Abstract
Background
Histamine H3 receptor (H3R) is associated with several neuropsychological diseases, and thus it is an important target involved in several CNS disorders, such as narcolepsy, attention deficit hyperactivity disorder and schizophrenia. Since QSAR modeling is a feasible approach to explain the role of the molecular substituents in the biological activity, it can help in improving the design of better H3R ligands for these conditions.
Methods
This article reviews papers previously published in literature to show the current status of the contribution from QSAR modeling to reach H3R antagonists/inverse agonists.
Results
Classical and 3D-QSAR models were retrieved, showing that the steric and hydrophobic properties of the H3R ligands are most important to reach good affinity.
Conclusion
Although QSAR methods are valuable to design better H3R antagonists/inverse agonists, pharmacokinetics should also be considered in future models to ensure good CNS penetration.
Keywords: QSAR, H3 receptor, H3R antagonists, H3R inverse agonists, neurodegenerative diseases, neuropsychiatric disorders, structure-activity relationship
1. INTRODUCTION
The histaminergic system in the CNS is mainly modulated by histamine, a biogenic amine involved in several pathophysiological effects. The effects of histamine are produced through interaction with histamine G protein coupled receptors (GPCRs). There are four known subtypes of histamine receptors, H1 to H4 (H1R-H4R), differing in localization and mechanism of cellular signaling [1].
The H3R was discovered in 1983 by Arrang and colleagues, by an experimental observation that H1R antagonists did not show any response in this target and the H2R antagonists exhibited variable affinities not correlated to the H2R affinities, suggesting considerable differences between the classic receptors and H3R [2, 3]. Peripherally, H3R can be found in nerve endings of the gastrointestinal tract and the heart [4]. However, the H3R is found predominantly in the central nervous system (CNS) and is highly expressed in the cerebral cortex, basal ganglia [5], hippocampus [6], nucleus accumbens and substantia nigra [7]. The H3R is located in the presynaptic neurons, where it acts both as autoreceptor (modulating the synthesis and release of histamine in histaminergic neurons), and heteroreceptor in non-histaminergic neurons, regulating release of other neurotransmitters such as acetylcholine, norepinephrine, dopamine and serotonin [8]. H3R has different signaling pathways such as, inhibition of Na+/H+ exchanger activity, modulation of the MAPK pathway, activation of the AKT/GSK-3β axis and activation of phospholipase A2 [9]. However, signal transduction by H3R is primarily mediated by Gi/o protein, leading to reduction in intracellular cAMP concentration and influx of calcium into neurons, so it acts as inhibitory controller of neurotransmitters release [10].
Nowadays, there are at least six human isoforms for H3R, although its exact physiological role is still unclear. However, the CNS distribution of each isoform is considerably different, possibly leading to different pharmacological profile [11]. In addition, there are important differences in the binding affinities of H3R antagonists among species that are attributable to differences in some amino acids. While first-generation H3R antagonists were generally more potent at rodent receptors (including imidazole and non-imidazole compounds), more recent non-imidazole compounds are much more potent at human receptors [12]. These differences must be considered during the design process, since it can lead to human activity profile far from the predicted by the animal receptors.
Considering the large expression of H3R in the CNS and its ability to control the release of several distinct neurotransmitters, H3R has become an interesting target for bioactive molecules to treat neurological and psychiatric disorders such as Alzheimer's (AD) and Parkinson’s (PD) disease, epilepsy, hyperactivity, attention deficit disorder and schizophrenia [1, 13].
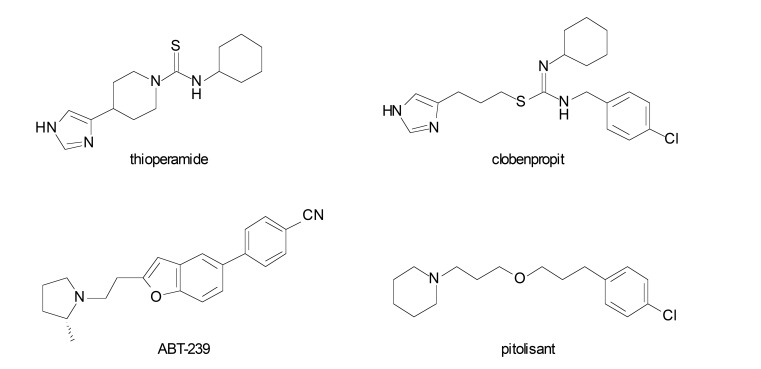
Since H3R acts through a negative feed-back mechanism, H3R antagonists/inverse agonists can be useful in increasing the release of neurotransmitters, thereby helping in the treatment of conditions involved in reducing neurotransmitter activity [14]. Recently, the H3R inverse agonist pitolisant or tripolisant (Wakix®, Fig. 1) has been approved in the United States and the European Union for the treatment of narcolepsy with or without cataplexy. In addition, pitolisant is also being evaluated in clinical trials with indications for treatment of other disorders such as schizophrenia and some types of dementia [15]. With regards to neurodegenerative diseases, pitolisant has shown promising results in clinical trials for PD, especially in improving the excessive daytime sleepiness in PD patients [16].
Fig. (1).
Selected H3R antagonists/inverse agonists from literature.
Several antagonists/inverse agonists have also demonstrated pro-cognitive activity in cognitive deficit models. Administration of H3R antagonists in hypothalamic tuberomammillary nucleus increased the release of acetylcholine, dopamine and norepinephrine into the prefrontal cortex [17] and may cause both improvement and increased cognitive functions [18-20]. H3R antagonists have also shown to elevate acetylcholine levels in cortex and hippocampus, enhancing memory. However, preclinical studies have shown that H3R antagonists activate signaling pathways that may improve cognitive efficacy and disease-modifying effects beyond symptomatic alleviation in AD. For instance, administration of ABT-239 (a H3R antagonist, Fig. 1) in normal mice showed increased cortical cAMP response element-binding (CREB) and glycogen synthase kinase 3β (GSK3β) phosphorylation, producing cognitive efficacy independently of increasing acetylcholine release [12]. In addition, some H3R antagonists such as ABT-239 and thioperamide (Fig. 1), demonstrated neuroprotective action in vitro and in vivo in neurotoxicity models [20].
