Abstract
Significance: The emerging connections between an increasing number of long noncoding RNAs (lncRNAs) and oncogenic hallmarks provide a new twist to tumor complexity.
Recent Advances: In the present review, we highlight specific lncRNAs that have been studied in relation to tumorigenesis, either as participants in the neoplastic process or as markers of pathway activity or drug response. These transcripts are typically deregulated by oncogenic or tumor-suppressing signals or respond to microenvironmental conditions such as hypoxia.
Critical Issues: Among these transcripts are lncRNAs sufficiently divergent between mouse and human genomes that may contribute to biological differences between species.
Future Directions: From a translational standpoint, knowledge about primate-specific lncRNAs may help explain the reason behind the failure to reproduce the results from mouse cancer models in human cell-based systems. Antioxid. Redox Signal. 29, 922–935.
Keywords: : cancer, non-coding RNA, metabolism, hypoxia, microenvironment
Introduction
Since the declaration of War on Cancer in 1971, increased efforts have been focused on characterizing the cellular and molecular changes associated with tumor progression and therapeutic response. Six key hallmarks have been highlighted to describe the mechanisms through which tumor cells proliferate, invade, and metastasize (39), and new emerging hallmarks are added as knowledge advances (40). Nevertheless, despite the progress in understanding the genetic programming of tumor cells, the overall decrease in mortality remains relatively modest. A major reason for therapeutic failure is tumor heterogeneity: advanced tumors being in fact a collection of dynamic subpopulations with different mutation spectra and vulnerabilities. Second, increasingly detailed molecular dissection of tumor-driving signals has revealed additional layers of complexity, with special attention being drawn by the expanding world of noncoding RNAs.
Over the past two decades, these cellular RNAs that translated into proteins have been gradually implicated in virtually all physiological, developmental, and disease processes, including cancer (6, 9, 25, 29, 57, 62). According to the classic dogma, RNA transcripts simply served as templates for protein synthesis (22), which led to decades of protein-centered research. However, successive waves of discovery identified multiple categories of functional noncoding transcripts, beginning with heterogeneous nuclear RNAs (45, 113), followed by introns (4, 5, 20), small nuclear RNAs (37, 38, 63, 81, 102), microRNAs (miRNAs) (61), and long noncoding RNAs (lncRNAs) (10). While the study of miRNAs dominated the first decade of the noncoding RNA revolution, in recent years, lncRNAs—generically defined as noncoding transcripts longer than 200 ribonucleotides—have moved to center stage.
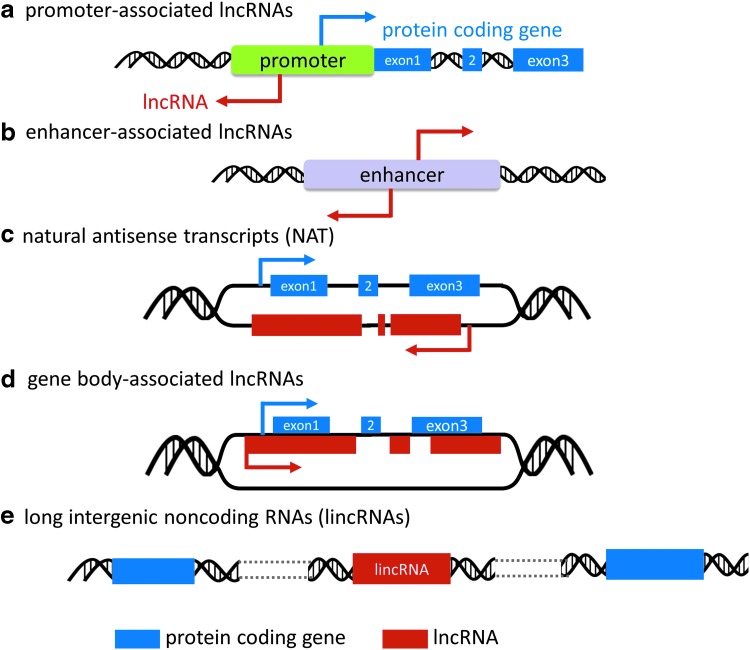
The completion of human genome project and the parallel progress in RNA sequencing technology have been instrumental for identification of thousands of lncRNA transcripts (14, 17, 55, 97). In the latest Human GENCODE release (version 26, October 2016, GRCh38, Ensembl 88), 15,787 genes originating 27,720 RNA locus transcripts are identified as lncRNA genes. While the genome size tends to increase during metazoan evolution toward increasingly complex life forms, the number of protein coding genes has remained relatively steady (105). In contrast, the number of noncoding elements, including lncRNAs, appears to have increased dramatically. As the term lncRNA is a generic designation based on size, additional classifications are required when dissecting their biological roles. A popular categorization is based on their genomic contexts (Fig. 1): (i) promoter-associated lncRNAs (Fig. 1a) (e.g., promoter of CDKN1A antisense DNA damage-activated RNA); (ii) enhancer-associated lncRNAs (Fig. 1b) (e.g., Evf2); (iii) natural antisense transcripts (NATs, Fig. 1c) (e.g., hypoxia-inducible factor 1 alpha antisense 2 [HIF1A-AS2]); (iv) gene body-associated (sense) lncRNAs (Fig.1d) (e.g., CCAAT/enhancer binding protein alpha - ecCEBPA); and (v) long intergenic ncRNAs (lincRNAs, Fig. 1e) (e.g., HOX transcript antisense RNA [HOTAIR], metastasis-associated lung adenocarcinoma transcript 1 [MALAT1]) (8, 101).
FIG. 1.
lncRNA classification. Based on genomic context, lncRNA can be classified into five categories: (a) promoter-associated lncRNAs, (b) enhancer-associated lncRNAs, (c) natural antisense transcript, (d) gene body-associated lncRNAs, and (e) intergenic lncRNAs. lncRNA, long noncoding RNA.
From a cancer perspective, the ever-increasing number of connections between lncRNAs and oncogenic hallmarks adds a new twist to tumor complexity. lncRNAs tend to be less conserved during evolution and their expression exhibits higher tissue specificity compared with PCGs. Therefore, detailed knowledge about cancer-associated lncRNAs may explain some differences between neoplastic cells derived from different tissues or different species.
Despite the fundamental difference with respect to protein coding ability, lncRNAs exhibit important similarities with PCGs, including chromatin marks at their promoters or enhancers (35). Furthermore, lncRNA genes are also transcribed by RNA polymerase II, spliced at canonical splicing sites, and some undergo polyadenylation (93). Similarly to coding genes, lncRNAs are regulated, positively and negatively, by complexes of transcription factors, coregulators and corepressors, from proximal promoters or enhancers. It is predictable therefore that transcription factors that drive proliferation and survival programs in normal or tumor cells also engage lncRNAs that regulate specific aspects of tumor biology.
In addition to cell-autonomous regulatory mechanisms, the tumor microenvironment has a significant impact in shaping the lncRNA landscape. The combination of stress factors, including oxygen and nutrient depletion, favors the selection of populations with increased ability to survive by rewiring their molecular networks, including metabolism, apoptotic responses, and proliferative programs. As discussed below, it is predictable that tumor microenvironment-regulated lncRNAs should impact this set of basic cell responses.
How can a ribonucleotide stretch affect the survival and proliferation of a neoplastic cell? lncRNAs have been shown to regulate gene expression at multiple levels (epigenetic, transcriptional, and post-transcriptional) through interaction with other biomolecules, such as proteins, regulatory DNA regions, and miRNAs (Fig. 2). Subcellular localization appears to be a major determinant for lncRNA interactions and therefore functions. In particular, nuclear lncRNAs modulate gene expression in cis or trans by interacting with transcriptional coregulators and chromatin remodeling complexes. As shown by Rinn and colleagues, ∼20% lncRNAs associate with the polycomb repressive complex 2 (PRC2), a multicomponent histone methyltransferase required for epigenetic silencing (56). A classic example is provided by HOTAIR, shown to reprogram PRC2 and LSD1-CoREST (lysine-specific demethylase 1 and REST corepressor 1 complex) occupancy within the homeobox D cluster (34, 108). Subsequent studies, however, indicated that the lncRNA-based PRC2 guiding model needs to be revised (23, 54). Furthermore, Portoso et al.'s recent results suggest that HOTAIR-PRC2 interactions are dispensable for HOTAIR-mediated transcriptional silencing (90).
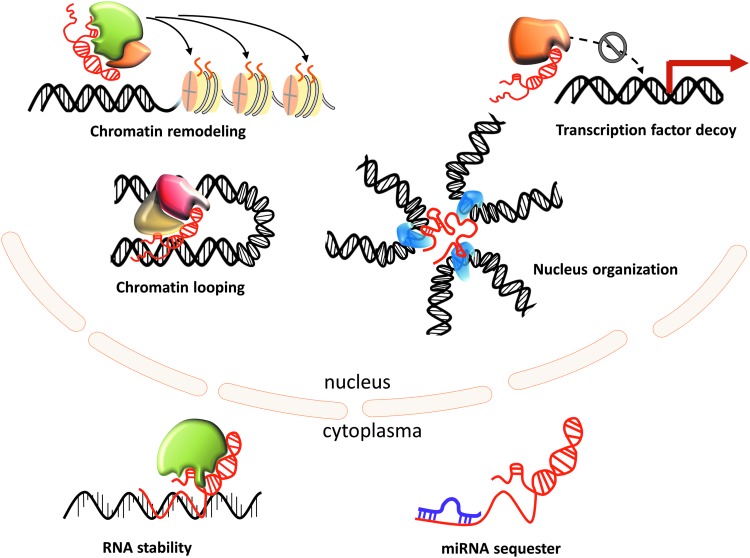
FIG. 2.
lncRNA functions. A diverse range of mechanisms have been described for lncRNA regulation of their targets depending on their subcellular localization: assembly and recruitment of chromatin-modifying complexes to their DNA targets in cis; some lncRNAs act as RNA decoys, tethering transcription factors away from their DNA targets by directly binding to them as target mimics; and guiding of the physical looping that occurs between enhancers and targeted promoters (enhancer lncRNAs). Many lncRNAs bind to various protein partners to regulate RNA splicing, degradation, and translation; others act as microRNA target site decoys miRNA, microRNA.
