Abstract
Abstract: Background
As the number of elderly persons increases, neurodegenerative diseases are becoming ubiquitous. There is currently a great need for knowledge concerning management of old-age neurodegenerative diseases; the most important of which are: Alzheimer’s disease, Parkinson’s disease, Amyotrophic Lateral Sclerosis, and Huntington’s disease.
Objective
To summarize the potential of computationally predicted molecules and targets against neurodegenerative diseases.
Method
Review of literature published since 1997 against neurodegenerative diseases, utilizing as keywords: in silico, Alzheimer’s disease, Parkinson’s disease, Amyotrophic Lateral Sclerosis ALS, and Huntington’s disease was conducted.
Results and Conclusion
Due to the costs associated with experimentation and current ethical law, performing experiments directly on living organisms has become much more difficult. In this scenario, in silico techniques have been successful and have become powerful tools in the search to cure disease. Researchers use the Computer Aided Drug Design pipeline which: 1) generates 3-dimensional structures of target proteins through homology modeling 2) achieves stabilization through molecular dynamics simulation, and 3) exploits molecular docking through large compound libraries. Next generation sequencing is continually producing enormous amounts of raw sequence data while neuroimaging is producing a multitude of raw image data. To solve such pressing problems, these new tools and algorithms are required. This review elaborates precise in silico tools and techniques for drug targets, active molecules, and molecular docking studies, together with future prospects and challenges concerning possible breakthroughs in Alzheimer’s, Parkinson’s, Amyotrophic Lateral Sclerosis, and Huntington’s disease.
Keywords: In silico analysis, computer aided drug design, alzheimer’s disease, parkinson’s disease, amyotrophic lateral sclerosis, and huntington ’s disease
1. Introduction
A better quality of life, and up-to-date individualized health care plans are major concerns of the current health care industry. Such measures have increased the individual’s lifespan, however old age diseases are now becoming more common. According to a 2010 survey by the US National Institute of Health (NIH); 8% (524 million people) of the world’s total population is at or above 65 years old, this group is anticipated to grow rapidly; up to 2 billion (16%) by 2050 [1]. Given this scenario, effective measures should be taken to cope with old age diseases. Degenerative nerve diseases are the most common diseases of old age. Many organizations and individual laboratories are working to cure neurodegenerative diseases, but there are still no available medicines that can cure neurological diseases effectively. The major hurdles to finding a potent cure are the complexity of the brain system, the availability of resources, and ethical restraints. However, advances in computational technology have enabled us to perform in silico experiments which are influencing our understanding of the brain tremendously [2-4].
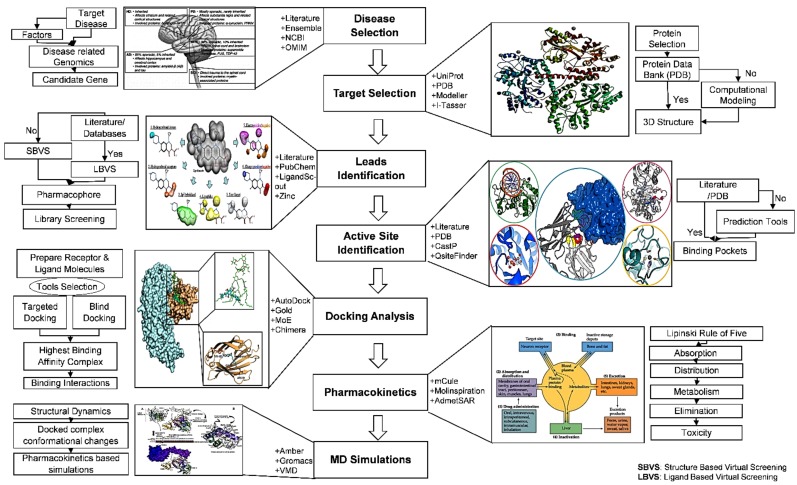
Bioinformatics, utilizing computational, mathematical and statistical approaches is a very recent emerging interdisciplinary science that solves biological problems [5]. Molecular docking is a computational technique that predicts the preferred binding orientations of one molecule (receptor) to another molecule (ligand) to form a firm or stable complex. Computational drug discovery or computer-aided drug design (CADD) is one of the principal approaches applied in drug discovery to reduce time and costs [5, 6]. CADD techniques involve sequence based drug design, ligand based drug design, and structure based drug design. The most used methodologies in CADD are target (protein) identification, molecular docking analyses, molecule design, quantitative and qualitative structure activity relationship, lead optimization, and ADMET among others, as depicted in Fig. (1). In silico approaches and bioinformatic analyses have been successful in both solving biological problems [5] and in designing numerous novel computer-aided molecules to fight neurological disorders, [7-11] and cancer [12-15]. Revealing protein structural information, using X-ray crystallography, Nuclear Magnetic Resonance (NMR), and Electron Microscopy is difficult and requires large resources and time [16-18]. Due to their abilities to build 3D protein structures based on already available homologous protein family information, computational methods are becoming more prominent [19-21]. Numerous servers and tools are available to model proteins through homology modeling techniques including I-Tasser, M4T, Phyre2, and Intfold2 [22-25].
Fig. (1).
CADD methodology comprises target selection, molecular docking studies, pharmacokinetics analysis, and MD simulations.
Using only protein sequence information, servers employing homology modeling build 3D models based on i) information in the Protein Data Bank (PDB) database, ii) the nature of the protein sequence, and iii) fine tuning of the model through either energy minimization or low level molecular dynamics simulation. The idea of homology modeling is based on templates of either X-ray, NMR, or Electron Microscopy structures. Often multiple templates are used for protein modeling based on sequence alignments of a particular query. Utilizing Newtonian equations, simulated Molecular Dynamics (MD) studies help to resolve protein structures [26-28]. During MD simulations, being based on force-fields, virtual systems are generated which allow independent structural repositioning of atoms within proteins in an attempt to get the native protein structure. MD simulation followed with molecular modeling has helped to solve many large bio-molecule structures [29, 30]. Virtual Screening (VS) is a technique to screen potent compounds from millions of compounds against disease targets [31-33]. VS can be implemented through ligand based or structure based methods. For VS, different databases can be used including ZINC [34, 35], ChemDB [36], ChEMBL [37, 38], Mcule [39], and recently, the Cresset BMD compound database [40]. The resulting compounds are docked with the target protein to check which molecule is most likely to bind and build suitable interactions with the target protein. For docking purposes, Autodock [41, 42], Autodock Vina [43], GOLD [44, 45], and other tools are available [20, 46-50].
Currently, there is a large gap between designing and implementing effective neuro-drugs. The old-age population time bomb is soon to explode, underscoring the immediate need for measures to cope with neurodegenerative diseases [51-53]. Progressive degeneration and death of nerve cells produces debilitating conditions; principally dementia and loss of movement control. This study attempts to summarize the published literature since 1997, on computational drug design against neurodegenerative diseases using the keywords: in silico, neurodegenerative diseases, Alzheimer’s Diseases (AD), Parkinson’s Disease (PD), Amyotrophic Lateral Sclerosis (ALS), Huntington’s Disease (HD), and CADD. This review attempts to recapitulate both current and novel drug targets in the principal neurodegenerative diseases (AD, PD, ALS, and HD) using in silico approaches (Table 1).
Table 1.
Summarized new and current drug targets in neurodegenerative diseases (AD, PD, ALS and HD) using in-silico approaches.
| Diseases | AD | PD | ALS | HD |
|---|---|---|---|---|
| Characterization | Death and malfunctioning of neurons |
Death of substantia nigra cells | Loss of upper and lower motor neurons | Degeneration of Nerve Cells |
| Risk Factors | Hyperphosphorylation of tau proteins FAD Mutations Presenilin-1 or -2, APP SAD Apo-ε, APOE4,Aging |
Poor lifestyle & Poor diet Abnormal expression SNCA, PARK2, PINK1, DJ1, LRRK2, ATP13A2 Blockage of Adenosine A2A receptors |
Mutations SODI, FIG4, FUS/TLS, TARDBP, ANG, VAPB PON1, Hetereochromatosis gene, Neurofilament heavy chain gene Repeat (GGGGCC) in C9ORF72 |
Mutations HTT, Higher repeats of CAG in HTT allele Interacting Proteins HTT-interacting protein 1 GRB2-like protein 3 Protein kinase C Postsynaptic density-95 FIP-2 |
| Targets for potential therapeutics | Acetylcholinesterase, N-methyl-D-aspartate receptor, Muscarinic and nicotinic ACh receptors, Tau proteins Beta-secretase enzymes |
Mutant LRRK2 SNCA Motif Mutated expression & mitochondria localization PINK1, PARK2, DJ1 Dopamine receptors |
Mutant SODI SODI oligomerization CASP-3, CASP-8 TDP-43, p38 MAPK Nav1.6 sodium channel |
Mutant HTT, Infant Testing HTT Interacting proteins Specificity protein 1 Nuclear receptor co-repressor, HTT-interacting protein 1, Postsynaptic density-95, FIP-2 |
| Computationally derived compounds for Neurodegenerative diseases |
AChE Compounds Flavonoid derivatives, macluraxanthone, kaempferol, rutin, quercetin, pyridopyrimidine, 6-chloro-pyridonepezil and pyridonepezil & piperazine derivatives, memantine antagonist of NMDA Triazolyl-amidine derivatives 3-substituted-1H-indoles, 1-benzyl-1,2,3,4-tetrahydro- b-carboline, and ifrenprodil, AF267B compound OM00-3 and OM99-2 of BACE1 Flavones & flavonol derivatives morin, kaempferol, quercetin, apigenin and myricentin |
LRRK2 kinase inhibitors pyrroloquinoline quinone Dopamine related compounds |
Antiglutamatergic compound Riluzole TCM Hesperidin and THSG Non-peptidyl natural Curcumin and Rosmarinic CK-1δ inhibitors Compound 20 and 24 ZINCPharmer 1, 2, 3, 16, 17, 18, 22,24,25,27,28 |
T1-11 from Chinese medicinal herb inhibiting transcription of adenosinergic pathway |
2. Alzheimer’s disease
AD is classified into two types; sporadic AD (SAD), and familial/early onset AD (FAD), and is considered the most common cause of dementia [54]. The gradual death and malfunctioning of neurons in the disease lead to loss of cognitive function and memory. AD is characterized by hyperphosphorylated microtubules linked with tau proteins forming neurofibrillary tangles within the cell, this with accumulation of amyloid (Ab) plaque around the neurons [55, 56]. The amyloidogenic pathway is exacerbated by mutations, and disturbs the normal pathway in which a-secretase acts on the membrane protein APP (amyloid precursor protein), followed by a harmful peptide synthesized by g-secretase. While in the amyloidogenic pathway, APP breakdown by b-secretase is followed by g-secretase and leads to Ab plaque formation whose key constituent is the (42 residue) Ab 42 [57, 58].
AD is a progressive neurodegenerative disorder characterized by diminished motor and cognitive functions, and progressive memory loss due to the demise of brain cells, which finally leads the patient to death [59]. AD progression can be classified in three stages; i) dementia, ii) mild cognitive impairment and iii) preclinical (no symptoms or signs) [59]. It was recently reported that more than 4.7 million people over 65 years old have AD in the USA [60]. It has also been predicted globally that one in 85 people will have AD in 2050 [61, 62].
Ab plaque and oligomers are potential synaptotoxins, alter intracellular Ca 3þ levels, inhibit mitochondrial activity, block proteasome functions [63, 64] and stimulate inflammatory processes [65, 66]. The above mentioned processes also contribute to neuron dysfunction.
Hyperphosphorylation of tau proteins result in neurofibrillary tangle accumulations in neurons [3]. Consequently, synaptic and biochemical communication between neurons is interrupted which gradually leads to cell death [67]. FAD is usually caused by autosomal dominant mutations in the presenilin-1 or -2 gene, or in APP [68, 69]. The majority AD cases are sporadic (SAD) [70].
The Apo-ε gene is considered a key high risk factor in SAD [71]. SAD has numerous risk factors including the APOE4 (apolipoprotein E4) allele and aging, which leads to gradual motor function loss, and cardiac and stroke disease [72, 73].
3. Targets in AD
3.1. Acetylcholinesterase
Brain cortex plasticity and certain forms of learning depend on the presence of ACh [74, 75]. The nerve fibers, during transmission of cholinergic release the neurotransmitter ACh which binds other cholinergic nerve fibers in designated receptors and conveys a message for response. The enzyme cholinesterase decreases ACh concentration by hydrolyzing the molecule [76]. Enzymes that bind to and inhibit cholinesterase result in increasing ACh synapse concentrations [77]. ACh accumulation causes continuous stimulations of glands and muscles; and potentiates parasympathetic activities including constrictions of bronchioles, respiratory tract mucus secretion, slowed heart rate, pupil constriction, vasodilatation, and increased tear, saliva, and sweat production [78].