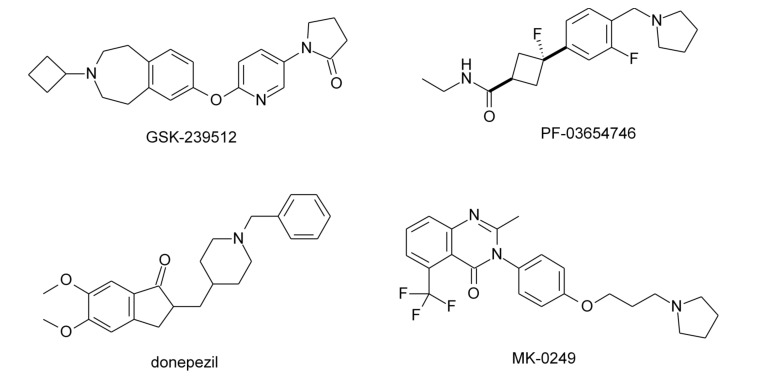
Clinical studies have shown that H3R antagonists may present therapeutic potential for neurodegenerative diseases. The H3R antagonist GSK-239512 (Fig. 2) was developed for the treatment of various types of dementia with cognitive impairment. In phase I trials, it was evaluated with indication for AD with mild to moderate symptoms [21] and in phase II studies with indication for schizophrenia [22]. PF-03654746 (Fig. 2) was evaluated for efficacy and safety in volunteers with excessive daytime sleepiness (EDS) associated with narcolepsy and presented significant improvement in symptoms, as pitolisant [23], and also completed phase I clinical trials in patients with schizophrenia, showing significant improvement in cognitive symptoms [24]. Other H3R antagonist, SAR-110894, was evaluated in phase II clinical trials for treatment of AD, in association with donepezil [25]. Finally, the compound MK-0249 was evaluated for the treatment of AD associated with cognitive deficits [26], and also in phase II studies for patients with paranoid schizophrenia (Fig. 2) [27].
Fig. (2).
Examples of H3R antagonists/inverse agonists evaluated in clinical studies.
In summary, numerous advantages have been observed for H3R antagonists/inverse agonists that may translate into promising AD and PD enhancing agents. Considering all these therapeutic potentials, H3R ligands are constantly a focus in search of new chemical entities, and thus the structural requirements necessary for appropriate interaction with the receptor and the design of more active compounds have been constantly reported. Considering this, the quantitative structure-activity relationships (QSAR) strategy can be highlighted. In addition to the appropriate pharmacodynamic requirements, the QSAR approach may also assist in predicting the pharmacokinetic profile of the designed molecules [28]. Pharmacokinetics is among the most important bottlenecks in the drug discovery, since several compounds that fail in the preclinical and clinical phases do not fulfill the ADME (absorption, distribution, metabolism and excretion) requirements or present inappropriate significant toxicity [20, 29].
In this paper, we performed a review of the main structural requirements to improve H3R antagonist/inverse agonist activity as suggested by quantitative structure-activity relationships (QSAR) models reported in literature. This data is quite important to guide a more rational design of new H3R ligands with better activity profile. The reports were raised in PubMed and Web of Science databases, using the terms “QSAR” and “H3 ligands” as keywords.
2. QSAR
Several H3R ligands were identified and evaluated as potent H3R antagonists and different approaches trying to explore structural determinants for H3R antagonistic activity. The QSAR approach is a feasible technique for identifying the important structural determinants to a defined biological activity. QSAR is a statistical model that has been successfully used in agrochemical, toxicological and environmental studies beyond drug discovery [30]. Accordingly, QSAR approach was also explored to determine H3R affinity, activity and selectivity.
The QSAR approach is based on the assumption that similar molecules have similar activities, and considering that similarity refers to chemical characteristics, the activity of a molecule is dependent of certain physicochemical properties, called as descriptors. In medicinal chemistry, this assumption is known as structure-activity relationships (SAR).
Through statistically validated mathematical models, the QSAR modeling allows to predict and quantify the SAR variables, present in a series of analogues with known quantitative activity, responsible for the affinity and activity in a specific target. In these mathematical models, the different structural, physicochemical and conformational properties of the molecules are expressed by structural descriptors (independent variables). The different types of descriptors are obtained experimentally or calculated in silico, and evaluated for correlation with previously known activity of the compounds (dependent variable) [31, 32].
The QSAR studies started with the work of Corwin Hansch group through the investigation of the role of hydrophobicity in the biological activity of compounds. In later works, they also added the influence of electronic and steric properties on the model. The earlier QSAR models are known as Hansch's analysis [33].
To present reliable results, it is necessary that these models should be statistically validated. Basically, the validations of the QSAR models are subjected to statistical model validation parameters such as degree of adjustment, degree of significance and degree of predictability [34]. The analysis of adjustment of the model can be performed by calculating the correlation coefficient (R), standard deviation (s) [31] and other parameters such as standard error (SE) of prediction (standard error, SEP) or estimated (standard error of estimate, SEE) and its deviation (standard deviation error in prediction, SDEP). The statistical significance of a model can be evaluated using, per example, the multiple determination coefficient (R2) and Fisher's test (F) [34]. The predictability can be determined by cross-validation processes, through evaluation of cross-correlation coefficient (Q2), leave-N-out, scrambling of dependent variable and also through prediction of the activity of a test set [32].
In addition to the classical QSAR modeling (2D-QSAR), QSAR studies can enhance the steric factors through a three-dimensional approach (3D-QSAR), such as comparative molecular field analysis (CoMFA) and comparative molecular similarity index analysis (CoMSIA). These approaches allow to obtain descriptors (such as Coulomb and Lennard-Jones potentials, hydrogen bonding and others) in a three-dimensional environment, to identify possible points of interaction of a ligand in the space, and possibly to the pharmacological target responsible for the biological activity [35].
As described, the QSAR modeling allows to obtain simple and fast models that can be useful in explaining the dependence of the activity on certain descriptors, and more importantly, in predicting the activity of new and non-tested compounds. However, as any statistical model, mispredictions can happen and it is important to understand the applicability domain of each model. Therefore, the use of dataset obtained by different methodologies or animal species, choice of redundant descriptors, employment of non-sense descriptors, inadequate number of variables in each model and others mistakes should be observed [33].
3. QSAR STUDIES ON H3R ANTAGONISTS
Table 1 presents the descriptors, with the meanings and nature, addressed throughout the review.
Table 1.
List of descriptors from reported QSAR models for H3R activities and their meaning.
