Summary
Susceptibility to the root‐knot nematode Meloidogyne incognita in plants is thought to be a complex trait based on multiple genes involved in cell differentiation, growth and defence. Previous genetic analyses of susceptibility to M. incognita have mainly focused on segregating dominant resistance genes in crops. It is not known if plants harbour significant genetic variation in susceptibility to M. incognita independent of dominant resistance.
To study the genetic architecture of susceptibility to M. incognita, we analysed nematode reproduction on a highly diverse set of 340 natural inbred lines of Arabidopsis thaliana with genome‐wide association mapping. We observed a surprisingly large variation in nematode reproduction among these lines.
Genome‐wide association mapping revealed four quantitative trait loci (QTLs) located on chromosomes 1 and 5 of A. thaliana significantly associated with reproductive success of M. incognita, none of which harbours typical resistance gene homologues. Mutant analysis of three genes located in two QTLs showed that the transcription factor BRASSINAZOLE RESISTANT1 and an F‐box family protein may function as (co‐)regulators of susceptibility to M. incognita in Arabidopsis.
Our data suggest that breeding for loss‐of‐susceptibility, based on allelic variants critically involved in nematode feeding, could be used to make crops more resilient to root‐knot nematodes.
Keywords: allelic variation, Arabidopsis thaliana, brassinosteroid signalling, F‐box protein, genome‐wide association, Meloidogyne incognita, susceptibility
Introduction
Polyphagous root‐knot nematodes significantly undermine agricultural productivity in major food crops worldwide (Jones et al., 2013). In a recent study on biotic risk factors of global food security, the tropical root‐knot nematode Meloidogyne incognita was ranked as the most invasive plant disease‐causing agent (Bebber et al., 2014). For decades, root‐knot nematode infestations have been controlled by applications of chemical pesticides. However, most pesticides against root‐knot nematodes face regulatory bans owing to their high human and environmental toxicity. The phasing‐out of chemical pesticides to root‐knot nematodes has significantly increased the global demand for nematode‐resistant crops. However, for only a few crops, such as tomato, prune, carrot and pepper, highly specific dominant resistance genes against root‐knot nematodes are available (Williamson & Kumar, 2006; Davies & Elling, 2015).
Two natural phenomena currently threaten the use of dominant resistances to root‐knot nematodes, the first of which is genetic selection for resistance‐breaking nematode populations. For instance, most of the commercial cultivars of tomato (Solanum lycopersicum) carry introgressions of the dominant Mi‐1.2 gene from the wild tomato species Solanum peruvianum, which in many areas is no longer able to confer high levels of resistance to several tropical root‐knot nematode species (e.g. M. incognita, Meloidogyne javanica and Meloidogyne arenaria; Castagnone‐Sereno et al., 2013). Findings of virulent field populations of M. incognita in tomato with the Mi‐1.2 gene are not a particularly recent development (Kaloshian et al., 1996; Semblat et al., 2000). However, their widespread dispersal across major tomato‐producing regions has lately turned them into a major concern for growers. Second, many known dominant resistances to tropical root‐knot nematodes are temperature sensitive and rising soil temperatures by global warming may render them ineffective (Jacquet et al., 2005).
Root‐knot nematodes are obligate biotrophs that feed for weeks on living cells within the vascular cylinder of the root of a host plant. Soil‐borne second‐stage juveniles (J2s) of M. incognita invade the roots at the transition zone close to the root tip. The J2s then migrate intercellularly through the root cortex towards the root meristem, where they enter the vascular cylinder from below. Inside the vascular cylinder the J2s establish a permanent feeding structure consisting of several giant nurse cells (Caillaud et al., 2008). For the initiation of these giant cells, the J2s redirect the differentiation and growth of vascular cells into large transfer cell‐like units. The exact molecular mechanisms underlying the cellular transformation of vascular parenchyma into giant cells are not well understood. However, it is clear that giant cell formation involves alterations in a wide range of fundamental molecular and cellular processes, including epigenetic control of gene expression, cell cycle regulation, plant cell wall modifications and cytoskeletal rearrangements (Kyndt et al., 2013). Prolonged feeding on giant cells enables the J2s to moult three times into the adult female stage. After a couple of weeks, adult female root‐knot nematodes produce offspring as an aggregate of eggs held together by a gelatinous matrix (Kyndt et al., 2013).
Giant cells are a polygenic trait of nematode‐infected plants, involving hundreds of different plant genes. Studies on giant cell‐enriched root tissue from Arabidopsis thaliana infected with M. incognita revealed > 3000 differentially regulated genes in a comparison with uninfected root tissue (Jammes et al., 2005). Similarly, c. 1000 genes appeared to be differentially regulated in giant cell‐enriched tissue of M. incognita at 21 d post‐inoculation in Arabidopsis compared to uninfected tissue (Fuller et al., 2007). A similar number of differentially expressed genes were identified in a comparison of microdissected giant cells and neighbouring vascular cells in Arabidopsis at 3 d post‐inoculation with the tropical root‐knot nematode M. javanica (Barcala et al., 2010). Although not all genes regulated in association with giant cell formation will be causally linked to this process, allelic variation in specific subsets of these genes may quantitatively affect the susceptibility of a host plant to root‐knot nematodes.
Quantitative traits can be mapped onto specific genome loci by exploiting linkage disequilibrium (LD) between allelic variants (i.e. single nucleotide polymorphisms (SNPs)) and a particular trait in a set of individuals. Genome‐wide association (GWA) mapping expands on this principle by studying associations between a large number of SNPs across a genome and complex traits within a sample of genetically diverse individuals from a natural population (Zhu et al., 2008). At present, the richest resources for GWA mapping between SNPs and complex traits in plants focus on large collections of natural inbred lines of Arabidopsis (Atwell et al., 2010; Cao et al., 2011; Weigel, 2012). Arabidopsis serves as a model organism to study plant responses to all kinds of abiotic and biotic stresses, including infections by root‐knot nematodes (Sijmons et al., 1991). Genome‐wide associations between allelic variants and responses to a variety of biotic and abiotic stresses have recently been mapped onto the genome of Arabidopsis (Kloth et al., 2012, 2016; Bac‐Molenaar et al., 2015; Davila Olivas et al., 2017). Moreover, multi‐trait genome‐wide association mapping has been used to reveal cross‐correlations between SNPs and resistances to different biotic and abiotic stresses in Arabidopsis, including parallels in responses to osmotic stress and root‐knot nematodes (Thoen et al., 2017).
In theory, plants could be made more resistant to nematode infections by selecting for less conducive allelic variants of genes that critically determine susceptibility (i.e. S‐genes; de Almeida Engler et al., 2005; van Schie & Takken, 2014). Given the problems with dominant resistance genes in food crops, we asked whether plants harbour significant natural variation in susceptibility to root‐knot nematodes, which is not related to dominant resistance. Here, we present the results of a GWA study of quantitative variation in susceptibility to the root‐knot nematode M. incognita in Arabidopsis. Our primary interest was to analyse allelic variation in genes that do not resemble major resistance gene homologs. For this, we chose to work with Arabidopsis, because previous research suggested that dominant resistance to M. incognita may be absent in this species (Niebel et al., 1994). In fact, it is not so likely that the resistance gene repertoire of Arabidopsis, with its native range in more temperate regions of Europe and Asia (Beck et al., 2008), has undergone extensive adaptations to tropical root‐knot nematodes (e.g. M. incognita). Natural Arabidopsis inbred lines are also particularly well suited for GWA mapping of disease susceptibility, because they allow for repeatedly phenotyping of genetically identical individuals in notoriously variable in vitro bioassays with nematodes. In total, we found eight SNPs in our GWA study to be significantly associated with the reproductive rate of M. incognita in 340 Arabidopsis lines. By using the predicted LD decay for the Arabidopsis genome, we aggregated the SNPs into four genomic regions, two of which we examined in more detail in this paper. Our data on the candidate genes in these loci demonstrate that the transcription factor BRASSINAZOLE RESISTANT‐1 and an F‐box family protein in Arabidopsis probably (co‐)regulate susceptibility to M. incognita.
Materials and Methods
Plant material
For genome‐wide association mapping, we used a population consisting of 340 natural inbred lines selected from a global HapMap collection of Arabidopsis thaliana (L.) Heynh. (http://bergelson.uchicago.edu/wp-content/uploads/2015/04/Justins-360-lines.xls). The homozygous T‐DNA insertion mutant lines Salk_052305C (hereafter referred to as gsp1‐1) and Salk_050274C (hereafter frni1‐1), the ethyl methanesulfonate (EMS)‐induced mutant line bzr1‐1D, and the BZR1:CFP gene fusion reporter line were obtained from the Nottingham Arabidopsis Stock Centre (Alonso et al., 2003). The bzr1‐1D and BZR1:CFP lines were originally described by Wang et al. (2002). The bzr1‐1D, BZR1:CFP, gsp1‐1 and frni1‐1 lines were all generated in the background of A. thaliana Col‐0.
The homozygosity of T‐DNA inserts was checked by PCR on genomic DNA isolated from leaf material (Holterman et al., 2006) of 12 seedlings using primer combinations as indicated in Supporting Information Table S1. The following conditions were used for PCR: 10 min at 94°C, 35 cycles of 30 s at 94°C, 1.5 min at 60°C and 1 min at 72°C, and a final incubation of 10 min at 72°C. The PCR amplification products were analysed by agarose gel electrophoresis.
