Abstract
Myotubular myopathy, or X-linked centronuclear myopathy, is a severe muscle disorder representing a significant burden for patients and their families. It is clinically characterized by neonatal and severe muscle weakness and atrophy. Mutations in the myotubularin (MTM1) gene cause myotubular myopathy, and no specific curative treatment is available. We previously found that dynamin 2 (DNM2) is upregulated in both Mtm1 knockout and patient muscle samples, whereas its reduction through antisense oligonucleotides rescues the clinical and histopathological features of this myopathy in mice. Here, we propose a novel approach targeting Dnm2 mRNA. We screened and validated in vitro and in vivo several short hairpin RNA (shRNA) sequences that efficiently target Dnm2 mRNA. A single intramuscular injection of AAV-shDnm2 resulted in long-term reduction of DNM2 protein level and restored muscle force, mass, histology, and myofiber ultrastructure and prevented molecular defects linked to the disease. Our results demonstrate a robust DNM2 knockdown and provide an alternative strategy based on reduction of DNM2 to treat myotubular myopathy.
Keywords: AAV, DNM2, shRNA, XLMTM, X-linked myotubular myopathy, MTM1, congenital myopathy, myotubularin, dynamin, centronuclear myopathy
Myotubular myopathy is a severe muscle disorder with no specific treatment. In this issue of Molecular Therapy, Tasfaout et al. (2018) show that intramuscular injections of adeno-associated virus expressing shRNA specific to Dnm2 reduce efficiently DNM2 level and restore muscle force, mass, histology, and myofiber ultrastructure in a mouse model.
Introduction
Centronuclear myopathies (CNMs) are a group of rare congenital disorders characterized clinically by severe muscle atrophy and generalized muscle weakness.1, 2 Muscle biopsies from affected patients present numerous myofibers with nuclei abnormally located at the center, without excessive regeneration. Different forms of CNM have been characterized to date. The most common and severe form is the X-linked CNM (XLCNM), also called myotubular myopathy.3 XLCNM patients present with severe hypotonia and respiratory distress at birth, strongly impacting on life expectancy. More than 200 mutations in MTM1 have been identified to cause XLCNM, most of them leading to a strong decrease or absence of the MTM1 protein.4, 5, 6, 7, 8, 9 MTM1 is a phosphoinositide phosphatase involved in key cellular processes, including endosomal trafficking,10 excitation contraction coupling,11, 12 intermediate filament organization,13 neuromuscular junction (NMJ) structure,14, 15 satellite cell proliferation,16 apoptosis,16, 17 and autophagy.18, 19 Mutations in DNM2 have been associated with a less severe autosomal dominant form.20, 21 It is characterized by moderate muscle weakness with late onset20; however, several neonatal cases have been reported with very severe phenotype. DNM2 encodes for dynamin 2, a mechanoenzyme implicated in endocytosis,22 exocytosis,23, 24 as well as cytoskeleton organization.25, 26, 27 Different in vitro studies have proposed CNM-causing DNM2 mutations increase dynamin 2 guanosine triphosphatase (GTPase) activity and oligomer stability, suggesting a gain of function of the mutated forms.28, 29 This hypothesis is sustained by the fact that overexpression of wild-type DNM2 in mice recapitulates a CNM-like phenotype.30, 31 To date, no effective and specific therapy is available for patients with CNM.
The Mtm1 knockout (Mtm1KO) mice present a severe muscular phenotype and have been extensively used to understand the physiopathology of XLCNM and to test therapeutic approaches.32, 33, 34, 35, 36, 37 They develop a severe progressive myopathy starting from week 3 that is clinically reflected by a severe muscle atrophy and weakness.32 They present a short lifespan and die between 1 and 3 months of age. Transversal muscle sections of Mtm1KO exhibit a typical XLCNM histology with predominance of hypotrophic and rounded fibers with centralized nuclei, abnormal organelle distribution, and aggregated proteins, including caveolin 3 (Cav3) and desmin.13, 36 In addition, and similarly to muscle biopsies from XLCNM patients, DNM2 was found to be upregulated at the symptomatic phase, pointing to DNM2 reduction as a therapeutic strategy.36
We have previously reported that reduction of DNM2 expression by genetic crossing rescues the early XLCNM lethality and ameliorates most CNM features, notably increased muscle force and correction of myofiber histology, indicating an epistasis between MTM1 and DNM2.36 These data have been recently confirmed using antisense oligonucleotides (ASOs) that target the nuclear pre-mRNA of Dnm2. Repeated injections of ASO targeting Dnm2 into Mtm1KO mice efficiently reduced the DNM2 protein level and revert the CNM features.35 However, such ASO strategy would require repeated injections in patients, potentially on a long-term basis, and an efficient biodistribution in patients to reach all muscles of the body.
Here, we tested another approach of reducing DNM2 expression based on AAV-short hairpin RNA (shRNA). Adeno-associated virus (AAV) vectors have been intensively used to re-express the defective gene in many monogenic disorders.38 In addition, an increasing number of studies showed their efficacy to repress gene expression when they encode for shRNA complementary to the target mRNA.39, 40, 41, 42 In the present study, we evaluated the potential of AAV-shRNA to knockdown DNM2 in mice. A single intramuscular injection of AAV-shDnm2 resulted in long-term reduction of DNM2 protein level and restored different CNM features. Our data validate the epistasis between Mtm1 and Dnm2 and propose an alternative approach to target DNM2 for therapeutic approaches.
Results
Design and In Vitro Validation of shRNA Targeting DNM2 Conserved Regions
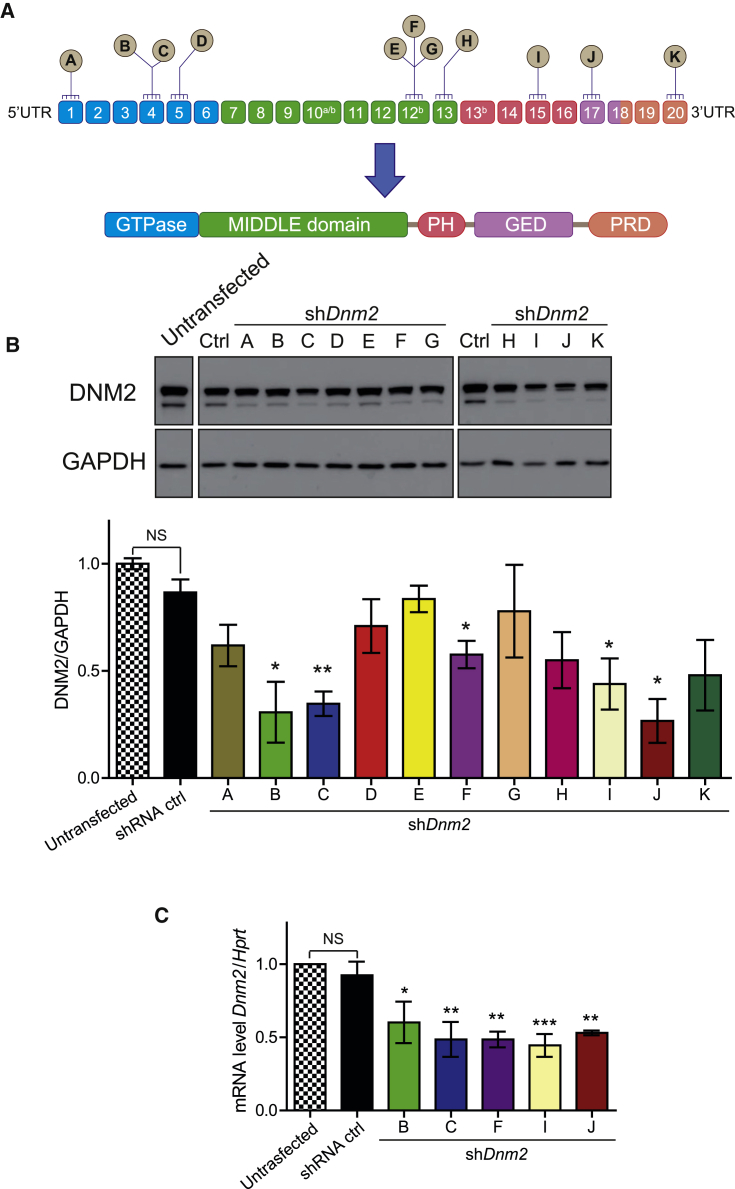
To target DNM2 mRNA for degradation, eleven highly conserved regions between the human and mouse DNM2 mRNA were selected to design the different shRNA sequences (Figure 1A; Table 1). All shRNA-Dnm2 sequences as well as a scrambled sequence (shRNA ctrl) had an identical nine-nucleotide hairpin or loop sequence.
Figure 1.
