Summary
Tritrophic interactions involving a biocontrol agent, a pathogen and a plant have been analyzed predominantly from the perspective of the biocontrol agent. We have conducted the first comprehensive transcriptomic analysis of all three organisms in an effort to understand the elusive properties of Pseudozyma flocculosa in the context of its biocontrol activity against Blumeria graminis f.sp. hordei as it parasitizes Hordeum vulgare.
After inoculation of P. flocculosa, the tripartite interaction was monitored over time and samples collected for scanning electron microscopy and RNA sequencing.
Based on our observations, P. flocculosa indirectly parasitizes barley, albeit transiently, by diverting nutrients extracted by B. graminis from barley leaves through a process involving unique effectors. This brings novel evidence that such molecules can also influence fungal–fungal interactions. Their release is synchronized with a higher expression of powdery mildew haustorial effectors, a sharp decline in the photosynthetic machinery of barley and a developmental peak in P. flocculosa. The interaction culminates with a collapse of B. graminis haustoria, thereby stopping P. flocculosa growth, as barley plants show higher metabolic activity.
To conclude, our study has uncovered a complex and intricate phenomenon, described here as hyperbiotrophy, only achievable through the conjugated action of the three protagonists.
Keywords: biocontrol agent, effectors, haustorium, hyperbiotrophy, powdery mildew, Pseudozyma flocculosa, tritrophic interaction, Ustilaginales
Introduction
Powdery mildew causes important losses to a wide range of agricultural and horticultural plants. Despite being recognized as one disease, over 650 powdery mildew species are known to occur on nearly 10 000 host species (Glawe, 2008). Powdery mildew fungi belong to the Erysiphales within the Ascomycota and are obligate biotrophic parasites. To establish infection, they depend on living cells where they form a specialized feeding structure, the haustorium, which invaginates the plasma membrane of the host epidermal cell. The interaction between the haustorium and the plant cell is tightly regulated so that the fungus can obtain resources from the plant without killing it (Chaudhari et al., 2014). Diversion of nutrients from the host to powdery mildew fungus leads to the production of mycelium, conidiophores and conidia, thus maintaining the development and asexual reproduction of the fungus. For the plant, the infection results in reduced photosynthesis, slower development and lower productivity.
The economic impact of powdery mildews has driven research in many areas and possibly none more than in the field of genetic control. The pioneering work of Moseman et al. (1964) was the first to investigate gene‐for‐gene interactions between isolates of cereal powdery mildew (Blumeria graminis) and varieties of barley and wheat. As a result, barley powdery mildew (B. graminis (DC.) Speer f. sp. hordei Marchal; B. graminis) is currently one of the best‐studied fungal diseases of plants in terms of the genetics of the host–pathogen interactions. Moreover, the B. graminis genome was the first to be released among powdery mildew species (Spanu et al., 2010), which led to the annotation of 6469 genes from which 491 were identified as candidate secreted effector proteins (CSEPs) (Pedersen et al., 2012). As in many plant–pathogen interactions, effector proteins are involved in disease development by, among other functions, suppressing host defense responses and affecting the host metabolism for the purpose of nutrient acquisition by the pathogen (Chaudhari et al., 2014). In spite of the overwhelming evidence linking CSEPs to the virulence arsenal of plant pathogens, very little is known about their functionality (Sonah et al., 2016). Out of the 491 putative CSEPs identified in silico in B. graminis, recent efforts have so far directly associated about a dozen candidate effectors to the virulence/aggressiveness of the pathogen (Pliego et al., 2013; Ahmed et al., 2015, 2016; Whigham et al., 2015).
Although B. graminis is widely recognized as a study model for powdery mildews, it is clear that its gene‐for‐gene interaction with barley is unique, this notion being reinforced by the fact that all 491 CSEPs are found only in Blumeria spp. Moreover, the majority of resistance genes that can be exploited in barley for powdery mildew control do not confer protection in other powdery mildew–plant interactions. For this reason, other research efforts have concentrated on the development of control methods that were more generic against powdery mildews. With growing concerns over the environmental impact of fungicides, an important body of work has concentrated on biological approaches. Many efforts have been directed toward the identification/isolation of natural enemies of powdery mildews that could be exploited as biocontrol agents (BCA) (Bélanger & Labbé, 2002; Kiss et al., 2010). Among the fungi and bacteria that have been reported to display an antagonistic activity against the Erysiphales, Pseudozyma flocculosa is arguably the most intriguing.
Pseudozyma flocculosa was first discovered and described in 1987 as an epiphytic yeast on powdery mildew‐infected clover leaves (Traquair et al., 1988). It was subsequently found to be a powerful antagonist of powdery mildews and its activity appears to extend to all and only members of the Erysiphales (Bélanger et al., 2012). Over the years, considerable resources have been invested in trying to decipher how P. flocculosa could specifically attack powdery mildews based on the principle that BCAs exert their activity through the manifestation of competition, parasitism, antibiosis or induced resistance (Whipps, 2001). In the case of P. flocculosa, in vitro bioassays, electron microscopy studies and chemical analyses have all pointed to a single mode of action: antibiosis. This conclusion was reinforced by the characterization and purification of an active molecule, flocculosin, that displayed powerful antimicrobial activity (Cheng et al., 2003; Mimee et al., 2005, 2009). This was further supported by the discovery of a complex gene cluster regulating the synthesis of flocculosin (Teichmann et al., 2011b). Intriguingly, flocculosin is nearly identical to ustilagic acid – a compound produced by Ustilago maydis under the control of a similar gene cluster (Teichmann et al., 2007, 2011a). However, the latter discovery, although confirming the classification of P. flocculosa within the Ustilaginales and its close link to smut pathogens, raised questions about the role of flocculosin in the biocontrol activity. If flocculosin indeed confers biocontrol activity against powdery mildews, what is the function of ustilagic acid in U. maydis considering that the fungus is a plant pathogen and does not act as BCA? In an effort to relate the production of flocculosin (or ustilagic acid) to biocontrol, Clément‐Mathieu et al. (2008) and Hammami et al. (2011) tested and compared P. flocculosa, U. maydis and other species of Pseudozyma with or without the ability to produce glycolipids. In all bioassays, only P. flocculosa was capable of antagonizing powdery mildews. The authors concluded that the biocontrol specificity of P. flocculosa could not be attributed to its ability to produce flocculosin alone, given that other similar organisms producing the same glycolipids were incapable of biocontrol. Rather, the results suggested that the particular properties of P. flocculosa, if modulated by flocculosin, were dependent on other factors stimulating the growth and development of the yeast‐like fungus in the presence of powdery mildew colonies.
Recently, P. flocculosa was fully sequenced and annotated and its genome compared to that of closely related Ustilaginales including U. maydis (Lefebvre et al., 2013). Interestingly, on the one hand, its genome was highly similar to that of the smut fungi, with the notable exception of the loss of a few key CSEPs reported to be involved in the virulence of U. maydis; this led to the conclusion that this accounted for the different lifestyles between the two fungi. On the other hand, the P. flocculosa genome contains a few unique features such as 200 CSEPs, and lytic enzymes that could explain its singular activity against powdery mildews. However, no evidence supporting this hypothesis has been found to date.
We posit that the specific antagonistic activity of P. flocculosa against powdery mildews might be deciphered by studying the simultaneous transcriptomic responses of all three organisms over the development of the tritrophic interaction P. flocculosa–B. graminis–H. vulgare. We report here that CSEPs produced by P. flocculosa are involved in its interaction against B. graminis, and that B. graminis responds by increasing quickly and transiently its virulence against the host, until it collapses. We conclude that the BCA exploits B. graminis as a conduit to the plant in a hitherto unsuspected strategy, we define here as hyperbiotrophy.
Materials and Methods
Pseudozyma flocculosa and powdery mildew‐infected plant material
Pseudozyma flocculosa (Traquair, Shaw and Jarvis) Boekhout and Traquair (DAOM 196992) was grown in flasks in YMPD medium (yeast extract, malt extract, peptone, and dextrose; yeast extract 3 g l−1, malt extract 3 g l−1, peptone water 5 g l−1, and dextrose 10 g l−1) following Hammami et al. (2008). The culture was harvested at four different times spanning different phases corresponding to germination (2 h), exponential growth (8 h), flocculosin production under nutrient depletion (18 h) and maturation of sporidia (30 h). Samples collected in vitro were used for comparison with P. flocculosa transcriptome dynamics during the tripartite bioassay.
Barley (Hordeum vulgare L. cv Foster) plants displaying high susceptibility to B. graminis were used for this study. Barley seedlings were grown in a glasshouse under semi‐controlled conditions (18 h 24°C : 6 h 18°C, day : night, light : dark cycle). Three‐week‐old seedlings were exposed to natural infection with B. graminis. The fungus was allowed to grow on the surface of leaves under moist conditions for 4–5 d until it covered c. 50% of the leaf surface.
Inoculation with P. flocculosa and sampling strategy
Diseased leaves were inoculated by spraying uniformly until runoff a suspension of P. flocculosa sporidia (c. 2 × 107 cells ml−1) obtained from a 3‐d‐old culture grown in YMPD broth. The solution was left to evaporate for 20 min and the plants were covered with plastic bags to maintain a relative humidity over 90%. Plants were sampled at 0, 2, 4, 8, 12, 24 and 36 h post‐inoculation (hpi) with P. flocculosa.