Acetylcholinesterase (AChE) inhibitors disrupt the cholinergic pathway in the basal forebrain and cerebral cortex which contribute to cognitive impairment in AD patients [79, 80]. Tacrine, rivastigmine, galantamine and donepezil are drugs that act as AChE inhibitors and are approved for symptomatic relief [81]. CADD has promoted progressive advancements in development and design of more potent drug target ligands against diseases. Besides the approved marketed AChE inhibitors, numerous new natural and modified synthetic compounds have been shown to present potent cholinesterase activity. Recently, a mini-review also summarized various approaches used to design and identify multi-targeted sites against AD [82].
Besides the catalytic site on AChE, the presence of a peripheral anionic site (PAS) has been implicated in promoting formation and localization of amyloid fibrils. Various novel flavonoid derivatives have been reported that can bind AChE at both sites, and provide promising results as compared to donepezil and rivastigmine for AChE activity inhibition [83]. Another four flavonoid derivatives; macluraxanthone, kaempferol, rutin, and quercetin were computationally and chemically tested for AChE inhibition, and the quercetin and macluraxanthone derivatives also showed potential inhibitory activity against cholinesterase [84].
Various novel modified carbamates have been synthesized and analyzed using in silico and in vitro approaches and have shown good inhibitory activity against AChE [85]. Pyridopyrimidine, a novel compound developed using in silico molecular docking studies, and confirmed with in vitro synthesis, inhibits AChE activity with more potency than the drug galantamine [86]. A hybrid of aminopyridine and donepezil (6-chloro-pyridonepezil and pyridonepezil) has been reported as a potential inhibitor of cholinesterase using in vitro and in silico analyses. These compounds inhibit both the PAS and catalytic sites [87, 88]. Several derivatives of piperazine were also reported as AChE inhibitors and a few acted as dual-site inhibitors [89]. Das et al., (2017) elucidated flavonoids with potential AChE inhibition which might be helpful in Alzheimer's disease management [90].
3.2. N-methyl-D-aspartate Receptor
In patients with AD, hyper-activation of N-methyl-D-aspartate (NMDA) glutamate type receptors causes continuous and excessive Ca2+ influx via receptor associated ion channels [91]. The transmission of glutamate-mediated synaptic signaling is significant for normal functioning of the nervous system where glutamate behaves as a vital excitatory neurotransmitter in the brain [92]. Hyperactivation of NMDA receptors with glutamate results in production of both free radicals and various enzymes that can contribute to neuronal cell death. During chronic and acute neurodegenerative disorders glutamate can be released inappropriately and is usually not eliminated properly due to metabolic energy disruption. Genetically compromised neurons cannot maintain ionic homeostasis in the absence of energy and become depolarized. Normal Mg2+ blockage of NMDA receptor coupled channels is relieved by this depolarization [93]. Therefore, it is supposed that excessive stimulation of glutamate receptors occurs during neurodegenerative symptoms and ischemia. An antagonist of NMDA receptors could be therapeutically beneficial in a number neurological disorders including neuropathic pain syndromes, dementia and strokes [94]. NMDA receptors are composed of various subunits such as the NR1, NR2A-D, NR3A or NR3B subunits. These subunits form in tetramers to compose the receptor and the composition of these subunits determines the parameters and pharmacology of the receptor-ion channel complex. Alternative splicing (NR1 and the other subunits) further contributes to the pharmacological properties of the receptor [95].
The only marketed NMDA receptor antagonist (memantine) presents rapid receptor blocking and unblocking activity [96]. Memantine is the only NMDA antagonist drug available on the market and there are numerous molecular docking studies in progress to a design active and novel ligands to target this receptor in AD.
Several novel ligands have been identified with successful molecular docking results including triazolyl-amidine derivatives [97], 3-substituted-1H-indoles [98], 1-benzyl-1,2,3,4-tetrahydro- b-carboline [99], 3-hydroxy-1H-quinazoline-2,4-dione derivatives [100] and others. Ifrenprodil and similar molecules act as potential inhibitors of the NR2B subunit of NMDA and were discovered in molecular docking studies [101, 102].
3.3. Muscarinic and Nicotinic ACh Receptors
Muscarinic receptors (mAChR) are ACh receptors present in the peripheral and central nervous system (CNS) that form G protein receptor complexes in cell membranes of neurons, and other cells. Muscarinic receptors are involved in learning, motor control, and memory management in the CNS. These receptors are divided into five different subtypes named M1 to M5 [103], and play different roles in the parasympathetic nervous system (PNS) acting as the principal end receptors for ACh released from postganglionic fibers. The M1 type of mAChR plays a key role in learning, memory management, and cognitive processing in the cerebral cortex and in the hippocampus as impaired by AD [104].
Cholinergic activation can restore cholinergic deficits that are a principle feature of AD. Several studies have been performed with muscarinic agonists that improved cognitive functions in patients, but were unable to complete trials due to co-activation of other non-specific subtypes [105]. Both α4β2 and α7 nicotinic receptor subtype expressing neurons have been observed as damaged in AD patients [106]. In this last decade, certain selective M1 subtype agonists such as AF292, AF267B, AF150 and AF102B of the AF series of drugs have been tested on AD patients. AF267B showed effective pharmacokinetics and with oral administration also penetrates the blood-brain barrier, and AF267B, AF102B and AF150(S) were reported to have decreased Aβ, and elevated non-amyloidogenic APP and neurotrophic effects [107].
Formation of AD amyloid decreases the receptor’s ability to transmit signals, resulting in decreased cholinergic activity. Activation of mAChR M1 can attenuate the pathological features of Alzheimer’s and restore cognitive functions. Certain mechanisms decrease hyper-phosphorylated tau and upregulate α-APP; and hypocholinergic effects result in Aβ formation [108]. An allosteric candidate of M1 (77-LH-28-1) from GlaxoSmithKline (Harlow, UK) presented both effective CNS penetration and pharmacological profile. VU0364572 and VU0357017, two M1 selective agonists from Vanderbilt Centre for Neuroscience Drug Discovery, (Nashville, TN, USA) were analyzed and tested on cell lines of animal models and showed potentially active behavior on several parameters. Certain agonists of M1 could not complete clinical trials [109]. Elan Pharmaceuticals developed an α7 nicotinic receptor agonist EVP-6124, terminating its phase II trial [110]. Awasthi et al., (2016) summarized the results of natural compound analyses utilizing computational approaches of therapeutic molecules effective in preventing Aβ plaque formation [111].
3.4. Tau Proteins
The microtubule linked Tau proteins have a key role in microtubule stability and assembly and are also involved in maintaining cell integrity. Primarily in axons, they are present in phosphorylated soluble form. In AD, hyperphosphorylation of tau proteins causes insoluble intracellular neurofibrillary tangles in neurons. Normal synaptic plasticity is disturbed causing neurodegenerative changes. Cdk-5 and glycogen synthase kinase (Tau kinase 1/GSK-3b) also have involvement in hyperphosphorylation of tau proteins [112, 113]. Thus, reducing hyperphosphorylation of tau proteins by inhibiting GSK-3b has been suggested as a potential therapeutic alternative against AD.
Several reputed pharmaceutical companies including GlaxoSmithKline (Harlow, UK), Eli Lilly (IN, USA), and Roche (Basel, Switzerland) have analyzed and tested numerous GSK-3b inhibitors. Maleimide derivatives including quinolones, oxadiazole, benzimidazoles, imidazopyridines and derivatives of pyrimidine thiazolidinedione are a few common compounds which presented potential activities during in silico analyses and were further analyzed by in vitro assay [114].
3.5. Beta-secretase Enzymes
To treat AD, β-secretase initiates amyloid beta generation and is considered a potential drug target to reduce cerebral levels of APP. APP is subject to degradation via non-amyloidogenic pathways or amyloidogenic pathways. Either β-secretase or α-secretase cleave APP, and γ-secretase processes the remaining membrane attached fragments [57]. Α- cleavage followed by γ-cleavage products are non-amyloidogenic and highly soluble [58]; β-secretase mediated cleavage produces Aβ. APP amyloidogenic cleavage results in APP intracellular domain synthesis that has the ability to change diverse cellular functions [115]. APP synthesis in neuronal cell bodies undergoes axonal transport being contained in transport vesicles. Presynaptic terminals secrete Aβ into the extracellular matrix, and fibrillary Aβ forms outside of the neurons.
The APP gene also has FAD mutations which either modulate γ-secretase activity to increase amyloidogenic Aβ42-Aβ40 or β-cleavage to α-cleavage [57]. The amyloid processing pathway makes BACE1 (β-sectetase/memapsin 2) a potential drug target. BACE1 is present in the intracellular acidic compartments, and is a type 1 transmembrane aspartyl protease. Its highest expression is found in neurons. Knockouts and over expression of BACE1 respectively decrease and increase production of Aβ [116]. There are two aspartic acid residues (Asp-32 and Asp-228) in the active site of BACE1 located in the hydrophobic cleft. A significant role is played by water molecules which help maintain enzymatic function and stability [116]. In silico studies have reported two first generation BACE1 inhibitors (OM00-3 and OM99-2) that behave like natural substrates [117].
Derivatives of aminoquinazoline, acyl-guanidine, hydroxyethylene (HE), aminoimidazole, carbinamine, and hydrox-yethyleneamine (HEA) are reported inhibitors [118]. Derivatives of synthetic coumarin were validated computationally as dual inhibitors of BACE1 and AChE [105, 106]. By employing molecular docking analyses, dual inhibitors of BACE1 and AChE have been reported for hydroxymethylcarbonyl, HEA and HE scaffolds. Interestingly, these two inhibitors revealed potential activity in cell based assays [121]. In other in silico analyses, flavones and flavonols, namely morin, kaempferol, quercetin, apigenin and myricetin have been reported as potential inhibitors of BACE1 [122]. Peptidomimetic inhibitors of BACE1 have a statin based structure with effective IC50 values and binding efficacy [123].
3.6. In vitro and in vivo Validation of Identified in silico Molecules
Derivatives of flavonoid have been validated in in vitro AChE rat studies, with better inhibitory activity than was observed for rivastigmine and certain inhibitory activities similar to donepezil [83]. Nordihydroguaiaretic acid is an effective phenolic lignin extracted from Larrea tridentates. This acid was observed as a cholinesterase inhibitor, having potential activity similar to marketed drugs and has an anti-aggregation effect on Aβ [124].
Numerous novel carbamate molecules have been designed and chemically synthesized against AChE and better inhibitory activity has been observed as compared to the currently available rivastigmine [85]. Certain derivatives of pyridopyrimidine have been reported to have about 2-2.5 times higher inhibitory effect against AChE as compared to galantamine, these were validated via in vitro enzyme assay [86]. Derivatives of pyridonepezil have been reported as effective and selective inhibitors of AChE when compared with the reference compound donepezil [87].
Derivatives of 4-hydroxycoumarin were also reported as significant inhibitors of AChE [125]. Novel derivatives of piperidine showed dual inhibitory activities against Aβ and AChE aggregation in in vitro assays [126]. Certain 6-chloro-pyridonepezils were also reported as dual inhibitors against PAS and the catalytic site of AChE [88]. Derivatives of peperzine also showed dual site inhibitory effects [89].
Derivatives of HE present dual inhibitory activities against both AChE and BACE1 [123]. Flavonoids, specifically quercetin and myricetin reveal potential BACE1 inhibitory effects. Extracellular Aβ concentration and neuronal BACE1 secretion showed significant reductions after quercetin and myricetin administrations [124].
Benzodiazepine modified molecules presented potential inhibition against BACE1 in a cell based assay [127]. Derivatives of HEA were reported as a significant inhibitor of BACE1 in a preclinical animal model [128]. Takeda Pharmaceuticals Japan developed a non-peptide novel BACE1 inhibitor (TAK-070) which significantly reduced the activity of Aβ in a mouse model [129].
Another compound against BACE1 from Merck & Co., Kenilworth, NJ, USA, MK-8931 is in clinical trial phase 3 [129]. Torrey Pines Therapeutics, Inc., CA, USA) developed AF267B M1, a muscarinic agonist to decrease levels of Aβ, it was shown to prevent aggregation in mouse and preclinical rabbit models. AF102B, a long-term treatment for AD patients decreased Aβ levels in cerebrospinal fluid [130].