In Vitro Validation of shRNA Targeting DNM2
(A) Location of shRNA-targeting sequences in Dnm2 mRNA. Exons are depicted as boxes, and exons 10a, 10b, 12b, and 13b are alternative exons. Below, the protein domains encompass a GTPase domain, a middle and GTP effector (GED) domains forming the stalk, a pleckstrin homology (PH), and a proline-rich (PRD) domain. (B) Representative western blot from HEK293T cells co-transfected with two plasmids is shown: one encoding human DNM2 and the second expressing the different shRNA. DNM2 protein levels were determined by densitometry and standardized to GAPDH. (C) Endogenous Dnm2 mRNA levels in shRNA-treated C2C12 mouse myoblast cells were determined by qRT-PCR and standardized to Hprt. Cells were electroporated with shRNA ctrl or shDnm2 (B, C, F, I, or J). n = 3 biological replicates per each group. Data represent an average of three independent experiments ± SEM. *p < 0.05; **p < 0.01 for shDnm2-treated versus shRNA ctrl-treated cells (ANOVA test).
Table 1.
Selected shRNA and Target Sequences
| shRNA | Target Sequence (Mouse) | Remarks | Top 3 of Potential Off-Target mRNA (Mouse) | Sequence Homology (%) |
|---|---|---|---|---|
| shRNA ctrl | GGGCTATCCCAACGCTATTAGT | no target | UTP23 small subunit processome component (Utp23) | 68 |
| Von Willebrand factor A domain containing 2 (Vwa2) | 63 | |||
| B cell CLL/lymphoma 11A (zinc finger protein) (Bcl11a) | 59 | |||
| shDnm2 A | AACCGCGGGATGGAAGAGCT | identical to human sequence | transmembrane protein 67 (Tmem67) | 70 |
| family with sequence similarity 206, member A (Fam206a) | 70 | |||
| protein phosphatase 1F (PP2C domain containing) (Ppm1f) | 70 | |||
| shDnm2 B | AACTTGACCCTCATCGACCTC | identical to human sequence | potassium voltage-gated channel, subfamily G, member 1 (Kcng1) | 76 |
| IKAROS family zinc finger 1 (Ikzf1) | 76 | |||
| DnaJ heat shock protein family (Hsp40) member C2 (Dnajc2) | 71 | |||
| shDnm2 C | AAGGACATGATCCTGCAGTTCAT | identical to human sequence | adaptor-related protein complex 3, delta 1 subunit (Ap3d1) | 78 |
| suppressor of glucose, autophagy-associated 1 (Soga1) | 65 | |||
| adhesion G protein-coupled receptor G2 (Adgrg2) | 60 | |||
| shDnm2 D | TCGGTGTCATCACCAAGCT | identical to human sequence | myosin IA (Myo1a) | 78 |
| ankyrin repeat domain 52 (Ankrd52) | 73 | |||
| zinc finger protein 407 (Zfp407) | 73 | |||
| shDnm2 E | TGCCAACTGTTTCTATACT | 1 nucleotide different to human (underlined) | protocadherin 17 (Pcdh17) | 73 |
| folliculin interacting protein 1 (Fnip1) | 73 | |||
| zeta-chain (TCR)-associated protein kinase (Zap70) | 73 | |||
| shDnm2 F | AACTGTTTCTATACTGAGGAG | 2 nucleotides different to human (underlined) | protocadherin 17 (Pcdh17) | 71 |
| tenascin R (Tnr) | 71 | |||
| ligand-dependent nuclear receptor corepressor-like (Lcorl) | 66 | |||
| shDnm2 G | TTTCTATACTGAGGAGCTGGT | 2 nucleotides different to human (underlined) | WNK lysine-deficient protein kinase 2 (Wnk2) | 71 |
| TRAF3 interacting protein 2 (Traf3ip2) | 66 | |||
| trans-2,3-enoyl-CoA reductase-like (Tecrl) | 66 | |||
| shDnm2 H | GCACGCAGCTGAACAAGAA | identical to human sequence | HGF-regulated tyrosine kinase substrate (Hgs) | 78 |
| two-pore segment channel 2 (Tpcn2) | 73 | |||
| interleukin-1 alpha (Il1a) | 73 | |||
| shDnm2 I | AAGAAGTACATGCTGCCACTGGA | 1 nucleotide different to human (underlined) | zinc finger, C3H1-type containing (Zfc3h1) | 69 |
| DENN/MADD domain containing 5A (Dennd5a) | 69 | |||
| small G protein signaling modulator 1 (Sgsm1) | 65 | |||
| shDnm2 J | AACACCTTCTCCATGGACCC | identical to human sequence | proteasome (prosome, macropain) 26S subunit, non-ATPase, 12 (Psmd12) | 75 |
| GTP cyclohydrolase 1 (Gch1) | 75 | |||
| acyl-CoA synthetase medium-chain family member 3 (Acsm3) | 70 | |||
| shDnm2 K | CCATTATCCGCCCAGCCGAGC | identical to human sequence | poly (ADP-ribose) polymerase family, member 6 (Parp6) | 66 |
| RAP2B, member of RAS oncogene family (Rap2b) | 66 | |||
| arylsulfatase i (Arsi) | 66 |
CoA, coenzyme A.
To evaluate the efficacy of shRNA to knockdown DNM2 protein level in vitro, HEK293T cells were co-transfected with a plasmid expressing human DNM2 and plasmids expressing the shDnm2 under the control of the mouse U6 promoter. DNM2 protein level was measured 72 hr post-transfection. A statistically significant decrease of DNM2 protein level was observed with five shDnm2 constructs (B, C, F, I, and J) out of the 11 shDnm2s tested (Figure 1B). In addition, selected shDnm2s elicited a strong decrease of the endogenous murine Dnm2 mRNA when electroporated into mouse C2C12 myoblasts (Figure 1C). However, protein level was unchanged 48 hr after electroporation, most likely due to longer stability of endogenous DNM2 (Figure S1). These results indicate that five shRNAs (B, C, F, I, and J) target efficiently both human and murine DNM2 mRNA.
