Abstract
Background
Neurodegenerative disorders (NDs) are diverse group of disorders characterized by escalating loss of neurons (structural and functional). The development of potential therapeutics for NDs presents an important challenge, as traditional treatments are inefficient and usually are unable to stop or retard the process of neurodegeneration. Computer-Aided Drug Design (CADD) has emerged as an efficient means of developing candidate drugs for the treatment of many disease types. Applications of CADD approach to drug discovery are progressing day by day. The recent tendency in drug design is to rationally design potent therapeutics with multi-targeting effects, higher efficacies, and fewer side effects, especially in terms of toxicity.
Methods
A wide literature search was performed for writing this review. An updated view on different types of NDs, their effect on human population and a brief introduction to CADD, various approaches involved in this technique, ranging from structural-based to ligand-based drug design has been discussed. The successful application of CADD approaches for the treatment of neurodegenerative disorders is also included in this review.
Results
In this review, we have briefly described about CADD and its use in the development of the therapeutic drug candidates against NDs. The successful applications, limitations and future prospects of this approach have also been discussed.
Conclusion
CADD can assist researchers studying interactions between drugs and receptors. We believe this review will be helpful for better understanding of CADD and its applications towards the discovery of new drug candidates against various fatal NDs.
Keywords: CADD, NDs, QSAR, molecular docking, homology modeling, SBDD, LBDD
1. BACKGROUND
The development of novel potential therapies for the treatment of NDs represents an important means of extending life span and quality of life in the elderly [1]. The development of potential therapeutics for NDs presents an important challenge, as traditional treatments are inefficient and usually are unable to stop or retard the process of neurodegeneration [2], which is highly complex and includes many neuropathological conditions and cognitive function losses, such as, memory and learning losses. NDs cause neuron loss and brain aging, which eventually lead to death. In fact, it has been estimated that more than 25% of global deaths and disabilities are caused by brain-associated disorders [3], such as, Alzheimer’s disease (AD), Parkinson’s disease (PD), Huntington’s disease (HD) and amyotrophic lateral sclerosis (ALS) [4, 5]. Available treatments are limited, and no truly effective drug is available for many NDs. Furthermore, the number of drugs approved is limited by the high failure rates of lead compounds in clinical trials [6].
Identifying novel, potential drugs for NDs is difficult using traditional approaches of drug discovery [7]. However, during the last decade, computers have been used to aid and accelerate the process of drug discovery, and this process is now referred to as computer-aided drug design (CADD) or computer-assisted molecular design (CAMD).
Computer-Aided Drug Design (CADD) emerged as an efficient means of identifying potential lead compounds and for aiding the developments of possible drugs for a wide range of diseases [8, 9]. Today, a number of computational approaches are being used to identify potential lead molecules from huge compound libraries.
Applications of CADD approach to drug discovery are progressing on a daily basis. The recent tendency in drug design is to rationally design potent therapeutics with
multi-targeting effects, higher efficacies, and fewer side effects, especially in terms of toxicity. In this review, we provide a brief introduction to CADD and include details of structure-based drug design (SBDD) and ligand-based drug design (LBDD), and their uses to identify potential drug candidates for NDs. In addition, we provide an up-to-date summary of the successes and limitations of CADD against NDs and discuss its future prospects.
2. COMPUTER AIDED DRUG DESIGN (CADD)
To introduce a new drug to the market is a costly affair that involves considerable time and money. The average time taken to discover/develop a drug is around 10-15 years and the cost stands at around US$ 800 million [10-12]. Not surprisingly, pharmaceutical companies focus on reducing development times and budgets without adversely affecting quality. In the 1990’s, a large number of developments were undertaken using combinatorial and high-throughput screening technologies, which accelerated drug discovery [13-15]. These technologies were widely adopted because they enabled the rapid synthesis and screening of large libraries, but unfortunately, no significant success was achieved and little progress toward the development of new molecular entities was made [16, 17].
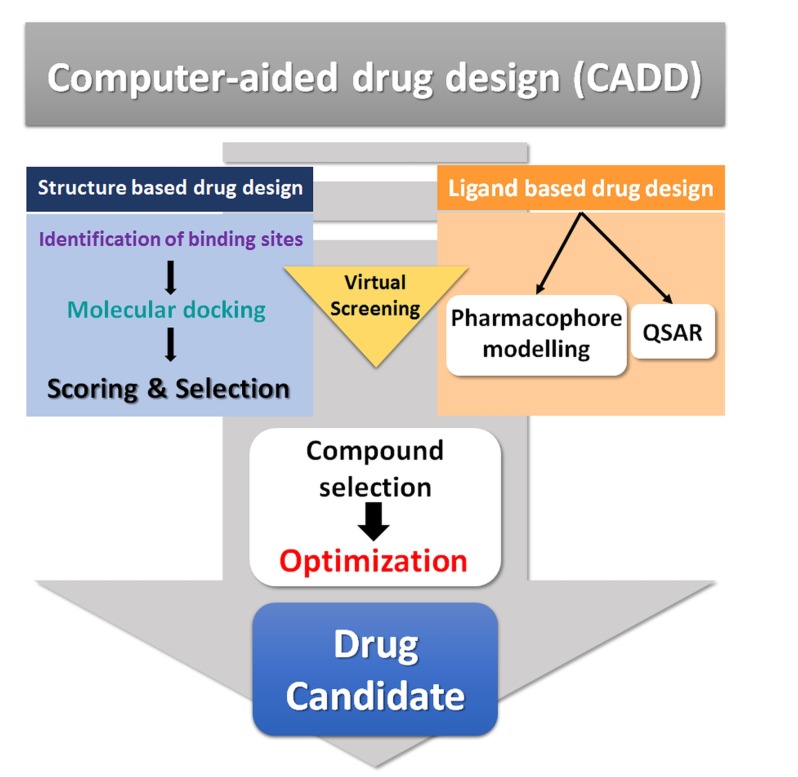
A combination of advanced computational techniques, biological science, and chemical synthesis was introduced to facilitate the discovery process, and this combinational approach enhanced the scale of discovery. Eventually, the term computer-aided drug design (CADD) was adopted for the use of computers in drug discovery [17, 18]. Advanced computational applications have been shown to be effective tools and notable successes have been achieved using these techniques. CADD is a specialized discipline, whereby different computational methods are used to simulate interactions between receptors and drugs in order to determine binding affinities [19]. However, the technique is not limited to studies of chemical interactions and binding affinity predictions, as it has many more applications ranging from the design of compounds with desired physiochemical properties to the management of digital repositories of compounds. An overview of CADD is provided in Fig. (1). CADD may be broadly categorized embracing both structure- and ligand-based drug design. Fig. (2) illustrates various approaches applied in CADD.
Fig. (1).
Overview of CADD process.
Fig. (2).
Various approaches applied in CADD.
Virtual screening (VS) is a computational technique used for screening large datasets of molecules, and has been successfully used to complement High Throughput Screening (HTS) for drug discovery [8, 20, 21]. The major aim of VS is to enable the rapid, cost-effective evaluation of huge virtual compound databases to screen for effective leads for synthesis and further study [22]. Virtual database screening can be applied to screen large libraries of compounds using various computational approaches to identify those entities likely to bind to a molecular target of interest [23, 24]. To a large extent, VS mitigates the problem of drug synthesis because it utilizes large libraries of pre-synthesized compounds.