| Descriptor | Meaning | Nature |
|---|---|---|
| MRI | Molecular redundancy index: indicates the capacity and symmetry of a molecule | Topological |
| Ip1 | Indicator parameter for the presence or absence of one benzene ring | - |
| EHOMO | Highest occupied molecular orbital energy | Thermodynamic |
| logDpH7.4 | Distribution coefficient in pH 7.4 | Hydrophobic |
| Mor19V | 3D atomic coordinates by the transform used in electron diffraction studies weighted by volume | 3D-Steric |
| Mor30M | 3D atomic coordinates by the transform used in electron diffraction studies weighted by mass | 3D-Steric |
| εFERMO | Frontier effective-for-reaction molecular orbital energy | Thermodynamic |
| ω | Electrophilicity index | Electronic |
| vsurf_DD13 | Distance between polar (H2O probe) and hydrophobic (DRY probe) groups to the receptor | 3D-Mixed |
| vsurf_Wp4 | Polarizability on the van der Waals surface | 3D-Mixed |
| φ | Dihedral angle | Steric |
| δ | Electron density | Electronic |
| E_stb | The bond stretch-bend cross-term potential energy descriptor calculated from stored 3D conformations | Thermodynamic |
| DRY | Interaction energy value of hydrophobic probe | 3D-Mixed |
| BCUT_SMR_0 | BCUT descriptors using atomic contribution to molar refractivity | Steric-electronic |
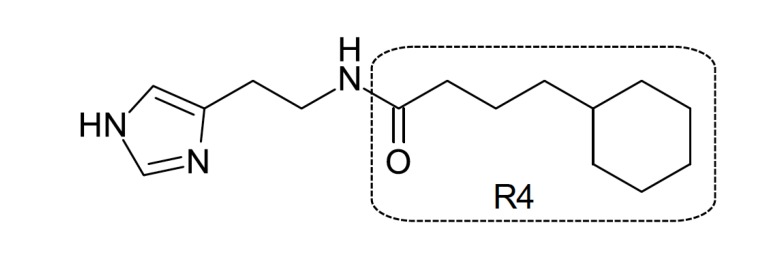
There are few reports on QSAR studies with H3R antagonists in literature. Among them, one of the sets evaluated was comprised of acylated histamine derivatives (pKi 5.7-7.3) previously reported by Stark et al. [36]. Agrawal and colleagues presented a classical QSAR model using topological descriptors and obtained through multiple regression analysis (MRA). The MRI descriptor was considered essential to attribute activity and to prediction power of the model. The addition of a qualitative indicator parameter (Ip1) in the presence of a benzene ring in R4 in the acylated histamine structure of the compounds (Fig. 3) also resulted in improvement in the obtained model. Thus, it was proposed that the presence of an aromatic ring in these kinds of compounds is essential to good activity [37]. However, the number of molecules used to build the models was relatively low (n = 13), leading to quite simple models. Since QSAR modeling is a ligand-based approach, representative number of molecules is indispensable to statistical significance. To avoid correlations by coincidence using QSAR approach, it is suggested to use at least 5 compounds per descriptor in the final model obtained through classical MRA [34]. The obtained statistical parameters for the best model were: SD = 0.2199, R = 0.9281, F = 12.430, R2 = 0.7921 and Q = 0.4221.
Fig. (3).
Acylated histamine derivative with H3R antagonist activity.
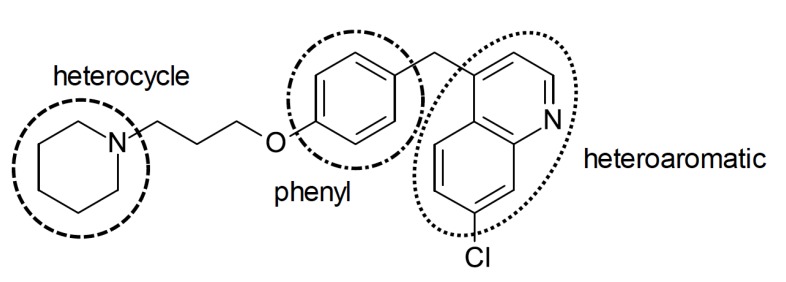
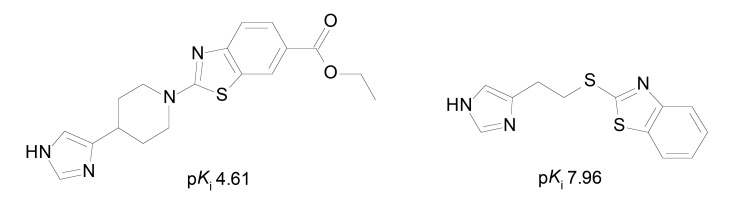
3D-QSAR models (CoMFA e CoMSIA) were built using a set of 144 H3R antagonists (118 in the training set, 26 in the test set) with general structure consisting on an heterocycle linked to a phenyl group and a heteroaromatic group (Fig. 4). The compounds were aligned based on the center of the heterocycle to obtain de CoMFA models. The 3D descriptors used were of electrostatic, hydrophobic and donor of hydrogen bonding nature, in addition to added descriptors such as HOMO and LUMO. The use of additional descriptors was important for the improvement of 3D-QSAR models (CoMFA: q2 = 0.721, r2 = 0.931, SEE = 0.236 e CoMSIA: q2 = 0.700, r2 = 0.921, SEE = 0.252) and better predictive capacity on the contribution of the descriptors in H3R affinity. The CoMFA results showed that the presence of bulky groups near the heterocycle and especially, near the phenyl group, is favorable for the activity. Moreover, electronegative and positively charged groups in the heterocycle and in the phenyl group, respectively, may increase the affinity for H3R. The CoMSIA models showed that hydrophilicity in the heteroaromatic region is favorable to activity, whereas in the heterocyclic region, the H3R affinity may be increased through hydrogen bonding acceptors and hydrophobic groups (Fig. 4) [38]. In fact, hydrophobic groups may constitute an additional point of interaction due to the presence of a lipophilic pocket between the TM3 and TM6 (transmembrane domains) in the H3R [39], and compound presented in Fig. (4) is an example of high activity compound.
Fig. (4).
Example of H3R ligand used by Chen [38] for CoMFA and CoMSIA studies.
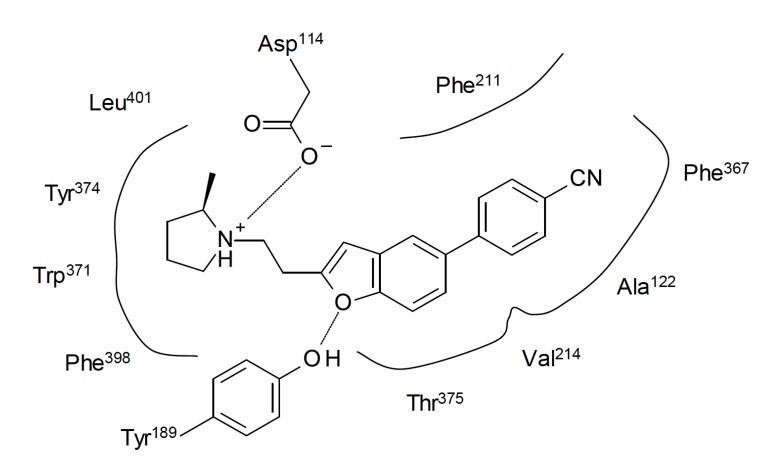
A set of 58 (44 in the training set) arylbenzofuran derivatives (pKi 7.2-10.1) previously reported by Gfesser et al. [40] including ABT-239 was also used to built QSAR models which could help to improve the affinity for both human and rat H3R [41]. The final model was fully (internal and external) validated. The model is dependent on four descriptors (EHOMO, logDpH7.4, Mor19V e Mor30M) to explore the structural requirements of this derivatives to achieve adequate human H3R affinity and presented relatively satisfactory statistics (r2 = 0.754, F = 40.7, SE = 0.317, q2LOO = 0.71). The negative correlation between the descriptor EHOMO and pKi indicates that lower EHOMO values lead to higher H3R affinity. The researchers observed that EHOMO was highly correlated to the electronic density in the aromatic system, suggesting that it can perform specific interactions in the receptor. Moreover, more hydrophilic substituents may lead to higher affinity molecules, as indicated by logDpH7.4. Molecular docking studies were used to explore and validate the obtained models. Human H3R homology model from the bovine rhodopsin structure was built and the compounds were docked using known interaction points between H3R and histamine. The selected interactions were between Asp114 (the most important in biogenic amines receptors) and the aliphatic NH2 group, and between Glu206 and the Nt imidazole ring of histamine. The results suggested that the compounds may interact through charge transfer between the phenyl or benzofuran rings of the ligands and the hydrophobic π system comprised by Ala122, Phe211, Val214, Trp371, Phe367, Phe398 and Leu401 amino acids (Fig. 5). The QSAR model also showed unfavorable correlation of lipophilicity with molecular mass (Mor30M) and positive correlation with molecular volume (Mor19V), suggesting that bulky hydrophilic substituents may improve the H3R affinity of the ligands [41]. It must be stressed that although hydrophilicity is favorable to H3R affinity, it may be detrimental to pharmacokinetics, since it is expected that the compounds reach the CNS and they must be capable of crossing the blood-brain barrier (BBB) indeed. In addition, more hydrophilic compounds can have poor oral absorption, leading to low bioavailability in vivo. Fortunately, all the compounds from the set were quite lipophilic, but a threshold for hydrophilicity of the substituents should be observed in the design of new molecules. The QSAR model to rat H3R affinity was even better (r2 = 0.840, F = 69.8, SE = 0.288, q2LOO = 0.81), however the descriptors present in both human and rat models were different, showing the importance of specie-related pKi value. In summary, when performing QSAR studies, species variability regarding pKi values must be considered as key information for prediction of affinity for H3R.