Efficient In Vivo Dnm2 Knockdown following a Single Intramuscular Injection of AAV shRNA
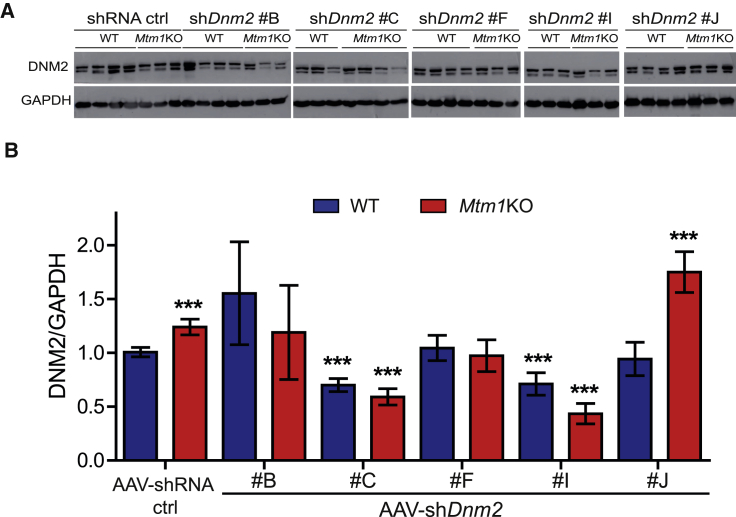
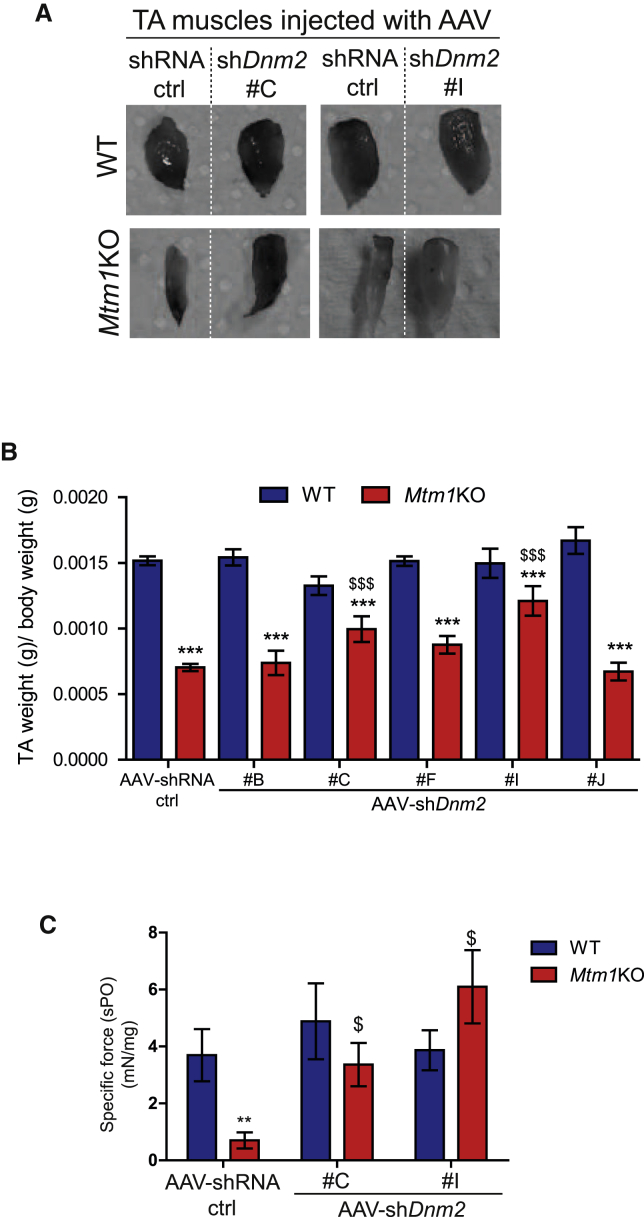
To test the in vivo efficacy of the five best shDnm2s, AAV serotype 2/9 vectors encoding the selected shDnm2 or shRNA ctrl were produced. AAV2/9 serotype efficiently transduces skeletal muscle.43 The AAV-shDnm2s (B, C, F, H, or I) were administrated locally into right tibialis anterior muscles (TAs) of 3-week-old wild-type (WT) or Mtm1KO mice at a dose of 1.2 × 1011 viral genome per TA while the same dose of AAV-shRNA ctrl was injected into the contralateral TA. Four weeks later, GFP expression was visualized in the injected TAs to ensure an effective viral transduction (Figure S2). TAs were weighed, and in situ muscular force as well as DNM2 protein level were measured. DNM2 protein levels in TA injected with AAV-shDnm2 B, F, and J were not reduced below TA injected with AAV-shRNA ctrl (Figures 2A and 2B). Accordingly, these muscles remain atrophic (Figures 3A, 3B, and S3). Conversely, muscles of WT or Mtm1KO injected with shDnm2 C or I presented a strong decrease of DNM2 protein level (Figures 2A and 2B). In addition, improvement of muscle mass and force were noted for Mtm1KO injected with shDnm2 C or I. Mtm1KO muscles injected with AAV-shDnm2 C or I were remarkably bigger and exhibit an increase of specific muscle force compared to the contralateral TA injected with shRNA ctrl (Figures 3A–3C and S3). Nevertheless, we noted that the absolute force of WT muscles injected with AAV was reduced when compared to those injected with NaCl, whereas muscle mass and histology were similar (Figure S4). This may indicate a deleterious effect of AAV injection and GFP expression specifically on muscle force.44, 45 Moreover, these results validate in vivo the efficiency of several shRNA sequences targeting Dnm2 mRNA and show that reducing DNM2 after a single injection of AAV-shDnm2 efficiently ameliorates muscle mass and force of the Mtm1KO XLCNM model.
Figure 2.
In Vivo Validation of shRNA Targeting Dnm2 following Intramuscular Injection
(A) Representative western blot of 7-week-old WT or Mtm1KO TAs injected with either AAV-shRNA ctrl or AAV-shDnm2 (B, C, F, I, or J). (B) DNM2 densitometries were quantified and standardized to the loading control GAPDH (n = 5–7 mice/group). ***p < 0.01 versus WT muscles treated with AAV-shRNA ctrl (ANOVA test).
Figure 3.
Positive Physiological Effects of AAV shRNA-Mediated Dnm2 Knockdown
(A) Photography of TAs from WT or Mtm1KO mice treated with AAV-shRNA ctrl or shDnm2 (C or I). (B) TA weight relative to body weight is shown (n = 6). (C) Specific muscle force of TA was measured after sciatic nerve stimulation. The specific muscle force was calculated by dividing the absolute force by TA weight (n = 5–7 mice/group). **p < 0.01; ***p < 0.001 versus WT TA treated with shRNA ctrl. $p < 0.05; $$$p < 0.01 Mtm1KO treated with AAV-shDnm2 versus Mtm1KO treated with AAV-shRNA ctrl (ANOVA test).
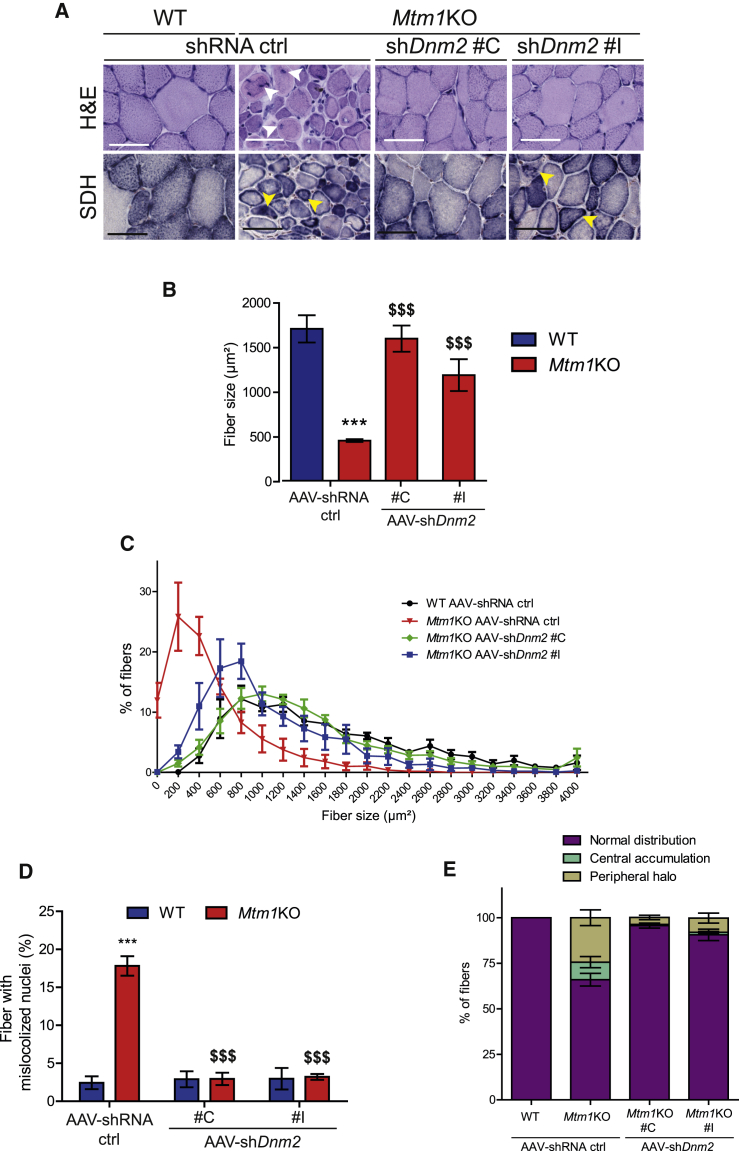
AAV shRNA-Mediated Knockdown of Dnm2 Rescues Mtm1KO Muscle Histology
To assess whether the functional amelioration of Mtm1KO muscles is linked to an improved muscle histology, transversal sections of TA 4 weeks post-injection were stained with H&E. Mtm1KO muscles treated with AAV-shRNA ctrl exhibited small and rounded fibers with a 3-fold reduction in fiber size compared to WT muscles (Figures 4A–4C). In addition, approximately 20% of the myofibers presented with abnormal internal localization of nuclei (Figure 4D). Succinate dehydrogenase (SDH) staining showed abnormal mitochondrial distribution with aggregated staining in the center or forming a halo at the periphery of the myofiber (Figures 4A and 4E). These abnormal histological features are reminiscent of the typical histology seen in XLCNM patients46 and were corrected in Mtm1KO TA when treated with two different AAV-shDnm2. Muscles injected with AAV-shDnm2 C or I showed a normalization of the myofiber size with nuclei positioned at the periphery (Figures 4A–4D). In addition, TAs injected with AAV-shDnm2 C exhibit a normal muscle histology similar to WT with normal mitochondrial distribution and a decreased number of fibers with central or peripheral accumulation of mitochondria (Figures 4A and 4E). Injection of shDnm2 I correlated with an increase of myofiber size and a strong improvement of mitochondria distribution; however, few myofibers still presented an intermediate size with mitochondrial oxidative staining in the center or forming a halo at the periphery (Figures 4A–4E). These results suggest only a slight difference in efficacy between shDnm2 C and I based on SDH staining and show overall a strong rescue of muscle histology following a single injection of two different AAV-shDnm2s.
Figure 4.