Nematode infection assays
Eggs of M. incognita were obtained by treating tomato roots infected with M. incognita (strain ‘Morelos’ from INRA, Sophia Antipolis, France) with 0.05% (v/v) NaOCl for 3 min. Roots were rinsed with tap water and the eggs were collected on a 25 μm sieve. Next, the eggs were incubated in a solution of 2.4 mM NaN3 for 20 min with shaking. Thereafter, the eggs were rinsed with tap water and incubated on a 25 μm sieve in a solution of 1.5 mg ml−1 gentamycin and 0.05 mg ml−1 nystatin in the dark at room temperature. Hatched juveniles were collected after 4 d and surface sterilized (0.16 mM HgCl2, 0.49 mM NaN3, 0.002% (v/v) Triton X‐100) for 10 min. After surface sterilization, the juveniles were rinsed three times with sterile tap water and transferred to 0.7% Gelrite solution (Duchefa Biochemie, Haarlem, the Netherland).
To generate cultures of Arabidopsis seedlings in vitro, seeds were vapour‐sterilized (in 0.7 M NaOCl and 1% HCl in tap water) for 5 h and transferred to a six‐well cell culture plate containing Murashige and Skoog (MS) medium with vitamins 4.7 g l−1 (Duchefa Biochemie), 58.5 mM sucrose and 5 g l−1 Gelrite (Duchefa Biochemie). The six‐well plates with seeds were incubated in the dark at 4°C for 3 d. Next, the seeds were allowed to germinate at 21°C under 16 h : 8 h, light : dark conditions. To determine the susceptibility of the 340 natural Arabidopsis inbred lines and the bzr1‐1D, gsp1‐1 and frni1‐1 mutant lines, 1‐wk‐old seedlings were manually transferred to wells in a new six‐well plate containing MS medium and incubated for another 7 d at 21°C under a 16 h : 8 h, light : dark regime. Each well contained only one seedling. Next, the seedlings were inoculated with 180 infective J2s of M. incognita per plant and incubated at 24°C in the dark.
To be able to count the number of infective juveniles in the bzr1‐1D, gsp1‐1 and frni1‐1 mutant lines at 7 d after inoculation, whole roots were stained with acid fuchsin. To this end, clean roots were first incubated in 16.8 mM NaOCl for 5 min, and thereafter in tap water for 10 min. Next, the roots were transferred into an acid fuchsin solution (0.2 M acid fuchsin and 0.8% glacial acetic acid in tap water) and heated in a microwave oven for 30 s. After cooling, roots were transferred to 40% glycerol and the number of juveniles was counted by visually inspecting the roots with a dissection microscope.
The number of egg masses per plant was counted 6 wk after inoculation by visually inspecting the roots with a dissection microscope. The natural inbred lines were screened in batches of 20 accessions, including Columbia‐0 (Col‐0) as a reference in each batch. Each inbred line was tested in at least four technical replicates. The average number of egg masses per plant, the standard error of the mean and the number of technical replicates (n) of each inbred line are summarized in Table S2. The data were analysed for narrow sense heritability and genome‐wide associations as described below.
To determine the susceptibility of the bzr1‐1D, gsp1‐1 and frni1‐1 mutant lines, the number of juveniles and egg masses per plant was statistically analysed using two‐way ANOVA and post‐hoc Tukey HSD test in R (v.3.0.2, http://www.r-project.org). Each line was tested in at least three independent experiments and 18 replicates per experiment. Both genotype and experiment number were used as factors to test for significance in the ANOVA.
To collect nematode‐infected roots for gene expression analysis by quantitative reverse transcription PCR (qRT‐PCR), freshly germinated 7‐d‐old seedlings were transferred to 12 cm square plates containing MS medium and placed vertically for a further 7 d at 21°C under a 16 h : 8 h, light : dark regime. Each plate contained four seedlings. Next, the seedlings were inoculated with 180 infective J2s of M. incognita per plant and incubated horizontally at 24°C in the dark. In parallel, seedlings in plates without juveniles were also incubated horizontally at 24°C in the dark to serve as uninfected controls. Furthermore, whole root systems of a subset of the seedlings were collected just before the inoculation with juveniles. Similarly, at 7 d after inoculation whole root systems were collected of inoculated and non‐inoculated seedlings. Root systems of 12 seedlings were aggregated to make one sample, which was snap frozen in liquid nitrogen and then stored at −80°C until further use. Three biological replicates were performed for each experiment.
Quantitative reverse transcription PCR
Expression analysis for a gene of interest was performed on the stored root samples produced during the nematode infection study. Whole root systems were cut from aerial parts of the seedlings and snap frozen in liquid nitrogen. Total RNA was isolated from whole roots of 12 14‐d‐old plants of gsp1‐1, bzr1‐1D, frni1‐1 and Col‐0 wild‐type. The frozen root systems were homogenized using TissueLyser (Qiagen) twice for 30 s. Total RNA was extracted from 100 mg of the homogenate with the Maxwell Plant RNA kit (Promega) using the Maxwell 16 Robot (Promega) according to the manufacturer's protocol. The amount of total RNA per sample was determined by an ND‐1000 spectrophotometer (Isogen Life Science, Utrecht, the Netherlands). First‐strand cDNA was synthesized from total RNA using the Superscript III First‐Strand synthesis system (Invitrogen) according to the manufacturer's protocol. Samples were analysed by quantitative PCR using Absolute SYBR Green Fluorescein mix (Thermo Fisher Scientific, Waltham, USA). cDNA matching A. thaliana elongation factor 1 alpha was amplified as a reference for constitutive expression using primers as indicated in Table S1 (Czechowski et al., 2004). To quantify the expression level for the gene of interest, specific gene primers were used (Table S1). For qRT‐PCR, 5 ng cDNA was used with the following conditions: 15 min at 95°C, 40 cycles of 30 s at 95°C, 30 s at 62°C and 30 s at 72°C, and a final incubation of 5 min at 72°C. The relative expression ratio between the gene of interest and the reference gene was calculated as described elsewhere (Pfaffl, 2001). This ratio was statistically analysed for significance with a two‐way ANOVA and post‐hoc Tukey HSD test in R (P < 0.05).
Root phenotypes
Arabidopsis seedlings were allowed to germinate and grow for 14 d on MS medium as described above. To determine the number of root tips and root length of the seedlings, the complete plants were transferred from the media onto a plastic tray with water. Next, the leaves of the seedlings were removed and the roots were spread out over the surface of the tray. A scan of the roots was made with a photo scanner (Epson Perfection V800). The scan was analysed to measure root length using WinRhizo package for Arabidopsis (WinRhizo pro2015; Regent Instruments Inc., Ville de Québec, Canada). The number of root tips was counted by visually inspecting the scan. Differences in the number of root tips and the root length per plant were statistically analysed for significance with a two‐way ANOVA and post‐hoc Tukey HSD test in R (P < 0.05).
Confocal microscopy of BZR1‐CFP
Seeds of the Arabidopsis BZR1:CFP reporter line and Col‐0 were vapour sterilized and incubated for 3 d at 4°C in the dark as described above. Next, 20 seeds were transferred to 12 cm square plates with MS media and placed vertically in a growth chamber at 21°C with a 16 h : 8 h, light : dark regime. After 5 d, the seedlings were inoculated with 25 surface‐sterilized juveniles of M. incognita per plant and placed vertically at 24°C in the dark. Three days after inoculation seedlings were transferred to a microscope slide and analysed with a Zeiss LSM 710 confocal microscope. Seedlings were incubated in 0.5 μg ml−1 propidium iodide in phosphate‐buffered saline to stain the plant cell walls. The emission spectra were set to 463–538 and 586–719 nm for cyan fluorescent protein (CFP) and propidium iodide, respectively. Non‐adjusted images were analysed with ImageJ, wherein the pixel intensity of the root area was compared to the background. Data of two independent experiments, including the analysis of 10 seedlings per experiment, were analysed with a two‐way ANOVA and post‐hoc Tukey HSD test in R. Images were enhanced in brightness for publication in print.
Narrow‐sense heritability
To estimate the amount of variation that can be explained by genome‐wide association mapping, we calculated the narrow‐sense heritability. For this, we used a mixed model approach using efficient mixed‐model association (EMMA) based on restricted maximum likelihood (REML) to estimate the variance components as described (Kang et al., 2008; Rockman et al., 2010). The kinship matrix was calculated using all 214 051 SNPs (Horton et al., 2012) and narrow sense heritability was calculated as
where V g is the genetic variance and V E is the residual variance, as estimated by REML (excluding SNPs with a frequency < 0.05 from the estimation).
Genome‐wide association mapping
Genotypic means of the egg mass data were used as input for the GWA mapping using 214 051 SNPs (Horton et al., 2012) using rrBLUP and the TAIR10 database (Yu et al., 2006; Endelman, 2011). First, a kinship matrix based on all SNPs was constructed to correct for population structure. Second, association mapping was done, excluding SNPs with a frequency < 0.05 from analysis. SNPs with a −log10(P) > 5.0 were considered significantly associated with phenotypic variance. To determine the false discovery rate at this threshold for significance, an empirical multiple testing threshold was calculated by permutation. Trait levels were randomly assigned to the genotypes, after which the association mapping was performed as described above. This procedure was repeated 1000 times resulting in a false discovery rate of 0.2 at −log10(P) > 5.0. To calculate how much of the total narrow sense heritability can be explained by significantly associating SNPs we used an additive linear model incorporating all SNPs in order to avoid a bias in SNPs capturing the same variation.