A significant proportion of lncRNAs are thought to act in cis by enhancing or, conversely, repressing the expression of nearby genes (89). How exactly these lncRNAs perform these functions remains a debated topic. Some of these noncoding transcripts may reorganize the local architecture of chromatin and stoichiometry of transcriptional complexes by specific RNA-protein interactions. Recently, however, Engreitz et al. (26) provided a surprising twist to the function of lncRNAs. Their results indicate that (at least in some cases) the lncRNA transcript itself is not critical for the regulation of a neighboring gene as long as there is active transcription of this noncoding locus. In other words, the sequence and interactions of the noncoding RNA product take a backseat to the actual process that generates it. Based on the large number of lncRNAs and the enormous diversity of contexts in which they function, it seems reasonable to assume that these mechanisms are not mutually exclusive.
A puzzling characteristic of lncRNAs is that most of them exhibit very low expression in a particular cell context, including tumors. Many are often considered transcriptional noise and tend to be discounted by arbitrarily set expression cutoff. For lncRNAs, however, low expression should not automatically be viewed as lack of significance as they may achieve biologically meaningful concentrations in specific subcellular compartments; for example, the physical looping mediated by a specific lncRNA bringing together an enhancer and a promoter (66, 89). Supporting evidence has been presented linking lncRNAs to the three-dimensional organization of the nucleus, such as paraspeckle formation or multichromosomal structure (21, 36). These highly specific and localized interactions may support therefore the compatibility between low expression and tissue specificity (84).
At the post-transcriptional level, lncRNAs have been shown to be involved in virtually all steps of RNA metabolism, including stability, processing, and decay. The upregulation of natural antisense (NAT) type of lncRNAs often affects gene expression on the opposite strand by generating RNA duplexes, either by transcript stabilization or degradation via RNA interference. Several other effects such as alternative splicing lncRNA-mediated RNA processing have been described as well as an mRNA degradation process called Staufen-mediated decay, which involves lncRNAs binding to the 3′UTR of Staufen-targeted genes (58, 59). Furthermore, lncRNAs can also directly bind proteins, mostly transcription factors, to disrupt their interaction with targeted DNA or other proteins (49).
We will now apply the interactions and functions summarized to the specific case of tumors and highlight how lncRNAs can be integrated in the network of classic neoplastic determinants.
Tumor Microenvironment and lncRNAs; the Effect of Hypoxia
It is estimated that more than half of solid tumors contain hypoxic regions (75) (11, 110) that represent sources of cells with aggressive phenotype and high resistance to therapy (42, 94, 98, 99). The imbalance between high oxygen consumption of fast proliferating tumor cells and impaired oxygen delivery due to abnormalities in tumor vasculature (30) triggers signaling pathways that regulate tumor cell survival, angiogenesis, metastasis, immune response, and metabolic reprogramming.
Hypoxia-mediated cellular response is primarily driven through the HIF pathway, a complex regulatory network, with multiple feedbacks and checkpoint signaling loops. HIF transcription factors are heterodimers comprising two subunits: an oxygen-sensitive α-subunit (hypoxia-inducible factor 1 alpha [HIF-1α], HIF-2α, and HIF-3α) and a constitutively expressed β-subunit (HIF-1β/ARNT, HIF-2β/ARNT2). While HIF-1β/ARNT is ubiquitously expressed, HIF-2β/ARNT2 is mainly localized in neural tissue and kidney. Three prolyl hydroxylases, EGLN 1–3/PHD 1–3, hydroxylate two proline residues of HIF-α (12, 27, 51), thus favoring the binding of von Hippel-Lindau tumor suppressor protein (pVHL) to HIF-α, which subsequently targets HIF-α for ubiquitination-mediated proteasomal degradation (52, 53, 82). Under hypoxic conditions, the activity of EGLN enzymes decreases, resulting in increased abundance of nonhydroxylated HIF-α subunits, which cannot be recognized by the pVHL complex, thus able to form an active transcriptional complex.
Active HIF regulates the transcription of hundreds of coding and noncoding genes. While a large set of miRNAs have been reported as hypoxia responsive (31), a smaller number of lncRNAs have been identified that respond with at least some consistency to oxygen availability. However, it should not be interpreted that miRNAs are generally more responsive to oxygen deprivation. First, miRNAs have been systematically investigated for a longer period of time (103). Second, multiple nomenclatures coexisted for lncRNAs and one could speculate that the same lncRNA may have been identified by different screens under different names. Finally, omission of nonpolyadenylated transcripts from library preparation may have led to the loss of hypoxia-responsive lncRNAs.
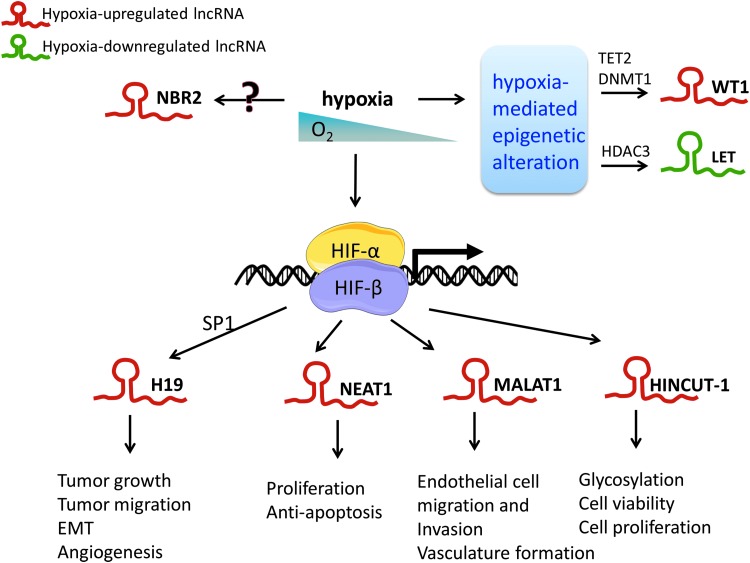
Overall, two types of lncRNA-HIF connections have been described: lncRNAs that are regulated in response to hypoxia and HIF signaling subsequently and lncRNAs that regulate HIF signaling (Fig. 3). Many hypoxia-inducible lncRNAs reported to date are direct transcriptional HIF targets (18, 19, 28, 79, 85, 86, 121, 124, 130). Chromatin immunoprecipitation sequencing studies of HIF-DNA binding have revealed that ∼30% of HIF binding sites are close to noncoding gene loci; correlations with hypoxic gene regulation revealed significant associations between HIF binding and transcription of lncRNA. Nevertheless, several lncRNAs such as nuclear paraspeckle assembly transcript 1 (NEAT1), MALAT1, HIF1A-AS2, imprinted maternally expressed lncRNA (H19), hypoxia-induced noncoding ultraconserved transcript 1 (HINCUT-1), and urothelial cancer-associated 1 have been identified as hypoxia-responsive lncRNAs (18, 19, 85, 86). A selection of hypoxia-regulated lncRNAs and their impact on tumor biology are summarized in Table 1.
FIG. 3.
Examples of hypoxia-regulated lncRNAs. lncRNAs can be regulated by hypoxia through direct or indirect manner. Some lncRNAs, such as NEAT1, MALAT1, and HINCUT-1, etc. have been identified as direct HIF targets. HIF is also involved in upregulation of H19, but most likely via an indirect signaling through SP1. Hypoxia-mediated epigenetic alterations also play important roles in the expression of some hypoxia-responsive lncRNAs such as lncRNA WT1 and LET. H19, imprinted maternally expressed lncRNA; HIF, hypoxia-inducible factor; HINCUT-1, hypoxia-induced noncoding ultraconserved transcript 1; LET, low expression in tumor; MALAT1, metastasis-associated lung adenocarcinoma transcript 1; NEAT1, nuclear paraspeckle assembly transcript 1; TET2, tet methylcytosine dioxygenase 2; WT1, Wilms' tumor 1.
Table 1.
Hypoxia-Regulated Long Noncoding RNAs and Their Role in Cancer
| lncRNA | Hypoxia response | Transcription regulation | Cancer type | Cancer impact | Refs. |
|---|---|---|---|---|---|
| H19 | Up | HIF/MYC/SP1 | HCC | Cell survival and proliferation | (13, 46, 73, 76–80, 87, 117, 122) |
| Bladder cancer | |||||
| CRC | Migration | ||||
| Esophageal cancer | Angiogenesis | ||||
| Breast cancer | EMT | ||||
| Gastric cancer | |||||
| HINCUT1/uc.475 | Up | HIF | Colon cancer | Cell proliferation | (28) |
| UCA1/CURD | Up | HIF | Bladder cancer | Cell proliferation | (121) |
| Migration and invasion | |||||
| Apoptosis | |||||
| NEAT1 | Up | HIF | Breast cancer | Cell survival | (18) |
| Apoptosis | |||||
| MALAT1 | Up | HIF | Breast cancer | Cell cycle | (18, 19, 85, 107) |
| Lung cancer | Angiogenesis | ||||
| Tumor metastasis | |||||
| lncRNA-NUTF2P3-001 | Up | HIF | Pancreatic cancer | Proliferation, invasion | (67) |
| HIF1A-AS2 | Up | HIF | Glioblastoma | Maintain mesenchymal glioblastoma stem-like cells in hypoxia niches | (7, 86) |
| Breast cancer | |||||
| SARCC | Up | HIF | RCC | Proliferation | (128) |
| ANRIL | Up | HIF | Osteosarcoma | Invasion, apoptosis | (114) |
| GAPLINC | Up | HIF | Gastric cancer | Proliferation | (70) |
| PVT1 | Up | HIF | Cervical cancer | Proliferation, apoptosis, migration, invasion, cisplatin cytotoxicity | (47, 50) |
| Gastric cancer | |||||
| NBR2 | Up | — | Breast cancer | Cell cycle | (72, 116) |
| Apoptosis/autophagy | |||||
| Metabolic checkpoint under energy stress | |||||
| EFNA3 lncRNA | Up | — | Breast cancer | Tumor metastasis | (32) |
| AK058003 | Up | — | Gastric cancer | Modulate DNA methylation at SNCG CpG island | (111) |
| Migration and invasion | |||||
| Tumor metastasis | |||||
| AK123072 | Up | — | Gastric cancer | Migration, invasion, metastasis | (125) |
| linc-ROR | Up | — | HCC | RNA sponge (miR145) | (106) |
| Upregulate HIF1A mRNA | |||||
| Cell survival | |||||
| HOTAIR | Up | HIF | Breast cancer | Cell viability | (34, 130) |
| Lung cancer | Invasion | ||||
| Apoptosis | |||||
| Metastasis | |||||
| lncRNA-LET | Down | Hypoxia-mediated epigenetic alteration: HDAC3 | Gallbladder cancer | Inhibit invasion and metastasis | (74, 123) |
| SCLC | |||||
| HCC | |||||
| CRC | |||||
| WT1 lncRNA | Up | Hypoxia-mediated epigenetic alteration: DNMT1, TET2 | AML | Modulate histone methylation at WT1 TSS | (83) |
AML, acute myeloid leukemia; ANRIL, CDKN2B antisense RNA 1; CRC, colorectal cancer; DNMT1, DNA methyltransferase 1; EMT, epithelial-to-mesenchymal transition; H19, imprinted maternally expressed lncRNA; HCC, hepatocellular carcinoma; HIF, hypoxia-inducible factor; HIF1A-AS2, hypoxia-inducible factor 1 alpha antisense 2; HINCUT-1, hypoxia-induced noncoding ultraconserved transcript 1; HOTAIR, HOX transcript antisense RNA; lncRNA, long noncoding RNA; MALAT1, metastasis-associated lung adenocarcinoma transcript 1; NBR2, neighbor of the BRCA1 gene 2; NEAT1, nuclear paraspeckle assembly transcript 1; PVT1, PVT1 plasmacytoma variant translocation 1 lncRNA; RCC, renal cell carcinoma; SARCC, suppressing androgen receptor in renal cell carcinoma; SCLC, squamous-cell lung cancer; TET2, tet methylcytosine dioxygenase 2; UCA1/CUDR, urothelial cancer-associated 1; WT1, Wilms' tumor 1.