Histology Amelioration of TA Treated with AAV shDnm2
(A) TA sections of WT or Mtm1KO mice treated with AAV-shRNA ctrl or shDnm2 (C or I) were stained with H&E and SDH to visualize fiber size, shape, nuclei positioning, and mitochondrial oxidative activity. Arrows show mislocalized nuclei or aggregated mitochondria (scale bars: 50 μm). (B) TA myofiber area was calculated on 300–400 fibers per sample (n = 5 mice/group). (C) Fiber size distribution of WT or Mtm1KO TAs according to size groups. Affected Mtm1KO TAs have more small fibers and less medium-size or large fibers. (D) Percentage of fibers with mislocalized nuclei was determined in 1,000 fibers (n = 5–6 mice/group). (E) Percentage of fibers with abnormal mitochondrial distribution was determined in 1,000 fibers (n = 5 mice/group). ***p < 0.001 versus WT TA treated with shRNA ctrl. $$$p < 0.01 Mtm1KO treated with AAV-shDnm2 versus Mtm1KO treated with AAV-shRNA ctrl (ANOVA test).
Dnm2 Reduction through AAV-shRNA Rescues Myofiber Organization in Mtm1KO Muscles
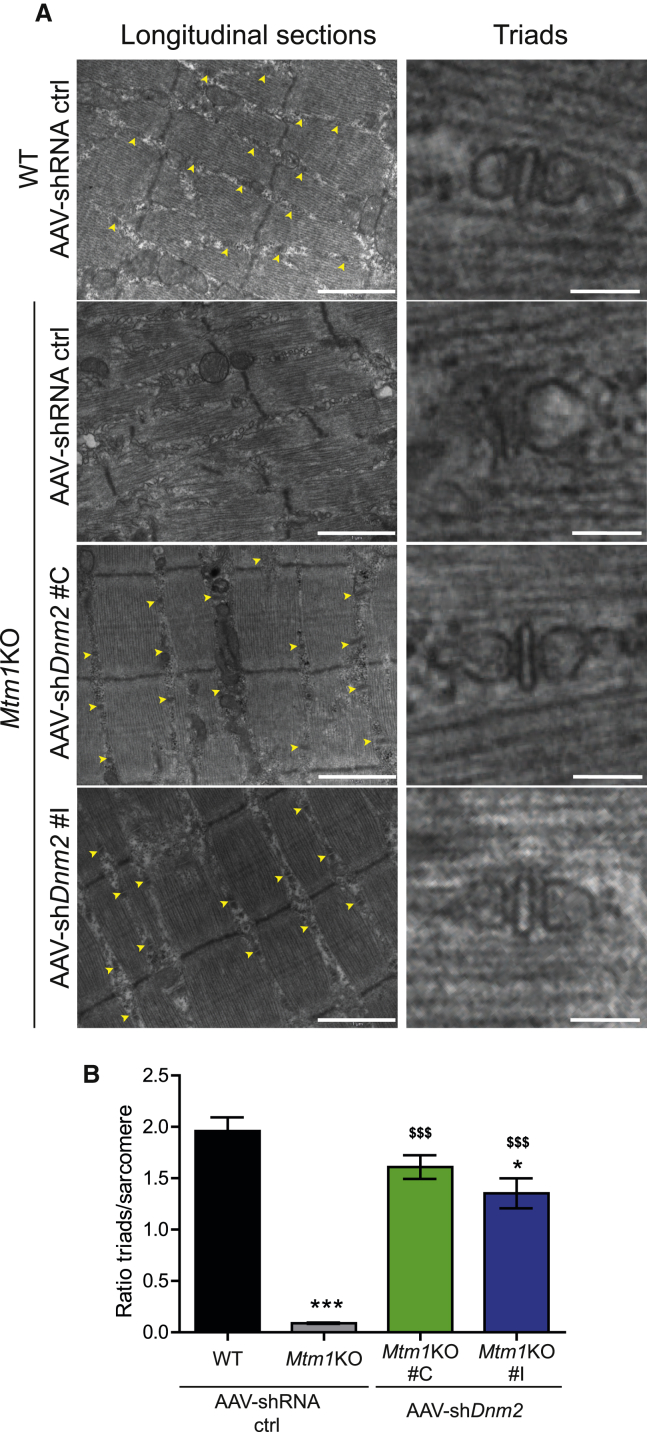
MTM1 deficiency was documented to induce disorganization of muscle fiber in several species.12, 32, 47 MTM1-deficient fibers present with misalignment of Z-line; mislocalization of organelles, including nuclei and mitochondria; and a strong defect in triad structure (formed by one T-tubule and two sarcoplasmic reticulum cisternae) implicated in excitation-contraction coupling. In agreement with the literature, misalignment of the Z-line was observed in the Mtm1KO TAs by transmission electronic microscopy (TEM) in this study (Figure 5A) and confirmed by immunolabeling of alpha-actinin (Z-line marker; Figure 6). Well-organized triads are seldom observed in this model (Figures 5A and 5B), and this finding parallels the perturbed localization of the junctional sarcoplasmic reticulum calcium channel RYR1 and caveolin 3 implicated in T-tubule biogenesis (Figure 6).
Figure 5.
Amelioration of Muscle Ultrastructure through AAV-shDnm2 Injections
(A) Electron microscopy was used to assess the general ultrastructure of myofibers and to count the triads formed (yellow arrows, scale bar: 1 μm for left panels and 100 nm for right panels). (B) Graph representing the number of triad well formed and localized per sarcomere is shown (n = 3 per group). *p < 0.05; ***p < 0.001 for Mtm1KO treated with AAV-shRNA ctrl or AAV-shDnm2 versus WT treated with AAV-shRNA ctrl. $$$p < 0.001 Mtm1KO treated with AAV-shDnm2 versus Mtm1KO treated with AAV-shRNA ctrl (ANOVA test).
Figure 6.
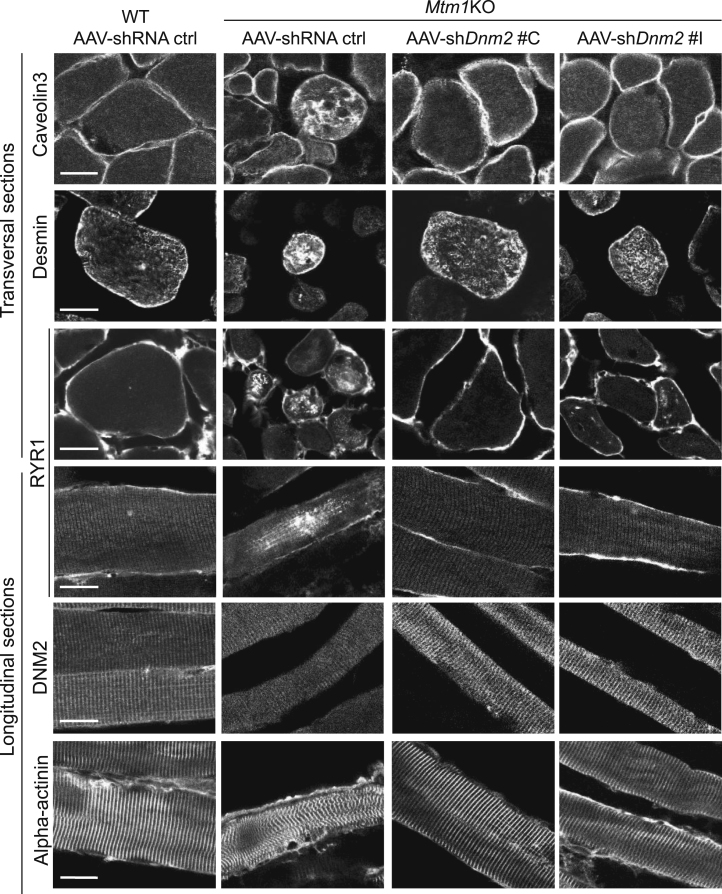
Improvement of Molecular Defects in Mtm1KO Mice Treated with AAV shDnm2
Immunolabeling of CAV3, desmin, RYR1, DNM2, and alpha-actinin was performed on 8 μm transversal or longitudinal sections. The staining was visualized using confocal microscopy (scale bar: 20 μm).
AAV-shDnm2 injection into Mtm1KO mice restored the disorganized ultrastructure observed in untreated mice. Using TEM, myofiber ultrastructure revealed a well-organized structure with increased ratio of normal and well-positioned triads in treated Mtm1KO mice (Figures 5A and 5B), equivalent to the rescue observed targeting Dnm2 systemically by ASO at higher doses.35 RYR1 and caveolin 3 were also correctly localized as in WT muscle (Figure 6). Furthermore, Z-line alignment and alpha-actinin labeling were also normal. DNM2 localized normally in both untreated and treated Mtm1KO muscles, in a transverse localization. In addition, the desmin aggregation observed in the Mtm1KO mice treated with AAV-shRNA ctrl was also rescued upon injection with AAV-shDnm2 (Figure 6).
Altogether, these findings underline a complete amelioration of the XLCNM-like phenotypes in the Mtm1KO model at the structural and molecular levels upon DNM2 reduction through AAV-shRNA treatment and fully correlate with the rescue in muscle mass and force.