4. Parkinson’s disease
Parkinson’s disease (PD) has become the second most common old age (roughly 60 years old), movement disorder disease [1]. The substantia nigra is the part of the brain which secretes dopamine and other neurotransmitters for communication with movement control centers [131]. When substantia nigra cells die, the amount of dopamine falls, which negatively affects movement control centers. The cause of substantia nigra cell death is still not fully understood, but it is believed to be due to additive genetic and environmental factors. In some cases, PD is genetic, yet it is mostly found to occur due to poor lifestyle and diet [1]. Patients gradually develop the disease, which first affects one side, and later the other side of the body, leading to trembling and stiffness of the jaw, hands, arms, legs, and trunk, causing slowness of movement, with poor balance and body coordination. In severe conditions, patients also face difficulties walking and chewing, and also develop depression and sleeping problems.
Studies have shown that expression abnormalities in SNCA, PARK2, PINK1, DJ1, LRRK2, and ATP13A2 genes disturb generation/reception in the dopamine pathway, yielding to PD [132]. Dopamine is received specifically by 5 dopamine receptors DRD1 – DRD5 [5, 133, 134]. Adenosine A2A receptor blockages, as expressed in basal ganglia are also involved in PD development [135-137].
4.1. LRRK2
The ubiquitously expressed gene LRRK2 has important functions in the region where dopaminergic neuronal degeneration starts in PD patients [138, 139]. More than 40 mutations are reported for the LRRK2 gene, and most of its biology is still poorly understood. The LRRK2 gene encodes a large 2527 amino acid transcript encoding a protein with multiple protein interactions and enzymatic domains [140, 141]. Many studies have been performed to uncover structural features of the LRRK2 protein including domain analyses and inhibitor design. LRRK2 kinase inhibitors are a hot topic of interest for pathological LRRK2 activity [142]. Aside from first generation inhibitor tools for LRRK2, computational design and docking has revealed the first successful potent inhibitor for mouse LRKK2. A panel of 160 cell permeable and ATP competitive kinase inhibitors used for LRRK2 dephosphorylation at serine cluster including Ser910/935/955/973 have shown positive results [1, 131, 141].
4.2. SNCA
The SNCA gene codes for the protein alpha-synuclein involved in neuropsychiatric pathology. There are a total of 5 exons for the SNCA gene encoding 140 amino acids [143]. Evolutionary studies have shown that amino acids ranging from 32 to 58 residues and having an N-terminal domain and degenerative amino acid motif “KTKEGV” have critical structural and functional implications derived through lineage specific substitutions under epistatic influence [144]. Homology models showed amphipathic helices as a characteristic structural SNCA feature. Pyrroloquinoline has been computationally identified as inhibiting SNCA derived plaques [145, 146].
4.3. PINK1, PARK2, and DJ1
The autosomal recessive “three musketeers” of neuroprotection for PD are the PINK1, PARK2, and DJ1 genes [147]. Much less work has been done on their three proteins and researchers are trying to figure out pathways and possible inhibition involved in their mutated expression. The PINK1 protein, composed of 581 amino acids was recently localized; having a conserved kinase domain facing the mitochondrial cytoplasm, and is thought to be composed of multiple mitochondrial targeting sequences [132]. The involvement of PINK1 in PD is suggested in three pathways; 1) incorrect phosphorylation and assembly of mitochondrial complexes affecting Na+ / K+ homeostasis, 2) activation of the apoptosis pathway, and 3) disturbance in mitochondrial localization of PARK2 1. PARK2 also termed as Parkin is an autosomal recessive PD gene encoding a 52 kDa protein. Much work has been performed to functionally characterize PARK2, but most of its pathway is still unclear. PARK2 is an E2 dependent E3 ubiquitin ligase with a role in sophisticated mitochondrial quality control pathways.
DJ-1 was first identified as an oncogene and later associated with familial PD; and localized in the mitochondria, has a role in neuroprotective activity [148]. DJ-1 forms as a single 20 kDa domain but its abnormal behavior allows altered mitochondrial morphological dynamics through increased Reactive Oxygen Species (ROS), which may still be rescued by cell permeable glutathione precursors, or by PRAK2 or PINK1 overexpression [149]. Even though its crystal structure was available a decade ago, the dynamics and understanding of the DJ-1 neuroprotective pathway is still little known [1, 147].
Dopamine receptors are PD treatment targets and dopamine related compounds can be directly provided to these receptors to enable proper function [150-152]. Such treatments are provided when no other treatment is available because it does not cure, but helps the neurons to perform their normal functions. In silico studies combined with in vitro studies have helped to discover many compounds that target certain dopamine receptors [5, 133, 152-156]. It is very difficult to target dopamine receptors inside the brain due to brain permeability barriers, thus mixed studies are focused currently on computational drug design, and their toxicity properties [157]. Due to the complexity of the brain function, it has become an ambitious task for researchers to design non-toxic or less-toxic dopamine related compounds [134, 158, 159].
5. Amyotrophic Lateral Sclerosis
ALS is a lethal neurodegenerative disease, characterized by the loss of both upper motor neurons in the motor cortex, and lower motor neurons in the spinal cord and brainstem. This adult onset disorder leads to muscle weakness and atrophy, twitching and convulsiveness [160]. Denervation of lower motor-muscle neurons, and axon retractions lead to the failure of axonal connections which develops as the signs and symptoms of ALS. At first, axon retraction is restored with collateral re-innervation and axons that appear less susceptible to degenerative events. However, on account of these newer less susceptible neurons, the compensation mechanism fails as the disease progresses. It has also been shown in animal models that the neuronal cell body dies only after the preliminary stage of axonal retraction and dysfunction [161, 162].
In humans, the particular timing of ALS events is less known, but the order was confirmed by autopsies in ALS patients who died early. ALS is conventionally categorized into sporadic ALS (SALS) and familial ALS (FALS), which are clinically similar. Incomplete penetration, partial family history and non-paternity are reasons for misclassification. SALS patients are not hereditarily affected, yet FALS is predominantly hereditary, being caused by mutations in a diverse set of genes [160]. Approximately, 90% of ALS patients present the sporadic form, the number of FALS causative genes is high, but the patho-mechanism is still unknown [163, 164].
Currently, there is no therapy for ALS other than the anti-glutamatergic compound riluzole, which is less effective in improving symptoms and only prolongs median survival by a few months [165]. There remains a great need to develop novel therapeutics for ALS and identify its underlying mechanisms. Although, most ALS cases are sporadic, the triggering mechanism is unknown which hinders target discovery and drug development for this neurodegenerative disorder [166, 167]. Clinical trials have been performed on more than 30 compounds, but most of these failed to provide the therapeutic benefits in ALS patients [168].
ALS etiology involves multiple genetic factors. However, increasing numbers of ALS linked genes are being identified. Superoxide dismutase type 1 (SODI) mutations are described as one of the key causes of ALS [167]. SODI mutations have been found in both FALS and SALS and extensively studied. Mutations in genes, including FIG4, FUS/TLS, TARDBP, ANG, VAPB and hexanucleotide repeat expansion (GGGGCC) in C9ORF72 are genetic factors responsible for the onset of ALS. In ALS, genetic association studies reveal other risk factors including insertions or deletions in paraoxonase-1 (PON1), neurofilament heavy chain genes, and hetereochromatosis genes (HFE) [163, 169].
5.1. SODI
SODI, is an antioxidant enzyme involved in detoxification of superoxide radicals becoming enzymatically active by binding zinc and copper ions, and engenders a highly conserved intra-molecular disulfide bond [170]. Variants in Cu/Zn binding to SODI can augment aggregation and reduce protein stability concomitant with ALS [171, 172]. Genetic linkage of SODI mutations in ALS has been demonstrated: including alanine 4 to valine (A4V) [173], histidine 46 to arginine (H46R) [174] and isoleucine 113 to threonine (I113T) [175]. A4V mutation is a frequent causative agent of ALS in the United States and accounts for about 50% of SODI-ALS patients. About, 80% Japanese familial ALS are affected by mutant H46R; and I113T mutation is also common in FALS. When designing novel ALS drugs, SODI stabilization and aggregation are targeted [176].
From Traditional Chinese Medicine (TCM), Huang et al., (2014) [176] conducted research to identify inhibitors of mutant SODI for ALS therapy. Through CADD, a mutant SODI and dopamine complex was utilized to scrutinize new lead compounds to inhibit aggregation. The world TCM database was employed to screen potential compounds with high-affinity binding to the active site of mutant SODI and validate the stability of the complex for binding through MD simulation assay. The TCM Database@Taiwan (61,000 compounds) and Chang’s laboratory were utilized to screen the database and perform the binding assay. Docking analyses revealed that 2, 3, 5, 4-tetrahydroxystilbene-2-O-𝛽-D glucoside (THSG) and hesperidin had higher binding affinities than dopamine. Glu100 was the common residue for each binding analysis. Root Mean Square Deviation (RMSD) value ranged from 0.16-0.24 nm indicating that all the protein ligand complexes were stable during 5000 ps simulations. Current research has concluded that hesperidin and THSG could be potential targets for ALS therapy to design novel drugs. Currently, there is no effective therapy available for SODI associated ALS and oligomerization of SODI might provide good targets to develop effective ALS therapeutics. In vitro, a SODI oligomerization model tested to screen 640 FDA approved drugs, recognized three effective chemical compound classes that inhibit oligomerization of SODI proteins. The oligomerization model might also be used to test computationally screened compounds to identify and develop effective cures for this devastating disease [166].
5.2. Caspase 3
Khan et al., (2015) [177] conducted in silico analyses to identify the role of caspase-3 in the regulation of multi-neurodegenerative disorders: namely, ALS, HD, AD and PD using an interaction network to identify natural potent non-peptidyl compounds against caspase-3. High caspase-3 activity has been identified in human ALS [178]. The excitatory amino acid transporter-2 (EAAT2) was cleaved by caspase-3 at a specific cytosolic C-terminal site, leading to significant transporter inhibition, Thus, it plays a vital role in ALS pathogenesis [179]. Most of the known inhibitors of caspase-3 are peptidyl in nature due to its proteolytic enzymatic activity for caspase-3 [180].
The STRING database exhibited a 0.506 interaction confidence score for caspase-3 in SODI, indicating strong evidence of interaction; a potential therapeutic target for neurodegenerative disorders including ALS. In current in silico research, a molecular docking study was carried out to identify compounds from plant-derived non-peptidyl natural inhibitors against proteolytic enzymes [177]. Curcumin and rosmarinic acid are most promising leads imitating the inhibitory effects of peptidyl inhibitors. The discovery of natural non-peptidyl inhibitors as viable drug candidates for multiple neurodegenerative disorders is significant. An interaction network including all of the causative proteins of neurodegenerative diseases including ALS and protein-protein interaction pathways revealed the importance of CASP-3 and CASP-8 in neurodegenerative disorders. They could be used as potential targets for effective ALS therapies [181].
5.3. Protein Kinase CK-1 Inhibitors
TAR DNA 43 binding protein (TDP-43) accumulation, a pathological hallmark in SALS, actually presents new therapeutic targets for drug development. Post-translational modification of TDP-43 might be the effect of up-regulated protein kinase CK-1 in affected neurons, such that phosphorylation of TDP-43 initiates the onset and development of ALS [182].
Salado et al., (2014) utilized a chemical-genetic approach, molecular docking, and cellular-based assays to discover and optimize potent CK-1δ inhibitors for TDP-43 proteinopathies and ALS. The initial screening involved chemically diverse heterocyclic compounds from an in-house chemical library, and a focalized subset of structures was then evaluated. Compound MR-3.15 with an IC50 value of 0.85 μM was further optimized in series of chemical reactions. Molecular docking studies were carried out on N-Benzothiazolyl-2-phenyl-acetamide derivatives with CK-1δ crystallographic structures, and a potent inhibitor (Compound 20) (IC50 = 23 nM) and (Compound 24) showed the lowest energy poses and the most populated clusters in ATP binding sites; these were evaluated further in cellular assays. They present the ability to cross the blood-brain barrier, making them good drug candidates and possibly useful as tools for novel drug development [183].
5.4. MAPK
Several cellular events including apoptosis, differentiation, oncogenesis, and mitogenesis are linked to the mitogen-activated protein kinases (MAPKs) family [182]. The p38 MAPK is associated with proinflammatory cytokine inhibition and is being studied as a therapeutic target in preclinical animal models of CNS diseases including ALS [184].
Pharmacophore generation, together with similarity-based virtual screening of more than 18.3 million compounds was executed to identify novel potential compounds able to reach phase 3 clinical trials. Twenty pharmacophore hypotheses were generated with linear binding modes and screening was conducted using the ChemBridge and ZINC (CNS) databases. The ZINCPharmer database, having 176 million conformers of 18.3 million compounds was also employed in another pharmacophore screening. Pharmacokinetic analysis scrutinized 11 compounds having (Top-5) drug like properties ranking in PharmMapper for p38α MAPK. Among the scrutinized compounds, similarity-based VS generated 4 compounds (compounds 24, 25, 27 and 28), together with 7 compounds (compounds 1, 2, 3, 16, 17, 18, and 22) obtained from pharmacophore based VS, which might be used as potential drug candidates after in vitro assays and quantitative structure-activity relationship studies [185].
