Abstract
Rilpivirine (RPV) and Etravirine (ETR) are approved second-generation non-nucleoside reverse transcriptase inhibitors (NNRTIs) for HIV treatment. There is a cross-resistance HIV mutation profile between first- and second-generation NNRTI drugs. We determined the prevalence of HIV-1 drug resistance mutations (DRMs) to RPV and ETR in Botswana. A total of 168 HIV-1 polymerase gene sequences from participants failing nevirapine (NVP)- or efavirenz (EFV)-containing regimens were analyzed for DRMs using the Stanford University HIV drug resistance database. Forty-one sequences were from an adult antiretroviral therapy (ART) study, the Tshepo study, and 127 from a prevention of mother-to-child transmission (PMTCT) study, the Mashi study, all conducted in Botswana. Prevalence of RPV and ETR highest DRM in the adult ART study (n = 41) were K101E (26.2%), E138A (23.8%), and Y181C (26.2%). The PMTCT cohort's (n = 127) high prevalence mutations were Y181C (15.7%), E138A (15%), and K101E (11%). A total of 42.9% and 3.2% of patients in the adult ART study and PMTCT study, respectively, had three or more NNRTI mutations at virologic failure. We identified HIV-1 mutations conferring resistance to RPV and ETR even though they have not been used in Botswana. Of concern was the high proportion of sequences from the adult ART study that displayed multiple DRMs; as the number of NNRTI mutations increases, the level of cross-resistance increases. It is plausible that patients displaying such profiles maybe at increased risk of failing second-generation NNRTI drugs, hence, calls for genotyping in patients with prior NVP or efavirenz exposure before prescription of RPV- or ETR-containing cART.
Keywords: : rilpivirine, etravirine, drug resistance, nevirapine, efavirenz
Introduction
Non-nucleoside reverse transcriptase inhibitors (NNRTIs) continue to be the mainstay of combination antiretroviral therapy (cART) especially in developing countries.1,2 The clinical use of first-generation NNRTIs has been limited by side effects, a low barrier to resistance, and broad cross-resistance.3 New second-generation NNRTIs, etravirine (ETR) and rilpivirine (RPV), have been recently approved for human immunodeficiency virus type 1 (HIV-1) treatment.4,5
ETR binds and adjusts to the changes within the non-nucleoside reverse transcriptase binding pocket to inhibit viral replication.6 The possession of a unique chemical structure of RPV that can also adapt to changes in the HIV-1 reverse transcriptase binding pocket possibly gives the drug an advantage over first-generation NNRTIs.7,8
A recent in vitro study focusing on the patterns of HIV-1 drug resistance mutations (DRMs) reported that, in the absence of other mutations, ETR and RPV are likely to select for the E138K as a major mutation.9 More than 12 DRMs have been reported for ETR and RPV across different drug resistance algorithms, studies, and databases.10 Two phase-three randomized trials comparing a regimen containing efavirenz (EFV) to an RPV-containing regimen in a treatment-naive population revealed a favorable safety profile with higher incidence of virologic failures in RPV than in EFV arm.11,12
Botswana was the first country in sub-Saharan Africa, in 2002, to offer cART to all HIV patients qualifying for treatment.13 The treatment program is considered a success as evidenced by high levels of an ART access and viral suppression rates.14 Like most HIV treatment programs in developing countries, NNRTIs have formed a cornerstone of the treatment and prevention of mother-to-child transmission (PMTCT) regimens. To our knowledge, there is currently no published data from Botswana on the prevalence of both ETR and RPV DRMs in ETR and RPV-naive patients. In this article, we report the prevalence of ETR and RPV DRMs in HIV-1 subtype C-infected patients who were on nevirapine (NVP)- or EFV-based cART.