Arguably the first reported hypoxia-inducible noncoding RNA is the transcript generated by the imprinted oncofetal gene H19 (77, 79, 80). While details remain to be elucidated, Wu et al. provided evidence that HIF-1, while required, is not the direct activator of H19 transcription. They proposed that HIF-1 activates SP1, which in turn activates the H19 promoter (117). Functionally, H19 has been shown to promote tumor growth and regulate anchorage-independent growth after hypoxia recovery (77, 79).
H19 exerts broader proneoplastic effects unrelated to its response to hypoxia. The H19 gene is highly expressed in common metastatic sites regardless of tumor primary origin. H19 enhances cell migration in vitro and stimulates tumor metastasis in vivo (95). In ovarian carcinoma cells, H19 overexpression is associated with chemoresistance and epithelial-to-mesenchymal transition phenotype (80). H19 knockdown leads to decreased expression of genes with antiapoptotic function, such as microphthalmia-associated transcription factor, immediate early response 3, protein kinase C, zeta, B cell CLL/lymphoma 3, and serine/threonine kinase 1, and upregulation of proapoptotic genes such as DNA damage-inducible transcript 3 also known as GADD153 (77). H19 also plays an important and multipronged role in tumor angiogenesis (77) by regulating the production of proangiogenic factors such as angiogenin, fibroblast growth factor 18, prolylcarboxypeptidase, tumor necrosis factor α-induced protein 1, calponin 2, and inhibitor of DNA binding 2.
Although much remains to be understood about how these complex regulatory effects are set in motion, H19 actions most likely involve a multitude of interactions, including proteins and RNA interactions. For example, H19 can modulate chromatin structure within the imprinted gene network through interaction with methyl-CpG-binding domain protein 1 (87). Other studies provided evidence that H19 functions as an endogenous miRNA sponge for let-7 tumor suppressor miRNAs (118).
Choudhry et al. reported NEAT1 and MALAT1 as the main lncRNAs induced in hypoxic MCF7 breast cancer cells (18). NEAT1 is predominantly controlled by HIF-2, rather than by HIF-1, and is involved in paraspeckle formation (21). One of the paraspeckle functions is to sequester hyperedited RNAs into the nucleus, thus impeding their translocation to the cytoplasm (3). Interestingly, elimination of these lncRNAs in the mouse embryo is compatible with life; therefore, it is conceivable that these transcripts play fine-tuning rather than essential roles in proliferation. However, in tumor cells, hypoxic induction of NEAT1 promotes proliferation and suppresses apoptosis (18). Protumorigenic roles have been described for MALAT1 as well. In the highly angiogenic neuroblastomas, upregulation of MALAT1 promotes endothelial cell migration, invasion, and vasculature formation through fibroblast growth factor 2 upregulation (107). In breast cancer, MALAT1 regulates critical processes such as tumor growth, differentiation, and metastasis (1). Genetic loss or antisense oligonucleotide (ASO)-mediated knockdown in MMTV-PyMT mouse mammary carcinoma models leads to gene expression alterations and splicing patterns of genes involved in pathogenesis, resulting in reduced branching morphogenesis in MMTV-PyMT- and Her2/neu-amplified tumor organoids, increased cell adhesion, and loss of migration (1). Many miRNAs, including miR-205, miR-200c, and miR-204, have been reported to interact with MALAT1 and thus contribute to its tumor-promoting mechanism in various cancer types (44, 65, 120). However, these studies often provide little significant molecular proof and require further validation.
HINCUTs are a family of lncRNAs that are transcribed from regions exhibiting extremely high conservation between human, rat, and mouse genomes (2). Our groups have shown that HINCUT-1 (originally termed uc.475) is an lncRNA transcribed as a retained intron of O-linked N-acetylglucosamine transferase (OGT) mRNA and is induced by hypoxia in an HIF-dependent manner (28). Although details are unclear, HINCUT-1 appears to play an important role in steady-state OGT expression and overall cellular glycosylation and its inactivation has detrimental effects on cell viability and proliferation (28).
Recently, suppressing androgen receptor in renal cell carcinoma (SARCC) lncRNA was reported as an HIF-2 target in clear cell renal carcinoma. The authors provide preliminary evidence that lncRNA-SARCC binds and destabilizes androgen receptor (AR), which results in suppression of AR/HIF-2α/c-MYC signaling (128).
Another lncRNA probably driven by HIF is plasmacytoma variant translocation 1 lncRNA, which appears to be a multifaceted player in cancer. On the one hand, it promotes cell migration and invasion, and on the other hand, it was shown to correlate with immune response stimulation in cervical cancer (50).
Neighbor of breast cancer 1 (BRCA1) gene 2 (NBR2) lncRNA is a transcript expressed in the opposite orientation from the bidirectional BRCA1 promoter that has recently been shown to regulate AMP-activated protein kinase under energy stress (72). Wiedmeier et al. have recently shown that NBR2 is induced under prolonged hypoxia in MCF7 cells, while BRCA1 is repressed. These results suggest that the two transcripts driven by the BRCA1 promoter are differentially regulated in response to hypoxia, although the regulatory element(s) required for induction of NBR2 appear to reside outside of the BRCA1 minimal promoter (116).
A rare case of lncRNA reported as downregulated in hypoxia is lncRNA-LET (74, 123), which exhibits the behavior of a tumor-suppressing element. In primary hepatocellular carcinoma, lncRNA-LET expression is inversely correlated with the prototypical hypoxia marker carbonic anhydrase 9, and experimentally, lncRNA-LET downregulation leads to hypoxia-induced cancer cell invasion in hepatocellular carcinoma cells (123). In a different tumor context, ectopic expression of lncRNA-LET leads to G0/G1 cell cycle arrest and induction of apoptosis under hypoxic conditions and suppresses gallbladder tumor growth in vivo (74).
Occasionally, hypoxia-dependent lncRNA regulation may occur through epigenetic regulators rather than direct HIF activation. In acute myeloid leukemia cells, induction of Wilms' Tumor 1 (WT1) lncRNA, an antisense-oriented lncRNA overlapping with intron 1 CpG island of the WT1 gene, appears to be the result of demethylation through hypoxia-regulated expression of DNA methyltransferase 1 and tet methylcytosine dioxygenase 2 (83).
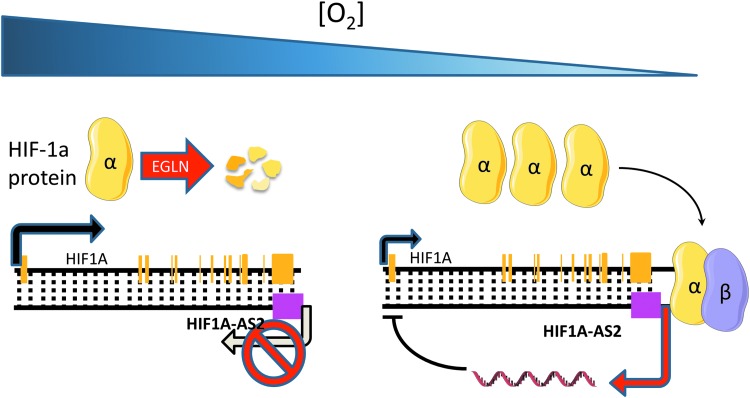
lncRNAs are not only direct targets of HIF transcriptional activation but also have been demonstrated to regulate the transcription of HIF genes themselves, through direct or indirect interactions. This mechanism creates complex signaling networks with positive and negative feedback loops that integrate multiple signaling pathways to control HIF response to hypoxia. HIF-1α antisense transcripts have long been known to be induced in response to hypoxia (7, 86) and have been shown to negatively regulate HIF expression by chromatin inactivation or mRNA degradation (7) (Fig. 4). More recently, in mesenchymal glioblastoma stem-like cells, HIF1A-AS2 was found to be the most significantly upregulated lncRNA, playing a protumorigenic role. The authors identified DExH-box helicase 9 and insulin-like growth factor 2-binding protein 2 proteins as major interactors of HIF1A-AS2 and this interaction in turn drives the expression of tumor-promoting downstream targets, in particular the high-mobility group AT-hook 1 (86). Preliminary evidence suggests that HIF1A-AS2 may be relevant in a broader context as its knockdown was found to inhibit gastric cancer cell proliferation (16).
FIG. 4.
HIF1A antisense transcript forms negative feedback loops to control HIF response to hypoxia. HIF1A-AS2 is an HIF-target lncRNA. Under hypoxia, HIF directly binds to the HIF1A-AS2 promoter, driving the expression of an antisense transcript to HIF1A, which in return negatively regulates HIF1A expression level as a negative feedback mechanism. HIF1A-AS2, hypoxia-inducible factor 1 alpha antisense 2.
A more recent, and incompletely understood, mechanism appears to involve HIF-2 and is based on HIF-2α promoter upstream transcript lncRNA. This is an lncRNA transcribed from the upstream of the HIF-2α promoter, which induces cis HIF-2α activation in osteosarcoma (112) and colorectal cancer (126).
