Summary
Local adaptation in tree species has been documented through a long history of common garden experiments where functional traits (height, bud phenology) are used as proxies for fitness. However, the ability to identify genes or genomic regions related to adaptation to climate requires the evaluation of traits that precisely reflect how and when climate exerts selective constraints.
We combine dendroecology with association genetics to establish a link between genotypes, phenotypes and interannual climatic fluctuations. We illustrate this approach by examining individual tree responses embedded in the annual rings of 233 Pinus strobus trees growing in a common garden experiment representing 38 populations from the majority of its range.
We found that interannual variability in growth was affected by low temperatures during spring and autumn, and by summer heat and drought. Among‐population variation in climatic sensitivity was significantly correlated with the mean annual temperature of the provenance, suggesting local adaptation. Genotype–phenotype associations using these new tree‐ring phenotypes validated nine candidate genes identified in a previous genetic–environment association study.
Combining dendroecology with association genetics allowed us to assess tree vulnerability to past climate at fine temporal scales and provides avenues for future genomic studies on functional adaptation in forest trees.
Keywords: adaptive capacity, climate change, common garden, dendroecology, local adaptation, needleleaf, temperate forests, tree rings
Short abstract
See also the Commentary on this article by https://doi.org/10.1111/nph.15094.
Introduction
In boreal and temperate ecosystems, a substantial increase in temperature and in the frequency of extreme events (e.g. severe droughts or heat waves) is projected for the upcoming century (IPCC, 2013). Poleward climatic niche shifts are expected (e.g. McKenney et al., 2014), therefore raising questions about the ability of trees to cope with those rapid changes (Price et al., 2013). Migration and long‐term genetic adaptation are expected to be too slow to keep pace with climatic niche shifts (Jump & Peñuelas, 2005; Aitken et al., 2008; Jump et al., 2009; Jezkova & Wiens, 2016). Nevertheless, populations may persist in their current location depending on species sensitivity and adaptive capacity to future environmental changes (Aubin et al., 2016). To better predict the fate of tree species, knowledge is needed about the climatic constraints affecting their growth/development and the key genetic and physiological mechanisms involved in the response to those constraints (Aubin et al., 2016; Urban et al., 2016). These knowledge gaps challenge the forecast of species’ responses to shifting climatic conditions and the implementation of forest management strategies such as assisted gene flow from pre‐adapted populations (Aitken & Whitlock, 2013). Without a genuine tree‐centered approach based on time‐series data to evaluate sensitivity to fluctuating environmental conditions, elucidating the genetic architecture of adaptation to climate throughout a tree's lifespan will remain difficult (Alberto et al., 2013).
Local adaptation in tree species has been documented through a long history of common garden experiments composed of georeferenced genetic material (provenances), where dendrometric traits such as diameter or height growth were used as proxies for fitness (Langlet, 1971; Alberto et al., 2013). The rapidity of climate change advocates the use of those existing common garden experiments (which have already been exposed to warming during the last few decades) to investigate responses to climate change (e.g. Rehfeldt et al., 1999; Sork et al., 2013). In combination with genomic approaches, common gardens represent a unique opportunity to decipher the genomic architecture of local adaptation, and to identify putative genes or genomic regions involved in tree species’ responses to climate (Lepais & Bacles, 2014; de Villemereuil et al., 2016).
Two genomic strategies are commonly used to search for genes involved in local adaptation to climate (Sork et al., 2013). The first is the genotype–environment association (GEA) method that looks for correlations between genetic markers and environmental variables from where populations/individuals originate (Joost et al., 2007; Coop et al., 2010). However, GEA does not provide information about the phenotypic traits controlled by the candidate genes. Another strategy is to test for associations between genotypes growing in a common environment and traits of interest (genotype–phenotype association; GPA). GPA has the advantage of establishing more direct links between genes and traits potentially under selection (e.g. Eckert et al., 2015). However, in tree species, the list of functional traits that closely reflect adaptation to climate is still rather limited and labor‐intensive to evaluate (for a review, see Aitken & Bemmels, 2016). In particular, assessment of dendrometric traits usually consists of punctual measurements (e.g. height or diameter at a certain age) that are probably the cumulative result of many different climatic events and constraints that occurred throughout the tree's lifespan. The use of innovative tools and methods to measure traits that reflect how and when climate exerts selective constraints (Rahaman et al., 2015; Urban et al., 2016) is thus needed to improve our understanding of species sensitivity and adaptive capacity to climate.
Through the investigation of interannual tree‐ring variations, dendroecology allows the quantification of climatic constraints that are exerted on trees. For decades, dendroecologists have developed methods to link climate with variations in wood anatomy characteristics (Fritts, 1976; Cook & Kairiūkštis, 1990; Schweingruber, 1996; Gessler et al., 2014; Housset et al., 2015; Girardin et al., 2016; Hartmann & Trumbore, 2016). Indeed, cambial activity is an integrative indicator that accounts for physiological mechanisms such as responses to drought stress, phenology of dormancy and resistance to freezing injuries. Those wood characteristics can be measured using nondestructive techniques to build a retrospective time‐series of growth‐related traits throughout a tree's lifespan. The influence of abiotic stress can be estimated by quantifying annual growth responses to punctual extreme climatic events (e.g. Montwé et al., 2016). Several recent reports have demonstrated the intraspecific genetic variation in growth response to punctual extreme climatic events (Montwé et al., 2016) or in cumulative growth responses to the average climate (over many years) in common garden experiments (McLane et al., 2011; Taeger et al., 2013; Leland et al., 2016). Most importantly, analysis of the relationships between growth time‐series and monthly climatic variables makes it possible to assess sensitivity to climatic constraints on a year‐to‐year basis. Therefore, it enables the identification of growth‐limiting factors and the time of the year (month or season) when trees are the most affected (Fritts, 1976; Girardin et al., 2012). Such tree‐ring traits reflecting sensitivity to climatic constraints have never been used as phenotypic traits in the context of quantitative genetics.
Here, we develop a new class of phenotypes that reflect responses to interannual climatic variability across trees’ lifespan. We then combine these new climate‐related phenotypes with association genetics to establish a link between the genotypes, phenotypes and climatic constraints affecting trees (Fig. 1). The present study demonstrates how tree‐ring traits derived from dendroecology can be used to identify and quantify climatic constraints influencing tree growth; study local adaptation in growth responses to such constraints; and identify and validate the genes putatively involved in such adaptation processes. We illustrate our approach using a Pinus strobus (eastern white pine, hereafter P. strobus) common garden experiment located in Quebec that includes provenances from a wide range of environments across the species’ distribution area in eastern North America. This species is representative of many temperate and boreal tree species for which the magnitude and speed of climate change are expected to induce widespread range shifts by 2060 (Joyce & Rehfeldt, 2013; Pedlar & McKenney, 2017). Local adaptation in P. strobus was made evident in provenance trial studies, which detected significant among‐population variation in classic quantitative traits (e.g. height, bud phenology: see Li et al., 1997; Joyce & Rehfeldt, 2013). More recently, GEA studies found several single nucleotide polymorphisms (SNPs) associated with climate (Nadeau et al., 2016; Rajora et al., 2016).
Figure 1.
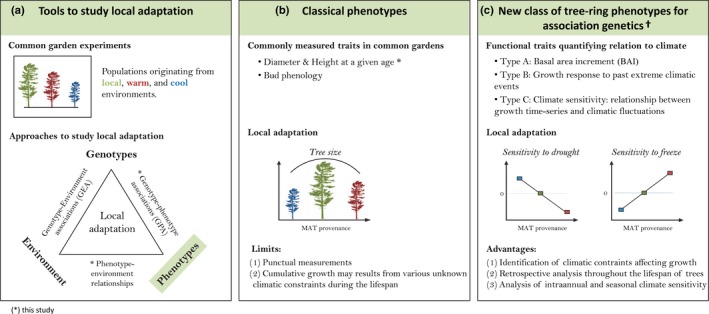
Tree‐ring trait assets in establishing the links between genotypes, phenotypes and environmental selection pressures acting on trees. (a) Tools to study local adaptation include common garden experiments and association genetics (triangle: adapted from Sork et al., 2013). (b) Commonly used phenotypes in dendroecology and forest genetics are based on dendrometric traits. (c) The use of tree‐ring phenotypes in association genetic studies allows fine temporal analysis of growth responses to past climatic events in addition to the identification of new candidate genes. †, Traits examined in the present study. Types A, B and C: please refer to the Materials and Methods section.
As a first step, a tree‐ring analysis was conducted to provide three different classes of phenotypic traits (Fig. 1). Traits included average annual ring width and density (hereafter type A traits), relative growth variation for a given year following an extreme climatic event (type B traits) and climatic sensitivity (CS) traits, that is, the strength of relationships between annual growth and climatic variation such as temperature and drought (type C traits). In a second step, those traits were correlated with mean annual temperature of the provenance (MATP) to search for local adaptation in growth responses to climate (e.g. genetic–environment clines). Third, GPA analyses were performed on those tree‐ring traits to identify putative loci involved in growth responses to climatic constraints and to further cross‐validate candidate genes that had been identified in a previous GEA study (Nadeau et al., 2016).
Materials and Methods
Biological material and experimental design
Throughout this paper, we will use the term ‘provenance’ when referring to the geographic and climatic origin of a population, and the term ‘population’ when referring to the trees grown from seed sampled at the provenance (one population per provenance; McLane et al., 2011). A list of definitions for abbreviations used throughout this paper is given in Table 1. We studied P. strobus L. trees from 38 provenances planted in a common garden experiment. Provenances were selected to cover the majority of the species’ range from Canada and the US (Fig. 2; Supporting Information Table S1) to maximize the representativity of the intraspecific genetic variability. Nadeau et al. (2015) identified two main genetic groups (southern and northern) probably resulting from separate glacial refugia. Thirty‐six provenances were part of the northern genetic group and two provenances were from the southern group (provenances 12 and 13). Seeds were collected between 1976 and 1986 in natural stands, stored at −18°C, germinated in a glasshouse and cultivated for 3 yr in a tree nursery before their establishment in the common garden experiment at the Valcartier arboretum (Quebec, Canada; 46.94886°N, 71.4962°W) in 1989. The experiment was designed as two randomized complete blocks, with up to n = 16 trees per seedlot and blocks arranged in four‐tree row plots. Spacing between trees was 2.0 × 1.5 m (thinned to 2.0 × 3.0 m after the 2002 growing season) in block 1, and 2.0 × 2.0 m in block 2. On average, three trees (SD = 0.96) per population were sampled in each block. A total of 233 trees were sampled, representing two to ten trees per population (mean = 6.13, SD = 1.51), depending on the population survival rate (Table S1).