The linkage disequilibrium between SNPs was calculated using correlation analysis. First, the SNPs were converted to binary traits (either 0 or 1), which was possible because the HapMap genetic map was constructed with SNPs with only two variants per site. Per two locations the correlation between SNPs was calculated by Pearson correlation (as provided by R). The squared correlations are reported because the direction of the correlation does not confer real information (as the conversion to binary was arbitrary).
Results
Quantitative variation in susceptibility to M. incognita
Arabidopsis has not been systematically analysed for intraspecific variation in susceptibility to M. incognita before. To investigate whether Arabidopsis harbours any significant quantitative variation in susceptibility to M. incognita, we tested seedlings of 340 natural inbred lines of the Arabidopsis HapMap population with nematode bioassays in vitro. These natural inbred lines were phenotypically screened for reproductive success of M. incognita in batches of 20 accessions, including Col‐0 as reference in each batch. Approximately 60 accessions were tested multiple times in different batches to monitor consistency across different batches. Six weeks after inoculation, the average number of egg masses of M. incognita on the 340 accessions ranged from five to 45 per plant (Fig. 1; Table S2). Inoculations with M. incognita on Col‐0 resulted on average in 12 egg masses per plant. Based on our extensive phenotype screening we concluded that Arabidopsis harbours large quantitative variation in susceptibility to M. incognita.
Figure 1.
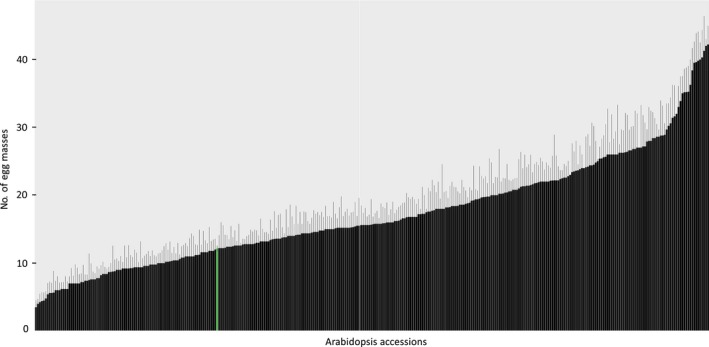
Quantitative variation in susceptibility of Arabidopsis thaliana to the root‐knot nematode Meloidogyne incognita. Average number of egg masses per plant including standard error of the mean on 340 natural inbred lines of Arabidopsis (accessions) at 6 wk after inoculation with 2nd stage juveniles of M. incognita. The green bar indicates the number of egg masses per plant for Col‐0, which was used as a reference throughout this study.
To estimate how much of the variance in reproductive success of M. incognita was caused by underlying genetic variation in the Arabidopsis lines, we calculated the narrow‐sense heritability. Using 214 051 SNPs as a basis for the genetic similarity, we estimated that 52% of the variation in susceptibility to M. incognita was attributable to additive genetic variation between the different Arabidopsis lines. We therefore decided to use our data set to identify genome‐wide associations between SNPs in Arabidopsis and susceptibility to M. incognita.
Four QTLs for susceptibility to M. incognita in Arabidopsis
To link allelic variation in Arabidopsis to reproductive success of M. incognita, we mapped genome‐wide associations underlying the number of egg masses per plant using linear mixed models (Yu et al., 2006; Endelman, 2011). Only SNPs with a minor allele frequency above 0.05 (199 252 SNPs) were included in the analysis. We identified significant associations between eight SNPs and the number of egg masses per plant 6 wk after inoculation with M. incognita in Arabidopsis (threshold for significance −log10(P) > 5; Fig. 2). Furthermore, by using an additive linear model incorporating all SNPs again, we calculated that 22% of the total variation in susceptibility of Arabidopsis can be linked to these eight SNPs.
Figure 2.
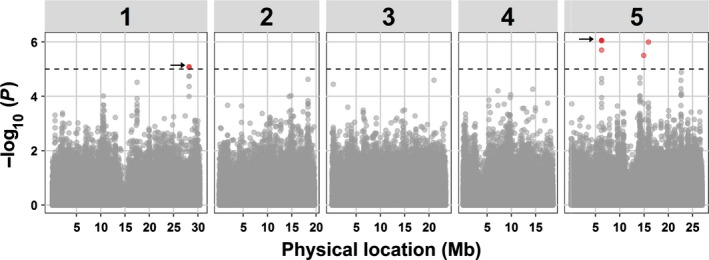
Manhattan plot of associations between 199 252 single nucleotide polymorphisms (SNPs) and the number of egg masses per plant of Meloidogyne incognita in Arabidopsis. Dashed horizontal line indicates threshold for significance in genome‐wide association mapping set at −log10(P) = 5. Red dots indicate the positions of eight significantly associated SNPs, of which five are overlapping and are indicated by the arrows. Numbers 1–5 in grey rectangles mark the five chromosomes of Arabidopsis.
LD in populations of natural inbred lines of Arabidopsis decays on average within 10 kb (Kim et al., 2007). Based on this predicted LD decay, we aggregated the eight SNPs into four QTLs located on two chromosomes (Table 1). We also analysed the specific LD between the eight significantly associated SNPs (Fig. S1). As expected, only low LD was observed between SNPs located in different QTLs (r 2 < 0.11). However, moderate LD was observed for the SNPs in QTL1 on chromosome 1 (r 2 = 0.55), while strong LD was observed for the four SNPs in QTL2 on chromosome 5 (r 2 > 0.99). The two SNPs marking QTLs 3 and 4 segregate independently (r 2 = 0.01). In conclusion, allelic variation in at least four genome locations is linked to quantitative variation in susceptibility to M. incognita in our population of Arabidopsis natural inbred lines.
Table 1.
Eight single nucleotide polymorphisms (SNPs) significantly associated with reproductive success of Meloidogyne incognita aggregate into four quantitative trait loci (QTLs) located on two chromosomes of Arabidopsis
| QTL | Chromosome | Position (bp) | SNPa | SNP frequencyb | −Log10(P)c | Effect size | SNP located in gene |
|---|---|---|---|---|---|---|---|
| 1 | 1 | 28 187 392 | C : G | 278 : 71 | 5.1 | 5.62 | At1G75080 |
| 28 188 151 | C : G | 102 : 247 | 5.1 | 5.10 | At1G75090 | ||
| 2 | 5 | 6 263 591 | A : T | 139 : 210 | 6.1 | 5.31 | At5G18780 |
| 6 263 577 | A : T | 139 : 210 | 6.1 | 5.31 | At5G18780 | ||
| 6 263 644 | A : T | 140 : 209 | 5.7 | 5.18 | At5G18780 | ||
| 6 263 678 | C : G | 139 : 210 | 6.1 | 5.31 | At5G18780 | ||
| 3 | 5 | 14 913 458 | A : T | 288 : 61 | 5.5 | 5.35 | At5G37540 |
| 4 | 5 | 15 904 331 | C : T | 290 : 59 | 6 | 4.47 | At5G39740 |
aPossible alleles for each SNP position. bFrequency of lines harbouring the SNP. cLevel of significance of the association of an individual SNP.
To further investigate the genetic architecture underlying the reproductive success of M. incognita in Arabidopsis, we focused on QTL1 and QTL2 located on chromosomes 1 and 5, respectively. QTL1 is marked by two significantly associated SNPs with moderate LD (markers Chr1.28187392 and Chr1.28188151). These two SNPs were located within 1 kb distance from each other (Table 1; Fig. 3a). SNP marker Chr1.28187392 is located in BRASSINAZOLE‐RESISTANT 1 (BZR1; AT1G75080) (Fig. 3a). The neighbouring SNP marker Chr1.28188151 is located in a predicted gene in complementary orientation encoding a putative DNA glycosylase superfamily protein (AT1G75090; hereafter named GSP1). Marker Chr1.28188151 was in strong LD (r 2 > 0.94) with three other markers at this locus (i.e. Chr1.28187959, Chr1.28187978 and Chr1.28188103), which were just below our threshold for significance in the GWA. Marker Chr1.28188103 was located in GSP1, while Chr1.28187959 and Chr1.28187978 were located in the regions where transcripts of BRZ1 and GSP1 overlap.
Figure 3.