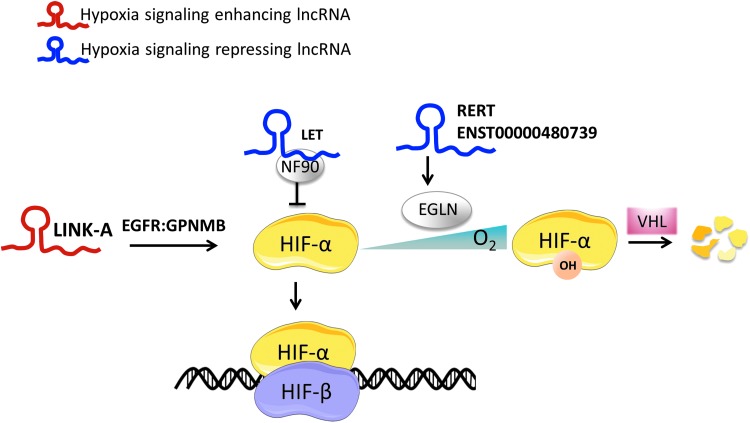
Several lncRNAs have been reported to regulate HIF signaling through indirect mechanisms (Fig. 5). Long intergenic noncoding RNA for kinase activation mediates heparin-binding epidermal growth factor-like growth factor-triggered, epidermal growth factor receptor: glycoprotein nonmetastatic melanoma protein B heterodimer-dependent HIF-1α phosphorylation leading to HIF-1α stabilization, HIF-1α-p300 interaction, and activation of hypoxic programs, including glycolysis under normal oxygen conditions in breast cancer (68). In pancreatic ductal adenocarcinoma, lncRNA ENST00000480739 has been demonstrated to increase the levels of endoplasmic reticulum lectin protein (104), which is known to increase the affinity between HIF-1α and EGLN hydroxylases, therefore leading to HIF-1α destabilization. Somewhat similarly, RAB4B-EGLN2 read-through lncRNA appears to suppress HIF-1α signaling through EGLN transcription activation (132).
FIG. 5.
Examples of lncRNAs that regulate hypoxia signaling. Several lncRNAs have been reported to regulate hypoxia signaling through indirect mechanisms. LINK-A stabilizes HIF-1α through an EGFR:GPNMB heterodimer-dependent HIF-1α phosphorylation and thus activates HIF-1α transcriptional programs. lncRNA LET reduces the protein levels of HIF-1α through its association with NF90. lncRNA RERT and ENST00000480739 downregulate hypoxia signaling through modulation of EGLN. EGFR, epidermal growth factor receptor; EGLN 1–3/PHD 1–3, prolyl hydroxylases 1–3; GPNMB, glycoprotein nmb; HIF-1α, hypoxia-inducible factor 1 alpha; LINK-A, long intergenic noncoding RNA for kinase activation; RERT, RAB4B-EGLN2 read-through lncRNA.
Other Cancer-Associated lncRNAs
Colon cancer-associated transcripts (CCATs), 1 (119) and 2 (69), are lncRNAs transcribed from the highly conserved 8q24 region that has been shown to enhance the transcription of MYC oncogene and promote cancer progression, invasion, and metastasis. CCAT1 is involved in maintaining chromatin looping between the MYC promoter and its enhancers in coordination with the CCCTC-binding factor. CCAT2 increases chromosomal instability through transcription factor 7-like 2-mediated transcriptional regulation. Thus, both CCAT1 and CCAT2 have been associated with increased risk of cancer and have been shown to regulate multiple molecular pathways to promote cell proliferation, metastasis, and cancer metabolism (96).
lincRNA-p21 is a p53 transcriptional target that has been shown to repress p53 transcriptional response through heterogeneous nuclear ribonucleoprotein K and trigger apoptosis (48). While in coordination with RNA-binding protein HuR, it inhibits the translation of p53 targets such as jun B proto-oncogene and catenin beta 1 (127). In nonsmall cell lung cancer, tumor samples with high lincRNA-p21 levels show higher microvascular density. lincRNA-p21 induces angiogenesis in vitro, while lincRNA-p21 inhibition leads to downregulation of angiogenesis-related genes, such as vascular endothelial growth factor A (15).
Prostate cancer-associated ncRNA transcript 1 (PCAT-1), although initially reported as a PCAT (91), has been described to associate with multiple types of cancers. PCAT-1 is a target of the PRC2 and represses the transcription of genes involved in cell proliferation, invasion, and metastasis.
CDKN2B antisense RNA 1 (ANRIL) is transcribed in the opposite direction from the INK4b-ARF-INK4a cluster and it is one of the most frequently altered lncRNAs in cancer. The molecular mechanisms through which ANRIL mediated cancer development and progression are still uncertain; however, it is hypothesized that aberrant expression levels of ANRIL may block the DNA damage response mechanism, leading to genomic instability. In addition, ANRIL promotes tumor cell proliferation by regulating target genes in trans. ANRIL promotes the epigenetically silencing of miR-99A/miR-449A, therefore upregulating mechanistic target of rapamycin and cyclin-dependent kinase 6/E2F transcription factor 1 pathways (129).
lncRNAs: Are Diagnostic and Therapeutic Applications Feasible?
As functional molecules, the lncRNA expression levels may serve as better prognostic and diagnostic indicators of diseases than mRNAs. Additional, their highly specific spatial and temporal expression signatures could lead to a more accurate disease diagnosis and classification. Potential applications of lncRNAs in clinical oncology have been proposed, such as diagnostic biomarkers and therapy response predictors. Prostate cancer-associated 3 (PCA3/DD3) lncRNA, for example, has already been tested in controlled clinical settings based on its much higher expression in prostate tumors compared with normal prostate and other tissues. However, based on the available data, PCA3/DD3 does not appear to be superior to the routinely used prostate-specific antigen (24, 60). Another potentially valuable marker may be HOTAIR, which was found to be upregulated dramatically in metastatic breast cancer tissue compared with normal breast tissue (34).
The therapeutic relevance of lncRNAs is currently under exploration, but critical hurdles need to be overcome. Due to their size, transduction of tumor suppressor lncRNAs necessitates delivery systems (e.g., viruses) that have yet to prove their value in clinical settings. On the other hand, oncogenic lncRNAs may be targetable with synthetic RNAs, such as siRNAs, ASOs, or miRNAs. While siRNA-mediated knockdown of cytoplasmic lncRNAs is highly efficient, targeting nuclear lncRNAs is more challenging. Thus, the ASO technology has been optimized to target nuclear lncRNAs for RNase H1-mediated RNA degradation. Promising in vivo results have been reported for several lncRNAs such as MALAT1 (1) and SAMSSON (64).
Another approach for lncRNA targeting may be based on lessons learned from the study of vault RNAs (vtRNAs) as mediators of multidrug resistance (33). It was shown that vtRNAs directly bind to chemotherapeutic agents, indicating that it would also be possible to design small molecules that interact with lncRNAs (33). vtRNAs are technically short RNAs, ranging from 80 to 90 nucleotides; however, examples of longer RNAs involved with drug interactions exist, such as aptamers (41, 43, 88, 115). Targeting transcripts the size of lncRNAs may appear challenging, but there is a precedent for fragmenting large ribonucleoprotein complexes into more manageable sizes. This strategy has been applied to design ligands for the expanded rCUG and rCAG repeats expressed in myotonic dystrophy type 1 that interact with Muscleblind-like 1 protein (92). Moreover, unbiased methods such as systematic evolution of ligands by exponential enrichment have the potential to be used to identify molecules that interact with lncRNAs (109).
The clustered regularly interspaced short palindromic repeat (CRISPR)/Cas9-based technologies have revolutionized the study of genetic reprogramming by being developed into a genome-wide editing tool with large applications, including noncoding transcriptome functionality (71, 131). However, CRISPR-Cas9-directed lncRNA genomic deletions do not necessarily induce repression of biological activity (100, 131). The CRISPR interference has been reported as a better approach for lncRNA functionality studies as this technology uses a nuclease-dead dCAS9-KRAB repressor fusion protein to repress gene transcription. This protein can be recruited by single-guided RNA pools to the lncRNA transcriptional start sites, and association with a specific phenotype (71) can be selected based on specific markers as a readout.
In conclusion, multidisciplinary approaches continue to provide critical insights into the involvement of lncRNAs in various aspects of cancer biology. While many lncRNAs already show significant potential as therapeutic targets or cancer biomarkers, transitioning from basic knowledge to viable clinical applications remains challenging. Future studies will need to clarify which, if any, lncRNAs are truly essential for cancer cell viability and to develop more efficient tools for their inactivation in clinical tumors. Furthermore, it would be highly impactful to identify lncRNAs that inform about tumor vulnerability to specific therapeutic agents, potentially in a defined genetic context.
Abbreviations Used
- AML
acute myeloid leukemia
- ANRIL
CDKN2B antisense RNA 1
- AR
androgen receptor
- ASO
antisense oligonucleotide
- BRCA1
breast cancer 1
- CCATs
colon cancer-associated transcripts
- CRC
colorectal cancer
- CRISPR
clustered regularly interspaced short palindromic repeat
- DNMT1
DNA methyltransferase 1
- EGFR
epidermal growth factor receptor
- EGLN 1–3/PHD 1–3
prolyl hydroxylases 1–3
- EMT
epithelial-to-mesenchymal transition
- GPNMB
glycoprotein nmb
- H19
imprinted maternally expressed lncRNA
- HCC
hepatocellular carcinoma
- HIF
hypoxia-inducible factor
- HIF-1α
hypoxia-inducible factor 1 alpha
- HIF1A-AS2
hypoxia-inducible factor 1 alpha antisense 2
- HINCUT-1
hypoxia-induced noncoding ultraconserved transcript 1
- HOTAIR
HOX transcript antisense RNA
- lincRNA
long intergenic ncRNA
- LINK-A
long intergenic noncoding RNA for kinase activation
- lncRNA
long noncoding RNA
- MALAT1
metastasis-associated lung adenocarcinoma transcript 1
- miRNA
microRNA
- NAT
natural antisense transcript
- NBR2
neighbor of the BRCA1 gene 2
- NEAT1
nuclear paraspeckle assembly transcript 1
- OGT
O-linked N-acetylglucosamine transferase
- PCA3/DD3
prostate cancer-associated 3
- PCAT-1
prostate cancer-associated ncRNA transcript 1
- PRC2
polycomb repressive complex 2
- pVHL
von Hippel-Lindau tumor suppressor protein
- PVT1
PVT1 plasmacytoma variant translocation 1 lncRNA
- RCC
renal cell carcinoma
- RERT
RAB4B-EGLN2 read-through lncRNA
- SARCC
suppressing androgen receptor in renal cell carcinoma
- SCLC
squamous-cell lung cancer
- TET2
tet methylcytosine dioxygenase 2
- UCA1/CUDR
urothelial cancer-associated 1
- VEGFA
vascular endothelial growth factor A
- vtRNA
vault RNA
- WT1
Wilms' tumor 1
Acknowledgments
Work in Dr. Ivan's laboratory is supported by a National Institutes of Health (NIH/NCI) grant R01CA155332; Dr. Calin's laboratory is supported by National Institutes of Health (NIH/NCATS) grant UH3TR00943-01 through the NIH Common Fund, Office of Strategic Coordination (OSC); the NIH/NCI grant 1 R01 CA182905-01; U54 grant—UPR/MDACC Partnership for Excellence in Cancer Research 2016 Pilot Project; CLL Moonshot Flagship project; and the Estate of C.G. Johnson, Jr.; and Dr. Tudoran's work is supported by the Romanian National Grant Program PN-II-RU-TE-2014-4-1984.