2.1. Structure-based Drug Design (SBDD)
Structure-based drug design utilizes protein three-dimensional (3D) structural information to design new biologically active molecules [25]. Thus, the identification of a target molecule and the determination of its structure is the main, initial step of SBDD [23, 26]. The identified target may be an enzyme associated with a disease of interest. Based on binding affinity determinations, potential compounds are determined which attenuates the activity of target by its inhibition. Thus, SBDD utilizes information about a biological target and identifies potentially new medications. As such SBDD constitutes a marked advancement in the computational techniques used in the biophysics, medicinal chemistry, statistics, biochemistry, and other fields [27]. Scientific advancements have resulted in a large number of techniques for predicting protein structures. These state-of-the-art technologies enable the determination of the structures of large numbers of proteins by using cryo-electron microscopy (EM), nuclear magnetic resonance (NMR), X-ray crystallography and computational methods like homology modeling and molecular dynamic (MD) simulation [28].
2.1.1. Homology Modeling
Determining the structure of a target molecule follows the identification of a specific drug target [29]. Despite the availability of advanced techniques, the structures of a large number of proteins have not been identified [30]. Homology modelling helps in this situation because it can be used to generate the structures of proteins on information available for similar proteins [31].
Structural information about an identified target is a prerequisite for SBDD, but the structures of several identified neurodegenerative drug targets have yet not been determined [32, 33]. A large number of studies have been conducted using the homology modelling approach to generate structures of identified target molecules. Structural information is also required to gain insights of protein activities. Dhanavade et al. generated the structure of cysteine protease, which degrades amyloid beta peptide, an important causative agent of Alzheimer’s disease [34]. Several in silico experiments have been conducted using the modeled structure of cysteine protease to investigate the nature of its binding site.
2.1.2. Molecular Docking
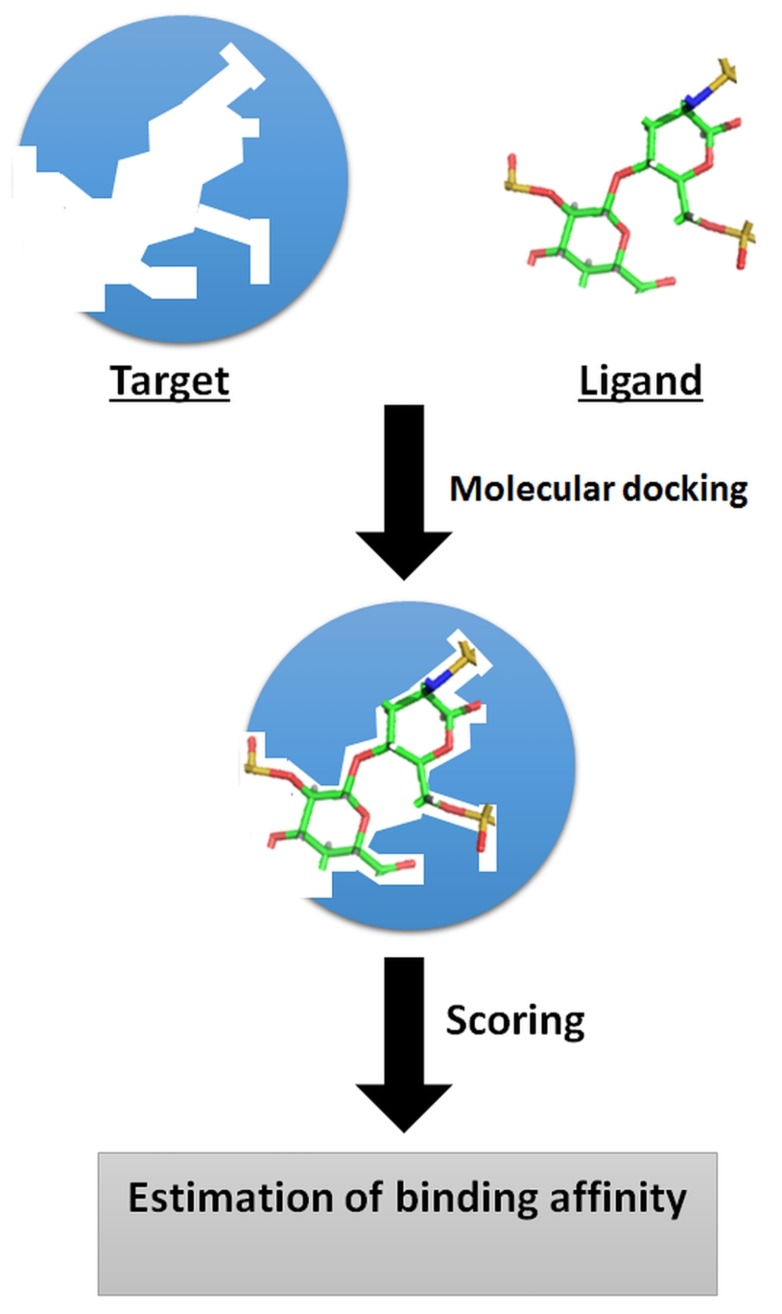
Molecular docking is a computational process widely used for rapidly predicting the binding modes and affinities of small molecules against their target molecules (usually proteins) [35, 36]. This in silico process has achieved a position of great importance in the drug discovery field [21, 36-38]. Molecular docking has emerged over the last two decades and is now considered an indispensable tool for CADD and in the structural biology field, and has been shown to be more efficient than traditional drug discovery methods. Molecular docking has been greatly facilitated by dramatic growth in computer power and the increasing availability of small molecule and protein databases. Fig. (3) illustrates the basic principle of molecular docking. Recent advancements in computer methods and access to 3D structural information of biological targets are set to increase the effectiveness of this technique and facilitate its large-scale application to studies of molecular interactions involved in ligand-protein binding. Generally, small molecules can be docked in three different ways, that is; (a) by rigid docking, where both target and ligands are treated as rigid entities; (b) by flexible docking, where both ligand and target are considered to be flexible; and (c) by flexible ligand docking, where the ligand is considered to be flexible and the target is considered rigid [39]. Many molecular docking programs have been developed during recent years, such as, AutoDock [40], Dock [41], FlexX [42], Glide [43], Gold [44], Surflex [45], ICM, and LigandFit [46], and been used successfully in many computer based drug discovery projects. Table 1 provides a list of major molecular docking tools in practice. Typically, the major goal of molecular docking is to identify ligands that bind most favorably within receptor binding sites and to determine its most energetically favored binding orientations (poses). The term “binding pose” is the orientation/confirmation of a ligand relative to its receptor. A binding pose either refers to a conformation of a ligand molecule within the binding site of its target protein which has been confirmed experimentally, or a computationally modelled hypothetical conformation. The search algorithm and the scoring function are two important components for determining protein-ligand interactions [47]. The search algorithm is responsible for searching different poses and conformations of a ligand within a given target protein and the scoring function estimates the binding affinities of generated poses, ranks them, and identifies the most favorable receptor/ligand binding modes [47, 48]. An ideal search algorithm should be fast and effective, and the scoring function must be capable of determining the physicochemical properties of molecules and the thermodynamics of interactions.
Fig. (3).
Basic principle of molecular docking.
Table 1.