5.5. Nav1.6 Sodium Channel
The Nav1.6 channel could serve as an important ALS target for design drug therapy. In silico analyses demonstrated the interaction of Riluzole with the Nav1.6 channel, Tyr-1787, Gln-1799 and Leu-1843 as key residues for drug discovery and development [166, 186].
6. Huntington’s disease
An inherited autosomal dominant old age disease, Huntington’s disease (HD), is another neurodegenerative disease which wastes away neuronal structures deep within brain, at first causing uncontrolled movements and balance problems, and later leading to more severe problems like the inability to walk, swallow, talk, think, and perceive, and affects emotions and stable memory (family members recognition) [187]. The available drugs manage some HD symptoms, but today’s drugs can neither cure nor slow down disease onset. HD is caused by one HTT gene present on chromosome 4. Transgenic mouse model studies have suggested that HD can develop through mutations in exon1 of the HTT gene [188]. HD can be diagnosed through blood sample analysis together with family history, Magnetic Resonance Imaging (MRI), and Computed Tomography (CT).
Normal alleles of HTT contain trinucleotide repeats (CAG repeats). Studies have shown that between 28 to 35 CAG repeats is normal and unassociated to HD. When exceeding 28 CAG repeats, it causes replication instability and CAG repeats may increase further. While 36–40 CAG repeats may cause abnormal protein behavior, 41 or more repeats signifies complete penetration, and symptoms begin to appear [187, 189, 190]. The higher replication rates in spermatogenesis as compared to oogenesis suggest that the HTT gene is mostly transferred paternally, a phenomenon called anticipation. It can be inferred from certain studies that the HTT gene presents dynamic behavior sync; even identical twins present differing HD onset ages, and different clinical symptoms [191]. There are several interacting proteins involved with HTT proteins, and interestingly each different protein interaction causes loss of the different type of neurons. Currently, known HTT protein interacting partners are HTT-interacting protein 1, SRC homology region 3-containing GRB2-like protein 3, protein kinase C, and casein kinase substrate in neurons 1, HTT-associated protein 1, postsynaptic density-95, FIP-2, specificity protein 1 and nuclear receptor co-repressor [192].
It is highly recommended to screen for HD in newborn babies so that management and care can be provided and lessen the effects of HD [193]. Since HTT mutations are genetic, and control is currently not possible; more efforts are being made to target HTT interacting proteins and minimize HD symptoms. In a recent study, T1-11 purified from a Chinese medicinal herb, and used for about 1500 years to treat brain-related problems, has been shown to provide beneficial effects for certain HD symptoms inhibiting transcription for the adenosinergic pathway [194].
7. Future Directions
Molecular docking studies are used to correctly estimate receptor binding to the ligand site and to accurately predict the binding strength. To elucidate potential molecules for treating neurodegenerative disorders, numerous compounds against both known and novel targets can be analyzed and designed with molecular docking. Multi-target inhibitors of neurodegenerative disorders can also be designed and analyzed. Currently, no effective treatments are available to cure or prevent neurodegenerative disorders but numerous approved drugs provide temporary and modest improvements. To directly target the causes of neurodegenerative disorders would constitute a rational approach. Numerous compounds
mentioned in this study have been withdrawn in different stages of drug design including clinical trials because of receptor protein non-specificity or ineffectiveness in human trials. In terms of structural accessibility and the blood-brain barrier, the brain is considered a most difficult organ. Thus, it is often less likely that molecules predicted in silico, in vivo and in vitro are effective in situ. In silico analyses using molecular modeling and molecular docking studies have been shown to be effective in novel drug design. Yet to design potential ligands against neurodegenerative disorders more funding for critical analysis is needed.
Next-generation sequencing has enabled researchers to sequence and compare inter- and intra-specific genomes. Genome Wide Association Studies (GWAS) are another tool that studies how genes are correlated with each other within a genome, and CRISPR-CAS technology is now reshaping our understanding and ability to solve genetic problems by specifically editing genes. Through CRISPR-CAS, in the near future trinucleotide repeats of HD may well be controlled to within supportable limits. Such methodologies in research are progressively being integrated to solve neurodegenerative disorders. Functional MRI (fMRI) technique is being used to identify active brain regions for specific tasks, whereas fMRI in-conjunction with pattern recognition analysis is being used for searching linked brain areas to analyze patients with Alzheimer’s and Parkinson’s diseases. Due to the high level of brain complexity; its functions and collateral effect trade-offs; it is still quite difficult to cure disease without effects spreading to other brain areas. Computational analyses offer a place to pre-test and simulate brain parts using various approaches, yet new techniques and algorithms are required which may help to design and cure neurological diseases with minimum trade-offs.
We are in the era of Big Data with scientists producing a gigantic amount of data by various means including bioinformatics. Usually, computational analyses give differing results for the same sample; this is due to versions of tools and algorithms utilized, parameters, environments, and other reasons. We should build a database that will have all the published in silico molecules against diseases. Using a single open access database where all published articles are submitted from the scientific community, from Big Data gathered together in one repository, similar analyses with minor parameter variations can be analyzed and meaningful results deduced.
Acknowledgements
The authors acknowledge Dr. Asif Mir Department of Bioinformatics and Biotechnology, International Islamic University of Islamabad, for his continuous guidance and support throughout the manuscript.
List of Abbreviations
- 3D
3-dimensional
- Ab
amyloid
- AChE
acetylcholinesterase
- AD
Alzheimer’s Diseases
- ADMET
Absorption, Distribution, Metabolism, Excretion and Toxicity
- ALS
Amyotrophic Lateral Sclerosis
- APP
amyloid precursor protein
- BACE1
β-sectetase/memapsin 2
- CADD
computer-aided drug design
- CNS
central nervous system
- CT
Computed Tomography
- EAAT2
Excitatory amino acid transporter-2
- FAD
familial/early onset AD
- FALS
familial ALS
- GWAS
Genome Wide Association Studies
- HD
Huntington’s disease
- mAChR
muscarinic receptors
- MAPKs
mitogen-activated protein kinases
- MD
Molecular Dynamics
- MRI
Magnetic Resonance Imaging
- NIH
National Institute of Health
- NMDA
N-methyl-D-aspartate
- NMR
Nuclear Magnetic Resonance
- PAS
peripheral anionic site
- PD
Parkinson’s Disease
- PDB
Protein Data Bank
- PNS
parasympathetic nervous system
- PON1
paraoxonase-1
- RMSD
Root Mean Square Deviation
- ROS
Reactive Oxygen Species
- SAD
sporadic AD
- SALS
sporadic ALS
- SODI
Superoxide dismutase type 1
- TCM
traditional Chinese medicine
- TDP-43
TAR DNA binding protein 43
- THSG
2, 3, 5, 4-tetrahydroxystilbene-2-O-𝛽-D glucoside
- VS
Virtual Screening
Consent for Publication
Not applicable.
Conflict of Interest
The authors declare no conflict of interest, financial or otherwise.
References
- 1.Sanders S., Zhang Z., Tang B. 2013.
- 2.Jntu K.V.R., Jntu L.P.R. A neural network based classification and diagnosis of brain hemorrhages.; 2009. [Google Scholar]
- 3.Lin J.S. Brain structures and mechanisms involved in the control of cortical activation and wakefulness, with emphasis on the posterior hypothalamus and histaminergic neurons. Sleep Med. Rev. 2000;4(5):471–503. doi: 10.1053/smrv.2000.0116. [DOI] [PubMed] [Google Scholar]
- 4.Navarrete A., van Schaik C.P., Isler K. Energetics and the evolution of human brain size. Nature. 2011;480(7375):91–93. doi: 10.1038/nature10629. [DOI] [PubMed] [Google Scholar]
- 5.Sehgal S., Khattak N., Mir A. Structural, phylogenetic and docking studies of D-amino acid oxidase activator (DAOA), a candidate schizophrenia gene. Theor. Biol. Med. Model. 2013;10(1):3. doi: 10.1186/1742-4682-10-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sehgal S.A. Pharmacoinformatics, adaptive evolution, and elucidation of Six novel compounds for Schizophrenia treatment by targeting DAOA (G72) Isoforms. BioMed Res. Int. 2017;2017:1–19. doi: 10.1155/2017/5925714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tahir R.A., Sehgal S.A. Pharmacoinformatics and molecular docking studies reveal potential novel compounds against Schizophrenia by target SYN II. Comb. Chem. High Throughput Screen. 2018;21:180213092018. doi: 10.2174/1386207321666. [DOI] [PubMed] [Google Scholar]
- 8.Sehgal S.A. Pharmacoinformatics and molecular docking studies reveal potential novel proline Dehydrogenase (PRODH) com-pounds for Schizophrenia inhibition. Med. Chem. Res. 2017;26(2):314–326. doi: 10.1007/s00044-016-1752-2. [DOI] [Google Scholar]
- 9.Sehgal S.A., Mannan S., Ali S. Pharmacoinformatic and molecular docking studies reveal potential novel antidepressants against neurodegenerative disorders by targeting HSPB8. Drug Des. Devel. Ther. 2016;10:1605. doi: 10.2147/DDDT.S101929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sehgal S.A., Mannan S., Kanwal S., Naveed I., Mir A. Adaptive evolution and elucidating the potential inhibitor against schizophrenia to target DAOA (G72) isoforms. Drug Des. Devel. Ther. 2015;9:3471. doi: 10.2147/DDDT.S63946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sehgal S.A., Hassan M., Rashid S. Pharmacoinformatics elucidation of potential drug targets against migraine to target ion channel protein KCNK18. Drug Des. Devel. Ther. 2014;8:571. doi: 10.2147/DDDT.S63096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sehgal S.A. Kanwal. S.; Tahir, R.A.; Khalid, Z.; Hammad, M.A. In silico elucidation of potential drug target sites of the Thumb Index Fold Protein, Wnt-8b. Trop. J. Pharm. Res. 2018;17(3):491–497. doi: 10.4314/tjpr.v17i3.15. [DOI] [Google Scholar]