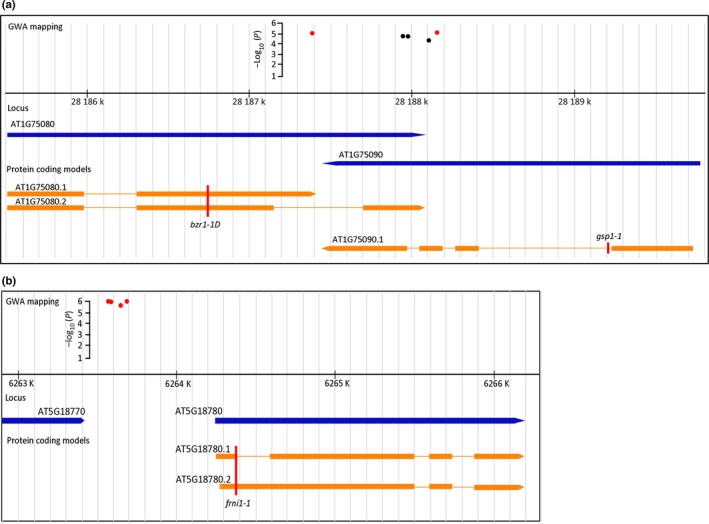
Overview of the genomic region harbouring QTL1 and QTL2 located on chromosome 1 and 5, respectively. (a) Genomic region of QTL1. The red dots represent the significantly associated single nucleotide polymorphisms (SNPs). The black dots represent three SNPs in strong linkage disequilibrium, but with −log10(P) scores below 5. The blue arrows represent two predicted genes (At1G75080 and At1G75090) in complementary orientation. Transcripts deriving from these genes are indicated in orange, with rectangles marking the protein coding exons. The red vertical line marked as bzr1‐1D indicates the position of the dominant ethyl methanesulfonate (EMS) mutation in the BZR1 gene. The red vertical line marked as gsp1‐1 indicates the position of the T‐DNA insert in a homozygous knockout mutant of GSP1. (b) Genomic region harbouring QTL2. The red dots represent the significantly associated SNPs. The blue arrows represent two predicted genes (At5G18770 and At5G18780) in similar orientation. Transcripts deriving from At5G18780 are indicated in orange, with rectangles marking the protein coding exons. The red vertical line marked as frni1‐1 indicates the position of the T‐DNA insert in a homozygous knockout mutant of FRNI1.
We used SNPs markers Chr1.28187392 and Chr1.28188151 to determine the most susceptible and the least susceptible haplotype for QTL1. Arabidopsis lines harbouring a C at Chr1.28187392 (n = 278) were less susceptible to M. incognita than those harbouring a G (n = 71). Similarly, lines harbouring a G at Chr1.28188151 (n = 247) were also less susceptible than those harbouring a C (n = 102). These polymorphisms occurred in four haplotype combinations: CC (n = 32), GC (n = 62), CG (n = 232) and GG (n = 2). Interestingly, lines with the most prevalent CG haplotype (e.g. Col‐0) were also the least susceptible to M. incognita, which could point to a selective advantage of this haplotype (Fig. S2).
Next, we focused on four significantly associated SNP markers with strong LD (i.e. Chr5.6263591, Chr5.6263577, Chr5.6263644 and Chr5.6263678) that mark QTL2 on chromosome 5. The SNPs are all located in an intergenic region c. 600 bp upstream of predicted gene At5G18780 (Fig. 3b). Two splice variants have been observed for At5G18780, both with unknown function. The protein encoded by At5G18780 is annotated as F‐box/Ribonuclease inhibitor‐like superfamily protein of 441 amino acids (hereafter FRNI1). The predicted topology of FRNI1 includes an amino terminal F‐box of 50 amino acids long (pfam 00646), seven leucine‐rich repeats with similarity to ribonuclease inhibitor 1 (RNI) and a carboxy terminal FBD domain (pfam08384) that is found in F‐box domain‐containing plant proteins.
BZR1 and FRNI1 (co‐)regulate reproductive success of M. incognita
To find further support for a role of BZR1, GSP1 and FRNI1 in susceptibility of Arabidopsis to M. incognita, we first assessed their expression levels in roots of infected and non‐infected seedlings using qRT‐PCR. Expression of the genes was determined in whole root systems collected at the time of inoculation and at 7 d after inoculation in infected and non‐infected plants. This set up allowed us to study the developmental regulation of the genes in young Arabidopsis seedlings, as well as their regulation in response to infection by M. incognita. BZR1, GSP1 and FRNI1 were all upregulated in non‐infected roots of Arabidopsis seedlings, as they developed in the 7 d after the time of inoculation (Fig. 4). Expression of both GSP1 and FRNI1 was significantly downregulated in nematode‐infected roots at the same time point after inoculation (P < 0.05). By contrast, infection by M. incognita did not alter the developmentally regulated expression of BZR1. As we used whole root systems, dilution effects could keep local changes in expression of BZR1 at the infection site below the detection limits of the qRT‐PCR. To address this concern, we also investigated the expression of BZR1 with confocal microscopy of the Arabidopsis BZR1:CFP reporter line at 3 d after inoculation with M. incognita (Fig. S3). Based on image analysis, we concluded that infections with M. incognita do not lead to significant changes in BZR1 expression at the infection site of the nematodes.
Figure 4.
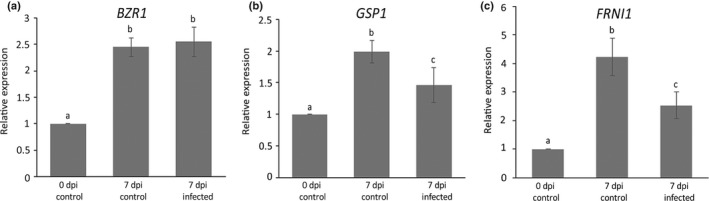
Relative expression of BZR1,GSP1 and FRNI1 in infected and noninfected roots of wild‐type Arabidopsis Col‐0 plants at 7 d after inoculation (dpi) with Meloidogyne incognita. The expression levels of (a) BZR1, (b) GSP1 and (c) FRNI1 are given as ratios relative to the expression levels of these genes at the time of inoculation. Data reflect gene expression levels in whole roots collected at the time of inoculation with M. incognita (0 dpi control), in whole roots collected at 7 d after mock‐inoculation (7 dpi control) and 7 d after inoculation with M. incognita (7 dpi infected). Bars represent average values based on three independent biological samples with three technical replicates per biological sample. Error bars represent ±SEM. Different lower‐case letters indicate statistical difference determined by ANOVA with post‐hoc Tukey HSD (P < 0.05).
To test if BZR1, GSP1 and FRNI1 are required for reproductive success of M. incognita, we challenged several Arabidopsis mutant lines with infective juveniles in a bioassay. Homozygous Arabidopsis T‐DNA knockout mutants of BZR1 have a lethal phenotype and could not be used to test the involvement of this gene in the reproductive success of M. incognita. Instead, we analysed the susceptibility of the dominant positive EMS mutant Arabidopsis line bzr1‐1D in our bioassays with M. incognita (Wang et al., 2002). Both the number of J2s of M. incognita per plant at 7 d after inoculation and the number of egg masses per plant at 6 wk after inoculation were significantly reduced on the bzr1‐1D mutant line compared to the wild‐type Arabidopsis plants (Fig. 5a). The bzr1‐1D mutant harbours a functional mutant allele of the BZR1 transcription factor that makes it insensitive to the brassinosteroid (BR) biosynthetic inhibitor brassinazole (Wang et al., 2002). Seedlings of the bzr1‐1D mutant typically show anomalous root architecture under specific light conditions (Wang et al., 2002), and this could affect susceptibility to nematode infections. Indeed, in our experimental set‐up the average total root length at the time of inoculation was significantly smaller in bzr1‐1D mutants as compared to wild‐type Col‐0 plants (Fig. 5c). More importantly, susceptibility of plants to root‐knot nematodes is known to depend on the number of available root tips at the time of inoculation. This parameter of root architecture was not significantly different between the bzr1‐1D mutant and the wild‐type Arabidopsis plants (Fig. 5b).
Figure 5.
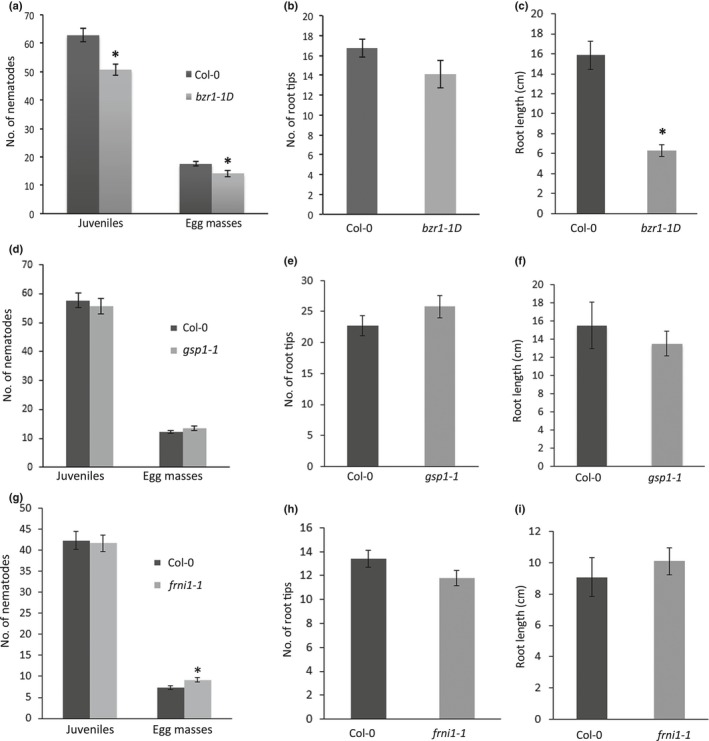
Susceptibility of a dominant positive ethyl methanesulfonate (EMS) mutant bzr1‐1D and homozygous T‐DNA insert mutants gsp1‐1 and frni1‐1 of Arabidopsis to Meloidogyne incognita. (a) Number of juveniles at 7 d after inoculation and egg masses per plant at 6 wk after inoculation on bzr1‐1D and wild‐type Arabidopsis plants. (b) Number of root tips and (c) total root length of seedlings of bzr1‐1D at the age of inoculation. (d) Number of juveniles at 7 d after inoculation and egg masses per plant at 6 wk after inoculation on gsp1‐1 and wild‐type Arabidopsis plants. (e) Number of root tips and (f) total root length of seedlings of gsp1‐1 at the age of inoculation. (g) Number of juveniles at 7 d after inoculation and egg masses per plant at 6 wk after inoculation on frni1‐1 and wild‐type Arabidopsis plants. (h) Number of root tips and (i) total root length of seedlings of frni1‐1 at the age of inoculation. (a, d, g) Bars reflect the averages and SEM of three independent experiments (n > 50). (b, c, e, f, h, i) Bars represent the mean ± SEM of three independent experiments (n > 12). Data were statistically tested for significance with ANOVA with post‐hoc Tukey HSD: *, P < 0.05.