List of major available molecular docking tools.
| S.No. | Program | Availability | Search Method | Refs. |
|---|---|---|---|---|
| 1. | AutoDock | Freely available | Genetic Algorithm/Monte Carlo | [40] |
| 2. | Gold | Paid | Genetic Algorithm | [44] |
| 3. | Glide | Paid | Monte Carlo | [43] |
| 4. | FlexX | Paid | Incremental construction | [42] |
| 5. | Dock | Freely available | Shape fitting (sphere sets) | [41] |
| 6. | LigandFit | Paid | Monte Carlo | [46] |
| 7. | FRED | Freely available | Shape fitting (Gaussian) | [91] |
| 8. | ICM | Paid | Monte Carlo | [92] |
| 9. | eHiTS | Paid | Incremental construction | [93] |
| 10. | Surflex-Dock | Paid | Incremental construction | [45] |
A large number of trials are being conducted to identify binding modes of ligands and selection of the most energetically favored poses. In order to achieve this, molecular docking tools are used to generate a set of different ligand binding poses and a scoring function is used to estimate the binding affinities of generated poses to identify the best binding mode. The energy change caused by ligand/receptor complex formation, is given by the Gibbs free energy (ΔG) and the binding constant (Kd) [49, 50]. The binding energy of a complex is predicted by evaluating physicochemical features involved in ligand-receptor binding, which include desolvation, intermolecular interactions, and entropic effects [51]. Sehgal et al. identified a number of compounds active against HSPB8 based on molecular docking results [52].
2.2. Ligand Based Drug Design (LBDD)
LBDD offers a general approach for elucidating relationships between the structural and physicochemical properties of compounds/ligands and their biological activities. This approach is applied when 3D structural information of a target protein is unavailable. In this process the available information of ligands and their biological activity is used for the development of new potential drug candidates. LBDD is widely used in pharmaceutical research, as more than 50% of approved drugs targeting membrane proteins (for which 3D structures are often not available, such as, GPCR). It is based on the assumption that compounds with similar structural features share common biological activities and interact/inhibit common target molecules [36, 37].
The representation of molecules is the basis of LBDD approach. Molecular descriptors are numerical values used to represent the structural and physicochemical properties of molecules [53, 54]. The molecular descriptor field is strikingly interdisciplinary and includes a number of different theories [55]. Active molecules are represented by the 0D-4D class of molecular descriptors [56]. Constitutional and count descriptors are 0D molecular descriptors, chemical fingerprints or lists of structural fragments, such as, SMILES and SLN, are 1D descriptors, graph invariants in which atoms are denoted as nodes and bonds as edges are 2D-descriptors, geometrical, WHIM and others are 3D descriptors, and those derived from CoMFA or DRID methods are classified as 4D descriptors [57]. Similarity searching is a key aspect of the LBDD method. This technique uses a known active compound as a query compound to find similar compounds and then rank compounds identified in a database. Based on this belief, structurally similar molecules exhibit similar biological activities and physicochemical properties. Numerical descriptors are applied and similarity coefficient is defined to quantify the degree of similarity (similarity/ dissimilarity). Fingerprint-based similarity or 2D similarity measures are widely used for similarity searching. A number of coefficients are applied in similarity searching with different fingerprint molecular databases (Cosine, Euclidian distance, Forbes, Tanimoto coefficients etc.).
The Tanimoto coefficient is most popular and widely accepted similarity index for binary variables, despite its well-documented size bias. It may be defined as:
where overlapab, overlapaa, and overlapbb are the volume overlaps of molecule a with molecule b, of molecule a with itself, and of molecule b with itself, respectively. Values of Tc range from 0 (no similarity) to 1 (a perfect match).
LBDD is generally categorized as Quantitative Structure Activity Relationship (QSAR) or pharmacophore modeling.
2.2.1. QSAR and its Role in Drug Discovery
The QSAR method and pharmacophore modeling are the most popular approaches to ligand-based drug design [58, 59]. QSAR methods are based on the belief that molecular structures are directly associated with biological activities, and thus, that molecular or structural variations alter biological activities. QSAR is defined as a process involving the construction of computational or mathematical models using chemometric techniques to identify significant correlations between a series of structures and functions [60]. For QSAR, the primary hypothesis is that “compounds with similar structural or physiochemical properties show similar activities”. To identify potential leads, a library of lead compounds with the desired biological activities is produced. A model is then developed to predict the quantitative relation between the structural and physico-chemical features of these compounds and their biological activities. A statistical model generated using such relations is then used to mathematically optimize the biological properties of sets of compounds and maximize relevant biological activities. QSAR is used to modify existing compounds and improve their activities, and has been widely used in drug discovery to improve existing drugs for NDs. A study conducted by Dong et al. successfully designed a new series of PDE-4 inhibitors using the QSAR approach [61]. Bhadoriya et al. successfully implemented QSAR to the discovery of more potent anti-Alzheimer’s agents [62]. In latter, QSAR studies were carried out on a series of 34 fused 5,6-bicyclic heterocycles to identify the structural characteristics needed to inhibit Aβ42.
2.3. Pharmacophore Modelling
A pharmacophore is an assembly (3D arrangement) of 'steric' and 'electronic' features required for optimal supramolecular interaction with a specific biological target structure and to prompt/block its biological response [63]. Ligand-based pharmacophore model generation is based on available information on the biological activities of compounds/ligands. A pharmacophore does not symbolize an actual molecule/ligand or real connection between functional groups, but rather provides an abstract description of molecular features that are vital for molecular interactions between molecules and macromolecular ligands.
Pharmacophore modeling is widely used to identify potential lead molecules quickly. During the recent era of drug design, many therapeutically potent and well accepted drug targets with unknown active site geometries have been identified. Pharmacophore modeling provides an efficient means of rapidly screening huge databases of compounds. The elucidation of common pharmacophore features is conducted by aligning conformational models and active compounds three dimensionally. A superimposition algorithm assembles training set compounds (3D structure) in the same position/arrangement of their respective chemical properties/features. Pharmacophoric features are positioned such that all/maximum compounds share a common chemical functionality. To refine a shared pharmacophore feature, information regarding inactive compounds can be included in the model generation process. A number of tools and software have been developed for pharmacophore development, such as, Phase, Catalyst/Discovery Studio, MOE, and LigandScout [64].
3. SUCCESSFUL CADD APPROACHES TO THE TREATMENT OF NEURODEGENERATIVE DISORDERS
The success of CADD has resulted in its being recognized as an important technique in the research and pharmaceutical fields. There are many examples of the successful application of CADD, but here we describe its successes with respect to the design of drugs for the treatment of NDs. Amyloid-β is an important therapeutic target in Alzheimer’s disease [65]. Chen et al. used an in silico approach to study a series of peptides against the fibrillar form of Aβ, and reported two highly active compounds [66]. These peptides were subsequently found to inhibit the neurotoxic effects of Aβ on neuroblastoma cells.
BACE-1 is an enzyme that has been reported to be essential for β-amyloid generation [67]. Research suggests inhibition of this enzyme stops the production of β-amyloid, and thus, prevents NDs like Alzheimer's disease [68]. This finding has made BACE-1 an important therapeutic target for NDs. During the last few years, several computational approaches have been used to study the structural behavior of BACE-1 and to design their inhibitors [69-71].
ROCK-I and NOX2 are among the most attractive potential therapeutic targets for several NDs [72-75]. Inhibition of these two enzymes constitutes treatment for neurological diseases like autism spectral disorder, Alzheimer, and fragile X syndrome. Alokam et al. reported the successful use of CADD to design dual inhibitors for these enzymes [76], by employing a combination of pharmacophores and using a molecular docking approach to identify chemical entities. In vitro validation of selected chemical entities demonstrated their inhibitory potentials against ROCK-I and NOX2.
HDAC6 is a member of the class IIb Histone deacetylases (HDACs) family and is usually found in cytosol in association with non-histone proteins [77, 78]. HDAC6 has been widely reported to be a crucial therapeutic drug target for several NDs [79-81]. The implementation of CADD has been reported to result in the design of a potential inhibitor of this enzyme. In one study conducted by Goracci et al., a virtual screening approach was used to identify potential inhibitors for HDAC6, and these were then subjected to in vitro testing. The results obtained showed inhibitors had low cytotoxicities, suggesting potential for drug development [82]. Several other reports have described the successful use of CADD in the NDs.