RNA extraction, libraries construction and sequencing
For the transcriptome analyses, leaves of four individual plant replicates of each treatment were collected, immediately frozen in liquid nitrogen and stored at −80°C.
Total RNA was extracted using Trizol™ reagent (Invitrogen) and RNeasy® Mini purification kit (Qiagen) including DNAse treatment as per manufacturer's instructions. Before labelling, RNA quality and concentration were checked by agarose gel electrophoresis, spectrophotometry (Nanodrop ND‐1000; NanoDrop Technologies, Wilmington, DE, USA) and ultimately, by an Agilent 2100 Bioanalyzer™ (Agilent Technologies, Palo Alto, CA, USA).
RNA‐seq libraries were generated using the TruSeq® RNA‐Seq Sample Preparation kit according to the manufacturer's protocol (Illumina Inc., San Diego, CA, USA). This kit included a polyA mRNA purification step. After quality control of the indexed cDNA libraries, multiplexed sequencing was carried out on an Illumina HiSeq™ 2000 platform by Genome Quebec (McGill University, Montréal, Canada). All of the primary sequencing data can be found in the DDBJ Sequence Read Archive (DRA) under the accession no. PSUB006820.
RNA‐Seq data analysis
Raw reads processing and high‐quality reads alignment to the reference genomes
Poly‐A, adaptor sequence contaminants and low‐quality bases (Q < 15) were trimmed from Illumina reads in FASTQ format using the RNA‐seq analysis tool of CLC Genomics Workbench v8 (CLC Bio, Aarhus, Denmark) before further processing. All cleaned reads > 40 bp in length were aligned to the P. flocculosa reference genome; the remaining unmapped reads were then aligned to B. graminis and H. vulgare reference genomes in that order. The following criteria were used to map the unique sequence reads: mismatch cost of 2; insert cost of 3; deletion cost of 3; minimum length fraction of 0.9 and minimum similarity fraction of 0.8. Raw read counts and mapping distribution for each condition are given in Supporting Information Table S1.
Gene expression, differential gene expression analyses and clustering
In order to estimate the expression levels, data were expressed as reads per kilobase per million mapped reads (RPKM) by calculating the mapped reads (total exon reads/total mapped reads in millions × exon length in kb) for each gene. After normalization, data were log2‐transformed (0.25 was added to each RPKM number to deal with zero counts) for differential expression analysis. Significant differences in gene expression were detected using the EdgeR algorithm implemented in the CLC Genomics Workbench that utilizes the Exact Test developed by Robinson & Smyth (2008). The P‐value threshold was determined by the false discovery rate (FDR) to account for multiple tests of significance. A FDR threshold ≤ 0.01 was adopted to judge the significance of the gene expression change throughout the tripartite bioassay. In order to identify general trends during the tripartite bioassay, a hierarchical clustering of features was also generated for each organism using normalized expression values (mean expression level was subtracted). Clusters with similar expression patterns based on Manhattan distances were identified. Given that we had more than three replicates for each treatment, quantitative PCR validation was deemed unnecessary (Fang & Cui, 2011).
Functional annotation and enrichment analyses
The web‐based AgriGO tool (Du et al., 2010) was used to obtain Gene Ontology (GO) annotations and to perform singular enrichment analyses (FDR P‐value ≤ 0.05) of genes of P. flocculosa, B. graminis and H. vulgare differentially expressed during the tripartite interaction.
Photosynthesis measurements
In order to validate some of the transcriptomic responses observed with the plant, photosynthesis measurements were conducted on the uppermost, fully expanded infected leaf of at least five plants at 0, 12, 24 and 36 hpi with P. flocculosa, as well as on five noninfected plants. All measurements were made between 9 h00 and 11 h00 am with a Li‐Cor 6400 portable system (Li‐Cor Inc., Lincoln, NE, USA). The internal LED light source was set at 500 μmol m−2 s−1 to ensure a constant uniform light across samples and leaf temperature was maintained at 25°C. In the cuvette, CO2 concentration was 383 ± 2 μmol mol−1 and relative humidity at 60% ± 1. Once steady‐state gas exchange rates were observed, net photosynthetic rate was recorded and thereafter normalized by the actual leaf surface area enclosed in the chamber.
Light and transmission electron microscopy observations
At 0, 12, 24, 36 and 48 hpi with P. flocculosa, 1‐cm2 leaf pieces were directly imaged with an Olympus SZ61 stereomicroscope (Olympus Tokyo, Tokyo, Japan) or collected for later observations. Controls consisted of infected leaves sprayed with water and kept under plastic bags.
For light microscopy and transmission electron microscopy (TEM) observations, segments of randomly selected plants of each treatment were fixed overnight in 2.5% glutaraldehyde, 4% formaldehyde in 100 mM sodium cacosylate buffer (pH 7.3) at 4°C and postfixed for 1 h with 1% osmium tetroxide before dehydration in a grade ethanol series and embedding in LR‐white resin.
Sections were cut at 1 μm thick, stained with toluidine blue and finally imaged with a ZEISS Axioscope microscope (Zeiss, Din Mills, Ontarion, Canada). For TEM imaging, ultrathin sections (100 nm) collected on nickel gris were stained with uranyl acetate and observed with a Jeol 1200 EX microscope (Jeol, Tokyo, Japan).
For scanning electron microscopy, leaf segments were immersed in 100% dry methanol for 10 min, followed by 2 × 30 min changes in 100% dry ethanol. Ethanol was then used for critical point drying of the tissue. Finally, samples were mounted on aluminium stubs, coated with c. 20 nm of gold‐palladium and imaged with a Jeol JSM6360LV Scanning Electron Microscope (Jeol) at 15 kV accelerating voltage.
Results
Dynamic changes in phenotypes of the P. flocculosa–B. graminis–H. vulgare interaction
In order to examine relevant and specific data, sampling of the tritrophic interaction P. flocculosa–B. graminis–H. vulgare was optimized to highlight both phenotypic and transcriptomic responses of all three interacting organisms. Preliminary experiments indicated that no distinct or uniform pattern emerged within the first 10 h following inoculation of P. flocculosa (results not shown).
At 12 hpi, an initial collapse of B. graminis conidial chains was observed, followed by the agglutination of conidia and conidiophores at 24 hpi, which coincided with a faint discoloration on the surface of the colonies, typical of P. flocculosa presence (Fig. 1a). By 36 hpi, B. graminis colonies were completely overgrown by P. flocculosa (Fig. 1a). At higher magnification under SEM, mycelial filaments of P. flocculosa were clearly visible throughout the B. graminis colonies at 12 hpi, a clear indication of its nascent developmental stage following germination (Fig. 1b); by 24 hpi, P. flocculosa mycelium had extensively colonized B. graminis structures and, as expected, no evidence of penetration or coiling could be observed; finally, after 36 h, the P. flocculosa had completely overtaken B. graminis and initiated the production of sporidia (Fig. 1b).
Figure 1.
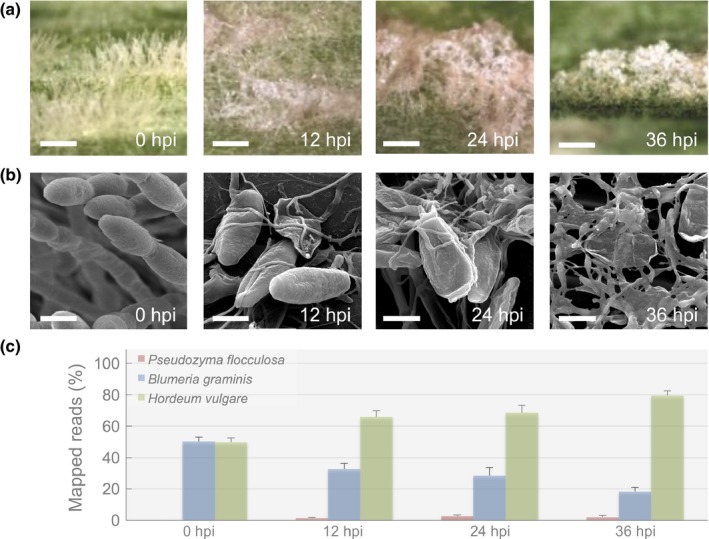
Rapid changes in phenotypes during the tripartite interaction Pseudozyma flocculosa–Blumeria graminis f.sp. hordei–Hordeum vulgare. (a) Optical stereomicroscopic observations of the antagonism of P. flocculosa on B. graminis colonies over time (bar, 400 μm). (b) Scanning electron microscopy observations of B. graminis colonies progressively invaded by P. flocculosa (bar, 10 μm). (c) Percentage of mapped reads for all three interacting organisms over different time points following P. flocculosa inoculation on B. graminis colonies. Each value is the mean ± SE (n ≥ 3).
In terms of raw transcriptomic data, an equal proportion of reads mapped to either B. graminis or H. vulgare reference genomes at 0 hpi (Fig. 1c). After inoculation with P. flocculosa, the proportion of reads that mapped onto B. graminis genome dropped to about one third of the total mapped reads (Fig. 1c). For P. flocculosa, reads were first detectable at 12 hpi and more than doubled at 24 hpi (1.3–2.9% total) (Fig. 1c), a time where the BCA appeared to be the most active (Fig. 1b). Interestingly, both reads from B. graminis and P. flocculosa decreased noticeably at 36 hpi (Fig. 1c) coinciding with the suppression of B. graminis (Fig. 1a,b).