A homozygous Arabidopsis T‐DNA mutant line was available for the GSP1 gene, which harbours an insert in the predicted first intron of the coding sequence of GSP1 (Fig. 3a). Expression of GSP1 was strongly reduced in roots of the gsp1‐1 mutant line, but not completely knocked‐out (Fig. S4a,b). Despite this reduction in gene expression, the number of egg masses per plant at 6 wk after inoculation was not significantly different in the gsp1‐1 mutant line, when compared to wild‐type plants (Fig. 5d). Similarly, the number of infective juveniles per plant at 7 d after inoculation was also not significantly different between the gsp1‐1 mutant and wild‐type plants. Furthermore, the root architecture of this mutant was not significantly different from wild‐type Arabidopsis plants (Fig. 5e,f).
BZR1 and GSP1 are located in antisense direction and their coding sequences partially overlap. BZR1 and GSP1 could therefore act as cis‐natural antisense pairs, which could lead to the formation of siRNA and thus transcript breakdown. To test this, we analysed the expression of GSP1 in bzr1‐1D and the expression of BZR1 in gsp1‐1. In a comparison with wild‐type Arabidopsis seedlings at 7 d after inoculation, the expression of GSP1 was not altered by the EMS mutation in bzr1‐1d, and vice versa the expression of BZR1 was not altered by the T‐DNA insert in gsp1‐1 (Fig. 6). Our data therefore showed that it is unlikely that the phenotype of the bzr1‐1D mutation arises through it actions on transcript levels of GSP1.
Figure 6.
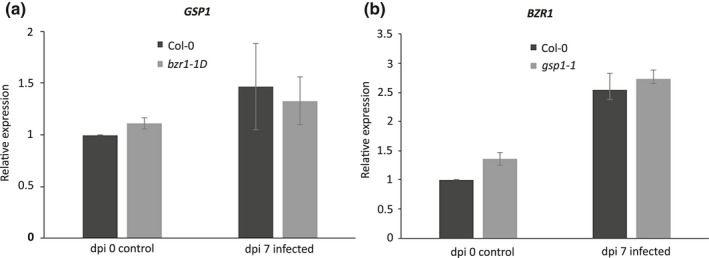
Mutation in the Arabidopsis bzr1‐1D line does not affect expression of neighbouring GSP1, and vice versa. (a) Relative expression of GSP1 in bzr1‐1D and wild‐type Arabidopsis plants at 7 d after inoculation (dpi) with Meloidogyne incognita. (b) Relative expression of BZR1 in gsp1‐1 and wild‐type Arabidopsis plants at 7 d after inoculation with M. incognita. Gene expression levels are determined by quantitative reverse transcription PCR and presented here as a ratio relative to the expression level in wild‐type Arabidopsis plants at the time of inoculation. Mars represent average values based on three independent biological samples with three technical replicates per biological sample. Error bars represent ± SEM.
A homozygous Arabidopsis knock‐out line was also available for the FRNI1 gene, which harbours a T‐DNA insert in the first predicted exon of FRNI1 (Fig. 3b). qRT‐PCR showed that the expression of FRNI1 was completely knocked‐out in roots of the frni1‐1 mutant line (Fig. S4c,d). The number of egg masses on roots of the frni1‐1 mutant line at 6 wk after inoculation was significantly higher as compared to the wild‐type Arabidopsis plants (Fig. 5g). By contrast, we observed no significant difference in the number of juveniles inside roots at 7 d after inoculation between frni1‐1 and wild‐type Arabidopsis plants. The root architecture of the frni1‐1 mutant was also not significantly different from wild‐type Arabidopsis plants (Fig. 5h,i). Altogether, we concluded that BZR1 and FRNI probably function as (co‐)regulators of reproductive success of M. incognita in Arabidopsis. Allelic variation in these genes may therefore contribute to quantitative variation in susceptibility to M. incognita in our collection of Arabidopsis lines. By contrast, despite its downregulation in association with nematode infections, allelic variation in GSP1 is less likely to be causal for quantitative variation in susceptibility of Arabidopsis to M. incognita.
BZR1 is a master regulator of both cell proliferation, differentiation and defence. As such it regulates cell elongation, which is evident from the reduced root growth phenotype of the bzr1‐1D mutant. Reduced cell growth may affect the expansion of nematode‐induced giant cells, but we could not exclude the possibility that the bzr1‐1D mutant is also affected in its ability to mount a defence response to M. incognita. We therefore analysed the expression of markers for salicylic acid‐ and jasmonic acid‐related defence responses (i.e. At2G14610 (PR1) and A5G44120 (PDF1.2)) and a marker for cellular expansion (i.e. At2G28950 (EXP6)) in nematode‐infected roots of the bzr1‐1D mutant line and wild‐type Arabidopsis. Surprisingly, the expression of PR1 was constitutively and highly upregulated at the time of inoculation and at 7 d after inoculation in both infected and non‐infected bzr1‐1D mutants when compared to wild‐type Arabidopsis plants (Fig. 7a). By contrast, the marker genes for jasmonic acid‐dependent defences and cellular expansion were not differentially regulated in bzr1‐1D and wild‐type Arabidopsis (Fig. 7b,c). The function of FRNI1 is not known, and we therefore conducted a similar marker gene experiment on nematode‐infected roots of the frni1‐1 mutant line. However, none of the marker genes was differentially regulated between frni1‐1 and wild‐type Arabidopsis plants (Fig. 7d–f).
Figure 7.
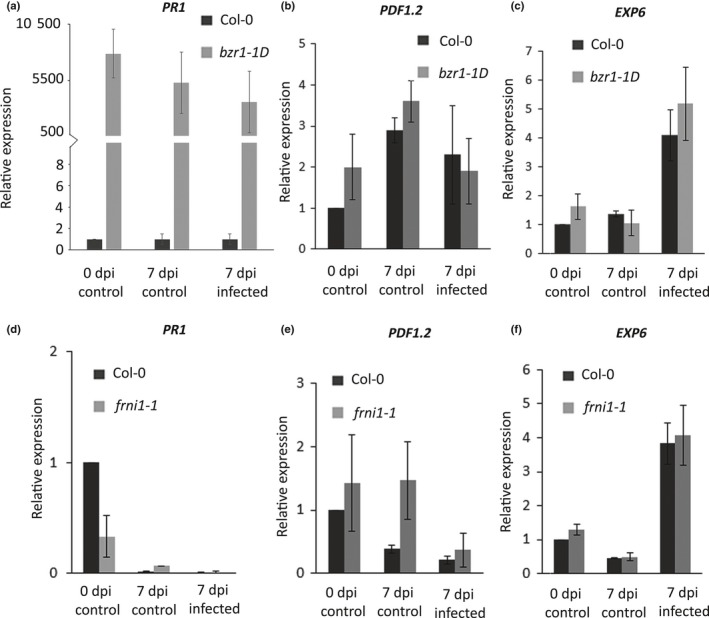
Differential expression of marker genes for salicylic acid‐ (PR1) and jasmonic acid‐ (PDF1.2) dependent defence responses and plant cell expansion (EXP6) in the bzr1‐1D and frni1‐1 mutant and wild‐type Arabidopsis plants. Relative expression levels of (a) PR1, (b) PDF1.2 and (c) EXP6 in bzr1‐1D and wild‐type Arabidopsis plants. Relative expression levels of (d) PR1, (e) PDF1.2 and (f) EXP6 in gsp1‐1 and wild‐type Arabidopsis plants. Gene expression levels are determined with quantitative reverse transcription PCR and presented here as a ratio relative to the expression level in wild‐type Arabidopsis plants at the time of inoculation. Data reflect expression in whole roots collected at the time of inoculation with Meloidogyne incognita (0 d post‐inoculation (dpi) control), in whole roots collected at 7 d after mock‐inoculation (7 dpi control) and 7 d after inoculation with M. incognita (7 dpi infected). Bars represent average values based on three independent biological samples with three technical replicates per biological sample. Error bars indicate SEM.
Discussion
Most genetic analyses of wild relatives of crop plant species have focused on identifying new sources of dominant resistance to nematodes, while largely disregarding natural variation in susceptibility. Here, we used GWA mapping to assess the genetic underpinnings of a large variation in susceptibility to the root‐knot nematode M. incognita in a population of natural inbred lines of Arabidopsis. By applying a −log10(P) score of 5 as a threshold for significance (corresponding to a false discovery rate of 0.2), we identified four QTLs in Arabidopsis associated with the number of egg masses of M. incognita at 6 wk post‐inoculation (Fig. 2). These four QTLs probably harbour allelic variants that are causally related to reproductive success of M. incognita on Arabidopsis.