4. Limitations
Despite a number of successful applications of CADD to modern drug design, it has its limitations. In particular, like any computer assisted hypothetical system results must be validated in actual systems, and many lead molecules identified using CADD have failed to exhibit desired activities in biological systems [83, 84]. Several parameters must be met before potential compound to be approved as potent lead/drug, as it has to pass several pharmacological criteria. In fact, an average of only 40% of lead/drug candidates passes the different phases of clinical trials and obtains approval for clinical use.
Any computational tool based on pre-defined algorithms and scripts has its limitations, and the computational tools/methods used in CADD, such as, molecular docking, virtual screening, QSAR, pharmacophore modeling, and molecular dynamics, have their own limitations [49, 85-88]. Furthermore, ADME and many toxicity prediction tools are not supported by solid experimental data, and many examples of the failure of these computational approaches can be found in the literature [89, 90].
To overcome limitations and improve accuracy in terms of predicting potent leads, regular updates of tools and algorithms are needed. Database reliability and high quality validated experimental molecules is to be developed and updated because many pharmacophores do not pass biological activity process due to non-availability of good quality data sets. Databases should contain detail data on genomics and proteomics, high quality sequence information, physicochemical properties, and structures.
Conclusion
In the present era of drug discovery, the application of CADD counts up the most important accountability, and provides computational tools and algorithms that save time, costs, and reduce the risk of detecting non-viable developmental leads. The discovery of a new lead/drug using recent CADD paradigms requires a systematic understanding of the molecular and pathological conditions induced by diseases. Early diagnosis of NDs remains a huge challenge for researchers and clinicians. However, CADD can assist researchers studying interactions between drugs and receptors. The pharmacoinformatic approach is being applied to modern drug discovery and is providing much basic knowledge regarding drug-receptor interactions. Novel technologies and computational algorithms are required to move the CADD approach forward, as new developments are likely to lead to tools for disease identification and the screening of potential lead compounds. The emerging field of neurological studies, which includes neuroproteomics and neurogenomics, may aid understanding of the neuronal alterations associated with NDs. Furthermore, the application of technologies associated with neuroproteomics, neurogenomics, and next generation sequencing, and genome wide association studies may result in the identification of novel therapeutic targets and ultimately improve our ability to treat NDs.
Acknowledgements
This work was supported by the grant K16281 awarded to Korea Institute of Oriental Medicine (KIOM) from Ministry of Education, Science and Technology (MEST), Republic of Korea.
List OF Abbreviations
- CADD
Computer-aided drug design
- QSAR
Quantitative structure activity relationship
- SBDD
Structure based drug design
- LBDD
Ligand based drug design
- NDs
Neurodegenerative diseases
Consent for Publication
Not applicable.
Conflict of Interest
The authors declare no conflict of interest, financial or otherwise.
References
- 1.Madonna R., Novo G., Balistreri C.R. Cellular and molecular basis of the imbalance between vascular damage and repair in ageing and age-related diseases: As biomarkers and targets for new treatments. Mech. Ageing Dev. 2016;159:22–30. doi: 10.1016/j.mad.2016.03.005. [http://dx.doi. org/10.1016/j.mad.2016.03.005]. [PMID: 26993150]. [DOI] [PubMed] [Google Scholar]
- 2.Anand R, Gill KD. 2014.
- 3.Silberberg D. The high impact of neurologic disorders in developing countries: the struggle for global recognition. Neurology. 2011;77(3):307–308. doi: 10.1212/WNL.0b013e3182285da9. [http://dx.doi.org/10.1212/WNL.0b013e3182285da9]. [PMID: 21768602]. [DOI] [PubMed] [Google Scholar]
- 4.Ke Z., Zhang X., Cao Z., Ding Y., Li N., Cao L., Wang T., Zhang C., Ding G., Wang Z., Xu X., Xiao W. Drug discovery of neurodegenerative disease through network pharmacology approach in herbs. Biomed. Pharmacother. 2016;78:272–279. doi: 10.1016/j.biopha.2016.01.021. [http:// dx.doi.org/10.1016/j.biopha.2016.01.021]. [PMID: 26898452]. [DOI] [PubMed] [Google Scholar]
- 5.Lausted C., Lee I., Zhou Y., Qin S., Sung J., Price N.D., Hood L., Wang K. Systems approach to neurodegenerative disease biomarker discovery. Annu. Rev. Pharmacol. Toxicol. 2014;54:457–481. doi: 10.1146/annurev-pharmtox-011613-135928. [http://dx.doi.org/10.1146/annurev-pharmtox-011613-135928]. [PMID: 24160693]. [DOI] [PubMed] [Google Scholar]
- 6.Morris G.P., Clark I.A., Vissel B. Inconsistencies and controversies surrounding the amyloid hypothesis of Alzheimer’s disease. Acta Neuropathol. Commun. 2014;2:135. doi: 10.1186/s40478-014-0135-5. [PMID: 25231068]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zeng H., Wu X. Alzheimer’s disease drug development based on computer-aided drug design. Eur. J. Med. Chem. 2016;121:851–863. doi: 10.1016/j.ejmech.2015.08.039. [http://dx.doi.org/10.1016/j.ejmech.2015.08.039]. [PMID: 26415837]. [DOI] [PubMed] [Google Scholar]
- 8.Baig M.H., Ahmad K., Roy S., Ashraf J.M., Adil M., Siddiqui M.H., Khan S., Kamal M.A., Provazník I., Choi I. Computer Aided Drug Design: Success and Limitations. Curr. Pharm. Des. 2016;22(5):572–581. doi: 10.2174/1381612822666151125000550. [http://dx.doi.org/10.2174/1381612822666 151125000550]. [PMID: 26601966]. [DOI] [PubMed] [Google Scholar]
- 9.Scotti L., Scotti M.T. Computer aided drug design studies in the discovery of secondary metabolites targeted against age-related neurodegenerative diseases. Curr. Top. Med. Chem. 2015;15(21):2239–2252. doi: 10.2174/1568026615666150610143510. [http://dx.doi.org/10.2174/1568026615666150610143510]. [PMID: 26059353]. [DOI] [PubMed] [Google Scholar]