- 13.Sehgal S., Tahir R., Shafique S., Hassan M., Rashid S. Molecular modeling and docking analysis of CYP1A1 associated with head and neck cancer to explore its binding regions. J. Theor. Comput. Sci. 2014;1(112):2. [Google Scholar]
- 14.Jamil F., Ali A., Sehgal S.A. Comparative modeling, molecular docking, and revealing of potential binding pockets of RASSF2; a candidate cancer gene. Interdiscip. Sci. 2016:1–10. doi: 10.1007/s12539-016-0145-z. [DOI] [PubMed] [Google Scholar]
- 15.Tahir R.A., Sehgal S.A., Khattak N.A., Khattak J.Z.K., Mir A. Tumor necrosis factor receptor superfamily 10B (TNFRSF10B): an insight from structure modeling to virtual screening for design drug against head and neck cancer. Theor. Biol. Med. Model. 2013;10(1):1. doi: 10.1186/1742-4682-10-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leuzinger W., Baker A.L., Acetylcholinesterase I. Large-scale purification, homogeneity, and amino Acid analysis. Proc. Natl. Acad. Sci. USA. 1967;57(2):446–451. doi: 10.1073/pnas.57.2.446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.MacArthur M.W., Laskowski R.A., Thornton J.M. Knowledge-based validation of protein structure coordinates derived by X-ray crystallography and NMR spectroscopy. Curr. Opin. Struct. Biol. 1994;4(5):731–737. doi: 10.1016/S0959-440X(94)90172-4. [DOI] [Google Scholar]
- 18.Sippl M. J. Recognition of errors in three-dimensional structures of proteins. 1993. [DOI] [PubMed]
- 19.Congreve M., Chessari G., Tisi D., Woodhead A.J. Recent developments in fragment-based drug discovery. J. Med. Chem. 2008;51(13):3661–3680. doi: 10.1021/jm8000373. [DOI] [PubMed] [Google Scholar]
- 20.Perola E., Walters W.P., Charifson P.S. A detailed comparison of current docking and scoring methods on systems of pharmaceutical relevance. Proteins. 2004;56(2):235–249. doi: 10.1002/prot.20088. [DOI] [PubMed] [Google Scholar]
- 21.Yuan Z., Bailey T. L., Teasdale R. D. Prediction of protein B-factor profiles. 2005. [DOI] [PubMed]
- 22.Cavasotto C.N., Phatak S.S. Homology modeling in drug discovery: current trends and applications. Drug Discov. Today. 2009;14(13):676–683. doi: 10.1016/j.drudis.2009.04.006. [DOI] [PubMed] [Google Scholar]
- 23.Eswar N., John B., Mirkovic N., Fiser A., Ilyin V.A., Pieper U., Stuart A.C., Marti-Renom M.A., Madhusudhan M.S., Yerkovich B. Tools for comparative protein structure modeling and analysis. Nucleic Acids Res. 2003;31(13):3375–3380. doi: 10.1093/nar/gkg543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martí-Renom M.A., Stuart A.C., Fiser A., Sánchez R., Melo F., Šali A. Comparative protein structure modeling of genes and genomes. Annu. Rev. Biophys. Biomol. Struct. 2000;29(1):291–325. doi: 10.1146/annurev.biophys.29.1.291. [DOI] [PubMed] [Google Scholar]
- 25.Wallner B., Elofsson A. All are not equal: a benchmark of different homology modeling programs. Protein Sci. 2005;14(5):1315–1327. doi: 10.1110/ps.041253405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Karplus M., Petsko G.A. Molecular dynamics simulations in biology. Nature. 1990;347(6294):631–639. doi: 10.1038/347631a0. [DOI] [PubMed] [Google Scholar]
- 27.Ogbonna N. Molecular dynamics simulation. Muizenberg. African Institute for Mathematical Sciences; 2004. [Google Scholar]
- 28.Snow C. D., Sorin E. J., Rhee Y. M., Pande V. S. How well can simulation predict protein folding kinetics and thermodynamiscs. 2005. [DOI] [PubMed]
- 29.Azam S.S., Mirza A.H. Role of thumb index fold in Wnt-4 protein and its dynamics through a molecular dynamics simulation study. J. Mol. Liq. 2014;198(0):313–321. doi: 10.1016/j.molliq.2014.07.007. [DOI] [Google Scholar]
- 30.Karplus M., McCammon J.A. Molecular dynamics simulations of biomolecules. Nat. Struct. Mol. Biol. 2002;9(9):646–652. doi: 10.1038/nsb0902-646. [DOI] [PubMed] [Google Scholar]
- 31.Reddy S., Pati S.P., Kumar P.P., Pradeep H.N., Sastry G.N. Virtual screening in drug discovery -- a computational perspective. Curr. Protein Pept. Sci. 2007;8(4):329–351. doi: 10.2174/138920307781369427. [DOI] [PubMed] [Google Scholar]
- 32.Schneider G. Virtual screening: an endless staircase? Nat. Rev. Drug Discov. 2010;9(4):273–276. doi: 10.1038/nrd3139. [DOI] [PubMed] [Google Scholar]
- 33.Zavodszky M.I., Rohatgi A., Van Voorst J.R., Yan H., Kuhn L.a. Scoring ligand similarity in structure-based virtual screening. J. Mol. Recognit. 2009;22(4):280–292. doi: 10.1002/jmr.942. [DOI] [PubMed] [Google Scholar]
- 34.Irwin J.J., Shoichet B.K. ZINC-a free database of commercially available compounds for virtual screening. J. Chem. Inf. Model. 2005;45(1):177–182. doi: 10.1021/ci049714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ma D-L., Chan D.S-H., Leung C-H. Molecular docking for virtual screening of natural product databases. Chem. Sci. (Camb.) 2011;2(9):1656–1665. doi: 10.1039/C1SC00152C. [DOI] [Google Scholar]
- 36.Tahir I., Ahmad M.N., Islam A.K.M.S., Arbain D. Virtual searching of dummy template for Sinenstein based on 2D molecular similarity using chemdb tool. Indones. J. Chem. 2012;12(3):217–222. [Google Scholar]
- 37.Bottegoni G., Favia A. D., Recanatini M., Cavalli A. The role of fragment-based and computational methods in polypharmacology. 2012. [DOI] [PubMed]
- 38.Schlessinger A., Geier E., Fan H., Irwin J.J., Shoichet B.K., Giacomini K.M., Sali A. Structure-based discovery of prescription drugs that interact with the norepinephrine transporter, NET. Proc. Natl. Acad. Sci. USA. 2011;108(38):15810–15815. doi: 10.1073/pnas.1106030108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mirza M.U., Mirza A.H., Ghori N-U-H., Ferdous S. Glycyrrhetinic acid and E. resveratroloside act as potential plant derived compounds against dopamine receptor D3 for Parkinson’s disease: a pharmacoinformatics study. Drug Des. Devel. Ther. 2015;9:187–198. doi: 10.2147/DDDT.S72794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rose S.T. Cheeseright finding better leads using molecular fields. Chem. Bio. News; 2008. [Google Scholar]
- 41.Goodsell D.S., Morris G.M., Olson A.J. Automated dock-ing of flexible ligands: applications of AutoDock. J. Mol. Recognit. 1996;9(1):1–5. doi: 10.1002/(sici)1099-1352(199601)9:1<1::aid-jmr241>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- 42.Morris G.M., Huey R., Olson A.J. 2008. [Google Scholar]
- 43.Trott O., Olson A.J. Auto Dock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010;31(2):455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Marcou G., Kellenberger E. Docking with GOLD. 2009.
- 45.Verdonk M.L., Cole J.C., Hartshorn M.J., Murray C.W., Taylor R.D. Improved protein-ligand docking using GOLD. Proteins. 2003;52(4):609–623. doi: 10.1002/prot.10465. [DOI] [PubMed] [Google Scholar]
- 46.Bissantz C., Folkers G., Rognan D. Protein-based virtual screening of chemical databases. 1. Evaluation of different docking/scoring combinations. J. Med. Chem. 2000;43(25):4759–4767. doi: 10.1021/jm001044l. [DOI] [PubMed] [Google Scholar]
- 47.Chen Z., Li H., Zhang Q., Bao X., Yu K., Luo X., Zhu W., Jiang H. Pharmacophore-based virtual screening versus docking-based virtual screening: a benchmark comparison against eight targets. Acta Pharmacol. Sin. 2009;30(12):1694–1708. doi: 10.1038/aps.2009.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Corbeil C.R., Englebienne P., Yannopoulos C.G., Chan L., Das S.K., Bilimoria D., L’heureux L., Moitessier N. Docking ligands into flexible and solvated macromolecules. 2. Development and application of fitted 1.5 to the virtual screening of potential HCV polymerase inhibitors. J. Chem. Inf. Model. 2008;48(4):902–909. doi: 10.1021/ci700398h. [DOI] [PubMed] [Google Scholar]
- 49.Warren G.L., Andrews C.W., Capelli A-M., Clarke B., LaLonde J., Lambert M.H., Lindvall M., Nevins N., Semus S.F., Senger S., Tedesco G., Wall I.D., Woolven J.M., Peishoff C.E., Head M.S. A critical assessment of docking programs and scoring functions. J. Med. Chem. 2006;49(20):5912–5931. doi: 10.1021/jm050362n. [DOI] [PubMed] [Google Scholar]
- 50.Yuriev E., Agostino M., Ramsland P.A. Challenges and advances in computational docking: 2009 in review. J. Mol. Recognit. 2011;24(2):149–164. doi: 10.1002/jmr.1077. [DOI] [PubMed] [Google Scholar]
- 51.Allan B.D. Amyloid β-peptide (1-42)-induced oxidative stress and neurotoxicity: Implications for neurodegeneration in Alzheimer’s disease brain. A review. Free Radic. Res. 2002;36(12):1307–1313. doi: 10.1080/1071576021000049890. [DOI] [PubMed] [Google Scholar]
- 52.Brinton R. Requirements of a brain selective estrogen: ad-vances and remaining challenges for developing a neuroserm. J. Alzheimers Dis. 2004;6:27–35. doi: 10.3233/jad-2004-6s607. [DOI] [PubMed] [Google Scholar]
- 53.Fratiglioni L., Wang H-X. Smoking and Parkinson’s and Alzheimer’s disease: review of the epidemiological studies. Behav. Brain Res. 2000;113(1-2):117–120. doi: 10.1016/S0166-4328(00)00206-0. [DOI] [PubMed] [Google Scholar]
- 54.Thies W., Bleiler L. 2013 Alzheimer’s disease facts and figures. Alzheimers Dement. 2013;9(2):208–245. doi: 10.1016/j.jalz.2013.02.003. [DOI] [PubMed] [Google Scholar]
- 55.Selkoe D. J. Alzheimer’s disease: genes, proteins, and therapy. 2001. [DOI] [PubMed]
- 56.Noble W., Hanger D.P., Miller C.C.J., Lovestone S. The importance of tau phosphorylation for neurodegenerative diseases. Front. Neurol. 2013;4:83. doi: 10.3389/fneur.2013.00083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang Y-w., Thompson R., Zhang H., Xu H. APP processing in Alzheimer’s disease. Mol. Brain. 2011;4(1):3. doi: 10.1186/1756-6606-4-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gazzaley A., Small S. A. 2011.
- 59.Bäckman L., Jones S., Berger A.K., Laukka E.J., Small B.J. Multiple cognitive deficits during the transition to Alzheimer’s disease. Int. J. Integr. Med. 2004;256:195–204. doi: 10.1111/j.1365-2796.2004.01386.x. [DOI] [PubMed] [Google Scholar]
- 60.Hebert L. E., Weuve J., Scherr P. A., Evans D. A. 2013.