References
- 1.Arun G, Diermeier S, Akerman M, Chang KC, Wilkinson JE, Hearn S, Kim Y, MacLeod AR, Krainer AR, Norton L, Brogi E, Egeblad M, and Spector DL. Differentiation of mammary tumors and reduction in metastasis upon Malat1 lncRNA loss. Genes Dev 30: 34–51, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bejerano G, Pheasant M, Makunin I, Stephen S, Kent WJ, Mattick JS, and Haussler D. Ultraconserved elements in the human genome. Science 304: 1321–1325, 2004 [DOI] [PubMed] [Google Scholar]
- 3.Ben-Zvi M, Amariglio N, Paret G, and Nevo-Caspi Y. F11R expression upon hypoxia is regulated by RNA editing. PLoS One 8: e77702, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berget SM. and Sharp PA. A spliced sequence at the 5′-terminus of adenovirus late mRNA. Brookhaven Symp Biol 12–20: 332–344, 1977 [PubMed] [Google Scholar]
- 5.Berk AJ. and Sharp PA. Sizing and mapping of early adenovirus mRNAs by gel electrophoresis of S1 endonuclease-digested hybrids. Cell 12: 721–732, 1977 [DOI] [PubMed] [Google Scholar]
- 6.Bernstein E, Caudy AA, Hammond SM, and Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 409: 363–366, 2001 [DOI] [PubMed] [Google Scholar]
- 7.Bertozzi D, Iurlaro R, Sordet O, Marinello J, Zaffaroni N, and Capranico G. Characterization of novel antisense HIF-1alpha transcripts in human cancers. Cell Cycle 10: 3189–3197, 2011 [DOI] [PubMed] [Google Scholar]
- 8.Bonasio R. and Shiekhattar R. Regulation of transcription by long noncoding RNAs. Annu Rev Genet 48: 433–455, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bracken CP, Gregory PA, Khew-Goodall Y, and Goodall GJ. The role of microRNAs in metastasis and epithelial-mesenchymal transition. Cell Mol Life Sci 66: 1682–1699, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brannan CI, Dees EC, Ingram RS, and Tilghman SM. The product of the H19 gene may function as an RNA. Mol Cell Biol 10: 28–36, 1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brown JM. and Giaccia AJ. The unique physiology of solid tumors: opportunities (and problems) for cancer therapy. Cancer Res 58: 1408–1416, 1998 [PubMed] [Google Scholar]
- 12.Bruick RK. and McKnight SL. A conserved family of prolyl-4-hydroxylases that modify HIF. Science 294: 1337–1340, 2001 [DOI] [PubMed] [Google Scholar]
- 13.Cai X. and Cullen BR. The imprinted H19 noncoding RNA is a primary microRNA precursor. RNA 13: 313–316, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, Oyama R, Ravasi T, Lenhard B, Wells C, Kodzius R, Shimokawa K, Bajic VB, Brenner SE, Batalov S, Forrest AR, Zavolan M, Davis MJ, Wilming LG, Aidinis V, Allen JE, Ambesi-Impiombato A, Apweiler R, Aturaliya RN, Bailey TL, Bansal M, Baxter L, Beisel KW, Bersano T, Bono H, Chalk AM, Chiu KP, Choudhary V, Christoffels A, Clutterbuck DR, Crowe ML, Dalla E, Dalrymple BP, de Bono B, Della Gatta G, di Bernardo D, Down T, Engstrom P, Fagiolini M, Faulkner G, Fletcher CF, Fukushima T, Furuno M, Futaki S, Gariboldi M, Georgii-Hemming P, Gingeras TR, Gojobori T, Green RE, Gustincich S, Harbers M, Hayashi Y, Hensch TK, Hirokawa N, Hill D, Huminiecki L, Iacono M, Ikeo K, Iwama A, Ishikawa T, Jakt M, Kanapin A, Katoh M, Kawasawa Y, Kelso J, Kitamura H, Kitano H, Kollias G, Krishnan SP, Kruger A, Kummerfeld SK, Kurochkin IV, Lareau LF, Lazarevic D, Lipovich L, Liu J, Liuni S, McWilliam S, Madan Babu M, Madera M, Marchionni L, Matsuda H, Matsuzawa S, Miki H, Mignone F, Miyake S, Morris K, Mottagui-Tabar S, Mulder N, Nakano N, Nakauchi H, Ng P, Nilsson R, Nishiguchi S, Nishikawa S, Nori F, Ohara O, Okazaki Y, Orlando V, Pang KC, Pavan WJ, Pavesi G, Pesole G, Petrovsky N, Piazza S, Reed J, Reid JF, Ring BZ, Ringwald M, Rost B, Ruan Y, Salzberg SL, Sandelin A, Schneider C, Schonbach C, Sekiguchi K, Semple CA, Seno S, Sessa L, Sheng Y, Shibata Y, Shimada H, Shimada K, Silva D, Sinclair B, Sperling S, Stupka E, Sugiura K, Sultana R, Takenaka Y, Taki K, Tammoja K, Tan SL, Tang S, Taylor MS, Tegner J, Teichmann SA, Ueda HR, van Nimwegen E, Verardo R, Wei CL, Yagi K, Yamanishi H, Zabarovsky E, Zhu S, Zimmer A, Hide W, Bult C, Grimmond SM, Teasdale RD, Liu ET, Brusic V, Quackenbush J, Wahlestedt C, Mattick JS, Hume DA, Kai C, Sasaki D, Tomaru Y, Fukuda S, Kanamori-Katayama M, Suzuki M, Aoki J, Arakawa T, Iida J, Imamura K, Itoh M, Kato T, Kawaji H, Kawagashira N, Kawashima T, Kojima M, Kondo S, Konno H, Nakano K, Ninomiya N, Nishio T, Okada M, Plessy C, Shibata K, Shiraki T, Suzuki S, Tagami M, Waki K, Watahiki A, Okamura-Oho Y, Suzuki H, Kawai J, Hayashizaki Y FA.NTOM Consortium, and RIKEN Genome Exploration Research Group and Genome Science Group (Genome Network Project Core Group). The transcriptional landscape of the mammalian genome. Science 309: 1559–1563, 2005 [DOI] [PubMed] [Google Scholar]
- 15.Castellano JJ, Navarro A, Vinolas N, Marrades RM, Moises J, Cordeiro A, Saco A, Munoz C, Fuster D, Molins L, Ramirez J, and Monzo M. LincRNA-p21 Impacts prognosis in resected non-small cell lung cancer patients through angiogenesis regulation. J Thorac Oncol 11: 2173–2182, 2016 [DOI] [PubMed] [Google Scholar]
- 16.Chen WM, Huang MD, Kong R, Xu TP, Zhang EB, Xia R, Sun M, De W, and Shu YQ. Antisense long noncoding RNA HIF1A-AS2 is upregulated in gastric cancer and associated with poor prognosis. Dig Dis Sci 60: 1655–1662, 2015 [DOI] [PubMed] [Google Scholar]
- 17.Cheng J, Kapranov P, Drenkow J, Dike S, Brubaker S, Patel S, Long J, Stern D, Tammana H, Helt G, Sementchenko V, Piccolboni A, Bekiranov S, Bailey DK, Ganesh M, Ghosh S, Bell I, Gerhard DS, and Gingeras TR. Transcriptional maps of 10 human chromosomes at 5-nucleotide resolution. Science 308: 1149–1154, 2005 [DOI] [PubMed] [Google Scholar]
- 18.Choudhry H, Albukhari A, Morotti M, Haider S, Moralli D, Smythies J, Schodel J, Green CM, Camps C, Buffa F, Ratcliffe P, Ragoussis J, Harris AL, and Mole DR. Tumor hypoxia induces nuclear paraspeckle formation through HIF-2alpha dependent transcriptional activation of NEAT1 leading to cancer cell survival. Oncogene 34: 4546, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Choudhry H, Schodel J, Oikonomopoulos S, Camps C, Grampp S, Harris AL, Ratcliffe PJ, Ragoussis J, and Mole DR. Extensive regulation of the non-coding transcriptome by hypoxia: role of HIF in releasing paused RNApol2. EMBO Rep 15: 70–76, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chow LT, Roberts JM, Lewis JB, and Broker TR. A map of cytoplasmic RNA transcripts from lytic adenovirus type 2, determined by electron microscopy of RNA:DNA hybrids. Cell 11: 819–836, 1977 [DOI] [PubMed] [Google Scholar]
- 21.Clemson CM, Hutchinson JN, Sara SA, Ensminger AW, Fox AH, Chess A, and Lawrence JB. An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol Cell 33: 717–726, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Crick FH. On protein synthesis. Symp Soc Exp Biol 12: 138–163, 1958 [PubMed] [Google Scholar]
- 23.Davidovich C, Zheng L, Goodrich KJ, and Cech TR. Promiscuous RNA binding by polycomb repressive complex 2. Nat Struct Mol Biol 20: 1250–1257, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Day JR, Jost M, Reynolds MA, Groskopf J, and Rittenhouse H. PCA3: from basic molecular science to the clinical lab. Cancer Lett 301: 1–6, 2011 [DOI] [PubMed] [Google Scholar]
- 25.Doi N, Zenno S, Ueda R, Ohki-Hamazaki H, Ui-Tei K, and Saigo K. Short-interfering-RNA-mediated gene silencing in mammalian cells requires Dicer and eIF2C translation initiation factors. Curr Biol 13: 41–46, 2003 [DOI] [PubMed] [Google Scholar]
- 26.Engreitz JM, Haines JE, Perez EM, Munson G, Chen J, Kane M, McDonel PE, Guttman M, and Lander ES. Local regulation of gene expression by lncRNA promoters, transcription and splicing. Nature 539: 452–455, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Epstein AC, Gleadle JM, McNeill LA, Hewitson KS, O'Rourke J, Mole DR, Mukherji M, Metzen E, Wilson MI, Dhanda A, Tian YM, Masson N, Hamilton DL, Jaakkola P, Barstead R, Hodgkin J, Maxwell PH, Pugh CW, Schofield CJ, and Ratcliffe PJ. C. elegans EGL-9 and mammalian homologs define a family of dioxygenases that regulate HIF by prolyl hydroxylation. Cell 107: 43–54, 2001 [DOI] [PubMed] [Google Scholar]