- 10.Pan S.Y., Zhou S.F., Gao S.H., Yu Z.L., Zhang S.F., Tang M.K., Sun J.N., Ma D.L., Han Y.F., Fong W.F., Ko K.M. New perspectives on how to discover drugs from herbal medicines: CAM’s outstanding contribution to modern therapeutics. Evid. Based Complement. Alternat. Med. 2013;2013:627375. doi: 10.1155/2013/627375. [http://dx. doi.org/10.1155/2013/627375]. [PMID: 23634172]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Paul S.M., Mytelka D.S., Dunwiddie C.T., Persinger C.C., Munos B.H., Lindborg S.R., Schacht A.L. How to improve R&D productivity: the pharmaceutical industry’s grand challenge. Nat. Rev. Drug Discov. 2010;9(3):203–214. doi: 10.1038/nrd3078. [http://dx.doi.org/10.1038/ nrd3078]. [PMID: 20168317]. [DOI] [PubMed] [Google Scholar]
- 12.Dickson M., Gagnon J.P. Key factors in the rising cost of new drug discovery and development. Nat. Rev. Drug Discov. 2004;3(5):417–429. doi: 10.1038/nrd1382. [http://dx.doi.org/10.1038/nrd1382]. [PMID: 15136789]. [DOI] [PubMed] [Google Scholar]
- 13.Clark R.L., Johnston B.F., Mackay S.P., Breslin C.J., Robertson M.N., Harvey A.L. The Drug Discovery Portal: a resource to enhance drug discovery from academia. Drug Discov. Today. 2010;15(15-16):679–683. doi: 10.1016/j.drudis.2010.06.003. [http://dx.doi.org/10.1016/j.drudis.2010.06.003]. [PMID: 20547242]. [DOI] [PubMed] [Google Scholar]
- 14.Entzeroth M., Flotow H., Condron P. Overview of high-throughput screening. 2009. [DOI] [PubMed] [Google Scholar]
- 15.Szymański P., Markowicz M., Mikiciuk-Olasik E. Adaptation of high-throughput screening in drug discovery-toxicological screening tests. Int. J. Mol. Sci. 2012;13(1):427–452. doi: 10.3390/ijms13010427. [http://dx.doi.org/ 10.3390/ijms13010427]. [PMID: 22312262]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lahana R. How many leads from HTS? Drug Discov. Today. 1999;4(10):447–448. doi: 10.1016/s1359-6446(99)01393-8. [http://dx.doi.org/10.1016/S1359-6446(99) 01393-8]. [PMID: 10481138]. [DOI] [PubMed] [Google Scholar]
- 17.Song C.M., Lim S.J., Tong J.C. Recent advances in computer-aided drug design. Brief. Bioinform. 2009;10(5):579–591. doi: 10.1093/bib/bbp023. [http:// dx.doi.org/10.1093/bib/bbp023]. [PMID: 19433475]. [DOI] [PubMed] [Google Scholar]
- 18.Veselovsky A.V., Zharkova M.S., Poroikov V.V., Nicklaus M.C. Computer-aided design and discovery of protein-protein interaction inhibitors as agents for anti-HIV therapy. SAR QSAR Environ. Res. 2014;25(6):457–471. doi: 10.1080/1062936X.2014.898689. [http://dx.doi.org/10.1080/1062936X. 2014.898689]. [PMID: 24716798]. [DOI] [PubMed] [Google Scholar]
- 19.Pârvu L. QSAR - a piece of drug design. J. Cell. Mol. Med. 2003;7(3):333–335. doi: 10.1111/j.1582-4934.2003.tb00235.x. [http://dx.doi.org/10.1111/j.1582-4934.2003.tb00235.x]. [PMID: 14594559]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim K.H., Kim N.D., Seong B.L. Pharmacophore-based virtual screening: a review of recent applications. Expert Opin. Drug Discov. 2010;5(3):205–222. doi: 10.1517/17460441003592072. [http://dx.doi.org/10.1517/17460441003592072]. [PMID: 22823018]. [DOI] [PubMed] [Google Scholar]
- 21.Sousa S.F., Cerqueira N.M., Fernandes P.A., Ramos M.J. Virtual screening in drug design and development. Comb. Chem. High Throughput Screen. 2010;13(5):442–453. doi: 10.2174/138620710791293001. [http://dx.doi.org/10. 2174/138620710791293001]. [PMID: 20236061]. [DOI] [PubMed] [Google Scholar]
- 22.Waszkowycz B., Perkins T.D.J., Sykes R.A., Li J. Large-scale virtual screening for discovering leads in the postgenomic era. IBM Syst. J. 2001;40(2):360. [http://dx.doi.org/10.1147/sj.402.0360]. [Google Scholar]
- 23.Lionta E., Spyrou G., Vassilatis D.K., Cournia Z. Structure-based virtual screening for drug discovery: principles, applications and recent advances. Curr. Top. Med. Chem. 2014;14(16):1923–1938. doi: 10.2174/1568026614666140929124445. [http://dx.doi.org/10.2174/1568026614666140929124445]. [PMID: 25262799]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shoichet B.K. Virtual screening of chemical libraries. Nature. 2004;432(7019):862–865. doi: 10.1038/nature03197. [http://dx.doi.org/10.1038/nature03197]. [PMID: 15602552]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lounnas V., Ritschel T., Kelder J., McGuire R., Bywater R.P., Foloppe N. Current progress in Structure-Based Rational Drug Design marks a new mindset in drug discovery. Comput. Struct. Biotechnol. J. 2013;5:e201302011. doi: 10.5936/csbj.201302011. [http://dx.doi.org/10.5936/csbj. 201302011]. [PMID: 24688704]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Anderson A.C. Structure-based functional design of drugs: from target to lead compound. Methods Mol. Biol. 2012;823:359–366. doi: 10.1007/978-1-60327-216-2_23. [http://dx.doi.org/10.1007/978-1-60327-216-2_23]. [PMID: 22081357]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Andricopulo A.D., Salum L.B., Abraham D.J. Structure-based drug design strategies in medicinal chemistry. Curr. Top. Med. Chem. 2009;9(9):771–790. doi: 10.2174/156802609789207127. [http://dx.doi.org/10.2174/156802609789207127]. [PMID: 19754394]. [DOI] [PubMed] [Google Scholar]
- 28.Goh B.C., Hadden J.A., Bernardi R.C., Singharoy A., McGreevy R., Rudack T., Cassidy C.K., Schulten K. Computational methodologies for real-space structural refinement of large macromolecular complexes. Annu. Rev. Biophys. 2016;45:253–278. doi: 10.1146/annurev-biophys-062215-011113. [http://dx.doi.org/10.1146/annurev-biophys-062215-011113]. [PMID: 27145875]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fang Y. Combining label-free cell phenotypic profiling with computational approaches for novel drug discovery. Expert Opin. Drug Discov. 2015;10(4):331–343. doi: 10.1517/17460441.2015.1020788. [http://dx.doi.org/10.1517/ 17460441.2015.1020788]. [PMID: 25727255]. [DOI] [PubMed] [Google Scholar]
- 30.Schwede T. Protein modeling: what happened to the “protein structure gap”? Structure. 2013;21(9):1531–1540. doi: 10.1016/j.str.2013.08.007. [http://dx.doi. org/10.1016/j.str.2013.08.007]. [PMID: 24010712]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vyas V.K., Ukawala R.D., Ghate M., Chintha C. Homology modeling a fast tool for drug discovery: current perspectives. Indian J. Pharm. Sci. 2012;74(1):1–17. doi: 10.4103/0250-474X.102537. [http://dx.doi.org/10.4103/ 0250-474X.102537]. [PMID: 23204616]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Eberini I., Daniele S., Parravicini C., Sensi C., Trincavelli M.L., Martini C., Abbracchio M.P. In silico identification of new ligands for GPR17: a promising therapeutic target for neurodegenerative diseases. J. Comput. Aided Mol. Des. 2011;25(8):743–752. doi: 10.1007/s10822-011-9455-8. [http://dx.doi.org/10.1007/s10822-011-9455-8]. [PMID: 21744154]. [DOI] [PubMed] [Google Scholar]