Transcriptome profiling of P. flocculosa
We exploited the extensive body of work on several aspects of P. flocculosa (Hammami et al., 2008; Lefebvre et al., 2013) to devise a strategy for the analysis and interpretation of transcriptome dynamics. In this regard, our analyses were greatly facilitated by the previous identification of several genes, or gene groups that were proposed to be relevant to its specific lifestyle.
Of the 6877 genes annotated in the P. flocculosa genome (Lefebvre et al., 2013), 91 ± 2% were expressed in vitro ≥ 1 RPKM in at least one of the four sampling times, and 94 ± 1% in biocontrol conditions (see Table S2 for detailed expression data). A total of 3207 differentially expressed genes (DEG) were identified (fold‐change ≥ ∣2∣ between two sampling times; FDR P‐value ≤ 0.01) in P. flocculosa during the course of the tripartite bioassay. In addition, many of the highly expressed genes by sporidia at 30 h in vitro, were also observed in planta at 36 hpi when P. flocculosa started producing sporidia (Table S2; Fig. 1b).
Pseudozyma flocculosa mode of action is unrelated to plant pathogen relatives
As many as 285 genes showed no expression in planta; these included both b locus‐encoded homeodomain proteins, pf03264 and pf03265, orthologs of genes involved in mating and subsequent pathogenicity in U. maydis. In addition, none of the 47 putative pathogenesis associated genes associated to smut fungi and found in P. flocculosa genome exhibited higher expression values in planta than in vitro (Table S3).
Flocculosin appears to have limited role in biocontrol activity
We analyzed the expression of all 11 genes involved in flocculosin synthesis, the antifungal molecule long associated with the biocontrol arsenal of P. flocculosa. All genes were weakly expressed compared to in vitro conditions in the early stage of interaction at 12 hpi (Fig 2a, S1). A generalized increase in expression was observed at 24 hpi, albeit at much lower levels than in vitro before receding to lower levels at 36 hpi.
Figure 2.
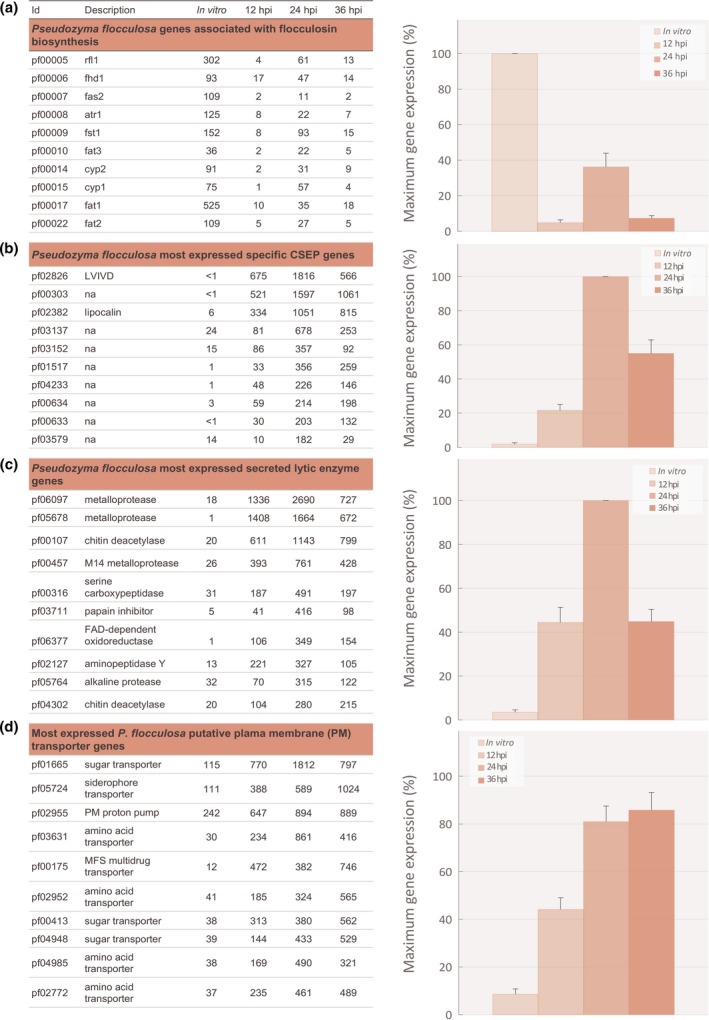
Transcriptome analysis of Pseudozyma flocculosa during the tripartite interaction P. flocculosa–Blumeria graminis f.sp. hordei–Hordeum vulgare. Transcriptome analysis P. flocculosa during the tripartitre interaction P. flocculosa–B. graminis–H. vulgare revealed: (a) the limited transcription of genes associated with flocculosin biosynthesis in P. flocculosa during the tripartite interaction; (b) the rapid induction of genes encoding candidate secreted effector proteins (CSEPs) specific to the biocontrol agent; (c) the rapid induction of genes encoding putative secreted lytic enzymes and (d) the rapid induction of genes encoding putative transmembrane transporters. Average reads per kilobase per million mapped reads (RPKM) data of genes of the flocculosin cluster and of genes significantly differentially expressed (FDR P‐value ≤ 0.01) during the tripartite interaction P. flocculosa–B. graminis–H. vulgare at 12, 24 and 36 h post‐inoculation (hpi) on B. graminis colonies compared to in vitro cultures (n ≥ 3) are given in the table on the left. On the right, histograms showing the average relative (%) cluster expression at each time point based on the highest level of expression for each gene as a measure to showcase the trend in expression dynamics. Each value is the mean ± SE (n = 10 genes).
Unique P. flocculosa CSEPs and other secreted proteins are involved in antagonism against B. graminis
Among the highest DEGs observed, many belonged to CSEPs reported to be unique to P. flocculosa (Lefebvre et al., 2013). From those CSEPs, we identified a group of 10 highly expressed genes whose analysis confirmed a similar pattern of expression (Figs 2b, S2). In fact, those effectors were highly expressed during contact with B. graminis, contrasting with a complete or near complete lack of expression in vitro. Expression increased over time to reach a peak at 24 hpi and subsided at 36 hpi when P. flocculosa had completely overtaken B. graminis (see Fig. 1). Of particular importance, three transcripts, pf02826, pf00303 and pf02382, had a differential fold‐change > 175 when compared to in vitro conditions.
Within the secretome, several lytic enzyme genes also were found to be strongly induced in presence of B. graminis (Figs 2c, S3). As with CSEPs above, their expression was consistently higher at 24 hpi. In particular, two metalloproteases, pf06097 and pf05678, showed the highest RPKM values of the group.
Importance of P. flocculosa transporters revealed in the interaction with B. graminis
Considering that transporter proteins play an important role in substrate acquisition, a particular emphasis was placed on their analysis. This revealed that several genes coding for sugar and amino acid transporters were upregulated during the rapid growth phase of the BCA compared to growth in vitro (Figs 2d, S4).
Transcriptome profiling of B. graminis
The transcriptomic response of B. graminis to P. flocculosa presence was broad: 1872 genes exhibited differential expression levels (Table S4; fold‐change ≥ ∣2∣ between two sampling times, FDR P‐value ≤ 0.01) throughout the sampling period including 1082 that were upregulated.
Generalized increase of transport system in B. graminis appears to benefit P. flocculosa
AgriGO singular enrichment analysis of the differentially expressed genes revealed the overrepresentation of GO terms: ‘transmembrane transporter function’, ‘membrane component’ and ‘carbohydrate metabolic process’ among the genes induced in presence of P. flocculosa (Fig. S5). These results were seemingly in contradiction with the phenotypes observed. Genes associated with B. graminis nutrient acquisition system were over‐stimulated in spite of a vegetative development obviously altered by the rapid growth of P. flocculosa (Figs 3a, S6). In fact, several sugar and amino acid transporters in B. graminis were found to be upregulated in synchrony with the dynamic regulation of similar transport genes of P. flocculosa (see Fig. 2d).
Figure 3.