- 28.Ferdin J, Nishida N, Wu X, Nicoloso MS, Shah MY, Devlin C, Ling H, Shimizu M, Kumar K, Cortez MA, Ferracin M, Bi Y, Yang D, Czerniak B, Zhang W, Schmittgen TD, Voorhoeve MP, Reginato MJ, Negrini M, Davuluri RV, Kunej T, Ivan M, and Calin GA. HINCUTs in cancer: hypoxia-induced noncoding ultraconserved transcripts. Cell Death Differ 20: 1675–1687, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fernandez-Valverde SL, Taft RJ, and Mattick JS. MicroRNAs in beta-cell biology, insulin resistance, diabetes and its complications. Diabetes 60: 1825–1831, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fukumura D. and Jain RK. Tumor microvasculature and microenvironment: targets for anti-angiogenesis and normalization. Microvasc Res 74: 72–84, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gee HE, Ivan C, Calin GA, and Ivan M. HypoxamiRs and cancer: from biology to targeted therapy. Antioxid Redox Signal 21: 1220–1238, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gomez-Maldonado L, Tiana M, Roche O, Prado-Cabrero A, Jensen L, Fernandez-Barral A, Guijarro-Munoz I, Favaro E, Moreno-Bueno G, Sanz L, Aragones J, Harris A, Volpert O, Jimenez B, and del Peso L. EFNA3 long noncoding RNAs induced by hypoxia promote metastatic dissemination. Oncogene 34: 2609–2620, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gopinath SC, Matsugami A, Katahira M, and Kumar PK. Human vault-associated non-coding RNAs bind to mitoxantrone, a chemotherapeutic compound. Nucleic Acids Res 33: 4874–4881, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, Wang Y, Brzoska P, Kong B, Li R, West RB, van de Vijver MJ, Sukumar S, and Chang HY. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 464: 1071–1076, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, Cabili MN, Jaenisch R, Mikkelsen TS, Jacks T, Hacohen N, Bernstein BE, Kellis M, Regev A, Rinn JL, and Lander ES. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature 458: 223–227, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hacisuleyman E, Goff LA, Trapnell C, Williams A, Henao-Mejia J, Sun L, McClanahan P, Hendrickson DG, Sauvageau M, Kelley DR, Morse M, Engreitz J, Lander ES, Guttman M, Lodish HF, Flavell R, Raj A, and Rinn JL. Topological organization of multichromosomal regions by the long intergenic noncoding RNA Firre. Nat Struct Mol Biol 21: 198–206, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hadjiolov AA, Venkov PV, and Tsanev RG. Ribonucleic acids fractionation by density-gradient centrifugation and by agar gel electrophoresis: a comparison. Anal Biochem 17: 263–267, 1966 [DOI] [PubMed] [Google Scholar]
- 38.Hamm J, Darzynkiewicz E, Tahara SM, and Mattaj IW. The trimethylguanosine cap structure of U1 snRNA is a component of a bipartite nuclear targeting signal. Cell 62: 569–577, 1990 [DOI] [PubMed] [Google Scholar]
- 39.Hanahan D. and Weinberg RA. The hallmarks of cancer. Cell 100: 57–70, 2000 [DOI] [PubMed] [Google Scholar]
- 40.Hanahan D. and Weinberg RA. Hallmarks of cancer: the next generation. Cell 144: 646–674, 2011 [DOI] [PubMed] [Google Scholar]
- 41.Hanson S, Berthelot K, Fink B, McCarthy JE, and Suess B. Tetracycline-aptamer-mediated translational regulation in yeast. Mol Microbiol 49: 1627–1637, 2003 [DOI] [PubMed] [Google Scholar]
- 42.Harris AL. Hypoxia—a key regulatory factor in tumour growth. Nat Rev Cancer 2: 38–47, 2002 [DOI] [PubMed] [Google Scholar]
- 43.Hermann T. and Patel DJ. Adaptive recognition by nucleic acid aptamers. Science 287: 820–825, 2000 [DOI] [PubMed] [Google Scholar]
- 44.Hirata H, Hinoda Y, Shahryari V, Deng G, Nakajima K, Tabatabai ZL, Ishii N, and Dahiya R. Long Noncoding RNA MALAT1 promotes aggressive renal cell carcinoma through Ezh2 and interacts with miR-205. Cancer Res 75: 1322–1331, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Holmes DS, Mayfield JE, Sander G, and Bonner J. Chromosomal RNA: its properties. Science 177: 72–74, 1972 [DOI] [PubMed] [Google Scholar]
- 46.Huang C, Cao L, Qiu L, Dai X, Ma L, Zhou Y, Li H, Gao M, Li W, Zhang Q, Han K, and Lv H. Upregulation of H19 promotes invasion and induces epithelial-to-mesenchymal transition in esophageal cancer. Oncol Lett 10: 291–296, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Huang T, Liu HW, Chen JQ, Wang SH, Hao LQ, Liu M, and Wang B. The long noncoding RNA PVT1 functions as a competing endogenous RNA by sponging miR-186 in gastric cancer. Biomed Pharmacother 88: 302–308, 2017 [DOI] [PubMed] [Google Scholar]
- 48.Huarte M, Guttman M, Feldser D, Garber M, Koziol MJ, Kenzelmann-Broz D, Khalil AM, Zuk O, Amit I, Rabani M, Attardi LD, Regev A, Lander ES, Jacks T, and Rinn JL. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell 142: 409–419, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hung T, Wang Y, Lin MF, Koegel AK, Kotake Y, Grant GD, Horlings HM, Shah N, Umbricht C, Wang P, Wang Y, Kong B, Langerod A, Borresen-Dale AL, Kim SK, van de Vijver M, Sukumar S, Whitfield ML, Kellis M, Xiong Y, Wong DJ, and Chang HY. Extensive and coordinated transcription of noncoding RNAs within cell-cycle promoters. Nat Genet 43: 621–629, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Iden M, Fye S, Li K, Chowdhury T, Ramchandran R, and Rader JS. The lncRNA PVT1 contributes to the cervical cancer phenotype and associates with poor patient prognosis. PLoS One 11: e0156274, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ivan M, Haberberger T, Gervasi DC, Michelson KS, Gunzler V, Kondo K, Yang H, Sorokina I, Conaway RC, Conaway JW, and Kaelin WG., Jr Biochemical purification and pharmacological inhibition of a mammalian prolyl hydroxylase acting on hypoxia-inducible factor. Proc Natl Acad Sci U S A 99: 13459–13464, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ivan M, Kondo K, Yang H, Kim W, Valiando J, Ohh M, Salic A, Asara JM, Lane WS, and Kaelin WG., Jr HIFalpha targeted for VHL-mediated destruction by proline hydroxylation: implications for O2 sensing. Science 292: 464–468, 2001 [DOI] [PubMed] [Google Scholar]
- 53.Jaakkola P, Mole DR, Tian YM, Wilson MI, Gielbert J, Gaskell SJ, von Kriegsheim A, Hebestreit HF, Mukherji M, Schofield CJ, Maxwell PH, Pugh CW, and Ratcliffe PJ. Targeting of HIF-alpha to the von Hippel-Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science 292: 468–472, 2001 [DOI] [PubMed] [Google Scholar]
- 54.Kaneko S, Son J, Shen SS, Reinberg D, and Bonasio R. PRC2 binds active promoters and contacts nascent RNAs in embryonic stem cells. Nat Struct Mol Biol 20: 1258–1264, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kapranov P, Cawley SE, Drenkow J, Bekiranov S, Strausberg RL, Fodor SP, and Gingeras TR. Large-scale transcriptional activity in chromosomes 21 and 22. Science 296: 916–919, 2002 [DOI] [PubMed] [Google Scholar]
- 56.Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D, Thomas K, Presser A, Bernstein BE, van Oudenaarden A, Regev A, Lander ES, and Rinn JL. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci U S A 106: 11667–11672, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kim DH, Villeneuve LM, Morris KV, and Rossi JJ. Argonaute-1 directs siRNA-mediated transcriptional gene silencing in human cells. Nat Struct Mol Biol 13: 793–797, 2006 [DOI] [PubMed] [Google Scholar]
- 58.Kretz M. TINCR, staufen1, and cellular differentiation. RNA Biol 10: 1597–1601, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kretz M, Siprashvili Z, Chu C, Webster DE, Zehnder A, Qu K, Lee CS, Flockhart RJ, Groff AF, Chow J, Johnston D, Kim GE, Spitale RC, Flynn RA, Zheng GX, Aiyer S, Raj A, Rinn JL, Chang HY, and Khavari PA. Control of somatic tissue differentiation by the long non-coding RNA TINCR. Nature 493: 231–235, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lee GL, Dobi A, and Srivastava S. Prostate cancer: diagnostic performance of the PCA3 urine test. Nat Rev Urol 8: 123–124, 2011 [DOI] [PubMed] [Google Scholar]
- 61.Lee RC, Feinbaum RL, and Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75: 843–854, 1993 [DOI] [PubMed] [Google Scholar]
- 62.Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Radmark O, Kim S, and Kim VN. The nuclear RNase III Drosha initiates microRNA processing. Nature 425: 415–419, 2003 [DOI] [PubMed] [Google Scholar]
- 63.Legrain P, Seraphin B, and Rosbash M. Early commitment of yeast pre-mRNA to the spliceosome pathway. Mol Cell Biol 8: 3755–3760, 1988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Leucci E, Vendramin R, Spinazzi M, Laurette P, Fiers M, Wouters J, Radaelli E, Eyckerman S, Leonelli C, Vanderheyden K, Rogiers A, Hermans E, Baatsen P, Aerts S, Amant F, Van Aelst S, van den Oord J, de Strooper B, Davidson I, Lafontaine DL, Gevaert K, Vandesompele J, Mestdagh P, and Marine JC. Melanoma addiction to the long non-coding RNA SAMMSON. Nature 531: 518–522, 2016 [DOI] [PubMed] [Google Scholar]
- 65.Li J, Wang J, Chen Y, Li S, Jin M, Wang H, Chen Z, and Yu W. LncRNA MALAT1 exerts oncogenic functions in lung adenocarcinoma by targeting miR-204. Am J Cancer Res 6: 1099–1107, 2016 [PMC free article] [PubMed] [Google Scholar]