Table 1.
Abbreviations used in this study
| Abbreviation | Definition |
|---|---|
| BAIi–j | Basal area increment from year i to j (mm2 yr−1) |
| avDensi–j | Average density from year i to j (kg m−³) |
| D21 | Diameter of 21‐yr‐old trees (m) |
| H21 | Height of 21‐yr‐old trees (m) |
| GEA | Genotype–environment association |
| GPA | Genotype–phenotype association |
| DAPC | Discriminant analysis of principal components |
| CSX – Y.Z (t) | Climatic sensitivity traits. Defined as the statistical associations between a growth metric X (residuals of BAI or avDens) and a climate variable Y (Temp for temperature; Prec for precipitation; Drought for drought code; Freeze for number of freezing days) in month Z (June of the year preceding growth (labelled t–1) to September of the year contemporaneous to growth (t)) |
| MAPp | Mean annual precipitation of provenance (mm) |
| MATp | Mean annual temperature of provenance (°C) |
| SNP | Single nucleotide polymorphism |
Figure 2.
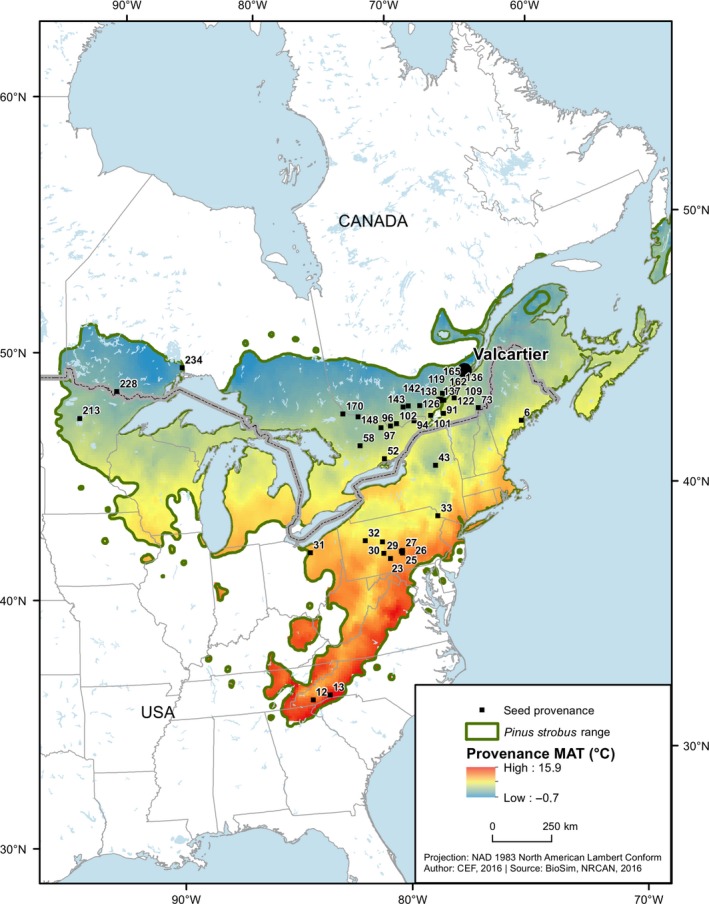
Location of the Valcartier common garden experiment and of provenances of sampled Pinus strobus. Mean annual temperature of each provenance (MAT p) averaged over 1993–2014 is superimposed on the range‐wide distribution of P. strobus.
Meteorological data
Monthly means of daily temperature (T), mean monthly precipitation (P) and monthly means of daily drought code (DC) time‐series at the Valcartier experimental site were obtained for the 1993–2014 period using the BioSIM v.10 software (Régnière & Bolstad, 1994). Daily temperature and precipitation data were interpolated from the eight closest weather stations, adjusted for elevation and location differentials with regional gradients, and averaged using inverse distance weighting (1/d 2, where d is distance). Monthly values were computed from the average of the daily quantities. DC is an index for the amount of moisture stored in deep organic layers and soils and takes into account snow arrival in autumn and thawing in spring based on modeled snow accumulation (Terrier et al., 2013). The drought season is defined as the snow‐free period, which ranges from c. May to October. DC is unitless and ranges from 0 to c. 800, with low values indicating low drought risk and high values indicating high drought risk. Collinearity amongst these different monthly climate variables (temperature, precipitation and drought) is generally low according to pairwise Spearman rank correlation analysis (Table S2). Mean annual temperature and precipitation of each provenance (MATp and MAPp, respectively) were also computed and averaged over the 1961–1990 period (i.e. the period corresponding to seed collection) using BioSIM v.10.
Tree‐ring data
At the end of October 2014, a 5‐mm‐diameter core was taken (from bark to pith) from each tree at 1.3 m above ground using a Pressler increment borer. Cores were kept frozen until being air‐dried, with no risk of developing mold. They were later sawn to a 1.7‐mm thickness longitudinally using a twin blade sleeve to obtain a smooth, thin surface. The same sample was used for dendrochronological and densitometric analyses. Annual rings of each core were visually cross‐dated with skeleton‐plots and pointer‐year identification under 4.5× magnification. Samples were then conditioned at a temperature of 20°C and relative humidity of 40% to maintain an equilibrium moisture content of 8%. For dendrochronological analyses, ring‐widths were measured from high‐resolution scanned images of the cores using the software Coo‐Recorder (Cybis, Saltsjöbaden, Sweden) at a precision of 10−2 mm. Cross‐dating was validated using the software CDendro (Cybis) and cofecha (Holmes, 1983). The ring‐width measurement series were converted into basal area increment (BAI) chronologies using the R library ‘dplR’ 1.6.3 (Bunn, 2008). Average annual BAI for the 1993–2014 period was calculated for each population from the raw data to obtain absolute levels of growth.
Two BAI metrics were used in downstream analyses: the raw – absolute BAI and standardized residual BAI scores. Eliminating the intrinsic age‐ and size‐related growth trends in BAI makes it possible to address annual growth variability independently of biological age or tree size. Here, raw BAI chronologies, which were not normally distributed, were transformed using a logarithmic scaling and detrended using modified generalized exponential functions (‘Hugershoff’ type; Warren, 1980) with the software arstan v.41d XP (Cook & Holmes, 1986). Additionally, an autoregressive model was applied to remove temporal autocorrelation in the time‐series, and residuals were extracted. Annual averages of these standardized residual BAI scores were computed for each population using a robust bi‐weight mean, which removes outliers attributable to disturbance or endogenous site factors (R library ‘dplR’ 1.6.3; Bunn, 2008). The mean interseries correlation (rbar) computed from all of the standardized residual tree‐level BAI time‐series (Wigley et al., 1984) was rbar = 0.80 (SD ± 0.17). The expressed population signal obtained for the residual tree‐level series was 0.99, which was higher than the commonly used threshold value (0.85) for growth–climate correlation analysis (Wigley et al., 1984).
The density of each thin core slice was measured using a Quintek X‐ray measuring system at Université Laval (Quebec, Canada). Cores were scanned in 0.04‐mm steps, producing high‐resolution density profiles. Average annual density was determined for each tree ring and the correct identification of annual ring borders was verified using the previously cross‐dated ring‐width measurements. Mean ring density was calculated for each population by averaging the annual ring density of all trees in a population for the 1993–2014 period. Then, for each core, average ring‐density time‐series were detrended using modified generalized exponential functions and temporal autocorrelation was removed as described above. Finally, annual averages of these standardized residual density scores were computed for each population as described above.
Three classes of tree‐ring traits derived using a dendroecological approach
Traits were classified according to the following nomenclature (A, B and C; see description in Fig. 1c): (A) mean xylem BAI (meanBAI1993–2014) and average density (avDens1993–2014) over the 1993–2014 period, corresponding to classic measures used in forestry; (B) growth variation for a specific year following an extreme climatic event; and (C) climatic sensitivity (CS) traits. Type A traits are long‐term averages that probably result from many climatic events or constraints on growth throughout a tree's lifespan, whereas type B and type C traits reflect growth responses to climate on a year‐to‐year basis. To illustrate type B traits, we analyzed the standardized residual BAI scores for the year 2003 (BAI2003) because we observed large among‐population variation during that year (Fig. 3), which could be related to lower temperatures observed in autumn 2002 and spring 2003 (Figs S1, S2). The standardized residual BAI2003 scores indicate the relative growth variation of a population in comparison to its average growth level. Therein, a value of BAI2003 < 1 indicates a lower growth in 2003 as compared with the population's average growth level for the 1993–2014 period or, conversely, a BAI2003 > 1 indicates a higher growth than average.
Figure 3.
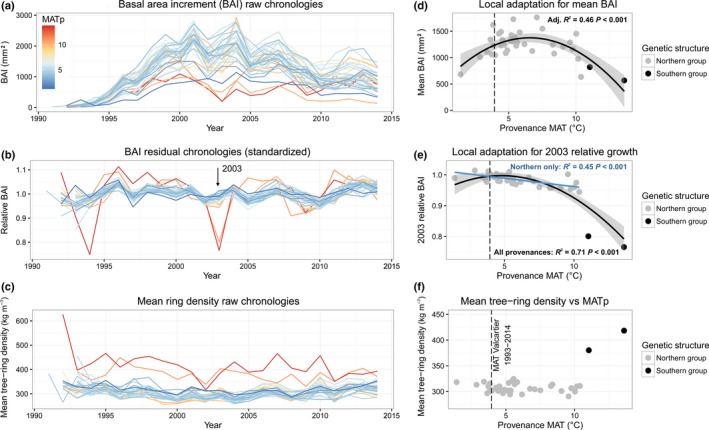
Tree‐ring phenotypes. (a) Annual raw–absolute basal area increment (BAI), (b) annual standardized residual BAI scores and (c) annual raw‐absolute tree‐ring density averaged by populations. The color gradient represents the mean annual temperature of the provenance (MAT p, °C) (red, warmer; blue, colder). The following relationships between MAT p and tree‐ring phenotypes of the populations are illustrated: (d) BAI averaged over 1993–2014, (e) relative growth variation in 2003 (BAI 2003) and (f) tree‐ring density averaged over 1993–2014. The genetic structure groups described by Nadeau et al. (2015) are represented by gray (northern group) and black (southern group) points. Significant relationships among all provenances suggesting local adaptation are represented by black lines. Significant relationships for the subset of provenances from the northern genetic structure group are represented by a blue line. The relationships were chosen according to a model selection procedure (Akaike information criterion, see the Materials and Methods section) between a linear model and a second‐order polynomial regression model. Only significant models are shown.