Materials and Methods
Study population
We analyzed HIV sequence data from ETR and RPV-related DRMs from a PMTCT study, the Mashi study15 and adult ART study, the Tshepo study,16,17 both conducted in Botswana between 2002 and 2007. The Mashi study was a PMTCT study, which enrolled pregnant HIV-infected women between June 2002 and August 2003. The Mashi study objective was to assess the equivalence of maternal single-dose NVP versus placebo, on a backdrop of both maternal and infant zidovudine (ZDV), in prevention of HIV transmission to infants. Some of the enrolment criteria used were that these women be ≥18 years old, between 33- and 35-week gestational age, and did not have known ZDV or NVP intolerance.18
We randomly selected 232 samples from the Mashi study participating mothers were genotyped for HIV DRMs at 1-month postpartum time point. A total of 127 of the maternal viral sequences with NNRTI resistance mutations were included in the current analysis.
The adult ART study (Tshepo study) was an open-label unblinded 3 × 2 × 2 factorial design study that compared six highly active ART regimen arms as used in the first-line treatment of HIV.16 The antiretroviral drugs used in the various treatment arms were ZDV, lamivudine (3TC), NVP, EFV, stavudine (d4T), and didanosine (ddI). The study evaluated efficacy, tolerability, and the development of drug resistance in a cohort of 650 adult participants. The treatment arms drug combinations compared were as follows: arm A (ZDV/3TC/NVP), arm B (ZDV/3TC/EFV), arm C (ZDV/ddI/NVP), arm D (ZDV/ddI/EFV), arm E (d4T/NVP/3TC), and arm F (d4T/3TC/EFV).16 As of August 21, 2007, 72 participants in Tshepo had virologic failure. From virologic failures with resistance, all 41 viral sequences with NNRTI resistance mutations were used for this analysis.
HIV genotypic drug resistance analysis
All specimens were genotyped using the HIV-1 Viroseq Genotyping System v2.0 (Celera Diagnostics, Alameda, CA) and sequences represented part of the polymerase gene (pol), covering the whole of HIV protease and the first 335 codons of the reverse transcriptase, as previously performed.19 Sequences were screened for hypermutation using the online Hypermut tool from Los Alamos (www.hiv.lanl.gov/content/sequence/HYPERMUT/hypermut.html). The protease and reverse transcriptase-recognized mutations were interpreted according to the Stanford HIV drug resistance database (http://hivdb.stanford.edu).
DRMs and interpretation
The list of ETR and RPV resistance mutations used in this study were as those recorded by previous studies.11,12,20 The following reverse transcriptase mutations were considered as resistance mutations for ETR: V90I, A98G, L100I/V, K101E/H/P, V106A/I/M, E138A/G/K/Q, V179D/E/F/I/L/M/T, Y181C/I/S/V, Y188C/H/L, G190A/C/E/Q/S/T/V, P225H, F227C, M230 L, and K238 N/T. For RPV, the following were considered as resistance mutations: V90I, L100I, K101E/P/T, V106A/I, V108I, E138A/G/K/Q/R, V179F/I/L, Y181C/I/V, Y188I, G190E, H221Y, F227C/L, and M230I/L.
HIV subtyping
Subtyping was conducted using the REGA HIV-1 Automated Subtyping Tool Version 3.0.21 Phylogenetic trees were reconstructed using Maximum Likelihood Phylogenetic analysis based on the General Time Reversible model implemented in MEGA 6. Subtypes were also confirmed with Stanford HIV drug resistance database analysis.22,23
Ethical considerations
The Mashi and Tshepo studies received ethical approval from the Health Research and Development Committee of the Ministry of Health in Botswana and the Office of Human Research Administration at the Harvard T.H. Chan School of Public Health in the United States of America. All the enrolled participants provided written informed consent.
Results
ETR and RPV resistance-associated mutation prevalence
Upon checking with the Stanford HIV drug resistance database, we did not identify any of the 168 sequences to have any hypermutations, so all the sequences were included in the analysis (Table 1).
Table 1.