Fig. (5).
Representation of a possible binding mode for ABT-239 in human H3R, as suggested by Dastmalchi et al. [41]. Lines represent hydrophobic interactions and dashed lines the hydrogen bonding.
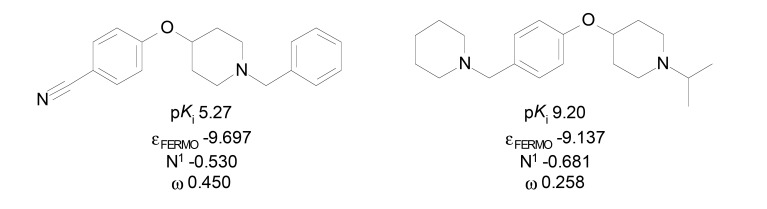
Da Costa and Trsic [14] reported QSAR models to a set of 28 4-phenoxypiperidine derivatives (pKi 5.27-9.20) previously published by Dvorak et al. (Fig. 6) [42]. Quantum-chemical descriptors were used to investigate the contribution of electronic characteristics in the binding affinity to H3R. The calculated descriptors have presented high correlation with the antagonist activity, classifying the compounds into two distinct groups with higher and lower activity by using hierarchical cluster analysis (HCA) and principal component analysis (PCA). The presence of εFERMO (frontier effective-for-reaction molecular orbital energies) in the model showed important statistic contribution (R2 = 0.927, F = 80.11, SEP = 0.141) with high prediction power (88%) of the pKi values. The descriptors HOMO and LUMO were also evaluated in some built models [43], however these descriptors showed poor correlation to the activity than εFERMO descriptor, possibly due to limitations of the HOMO and LUMO model of electron transfer. The model showed that molecules with high nucleophilic characteristic may have higher H3R affinity, since εFERMO value showed favorable contribution to pKi. On the other hand, the electrophilicity (ω descriptor) showed unfavorable contribution to the affinity. In addition, the authors demonstrated that higher electronic density in the piperidine nitrogen atom (N1) showed higher activity (pKi), showing importance of such atom for interaction with the receptor.
Fig. (6).
Low (left) and high (right) affinity molecules present in the set studied by da Costa and Trsic [14].
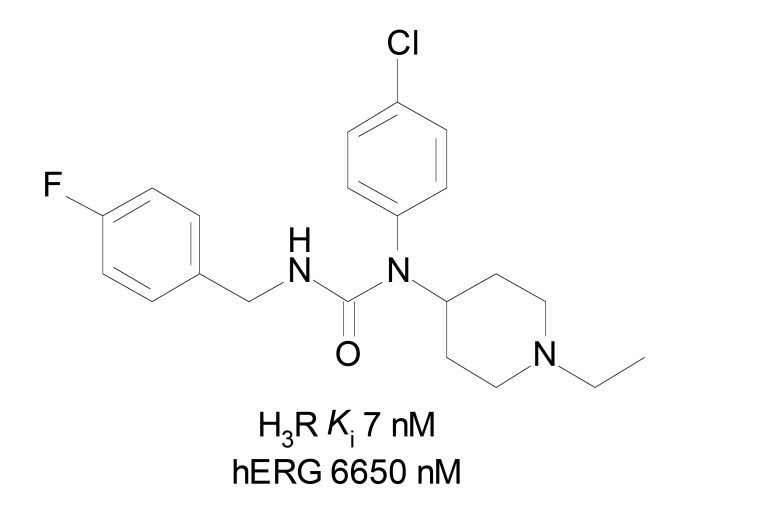
Several H3R ligands show the imidazole ring present in histamine. Imidazole nitrogen interacts with H3R through the proton transfer [38], being crucial for the activation of H3R. Since the blockade of the receptor is more relevant than activation to improve neurodegenerative diseases, this feature is undesirable to the development of H3R ligands with good pharmacological profile. Piperidinylurea derivatives (Fig. 7) from a set of 15 compounds were employed in QSAR studies. The activity was evaluated as pIC50, and the biparametric QSAR models were validated by different techniques, presenting good statistical quality (R = 0.8820, R2 = 0.7780, AdjR2 = 0.7409, F(2,12,0.05) = 21.0210, SEE = 0.2988, t(12,0.0005) = 13.7890, X2(0.05) = 0.1438 and p = 0.0000). The negative correlation of the vsurf_DD13 and vsurf_Wp4 values indicated that small distance between polar/hydrophobic groups and the atom of the target molecule, as well as superficial hydrophobicity of the compounds, are favorable to H3R antagonistic activity [44]. Moreover, considering these compounds also have considerable human ether-a-go-go-related gene (hERG) channel blocking activity and that the properties involved in hERG activity are quite different from those involved in H3R activity, the obtained model can be considered quite useful in improving H3R affinity with lower hERG blockade activity. The hERG channel blockade is involved in dangerous cardiac arrhythmias which means high toxicity to humans, and thus must be avoided. In summary, lower distance between polar and aromatic/hydrophobic groups should be small to avoid hERG activity, as well as more hydrophobic molecules can have better H3R affinity than hERG activity. Polar groups on the surface area also increase hERG activity.
Fig. (7).
Example of molecule with high H3R affinity and poor hERG activity.