CS (type C) traits were determined as follows. For each population, correlation analyses were computed between the annual standardized residual BAI scores, averaged at the population level, and the matrix of monthly climatic data (from 1993 to 2014) interpolated at the location of the common garden (Biondi & Waikul, 2004). Long‐term trends in climatic series were linearly detrended to obtain unbiased data of interannual climatic variations. Correlation analyses were performed for the period spanning from June of the year preceding ring formation to September of the year concurrent with ring formation to account for lagged effects from year to year (e.g. previous autumn freezing injury). The coefficients of correlation between growth metrics and climatic variables hereafter constitute the CS traits. Significance in correlation analyses was assessed using bootstrapped randomization (n = 1000 simulations): when the 95% confidence interval included zero, the null hypothesis of ‘no relationship’ could not be rejected. CS traits were also calculated at the individual‐tree level to be used as phenotypes in GPAs (see below). As is routinely done in dendroecology, a response function analysis was also used as a complement to quantify the influence of climate on tree growth (Cook & Kairiūkštis, 1990). This analysis is a multiple regression technique that uses the principal components of monthly climatic data to estimate growth–climate associations. This complementary analysis, made in support of the correlation analysis, controls for collinearity of monthly climate predictors; results are presented in the Supporting Information section and are not discussed herein (Figs S3, S4). All analyses were conducted using the R library ‘treeclim’ 1.0.11 (Zang & Biondi, 2015). CS traits of the average density metric were also determined by repeating all of the above analyses on the annual standardized residual density scores, averaged at the population level.
Local adaptation in growth responses to climate
We screened tree‐ring traits (type A, B and C) exhibiting among‐population variation that could be correlated with the climate of the provenance, an indication of local adaptation in growth responses to climate (Sork et al., 2013). We used MATp as an indicator because it was highly correlated with the number of growing degree‐days above 5°C (r = 0.97), which was found to be the best predictor of genetic variation in growth potential among P. strobus populations (Joyce & Rehfeldt, 2013). MATp is correlated with MAPp in our study system (Spearman r = 0.42, with P = 0.030 and df = 24, taking into account spatial autocorrelation; Dutilleul, 1993), and hence it also partially reflects a moisture availability gradient. First, Mantel tests were conducted to determine if similarities between population‐averaged time‐series of standardized residual BAI scores, as measured using pairwise correlations between time‐series, and geophysical characteristics of the provenances, were related (‘mantel.rtest’ function in the ade4 R package; Dray & Dufour, 2007). Next, for type A and B traits, the relationship with provenance MATp was tested using both linear and second‐degree polynomial regressions. We tested for the effect of block sample size by including a covariate in these models, which was defined as the sample size ratio of block 1 vs block 2; the effect of the covariate was not significant. Model selection was based on the Akaike information criterion (Burnham & Anderson, 2010). The null hypothesis (H 0) was ‘no relationship with MATp’. Regarding the effect of MATp on CS traits (type C), a linear regression model provided the best fit. To reduce the risk of generating spurious results, we only tested the CS traits for which growth–climate correlations were significant in at least five populations. This threshold was arbitrarily set to allow sufficient interpopulation variance in the CS traits for the detection of genetic associations.
Genetic data acquisition
For genotyping, DNA from each of the 233 sampled trees was extracted from foliage using the protocol described by Nadeau et al. (2015). Leaf sampling was conducted independently from the sampling of Nadeau et al. (2015, 2016): only 21 out of 133 populations sampled in Nadeau et al. (2015, 2016) are common to both datasets (16%), and samples within populations are largely different (c. 4% in common). The 233 trees were genotyped on the same set of 153 SNP markers as in Nadeau et al. (2015) previously developed in a Sequenom iPlex Gold MassARRAY (Agena Bioscience, San Diego, CA, USA). This array included SNPs from 52 candidate genes for growth and phenology previously identified in Picea glauca and 68 noncandidate genes randomly distributed across the genome (for more details see Nadeau et al., 2015, 2016). Using this array and an independent set of populations distributed across the range of P. strobus, Nadeau et al. (2016) identified 26 candidate SNPs for local adaptation using GEAs and F ST outlier tests. In this study, eight trees and six SNPs failed during the genotyping step and were discarded from subsequent analyses. SNPs with a minor allele frequency < 0.03 were also discarded. This resulted in 225 trees and 128 SNPs successfully genotyped (SNP genotyping call rate: min. = 90.7%, mean = 98.9%), including 21 of the candidate SNPs identified by Nadeau et al. (2016). All of the 21 candidate SNPs were detected by at least one GEA method, and two SNPs were F ST outliers (Nadeau et al., 2016; Table S3). No trees were discarded based on the genotyping call rate (min. = 78.9%; mean = 98.9%). GenBank accession numbers of the genes containing the 128 SNPs are given in Table S3.
Genotype–phenotype associations
We searched for genes involved in growth‐sensitivity to climate by performing GPA analyses on tree‐ring traits. GPAs were tested at the individual‐tree level for the 128 SNPs using tassel v.5.0 software (Yu et al., 2006; Zhang et al., 2010). We only tested the tree‐ring traits that showed a highly significant correlation with MATp at the population level (P < 0.01, see Results). tassel tests for associations between genotypes and phenotypes for each SNP–trait combination. The full linear mixed model used can be expressed as follows:
where y is the vector of observations for a tree‐ring trait (i.e. the phenotypes), β is the vector of fixed block effects, α is the vector of fixed SNP (genotypes) effects, v is the vector of fixed population structure effects (described below), μ is a vector of additive or ‘background’ polygenic effects, which takes into account relatedness among individuals, and is the vector of random residuals. X, S, Q and Z are incidence matrices relating y to β, α, v and μ, respectively. Random additive effects and errors are assumed to be normally distributed with μ ~ N(0, 2K ) and ϵ ~ N(0,I ), where is the additive genetic variance, is the residual variance, K is the kinship matrix and I is the identity matrix. The kinship matrix K was computed from the 128 SNPs using the method of Ritland (1996) implemented in the program SPAGeDi v1.5a (Hardy & Vekemans, 2002).
Population structure can create spurious associations between genotypes and phenotypes (false positives). Nevertheless, when population structure covaries with adaptive genetic variation as in P. strobus (Nadeau et al., 2016), over‐adjustment for population structure may decrease power and result in false negatives (Sork et al., 2013). Therefore, three models were tested with tassel: no correction (i.e. omitting the population structure and kinship effects); correction for population structure (Q model, i.e. omitting the kinship effects); and correction for population structure and for kinship (Q + K model, i.e. full model). To obtain the population membership matrix (Q), we performed a discriminant analysis of principal components (DAPC; Jombart et al., 2010) using all 225 genotyped trees from this study and 821 additional trees (133 provenances) sampled across the entire range and genotyped on the same set of 128 SNPs taken from Nadeau et al. (2015). DAPC maximizes among‐population genetic variation while minimizing within‐group genetic variation. Therefore, this method is well suited for GPA analyses because population structure is removed by including the group memberships identified by DAPC as covariates in the tassel model (Jombart et al., 2010). A complete description of the DAPC method can be found in Fig. S5. We used the membership probabilities for k = 3 clusters (k − 1 = 2 covariates) as the Q matrix to correct for population structure in the tassel analyses (Fig. S5b). The three clusters detected were similar to those in Nadeau et al. (2015): a southern and a northern group with weak west to east differentiation (Fig. S5c). To summarize results from the three models (no correction, Q and Q + K), each SNP–phenotype association was classified as follows: ‘very likely’ when significant (P < 0.05) for all three models; ‘likely’ when significant for two out of the three models; ‘uncertain’ when significant for only one model; and ‘no evidence’ when not significant in the three models (Girardin et al., 2016). While summarizing results across models may help controlling for errors, the likelihood of false discoveries is significantly raised under multiple testing. Because we tested a small number of SNPs (128), we preferred to reduce the risk of false negatives (i.e. not detecting true associations) by using P‐values that were not corrected for multiple testing. Instead, the cross‐validation of the SNPs detected by tassel with the 21 candidate SNPs found in Nadeau et al. (2016), which used independent samples and different methods, should reduce false positives and make a stronger case for those candidate genes. GPAs excluding the two southern group populations (n = 221) gave similar results (Table S4).
To look for functional categories, genes containing SNPs involved in significant GPAs were annotated using blastx against the RefSeq database (https://www.ncbi.nlm.nih.gov/refseq) using a threshold E‐value of 1 e−10. Given that two P. strobus sequences had no significant hit on the RefSeq database, blast analyses were performed against the P. glauca gene catalog (tblastx, E‐value of 10−10; Gcat 3.3; Rigault et al., 2011) and the P. strobus transcriptome (blastn, E‐value of 10−20; Ingo Ensminger (University of Toronto), personal communication). Complementary information regarding their putative function was then deduced from P. glauca and P. strobus best‐ortholog TAIR annotations (TAIR, Arabidopsis thaliana database, https://www.arabidopsis.org/index.jsp). SNP annotations (synonymous, nonsynonymous) were deduced as previously described by Nadeau et al. (2016).
Results
High variability was observed for annual averages of raw‐absolute BAI among P. strobus populations (Fig. 3a). Globally, time‐series of population‐averaged standardized residual BAI scores were well intercorrelated; the mean of the pairwise correlations was 0.66. Yet, the values of these pairwise correlations tended to be higher when computed between provenances that had smaller geographic distances (Mantel test observed correlation r = −0.30, P = 0.019) and smaller MATp differences (r = −0.39, P = 0.003).