Demographic Data in the Tshepo and Mashi Patients Who Were Evaluated for Drug Resistance Mutations
| Tshepo | Mashi | |
|---|---|---|
| Gender N (%) | ||
| Female | 27 (65.9) | 127 (100) |
| Male | 14 (34.1) | 0 (0) |
| Age in years, median (IQR) | 33.4 (29.2, 40.5) | 24.5 (21, 28.3) |
| CD4+ T cells median, cells/μL (IQR) | 269 (179, 411) | 440 (357.5, 664.3) |
| Viral load median, copies/mL (IQR) | 9860 (5290, 42300) | 7195 (3430, 26595) |
IQR, interquartile range.
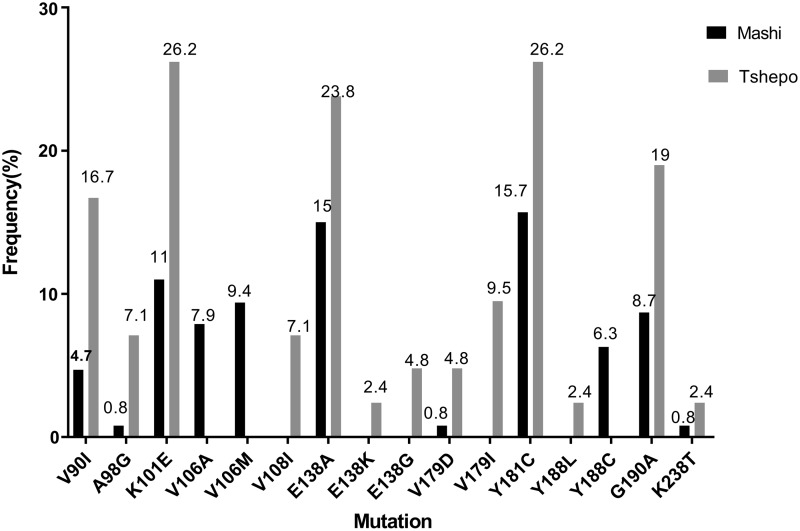
The Y181C mutation was the most prevalent mutation at 15.7% in the PMTCT study (Mashi study), followed by E138A at 15%. The K101E mutation occurred at 11%, whereas the rest of the other mutations observed were below a prevalence of 10% (Fig. 1). Within the Tshepo study (the adult antiretroviral study), patients were randomized to six different combinations of ART, each of these either containing NVP or EFV. The most prevalent mutations in the adult ART study were the Y181C and K101E (both at 26.2%) followed by the E138A (23.8%) (Fig. 1).
FIG. 1.
Prevalence of ETR and/or RPV DRMs at 1-month postpartum in a prevention of mother-to-child transmission of HIV study (Mashi study, N = 127) and at failure in the adult antiretroviral study (Tshepo study, N = 41) in Botswana. DRM, drug resistance mutation; ETR, etravirine; RPV, rilpivirine.
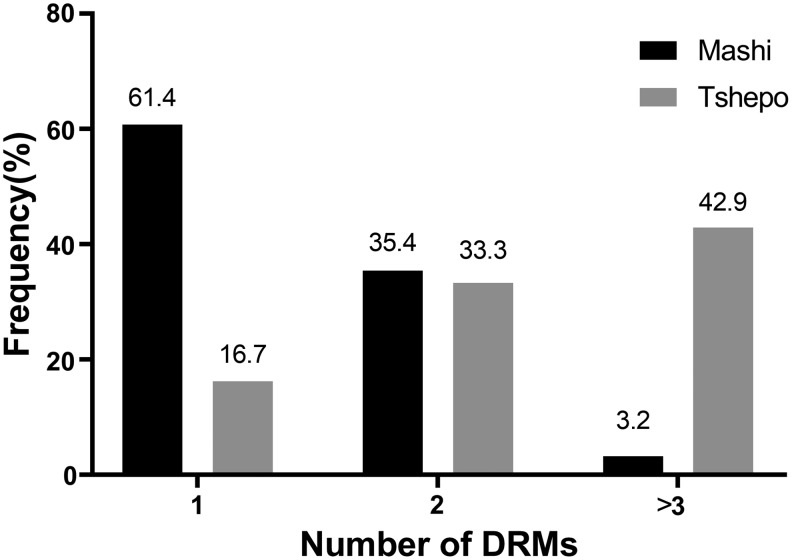
The proportion of Mashi genotypes that had 1, 2, and 3 DRMs was 61.4%, 35.4%, and 3.2%, respectively (Fig. 2). Tshepo genotypes that had 1 DRM and 2 DRMs were 16.7% and 33.3%, respectively. Three or more DRMs were found in 42.9% of the Tshepo genotypes.