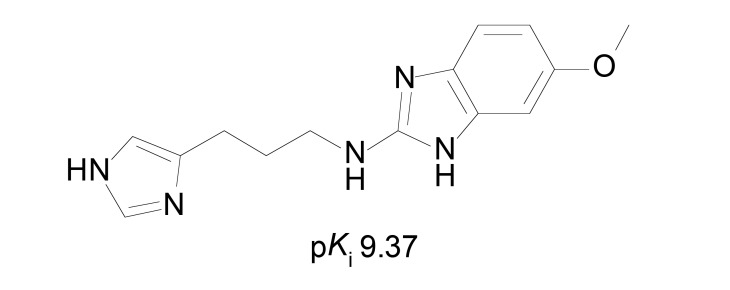
The role of lipophilicity (logP) and basicity (pKa) were evaluated with a set of 11 2-aminobenzimidazole (Fig. 8) derivatives [45]. It is important to stress that lipophilicity also increases the binding to plasma proteins, affecting the distribution of the compounds in vivo. The pKa can also be correlated to ionization state, that can influence in both pharmacokinetics and receptor affinity. The pKi values of compounds (pKi 6.91-9.37) were measured through binding assays in rat receptors from cerebral cortex synaptosomes. The obtained QSAR models were not considered statistically satisfactory by using classical MRA. When using the partial least square (PLS) analysis, better but not sufficient model was obtained. Although the QSAR data was not conclusive, some SAR information regarding these compounds was obtained. Protonation of 2-aminobenzimidazole nucleus is unfavorable to basicity, suggesting these compounds may interact in the neutral form with the receptor. In addition, the lipophilicity given by the substituent in the benzimidazole nucleus is favorable to the H3R affinity. The work also suggests that optimal lipophilicity is around logP 2.4, as indicated by the second-order descriptor logP2 [45].
Fig. (8).
Potent aminobenzimidazole derivative from Mor et al. [45].
In another work, a set of 38 H3R antagonists (pKi 4.05-9.89) was evaluated through 3D-QSAR (CoMFA and CoMSIA methods) which employs steric, electrostatic, lipophilic, hydrogen bonding acceptor and hydrogen bonding donor descriptors. The compounds had as common structure a polar heterocycle group (containing an imidazole or thiazole group) attached to an imidazole ring by an alkyl spacer. Initially the models were built using the imidazole, the terminal apolar group and a hydrogen-bond donor group of thioperamide in the alignment. However, this led to poor models, suggesting the role of hydrogen-bond is different. Using the nitrogen atom with a lone electron pair present in all the compounds instead of the donor group, the models improved significantly and then this alignment was used. The best model was obtained (CoMSIA: n = 38, LVs = 3, Q2 = 0.85, SDEP = 0.45, R2 = 0.91, s = 0.38) using only the steric field as descriptor, proving to be determinant for H3R affinity. It is possible to verify that increasing volume in the polar region of the heterocycle can improve the affinity, but the substitution in the benzene ring with bulky groups should be avoided (Fig. 9). In addition, the presence of donor groups and hydrogen bonding in the polar region of the molecule did not demonstrate statistical significance in the H3R affinity [46]. It is interesting that ABT-239 (Fig. 1) is quite similar to the compounds used in this set, but it presents a bulky 4-cyanobenzene group in this prohibited region and also present high affinity. Maybe benzofurans can be considered outliers to this model, but it limits its application domain.
Fig. (9).
Examples of low (left) and high (right) affinity compounds used by Rivara et al. [46] in the 3D-QSAR models.
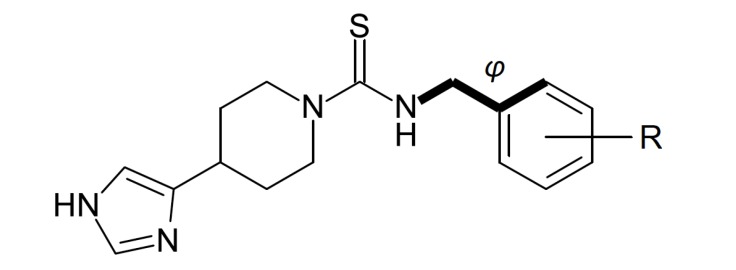
A set of monosubstituted benzyl analogues of thioperamide (Fig. 10) was synthesized and also evaluated through classical QSAR modeling to obtain better tools for positron emission tomography (PET) and single photon emission computed tomography (SPECT) applications, and halogens are suitable atoms to reach this objective [47]. In this work, the intrinsic activity (pA2 value) was used as dependent variable for H3R activity. The synthesized thioperamide analogues have shown clear influence of the substituent in the benzyl group in the activity, correlated to steric and electronic factors. To evaluate this quantitatively, steric (the dihedral angle φ) and electronic (the electron density δ) descriptors were calculated. The best model (n = 13, r = 0.93, s = 0.28, F = 31.57) exhibited influence of both descriptors. The angle between the phenyl group and the isothiourea group (φ) showed minor contribution to explain the activity, whereas the electron density of the substituted carbon of the phenyl ring (δ) was highly correlated to the pA2 values. The results suggest that substituents which lead to more negative charge on the carbon linked to them (i.e. when linked to iodine) may have better affinity for H3R, and ortho-substitution can also improve the activity due to its steric influence on φ. Although the model is limited due to the small size of the dataset, the residual values from the predicted activities were quite low, and thus the model brought important information allowing some preliminary evaluation to this set of compounds.
Fig. (10).
General structure for the set used by Windhorst et al. [47].
H3R has considerable homology with H4R (~ 37% total, 68% within TM) [48], and so obtaining molecules with selectivity towards each is a hard task. Due to the similarity between them, it is likely that several compounds that bind at one receptor may have considerable affinity for the other. The proposed pharmacophore model for H3R ligands may also be applicable for H4R ligands [1], making it difficult to achieve selective compounds. In fact, several ligands designed to bind to H3R present considerable affinity for H4R, such as thioperamide and clobenpropit (Fig. 1). Lim et al. [48] used a set of 22 clobenpropit analogues (pKi 6.5–8.6) to determine the main characteristics that play the role in the affinity for both H3R and H4R. Electronic, hydrophobic and steric descriptors were calculated and used to search valid QSAR models. Two final models were obtained, each for H3R affinity (r = 0.982, R2 = 0.964, S = 0.099, F4,17 = 115.091, F5%,4,17 = 2.965, q2 = 0.946) and for H4R affinity (r = 0.946, R2 = 0.894, S = 0.166, F4,17 = 35.994, F5%,4,17 = 2.965, q2 = 0.801). It was verified that the descriptors which correlate with the affinity for H3R also show high correlation with H4R affinity and vice-versa, proving that it is really difficult to design clobenpropit analogues with considerable selectivity towards either receptors. The only descriptor with positive correlation to H4R affinity that had shown inverse correlation with H3R affinity was the energy of bond stretch-bend (E_stb), although its contribution alone is not sufficient to achieve such objective. Lower E_stb values may favor affinity for H4R without increasing the affinity for H3R [48].