CS traits
Analysis of interannual variability of population‐averaged standardized residual BAI and density scores in relation to climatic fluctuations enables the identification of climatic constraints acting upon ring formation. The coefficients of correlation between growth metrics and the climatic variables hereafter constitute the CS traits (type C; Figs 4, 5; see Figs S3, S4 for response function analyses). The positive influence of a lengthening of the growing season on BAI was made evident by the positive correlations between population averages of standardized residual BAI scores (1993–2010) and October temperatures during the autumn of the year preceding ring formation (CSBAI–Temp.Oct(t−1)), and with May temperature of the year concurrent with ring formation (CSBAI–Temp.May(t)) (Fig. 4a). Furthermore, standardized residual BAI scores were negatively correlated with the numbers of freezing days occurring in October of the year preceding growth (CSBAI–Freeze.Oct(t−1)) and in June of the year of ring formation (CSBAI–Freeze.Jun(t)) (Fig. S6a). In addition, the effect of heat stress on BAI was revealed by the negative correlations between standardized residual BAI scores and July temperature of the current growth year (CSBAI–Temp.Jul(t); Fig. 4a). Negative correlations with July and August DCs (CSBAI–Drought.Jul(t) and CSBAI‐Drought.Aug(t)) were also observed, highlighting the influence of summer drought stress on BAI (Fig. 5a). In parallel, standardized residual BAI scores were positively correlated with current‐year July precipitation (CSBAI–Prec.Jul(t); Fig. S7a). Individual‐tree level CS traits were consistent with the CS traits computed at the population level (visual inspection, Fig. S8). As for ring density, a negative effect of summer drought during the year preceding growth was detected (Fig. S9). Tree‐ring density was positively influenced by high temperatures during the growing season (May, June and July; Fig. S10).
Figure 4.
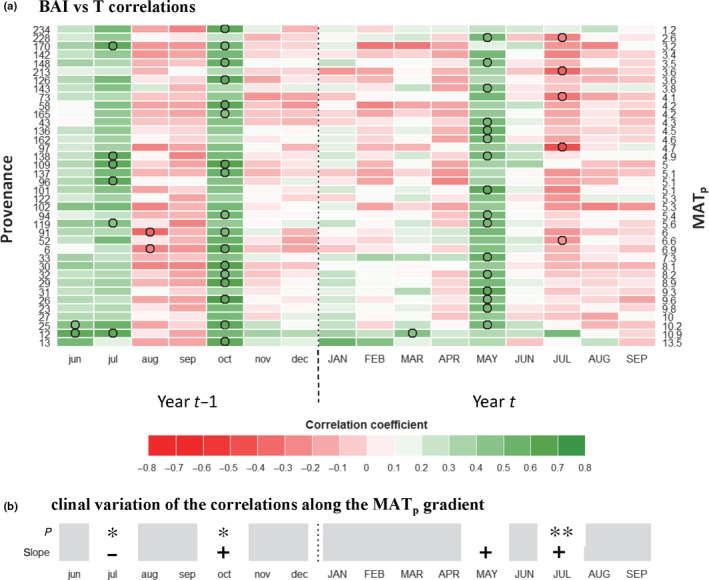
Climatic sensitivity (CS) traits: basal area increment (BAI) and monthly temperature (T). (a) Bootstrapped correlation coefficients were computed between Pinus strobus standardized residuals of BAI scores (from Fig. 3b) and monthly mean temperature at the Valcartier common garden experiment (Quebec, Canada) over the 1993–2014 period. Populations (rows) are ordered by increasing mean annual temperature of the provenance (MAT p; right axis labels). Months in capital letters represent the current year of ring formation; months in lower case represent climate variables during the year preceding ring formation. Significant coefficients are represented by circles. (b) Test for clinal variations in correlation coefficients along the MAT p gradient: P‐value and slope sign of a regression against MAT p. Significance: *, P < 0.05; **, P < 0.01.
Figure 5.
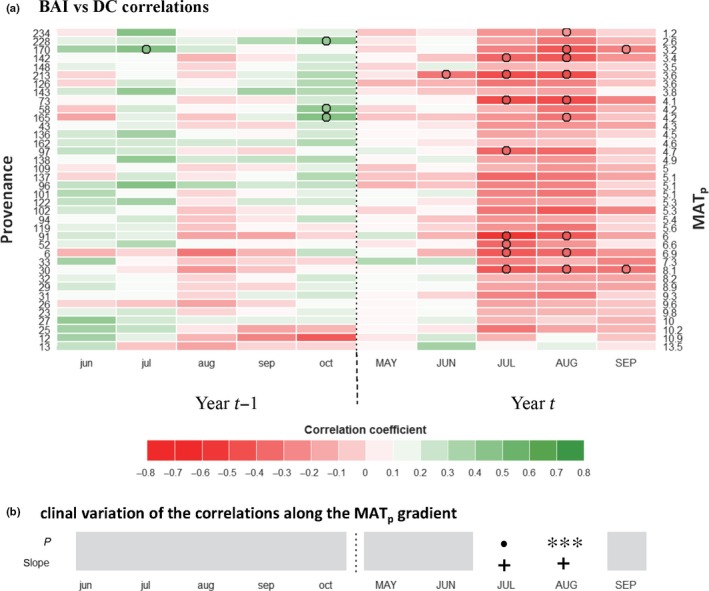
Climatic sensitivity (CS) traits: basal area increment (BAI) and monthly drought code (DC). (a) Bootstrapped correlation coefficients were computed between Pinus strobus BAI standardized residuals of BAI scores and the monthly DC at the Valcartier common garden experiment (Quebec, Canada) over the 1993–2014 period. (b) Test for clinal variations in correlation coefficients along the MAT p gradient: P‐value and slope sign of a regression against MAT p. Significance: •, P < 0.1; ***, P < 0.001.
Local adaptation in tree‐ring traits
The raw BAI averaged by population over the entire period (absolute meanBAI1993–2014, type A trait), expressed as a function of MATp, exhibited a second‐order curve (Fig. 3d; adj R 2 = 0.46, P < 0.001). This revealed a high degree of variation among populations and suggested local adaptation to the mean annual temperature of the provenance. According to that model, the best‐performing population was nonlocal. The MATp of the population that had the highest growth performance was 6.6°C, which is higher than the MAT in the Valcartier common garden experiment (3.95°C). Populations from both warmer and colder provenances exhibited lower growth. No significant relationship was observed between absolute avDens1993–2014 and MATp (Fig. 3c). However, note that the two populations from the southern group have a much higher tree‐ring density (Fig. 3f).
Among‐population variation was also observed for climate‐related tree‐ring traits (types B and C). The reduction in growth in 2003 (BAI2003, type B trait) was stronger for the populations from warmer provenances and absent in some cold‐climate provenances (Fig. 3e). Growth in 2003 was particularly reduced for the two southernmost provenances, corresponding to the southern genetic group. Among‐population genetic variation was found for CS traits using growth–climate correlations (type C traits). Among traits related to heat and drought stresses, CSBAI–Temp.Jul(t) (Fig. 4) and CSBAI–Drought.Aug(t) (Fig. 5) were positively correlated with MATp, and CSBAI–Prec.Jul(t) (Fig. S7) was negatively correlated with MATp. Among traits reflecting cold tolerance, CSBAI–Freeze.Oct(t−1) and CSBAI–Freeze.Jun(t) (Fig. S6) were negatively correlated with MATp, whereas CSBAI–Temp.Oct(t−1) (Fig. 4) was positively correlated with MATp.
CS and genotype–phenotype associations
We searched for genes related to local adaptation in growth responses to climate by testing those tree‐ring traits that showed a significant (P < 0.01) correlation with MATp: meanBAI1993–2014 (type A), BAI2003 (type B) and five CS traits (type C): CSBAI–Drought.Aug(t), CSBAI–Prec.Jul(t), CSBAI–Temp.Jul(t), CSBAI–Freeze.Oct(t−1) and CSBAI–Freeze.Jun(t). Diameter at breast height (DBH21) and height measured at age 21 (H21), referred to as dendrometric phenotypic traits in the present study, were also tested. GPA analysis revealed significant associations (P < 0.05) with dendrometric traits, as well as with the three types of tree‐ring traits for both candidate and noncandidate SNPs (Table S3). For dendrometric traits (DBH21 and H21) and type A tree‐ring traits (meanBAI1993–2014), the proportion of significant SNPs was similar between candidate and noncandidate SNPs (Table S5). For all the climate‐related type B (BAI2003) and type C traits (CS traits), we found a greater proportion of significant candidate SNPs as compared to noncandidate SNPs, although the proportion of SNPs classified as very likely was greater for noncandidate SNPs for CSBAI–Prec.Jul(t) and CSBAI‐FreezeOct(t−1).
Among the 21 candidate SNPs tested (Nadeau et al., 2016), 12 were confirmed in this study (10 genes; Fig. 6; Table 2): three SNPs (three genes) were significantly associated with dendrometric traits DBH21 and H21, while 11 SNPs (nine genes) were associated with tree‐ring traits (types A, B and C). As many as 10 SNPs (eight genes) were associated with the climate‐related type B and type C traits (seven SNP–trait associations classified as very likely, five likely and eight uncertain). Traits reflecting responses to specific freezing events (BAI2003, CSBAI–Freeze.Oct(t−1), and CSBAI–Freeze.Jun(t)) were associated with SNPs G‐001, G‐004, M‐016, M‐017, N‐029, N‐033, N‐040 and O‐002. CS traits reflecting heat and drought tolerance (CSBAI–Temp.Jul(t), CSBAI–Prec.Jul(t), CSBAI–Drought.Aug(t)) were associated with SNPs G‐014, M‐015, M‐016, N‐029, N‐033 and N‐040.
Figure 6.
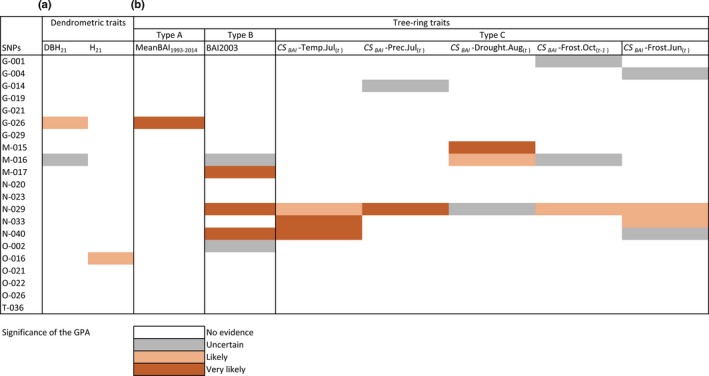
Tests of genotype–phenotype associations (GPAs) between traits and candidate loci from a previous genotype–environment association (GEA) analysis (Nadeau et al., 2016): (a) dendrometric traits (DBH 21 and H21 = diameter at breast height and height at 21 yr), and (b) tree‐ring traits of types A, B and C. For each SNP–trait pair, the association was tested with different levels of corrections: without correction, with correction for the genetic structure (Q), and with correction for both genetic structure and kinship (Q + K). The significance of the associations was classified as very likely (when significant for three levels of correction), likely (when significant for two levels) or uncertain (when significant for only one level of correction). BAI, basal area increment; CS, climate sensitivity trait; Temp, temperature; Prec, precipitation; Drought, drought code; Freeze, number of freezing days during the month.