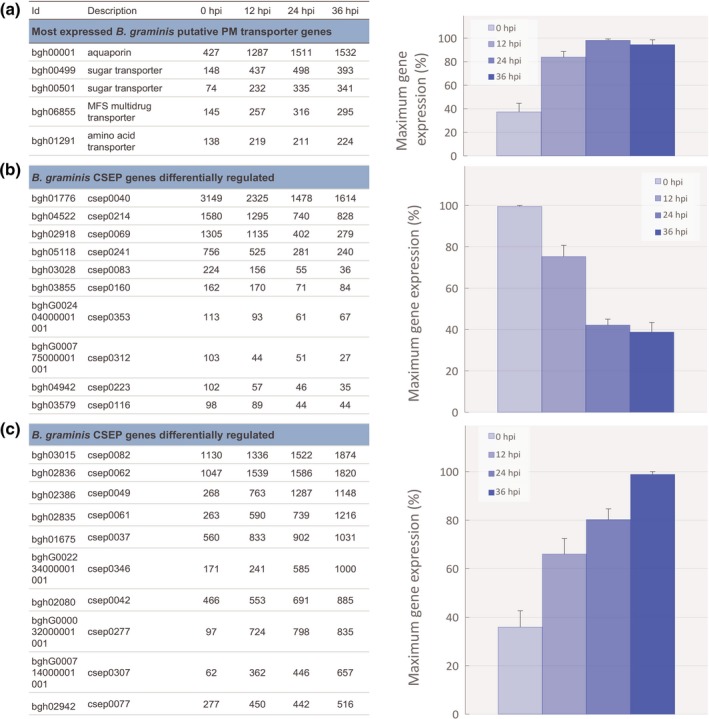
Transcriptome analysis of Blumeria graminis f.sp. hordei during the tripartite interaction Pseudozyma flocculosa–B. graminis–Hordeum vulgare. Transcriptome analysis B. graminis during the tripartitre interaction P. flocculosa–B. graminis–H. vulgare revealed: (a) the increasing transcription pattern of genes encoding putative transmembrane transporters; (b) the decreasing expression pattern of conidia‐ and hyphae‐specific candidate secreted effector protein (CSEP) genes and (c) the increasing expression pattern of haustoria‐specific CSEP genes. Average reads per kilobase per million mapped reads (RPKM) data of genes significantly differentially expressed (FDR P‐value ≤ 0.01) during the tripartite interaction P. flocculosa–B. graminis–H. vulgare at 0, 12, 24 and 36 h post‐inoculation (hpi) on B. graminis colonies (n ≥ 3) are given in the table on the left. On the right, histogram showing the average relative (%) cluster expression at each time point based on the highest level of expression for each gene as a measure to showcase the trend in expression dynamics. Each value is the mean ± SE (n = number of genes presented in the table on the left).
Unexpected patterns of CSEP expression are observed in B. graminis
Several CSEPs were identified in the sequencing and annotation of B. graminis genome (Spanu et al., 2010). These were implicated in the virulence of the pathogen. For this reason, they were emphasized in our analyses. Based on cluster analysis and expression dynamics, two clear and distinct patterns emerged among the most expressed CSEPs.
In a first group of CSEPs, all reported as encoding proteins produced by conidia and hyphae (Pedersen et al., 2012), we observed a declining level of expression in B. graminis colonies following P. flocculosa inoculation, a pattern well‐aligned with the early degradation of B. graminis mycelium and conidia (Figs 3b, S7). In addition, another 15 CSEPs were similarly repressed in the presence of P. flocculosa (FDR P‐value ≤ 0.01; Table S4).
The second group had arguably the most counterintuitive pattern. Based on cluster analysis, 112 CSEPs were found to have an increased expression over time (FDR P‐value ≤ 0.01) in spite of the obvious stress imposed by the presence of P. flocculosa. Analysis of the ten most expressed ones revealed that they were all of haustorial origin (Bindschedler et al., 2009, 2011), and that they steadily increased over the base level maintaining infection with barley (Figs 3d, S8), even when ectotrophic structures were physically destroyed by P. flocculosa (Fig. 1).
Profiling of Hordeum vulgare transcriptome and physiology
A tale of two pathogens?
Photosynthesis deregulation is one of the most reliable markers of powdery mildew presence on a plant. At the transcriptomic level, ‘Photosynthesis’, ‘secondary metabolic process’ and ‘response to stimulus’ were the most significantly over‐represented GO terms (Fig. S9) among the 1259 H. vulgare DEGs observed in response to P. flocculosa inoculation (Table S5; fold‐change ≥ ∣2∣ between two sampling times, FDR P‐value ≤ 0.01). Among other common and important DEGs in barley, many defense‐related proteins were observed within highly expressed genes (Fig. S10; Table S5). Their expression increased over time consistent with the phenotype of a plant fighting off infection throughout the experimental period. Most importantly, the pattern of expression of photosynthesis‐related genes showed a sharp reduction over time that culminated at 24 hpi before going back up at 36 hpi (Figs 4a, S11).
Figure 4.
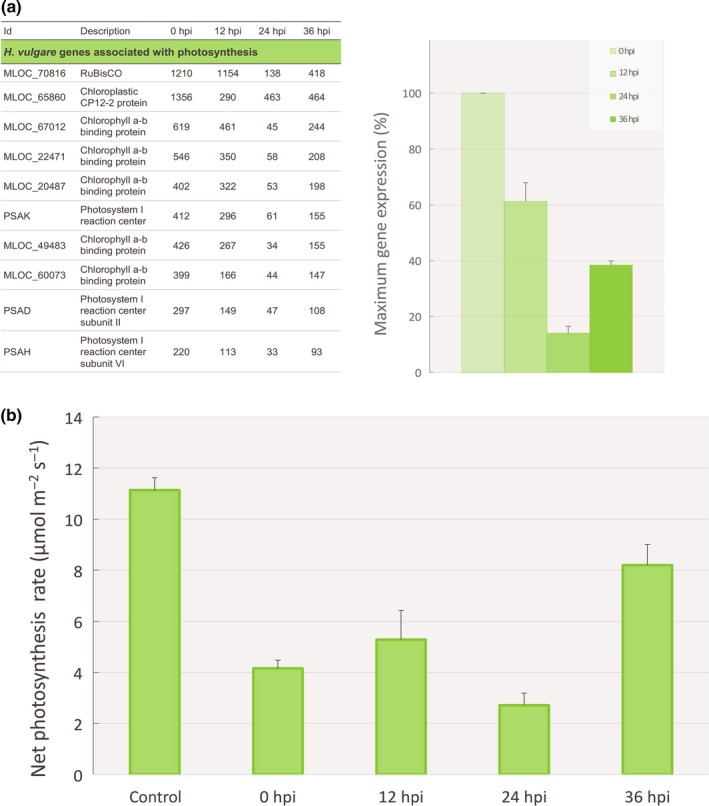
Effect of Pseudozyma flocculosa on the expression profile of the most expressed photosynthesis‐related genes and on net photosynthesis rate in Hordeum vulgare during the tripartite interaction P. flocculosa–Blumeria graminis f.sp. hordei–H. vulgare. (a) Average reads per kilobase per million mapped reads (RPKM) data of the most expressed photosynthesis‐related genes of H. vulgare significantly differentially expressed (FDR P‐value ≤ 0.01) during the tripartite interaction P. flocculosa–B. graminis–H. vulgare at 0, 12, 24 and 36 h post‐inoculation (hpi) on B. graminis colonies (n ≥ 3) are given in the table on the left. The histogram shows the average relative (%) cluster expression at each time point based on the highest level of expression for each gene as a measure to showcase the trend in expression dynamics. Each value is the mean ± SE (n = 10 genes). (b) Net photosynthesis rate (μmol m−2 s−1) of H. vulgare leaves during the tripartite interaction. Each value is the mean ± SE (n ≥ 5 plants).
Photosynthesis measurements
Based on barley transcriptomic responses, photosynthetic rates were measured on plants subjected to the same treatment to determine if the fungal interactions influenced the plants reaction. A pattern of declining photosynthetic rate of leaves was observed up to 24 hpi, followed by a resurgence in the following 12 h (36 hpi; Fig. 4b), in line with the reduced activity of both B. graminis and P. flocculosa.
Light microscopy and TEM observations of the interaction
Light and transmission electron microscopy observations throughout the tripartite interaction yielded phenotypes that supported the transcriptomes of the three organisms (Fig. 5). Compared with the control (Fig. 5a), haustoria in barley cells presented digitations seemingly unaltered whereas a few ectotrophic hyphae were devoid of cytoplasm, 12 h after P. flocculosa inoculation (Fig. 5b). At 24 hpi, surface conidia and hyphae had clear signs of degradation, whereas haustorial bodies started to show evidence of alteration along with a thicker extra‐haustorial matrix (Fig. 5c). At 36 hpi, a clear layer of dead hyphae was observed on the leaf surface and, when still integral, haustorial structures were convoluted and sometimes collapsed (Fig. 5d).
Figure 5.