The ability of Arabidopsis to support reproduction of root‐knot nematodes is thought to be a complex trait involving many different plant genes (Jammes et al., 2005; Dubreuil et al., 2007; Barcala et al., 2010; Portillo et al., 2013; Cabrera et al., 2014). Given the anticipated complexity of this trait, the number of QTLs significantly associated with reproduction of M. incognita in our GWA study was relatively small. It is possible that the genetic architecture underlying susceptibility of Arabidopsis to M. incognita is much simpler than previously thought. However, it should be noted that only 22% of the observed phenotypic variance can be linked to the four QTLs identified our GWA study. This could indicate the presence of an abundance of rare alleles (with minor allele frequencies below 0.05), which are capturing most of the variation but were excluded from GWA mapping. Alternatively, many QTLs for susceptibility to M. incognita in the Arabidopsis genome may have small effect sizes that cannot be detected with the resolution of our bioassays. In either case, our GWA study probably underestimates the complexity of the genetic architecture of susceptibility to M. incognita in Arabidopsis.
Others have investigated the genetic architecture of responses to biotic stresses in Arabidopsis by accepting a less stringent threshold for significant associations in GWA mapping (e.g. −log10(P) > 4; El‐Soda et al., 2015; Kloth et al., 2016; Kooke et al., 2016; Davila Olivas et al., 2017). Similarly relaxing the stringency in our analysis would result in significant associations between 36 SNPs (located within 19 QTLs) and reproduction of M. incognita on Arabidopsis. Lowering the threshold for significance in the GWA mapping may thus reveal more common alleles with smaller effect sizes in our population of Arabidopsis lines (Korte & Farlow, 2013; Kooke et al., 2016). However, this would also raise the false discovery rate to 60%, which would reduce the chances of identifying causal genes in follow‐up studies.
To assess whether our GWA study (using stringent criteria) can help to identify genes involved in susceptibility of Arabidopsis to M. incognita, we focused on two SNPs located in QTL1 on chromosome 1 (Fig. 3). Chr1.28187392 showed moderate LD (r 2 = 0.55) with Ch1.28188151, while LD seems to rapidly decay with SNPs directly flanking Chr1.28187392 and Chr1.28188151. Based on the locations of the two significant SNPs and the predicted LD decay in this region, we concluded that BZR1 and GSP1 were the only two candidates in this region that could contribute to the variance in susceptibility of Arabidopsis to M. incognita.
Our infection assays with the dominant positive bzr1‐1D mutant line showed that BZR1 probably acts as a rate‐limiting factor in the reproductive success of M. incognita in Arabidopsis (Fig. 5). The number of juveniles inside seedlings during the early stages of parasitism and the number of egg masses at 6 wk post‐inoculation was consistently smaller on the bzr1‐1D mutant when compared to wild‐type Col‐0. BZR1 is constitutively active in the bzr1‐1D mutant line, which simulates the accumulation of BR (Wang et al., 2002). Under specific light conditions the dominant bzr1‐1D mutation results in an anomalous root architecture. In our nematode infection assays the number of root tips at the time of inoculation was not significantly different between the bzr1‐1D mutant line and wild‐type Col‐0. This is important because the invasion of Arabidopsis by M. incognita occurs only in the transition zone close to root tips (Sijmons et al., 1991). However, the reduced total root length of the bzr1‐1D mutant could point to defects in cell growth, which may affect the expansion of nematode‐induced giant cells.
The transcription factor BZR1 is at the end of a signalling cascade which is activated by the BRI1/BAK1 co‐receptor complex upon detection of brassinolide (Jaillais & Vert, 2016). The activation of BR signalling in Arabidopsis results in the dephosphorylation and translocation to the nucleus of BZR1, where it binds to DNA and specifically activates or represses the expression of almost 1000 genes (Sun et al., 2010; Di Rubbo et al., 2011). BR signalling plays a crucial role in determining cell growth by promoting elongation of differentiated cells, but also by regulating the transition between cell cycle progression and cell differentiation (Jaillais & Vert, 2016). Aberrant progression through the mitotic cell cycle, extensive cell elongation and expansion are all considered essential steps in the ontogeny of giant cells in nematode‐infected roots of A. thaliana (de Almeida Engler & Gheysen, 2013; Kyndt et al., 2013; Vieira et al., 2013). The outcome of BR signalling in roots is cell type and position specific, but generally antagonises the effect of auxin (Chaiwanon & Wang, 2015). BZR1 regulates the expression of several genes related to auxin biosynthesis and signalling (Sun et al., 2010). For instance, BZR1 directly represses the expression of PIN auxin efflux carriers involved in directing polar auxin transport towards root tips (i.e. PIN3 and PIN4; Feraru & Friml, 2008; Sun et al., 2010; Vragović et al., 2015). Recently, it was shown that development of M. incognita is hampered on Arabidopsis knockout mutants of PIN3 and PIN4 (Kyndt et al., 2016). PIN3 and PIN4 are thought to be involved in redirecting the flow of auxin during giant cell formation. Similarly, BZR1 regulates the expression of genes involved in plant cell wall plasticity, which is a fundamental requirement for cell growth but also for the expansion of giant cells (e.g. EXPA1; Jammes et al., 2005; Sun et al., 2010).
Furthermore, BZR1 also regulates the trade‐off between growth and immunity, which may explain the constitutive upregulation of PR1 that we observed in the bzr1‐1D mutant (Lozano‐Duran et al., 2013; Lozano‐Durán & Zipfel, 2015). The loss of susceptibility to M. incognita in the bzr1‐1D mutant could therefore also reflect alterations in basal immunity of Arabidopsis seedlings. This latter scenario would be in agreement with the recently observed enhanced resistance of transgenic Lotus japonicus plants ectopically expressing the Arabidopsis bzr1‐1D allele to feeding by onion thrips (Miyaji et al., 2014). Altogether, we conclude that BZR1 probably (co‐)regulates susceptibility to M. incognita in A. thaliana through its role in plant cell growth, basal defence or both. Allelic variation in BZR1 could therefore be casual for some of the observed variance in reproductive success of M. incognita in our population of Arabidopsis lines.
The second SNP marker significantly associated with the number of egg masses of M. incognita per plant in QTL1 was located in the first exon of GSP1, a putative DNA glycosylase superfamily protein. The function of GSP1 has not been studied before, but based on sequence homology it is predicted to be involved in base‐excision repair of DNA (Manova & Gruszka, 2015). Despite the fact that GSP1 is strongly downregulated upon infection by M. incognita at 7 d after inoculation, the homozygous knockdown mutant Arabidopsis line of GSP1 in the Col‐0 background showed no altered susceptibility to M. incognita. This suggests that GSP1 is regulated in association with, but not required for, reproduction of M. incognita in Arabidopsis. However, it should be noted that the Arabidopsis Col‐0 line carries the nonsusceptible haplotype (i.e. CG) for this locus, and that a knockdown by the T‐DNA insert in gsp1‐1 may therefore not lead to further reduction in susceptibility. In conclusion, we have found no evidence suggesting that allelic variation in GSP1 significantly contributes to the variance in susceptibility of Arabidopsis to M. incognita.
Four co‐segregating SNPs marking QTL2 on chromosome 5 pointed at FRNI1 as a co‐regulator of susceptibility of Arabidopsis to M. incognita. The SNP markers are located in the putative regulatory region upstream of the predicted coding sequence of FRNI1, where they might affect expression levels of this gene. Unlike BZR1, FRNI1 is strongly downregulated in nematode‐infected roots of wild‐type Arabidopsis Col‐0 plants. Alterations in FRNI1 expression are therefore likely to affect the susceptibility of Arabidopsis to M. incognita. In fact, our data showed that the compete loss of FRNI1 expression in the frni1‐1 knockout mutant resulted in a small but significant increase in the number of egg masses per plant. So far, no function has been ascribed to FRNI1, but its architecture as an F‐box‐like and RNI‐like protein suggests that it might be involved in protein–protein interactions. More specifically, the F‐box is defined as a component of the E3 ubiquitin ligase complex SCF (Skp1‐cullin‐F‐box protein ligase), which targets proteins to the 26S proteasome for degradation (Lechner et al., 2006). F‐box‐containing proteins are involved in many cellular process in plants, including hormone signalling and defence responses. The lack of differential expression of PR1, PDF1.2 and EXP6 in nematode‐infected roots of frni1‐1 mutant and wild‐type Arabidopsis plants offered no clue as to whether FRNI1 co‐regulates susceptibility by affecting defence, development or both. Further investigations are therefore needed to shed light on the function of FRNI1 in Arabidopsis.
The main objective of this study was to explore the natural variation in susceptibility to M. incognita of Arabidopsis, which is thought to lack dominant resistance genes to this nematode species. In our phenotype screening of the Arabidopsis lines we observed an unexpected large variation in reproductive success of M. incognita. Extensive variation in susceptibility was also observed within a smaller set of 45 Arabidopsis inbred lines challenged with the northern root‐knot nematode Meloidogyne hapla (Boiteux et al., 1999). As the natural distribution of M. hapla and A. thaliana in temperate regions may have overlapped, this variation could be partly based on segregating major resistance genes. We found no evidence by GWA mapping that the large phenotypic variance in susceptibility to M. incognita is based on the presence of segregating dominant resistance genes linked to any of the QTLs. By contrast, GWA mapping of susceptibility to Meloidogyne graminicola in rice cultivars identified 11 genomic loci, at least one of which harbours major resistance gene homologues (Dimkpa et al., 2016). Our data thus indicate that plants could be made more resistant to infections of root‐knot nematodes by selecting unfavourable alleles of S‐genes that are essential for giant cell initiation, expansion and maintenance. However, it remains to be investigated if these loss‐of‐susceptibility alleles can be exploited by plant breeders to improve the resilience of crops without experiencing undesirable pleiotropic effects on other agronomically important traits.