Table 2.
Annotation of the candidate single nucleotide polymorphisms (SNPs) associated with dendrometric and tree‐ring traits
| Trait type | SNP | Gene | SNP annotationa | Annotationb | Traits pointed out by GPA | Known function | Known response to abiotic stresses |
|---|---|---|---|---|---|---|---|
| Dendrometric | O‐016 | 2_6731_01 | NS | SLY1 gene, F‐box family protein | H21 | Positive regulator of GA signaling in Arabidopsis thaliana (Ariizumi et al., 2011) | GA3 in A. thaliana (Ariizumi et al., 2011) |
| Dendrometric, A | G‐026 | GQ0206.B3_C13 | NS | Hypothetical protein ACMD2_05917 (Ananas comosus) | DBH21, meanBAI1993–2014 | – | – |
| B | M‐017 | 0_8844_01 | Intron | Galacturonosyltransferase (GAUT) 14 | Tolerance to cold: BAI2003 | Carbohydrate metabolism and cell wall pectin biosynthetic processes in A. thaliana (Sterling et al., 2006; Wang et al., 2011) | Cold in conifers (Bannister & Neuner, 2001) |
| B | O‐002 | 0_8844_01 | S | Galacturonosyltransferase (GAUT) 14 | Tolerance to cold: BAI2003 | Carbohydrate metabolism and cell wall pectin biosynthetic processes in A. thaliana (Sterling et al., 2006; Wang et al., 2011) | Cold in conifers (Bannister & Neuner, 2001) |
| C | G‐004 | GQ0033.TB_H23 | Intron | Endoglucanase 25‐like | Tolerance to cold: CSBAI–Frost.Jun(t) | Carbohydrate metabolism and cellulose synthesis (A. thaliana, Picea glauca; Maloney, 2010) | Diverse abiotic stresses in herbaceous plants (Wang et al., 2016) |
| C | G‐001 | GQ0015.B3.r_B10 | Intron | Vacuolar‐processing enzyme beta‐isozyme 1‐like | Tolerance to cold: CSBAI–Frost.Oct(t−1) | Storage protein accumulation (Gruis et al., 2004) | Stress‐induced programmed cell death process (Hatsugai et al., 2015) |
| C | G‐014 | GQ0081.BR.1_D09 | NS | Plastid movement impaired 1‐related 1 (PMIR1) | Tolerance to drought: CSBAI–Prec.Jul(t) | Chloroplast photorelocation movement (Suetsugu et al., 2015) | Drought in A. thaliana (Rojas‐Pierce et al., 2014) |
| C | N‐033 | 0_7001_01 | NS | NADPH‐dependent diflavin oxidoreductase 1‐like isoform X3 |
Tolerance to cold: CSBAI–Frost.Jun(t)
Tolerance to heat: CSBAI–Temp.Jul(t) |
Modulation of plant growth and development in higher plants (Lodeyro et al., 2012; Karlusich et al., 2014) | Diverse abiotic stresses using transgenic plants (Lodeyro et al., 2012) |
| C | M‐015 | 0_8683_01 | S | Serine/threonine‐protein kinase At1g18390 | Tolerance to drought: CSBAI–Drought.Aug(t) | Signaling and plant defense (Afzal et al., 2008) | Diverse abiotic stresses in herbaceous plants (Afzal et al., 2008) |
| Dendrometric, B, C | M‐016 | 0_8683_01 | NS | Serine/threonine‐protein kinase At1g18390 |
DBH21
Tolerance to cold: BAI2003, CSBAI–Frost.Oct(t−1) Tolerance to drought: CSBAI–Drought.Aug(t) |
Signaling and plant defense (Afzal et al., 2008) | Diverse abiotic stresses in herbaceous plants (Afzal et al., 2008) |
| B, C | N‐029 | 0_6047_02 | Not annotated | Basic helix–loop–helix (bHLH) DNA‐binding superfamily protein |
Tolerance to cold: BAI2003, CSBAI–Frost.Oct(t−1), CSBAI–Frost.Jun(t)
Tolerance to heat: CSBAI–Temp.Jul(t) Tolerance to drought: CSBAI–Prec.Jul(t), CSBAI–Drought.Aug(t) |
Anthocyanin biosynthesis, cell proliferation and differentiation (Yamada et al., 2011; Xiang et al., 2015) | Diverse abiotic stresses (Ji et al., 2016) |
| B, C | N‐040 | 2_4107_01 | S | Thylakoid lumenal 19 kDa protein, chloroplastic |
Tolerance to cold: BAI2003, CSBAI–Frost.Jun(t)
Tolerance to heat: CSBAI–Temp.Jul(t) |
Photosynthetic electron transfer (Ishihara et al., 2007) | Diverse abiotic stresses in herbaceous plants (Gururani et al., 2015) |
Known functions and responses to abiotic stresses of homologous genes in other plant species are reported in the two right‐hand columns of the table.
–, No information reported.
S, synonymous SNP; NS, nonsynonymous SNP.
The most informative annotation has been selected among the 10 top hits for the GQ0081.BR.1_D09 gene.
The annotation of the 12 candidate SNPs confirmed in this study revealed a good match with the known functions of homologous genes in other plant models (Table 2). Eight SNPs were located in coding regions and three were located in a noncoding region (intron). Among those located in coding regions, three SNPs were identified as synonymous and five were identified as nonsynonymous. All the putative gene products had functions ranging from transcription factor activity to regulation of signaling molecules.
Discussion
This study illustrates how the combined analysis of tree‐ring traits derived from dendroecology and genotypes from a common garden experiment can contribute to understanding the complex relationship between genotypes, phenotypes (growth responses embedded in the annual rings) and interannual climatic fluctuations. Indeed, the new class of phenotypes obtained provided useful information to identify climatic constraints for a given tree species. We illustrated this approach by an examination of the CS traits shaping the growth of an important conifer in eastern North America's temperate forests. We quantified the sensitivity of populations to climatic‐related constraints across their lifespan and identified the critical times of the year (or previous year) when those constraints affected growth. We then highlighted the clinal variation along MATp for those traits. This approach made it possible to identify candidate genes for local adaptation in growth responses to climatic variability, and to cross‐validate candidate loci obtained from an independent GEA study.
Tree sensitivity to climate
The dendroclimatic analysis pointed to traits reflecting CS for each population (type C traits). At our study site, growth was mainly limited by very low/freezing temperature during the dehardening phase in spring and cold acclimation phase in autumn, when trees are probably the most vulnerable (probably causing freeze injuries; Howe et al., 2003); and by summer heat and drought stress.
The positive influence of a longer growing season on growth is shown by the positive effect of previous October and current May temperatures (Fig. 4), as well as the negative effect of previous October and current June number of days below 0°C (Fig. S6). Warm spring conditions result in an earlier cessation of dormancy and earlier cambium reactivation, which leads to a longer growth period (Begum et al., 2013). Similarly, a warm autumn during the year before ring formation translates into more time available for accumulation of nonstructural carbohydrate reserves (Hartmann & Trumbore, 2016) and for the division of leaf primordia meristematic cells in buds (Fritts, 1976; Macey & Arnott, 1986). This influence of temperature in combination with photoperiod on the growth of the tree populations under study could be confirmed by studying bud and cambium phenology (e.g. Perrin et al., 2017).
Growth was limited by summer heat or drought, with a negative correlation with July temperature (Fig. 4) and July and August drought severity (Fig. 5), and a positive correlation with July precipitation (Fig. S7). Warm summers can have a direct negative effect on growth by increasing the ratio of carbohydrates allocated to maintenance respiration, while hydric stress reduces both water intake and photosynthesis through stomata closure (Hartmann & Trumbore, 2016). The correlations with climate that we found for this common garden were consistent with those reported for natural stands in other regions (e.g. Marchand & Filion, 2011; Chhin et al., 2013; Girardin et al., 2016).
The study of tree rings makes it possible to precisely determine the influence of yearly climatic anomalies on growth during a tree's life cycle; this information cannot be obtained from conventional approaches used in tree genetics, such as the study of tree height or diameter at a given age. It also has the advantage of accounting for time‐lagged effects of climate, which is particularly relevant for conifer species showing a determined growth pattern. For instance, autumn temperatures may influence the timing of budburst of the subsequent year, which in turn can impact tree fitness (Heide, 2003).
Tree adaptation potential to climate change
Comparing the populations’ responses to climate (type B and type C traits) in the common garden revealed existing genetic variation for tolerance to freeze and drought. The positive influence of a warm autumn during the year preceding ring formation (CSBAI–Temp.Oct(t−1)) was stronger for populations from southernmost provenances and significantly increased with MATp (Fig. 4a,b). Furthermore, the negative effect of early autumn freeze (CSBAI–Freeze.Oct(t−1)) and late spring freeze (CSBAI–Freeze.Jun(t)) was greater for warm provenances (Fig. S6). The timing of bud set and bud flush are two important traits involved in the adaptation to local climate that are known to be under strong genetic control in forest trees (e.g. Billington & Pelham, 1991; Savolainen et al., 2004; Bennie et al., 2010; McKown et al., 2014). Populations from colder provenances have probably evolved to set buds and enter dormancy earlier, thereby minimizing the risk of early autumn freeze injuries. Clinal variations in cold hardiness were also reported for P. strobus (Lu et al., 2003), thus supporting our findings that warmer provenances were more likely to be impacted by freeze injuries and to exhibit reduced growth. Genetic variation related to cold hardiness was further demonstrated by among‐provenance variations in the standardized residual growth of 2003 (BAI2003) following lower than average temperatures (e.g. −2°C) during autumn 2002 and spring 2003 (type B traits). The relative growth reduction following this event was significantly more pronounced for populations from warmer provenances (Fig. 3e), with the two southern group provenances exhibiting a very severe growth reduction.