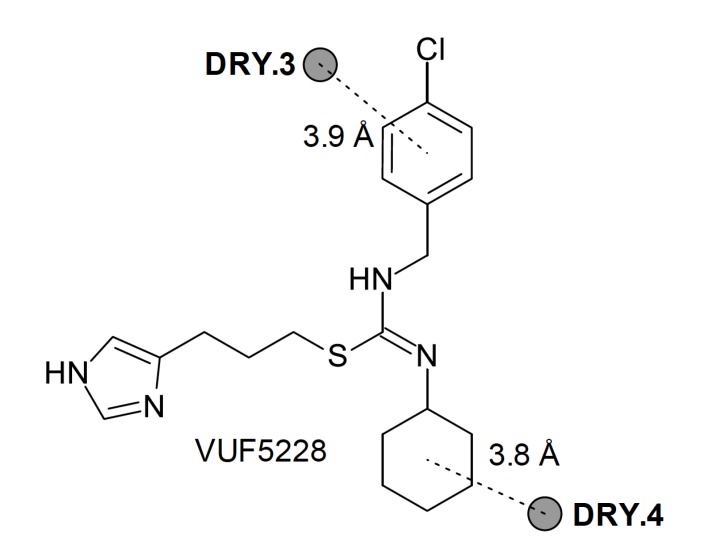
Finally, another study aiming determinants of selectivity was also performed [49]. In this work, QSAR models were constructed for a series of 14 compounds derived from clobenpropit with an additional lipophilic moiety linked to the thiourea group. The authors mixed classical and 3D-QSAR models, as well as data obtained from docking of clobenpropit and VUF5228 in both H3R and H4R. The obtained models showed that the higher topological diameter increases the affinity for H3R, corroborating with data from other QSAR models, and also BCUT_SMR_0 descriptor (r = 0.900, R2 = 0.810, S = 0.312, F2,11 = 23.454, F5%,2,11 = 3.982, q2 = 0.699). The mixed classical and 3D model (r = 0.955, R2 = 0.912, S = 0.277, F2,11 = 57.249, F5%,2,11 = 3.982, q2 = 0.862) furnished some information regarding selectivity towards H3R, by using the difference of activity between both receptors (ΔpKi = pKi H3R - pKi H4R). The selectivity can be explained by 3D interaction probes which may represent different interaction points in the receptors (represented by compound VUF5228, Fig. 11).
Fig. (11).
Schematic representation of 3D descriptors which determine the H3R selectivity over H4R.
These probes (DRY) represent hydrophobic interactions with 4-chlorophenyl group and the additional cyclohexyl group, and these groups led to higher H3R selectivity. By combining the results from 3D QSAR with data from homology modeling of H4R, the result suggests that Asn4.57 and Glu5.46 residues are determinants of H4R selectivity. However, the 3D models are not useful in explaining which substitution pattern may lead to higher H3R selectivity, although the additional cyclohexyl led to higher affinity for H3R.
CONCLUSION
Many researchers have been directing efforts to obtain models that explain how molecules can be modified to achieve higher H3R affinity, antagonistic activity and selectivity over other targets. Among the several reports in literature presenting compounds which were designed and evaluated through SAR analysis, mainly piperidine, benzofuran, piperidinylurea and imidazole series were deeply studied through QSAR approaches. The data from these reports suggest that the steric and hydrophobic factors play the most important role in the H3R activity. Important features regarding the interaction with H3R have been described, but it must be adequately explored to improve both the affinity and the selectivity. However, considering the wide range of compounds reported, it is noted that the number of QSAR studies in the literature is still scarce. In addition, with some exceptions, such models focus only on the pharmacodynamic aspect of the biological activity. Considering that the H3R is mainly found on CNS, pharmacokinetic and toxicological aspects such as BBB penetration, metabolic stability, distribution coefficients and promiscuity should be used as dependent variable in QSAR studies. Pharmacokinetics is among the main bottlenecks to reach clinical trials, and it must be predicted in the earlier stages of drug discovery process to maximize the chances to reach the patient bedside.
Consent for Publication
Not applicable.
Acknowledgements
Declared none.
Conflict of Interest
The authors declare no conflict of interest, financial or otherwise.
REFERENCES
- 1.Corrêa M.F., dos Santos Fernandes J.P. Histamine H4 receptor ligands: future applications and state of art. Chem. Biol. Drug Des. 2015;85(4):461–480. doi: 10.1111/cbdd.12431. [http://dx.doi.org/10.1111/cbdd.12431]. [PMID: 25228262]. [DOI] [PubMed] [Google Scholar]
- 2.Arrang J.M., Garbarg M., Schwartz J.C. Auto-inhibition of brain histamine release mediated by a novel class (H3) of histamine receptor. Nature. 1983;302(5911):832–837. doi: 10.1038/302832a0. [http://dx.doi.org/10. 1038/302832a0]. [PMID: 6188956]. [DOI] [PubMed] [Google Scholar]
- 3.Arrang J.M., Garbarg M., Lancelot J.C., Lecomte J.M., Pollard H., Robba M., Schunack W., Schwartz J.C. Highly potent and selective ligands for histamine H3-receptors. Nature. 1987;327(6118):117–123. doi: 10.1038/327117a0. [http://dx.doi.org/10.1038/327117a0]. [PMID: 3033516]. [DOI] [PubMed] [Google Scholar]
- 4.Ramos-Jiménez J., Garduño-Torres B., Arias-Montaño J.A. Histamina y comunicación intercelular: 99 años de historia. Rev Biomed. 2009;20:100–126. [Google Scholar]
- 5.Martinez-Mir M.I., Pollard H., Moreau J., Arrang J.M., Ruat M., Traiffort E., Schwartz J.C., Palacios J.M. Three histamine receptors (H1, H2 and H3) visualized in the brain of human and non-human primates. Brain Res. 1990;526(2):322–327. doi: 10.1016/0006-8993(90)91240-h. [http://dx.doi. org/10.1016/0006-8993(90)91240-H]. [PMID: 1979518]. [DOI] [PubMed] [Google Scholar]
- 6.Drutel G., Peitsaro N., Karlstedt K., Wieland K., Smit M.J., Timmerman H., Panula P., Leurs R. Identification of rat H3 receptor isoforms with different brain expression and signaling properties. Mol. Pharmacol. 2001;59(1):1–8. [http://dx.doi.org/10. 1124/mol.59.1.1]. [PMID: 11125017]. [PubMed] [Google Scholar]
- 7.Brown R.E., Stevens D.R., Haas H.L. The physiology of brain histamine. Prog. Neurobiol. 2001;63(6):637–672. doi: 10.1016/s0301-0082(00)00039-3. [http://dx.doi. org/10.1016/S0301-0082(00)00039-3]. [PMID: 11164999]. [DOI] [PubMed] [Google Scholar]
- 8.Gemkow M.J., Davenport A.J., Harich S., Ellenbroek B.A., Cesura A., Hallett D. The histamine H3 receptor as a therapeutic drug target for CNS disorders. Drug Discov. Today. 2009;14(9-10):509–515. doi: 10.1016/j.drudis.2009.02.011. [http://dx.doi.org/10.1016/j.drudis.2009.02.011]. [PMID: 19429511]. [DOI] [PubMed] [Google Scholar]