- 66.Li W, Notani D, Ma Q, Tanasa B, Nunez E, Chen AY, Merkurjev D, Zhang J, Ohgi K, Song X, Oh S, Kim HS, Glass CK, and Rosenfeld MG. Functional roles of enhancer RNAs for oestrogen-dependent transcriptional activation. Nature 498: 516–520, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Li X, Deng SJ, Zhu S, Jin Y, Cui SP, Chen JY, Xiang C, Li QY, He C, Zhao SF, Chen HY, Niu Y, Liu Y, Deng SC, Wang CY, and Zhao G. Hypoxia-induced lncRNA-NUTF2P3-001 contributes to tumorigenesis of pancreatic cancer by derepressing the miR-3923/KRAS pathway. Oncotarget 7: 6000–6014, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lin A, Li C, Xing Z, Hu Q, Liang K, Han L, Wang C, Hawke DH, Wang S, Zhang Y, Wei Y, Ma G, Park PK, Zhou J, Zhou Y, Hu Z, Zhou Y, Marks JR, Liang H, Hung MC, Lin C, and Yang L. The LINK-A lncRNA activates normoxic HIF1alpha signalling in triple-negative breast cancer. Nat Cell Biol 18: 213–224, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ling H, Spizzo R, Atlasi Y, Nicoloso M, Shimizu M, Redis RS, Nishida N, Gafa R, Song J, Guo Z, Ivan C, Barbarotto E, De Vries I, Zhang X, Ferracin M, Churchman M, van Galen JF, Beverloo BH, Shariati M, Haderk F, Estecio MR, Garcia-Manero G, Patijn GA, Gotley DC, Bhardwaj V, Shureiqi I, Sen S, Multani AS, Welsh J, Yamamoto K, Taniguchi I, Song MA, Gallinger S, Casey G, Thibodeau SN, Le Marchand L, Tiirikainen M, Mani SA, Zhang W, Davuluri RV, Mimori K, Mori M, Sieuwerts AM, Martens JW, Tomlinson I, Negrini M, Berindan-Neagoe I, Foekens JA, Hamilton SR, Lanza G, Kopetz S, Fodde R, and Calin GA. CCAT2, a novel noncoding RNA mapping to 8q24, underlies metastatic progression and chromosomal instability in colon cancer. Genome Res 23: 1446–1461, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Liu L, Zhao X, Zou H, Bai R, Yang K, and Tian Z. Hypoxia promotes gastric cancer malignancy partly through the HIF-1alpha dependent transcriptional activation of the long non-coding RNA GAPLINC. Front Physiol 7: 420, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Liu SJ, Horlbeck MA, Cho SW, Birk HS, Malatesta M, He D, Attenello FJ, Villalta JE, Cho MY, Chen Y, Mandegar MA, Olvera MP, Gilbert LA, Conklin BR, Chang HY, Weissman JS, and Lim DA. CRISPRi-based genome-scale identification of functional long noncoding RNA loci in human cells. Science 355, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Liu X, Xiao ZD, Han L, Zhang J, Lee SW, Wang W, Lee H, Zhuang L, Chen J, Lin HK, Wang J, Liang H, and Gan B. LncRNA NBR2 engages a metabolic checkpoint by regulating AMPK under energy stress. Nat Cell Biol 18: 431–442, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Luo M, Li Z, Wang W, Zeng Y, Liu Z, and Qiu J. Long non-coding RNA H19 increases bladder cancer metastasis by associating with EZH2 and inhibiting E-cadherin expression. Cancer Lett 333: 213–221, 2013 [DOI] [PubMed] [Google Scholar]
- 74.Ma MZ, Kong X, Weng MZ, Zhang MD, Qin YY, Gong W, Zhang WJ, and Quan ZW. Long non-coding RNA-LET is a positive prognostic factor and exhibits tumor-suppressive activity in gallbladder cancer. Mol Carcinog 54: 1397–1406, 2015 [DOI] [PubMed] [Google Scholar]
- 75.Manoochehri Khoshinani H, Afshar S, and Najafi R. Hypoxia: a double-edged sword in cancer therapy. Cancer Invest 34: 536–545, 2016 [DOI] [PubMed] [Google Scholar]
- 76.Matouk I, Raveh E, Ohana P, Lail RA, Gershtain E, Gilon M, De Groot N, Czerniak A, and Hochberg A. The increasing complexity of the oncofetal h19 gene locus: functional dissection and therapeutic intervention. Int J Mol Sci 14: 4298–4316, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Matouk IJ, DeGroot N, Mezan S, Ayesh S, Abu-lail R, Hochberg A, and Galun E. The H19 non-coding RNA is essential for human tumor growth. PLoS One 2: e845, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Matouk IJ, Halle D, Gilon M, and Hochberg A. The non-coding RNAs of the H19-IGF2 imprinted loci: a focus on biological roles and therapeutic potential in lung cancer. J Transl Med 13: 113, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Matouk IJ, Mezan S, Mizrahi A, Ohana P, Abu-Lail R, Fellig Y, Degroot N, Galun E, and Hochberg A. The oncofetal H19 RNA connection: hypoxia, p53 and cancer. Biochim Biophys Acta 1803: 443–451, 2010 [DOI] [PubMed] [Google Scholar]
- 80.Matouk IJ, Raveh E, Abu-lail R, Mezan S, Gilon M, Gershtain E, Birman T, Gallula J, Schneider T, Barkali M, Richler C, Fellig Y, Sorin V, Hubert A, Hochberg A, and Czerniak A. Oncofetal H19 RNA promotes tumor metastasis. Biochim Biophys Acta 1843: 1414–1426, 2014 [DOI] [PubMed] [Google Scholar]
- 81.Maxwell ES. and Fournier MJ. The small nucleolar RNAs. Annu Rev Biochem 64: 897–934, 1995 [DOI] [PubMed] [Google Scholar]
- 82.Maxwell PH, Wiesener MS, Chang GW, Clifford SC, Vaux EC, Cockman ME, Wykoff CC, Pugh CW, Maher ER, and Ratcliffe PJ. The tumour suppressor protein VHL targets hypoxia-inducible factors for oxygen-dependent proteolysis. Nature 399: 271–275, 1999 [DOI] [PubMed] [Google Scholar]
- 83.McCarty G. and Loeb DM. Hypoxia-sensitive epigenetic regulation of an antisense-oriented lncRNA controls WT1 expression in myeloid leukemia cells. PLoS One 10: e0119837, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Mele M. and Rinn JL. “Cat's Cradling” the 3D genome by the act of LncRNA transcription. Mol Cell 62: 657–664, 2016 [DOI] [PubMed] [Google Scholar]
- 85.Michalik KM, You X, Manavski Y, Doddaballapur A, Zornig M, Braun T, John D, Ponomareva Y, Chen W, Uchida S, Boon RA, and Dimmeler S. Long noncoding RNA MALAT1 regulates endothelial cell function and vessel growth. Circ Res 114: 1389–1397, 2014 [DOI] [PubMed] [Google Scholar]
- 86.Mineo M, Ricklefs F, Rooj AK, Lyons SM, Ivanov P, Ansari KI, Nakano I, Chiocca EA, Godlewski J, and Bronisz A. The long non-coding RNA HIF1A-AS2 facilitates the maintenance of mesenchymal glioblastoma stem-like cells in hypoxic niches. Cell Rep 15: 2500–2509, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Monnier P, Martinet C, Pontis J, Stancheva I, Ait-Si-Ali S, and Dandolo L. H19 lncRNA controls gene expression of the imprinted gene network by recruiting MBD1. Proc Natl Acad Sci U S A 110: 20693–20698, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Nahvi A, Sudarsan N, Ebert MS, Zou X, Brown KL, and Breaker RR. Genetic control by a metabolite binding mRNA. Chem Biol 9: 1043, 2002 [DOI] [PubMed] [Google Scholar]
- 89.Orom UA, Derrien T, Beringer M, Gumireddy K, Gardini A, Bussotti G, Lai F, Zytnicki M, Notredame C, Huang Q, Guigo R, and Shiekhattar R. Long noncoding RNAs with enhancer-like function in human cells. Cell 143: 46–58, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Portoso M, Ragazzini R, Brencic Z, Moiani A, Michaud A, Vassilev I, Wassef M, Servant N, Sargueil B, and Margueron R. PRC2 is dispensable for HOTAIR-mediated transcriptional repression. EMBO J 36: 981–994, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Prensner JR, Iyer MK, Balbin OA, Dhanasekaran SM, Cao Q, Brenner JC, Laxman B, Asangani IA, Grasso CS, Kominsky HD, Cao X, Jing X, Wang X, Siddiqui J, Wei JT, Robinson D, Iyer HK, Palanisamy N, Maher CA, and Chinnaiyan AM. Transcriptome sequencing across a prostate cancer cohort identifies PCAT-1, an unannotated lincRNA implicated in disease progression. Nat Biotechnol 29: 742–749, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Pushechnikov A, Lee MM, Childs-Disney JL, Sobczak K, French JM, Thornton CA, and Disney MD. Rational design of ligands targeting triplet repeating transcripts that cause RNA dominant disease: application to myotonic muscular dystrophy type 1 and spinocerebellar ataxia type 3. J Am Chem Soc 131: 9767–9779, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Quinn JJ. and Chang HY. Unique features of long non-coding RNA biogenesis and function. Nat Rev Genet 17: 47–62, 2016 [DOI] [PubMed] [Google Scholar]
- 94.Rankin EB. and Giaccia AJ. The role of hypoxia-inducible factors in tumorigenesis. Cell Death Differ 15: 678–685, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Raveh E, Matouk IJ, Gilon M, and Hochberg A. The H19 long non-coding RNA in cancer initiation, progression and metastasis—a proposed unifying theory. Mol Cancer 14: 184, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Redis RS, Vela LE, Lu W, Ferreira de Oliveira J, Ivan C, Rodriguez-Aguayo C, Adamoski D, Pasculli B, Taguchi A, Chen Y, Fernandez AF, Valledor L, Van Roosbroeck K, Chang S, Shah M, Kinnebrew G, Han L, Atlasi Y, Cheung LH, Huang GY, Monroig P, Ramirez MS, Catela Ivkovic T, Van L, Ling H, Gafa R, Kapitanovic S, Lanza G, Bankson JA, Huang P, Lai SY, Bast RC, Rosenblum MG, Radovich M, Ivan M, Bartholomeusz G, Liang H, Fraga MF, Widger WR, Hanash S, Berindan-Neagoe I, Lopez-Berestein G, Ambrosio AL, Gomes Dias SM, and Calin GA. Allele-specific reprogramming of cancer metabolism by the long non-coding RNA CCAT2. Mol Cell 61: 520–534, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Rinn JL, Euskirchen G, Bertone P, Martone R, Luscombe NM, Hartman S, Harrison PM, Nelson FK, Miller P, Gerstein M, Weissman S, and Snyder M. The transcriptional activity of human chromosome 22. Genes Dev 17: 529–540, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Schindl M, Schoppmann SF, Samonigg H, Hausmaninger H, Kwasny W, Gnant M, Jakesz R, Kubista E, Birner P, Oberhuber G, Austrian B, and Colorectal Cancer Study Group. Overexpression of hypoxia-inducible factor 1alpha is associated with an unfavorable prognosis in lymph node-positive breast cancer. Clin Cancer Res 8: 1831–1837, 2002 [PubMed] [Google Scholar]