- 61.Brookmeyer R., Johnson E., Ziegler-Graham K., Arrighi H.M. Forecasting the global burden of Alzheimer’s disease. Alzheimers Dement. 2007;3(3):186–191. doi: 10.1016/j.jalz.2007.04.381. [DOI] [PubMed] [Google Scholar]
- 62.Niedowicz M. D.; Nelson, P.T.; Murphy, M.P. Alzheimer’s Disease: pathological mechanisms and recent insights. Curr. Neuropharmacol. 2011;9(4):674–684. doi: 10.2174/157015911798376181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lacor P. N., Buniel M. C., Furlow P. W., Clemente A. S., Velasco P. T., Wood M., Viola K. L., Klein W. L. Abeta oligomer-induced aberrations in synapse composition, shape, and density provide a molecular basis for loss of connectivity in Alzheimer’s disease. 2007. [DOI] [PMC free article] [PubMed]
- 64.Yankner B. A., Duffy L. K., Kirschner D. A. Neurotrophic and neurotoxic effects of amyloid beta protein: reversal by tachykinin neuropeptides. 1990. [DOI] [PubMed]
- 65.Shovlin C., Haslett C.L.J. The molecular and cellular basis of disease. Davidson’s Princ. Pract. Med. (Barc.) 1999;2(4):1–56. doi: 10.5582/irdr.2013.v2.4.115. [DOI] [Google Scholar]
- 66.Wenk G. Neuropathologic changes in Alzheimer’s disease. J. Clin. Psychiatry. 2003;64(64) Suppl. 9:7–10. [PubMed] [Google Scholar]
- 67.Chun W., Johnson G. The role of tau phosphorylation and cleavage in neuronal cell death. Front. Biosci. 2007;12:733–756. doi: 10.2741/2097. [DOI] [PubMed] [Google Scholar]
- 68.Waring S. C., Rosenberg R. N. Genome-Wide Association Studies in Alzheimer Disease. Arch. Neurol. 2008;65(3):353–356. doi: 10.1001/archneur.65.3.329. [DOI] [PubMed] [Google Scholar]
- 69.Shioi J., Georgakopoulos A., Mehta P., Kouchi Z., Litterst C.M., Baki L., Robakis N.K. FAD mutants unable to increase neurotoxic Aβ 42 suggest that mutation effects on neurodegeneration may be independent of effects on Aβ. J. Neurochem. 2007;101(3):674–681. doi: 10.1111/j.1471-4159.2006.04391.x. [DOI] [PubMed] [Google Scholar]
- 70.Gazzaley A., Small S.a. Alzheimer’s disease. Network. 2011;22(1-4):173–185. doi: 10.3109/0954898X.2011.638696. [DOI] [PubMed] [Google Scholar]
- 71.Fassbender K., Masters C., Beyreuther K. Alzheimer’s disease: Molecular concepts and therapeutic targets. Naturwissenschaften. 2001;88(6):261–267. doi: 10.1007/s001140100237. [DOI] [PubMed] [Google Scholar]
- 72.Strittmatter W.J., Saunders A.M., Schmechel D., Pericak-Vance M., Enghild J., Salvesen G.S., Roses A.D. Apolipoprotein E: high-avidity binding to beta-amyloid and increased frequency of type 4 allele in late-onset familial Alzheimer disease. Proc. Natl. Acad. Sci. USA. 1993;90(5):1977–1981. doi: 10.1073/pnas.90.5.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mahley R.W., Weisgraber K.H., Huang Y. Apolipoprotein E4: a causative factor and therapeutic target in neuropathology, including Alzheimer’s disease. Proc. Natl. Acad. Sci. USA. 2006;103(15):5644–5651. doi: 10.1073/pnas.0600549103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lane R.M., Potkin S.G., Enz A. Targeting acetylcholines-terase and butyrylcholinesterase in dementia. Int. J. Neuropsychopharmacol. 2006;9(1):101–124. doi: 10.1017/S1461145705005833. [DOI] [PubMed] [Google Scholar]
- 75.Thompson P.A., Wright D.E., Counsell C.E., Zajicek J. Statistical analysis, trial design and duration in Alzheimer’s disease clinical trials: A review. Int. Psychogeriatr. 2012;24(5):689–697. doi: 10.1017/S1041610211001116. [DOI] [PubMed] [Google Scholar]
- 76.Turner P.R., O’Connor K., Tate W.P., Abraham W.C. Roles of amyloid precursor protein and its fragments in regulating neural activity, plasticity and memory. Prog. Neurobiol. 2003;70(1):1–32. doi: 10.1016/S0301-0082(03)00089-3. [DOI] [PubMed] [Google Scholar]
- 77.Čolović M.B. Krstić, D. Z.; Lazarević-Pašti, T. D.; Bondžić, A. M.; Vasić, V. M. Acetylcholinesterase inhibitors: Pharmacology and toxicology. Curr. Neuropharmacol. 2013;11(3):315–335. doi: 10.2174/1570159X11311030006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Giacobini E. Cholinesterase inhibitors: new roles and therapeutic alternatives. 2004. [DOI] [PubMed]
- 79.Birks J. Cholinesterase inhibitors for Alzheimer’s disease. Cochrane Database Syst. Rev. 2006;(1):CD005593. doi: 10.1002/14651858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Teipel S., Heinsen H., Amaro E., Grinberg L.T., Krause B., Grothe M. Cholinergic basal forebrain atrophy predicts amyloid burden in Alzheimer’s disease. Neurobiol. Aging. 2014;35(3):482–491. doi: 10.1016/j.neurobiolaging.2013.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Armstrong R.A. What causes Alzheimer’s disease? Folia Neuropathol. 2013;51(3):169–188. doi: 10.5114/fn.2013.37702. [DOI] [PubMed] [Google Scholar]
- 82.Ambure P., Roy K. CADD Modeling of multi-target drugs against Alzheimer’s disease. Curr. Drug Targets. 2017;18(5):522–533. doi: 10.2174/1389450116666150907104855. [DOI] [PubMed] [Google Scholar]
- 83.Jacobsen J.S., Reinhart P., Pangalos M.N. Current concepts in therapeutic strategies targeting cognitive decline and disease modification in Alzheimer’s disease. NeuroRx. 2005;2(4):612–626. doi: 10.1602/neurorx.2.4.612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sheng R., Lin X., Zhang J., Chol K.S., Huang W., Yang B., He Q., Hu Y. Design, synthesis and evaluation of flavonoid derivatives as potent AChE inhibitors. Bioorg. Med. Chem. 2009;17(18):6692–6698. doi: 10.1016/j.bmc.2009.07.072. [DOI] [PubMed] [Google Scholar]
- 85.Khan M.T.H., Orhan I., Şenol F.S., Kartal M., Şener B., Dvorská M., Šmejkal K., Šlapetová T. Cholinesterase in-hibitory activities of some flavonoid derivatives and chosen xanthone and their molecular docking studies. Chem. Biol. Interact. 2009;181(3):383–389. doi: 10.1016/j.cbi.2009.06.024. [DOI] [PubMed] [Google Scholar]
- 86.Roy K.K., Tota S., Tripathi T., Chander S., Nath C., Saxena A.K. Lead optimization studies towards the discovery of novel carbamates as potent AChE inhibitors for the potential treatment of Alzheimer’s disease. Bioorg. Med. Chem. 2012;20(21):6313–6320. doi: 10.1016/j.bmc.2012.09.005. [DOI] [PubMed] [Google Scholar]
- 87.Basiri A., Murugaiyah V., Osman H., Kumar R.S., Kia Y., Ali M.A. Microwave assisted synthesis, cholinesterase enzymes inhibitory activities and molecular docking studies of new pyridopyrimidine derivatives. Bioorg. Med. Chem. 2013;21(11):3022–3031. doi: 10.1016/j.bmc.2013.03.058. [DOI] [PubMed] [Google Scholar]
- 88.Samadi A., Estrada M., Pérez C., Rodríguez-Franco M.I., Iriepa I., Moraleda I., Chioua M., Marco-Contelles J. Pyri-donepezils, new dual AChE inhibitors as potential drugs for the treatment of Alzheimer’s disease: Synthesis, biological as-sessment, and molecular modeling. Eur. J. Med. Chem. 2012;57:296–301. doi: 10.1016/j.ejmech.2012.09.030. [DOI] [PubMed] [Google Scholar]
- 89.Samadi A., Revenga M.D.L.F., Pérez C., Iriepa I., Morale-da I., Rodríguez-Franco M.I., Marco-Contelles J. Synthesis, pharmacological assessment, and molecular modeling of 6-chloro-pyridonepezils: New dual AChE inhibitors as potential drugs for the treatment of Alzheimer’s disease. Eur. J. Med. Chem. 2013;67:64–74. doi: 10.1016/j.ejmech.2013.06.021. [DOI] [PubMed] [Google Scholar]
- 90.Das S., Laskar M.A., Sarker S.D., Choudhury M.D., Choudhury P.R., Mitra A., Jamil S., Lathiff S.M.A., Abdullah S.A., Basar N. Prediction of Anti‐Alzheimer’s activity of flavonoids targeting acetylcholinesterase in silico. Phytochem. Anal. 2017;28(4):324–331. doi: 10.1002/pca.2679. [DOI] [PubMed] [Google Scholar]
- 91.Varadaraju K.R., Kumar J.R., Mallesha L., Muruli A., Mohana K.N.S., Mukunda C.K., Sharanaiah U. Virtual screening and biological evaluation of piperazine derivatives as human acetylcholinesterase inhibitors. Int. J. Alzheimers Dis. 2013;2013 doi: 10.1155/2013/653962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lipton S.A. The molecular basis of memantine action in Alzheimer’s disease and other neurologic disorders: low-affinity, uncompetitive antagonism. Curr. Alzheimer Res. 2005;2(2):155–165. doi: 10.2174/1567205053585846. [DOI] [PubMed] [Google Scholar]
- 93.Glycosylation D., Reduction B., Mg A.E., Mg B.I. The glutamate receptor ion channels. Pharmacology. 1999;51(1) [Google Scholar]
- 94.Zhou Y., Danbolt N.C. Glutamate as a neurotransmitter in the healthy brain. J. Neural Transm. (Vienna) 2014;121(8):799–817. doi: 10.1007/s00702-014-1180-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Danysz W., Parsons C.G. Alzheimer’s disease, β-amyloid, glutamate, NMDA receptors and memantine--searching for the connections. Br. J. Pharmacol. 2012;167(2):324–352. doi: 10.1111/j.1476-5381.2012.02057.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Paoletti P., Neyton J. NMDA receptor subunits: function and pharmacology. Curr. Opin. Pharmacol. 2007;7(1):39–47. doi: 10.1016/j.coph.2006.08.011. [DOI] [PubMed] [Google Scholar]
- 97.Gitto R., De Luca L., Ferro S., Russo E., De Sarro G., Chisari M., Ciranna L., Alvarez-Builla J., Alajarin R., Buemi M.R. Synthesis, modelling and biological characterization of 3-substituted-1H-indoles as ligands of GluN2B-containing N-methyl-D-aspartate receptors. Bioorg. Med. Chem. 2014;22(3):1040–1048. doi: 10.1016/j.bmc.2013.12.040. [DOI] [PubMed] [Google Scholar]
- 98.Espinoza-Moraga M., Caballero J., Gaube F., Winckler T., Santos L.S. 1-Benzyl-1,2,3,4-Tetrahydro-β-Carboline as channel blocker of N-Methyl-d-Aspartate receptors. Chem. Biol. Drug Des. 2012;79(4):594–599. doi: 10.1111/j.1747-0285.2012.01317.x. [DOI] [PubMed] [Google Scholar]
- 99.Colotta V., Lenzi O., Catarzi D., Varano F., Squarcialupi L., Costagli C., Galli A., Ghelardini C., Pugliese A.M., Ma-raula G., Coppi E., Pellegrini-Giampietro D.E., Pedata F., Sabbadin D., Moro S. 3-Hydroxy-1H-quinazoline-2,4-dione derivatives as new antagonists at ionotropic glutamate receptors: Molecular modeling and pharmacological studies. Eur. J. Med. Chem. 2012;54:470–482. doi: 10.1016/j.ejmech.2012.05.036. [DOI] [PubMed] [Google Scholar]
- 100.Parsons C.G., Danysz W., Dekundy A., Pulte I. Memantine and cholinesterase inhibitors: complementary mechanisms in the treatment of Alzheimer’s disease. Neurotox. Res. 2013;24(3):358–369. doi: 10.1007/s12640-013-9398-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Krueger B.A., Weil T., Schneider G. Comparative virtual screening and novelty detection for NMDA-GlycineB antagonists. J. Comput. Aided Mol. Des. 2009;23(12):869–881. doi: 10.1007/s10822-009-9304-1. [DOI] [PubMed] [Google Scholar]
- 102.Gitto R., De Luca L., Ferro S., Occhiuto F., Samperi S., De Sarro G., Russo E., Ciranna L., Costa L., Chimirri A. Computational studies to discover a new NR2B/NMDA receptor antagonist and evaluation of pharmacological profile. ChemMedChem. 2008;3(10):1539–1548. doi: 10.1002/cmdc.200800124. [DOI] [PubMed] [Google Scholar]
- 103.Caulfield M.P., Birdsall N.J. International Union of Pharmacology. XVII. Classification of muscarinic acetylcholine receptors. . Pharmacol. Rev. 1998;50(2):279–290. [PubMed] [Google Scholar]
- 104.Jiang S., Li Y., Zhang C., Zhao Y., Bu G., Xu H., Zhang Y.W. M1 muscarinic acetylcholine receptor in Alzheimer’s disease. Neurosci. Bull. 2014;30(2):295–307. doi: 10.1007/s12264-013-1406-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Buckingham S.S.D., Jones A.K.A., Brown L.A., Sattelle D.B. Nicotinic acetylcholine receptor signalling: roles in Alzheimer’s disease and amyloid neuroprotection. Pharmacology. 2009;61(1):39–61. doi: 10.1124/pr.108.000562.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Davie B.J., Christopoulos A., Scammells P.J. Development of M1 mAChR allosteric and bitopic ligands: Prospective therapeutics for the treatment of cognitive deficits. ACS Chem. Neurosci. 2013;4(7):1026–1048. doi: 10.1021/cn400086m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Melancon B.J., Tarr J.C., Panarese J.D., Wood M.R., Lindsley C.W. Allosteric modulation of the M1 muscarinic acetylcholine receptor: Improving cognition and a potential treatment for schizophrenia and Alzheimer’s disease. Drug Discov. Today. 2013;18(23-24):1185–1199. doi: 10.1016/j.drudis.2013.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Moebius H., Loewen G., Dgetluck N., Hilt D. 2015.