FIG. 2.
Number of ETR and/or RPV DRMs in analyzed HIV-1 genotypes of patients failing a nevirapine or efavirenz-containing regimen in Tshepo (n = 41) and Mashi (n = 127) studies.
The proportions of analyzed genotypes that had the NNRTI resistance mutations also carried the K103 N mutation at frequencies of 62.2% and 17.3% for Mashi and Tshepo, respectively, (data not shown).
NNRTI mutation patterns in patients with more than one DRM
The only distinguished common pattern of NNRTI resistance in the Tshepo study was the concurrent appearance of the K101E and G190A (7.3%) in three patients. Six common NNRTI DRM patterns were observed in the Mashi study, and they were as follows: K101E + E138A (3.1%), K101E + K103 N (4.7%), K103 N + G190A (6.3%), K103 N + V106A (6.3%), K103 N + Y181C (5.5%), and K103 N + Y188C (3.1%).
HIV subtyping
All the 168 analyzed sequences for the two studies were HIV-1 subtype C.
Discussion
To our knowledge, this is the first study to show the prevalence of mutations conferring resistance to two recently approved second-generation NNRTIs, ETR and RPV, in HIV-1 subtype C-infected patients in Botswana. Two patient cohorts were used: that of women who used NVP-containing treatment for PMTCT of HIV and also that of patients failing NVP or EFV-containing cART. The lack of data pertaining to DRMs to the second-generation NNTRIs in Botswana and elsewhere warrants research such as reported by this article. Botswana is yet to introduce ETR and RPV, therefore, our study is timely and could inform future prescription practices of such drugs for HIV-infected patients in the country.
The most prevalent mutation was Y181C. This mutation causes high level resistance to NVP and intermediate level resistance to both ETR and RPV and was determined in 26.2% of the genotypes. The observed Y181C mutation prevalence in this article is higher than that of other NNRTI-treated HIV-1 subtype C patients from various studies (based on analysis of 5553 HIV-1 subtype C viral isolates).23 The E138A mutation that leads to reduced susceptibility to both ETR and RPV was found at prevalence of 23.8% and 15% in the Tshepo and Mashi studies, respectively, values that are higher than expected in HIV-1 subtype C NNRTI-experienced patients (6.4%).23 Recent research has demonstrated that the E138A substitution mutation occurs more frequently in HIV-1 subtype C viruses than subtype B viruses, which is of concern since this virus clade forms almost half of all HIV-1 infections globally.24,25
The mutations occurring at codon 138 of the HIV reverse transcriptase have been associated with selection by the human leukocyte antigen B18 (HLA-B*18)-restricted cytotoxic T lymphocytes.26 This implies that the development of the E138A mutation depends on host genetics, however, this observation for this particular mutation has not been verified elsewhere, and thus provides an opportunity to conduct such research in our HIV-1-infected population. The HLA is known to be influential in the evolution and diversification of HIV-1 within individuals and populations, and also that positive selection of HIV escape mutations increases its chances for survival.27,28
The observed prevalence of 26.2% for the K101E mutation in the Tshepo cohort and 11% in Mashi cohort is also high compared with expected frequencies (7.8%) within HIV-1 subtype C NNRTI-experienced populations. The K101E mutation confers intermediate resistance to RPV and NVP, whereas it leads to low resistance for ETR and EFV.