Local adaptation in response to drought was suggested by the correlation between MATp and among‐population variation of CSBAI–Temp.Jul(t) (Fig. 4b), CSBAI–Drought.Aug(t) (Fig. 5b) and CSBAI–Prec.Jul(t) (Fig. S7). Provenances from warmer locations were less sensitive to summer drought in the common garden, suggesting that those provenances were more adapted to droughts (in agreement with Savva et al., 2002; McLane et al., 2011; Taeger et al., 2013; Montwé et al., 2016). Our study demonstrates the great potential of using CS derived from tree rings sampled in common gardens to investigate trees’ adaptive capacity to climate change, especially when testing provenances for suitability in future warming climates.
Genetic basis of climate sensitivity
GPA analysis conducted on a combination of dendrometric and tree‐ring traits allowed the detection of significant associations for both candidate and noncandidate SNPs from Nadeau et al. (2016). While the noncandidate SNPs may also contain relevant genes for responses to climate, our main goal was to see whether we could confirm those candidate SNPs that had previously been detected in genetic–environment associations (Nadeau et al., 2016) and gain further insights about their putative roles in adaptation to climate. For climate‐related traits (types B and C), we found a greater proportion of significant SNPs in candidate than in noncandidate SNPs. In addition, the independent validation of candidate SNPs from a previous study using different samples and different methods should reduce false positives and so we further discuss the candidate genes here. GPA analyses confirmed 12 of the 21 candidate SNPs (six very likely, two likely, four uncertain), and 10 of them (eight genes) were associated with climate‐related traits (types B and C). The five strongest candidate SNPs in Nadeau et al. (2016; namely N‐029, G‐014, M‐015, M‐016 and M‐017), which had previously been detected by at least two GEA or F ST outlier methods, were also significantly associated with climate‐related tree‐ring traits in this study. Although false positives are possible, many loci were associated with several traits and with different levels of correction for population structure and kinship, thus increasing our confidence in the results.
The associations between candidate SNPs and type B and C tree‐ring traits provided information about the climatic constraints affecting P. strobus growth, and thus helped bridge the gap between the genotypes, the phenotypes and the environmental selection pressures. Taken together, our results suggest that the putative functions obtained for the genes detected in GPA associations were plausible, because information found in the literature supports the established associations between the candidate SNPs and the measured traits reflecting either drought tolerance, freeze tolerance or both. Several SNPs were associated with both freeze and drought tolerance traits (three very likely and one likely; Fig. 6). Among those, SNP N‐029 was located within a bHlH (basic helix–loop–helix) transcription factor, which is known to confer tolerance to multiple abiotic stresses in higher plants (Table 2). Similarly, the gene containing SNP N‐040 codes for a thylakoid luminal protein involved in photosystem II, one of the most susceptible components of the photosynthetic machinery that bears the brunt of abiotic stresses. Other SNPs were specifically associated with either freeze or drought tolerance traits. Interestingly, the GAUT (SNPs M‐017, very likely; O‐002, uncertain) and the endoglucanase genes (SNP G‐004, uncertain), which are involved in carbohydrate metabolism, were specifically associated with growth responses to freezing events (Table 2). By preserving and stabilizing cell structures, carbohydrates associated with the cell wall can enhance freezing tolerance by controlling water movement and ice formation under cold conditions (Tenhaken, 2015). In addition, one SNP (G‐014; uncertain), exclusively associated with drought tolerance traits (Table 2), encoded a PLASTID MOVEMENT IMPAIRED1 (PMI1), a protein that mediates the abscisic acid (ABA)‐dependent pathway under drought conditions in Arabidopsis thaliana (Rojas‐Pierce et al., 2014). Contrary to cold‐responsive genes, many water‐stress‐inducible genes are upregulated by ABA under drought conditions (Yamaguchi‐Shinozaki & Shinozaki, 2006). Further gene expression and functional studies aimed at identifying coexpressed gene networks involved in specific biological processes, such as freeze tolerance, would be needed to determine the exact role of those genes in P. strobus and in other conifers (for a review, see Serin et al., 2016). Multi‐SNP GPA models (e.g. Segura et al., 2012) should also be tested given that highly metric traits such as growth are probably controlled by a large number of interacting genes, which was beyond the scope of this study given the small number of SNPs tested.
Conclusion
Accurately determining the periods of the year when trees are most vulnerable to abiotic and biotic factors is crucial to understanding the genetic architecture of local adaption. In this paper, we showed that tree‐ring traits measured in common garden experiments offer great potential to unravel local adaptation in forest trees through a fine‐scale, retrospective and dynamic view of tree responses to past environmental variations. For the first time, we proposed using a metric (i.e. climate sensitivity traits; CS) that incorporates both the phenotypic trait under study (i.e. growth or wood density) and the response to climate in association genetic studies. We identified important climatic constraints and the critical times of the year when those constraints affect growth, information that cannot be derived from classic measurements such as height and DBH. We believe that this novel approach will provide a valuable contribution to the study of differences in climate sensitivity between provenances in common garden experiments, and more generally to examine forest adaptation to environmental changes.
Author contributions
J.M.H., S.N., C.D., I.D., P.L., N.I. and M.P.G. planned and designed the research; J.M.H., S.N., M.P.G. and C.D. performed the experiments and analyzed the data; J.M.H., S.N., C.D., I.D., P.L., N.I. and M.P.G. wrote the manuscript. J.M.H. and S.M. contributed equally to this work.
Supporting information
Please note: Wiley Blackwell are not responsible for the content or functionality of any Supporting Information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
Fig. S1 Monthly mean temperature, drought code, minimum temperature and precipitation in 2002 and 2003 at Valcartier, compared to interannual variability during the 1993–2014 period.
Fig. S2 Spring mean temperature and August drought code in 2003, and number of days with minimum temperature below zero in October 2002 at Valcartier, compared to interannual variability in standardized residual BAI scores.
Fig. S3 Correlation analysis for climatic sensitivity traits basal area increment and monthly temperature.
Fig. S4 Correlation analysis for climatic sensitivity traits basal area increment and monthly drought code.
Fig. S5 Genetic structure assessed through a discriminant analysis of principal components (DAPC).
Fig. S6 Correlation analysis for climatic sensitivity traits basal area increment and monthly number of days below 0°C.
Fig. S7 Correlation analysis for climatic sensitivity traits basal area increment and monthly precipitation.
Fig. S8 Comparison of growth–climate relationships at the provenance and at the individual levels.
Fig. S9 Correlation analysis for climatic sensitivity traits ring average density and monthly drought code.
Fig. S10 Correlation analysis for climatic sensitivity traits ring density and monthly mean temperature.
Table S1 Summary of provenance location, mean annual temperature of provenance, mean annual precipitation and number of sampled trees per provenance
Table S2 Spearman rank correlations between monthly climate variables
Table S5 Comparisons between the number of SNPs detected in GPAs with tassel between candidate and noncandidate SNPs
Table S3 Detailed results of the genotype–phenotype association (GPA) analysis with tassel
Table S4 Detailed results of the genotype–phenotype association (GPA) analysis with tassel when excluding the two populations from the southern group
Acknowledgements
This work was made possible thanks to the financial and in‐kind support provided by the Canadian Forest Service and the Canadian Wood Fibre Centre of Natural Resources Canada, and funding from the Natural Sciences and Engineering Research Council of Canada (NSERC; Discovery Grant to M.P.G.). We thank Christine Simard, Philippe Labrie, David Gervais, Daniel Plourde, Eric Dussault, Jean‐François Légaré, and Marie‐Claude Gros‐Louis for their valuable contributions to field and laboratory work, and Manuel Lamothe, Claude Bomal and Julie Godbout for methodological discussions. We are grateful to Sébastien Clément and Ilga Porth for useful comments on a previous version of the manuscript and to Isabelle Lamarre for technical English editing. Finally, we would like to thank Dr Andrew Eckert and two anonymous reviewers for their insights and constructive comments.
See also the Commentary on this article by https://doi.org/10.1111/nph.15094.
Contributor Information
Nathalie Isabel, Email: nathalie.isabel@canada.ca.