- 109.Balaraman Y. Limaye, a R.; Levey, a I.; Srinivasan, S. Glycogen synthase kinase 3beta and Alzheimer’s disease: patho-physiological and therapeutic significance. Cell. Mol. Life Sci. 2006;63(11):1226–1235. doi: 10.1007/s00018-005-5597-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Uehara F., Shoda A., Aritomo K., Fukunaga K., Watanabe K., Ando R., Shinoda M., Ueno H., Kubodera H., Sunada S. 6-(4-Pyridyl) pyrimidin-4 (3H)-ones as CNS penetrant glycogen synthase kinase-3β inhibitors. Bioorg. Med. Chem. Lett. 2013;23(24):6928–6932. doi: 10.1016/j.bmcl.2013.09.021. [DOI] [PubMed] [Google Scholar]
- 111.Awasthi M., Singh S., Pandey V.P., Dwivedi U.N. Alzheimer’s disease: An overview of amyloid beta dependent pathogenesis and its therapeutic implications along with in silico approaches emphasizing the role of natural products. J. Neurol. Sci. 2016;•••:56–271. doi: 10.1016/j.jns.2016.01.008. [DOI] [PubMed] [Google Scholar]
- 112.Hanger D.P., Anderton B.H., Noble W. Tau phosphorylation: the therapeutic challenge for neurodegenerative disease. Trends Mol. Med. 2009;15(3):112–119. doi: 10.1016/j.molmed.2009.01.003. [DOI] [PubMed] [Google Scholar]
- 113.Hernandez F., Avila J. Tauopathies. Cell. Mol. Life Sci. 2007;64(17):2219–2233. doi: 10.1007/s00018-007-7220-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Saido T.C. Metabolism of amyloid β peptide and pathogenesis of Alzheimer’s disease. Proc. Jpn. Acad., Ser. B, Phys. Biol. Sci. 2013;89(7):321–339. doi: 10.2183/pjab.89.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Vassar R., Kandalepas P.C. The β-secretase enzyme BACE1 as a therapeutic target for Alzheimer’s disease. Alzheimers Res. Ther. 2011;3(3):20. doi: 10.1186/alzrt82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Mancini F., De Simone A., Andrisano V. Beta-secretase as a target for Alzheimer’s disease drug discovery: An overview of in vitro methods for characterization of inhibitors. Anal. Bioanal. Chem. 2011;400(7):1979–1996. doi: 10.1007/s00216-011-4963-x. [DOI] [PubMed] [Google Scholar]
- 117.Ghosh A.K., Gemma S., Tang J. Beta-secretase as a therapeutic target for Alzheimer’s disease. Neurotherapeutics. 2008;5(3):399–408. doi: 10.1515/revneuro-2016-0023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Ghosh A.K. Margherita. Brindisi, and J. T. Developing β-secretase inhibitors for treatment of Alzheimer’s disease. J. Neurochem. 2012;120:71–83. doi: 10.1111/j.1471-4159.2011.07476.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Piazzi L., Cavalli A., Colizzi F., Belluti F., Bartolini M., Mancini F., Recanatini M., Andrisano V., Rampa A. Multitarget-directed coumarin derivatives: hAChE and BACE1 inhibitors as potential anti-Alzheimer compounds. Bioorg. Med. Chem. Lett. 2008;18(1):423–426. doi: 10.1016/j.bmcl.2007.09.100. [DOI] [PubMed] [Google Scholar]
- 120.Zhu Y., Xiao K., Ma L., Xiong B., Fu Y., Yu H., Wang W., Wang X., Hu D., Peng H. Design, synthesis and biological evaluation of novel dual inhibitors of acetylcholinesterase and β-secretase. . Bioorg. Med. Chem. 2009;17(4):1600–1613. doi: 10.1016/j.bmc.2008.12.067. [DOI] [PubMed] [Google Scholar]
- 121.Shimmyo Y., Kihara T., Akaike A., Niidome T., Sugimoto H. Flavonols and flavones as BACE-1 inhibitors: structure–activity relationship in cell-free, cell-based and in silico studies reveal novel pharmacophore features. Biochim. Biophys. Acta, Gen. Subj. 2008;1780(5):819–825. doi: 10.1016/j.bbagen.2008.01.017. [DOI] [PubMed] [Google Scholar]
- 122.Zuo Z., Luo X., Zhu W., Shen J., Shen X., Jiang H., Chen K. 2005.
- 123.Remya C., Dileep K.V., Tintu I., Variyar E.J., Sadasivan C. In vitro inhibitory profile of NDGA against AChE and its in silico structural modifications based on ADME profile. J. Mol. Model. 2013;19(3):1179–1194. doi: 10.1007/s00894-012-1656-0. [DOI] [PubMed] [Google Scholar]
- 124.Razavi S. F., Khoobi M., Nadri H., Sakhteman A., Moradi A., Emami S., Foroumadi A., Shafiee A. 2013.
- 125.Kwon Y.E., Park J.Y., No K.T., Shin J.H., Lee S.K., Eun J.S., Yang J.H., Shin T.Y., Kim D.K., Chae B.S. Synthesis, in vitro assay, and molecular modeling of new piperidine derivatives having dual inhibitory potency against acetylcholinesterase and Aβ 1–42 aggregation for Alzheimer’s disease therapeutics. Bioorg. Med. Chem. 2007;15(20):6596–6607. doi: 10.1016/j.bmc.2007.07.003. [DOI] [PubMed] [Google Scholar]
- 126.Butini S., Gabellieri E., Brindisi M., Casagni A., Guarino E., Huleatt P.B., Relitti N., La Pietra V., Marinelli L., Giustiniano M. Novel peptidomimetics as BACE-1 inhibitors: Synthesis, molecular modeling, and biological studies. Bioorg. Med. Chem. Lett. 2013;23(1):85–89. doi: 10.1016/j.bmcl.2012.11.011. [DOI] [PubMed] [Google Scholar]
- 127.Truong A.P., Tóth G., Probst G.D., Sealy J.M., Bowers S., Wone D.W., Dressen D., Hom R.K., Konradi A.W., Sham H.L. Design of an orally efficacious hydroxyethylamine (HEA) BACE-1 inhibitor in a preclinical animal model. Bioorg. Med. Chem. Lett. 2010;20(21):6231–6236. doi: 10.1016/j.bmcl.2010.08.102. [DOI] [PubMed] [Google Scholar]
- 128.Fukumoto H., Takahashi H., Tarui N., Matsui J., Tomita T., Hirode M., Sagayama M., Maeda R., Kawamoto M., Hirai K. A noncompetitive BACE1 inhibitor TAK-070 ameliorates Aβ pathology and behavioral deficits in a mouse model of Alzheimer’s disease. J. Neurosci. 2010;30(33):11157–11166. doi: 10.1523/JNEUROSCI.2884-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Kumar A., Nisha C.M., Silakari C., Sharma I., Anusha K., Gupta N., Nair P., Tripathi T., Kumar A. Current and novel therapeutic molecules and targets in Alzheimer’s disease. J. Formos. Med. Assoc. 2016;115(1):3–10. doi: 10.1016/j.jfma.2015.04.001. [DOI] [PubMed] [Google Scholar]
- 130.Potter P.E. Investigational medications for treatment of patients with Alzheimer disease. J. Am. Osteopath. Assoc. 2010;110(9) Suppl. 8:S27–S36. [PubMed] [Google Scholar]
- 131.Choi H.G., Zhang J., Deng X., Hatcher J.M., Patricelli M.P., Zhao Z., Alessi D.R., Gray N.S. Brain penetrant LRRK2 inhibitor. ACS Med. Chem. Lett. 2012;3(8):658–662. doi: 10.1021/ml300123a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Cardona F., Sánchez-Mut J.V., Dopazo H., Pérez-Tur J. Phylogenetic and in silico structural analysis of the Parkinson disease-related kinase PINK1. Hum. Mutat. 2011;32(4):369–378. doi: 10.1002/humu.21444. [DOI] [PubMed] [Google Scholar]
- 133.Jatana N., Thukral L., Latha N. Structure and dynamics of DRD4 bound to an agonist and an antagonist using in silico approaches. Proteins. 2015;83(5):867–880. doi: 10.1002/prot.24716. [DOI] [PubMed] [Google Scholar]
- 134.Singh V., Somvanshi P. Homology modeling of adenosine A2A receptor and molecular docking for exploration of ap-propriate potent antagonists for treatment of Parkinson’s disease. Curr. Aging Sci. 2009;2(2):127–134. doi: 10.2174/1874609810902020127. [DOI] [PubMed] [Google Scholar]
- 135.Michielan L., Bacilieri M., Schiesaro A., Bolcato C., Pas-torin G., Spalluto G., Cacciari B., Klotz K.N., Kaseda C., Moro S. Linear and nonlinear 3D-QSAR approaches in tandem with ligand-based homology modeling as a computation-al strategy to depict the Pyrazolo-Triazolo-Pyrimidine antagonists binding site of the human adenosine A2A receptor. J. Chem. Inf. Model. 2008;48(2):350–363. doi: 10.1021/ci700300w. [DOI] [PubMed] [Google Scholar]
- 136.Kim S-K., Gao Z-G., Van Rompaey P., Gross A.S., Chen A., Van Calenbergh S., Jacobson K.A. Modeling the adenosine receptors: Comparison of the binding domains of A2A agonists and antagonists. J. Med. Chem. 2003;46(23):4847–4859. doi: 10.1021/jm0300431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.McRobb F.M., Capuano B., Crosby I.T., Chalmers D.K., Yuriev E. Homology modeling and docking evaluation of Aminergic G protein-coupled receptors. J. Chem. Inf. Model. 2010;50(4):626–637. doi: 10.1021/ci900444q. [DOI] [PubMed] [Google Scholar]
- 138.Chen H., Chan B.K., Drummond J., Estrada A.A., Gunzner-Toste J., Liu X., Liu Y., Moffat J., Shore D., Sweeney Z.K. Discovery of selective LRRK2 inhibitors guided by computational analysis and molecular modeling. J. Med. Chem. 2012;55(11):5536–5545. doi: 10.1021/jm300452p. [DOI] [PubMed] [Google Scholar]
- 139.Ray S., Liu M. Current understanding of LRRK2 in Parkinson’s disease: biochemical and structural features and inhibitor design. 2012. [DOI] [PMC free article] [PubMed]
- 140.Mata I.F., Wedemeyer W.J., Farrer M.J., Taylor J.P., Gallo K.A. LRRK2 in Parkinson’s disease: protein domains and functional insights. Trends Neurosci. 2006;29(5):286–293. doi: 10.1016/j.tins.2006.03.006. [DOI] [PubMed] [Google Scholar]
- 141.Vancraenenbroeck R., De Raeymaecker J., Lobbestael E., Gao F., De Maeyer M., Voet A., Baekelandt V., Taymans J-M. In silico, in vitro and cellular analysis with a kinome-wide inhibitor panel correlates cellular LRRK2 dephosphorylation to inhibitor activity on LRRK2. Front. Mol. Neurosci. 2014;7:51. doi: 10.3389/fnmol.2014.00051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Lee J.Y., Kim Y. Comparative homology modeling and ligand docking study of human catechol-O-methyltransferase for antiparkinson drug design. Bull. Korean Chem. Soc. 2005;26(11):1695–1700. [Google Scholar]
- 143.Siddiqui I.J., Pervaiz N., Abbasi A.A. The Parkinson disease gene SNCA: Evolutionary and structural insights with pathological implication. Sci. Rep. 2016;6:24475. doi: 10.1038/srep24475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Ganguli S., Datta A. 2010.