The risks of NNRTIs cross-resistance involving ETR and RPV in the Mashi cohort remained low as was evident from the low proportions (3.2%) of genotypes with three or more DRMs shown in Figure 2. However, risk for the Tshepo cohort was higher as the proportion of genotypes with 2 and ≥3 DRMs was 33.3% and 42.9%, respectively. Interpretation of these results should be done keeping in mind that none of the patients whose data were analyzed here was ever exposed to either ETR or RPV. Furthermore, ETR and RPV share an almost similar resistance profile, as well as, to some extent, with the first-generation NNRTIs.
A small number of patients (3 of 41) with a similar pattern of NNRTI resistance carried varying nucleoside reverse transcriptase inhibitors (NRTI) genotypes (data not shown), which follows an observation of lack of association between a unique thymidine analogue mutation genotype noted by Novitsky et al.29 in the same cohort we studied. The two NNRTI mutations (K101E and G190A) described for the mentioned three patients imply that if these patients were to be switched to second-generation NNRTIs at the time of failure, chances of further failure would be high, although with intermediate resistance to ETR. We are limited in this case to state possible influence of the drug–drug interaction(s) on mutational outcome. The other limiting factor for such a conclusion is the number of patients identified as having the observed patterns of NNRTI resistance in Tshepo. Associations of mutations across regimens (NNRTIs and NRTIs) were not made for Mashi because of homogeneity of treatment in this study.
A limitation to this study could be the possible underestimation of DRMs by the method that was used for genotyping, namely the Viroseq HIV-1 genotyping system. The system has been shown to have a higher failure rate for drug resistance testing in the nonsubtype B HIV-1 viruses. The difficulty with amplification by some of the Viroseq sequencing primers leads to the generation of single-strand sequences, which, in turn, makes calling of ambiguous bases a challenge.30 Higher failure rates of the Viroseq sequencing primers ranged from 3% to 76% in non-Subtype B viruses in Central Africa.31Utilization of more sensitive assays such as deep sequencing assays and allele-specific PCR and other assays that address HIV-1 genetic variability could assist in such instances.
To our knowledge, this is the first report on the prevalence of DRMs to second-generation NNRTIs in Botswana. The data provide a guide into the expected drug resistance profile for patients who have had prior exposure to either NVP- or EFV-containing regimens. It is hoped that findings of this study would influence prescription practices and policy regarding the use of these second-generation NNRTIs in the country. It is thus recommended that, before initiating or switching to newer antiretroviral drugs, drug resistance genotyping should be performed to avoid issuing drugs that might not be effective to patients with respective drug resistant viral variants.
Sequence Data
GenBank accession numbers for the sequences utilized in this study are MF278361 to MF278528
Acknowledgments
We are grateful to the study participants of both the Mashi and Tshepo studies. We would also like to thank the principal investigators of these studies for permitting usage of data collected during these trials. This work was supported by the Southern Africa Consortium for Research Excellence (grant no. 087537/F/08/Z) from Wellcome Trust. T.D and S.G are partly funded by Human Health and Heredity H3ABioNet (grant number 1 U41HG006941-01) from NIH. S.G is partially supported through the Sub-Saharan African Network for TB/HIV Research Excellence (SANTHE), a DELTAS Africa Initiative [grant no. DEL-15-006]. The DELTAS Africa Initiative is an independent funding scheme of the African Academy of Sciences (AAS)'s Alliance for Accelerating Excellence in Science in Africa (AESA) and supported by the New Partnership for Africa's Development Planning and Coordinating Agency (NEPAD Agency) with funding from the Wellcome Trust [grant no. 107752/Z/15/Z] and the U.K. government. The views expressed in this publication are those of the author(s) and not necessarily those of AAS, NEPAD Agency, Wellcome Trust, or the U.K. government.
Author Disclosure Statement
The authors declare no conflict of interest.