Discussion
Myotubular myopathy is the most severe form of CNM with highest incidence, and no available effective treatment exists. Among the different therapeutic approaches that were tested in preclinical trials on animal models, few showed significant histological and functional improvements. Here, we provide evidence showing that a single injection of AAV-shDnm2 rescues the CNM phenotype of Mtm1KO mice at the functional, histological, and ultrastructural level when analyzed four weeks after injection. Molecular defects were greatly ameliorated with a strong improvement of the localization of desmin, caveolin 3, and RYR1. The structural alterations of the myofibers were rescued, including normalization of mitochondria and nuclei positioning and triad shape and distribution. Furthermore, the different histological hallmarks of CNM were absent upon AAV-shDnm2 injection, with an increase in myofiber size and decrease in frequency of fibers with mislocalized nuclei. These effects correlated with a normalization of muscle mass and force. Nonetheless, muscles injected with AAV showed a reduction in general absolute muscle force. This could be due to GFP overexpression, as previous studies showed that GFP expression impairs contractile activity of the muscle by affecting actin-myosin interaction.44, 45
Our results demonstrate that reduction of DNM2 level, through local injection of AAV-shDnm2, prevents the disease progression. These data confirmed the therapeutic potential of DNM2 knockdown and are concordant with those obtained with ASO targeting Dnm2 or genetic reduction of Dnm2.35, 36 Furthermore, it is noteworthy that, using this approach, we almost fully rescued the CNM phenotype targeting DNM2 in one muscle alone, despite the severe phenotype still observed in other muscles of the mouse. DNM2 protein levels are increased in muscles from both XLCNM patients and Mtm1KO mice at symptomatic ages. The fact that reducing DNM2 level promotes disease prevention strongly supports the observed increase in DNM2 as a significant cause of the disease initiation and/or progression. This finding is sustained by the fact that overexpression of DNM2 in WT mice by transgenesis or AAV injections creates a CNM phenotype.30, 31 Moreover, in vitro experiments showed that DNM2 mutations leading to a dominant CNM form increase its GTPase activity and oligomerization stability.28, 29 We thus hypothesize that either increase in DNM2 protein level upon impairment of MTM1 or increase in DNM2 activity following mutations trigger the CNM phenotypes in XLCNM or ADCNM, respectively.
The present results confirm the epistasis between Mtm1 and Dnm2 and place DNM2 downstream of MTM1 in the pathological pathway causing CNM. To date, the functional link between MTM1 and DNM2 and the molecular mechanism underlying this rescue remain unclear. However, MTM1 deficiency was reported to cause an alteration of autophagy and protein accumulation.18, 19 One could hypothesize that autophagy defects lead to imbalance between transcription and clearance of DNM2, causing its accumulation and increased overall activity. Alternatively, MTM1 might somehow regulate DNM2 transcription or translation. However, this is less likely, as Dnm2 mRNA is not elevated in Mtm1KO mice.36 In addition, one cannot exclude that MTM1 directly regulates DNM2 activity or transport in addition to its expression.
Here, we validated several shRNA sequences targeting Dnm2 mRNA in vitro and in vivo for efficient DNM2 knockdown. In particular, we identified shRNA sequences partially (I) or fully (C) improving the molecular, cellular, and physiological phenotypes. These data provide the proof of concept that targeting Dnm2 mRNA for degradation using AAV as a delivery method is efficient to ameliorate XLCNM, thus validating a novel approach to tackle this disease. Compared to the recently reported ASO targeting nuclear Dnm2 pre-mRNA, shRNA targets the cytoplasmic mRNA, and both approaches efficiently decrease DNM2 level. Moreover, AAVs have two main advantages: they can have a good muscle tropism and they can sustain long-term expression following a single injection. Indeed, tissue specificity can be further achieved by modulating the AAV serotype and the promoter controlling the expression of the transgene. Numerous AAV serotypes have been identified, and several showed a strong skeletal muscle tropism.48 In addition, several promoter and regulatory cassettes have been reported to have a high activity in skeletal muscle.49 Together, the combination of these two elements would largely restrict the delivery of shRNA to skeletal muscles, which may improve the efficacy while decreasing the required dose and minimizing potential side effects related to reduction of DNM2 in other organs. Of note, reduction of DNM2 by 50% through transgenesis or ASO injection did not induce a detectable toxicity.35, 36
Many studies showed the potential of AAV to maintain a long-term and robust transgene expression,50, 51, 52, 53 which implies avoiding multiple injections that may trigger immune reaction against these vectors. Moreover, several reports claim the safety of recombinant AAV vector. Unlike other viral vectors, they do not integrate the genome of the host cell, and they elicit a mild and manageable immune response.52, 53, 54 However, they represent some limitations. Following their transduction into host cells, AAVs are mostly present as non-integrated episomes. Loss of the transgene expression could be expected due to normal muscle turnover. In addition, it has been reported that AAVs do not transduce satellite cells, which are multipotent cells able to differentiate and give rise to myofibers.55 Thus, re-administration of these vectors after the initial infusion might be needed. Furthermore, human administration of AAV requires a large amount and high-titer production, which represent a manufacturing challenge and costly process. Also, expression of shRNA to downregulate DNM2 level using ubiquitous RNA polymerase III promoters (like U6 or H1) might have some side effects in non-muscle tissues. These promoters do not provide a spatial or temporal control, and they show a high transcription activity in some cell types causing microRNA processing factors (Exportin-5 and RNA-induced silencing complex [RISC]) saturation, which leads to cytotoxicity and tissue damage.56, 57, 58 Transcription of shRNA from polymerase II promoters could be an alternative for effective and safer RNAi therapeutics.59
In conclusion, this study shows once again the usefulness of Mtm1KO as a bio-assay for screening of therapeutic agents and provides the first proof of principle that using AAV encoding for shDnm2 in this XLCNM mouse model shows a strong and robust reduction of DNM2 protein level following a single injection. These shRNA-targeting Dnm2s exhibit a strong affinity to human DNM2 transcript as shown in vitro (Figure 1), which might be relevant for human application in future clinical trials.
Materials and Methods
Cell Transfection
HEK293Ts were co-transfected by plasmid encoding human DNM2 and another plasmid expressing either shRNA ctrl or different shDnm2 using Lipofectamine 2000 and following the manufacturer’s instructions. Cells were seeded in 6-well plates. At 80% of confluency, a mixture of Lipofectamine reagent and opti-minimum essential medium (MEM) medium containing 2 μg of plasmid expressing human DNM2 and 2 μg of plasmid encoding the different shRNA was incubated for 4 hr. Then, the mixture was removed and replaced by DMEM containing 20% fetal calf serum (FCS). The transfected cells were incubated at 37°C for 72 hr before harvesting for protein analysis.
C2C12 mouse myoblast were electroporated with plasmids encoding the different shRNA candidates using Amaxa nucleofector2B Kit V and following the manufacturer’s instructions. Briefly, cells were trypsinized and 1 × 106 cells per sample were gently centrifuged at 90 g for 10 min at room temperature. Cell pellet was resuspended in 100 μL of the solution provided in the kit. Then, 2 μg of plasmid expressing shRNA ctrl or different shDnm2s were added to the cell suspension and transferred immediately to an electroporation cuvette. Electroporation was performed using the B-032 program of Nucleofector I Device. The electroporated cells were incubated for 48 hr in DMEM medium containing 20% FCS at 37°C before harvesting the electroporated cells for protein and RNA analysis.
Animals
Animal experiments were conducted on male Mtm1KO or WT 129SvPAS mice according to the French and European legislation on animal care and experimentation. Protocol nos. Com’Eth IGBMC-ICS 2012-132, 2013-034, and 2016-5453 were approved by the Institutional Ethics Committee. Mice were housed in a temperature-controlled room and ventilated cages with 12 hr day light/darkness cycle and given free access to food.
Intramuscular Injection of AAV
3-week-old male Mtm1KO or WT was weighed and then an anesthetic solution of ketamine 20 mg/mL and xylazine 0.4% at 5 μL/g of body weight was administrated by intraperitoneal injection. Right TAs were injected with 1.2 × 1011 vg/TA in 20 μL of AAV2/9 expressing shDnm2 (either B, C, F, I, or J), and the same dose of AAV2/9 expressing shRNA ctrl was injected in the contralateral TA. As an additional control, 20 μL of NaCl 0.9% was administrated into WT TA. At 7 weeks of age, mice were humanely killed by CO2 suffocation followed by cervical dislocation. The TAs were dissected and weighed. They were snap frozen in liquid nitrogen-cooled isopentane and stored at −80°C for H&E and SDH histology analysis as well as protein extraction and quantification. For immunostaining, TAs were stored in paraformaldehyde (PFA) 4% for 24 hr, and then they were transferred to sucrose 30% overnight and stored at 4°C.
Immunostaining of Muscle Sections
8 μm transversal or longitudinal cryosections of TA were stained with antibodies against CAV3 (Santa Cruz Biotechnology N-18 sc-7665; 1:500), RYR1 (34C abcam2668; 1:500), desmin (Santa Cruz sc-7559; 1:500), DNM2 (homemade antibody R2680; 1:500), and alpha-actinin (anti-α-actinin sarcomeric clone EA-53; 1:500). Images were taken with a Leica SP8-UV confocal microscope.