Martin P. Girardin, Email: martin.girardin@canada.ca
References
- Afzal AJ, Wood AJ, Lightfoot DA. 2008. Plant receptor‐like serine threonine kinases: roles in signaling and plant defense. Molecular Plant‐Microbe Interactions 21: 507–517. [DOI] [PubMed] [Google Scholar]
- Aitken SN, Bemmels JB. 2016. Time to get moving: assisted gene flow of forest trees. Evolutionary Applications 9: 271–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aitken SN, Whitlock MC. 2013. Assisted gene flow to facilitate local adaptation to climate change. Annual Review of Ecology, Evolution, and Systematics 44: 367–388. [Google Scholar]
- Aitken SN, Yeaman S, Holliday JA, Wang T, Curtis McLane S. 2008. Adaptation, migration or extirpation: climate change outcomes for tree populations. Evolutionary Applications 1: 95–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alberto FJ, Aitken SN, Alía R, González‐Martínez SC, Hänninen H, Kremer A, Lefèvre F, Lenormand T, Yeaman S, Whetten R et al 2013. Potential for evolutionary responses to climate change – evidence from tree populations. Global Change Biology 19: 1645–1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ariizumi T, Lawrence PK, Steber CM. 2011. The role of two F‐box proteins, SLEEPY1 and SNEEZY, in Arabidopsis gibberellin signaling. Plant Physiology 155: 765–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aubin I, Munson AD, Cardou F, Burton PJ, Isabel N, Pedlar JH, Paquette A, Taylor AR, Delagrange S, Kebli H et al 2016. Traits to stay, traits to move: a review of functional traits to assess sensitivity and adaptive capacity of temperate and boreal trees to climate change. Environmental Reviews 24: 164–186. [Google Scholar]
- Bannister P, Neuner G. 2001. Frost resistance and the distribution of conifers In: Bigras FJ, Colombo SJ, eds. Conifer cold hardiness. Dordrecht, the Netherlands: Springer, 3–21. [Google Scholar]
- Begum S, Nakaba S, Yamagishi Y, Oribe Y, Funada R. 2013. Regulation of cambial activity in relation to environmental conditions: understanding the role of temperature in wood formation of trees. Physiologia Plantarum 147: 46–54. [DOI] [PubMed] [Google Scholar]
- Bennie J, Kubin E, Wiltshire A, Huntley B, Baxter R. 2010. Predicting spatial and temporal patterns of bud‐burst and spring frost risk in north‐west Europe: the implications of local adaptation to climate. Global Change Biology 16: 1503–1514. [Google Scholar]
- Billington HL, Pelham J. 1991. Genetic variation in the date of budburst in Scottish birch populations: implications for climate change. Functional Ecology 5: 403–409. [Google Scholar]
- Biondi F, Waikul K. 2004. DENDROCLIM2002: a C++ program for statistical calibration of climate signals in tree‐ring chronologies. Computers & Geosciences 30: 303–311. [Google Scholar]
- Bunn AG. 2008. A dendrochronology program library in R (dplR). Dendrochronologia 26: 115–124. [Google Scholar]
- Burnham KP, Anderson DR. 2010. Model selection and multi‐model inference: a practical information‐theoretic approach. Dordrecht, the Netherlands: Springer. [Google Scholar]
- Chhin S, Chumack K, Dahl T, David ET, Kurzeja P, Magruder M, Telewski FW. 2013. Growth–Climate relationships of Pinus strobus in the floodway versus terrace Forest along the banks of the Red Cedar River, Michigan. Tree‐Ring Research 69: 37–47. [Google Scholar]
- Cook E, Kairiūkštis L. 1990. Methods of dendrochronology: applications in the environmental sciences. Dordrecht, the Netherlands: Kluwer Academic. [Google Scholar]
- Cook ER, Holmes R. 1986. Guide for computer program ARSTAN. Tucson, AZ, USA: Laboratory of Tree‐Ring Research, University of Arizona. [Google Scholar]
- Coop G, Witonsky D, Rienzo AD, Pritchard JK. 2010. Using environmental correlations to identify loci underlying local adaptation. Genetics 185: 1411–1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dray S, Dufour AB. 2007. The ade4 package: implementing the duality diagram for ecologists. Journal of Statistical Software 22: 1–20. [Google Scholar]
- Dutilleul P. 1993. Modifying the t‐test for assessing the correlation between two spatial processes. Biometrics 49: 305–314. [Google Scholar]
- Eckert AJ, Maloney PE, Vogler DR, Jensen CE, Mix AD, Neale DB. 2015. Local adaptation at fine spatial scales: an example from sugar pine (Pinus lambertiana, Pinaceae). Tree Genetics & Genomes 11: 42. [Google Scholar]
- Fritts HC. 1976. Tree rings and climate. London, UK: Academic Press. [Google Scholar]
- Gessler A, Ferrio JP, Hommel R, Treydte K, Werner RA, Monson RK. 2014. Stable isotopes in tree rings: towards a mechanistic understanding of isotope fractionation and mixing processes from the leaves to the wood. Tree Physiology 34: 796–818. [DOI] [PubMed] [Google Scholar]
- Girardin MP, Bouriaud O, Hogg EH (Ted), Kurz WA, Zimmermann NE, Metsaranta J, de Jong R, Frank DC, Esper J, Büntgen U et al 2016. No growth stimulation of Canada's boreal forest under half‐century of combined warming and CO2 fertilization. Proceedings of the National Academy of Sciences, USA 113: E8406–E8414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girardin MP, Guo XJ, Bernier PY, Raulier F, Gauthier S. 2012. Changes in growth of pristine boreal North American forests from 1950 to 2005 driven by landscape demographics and species traits. Biogeosciences 9: 2523–2536. [Google Scholar]
- Gruis D, Schulze J, Jung R. 2004. Storage protein accumulation in the absence of the vacuolar processing enzyme family of cysteine proteases. Plant Cell 16: 270–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gururani MA, Venkatesh J, Tran LSP. 2015. Regulation of photosynthesis during abiotic stress‐induced photoinhibition. Molecular Plant 8: 1304–1320. [DOI] [PubMed] [Google Scholar]
- Hardy OJ, Vekemans X. 2002. spagedi: a versatile computer program to analyse spatial genetic structure at the individual or population levels. Molecular Ecology Notes 2: 618–620. [Google Scholar]
- Hartmann H, Trumbore S. 2016. Understanding the roles of nonstructural carbohydrates in forest trees – from what we can measure to what we want to know. New Phytologist 211: 386–403. [DOI] [PubMed] [Google Scholar]
- Hatsugai N, Yamada K, Goto‐Yamada S, Hara‐Nishimura I. 2015. Vacuolar processing enzyme in plant programmed cell death. Frontiers in Plant Science 6: 234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heide OM. 2003. High autumn temperature delays spring bud burst in boreal trees, counterbalancing the effect of climatic warming. Tree Physiology 23: 931–936. [DOI] [PubMed] [Google Scholar]
- Holmes RL. 1983. Computer‐assisted quality control in tree‐ring dating and measurement. Tree‐Ring Bulletin 43: 69–78. [Google Scholar]
- Housset JM, Girardin MP, Baconnet M, Carcaillet C, Bergeron Y. 2015. Unexpected warming‐induced growth decline in Thuja occidentalis at its northern limits in North America. Journal of Biogeography 42: 1233–1245. [Google Scholar]
- Howe GT, Aitken SN, Neale DB, Jermstad KD, Wheeler NC, Chen TH. 2003. From genotype to phenotype: unraveling the complexities of cold adaptation in forest trees. Canadian Journal of Botany 81: 1247–1266. [Google Scholar]
- IPCC . 2013. Climate change 2013: the physical science basis. Contribution of Working Group I to the Fifth Assessment Report of the IPCC. Cambridge, UK: Cambridge University Press. [Google Scholar]
- Ishihara S, Takabayashi A, Ido K, Endo T, Ifuku K, Sato F. 2007. Distinct functions for the two PsbP‐like proteins PPL1 and PPL2 in the chloroplast thylakoid lumen of Arabidopsis. Plant Physiology 145: 668–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jezkova T, Wiens JJ. 2016. Rates of change in climatic niches in plant and animal populations are much slower than projected climate change. Proceedings of the Royal Society of London B 283: 2016–2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji X, Nie X, Liu Y, Zheng L, Zhao H, Zhang B, Huo L, Wang Y. 2016. A bHLH gene from Tamarix hispida improves abiotic stress tolerance by enhancing osmotic potential and decreasing reactive oxygen species accumulation. Tree Physiology 36: 193–207. [DOI] [PubMed] [Google Scholar]
- Jombart T, Devillard S, Balloux F. 2010. Discriminant analysis of principal components: a new method for the analysis of genetically structured populations. BMC Genetics 11: 94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joost S, Bonin A, Bruford MW, Després L, Conord C, Erhardt G, Taberlet P. 2007. A spatial analysis method (SAM) to detect candidate loci for selection: towards a landscape genomics approach to adaptation. Molecular Ecology 16: 3955–3969. [DOI] [PubMed] [Google Scholar]
- Joyce DG, Rehfeldt GE. 2013. Climatic niche, ecological genetics, and impact of climate change on eastern white pine (Pinus strobus L.): guidelines for land managers. Forest Ecology and Management 295: 173–192. [Google Scholar]
- Jump AS, Mátyás C, Peñuelas J. 2009. The altitude‐for‐latitude disparity in the range retractions of woody species. Trends in Ecology & Evolution 24: 694–701. [DOI] [PubMed] [Google Scholar]
- Jump AS, Peñuelas J. 2005. Running to stand still: adaptation and the response of plants to rapid climate change. Ecology Letters 8: 1010–1020. [DOI] [PubMed] [Google Scholar]
- Karlusich JJP, Lodeyro AF, Carrillo N. 2014. The long goodbye: the rise and fall of flavodoxin during plant evolution. Journal of Experimental Botany 65: 5161–5178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langlet O. 1971. Two hundred years genecology. Taxon 20: 653–721. [Google Scholar]
- Leland C, Hom J, Skowronski N, Ledig FT, Krusic PJ, Cook ER, Martin‐Benito D, Martin‐Fernandez J, Pederson N. 2016. Missing rings, synchronous growth, and ecological disturbance in a 36‐year pitch pine (Pinus rigida) provenance study. PLoS ONE 11: e0154730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepais O, Bacles CFE. 2014. Two are better than one: combining landscape genomics and common gardens for detecting local adaptation in forest trees. Molecular Ecology 23: 4671–4673. [DOI] [PubMed] [Google Scholar]
- Li P, Beaulieu J, Daoust G, Plourde A. 1997. Patterns of adaptive genetic variation in eastern white pine (Pinus strobus) from Québec. Canadian Journal of Forest Research 27: 199–206. [Google Scholar]
- Lodeyro AF, Ceccoli RD, Pierella Karlusich JJ, Carrillo N. 2012. The importance of flavodoxin for environmental stress tolerance in photosynthetic microorganisms and transgenic plants. Mechanism, evolution and biotechnological potential. FEBS Letters 586: 2917–2924. [DOI] [PubMed] [Google Scholar]
- Lu P, Joyce DG, Sinclair RW. 2003. Geographic variation in cold hardiness among eastern white pine (Pinus strobus L.) provenances in Ontario. Forest Ecology and Management 178: 329–340. [Google Scholar]
- Macey DE, Arnott JT. 1986. The effect of moderate moisture and nutrient stress on bud formation and growth of container‐grown white spruce seedlings. Canadian Journal of Forest Research 16: 949–954. [Google Scholar]
- Maloney V. 2010. Function, functional conservation and interactions of the membrane‐bound endo‐1,4‐beta‐glucanases orthologous to Korrigan. Retrospective Theses and Dissertations, 1919–2007. doi: 10.14288/1.0069960. [WWW document] URL http://hdl.handle.net/2429/24242 [accessed 18 December 2017]. [DOI]