- 33.Cavasotto C.N. Homology models in docking and high-throughput docking. Curr. Top. Med. Chem. 2011;11(12):1528–1534. doi: 10.2174/156802611795860951. [http:// dx.doi.org/10.2174/156802611795860951]. [PMID: 21510834]. [DOI] [PubMed] [Google Scholar]
- 34.Dhanavade M.J., Jalkute C.B., Barage S.H., Sonawane K.D. Homology modeling, molecular docking and MD simulation studies to investigate role of cysteine protease from Xanthomonas campestris in degradation of Aβ peptide. Comput. Biol. Med. 2013;43(12):2063–2070. doi: 10.1016/j.compbiomed.2013.09.021. [http://dx.doi.org/10.1016/j.compbiomed.2013. 09.021]. [PMID: 24290922]. [DOI] [PubMed] [Google Scholar]
- 35.Meng X.Y., Zhang H.X., Mezei M., Cui M. Molecular docking: a powerful approach for structure-based drug discovery. Curr Comput Aided Drug Des. 2011;7(2):146–157. doi: 10.2174/157340911795677602. [http://dx.doi.org/ 10.2174/157340911795677602]. [PMID: 21534921]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sousa S.F., Ribeiro A.J., Coimbra J.T., Neves R.P., Martins S.A., Moorthy N.S., Fernandes P.A., Ramos M.J. Protein-ligand docking in the new millennium--a retrospective of 10 years in the field. Curr. Med. Chem. 2013;20(18):2296–2314. doi: 10.2174/0929867311320180002. [http://dx.doi. org/10.2174/0929867311320180002]. [PMID: 23531220]. [DOI] [PubMed] [Google Scholar]
- 37.Manly C.J., Chandrasekhar J., Ochterski J.W., Hammer J.D., Warfield B.B. Strategies and tactics for optimizing the Hit-to-Lead process and beyond--a computational chemistry perspective. Drug Discov. Today. 2008;13(3-4):99–109. doi: 10.1016/j.drudis.2007.10.019. [http://dx.doi.org/10.1016/ j.drudis.2007.10.019]. [PMID: 18275907]. [DOI] [PubMed] [Google Scholar]
- 38.Huang S.Y., Zou X. Advances and challenges in protein-ligand docking. Int. J. Mol. Sci. 2010;11(8):3016–3034. doi: 10.3390/ijms11083016. [http://dx.doi. org/10.3390/ijms11083016]. [PMID: 21152288]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mohan V., Gibbs A.C., Cummings M.D., Jaeger E.P., DesJarlais R.L. Docking: successes and challenges. Curr. Pharm. Des. 2005;11(3):323–333. doi: 10.2174/1381612053382106. [http://dx.doi.org/10.2174/1381612053382106]. [PMID: 15723628]. [DOI] [PubMed] [Google Scholar]
- 40.Morris G.M., Goodsell D.S., Huey R., Olson A.J. Distributed automated docking of flexible ligands to proteins: parallel applications of AutoDock 2.4. J. Comput. Aided Mol. Des. 1996;10(4):293–304. doi: 10.1007/BF00124499. [http://dx.doi.org/10.1007/BF00124499]. [PMID: 8877701]. [DOI] [PubMed] [Google Scholar]
- 41.Ewing T.J., Makino S., Skillman A.G., Kuntz I.D. DOCK 4.0: search strategies for automated molecular docking of flexible molecule databases. J. Comput. Aided Mol. Des. 2001;15(5):411–428. doi: 10.1023/a:1011115820450. [http://dx.doi.org/10.1023/A:1011115820450]. [PMID: 11394736]. [DOI] [PubMed] [Google Scholar]
- 42.Kramer B., Rarey M., Lengauer T. Evaluation of the FLEXX incremental construction algorithm for protein-ligand docking. Proteins. 1999;37(2):228–241. doi: 10.1002/(sici)1097-0134(19991101)37:2<228::aid-prot8>3.0.co;2-8. [http://dx.doi.org/10.1002/ (SICI)1097-0134(19991101)37:2<228:AID-PROT8>3.0.CO;2-8]. [PMID: 10584068]. [DOI] [PubMed] [Google Scholar]
- 43.Friesner R.A., Banks J.L., Murphy R.B., Halgren T.A., Klicic J.J., Mainz D.T., Repasky M.P., Knoll E.H., Shelley M., Perry J.K., Shaw D.E., Francis P., Shenkin P.S. Glide: a new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. J. Med. Chem. 2004;47(7):1739–1749. doi: 10.1021/jm0306430. [http://dx.doi.org/10.1021/jm0306430]. [PMID: 15027865]. [DOI] [PubMed] [Google Scholar]
- 44.Verdonk M.L., Cole J.C., Hartshorn M.J., Murray C.W., Taylor R.D. Improved protein-ligand docking using GOLD. Proteins. 2003;52(4):609–623. doi: 10.1002/prot.10465. [http://dx.doi.org/10.1002/prot.10465]. [PMID: 12910460]. [DOI] [PubMed] [Google Scholar]
- 45.Jain A.N. Surflex: fully automatic flexible molecular docking using a molecular similarity-based search engine. J. Med. Chem. 2003;46(4):499–511. doi: 10.1021/jm020406h. [http://dx.doi.org/10.1021/jm020406h]. [PMID: 12570372]. [DOI] [PubMed] [Google Scholar]
- 46.Venkatachalam C.M., Jiang X., Oldfield T., Waldman M. LigandFit: a novel method for the shape-directed rapid docking of ligands to protein active sites. J. Mol. Graph. Model. 2003;21(4):289–307. doi: 10.1016/s1093-3263(02)00164-x. [http://dx.doi.org/10.1016/S1093-3263(02)00164-X]. [PMID: 12479928]. [DOI] [PubMed] [Google Scholar]
- 47.Du X., Li Y., Xia Y.L., Ai S.M., Liang J., Sang P., Ji X.L., Liu S.Q. Insights into Protein-Ligand Interactions: Mechanisms, Models, and Methods. Int. J. Mol. Sci. 2016;17(2):E144. doi: 10.3390/ijms17020144. [http:// dx.doi.org/10.3390/ijms17020144]. [PMID: 26821017]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sousa S.F., Fernandes P.A., Ramos M.J. Protein-ligand docking: current status and future challenges. Proteins. 2006;65(1):15–26. doi: 10.1002/prot.21082. [http://dx.doi.org/10.1002/prot.21082]. [PMID: 16862531]. [DOI] [PubMed] [Google Scholar]
- 49.Ferreira L.G., Dos Santos R.N., Oliva G., Andricopulo A.D. Molecular docking and structure-based drug design strategies. Molecules. 2015;20(7):13384–13421. doi: 10.3390/molecules200713384. [http://dx.doi.org/10.3390/ molecules200713384]. [PMID: 26205061]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Foloppe N., Hubbard R. Towards predictive ligand design with free-energy based computational methods? Curr. Med. Chem. 2006;13(29):3583–3608. doi: 10.2174/092986706779026165. [http://dx.doi.org/10.2174/092986706779026165]. [PMID: 17168725]. [DOI] [PubMed] [Google Scholar]
- 51.Jain A.N. Scoring functions for protein-ligand docking. Curr. Protein Pept. Sci. 2006;7(5):407–420. doi: 10.2174/138920306778559395. [http://dx.doi.org/10.2174/ 138920306778559395]. [PMID: 17073693]. [DOI] [PubMed] [Google Scholar]