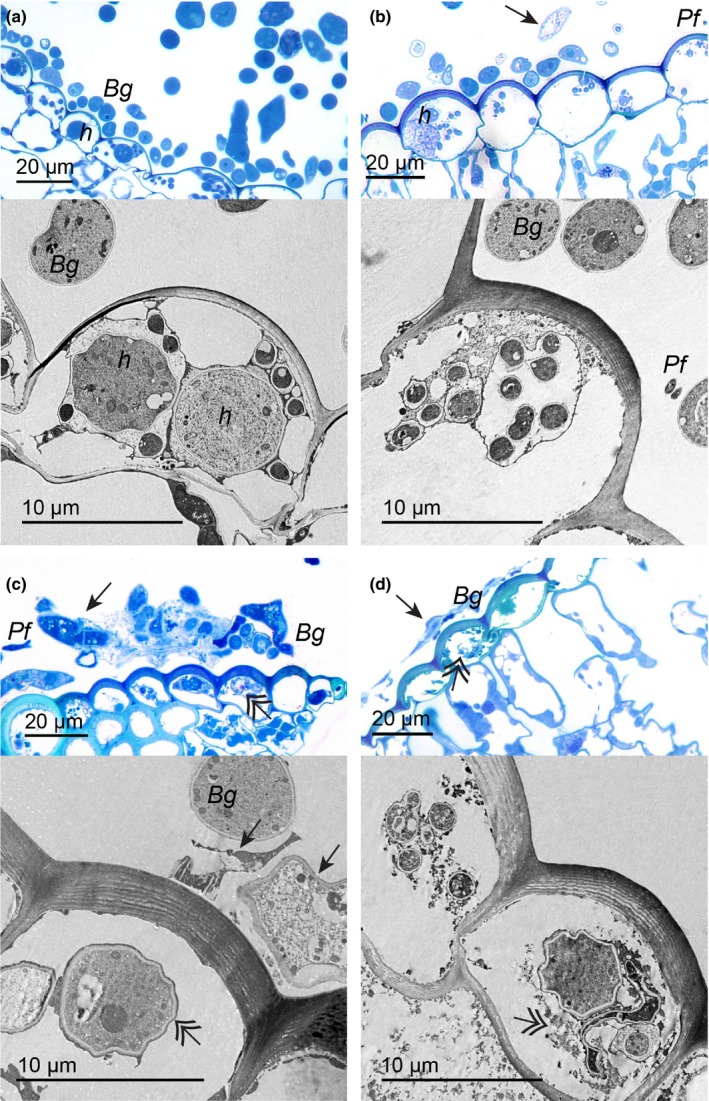
Light and transmission electron microscopy (TEM) of the interaction Pseudozyma flocculosa–Blumeria graminis f.sp. hordei–Hordeum vulgare reveal phenotypic responses supporting transcriptomic results. (a) Light and TEM. Healthy surface hyphae and conidia (Bg), and haustoria (h) of B. graminis are visible in barley cells before inoculation with P. flocculosa (0 hpi). (b) At 12 hpi, presence of P. flocculosa (Pf) as well as B. graminis empty hyphae are observed on the surface (arrows; light), whereas haustoria and digitations are seemingly healthy (TEM). (c) At 24 hpi, surface hyphae and conidia are clearly degraded (arrow; light), in barley cells, the haustoria also show signs of degradation (double arrow; TEM) along with a thicker extra‐haustorial matrix. (d) At 36 hpi, a denser layer of dead hyphae is evident on the leaf surface (arrow; light), and haustorial structures are convoluted when not completely plasmolyzed (double arrow; light and TEM).
Discussion
Historically, the interaction between a plant pathogen and a biocontrol agent (BCA) has been described as one of competition, antibiosis and/or parasitism based on 1D or 2D analyses (Paulitz & Bélanger, 2001). We present here the first comprehensive transcriptomic responses of a tritrophic interaction, BCA–powdery mildew–plant, highlighting unique and unsuspected interdependent reactions at each trophic level.
Since its discovery, Pseudozyma flocculosa has always been described as a superior antagonist because of its faster mode of action than other reported BCAs of powdery mildews such as Ampelomyces quisqualis or Tilletiopsis spp. (Hajlaoui & Bélanger, 1993; Dik et al., 1998). In our experiments, we reproduced the expected phenotypic responses where powdery mildew structures were quickly colonized and killed within a period of 24 h. For its part, P. flocculosa development was visible within 12 h post‐inoculation (hpi), peaked at 24 hpi and started receding at 36 hpi when powdery mildew colonies were seemingly saturated. This pattern is consistent with previous reports where longer bioassays have further shown that P. flocculosa populations are reduced to epiphytic levels once powdery mildew colonies are dead or absent (Neveu et al., 2007; Clément‐Mathieu et al., 2008; Hammami et al., 2011). Interestingly, the transcript proportions of both P. flocculosa and Blumeria graminis matched the observed phenotypes over time.
Comparative genomics has recently highlighted the remarkable similarity between P. flocculosa and smut fungi belonging to the Ustilaginomycetes (Lefebvre et al., 2013) but could only speculate about the factors that brought about such divergent lifestyles. In Ustilago maydis, the mating factor bE/bW heterodimer acts as the central regulator for pathogenicity (Romeis et al., 2000). In the course of our bioassays, the two P. flocculosa bE/bW orthologs were never transcribed, nor were numerous downstream genes or the putative pheromone precursor and receptors, which are determinant for the sexual reproduction and the development of the dikaryotic pathogenic state in plant pathogens. Together with the loss of key effectors for pathogenicity (Lefebvre et al., 2013), these results confirm that P. flocculosa has indeed lost the ability to be a plant pathogen in the same category as smut fungi.
Ever since its discovery, P. flocculosa has been described as a biocontrol agent of powdery mildews acting through antibiosis, the latter mode of action being entirely related to its ability to produce an unusual glycolipid through the activation of a conserved cluster of genes also found in the U. maydis genome (Teichmann et al., 2011a; Bélanger et al., 2012). Despite its potent antifungal properties (Mimee et al., 2005), the role of this glycolipid, flocculosin, in the biocontrol activity of P. flocculosa was recently questioned because its synthesis did not appear to be synchronized with P. flocculosa development on powdery mildews (Marchand et al., 2009; Hammami et al., 2011). In the present study, the 11 genes regulating flocculosin synthesis were strongly expressed in vitro but to a much lesser extent during the tripartite bioassay, especially at 12 hpi at a time where one would expect an early release facilitating access to powdery mildew colonies. Therefore, transcriptomic data support the notion that flocculosin is not a key factor in the biocontrol activity of P. flocculosa, namely in the context of its specificity to powdery mildews. Given its link to ustilagic acid produced by U. maydis and other related Ustilaginales (Kulakovskaya et al., 2005; Teichmann et al., 2007), conservation of the trait among those fungi appears more likely related to protection of an ecological niche on the leaf surface during the epiphytic stage.
Beyond the common genomic features, Lefebvre et al. (2013) reported several unique genes in P. flocculosa, including > 200 candidate secreted effector proteins (CSEPs) that we targeted to start analyzing transcripts of potential interest. The notion that some of them could be important for the antagonistic activity of P. flocculosa raised interesting possibilities because this phenomenon, although widely described in plant–pathogen interactions, has never been reported in fungal–fungal interactions. It was therefore fascinating to observe that several P. flocculosa CSEPs were among the most transcribed genes during the tripartite interaction. Their pattern of expression also was consistent with the biocontrol activity of P. flocculosa, thereby supporting their role as potential virulence factors against B. graminis. As a matter of fact, the repertoire of species‐specific candidate effectors in P. flocculosa is nearly three times as great compared to its smut relatives (U. maydis, U. hordei or Sporisorium reilianum; Lefebvre et al., 2013), thus suggesting different functions. Indeed, on the one hand, no orthologs of U. maydis effectors, actually characterized for their essential role in plant pathogenicity, were expressed in P. flocculosa. These include Pep1 (pf02348), Cmu1 (pf04511) or the Pit cluster (pf04022, pf04023) (Djamei et al., 2011; Doehlemann et al., 2011). On the other hand, several CSEPs, namely pf02826, pf00303 and pf02382, were highly and exclusively expressed during contact with powdery mildew structures, a result providing strong evidence that this fungal–fungal interaction is governed by effector proteins. For instance, the presence of a lipocalin PFAM domain on pf02382 is of particular interest. Lipocalins are secreted proteins that bind and transport small molecules, which suggests that pf02382 may be involved in the dissemination, sequestration of various compounds or also the capture for its own benefit of molecules such as iron‐charged siderophores (Fischbach et al., 2006).
Our results also revealed that the expression of many transcripts coding for lytic enzymes was specifically elicited following contact with B. graminis. On the one hand, although lytic enzymes have often been associated with direct parasitism in some BCAs (Whipps, 2001), this expression is unlikely to be related to this mode of action because the cycle of P. flocculosa is too short and no penetration of P. flocculosa inside powdery mildew structures has ever been observed (Hajlaoui & Bélanger, 1993; Bélanger et al., 2012). On the other hand, pf00107 and pf04302 proteins could potentially play a major role in rapid cell wall permeation as suggested for a chitinase of Trichoderma harzianum (Zeilinger et al., 1999). In addition, similar to the ones observed here, a secreted metallopeptidase with antifungal activity has been recently characterized as an essential component of T. guizhouense's ability to parasite the causative agent of banana wild disease (Zhang et al., 2016). Metalloproteases such as pf06097, pf05678 and pf00457 are also reported to be involved in the breakdown of nutrient sources from host cells (Druzhinina et al., 2012).
The previous observations are consequent with a pattern of nutrient acquisition and accordingly, major changes in the transmembrane transport system of P. flocculosa were noted. For instance, contact with B. graminis clearly triggered an accumulation of sugar and amino‐acid transporter transcripts in a manner reminiscent of plant surface signals inducing U. maydis transporters to provide the pathogen with resources diverted from the plant metabolism (Lanver et al., 2014). Interestingly, the most expressed sugar transporter gene, pf01665, is a close homolog of the U. maydis Hxt1 transporter required for virulence (Schuler et al., 2015). In the rust fungus Uromyces fabae, HXT1 is a haustorium‐specific protein involved in sugar efflux from the infected plant tissue to the pathogen (Voegele et al., 2001). The expression profile of pf03631, a Gap1 general amino‐acid permease homolog, mirrors Hxt1 in this study. In Saccharomyces cerevisiae, GAP1 is the major transporter of amino acid for use as a nitrogen source (Magasanik & Kaiser, 2002; Donaton et al., 2003). Considering their expression levels at 24 hpi, pf01665 and pf03631 are likely to have similar functions in P. flocculosa, whereby these specific permeases with putative sensor functions may help to assimilate nutrients directly from B. graminis.