- 145.Kuldeep S., Satpal B.S. Virtual screening approach of drug design for Parkinson’s disease. Int. J. Pharma Bio Sci. 2013;4(2):821–830. [Google Scholar]
- 146.Jeyam M., Raj K. G. R.; Poornima, V.; Sharanya, M. Molecular understanding and Insilico validation of traditional medicines for Parkinson’s disease. Asian J. Pharm. Clin. Res. 2012;5(4):125–128. [Google Scholar]
- 147.Trempe J.F., Fon E.A. Structure and function of Parkin, PINK1, and DJ-1, the three musketeers of neuroprotection. Front. Neurol. 2013;4:1–11. doi: 10.3389/fneur.2013.00038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Ansari M.Z. Computational approach towards targeting aggregate formation in Synucleinopathies. Proteom. Bioinform. 2015;2(6) doi: 10.15406/mojpb.2015.02.00066. [DOI] [Google Scholar]
- 149.Dawson T. M. Molecular Pathways of neurodegeneration in Parkinson’s disease. 2003. [DOI] [PubMed]
- 150.Platania C.B.M., Salomone S., Leggio G.M., Drago F., Bucolo C. Homology modeling of dopamine D2 and D3 receptors: Molecular dynamics refinement and docking evaluation. PLoS One. 2012;7(9):e44316. doi: 10.1371/journal.pone.0044316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Hobrath J.V., Wang S. Computational elucidation of the structural basis of ligand binding to the Dopamine 3 receptor through docking and homology modeling. J. Med. Chem. 2006;49(15):4470–4476. doi: 10.1021/jm0501634. [DOI] [PubMed] [Google Scholar]
- 152.Khoddami M., Nadri H., Moradi A., Sakhteman A. Homology modeling, molecular dynamic simulation, and docking based binding site analysis of human dopamine (D4) receptor. J. Mol. Model. 2015;21(2):36. doi: 10.1007/s00894-015-2579-3. [DOI] [PubMed] [Google Scholar]
- 153.Kortagere S., Cheng S-Y., Antonio T., Zhen J., Reith M.E.A., Dutta A.K. Interaction of novel hybrid compounds with the D3 dopamine receptor: Site-directed mutagenesis and homology modeling studies. Biochem. Pharmacol. 2011;81(1):157–163. doi: 10.1016/j.bcp.2010.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Varady J., Wu X., Fang X., Min J., Hu Z., Levant B., Wang S. Molecular modeling of the Three-Dimensional structure of dopamine 3 (D3) subtype receptor: Discovery of novel and potent D3 ligands through a hybrid pharmacophore-and structure-based database searching approach. J. Med. Chem. 2003;46(21):4377–4392. doi: 10.1021/jm030085p. [DOI] [PubMed] [Google Scholar]
- 155.Simpson M.M., Ballesteros J.A., Chiappa V., Chen J., Suehiro M., Hartman D.S., Godel T., Snyder L.A., Sakmar T.P., Javitch J.A. Dopamine D4/D2 receptor selec-tivity is determined by A divergent aromatic microdomain contained within the second, third, and seventh membrane-spanning segments. Mol. Pharmacol. 1999;56(6):1116–1126. doi: 10.1124/mol.56.6.1116. [DOI] [PubMed] [Google Scholar]
- 156.Indarte M., Madura J.D., Surratt C.K. Dopamine trans-porter comparative molecular modeling and binding site pre-diction using the LeuTAa leucine transporter as a template. Proteins. 2008;70(3):1033–1046. doi: 10.1002/prot.21598. [DOI] [PubMed] [Google Scholar]
- 157.Harris F., Pierpoint L. Photodynamic therapy based on 5-aminolevulinic acid and its use as an antimicrobial agent. Med. Res. Rev. 2012;29(6):1292–1327. doi: 10.1002/med. [DOI] [PubMed] [Google Scholar]
- 158.Pradeep H., Rajanikant G.K. Computational Prediction of a Putative binding site on Drp1: Implications for Antiparkinsonian therapy. J. Chem. Inf. Model. 2014;54(7):2042–2050. doi: 10.1021/ci500243h. [DOI] [PubMed] [Google Scholar]
- 159.Jimenez-Botello L.C., Soriano-Ursúa M.A., Deeb O., Segura-Cabrera A., Martínez-Archundia M., Correa-Basurto J. Homology modeling, molecular dynamics and docking simulations of rat A2A receptor: A three-dimensional model validation under QSAR studies. Chemistry. 2006:1. [Google Scholar]
- 160.Robberecht W., Philips T. The changing scene of amyo-trophic lateral sclerosis. Nat. Rev. Neurosci. 2013;14(4):1–17. doi: 10.1038/nrn3430. [DOI] [PubMed] [Google Scholar]
- 161.Saxena S., Caroni P. Selective neuronal vulnerability in neurodegenerative diseases: from stressor thresholds to degeneration. Neuron. 2011;71(1):35–48. doi: 10.1016/j.neuron.2011.06.031. [DOI] [PubMed] [Google Scholar]
- 162.Fischer L.R., Culver D.G., Tennant P., Davis A.A., Wang M., Castellano-Sanchez A., Khan J., Polak M.A., Glass J.D. Amyotrophic lateral sclerosis is a distal axonopathy: Evidence in mice and man. Exp. Neurol. 2004;185(2):232–240. doi: 10.1016/j.expneurol.2003.10.004. [DOI] [PubMed] [Google Scholar]
- 163.Andersen P.M., Al-Chalabi A. Clinical genetics of amyo-trophic lateral sclerosis: what do we really know? Nat. Rev. Neurol. 2011;7(11):603–615. doi: 10.1038/nrneurol.2011.150. [DOI] [PubMed] [Google Scholar]
- 164.Renton A.E., Chiò A., Traynor B.J. State of play in amyo-trophic lateral sclerosis genetics. Nat. Neurosci. 2014;17(1):17–23. doi: 10.1038/nn.3584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Genç B., Özdinler P.H. Moving forward in clinical trials for ALS: Motor neurons lead the way please. Drug Discov. Today. 2014;19(4):441–449. doi: 10.1016/j.drudis.2013.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Anzai I., Toichi K., Tokuda E., Mukaiyama A., Akiyama S., Furukawa Y. Screening of drugs Inhibiting In vitro Oli-gomerization of Cu/Zn-Superoxide dismutase with a mutation causing Amyotrophic lateral sclerosis. Front. Mol. Biosci. 2016;3:40. doi: 10.3389/fmolb.2016.00040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Ferraiuolo L., Kirby J., Grierson A.J., Sendtner M., Shaw P.J. Molecular pathways of motor neuron injury in amyotrophic lateral sclerosis. Nat. Rev. Neurol. 2011;7(11):616. doi: 10.1038/nrneurol.2011.152. [DOI] [PubMed] [Google Scholar]
- 168.Garbuzova-Davis S., Thomson A., Kurien C., Shytle R.D., Sanberg P.R. Potential new complication in drug therapy development for amyotrophic lateral sclerosis. Expert Rev. Neurother. 2016;16(12):1397–1405. doi: 10.1080/14737175.2016.1207530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.van den Berg L.H., Kiernan M.C. Clinical diagnosis and management of amyotrophic lateral sclerosis. Nat. Rev. Neurol. 2011;7(11):639–649. doi: 10.1038/nrneurol.2011.153. [DOI] [PubMed] [Google Scholar]
- 170.Furukawa Y., Torres A.S., O’Halloran T.V. Oxygen-induced maturation of SOD1: a key role for disulfide formation by the copper chaperone CCS. EMBO J. 2004;23(14):2872–2881. doi: 10.1038/sj.emboj.7600276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Wang L., Popko B., Roos R.P. An enhanced integrated stress response ameliorates mutant SOD1-induced ALS. Hum. Mol. Genet. 2014;23(10):2629–2638. doi: 10.1093/hmg/ddt658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Brasil A.A., Belati A., Mannarino S.C., Panek A.D., Eleu-therio E.C.A., Pereira M.D. The involvement of GSH in the activation of human Sod1 linked to FALS in chronologically aged yeast cells. FEMS Yeast Res. 2013;13(5):433–440. doi: 10.1111/1567-1364.12045. [DOI] [PubMed] [Google Scholar]
- 173.Ip P., Mulligan V.K., Chakrabartty A. ALS-causing SOD1 mutations promote production of copper-deficient misfolded species. J. Mol. Biol. 2011;409(5):839–852. doi: 10.1016/j.jmb.2011.04.027. [DOI] [PubMed] [Google Scholar]
- 174.Yoshii Y., Otomo A., Pan L., Ohtsuka M., Hadano S. Loss of glial fibrillary acidic protein marginally accelerates disease progression in a SOD1 H46R transgenic mouse model of ALS. Neurosci. Res. 2011;70(3):321–329. doi: 10.1016/j.neures.2011.03.006. [DOI] [PubMed] [Google Scholar]
- 175.Honjo Y., Kaneko S., Ito H., Horibe T., Nagashima M., Nakamura M., Fujita K., Takahashi R., Kusaka H., Kawa-kami K. Protein disulfide isomerase-immunopositive inclu-sions in patients with amyotrophic lateral sclerosis. Amyotroph. Lateral Scler. 2011;2968:1–7. doi: 10.3109/17482968.2011.594055. [DOI] [PubMed] [Google Scholar]
- 176.Huang H.J., Chang T.T., Chen H.Y., Chen C.Y.C. Finding inhibitors of mutant superoxide dismutase-1 for amyotrophic lateral sclerosis therapy from traditional Chinese medicine. Evidence-based complement. Altern. Med. 2014;2014:156276. doi: 10.1155/2014/156276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Khan S., Ahmad K., Alshammari E., Adnan M., Baig M.H., Lohani M., Somvanshi P., Haque S. Implication of caspase-3 as a common therapeutic target for multineurodegenerative disorders and its inhibition using nonpeptidyl natural compounds. BioMed Res. Int. 2015;2015:379817. doi: 10.1155/2015/379817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Martin L.J., Price A.C., Kaiser A., Shaikh A.Y., Liu Z. Mechanisms for neuronal degeneration in amyotrophic lateral sclerosis and in models of motor neuron death. Int. J. Mol. Med. 2000;5(1):3–13. doi: 10.3892/ijmm.5.1.3. [Review]. [DOI] [PubMed] [Google Scholar]
- 179.Boston-Howes W., Gibb S.L., Williams E.O., Pasinelli P., Brown R.H., Trotti D. Caspase-3 cleaves and inactivates the glutamate transporter EAAT2. J. Biol. Chem. 2006;281(20):14076–14084. doi: 10.1074/jbc.M600653200. [DOI] [PubMed] [Google Scholar]
- 180.Friedlander R.M. Apoptosis and caspases in neurodegenerative diseases. N. Engl. J. Med. 2003;348(14):1365–1375. doi: 10.1056/NEJMra022366. [DOI] [PubMed] [Google Scholar]
- 181.Ahmad K., Baig M.H., Gupta G.K., Kamal M.A., Pathak N., Choi I. Identification of common therapeutic targets for selected neurodegenerative disorders: An in silico approach. J. Comput. Sci. 2015;17:292–306. [Google Scholar]
- 182.Zhang Y-Y., Mei Z-Q., Wu J-W., Wang Z-X. Enzymatic activity and substrate specificity of mitogen-activated protein kinase p38alpha in different phosphorylation states. J. Biol. Chem. 2008;283(39):26591–26601. doi: 10.1074/jbc.M801703200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Salado I.G., Redondo M., Bello M.L., Perez C.n., Liachko N.F., Kraemer B.C., Miguel L., Lecourtois M., Gil C., Martinez A. Protein kinase CK-1 inhibitors as new potential drugs for amyotrophic lateral sclerosis. J. Med. Chem. 2014;57(6):2755–2772. doi: 10.1021/jm500065f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Chico L.K., Van Eldik L.J., Watterson D.M. Targeting protein kinases in central nervous system disorders. Nat. Rev. Drug Discov. 2009;8(11):892–909. doi: 10.1038/nrd2999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Pinsetta F.R., Taft C.A., Silva C.H.T.P. Rational design of novel potential p38α MAPK inhibitors with drug-like proper- ties using pharmacophore and similarity-based virtual screening procedures. Curr. Bioact. Compd. 2013;9(1):3–13. doi: 10.2174/1573407211309010002. [DOI] [Google Scholar]
- 186.Sierra B. O.; Gonzalez, J.; Capani, F.; Barreto, G. E. In silico docking reveals possible Riluzole binding sites on Nav1.6 sodium channel: Implications for amyotrophic lateral sclerosis therapy. J. Theor. Biol. 2012;315:53–63. doi: 10.1016/j.jtbi.2012.09.004. [DOI] [PubMed] [Google Scholar]
- 187.Walker F.O. Huntington’s disease. Lancet. 2007;369(9557):218–228. doi: 10.1016/S0140-6736(07)60111-1. [DOI] [PubMed] [Google Scholar]
- 188.Nasir J., Floresco S.B., O’Kusky J.R., Diewert V.M., Richman J.M., Zeisler J., Borowski A., Marth J.D., Phil-lips A.G., Hayden M.R. Targeted disruption of the Hunting-ton’s disease gene results in embryonic lethality and behavioral and morphological changes in heterozygotes. Cell. 1995;81(5):811–823. doi: 10.1016/0092-8674(95)90542-1. [DOI] [PubMed] [Google Scholar]
- 189.Yuan J., Yankner B.A. Apoptosis in the nervous system. Nature. 2000;407(6805):802–809. doi: 10.1038/35037739. [DOI] [PubMed] [Google Scholar]
- 190.MacDonald M.E., Ambrose C.M., Duyao M.P., Myers R.H., Lin C., Srinidhi L., Barnes G., Taylor S.A., James M., Groot N. A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington’s disease chromosomes. Cell. 1993;72(6):971–983. doi: 10.1016/0092-8674(93)90585-e. [DOI] [PubMed] [Google Scholar]
- 191.Steffan J.S., Kazantsev A., Spasic-Boskovic O., Greenwald M., Zhu Y.Z., Gohler H., Wanker E.E., Bates G.P., Housman D.E., Thompson L.M. The Huntington’s disease protein interacts with p53 and CREB-binding protein and re-presses transcription. Proc. Natl. Acad. Sci. USA. 2000;97(12):6763–6768. doi: 10.1073/pnas.100110097\r100110097. [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Harjes P., Wanker E.E. The hunt for huntingtin function: interaction partners tell many different stories. Trends Biochem. Sci. 2003;28(8):425–433. doi: 10.1016/S0968-0004(03)00168-3. [DOI] [PubMed] [Google Scholar]
- 193.Conforti P., Zuccato C., Gaudenzi G., Ieraci A., Camnasio S., Buckley N. J., Mutti C., Cotelli F., Contini A., Cattaneo E. Binding of the repressor complex REST-mSIN3b by small molecules restores neuronal gene transcription in Hunting-ton’s disease models. 2013. [DOI] [PubMed]
- 194.Huang N-K., Lin J-H., Lin J-T., Lin C-I., Liu E.M., Lin C-J., Chen W-P., Shen Y-C., Chen H-M., Chen J-B. A new drug design targeting the adenosinergic system for Huntington’s disease. PLoS One. 2011;6(6):e20934. doi: 10.1371/journal.pone.0020934. [DOI] [PMC free article] [PubMed] [Google Scholar]