References
- 1.Riddler SA, Haubrich R, DiRienzo AG, et al. : Class-sparing regimens for initial treatment of HIV-1 infection. N Engl J Med 2008;358:2095–2106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ministry of Health Botswana: Botswana National HIVand AIDS National Treatment Guidelines Ministry of Health, Gaborone, 2012 [Google Scholar]
- 3.Anta L, Llibre JM, Poveda E, et al. : Rilpivirine resistance mutations in HIV patients failing non-nucleoside reverse transcriptase inhibitor-based therapies. AIDS 2013;27:81–85 [DOI] [PubMed] [Google Scholar]
- 4.FDA notifications. Rilpivirine approved for HIV treatment. AIDS Alert 2011;26:82–84 [PubMed] [Google Scholar]
- 5.FDA approves new HIV drug after priority review. U.S. Food and Drug Administration. AIDS Patient Care STDs 2008;22:159–161 [DOI] [PubMed] [Google Scholar]
- 6.Das K, Jain B, Patel HS: Nile blue in Triton-X 100/benzene-hexane reverse micelles: A fluorescence spectroscopic study. Spectrochim Acta A Mol Biomol Spectrosc 2004;60:2059–2064 [DOI] [PubMed] [Google Scholar]
- 7.Xu HT, Colby-Germinario SP, Huang W, et al. : Role of the K101E substitution in HIV-1 reverse transcriptase in resistance to rilpivirine and other non-nucleoside reverse transcriptase inhibitors. Antimicrob Agents Chemother 2013;57:5649–5657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Das K, Bauman JD, Clark AD Jr, et al. : High-resolution structures of HIV-1 reverse transcriptase/TMC278 complexes: Strategic flexibility explains potency against resistance mutations. Proc Natl Acad Sci U S A 2008;105:1466–1471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Asahchop EL, Wainberg MA, Oliveira M, et al. : Distinct resistance patterns to etravirine and rilpivirine in viruses containing non-nucleoside reverse transcriptase inhibitor mutations at baseline. AIDS 2013;27:879–887 [DOI] [PubMed] [Google Scholar]
- 10.Wensing AM, Calvez V, Gunthard HF, et al. : Update of the drug resistance mutations in HIV-1. Top Antivir Med 2017;2017:132–133 [PMC free article] [PubMed] [Google Scholar]
- 11.Molina JM, Cahn P, Grinsztejn B, et al. : Rilpivirine versus efavirenz with tenofovir and emtricitabine in treatment-naive adults infected with HIV-1 (ECHO): A phase 3 randomised double-blind active-controlled trial. Lancet 2011;378:238–246 [DOI] [PubMed] [Google Scholar]
- 12.Cohen CJ, Andrade-Villanueva J, Clotet B, et al. : Rilpivirine versus efavirenz with two background nucleoside or nucleotide reverse transcriptase inhibitors in treatment-naive adults infected with HIV-1 (THRIVE): A phase 3, randomised, non-inferiority trial. Lancet 2011;378:229–237 [DOI] [PubMed] [Google Scholar]
- 13.UNAIDS: UNAIDS Report on the Global AIDS Epidemic | 2010. Joint United Nations Programme on HIV/AIDS (UNAIDS), Geneva, Switzerland, 2010 [Google Scholar]
- 14.Gaolathe T, Wirth KE, Holme MP, et al. : Botswana's progress toward achieving the 2020 UNAIDS 90-90-90 antiretroviral therapy and virological suppression goals: A population-based survey. Lancet HIV 2016;3:e221–e230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thior I, Lockman S, Smeaton LM, et al. : Breastfeeding plus infant zidovudine prophylaxis for 6 months vs formula feeding plus infant zidovudine for 1 month to reduce mother-to-child HIV transmission in Botswana: A randomized trial: The Mashi Study. JAMA 2006;296:794–805 [DOI] [PubMed] [Google Scholar]