When looking into the B. graminis transcriptomic profile following inoculation with P. flocculosa, several responses were seemingly incongruent with the observed interaction phenotypes. Although B. graminis was clearly being overwhelmed by P. flocculosa and its external structures plasmolysed, several transmembrane transporters like the haustorial‐specific sugar transporter bgh00499, known to be involved in pathogenesis (Bindschedler et al., 2011; Hacquard et al., 2013), exhibited an increase in expression as early as 12 hpi to a level that was maintained throughout the interaction. Considering the rapid development of P. flocculosa over that time period and a concomitant increase in transporters activity in both fungi, these results could suggest a diversion of nutrients extracted by B. graminis from barley for the benefit of P. flocculosa.
The expression dynamics of B. graminis CSEPs was also very informative with respect to their functionality. In essence, two distinct patterns of expression were observed and converged toward different roles. In a first group, there was a regular downward trend over time following P. flocculosa inoculation. This pattern is at first predictable because a reduction in expression should be synchronized with B. graminis being antagonized. Among the CSEP genes found in this group, two of them, csep0040 and csep0214 (Pedersen et al., 2012), have been associated with conidia, whereas the other ones encode secreted proteins of the mycelium. By contrast, several CSEPs were found to have an increasing pattern of expression over time. Interestingly, most of these CSEPs, including the highly expressed csep0082 and csep0062, are haustoria specific (Bindschedler et al., 2009, 2011) and contribute to the virulence of B. graminis. There is a counterintuitive quality to this because it is likely that this would affect the delicate balance inherent to biotrophy established between B. graminis and barley cells. A possible explanation is that P. flocculosa exploits the B. graminis pathogenic machinery and exacerbates its virulence, albeit for a short period of time, for its own benefit.
The transcriptome profiling of barley plantss also revealed patterns of gene expression that suggested the interaction between P. flocculosa and B. graminis was in fact greatly influenced by the plant. For instance, it is well known that powdery mildew infection results in reduction of plant photosynthesis (Higgins et al., 1985; Scholes et al., 1994; Chain et al., 2009; Chandran et al., 2010). As such, one would expect that P. flocculosa antagonism of B. graminis would quickly alleviate the stress exerted by the pathogen onto barley leaves. However, physiological data and analysis of genes related to photosynthesis revealed a pattern that suggested otherwise. Indeed, we observed a sharp downregulation of photosynthetic genes concomitant with P. flocculosa rapid development within the first 24 h, followed by an upregulation at 36 hpi when the activity of P. flocculosa receded. This would indicate, in line with earlier observations, that P. flocculosa actually exploits B. graminis as a conduit to extract nutrients from the plant.
Taken together, these findings have uncovered a novel mode of action within the antagonistic repertoire of biocontrol agents that we describe here as hyperbiotrophy. Indeed, against all expectations, the ultimate host target of P. flocculosa, appears to be the plant, a parasitism that is achieved through the powdery mildew pathogen. In Fig. 6, we propose a model that explains how the interactions between the three organisms lead to this rare form of parasitism. Upon contact with B. graminis, P. flocculosa releases an unique set of effectors, which in turn activates the virulence of B. graminis through an exacerbated production of haustorial effectors and deregulates its interaction with barley. This interaction accentuates the acquisition of nutrients from the plant, as shown by the increased stress on its photosynthetic ability, nutrients that are, in all likelihood, diverted toward P. flocculosa because the latter develops rapidly during this period whereas the ectotrophic structures of B. graminis become progressively destroyed. This process, as observed here and in other reports (Hammami et al., 2011), is very transient and lasts approximately only 24 h, its demise being linked with the collapse of B. graminis (see Figs 1, 5, 6). Incidentally, this model explains why P. flocculosa populations will be reduced to epiphytic levels in the hours following the interruption of the conduit between the plant and the pathogen (Hajlaoui et al., 1992; Hammami et al., 2011). This model is also coherent with previous observations showing that P. flocculosa is unable to grow on detached powdery mildew spores, or on the plant alone, and is specific to powdery mildew pathogens. The latter conclusion had puzzled scientists because flocculosin (and ustilagic acid), presumed to be the active principle of P. flocculosa biocontrol activity for a long time, was toxic against a wide array of fungi (Mimee et al., 2005; Teichmann et al., 2007). Future efforts should concentrate on assessing the functionality of the salient genes of P. flocculosa and B. graminis, a challenge currently hampered by the fact that neither of the fungi is amenable to transformation (Bélanger et al., 2012; Yan et al., 2012).
Figure 6.
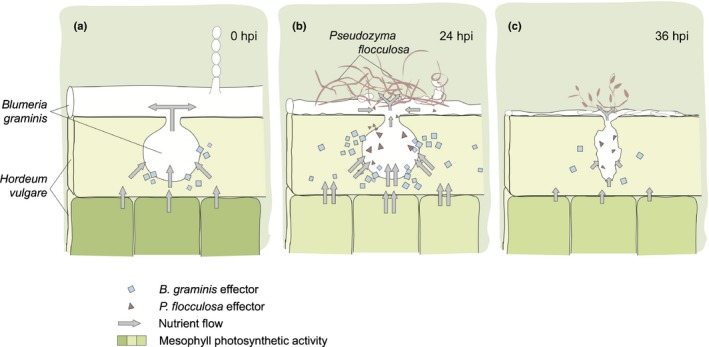
Proposed model of hyperbiotrophy in the tritrophic interaction Pseudozyma flocculosa–Blumeria graminis f.sp. hordei–Hordeum vulgare. (a) A biotrophic interaction is established between B. graminis and H. vulgare through the formation of a haustorium within the epidermal cells, and the release of effectors preventing defense reactions by the plant, and diverting nutrients from the plant to the pathogen (Chaudhari et al., 2014). (b) Upon contact with B. graminis structures, P. flocculosa overwhelms the pathogen colonies within 24 h, a phenomenon synchronized with the higher expression of transporters and unique P. flocculosa effectors, which in turn exacerbates the virulence of B. graminis by overstimulating its expression of effectors and the extraction of nutrients from H. vulgare cells to the profit of P. flocculosa. This hyperbiotrophic activity affects the plant transiently by further reducing gene expression linked to photosynthesis. (c) Deregulation of the delicate biotrophic balance between B. graminis and H. vulgare leads to the decline of the haustorium activity and its eventual collapse, thereby reducing the flow of nutrients toward P. flocculosa and halting its growth within 36 h, and at the same time allowing the plant to start recovering through increased photosynthetic activity.
In conclusion, our transcriptomic analysis of the tripartite interaction P. flocculosa–B. graminis–H. vulgare has uncovered a complex phenomenon of hyperbiotrophy, the first such description for a biocontrol agent. Based on our observations, P. flocculosa would actually parasitize the plant, albeit transiently, by diverting nutrients extracted by B. graminis through a process involving the release of unique effectors, bringing evidence that such molecules can also influence fungal–fungal interactions.
Author contributions
J.L. carried out physiological data measurements and transcriptomic analyses, participated in the interpretation of data and drafted the manuscript; J.L., C.L. and R.R.B. carried out optical microscopy and transmission electronic microscopic observations; G.B.R., F.L. and C.L. carried out sample collection, RNA‐Seq experiments and transcriptomic analyses; C.L. and P.D.S. were involved in interpretation of data and drafting of the manuscript; and R.R.B. conceived the study, participated in interpretation of data and writing of the article. All authors have read and approved the final manuscript.
Supporting information
Please note: Wiley Blackwell are not responsible for the content or functionality of any Supporting Information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
Fig. S1 Transcriptomic profiles of genes associated with flocculosin biosynthesis in Pseudozyma flocculosa during the tripartite interaction P. flocculosa–Blumeria graminis f.sp. hordei–Hordeum vulgare.
Fig. S2 Transcriptomic profiles of the most expressed CSEPs genes specific to Pseudozyma flocculosa during the tripartite interaction P. flocculosa–Blumeria graminis f.sp. hordei–Hordeum vulgare.
Fig. S3 Transcriptomic profiles of secreted lytic enzyme genes in Pseudozyma flocculosa during the tripartite interaction P. flocculosa–Blumeria graminis f.sp. hordei–Hordeum vulgare.
Fig. S4 Transcriptomic profiles of transporter genes in Pseudozyma flocculosa during the tripartite interaction P. flocculosa–Blumeria graminis f.sp. hordei–Hordeum vulgare.
Fig. S5 Gene ontology (GO) enrichment of Blumeria graminis differentially expressed genes during the tripartite interaction.
Fig. S6 Transcriptomic profiles of transporter genes in Blumeria graminis during the tripartite interaction Pseudozyma flocculosa–B. graminis f.sp. hordei–Hordeum vulgare.
Fig. S7 Transcriptomic profiles of conidia‐ and hyphae‐specific CSEPs in Blumeria graminis during the tripartite interaction Pseudozyma flocculosa–B. graminis f.sp. hordei–Hordeum vulgare.
Fig. S8 Transcriptomic profiles of haustoria‐specific CSEPs in Blumeria graminis during the tripartite interaction Pseudozyma flocculosa‐B. graminis f.sp. hordei–Hordeum vulgare.
Fig. S9 Gene ontology (GO) enrichment of Hordeum vulgare differentially expressed genes during the tripartite interaction.
Fig. S10 Transcriptomic profiles of photosynthesis‐related genes in Hordeum vulgareduring the tripartite interaction Pseudozyma flocculosa–Blumeria graminis f.sp. hordei–H. vulgare.