- 9.Bongers G., Bakker R.A., Leurs R. Molecular aspects of the histamine H3 receptor. Biochem. Pharmacol. 2007;73(8):1195–1204. doi: 10.1016/j.bcp.2007.01.008. [http://dx.doi.org/10.1016/j.bcp.2007.01.008]. [PMID: 17276412]. [DOI] [PubMed] [Google Scholar]
- 10.Tiligada E., Zampeli E., Sander K., Stark H. Histamine H3 and H4 receptors as novel drug targets. Expert Opin. Investig. Drugs. 2009;18(10):1519–1531. doi: 10.1517/14728220903188438. [http://dx.doi.org/10.1517/14728220903188438]. [PMID: 19758107]. [DOI] [PubMed] [Google Scholar]
- 11.Bakker R.A. Histamine H3-receptor isoforms. Inflamm. Res. 2004;53(10):509–516. doi: 10.1007/s00011-004-1286-9. [http://dx.doi.org/10.1007/s00011-004-1286-9]. [PMID: 15597144]. [DOI] [PubMed] [Google Scholar]
- 12.Brioni J.D., Esbenshade T.A., Garrison T.R., Bitner S.R., Cowart M.D. Discovery of histamine H3 antagonists for the treatment of cognitive disorders and Alzheimer’s disease. J. Pharmacol. Exp. Ther. 2011;336(1):38–46. doi: 10.1124/jpet.110.166876. [http://dx.doi.org/10.1124/jpet.110. 166876]. [PMID: 20864505]. [DOI] [PubMed] [Google Scholar]
- 13.Leurs R., Bakker R.A., Timmerman H., de Esch I.J. The histamine H3 receptor: from gene cloning to H3 receptor drugs. Nat. Rev. Drug Discov. 2005;4(2):107–120. doi: 10.1038/nrd1631. [http://dx.doi.org/10.1038/ nrd1631]. [PMID: 15665857]. [DOI] [PubMed] [Google Scholar]
- 14.da Costa E.B., Trsic M. A quantum chemical study on a set of non-imidazole H3 antihistamine molecules. J. Mol. Graph. Model. 2010;28(7):657–663. doi: 10.1016/j.jmgm.2010.01.003. [http://dx.doi.org/10.1016/j.jmgm.2010.01. 003]. [PMID: 20138791]. [DOI] [PubMed] [Google Scholar]
- 15.Syed Y.Y. Pitolisant: First global approval. Drugs. 2016;76(13):1313–1318. doi: 10.1007/s40265-016-0620-1. [http://dx.doi.org/10.1007/s40265-016-0620-1]. [PMID: 27438291]. [DOI] [PubMed] [Google Scholar]
- 16.Schwartz J.C. The histamine H3 receptor: from discovery to clinical trials with pitolisant. Br. J. Pharmacol. 2011;163(4):713–721. doi: 10.1111/j.1476-5381.2011.01286.x. [http://dx.doi.org/10.1111/j.1476-5381.2011.01286.x]. [PMID: 21615387]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Passani M.B., Blandina P. Histamine receptors in the CNS as targets for therapeutic intervention. Trends Pharmacol. Sci. 2011;32(4):242–249. doi: 10.1016/j.tips.2011.01.003. [http://dx.doi.org/10.1016/j.tips.2011.01.003]. [PMID: 21324537]. [DOI] [PubMed] [Google Scholar]
- 18.Fox G.B., Pan J.B., Radek R.J., Lewis A.M., Bitner R.S., Esbenshade T.A., Faghih R., Bennani Y.L., Williams M., Yao B.B., Decker M.W., Hancock A.A. Two novel and selective nonimidazole H3 receptor antagonists A-304121 and A-317920: II. In vivo behavioral and neurophysiological characterization. J. Pharmacol. Exp. Ther. 2003;305(3):897–908. doi: 10.1124/jpet.102.047241. [http://dx.doi.org/ 10.1124/jpet.102.047241]. [PMID: 12606600]. [DOI] [PubMed] [Google Scholar]
- 19.Howard H.R. Agents for attention-deficit hyperactivity disorder – an update. Expert Opin. Ther. Pat. 2004;14(7):983–1008. [http:// dx.doi.org/10.1517/13543776.14.7.983]. [Google Scholar]
- 20.Sadek B., Saad A., Sadeq A., Jalal F., Stark H. Histamine H3 receptor as a potential target for cognitive symptoms in neuropsychiatric diseases. Behav. Brain Res. 2016;312:415–430. doi: 10.1016/j.bbr.2016.06.051. [http:// dx.doi.org/10.1016/j.bbr.2016.06.051]. [PMID: 27363923]. [DOI] [PubMed] [Google Scholar]
- 21. https://clinicaltrials.gov
- 22. https://clinicaltrials.gov
- 23. https://clinicaltrials.gov
- 24. https://clinicaltrials.gov
- 25. https://clinicaltrials.gov
- 26. https://clinicaltrials.gov
- 27. https://clinicaltrials.gov
- 28.Lill M.A. Multi-dimensional QSAR in drug discovery. Drug Discov. Today. 2007;12(23-24):1013–1017. doi: 10.1016/j.drudis.2007.08.004. [http://dx.doi.org/10.1016/ j.drudis.2007.08.004]. [PMID: 18061879]. [DOI] [PubMed] [Google Scholar]
- 29.Keller T.H., Pichota A., Yin Z. A practical view of ‘druggability’. Curr. Opin. Chem. Biol. 2006;10(4):357–361. doi: 10.1016/j.cbpa.2006.06.014. [http://dx.doi. org/10.1016/j.cbpa.2006.06.014]. [PMID: 16814592]. [DOI] [PubMed] [Google Scholar]
- 30.Roy K., Kar S. Understanding the basics of QSAR for applications in phar-maceutical sciences and risk assessment. 1st ed. Amsterdam: Academic press; 2015. [Google Scholar]
- 31.Ferreira M.M.C., Montanari C.A., Gaudio A.C. Seleção de variáveis em QSAR. Quim. Nova. 2002;25(3):439–448. [http:// dx.doi.org/10.1590/S0100-40422002000300017]. [Google Scholar]
- 32.Martins J.P.A., Ferreira M.M.C. QASR modeling: um novo pacote computacional open source para gerar e validar modelos QSAR. Quim. Nova. 2013;36(4):554–560. [http://dx.doi.org/10.1590/ S0100-40422013000400013]. [Google Scholar]
- 33.Cherkasov A., Muratov E.N., Fourches D., Varnek A., Baskin I.I., Cronin M., Dearden J., Gramatica P., Martin Y.C., Todeschini R., Consonni V., Kuz’min V.E., Cramer R., Benigni R., Yang C., Rathman J., Terfloth L., Gasteiger J., Richard A., Tropsha A. QSAR modeling: where have you been? Where are you going to? J. Med. Chem. 2014;57(12):4977–5010. doi: 10.1021/jm4004285. [http://dx. doi.org/10.1021/jm4004285]. [PMID: 24351051]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gaudio A.C., Zandonade E. Proposição, validação e análise dos modelos que correlacionam estrutura química e atividade biológica. Quim. Nova. 2001;24(5):658–671. [http://dx.doi.org/10.1590/ S0100-40422001000500013]. [Google Scholar]