- 16.Wester CW, Thomas AM, Bussmann H, et al. : Non-nucleoside reverse transcriptase inhibitor outcomes among combination antiretroviral therapy-treated adults in Botswana. AIDS 2010;24(Suppl 1):S27–S36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bussmann H, Wester CW, Thomas A, et al. : Response to zidovudine/didanosine-containing combination antiretroviral therapy among HIV-1 subtype C-infected adults in Botswana: Two-year outcomes from a randomized clinical trial. J Acquir Immune Defic Syndr 2009;51:37–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shapiro RL, Thior I, Gilbert PB, et al. : Maternal single-dose nevirapine versus placebo as part of an antiretroviral strategy to prevent mother-to-child HIV transmission in Botswana. AIDS 2006;20:1281–1288 [DOI] [PubMed] [Google Scholar]
- 19.Doualla-Bell F, Gaseitsiwe S, Ndungu T, et al. : Mutations and polymorphisms associated with antiretroviral drugs in HIV-1C-infected African patients. Antivir Chem Chemother 2004;15:189–200 [DOI] [PubMed] [Google Scholar]
- 20.Anta L, Blanco JL, Llibre JM, et al. : Resistance to the most recent protease and non-nucleoside reverse transcriptase inhibitors across HIV-1 non-B subtypes. J Antimicrob Chemother 2013;68:1994–2002 [DOI] [PubMed] [Google Scholar]
- 21.Pineda-Pena AC, Faria NR, Imbrechts S, et al. : Automated subtyping of HIV-1 genetic sequences for clinical and surveillance purposes: Performance evaluation of the new REGA version 3 and seven other tools. Infect Genet Evol 2013;19:337–348 [DOI] [PubMed] [Google Scholar]
- 22.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S: MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 2013;30:2725–2729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stanford University. HIV Drug Resistance Database. Available at http://hivdb.stanford.edu/cgi-bin/MutPrevBySubtypeRx.cgi, accessed March 24, 2018.
- 24.Sluis-Cremer N, Jordan MR, Huber K, et al. E138A in HIV-1 reverse transcriptase is more common in subtype C than B: Implications for rilpivirine use in resource-limited settings. Antiviral Res 2014;107:31–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Osmanov S, Pattou C, Walker N, Schwardlander B, Esparza J: Estimated global distribution and regional spread of HIV-1 genetic subtypes in the year 2000. J Acquir Immune Defic Syndr 2002;29:184–190 [DOI] [PubMed] [Google Scholar]
- 26.Gatanaga H, Murakoshi H, Hachiya A, et al. : Naturally selected rilpivirine-resistant HIV-1 variants by host cellular immunity. Clin Infect Dis 2013;57:1051–1055 [DOI] [PubMed] [Google Scholar]
- 27.Rambaut A, Posada D, Crandall KA, Holmes EC: The causes and consequences of HIV evolution. Nat Rev Genet 2004;5:52–61 [DOI] [PubMed] [Google Scholar]
- 28.Carlson JM, Le AQ, Shahid A, Brumme ZL: HIV-1 adaptation to HLA: A window into virus–host immune interactions. Trends Microbiol 2015;23:212–224 [DOI] [PubMed] [Google Scholar]
- 29.Novitsky V, Wester CW, DeGruttola V, et al. : The reverse transcriptase 67N 70R 215Y genotype is the predominant TAM pathway associated with virologic failure among HIV type 1C-infected adults treated with ZDV/ddI-containing HAART in southern Africa. AIDS Res Hum Retroviruses 2007;23:868–878 [DOI] [PubMed] [Google Scholar]
- 30.Jagodzinski LL, Cooley JD, Weber M, Michael NL: Performance characteristics of human immunodeficiency virus type 1 (HIV-1) genotyping systems in sequence-based analysis of subtypes other than HIV-1 subtype B. J Clin Microbiol 2003;41:998–1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mouafo LC, Pere H, Ndjoyi-Mbiguino A, et al. : LETTER to the EDITOR performance of the ViroSeq(R) HIV-1 genotyping system v2.0 in central Africa. Open AIDS J 2015;9:9–13 [DOI] [PMC free article] [PubMed] [Google Scholar]