- 99.Semenza GL. Intratumoral hypoxia, radiation resistance, and HIF-1. Cancer Cell 5: 405–406, 2004 [DOI] [PubMed] [Google Scholar]
- 100.Shalem O, Sanjana NE, Hartenian E, Shi X, Scott DA, Mikkelsen TS, Heckl D, Ebert BL, Root DE, Doench JG, and Zhang F. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science 343: 84–87, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Sigova AA, Mullen AC, Molinie B, Gupta S, Orlando DA, Guenther MG, Almada AE, Lin C, Sharp PA, Giallourakis CC, and Young RA. Divergent transcription of long noncoding RNA/mRNA gene pairs in embryonic stem cells. Proc Natl Acad Sci U S A 110: 2876–2881, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Singh R. and Reddy R. Gamma-monomethyl phosphate: a cap structure in spliceosomal U6 small nuclear RNA. Proc Natl Acad Sci U S A 86: 8280–8283, 1989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.St. Laurent G, Wahlestedt C, and Kapranov P. The landscape of long noncoding RNA classification. Trends Genet 31: 239–251, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Sun YW, Chen YF, Li J, Huo YM, Liu DJ, Hua R, Zhang JF, Liu W, Yang JY, Fu XL, Yan T, Hong J, and Cao H. A novel long non-coding RNA ENST00000480739 suppresses tumour cell invasion by regulating OS-9 and HIF-1alpha in pancreatic ductal adenocarcinoma. Br J Cancer 111: 2131–2141, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Taft RJ, Pheasant M, and Mattick JS. The relationship between non-protein-coding DNA and eukaryotic complexity. Bioessays 29: 288–299, 2007 [DOI] [PubMed] [Google Scholar]
- 106.Takahashi K, Yan IK, Haga H, and Patel T. Modulation of hypoxia-signaling pathways by extracellular linc-RoR. J Cell Sci 127: 1585–1594, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Tee AE, Liu B, Song R, Li J, Pasquier E, Cheung BB, Jiang C, Marshall GM, Haber M, Norris MD, Fletcher JI, Dinger ME, and Liu T. The long noncoding RNA MALAT1 promotes tumor-driven angiogenesis by up-regulating pro-angiogenic gene expression. Oncotarget 7: 8663–8675, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F, Shi Y, Segal E, and Chang HY. Long noncoding RNA as modular scaffold of histone modification complexes. Science 329: 689–693, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Tsai MC, Spitale RC, and Chang HY. Long intergenic noncoding RNAs: new links in cancer progression. Cancer Res 71: 3–7, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Vaupel P. and Mayer A. Hypoxia in cancer: significance and impact on clinical outcome. Cancer Metastasis Rev 26: 225–239, 2007 [DOI] [PubMed] [Google Scholar]
- 111.Wang Y, Liu X, Zhang H, Sun L, Zhou Y, Jin H, Zhang H, Zhang H, Liu J, Guo H, Nie Y, Wu K, Fan D, Zhang H, and Liu L. Hypoxia-inducible lncRNA-AK058003 promotes gastric cancer metastasis by targeting gamma-synuclein. Neoplasia 16: 1094–1106, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Wang Y, Yao J, Meng H, Yu Z, Wang Z, Yuan X, Chen H, and Wang A. A novel long non-coding RNA, hypoxia-inducible factor-2alpha promoter upstream transcript, functions as an inhibitor of osteosarcoma stem cells in vitro. Mol Med Rep 11: 2534–2540, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Warner JR, Soeiro R, Birnboim HC, Girard M, and Darnell JE. Rapidly labeled HeLa cell nuclear RNA. I. Identification by zone sedimentation of a heterogeneous fraction separate from ribosomal precursor RNA. J Mol Biol 19: 349–361, 1966 [DOI] [PubMed] [Google Scholar]
- 114.Wei X, Wang C, Ma C, Sun W, Li H, and Cai Z. Long noncoding RNA ANRIL is activated by hypoxia-inducible factor-1alpha and promotes osteosarcoma cell invasion and suppresses cell apoptosis upon hypoxia. Cancer Cell Int 16: 73, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 115.Werstuck G. and Green MR. Controlling gene expression in living cells through small molecule-RNA interactions. Science 282: 296–298, 1998 [DOI] [PubMed] [Google Scholar]
- 116.Wiedmeier JE, Ohlrich A, Chu A, Rountree MR, and Turker MS. Induction of the long noncoding RNA NBR2 from the bidirectional BRCA1 promoter under hypoxic conditions. Mutat Res 796: 13–19, 2017 [DOI] [PubMed] [Google Scholar]
- 117.Wu W, Hu Q, Nie E, Yu T, Wu Y, Zhi T, Jiang K, Shen F, Wang Y, Zhang J, and You Y. Hypoxia induces H19 expression through direct and indirect Hif-1alpha activity, promoting oncogenic effects in glioblastoma. Sci Rep 7: 45029, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Xia T, Liao Q, Jiang X, Shao Y, Xiao B, Xi Y, and Guo J. Long noncoding RNA associated-competing endogenous RNAs in gastric cancer. Sci Rep 4: 6088, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Xiang JF, Yin QF, Chen T, Zhang Y, Zhang XO, Wu Z, Zhang S, Wang HB, Ge J, Lu X, Yang L, and Chen LL. Human colorectal cancer-specific CCAT1-L lncRNA regulates long-range chromatin interactions at the MYC locus. Cell Res 24: 513–531, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Xiao H, Tang K, Liu P, Chen K, Hu J, Zeng J, Xiao W, Yu G, Yao W, Zhou H, Li H, Pan Y, Li A, Ye Z, Wang J, Xu H, and Huang Q. LncRNA MALAT1 functions as a competing endogenous RNA to regulate ZEB2 expression by sponging miR-200s in clear cell kidney carcinoma. Oncotarget 6: 38005–38015, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Xue M, Li X, Li Z, and Chen W. Urothelial carcinoma associated 1 is a hypoxia-inducible factor-1alpha-targeted long noncoding RNA that enhances hypoxic bladder cancer cell proliferation, migration, and invasion. Tumour Biol 35: 6901–6912, 2014 [DOI] [PubMed] [Google Scholar]
- 122.Yang C, Tang R, Ma X, Wang Y, Luo D, Xu Z, Zhu Y, and Yang L. Tag SNPs in long non-coding RNA H19 contribute to susceptibility to gastric cancer in the Chinese Han population. Oncotarget 6: 15311–15320, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Yang F, Huo XS, Yuan SX, Zhang L, Zhou WP, Wang F, and Sun SH. Repression of the long noncoding RNA-LET by histone deacetylase 3 contributes to hypoxia-mediated metastasis. Mol Cell 49: 1083–1096, 2013 [DOI] [PubMed] [Google Scholar]
- 124.Yang F, Zhang H, Mei Y, and Wu M. Reciprocal regulation of HIF-1alpha and lincRNA-p21 modulates the Warburg effect. Mol Cell 53: 88–100, 2014 [DOI] [PubMed] [Google Scholar]
- 125.Yang Z, Wang R, Zhang T, and Dong X. Hypoxia/lncRNA-AK123072/EGFR pathway induced metastasis and invasion in gastric cancer. Int J Clin Exp Med 8: 19954–19968, 2015 [PMC free article] [PubMed] [Google Scholar]
- 126.Yao J, Li J, Geng P, Li Y, Chen H, and Zhu Y. Knockdown of a HIF-2alpha promoter upstream long noncoding RNA impairs colorectal cancer stem cell properties in vitro through HIF-2alpha downregulation. Onco Targets Ther 8: 3467–3474, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Yoon JH, Abdelmohsen K, Srikantan S, Yang X, Martindale JL, De S, Huarte M, Zhan M, Becker KG, and Gorospe M. LincRNA-p21 suppresses target mRNA translation. Mol Cell 47: 648–655, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Zhai W, Sun Y, Jiang M, Wang M, Gasiewicz TA, Zheng J, and Chang C. Differential regulation of lncRNA-SARCC suppresses VHL-mutant RCC cell proliferation yet promotes VHL-normal RCC cell proliferation via modulating androgen receptor/HIF-2alpha/C-MYC axis under hypoxia. Oncogene 35: 4866–4880, 2016 [DOI] [PubMed] [Google Scholar]
- 129.Zhang EB, Kong R, Yin DD, You LH, Sun M, Han L, Xu TP, Xia R, Yang JS, De W, and Chen J. Long noncoding RNA ANRIL indicates a poor prognosis of gastric cancer and promotes tumor growth by epigenetically silencing of miR-99a/miR-449a. Oncotarget 5: 2276–2292, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Zhou C, Ye L, Jiang C, Bai J, Chi Y, and Zhang H. Long noncoding RNA HOTAIR, a hypoxia-inducible factor-1alpha activated driver of malignancy, enhances hypoxic cancer cell proliferation, migration, and invasion in non-small cell lung cancer. Tumour Biol 36: 9179–9188, 2015 [DOI] [PubMed] [Google Scholar]
- 131.Zhu S, Li W, Liu J, Chen CH, Liao Q, Xu P, Xu H, Xiao T, Cao Z, Peng J, Yuan P, Brown M, Liu XS, and Wei W. Genome-scale deletion screening of human long non-coding RNAs using a paired-guide RNA CRISPR-Cas9 library. Nat Biotechnol 34: 1279–1286, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Zhu Z, Gao X, He Y, Zhao H, Yu Q, Jiang D, Zhang P, Ma X, Huang H, Dong D, Wan J, Gu Z, Jiang X, Yu L, and Gao Y. An insertion/deletion polymorphism within RERT-lncRNA modulates hepatocellular carcinoma risk. Cancer Res 72: 6163–6172, 2012 [DOI] [PubMed] [Google Scholar]