Author contributions
J.B., A.G., G.S., M.D., J.H. and S.W. conceived and designed the experiments. S.W., M.G.S., M.E.P.O., C.v.S., O.C.A.S. and J.L.L‐T. performed the experiments. S.W., M.G.S., J.L.L‐T., J.E.K., A.G., J.B. and G.S. analysed the data. S.W., M.G.S., J.E.K., A.G., J.B. and G.S. wrote the article. All authors edited and approved the final article.
Supporting information
Please note: Wiley Blackwell are not responsible for the content or functionality of any Supporting Information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
Fig. S1 Linkage disequilibrium (LD) between eight SNPs in Arabidopsis significantly associated with the number of egg masses of Meloidogyne incognita per plant.
Fig. S2 Haplotype‐specific susceptibility of Arabidopsis to Meloidogyne incognita.
Fig. S3 Expression of BZR1:CFP fusion protein under the control of the endogenous BZR1 promoter sequence in roots of Arabidopsis seedlings infected with Meloidogyne incognita.
Fig. S4 Strongly reduced expression of GSP1 and FRNI1 in the homozygous knockout Arabidopsis line gsp1‐1 and frni1‐1, respectively.
Table S1 Primers used for RT‐PCR
Table S2 Number of egg masses of Meloidogyne incognita per plant on 340 natural inbred lines of Arabidopsis thaliana at 6 wk after inoculation
Acknowledgements
We thank Willem Kruijer for his support with GWA mapping, and Hein Overmars and Anna Finkers‐Tomczak for help with setting up the nematode bioassays. We are also grateful for the financial support provided by the Netherlands Organization for Scientific Research (NWO, Perspectives Programme ‘Learning from Nature to Protect Crops’; STW grant 10997).
References
- de Almeida Engler J, Favery B, Engler G, Abad P. 2005. Loss of susceptibility as an alternative for nematode resistance. Current Opinion in Biotechnology 16: 112–117. [DOI] [PubMed] [Google Scholar]
- de Almeida Engler J, Gheysen G. 2013. Nematode‐induced endoreduplication in plant host cells: why and how? Molecular Plant‐Microbe Interactions 26: 17–24. [DOI] [PubMed] [Google Scholar]
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R et al 2003. Genome‐wide insertional mutagenesis of Arabidopsis thaliana . Science 301: 653–657. [DOI] [PubMed] [Google Scholar]
- Atwell S, Huang YS, Vilhjalmsson BJ, Willems G, Horton M, Li Y, Meng D, Platt A, Tarone AM, Hu TT et al 2010. Genome‐wide association study of 107 phenotypes in Arabidopsis thaliana inbred lines. Nature 465: 627–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bac‐Molenaar JA, Fradin EF, Becker FFM, Rienstra JA, van der Schoot J, Vreugdenhil D, Keurentjes JJB. 2015. Genome‐wide association mapping of fertility reduction upon heat stress reveals developmental stage‐specific QTLs in Arabidopsis thaliana . Plant Cell 27: 1857–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barcala M, Garcia A, Cabrera J, Casson S, Lindsey K, Favery B, Garcia‐Casado G, Solano R, Fenoll C, Escobar C. 2010. Early transcriptomic events in microdissected Arabidopsis nematode‐induced giant cells. Plant Journal 61: 698–712. [DOI] [PubMed] [Google Scholar]
- Bebber DP, Holmes T, Gurr SJ. 2014. The global spread of crop pests and pathogens. Global Ecology and Biogeography 23: 1398–1407. [Google Scholar]
- Beck JB, Schmuths H, Schaal BA. 2008. Native range genetic variation in Arabidopsis thaliana is strongly geographically structured and reflects Pleistocene glacial dynamics. Molecular Ecology 17: 902–915. [DOI] [PubMed] [Google Scholar]
- Boiteux LS, Fonseca MEN, Simon PW. 1999. Host status and reaction of Arabidopsis thaliana ecotypes to infection by the northern root‐knot nematode (Meloidogyne hapla). Plant Breeding 118: 355–358. [Google Scholar]
- Cabrera J, Barcala M, Fenoll C, Escobar C. 2014. Transcriptomic signatures of transfer cells in early developing nematode feeding cells of Arabidopsis focused on auxin and ethylene signaling. Frontiers in Plant Science 5: 107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caillaud M‐C, Dubreuil G, Quentin M, Perfus‐Barbeoch L, Lecomte P, de Almeida Engler J, Abad P, Rosso M‐N, Favery B. 2008. Root‐knot nematodes manipulate plant cell functions during a compatible interaction. Journal of Plant Physiology 165: 104–113. [DOI] [PubMed] [Google Scholar]
- Cao J, Schneeberger K, Ossowski S, Gunther T, Bender S, Fitz J, Koenig D, Lanz C, Stegle O, Lippert C et al 2011. Whole‐genome sequencing of multiple Arabidopsis thaliana populations. Nature Genetics 43: 956–963. [DOI] [PubMed] [Google Scholar]
- Castagnone‐Sereno P, Danchin EGJ, Perfus‐Barbeoch L, Abad P. 2013. Diversity and evolution of root‐knot nematodes, genus Meloidogyne: new insights from the genomic era. Annual Review of Phytopathology 51: 203–220. [DOI] [PubMed] [Google Scholar]
- Chaiwanon J, Wang Z‐Y. 2015. Spatiotemporal brassinosteroid signaling and antagonism with auxin pattern stem cell dynamics in Arabidopsis roots. Current Biology 25: 1031–1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czechowski T, Bari RP, Stitt M, Scheible W‐R, Udvardi MK. 2004. Real‐time RT‐PCR profiling of over 1400 Arabidopsis transcription factors: unprecedented sensitivity reveals novel root‐ and shoot‐specific genes. Plant Journal 38: 366–379. [DOI] [PubMed] [Google Scholar]
- Davies LJ, Elling AA. 2015. Resistance genes against plant‐parasitic nematodes: a durable control strategy? Nematology 17: 249–263. [Google Scholar]
- Davila Olivas NH, Kruijer W, Gort G, Wijnen CL, van Loon JJ, Dicke M. 2017. Genome‐wide association analysis reveals distinct genetic architectures for single and combined stress responses in Arabidopsis thaliana . New Phytologist 213: 838–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Rubbo S, Irani NG, Russinova E. 2011. PP2A phosphatases: the “On‐Off” regulatory switches of brassinosteroid signaling. Science Signaling 4: pe25. [DOI] [PubMed] [Google Scholar]
- Dimkpa SO, Lahari Z, Shrestha R, Douglas A, Gheysen G, Price AH. 2016. A genome‐wide association study of a global rice panel reveals resistance in Oryza sativa to root‐knot nematodes. Journal of Experimental Botany 67: 1191–1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubreuil G, Magliano M, Deleury E, Abad P, Rosso MN. 2007. Transcriptome analysis of root‐knot nematode functions induced in the early stages of parasitism. New Phytologist 176: 426–436. [DOI] [PubMed] [Google Scholar]
- El‐Soda M, Kruijer W, Malosetti M, Koornneef M, Aarts MGM. 2015. Quantitative trait loci and candidate genes underlying genotype by environment interaction in the response of Arabidopsis thaliana to drought. Plant, Cell & Environment 38: 585–599. [DOI] [PubMed] [Google Scholar]
- Endelman JB. 2011. Ridge regression and other kernels for genomic selection with R package rrBLUP. Plant Genome 4: 250–255. [Google Scholar]
- Feraru E, Friml J. 2008. PIN polar targeting. Plant Physiology 147: 1553–1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller VL, Lilley CJ, Atkinson HJ, Urwin PE. 2007. Differential gene expression in Arabidopsis following infection by plant‐parasitic nematodes Meloidogyne incognita and Heterodera schachtii . Molecular Plant Pathology 8: 595–609. [DOI] [PubMed] [Google Scholar]
- Holterman M, van der Wurff A, van den Elsen S, van Megen H, Bongers T, Holovachov O, Bakker J, Helder J. 2006. Phylum‐wide analysis of SSU rDNA reveals deep phylogenetic relationships among nematodes and accelerated evolution toward crown clades. Molecular Biology and Evolution 23: 1792–1800. [DOI] [PubMed] [Google Scholar]
- Horton MW, Hancock AM, Huang YS, Toomajian C, Atwell S, Auton A, Muliyati NW, Platt A, Sperone FG, Vilhjalmsson BJ et al 2012. Genome‐wide patterns of genetic variation in worldwide Arabidopsis thaliana accessions from the RegMap panel. Nature Genetics 44: 212–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacquet M, Bongiovanni M, Martinez M, Verschave P, Wajnberg E, Castagnone‐Sereno P. 2005. Variation in resistance to the root‐knot nematode Meloidogyne incognita in tomato genotypes bearing the Mi gene. Plant Pathology 54: 93–99. [Google Scholar]
- Jaillais Y, Vert G. 2016. Brassinosteroid signaling and BRI1 dynamics went underground. Current Opinion in Plant Biology 33: 92–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jammes F, Lecomte P, de Almeida‐Engler J, Bitton F, Martin‐Magniette M‐L, Renou JP, Abad P, Favery B. 2005. Genome‐wide expression profiling of the host response to root‐knot nematode infection in Arabidopsis. Plant Journal 44: 447–458. [DOI] [PubMed] [Google Scholar]