- 52.Sehgal S.A., Mannan S., Ali S. Pharmacoinformatic and molecular docking studies reveal potential novel antidepressants against neurodegenerative disorders by targeting HSPB8. Drug Des. Devel. Ther. 2016;10:1605–1618. doi: 10.2147/DDDT.S101929. [http://dx.doi.org/10.2147/DDDT. S101929]. [PMID: 27226709]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Guha R., Willighagen E. A survey of quantitative descriptions of molecular structure. Curr. Top. Med. Chem. 2012;12(18):1946–1956. doi: 10.2174/156802612804910278. [http://dx.doi.org/10.2174/156802612804910278]. [PMID: 23110530]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kombo D.C., Tallapragada K., Jain R., Chewning J., Mazurov A.A., Speake J.D., Hauser T.A., Toler S. 3D molecular descriptors important for clinical success. J. Chem. Inf. Model. 2013;53(2):327–342. doi: 10.1021/ci300445e. [http://dx.doi.org/10.1021/ci300445e]. [PMID: 23244494]. [DOI] [PubMed] [Google Scholar]
- 55.Mauri A., Consonni V., Pavan M., Todeschini R. Dragon software: An easy approach to molecular descriptor calculations. Match (Mulh.) 2006;56(2):237–248. [Google Scholar]
- 56.Andrade C.H., Pasqualoto K.F., Ferreira E.I., Hopfinger A.J. 4D-QSAR: perspectives in drug design. Molecules. 2010;15(5):3281–3294. doi: 10.3390/molecules15053281. [http://dx.doi.org/10.3390/molecules15053281]. [PMID: 20657478]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Myint K.Z., Xie X.Q. Recent advances in fragment-based QSAR and multi-dimensional QSAR methods. Int. J. Mol. Sci. 2010;11(10):3846–3866. doi: 10.3390/ijms11103846. [http://dx.doi.org/10.3390/ijms11103846]. [PMID: 21152304]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Acharya C., Coop A., Polli J.E., Mackerell A.D., Jr Recent advances in ligand-based drug design: relevance and utility of the conformationally sampled pharmacophore approach. Curr Comput Aided Drug Des. 2011;7(1):10–22. doi: 10.2174/157340911793743547. [http://dx.doi.org/10.2174/ 157340911793743547]. [PMID: 20807187]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yang S.Y. Pharmacophore modeling and applications in drug discovery: challenges and recent advances. Drug Discov. Today. 2010;15(11-12):444–450. doi: 10.1016/j.drudis.2010.03.013. [http://dx.doi.org/10.1016/j.drudis.2010. 03.013]. [PMID: 20362693]. [DOI] [PubMed] [Google Scholar]
- 60.Karelson M., Lobanov V.S., Katritzky A.R. Quantum-Chemical Descriptors in QSAR/QSPR Studies. Chem. Rev. 1996;96(3):1027–1044. doi: 10.1021/cr950202r. [http://dx.doi.org/10.1021/cr950202r]. [PMID: 11848779]. [DOI] [PubMed] [Google Scholar]
- 61.Dong X., Zheng W. A new structure-based QSAR method affords both descriptive and predictive models for phosphodiesterase-4 inhibitors. Curr. Chem. Genomics. 2008;2:29–39. doi: 10.2174/1875397300802010029. [http://dx.doi. org/10.2174/1875397300802010029]. [PMID: 20161841]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bhadoriya K.S., Sharma M.C., Sharma S., Jain S.V., Avchar M.H. An approach to design potent anti-Alzheimer’s agents by 3D-QSAR studies on fused 5, 6-bicyclic heterocycles as γ-secretase modulators using kNN–MFA methodology. Arab. J. Chem. 2014;7(6):924–935. [http://dx.doi.org/10.1016/j.arabjc.2013.02.002]. [Google Scholar]
- 63.Kaserer T., Beck K.R., Akram M., Odermatt A., Schuster D. Pharmacophore models and pharmacophore-based virtual screening: Concepts and applications exemplified on hydroxysteroid dehydrogenases. Molecules. 2015;20(12):22799–22832. doi: 10.3390/molecules201219880. [http:// dx.doi.org/10.3390/molecules201219880]. [PMID: 26703541]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Liao C., Sitzmann M., Pugliese A., Nicklaus M.C. Software and resources for computational medicinal chemistry. Future Med. Chem. 2011;3(8):1057–1085. doi: 10.4155/fmc.11.63. [http://dx.doi.org/10.4155/fmc.11.63]. [PMID: 21707404]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Selkoe D.J. Amyloid beta-protein and the genetics of Alzheimer’s disease. J. Biol. Chem. 1996;271(31):18295–18298. doi: 10.1074/jbc.271.31.18295. [http://dx.doi. org/10.1074/jbc.271.31.18295]. [PMID: 8756120]. [DOI] [PubMed] [Google Scholar]
- 66.Chen D., Martin Z.S., Soto C., Schein C.H. Computational selection of inhibitors of Abeta aggregation and neuronal toxicity. Bioorg. Med. Chem. 2009;17(14):5189–5197. doi: 10.1016/j.bmc.2009.05.047. [http://dx.doi.org/ 10.1016/j.bmc.2009.05.047]. [PMID: 19540126]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Fu H., Li W., Luo J., Lee N.T., Li M., Tsim K.W., Pang Y., Youdim M.B., Han Y. Promising anti-Alzheimer’s dimer bis(7)-tacrine reduces beta-amyloid generation by directly inhibiting BACE-1 activity. Biochem. Biophys. Res. Commun. 2008;366(3):631–636. doi: 10.1016/j.bbrc.2007.11.068. [http://dx.doi.org/10.1016/j.bbrc.2007.11.068]. [PMID: 18039469]. [DOI] [PubMed] [Google Scholar]
- 68.Jiang Y., Mullaney K.A., Peterhoff C.M., Che S., Schmidt S.D., Boyer-Boiteau A., Ginsberg S.D., Cataldo A.M., Mathews P.M., Nixon R.A. Alzheimer’s-related endosome dysfunction in Down syndrome is Abeta-independent but requires APP and is reversed by BACE-1 inhibition. Proc. Natl. Acad. Sci. USA. 2010;107(4):1630–1635. doi: 10.1073/pnas.0908953107. [http://dx.doi.org/10.1073/pnas.0908953107]. [PMID: 20080541]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kacker P., Bottegoni G., Cavalli A. Computational methods in the discovery and design of BACE-1 inhibitors. Curr. Med. Chem. 2012;19(36):6095–6111. [http://dx.doi.org/10.2174/0929867311 209066095]. [PMID: 23072352]. [PubMed] [Google Scholar]
- 70.Rueeger H., Lueoend R., Rogel O., Rondeau J.M., Möbitz H., Machauer R., Jacobson L., Staufenbiel M., Desrayaud S., Neumann U. Discovery of cyclic sulfone hydroxyethylamines as potent and selective β-site APP-cleaving enzyme 1 (BACE1) inhibitors: structure-based design and in vivo reduction of amyloid β-peptides. J. Med. Chem. 2012;55(7):3364–3386. doi: 10.1021/jm300069y. [http://dx.doi. org/10.1021/jm300069y]. [PMID: 22380629]. [DOI] [PubMed] [Google Scholar]
- 71.Ju Y., Li Z., Deng Y., Tong A., Zhou L., Luo Y. Identification of Novel BACE1 Inhibitors by Combination of Pharmacophore Modeling, Structure-Based Design and In Vitro Assay. Curr Comput Aided Drug Des. 2016;12(1):73–82. doi: 10.2174/1573409912666160222113103. [http://dx.doi.org/ 10.2174/1573409912666160222113103]. [PMID: 26899408]. [DOI] [PubMed] [Google Scholar]
- 72.Labandeira-Garcia J.L., Rodríguez-Perez A.I., Villar-Cheda B., Borrajo A., Dominguez-Meijide A., Guerra M.J. Rho Kinase and Dopaminergic Degeneration: A Promising Therapeutic Target for Parkinson’s Disease. Neuroscientist. 2015;21(6):616–629. doi: 10.1177/1073858414554954. [http://dx.doi.org/10.1177/1073858414554954]. [PMID: 25323761]. [DOI] [PubMed] [Google Scholar]
