. The number of pairwise comparisons used to calculate each value appears in brackets.
To determine whether relatedness among males and females might play a role in community structure, for all 3 communities the proportion of pairwise comparisons of all community members with relatedness levels equivalent or greater than that of half sibs (R = 0.195) was examined. In each community, the proportion of relationships exceeding this value was low (North, Middle, South; 5.9%, 7.6%, 6.3%; mean 6.2%). Most of the relationships were in fact between males and females (on average 3.4%) with a lesser proportion contributed by male–male (1.4%) or female–female (1.4%) relationships. Values for these within-community comparisons would be expected to be higher than those derived from intercommunity comparisons, and all intercommunity comparisons did indeed yield very low proportions of relationships exceeding R = 0.195 (North:Middle 0.5%; North:South 1.1%; Middle:South 2.2%, mean 1.2%). The difference between the means for the intra- and intercommunity comparisons is significant (P < 0.001), confirming that chimpanzees have more relatives within their home community than outside, despite males and females having on average equally low within-community relatedness levels.
For comparison, the data from a recent study of genetic relationships in an East African chimpanzee community, namely the Kasakela community at Gombe (3), were similarly analyzed. The dataset was restricted to those genotypes for which the authors reported replication of results at the level recommended for reliable genotyping and hence omitted 8 of the 39 individuals for which fewer than half of the loci were typed. The R values produced by analysis of these data are underestimates of the degree of relatedness because, as a result of the limited sample size, the allele frequencies were calculated from data that includes related individuals (Fig. 2). As in the Taï communities, the average R values for the males (R = −0.223) and females (r = −0.0823) at Gombe do not significantly differ (P > 0.51), and the average level of relatedness is low.
Discussion
An assessment of the paternity of 41 chimpanzees living in three contiguous communities in the Taï National Park revealed only one case of probable EGP as well as 34 assignments of paternity to within-group fathers. By considering only offspring for which all potential fathers were analyzed, we derive a frequency of EGP of 7%. This depiction of the chimpanzee community as the main reproductive unit is in stark contrast to earlier results from one of the same communities, from which a rate of ≈50% EGP was inferred (8). In fact, the results of this and the previous study are highly inconsistent, with disagreement on the paternity assignments for 7 of the 11 offspring compared. This inconsistency indicates that one or both of the studies produced inaccurate data. It has long been recognized that particular care must be taken to ensure the accuracy of genotypes produced by using low-concentration template DNA such as that typically obtained from noninvasive samples (23, 36). The fundamental problem is thought to be “allelic dropout,” the amplification of only one of two alleles at a heterozygous locus, thus producing a falsely homozygous result. We conclude from our direct comparison of results for 33 individuals at the vWF locus as well as from 4 mother–offspring mismatches in the original data that although Gagneux et al. used fewer repetitions than is recommended to control for allelic dropout (37), only some of the error could actually have been caused by allelic dropout. Other sources of error must have been present, including but not limited to contamination, artifactual “stutter” bands, and sample mix-up. Random inaccurate genotyping is more likely to lead to false exclusions rather than false assignments, and thus in that study a high rate of unassigned paternity resulted in a finding of a high rate of EGP.
The difficulty of producing accurate results from noninvasive samples has resulted in a scarcity of comprehensive microsatellite genotyping studies of wild animal populations (38), and the lack of consistent application of criteria for reliability means that critical evaluation of even published studies is necessary. A recent reanalysis of the genetic relationships of the chimpanzees of the Kasakela community at Gombe has verified one of the two paternity assignments but also shown that three paternity exclusions made by Morin et al. (22) were incorrect (3). Also, the data from the new study of the Gombe chimpanzees itself contains a substantial percentage (22%) of genotypes that are noted as not being confirmed to the recommended extent, whereas the paternity assignments for 3 of the 14 offspring were inferred despite the presence of a mismatch between the putative father and the offspring (see ref. 3; Table 3). Similarly, it was also mentioned in this article that reanalysis of the original Taï data (9), using a likelihood-based method of paternity assignment (31), which does not require genotype compatibility between father and offspring produced assignments for six of the seven EGP offspring (3). However, the new paternity assignments proposed all involved mismatches at two to four loci, rendering the accompanying calculation of paternity exclusion probabilities uninformative. A different measure of confidence in the assignments was calculated by comparisons of the likelihoods of the two most probable fathers, but values were low when unsampled extra-group males were included in the analysis (3). Noninvasive samples remain a viable means for genetic analysis of wild animal populations. Studies using microsatellites, however, clearly need to conform to recommended guidelines for accuracy by using DNA quantification and multiple verification of results (24), as well as use appropriate methods of data analysis.
The low rate of 7% EGP in chimpanzees found here is consistent with observational data in which extra-group copulations have been noted at a frequency of between 1% (Taï) and 13% (Gombe) (10, 11). Interestingly, paternity assignment of 14 offspring at Gombe was all to within-group males despite the higher level of extra-group copulations in this community (3). More comparative data are needed, but at this point, it seems that the community represents the main reproductive unit in chimpanzees.