Fig. S11 Transcriptomic profiles of the most expressed genes in Hordeum vulgare during the tripartite interaction Pseudozyma flocculosa–Blumeria graminis f.sp. hordei–H. vulgare.
Table S1 Summary of RPKM values obtained from the in vitro experiment and the tripartite bioassay
Table S2 Summary of RNAseq data of Pseudozyma flocculosa mapped reads during the tripartite bioassay
Table S3 RPKM data of orthologs of Pseudozyma flocculosa genes related to filament formation and pathogenic development
Table S4 Summary of RNAseq data of Blumeria graminis mapped reads during the tripartite bioassay
Table S5 Summary of RNAseq data of Hordeum vulgare mapped reads during the tripartite bioassay
Acknowledgements
We thank Bastien Jadot for technical assistance, and Jérôme Laroche and Brian Boyle from Institut de biologie integrative et des systèmes (IBIS) for help and support in bioinformatics. This work was supported by grants from the Natural Sciences and Engineering Research Council of Canada (NSERC), le Fonds de recherche du Québec – Nature et technologies (FRQNT) and the Canada Research Chairs Program to R.R.B.
References
- Ahmed AA, Pedersen C, Schultz‐Larsen T, Kwaaitaal M, Jørgensen HJ, Thordal‐Christensen H. 2015. The barley powdery mildew candidate secreted effector protein CSEP0105 inhibits the chaperone activity of a small heat shock protein. Plant Physiology 168: 321–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed AA, Pedersen C, Thordal‐Christensen H. 2016. The barley powdery mildew effector candidates CSEP0081 and CSEP0254 promote fungal infection success. PLoS ONE 11: e0157586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bélanger RR, Labbé C. 2002. Control of powdery mildews without chemicals: prophylactic and biological alternatives for horticultural crops In: Bélanger RR, Bushnell WR, Dik AJ, Carver TLW, eds. The powdery mildews, a comprehensive treatise. St Paul, MN, USA: APS Press, 256–267. [Google Scholar]
- Bélanger RR, Labbé C, Lefebvre F, Teichmann B. 2012. Mode of action of biocontrol agents: all that glitters is not gold. Canadian Journal of Plant Pathology 34: 469–478. [Google Scholar]
- Bindschedler LV, Burgis TA, Mills DJS, Ho JTC, Cramer R, Spanu PD. 2009. In planta proteomics and proteogenomics of the biotrophic barley fungal pathogen Blumeria graminis f. sp. hordei . Molecular & Cellular Proteomics 8: 2368–2381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bindschedler LV, McGuffin LJ, Burgis TA, Spanu PD, Cramer R. 2011. Proteogenomics and in silico structural and functional annotation of the barley powdery mildew Blumeria graminis f. sp. hordei . Methods 54: 432–441. [DOI] [PubMed] [Google Scholar]
- Chain F, Côté‐Beaulieu C, Belzile F, Menzies JG, Bélanger RR. 2009. A comprehensive transcriptomic analysis of the effect of silicon on wheat plants under control and pathogen stress conditions. Molecular Plant–Microbe Interactions Journal 22: 1323–1330. [DOI] [PubMed] [Google Scholar]
- Chandran D, Inada N, Hather G, Kleindt CK, Wildermuth MC. 2010. Laser microdissection of Arabidopsis cells at the powdery mildew infection site reveals site‐specific processes and regulators. Proceedings of the National Academy of Sciences, USA 107: 460–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhari P, Ahmed B, Joly DL, Germain H. 2014. Effector biology during biotrophic invasion of plant cells. Virulence 5: 703–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y, McNally DJ, Labbé C, Voyer N, Belzile F, Bélanger RR. 2003. Insertional mutagenesis of a fungal biocontrol agent led to discovery of a rare cellobiose lipid with antifungal activity. Applied and Environmental Microbiology 69: 2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clément‐Mathieu G, Chain F, Marchand G, Bélanger RR. 2008. Leaf and powdery mildew colonization by glycolipid‐producing Pseudozyma species. Fungal Ecology 1: 69–77. [Google Scholar]
- Dik AJ, Verhaar MA, Bélanger RR. 1998. Comparison of three biological control agents against cucumber powdery mildew (Sphaeroteca fuliginea) in semi‐commercial‐scale glasshouse trials. European Journal of Plant Pathology 104: 413–423. [Google Scholar]
- Djamei A, Schipper K, Rabe F, Ghosh A, Vincon V, Kahnt J, Osorio S, Tohge T, Fernie AR, Feussner I et al 2011. Metabolic priming by a secreted fungal effector. Nature 478: 395–398. [DOI] [PubMed] [Google Scholar]
- Doehlemann G, Reissmann S, Aßmann D, Fleckenstein M, Kahmann R. 2011. Two linked genes encoding a secreted effector and a membrane protein are essential for Ustilago maydis‐induced tumour formation. Molecular Microbiology 81: 751–766. [DOI] [PubMed] [Google Scholar]
- Donaton MC, Holsbeeks I, Lagatie O, Van Zeebroeck G, Crauwels M, Winderickx J, Thevelein JM. 2003. The Gap1 general amino acid permease acts as an amino acid sensor for activation of protein kinase A targets in the yeast Saccharomyces cerevisiae . Molecular Microbiology 50: 911–929. [DOI] [PubMed] [Google Scholar]
- Druzhinina IS, Shelest E, Kubicek CP. 2012. Novel traits of Trichoderma predicted through the analysis of its secretome. FEMS Microbiology Letters 337: 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Z, Zhou X, Ling Y, Zhang Z, Su Z. 2010. AgriGO: a GO analysis toolkit for the agricultural community. Nucleic Acids Research 38: W64–W70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang Z, Cui X. 2011. Design and validation issues in RNA‐seq experiments. Briefings in Bioinformatic 12: 280–287. [DOI] [PubMed] [Google Scholar]
- Fischbach MA, Lin H, Zhou L, Yu Y, Abergel RJ, Liu DR, Raymond KN, Wanner BL, Strong RK, Walsh CT et al 2006. The pathogen‐associated iroA gene cluster mediates bacterial evasion of lipocalin 2. Proceedings of the National Academy of Sciences, USA 103: 16502–16507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glawe DA. 2008. The powdery mildews: a review of the world's most familiar (yet poorly known) plant pathogens. Annual Review of Phytopathology 46: 27–51. [DOI] [PubMed] [Google Scholar]
- Hacquard S, Kracher B, Maekawa T, Vernaldi S, Schulze‐Lefert P, Ver Loren van Themaat E. 2013. Mosaic genome structure of the barley powdery mildew pathogen and conservation of transcriptional programs in divergent hosts. Proceedings of the National Academy of Sciences, USA 110: 2219–2228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajlaoui MR, Bélanger RR. 1993. Antagonism of the yeast‐like phylloplane fungus Sporothrix flocculosa against Erysiphe graminis var. tritici . Biocontrol Science and Technology 3: 427–434. [Google Scholar]
- Hajlaoui MR, Benhamou N, Bélanger RR. 1992. Cytochemical study of the antagonistic activity of Sporothrix flocculosa on rose powdery mildew, Sphaerotheca pannosa var. rosae . Phytopathology 82: 583–589. [Google Scholar]
- Hammami W, Labbé C, Chain F, Mimee B, Bélanger RR. 2008. Nutritional regulation and kinetics of flocculosin synthesis by Pseudozyma flocculosa . Applied Microbiology and Biotechnology 80: 307–315. [DOI] [PubMed] [Google Scholar]
- Hammami W, Qiroga Castro C, Rémus‐Borel W, Labbé C, Bélanger RR. 2011. Ecological basis of the interaction between Pseudozyma flocculosa and powdery mildew fungi. Applied and Environmental Microbiology 77: 926–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins CM, Manners JM, Scott KJ. 1985. Decrease in three messenger RNA species coding for chloroplast proteins in leaves of barley infected with Erysiphe graminis f.sp. hordei . Plant Physiology 78: 891–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss L, Pintye A, Zséli G, Jankovics T, Szentiványi O, Hafez YM, Cook RT. 2010. Microcyclic conidiogenesis in powdery mildews and its association with intracellular parasitism by Ampelomyces. European Journal of Plant Pathology 126: 445–451. [Google Scholar]
- Kulakovskaya TV, Shashkov AS, Kulakovskaya EV, Golubev WI. 2005. Ustilagic acid secretion by Pseudozyma fusiformata strains. FEMS Yeast Research 5: 919–923. [DOI] [PubMed] [Google Scholar]
- Lanver D, Berndt P, Tollot M, Naik V, Vranes M, Warmann T, Munch K, Rossel N, Kahmann R. 2014. Plant surface cues prime Ustilago maydis for biotrophic development. PLoS Pathogens 10: e1004272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefebvre F, Joly DL, Labbé C, Teichmann B, Linning R, Belzile F, Bakkeren G, Bélanger RR. 2013. The transition from a phytopathogenic smut ancestor to an anamorphic biocontrol agent deciphered by comparative whole‐genome analysis. Plant Cell 25: 1946–1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magasanik B, Kaiser CA. 2002. Nitrogen regulation in Saccharomyces cerevisiae . Gene 290: 1–18. [DOI] [PubMed] [Google Scholar]