Muscle Histology
8 μm transversal cryosections were prepared and stained by H&E or SDH. Entire muscle sections were imaged with the Hamamatsu NanoZoomer 2HT slide-scanner. The percentage of TA fibers with mislocalized (centralized or internalized) nuclei or abnormal mitochondrial distribution was counted using the cell counter plugin in Fiji image analysis software. The fiber area was measured using the Fiji software.
TA Contractile Properties
TA contraction properties were evaluated in situ after sciatic nerve stimulation using Complete1300A Mouse Test System from Aurora. Briefly, mice were anesthetized with intraperitoneal (i.p.) injection of pentobarbital (60 mg/kg) and maintained under deep anesthesia during the experiment. The distal tendon of TA was detached and tied to the isometric transducer using a silk ligature. The knee and foot were fixed with clamps. All isometric contraction measurements were made at an initial muscle length of L0 (length at which maximal tension was obtained during the twitch). Then, sciatic nerves were stimulated by pulses of 50– 200 Hz, and the absolute maximal force was measured. The specific maximal force was determined by dividing the absolute muscle force on the TA weight.
Protein Extraction and Western Blot
TA cryosections or transfected HEK293T cells were lysed in radio immunoprecipitation assay (RIPA) buffer supplemented with PMSF 1 mM and complete mini EDTA-free protease inhibitor cocktail (Roche Diagnostic). Protein concentrations were determined with the Bio-Rad Protein Assay Kit. Samples were denaturated at 95°C for 5 min. Then, 20 μg of protein was loaded in buffer containing 50 mM Tris-HCl, 2% SDS, and 10% glycerol, separated in 10% SDS-PAGE electrophoretic gel, and transferred on nitrocellulose membrane for 1.5 hr at 200 mA. Membranes were blocked for 2 hr in Tris-buffered saline (TBS) containing 5% non-fat dry milk and 0.1% Tween20 before an incubation for 2 hr with primary rabbit polyclonal antibodies against DNM2 (homemade antibody R2865; 1:500) or mouse antibody against GAPDH (1:100,000) diluted in blocking buffer containing 5% milk. Secondary antibodies coupled to horseradish peroxidase were goat anti-rabbit (for DNM2; 1:10,000) or goat anti-mouse (for GAPDH; 1:10,000) and were incubated overnight. Nitrocellulose membranes were visualized in Amersham Imager 600.
Production and Purification of rAAV
Recombinant adeno-associated virus (rAAV)2/9 vectors were generated by a triple transfection of AAV-293 cell line with pAAV2 insert containing either shRNA control (scrambled sequence) or shDnm2 (B, C, F, I, and J) under the control of the mouse U6 promoter, a GFP gene under the control of cytomegalovirus (CMV) promoter as a reporter and flanked by serotype-2 inverted terminal repeats, pXR1 containing rep and cap genes of AAV serotype-9, and pHelper encoding the adenovirus helper functions. Cell lysates were subjected to 3 rounds of freeze/thaw and then treated with 50 U/mL Benzonase (Sigma) for 30 min at 37°C and clarified by centrifugation. Viral vectors were purified by iodixanol gradient ultracentrifugation followed by dialysis and concentration against Dulbecco’s phosphate-buffered saline (DPBS) using centrifugal filters (Amicon Ultra-15 Centrifugal Filter Devices 30K). Physical particles were quantified by real-time PCR, and titers are expressed as viral genomes per mL (vg/mL).
RNA Extraction and qRT-PCR
Total RNA was isolated from transfected HEK293T or C2C12 cells using TRIzol reagent according to the manufacturer’s instruction (Invitrogen, UK). RT-PCR was carried out on 1–1.5 μg aliquot using SuperscriptII Reverse Transcriptase (Thermo Fisher Scientific). qRT-PCR was performed in Lightcycler 480 (Roche) using Dnm2 (F): CCAACAAAGGCATCTCCCCT, Dnm2 (R): TGGTGAGTAGACCCGAAGGT, Hprt(F): GTAATGATCAGTCAACGGGGG-AC, and Hprt (R): CCAGCAAGCTTGCAACCTTAACCA mixed in SybrGreen (QIAGEN).
Statistical Analysis
For in vitro studies, n = 3 biological replicates. Bar charts depict mean ± SEM. For in vivo studies, n = 5–8 were used. Curves and graphs were made using GraphPad Prism software. Difference between groups was analyzed by t test or two-way ANOVA followed by post hoc Bonferroni.
Author Contributions
Conceptualization, B.S.C. and J.L.; Investigation, H.T.; Resources, V.M.L., C.K., P.K., N.M., and D.B.; Writing – Original Draft, H.T.; Writing – Review & Editing, B.S.C. and J.L.
Conflicts of Interest
H.T., J.L., and B.S.C. are inventors of a patent on targeting DNM2 for the treatment of centronuclear myopathies. J.L. and B.S.C. are scientific advisors for Dynacure.
Acknowledgments
We thank Pascal Kessler, William Magnant, and Olivia Wendling for excellent technical assistance and Julie Johnston (Penn Vector Core at University of Pennsylvania) for providing pAAV2/9 plasmid. This study was supported by ANR-10-LABX-0030-INRT, a French state fund managed by the Agence Nationale de la Recherche under the frame program Investissements d’Avenir ANR-10-IDEX-0002-02, ANR-14-CE12-0009, Myotubular Trust and Sparks the Children’s Medical Research Charity, Association Française contre les Myopathies (AFM 20323), and the SATT Alsace Conectus. H.T. was supported by a MRT fellowship.
Footnotes
Supplemental Information includes four figures and can be found with this article online at https://doi.org/10.1016/j.ymthe.2018.02.008.
Contributor Information
Jocelyn Laporte, Email: jocelyn@igbmc.fr.
Belinda S. Cowling, Email: belinda@igbmc.fr.
Supplemental Information
References
- 1.Romero N.B. Centronuclear myopathies: a widening concept. Neuromuscul. Disord. 2010;20:223–228. doi: 10.1016/j.nmd.2010.01.014. [DOI] [PubMed] [Google Scholar]
- 2.Jungbluth H., Wallgren-Pettersson C., Laporte J. Centronuclear (myotubular) myopathy. Orphanet J. Rare Dis. 2008;3:26. doi: 10.1186/1750-1172-3-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Spiro A.J., Shy G.M., Gonatas N.K. Myotubular myopathy. Persistence of fetal muscle in an adolescent boy. Arch. Neurol. 1966;14:1–14. doi: 10.1001/archneur.1966.00470070005001. [DOI] [PubMed] [Google Scholar]
- 4.Laporte J., Hu L.J., Kretz C., Mandel J.L., Kioschis P., Coy J.F., Klauck S.M., Poustka A., Dahl N. A gene mutated in X-linked myotubular myopathy defines a new putative tyrosine phosphatase family conserved in yeast. Nat. Genet. 1996;13:175–182. doi: 10.1038/ng0696-175. [DOI] [PubMed] [Google Scholar]
- 5.Laporte J., Kress W., Mandel J.L. Diagnosis of X-linked myotubular myopathy by detection of myotubularin. Ann. Neurol. 2001;50:42–46. doi: 10.1002/ana.1033. [DOI] [PubMed] [Google Scholar]
- 6.Buj-Bello A., Biancalana V., Moutou C., Laporte J., Mandel J.L. Identification of novel mutations in the MTM1 gene causing severe and mild forms of X-linked myotubular myopathy. Hum. Mutat. 1999;14:320–325. doi: 10.1002/(SICI)1098-1004(199910)14:4<320::AID-HUMU7>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 7.Laporte J., Biancalana V., Tanner S.M., Kress W., Schneider V., Wallgren-Pettersson C., Herger F., Buj-Bello A., Blondeau F., Liechti-Gallati S., Mandel J.L. MTM1 mutations in X-linked myotubular myopathy. Hum. Mutat. 2000;15:393–409. doi: 10.1002/(SICI)1098-1004(200005)15:5<393::AID-HUMU1>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 8.Tsai T.C., Horinouchi H., Noguchi S., Minami N., Murayama K., Hayashi Y.K., Nonaka I., Nishino I. Characterization of MTM1 mutations in 31 Japanese families with myotubular myopathy, including a patient carrying 240 kb deletion in Xq28 without male hypogenitalism. Neuromuscul. Disord. 2005;15:245–252. doi: 10.1016/j.nmd.2004.12.005. [DOI] [PubMed] [Google Scholar]
- 9.Tosch V., Vasli N., Kretz C., Nicot A.S., Gasnier C., Dondaine N., Oriot D., Barth M., Puissant H., Romero N.B. Novel molecular diagnostic approaches for X-linked centronuclear (myotubular) myopathy reveal intronic mutations. Neuromuscul. Disord. 2010;20:375–381. doi: 10.1016/j.nmd.2010.03.015. [DOI] [PubMed] [Google Scholar]