Most chimpanzee communities are characterized as male-bonded because of the presence of strong social bonds among community males (17, 18). It has been suggested that this is the result of a combination of male philopatry and within-group reproduction resulting in higher average levels of genetic relatedness among community males than among community females (12). Although this study detected somewhat elevated average relatedness levels in males as compared with females in the three Taï communities and in the Gombe community (Fig. 2), this difference does not seem to be significant. The results of the tests can be summarized as (i) on average, males were not found to be significantly more related than females in the same community, (ii) the proportion of pairs that rejected the hypothesis of unrelatedness (R = 0) was 10–12% for males and 0–14% for females, and (iii) the proportion of individual comparisons compatible with R at the level of half-siblings or higher was low (5–10% for males and 4–14% for females). As mentioned previously, the original study of the Gombe chimpanzees (12, 22), suggesting that, on average, group males had the same level of relatedness as half-siblings, contained data that can now be recognized as flawed, making inferences based on that data untenable. The lack of higher male than female relatedness can plausibly be attributed to several factors. One is the introduction of new genes into the community each generation by the immigration of adolescent females from other communities, along with the emigration of adolescent females born in the community. Hence, mothers of all offspring are expected to be unrelated. Fathers of offspring primarily originate from within the community, but our results show that the collection of community offspring is fathered by a variety of individuals (see Table 1). Accordingly, individual males are typically unable to monopolize reproductive opportunities, while in addition, some offspring have EGP.
Although short-term mutualistic benefits as well as long-term social relationships are more likely to explain the basis for cooperative male behaviors such as hunting and meat-sharing (39), the presence of a greater number of related individuals within as compared with between communities shown here may indicate that kin selection theory can to some extent explain the typically hostile character of interactions between communities. Evidence presented here, supporting the concept of a chimpanzee society bonded through male–female relationships, implies that the use of chimpanzee society as the exemplar of the hypothesized male kin-bonded society of the common ancestor of chimpanzees and humans may require reconsideration.
Supplementary Material
Acknowledgments
We thank the Ivorian authorities for long-term support, especially the Ministry of the Environment and Forests as well as the Ministry of Research, the directorship of the Taï National Park, and the Swiss Research Center in Abidjan. Chimpanzee samples were collected by many members of the Taï chimpanzee project and we thank all of them for making this study possible. We thank A. Abraham and V. Jaenicke for technical assistance; K. Isler for sampling assistance; B. Bradley, E. Geffen, P. Morin, S. Pääbo, and M. Stoneking for helpful discussion; and the reviewers for useful comments. The Max Planck Society and the Swiss National Science Foundation supported this work.
Abbreviations
- EGP
extra-group paternity
- vWF
von Willebrand factor
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Altmann J, Alberts S C, Haines S A, Dubach J, Muruthi P, Coote T, Geffen E, Cheesman D J, Mututua R S, Saiyalel S N, et al. Proc Natl Acad Sci USA. 1996;93:5797–5801. doi: 10.1073/pnas.93.12.5797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Launhardt K, Borries C, Hardt C, Epplen J T, Winkler P. Anim Behav. 2001;61:53–64. doi: 10.1006/anbe.2000.1590. [DOI] [PubMed] [Google Scholar]
- 3.Constable J L, Ashley M V, Goodall J, Pusey A E. Mol Ecol. 2001;10:1279–1300. doi: 10.1046/j.1365-294x.2001.01262.x. [DOI] [PubMed] [Google Scholar]
- 4.Petrie M, Kempenaers B. Trends Ecol Evol. 1998;13:52–58. doi: 10.1016/s0169-5347(97)01232-9. [DOI] [PubMed] [Google Scholar]
- 5.Worthington Wilmer J, Allen P J, Pomeroy P P, Twiss S D, Amos W. Mol Ecol. 1999;8:1417–1429. doi: 10.1046/j.1365-294x.1999.00705.x. [DOI] [PubMed] [Google Scholar]