- 35.Potemkin V., Grishina M. Principles for 3D/4D QSAR classification of drugs. Drug Discov. Today. 2008;13(21-22):952–959. doi: 10.1016/j.drudis.2008.07.006. [http://dx.doi.org/10.1016/j.drudis.2008.07.006]. [PMID: 18721896]. [DOI] [PubMed] [Google Scholar]
- 36.Stark H., Lipp R., Arrang J.M., Garbarg M., Schwartz J.C., Schunack W. Acylated and alkylated histamine derivatives as new histamine H3-receptor antagonists. Eur. J. Med. Chem. 1994;29(9):695–700. [http://dx.doi.org/10.1016/0223-5234(94)90031-0]. [Google Scholar]
- 37.Agrawal V.K., Khadikar P.V. QSAR studies on acylated histamine derivatives. Bioorg. Med. Chem. 2001;9(11):2787–2792. doi: 10.1016/s0968-0896(01)00147-x. [http://dx.doi.org/10.1016/S0968-0896(01)00147-X]. [PMID: 11597458]. [DOI] [PubMed] [Google Scholar]
- 38.Chen H.F. Computational study of histamine H3-receptor antagonist with support vector machines and three dimension quantitative structure activity relationship methods. Anal. Chim. Acta. 2008;624(2):203–209. doi: 10.1016/j.aca.2008.06.048. [http://dx.doi.org/10.1016/j.aca.2008.06.048]. [PMID: 18706326]. [DOI] [PubMed] [Google Scholar]
- 39.Kim S.K., Fristrup P., Abrol R., Goddard W.A., III Structure-based prediction of subtype selectivity of histamine H3 receptor selective antagonists in clinical trials. J. Chem. Inf. Model. 2011;51(12):3262–3274. doi: 10.1021/ci200435b. [http://dx.doi.org/10.1021/ci200435b]. [PMID: 22035233]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gfesser G.A., Faghih R., Bennani Y.L., Curtis M.P., Esbenshade T.A., Hancock A.A., Cowart M.D. Structure-activity relationships of arylbenzofuran H3 receptor antagonists. Bioorg. Med. Chem. Lett. 2005;15(10):2559–2563. doi: 10.1016/j.bmcl.2005.03.047. [http://dx.doi.org/10.1016/ j.bmcl.2005.03.047]. [PMID: 15863316]. [DOI] [PubMed] [Google Scholar]
- 41.Dastmalchi S., Hamzeh-Mivehroud M., Ghafourian T., Hamzeiy H. Molecular modeling of histamine H3 receptor and QSAR studies on arylbenzofuran derived H3 antagonists. J. Mol. Graph. Model. 2008;26(5):834–844. doi: 10.1016/j.jmgm.2007.05.002. [http://dx.doi.org/10.1016/j.jmgm. 2007.05.002]. [PMID: 17561422]. [DOI] [PubMed] [Google Scholar]
- 42.Dvorak C.A., Apodaca R., Barbier A.J., Berridge C.W., Wilson S.J., Boggs J.D., Xiao W., Lovenberg T.W., Carruthers N.I. 4-phenoxypiperidines: potent, conformationally restricted, non-imidazole histamine H3 antagonists. J. Med. Chem. 2005;48(6):2229–2238. doi: 10.1021/jm049212n. [http://dx.doi.org/10.1021/jm049212n]. [PMID: 15771465]. [DOI] [PubMed] [Google Scholar]
- 43.Arroio A., Honório K.M., da Silva A.B.F. Propriedades químico-quânticas empregadas em estudos das relações estrutura-atividade. Quim. Nova. 2010;33(3):694–699. [http://dx.doi.org/10.1590/ S0100-40422010000300037]. [Google Scholar]
- 44.Moorthy N.S., Ramos M.J., Fernandes P.A. QSAR and pharmacophore analysis of a series of piperidinyl urea derivatives as HERG blockers and H3 antagonists. Curr. Drug Discov. Technol. 2013;10(1):47–58. doi: 10.2174/157016313804998889. [PMID: 22564166]. [DOI] [PubMed] [Google Scholar]
- 45.Mor M., Bordi F., Silva C., Rivara S., Zuliani V., Vacondio F., Rivara M., Barocelli E., Bertoni S., Ballabeni V., Magnanini F., Impicciatore M., Plazzi P.V. Synthesis, biological activity, QSAR and QSPR study of 2-aminobenzimidazole derivatives as potent H3-antagonists. Bioorg. Med. Chem. 2004;12(4):663–674. doi: 10.1016/j.bmc.2003.11.030. [http://dx.doi.org/10.1016/j.bmc.2003.11.030]. [PMID: 14759727]. [DOI] [PubMed] [Google Scholar]
- 46.Rivara S., Mor M., Bordi F., Silva C., Zuliani V., Vacondio F., Morini G., Plazzi P.V., Carrupt P.A., Testa B. Synthesis and three-dimensional quantitative structure-activity relationship analysis of H3 receptor antagonists containing a neutral heterocyclic polar group. Drug Des. Discov. 2003;18(2-3):65–79. [http://dx.doi. org/10.3109/10559610290249539]. [PMID: 14675944]. [PubMed] [Google Scholar]
- 47.Windhorst A.D., Timmerman H., Worthington E.A., Bijloo G.J., Nederkoorn P.H., Menge W.M., Leurs R., Herscheid J.D. Characterization of the binding site of the histamine H(3) receptor. 2. Synthesis, in vitro pharmacology, and QSAR of a series of monosubstituted benzyl analogues of thioperamide. J. Med. Chem. 2000;43(9):1754–1761. doi: 10.1021/jm981106w. [http://dx.doi.org/10.1021/jm981106w]. [PMID: 10794692]. [DOI] [PubMed] [Google Scholar]
- 48.Lim H.D., Istyastono E.P., van de Stolpe A., Romeo G., Gobbi S., Schepers M., Lahaye R., Menge W.M., Zuiderveld O.P., Jongejan A., Smits R.A., Bakker R.A., Haaksma E.E., Leurs R., de Esch I.J. Clobenpropit analogs as dual activity ligands for the histamine H3 and H4 receptors: synthesis, pharmacological evaluation, and cross-target QSAR studies. Bioorg. Med. Chem. 2009;17(11):3987–3994. doi: 10.1016/j.bmc.2009.04.007. [http://dx.doi.org/10.1016/j.bmc.2009.04.007]. [PMID: 19414267]. [DOI] [PubMed] [Google Scholar]
- 49.Istyastono E.P., Nijmeijer S., Lim H.D., van de Stolpe A., Roumen L., Kooistra A.J., Vischer H.F., de Esch I.J.P., Leurs R., de Graaf C. Molecular determinants of ligand binding modes in the histamine H(4) receptor: linking ligand-based three-dimensional quantitative structure-activity relationship (3D-QSAR) models to in silico guided receptor mutagenesis studies. J. Med. Chem. 2011;54(23):8136–8147. doi: 10.1021/jm201042n. [http://dx.doi.org/10.1021/jm201042n]. [PMID: 22003888]. [DOI] [PubMed] [Google Scholar]