- 10.Tsujita K., Itoh T., Ijuin T., Yamamoto A., Shisheva A., Laporte J., Takenawa T. Myotubularin regulates the function of the late endosome through the gram domain-phosphatidylinositol 3,5-bisphosphate interaction. J. Biol. Chem. 2004;279:13817–13824. doi: 10.1074/jbc.M312294200. [DOI] [PubMed] [Google Scholar]
- 11.Al-Qusairi L., Weiss N., Toussaint A., Berbey C., Messaddeq N., Kretz C., Sanoudou D., Beggs A.H., Allard B., Mandel J.L. T-tubule disorganization and defective excitation-contraction coupling in muscle fibers lacking myotubularin lipid phosphatase. Proc. Natl. Acad. Sci. USA. 2009;106:18763–18768. doi: 10.1073/pnas.0900705106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dowling J.J., Vreede A.P., Low S.E., Gibbs E.M., Kuwada J.Y., Bonnemann C.G., Feldman E.L. Loss of myotubularin function results in T-tubule disorganization in zebrafish and human myotubular myopathy. PLoS Genet. 2009;5:e1000372. doi: 10.1371/journal.pgen.1000372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hnia K., Tronchère H., Tomczak K.K., Amoasii L., Schultz P., Beggs A.H., Payrastre B., Mandel J.L., Laporte J. Myotubularin controls desmin intermediate filament architecture and mitochondrial dynamics in human and mouse skeletal muscle. J. Clin. Invest. 2011;121:70–85. doi: 10.1172/JCI44021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Robb S.A., Sewry C.A., Dowling J.J., Feng L., Cullup T., Lillis S., Abbs S., Lees M.M., Laporte J., Manzur A.Y. Impaired neuromuscular transmission and response to acetylcholinesterase inhibitors in centronuclear myopathies. Neuromuscul. Disord. 2011;21:379–386. doi: 10.1016/j.nmd.2011.02.012. [DOI] [PubMed] [Google Scholar]
- 15.Dowling J.J., Joubert R., Low S.E., Durban A.N., Messaddeq N., Li X., Dulin-Smith A.N., Snyder A.D., Marshall M.L., Marshall J.T. Myotubular myopathy and the neuromuscular junction: a novel therapeutic approach from mouse models. Dis. Model. Mech. 2012;5:852–859. doi: 10.1242/dmm.009746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lawlor M.W., Alexander M.S., Viola M.G., Meng H., Joubert R., Gupta V., Motohashi N., Manfready R.A., Hsu C.P., Huang P. Myotubularin-deficient myoblasts display increased apoptosis, delayed proliferation, and poor cell engraftment. Am. J. Pathol. 2012;181:961–968. doi: 10.1016/j.ajpath.2012.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lawlor M.W., Viola M.G., Meng H., Edelstein R.V., Liu F., Yan K., Luna E.J., Lerch-Gaggl A., Hoffmann R.G., Pierson C.R. Differential muscle hypertrophy is associated with satellite cell numbers and Akt pathway activation following activin type IIB receptor inhibition in Mtm1 p.R69C mice. Am. J. Pathol. 2014;184:1831–1842. doi: 10.1016/j.ajpath.2014.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Al-Qusairi L., Prokic I., Amoasii L., Kretz C., Messaddeq N., Mandel J.L., Laporte J. Lack of myotubularin (MTM1) leads to muscle hypotrophy through unbalanced regulation of the autophagy and ubiquitin-proteasome pathways. FASEB J. 2013;27:3384–3394. doi: 10.1096/fj.12-220947. [DOI] [PubMed] [Google Scholar]
- 19.Fetalvero K.M., Yu Y., Goetschkes M., Liang G., Valdez R.A., Gould T., Triantafellow E., Bergling S., Loureiro J., Eash J. Defective autophagy and mTORC1 signaling in myotubularin null mice. Mol. Cell. Biol. 2013;33:98–110. doi: 10.1128/MCB.01075-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bitoun M., Maugenre S., Jeannet P.Y., Lacène E., Ferrer X., Laforêt P., Martin J.J., Laporte J., Lochmüller H., Beggs A.H. Mutations in dynamin 2 cause dominant centronuclear myopathy. Nat. Genet. 2005;37:1207–1209. doi: 10.1038/ng1657. [DOI] [PubMed] [Google Scholar]
- 21.Böhm J., Biancalana V., Dechene E.T., Bitoun M., Pierson C.R., Schaefer E., Karasoy H., Dempsey M.A., Klein F., Dondaine N. Mutation spectrum in the large GTPase dynamin 2, and genotype-phenotype correlation in autosomal dominant centronuclear myopathy. Hum. Mutat. 2012;33:949–959. doi: 10.1002/humu.22067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Warnock D.E., Baba T., Schmid S.L. Ubiquitously expressed dynamin-II has a higher intrinsic GTPase activity and a greater propensity for self-assembly than neuronal dynamin-I. Mol. Biol. Cell. 1997;8:2553–2562. doi: 10.1091/mbc.8.12.2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Di A., Nelson D.J., Bindokas V., Brown M.E., Libunao F., Palfrey H.C. Dynamin regulates focal exocytosis in phagocytosing macrophages. Mol. Biol. Cell. 2003;14:2016–2028. doi: 10.1091/mbc.E02-09-0626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Arneson L.N., Segovis C.M., Gomez T.S., Schoon R.A., Dick C.J., Lou Z., Billadeau D.D., Leibson P.J. Dynamin 2 regulates granule exocytosis during NK cell-mediated cytotoxicity. J. Immunol. 2008;181:6995–7001. doi: 10.4049/jimmunol.181.10.6995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McNiven M.A., Kim L., Krueger E.W., Orth J.D., Cao H., Wong T.W. Regulated interactions between dynamin and the actin-binding protein cortactin modulate cell shape. J. Cell Biol. 2000;151:187–198. doi: 10.1083/jcb.151.1.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mooren O.L., Kotova T.I., Moore A.J., Schafer D.A. Dynamin2 GTPase and cortactin remodel actin filaments. J. Biol. Chem. 2009;284:23995–24005. doi: 10.1074/jbc.M109.024398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yamada H., Takeda T., Michiue H., Abe T., Takei K. Actin bundling by dynamin 2 and cortactin is implicated in cell migration by stabilizing filopodia in human non-small cell lung carcinoma cells. Int. J. Oncol. 2016;49:877–886. doi: 10.3892/ijo.2016.3592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kenniston J.A., Lemmon M.A. Dynamin GTPase regulation is altered by PH domain mutations found in centronuclear myopathy patients. EMBO J. 2010;29:3054–3067. doi: 10.1038/emboj.2010.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang L., Barylko B., Byers C., Ross J.A., Jameson D.M., Albanesi J.P. Dynamin 2 mutants linked to centronuclear myopathies form abnormally stable polymers. J. Biol. Chem. 2010;285:22753–22757. doi: 10.1074/jbc.C110.130013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cowling B.S., Toussaint A., Amoasii L., Koebel P., Ferry A., Davignon L., Nishino I., Mandel J.L., Laporte J. Increased expression of wild-type or a centronuclear myopathy mutant of dynamin 2 in skeletal muscle of adult mice leads to structural defects and muscle weakness. Am. J. Pathol. 2011;178:2224–2235. doi: 10.1016/j.ajpath.2011.01.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu N., Bezprozvannaya S., Shelton J.M., Frisard M.I., Hulver M.W., McMillan R.P., Wu Y., Voelker K.A., Grange R.W., Richardson J.A. Mice lacking microRNA 133a develop dynamin 2–dependent centronuclear myopathy. J. Clin. Invest. 2011;121:3258–3268. doi: 10.1172/JCI46267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Buj-Bello A., Laugel V., Messaddeq N., Zahreddine H., Laporte J., Pellissier J.F., Mandel J.L. The lipid phosphatase myotubularin is essential for skeletal muscle maintenance but not for myogenesis in mice. Proc. Natl. Acad. Sci. USA. 2002;99:15060–15065. doi: 10.1073/pnas.212498399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Childers M.K., Joubert R., Poulard K., Moal C., Grange R.W., Doering J.A., Lawlor M.W., Rider B.E., Jamet T., Danièle N. Gene therapy prolongs survival and restores function in murine and canine models of myotubular myopathy. Sci. Transl. Med. 2014;6:220ra10. doi: 10.1126/scitranslmed.3007523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Buj-Bello A., Fougerousse F., Schwab Y., Messaddeq N., Spehner D., Pierson C.R., Durand M., Kretz C., Danos O., Douar A.M. AAV-mediated intramuscular delivery of myotubularin corrects the myotubular myopathy phenotype in targeted murine muscle and suggests a function in plasma membrane homeostasis. Hum. Mol. Genet. 