- Jones JT, Haegeman A, Danchin EGJ, Gaur HS, Helder J, Jones MGK, Kikuchi T, Manzanilla‐López R, Palomares‐Rius JE, Wesemael WML et al 2013. Top 10 plant‐parasitic nematodes in molecular plant pathology. Molecular Plant Pathology 14: 946–961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaloshian I, Williamson VM, Miyao G, Lawn DA, Westerdahl BB. 1996. ‘Resistance‐breaking’ nematodes identified in California tomatoes. California Agriculture 50: 18–19. [Google Scholar]
- Kang HM, Zaitlen NA, Wade CM, Kirby A, Heckerman D, Daly MJ, Eskin E. 2008. Efficient control of population structure in model organism association mapping. Genetics 178: 1709–1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Plagnol V, Hu TT, Toomajian C, Clark RM, Ossowski S, Ecker JR, Weigel D, Nordborg M. 2007. Recombination and linkage disequilibrium in Arabidopsis thaliana . Nature Genetics 39: 1151–1155. [DOI] [PubMed] [Google Scholar]
- Kloth KJ, Thoen MPM, Bouwmeester HJ, Jongsma MA, Dicke M. 2012. Association mapping of plant resistance to insects. Trends in Plant Science 17: 311–319. [DOI] [PubMed] [Google Scholar]
- Kloth KJ, Wiegers GL, Busscher‐Lange J, van Haarst JC, Kruijer W, Bouwmeester HJ, Dicke M, Jongsma MA. 2016. AtWRKY22 promotes susceptibility to aphids and modulates salicylic acid and jasmonic acid signalling. Journal of Experimental Botany 67: 3383–3396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kooke R, Kruijer W, Bours R, Becker F, Kuhn A, van de Geest H, Buntjer J, Doeswijk T, Guerra J, Bouwmeester H et al 2016. Genome‐wide association mapping and genomic prediction elucidate the genetic architecture of morphological traits in Arabidopsis. Plant Physiology 170: 2187–2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korte A, Farlow A. 2013. The advantages and limitations of trait analysis with GWAS: a review. Plant Methods 9: 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyndt T, Goverse A, Haegeman A, Warmerdam S, Wanjau C, Jahani M, Engler G, de Almeida Engler J, Gheysen G. 2016. Redirection of auxin flow in Arabidopsis thaliana roots after infection by root‐knot nematodes. Journal of Experimental Botany 67: 4559–4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyndt T, Vieira P, Gheysen G, Almeida‐Engler J. 2013. Nematode feeding sites: unique organs in plant roots. Planta 238: 807–818. [DOI] [PubMed] [Google Scholar]
- Lechner E, Achard P, Vansiri A, Potuschak T, Genschik P. 2006. F‐box proteins everywhere. Current Opinion in Plant Biology 9: 631–638. [DOI] [PubMed] [Google Scholar]
- Lozano‐Duran R, Macho AP, Boutrot F, Segonzac C, Somssich IE, Zipfel C. 2013. The transcriptional regulator BZR1 mediates trade‐off between plant innate immunity and growth. eLife 2: e00983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozano‐Durán R, Zipfel C. 2015. Trade‐off between growth and immunity: role of brassinosteroids. Trends in Plant Science 20: 12–19. [DOI] [PubMed] [Google Scholar]
- Manova V, Gruszka D. 2015. DNA damage and repair in plants – from models to crops. Frontiers in Plant Science 6: 885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyaji T, Yamagami A, Kume N, Sakuta M, Osada H, Asami T, Arimoto Y, Nakano T. 2014. Brassinosteroid‐related transcription factor BIL1/BZR1 increases plant resistance to insect feeding. Bioscience Biotechnology and Biochemistry 78: 960–968. [DOI] [PubMed] [Google Scholar]
- Niebel A, Barthels N, deAlmeida‐Engler J , Karimi M, Vercauteren I, Montagu MV, Gheysen G. 1994. Arabidopsis thaliana as a model host plant to study molecular interactions with root‐knot and cyst nematodes In: Lamberti F, De Giorgi C, Bird DM, eds. Advances in molecular plant nematology. Boston, MA, USA: Springer US, 161–170. [Google Scholar]
- Pfaffl MW. 2001. A new mathematical model for relative quantification in real‐time RT‐PCR. Nucleic Acids Research 29: e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portillo M, Cabrera J, Lindsey K, Topping J, Andres MF, Emiliozzi M, Oliveros JC, Garcia‐Casado G, Solano R, Koltai H et al 2013. Distinct and conserved transcriptomic changes during nematode‐induced giant cell development in tomato compared with Arabidopsis: a functional role for gene repression. New Phytologist 197: 1276–1290. [DOI] [PubMed] [Google Scholar]
- Rockman MV, Skrovanek SS, Kruglyak L. 2010. Selection at linked sites shapes heritable phenotypic variation in C. elegans . Science 330: 372–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Schie CCN, Takken FLW. 2014. Susceptibility genes 101: how to be a good host. Annual Review of Phytopathology 52: 551–581. [DOI] [PubMed] [Google Scholar]
- Semblat JP, Bongiovanni M, Wajnberg E, Dalmasso A, Abad P, Castagnone‐Sereno P. 2000. Virulence and molecular diversity of parthenogenetic root‐knot nematodes, Meloidogyne spp. Heredity 84: 81–89. [DOI] [PubMed] [Google Scholar]
- Sijmons PC, Grundler FMW, von Mende N, Burrows PR, Wyss U. 1991. Arabidopsis thaliana as a new model host for plant‐parasitic nematodes. Plant Journal 1: 245–254. [Google Scholar]
- Sun Y, Fan X‐Y, Cao D‐M, Tang W, He K, Zhu J‐Y, He J‐X, Bai M‐Y, Zhu S, Oh E et al 2010. Integration of brassinosteroid signal transduction with the transcription network for plant growth regulation in Arabidopsis. Developmental Cell 19: 765–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoen MP, Davila Olivas NH, Kloth KJ, Coolen S, Huang PP, Aarts MG, Bac‐Molenaar JA, Bakker J, Bouwmeester HJ, Broekgaarden C et al 2017. Genetic architecture of plant stress resistance: multi‐trait genome‐wide association mapping. New Phytologist 213: 1346–1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira P, Escudero C, Rodiuc N, Boruc J, Russinova E, Glab N, Mota M, De Veylder L, Abad P, Engler G et al 2013. Ectopic expression of Kip‐related proteins restrains root‐knot nematode‐feeding site expansion. New Phytologist 199: 505–519. [DOI] [PubMed] [Google Scholar]
- Vragović K, Sela A, Friedlander‐Shani L, Fridman Y, Hacham Y, Holland N, Bartom E, Mockler TC, Savaldi‐Goldstein S. 2015. Translatome analyses capture of opposing tissue‐specific brassinosteroid signals orchestrating root meristem differentiation. Proceedings of the National Academy of Sciences, USA 112: 923–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ZY, Nakano T, Gendron J, He J, Chen M, Vafeados D, Yang Y, Fujioka S, Yoshida S, Asami T et al 2002. Nuclear‐localized BZR1 mediates brassinosteroid‐induced growth and feedback suppression of brassinosteroid biosynthesis. Developmental Cell 2: 505–513. [DOI] [PubMed] [Google Scholar]
- Weigel D. 2012. Natural variation in Arabidopsis: from molecular genetics to ecological genomics. Plant Physiology 158: 2–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson VM, Kumar A. 2006. Nematode resistance in plants: the battle underground. Trends in Genetics 22: 396–403. [DOI] [PubMed] [Google Scholar]
- Yu J, Pressoir G, Briggs WH, Vroh Bi I, Yamasaki M, Doebley JF, McMullen MD, Gaut BS, Nielsen DM, Holland JB et al 2006. A unified mixed‐model method for association mapping that accounts for multiple levels of relatedness. Nature Genetics 38: 203–208. [DOI] [PubMed] [Google Scholar]
- Zhu C, Gore M, Buckler ES, Yu J. 2008. Status and prospects of association mapping in plants. Plant Genome 1: 5–20. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Please note: Wiley Blackwell are not responsible for the content or functionality of any Supporting Information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
Fig. S1 Linkage disequilibrium (LD) between eight SNPs in Arabidopsis significantly associated with the number of egg masses of Meloidogyne incognita per plant.
Fig. S2 Haplotype‐specific susceptibility of Arabidopsis to Meloidogyne incognita.
Fig. S3 Expression of BZR1:CFP fusion protein under the control of the endogenous BZR1 promoter sequence in roots of Arabidopsis seedlings infected with Meloidogyne incognita.
Fig. S4 Strongly reduced expression of GSP1 and FRNI1 in the homozygous knockout Arabidopsis line gsp1‐1 and frni1‐1, respectively.
Table S1 Primers used for RT‐PCR
Table S2 Number of egg masses of Meloidogyne incognita per plant on 340 natural inbred lines of Arabidopsis thaliana at 6 wk after inoculation