- Marchand G, Rémus‐Borel W, Chain F, Hammami W, Belzile F, Bélanger RR. 2009. Identification of genes potentially involved in the biocontrol activity of Pseudozyma flocculosa . Phytopathology 99: 1142–1149. [DOI] [PubMed] [Google Scholar]
- Mimee B, Labbé C, Pelletier R, Bélanger RR. 2005. Antifungal activity of flocculosin, a novel glycolipid isolated from Pseudozyma flocculosa . Antimicrobial Agents and Chemotherapy 49: 1597–1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimee B, Pelletier R, Bélanger RR. 2009. In vitro antibacterial activity and antifungal mode of action of flocculosin, a membrane‐active cellobiose lipid. Journal of Applied Microbiology 107: 989–996. [DOI] [PubMed] [Google Scholar]
- Moseman JG, Scharen AL, Greely LW. 1964. Propagation of Erysiphe graminis f. sp. tritici on barley and Erysiphe graminis f. sp. hordei on wheat. Phytopathology 55: 92–96. [Google Scholar]
- Neveu B, Labbé C, Bélanger RR. 2007. GFP technology for the study of biocontrol agents in tritrophic interactions: a case study with Pseudozyma flocculosa . Journal of Microbiological Methods 68: 275–281. [DOI] [PubMed] [Google Scholar]
- Paulitz TC, Bélanger RR. 2001. Biological control in greenhouse systems. Annual Review of Phytopathology 39: 103–133. [DOI] [PubMed] [Google Scholar]
- Pedersen C, Ver Loren van Themaat E, McGuffin LJ, Abbott JC, Burgiss TA, Barton G, Spanu PD. 2012. Structure and evolution of barley powdery mildew effector candidates. BMC Genomics 13: 694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pliego C, Nowara D, Bonciani G, Gheorghe DM, Xu R, Surana P, Whigham E, Nettleton D, Bogdanove AJ, Wise RP et al 2013. Host‐induced gene silencing in barley powdery mildew reveals a class of ribonuclease‐like effectors. Molecular Plant–Microbe Interactions 26: 633–642. [DOI] [PubMed] [Google Scholar]
- Robinson MD, Smyth GK. 2008. Small‐sample estimation of negative binomial dispersion, with applications to SAGE data. Biostatistics 9: 321–332. [DOI] [PubMed] [Google Scholar]
- Romeis T, Brachmann A, Kahmann R, Kämper J. 2000. Identification of a target gene for the bE–bW homeodomain protein complex in Ustilago maydis . Molecular Microbiology 37: 54–66. [DOI] [PubMed] [Google Scholar]
- Scholes JD, Lee PJ, Horton P, Lewis DH. 1994. Invertase understanding changes in the photosynthetic and carbohydrate‐metabolism of barley leaves infected with powdery mildew. New Phytologist 126: 213–222. [Google Scholar]
- Schuler D, Wahl R, Wippel K, Vranes M, Munsterkotter M, Sauer N, Kamper J. 2015. Hxt1, a monosaccharide transporter and sensor required for virulence of the maize pathogen Ustilago maydis . New Phytologist 206: 1086–1100. [DOI] [PubMed] [Google Scholar]
- Sonah H, Deshmukh RK, Bélanger RR. 2016. Computational prediction of effector proteins in fungi: opportunities and challenges. Frontiers in Plant Science 7: 126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spanu PD, Abbott JC, Amselem J, Burgis TA, Soanes DM, Stüber K, Ver Loren van Themaat E, Brown JK, Butcher SA, Gurr SJ et al 2010. Genome expansion and gene loss in powdery mildew fungi reveal tradeoffs in extreme parasitism. Science 330: 1543–1546. [DOI] [PubMed] [Google Scholar]
- Teichmann B, Labbé C, Lefebvre F, Bölker M, Linne U, Bélanger RR. 2011a. Identification of a biosynthesis gene cluster for flocculosin a cellobiose lipid produced by the biocontrol agent Pseudozyma flocculosa . Molecular Microbiology 79: 1483–1495. [DOI] [PubMed] [Google Scholar]
- Teichmann B, Lefebvre F, Labbé C, Bölker M, Linne U, Bélanger RR. 2011b. Beta hydroxylation of glycolipids from Ustilago maydis and Pseudozyma flocculosa by an NADPH‐dependent β‐hydroxylase. Applied and Environmental Microbiology 77: 7823–7829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teichmann B, Linne U, Hewald S, Marahiel MA, Bölker M. 2007. A biosynthetic gene cluster for a secreted cellobiose lipid with antifungal activity from Ustilago maydis . Molecular Microbiology 66: 525–533. [DOI] [PubMed] [Google Scholar]
- Traquair JA, Shaw LA, Jarvis WR. 1988. New species of Stephanoascus with Sporothrix anamorphs. Canadian Journal of Botany 66: 926–933. [Google Scholar]
- Voegele RT, Struck C, Hahn M, Mendgen K. 2001. The role of haustoria in sugar supply during infection of broad bean by the rust fungus Uromyces fabae . Proceedings of the National Academy of Sciences, USA 98: 8133–8138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whigham E, Qi S, Mistry D, Surana P, Xu R, Fuerst GS, Pliego C, Bindschedler LV, Spanu P, Dickerson JA et al 2015. Broadly conserved fungal effector BEC1019 suppresses host cell death and enhances pathogen virulence in powdery mildew of barley (Hordeum vulgare L.). Molecular Plant‐Microbe Interactions 28: 968–983. [DOI] [PubMed] [Google Scholar]
- Whipps JM. 2001. Microbial interactions and biocontrol in the rhizosphere. Journal of Experimental Botany 52: 487–551. [DOI] [PubMed] [Google Scholar]
- Yan L, Chen Y, Yang Q, Ma Z. 2012. Heterologous expression of the CYP51 gene of the obligate fungus Blumeria graminis in the necrotrophic fungus Botrytis cinerea . Journal of Eukaryotic Microbiology 59: 88–92. [DOI] [PubMed] [Google Scholar]
- Zeilinger S, Galhaup C, Payer K, Woo SL, Mach RL, Fekete C, Lorito M, Kubicek CP. 1999. Chitinase gene expression during mycoparasitic interaction of Trichoderma harzianum with its host. Fungal Genetics and Biology 26: 131–140. [DOI] [PubMed] [Google Scholar]
- Zhang J, Bayram Akcapinar G, Atanasova L, Rahimi MJ, Przylucka A, Yang D, Kubicek CP, Zhang R, Shen Q, Druzhinina IS. 2016. The neutral metallopeptidase NMP1 of Trichoderma guizhouense is required for mycotrophy and self‐defence. Environmental Microbiology 18: 580–597. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Please note: Wiley Blackwell are not responsible for the content or functionality of any Supporting Information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
Fig. S1 Transcriptomic profiles of genes associated with flocculosin biosynthesis in Pseudozyma flocculosa during the tripartite interaction P. flocculosa–Blumeria graminis f.sp. hordei–Hordeum vulgare.
Fig. S2 Transcriptomic profiles of the most expressed CSEPs genes specific to Pseudozyma flocculosa during the tripartite interaction P. flocculosa–Blumeria graminis f.sp. hordei–Hordeum vulgare.
Fig. S3 Transcriptomic profiles of secreted lytic enzyme genes in Pseudozyma flocculosa during the tripartite interaction P. flocculosa–Blumeria graminis f.sp. hordei–Hordeum vulgare.
Fig. S4 Transcriptomic profiles of transporter genes in Pseudozyma flocculosa during the tripartite interaction P. flocculosa–Blumeria graminis f.sp. hordei–Hordeum vulgare.
Fig. S5 Gene ontology (GO) enrichment of Blumeria graminis differentially expressed genes during the tripartite interaction.
Fig. S6 Transcriptomic profiles of transporter genes in Blumeria graminis during the tripartite interaction Pseudozyma flocculosa–B. graminis f.sp. hordei–Hordeum vulgare.
Fig. S7 Transcriptomic profiles of conidia‐ and hyphae‐specific CSEPs in Blumeria graminis during the tripartite interaction Pseudozyma flocculosa–B. graminis f.sp. hordei–Hordeum vulgare.
Fig. S8 Transcriptomic profiles of haustoria‐specific CSEPs in Blumeria graminis during the tripartite interaction Pseudozyma flocculosa‐B. graminis f.sp. hordei–Hordeum vulgare.
Fig. S9 Gene ontology (GO) enrichment of Hordeum vulgare differentially expressed genes during the tripartite interaction.
Fig. S10 Transcriptomic profiles of photosynthesis‐related genes in Hordeum vulgareduring the tripartite interaction Pseudozyma flocculosa–Blumeria graminis f.sp. hordei–H. vulgare.
Fig. S11 Transcriptomic profiles of the most expressed genes in Hordeum vulgare during the tripartite interaction Pseudozyma flocculosa–Blumeria graminis f.sp. hordei–H. vulgare.
Table S1 Summary of RPKM values obtained from the in vitro experiment and the tripartite bioassay
Table S2 Summary of RNAseq data of Pseudozyma flocculosa mapped reads during the tripartite bioassay
Table S3 RPKM data of orthologs of Pseudozyma flocculosa genes related to filament formation and pathogenic development
Table S4 Summary of RNAseq data of Blumeria graminis mapped reads during the tripartite bioassay
Table S5 Summary of RNAseq data of Hordeum vulgare mapped reads during the tripartite bioassay