- 73.Mueller B.K., Mack H., Teusch N. Rho kinase, a promising drug target for neurological disorders. Nat. Rev. Drug Discov. 2005;4(5):387–398. doi: 10.1038/nrd1719. [http://dx.doi.org/10.1038/nrd1719]. [PMID: 15864268]. [DOI] [PubMed] [Google Scholar]
- 74.Nayernia Z., Jaquet V., Krause K.H. New insights on NOX enzymes in the central nervous system. Antioxid. Redox Signal. 2014;20(17):2815–2837. doi: 10.1089/ars.2013.5703. [http://dx.doi.org/10.1089/ars.2013.5703]. [PMID: 24206089]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sorce S., Krause K.H. NOX enzymes in the central nervous system: from signaling to disease. Antioxid. Redox Signal. 2009;11(10):2481–2504. doi: 10.1089/ars.2009.2578. [http://dx.doi.org/10.1089/ars.2009.2578]. [PMID: 19309263]. [DOI] [PubMed] [Google Scholar]
- 76.Alokam R., Singhal S., Srivathsav G.S., Garigipati S., Puppala S., Sriram D., Perumal Y. Design of dual inhibitors of ROCK-I and NOX2 as potential leads for the treatment of neuroinflammation associated with various neurological diseases including autism spectrum disorder. Mol. Biosyst. 2015;11(2):607–617. doi: 10.1039/c4mb00570h. [http://dx. doi.org/10.1039/C4MB00570H]. [PMID: 25465055]. [DOI] [PubMed] [Google Scholar]
- 77.Boyault C., Zhang Y., Fritah S., Caron C., Gilquin B., Kwon S.H., Garrido C., Yao T.P., Vourc’h C., Matthias P., Khochbin S. HDAC6 controls major cell response pathways to cytotoxic accumulation of protein aggregates. Genes Dev. 2007;21(17):2172–2181. doi: 10.1101/gad.436407. [http://dx.doi.org/10.1101/gad.436407]. [PMID: 17785525]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Xu W.S., Parmigiani R.B., Marks P.A. Histone deacetylase inhibitors: molecular mechanisms of action. Oncogene. 2007;26(37):5541–5552. doi: 10.1038/sj.onc.1210620. [http://dx.doi.org/10.1038/sj.onc.1210620]. [PMID: 17694093]. [DOI] [PubMed] [Google Scholar]
- 79.Gray S.G. Targeting histone deacetylases for the treatment of Huntington’s disease. CNS Neurosci. Ther. 2010;16(6):348–361. doi: 10.1111/j.1755-5949.2010.00184.x. [http://dx.doi.org/10.1111/j.1755-5949.2010.00184.x]. [PMID: 20642797]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Saha R.N., Pahan K. HATs and HDACs in neurodegeneration: a tale of disconcerted acetylation homeostasis. Cell Death Differ. 2006;13(4):539–550. doi: 10.1038/sj.cdd.4401769. [http://dx.doi.org/10.1038/sj.cdd.4401769]. [PMID: 16167067]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tang H., Wang X.S., Huang X.P., Roth B.L., Butler K.V., Kozikowski A.P., Jung M., Tropsha A. Novel inhibitors of human histone deacetylase (HDAC) identified by QSAR modeling of known inhibitors, virtual screening, and experimental validation. J. Chem. Inf. Model. 2009;49(2):461–476. doi: 10.1021/ci800366f. [http://dx.doi.org/10. 1021/ci800366f]. [PMID: 19182860]. [DOI] [PubMed] [Google Scholar]
- 82.Goracci L., Deschamps N., Randazzo G.M., Petit C., Dos Santos Passos C., Carrupt P.A., Simões-Pires C., Nurisso A. A Rational Approach for the Identification of Non-Hydroxamate HDAC6-Selective Inhibitors. Sci. Rep. 2016;6:29086. doi: 10.1038/srep29086. [http://dx.doi. org/10.1038/srep29086]. [PMID: 27404291]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Schneider G. Virtual screening: an endless staircase? Nat. Rev. Drug Discov. 2010;9(4):273–276. doi: 10.1038/nrd3139. [http://dx.doi.org/10.1038/ nrd3139]. [PMID: 20357802]. [DOI] [PubMed] [Google Scholar]
- 84.Verkhivker G.M., Bouzida D., Gehlhaar D.K., Rejto P.A., Arthurs S., Colson A.B., Freer S.T., Larson V., Luty B.A., Marrone T., Rose P.W. Deciphering common failures in molecular docking of ligand-protein complexes. J. Comput. Aided Mol. Des. 2000;14(8):731–751. doi: 10.1023/a:1008158231558. [http://dx.doi.org/10.1023/A:1008158231558]. [PMID: 11131967]. [DOI] [PubMed] [Google Scholar]
- 85.Cheatham T.E., III, Young M.A. Molecular dynamics simulation of nucleic acids: successes, limitations, and promise. Biopolymers. 2000-2001;56(4):232–256. doi: 10.1002/1097-0282(2000)56:4<232::AID-BIP10037>3.0.CO;2-H. [http://dx.doi.org/10.1002/1097-0282 (2000)56:4<232:AID-BIP10037>3.0.CO;2-H]. [PMID: 11754338]. [DOI] [PubMed] [Google Scholar]
- 86.Klebe G. Virtual ligand screening: strategies, perspectives and limitations. Drug Discov. Today. 2006;11(13-14):580–594. doi: 10.1016/j.drudis.2006.05.012. [http://dx.doi.org/10.1016/j.drudis.2006.05.012]. [PMID: 16793526]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Korb O., Olsson T.S., Bowden S.J., Hall R.J., Verdonk M.L., Liebeschuetz J.W., Cole J.C. Potential and limitations of ensemble docking. J. Chem. Inf. Model. 2012;52(5):1262–1274. doi: 10.1021/ci2005934. [http://dx.doi.org/10.1021/ci2005934]. [PMID: 22482774]. [DOI] [PubMed] [Google Scholar]
- 88.MacDonald D., Breton R., Sutcliffe R., Walker J. Uses and limitations of quantitative structure-activity relationships (QSARs) to categorize substances on the Canadian domestic substance list as persistent and/or bioaccumulative, and inherently toxic to non-human organisms. SAR QSAR Environ. Res. 2002;13(1):43–55. doi: 10.1080/10629360290002082. [http://dx.doi.org/10.1080/10629360290002082]. [PMID: 12074391]. [DOI] [PubMed] [Google Scholar]
- 89.Blomme E.A., Will Y. Toxicology Strategies for Drug Discovery: Present and Future. Chem. Res. Toxicol. 2016;29(4):473–504. doi: 10.1021/acs.chemrestox.5b00407. [http:// dx.doi.org/10.1021/acs.chemrestox.5b00407]. [PMID: 26588328]. [DOI] [PubMed] [Google Scholar]
- 90.van de Waterbeemd H., Gifford E. ADMET in silico modelling: towards prediction paradise? Nat. Rev. Drug Discov. 2003;2(3):192–204. doi: 10.1038/nrd1032. [PMID: 12612645]. [DOI] [PubMed] [Google Scholar]
- 91.McGann M. FRED and HYBRID docking performance on standardized datasets. J. Comput. Aided Mol. Des. 2012;26(8):897–906. doi: 10.1007/s10822-012-9584-8. [http://dx.doi.org/10.1007/s10822-012-9584-8]. [PMID: 22669221]. [DOI] [PubMed] [Google Scholar]
- 92.Neves M.A., Totrov M., Abagyan R. Docking and scoring with ICM: the benchmarking results and strategies for improvement. J. Comput. Aided Mol. Des. 2012;26(6):675–686. doi: 10.1007/s10822-012-9547-0. [http://dx.doi.org/ 10.1007/s10822-012-9547-0]. [PMID: 22569591]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ravitz O., Zsoldos Z., Simon A. Improving molecular docking through eHiTS' tunable scoring function. J. Comput. Aided Mol. Des. 2011;25(11):1033–1051. doi: 10.1007/s10822-011-9482-5. [http://dx.doi.org/10.1007/s10822-011-9482-5]. [PMID: 22076470]. [DOI] [PubMed] [Google Scholar]