- 6.Heinze J, Keller L. Trends Ecol Evol. 2000;15:508–512. doi: 10.1016/s0169-5347(00)01995-9. [DOI] [PubMed] [Google Scholar]
- 7.Zamudio K R, Sinervo E. Proc Natl Acad Sci USA. 2000;97:14427–14432. doi: 10.1073/pnas.011544998. . (First Published December 5, 2000; 10.1073/pnas.011544998) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gagneux P, Woodruff D S, Boesch C. Nature (London) 1997;387:358–359. doi: 10.1038/387358a0. [DOI] [PubMed] [Google Scholar]
- 9.Gagneux P, Boesch C, Woodruff D. Anim Behav. 1999;57:19–32. doi: 10.1006/anbe.1998.0972. [DOI] [PubMed] [Google Scholar]
- 10.Boesch C, Boesch-Achermann H. The Chimpanzees of the Taï Forest. Oxford: Oxford Univ. Press; 2000. [Google Scholar]
- 11.Goodall J. The Chimpanzees of Gombe. Cambridge, MA: Harvard Univ. Press; 1986. [Google Scholar]
- 12.Morin P A, Moore J J, Chakraborty R, Jin L, Goodall J, Woodruff D S. Science. 1994;265:1193–1201. doi: 10.1126/science.7915048. [DOI] [PubMed] [Google Scholar]
- 13.Pusey A, Williams J, Goodall J. Science. 1997;277:828–830. doi: 10.1126/science.277.5327.828. [DOI] [PubMed] [Google Scholar]
- 14.Hamilton W D. Am Nat. 1963;97:354–356. [Google Scholar]
- 15.Maynard Smith J. Nature (London) 1964;201:1145–1147. [Google Scholar]
- 16.Wrangham R King College Sociobiology Group, editors. Current Problems in Sociobiology. Cambridge, U.K.: Cambridge Univ. Press; 1982. pp. 269–289. [Google Scholar]
- 17.Foley R A. In: Comparative Socioecology. Standen V, Foley R A, editors. Oxford: Blackwell; 1989. [Google Scholar]
- 18.Wrangham R. In: The Evolution of Human Behavior: Primate Models. Kinzey W, editor. Albany: State Univ. New York Press; 1987. [Google Scholar]
- 19.Nishida T, Takasaki H, Takahata Y. In: The Chimpanzees of the Mahale Mountains. Nishida T, editor. Tokyo: Univ. Tokyo Press; 1990. pp. 63–97. [Google Scholar]
- 20.Mitani J, Watts D. Am J Phys Anthropol. 1999;109:439–454. doi: 10.1002/(SICI)1096-8644(199908)109:4<439::AID-AJPA2>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 21.Tutin C E G. Behav Ecol Sociobiol. 1979;6:29–38. [Google Scholar]
- 22.Morin P A, Wallis J, Moore J J, Woodruff D S. Mol Ecol. 1994;3:469–478. doi: 10.1111/j.1365-294x.1994.tb00125.x. [DOI] [PubMed] [Google Scholar]
- 23.Taberlet P, Griffin S, Goossens B, Questiau S, Manceau V, Escaravage N, Waits L P, Bouvet J. Nucleic Acids Res. 1996;24:3189–3194. doi: 10.1093/nar/24.16.3189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morin P A, Chambers K E, Boesch C, Vigilant L. Mol Ecol. 2001;10:1835–1844. doi: 10.1046/j.0962-1083.2001.01308.x. [DOI] [PubMed] [Google Scholar]
- 25.Herbinger I, Boesch C, Rothe H. Int J Primatol. 2001;22:143–167. [Google Scholar]
- 26.Kalmar T, Bachrati C Z, Marcsik A, Rasko I. Nucleic Acids Res. 2000;28:E67. doi: 10.1093/nar/28.12.e67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vigilant L. Biol Chem. 1999;380:1329–1331. doi: 10.1515/BC.1999.169. [DOI] [PubMed] [Google Scholar]
- 28.Bradley B J, Boesch C, Vigilant L. Cons Genet. 2000;1:289–292. [Google Scholar]
- 29.Sullivan K M, Mannucci A, Kimpton C P, Gill P. BioTechniques. 1993;15:636–641. [PubMed] [Google Scholar]
- 30.Bradley B J, Chambers K, Vigilant L. Cons Genet. 2001;2:179–181. [Google Scholar]
- 31.Marshall T C, Slate J, Kruuk L E, Pemberton J M. Mol Ecol. 1998;7:639–655. doi: 10.1046/j.1365-294x.1998.00374.x. [DOI] [PubMed] [Google Scholar]
- 32.Queller D C, Goodnight K F. Evolution (Lawrence, Kans) 1989;43:258–275. doi: 10.1111/j.1558-5646.1989.tb04226.x. [DOI] [PubMed] [Google Scholar]
- 33.Goodnight K F, Queller D C. Mol Ecol. 1999;8:1231–1234. [Google Scholar]
- 34.Alberts S C. Proc R Soc London Ser B. 1999;266:1501–1506. doi: 10.1098/rspb.1999.0807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chapais B, Savard L, Gauthier C. Behav Ecol Sociobiol. 2001;49:493–502. [Google Scholar]
- 36.Navidi W, Arnheim N, Waterman M S. Am J Hum Genet. 1992;50:347–359. [PMC free article] [PubMed] [Google Scholar]
- 37.Gagneux P, Boesch C, Woodruff D S. Mol Ecol. 1997;6:861–868. doi: 10.1111/j.1365-294x.1997.tb00140.x. [DOI] [PubMed] [Google Scholar]
- 38.Taberlet P, Waits L P, Luikart G. Trends Ecol Evol. 1999;14:323–327. doi: 10.1016/s0169-5347(99)01637-7. [DOI] [PubMed] [Google Scholar]
- 39.Boesch C. Anim Behav. 1994;48:653–667. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.