- Marchand N, Filion L. 2011. False rings in the white pine (Pinus strobus) of the Outaouais Hills, Québec (Canada), as indicators of water stress. Canadian Journal of Forest Research 42: 12–22. [Google Scholar]
- McKenney DW, Pedlar JH, Lawrence K, Papadopol P, Campbell K, Hutchinson MF. 2014. Change and evolution in the plant hardiness zones of Canada. BioScience 64: 341–350. [Google Scholar]
- McKown AD, Guy RD, Klápště J, Geraldes A, Friedmann M, Cronk QC, El‐Kassaby YA, Mansfield SD, Douglas CJ. 2014. Geographical and environmental gradients shape phenotypic trait variation and genetic structure in Populus trichocarpa . New Phytologist 201: 1263–1276. [DOI] [PubMed] [Google Scholar]
- McLane SC, Daniels LD, Aitken SN. 2011. Climate impacts on lodgepole pine (Pinus contorta) radial growth in a provenance experiment. Forest Ecology and Management 262: 115–123. [Google Scholar]
- Montwé D, Isaac‐Renton M, Hamann A, Spiecker H. 2016. Drought tolerance and growth in populations of a wide‐ranging tree species indicate climate change risks for the boreal north. Global Change Biology 22: 806–815. [DOI] [PubMed] [Google Scholar]
- Nadeau S, Godbout J, Lamothe M, Gros‐Louis M‐C, Isabel N, Ritland K. 2015. Contrasting patterns of genetic diversity across the ranges of Pinus monticola and P. strobus: a comparison between eastern and western North American postglacial colonization histories. American Journal of Botany 102: 1342–1355. [DOI] [PubMed] [Google Scholar]
- Nadeau S, Meirmans PG, Aitken SN, Ritland K, Isabel N. 2016. The challenge of separating signatures of local adaptation from those of isolation by distance and colonization history: the case of two white pines. Ecology and Evolution 6: 8649–8664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedlar JH, McKenney DW. 2017. Assessing the anticipated growth response of northern conifer populations to a warming climate. Scientific Reports 7: 43881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrin M, Rossi S, Isabel N. 2017. Synchronisms between bud and cambium phenology in black spruce: early‐flushing provenances exhibit early xylem formation. Tree Physiology 37: 593–603. [DOI] [PubMed] [Google Scholar]
- Price DT, Alfaro RI, Brown KJ, Flannigan MD, Fleming RA, Hogg EH, Girardin MP, Lakusta T, Johnston M, McKenney DW et al 2013. Anticipating the consequences of climate change for Canada's boreal forest ecosystems. Environmental Reviews 21: 322–365. [Google Scholar]
- Rahaman MM, Chen D, Gillani Z, Klukas C, Chen M. 2015. Advanced phenotyping and phenotype data analysis for the study of plant growth and development. Frontiers in Plant Science 6: 619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajora OP, Eckert AJ, Zinck JW. 2016. Single‐locus versus multilocus patterns of local adaptation to climate in eastern white pine (Pinus strobus, Pinaceae). PLoS ONE 11: e0158691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Régnière J, Bolstad P. 1994. Statistical simulation of daily air temperature patterns in Eastern North America to forecast seasonal events in insect pest management. Environmental Entomology 23: 1368–1380. [Google Scholar]
- Rehfeldt GE, Ying CC, Spittlehouse DL, Hamilton DA Jr. 1999. Genetic responses to climate in Pinus contorta: niche breadth, climate change, and reforestation. Ecological Monographs 69: 375–407. [Google Scholar]
- Rigault P, Boyle B, Lepage P, Cooke JEK, Bousquet J, MacKay JJ. 2011. A white spruce gene catalog for conifer genome analyses. Plant Physiology 157: 14–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritland K. 1996. Estimators for pairwise relatedness and individual inbreeding coefficients. Genetics Research 67: 175–185. [Google Scholar]
- Rojas‐Pierce M, Whippo CW, Davis PA, Hangarter RP, Springer PS. 2014. PLASTID MOVEMENT IMPAIRED1 mediates ABA sensitivity during germination and implicates ABA in light‐mediated chloroplast movements. Plant Physiology and Biochemistry 83: 185–193. [DOI] [PubMed] [Google Scholar]
- Savolainen O, Bokma F, García‐Gil R, Komulainen P, Repo T. 2004. Genetic variation in cessation of growth and frost hardiness and consequences for adaptation of Pinus sylvestris to climatic changes. Forest Ecology and Management 197: 79–89. [Google Scholar]
- Savva Y, Schweingruber FH, Milyutin LI, Vaganov E. 2002. Genetic and environmental signals in tree rings from different provenances of Pinus sylvestris L. planted in the southern taiga, central Siberia. Trees – Structure and Function 16: 313–324. [Google Scholar]
- Schweingruber FH. 1996. Tree rings and environment: dendroecology. Bern, Switzerland: Paul Haupt AG. [Google Scholar]
- Segura V, Vilhjálmsson BJ, Platt A, Korte A, Seren Ü, Long Q, Nordborg M. 2012. An efficient multi‐locus mixed‐model approach for genome‐wide association studies in structured populations. Nature Genetics 44: 825–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serin EAR, Nijveen H, Hilhorst HWM, Ligterink W. 2016. Learning from Co‐expression Networks: Possibilities and Challenges. Frontiers in Plant Science 7: 444. doi: 10.3389/fpls.2016.00444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sork VL, Aitken SN, Dyer RJ, Eckert AJ, Legendre P, Neale DB. 2013. Putting the landscape into the genomics of trees: approaches for understanding local adaptation and population responses to changing climate. Tree Genetics & Genomes 9: 901–911. [Google Scholar]
- Sterling JD, Atmodjo MA, Inwood SE, Kolli VSK, Quigley HF, Hahn MG, Mohnen D. 2006. Functional identification of an Arabidopsis pectin biosynthetic homogalacturonan galacturonosyltransferase. Proceedings of the National Academy of Sciences, USA 103: 5236–5241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suetsugu N, Higa T, Kong S‐G, Wada M. 2015. PLASTID MOVEMENT IMPAIRED1 and PLASTID MOVEMENT IMPAIRED1‐RELATED1 mediate photorelocation movements of both chloroplasts and nuclei. Plant Physiology 169: 1155–1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taeger S, Zang C, Liesebach M, Schneck V, Menzel A. 2013. Impact of climate and drought events on the growth of Scots pine (Pinus sylvestris L.) provenances. Forest Ecology and Management 307: 30–42. [Google Scholar]
- Tenhaken R. 2015. Cell wall remodeling under abiotic stress. Frontiers in Plant Science 5: 771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terrier A, Girardin MP, Périé C, Legendre P, Bergeron Y. 2013. Potential changes in forest composition could reduce impacts of climate change on boreal wildfires. Ecological Applications 23: 21–35. [DOI] [PubMed] [Google Scholar]
- Urban MC, Bocedi G, Hendry AP, Mihoub J‐B, Pe'er G, Singer A, Bridle JR, Crozier LG, Meester LD, Godsoe W et al 2016. Improving the forecast for biodiversity under climate change. Science 353: aad8466. [DOI] [PubMed] [Google Scholar]
- de Villemereuil P, Gaggiotti OE, Mouterde M, Till‐Bottraud I. 2016. Common garden experiments in the genomic era: new perspectives and opportunities. Heredity 116: 249–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, McFarlane HE, Persson S. 2016. The impact of abiotic factors on cellulose synthesis. Journal of Experimental Botany 67: 543–552. [DOI] [PubMed] [Google Scholar]
- Wang X, Piao S, Ciais P, Li J, Friedlingstein P, Koven C, Chen A. 2011. Spring temperature change and its implication in the change of vegetation growth in North America from 1982 to 2006. Proceedings of the National Academy of Sciences, USA 108: 1240–1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren WG. 1980. On removing the growth trend from dendrochronological data. Tree‐Ring Bulletin 40: 35–44. [Google Scholar]
- Wigley TML, Briffa KR, Jones PD. 1984. On the average value of correlated time series, with applications in dendroclimatology and hydrometeorology. Journal of Climate and Applied Meteorology 23: 201–213. [Google Scholar]
- Xiang L, Liu X, Li X, Yin X, Grierson D, Li F, Chen K. 2015. A novel bHLH transcription factor involved in regulating anthocyanin biosynthesis in chrysanthemums (Chrysanthemum morifolium Ramat.). PLoS ONE 10: e0143892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada Y, Koyama T, Sato F. 2011. Basic helix‐loop‐helix transcription factors and regulation of alkaloid biosynthesis. Plant Signaling & Behavior 6: 1627–1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi‐Shinozaki K, Shinozaki K. 2006. Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annual Review of Plant Biology 57: 781–803. [DOI] [PubMed] [Google Scholar]
- Yu J, Pressoir G, Briggs WH, Vroh Bi I, Yamasaki M, Doebley JF, McMullen MD, Gaut BS, Nielsen DM, Holland JB et al 2006. A unified mixed‐model method for association mapping that accounts for multiple levels of relatedness. Nature Genetics 38: 203–208. [DOI] [PubMed] [Google Scholar]
- Zang C, Biondi F. 2015. treeclim: an R package for the numerical calibration of proxy‐climate relationships. Ecography 38: 431–436. [Google Scholar]
- Zhang Z, Ersoz E, Lai C‐Q, Todhunter RJ, Tiwari HK, Gore MA, Bradbury PJ, Yu J, Arnett DK, Ordovas JM et al 2010. Mixed linear model approach adapted for genome‐wide association studies. Nature Genetics 42: 355–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Please note: Wiley Blackwell are not responsible for the content or functionality of any Supporting Information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
Fig. S1 Monthly mean temperature, drought code, minimum temperature and precipitation in 2002 and 2003 at Valcartier, compared to interannual variability during the 1993–2014 period.
Fig. S2 Spring mean temperature and August drought code in 2003, and number of days with minimum temperature below zero in October 2002 at Valcartier, compared to interannual variability in standardized residual BAI scores.
Fig. S3 Correlation analysis for climatic sensitivity traits basal area increment and monthly temperature.
Fig. S4 Correlation analysis for climatic sensitivity traits basal area increment and monthly drought code.
Fig. S5 Genetic structure assessed through a discriminant analysis of principal components (DAPC).
Fig. S6 Correlation analysis for climatic sensitivity traits basal area increment and monthly number of days below 0°C.
Fig. S7 Correlation analysis for climatic sensitivity traits basal area increment and monthly precipitation.
Fig. S8 Comparison of growth–climate relationships at the provenance and at the individual levels.
Fig. S9 Correlation analysis for climatic sensitivity traits ring average density and monthly drought code.
Fig. S10 Correlation analysis for climatic sensitivity traits ring density and monthly mean temperature.
Table S1 Summary of provenance location, mean annual temperature of provenance, mean annual precipitation and number of sampled trees per provenance
Table S2 Spearman rank correlations between monthly climate variables
Table S5 Comparisons between the number of SNPs detected in GPAs with tassel between candidate and noncandidate SNPs
Table S3 Detailed results of the genotype–phenotype association (GPA) analysis with tassel
Table S4 Detailed results of the genotype–phenotype association (GPA) analysis with tassel when excluding the two populations from the southern group