2008;17:2132–2143. doi: 10.1093/hmg/ddn112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tasfaout H., Buono S., Guo S., Kretz C., Messaddeq N., Booten S., Greenlee S., Monia B.P., Cowling B.S., Laporte J. Antisense oligonucleotide-mediated Dnm2 knockdown prevents and reverts myotubular myopathy in mice. Nat. Commun. 2017;8:15661. doi: 10.1038/ncomms15661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cowling B.S., Chevremont T., Prokic I., Kretz C., Ferry A., Coirault C., Koutsopoulos O., Laugel V., Romero N.B., Laporte J. Reducing dynamin 2 expression rescues X-linked centronuclear myopathy. J. Clin. Invest. 2014;124:1350–1363. doi: 10.1172/JCI71206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sabha N., Volpatti J.R., Gonorazky H., Reifler A., Davidson A.E., Li X., Eltayeb N.M., Dall’Armi C., Di Paolo G., Brooks S.V. PIK3C2B inhibition improves function and prolongs survival in myotubular myopathy animal models. J. Clin. Invest. 2016;126:3613–3625. doi: 10.1172/JCI86841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Samulski R.J., Muzyczka N. AAV-mediated gene therapy for research and therapeutic purposes. Annu. Rev. Virol. 2014;1:427–451. doi: 10.1146/annurev-virology-031413-085355. [DOI] [PubMed] [Google Scholar]
- 39.Koornneef A., Maczuga P., van Logtenstein R., Borel F., Blits B., Ritsema T., van Deventer S., Petry H., Konstantinova P. Apolipoprotein B knockdown by AAV-delivered shRNA lowers plasma cholesterol in mice. Mol. Ther. 2011;19:731–740. doi: 10.1038/mt.2011.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Grimm D., Pandey K., Kay M.A. Adeno-associated virus vectors for short hairpin RNA expression. Methods Enzymol. 2005;392:381–405. doi: 10.1016/S0076-6879(04)92023-X. [DOI] [PubMed] [Google Scholar]
- 41.Yang Q., Tang Y., Imbrogno K., Lu A., Proto J.D., Chen A., Guo F., Fu F.H., Huard J., Wang B. AAV-based shRNA silencing of NF-κB ameliorates muscle pathologies in mdx mice. Gene Ther. 2012;19:1196–1204. doi: 10.1038/gt.2011.207. [DOI] [PubMed] [Google Scholar]
- 42.Saal K.A., Koch J.C., Tatenhorst L., Szegő E.M., Ribas V.T., Michel U., Bähr M., Tönges L., Lingor P. AAV.shRNA-mediated downregulation of ROCK2 attenuates degeneration of dopaminergic neurons in toxin-induced models of Parkinson’s disease in vitro and in vivo. Neurobiol. Dis. 2015;73:150–162. doi: 10.1016/j.nbd.2014.09.013. [DOI] [PubMed] [Google Scholar]
- 43.Koo T., Malerba A., Athanasopoulos T., Trollet C., Boldrin L., Ferry A., Popplewell L., Foster H., Foster K., Dickson G. Delivery of AAV2/9-microdystrophin genes incorporating helix 1 of the coiled-coil motif in the C-terminal domain of dystrophin improves muscle pathology and restores the level of α1-syntrophin and α-dystrobrevin in skeletal muscles of mdx mice. Hum. Gene Ther. 2011;22:1379–1388. doi: 10.1089/hum.2011.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Agbulut O., Huet A., Niederländer N., Puceat M., Menasché P., Coirault C. Green fluorescent protein impairs actin-myosin interactions by binding to the actin-binding site of myosin. J. Biol. Chem. 2007;282:10465–10471. doi: 10.1074/jbc.M610418200. [DOI] [PubMed] [Google Scholar]
- 45.Agbulut O., Coirault C., Niederländer N., Huet A., Vicart P., Hagège A., Puceat M., Menasché P. GFP expression in muscle cells impairs actin-myosin interactions: implications for cell therapy. Nat. Methods. 2006;3:331. doi: 10.1038/nmeth0506-331. [DOI] [PubMed] [Google Scholar]
- 46.Lawlor M.W., Beggs A.H., Buj-Bello A., Childers M.K., Dowling J.J., James E.S., Meng H., Moore S.A., Prasad S., Schoser B., Sewry C.A. Skeletal muscle pathology in X-linked myotubular myopathy: review with cross-species comparisons. J. Neuropathol. Exp. Neurol. 2016;75:102–110. doi: 10.1093/jnen/nlv020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Beggs A.H., Böhm J., Snead E., Kozlowski M., Maurer M., Minor K., Childers M.K., Taylor S.M., Hitte C., Mickelson J.R. MTM1 mutation associated with X-linked myotubular myopathy in Labrador Retrievers. Proc. Natl. Acad. Sci. USA. 2010;107:14697–14702. doi: 10.1073/pnas.1003677107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zincarelli C., Soltys S., Rengo G., Rabinowitz J.E. Analysis of AAV serotypes 1-9 mediated gene expression and tropism in mice after systemic injection. Mol. Ther. 2008;16:1073–1080. doi: 10.1038/mt.2008.76. [DOI] [PubMed] [Google Scholar]
- 49.Salva M.Z., Himeda C.L., Tai P.W., Nishiuchi E., Gregorevic P., Allen J.M., Finn E.E., Nguyen Q.G., Blankinship M.J., Meuse L. Design of tissue-specific regulatory cassettes for high-level rAAV-mediated expression in skeletal and cardiac muscle. Mol. Ther. 2007;15:320–329. doi: 10.1038/sj.mt.6300027. [DOI] [PubMed] [Google Scholar]
- 50.Rivera V.M., Gao G.P., Grant R.L., Schnell M.A., Zoltick P.W., Rozamus L.W., Clackson T., Wilson J.M. Long-term pharmacologically regulated expression of erythropoietin in primates following AAV-mediated gene transfer. Blood. 2005;105:1424–1430. doi: 10.1182/blood-2004-06-2501. [DOI] [PubMed] [Google Scholar]
- 51.Husain T., Passini M.A., Parente M.K., Fraser N.W., Wolfe J.H. Long-term AAV vector gene and protein expression in mouse brain from a small pan-cellular promoter is similar to neural cell promoters. Gene Ther. 2009;16:927–932. doi: 10.1038/gt.2009.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Penaud-Budloo M., Le Guiner C., Nowrouzi A., Toromanoff A., Chérel Y., Chenuaud P., Schmidt M., von Kalle C., Rolling F., Moullier P., Snyder R.O. Adeno-associated virus vector genomes persist as episomal chromatin in primate muscle. J. Virol. 2008;82:7875–7885. doi: 10.1128/JVI.00649-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nathwani A.C., Tuddenham E.G., Rangarajan S., Rosales C., McIntosh J., Linch D.C., Chowdary P., Riddell A., Pie A.J., Harrington C. Adenovirus-associated virus vector-mediated gene transfer in hemophilia B. N. Engl. J. Med. 2011;365:2357–2365. doi: 10.1056/NEJMoa1108046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nowrouzi A., Penaud-Budloo M., Kaeppel C., Appelt U., Le Guiner C., Moullier P., von Kalle C., Snyder R.O., Schmidt M. Integration frequency and intermolecular recombination of rAAV vectors in non-human primate skeletal muscle and liver. Mol. Ther. 2012;20:1177–1186. doi: 10.1038/mt.2012.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Arnett A.L., Konieczny P., Ramos J.N., Hall J., Odom G., Yablonka-Reuveni Z., Chamberlain J.R., Chamberlain J.S. Adeno-associated viral (AAV) vectors do not efficiently target muscle satellite cells. Mol. Ther. Methods Clin. Dev. 2014;1:14038. doi: 10.1038/mtm.2014.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yi R., Doehle B.P., Qin Y., Macara I.G., Cullen B.R. Overexpression of exportin 5 enhances RNA interference mediated by short hairpin RNAs and microRNAs. RNA. 2005;11:220–226. doi: 10.1261/rna.7233305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hutvágner G., Simard M.J., Mello C.C., Zamore P.D. Sequence-specific inhibition of small RNA function. PLoS Biol. 2004;2:E98. doi: 10.1371/journal.pbio.0020098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Grimm D., Streetz K.L., Jopling C.L., Storm T.A., Pandey K., Davis C.R., Marion P., Salazar F., Kay M.A. Fatality in mice due to oversaturation of cellular microRNA/short hairpin RNA pathways. Nature. 2006;441:537–541. doi: 10.1038/nature04791. [DOI] [PubMed] [Google Scholar]
- 59.Giering J.C., Grimm D., Storm T.A., Kay M.A. Expression of shRNA from a tissue-specific pol II promoter is an effective and safe RNAi therapeutic. Mol. Ther. 2008;16:1630–1636. doi: 10.1038/mt.2008.144. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.